Abstract
The introduction of antiretroviral therapy (ART) and highly active antiretroviral therapy (HAART) has transformed human immunodeficiency virus (HIV)-1 into a chronic, well-managed disease. However, these therapies do not eliminate all infected cells from the body despite suppressing viral load. Viral rebound is largely due to the presence of cellular reservoirs which support long-term persistence of HIV-1. A thorough understanding of the HIV-1 reservoir will facilitate the development of new strategies leading to its detection, reduction, and elimination, ultimately leading to curative therapies for HIV-1. Although immune cells derived from lymphoid and myeloid progenitors have been thoroughly studied as HIV-1 reservoirs, few studies have examined whether mesenchymal stromal/stem cells (MSCs) can assume this function. In this review, we evaluate published studies which have assessed whether MSCs contribute to the HIV-1 reservoir. MSCs have been found to express the receptors and co-receptors required for HIV-1 entry, albeit at levels of expression and receptor localisation that vary considerably between studies. Exposure to HIV-1 and HIV-1 proteins alters MSC properties in vitro, including their proliferation capacity and differentiation potential. However, in vitro and in vivo experiments investigating whether MSCs can become infected with and harbour latent integrated proviral DNA are lacking. In conclusion, MSCs appear to have the potential to contribute to the HIV-1 reservoir. However, further studies are needed using techniques such as those used to prove that cluster of differentiation (CD)4+ T cells constitute an HIV-1 reservoir before a reservoir function can definitively be ascribed to MSCs.
Graphical abstract
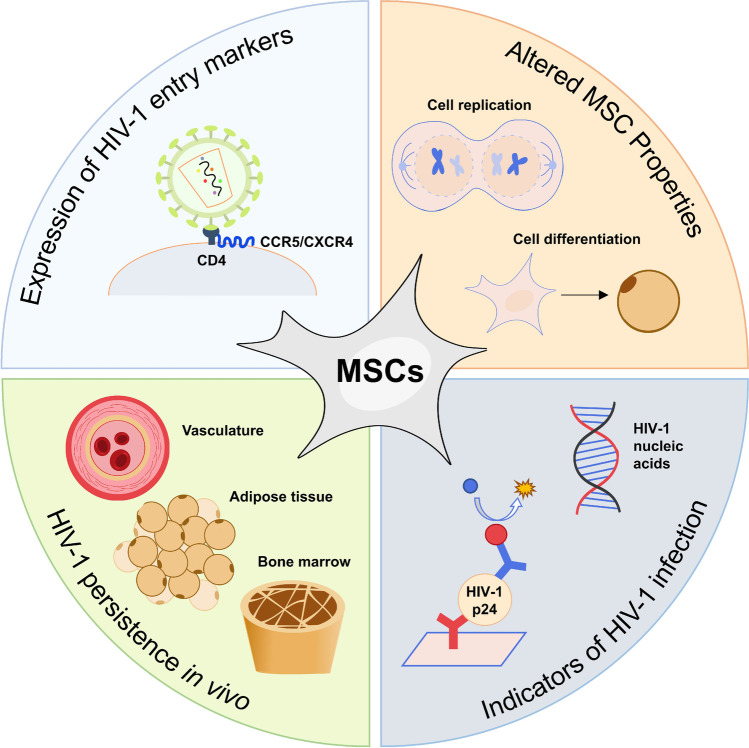
MSCs may contribute to HIV-1 persistence in vivo in the vasculature, adipose tissue, and bone marrow by being a reservoir for latent HIV-1. To harbour latent HIV-1, MSCs must express HIV-1 entry markers, and show evidence of productive or latent HIV-1 infection. The effect of HIV-1 or HIV-1 proteins on MSC properties may also be indicative of HIV-1 infection.
Keywords: Human immunodeficiency virus (HIV)-1, mesenchymal stromal/stem cells (MSCs), cellular reservoirs, receptors/co-receptors, HIV-1 proteins, HIV-1 latency, stem cells, infection, viral rebound
Introduction
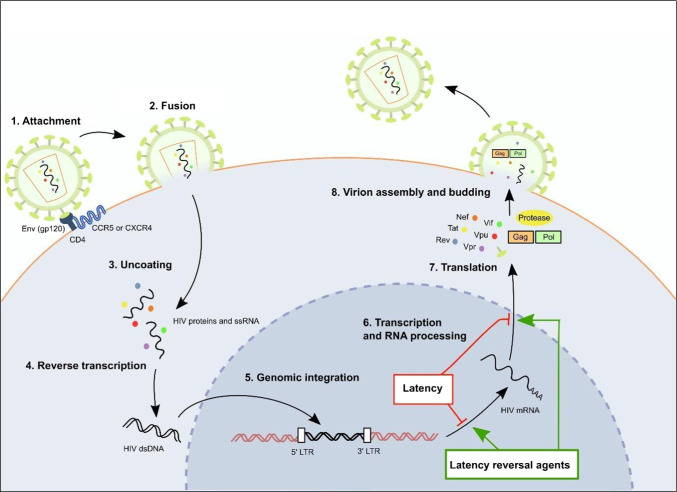
Human immunodeficiency virus (HIV)-1 primarily targets and infects cluster of differentiation (CD)4+ T cells, leading to compromised immune function and leaving the host vulnerable to opportunistic infections. Without treatment, immune function is gradually reduced and the individual advances to the final and most severe stage of the disease, acquired immune deficiency syndrome (AIDS) [1]. HIV-1 uses the machinery of CD4+ T cells to multiply and spread through the body. Entry of HIV-1 into CD4+ T cells occurs through binding of the HIV-1 envelope (Env) protein, gp120, to CD4, the primary cell surface receptor on the host cell (Fig. 1) [2, 3]. Subsequent co-receptor binding to C-C-motif chemokine receptor type 5 (CCR5) for R5-tropic viruses, to C-X-C-motif chemokine receptor type 4 (CXCR4) for X4-tropic viruses or to both co-receptors for R5X4 dual tropic viruses, results in fusion of the viral and CD4+ T cell membranes allowing transfer of the viral genome into the host cell [4, 5]. Inside the host cell, HIV-1 ribonucleic acid (RNA) is reverse transcribed into HIV-1 proviral deoxyribonucleic acid (DNA). Proviral DNA enters the host cell nucleus where it is integrated into the host cell genome. Upon integration, host RNA polymerase II enzymes transcribe the proviral DNA to form messenger RNA (mRNA). Viral mRNA is then translated by host cell translation machinery to form viral proteins. Viral proteins and transcribed viral single-stranded RNA (ssRNA) molecules move to the cell surface where they assemble to form immature, non-infectious virions. These virions bud from the host cell plasma membrane and undergo a protease-mediated maturation process to form mature, infectious virions that contain fully functional reverse transcriptase and integrase enzymes [6–8]. Refer to Fig. 1 for a detailed depiction of the HIV-1 life cycle.
Fig. 1.
HIV-1 life cycle and latency. To enter host cells, the HIV-1 gp120 Env protein binds to the host CD4 receptor and CCR5/CXCR4 co-receptors (1). Once bound, the HIV-1 viral envelope fuses with the host membrane and uncoats, releasing HIV-1 proteins and ssRNA into the host cell cytoplasm (2-3). HIV-1 ssRNA is reverse-transcribed into the dsDNA proviral genome using HIV-1 reverse transcriptase (4) and can integrate into the dsDNA host genome in the nucleus (5). Host genomic DNA containing the integrated HIV-1 transcript can then undergo transcription, RNA processing and translation to produce viral proteins, which assemble with viral ssRNA to form mature, infectious HIV-1 virions (6-8). Alternatively, interference with gene expression through mechanisms such as epigenetic silencing and/or post-translational modifications, can cause the replication-competent provirus to remain latent within the host cell. Certain chemical compounds, termed latency reversal agents, can reverse silencing mechanisms to reactivate latent HIV-1. Abbreviations: CCR5: C-C-motif chemokine receptor type 5; CD4: cluster of differentiation type 4; CXCR4: C-X-C-motif chemokine receptor type 4; dsDNA: double stranded DNA; Env: HIV-1 envelope protein; Gag: HIV-1 group specific antigen; gp120: HIV-1 glycoprotein 120; LTR: long terminal repeat; mRNA: messenger ribonucleic acid; Nef: HIV-1 negative factor protein; Pol: HIV-1 polymerase; Rev: HIV-1 response element protein; ssRNA: single-stranded RNA; Tat: HIV-1 transactivating protein; Vif: HIV-1 viral infectivity factor protein; Vpr: HIV-1 viral protein R; Vpu: HIV-1 viral protein U
HIV-1 persists in cells where it can adopt two different functional states, namely productive/replicative and latent [9]. The introduction of antiretroviral therapy (ART) and highly active antiretroviral therapy (HAART) has transformed HIV-1 into a chronic, well-managed disease by effectively reducing viral load and the effects of HIV-1 on the immune system. ART or HAART primarily target the productive/replicative state by reducing and/or inhibiting HIV-1 replication. However, these therapies do not eliminate all infected cells from the body despite suppressing viral load. To achieve effective viral suppression, a treatment adherence rate of 95% is recommended [10, 11]. Viral rebound is largely caused by HIV-1 latency, which is defined as a non-productive, reversible state of HIV-1 infection characterised by no or low-level expression of viral proteins and a lack of viral replication. Some studies have also expanded the definition of latent HIV-1 to include transcription- and translation-competent proviruses, which are not fully replication-competent, but produce viral mRNA and proteins [12, 13]. Long term persistence of HIV-1 is supported by infected cellular compartments harbouring latent HIV-1, termed cellular reservoirs [14, 15].
Due to a multitude of factors, including epigenetic regulation and/or silencing, HIV-1 reservoirs exhibit low-level or zero HIV-1 gene expression in patients receiving optimal ART [16, 17]. However, gene expression and viral replication resume following treatment interruption; thus, HIV-1 reservoirs constitute a challenge to the complete eradication of the virus from infected patients. Latency reversal agents (LRA) can be used to reactivate gene expression of latent HIV-1, rendering the virus vulnerable to elimination by the host immune system and ART, an approach labelled “shock and kill”. Additionally, LRAs can be used to distinguish between latently infected cells and cells either uninfected with HIV-1 or refractory to HIV-1 infection [18]. Complete eradication of the virus is difficult, since mechanisms contributing to HIV-1 latency are not fully understood and LRAs must effectively eliminate HIV-1 from a variety of cell types and tissues to be effective [19]. Figure 1 outlines the effect that latency and LRAs have on HIV-1 gene expression during the HIV life cycle.
A thorough understanding of the HIV-1 reservoir will facilitate the development of new strategies to detect, reduce and eliminate the reservoir, ultimately leading to HIV-1 curative therapies. Although immune cells derived from lymphoid and myeloid progenitors have been thoroughly studied as HIV-1 reservoirs, few studies have examined whether mesenchymal stromal/stem cells (MSCs) can assume this function [20]. MSCs are a heterogeneous population of cells containing a subset of non-hematopoietic multipotent adult stem cells. Although originally isolated from bone marrow, MSCs have been found to reside in virtually all post-natal human tissues and organs [21, 22]. MSCs are easily attainable, can be isolated from various tissues, and show promise therapeutically due to their differentiation capacity and ability to activate endogenous progenitor cells through paracrine signalling [23]. Furthermore, the ability of MSCs to migrate to sites of inflammation throughout the body, including the central nervous system (CNS), could facilitate viral dissemination if MSCs are indeed an additional HIV-1 reservoir [22, 24].
Many individuals infected with HIV-1 while receiving HAART still manifest clinical features such as reduced bone mineral density, altered lipid metabolism (lipodystrophy, dyslipidemia) and haematological abnormalities [25–27]. This was initially thought to be the result of ART toxicity; however, their occurrence in ART naïve HIV-infected people suggests that the virus plays a role in driving these abnormalities [28, 29]. Adipose tissue has been suggested to be a reservoir for HIV-1 [30, 31]. Adipocytes express HIV-1 entry receptors and co-receptors [32] and can be infected in vitro with HIV-1 [33], but the presence of HIV-1 in these cells has not been reported [34, 35]. Adipose tissue is not only involved in metabolic homeostasis but also contains MSCs that support bone and lipid homeostasis and that can differentiate into osteoblasts and adipocytes [36, 37]. Infection of MSCs in the bone marrow or adipose tissue niche by HIV-1 could explain these comorbidities. To examine whether MSCs contribute to the HIV-1 reservoir, this review evaluates studies addressing HIV-1 receptor and co-receptor expression on MSCs, and whether these cells can become infected with and harbour the virus.
HIV-1 Reservoirs
Lymphoid Progenitors
CD4+ T cells, which arise from lymphoid progenitors, represent the largest and best characterised HIV-1 reservoir. Resting CD4+ T memory cells were initially regarded as the main T cell population harbouring latent HIV-1 [38–41]. However, viral latency has also been observed in activated and proliferating [42, 43] as well as in regulatory CD4+ T cells [44, 45]. When infected T cells undergo clonal expansion, the integrated proviral genome is transferred to daughter cells [46–49]. In lymph nodes, follicular helper T cells have been shown to contribute to the viral reservoir [50, 51]. CD4+ resident memory T cells from ART-treated women whose plasma viral load was undetectable for up to 10 years, have been identified in cervical tissues; this may constitute another cellular reservoir [52]. Considering the wide distribution of resident CD4+ memory T cells in tissues throughout the body, this cell subset may contribute to persisting HIV-1 reservoirs [52, 53]. Notably, CD4+ T cells undergoing an effector-to-memory cell type transition have temporarily upregulated CCR5 expression and downregulated global gene expression, making them ideal for harbouring latent HIV-1 [54].
Myeloid Progenitors
Cells derived from myeloid progenitors, namely, macrophages, monocytes, basophils, and mast cells, can express HIV-1 receptors and co-receptors [55–60]. Despite this observation, the contribution of these cells to the HIV-1 reservoir is not fully understood.
Although macrophages exhibit lower CD4 expression than CD4+ T cells, macrophage tropic viruses can adapt to this low level of expression to infect macrophages [61, 62]. The self-renewal capabilities of tissue-resident macrophage populations makes them ideal for harbouring latent HIV-1 [63, 64]. Minor populations of HIV-1 isolated from patients undergoing ART interruption are macrophage-tropic and express macrophage markers indicative of macrophage origin [65]. For instance, a study using penile tissue from ART-suppressed individuals recovered replication-competent HIV-1 from urethral macrophages [66]. This suggests that macrophages can form a latent HIV-1 reservoir in patients undergoing ART, with re-activation of the virus following treatment interruption, although further studies are needed to confirm this.
