Abstract
De-oiled rice bran (DORB) is a potentially useful by-product of the rice bran oil industry. DORB may prove to be an important protein source, and also contains many other micronutrients. This study has the principal aim of optimizing the process of DORB protein hydrolysate preparation, and then testing the hydrolysate to determine its lipase inhibitory activity. DORB underwent hydrolysis using Alcalase® and response surface methodology (RSM). The resulting degree of hydrolysis (DH) was then monitored along with the extent of any lipase inhibitory activity. The optimum levels of lipase inhibition were obtained at a temperature of 49.88 °C, a duration of 150.43 minutes, and 1.53% Alcalase® used for the sample 5% (w/v) solution. In these conditions, the DH value was 35.65%, and the IC50 value for lipase inhibitory activity was 2.84 μg mL−1. Five ranges of different molecular weights were obtained via fractionation, whereupon it was determined that the highest level of inhibitory activity was achieved by the <0.65 kDa fraction. This fraction was then further purified via RP-HPLC, and the resulting peak had a retention time of 21.75 minutes (F2 sub-fraction) and exhibited high lipase inhibitory activity. Mass spectrometry was used to determine the amino acid sequence for this peak, identified as FYLGYCDY. This particular peptide is categorized as bitter, with a non-toxic profile, and having poor water solubility. The synthesized form of this peptide showed lipase inhibitory activity measured by an IC50 value of 0.47 ± 0.02 μM. The Lineweaver–Burk plot revealed that FYLGYCDY is a non-competitive inhibitor, while analysis of the docking results provided details of the FYLGYCDY peptide binding site with the porcine pancreatic lipase (PPL) complex, which is a competitive type. It can be inferred from these findings that DORB may prove a useful raw material source for the production of anti-obesity peptides which might enhance the therapeutic and commercial performance of functional foods and healthcare products.
De-oiled rice bran (DORB) is a potentially useful by-product of the rice bran oil industry.
1. Introduction
Obesity is becoming increasingly prevalent as a consequence of modern lifestyles which couple high intake levels of unhealthy food with a lack of physical activity.1,2 It is widely understood to be linked to a range of chronic diseases including hypertension, hyperlipidemia, type II diabetes, coronary heart disease, and various forms of cancer.3–5 One technique for treating obesity is to block lipid digestion and absorption in the gastrointestinal tract, accompanied by obstruction of new fat tissue development. Lipid metabolism is a process in which pancreatic lipase is the vital enzyme required to digest triglycerides. Triglycerides in the diet undergo hydrolysis by pancreatic lipase to form monoglycerides and free fatty acids, along with other small molecules which can be absorbed by the intestines where they resynthesize triglycerides, eventually resulting in obesity. For this reason, one key strategy to eliminate obesity is to inhibit this lipase activity, which can be tested in vitro. A number of different techniques to address and reduce obesity are understood, including the use of natural and synthetic pancreatic lipase inhibitors, which are able to block intestinal absorption of lipids.6–8 Studies conducted in animals have shown that an important tea catechin, (−)-epigallocatechin-3-gallate,9 along with polyphenol extracts from black tea,10 can stop obesity resulting from a diet containing high levels of fat through pancreatic lipase inhibition. One drug, orlistat, is a pancreatic inhibitor which is widely used as a means of limiting obesity.11 Nevertheless, they may cause side effects when used long-term.
According to recent developments in the functional food market, proteins are a significant driving force, providing energy and essential amino acids that are helpful to human health. The enzymatic hydrolysis mechanism generates different sizes of peptides, and also reducing unwanted side reactions. The nutritional and functional qualities of enzymatic protein hydrolysates allow them to play a role in both clinical practice and as protein supplements.12–15 It has been reported that enzymatic protein hydrolysates obtained from plants including rice,16 corn gluten,17 soy,18 potatoes,19 lemon basil seeds,20 and rice bran21 present bioactive properties which can act against oxidation and hypertension, and provide antithrombotic and hypocholesterolemic activity. Besides, plant protein hydrolysates are also rich sources of both proteins and other bioactive compounds, and can therefore be used as functional ingredients in nutritional food products.22
Rice is the most important of Thailand's agricultural crops, but paddy rice also yields a by-product, rice bran, at a rate of around 10%. The subject of this study, de-oiled rice bran (DORB), is a by-product obtained when rice bran oil is extracted from whole rice bran. Following the extraction of the oil, up to 20% of the protein will still be present in the DORB, which now serves as the main source of protein. As a result of its high protein content, rice bran offers has good potential as a source of beneficial hydrolysates. Useful bioactive properties have already been reported in rice bran proteins by a number of researchers, indicating the high value available from this particular by-product.23–26 There are a number of useful properties attributed to rice bran protein hydrolysates, including anti-oxidant, anti-diabetic, and anti-inflammatory activity.26 Due to the extensive disulfide bonds and compact agglomeration, rice bran proteins are insoluble in all common solvents.27 Most of the protein from rice bran is extracted using alkali. This processing method also results in protein denaturation and hydrolysis, as well as an increase in Maillard reaction, and causes increasing precipitation of non-protein material. Therefore, enzymatic methods have been applied as a method of enhancing rice bran protein solubility, yielding a wide variety of peptide hydrolysates.28
In this study, hydrolysis of DORB was performed using Alcalase® so as to derive a set of lipase inhibitory fractions. Response surface methodology (RSM) was used to determine optimal hydrolysis conditions, taking into consideration hydrolysis temperature, pH, and E/S ratio. The purification of lipase inhibitory peptides was performed using ultrafiltration along with RP-HPLC (reversed-phase high-performance liquid chromatography). Peptide sequences were established using an LC-MS/MS quadrupole time-of-flight (Q-TOF) mass spectrometer LC-MS/MS, while inhibition mode was evaluated with Lineweaver–Burk plots. In the last step, DORB peptides which showed effective activity against obesity had their molecular docking examined to assess the relationship between lipase and peptides, and the nature of the molecular inhibitory mechanism. This would provide useful information to guide the preparation of suitable peptides to counter obesity.
2. Materials and methods
2.1. Biomaterials and chemicals
Thai Edible Oil Co., Ltd (Bangkok, Thailand) supplied the DORB (Oryza sativa), while Novo (Novo Industries, Bagsvaerde, Denmark) provided the Alcalase®, stored at 4 °C until required and offering activity of 3.018 U mL−1 from Bacillus licheniformis. Pancreatic lipase (EC 3.1.1.3) from porcine pancreas type II, gum arabic (Acacia gum), p-nitrophenyl palmitate, t-octylphenoxypoly-ethoxyethanol (Triton X 100), 99% o-phthalaldehyde (OPA), bovine serum albumin (BSA), dimethyl sulfoxide (DMSO), and 99% trifluoroacetic acid were obtained from Sigma-Aldrich (St. Louis, MO, USA). Ajax Finechem (Tauren Point NSW, Australia) provided potassium dihydrogen orthophosphate (KH2PO4), di-potassium hydrogen orthophosphate (K2HPO4), sodium hydroxide (NaOH), sodium chloride (NaCl), and sodium tetraborate (Na2[B4O5(OH)4]·8H2O), while 8% phosphoric acid was obtained from Avantor™ Performance Materials (St. Broad, Phillipsburg, USA). March KGaA (St. Concord, Billerica MA, USA) supplied tris-base, Molecular Biology Grade, 37% hydrochloric acid (HCl), l-serine, dithiothreitol (DTT), sodium dodecyl sulfate (SDS), 2-propanol, and 95% ethanol. Coomassie Brilliant Blue G was supplied by USB Corporation (Cleveland, OH, USA). HPLC grade methanol (MeOH) was purchased from Merck KGaA (Merck KGaA, Darmstadt, Germany) while HPLC grade Acetonitrile was obtained from Fisher Scientific Korea Ltd (St. Gwangpyeong-ro, Gangnam-Gun, Seoul). All chemicals used in the course of the research were of analytical grade.
2.2. Preparation of DORB powder
The preparation of DORB followed the approach recommended by Saisavoey et al.21 albeit with minor adjustments. A fine powder of DORB was first produced using a grinder. This powder was then sieved using a mesh of 90 μm to ensure that the surface of the powder would be ready to carry out protein hydrolysis. Once prepared, the sieved DORB powder was held until required in a desiccator at room temperature, which was itself stored in a vacuum-sealed polypropylene bag.
2.3. Proximate analysis
The standardized approach of the Association of Official Analytical Chemists (AOAC) was employed to establish nutritional composition. In the first step, the DORB powder was placed in a crucible in an oven for 6 hours at a temperature of 105 °C, before being allowed to cool in a desiccator. The moisture content of the samples was then assessed by weighing (925.09 and 926.08).29 Petroleum ether extraction was then conducted in Soxhlet apparatus for 1 hour in order to measure the fat content (AOAC 2003.06).30 The ash content of the sample was determined by kindling the sample overnight using a muffle furnace at 550 °C (AOAC 923.03).31 Protein content was then determined using the Kjeldahl method and the application of a 6.25 nitrogen conversion factor (AOAC 968.06 and 992.15).32 Crude fiber content was assessed in line with AOAC 962.09.33 In each case, analysis was performed in triplicate. Using the data obtained, carbohydrate content was then determined by subtracting the amounts of fat, crude protein, moisture, ash, and fiber from total sample weight. Energy values of different date varieties were determined by considering the fat, carbohydrate, and crude protein content through the application of a formula devised by Crisan and Sands: energy value (kcal per 100 g) = (2.62 × % protein) + (8.37 × % fat) + (4.2 × % carbohydrate).34
2.4. Initial experimentation for DORB hydrolysates via enzymatic hydrolysis
The initial investigation of the DORB protein hydrolysate via single-factor analysis used a range of temperatures from 30 to 60 °C, a range of processing times from 1 to 4 hours, and concentrations of Alcalase® from 0.5 to 2.5%. The hydrolysis process involved the use of 2.5 g of sieved DORB powder, which then underwent suspension in 50 mL of 20 mM phosphate buffer at a pH of 8.0 prior to incubation in a shaker operating at 150 rpm. The process was ended by heating in a water bath up to a temperature of 90 °C for 20 minutes. Centrifugation was then carried out for 30 minutes at 15 900×g. Protein hydrolysate was the resulting supernatant, which was recovered and stored under refrigerated conditions at −20 °C until required. Analysis was then performed to determine degree of hydrolysis and level of pancreatic lipase inhibition. For each factor, an optimal value was then selected for response surface methodology.
2.5. DORB hydrolysate optimization via RSM
To optimize the preparation of the DORB hydrolysate, CCD (central composite design) was used to design the experiments and carry out statistical analysis. RSM testing design permitted analysis of a number of different independent variables: temperature, enzyme, and duration. Five levels were coded accordingly as −1.68, −1, 0, +1, and +1.68 using Design Expert software (Version 11, Stat-Ease, Inc., USA). Response variables to be optimized were the degree of hydrolysis and lipase inhibitory activity. RSM was capable of determining the effects of independent variables (temperature, X1; enzyme, X2; and time, X3) upon DH and lipase inhibitory activity (Y1 and Y2). A five-level and three-factor CCD allowed assessment during preliminary experiments of temperature effects on hydrolysis in the range of 41.95 °C to 48.4 °C, concentration of Alcalase® effects in the range of 0.5% to 2.5%, and incubation time effects in the range of 79.09 to 280.91 minutes. Overall, the design allowed the testing of 20 different combinations in optimizing response values during protein hydrolysis. Stat-Ease software (Design Expert version 11.0.5 Trial) was employed to design the experiment and conduct statistical analysis.
Hydrolysis was conducted through the addition of different Alcalase® concentrations to a suspension containing 2.5 g of DORB powder in 50 mL of 20 mM phosphate buffer at a pH of 8.0, and shaken in an incubator at 150 rpm for varying durations and at varying temperatures. Following hydrolysis under the specific conditions for variables set according to experimental design, the resulting protein hydrolysate solutions were heated to 90 °C for 20 minutes using boiling water in order to terminate enzymatic hydrolysis reactions via the deactivation of protease. Centrifugation was then performed at 15 900×g for 30 minutes. The supernatant was then placed in storage until required, at a temperature of 4 °C.
