Abstract
As a class of enzymes, esterases have been investigated for decades and have found use in industrial processes, synthetic organic chemistry, and elsewhere. Esters are functional groups composed of an alcohol moiety and a carboxylic acid moiety. Although much work has explored the influence of the carboxyl moiety of an ester on its susceptibility to esterases, little work has explored the influence of the alcohol moiety. Here, we describe an in vitro methodology to explore the influence of changing the alcohol moiety of an ester on its enzymatic hydrolysis, including strategies for analyzing such data. We then describe leveraging data from these assays to develop targeted antimicrobial prodrugs that activate in certain species due to the discriminatory activity of species-specific esterases. We envisage the potential of genomics and machine learning to further these efforts. Finally, we anticipate the potential future uses of these ideas, including developing targeted anti-cancer compounds.
Keywords: antimicrobial compounds, data visualization, esterases, genomics, prodrugs, targeted therapeutics
1. INTRODUCTION
Esterases (EC 3.1.1.1) catalyze the hydrolysis of an ester bond to form a carboxylic acid and an alcohol. These enzymes are one of the many subgroups of α,β-hydrolase-fold enzymes (Satoh and Hosokawa, 2006). Found in animals, plants, and microorganisms, esterases likely evolved to detoxify xenobiotic compounds but have found applications ranging from laboratory-scale organic syntheses to industrial-scale processing of fats and oils to enabling chiral syntheses of active pharmaceutical ingredients (Panda and Gowrishankar, 2005). Their substrate promiscuity, though useful, is poorly understood and remains an area of active interest (Devamani et al., 2016; Martínez-Martínez et al., 2018).
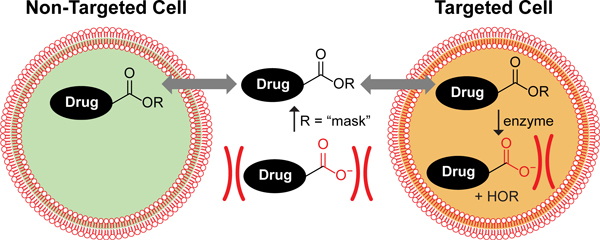
Beyond industrial and synthetic applications, understanding the substrate promiscuity of human carboxylesterases is especially important because of their activation of ester prodrugs. The ester prodrug strategy (Figure 1), in which a poorly bioavailable pharmacophore containing a charged carboxylate moiety is converted to an uncharged, potentially more bioavailable ester moiety, is the most common of prodrug strategies (Testa and Mayer, 2003; Lavis, 2008). The strategy has been applied to many drugs, including the antiviral oseltamivir (Tamiflu®), the antihypertensive enalapril (Vasotec®), and the thrombin inhibitor dabigatran etexilate (Pradaxa®), among others (Huttunen et al., 2011). The oseltamivir ester provides an interesting case study of ester prodrugs: the drug candidate was changed from a carboxylate drug to an ester prodrug near the end of its development, as the permeability was deemed to be too low. Oseltamivir contains a simple ethyl ester. That prodrug approach is common because many ethyl esters are readily cleaved by human carboxylesterase 1, and cleavage produces a nontoxic product, ethanol (Rautio et al., 2017). Intriguingly, however, later studies demonstrated oseltamivir is not cleaved by some variants of human carboxylesterase 1 that are present in the population, rendering the prodrug ineffective in some individuals (Zhu and Markowitz, 2009).
Figure 1.
The ester prodrug concept. Carboxylate-containing drugs are poorly permeable due to Coulombic repulsion. Masking the negative charge makes the drug more readily permeable in a nonspecific fashion. The active drug is revealed only in cells that contain an enzyme capable of hydrolyzing the ester bond to reveal the active moiety. In a targeted prodrug strategy, only cells that contain such an esterase would incur the effects of the drug.
The oseltamivir story intrigued us because ethyl esters were generally viewed as certain to be cleaved in human cells. The resistance of an ethyl ester to hydrolysis when faced with a naturally occurring variant of human carboxylesterase 1 caused us to wonder whether the alcohol moiety of the ester could significantly impact the ability of esterases to act on an ester prodrug. We learned that this impact had not been characterized thoroughly. Moreover, whereas many esterase probes are known that enable high-throughput screening of esterase activity, the vast majority rely on a phenolic chromophore that fluoresces upon hydrolysis from a variable acyl group (Lavis et al., 2006; Chyan and Raines, 2018). In these probes, the alcohol portion cannot be altered without impinging upon the fluorescent response. Prior to our work, few probes had been described wherein the acyl group is the nascent chromophore and the alcohol group is variable (Janes et al., 1998; Ueno et al., 2004; Tremblay et al., 2007; Kim et al., 2015).
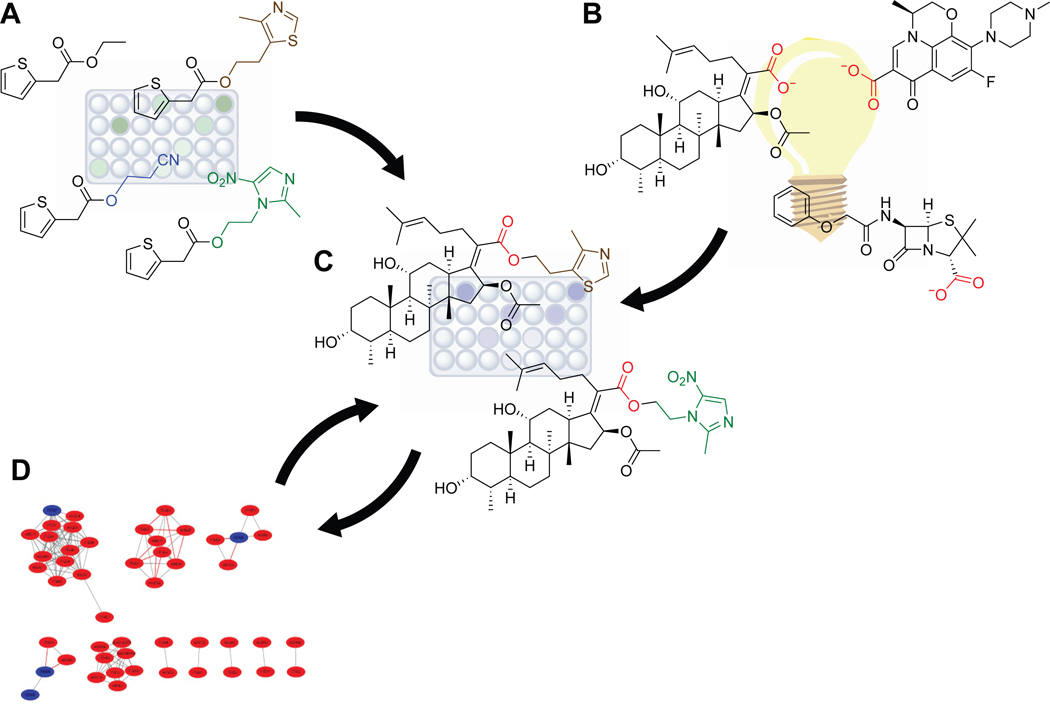
The idea of exploring how changes of the alcohol moiety in an ester prodrug could modulate esterase response across esterase types intrigued us, and we further wondered if knowledge of esterase cleavage patterns in different mammalian cell types or bacterial species could enable the creation of targeted ester prodrugs wherein the ester itself was responsible for the specificity of the prodrug, as shown in Figure 1. We reasoned that if we could establish esterase cleavage patterns of a given acyl group with varied alcohol moieties in vitro, we could then use that information to create ester prodrugs and test their impact in vivo on the desired cells. Furthermore, if we could establish through additional studies which esterases are responsible for the observed ester hydrolysis, then we could use genome sequences and bioinformatic analyses to predict what other organisms or cell types might cleave the developed esters and therefore be susceptible to the designed prodrugs. We saw these approaches as mutually supportive (Figure 2).