Classical monocytes (CD14+ CD16−), which comprise the majority of the circulating monocyte pool, are derived from the bone marrow and circulate in the bloodstream before migrating into surrounding tissues [67]. Migration of classical monocytes to the tissues is initiated during both steady-state and inflammatory conditions to produce dendritic cells or macrophage progeny [67, 68]. Despite being permissive to HIV-1 infection, circulating classical monocytes do not represent a stable reservoir since they have a short half-life and undergo apoptosis after approximately 24 hrs if migration into surrounding tissues has not taken place [67, 69–71]. Although infected classical monocytes do not technically represent an HIV-1 reservoir, they can cross the blood-brain barrier and differentiate into macrophages capable of transferring HIV-1 to astrocytes, microglia, and dendritic cells within the CNS, which form a prominent HIV-1 reservoir due to their longevity [72–77].
The role of other myeloid progenitor-derived cells such as basophils and mast cells in HIV-1 persistence is unclear. Some studies have shown that basophils and mast cells can express HIV-1 receptors and co-receptors on the cell surface [58–60]. Additionally, mast cells express cell surface HIV-1 attachment factors, such as heparan sulfate proteoglycan (HSPG) and α4β7 integrin [78]. Expression of HIV-1 receptors/co-receptors and/or attachment factors suggests that basophils and mast cells are susceptible to HIV-1 and may play a supporting role in the formation of HIV-1 reservoirs by disseminating the virus and trans-infecting susceptible CD4+ T cells [58–60, 78].
Hematopoietic Stem/Progenitor Cells
The self-renewal capabilities of haematopoietic stem/progenitor cells (HSPCs) make them ideal HIV-1 reservoir candidates. The jury is still out as to whether HSPCs can become infected with HIV-1 and function as an HIV-1 reservoir [79]. Some studies claim that HSPCs can become infected with HIV and contribute to an HIV-1 reservoir [80, 81], whereas others were unable to find evidence of HIV infection in HSPCs [82, 83]. However, research is starting to identify specific HSPC populations that express sufficient levels of CD4, CCR5 and CXCR4 to become infected with HIV-1 [84–88]. A relatively recent study that screened for integrated HIV-1 provirus found the presence of productive and latent infection in freshly isolated HSPCs from HIV-1-infected donors receiving ART. They reported that specific HSPC populations can be infected with and produce HIV-1 and harbour latent proviral genomes [84]. The role of HSPCs in HIV persistence and the identification of HSPC subpopulations that serve as HIV reservoirs needs to be clarified before HSPCs can be regarded as long-lived viral reservoirs. In addition, HSPCs infected with and/or harbouring latent HIV-1 may play a role in haematological co-morbidities observed in HIV-1 infected individuals; however, these co-morbidities could also result from the indirect effect of HIV-1 on other cell types [25, 81, 89, 90].
Techniques Used to Detect HIV-1 Reservoirs
Detecting Viral RNA/DNA and Genomic Integration
Polymerase chain reaction (PCR) techniques such as reverse transcription PCR (RT-PCR), quantitative PCR (qPCR), reverse transcription, quantitative PCR (RT-qPCR) and digital droplet PCR (ddPCR) are commonly used to detect the presence of HIV-1 RNA or DNA in host cells upon infection [64, 91]. These techniques can also be used to test for the integration of HIV-1 DNA into the human genome. RT-PCR and RT-qPCR measure levels of viral RNA in a non-quantitative and quantitative manner, respectively, before reverse-transcription to proviral DNA, while quantitative PCR (qPCR) and ddPCR measure levels of viral DNA either before or after integration into the host genome [91]. These techniques are cost-effective but can overestimate viral titre, since not all HIV-1 proviruses are replication-competent [64]. Additionally, the above-mentioned techniques are often not specific for integrated HIV-1 [64]. Nested Alu-gag PCR, a technique which combines PCR detection of HIV-1 gag gene sequences with detection of repetitive Alu sequences in the human host, can be employed to quantify integrated DNA to provide a more accurate estimation [92]. Additionally, to decrease the overestimation that often occurs with conventional PCR techniques, a newly introduced Q4PCR technique targets four conserved regions of the HIV-1 genome in an approach that combines both qPCR and next-generation sequencing (NGS) [43, 93].
Detecting Productive Infection
Conventional and digital p24 enzyme-linked immunosorbent assay (ELISA) methods are used to detect productive HIV-1 infection by quantitatively measuring levels of the HIV-1 capsid protein (p24) in plasma, serum, or cell culture supernatant [94]. An estimation of the productive viral load can be calculated from p24 levels; however, defective proviruses can still produce p24 proteins, leading to an overestimation of infectious viral levels [95]. A recent technique, the Tat/rev induced limiting dilution assay (TILDA), has been introduced to avoid overestimation of the productive viral titre. TILDA reduces productive viral titre overestimation by measuring the levels of viral Tat/rev multiply-spliced RNA (msRNA) [96]. Tat/rev msRNA is transcribed in productively but not latently infected cells unless these cells are activated with LRAs. The TILDA assay utilises this principle to measure the frequency of productively infected cells [96]. TILDA is accurate and sensitive; however, viral reservoir amounts can still be overestimated due to the occurrence of defective transcripts [70].
Detecting Latent Infection
To measure the size of the latent reservoir, the TILDA assay can be used on cells that have been exposed to LRAs such as phytohemagglutinin (PHA) which reactivates gene expression of latent HIV-1 [70]. The TILDA assay can become a powerful tool for determining the percentage of productively and latently infected cells when combined with LRAs [70]. Full-length individual proviral sequencing (FLIPS), an alternative for estimating the viral load of latent reservoirs, utilises NGS to detect near full-length proviral sequences [97]. FLIPS determines the proportion of replication-competent virus by identifying if there are defects within integrated HIV-1 proviral sequences that would potentially render the virus incapable of replication [97]. Using this principle, FLIPS can be used to detect replication-competent latent virus lying dormant within cells. This technique is time-consuming but gives highly accurate results; however, errors may occur during NGS library construction [43, 64].
HIV-1 quantitative viral outgrowth assays (qVOAs) in conjunction with p24 ELISA are considered to be the gold standard for measuring replication-competent latent HIV-1 [43]. Initial steps involve activation of infected cells with a mitogen and co-culturing these cells in serial dilution with PHA-activated, uninfected peripheral blood mononuclear cells (PBMCs) [98]. These steps induce reactivation and expansion of latent HIV-1, which can then be quantified using p24 ELISA techniques which provides an estimated size of the viral reservoir [99]. Table 1 summarises the different techniques along with relevant studies used to detect HIV-1 integration, infection, and latency in CD4+ T cells and where available, in MSCs.
Table 1.
Studies detecting HIV-1 infection and latency in CD4+ T cells and MSCs
| Technique | Purpose | Target | Technique used on activated cells*? | Relevant studies using CD4+ T cells to establish techniques | Relevant studies using MSCs (Papers discussed in this review) |
|---|---|---|---|---|---|
| PCR | Presence of viral DNA in cells exposed to HIV-1 | HIV-1 specific DNA such as gag | No |
BM-MSCs: [100] Vessel wall-derived MSCs: [101] Adipose cells/preadipocytes: [32] |
|
| qPCR | Genomic integration of HIV-1 into the host genome | Conserved genomic regions of HIV-1 DNA, including pol, gag, tat and/or episomal 2-LTR circles | No | [104–110] | ASCs: [111] |
| ddPCR | Conserved genomic regions of HIV-1 DNA, including pol and gag, and/or episomal 2-LTR circles | No | [112–114] | ||
| Nested qPCR | Alu elements in the human genome and HIV-1 gag and/or LTR DNA regions | No | [92, 115, 116] |
Vessel wall-derived MSCs: [101] BM-MSCs: [117] |
|
| Q4PCR | HIV-1 packaging signal, gag, pol, and env DNA regions | No | [93] | ||
| ELISA (conventional and digital) | Productive infection | p24 | No | [118–121] |
Vessel wall-derived MSCs: [101] ASCs: [111] Adipose cells/preadipocytes: [32] |
| TILDA | Tat and rev multiply spliced RNA | Yes or no | [96] | ||
| Latent infection | |||||
| FLIPS | Full-length HIV-1 genomic DNA | No | [124] | ||
| qVOA and ELISA | p24 | Yes | [125–128] |
*Activated with latency reversal agents, such as PHA and PMA
Abbreviations: ASCs Adipose-derived mesenchymal stromal/stem cells, (BM)-MSCs (Bone marrow-derived) mesenchymal stromal/stem cells, ddPCR digital droplet polymerase chain reaction, DNA deoxyribonucleic acid, ELISA enzyme-linked immunosorbent assay, env HIV-1 envelope protein, FLIPS full-length individual proviral sequencing, gag HIV-1 group specific antigen, LTR long terminal repeat, MSCs mesenchymal stromal/stem cells, PHA phytohaemagglutinin A, PMA phorbol 12-myristate 13-acetate, pol HIV-1 polymerase, PCR polymerase chain reaction, qPCR quantitative PCR, Q4PCR combined quadruplex qPCR, qVOA quantitative viral outgrowth assay, rev HIV-1 response element protein, RNA ribonucleic acid, tat HIV-1 transactivating protein, TILDA Tat/rev induced limiting dilution assay
Mesenchymal Stromal/Stem Cells
Adult stem cells maintain tissue homeostasis by affecting tissue specific turnover and repair. The fate of stem cells is controlled by their specialised microenvironment, referred to as the stem cell niche, through cell-to-cell interactions and molecular signalling [129]. Increasing interest in MSCs has led to inconsistencies regarding their nomenclature. To clarify the terminology, the International Society for Cellular Therapy (ISCT) proposed that fibroblast-like plastic adherent cells, regardless of tissue origin, be named multipotent mesenchymal stromal cells, while reserving the term mesenchymal stem cell for a subset of cells that clearly demonstrate stem cell properties [130, 131].
Sources of MSCs
MSCs are commonly isolated from the bone marrow [132] or adipose tissue [133, 134]; however, MSCs can also be isolated from neonatal-derived tissues such as umbilical cord blood (UCB) [135] and Wharton’s Jelly [136, 137], and from vessel walls [138, 139]. The International Federation for Adipose Therapeutics and Science (IFATS) has proposed the use of the term “adipose-derived (mesenchymal) stromal cells” (ASCs) to identify the plastic-adherent, multipotent cell population isolated from adipose tissue [140]. MSCs isolated from bone marrow are referred to as bone marrow-derived MSCs (BM-MSCs).
In Vitro Characterisation of MSCs
Stromal cells are classified as MSCs in vitro if they meet the following minimal criteria proposed by the ISCT: they need to 1) be plastic adherent in culture; 2) display a specific immunophenotype, with ≥95% of the MSC population being positive for CD105, CD73 and CD90 and ≤2% expressing CD45, CD34, CD14 or CD11b, CD79α or CD19 and HLA-DR; and 3) show multilineage differentiation capacity into adipocytes, osteoblasts and chondroblasts [141]. In 2013, IFATS and ISCT made a joint statement to define stromal cells specifically from adipose tissue [140]. They proposed the use of multi-colour flow cytometry analysis to confirm the phenotype of ASCs along with qualitative and quantitative evaluation of differentiation capacity. They defined ASCs as being positive for the cell surface markers CD90, CD73, CD29, CD105/CD13 and CD44 and negative for CD45 and CD31. Furthermore, they suggested that by including CD36 and CD106, MSCs from bone marrow and adipose tissue could be distinguished from one another: BM-MSCs are positive for CD106 and negative for CD36, whereas ASCs are negative for CD106 and positive for CD36.
In Vivo Characterisation of MSCs
The in vivo localisation, identification, and role of MSCs in their multiple anatomical locations is still poorly characterised. Whether they constitute a specific homogenous cell type, or a heterogeneous population, still needs to be elucidated. In search of the in vivo identity of MSCs, it has been postulated that MSCs exist throughout the body as pericytes due to their perivascular location [142, 143]. In the bone marrow niche, MSCs support haematopoiesis through the secretion of cytokines, growth factors and chemokines, and participate in bone remodelling and metabolism [144–146]. In the adipose niche, ASCs reside in the stromal vascular fraction (SVF) as multipotent precursor cells involved in adipocyte turnover [147, 148].
MSCs Express Receptors Required for HIV-1 Entry In Vitro
Different techniques can be used to detect, quantify, and characterise cell surface receptors [149]. Cell surface receptors are proteins, and thus they are synthesised via the same process as other proteins; DNA coding for the receptor is transcribed to mRNA, followed by translation of mRNA to the protein product. The detection of receptors can be performed at the mRNA level and at the protein level. RT-PCR/RT-qPCR detects mRNA levels as an indication of receptor transcription; however, the amount of mRNA does not necessarily correlate with protein production due to translational control, amongst other mechanisms [150, 151]. Protein expression of cell-surface or internalised receptors can be evaluated using immunostaining techniques or binding assays. Immunostaining techniques detect increased receptor-protein levels on sections of biological tissues using immunohistochemistry (IHC) or on intact cells using immunocytochemistry (ICC), whereas binding assays, such as flow cytometry, detect the presence of cell-surface receptors on live cells. These techniques use labelled (e.g., with a fluorescent dye) antibodies that specifically recognise the receptor.
It is important to understand whether MSCs express the CD4 receptor and CCR5/CXCR4 co-receptors required for HIV-1 entry before evaluating whether they can harbour latent HIV-1. Entry of HIV-1 into a host cell requires binding of HIV-1 to the CD4 receptor followed by CCR5 and/or CXCR4 co-receptor binding. Early during HIV-1 infection, CCR5 is predominantly utilised for HIV-1 entry. As infection progresses, co-receptor tropism changes can occur, increasing the frequency of CXCR4 utilisation [152]. Expression of CD4, CXCR4 and CCR5 mRNA by MSCs has been observed in vitro [101, 102, 111, 117, 153–161]. However, the reported level of expression and localisation of these receptors/co-receptors is inconsistent.
Expression of CD4
Using RT-qPCR, Cotter et al. found that HIV-1-exposed and unexposed human BM-MSCs express CD4 at the mRNA level [117]. CD4 glycoproteins were also detectable on the cell surface using ICC. In contrast, studies by Scadden et al. and Gibellini et al. failed to detect CD4 on the surface of human BM-MSCs and vessel wall-derived MSCs by flow cytometry [101, 102]. However, when the cells were fixed and permeabilised, intracellular CD4 was detected by flow cytometry in 20% of MSCs [101]. Additionally, CD4 mRNA was detectable in these MSCs using PCR and RT-qPCR techniques. The authors concluded that while CD4 was mainly expressed intracellularly, the very low levels expressed extracellularly may have been below the detection limit of flow cytometry [101]. Based on the above-mentioned findings, human MSCs appear to predominantly express CD4 intracellularly; however, additional studies are needed to address discrepancies in cell surface CD4 detection when using different experimental techniques, such as flow cytometry and ICC.