2.6. DH determination
An approach explained by Nielsen et al.35 was slightly adjusted and used to find the degree of hydrolysis of the DORB protein. Following hydrolysis of the DORB protein produced under each experimental condition, 400 mL of the supernatant was introduced to 3 mL of OPA reagent and mixed. The resulting mixture underwent precisely 2 minutes of room temperature incubation before detection using a spectrophotometer at 340 nm. This process was carried out in triplicate. Measurement of hydrolyzed proteins was based on the reaction occurring between OPA and amino acid groups when β-mercaptoethanol was present to form a colored compound which could be detected at 340 nm. The standard employed was serine, and the definition for DH held that it represented the percentage of cleaved peptide bonds, calculated using eqn (1):
| DH (%) = [((B × Nb)/MP) × (1/α) × (1/htot)] × 100 | 1 |
in which B indicates base consumption (mL), Nb represents base normality, MP indicates protein mass (g), α shows the average degree of dissociation for α -NH2 groups, and htot represents the overall number of peptide bonds contained within the protein substrate (8.0 meqv g−1 food protein).
2.7. Inhibition of pancreatic lipase
To assess pancreatic lipase inhibition, assay followed the approach set out by Adisakwattana et al.36 The test made use of 0.5 mg mL−1 of PPL (porcine pancreatic lipase) in 0.061 M Tris–HCl buffer at a pH of 8.0. For the substrate, p-nitrophenyl palmitate (p-NPP) was dissolved in 10 mL isopropanol in the form of a 1.66 mM substrate, which was then added to the Tris–HCl buffer containing gum arabic as an emulsifier and Triton X-100 as a surfactant. In the experimental phase, a 96-well plate was used to hold 50 μL as the sample solution, 25 μL of PPL solution, and 75 μL of p-NPP solution, while the blank sample in the absence of PPL instead contained Tris–HCl buffer. The negative control involved altering the sample to 50 μL of Tris–HCl buffer, and orlistat was used as a positive control. After 30 minutes of incubation, the reaction solution was then tested via spectrophotometer to evaluate the absorbance of released p-nitrophenol at 410 nm. The eqn (2) given below was then used to calculate lipase inhibitory percentage:
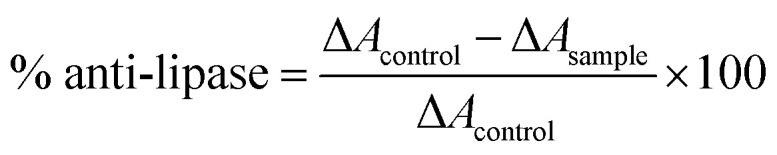 |
2 |
in which ΔAcontrol represents absorbance change at 410 nm with no test sample (Acontrol − Ablank of control), and ΔAsample indicates absorbance change at 410 nm in the presence of the test sample (Asample − Ablank of sample).
2.8. Soluble protein evaluation
The soluble protein concentration was determined using the previously described method (Bradford 1976).37 To perform the test, 200 μL of Bradford reagent working solution was added to 20 μL of DORB hydrolysate in a 96-well plate, prior to a room temperature incubation period of not less than 2 minutes, and not more than one hour. A spectrophotometer was then used to measure absorbance at 595 nm, while the concentration of proteins in the DORB hydrolysate can be specified through comparisons with a BSA of protein standard curve. This process was carried out in triplicate.
2.9. Fractionation and purification of lipase inhibitory peptides
2.9.1. Ultrafiltration
Fractionation was performed to isolate lipase inhibitory peptides, by first taking a sample of 800 mL of DORB protein hydrolysate solution and fractionating through the use of Minimate™ tangential flow filtration capsules and Omega™ membranes (Pall Life Sciences, Ann Arbor, USA). Four ultrafiltration membranes were used, allowing the application of four different molecular weight cut-off values at 10 kDa, 5 kDa, 3 kDA, and 0.65 kDa. The first step allowed the material to be separated using 10 kDa membrane, whereby the retentate served as the >10 kDa fraction, and the permeate underwent subsequent filtration using the 5 kDa membrane. This process was repeated until five separate fractions had been obtained: >10 kDa, 5–10 kDa, 3–5 kDa, 0.65–3 kDa, and <0.65 kDa. These fractions were placed into storage at 4 °C until required. Measurements of protein content and pancreatic lipase inhibitory activity were obtained, and the fraction which offered the greatest potential for the inhibition of pancreatic lipase was selected for further concentration via seep-vacuum before storage at −20 °C prior to the next step of purification.
2.9.2. RP-HPLC
Reversed-phase HPLC (RP-HPLC) was used in this study to purify lipase inhibitory peptides using a Luna C18 column (Shimpak, 250 × 46 mm, Luna SU; Phenomenex, Torrance, CA, USA). To perform these procedures, once the lipase inhibitory peptide had undergone ultrafiltration to obtain the selected fraction, 1.00 mL of this sample was further filtered through a 0.45 μm membrane (NYL Whatman membrane, GE, Buckinghamshire, UK). It was then introduced into the column, whereby separation was achieved by the HPLC system via gradient elution of solvent A (0.10% (v/v) trifluoroacetic acid (TFA) in DDI (distilled de-ionized) water) and solvent B (0.05% (v/v) TFA in acetonitrile). A linear gradient was used for elution, starting at 88% solvent A: 12% solvent B. From this point, the proportion of solvent A declined as the proportion of solvent B increased for a period of 50 minutes at a flow rate of 0.70 mL min−1. Eluent absorbance was measured at 280 nm using a UV detection approach, while ChromQuest Software (Thermo Fisher Scientific, San Jose, CA, USA) was used to perform chromatographic analysis. Identifiable high and clear peaks were gathered, and a speed vacuum concentrator was used to raise concentrations before lipase inhibitory activity was tested. The most effective fraction was then selected and freeze dried, whereupon peptide sequence could be determined through mass spectrometry.
2.10. Amino acid sequence identification via LC-ESI-Q-TOF-MS/MS
Characterization of the amino acid sequences of the lipase inhibitory peptide isolated using RP-HPLC was performed using a Q-TOF mass spectrometer along with an electrospray ionization source mass spectrometer (Model Amazon SL, Bruker, Germany). When calibrated for the initial step, ESI-Q-TOF aimed to determine those peptide chains with a mass ranging from 50 to 1200 m/z, whereupon the collected data were assessed via de novo sequencing, which involves the analysis of mass differences arising between fragment ion pairs, allowing the mass of amino acid residues to be subsequently calculated for the peptide chain. The determination of that residue thus permits identification.
Having identified the peptide sequence, the next step was to examine the profiles further to determine the extent of lipase inhibitory activity which would be linked to the synthetic peptide which could potentially serve as a biological agent. This information is available from the BIOPEP database (http://www.uwm.edu.pl/biochemia/index.php/en/biopep). The toxicity level of lipase inhibitory peptide can also be discovered using ToxinPred (http://crdd.osdd.net/raghava/toxinpred/) to determine the suitability of the peptide for use as an ingredient in functional food products. SVM (support vector machine) analysis was used for predictions in order to separate the peptides into those which were toxic and those which were considered safe, by setting a threshold of 0.00. In addition, the BIOPEP database was used to make predictions of the taste qualities of the selected lipase inhibitory peptide, since amino acids and peptides can have a significant influence on the taste of functional foods.
2.11. Synthesis of peptides
The DORB hydrolysate-derived lipase inhibitory peptide which was found to offer the greatest potential can by synthesized chemically using Fmoc solid-phase synthesis. This is performed by Biotech Bioscience & Technology Co., Ltd, in Shanghai, China, making use of an Applied Biosystems Model 433A Synergy peptide synthesizer. The purity of this lipase inhibitory peptide underwent analysis for verification using the MS system (Thermo Mod. Finnigan™ LXQ™) linked to a Surveyor HPLC. For this synthetic peptide, the lipase inhibitory peptide sequence was shown to achieve at least 98% purity.
2.12. Kinetics evaluation
Lineweaver–Burk plots allowed the identification of the inhibitory mechanisms used by the synthesized peptide FYLGYCDY. The Michaelis equation was used to determine kinetic parameters, including Km, Vmax, and Ki, while the measurement of lipase inhibitory activity was performed using differing substrate concentrations (0.07, 0.1, 0.13, and 0.16 mM) as well as varying synthesized peptide concentrations (0, 0.2, 0.5, and 0.8 mM). The substrate concentration reciprocal was then used as Km on the x-axis for linear regression analysis of Lineweaver–Burk plots, while the reciprocal of the absorbed substrate product at 410 nm served as Vmax on the y-axis. From these data it was then possible to establish the value of Ki, the inhibitor constant.
2.13. FYLGYCDY molecular docking at the lipase binding site
Discovery Studio 2019 Software was employed to produce a 3D FYLGYCDY peptide model. The molecular docking study made use of a 3D crystal structure of a PPL complex (PDB: 1ETH) to serve as the receptor, obtained online from the RCSB PDB Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). Orlistat, which was used as the positive control, was obtained online from the PubChem substance and compound databases (https://pubchem.ncbi.nlm.nih.gov). The study of molecular docking involving FYLGYCDY, orlistat, and PPL was performed using GOLD 5.7.1 software. Before docking takes place, it is necessary to first eliminate all heteroatoms from the PPL model, such as water molecules which are not needed, and ligands of the inhibitor. Polar hydrogens, de-oiled rice bran peptides, and orlistat were then added to the PPL model. A number of different scoring functions were then employed to carry out evaluations, including ChemPLP, ChemScore, GoldScore, ASP, and User Defined Score. Docking scores and measurements of binding energy were employed to determine the ideal pose for each residue. This helped in achieving a better understanding of de-oiled rice bran peptide binding to PPL and molecular docking of ligands lipase inhibitory peptide, as well as orlistat binding to the PPL receptor. Discovery Studio 19 software was employed to investigate the different interactions which arose between FYLGYCDY, PPL and orlistat. These interactions involved various forces, including electrostatic, van der Waals, hydrophilic, hydrophobic, and coordination forces. The optimal inhibitor peptide pose was also identified involving access to the active PPL site, while catalytic PPL sites included Ser153, Asp177, and His264, whereas substrate-binding sites included Phe78, His152, and Phe216.38
2.14. Statistical analysis
Design Expert was used to analyze the dependent and independent variables applied in the experimental, aiming to optimize preparation conditions for the DORB hydrolysate. Variables were measured in triplicate for all experimental results and presented in the form of mean ± standard deviation before analysis using the SPSS 11.5 statistical package. Data were analyzed with one-way analysis of variance (ANOVA) and least significant difference (LSD) test, with a significance level of 0.05. The best result obtained in terms of cost and convenience was then analyzed further for validation and to carry out further optimization.
3. Results and discussion
3.1. Proximate composition
By establishing the basic composition of a product in terms of its fat, fiber, moisture, ash, carbohydrates, and protein, it is possible to determine the nutritional value of that product, albeit on the understanding that the values may be altered when the proportions of materials used in creating the product are changed. Table 1 presents information concerning the chemical composition of DORB. DORB is typically the second by-product obtained from rice bran when it is used for oil extraction purposes. As a consequence of the oil removal, the relative quantities of nutrients in DORB may change, usually resulting in greater proportions of ash, carbohydrates, crude proteins, and fiber. DORB has been shown to have moisture content of just 6.72%, presenting a storage challenge given that the safe level of moisture content to store processed rice is 14%, while 12% is considered reasonable for storage over the longer term if some nutrients are to be maintained.39 Among the nutrients will be dietary fiber, which is found in cereal by-products, one of which is rice bran. As understanding of its health benefits grows, foods which are rich in fiber are increasingly promoted to consumers. There is very strong and growing demand for foods which offer added dietary fiber,40 so DORB makes an excellent choice, reaching as much as 13.94% dietary fiber.
Proximate DORB powder compositiona.
| Parameter | Proximate in dry weight (% w/w) |
|---|---|
| Moisture | 6.72 ± 0.11 |
| Fat | 1.44 ± 0.05 |
| Fiber | 6.11 ± 0.06 |
| Ash | 13.57 ± 0.12 |
| Carbohydrate | 59.20 ± 0.24 |
| Protein | 19.68 ± 0.30 |
Values are expressed as mean ± standard deviation and tests were performed in triplicate.