Figure 2.
Scheme for the development of targeted ester prodrugs. (A) The in vitro 2TA–terbium luminescence assay is used to identify alcohol moieties that modulate esterase activity in desired cell lysates. (B) Candidate carboxylate compounds are unearthed through literature mining, machine learning, or other approaches. (C) Carboxylate compounds are condensed with the alcohol compounds of interest and their activity against target and off-target cell lines assessed using a viability assay. (D) Genomic and bioinformatic analyses are used to determine which esterase(s) present in the target strain(s) are responsible for the observed cleavage. These data further suggest which strains are susceptible to the compounds and which strains are not. Additional viability assays can then be employed to confirm conclusions.
With this framework in mind, we first developed an analytical assay that allowed for the in vitro interrogation of esterase cleavage of a select acyl group with a varied alcohol moiety (Hetrick et al., 2019). Employing this methodology, we saw marked differences in cleavage patterns among Bacillus subtilis, Mycolycibacterium smegmatis (basionym Mycobacterium smegmatis), and Escherichia coli. Using a combination of viability assays, bioinformatics, and genetic manipulation of E. coli, we showed that a carboxylate-containing pharmacophore could indeed be transformed into a targeted drug through judicious choice of the alcohol moiety (Hetrick et al., 2021). With this proof-of-concept in hand, we report here on our methodology for measuring esterase activity with a varied alcohol in a high-throughput screen, assessing the impact of the ester prodrug candidates on target and non-target cell types, and determining and confirming the candidate esterase responsible for the cleavage, as well as on future prospects for this strategy.
2. Esterase Activity Assays with a Variable Alcohol Component
2.1. Fluorophore design considerations
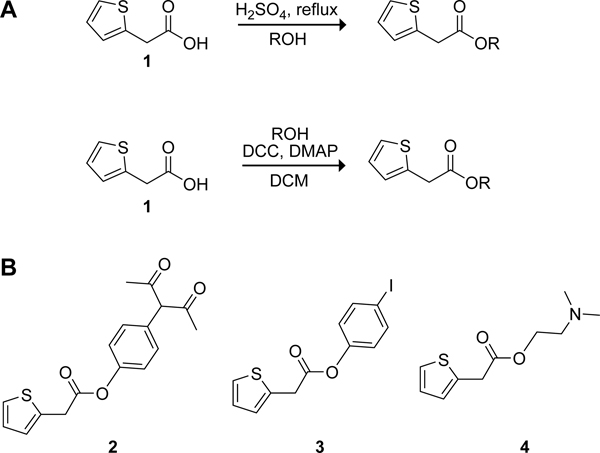
Esterase probes have been reviewed elsewhere (Chyan and Raines, 2018; Singh et al., 2021). As noted above, a myriad of probes have an invariable alcohol portion but few have a variable alcohol portion. Our terbium(III) luminescence-based probe offers the advantage of employing the commercially available 2-thiopheneacetic acid (2TA) as the acyl group. 2TA is small and, aside from its carboxyl group, relatively inert. Condensation of 2TA and simple alcohols can be achieved through Fischer esterification (Figure 3A).(Fischer and Speier, 1895) More complex esters can be formed through the activation of the 2TA carboxyl group with dicyclohexylcarbodiimide and the use of catalytic amounts of 4-dimethylaminopyridine (DMAP). Typically, purification by chromatography on silica gel delivers the desired product in good yields.
Figure 3.
Scheme for the synthesis of 2TA esters for the in vitro 2TA–terbium luminescence assay. (A) 2-Thiopheneacetic acid (1) is readily converted into esters through condensation with alcohols either via boiling in the alcohol with sulfuric acid (Fischer esterification) or through the use of DCC and DMAP. (B) Compounds that influence the luminescent response of ester cleavage.
The turn-on effect of the 2TA-based probes is dependent on the liberated carboxylate coordinating terbium(III). Certain esters are, however, also capable of such coordination prior to and/or after hydrolysis. For example, 2TA esters containing primary or secondary amino groups might coordinate terbium(III) while still in the ester form and thereby alter the resulting luminescence (Figure 3B). Thus, prior to commencing assays, we advise assessing the sensitivity of a synthesized probe by the addition of pig liver esterase (PLE) or another esterase to ensure that adequate discrimination is evident between hydrolyzed and non-hydrolyzed probes. As examples, we observed a decrease in luminescence for the diketophenyl ester 2 when hydrolyzed, no change for iodophenyl ester 3, and a minor change for dimethylamino-containing ester 4. In probes in which the ester is observed to chelate terbium(III), we suggest assessing the linear range of the method, which can differ significantly from the range of the free acid in the presence of a non-coordinating alcohol.
Another consideration in experimental design is determining the range of alcohol functionalities to include in a library. Aspects including chain length, steric bulk, and logP value should be considered. Other factors that can impact esterase cleavability include the number of sp3 atoms, polar surface area, logS values, and number of heteroatoms.
The toxicity of the alcohol is another consideration. Alcohols can be assessed for potential toxicity through in silico prescreening with the ToxAlert database (Sushko et al., 2012). In addition, candidate alcohols that are already Generally Recognized as Safe (GRAS) according to the United States Food and Drug Administration (FDA) provide a set that serves as a good starting point. Finally, known therapeutic compounds could also be wise to include, depending on the desired effect of the prodrug. Such compounds can be identified by using databases described in Section 3.2.