Expression of CCR5
Contradictory results have been reported regarding CCR5 expression on human BM-MSCs. Sordi et al. and Karnoub et al. failed to detect CCR5 mRNA and protein on human BM-MSCs [156, 162]; whereas other studies suggest that BM-MSCs are capable of both CCR5 mRNA and protein expression, albeit at varying levels between studies [117, 160, 163, 164]. Similar inconsistencies in CCR5 expression have been observed in murine BM-MSCs. Some studies reported low or negligible expression of cell surface CCR5 on murine BM-MSCs by flow cytometry [164, 165], while Alexeev et al. reported detectable expression [159]. A thorough study on murine BM-MSCs demonstrated expression of CCR5 mRNA and localisation of the CCR5 co-receptor on the cell surface and in the cytoplasm using a combination of RT-PCR, flow cytometry and ICC, respectively [166]. Further insight into the CCR5 expression profiles of human BM-MSCs is needed and should use a similar approach to that of Ji et al., whereby complementary techniques are used to verify CCR5 1) mRNA expression, 2) protein expression and 3) localisation of the co-receptor within the cell [166].
A paucity of research exists on CCR5 expression profiles of MSCs and MSC-like cells isolated from sources other than bone marrow, namely dermal MSCs, vessel wall-derived MSCs and ASCs. A single pilot study on human dermal MSCs found by flow cytometry that the cells had negligible CCR5 cell surface expression [167]. On the other hand, RT-qPCR and flow cytometry analysis of human vessel wall-derived MSCs indicated that these cells were capable of CCR5 mRNA and cell surface protein expression [101]. Hazan et al., Maurin et al., and Munier et al. studied the expression profiles of preadipocytes [32–34]. The methods used to isolate these preadipocytes were similar to those used to isolate ASCs [133, 134]. Thus, the cells they classified as preadipocytes may have contained a subset of ASCs. Maurin et al. demonstrated the presence of CCR5 mRNA by RT-PCR and cell surface CCR5 by ICC, while Hazan et al. and Munier et al. found low or undetectable levels of CCR5. A study by Kroeze et al. using ASCs characterised according to ISCT guidelines, demonstrated negligible expression of cell surface CCR5 [167]. Additional studies are needed to confirm whether characterised ASCs also express CCR5 mRNA and whether co-receptor expression can be detected intracellularly in addition to being located on the cell surface.
Expression of CXCR4
According to several studies, 26-96% of human BM-MSCs express CXCR4 on the cell surface [153, 154, 156, 164, 168, 169]. In contrast, Wynn et al. and Shi et al. found respectively that 83-98% and 95.9% of human BM-MSCs expressed CXCR4 intracellularly, with less than 1% of cells displaying extracellular expression [155, 169]. Inconsistencies in results may be due to different experimental techniques and culturing conditions, since extracellular CXCR4 has been shown to decrease with passage number in MSCs [153, 159, 170]. Depletion of cytokines, chemotactic and growth factors during cell culture has been proposed as a possible reason for the loss of extracellular CXCR4 [169]. The interaction of CXCR4 with cyclic AMP, prostaglandin E2 and nitric oxide mediators, interleukin (IL)-3, IL-6, IL-7, and stem cell factor (SCF) cytokines, and growth factors such as hepatocyte growth factor (HGF), commonly occur in vivo, inducing expression and cell surface localisation of the co-receptor [170–173].
Interactions of CXCR4 with its ligand, stromal-derived factor 1 (SDF-1), additionally play a large, dynamic role in the expression and cellular localisation of CXCR4. The SDF-1-CXCR4 axis is crucial for MSC chemokine-mediated migration to sites of injury [154, 155, 170, 174]. SDF-1-induced chemokine signals increase expression of CXCR4 on the surface of murine MSCs [158]. Upon binding SDF-1, MSC CXCR4 expression is decreased, and the co-receptor is internalised through endocytosis or micropinocytosis [157, 175]. CXCR4 is then internally degraded by lysosomes or is recycled back to the cell surface [175, 176].
Effect of HIV-1 on Expression Of Entry Receptors
Whether exposure to HIV-1 or HIV-1 proteins can enhance expression and cell surface localisation of HIV-1 receptors/co-receptors on MSCs is an area of active research. The interaction of HIV-1 gp120 with CXCR4 is of particular interest, since SDF-1 and gp120 both bind to CXCR4, albeit at non-intersecting N-terminal regions [172, 177–181]. An early study by Tarasova et al. found that gp120, like SDF-1, has a high affinity for CXCR4 and can cause irreversible co-receptor internalisation [182]. Other studies further discovered that the internalisation of CXCR4, as the result of gp120 binding, reduced SDF-1-CXCR4 interactions and migration of B cells, CD4+ T cells and monocytes [183–185]. However, this may not be the case with MSCs. Recently, Li et al. found that exposure of MSCs to 100 ng/mL X4- and R5-tropic HIV-1 gp120 glycoproteins increased external expression of CXCR4 by approximately 2-3-fold as measured by flow cytometry, and enhanced MSC migration [154]. This study suggested that exposure of MSCs to gp120 may prime HIV-1 to undergo a R5- to X4-tropism change by increasing the expression of CXCR4 on the MSC cell surface [154]. If this theory is valid, it would be advantageous for HIV-1 to not only increase cell surface expression/localisation of the CXCR4 co-receptor, but also the CD4 receptor, since HIV-1 entry is initiated by sequential binding to CD4 followed by CXCR4/CCR5. A study by Cotter et al. has shown that this is plausible, as MSCs exposed to high viral load (HVL; 100 000 to 150 000 copies/mL) HIV-1 sera for at least 72 hrs increased expression of extracellular CD4, as detected by ICC [117]. However, further studies are required to assess the validity of this hypothesis and to unravel the effects of HIV-1 and HIV-1 proteins on MSC receptor/co-receptor expression and localisation, particularly in vivo. Table 2 outlines studies examining expression of the HIV-1 receptors/co-receptors by human MSCs, and includes the experimental techniques used for detection of cell surface and intracellular expression.
Table 2.
HIV-1 receptor and co-receptor expression in cultured human MSCs
| MSC source | Receptor/co- receptor | Technique | Outcome | Reference | |
|---|---|---|---|---|---|
| Intracellular | Extracellular | ||||
| BM-MSCs | CD4 | RT-qPCR | Immunocytochemistry on HIV-1 exposed/unexposed cells |
Detection of mRNA. Cell surface expression, which increased with HIV-1 exposure. |
[117] |
| PCR | Flow cytometry |
Detection of mRNA. Failure to detect cell surface expression. |
[102] | ||
| CCR5 | Immunocytochemistry | Low expression on cell surface. | [117] | ||
| RT-PCR | Flow cytometry | Failure to detect mRNA and cell surface expression. | [156] | ||
| RT-qPCR | Immunocytochemistry |
Detection of mRNA. Cell surface expression. |
[160] | ||
| Immunocytochemistry | Failure to detect cell surface expression. | [162] | |||
| Immunocytochemistry | High expression in cells. | [163] | |||
| Flow cytometry | Cell surface expression on approximately 78% of cells. | [165] | |||
| CXCR4 | RT-PCR | Flow cytometry |
Failure to detect mRNA. Cell surface expression on 1% of cells. Intracellular expression in 83-98% of cells. |
[155] | |
| RT-PCR | Flow cytometry |
Detection of mRNA. Cell surface expression on approximately 26% of cells. |
[156] | ||
| RT-PCR | Flow cytometry |
Detection of mRNA. Cell surface expression on 43% of cells. |
[153] | ||
| RT-qPCR | Immunocytochemistry |
Detection of mRNA. Cell surface expression. |
[160] | ||
| Immunocytochemistry | Cell surface expression. | [117] | |||
| RT-qPCR | Flow cytometry on 100 ng/mL gp120 treated/untreated cells | Detection of mRNA. Cell surface expression on 84% of cells. Treatment with gp120 upregulated surface expression by 2-3-fold. | [154] | ||
| RT-qPCR | Flow cytometry |
Low levels of mRNA. Cell surface expression on approximately 2% of cells. |
[186] | ||
| RT-qPCR | Flow cytometry |
Detection of mRNA. Cell surface expression on approximately 29.8% of cells. |
[168] | ||
| RT-qPCR and flow cytometry | Flow cytometry |
Detection of mRNA. Intracellular expression in 95.9% of cells. Cell surface expression on 0.1% of cells. |
[169] | ||
| Flow cytometry | Cell surface expression on approximately 96% of cells. | [164] | |||
| Preadipocytes (may contain a subset of ASCs) | CD4 | RT-PCR | Detection of mRNA. | [32] | |
| RT-PCR | Immunocytochemistry |
Detection of mRNA. Failure to detect cell surface expression. |
[33] | ||
| RT-PCR | Failure to detect mRNA. | [34] | |||
| CCR5 | RT-PCR | Low levels of mRNA. | [32] | ||
| RT-PCR | Immunocytochemistry |
Detection of mRNA. Cell surface expression. |
[33] | ||
| RT-PCR | Failure to detect mRNA. | [34] | |||
| CXCR4 | RT-PCR | High levels of mRNA. | [32] | ||
| RT-PCR | Immunocytochemistry |
Detection of mRNA. Cell surface expression. |
[33] | ||
| RT-PCR | Detection of mRNA. | [34] | |||
| ASCs | CCR5 | Flow cytometry | Negligible cell surface expression. | [167] | |
| CXCR4 | Flow cytometry | Cell surface expression on approximately 1.21% of cells. | [167] | ||
| Dermal MSCs | CCR5 | Flow cytometry | Negligible cell surface expression. | [167] | |
| CXCR4 | Flow cytometry | Cell surface expression on approximately 1.31% of cells. | [167] | ||
| Vessel wall-derived MSCs | CD4 | RT-qPCR and flow cytometry | Flow cytometry |
Detection of mRNA. Intracellular expression in 20% of cells. Failure to detect cell surface expression. |
[101] |
| CCR5 | RT-qPCR | Flow cytometry |
Detection of mRNA. High levels of expression on cell surface. |
||
| CXCR4 | RT-qPCR | Flow cytometry |
Detection of mRNA. High levels of expression on cell surface. |
||
| Fetal blood MSCs | CXCR4 | RT-PCR and flow cytometry | Flow cytometry | Detection of mRNA. Cell surface expression on 23-25% of cells. Intracellular expression in 78-83% of cells. | [158] |
| Fetal BM-MSCs | CXCR4 | RT-qPCR and flow cytometry | Flow cytometry | Low levels of mRNA. Cell surface expression on 3.8% of cells. Intracellular expression in 50-90% of cells. | [157] |
Abbreviations: ASCs adipose-derived mesenchymal stromal/stem cells, (BM)-MSCs (Bone marrow-derived) mesenchymal stromal/stem cells, CCR5 C-C-motif chemokine receptor type 5, CD4 cluster of differentiation type 4, CXCR4 C-X-C-motif chemokine receptor type 4, gp120 HIV-1 glycoprotein 120, mRNA messenger ribonucleic acid, MSCs mesenchymal stromal/stem cells, PCR polymerase chain reaction, RT-PCR reverse transcription PCR, RT-qPCR reverse transcription quantitative PCR, qPCR quantitative PCR
In Vitro Infection of MSCs with HIV-1
Since MSCs express CD4, CXCR4 and CCR5 receptors/co-receptors that are required for HIV-1 infection [117], studies in vitro have sought to establish whether MSCs could be infected with the virus. A limited number of studies can be found in the literature investigating the infectability of MSCs by HIV-1. Early studies focused on BM-derived stromal cells and the effect HIV-1 had on these cells. Recent studies have focused more on the effect of HIV-1 and HIV-1 proteins on MSC characteristics which will be described in the subsequent section.
Early Experiments
In the early 1990’s, Scadden et al. reported that primary human BM-derived stromal cells are susceptible to HIV infection [102]. Since these cells were characterised as being free of monocyte/macrophage or endothelial cell contamination, they were referred to as BM stromal fibroblasts. MSCs are morphologically indistinguishable from fibroblasts and share surface immunophenotype, proliferation and differentiation capacity. It has thus been proposed that MSCs may originate from a fibroblast lineage [187]. The BM stromal fibroblasts isolated and investigated by Scadden et al. may be representative of the cells we label as BM-MSCs today. Cultures of primary BM stromal fibroblasts were positive for p24 antigen when exposed to X4-tropic HIV-1IIIB. When co-cultured with either H9 lymphoid or KG-1 myeloid cells, the BM stromal fibroblasts were capable of transferring HIV to lymphoid or myeloid cells. Using PCR, HIV-1 DNA was detectable for up to 4 weeks post-infection. However, following exposure to R5-tropic HIV-1Ba-L or HIV-1RJ9533 strains, the authors could not detect p24 antigen or viral DNA by PCR, indicating that exposure to HIV-1 failed to result in HIV-1 infection.
Canque et al. investigated whether primary human BM-derived stromal cells (containing a mixture of stromal cells, fibroblasts, endothelial cells, and macrophages) are susceptible to HIV [103]. Once the BM-derived stromal cultures reached confluency, non-adherent cells were removed and the cultures exposed overnight to either R5-tropic HIV-1Ada, HIV-1Ba-L and HIV-1JR-FL strains or X4-tropic HIV-1LAI and HIV-1MN strains. BM-derived stromal cells were positive for p24 antigen when exposed to R5-tropic HIV-1 strains but failed to produce detectable p24 when exposed to X4 tropic HIV-1 stains. To identify which subset of BM-derived stromal cells was producing virus, ICC was used and identified macrophages (CD14+CD68+CD45+TE7-) as being responsible for the detected p24 expression (Kal-1+) whereas fibroblasts (CD14-CD68loCD45-TE7-) were p24-negative (Kal-1-). In contrast to Scadden et al. who reported that stromal fibroblasts supported HIV-1IIIB infection and replication, Canque et al. showed that BM-derived stromal cells depleted of macrophages did not produce p24 when exposed to X4-tropic HIV-1LAI or R5-tropic HIV-1Ba-L. However viral DNA (123 bp, HIV-1 pol) could be detected and coculture with PHA-treated lymphocytes could rescue viral levels only in HIV-1Ba-L exposed macrophage depleted BM-derived stromal cells. Exposure to higher HIV-1LAI virus titres resulted in low levels of p24 which could mean that there is a correlation between infectability and virus titre. Marandin et al. confirmed the studies by Canque et al. showing that productive infection could be established in BM-derived stromal cells by p24 expression when exposed to R5-tropic but not X4-tropic HIV-1 strains [123].