Ash levels can provide critical information about the presence of inorganic minerals which play key roles in nutrition. These can be seen in Table 1. When a food sample is tested for ash content, this gives useful insights into the extent of any mineral elements in that sample.41 In DORB, the ash content was reported to be 13.57%, while in rice bran, levels up to 13% have been recorded. In rice husks, ash content is affected by climate and location, as well as rice type. The quality of milled rice is partly determined by ash content, since the extent of milling which will be carried out on the rice grain will depend on ash and crude fat content, whiteness, and polished rice yield.42 DORB can have carbohydrate content reaching as high as 59.20%, possibly as a result of the starchy endosperm ridges which are present, along with broken pieces which are produced during the final milling stage, which eliminates the last traces of the bran layer along with the kernel endosperm.43 Extracted lipid content will also be lower. Rice bran carbohydrates take various forms and typically contain celluloses, hemicelluloses, starch, polysaccharides, oligosaccharides, and various sugars. Rice bran contains a number of water-soluble polysaccharides such as hemicelluloses, oligosaccharides as well as non-starchy polysaccharides and some phytochemicals which have water-soluble properties.44 The conformational structure of plant polysaccharides is highly specific, and leads to the generation of anti-inflammatory cytokines while preventing pro-inflammatory cytokines. Plant polysaccharides also have the ability serve as immunomodulators via both innate and cell-mediated immunity, and via the interactions they have with monocytes, macrophages, T-cells, and polymorphonuclear lymphocytes.45
DORB has a protein content of approximately 19.68%. RBP are mostly high nutrient conservation protein, such as globulin, glutein, albumin, and prolamin, and their general properties tend to match those of other cereal proteins. The greatest nutritional value is found in proteins derived from rice bran, which are often hypoallergenic, and for this reason proteins which might be obtained from rice bran may be very valuable.46 Furthermore, the RBP amino acid profile is excellent due to the abundance of essential amino acids, one of which is lysine, which in cereal grains acts as a limiting amino acid. The easy digestibility of RBP allows its use as a food product ingredient. Since rice bran is rich in protein, it offers great potential as a source of protein hydrolysates. In addition, proteins and peptides obtained from rice bran are already well known to be effective against certain health conditions and can also work well as nutraceuticals.47–49 The preparation of these bio-functional peptides from rice bran typically involves enzymatic hydrolysis. RBP are known to serve as antioxidants and also offer immunomodulatory and antihypertensive properties. They can also inhibit cancer and ACE (angiotensin-converting enzyme) as well as providing inflammatory cytokine signaling. RBP can also improve food matrix physicochemical stability as well as exhibiting viscoelastic properties, cake properties, and enzymatic browning inhibitory activity.24 The impact of food products on health can be assessed partly through energy value, which also provides important information to consumers who are particularly sensitive to the health qualities of the product. Food energy values indicate the extent to which the body can use the food as a fuel, and provide information concerning the chemical energy potential of the organic compound bonds in food as well as the extent of the fat, protein, and carbohydrate along with minor content including organic acids.50 This study sought to arrive at a figure for DORB energy value, which amounted to 328.48 kcal per 100 g.
3.2. Optimization of the DORB protein hydrolysis process
3.2.1. Preliminary evaluation
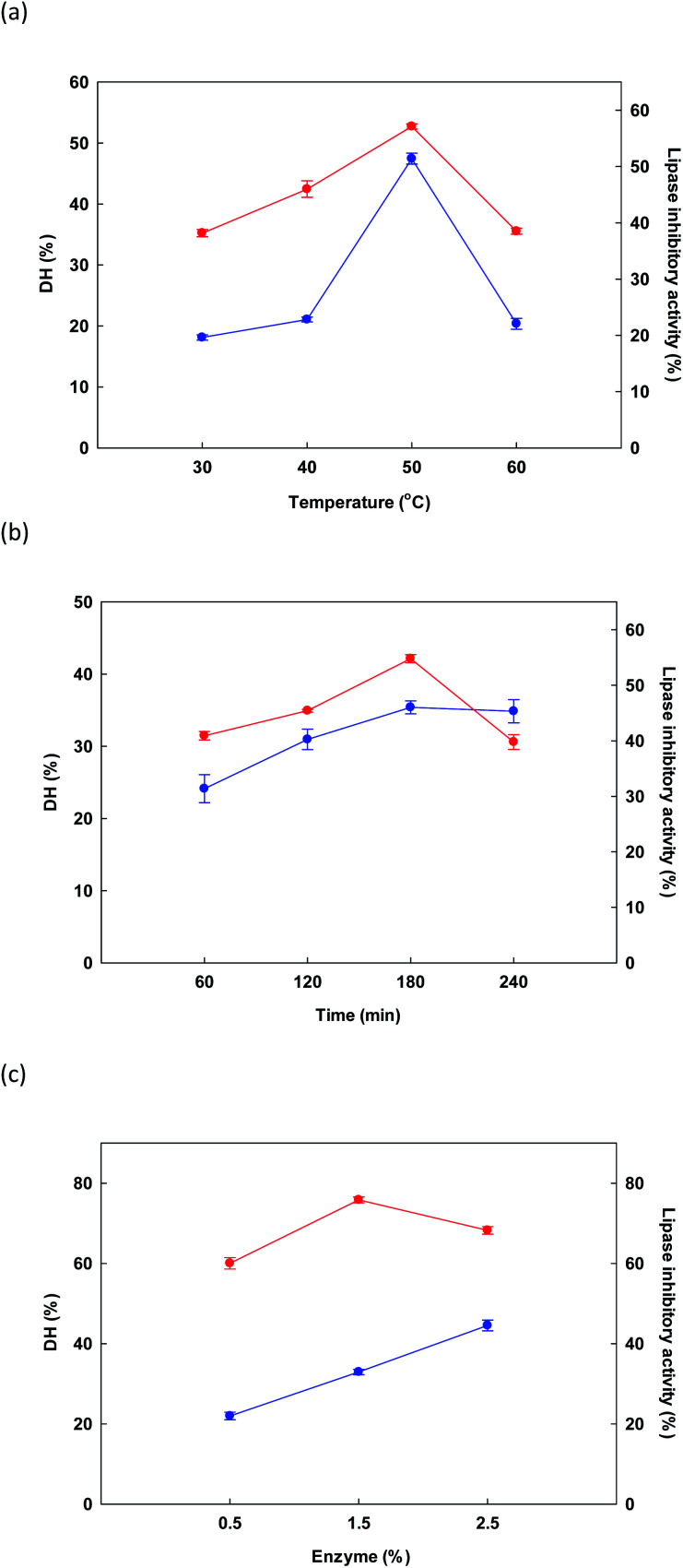
In initial testing, the influence of temperature on the DH and lipase inhibitory activity of the protein hydrolysate derived from DORB protein was measured while holding time at 180 minutes and enzyme concentration at 1.5% Alcalase® for a sample solution of 5% w/v. Fig. 1a presents outcomes, revealing that as the temperature increased from 30 °C to 50 °C, both DH and lipase inhibition increased, reaching a maximum level in both cases at 50 °C, before starting to decline thereafter. In similar research, Gao et al.51 studied the influence of temperature on DH and ACE inhibitory activity in cottonseed protein hydrolysis. When temperatures were high, denaturation of the protein took place along with the resolution of incomplete enzymatic hydrolysis, thus causing a drop in DH and ACE inhibition. In this study, it was confirmed that the effect of temperature upon DORB protein hydrolysis was significant at the 0.05 level, while the optimal temperature to be used as RSM central point was 50 °C.
Fig. 1. Initial examination of the influence of changes in different factors: temperature (a), time (b) and enzyme (c) for the reaction upon DH ( ) and lipase inhibitory activity (
) and lipase inhibitory activity ( ).
).
In Fig. 1b, the influence on DH and lipase inhibition of the various hydrolysis times is shown, including 60, 120, 180, and 240 minutes. For hydrolysis experiments, temperature was 50 °C while Alcalase® concentration was held at 1.5%. As time increased from 50 to 180 minutes, response variables of DH and lipase inhibition also rose, reaching a maximum at 180 minutes. A decline was then observed at longer time periods. Liu et al.52 reported similar findings when evaluating the DH of fish protein during enzymatic hydrolysis, where DH climbed notably as the time period increased from zero to 8 hours, whereupon it reached an equilibrium, or underwent a very small decrease. Lipase inhibition results showed a similar pattern. Our findings confirmed that the influence of time was significant in the context of DORB protein hydrolysis (p < 0.05). On the basis of these experiments, the selected RSM central point was 180 minutes.
Finally, in Fig. 1c the influence on DH and lipase inhibition is shown for different enzyme concentrations in the range of 0.5% to 2.5%. Temperature was fixed at 50 °C while duration was 180 minutes. As enzyme concentration increased, DH also increased, finally reaching 44.55 ± 1.34% at the maximum 2.5% enzyme concentration. However, for lipase inhibition, the best outcome occurred at a concentration of 1.5%, while increasing the concentration further caused the hydrolyzed peptides to be shorter, and thus less effective for the inhibition of lipase.52 On the basis of cost, as well as the importance of lipase inhibition, the concentration of 1.5% was therefore selected to serve as the central point for RSM, as shown in Table 2.
Independent variables and CCD coded levels for the hydrolysis of DORB protein.
| Independent variables | Code levels | ||||
|---|---|---|---|---|---|
| −1.68 | −1 | 0 | 1 | 1.68 | |
| X 1: temperature (°C) | 41.59 | 45.00 | 50.00 | 55.00 | 58.41 |
| X 2: E/S (% w/v) | 0.50 | 0.90 | 1.50 | 2.10 | 2.50 |
| X 3: hydrolysis time (min) | 79.09 | 120.00 | 180.00 | 240.00 | 280.91 |
3.2.2. RSM fitting
CCD (central composite design) was used to evaluate experimental design and conduct statistical analysis concerning lipase inhibitory peptide production. The three independent variables were temperature, enzyme, and duration, while the five coded levels employed were designated as −1.68, −1, 0, +1, and +1.68, whereby the central value (0) was taken from a preliminary evaluation of variables which can be seen in Table 2. In all, the experiments were performed in triplicate for 20 different variable combinations at different coded levels in line with CCD outline in order to optimize DH and lipase inhibition (Y1 and Y2 respectively) through RSM. These two responses were in the range of 25.03–39.79% for DH and 2.02–19.68 (IC50; μg mL−1) for lipase inhibition. It is necessary, however, for the relationship between the predicted and experimental responses to fall within an acceptable range, not exceeding 5% (Table 3).
Independent variable levels for temperature (X1), enzyme (X2), and time (X3), with tested values of the two response variables: % DH (Y1), and lipase inhibition (IC50; μg mL−1) (Y2)a.
| Run | Space type | Coded levels | Independent variables | Response | |||||
|---|---|---|---|---|---|---|---|---|---|
| X 1 | X 2 | X 3 | X 1 | X 2 | X 3 | Y 1 | Y 2 | ||
| 1 | Factorial | −1 | −1 | −1 | 45.00 | 0.90 | 120.00 | 25.03 | 9.77 |
| 2 | Factorial | +1 | −1 | −1 | 55.00 | 0.90 | 120.00 | 30.10 | 14.33 |
| 3 | Factorial | −1 | +1 | −1 | 45.00 | 2.10 | 120.00 | 31.23 | 11.00 |
| 4 | Factorial | +1 | +1 | −1 | 55.00 | 2.10 | 120.00 | 33.91 | 6.30 |
| 5 | Factorial | −1 | −1 | +1 | 45.00 | 0.90 | 240.00 | 30.56 | 7.06 |
| 6 | Factorial | +1 | −1 | +1 | 55.00 | 0.90 | 240.00 | 31.74 | 14.69 |
| 7 | Factorial | −1 | +1 | +1 | 45.00 | 2.10 | 240.00 | 36.13 | 19.25 |
| 8 | Factorial | +1 | +1 | +1 | 55.00 | 2.10 | 240.00 | 36.47 | 19.68 |
| 9 | Axial | −1.68 | 0 | 0 | 41.59 | 1.50 | 180.00 | 29.34 | 15.92 |
| 10 | Axial | +1.68 | 0 | 0 | 58.41 | 1.50 | 180.00 | 34.64 | 18.01 |
| 11 | Axial | 0 | −1.68 | 0 | 50.00 | 0.50 | 180.00 | 27.83 | 9.19 |
| 12 | Axial | 0 | +1.68 | 0 | 50.00 | 2.50 | 180.00 | 39.79 | 12.50 |
| 13 | Axial | 0 | 0 | −1.68 | 50.00 | 1.50 | 79.09 | 30.15 | 7.31 |
| 14 | Axial | 0 | 0 | +1.68 | 50.00 | 1.50 | 280.91 | 34.93 | 12.98 |
| 15 | Center | 0 | 0 | 0 | 50.00 | 1.50 | 180.00 | 34.54 | 3.02 |
| 16 | Center | 0 | 0 | 0 | 50.00 | 1.50 | 180.00 | 35.24 | 3.83 |
| 17 | Center | 0 | 0 | 0 | 50.00 | 1.50 | 180.00 | 34.36 | 4.12 |
| 18 | Center | 0 | 0 | 0 | 50.00 | 1.50 | 180.00 | 34.51 | 3.46 |
| 19 | Center | 0 | 0 | 0 | 50.00 | 1.50 | 180.00 | 33.07 | 3.65 |
| 20 | Center | 0 | 0 | 0 | 50.00 | 1.50 | 180.00 | 34.89 | 3.89 |
All tests were performed in triplicate.