2.2. Assay design and execution
As with all enzymatic assays, a plan considering the many factors that can influence the results is essential. (For an excellent description of general enzymatic assay considerations, see: (Bisswanger, 2014)). Our assay has, however, some additional unique considerations. Unlike a classic fluorogenic assay, wherein a chemical change reveals a latent, inherent fluorescence, our assay relies on the coordination of a revealed carboxylate moiety to terbium(III). The carboxylate 2TA serves as an “antenna” that transfers excitation energy from 2TA to the terbium atom, creating a luminescent response. The luminescent response requires formation of a coordination complex that is composed of multiple components: a shell of Triton X-100, a synergic agent tri-n-octylphosphine oxide (TOPO), and 2TA/terbium(III) in a ratio of 3:2, where each 2TA molecule likely bridges two terbium atoms. We have established a robust range with respect to Triton X-100 concentration, HEPES–NaOH concentration, TOPO concentration, terbium(III) concentration, and pH, thus ensuring that small changes in these factors will not materially impact the readout. Nonetheless, additional components present in the assay mixture, such as those introduced when adding enzyme or cell preparations, can impact the sensitization complex and should be assessed with care prior to deploying the assay on a large scale.
Given these noted sensitivities, the following practical steps should be taken to assay esterase activity.
A 2× assay buffer mix should be prepared to allow for the one-step addition of all assay components. The 2× assay buffer is composed of 10 mM HEPES–NaOH buffer, pH 7.4, containing TbCl3 (8 mM), Triton X-100 (0.2% w/v), and TOPO (700 µM). (Note that TOPO is sparingly soluble in aqueous solutions and hence must be added at the time or after the addition of the Triton X-100; sonication might also be required to achieve TOPO dissolution.)
When using a standard curve to calibrate the concentration of liberated 2TA, it is essential that the standard curve be prepared similarly to the sample curve. For example, if an alcohol interferes with the formation or intensity of the terbium complex, standard curves should be prepared that mimic exactly the hydrolyzed 2TA ester, e.g., the standard for a half-hydrolyzed sample of a 100 µM 2TA ethyl ester should include 50 µM 2TA and 50 µM ethanol.
A blank containing all assay components except the ester substrate should be monitored throughout the assay to correct for any drift effects.
The standard curve should be generated at the same time as the sample curve and monitored throughout the time course of the assay.
Care should be taken to ensure that all components that will be added to the assay and the assay plate itself are at the same temperature prior to the start of the assay. Finally, when preparing the assay in a microplate, it is advisable to use a multichannel pipette to ensure that all components are added swiftly and at nearly the same time.
To acquire data, our assay requires excitation at 285 nm, monitoring emission at 547 nm, and the use of an instrument that allows for time-resolved fluorescence (TRF), which is essential for taking advantage of the slow luminescence decay of lanthanide complexes to achieve a sufficient signal-to-noise ratio (Thibon and Pierre, 2009). The assay is ideal for use in a microplate reader such that replicate samples and the standard calibration curve(s) can be monitored nearly simultaneously. Data collection should be continuous. Optimal time points will depend on the amount of enzyme added and the speed of the reaction but should be spaced closely together to generate a clean and well-characterized progress curve. Typically, data points can be acquired every 1–3 minutes.
Assays using purified enzyme have the fewest confounding contaminates and thus the highest signal-to-noise ratio. When preparing for such assays, the enzyme should be added last to initiate the reaction, reaction mixtures mixed thoroughly, and emission monitored as soon as possible after mixing is complete. The enzyme should be added to the standard curve solution as well because of its potential influence on the luminescent response, and all solutions should be equilibrated to the same temperature prior to mixing. In addition, we have observed that if the plate reader does not have a cooling function, then the ambient temperature in the reader increases as the assay progresses due to mechanical heat generated by the apparatus. Accordingly, monitoring the temperature could be important. Many esterases, such as human carboxylesterases, have established buffer systems and their kinetic values could differ markedly in our assay conditions; hence, it could be necessary to use a reference compound such as p-nitrophenyl acetate to compare kinetics under these conditions to historical data (Anderson et al., 1994).
Assays can also be performed with a whole-cell lysate. Such assays are an effective means to compare the relative esterase activity of many substrates. We have attempted to use the assay with Luria–Bertani (LB) medium, Middlebrook 7H9 broth, and Dulbecco’s modified Eagle’s medium (DMEM) but have found that components in the cell medium deplete the signal from the assay too much for reliable measurements. Hence, when using cell lysate with either mammalian or bacterial cell lines, cells must first be pelleted, washed rapidly with a low ionic strength buffer or distilled water, pelleted immediately again, gently resuspended in distilled water (generally 1/10th of the original volume), and added rapidly to the assay to initiate. Because the assay contains the Triton X-100 surfactant above its critical micellar concentration, most cell types will rapidly lyse when combined with the assay buffer, thereby liberating any intracellular ester for action on the substrate after thorough mixing. Likewise, the washing step will remove secreted esterases, and their activity will not contribute to the data.
Comparisons of esterase activity within a cell type or bacterial species requires using the same cell preparation with all substrates. The standard curve should also include the lysate, which can impact the data significantly. A reference ester should be included for each cell preparation because a wide variety of factors can influence the esterase activity. We recommend using the 2TA-phenylester as a standard because it is usually hydrolyzed to completion, providing a convenient progress curve to use to normalize esterase results. When comparing esterase activity across different cell types or bacterial species, the activity can be normalized against a measure of the amount of added cell material. Absorbance at 280 nm or pellet mass or volume are potential crude measures, though total protein content as determined with a bicinchoninic (BCA) assay is a more robust means (Smith et al., 1985).