Experiments by Bahner et al. demonstrated that incubation of BM-derived stromal cell monolayers with R5-tropic HIV-1JR-FL at an MOI of 1 could initiate a productive low level viral infection which could not be achieved at lower MOIs (MOI < 1) [122]. The low-level infection was detectable by p24 antigen ELISA 3 weeks after inoculation and could be amplified by co-culturing the BM-derived stromal cells with monocytes, suggesting that the BM-derived stromal cells were infected with HIV-1 [122]. In contrast, co-culturing with stromal cells incapable of supporting HIV-1 infection, such as murine stromal cells, did not rescue viral levels [122]. These experiments show that BM-derived stromal cells can be infected in vitro with HIV-1 at high MOI, but that the level of virus being produced is low. Low level expression of viral proteins and viral replication could be an indicator of viral persistence through latency [188]; however, identification of latency was not the focus of this paper and was not evaluated further.
Research by Wang et al. supported the findings of Bahner et al. [100]. The authors exposed BM-derived stromal cells to dual-tropic HIV-1RF (MOI = 1) 24 hrs post-culturing for a week. The viral levels were approximated post-exposure by quantifying p24 on a weekly basis for 2-3 weeks using ELISA. To amplify HIV-1, 2- to 3-week post-exposure BM-derived stromal cells were co-cultured with uninfected T cells and p24 levels were measured using ELISA after a week of co-culturing. Negligible or low levels of p24 were detectable in the BM-derived stromal cells without co-culture, while stromal cells co-cultured with T cells demonstrated amplification of viral levels for up to 3-4 weeks post-infection. PCR was performed to test for the presence of HIV-1 proviral DNA within the BM-derived stromal cells. HIV-1 DNA products of 115 bp were detected. These results collectively suggested viral persistence in the BM-derived stromal cells and transmission to T cells. To test which BM stromal subpopulations harboured HIV-1 DNA, the authors performed in situ hybridisation using an HIV-1 DNA fragment as a probe. Stromal subpopulations which tested positive for HIV-1 DNA also stained positive for CD83, a dendritic cell/dendritic precursor marker detected using immunostaining methods. Thus, the authors concluded that CD83+ subpopulations may be the cell type in BM-derived stromal cells responsible for harbouring HIV-1.
The early studies by Scadden, Canque, Bahner, Wang and colleagues, which utilised adherent BM-derived stromal cells for experiments, seemed to indicate that BM-MSCs could be infected with HIV-1 [100, 102, 103, 122]. However, the studies did not pinpoint which BM-derived stromal cells were infected by confirming the immunophenotype of the infected cells, as this was not considered necessary at the time. Stroma isolated from bone marrow may contain fibroblasts, macrophages, adipocytes, and endothelial cells, in addition to MSCs. These cells can have overlapping characteristics. For instance, CD83+ stromal cell fractions can contain BM-MSCs and dendritic cells, since both cell types are semi-adherent and express CD83 [189]. Therefore, immunophenotyping using techniques such as flow cytometry is now considered to be an important approach for defining stromal cell sub-populations [140, 141].
A study by Fazeley et al. evaluated the infection of primary human placental fibroblasts with HIV-1 [190]. The isolated cells were classified as mesenchymal due to their expression of vimentin (mesenchymal cells) and lack of factor-VIII (endothelial cells) and CD14 (monocytes/macrophages) expression. MSCs have also been found to reside in different perinatal tissues such as placental tissues (amniotic membrane, chorionic plate, and decidua parietalis) and the umbilical cord [191]. It can thus be postulated that the placental fibroblasts evaluated by Fazeley may have contained a subset of the cells we know today as MSCs. The authors reported that placental fibroblast infection by HIV-1Lai and HIV-1IIIB could only be detected by rescuing virus with susceptible target cells, which is suggestive of latent infection.
Other early studies were also interested in adipose cells as possible targets for HIV-1 [32–34]. These studies however did not analyse MSCs/ASCs per se but evaluated preadipocytes isolated and differentiated from human subcutaneous adipose tissue. The methods used to isolate these preadipocytes [32, 33] are similar to those used to isolate ASCs [133, 134]. Thus, the cells they classified as preadipocytes may have contained a subset of ASCs. Hazan et al. isolated preadipocytes from adipose tissue harvested from HIV-1 seronegative donors, and investigated whether these adipose cells supported HIV-1 entry [32]. Primary human preadipocytes exposed to HIV-1 X4- and R5-trophic viruses had low p24 levels. Additionally, analysis of genomic DNA from preadipocytes 10-15 days post HIV-1 exposure resulted in the detection viral DNA specific for the HIV-1 gag region. Taken together, preadipocytes appear to be susceptible to HIV-1 infection; however viral integration and expression was not evaluated. The low-level viral production may be indicative of an infectious replication process, but whether this was efficient viral production was not clearly demonstrated. Reassessment of these experiments by the same authors concluded that adipose tissue cannot be infected with HIV-1 in vitro since HIV-1 receptor and co-receptor levels did not permit entry [34]. In addition, adipose cells obtained from donor biopsies failed to show high levels of HIV-1 receptor and co-receptor expression, which may be the limiting factor for efficient viral entry. They further propose that once the entry step is bypassed, viral replication can occur efficiently in adipose cells, but that this is likely to be a rare event. Maurin et al. performed similar experiments on adipose cells from human subcutaneous adipose tissue [33]. They demonstrated that inefficient HIV-1 replication in adipose cells was due to a lack of activation of adipocyte-signalling pathways. HIV-1 was able to infect human adipocytes in vitro and HIV-1 genes were expressed upon stimulation with pro-inflammatory cytokines. It will be important to repeat the experiments performed by Hazan, Maurin and colleagues using cells characterised as ASCs according to the criteria set out by ISCT and IFATS, to determine whether ASCs can indeed be infected with HIV-1 and contribute to the HIV-1 reservoir. Consensus regarding the expression of CD4, CCR5 and CXCR4 on adipose cells (primary culture preadipocytes, differentiated adipocytes, adipose tissue biopsies etc) has not been reached.
Recent Experiments
More recent in vitro HIV-1 infection studies using MSCs phenotypically identified by flow cytometry have generated contrasting results. Gibellini et al. found that undifferentiated, vessel wall-derived MSCs display evidence of infection when exposed to classical laboratory HIV-1 strains HIV-1lIIb (X4-tropic) and HIV-1ada (R5-tropic) [101]. HIV-1 proviral DNA for both strains were detectable in vessel wall-derived MSCs by PCR for HIV-1 gag (142 bp). Host cell integrated HIV-1 DNA (100 bp) was also detectable by nested Alu-gag PCR [101]. In support of this observation, exposure of these vessel wall-derived MSCs to HIV-1 was found to increase apoptosis, which was attenuated following treatment with CD4 antagonists, suggesting possible HIV-1 CD4 receptor utilisation [101]. HIV-1 p24 levels were analysed by ELISA to determine the presence of replicative HIV-1, and were found to be low and decreasing, suggesting possible low-level infection [101].
Cotter et al. observed results similar to those of Gibellini et al. [117]. They detected HIV-1 products (100 bp) using nested Alu-gag PCR which is indicative of viral integration, both in DNA from HIV-1 positive patients and DNA extracted from HVL-exposed BM-MSCs. Exposure of BM-MSCs to HVL HIV-1 positive sera for 72 hrs also altered MSC properties, which was no longer present upon treatment of the cells with antiretroviral drugs or CD4 antagonists. The authors concluded that the effect of HIV-1 on BM-MSCs may be due to infection and integration into the host genome. In subsequent experiments, the authors could not verify a productive infection of HIV-1 exposed BM-MSCs, since viral p24 and Tat proteins were undetectable by ELISA and ICC, respectively. However, the authors could detect Rev, the first protein to be translated in the HIV genome, by ICC. This partial transcription of viral proteins may have been due to a latent infection.
Nazari Shafti et al. found that undifferentiated ASCs exposed to low levels of HIV-1 (MOI of 0.1) had negligible p24 levels in the culture supernatant, and expression of Gag and Tat HIV-1 proteins was not evident in ASCs [111]. Productive infection was not detected in HIV-1 exposed ASCs, whereas ASCs that underwent differentiation into cells with hematopoietic attributes, referred to as hematopoietic differentiated (HD) cells, and then exposed to HIV-1, presented with detectable p24 levels, Gag and Tat mRNA, all indicative of productive infection. However, HD cells derived from ASCs pre-exposed to HIV-1 failed to show evidence of productive infection [111].
Effect of HIV-1 on MSC Properties
HIV-1 infected individuals display bone and lipid toxicities; however, the exact mechanism underlying these toxicities is not fully understood. Interest has arisen in determining the effects of HIV-1 on MSCs and whether bone and lipid toxicities in HIV-1 positive individuals might result from altered MSC function. MSCs are involved in lipid metabolism and bone mineralisation and can differentiate into adipocytes and osteoblasts in vitro [192]. The formation of fat and bone is inversely related [193, 194] and is controlled by an antagonistic balance between peroxisome proliferator-activated receptor gamma (PPARγ, which induces adipogenesis) and the runt-related transcription factor-2 (RUNX-2, which drives osteogenesis) [195]. The effect of various HIV-1 proteins, such as Tat, gp120, p55-gag, Rev and Nef on MSC properties has been investigated and will be discussed in detail below. Table 3 summarises this information and highlights the lack of studies investigating the effects of HIV-1 proteins on ASCs.
Table 3.
MSC characteristics affected by HIV-1 infection and/or exposure to HIV-1 proteins in vitro
| MSC source | Treatment | Characteristic | Outcome | Proposed mechanism | Reference |
|---|---|---|---|---|---|
| Vessel wall-derived MSCs |
HIV-1 strains: HIV-1IIIb HIV-1ada HIV-1 gp120 protein |
Apoptosis | Induced apoptosis in sub-confluent MSCs but failed to induce apoptosis during differentiation. | Direct interaction of gp120 and CD4 with the cell membrane led to apoptosis. Blocking gp120 or CD4 reversed the activation of apoptosis. | [101] |
| Adipogenic differentiation | Enhanced adipogenesis. | Upregulates expression of C/EBP β, C/EBP δ, adipsin and PPARγ mRNA. | |||
| Endothelial cell differentiation | Inhibited endothelial differentiation. | Downregulates expression of endothelial markers vWF, Flt-1 and KDR (as detected by flow cytometry and RT-qPCR). | |||
| Vessel wall-derived MSCs | HIV-1 Tat protein | Apoptosis | Induced apoptosis in sub-confluent MSCs with high Tat concentration but failed to induce apoptosis during differentiation. | Tat modulates cell proliferation and survival by inducing pro-apoptotic or anti-apoptotic responses that are dependent on Tat concentration. | [196] |
| Adipogenic differentiation | Enhanced adipogenesis. | Upregulates expression of C/EBP β, C/EBP δ, and PPARγ mRNA. | |||
| Endothelial cell differentiation | Inhibited endothelial differentiation. | Downregulates expression of endothelial markers vWF, Flt-1 and KDR. | |||
| BM-MSCs | HIV-1 Rev and p55-gag protein | Osteogenic differentiation |
Rev: pro-osteogenic effect. p55-gag: decreased osteogenesis. |
Rev increases calcium deposition and ALP activity. p55-gag decreases calcium deposition and ALP activity by downregulating BMP-2 and osteocalcin secretion and RUNX-2 expression. | [197] |
| BM-MSCs | HIV-1 p55-gag and Rev protein | Osteogenic differentiation |
p55-gag: reduced overall level of osteogenesis. Rev: increased overall rate of mineralisation. |
p55-gag leads to an earlier increase in CTGF levels, RUNX-2 activity and BMP-2 secretion. Rev reduces BMP-2 secretion, RUNX-2 activity, CTGF levels and ALP activity. | [198] |
| BM-MSCs | HIV-1 Tat and Nef protein | Cell senescence | Reduced proliferative activity and induced senescence. | Results from increased oxidative stress and mitochondrial dysfunction. Tat induces early increase in NF-κB activity and cytokine/ chemokine secretion, and these effects are prevented with an NF-κB inhibitor. Nef induces early inhibition of autophagy, and this effect is reversed with an autophagy inducer. | [199] |
| Osteogenic differentiation | Decreased osteogenic differentiation. | Lower level of RUNX-2 protein, decreased RUNX-2 mRNA expression and decreased level of osteocalcin secretion. | |||
| BM-MSCs | HIV-1 p55-gag protein | Senescence | Inhibited proliferation and induced senescence. | Not reported. | [200] |
| Hematopoietic supportive function | Reduced HSC colony forming capability, HSC expansion and p55-gag BM-MSCs did not express TPO and HGF (by RT-qPCR and ELISA) and had decreased expression for Flt3L, SCF, IL-7, and IL-8 cytokines. | BM-MSC senescence and decreased production of cytokines impacts HSC homeostasis. | |||
| BM-MSCs |
HIV-1 Tat protein Transfected with HIV-1 tat mRNA |
Hematopoietic supportive function | Reduced expansion and CFU forming capacity of HSCs co-cultured with BM-MSC. BM-MSCs lost their ability to assist hematopoietic recovery after co-transplantation of HSCs and BM-MSCs in vivo. Decreased expression of hematopoietic cytokines (TPO, Flt3L, SCF, IL-7 and IL-8) by BM-MSCs. | Tat protein inhibits hematopoietic support function of BM-MSCs by reducing hematopoietic cytokine expression by BM-MSCs possibly induced by BM-MSC senescence. | [201] |
| BM-MSCs | HIV-1 gp120 protein | Cell migration | Enhanced cell migration of MSCs in response to SDF-1. | Upregulates CXCR4 (protein and mRNA expression as well as cell surface expression), leading to amplification of the FAK-Paxillin and ERK 1/2 signalling pathways. | [154] |
| ASCs | HIV-1 Tat and Nef protein | Proliferation and ECM production | Induced profibrotic phenotype. Increased mRNA and protein levels of collagen 1-α1. | Increases secretion of fibronectin and TGF-β1 and expression of the myofibroblast marker, αSMA. | [202] |
| Adipogenesis and ECM production | Nef, but not Tat decreases cellular lipid accumulation. Nef induced collagen 1-α2 expression and both HIV-1 proteins induced collagen 6-α1 expression. | Downregulates expression of PPARγ and FABP4 (protein and/or mRNA). | |||
| BM-MSCs |
HIV-1 virus HIV-1 Tat protein |
Clonogenic ability and developmental potential | Suppressed the proliferation and differentiation of MSC-derived colony formation. | Increases levels of inflammatory cytokines being released. | [100] |
Abbreviations: (BM)-MSCs (Bone marrow-derived) mesenchymal stromal/stem cells, ASCs adipose-derived mesenchymal stromal/stem cells, HSCs hematopoietic stem cells, TPO thrombopoietin, Flt3L Flt-3 ligand, SCF stem cell factor. IL interleukin, HGF hepatocyte growth factor, BMP-2 bone morphogenic protein-2, RUNX-2 Runt-related transcription factor 2, PPARγ Peroxisome proliferator-activated receptor gamma, CTGF Connective tissue growth factor, ALP alkaline phosphatase, vWF von Willebrand factor, Flt-1 Fms related receptor tyrosine kinase-1, KDR kinase insert domain, C/EBP β or δ CCAAT/enhanced-binding protein beta or delta, FABP4 Fatty acid-binding protein 4, ERK 1/2 Extracellular signal-regulated kinase 1/2, FAK Focal adhesion kinase, αSMA alpha smooth muscle actin, TGF-β1 Transforming growth factor beta 1, CXCR4 CXC-chemokine receptor 4, NFκB Nuclear factor kappa-light-chain-enhancer of activated B cells, CFU colony forming unit, RT-qPCR reverse transcription quantitative PCR, gp120 HIV-1 glycoprotein 120
Viability, Proliferation, And Differentiation Capacity
In an ex vivo model, cultured human BM-MSCs were exposed to serum from uninfected donors as well as treatment naïve HIV-1 positive patients with a HVL (100 000-150 000 copies/mL) or a low viral load (LVL; 200-4 000 copies/mL) for 72 hr, before being differentiated into adipogenic or osteogenic lineages [117]. The authors found that exposure to HIV-1 patient sera favoured pro-adipogenic MSC differentiation and gene expression in a viral-load specific manner, and that this was driven by Tat. The effect of HVL sera on MSCs was attenuated by reverse transcriptase-inhibiting compounds and CD4 antagonists.
