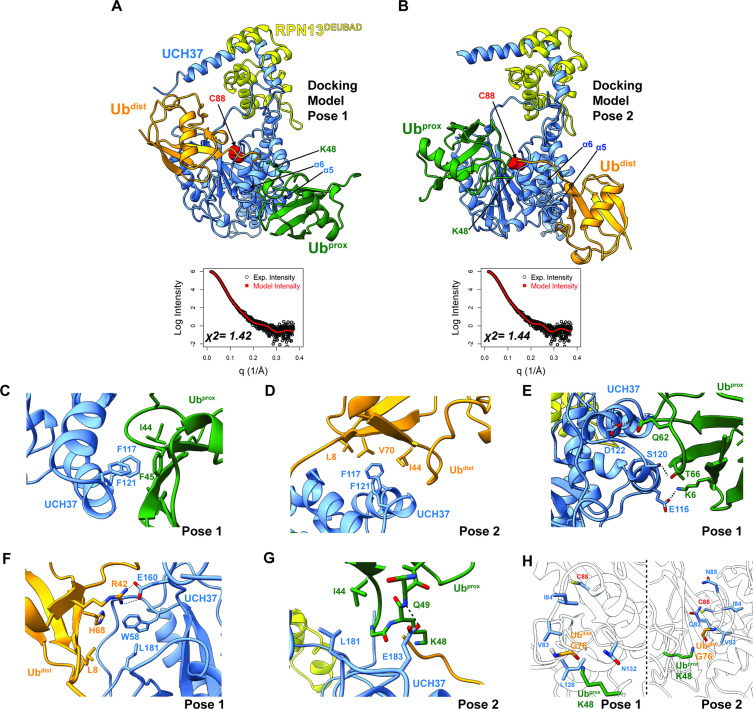
Figure 4. Docking models of the K48 di-ubiquitin (Ub):UCH37•RPN13DEUBAD complex.
(A–B) HADDOCK docking models show two poses corresponding to the interaction between K48 di-Ub and UCH37 along with their fit to experimental small-angle X-ray scattering data of the K48 di-Ub:UCH37C88A•RPN13DEUBAD complex. The goodness of fit to the experimental intensity is represented by χ2 values. In the first pose (A), the proximal Ub (Ubprox; green) interacts with α5–6 of UCH37. In the second pose (B), the distal Ub (Ubdist; orange) interacts with α5–6 of UCH37. (C) Residues highlighting the interaction between the aromatic-rich region of UCH37 α5–6 and the I44 patch of Ubprox in pose 1 (A). (D) Residues highlighting the interaction between the aromatic-rich region of UCH37 α5–6 and the I44 patch of Ubdist in pose 2 (B). (E) Polar contacts between Ubprox and UCH37 α5–6 in pose 1 (A). (F) Contacts between Ubdist and residues of UCH37 located outside the α5–6 motif in pose 1 (A). (G) Contacts between Ubprox and residues of UCH37 located outside the α5–6 motif in pose 2 (B). (H) The relative location of active site and the scissile, K48 isopeptide bond in molecular docking poses 1 and 2. In pose 1, residues of α6 and the loop leading into the catalytic Cys (C88) form a barrier for the isopeptide bond. In pose 2, only Q82 of UCH37 blocks the K48 isopeptide bond.