2.3. Fitting continuous assay data
As noted above, the assay generates continuous data and allows for the facile monitoring of replicate preparations. Including an appropriately prepared standard curve as well as an enzyme-containing blank—also monitored continuously at the same time as the sample—allows for the accurate conversion of the instrument output (relative fluorescence units or RFU) into concentration of 2TA. Processing of the RFU data is achieved using eq 1:
| (1) |
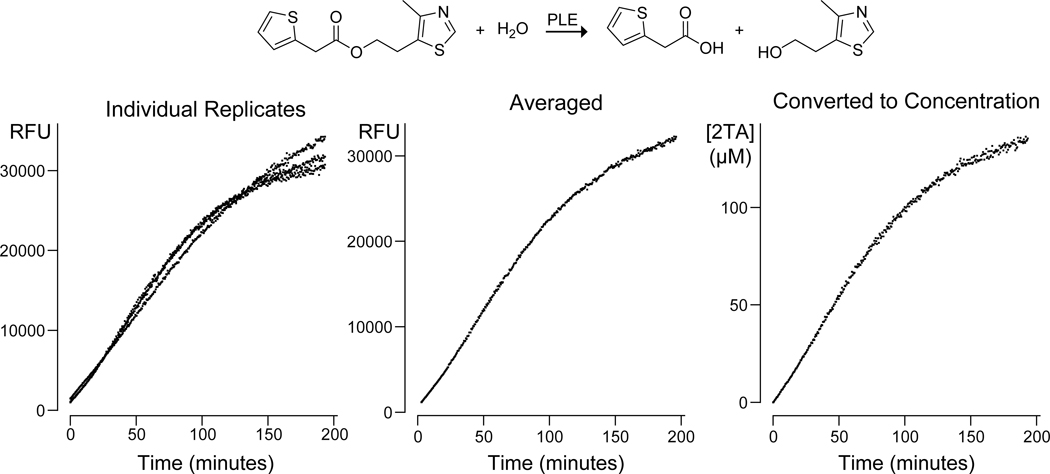
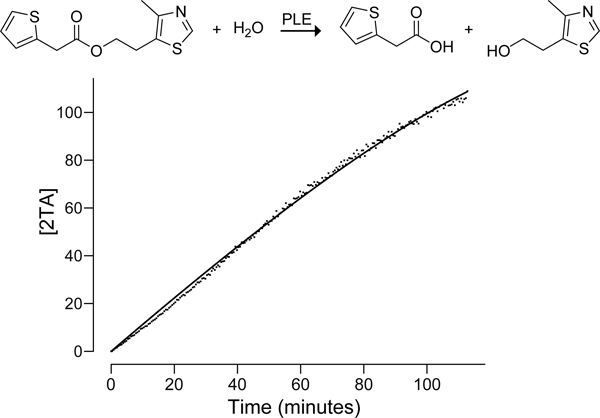
In eq 1, the slope at time t is determined from the fluorescence response of a standard curve consisting of 3 or more points. This slope is thus determined at each point that the sample is measured, thereby making the calculation of concentration robust to instrument drift, temperature variations, or other factors that could influence the measurement. The background term is assessed from three or more measurements of the luminescence of a given well immediately prior to adding the enzyme. An example of data analysis with eq 1 is shown in Figure 4.
Figure 4.
Graphs shows raw individual replicates, averaged replicate, and concentration results for the hydrolysis of the 2TA–sulfurylester by pig liver esterase (PLE). The raw data as well as the R code for creating the graphs is available at https://github.com/hetrickk/Plots_for_MIE.
With the data transformed into 2TA concentration, progress curves can be used to determine Michaelis–Menten parameters for catalysis by a purified enzyme. The classic initial rates method can be employed, where multiple substrate concentrations are measured in triplicate and the initial rate (where <10% of the substrate is consumed) is determined. A plot of the initial rate V0 against the initial substrate concentration [S]0 can then be fitted to the Michaelis–Menten equation (eq 2) to determine KM and Vmax = kcat[E].
| (2) |
Fitting the integrated Michaelis–Menten equation offers several advantages compared to the classic methodology (Liao et al., 2003; Goličnik, 2012). The integrated Michaelis–Menten equation (eq 3) fits to an individual progress curve itself rather than to values derived from the progress curve, thus removing a data-processing step that can add error. Fitting the classic equation also requires acquiring many progress curves, particularly those where the initial substrate concentration [S]0 is less than the KM value to ensure an accurate fit, which can be quite difficult to acquire because at low substrate concentrations the initial rate decreases quickly. On the other hand, fitting the integrated Michaelis–Menten equation (eq 3) requires only a few runs and is best accomplished where [S]0 is at least twice the KM value, which generally ensures a more accurate measurement. Additionally, progress curves should be fitted up to 80% of substrate consumption.
| (3) |
In eq 3, [P] is the product concentration at time t (which in our assay corresponds to the concentration of 2TA), and W represents the LambertW function (Corless et al., 1996). A commercial program, DynaFit (Kuzmič, 2009), is available and can perform the fit. A MatLab program is also available (Gutheil et al., 1994), and a further software package called the interactive continuous enzyme kinetics analysis tool (ICEKAT) provides an interactive environment that can tie the integrated equation back to the initial rates method (Olp et al., 2020). In an uncomplicated system such as one using purified PLE, we were able to fit progress curves easily with R software (R Core Team, 2014) using the script available at https://github.com/hetrickk/Plots_for_MIE (Figure 5). Executing this analysis in R produces the estimates of KM (termed “K” in R) and Vmax (termed “V” in R) as shown:
Figure 5.
Graph of the fitted progress curve for the hydrolysis of the 2TA–sulfurylester by pig liver esterase (PLE). Raw data and the script for making this plot and fit are available at https://github.com/hetrickk/Plots_for_MIE. Note that the progress curve data is saved as comma-separated values (csv).
Formula: P ∼ s − K * lambertW0(s/K * exp((s − V * t * 60)/K))
Parameters:
Estimate Std. Error t value Pr(>|t|)
K 2.792e+01 1.718e+00 16.26 <2e−16 ***
V 2.281e−02 3.466e−04 65.82 <2e−16 ***
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.508 on 226 degrees of freedom
Number of iterations to convergence: 14
Achieved convergence tolerance: 1.49e−08
Note that in the latest release of R, the Rcpp package (a required package for the lamW package) often generates the error “‘Rcpp_precious_remove’ not provided by package ‘Rcpp’”. This error can be resolved by restarting R, reinstalling Rcpp using install.packages(“Rcpp”), then proceeding with the code. Additionally, note that t is multiplied by 60 in the formula in R to convert t from minutes to seconds to yield a Vmax in s−1.
3. Antimicrobial prodrug development
3.1. Strain choice and considerations
Although we envision our approach as being applicable to many cell types, we first applied the ester prodrug targeting approach to narrowing the spectrum of a known antimicrobial compound. We took advantage of the knowledge that E. coli DH10B has low esterase activity (Antonczak et al., 2009) whereas M. smegmatis mc2155 (Tallman et al., 2016; Bassett et al., 2018) and B. subtilis OI1085 have significantly higher esterase activity (Eggert et al., 2000). We used these well-established and stark esterase differences to enable a proof-of-concept of our targeted ester prodrug approach. In our proof-of-concept, we demonstrated that the sulfurol ester of trans-3-(4-chlorobenzoyl)acrylic acid was a more potent antimicrobial against species harboring an esterase capable of cleaving it to the carboxylate form.