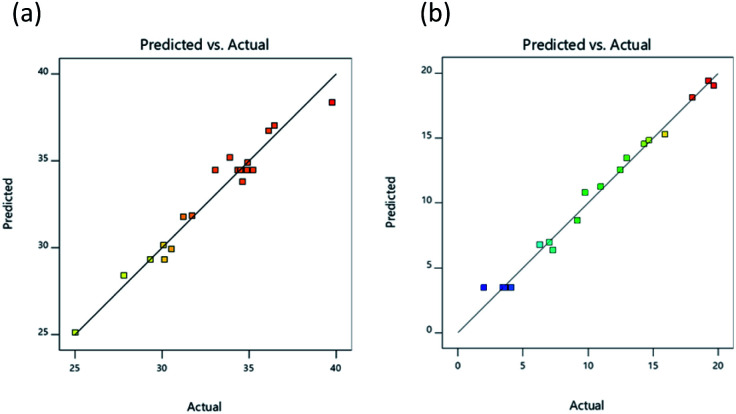
The 2D response graph presented in Fig. 2 shows the actual and predicted values for responses, with actual values illustrated on a straight line of the predicted value (Fig. 2a and b). Results confirmed the suitability of the independent variables for both of the response surface models. A summary of ANOVA (analysis of variance) results investigating regression models for the experiments testing DH and IC50 was provided in Design Expert 11 software and can be observed in Table 4. ANOVA results were shown to be significant (p < 0.05) for DH, taking into consideration the linear, interaction, and quadratic terms represented respectively by (X1, X2, and X3), (X1X3), and (X12, X32), as indicted by regression coefficients, while there was no significant influence exerted upon DH by interaction terms (X1X2 and X2X3) or quadratic term (X22). For an IC50 value of lipase inhibition, ANOVA results showed significance (p < 0.05) for linear, interaction, and quadratic terms represented respectively by (X1, X2, and X3), (X1X2, X1X3, and X2X3), and (X12, X22, and X32).
Fig. 2. Relationship between the experimental and predicted values for DH (a), and lipase inhibitory activity (IC50; μg mL−1) (b).
Analysis of variance (ANOVA) for DH (Y1), and IC50 of lipase inhibition (Y2) via the response surface quadratic model.
| Source | df | Y 1 | Y 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sum of square | Mean square | F-value | P-value | Sum of square | Mean square | F-value | P-value | ||
| Model | 9 | 211.46 | 23.50 | 24.59 | <0.0001a | 619.70 | 68.86 | 107.72 | <0.0001a |
| X 1 | 1 | 24.21 | 24.21 | 25.34 | 0.0005a | 9.58 | 9.58 | 14.99 | 0.0031a |
| X 2 | 1 | 119.66 | 119.66 | 125.23 | <0.0001a | 18.60 | 18.60 | 29.10 | 0.0003a |
| X 3 | 1 | 37.63 | 37.63 | 39.38 | <0.0001a | 60.83 | 60.83 | 95.17 | <0.0001a |
| X 1 X 2 | 1 | 1.30 | 1.30 | 1.36 | 0.2698 | 33.88 | 33.88 | 53.01 | <0.0001a |
| X 1 X 3 | 1 | 4.85 | 4.85 | 5.08 | 0.0479a | 8.39 | 8.39 | 13.13 | 0.0047a |
| X 2 X 3 | 1 | 0.01 | 0.01 | 0.01 | 0.9185 | 71.90 | 71.90 | 112.48 | <0.0001a |
| X 1 2 | 1 | 15.28 | 15.28 | 15.99 | 0.0025a | 316.33 | 316.33 | 494.90 | <0.0001a |
| X 2 2 | 1 | 2.15 | 2.15 | 2.25 | 0.1645 | 91.64 | 91.64 | 143.37 | <0.0001a |
| X 3 2 | 1 | 10.05 | 10.05 | 10.52 | 0.0088a | 74.49 | 74.49 | 116.54 | <0.0001a |
| Residual | 10 | 9.55 | 9.56 | 6.39 | 0.6392 | ||||
| Lack of fit | 5 | 6.81 | 1.36 | 2.49 | 0.1701 | 3.69 | 0.7388 | 1.39 | 0.3694 |
| Pure error | 5 | 2.74 | 0.55 | 2.70 | 0.5396 | ||||
| Cor total | 19 | 221.01 | 626.09 | ||||||
| Std dev. | 0.9775 | 0.7995 | |||||||
| R 2 | 0.9568 | 0.9898 | |||||||
| Adj R2 | 0.9179 | 0.9806 | |||||||
Indicates those variables which significantly influence the response variables (p < 0.05).
The p-values for both regression models were lower than 0.01, so the models can be considered significant. The p-values for lack of fit for the two models, however, exceeded 0.05 and hence were not significant. It can therefore be concluded that the models were suitably adjusted to responses. Model suitability can also be evaluated by coefficient of determination (adjusted R2) which was 0.9179, suggesting that 91.79% of response variability for DH could be explained by the model, while for lipase inhibition, the adjusted R2 value was 0.9806, thus demonstrating that 98.06% of response variability was explained by the model. In the case of our own R2 value, the models for both DH and lipase inhibition had values approaching 1.00, at 0.9568 and 0.9898 respectively, from which it can be inferred that the models fit well with experimental data. The response surface models which were developed had appropriate relationships involving independent variables constructed through the use of the regression coefficients for linear, interaction, and quadratic terms, along with a second-order polynomial model. Eqn (3) was used to calculate response for DH while lipase inhibition response was calculated with eqn (4):
| Y1 = −137.32471 + 5.0544X1 + 14.70023X2 + 0.239475X3 − 0.134583X1X2 − 0.002596X1X3 + 0.001007X2X3 − 0.04119X12 − 1.07299X22 − 0.000232X32 | 3 |
| Y2 = 492.32703 − 18.15846X1 + 0.240212X2 − 0.487773X3 − 0.685979X1X2 + 0.003414X1X3 + 0.083274X2X3 + 0.187405X12 + 7.0048X22 + 0.000632X32 | 4 |
In these equations, Y1 is DH prediction, Y2 is IC50 prediction, and X1, X2, and X3 represent temperature, enzyme, and time, respectively, as independent variables. Optimal levels for independent variables are those which generate the maximum DH value combined with minimum value for IC50.
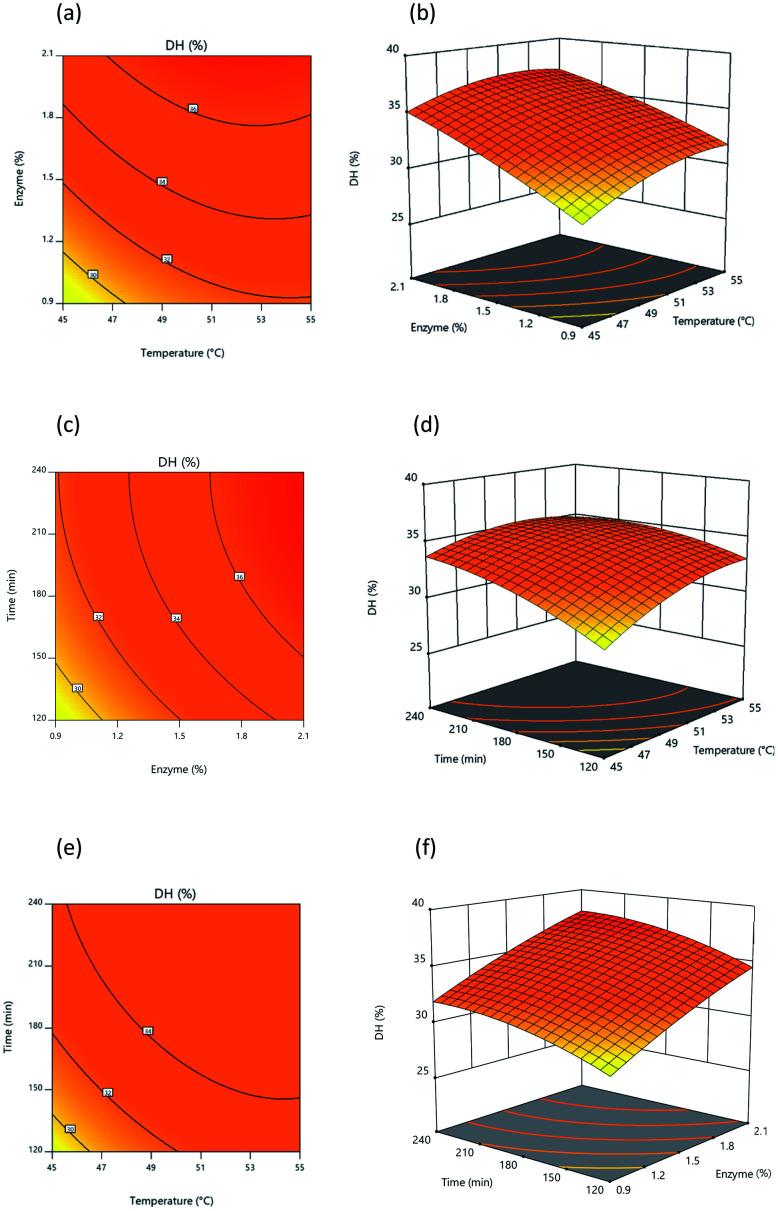
3.2.3. Analysis of response surface and contour plots
Fig. 3 and 4 present 3D surface response plots and associated contour plots which show the effects of independent variables on response variables. The 3D response surface plots for both DH and lipase inhibition were developed by plotting response variables on the z-axis (Y1 and Y2) against pairs of independent variables on the x- and y-axis, while the third variable is held constant using the central point values determined earlier. Fig. 3 makes it apparent that DH tends to increase as a consequence of interactions among the three independent variables. Fig. 3a and c present response surface plots which show mound-shaped patterns indicative of mutual interactions between temperature (X1) and enzyme (X2) and between enzyme (X2) and time (X3). By increasing the enzyme concentration used for hydrolysis, DH rises, but where an increase in time and temperature is recorded, DH rises initially before declining. Response surface (Y1) results were similar to those outlined by Hanmoungjai et al.53 and Montilha et al.54 who studied the hydrolysis of proteins using Alcalase®. Meanwhile, Fig. 3b confirms that the interaction between temperature (X1) and time (X3) influences DH in the manner of a convex plot, because as the independent variables rise, DH rises at first then falls. It is apparent that during hydrolysis of the protein, time and temperature interact together as enzyme concentration increases to produce a higher DH. Time and temperature have no effect, as the enzyme is still able to hydrolyze the protein, though time and temperature also have no adverse effects.
Fig. 3. Response surface and contour plot illustrating the influence of: (a) and (b) temperature and percentage of enzyme, (c) and (d) temperature and time, (e) and (f) percentage of enzyme and time upon DH.