In vitro, HIV-1 exposure suppressed the proliferative, colony-forming and differentiation capacity of BM-MSCs, possibly through increased expression of inflammatory cytokines [100]. The HIV-1 proteins p55-gag and gp120 reduced osteogenesis in human osteoblast cells, while Rev and p55-gag dysregulated osteogenesis in BM-MSCs [197]. In a follow-up study, exposure to Rev increased the overall rate of mineralisation whereas p55-gag had the opposite effect, reducing osteogenesis [198]. The investigators found that p55-gag and Rev could significantly alter MSC osteogenesis by altering key signalling events during the differentiation process (alkaline phosphatase (ALP) activity, calcium deposition, bone morphogenic protein- 2 (BMP-2), RUNX-2, connective tissue growth factor (CTGF)).
In human vessel wall-derived MSCs, HIV-1 and recombinant gp120 [101] and Tat [196] increased MSC apoptosis and regulated their differentiation capacity. Exposure to either gp120 or Tat increased adipogenesis by upregulating PPARγ activity, whereas endothelial differentiation was impaired by downregulating expression of endothelial markers Von Willebrand factor (vWF), Fms related receptor tyrosine kinase-1 (Flt-1) and kinase insert domain (KDR). Tat and Nef also reduced the proliferative activity of human BM-MSCs, promoting senescence and altering osteoblastic differentiation [199]. The investigators explored the mechanism of the Tat and Nef effects and found that Tat induced senescence via NF-κB pathway activation, resulting in oxidative stress, while Nef induced senescence by inhibiting autophagy. The reduced ability of BM-MSCs treated long term with Tat and/or Nef to differentiate into the osteogenic lineage correlated with decreased levels of RUNX-2 protein and mRNA expression, and osteocalcin secretion. Using an HIV-1 mouse model, BM-MSCs isolated from Tg26 HIV-1 transgenic mice showed reduced proliferation, osteogenic differentiation, function (impaired expression of multiple cytokines and chemokines) and therapeutic potential compared to BM-MSCs from healthy mice [203]. In vitro, Tat and Nef induced a profibrotic phenotype in proliferating ASCs by upregulating production of fibronectin, TGF-β1, alpha smooth muscle actin (αSMA), collagen 1-α2 and collagen 6-α1 [202]. Nef altered the adipogenic differentiation capacity of ASCs by reducing cellular lipid accumulation and expression of adipogenic markers PPARγ and fatty acid binding protein 4 (FABP4) [202].
Hematopoietic Supportive Functions
BM-MSCs are known for their hematopoietic supportive functions; however, resident MSCs could be impaired by HIV-1 proteins released by infected cells leading to the well-known cytopenia’s found in HIV-1 patients. It has been suggested that p55-gag and Tat may influence the hematopoietic supportive function of MSCs in vitro [200, 201]. HSPCs cultured with BM-MSCs exposed to p55-gag had fewer living cells and fewer colony forming units compared to HSPCs cultured with unexposed BM-MSCs [200]. BM-MSCs treated with p55-gag also showed reduced proliferation and higher senescence rates in culture. These studies postulate that senescence in BM-MSC impaired their ability to support HSPC proliferation and survival. BM-MSCs exposed to Tat displayed a reduced ability to support expansion of HSPCs in vitro. In vivo, Tat exposed BM-MSCs failed to support hematopoietic reconstitution after co-transplantation with HSPCs in sublethal-dose-irradiated NOD/SCID mice compared to HSPCs transplanted with unexposed BM-MSCs [201].
Discussion
MSCs reside in the bone marrow and in adipose tissue, both of which are sites of HIV-1 infection [31, 143, 204, 205]. The proximity of MSCs to HIV-1 susceptible cells in vivo, their haematopoietic supportive functions, involvement in regulating adipogenesis and osteogenesis, ability to home to sites of inflammation and/or tissue injury, and ability to modulate the immune response, make them a potential cell reservoir for HIV-1. The clinical application of MSCs is gaining momentum and they are being evaluated in clinical trials for musculoskeletal defects, immune system disorders and myocardial infarction, amongst others [206]. There is a need to understand whether MSCs can become infected with and harbour HIV-1 before they can be safely administered clinically. Treatment with MSCs to improve host immune reconstitution outcomes in HIV-1 positive patients treated with HAART who are immune non-responders (INRs), has been proposed [207, 208]. It would be imperative to understand the interaction of MSCs and HIV if we are to treat HIV-1 patients with MSCs. If HIV-1 negatively affects MSC properties and if MSCs can harbour HIV-1, then we cannot expect them to have an effective therapeutic effect.
The experiments described in this review focused mainly on whether MSCs could become infected with HIV-1 and how this affected their characteristics such as proliferation, differentiation capacity and hematopoietic supportive functions. These experiments fell short of being able to prove whether the low productive infection detected in MSCs was indeed due to latent infection. There is a need for additional research focused on specific areas, such as whether (i) MSCs from different sources (bone marrow, adipose tissue etc) express HIV-1 entry receptors and co-receptors at levels sufficiently high enough in vivo, ex vivo and in vitro to allow for active HIV-1 entry into these cells; (ii) MSCs support HIV-1 integration and not only HIV-1 DNA presence; (iii) MSCs allow productive and/or latent infection to occur; and (iv) MSCs can be classified as an additional HIV-1 reservoir by proving the presence and determining the size of this latent reservoir.
The levels of expression of the HIV-1 CD4 receptor and CCR5 and CXCR4 co-receptors are a limiting factor for HIV-1 entry and directly correlate with the level of productive or latent infection [209–211]. Cell surface markers such as sole markers (used to select for or purify MSCs from their in vivo environment) and stemness markers (used to identify a subset of MSCs with high fibroblastic colony-forming units (CFU-Fs) and differentiation potential) are not stably expressed on MSCs and vary between MSCs from different sources and within MSC sub-populations [212]. For example, the presence of markers Stro-1, CD271, SSEA-4 and CD146 differs between MSCs from various sources [212]. The expression of CD4, CCR5 and CXCR4 receptors/co-receptors on MSCs is no exception. HIV-1 receptors/co-receptors are detectable at the mRNA and protein levels in MSCs; however, expression levels differ between MSC sub-populations [101, 117, 153–155, 159–161, 194]. It is not yet known whether MSCs consistently express HIV-1 receptors/co-receptors on the cell surface at levels sufficient to allow for HIV-1 infection.
To conclusively confirm the presence or absence of HIV-1 entry receptors and co-receptors on MSCs, it will be imperative to understand the strengths and limitations of the techniques used when interpreting the data. Every method requires adequate controls to be included so that false-positive and false-negative outcomes can be excluded. mRNA expression data on its own is not sufficient and must be related to protein expression on the cell surface. In addition, further analysis is needed to determine whether detected mRNA levels are sufficient for protein synthesis, surface expression and functionality of the receptor. Ultimately mRNA and cell surface receptor expression data will be needed alongside functional experiments where cells are exposed to HIV-1 and infection is evaluated in order to conclude whether MSCs express functional receptors/co-receptors at a level sufficient to allow for HIV-1 entry. MSCs should also be evaluated before and after HIV-1 exposure since cell-surface receptors may only become expressed on the cell surface at detectable levels with specific signalling.
In certain HIV-1 susceptible cell types, interaction with HIV-1 or HIV-1 proteins can cause an up- or downregulation of HIV-1 receptor/co-receptor expression. For instance, CD4+ T cells from HIV-1 positive patients show upregulated CCR5 expression, and CD4+ and CD8+ T cells as well as CD14+ monocytes had downregulated CXCR4 expression compared to cells from HIV-1 negative patients [213]. The authors proposed that the upregulation of CCR5 resulted in a favourable environment for infection by R5-tropic HIV-1, the dominant viral strain type during early stages of infection. Similarly, the combined actions of HIV-1 proteins, Nef and Vpu, released post- HIV-1 infection, caused decreased cell-surface expression of CD4 on T cells, resulting in higher levels of infection [214–217]. Decreasing CD4 expression seems counterintuitive since CD4 is required for viral entry; however, it has been shown that high levels of CD4 can interfere with HIV-1 infectivity [214, 215, 218]. Although MSCs do not appear to consistently express HIV-1 receptors/co-receptors, MSCs may become primed to express elevated levels of these receptors, which is conducive to HIV-1 infection upon interaction with the virus [154].
The HIV-1 gp120 protein may be involved in priming MSCs to permit HIV-1 infection. As gp120 is shed from the viral membrane, it accumulates in tissues, such as lymphoid tissues [219]. Its presence induces apoptosis and alters the immune response to the virus, impeding viral clearance in CD34+ HSPCs and CD4+ T cells [220, 221]. Exposure to gp120 upregulates the expression of CXCR4 on MSCs, elicits apoptosis in sub-confluent MSCs, and alters their in vitro differentiation capacity by enhancing adipogenesis and inhibiting endothelial cell differentiation [101, 154]. This suggests that MSCs express HIV-1 receptors/co-receptors on the cell surface at levels that is sufficient to allow for the interaction of gp120 with MSCs and to initiate cell death pathways in these cells [101].
The transcriptome, immunophenotype and differentiation capacity of MSCs changes during in vitro culturing, and continues to change with increasing passage number [212, 222]. Additionally, enzymatic detachment of MSCs during cell culture passaging may alter or remove receptors on the cell surface, changing the observed MSC receptor expression profile [164]. Thus, the HIV-1 receptor/co-receptor expression profile in MSCs may be the consequence of cell culture conditions and needs to be evaluated in vivo or on the initial isolated cell population prior to culturing using flow cytometry or immuno-staining [223]. Expression may also vary based on tissue localisation, making it difficult to compare studies using MSCs from different sources [224–227]. To circumnavigate these challenges, MSCs, freshly harvested from ART-receiving or treatment naïve HIV-1 positive patients, should be evaluated for HIV-1 receptor and co-receptor expression prior to culturing. Affan et al. used freshly harvested or cultured synovial MSCs from tissue biopsies to confirm differences in phenotype between the cells [228]. They found that the surface marker expression of freshly harvested MSCs was distinct from that of cultured cells, emphasising the need for experiments using freshly harvested cells.
Whether MSCs can also become infected with, and harbour HIV-1 in vivo, remains to be clearly established. The viral capsid protein p24 is an early virologic biomarker of HIV-1 infection and is mostly used for diagnostic purposes during acute infection [229]. HIV-1 research in vitro uses p24 ELISA to detect productive infection. On the other hand, qVOA in combination with p24 ELISA and LRAs can be used as an indicator of viral reactivation to measure the latent HIV-1 reservoir [230]. The papers discussed in this review reported low and sometimes undetectable levels of p24 in HIV-1 exposed MSCs [32–34, 100–103, 111, 117, 122]. It is however difficult to conclude whether latent infection might account for low and undetectable p24 levels. In some studies, p24 levels in culture supernatant were measured over 4 weeks, while in other studies, levels were only measured at 1- or 2-week time-points [100, 117]. Also, MOIs of between 0.01 to 1 were used for in vitro infection studies. This is important since the time required for cells to become infected decreases with increasing MOI [231, 232]. Cells exposed to a lower MOI of 0.01 to 0.1 would have required more time to reach a detectable level of infection [231, 232]. In addition to the MOI, it is difficult to determine whether the virus used for these experiments was infectious and replication-competent, since viral propagation in a laboratory setting may introduce mutations which render HIV-1 defective [233, 234]. Co-culture experiments of HIV-1 exposed BM-derived stromal cells with cells susceptible to HIV-1 infection, such as lymphoid and myeloid cells [102], PHA-treated lymphocytes [103], monocytes [122] and T cells [100], was able to amplify viral levels. This viral amplification suggests that although HIV-1 exposed BM-derived stromal cells did not show productive infection, they may have been latently infected since they were able to infect susceptible cells. A low p24 level may imply that MSCs (i) cannot be efficiently infected with HIV-1; (ii) cannot produce HIV-1 virus or produce defective virus; (iii) harbour latent virus; or that (iv) the virus concentration used to infect MSCs was too low. Future experiments need to be carefully planned and include steps to evaluate the reason for low and undetectable p24 levels.