One of the key experiments in this study was using a bioinformatic analysis of the carboxylesterases present in E. coli DH10B, B. subtilis 168, and M. smegmatis mc2155 to identify which carboxylesterase family is likely responsible for the cleavage of trans-3-(4-chlorobenzoyl)acrylic acid. To achieve this goal, we used the NCBI protein database to identify all of the annotated carboxylesterases present in the three species. We then downloaded the amino acid sequences of all identified carboxylesterases and used the Enzyme Function Institute’s Enzyme Similarity Tool (EFI–EST) to determine the e-value connecting each individual enzyme sequence to each other enzyme sequence, that is, to perform an all-by-all BLAST (Gerlt et al., 2015). Note that the e-value, or expect value, is related to the number of hits of similarly strong alignment that one would expect to find in the database by random chance. For example, if two enzymes are found by BLAST to have an e-value of 15, then one would expect that 15 hits would be found in the database that appear to be similarly related but are actually just related by chance. On the other hand, if two enzymes have an e-value of 0.00000001 (or 10−9), then it is extremely unlikely that such a strong hit would be found by random chance. In short, the lower the e-value, the more closely related are the two enzymes. Analysis of the network generated with the EFI–EST using Cytoscape software allowed us to pinpoint a cluster of esterases that contained members from M. smegmatis and B. subtilis but not E. coli, and further experiments successfully confirmed that enzymes in this cluster were indeed responsible for cleaving the sulfurol ester. For a complete tutorial on using the EFI–EST, see: https://efi.igb.illinois.edu/efi-est/tutorial.php.
Useful as this bioinformatic analysis was in identifying an esterase that accepts the sulfurol ester as a substrate, it is potentially of even greater use in pointing towards a more sophisticated strain selection process than literature mining. Genomic data is abundantly available, as many pathogenic and commensal bacterial genomes have known sequences. The carboxylic ester hydrolase enzymes (CASTLE) database has gathered a wealth of carboxylesterase sequences into a single, freely available database that can be downloaded in its entirety upon request from the authors (Chen et al., 2016). This database can be used to map out carboxylesterase sequence-space by species, yielding knowledge of esterase families that are unique in certain pathogenic species, such as M. tuberculosis, but absent in commensal bacteria. This or other genomic-based approaches will allow future work to select species harboring esterases of particular interest, thereby enhancing the applicability of discovered targeted prodrugs.
3.2. Compound selection
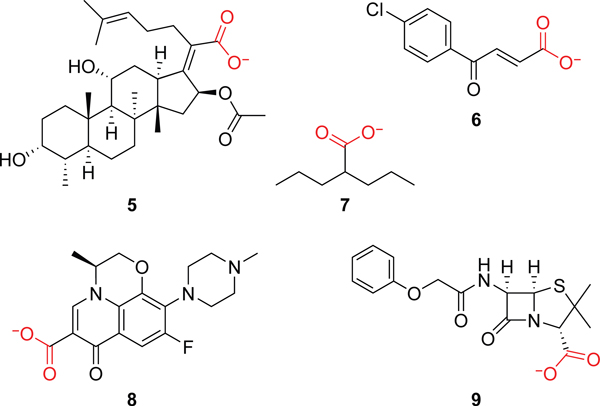
A carboxylate moiety is present in many major classes of antimicrobial compounds, including β–lactams, quinolones, many lipopeptides, and others. These drugs tend to have a broad spectrum and hence offer excellent opportunities for turning into targeted prodrugs via esterification. Additional compounds can be mined from online databases, such as Drugbank (https://go.drugbank.com/), ChEMBL (https://www.ebi.ac.uk/chembl/), and PubChem (https://pubchem.ncbi.nlm.nih.gov/). Drugbank is a particularly useful resource since it can be downloaded as an XML file, parsed either in R using the package dbparser or in Python using the ElementTree XML API, and queried to discover carboxylate-containing compounds. This approach also allows a user to easily identify other chemical moieties of interest, including potential alcohol components, and quickly learn what is known about safety and toxicology. Using these approaches, we identified fusidic acid (5), trans-3-(4-chlorobenzoyl)acrylic acid (6), valproic acid (7), levofloxacin (8), and pencillin V (9) as carboxylate-containing antimicrobials of particular interest (see Figure 6).
Figure 6.
Carboxylate-containing compounds of interest for the ester prodrug strategy.
In addition to identifying compounds that have been tested previously for antimicrobial activity, new computational efforts allow for discovery of novel, uncharacterized compounds. Recently, sophisticated machine-learning approaches allowed the de novo discovery of a potent antimicrobial compound (Stokes et al., 2020). These approaches will doubtless yield many results in the coming decades. Nevertheless, we have discovered that some leads can be generated using just the computational power of a laptop. The above databases all contain numerous drug-like compounds. Separation of these small-molecule drugs into antimicrobial compounds and non-antimicrobial compounds can be done readily and then used as a training set for a simple classification algorithm called support vector machine (SVM) (Amarappa and Sathyanarayana, 2014). This model can then be used to sort in silico databases of theoretical chemical compounds, such as GDB17 (Ruddigkeit et al., 2012), into compounds that might be antimicrobial and those that are unlikely to be antimicrobial.
For the experimental condensation of these target acids with alcohols, we have found no single method that is effective for accessing all esters. Fischer esterification is a straightforward procedure, but many carboxylates contain functionality that will not survive these harsh conditions. DMAP and DCC is another effective option, though Mukaiyama’s reagent, 2-chloro-1-methylpyridinium iodide, is perhaps the most general and effective means to accomplish ester synthesis (Novosjolova, 2012).
3.3. Viability assay
With microbial strains of interest and ester compounds in-hand, viability assays can be rapidly deployed to assess the potency of the esters. We adapted the resazurin dye assay to assess viability, as has been done previously (Fleck et al., 2014). Resazurin dye in the presence of live cells is reduced by NADH to resorufin, which generates a fluorescent response (although it should be noted that some cell types can reduce it further to dihydroresorufin—a cytotoxic and nonfluorescent molecule) (Prabst et al., 2017). We have used the commercially available alamarBlue™ formulation, loaded to 10% of the total volume of a well, to assess the viability of B. subtilis, M. smegmatis, and E. coli cells over 12 hours or more. Assay conditions can be optimized as described elsewhere (Kim and Jang, 2018). For rapid assay deployment, only a few key factors need consideration. First, the number of bacterial cells added to each well directly impacts the time course of the assay and its sensitivity. Starting with low bacterial cell density will yield a sensitive assay but could require many hours of fluorescence monitoring. Starting with higher bacterial cell-loading in each well yields a shorter assay time, because cell growth will reach saturation more quickly, but reduces sensitivity to smaller perturbations in viability. We found that diluting an overnight bacterial culture to an OD600 nm between 0.02 and 0.05 was generally appropriate. Additionally, we found that each bacterial preparation demonstrated slightly different growth kinetics, possibly due to variations in media preparation or temperature. Hence, replicates of untreated bacterial cells must be included with each cell preparation. Likewise, medium plus dye control replicates are essential to ensure there are no contaminating bacterial species present. As we performed these assays with 100 µL of medium in a 96-well plate held at 37 °C with shaking, applying a polyfilm cover to each plate is necessary to minimize evaporation of medium during longer assays. Finally, each well should be prepared in triplicate, and bacterial cells should be the last component added to each well.