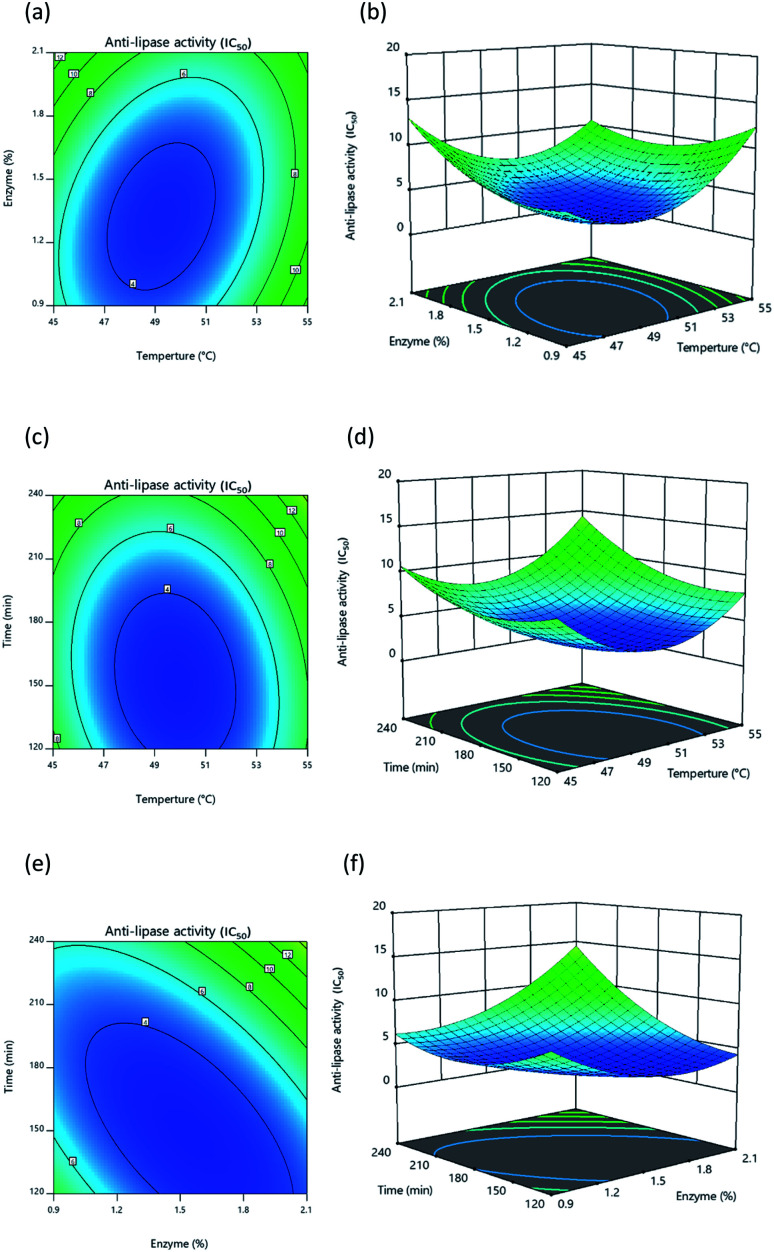
Fig. 4. Response surface and contour plot illustrating the influence of: (a) and (b) temperature and percentage of enzyme, (c) and (d) temperature and time, (e) and (f) percentage of enzyme and time upon lipase inhibition.
Once protein hydrolysates had been obtained under the 20 different conditions, they were tested to determine their lipase inhibitory activity in order to develop response surface plots, which matched those defined for optimal conditions, showing convex shapes as presented in Fig. 4a–4c. Increasing the independent variables during hydrolysis results in an initial increase of lipase inhibition followed later by a decline. It is apparent, however, that the lipase inhibition from protein hydrolysates will be affected in part by the way the three independent variables interacting with each other. In general, it is typically found that response surfaces that maximize yield provide the greatest level of biological activity; for instance, according to Ahmad et al.55 high yields of Artocarpus altilis extract also exhibit heightened DPPH scavenging activity. However, in this research, maximized protein yield did not result in the highest level of lipase inhibition, thus matching the findings of Baharuddin et al.56 It is advisable, therefore, to focus primarily upon regression models for the IC50 value indicating lipase inhibition, since this will show hydrolyzed protein size, which is a more important factor in lipase inhibition.
3.2.4. Validation test
Analysis of the three independent variables and two response variables allows optimal conditions to be determined through the use of Design Expert 11 software. To carry out this process, the DH response variable was set to be maximized while the independent variables remained within their specified ranges. The independent variables of temperature, enzyme, and duration used in RSM analysis were assessed in order to determine the degree of importance of each. This was determined by considering the differences between response variables and independent variables where a high value is indicative of greater importance for that particular variable. In the case of lipase inhibition, the RSM had an IC50 value set to be minimized, since that would be indicative of the greatest level of activity. This resulted in predicted independent values of 49.88 °C, 150.43 minutes, and 1.53% Alcalase® for a sample solution of 5% w/v. These parameters were predicted to give a DH value of 36.11% while the value for IC50 was 2.96 μg mL−1. Model validation experiments were carried out in triplicate under optimal conditions. Experimental results obtained were 35.65%, with an error of 1.29%, and 2.84 μg mL−1, with an error of 4.23%. These findings were very similar to the predicted values.
3.3. Purification of the lipase inhibitory peptides
3.3.1. Ultrafiltration
Lipase inhibition is significantly influenced by peptide molecular weight. To test this, ultrafiltration allows the enrichment of bioactive peptides and separation of the crude protein hydrolysates. In the first step, a 10 kDa filtration membrane is employed to remove the fraction >10 kDa. Then in the next steps, further membranes are used at 5 kDa, 3 kDa, and 0.65 kDa. Table 5 shows that the highest level of lipase inhibitory activity was displayed by the fraction <0.65 kDa, achieving an IC50 value of 3.5097 μg mL−1. This result matched the findings of O'Keeffe et al.57 who studied the ACE inhibition achieved by antioxidant peptides derived from whey proteins. Meanwhile, the work of Zhang et al.58 examined the lipase inhibitory activity of Chlorella pyenoidose proteins after enzymatic hydrolysis with Alcalase® and fractionation using a 5 kDa ultrafiltration membrane, which produced two fractions: >5 kDa and <5 kDa. It was found that the fraction <5 kDa produced far better pancreatic lipase inhibitory activity than the larger peptide size, as 75.73%. In another study, RBP underwent hydrolysis using trypsin before ultrafiltration using a pair of membranes at 4 kDa and 6 kDa to create three fractions of different molecular weights: <4 kDa, 4–6 kDa, and >6 kDa. The smallest fraction, <4 kDa, showed the greatest antioxidant and ACE inhibitory activity.59 This offers further evidence off the important role played by molecular weight in lipase inhibition, since smaller peptides are more easily absorbed by the human body. Accordingly, further separation was carried out on the <0.65 kDa fraction to isolate the pure peptide.
Lipase inhibition measured by IC50 values for the five fractions obtained using the ultrafiltration membranes for the DORB hydrolysate derived from Alcalase® hydrolysis.
| Molecular weight (kDa) | Lipase inhibitory activitya (IC50; μg mL−1) |
|---|---|
| Crude protein | 2.8360 ± 0.2022b |
| >10 | 35.4567 ± 0.3009e |
| 5–10 | 19.2533 ± 0.3523c |
| 3–5 | 6.2967 ± 0.2571b |
| 0.65–3 | 20.1750 ± 0.4171d |
| <0.65 | 3.5097 ± 0.1729a |
All results are presented in the form of mean ± SD and tests are performed in triplicate.
3.3.2. RP-HPLC
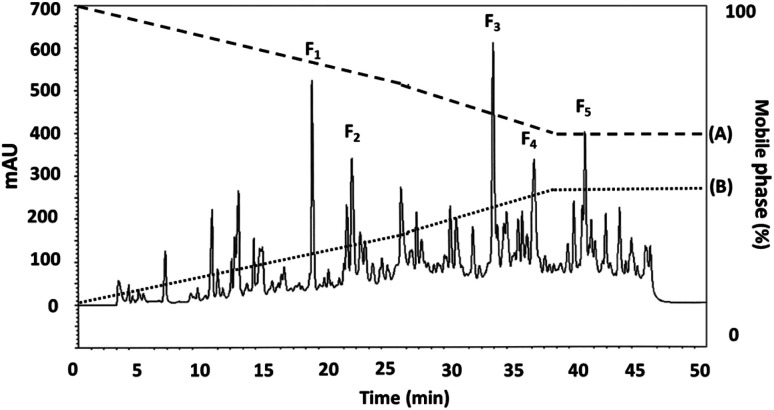
RP-HPLC offers a means of further separating smaller proteins after ultrafiltration. Where the compounds involved have differing hydrophobicity, hydrophilic peptides can initially be eluted, followed by the fractions which offer only moderate hydrophilic characteristics, while peptides which are hydrophobic, or offer non-polar attributes will be eluted last. The DORB protein fraction which proved to be most effective in lipase inhibition (0.65 kDa) was then selected for further fractionation using RP-HPLC. The resulting elution profile can be seen in Fig. 5. Taken in order, the F1 sub-fraction showed very weak lipase inhibitory activity, so the F2 sub-fraction was the first to produce a measurable IC50 value at 0.9109 μg mL−1 for lipase inhibition, having undergone elution at 21.75 minutes. The F3 sub-fraction was shown to be moderately hydrophilic with lipase inhibition at 0.0297 μg mL−1 after elution at 32.97 minutes. The F4 and F5 sub-fractions were found to be hydrophobic, with IC50 values in the range of 0.0419 and 0.0415 μg mL−1 respectively, following elution at 36.25 and 40.25 minutes. While peaks exhibited similar protein concentrations, those for F1, F3, F4, and F5 had a lower proportion displaying lipase inhibitory activity, and although protein concentrations were higher than for the other fractions presented in Table 6, so IC50 values can be calculated to equal F2. It is therefore the case that hydrophobicity affects the lipase inhibition capabilities of the peptides. These findings are confirmed by Jafar et al.,60 whose work involved whey protein peptide digested with pepsin, reporting that the separation of compounds using RP-HPLC revealed that those which were moderately hydrophobic exhibited the highest level of pancreatic lipase inhibition. In the case of the F2 sub-fraction peptides obtained from DORB hydrolysate, the gradient elution of the solvent was moderate, and the sub-fraction lay in the range of both hydrophilic and hydrophobic groups. For this reason, F2 was associated with further sequencing by LC-ESI-Q-TOF-MS/MS.
Fig. 5. The RP-HPLC profile for the DORB protein hydrolysate active fraction (<0.65 kDa) separated using eluent A: 0.1% trifluoroacetic acid (TFA), and eluent B: 70% acetonitrile.
RP-HPLC profile for the <0.65 kDa active fraction derived from DORB protein hydrolysate.
| Fraction | Retention time (min) | Lipase inhibitory activitya (IC50; μg mL−1) |
|---|---|---|
| F 1 | 18.54 | NCb |
| F 2 | 21.75 | 0.9109 ± 0.0272 |
| F 3 | 32.97 | NC |
| F 4 | 36.25 | NC |
| F 5 | 40.25 | NC |
All results are presented in the form of mean ± SD and tests are performed in triplicate.
NC – not calculable.