Integration of HIV-1 proviral DNA into host genomic DNA is an important indicator of infection alongside the measurement of viral p24 production. Integrated HIV-1 DNA may also be an indicator of latency in cells that fail to produce virus unless stimulated with LRAs [235]. To detect the presence of HIV-1 DNA in HIV-1 exposed MSCs, conventional and nested Alu-gag PCR techniques were used in several studies cited in this review. Nested Alu-gag PCR is a highly specific technique used to detect integrated HIV-1 DNA, unlike conventional PCR which cannot differentiate between DNA that exists freely within the cell and DNA integrated into the host genome. Conventional PCR experiments were utilised to test for the presence of HIV-1 DNA in BM-derived stromal cells [100, 102], vessel wall-derived MSCs [101] and BM-MSCs [117], and amplified HIV-1 gene products were detected in all the MSCs evaluated. However, only 2 studies employed nested Alu-gag PCR, and identified the presence of integrated HIV-1 gene products [101, 117]. The presence of integrated HIV-1 gene products in MSCs in addition to low or undetectable levels of p24 found in the above-mentioned experiments, suggests that MSCs may harbour latent HIV-1 [32–34, 101–103, 111, 117, 122, 189]. When designing experiments to detect the presence of integration, researchers should focus on targeting conserved HIV-1 DNA regions which are crucial for HIV-1 replication and survival, such as gag and pol, to limit the chance of detecting defective HIV-1 provirus [236]. This approach is utilised by the Q4PCR technique, which targets four conserved HIV-1 DNA regions [93].
Although various techniques have been employed to confirm that CD4+ T cells contribute to the HIV-1 reservoir (Table 1), only a limited number of these techniques have been used to investigate MSCs. To unequivocally assess whether MSCs are infectable and contribute to the HIV-1 latent reservoir, in-depth studies investigating HIV-1 integration into MSCs, and the infection state of these cells are needed. Additionally, there is a need to compare the HIV-1 production rates between MSCs and the well described rates seen in T cells. Since MSCs are present in known HIV-1 reservoir sites, there is a possibility that MSCs do not become latently infected themselves, but instead contribute to the persistence of HIV-1 by transmitting the virus to or interacting with latently infected cells. A recent study supports this hypothesis and found that MSCs migrate towards latently infected macrophages and T-helper cells, and thereby increase the latency reactivation potential of these cells as quantified by p24 production [237].
To ensure that results are comparable, future research examining the productive or latent infectability of MSCs should address the need for consistency between HIV-1 strains and MSC sources used. Infection experiments performed to date have utilised different strains and tropisms of HIV-1 [100–103, 111, 117, 122, 123]. A study with 11 different strains of HIV-1 highlighted differences in infectivity between the different strains due to differences in HIV-1 Env membrane stoichiometry which affects entry into host cells [238]. Thus, differences in infectivity observed between experiments in this review may be due to the use of different HIV-1 strains. It will also be important to ensure that the same MSC types are compared in infection experiments. Contrasting results were generated regarding the infectability of fibroblast-like and macrophage-like BM-derived stromal cells using R5- and X4-tropic HIV-1 [102, 103]. Fibroblast-like BM-derived stromal cells showed signs of infection by X4-tropic HIV-1 based on detection of p24; however, these cells showed no evidence of infection by R5-tropic HIV-1. The opposite trend was noted with macrophage-like BM-derived stromal cells, which only demonstrated evidence of infection upon exposure to R5-tropic but not X4-tropic HIV-1. It is likely that the differences in susceptibility of these BM-derived stromal cells to infection may have been due to the nature of the cells used. R5- and X4-tropic HIV-1 are known to predominantly target macrophages and T-cells, respectively, amongst other cell types [239, 240].
Although there is not enough data to confirm whether MSCs represent an additional HIV-1 reservoir, they appear to be affected by HIV-1 and its proteins. The studies discussed in this review have reported that HIV-1 proteins, Tat, Nef, gp120, Rev and p55-gag alter basic biological characteristics of MSCs such as self-renewal, proliferation, viability, senescence, apoptosis, colony forming and in vitro differentiation capacity [100, 101, 117, 197–203]. Thus, HIV-1 appears to alter MSC properties in vitro, but whether this is the case in vivo needs to be evaluated. Differentiation capacity into the adipogenic and osteogenic lineages was also affected by HIV-1 infection and/or HIV-1 proteins [101, 196–199, 202]. It therefore seems plausible that the effects of HIV-1 infection on endogenous MSCs could in part contribute to the lipid and bone toxicities reported in patients. Additional experiments are needed to identify the cause of these changes in MSC properties. Are these changes due to direct infection with HIV-1, interactions with HIV-1 proteins, or from interactions and signalling from other infected cells? MSC therapies in HIV-1 patients may not produce a beneficial effect, which may be due to the deleterious effects of HIV-1 on the cells in vivo and not necessarily because they do not work for the specific indications.
Conclusion
The papers reviewed do not provide enough data to verify whether MSCs can become infected with and harbour HIV-1. Additionally, the effect of HIV-1 and HIV-1 proteins on MSC properties such as self-renewal, proliferation, viability, senescence, apoptosis, colony forming and differentiation capacity were well described in vitro but not verified in vivo. Although MSCs appear to be capable of expressing HIV-1 entry receptors and co-receptors in vitro, there is no consensus on the level of intracellular and extracellular expression. Additionally, the results generated in vitro are not necessarily representative of what occurs in vivo.
The persistent presence of integrated viral nucleic acids and low levels of p24 in cell culture supernatant suggest that MSCs exposed to HIV-1 may be latently infected. However, due to lack of in vivo confirmation, low-level productive infection of MSCs or the possibility of MSCs being refractory to HIV-1 infection in vivo cannot be ruled out. Therefore, future experiments should evaluate MSCs directly after isolation from HIV-1 infected individuals instead of MSCs exposed to HIV-1 in vitro, to determine the infectability of these cells.
There is a paucity of experiments focusing on MSCs from adipose tissue in the articles evaluated in this review. ASCs may be one of the MSC cell types contributing to the reservoir if they are susceptible to HIV infection. Thus, there is a need to examine whether ASCs can harbour integrated HIV-1 DNA and compare the results to BM-MSCs. The studies described in the review failed to prove unequivocally that MSCs are an additional HIV-1 reservoir. To conclusively determine whether MSCs may indeed serve as an additional reservoir for HIV-1, it will be necessary to employ the techniques mentioned in Table 1, which have successfully proven that CD4+ T cells function as a reservoir of latent HIV-1.
Acknowledgements
We would like to acknowledge the assistance of Fiona A van Vollenstee, Marnie Potgieter and Tebogo Maborwa during the early stages of the preparation of this review.
Code Availability
Not applicable
Authors' Contributions
Karlien Kallmeyer: Study design, collection and/or assembly of data and interpretation thereof, manuscript writing, final approval of manuscript
Megan A. Ryder: Study design, collection and/or assembly of data and interpretation thereof, manuscript writing, final approval of manuscript
Michael S. Pepper: Conception and design, fundraising, data interpretation, manuscript writing, final approval of manuscript
Funding
This work was funded by the South African Medical Research Council (Extramural Unit for Stem Cell Research and Therapy and University Flagship grants) and the University of Pretoria through the Institute for Cellular and Molecular Medicine.
Data Availability
Not applicable
Declarations
Conflicts of Interest
The authors have no conflicts of interest to disclose.
Ethics Approval
Not applicable
Consent to Participate
Not applicable
Consent for Publication
Not applicable
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Simon V, Ho DD, Abdool Karim Q. HIV/AIDS epidemiology, pathogenesis, prevention, and treatment. Lancet. 2006;368(9534):489–504. doi: 10.1016/S0140-6736(06)69157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koppensteiner H, Brack-Werner R, Schindler M. Macrophages and their relevance in Human Immunodeficiency Virus Type I infection. Retrovirology. 2012;9(1):82. doi: 10.1186/1742-4690-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maddon PJ, Dalgleish AG, McDougal JS, Clapham PR, Weiss RA, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47(3):333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 4.Berger, E. A., Doms, R. W., Fenyo, E. M., Korber, B. T., Littman, D. R., Moore, J. P., ... Weiss, R. A. (1998). A new classification for HIV-1. Nature, 391(6664), 240. 10.1038/34571 [DOI] [PubMed]
- 5.Wilen CB, Tilton JC, Doms RW. HIV: cell binding and entry. Cold Spring Harbour Perspectives in Medicine. 2012;2(8):a006866. doi: 10.1101/cshperspect.a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gulnik S, Erickson JW, Xie D. HIV protease: enzyme function and drug resistance. Vitamins and Hormones. 2000;58:213–256. doi: 10.1016/s0083-6729(00)58026-1. [DOI] [PubMed] [Google Scholar]
- 7.Briggs JA, Wilk T, Welker R, Krausslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. The EMBO Journal. 2003;22(7):1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundquist WI, Kräusslich H-G. HIV-1 assembly, budding, and maturation. Cold Spring Harbor Perspectives in Medicine. 2012;2(7):a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Picado J, Deeks SG. Persistent HIV-1 replication during antiretroviral therapy. Current Opinion in HIV and AIDS. 2016;11(4):417–423. doi: 10.1097/COH.0000000000000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Lee E, Park B-J, Bang JH, Lee JY. Adherence to antiretroviral therapy and factors affecting low medication adherence among incident HIV-infected individuals during 2009–2016: a nationwide study. Scientific Reports. 2018;8(1):1–8. doi: 10.1038/s41598-018-21081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conway JM, Perelson AS, Li JZ. Predictions of time to HIV viral rebound following ART suspension that incorporate personal biomarkers. PLoS Computational Biology. 2019;15(7):e1007229. doi: 10.1371/journal.pcbi.1007229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baxter AE, O'Doherty U, Kaufmann DE. Beyond the replication-competent HIV reservoir: transcription and translation-competent reservoirs. Retrovirology. 2018;15(1):18. doi: 10.1186/s12977-018-0392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avettand-Fenoel, V., Hocqueloux, L., Ghosn, J., Cheret, A., Frange, P., Melard, A., ... Rouzioux, C. (2016). Total HIV-1 DNA, a Marker of Viral Reservoir Dynamics with Clinical Implications. Clinical Microbiology Reviews, 29(4), 859-880. 10.1128/CMR.00015-16 [DOI] [PMC free article] [PubMed]
- 14.Svicher V, Ceccherini-Silberstein F, Antinori A, Aquaro S, Perno CF. Understanding HIV compartments and reservoirs. Current HIV/AIDS Reports. 2014;11(2):186–194. doi: 10.1007/s11904-014-0207-y. [DOI] [PubMed] [Google Scholar]
- 15.Churchill MJ, Deeks SG, Margolis DM, Siliciano RF, Swanstrom R. HIV reservoirs: what, where and how to target them. Nature Reviews Microbiology. 2016;14(1):55. doi: 10.1038/nrmicro.2015.5. [DOI] [PubMed] [Google Scholar]
- 16.Khoury, G., Darcis, G., Lee, M. Y., Bouchat, S., Van Driessche, B., Purcell, D. F., & Van Lint, C. (2018). The molecular biology of HIV latency. In: HIV Vaccines and Cure (pp. 187-212). Springer. [DOI] [PubMed]
- 17.Karn J, Stoltzfus CM. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harbour Perspectives in Medicine. 2012;2(2):a006916. doi: 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deeks SG. HIV: Shock and kill. Nature. 2012;487(7408):439–440. doi: 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- 19.Barton, K., Hiener, B., Winckelmann, A., Rasmussen, T. A., Shao, W., Byth, K., ... Palmer, S. (2016). Broad activation of latent HIV-1 in vivo. Nature Communications, 7(1), 12731. 10.1038/ncomms12731 [DOI] [PMC free article] [PubMed]
- 20.Granick JL, Simon SI, Borjesson DL. Hematopoietic stem and progenitor cells as effectors in innate immunity. Bone Marrow Research. 2012;2012:165107. doi: 10.1155/2012/165107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedenstein AJ, Petrakova KV, Kurolesova AI, Frolova GP. Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation. 1968;6(2):230–247. doi: 10.1097/00007890-196803000-00009. [DOI] [PubMed] [Google Scholar]
- 22.da Silva Meirelles L, Chagastelles PC, Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. Journal of Cell Science. 2006;119(Pt 11):2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 23.Ankrum J, Karp JM. Mesenchymal stem cell therapy: Two steps forward, one step back. Trends in Molecular Medicine. 2010;16(5):203–209. doi: 10.1016/j.molmed.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azari MF, Mathias L, Ozturk E, Cram DS, Boyd RL, Petratos S. Mesenchymal stem cells for treatment of CNS injury. Current Neuropharmacology. 2010;8(4):316–323. doi: 10.2174/157015910793358204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durandt, C., Potgieter, J. C., Mellet, J., Herd, C., Khoosal, R., Nel, J. G., ... Pepper, M. S. (2019). HIV and haematopoiesis. South African Medical Journal, 109(8b), 40-45. 10.7196/SAMJ.2019.v109i8b.13829 [DOI] [PubMed]