As noted above, comparison of results between different preparations of bacteria on different days can be difficult, particularly when the desired differences are subtle. To accomplish such a comparison, the bacteria-only control growth curves from each preparation can be fitted readily with R software to Richards’s logistic growth curve (eq 4) to determine the inflection point (Richards, 1959; Zwietering et al., 1990). Using the inflection point, the time axes can be adjusted such that the bacteria-only controls overlap exactly, thereby enabling direct comparisons.
| (4) |
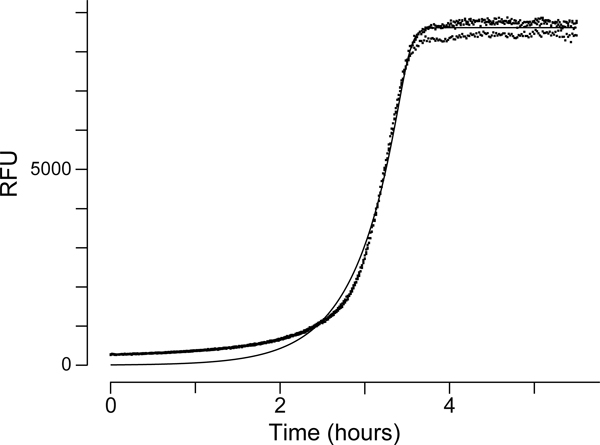
In eq 4, a is the upper asymptote, v is the parameter affecting near which asymptote the maximum growth occurs, k is the growth rate, l is the point of inflection on the abscissa, and t is the time (in minutes) plotted on the ordinate. Raw data and an example curve fit are available at https://github.com/hetrickk/Plots_for_MIE and shown in Figure 7.
Figure 7.
Fit of a growth curve of E. coli in CAMHB.
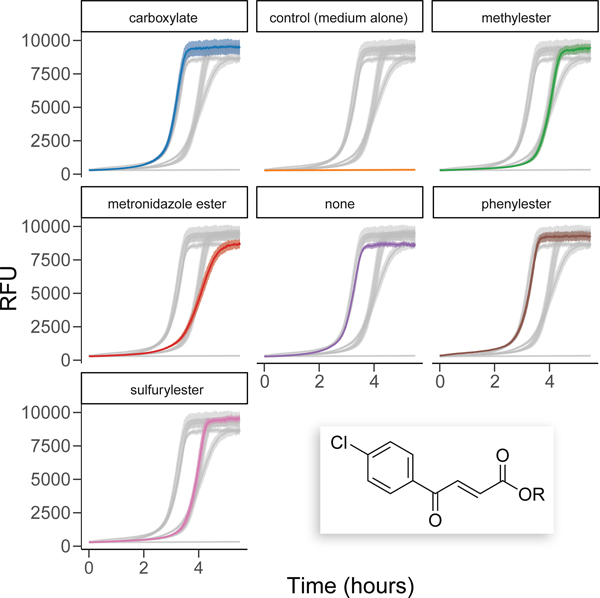
Plotting viability curves in R also allows for the easy inclusion of error ribbons to visualize standard error, standard deviation, or confidence interval. Finally, the ggplot2 and gghighlight packages allow for easy visual distinction between different assay conditions. The code for making such plots is available at https://github.com/hetrickk/Plots_for_MIE, and an example plot is shown in Figure 8.
Figure 8.
Example plot of viability assays using gghighlight to generate a visual comparison of the viability profiles of select esters of trans-3-(4-chlorobenzoyl)acrylic acid (inset).
3.4. Verification of esterase specificity
If differential activity is observed in viability assays for a set ester, then it is essential to verify that the ester is indeed cleaved by the targeted bacterial cells. We have used extraction and liquid chromatography–mass spectrometry (LC–MS) to accomplish this aim. A culture of the bacterium of interest was grown overnight, and a 500-µL aliquot was removed and lysed using sonication. The ester of interest was added to this lysate to a final concentration of 1 mM. As a control, the ester of interest was added to medium alone at the same concentration. Both the control and the sample were incubated at 37 °C for two hours. Dichloromethane (500 µL) was added to extract the ester and its hydrolysis products, and the dichloromethane layer was dried under a stream of N2(g). The dried residue was then resuspended in 75 µL of 1:1 acetonitrile/water and analyzed with LC–MS.
Semi-quantitative analysis can be readily achieved if at least two of the ester, alcohol, or carboxylate compounds can be extracted and observed with LC–MS. In our case, the percent of the sulfurol ester of trans-3-(4-chlorobenzoyl)acrylate that had been hydrolyzed could be determined because the alcohol hydrolysis product, sulfurol, and the ester itself could be observed in the LC–MS trace, thus allowing comparison of the ion count of these two peaks within a sample. Comparison of this ratio to the ratio of a set of standards that mimicked hydrolysis ranging from 0–100% hydrolyzed, prepared using extraction from the sample medium in the same manner as the sample and that were injected in the same LC–MS run, allowed for semi-quantitation of hydrolysis. The data provided clear evidence that the ester was indeed hydrolyzed in the presence of M. smegmatis or B. subtilis lysate but not in the presence of E. coli lysate. We were unable to observe the carboxylate reliably. If two of the three components cannot be readily observed, it might be possible to spike the sample with an internal standard compound and estimate the percent hydrolysis through a comparison with the internal standard.
Having established that the ester is indeed hydrolyzed in the presence of the bacterium, it is next useful to identify the esterase responsible for its hydrolysis. Bioinformatics is a key enabler for determining which esterase could be responsible for the observed hydrolysis, and using the EFI–EST tool is a powerful methodology for accomplishing this purpose. Once candidate esterases have been identified, they can be verified through cloning and expressing the candidate esterase in E. coli DH10B cells, as these cells have virtually no esterase activity. (Although, we note that this strain does not express T7 RNA polymerase and thus requires that either a different promoter is associated with the esterase or T7 RNA polymerase is co-expressed through addition of a plasmid containing the polymerase, such as pCS6 (Schmidt et al., 2012).) Having cloned the candidate esterase into E. coli, repeating viability and LC–MS studies from above is an effective means of verifying that the esterase is indeed capable of hydrolyzing the ester. Note that including E. coli transformed with pCS6 and an empty plasmid is an essential negative control in these studies.