3.4. Identification of peptides and their properties in F2 sub-fractions using quadrupole time-of-flight (Q-TOF) liquid chromatography-tandem mass spectrometry (LC-MS/MS)
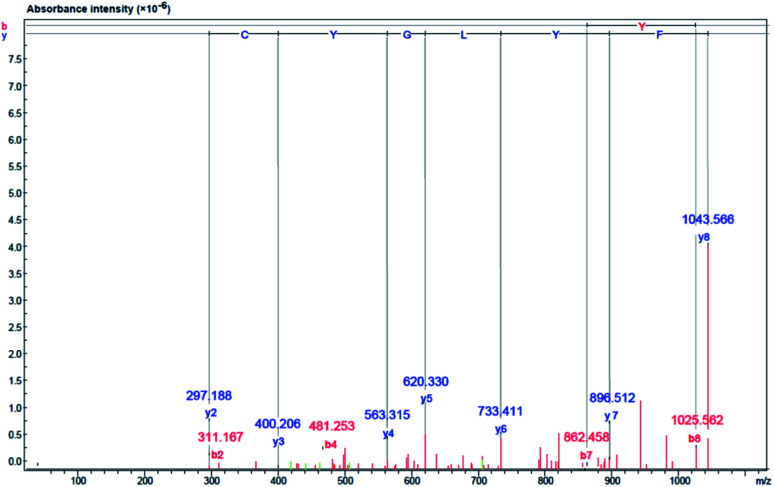
The amino acid sequences of the lipase inhibitory peptide in the RP-HPLC fraction (F2) were identified using LC-ESI-Q-TOF-MS/MS de novo sequencing as Phe–Tyr–Leu–Gly–Tyr–Cys–Asp–Tyr (FYLGYCDY), as shown in Fig. 6. Molecular weight was determined as 1043.15 Da. The homologous region for FYLGYCDY was sought through the NCBI GenBank on the basis of the Oryzeae genus, which offered a similar sequence to the hypothesized protein EE612_044279 of O. sativa (accession number KAB8108513.1), U-box domain-containing protein 19 of O. sativa Japonica (accession number XP_015649021.1), and hypothetical protein OsI_29238 of O. sativa Indica (accession number EAZ06993.1). Database sequences had an E-value of 6.6, sequence score of 23.10%, with 63% equal identities (7/11). The FYLGYCDY peptide underwent synthesis as the database specified, resulting in production of the purified peptide. This synthesized peptide had an IC50 value of 0.47 ± 0.02 μM. The Innovagen server provides data indicating that the FYLGYCDY peptide offered solubility lower than 0.10 mg mL−1, attaining the categorization of ‘undissolved’, so the solution for the peptide will be less than 10 mg mL−1 of gross peptide concentration in dimethyl sulfoxide (DMSO). Data can be seen in Table 7. The key properties of lipase inhibitory peptides include chain length and hydrophobicity; after examination of a large number of peptide sequences, it is clear that the best inhibitors of lipase offer short chains comprising just 2–16 amino acids. It is easier for these short peptides to enter the enzyme active site.61 It is also easier for such short peptides to enter the bloodstream without any alteration to their general properties.62
Fig. 6. Identification of amino acid sequence and lipase inhibitor peptide molecular mass following purification of DORB hydrolysate (F2 from RP-HPLC). Mass fragmentation spectrum of FYLGYCDY peptide.
FYLGYCDY peptide properties.
| Synthesized peptides | Solubility in watera | Hydrophobicityb (%) | Potential biological activityd | Toxicity prfile (SVM score)c | Sensory characteristics |
|---|---|---|---|---|---|
| FYLGYCDY | Poor | 25.0 | GY (ACE inhibitor and DPP IV inhibitor), FY (ACE inhibitor), LG (ACE inhibitor), DY (ACE inhibitor), YL (neuropeptide, DPP IV inhibitor and DPP-III inhibitor), LGY (immune-stimulating), LGY (regulating), YLGY (ACE inhibitor and antioxidative) | Non-toxic (−0.44) | GY (bitter), LG (bitter), FY (bitter), DY (bitter), YL (bitter) |
Solubility data for the peptide comes from the Innovagen server (http://www.innovagen.com/proteomics-tools).
Peptide properties were calculated using the Peptide 2.0 server (http://www.peptide2.com).
Analysis of the peptide toxicity was performed using the ToxinPred server (http://crdd.osdd.net/raghava/toxinpred/).
Sensory attributes and potential biological properties of the peptide are obtained from the BIOPEP database (http://www.uwm.edu.pl/biochemia/index.php/en/biopep).
The BIOPEP database (http://www.uwm.edu.pl/biochemia/index.php/en/biopep) contains data on the properties of many of the peptides which are understood to offer potentially useful bioactive properties. To determine the likely properties of the FYLGYCDY peptide it is possible to examine similar sequences in the database, which are shown in Table 7. The investigated peptide sequences which showed bioactivity were GY (location 4–5), FY (location 1–2), LG (location 3–4), DY (location 7–8), and YLGY (location 2–5), all demonstrating inhibitory qualities. The peptide which serves to regulate the flow of ions was DY (location 7–8), while the peptide controlling the phosphoinositol metabolism was YLGY (location 2–5). Meanwhile, YL (location 2–3) is a neuropeptide of anxiolytic, a dipeptidyl peptidase IV inhibitor (DPP IV inhibitor), and a DPP-III inhibitor, while GY (location 4–5) is also a dipeptidyl peptidase IV inhibitor. LGY (location 3–5) demonstrated immunostimulatory activity while YLGY (location 2–5) showed antioxidative activity. There are certain concerns which remain in the context of using peptides to develop therapeutic products or functional foods, and one is the issue of toxicity.63 FYLGYCDY peptide, however, was obtained from plant-based proteins, and it is possible to evaluate its toxicity using the ToxinPred server (http://crdd.osdd.net/raghava/toxinpred/), which allows for predictions to be made which suggest that FYLGYCDY is not toxic and has SVM scores below zero, which can be seen in Table 7. It is therefore feasible for this lipase inhibitory peptide to undergo further development, but evaluations must be carried out both in vitro and in vivo prior to human use, in order to ensure that toxicity problems are avoided. Another important consideration for peptides and amino acids used as food ingredients is that of sensory properties, and in particular taste, which is the consequence of certain chemical structures. The sensory attributes of peptides can be predicted using data from the BIOPEP database by applying the procedures outlined by Iwaniak et al.64 It can be concluded that the taste of the FYLGYCDY peptide would be bitter, as Table 7 indicates.
3.5. Inhibition kinetic of the lipase inhibitory peptide
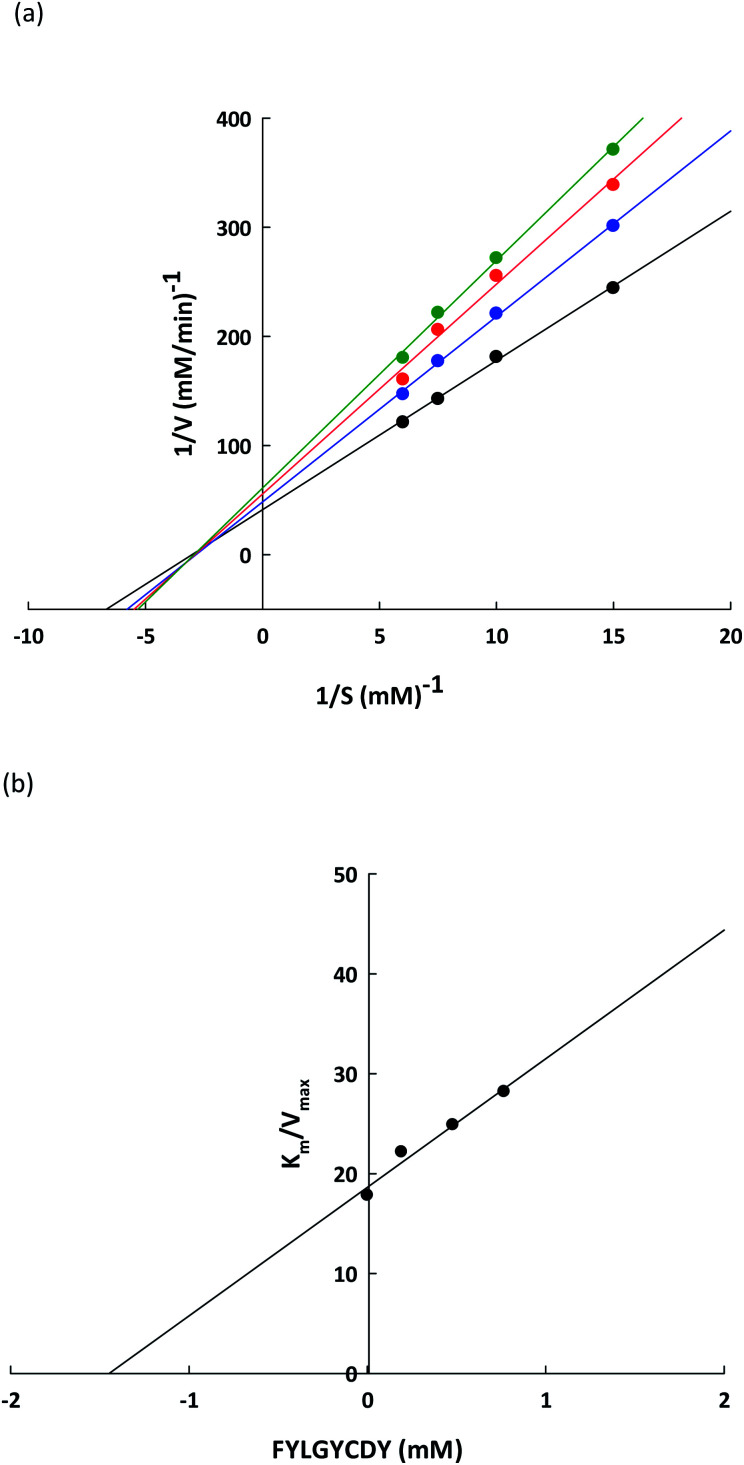
To verify the lipase inhibitory mechanism, a Lineweaver–Burk plot of the FYLGYCDY peptide derived from the DORB protein was analyzed. Analysis of the inhibition kinetics employed varying p-NPP concentrations as a substrate along with a fixed amount and concentration of the peptide. Fig. 7a shows that this peptide produces generally non-competitive inhibition. The four straight lines obtained had an intersection at one point on the 1/S axis and the value for Km was 0.6229 ± 0.1170 mM, while maximum velocity (Vmax) for the differing peptide concentrations along with the slopes of the straight lines were different. This occurred because an increase in peptide concentration resulted in a decline in the enzymatic reaction efficiency of the substrate. This is apparent in Table 8. Meanwhile, Ki represents the inhibition constant, which can be seen in Fig. 7b, revealing the bond strength between lipase and peptide, whereby a lower value is associated with higher bond strength. For FYLGYCDY, the value for Ki was 1.4493 ± 0.0961 mM, which is notable in peptides exhibiting non-competitive inhibition. The FYLGYCDY peptide therefore works by lowering the number of functional enzymes capable of reacting with the substrate. These findings are similar to those of Liu et al.,52 whose work on fish protein hydrolysates showed that the PPL inhibition mechanism was of a non-competitive inhibitor type, thus limiting the ability of the enzyme to generate efficient catalysis. It has also been reported that another non-competitive inhibitor for PPL is astaxanthin, which is able to block the channel to the enzyme catalytic site, thus delaying entrance to the substrate.
Fig. 7. (a) Lineweaver–Burk plots for the FYLGYCDY lipase inhibitor obtained from DORB protein hydrolysates. Reactions were carried out at varying inhibitor concentrations: no inhibitor (●), 0.2 mM of peptide ( ), 0.5 mM of peptide (
), 0.5 mM of peptide ( ), and 0.8 mM of peptide (
), and 0.8 mM of peptide ( ). 1/V and 1/[S] are reciprocals of velocity and substrate. (b) Dixon plot measuring the inhibitor constant (Ki) for the FYLGYCDY peptide.
). 1/V and 1/[S] are reciprocals of velocity and substrate. (b) Dixon plot measuring the inhibitor constant (Ki) for the FYLGYCDY peptide.
Kinetics parameters for PPL-catalyzed reactions at different peptide concentrationsa.
| Kinetic parameter | Control | FYLGYCDY (mM) | ||
|---|---|---|---|---|
| 0.2 | 0.5 | 0.8 | ||
| K M (mM) | 0.6229 ± 0.1170 | 0.6229 ± 0.1170 | ||
| V max (mM min−1) | 0.0352 ± 0.0039 | 0.0282 ± 0.0023 | 0.0252 ± 0.0030 | 0.0222 ± 0.0027 |
| K i (mM) | 1.4493 ± 0.0961 | |||
All results are presented in the form of mean ± SD and tests are performed in triplicate.