- 26.McComsey, G. A., Tebas, P., Shane, E., Yin, M. T., Overton, E. T., Huang, J. S., ... Brown, T. T. (2010). Bone disease in HIV infection: a practical review and recommendations for HIV care providers. Clinical Infectious Diseases, 51(8), 937-946. 10.1086/656412 [DOI] [PMC free article] [PubMed]
- 27.Feeney ER, Mallon PW. HIV and HAART-Associated Dyslipidemia. Open Cardiovascular Medical Journal. 2011;5:49–63. doi: 10.2174/1874192401105010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruera D, Luna N, David DO, Bergoglio LM, Zamudio J. Decreased bone mineral density in HIV-infected patients is independent of antiretroviral therapy. AIDS. 2003;17(13):1917–1923. doi: 10.1097/00002030-200309050-00010. [DOI] [PubMed] [Google Scholar]
- 29.Koethe, J. R., Lagathu, C., Lake, J. E., Domingo, P., Calmy, A., Falutz, J., ... Capeau, J. (2020). HIV and antiretroviral therapy-related fat alterations. Nature Reviews Disease Primers, 6(1), 48. 10.1038/s41572-020-0181-1 [DOI] [PubMed]
- 30.Damouche, A., Lazure, T., Avettand-Fenoel, V., Huot, N., Dejucq-Rainsford, N., Satie, A. P., ... Bourgeois, C. (2015). Adipose Tissue Is a Neglected Viral Reservoir and an Inflammatory Site during Chronic HIV and SIV Infection. PLoS Pathogens, 11(9), e1005153. 10.1371/journal.ppat.1005153 [DOI] [PMC free article] [PubMed]
- 31.Couturier, J., Suliburk, J. W., Brown, J. M., Luke, D. J., Agarwal, N., Yu, X., ... Lewis, D. E. (2015). Human adipose tissue as a reservoir for memory CD4+ T cells and HIV. AIDS, 29(6), 667-674. 10.1097/QAD.0000000000000599 [DOI] [PMC free article] [PubMed]
- 32.Hazan, U., Romero, I. A., Cancello, R., Valente, S., Perrin, V., Mariot, V., ... Pietri-Rouxel, F. (2002). Human adipose cells express CD4, CXCR4, and CCR5 [corrected] receptors: a new target cell type for the immunodeficiency virus-1? The FASEB Journal, 16(10), 1254-1256. 10.1096/fj.01-0947fje [DOI] [PubMed]
- 33.Maurin T, Saillan-Barreau C, Cousin B, Casteilla L, Doglio A, Penicaud L. Tumor necrosis factor-alpha stimulates HIV-1 production in primary culture of human adipocytes. Experimental Cell Research. 2005;304(2):544–551. doi: 10.1016/j.yexcr.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Munier S, Borjabad A, Lemaire M, Mariot V, Hazan U. In vitro infection of human primary adipose cells with HIV-1: a reassessment. AIDS. 2003;17(17):2537–2539. doi: 10.1097/00002030-200311210-00019. [DOI] [PubMed] [Google Scholar]
- 35.Dupin, N., Buffet, M., Marcelin, A. G., Lamotte, C., Gorin, I., Ait-Arkoub, Z., ... Peytavin, G. (2002). HIV and antiretroviral drug distribution in plasma and fat tissue of HIV-infected patients with lipodystrophy. AIDS, 16(18), 2419-2424. 10.1097/00002030-200212060-00006 [DOI] [PubMed]
- 36.Souza-Moreira L, Soares VC, Dias S d SG, Bozza PT. Adipose-derived Mesenchymal Stromal Cells Modulate Lipid Metabolism and Lipid Droplet Biogenesis via AKT/mTOR–PPARγ Signalling in Macrophages. Scientific Reports. 2019;9(1):1–11. doi: 10.1038/s41598-019-56835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu L, Yin C, Zhao F, Ali A, Ma J, Qian A. Mesenchymal Stem Cells: Cell Fate Decision to Osteoblast or Adipocyte and Application in Osteoporosis Treatment. International Journal of Molecular Sciences. 2018;19(2):360. doi: 10.3390/ijms19020360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chun, T. W., Stuyver, L., Mizell, S. B., Ehler, L. A., Mican, J. A., Baseler, M., ... Fauci, A. S. (1997). Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proceedings of the National Academy of Sciences of the United States of America, 94(24), 13193-13197. 10.1073/pnas.94.24.13193 [DOI] [PMC free article] [PubMed]
- 39.Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(15):8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finzi, D., Hermankova, M., Pierson, T., Carruth, L. M., Buck, C., Chaisson, R. E., ... Siliciano, R. F. (1997). Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science, 278(5341), 1295-1300. 10.1126/science.278.5341.1295 [DOI] [PubMed]
- 41.Finzi, D., Blankson, J., Siliciano, J. D., Margolick, J. B., Chadwick, K., Pierson, T., ... Siliciano, R. F. (1999). Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nature Medicine, 5(5), 512-517. 10.1038/8394 [DOI] [PubMed]
- 42.Chavez L, Calvanese V, Verdin E. HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells. PLoS Pathogens. 2015;11(6):e1004955. doi: 10.1371/journal.ppat.1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Falcinelli SD, Ceriani C, Margolis DM, Archin NM. New Frontiers in Measuring and Characterizing the HIV Reservoir. Frontiers in Microbiology. 2019;10(2878):2878. doi: 10.3389/fmicb.2019.02878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rocco J, Mellors JW, Macatangay BJ. Regulatory T cells: the ultimate HIV reservoir? Journal of Virus Eradication. 2018;4(4):209–214. doi: 10.1016/S2055-6640(20)30305-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tran, T. A., de Goer de Herve, M. G., Hendel-Chavez, H., Dembele, B., Le Nevot, E., Abbed, K., ... Taoufik, Y. (2008). Resting regulatory CD4 T cells: a site of HIV persistence in patients on long-term effective antiretroviral therapy. PLoS One, 3(10), e3305. 10.1371/journal.pone.0003305 [DOI] [PMC free article] [PubMed]
- 46.Kuo HH, Lichterfeld M. Recent progress in understanding HIV reservoirs. Current Opinion in HIV and AIDS. 2018;13(2):137–142. doi: 10.1097/COH.0000000000000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee, G. Q., Orlova-Fink, N., Einkauf, K., Chowdhury, F. Z., Sun, X., Harrington, S., ... Lichterfeld, M. (2017). Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. Journal of Clinical Investigation, 127(7), 2689-2696. 10.1172/JCI93289 [DOI] [PMC free article] [PubMed]
- 48.Hosmane, N. N., Kwon, K. J., Bruner, K. M., Capoferri, A. A., Beg, S., Rosenbloom, D. I., ... Siliciano, R. F. (2017). Proliferation of latently infected CD4(+) T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. Journal of Experimental Medicine, 214(4), 959-972. 10.1084/jem.20170193 [DOI] [PMC free article] [PubMed]
- 49.Bui, J. K., Sobolewski, M. D., Keele, B. F., Spindler, J., Musick, A., Wiegand, A., ... Mellors, J. W. (2017). Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathogens, 13(3), e1006283. 10.1371/journal.ppat.1006283 [DOI] [PMC free article] [PubMed]
- 50.Banga, R., Procopio, F. A., Noto, A., Pollakis, G., Cavassini, M., Ohmiti, K., ... Perreau, M. (2016). PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nature Medicine, 22(7), 754-761. 10.1038/nm.4113 [DOI] [PubMed]
- 51.Perreau, M., Savoye, A. L., De Crignis, E., Corpataux, J. M., Cubas, R., Haddad, E. K., ... Pantaleo, G. (2013). Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. Journal of Experimental Medicine, 210(1), 143-156. 10.1084/jem.20121932 [DOI] [PMC free article] [PubMed]
- 52.Cantero-Perez, J., Grau-Exposito, J., Serra-Peinado, C., Rosero, D. A., Luque-Ballesteros, L., Astorga-Gamaza, A., ... Genesca, M. (2019). Resident memory T cells are a cellular reservoir for HIV in the cervical mucosa. Nature Communications, 10(1), 4739. 10.1038/s41467-019-12732-2 [DOI] [PMC free article] [PubMed]
- 53.Kumar, B. V., Ma, W., Miron, M., Granot, T., Guyer, R. S., Carpenter, D. J., ... Farber, D. L. (2017). Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Reports, 20(12), 2921-2934. 10.1016/j.celrep.2017.08.078 [DOI] [PMC free article] [PubMed]
- 54.Shan, L., Deng, K., Gao, H., Xing, S., Capoferri, A. A., Durand, C. M., ... Siliciano, R. F. (2017). Transcriptional Reprogramming during Effector-to-Memory Transition Renders CD4(+) T Cells Permissive for Latent HIV-1 Infection. Immunity, 47(4), 766-775 e763. 10.1016/j.immuni.2017.09.014 [DOI] [PMC free article] [PubMed]
- 55.Tuttle DL, Harrison JK, Anders C, Sleasman JW, Goodenow MM. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage susceptibility to infection by human immunodeficiency virus type 1. Journal of Virology. 1998;72(6):4962–4969. doi: 10.1128/JVI.72.6.4962-4969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pesenti E, Pastore C, Lillo F, Siccardi AG, Vercelli D, Lopalco L. Role of CD4 and CCR5 levels in the susceptibility of primary macrophages to infection by CCR5-dependent HIV type 1 isolates. AIDS Research and Human Retroviruses. 1999;15(11):983–987. doi: 10.1089/088922299310494. [DOI] [PubMed] [Google Scholar]
- 57.Naif, H. M., Li, S., Alali, M., Sloane, A., Wu, L., Kelly, M., ... Cunningham, A. L. (1998). CCR5 expression correlates with susceptibility of maturing monocytes to human immunodeficiency virus type 1 infection. Journal of Virology, 72(1), 830-836. 10.1128/JVI.72.1.830-836.1998 [DOI] [PMC free article] [PubMed]
- 58.Li, Y., Li, L., Wadley, R., Reddel, S. W., Qi, J. C., Archis, C., ... Kouts, S. (2001). Mast cells/basophils in the peripheral blood of allergic individuals who are HIV-1 susceptible due to their surface expression of CD4 and the chemokine receptors CCR3, CCR5, and CXCR4. Blood, The Journal of the American Society of Hematology, 97(11), 3484-3490. [DOI] [PubMed]
- 59.Sundstrom, J. B., Ellis, J. E., Hair, G. A., Kirshenbaum, A. S., Metcalfe, D. D., Yi, H., ... Ansari, A. A. (2007). Human tissue mast cells are an inducible reservoir of persistent HIV infection. Blood, 109(12), 5293-5300. 10.1182/blood-2006-11-058438 [DOI] [PMC free article] [PubMed]
- 60.Qi, J. C., Stevens, R. L., Wadley, R., Collins, A., Cooley, M., Naif, H. M., ... Krilis, S. A. (2002). IL-16 regulation of human mast cells/basophils and their susceptibility to HIV-1. Journal of Immunology, 168(8), 4127-4134. 10.4049/jimmunol.168.8.4127 [DOI] [PubMed]
- 61.Walter BL, Wehrly K, Swanstrom R, Platt E, Kabat D, Chesebro B. Role of low CD4 levels in the influence of human immunodeficiency virus type 1 envelope V1 and V2 regions on entry and spread in macrophages. Journal of Virology. 2005;79(8):4828–4837. doi: 10.1128/JVI.79.8.4828-4837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joseph SB, Arrildt KT, Swanstrom AE, Schnell G, Lee B, Hoxie JA, Swanstrom R. Quantification of entry phenotypes of macrophage-tropic HIV-1 across a wide range of CD4 densities. Journal of Virology. 2014;88(4):1858–1869. doi: 10.1128/JVI.02477-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roszer T. Understanding the Biology of Self-Renewing Macrophages. Cells. 2018;7(8):103. doi: 10.3390/cells7080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wong ME, Jaworowski A, Hearps AC. Corrigendum: The HIV Reservoir in Monocytes and Macrophages. Frontiers in Immunology. 2019;10:2517. doi: 10.3389/fimmu.2019.02517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andrade, V. M., Mavian, C., Babic, D., Cordeiro, T., Sharkey, M., Barrios, L., ... Llano, A. (2020). A minor population of macrophage-tropic HIV-1 variants is identified in recrudescing viremia following analytic treatment interruption. Proceedings of the National Academy of Sciences, 117(18), 9981-9990. [DOI] [PMC free article] [PubMed]
- 66.Ganor, Y., Real, F., Sennepin, A., Dutertre, C. A., Prevedel, L., Xu, L., ... Bomsel, M. (2019). HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nature Microbiology, 4(4), 633-644. 10.1038/s41564-018-0335-z [DOI] [PubMed]
- 67.Patel, A. A., Zhang, Y., Fullerton, J. N., Boelen, L., Rongvaux, A., Maini, A. A., ... Yona, S. (2017). The Fate and Lifespan of Human Monocyte Subsets in Steady State and Systemic Inflammation. The Journal of Experimental Medicine, 214(7), 1913-1923. 10.1084/jem.20170355 [DOI] [PMC free article] [PubMed]
- 68.Italiani, P., & Boraschi, D. (2014). From monocytes to M1/M2 macrophages: Phenotypical vs. functional differentiation. Frontiers in Immunology, 5(514). 10.3389/fimmu.2014.00514 [DOI] [PMC free article] [PubMed]
- 69.Veenstra M, Leon-Rivera R, Li M, Gama L, Clements JE, Berman JW. Mechanisms of CNS Viral Seeding by HIV(+) CD14(+) CD16(+) Monocytes: Establishment and Reseeding of Viral Reservoirs Contributing to HIV-Associated Neurocognitive Disorders. mBio. 2017;8(5):e01280–e01217. doi: 10.1128/mBio.01280-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Monie TP. A Snapshot of the Innate Immune System. In: Monie TP, editor. The Innate Immune System. Academic Press; 2017. pp. 1–40. [Google Scholar]
- 71.van Furth R, Cohn ZA. The Origin and Kinetics of Mononuclear Phagocytes. Journal of Experimental Medicine. 1968;128(3):415–435. doi: 10.1084/jem.128.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Al-Harti L, Joseph J, Nath A. Astrocytes as an HIV CNS reservoir: highlights and reflections of an NIMH-sponsored symposium. Journal of Neurovirology. 2018;24(6):665–669. doi: 10.1007/s13365-018-0691-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Narasipura SD, Kim S, Al-Harthi L. Epigenetic regulation of HIV-1 latency in astrocytes. Journal of Virology. 2014;88(5):3031–3038. doi: 10.1128/jvi.03333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carroll-Anzinger D, Kumar A, Adarichev V, Kashanchi F, Al-Harthi L. Human immunodeficiency virus-restricted replication in astrocytes and the ability of gamma interferon to modulate this restriction are regulated by a downstream effector of the Wnt signaling pathway. Journal of Virology. 2007;81(11):5864–5871. doi: 10.1128/JVI.02234-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wallet, C., De Rovere, M., Van Assche, J., Daouad, F., De Wit, S., Gautier, V., ... Schwartz, C. (2019). Microglial Cells: The Main HIV-1 Reservoir in the Brain. Frontiers in Cellular and Infection Microbiology, 9, 362. 10.3389/fcimb.2019.00362 [DOI] [PMC free article] [PubMed]