4. Prospectus
Our current work has shown that the simple transformation of a carboxylate to an ester can improve not only the permeability but also the specificity of an antibiotic. In doing so, we have demonstrated that the combination of an in vitro esterase specificity assay to identify alcohol moieties of interest, cheminformatic and (potentially) machine-learning approaches to identify parent antimicrobial carboxylic acids, and a viability assay to assess the impact of novel esters can be a powerful engine for designing targeted ester prodrugs. Bioinformatics and genomics further illuminate and inform all of these endeavors, as illustrated in Figure 2. Going forward, we envision this approach leading to a host of targeted antimicrobial compounds.
This approach is in no way limited to esters or antimicrobial compounds alone. Knowledge of unique bacterial enzymes is ever-increasing (Veprinskiy et al., 2017; Rodríguez Benítez and Narayan, 2019), and many of these transformations, particularly those unique to a small subset of bacteria, could be leveraged to create prodrugs that are unmasked in novel chemical ways and in desired bacteria alone. Methylation or demethylation, reductions, oxidations, halogenations, and a host of other transformations could all be leveraged. As an example of the potential of other transformations, the tuberculosis-treating compound pyrazinamide was used therapeutically long before it was discovered that pyrazinamide is actually a prodrug that is converted to pyrazinoic acid by a pyrazinamidase that is found in Mycobacterium tuberculosis. Moreover, unique enzymatic environments are not, in fact, unique to different bacterial species. Cancer tumor cells exhibit a unique enzymatic environment relative to normal cells (Herling et al., 2011), and that could be explored in a similar fashion to reduce the toxicity and improve the potency of cancer drugs such as 6-diazo-5-oxo-l-norleucine, which has proven to be too toxic to be deployed clinically (Lemberg et al., 2018). Ultimately, we expect this general approach at the interface of genomics, biology, and chemistry will be a uniquely fertile ground for the discovery of targeted therapeutic prodrugs.
Acknowledgments
This work was supported by the Merkin Institute for Transformative Technologies in Healthcare. Additional support was provided by Grants R21 GM135780 and R01 GM044783 (NIH).
References
- Amarappa S, and Sathyanarayana SV (2014). Data classification using support vector machine (SVM), a simplified approach. Int. J. Electron. Comput. Sci. Eng. 3, 435–445. [Google Scholar]
- Anderson J, Byrne T, Woelfel KJ, Meany JE, Spyridis GT, and Pocker Y (1994). The hydrolysis of p-nitrophenyl acetate: A versatile reaction to study enzyme kinetics. J. Chem. Educ. 71, 715–718. [Google Scholar]
- Antonczak AK, Simova Z, and Tippmann EM (2009). A critical examination of Escherichia coli esterase activity. J. Biol. Chem. 284, 28795–28800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett B, Waibel B, White A, Hansen H, Stephens D, Koelper A, Larsen EM, Kim C, Glanzer A, Lavis LD, Hoops GC, and Johnson RJ (2018). Measuring the global substrate specificity of mycobacterial serine hydrolases using a library of fluorogenic ester substrates. ACS Infect. Dis. 4, 904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisswanger H (2014). Enzyme assays. Perspect. Sci. 1, 41–55. [Google Scholar]
- Chen Y, Black DS, and Reilly PJ (2016). Carboxylic ester hydrolases: Classification and database derived from their primary, secondary, and tertiary structures. Protein Sci. 25, 1942–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyan W, and Raines RT (2018). Enzyme-activated fluorogenic probes for live-cell and in vivo imaging. ACS Chem. Biol. 13, 1810–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corless RM, Gonnet GH, Hare DEG, Jeffrey DJ, and Knuth DE (1996). On the Lambert W function. Adv. Comput. Math. 5, 329–359. [Google Scholar]
- Devamani T, Rauwerdink AM, Lunzer M, Jones BJ, Mooney JL, Tan MA, Zhang ZJ, Xu JH, Dean AM, and Kazlauskas RJ (2016). Catalytic promiscuity of ancestral esterases and hydroxynitrile lyases. J. Am. Chem. Soc. 138, 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert T, Pencreac’h G, Verger R, and Jaeger K-E (2000). A novel extracellular esterase from Bacillus subtilis and its conversion to a monoacylglycerol hydrolase. Eur. J. Biochem. 267, 6459–6469. [DOI] [PubMed] [Google Scholar]
- Fischer E, and Speier A (1895). Darstellung der Ester. Chem. Ber. 28, 3252–3258. [Google Scholar]
- Fleck LE, North EJ, Lee RE, Mulcahy LR, Casadei G, and Lewis K (2014). A screen for and validation of prodrug antimicrobials. Antimicrob. Agents Chemother. 58, 1410–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, and Whalen KL (2015). Enzyme function initiative–enzyme similarity tool (EFI–EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta Proteins Proteom. 1854, 1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goličnik M (2012). On the Lambert W function and its utility in biochemical kinetics. Biochem. Eng. J. 63, 116–123. [Google Scholar]
- Gutheil WG, Kettner CA, and Bachovchin WW (1994). Kinlsq: A program for fitting kinetics data with numerically integrated rate equations and its application to the analysis of slow, tight-binding inhibition data. Anal. Biochem. 223, 13–20. [DOI] [PubMed] [Google Scholar]
- Herling A, König M, Bulik S, and Holzhütter H-G (2011). Enzymatic features of the glucose metabolism in tumor cells. FEBS J. 278, 2436–59. [DOI] [PubMed] [Google Scholar]
- Hetrick KJ, Aguilar Ramos MA, and Raines RT (2019). Terbium(III) luminescence-based assay for esterase activity. Anal. Chem. 91, 8615–8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetrick KJ, Aguilar Ramos MA, and Raines RT (2021). Endogenous enzymes enable antimicrobial activity. ACS Chem. Biol. 16, 800–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttunen KM, Raunio H, and Rautio J (2011). Prodrugs—from serendipity to rational design. Pharmacol. Rev. 63, 750–771. [DOI] [PubMed] [Google Scholar]
- Janes LE, Lowendahl AC, and Kazlauskas RJ (1998). Quantitative screening of hydrolase libraries using pH indicators: Identifying active and enantioselective hydrolases. Chem.—Eur. J. 4, 2324–2331. [Google Scholar]