3.6. FYLGYCDY molecular docking at the lipase binding site
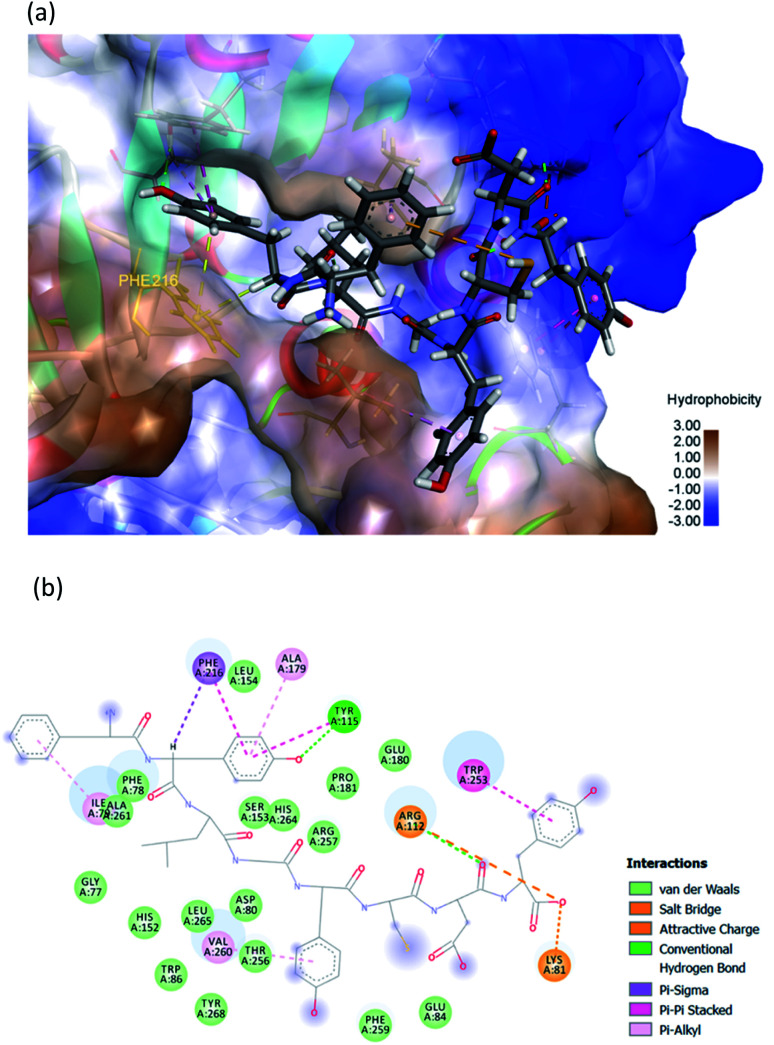
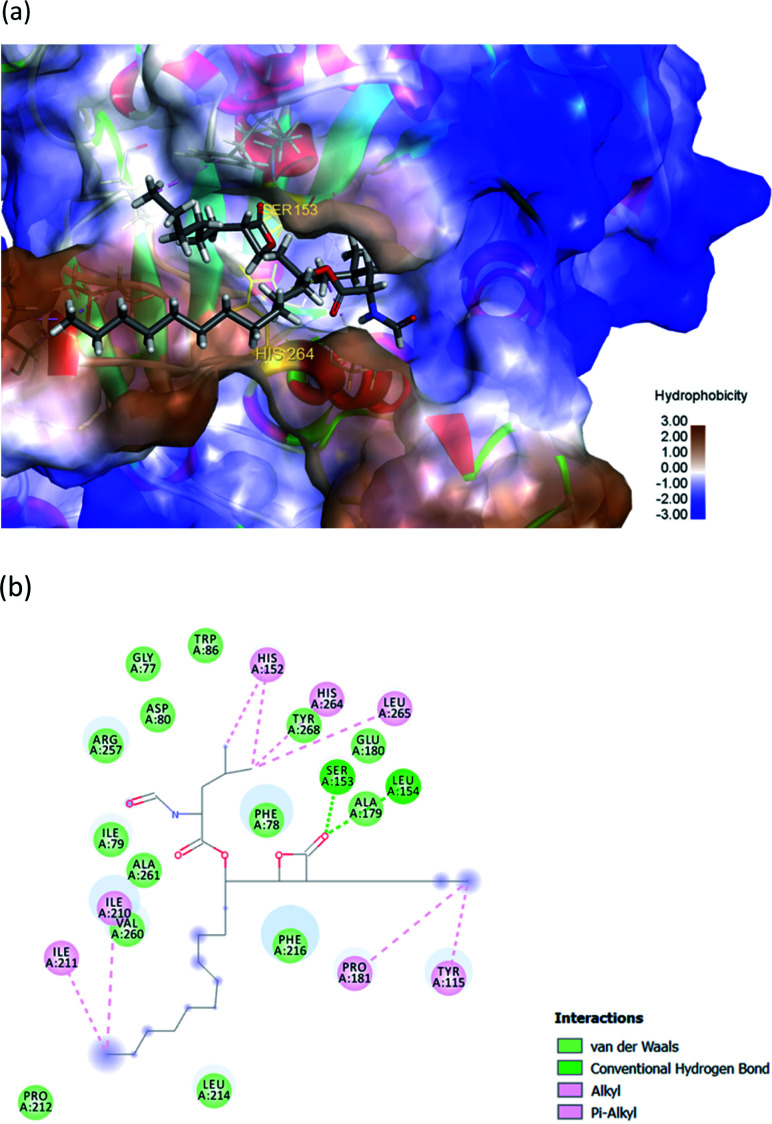
The hydrophobic peptide FYLGYCDY underwent docking with the crystal PPL structure (PDB: 1ETH) using GOLD 5.7.1 software to examine the interactions of the peptide (Fig. 8) binding with lipase and draw comparisons with orlistat which served as a positive control (Fig. 9). This process is shown in Fig. 8, which reveals that the Tyr amino acid, categorized as hydrophobic and located in position two of the inhibitor peptide, is capable of binding to the Phe216 of PPL.65 The hydrogen which forms part of Tyr2 serves as the donor atom, and Phe216 of PPL is the acceptor atom in the context of Pi–Sigma interaction. Phe216 acted as the substrate-binding site of the lipase, while Ser153, Asp177, and His264 acted as the PPL catalytic sites, whereby the inhibitor peptide did not bind to those catalytic sites but instead was under the influence of van der Waals forces. Table 9 shows this particular competitive type of molecular docking inhibition in the case of this peptide. It has been shown that hydrophobic peptides derived from the enzymatic hydrolysis of C. pryenoidose,58 with a greater molecular weight than the peptide which is the focus of this study, may bind to the PPL catalytic sites as a result of Pi-cation and conventional hydrogen bond interaction. Fig. 9 shows the properties of orlistat, which is designed to inhibit PPL as a mainly competitive inhibitor, when it binds to PPL at the principal catalytic sites, whereby Ser153 and His264 of the PPL complex by H-donor of Ser153 bind to the β-lactone ring of orlistat, while His264 binds to the side-chain hydrogen bond of the amino ester of orlistat, respectively binding through a conventional hydrogen bond and Pi–alkyl interaction.64 The evaluation was based on the different scoring functions for docking, the best score for which came from ChemPLP. The inhibitor peptide had a docking score of 122.54 for binding on PPL while orlistat achieved a docking score of 101.44. It therefore appears that the peptide would be able to inhibit the PPL function more effectively than orlistat.
Fig. 8. (a) 3D diagram of expected interactions between FYLGYCDY and the PPL complex. (b) 2D diagram of expected interactions between FYLGYCDY and the PPL molecule. Images produced using Discovery Studio 2019 software.
Fig. 9. (a) 3D diagram of expected interactions between orlistat and the PPL complex. (b) 2D diagram of expected interactions between orlistat and the PPL molecule. Images produced using Discovery Studio 2019 software.
Potential sites for binding between the FYLGYCDY bioactive peptide and PPL based on Discovery Studio 2019 software.
| Amino acid | Category | Donor atom | Acceptor atom | Interaction | Distance (Å) |
|---|---|---|---|---|---|
| Phe1 | Hydrophobic | Phe1 (Pi-orbitals) | Ile79 (alkyl) | Pi–alkyl | 3.9113 |
| Tyr2 | Hydrophobic | Tyr2 (Pi-orbitals) | Ala179 (alkyl) | Pi–alkyl | 5.0570 |
| Hydrophobic | Tyr2 (Pi-orbitals) | Phe216 (Pi-orbitals) | Pi–Pi stacked | 4.5563 | |
| Hydrophobic | Tyr2 (Pi-orbitals) | Try115 (Pi-orbitals) | Pi–Pi stacked | 4.4097 | |
| Hydrophobic | Tyr2 (C–H) | Phe216 (Pi-orbitals) | Pi–sigma | 2.7882 | |
| Hydrogen bond | Tyr115 (H-donor) | Tyr2 (H-acceptor) | Conventional hydrogen bond | 2.0815 | |
| Try5 | Hydrophobic | Try5 (Pi-orbitals) | Val260 (alkyl) | Pi–alkyl | 5.2900 |
| Asp7 | Hydrogen bond | Arg112 (H-donor) | Asp7 (H-acceptor) | Conventional hydrogen bond | 2.0172 |
| Try8 | Hydrogen bond | Try8 (Pi-orbitals) | Trp253 (Pi-orbitals) | Pi–Pi stacked | 3.9038 |
| Electrostatic | Arg112 (positive) | Try8 (negative) | Attractive charge | 3.8041 | |
| Hydrogen bond | Lys81 (H-donor; positive) | Try8 (H-acceptor; negative) | Salt bridge; attractive charge | 1.7087 | |
| Hydrophobic | Lrp253 (Pi-orbitals) | Try8 (Pi-orbitals) | Pi–Pi stacked | 4.1226 |
4. Conclusions
This study successfully identified a peptide which offered lipase inhibitory activity and which could be obtained from DORB proteins through the use of Alcalase®. In order to optimize the influence of temperature, time, and enzyme upon the degree of hydrolysis and lipase inhibitory activity as indicated by IC50 values, RSM was employed with CCD. To achieve the greatest level of lipase inhibitory activity from the hydrolysate, the necessary conditions were 49.88 °C, 150.43 minutes, and 1.53% enzyme for sample solutions of 5% w/v. Under these conditions, the DH value was 35.65%, and the IC50 value was 2.84 μg mL−1. The hydrolysate which offered the best performance in terms of lipase inhibition then underwent fractionation to obtain fractions bound by different molecular weight cut-offs, with the MW < 0.65 kDa proving to be the most effective inhibitor of lipase, having achieved an IC50 value of 3.51 μg mL−1. Following fractionation using ultrafiltration techniques, as well as purification using RP-HPLC, and peptide identification via LC-MS/MS, the lipase inhibitory peptide Phe–Tyr–Leu–Gly–Tyr–Cys–Asp–Tyr (FYLGYCDY) was identified, with an IC50 value of 0.47 μM and a molecular weight of 1043.15 Da. The peptide FYLGYCDY was shown to have the capacity to inhibit pancreatic lipase, and according to Lineweaver–Burk plots is a non-competitive inhibitor. However, molecular docking examination which aimed to predict the binding site for the peptide FYLGYCDY with the PPL complex showed a competitive binding type. From the results obtained, the peptide demonstrated the capacity to block the channel to the enzyme catalytic site, thus delaying the entrance of the substrate. The lipase inhibitory capacity of the DORB protein hydrolysate can function as a bioactive peptide, especially to serve as anti-obesity agents. This peptide, however, should also undergo further study in terms of its cytotoxicity and allergenicity in order that it might in future be used in the food or pharmaceutical sectors.
Funding
Sincere thanks are expressed to we acknowledge the financial support from the grant for research: the Research Assistantship Fund, Faculty of Science, Chulalongkorn University (RAF_2562_003), the Thailand Science Research and Innovation (TSRI) Fund (CU_FRB640001_01_61_2), and the Center of Excellence on Medical Biotechnology (CEMB), S&T Postgraduate Education and Research Development Office (PERDO), Office of Higher Education Commission (OHEC), Thailand (SN-63-009-01) for providing the financial support for this research in recognition of the financial assistance offered which enabled this research study to be completed.
Conflicts of interest
The authors have no conflict of interest. This publication has not been submitted earlier in any journal and is not being considered for publication elsewhere. All of the authors, including the corresponding authors, have read and approved the final submitted manuscript.
Supplementary Material
Acknowledgments
The researchers are also grateful to the Institute of Biotechnology and Genetic Engineering at Chulalongkorn University for the facilities which they made available during the course of the study. The authors were grateful to Dr Benjamin Richard Poole, Department of English, Faculty of Art, Chulalongkorn University for reviewing this manuscript.