- Kim HJ, and Jang S (2018). Optimization of a resazurin-based microplate assay for large-scale compound screenings against Klebsiella pneumoniae. 3 Biotech. 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim H, Choi Y, and Kim Y (2015). A new strategy for fluorogenic esterase probes displaying low levels of non-specific hydrolysis. Chem. Eur. J. 21, 9645–9649. [DOI] [PubMed] [Google Scholar]
- Kuzmič P (2009). DynaFit—a software package for enzymology. Methods Enzymol. 467, 247–280. [DOI] [PubMed] [Google Scholar]
- Lavis LD (2008). Ester bonds in prodrugs. ACS Chem. Biol. 3, 203–206. [DOI] [PubMed] [Google Scholar]
- Lavis LD, Chao T-Y, and Raines RT (2006). Fluorogenic label for biomolecular imaging. ACS Chem. Biol. 1, 252–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberg KM, Vornov JJ, Rais R, and Slusher BS (2018). We’re not “DON” yet: Optimal dosing and prodrug delivery of 6-diazo-5-oxo-L-norleucine. Mol. Cancer Ther. 17, 1824–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao F, Tian K-C, Yang X, Zhou Q-X, Zeng Z-C, and Zuo Y-P (2003). Kinetic substrate quantification by fitting the enzyme reaction curve to the integrated Michaelis–Menten equation. Anal. Bioanal. Chem. 375, 756–762. [DOI] [PubMed] [Google Scholar]
- Martínez-Martínez M, Coscolín C, Santiago G, Chow J, Stogios PJ, Bargiela R, Gertler C, Navarro-Fernández J, Bollinger A, Thies S, Méndez-Garcia C, Popovic A, Brown G, Chernikova TN, García-Moyano A, Bjerga GEK, Pírez-Garcia P, Hai T, Del Pozo MV, Stokke R, Steen IH, Cui H, Xu X, Nocek BP, Alcaide M, Distaso M, Mesa V, Peláez AI, Sánchez J, Buchholz PCF, Pleiss J, Fernández-Guerra A, Glöckner FO, Golyshina OV, Yakimov MM, Savchenko A, Jaeger K-E, Yakunin AF, Streit WR, Golyshin PN, Guallar V, Ferrer M, and The Inmare Consortium (2018). Determinants and prediction of esterase substrate promiscuity patterns. ACS Chem. Biol. 13, 225–234. [DOI] [PubMed] [Google Scholar]
- Novosjolova I (2012). The Mukaiyama reagent: An efficient condensation agent. Synlett. 24, 135–136. [Google Scholar]
- Olp MD, Kalous KS, and Smith BC (2020). ICEKAT: An interactive online tool for calculating initial rates from continuous enzyme kinetic traces. BMC Bioinformatics. 21, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda T, and Gowrishankar BS (2005). Production and applications of esterases. Appl. Microbiol. Biotechnol. 67, 160–169. [DOI] [PubMed] [Google Scholar]
- Prabst K, Engelhardt H, Ringgeler S, and Hubner H (2017). Basic colorimetric proliferation assays: MTT, WST, and resazurin. Methods Mol. Biol. 1601, 1–17. [DOI] [PubMed] [Google Scholar]
- R Core Team (2014). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rautio J, Karkkainen J, and Sloan KB (2017). Prodrugs—Recent approvals and a glimpse of the pipeline. Eur. J. Pharm. Sci. 109, 146–161. [DOI] [PubMed] [Google Scholar]
- Richards FJ (1959). A flexible growth function for empirical use. J. Exp. Bot. 10, 290–300. [Google Scholar]
- Rodríguez Benítez A, and Narayan ARH (2019). Frontiers in biocatalysis: Profiling function across sequence space. ACS Cent. Sci. 5, 1747–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddigkeit L, van Deursen R, Blum LC, and Reymond JL (2012). Enumeration of 166 billion organic small molecules in the chemical universe database GDB-17. J. Chem. Inf. Model. 52, 2864–2875. [DOI] [PubMed] [Google Scholar]
- Satoh T, and Hosokawa M (2006). Structure, function and regulation of carboxylesterases. Chem. Biol. Interact. 162, 195–211. [DOI] [PubMed] [Google Scholar]
- Schmidt CM, Shis DL, Nguyen-Huu TD, and Bennett MR (2012). Stable maintenance of multiple plasmids in E. coli using a single selective marker. ACS Synth. Biol. 1, 445–450. [DOI] [PubMed] [Google Scholar]
- Singh A, Gao M, and Beck MW (2021). Human carboxylesterases and fluorescent probes to image their activity in live cells. RSC Med. Chem. 12, 1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, and Klenk DC (1985). Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85. [DOI] [PubMed] [Google Scholar]
- Stokes JM, Yang K, Swanson K, Jin W, Cubillos-Ruiz A, Donghia NM, MacNair CR, French S, Carfrae LA, Bloom-Ackermann Z, Tran VM, Chiappino-Pepe A, Badran AH, Andrews IW, Chory EJ, Church GM, Brown ED, Jaakkola TS, Barzilay R, and Collins JJ (2020). A deep learning approach to antibiotic discovery. Cell. 180, 688–702 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sushko I, Salmina E, Potemkin VA, Poda G, and Tetko IV (2012). ToxAlerts: A web server of structural alerts for toxic chemicals and compounds with potential adverse reactions. J. Chem. Inf. Model. 52, 2310–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallman KR, Levine SR, and Beatty KE (2016). Profiling esterases in Mycobacterium tuberculosis using far-red fluorogenic substrates. ACS Chem. Biol. 11, 1810–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa B, and Mayer JM (2003). Hydrolysis in Drug and Prodrug Metabolism. Verlag Helvetica Chimica Acta, Zürich, Switzerland, pp. 419–534. [Google Scholar]
- Thibon A, and Pierre VC (2009). Principles of responsive lanthanide-based luminescent probes for cellular imaging. Anal. Bioanal. Chem. 394, 107–120. [DOI] [PubMed] [Google Scholar]
- Tremblay MS, Halim M, and Sames D (2007). Cocktails of Tb3+ and Eu3+ complexes: A general platform for the design of ratiometric optical probes. J. Am. Chem. Soc. 129, 7570–7577. [DOI] [PubMed] [Google Scholar]
- Ueno T, Urano Y, Setsukinai K. i., Takakusa H, Kojima H, Ohkubo K, Fukuzumi S, and Nagano T (2004). Rational principles for modulating fluorescence properties of fluorescein. J. Am. Chem. Soc. 126, 14079–14085. [DOI] [PubMed] [Google Scholar]
- Veprinskiy V, Heizinger L, Plach MG, and Merkl R (2017). Assessing in silico the recruitment and functional spectrum of bacterial enzymes from secondary metabolism. BMC Evol. Biol. 17, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu HJ, and Markowitz JS (2009). Activation of the antiviral prodrug oseltamivir is impaired by two newly identified carboxylesterase 1 variants. Drug Metab. Dispos. 37, 264–267. [DOI] [PubMed] [Google Scholar]
- Zwietering MH, Jongenburger I, Rombouts FM, and van’t Riet K (1990). Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56, 1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]