References
- Hurt R. T. Kulisek C. Buchanan L. A. McClave S. A. Gastroenterol. Hepatol. 2010;6:780–792. [PMC free article] [PubMed] [Google Scholar]
- Mitchell N. S. Catenacci V. A. Wyatt H. R. Hill J. O. Psychiatr. Clin. 2011;34:717–732. doi: 10.1016/j.psc.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi-Sunyer X. Postgrad. Med. 2009;121:21–33. doi: 10.3810/pgm.2009.11.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Melean C. M. Somers V. K. Rodriguez-Escudero J. P. Singh P. Sochor O. Llano E. M. Lopez-Jimenez F. Curr. Atheroscler. Rep. 2013;15:364. doi: 10.1007/s11883-013-0364-2. [DOI] [PubMed] [Google Scholar]
- Smith K. B. Smith M. S. Prim. Care. 2016;43:121–135. doi: 10.1016/j.pop.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Birari R. B. Bhutani K. K. Drug Discov. Today. 2007;12:879–889. doi: 10.1016/j.drudis.2007.07.024. [DOI] [PubMed] [Google Scholar]
- Yun J. W. Phytochemistry. 2010;71:1625–1641. doi: 10.1016/j.phytochem.2010.07.011. [DOI] [PubMed] [Google Scholar]
- de la Garza A. L. Milagro F. I. Boque N. Campión J. Martínez J. A. Planta Med. 2011;77:773–785. doi: 10.1055/s-0030-1270924. [DOI] [PubMed] [Google Scholar]
- Grove K. A. Sae-tan S. Kennett M. J. Lambert J. D. Obesity. 2012;20:2311–2313. doi: 10.1038/oby.2011.139. [DOI] [PubMed] [Google Scholar]
- Uchiyama S. Taniguchi Y. Saka A. Yoshida A. Yajima H. Nutrition. 2011;27:287–292. doi: 10.1016/j.nut.2010.01.019. [DOI] [PubMed] [Google Scholar]
- Ballinger A. Peikin S. R. Eur. J. Pharmacol. 2002;440:109–117. doi: 10.1016/S0014-2999(02)01422-X. [DOI] [PubMed] [Google Scholar]
- López-Barrios L. Gutiérrez-Uribe J. A. Serna-Saldívar S. O. J. Food Sci. 2014;79:R273–R283. doi: 10.1111/1750-3841.12365. [DOI] [PubMed] [Google Scholar]
- Saadi S. Saari N. Anwar F. Abdul Hamid A. Ghazali H. M. Biotechnol. Adv. 2015;33:80–116. doi: 10.1016/j.biotechadv.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Nasri M. Adv. Food Nutr. Res. 2017;81:109–159. doi: 10.1016/bs.afnr.2016.10.003. [DOI] [PubMed] [Google Scholar]
- Wang Y. Huang Q. Kon D. Xu P. Mini Rev. Med. Chem. 2018;18:1524–1535. doi: 10.2174/1389557518666180424110754. [DOI] [PubMed] [Google Scholar]
- Li G. H. Qu M. R. Wan J. Z. You J. M. Asia Pac. J. Clin. Nutr. 2007;16:275–280. [PubMed] [Google Scholar]
- Zhuang H. Tang N. Yuan Y. J. Funct. Foods. 2013;5:1810–1821. doi: 10.1016/j.jff.2013.08.013. [DOI] [Google Scholar]
- Zhong F. Zhang X. Ma J. Shoemaker C. F. Food Res. Int. 2007;40:756–762. doi: 10.1016/j.foodres.2007.01.005. [DOI] [Google Scholar]
- Zhang M. Mu T. H. Sun M. J. J. Funct. Foods. 2014;7:191–200. doi: 10.1016/j.jff.2014.02.012. [DOI] [Google Scholar]
- Kheeree N. Sangtanoo P. Srimongkol P. Saisavoey T. Reamtong O. Choowongkomon K. Karnchanatat A. Food Funct. 2020;11:8161–8178. doi: 10.1039/D0FO01240H. [DOI] [PubMed] [Google Scholar]
- Saisavoey T. Sangtanoo P. Reamtong O. Karnchanatat A. J. Food Biochem. 2016;40:731–740. doi: 10.1111/jfbc.12266. [DOI] [Google Scholar]
- Wouters A. G. B. Rombouts I. Fierens E. Brijs K. Delcour J. A. Compr. Rev. Food Sci. Food Saf. 2016;15:786–800. doi: 10.1111/1541-4337.12209. [DOI] [PubMed] [Google Scholar]
- Gul K. Yousuf B. Singh A. K. Singh P. Wani A. A. Bioact. Carbohydr. Diet. Fibre. 2015;6:24–30. doi: 10.1016/j.bcdf.2015.06.002. [DOI] [Google Scholar]
- Park H. Y. Lee K. W. Choi H. D. Food Funct. 2017;22:935–943. doi: 10.1039/C6FO01763K. [DOI] [PubMed] [Google Scholar]
- Sohail M. Rakha A. Butt M. S. Iqbal M. J. Rashid S. Crit. Rev. Food Sci. Nutr. 2017;22:3771–3780. doi: 10.1080/10408398.2016.1164120. [DOI] [PubMed] [Google Scholar]
- Liu Y. Q. Strappe P. Shang W. T. Zhou Z. K. Crit. Rev. Food Sci. Nutr. 2019;59:349–356. doi: 10.1080/10408398.2017.1374923. [DOI] [PubMed] [Google Scholar]
- Tang S. Hettiarachchy N. S. Shellhammer T. H. J. Agric. Food Chem. 2002;50:7444–7448. doi: 10.1021/jf025771w. [DOI] [PubMed] [Google Scholar]
- Fabian C. Ju Y. H. Crit. Rev. Food Sci. Nutr. 2011;51:816–827. doi: 10.1080/10408398.2010.482678. [DOI] [PubMed] [Google Scholar]
- Official Methods of Analysis of AOAC International. 18th ed. Methods 925.09 and 926.08, AOAC International, Gaithersburg, MD, USA, 2005 [Google Scholar]
- Official Methods of Analysis of AOAC International. 18th ed. Methods 2003.06, AOAC International, Gaithersburg, MD, USA, 2005 [Google Scholar]
- Official Methods of Analysis of AOAC International. 18th ed. Methods 923.03, AOAC International, Gaithersburg, MD, USA, 2005 [Google Scholar]
- Official Methods of Analysis of AOAC International. 18th ed. Methods 968.06 and 992.15, AOAC International, Gaithersburg, MD, USA, 2005 [Google Scholar]
- Official Methods of Analysis of AOAC International. 18th ed. Methods 962.09, AOAC International, Gaithersburg, MD, USA, 2005 [Google Scholar]
- Crisan E. V. and Sands A., Nutritional Value. Academic Press, New York, 1978, pp. 137–168 [Google Scholar]
- Nielsen P. Petersen D. Dambmann C. J. Food Sci. 2001;66:642–646. doi: 10.1111/j.1365-2621.2001.tb04614.x. [DOI] [Google Scholar]
- Adisakwattana S. Moonrat J. Srichairat S. Chanasit C. Tirapongporn H. Chanathong B. Ngamukote S. Mauml K. Sapwarobol S. J. Med. Plants Res. 2010;4:2113–2120. [Google Scholar]
- Bradford M. M. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Hermoso J. Pignol D. Kerfelec B. Crenon I. Chapus C. Fontecilla-Camps J. C. J. Biol. Chem. 1996;271:18007–18016. doi: 10.1074/jbc.271.30.18007. [DOI] [PubMed] [Google Scholar]
- Genkawa T. Uchino T. Inoue A. Tanaka F. Hamanaka D. Biosyst. Eng. 2008;99:515–522. doi: 10.1016/j.biosystemseng.2007.12.011. [DOI] [Google Scholar]
- Patel S. J. Funct. Foods. 2015;14:255–269. doi: 10.1016/j.jff.2015.02.010. [DOI] [Google Scholar]
- Edeogu C. O. Ezeonu F. C. Okaka A. N. C. Ekuma C. E. Elom S. O. Int. J. Biotechnol. Biochem. 2007;3:1–8. doi: 10.3844/ajbbsp.2007.1.9. [DOI] [Google Scholar]
- Kalpanadevi C. Singh V. Subramanian R. J. Food Sci. Technol. 2018;55:2259–2269. doi: 10.1007/s13197-018-3143-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd B. J. Siebenmorgen T. J. Beers K. W. Cereal Chem. 2000;77:551–555. doi: 10.1094/CCHEM.2000.77.5.551. [DOI] [Google Scholar]
- Yamagishi T. Tsuboi T. Kikuchi K. Cereal Chem. 2003;80:5–8. doi: 10.1094/CCHEM.2003.80.1.5. [DOI] [Google Scholar]
- Shi L. Int. J. Biol. Macromol. 2016;92:37–48. doi: 10.1016/j.ijbiomac.2016.06.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amagliani L. O'Regan J. Kelly A. L. O'Mahony J. A. Trends Food Sci. Technol. 2017;64:1–12. doi: 10.1016/j.tifs.2017.01.008. [DOI] [Google Scholar]
- Sereewatthanawut I. Prapintip S. Watchiraruji K. Goto M. Sasaki M. Shotipruk A. Bioresour. Technol. 2008;99:555–561. doi: 10.1016/j.biortech.2006.12.030. [DOI] [PubMed] [Google Scholar]
- Fabian C. Ju Y. H. Crit. Rev. Food Sci. Nutr. 2011;51:816–827. doi: 10.1080/10408398.2010.482678. [DOI] [PubMed] [Google Scholar]
- Piotrowicz I. B. B. Garcés-Rimón M. Moreno-Fernández S. Aleixandre A. Salas-Mellado M. Miguel-Castro M. Foods. 2020;9:812. doi: 10.3390/foods9060812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic I. Bossink B. A. G. van der Sijde P. C. Sustainability. 2019;11:7197. doi: 10.3390/su11247197. [DOI] [Google Scholar]
- Gao D. Zhang F. Ma Z. Chen S. Ding G. Tian X. Feng R. Int. J. Food Prop. 2019;22:1296–1309. doi: 10.1080/10942912.2019.1640735. [DOI] [Google Scholar]
- Liu L. Wang Y. Peng C. Wang J. Int. J. Mol. Sci. 2013;14:3124–3139. doi: 10.3390/ijms14023124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanmoungjai P. Pyle D. L. Niranjan K. J. Am. Oil Chem. Soc. 2001;78:817–821. doi: 10.1007/s11746-001-0348-2. [DOI] [Google Scholar]
- Montilha M. Sbroggio M. Figueiredo V. Ida E. Kurozawa L. Int. Food Res. J. 2007;24:1067–1074. [Google Scholar]
- Ahmad M. N. Karim N. U. Normaya E. Piah B. M. Iqbal A. Bulat K. H. K. Sci. Rep. 2020;10:1–14. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharuddin N. A. Halim N. R. A. Sarbon N. M. Int. Food Res. J. 2016;23:1424–1431. [Google Scholar]
- O'Keeffe M. B. Conesa C. FitzGerald R. J. Int. J. Food Sci. Technol. 2017;52:1751–1759. doi: 10.1111/ijfs.13448. [DOI] [Google Scholar]
- Zhang R. Chen J. Mao X. Qi P. Zhang X. Molecules. 2019;24:3527. doi: 10.3390/molecules24193527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Chen H. Fu X. Li S. Wei J. LWT–Food Sci. Technol. 2017;75:93–99. doi: 10.1016/j.lwt.2016.08.047. [DOI] [Google Scholar]
- Jafar S. Kamal H. Mudgil P. Hassan H. M. Maqsood S. LWT–Food Sci. Technol. 2018;98:212–218. doi: 10.1016/j.lwt.2018.08.024. [DOI] [Google Scholar]
- Mudgil P. Kamal H. Yuen G. C. Maqsood S. Food Chem. 2018;259:46–54. doi: 10.1016/j.foodchem.2018.03.082. [DOI] [PubMed] [Google Scholar]
- Aluko R. E. Annu. Rev. Food Sci. Technol. 2015;6:235–262. doi: 10.1146/annurev-food-022814-015520. [DOI] [PubMed] [Google Scholar]
- Gupta S. Kapoor P. Chaudhary K. Gautam A. Kumar R. Raghava G. P. S. PLoS One. 2013;8:e73957. doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaniak A. Minkiewicz P. Darewicz M. Sieniawski K. Starowicz P. Food Res. Int. 2016;85:155–161. doi: 10.1016/j.foodres.2016.04.031. [DOI] [PubMed] [Google Scholar]
- Hermoso J. Pignol D. Kerfelec B. Crenon I. Chapus C. Fontecilla-Camps J. C. J. Biol. Chem. 1996;271:18007–18016. doi: 10.1074/jbc.271.30.18007. [DOI] [PubMed] [Google Scholar]