- 76.Alvarez-Carbonell D, Ye F, Ramanath N, Dobrowolski C, Karn J. The Glucocorticoid Receptor Is a Critical Regulator of HIV Latency in Human Microglial Cells. Journal of Neuroimmune Pharmacology. 2019;14(1):94–109. doi: 10.1007/s11481-018-9798-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li G, Ma J, Yu H, Bi W, Wu Y, Cheng L, Su L. Plasmacytoid dendritic cells contribute to HIV diseases and viral persistence via IDO-dependent mechanisms. The Journal of Immunology. 2019;202(1 Supplement):75.71. [Google Scholar]
- 78.Jiang, A.-P., Jiang, J.-F., Wei, J.-F., Guo, M.-G., Qin, Y., Guo, Q.-Q., ... Veazey, R. S. (2016). Human mucosal mast cells capture HIV-1 and mediate viral trans-infection of CD4+ T cells. Journal of Virology, 90(6), 2928-2937. [DOI] [PMC free article] [PubMed]
- 79.Tsukamoto T. Hematopoietic Stem/Progenitor Cells and the Pathogenesis of HIV/AIDS. Frontiers in Cellular and Infection Microbiology. 2020;10:60. doi: 10.3389/fcimb.2020.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carter, C. C., McNamara, L. A., Onafuwa-Nuga, A., Shackleton, M., Riddell, J. T., Bixby, D., ... Collins, K. L. (2011). HIV-1 Utilizes the CXCR4 Chemokine Receptor to Infect Multipotent Hematopoietic Stem and Progenitor Cells. Cell Host & Microbe, 9(3), 223-234. 10.1016/j.chom.2011.02.005 [DOI] [PMC free article] [PubMed]
- 81.Nixon CC, Vatakis DN, Reichelderfer SN, Dixit D, Kim SG, Uittenbogaart CH, Zack JA. HIV-1 infection of hematopoietic progenitor cells in vivo in humanized mice. Blood. 2013;122(13):2195–2204. doi: 10.1182/blood-2013-04-496950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Durand, C. M., Ghiaur, G., Siliciano, J. D., Rabi, S. A., Eisele, E. E., Salgado, M., ... Siliciano, R. F. (2012). HIV-1 DNA Is Detected in Bone Marrow Populations Containing CD4+ T Cells but Is Not Found in Purified Cd34+ Hematopoietic Progenitor Cells in Most Patients on Antiretroviral Therapy. Journal of Infectious Diseases, 205(6), 1014-1018. 10.1093/infdis/jir884 [DOI] [PMC free article] [PubMed]
- 83.Josefsson, L., Eriksson, S., Sinclair, E., Ho, T., Killian, M., Epling, L., ... Palmer, S. (2012). Hematopoietic Precursor Cells Isolated from Patients on Long-Term Suppressive HIV Therapy Did Not Contain HIV-1 DNA. Journal of Infectious Diseases, 206(1), 28-34. 10.1093/infdis/jis301 [DOI] [PMC free article] [PubMed]
- 84.Sebastian, N. T., Zaikos, T. D., Terry, V., Taschuk, F., McNamara, L. A., Onafuwa-Nuga, A., ... Bixby, D. (2017). CD4 is expressed on a heterogeneous subset of hematopoietic progenitors, which persistently harbor CXCR4 and CCR5-tropic HIV proviral genomes in vivo. PLoS Pathogens, 13(7), e1006509. [DOI] [PMC free article] [PubMed]
- 85.Louache F, Debili N, Marandin A, Coulombel L, Vainchenker W. Expression of CD4 by human hematopoietic progenitors. Blood. 1994;84(10):3344–3355. doi: 10.1182/blood.V84.10.3344.3344. [DOI] [PubMed] [Google Scholar]
- 86.Zauli, G., Furlini, G., Vitale, M., Re, M. C., Gibellini, D., Zamai, L., ... La Placa, M. (1994). A subset of human CD34+ hematopoietic progenitors express low levels of CD4, the high-affinity receptor for human immunodeficiency virus-type 1. Blood, 84(6), 1896-1905. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/7521692 [PubMed]
- 87.Ruiz, M. E., Cicala, C., Arthos, J., Kinter, A., Catanzaro, A. T., Adelsberger, J., ... Fauci, A. S. (1998). Peripheral blood-derived CD34+ progenitor cells: CXC chemokine receptor 4 and CC chemokine receptor 5 expression and infection by HIV. Journal of Immunology, 161(8), 4169-4176. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9780190 [PubMed]
- 88.Banda, N. K., Simon, G. R., Sipple, J. D., Terrell, K. L., Archer, P., Shpall, E. J., ... Harrison, G. S. (1999). Depletion of CD34+ CD4+ cells in bone marrow from HIV-1-infected individuals. Biology of Blood and Marrow Transplantation, 5(3), 162-172. 10.1053/bbmt.1999.v5.pm10392962 [DOI] [PubMed]
- 89.Marandin A, Katz A, Oksenhendler E, Tulliez M, Picard F, Vainchenker W, Louache F. Loss of primitive hematopoietic progenitors in patients with human immunodeficiency virus infection. Blood. 1996;88(12):4568–4578. doi: 10.1182/blood.V88.12.4568.bloodjournal88124568. [DOI] [PubMed] [Google Scholar]
- 90.Costantini, A., Giuliodoro, S., Mancini, S., Butini, L., Regnery, C. M., Silvestri, G., ... Montroni, M. (2006). Impaired in-vitro growth of megakaryocytic colonies derived from CD34 cells of HIV-1-infected patients with active viral replication. AIDS, 20(13), 1713-1720. 10.1097/01.aids.0000242817.88086.8c [DOI] [PubMed]
- 91.Wang Z, Simonetti FR, Siliciano RF, Laird GM. Measuring Replication Competent HIV-1: Advances and Challenges in Defining the Latent Reservoir. Retrovirology. 2018;15(1):21. doi: 10.1186/s12977-018-0404-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liszewski MK, Yu JJ, O'Doherty U. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods. 2009;47(4):254–260. doi: 10.1016/j.ymeth.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gaebler, C., Lorenzi, J. C., Oliveira, T. Y., Nogueira, L., Ramos, V., Lu, C.-L., ... Caskey, M. (2019). Combination of quadruplex qPCR and next-generation sequencing for qualitative and quantitative analysis of the HIV-1 latent reservoir. Journal of Experimental Medicine, 216(10), 2253-2264. [DOI] [PMC free article] [PubMed]
- 94.Wehrly K, Chesebro B. P24 Antigen Capture Assay for Quantification of Human Immunodeficiency Virus Using Readily Available Inexpensive Reagents. Methods. 1997;12(4):288–293. doi: 10.1006/meth.1997.0481. [DOI] [PubMed] [Google Scholar]
- 95.Imamichi, H., Smith, M., Adelsberger, J. W., Izumi, T., Scrimieri, F., Sherman, B. T., ... Lane, H. C. (2020). Defective HIV-1 Proviruses Produce Viral Proteins. Proceedings of the National Academy of Sciences, 117(7), 3704. 10.1073/pnas.1917876117 [DOI] [PMC free article] [PubMed]
- 96.Procopio, F. A., Fromentin, R., Kulpa, D. A., Brehm, J. H., Bebin, A. G., Strain, M. C., ... Chomont, N. (2015). A Novel Assay to Measure the Magnitude of the Inducible Viral Reservoir in HIV-infected Individuals. EBioMedicine, 2(8), 874-883. 10.1016/j.ebiom.2015.06.019 [DOI] [PMC free article] [PubMed]
- 97.Hiener, B., Eden, J. S., Horsburgh, B. A., & Palmer, S. (2018). Amplification of Near Full-length HIV-1 Proviruses for Next-Generation Sequencing. Journal of Visualized Experiments, 140. 10.3791/58016 [DOI] [PMC free article] [PubMed]
- 98.Bosque A, Planelles V. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood. 2009;113(1):58–65. doi: 10.1182/blood-2008-07-168393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fun A, Mok HP, Wills MR, Lever AM. A highly reproducible quantitative viral outgrowth assay for the measurement of the replication-competent latent HIV-1 reservoir. Science Reports. 2017;7:43231. doi: 10.1038/srep43231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang L, Mondal D, La Russa VF, Agrawal KC. Suppression of clonogenic potential of human bone marrow mesenchymal stem cells by HIV type 1: putative role of HIV type 1 tat protein and inflammatory cytokines. AIDS Research and Human Retroviruses. 2002;18(13):917–931. doi: 10.1089/088922202760265597. [DOI] [PubMed] [Google Scholar]
- 101.Gibellini, D., Alviano, F., Miserocchi, A., Tazzari, P. L., Ricci, F., Clo, A., ... Re, M. C. (2011). HIV-1 and recombinant gp120 affect the survival and differentiation of human vessel wall-derived mesenchymal stem cells. Retrovirology, 8(1), 40. 10.1186/1742-4690-8-40 [DOI] [PMC free article] [PubMed]
- 102.Scadden, D. T., Zeira, M., Woon, A., Wang, Z., Schieve, L., Ikeuchi, K., ... Groopman, J. E. (1990). Human immunodeficiency virus infection of human bone marrow stromal fibroblasts. Blood, 76(2), 317-322. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/1695109 [PubMed]
- 103.Canque B, Marandin A, Rosenzwajg M, Louache F, Vainchenker W, Gluckman JC. Susceptibility of Human Bone Marrow Stromal Cells to Human Immunodeficiency Virus (HIV) Virology. 1995;208(2):779–783. doi: 10.1006/viro.1995.1211. [DOI] [PubMed] [Google Scholar]
- 104.Casabianca A, Gori C, Orlandi C, Forbici F, Federico Perno C, Magnani M. Fast and sensitive quantitative detection of HIV DNA in whole blood leucocytes by SYBR green I real-time PCR assay. Molecular and Cellular Probes. 2007;21(5-6):368–378. doi: 10.1016/j.mcp.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 105.Malnati, M. S., Scarlatti, G., Gatto, F., Salvatori, F., Cassina, G., Rutigliano, T., ... Lusso, P. (2008). A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nature Protocols, 3(7), 1240-1248. 10.1038/nprot.2008.108 [DOI] [PubMed]
- 106.Mbisa JL, Delviks-Frankenberry KA, Thomas JA, Gorelick RJ, Pathak VK. Real-time PCR analysis of HIV-1 replication post-entry events. Methods in Molecular Biology. 2009;485:55–72. doi: 10.1007/978-1-59745-170-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van der Sluis, R. M., van Montfort, T., Centlivre, M., Schopman, N. C., Cornelissen, M., Sanders, R. W., ... Pollakis, G. (2013). Quantitation of HIV-1 DNA with a sensitive TaqMan assay that has broad subtype specificity. Journal of Virological Methods, 187(1), 94-102. 10.1016/j.jviromet.2012.09.019 [DOI] [PubMed]
- 108.Casabianca, A., Orlandi, C., Canovari, B., Scotti, M., Acetoso, M., Valentini, M., ... Magnani, M. (2014). A real time PCR platform for the simultaneous quantification of total and extrachromosomal HIV DNA forms in blood of HIV-1 infected patients. PLoS One, 9(11), e111919. 10.1371/journal.pone.0111919 [DOI] [PMC free article] [PubMed]
- 109.Vandergeeten, C., Fromentin, R., Merlini, E., Lawani, M. B., DaFonseca, S., Bakeman, W., ... Chomont, N. (2014). Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. Journal of Virology, 88(21), 12385-12396. 10.1128/JVI.00609-14 [DOI] [PMC free article] [PubMed]
- 110.Rutsaert, S., De Spiegelaere, W., Van Hecke, C., De Scheerder, M. A., Kiselinova, M., Vervisch, K., ... Vandekerckhove, L. (2018). In-depth validation of total HIV-1 DNA assays for quantification of various HIV-1 subtypes. Science Reports, 8(1), 17274. 10.1038/s41598-018-35403-6 [DOI] [PMC free article] [PubMed]
- 111.Nazari-Shafti, T. Z., Freisinger, E., Roy, U., Bulot, C. T., Senst, C., Dupin, C. L., ... Izadpanah, R. (2011). Mesenchymal stem cell derived hematopoietic cells are permissive to HIV-1 infection. Retrovirology, 8(1), 3. 10.1186/1742-4690-8-3 [DOI] [PMC free article] [PubMed]
- 112.Henrich TJ, Gallien S, Li JZ, Pereyra F, Kuritzkes DR. Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR. Journal of Virological Methods. 2012;186(1-2):68–72. doi: 10.1016/j.jviromet.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Strain, M. C., Lada, S. M., Luong, T., Rought, S. E., Gianella, S., Terry, V. H., ... Richman, D. D. (2013). Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One, 8(4), e55943. 10.1371/journal.pone.0055943 [DOI] [PMC free article] [PubMed]
- 114.Eriksson, S., Graf, E. H., Dahl, V., Strain, M. C., Yukl, S. A., Lysenko, E. S., ... Siliciano, J. D. (2013). Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathogens, 9(2), e1003174. 10.1371/journal.ppat.1003174 [DOI] [PMC free article] [PubMed]
- 115.O'Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. Journal of Virology. 2002;76(21):10942–10950. doi: 10.1128/jvi.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brussel A, Sonigo P. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. Journal of Virology. 2003;77(18):10119–10124. doi: 10.1128/jvi.77.18.10119-10124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cotter EJ, Chew N, Powderly WG, Doran PP. HIV type 1 alters mesenchymal stem cell differentiation potential and cell phenotype ex vivo. AIDS Research and Human Retroviruses. 2011;27(2):187–199. doi: 10.1089/aid.2010.0114. [DOI] [PubMed] [Google Scholar]
- 118.Nishanian P, Huskins KR, Stehn S, Detels R, Fahey JL. A simple method for improved assay demonstrates that HIV p24 antigen is present as immune complexes in most sera from HIV-infected individuals. Journal of Infectious Diseases. 1990;162(1):21–28. doi: 10.1093/infdis/162.1.21. [DOI] [PubMed] [Google Scholar]
- 119.Chang, L., Song, L., Fournier, D. R., Kan, C. W., Patel, P. P., Ferrell, E. P., ... Wilson, D. H. (2013). Simple diffusion-constrained immunoassay for p24 protein with the sensitivity of nucleic acid amplification for detecting acute HIV infection. Journal of Virological Methods, 188(1-2), 153-160. 10.1016/j.jviromet.2012.08.017 [DOI] [PubMed]
- 120.Cabrera C, Chang L, Stone M, Busch M, Wilson DH. Rapid, Fully Automated Digital Immunoassay for p24 Protein with the Sensitivity of Nucleic Acid Amplification for Detecting Acute HIV Infection. Clinical Chemistry. 2015;61(11):1372–1380. doi: 10.1373/clinchem.2015.243287. [DOI] [PubMed] [Google Scholar]
- 121.Passaes, C. P. B., Bruel, T., Decalf, J., David, A., Angin, M., Monceaux, V., ... Saez-Cirion, A. (2017). Ultrasensitive HIV-1 p24 assay detects single infected cells and differences in reservoir induction by latency reversal agents. Journal of Virology, 91(6), e02296-02216. 10.1128/JVI.02296-16 [DOI] [PMC free article] [PubMed]
- 122.Bahner I, Kearns K, Coutinho S, Leonard EH, Kohn DB. Infection of human marrow stroma by human immunodeficiency virus-1 (HIV-1) is both required and sufficient for HIV-1–induced hematopoietic suppression in vitro: demonstration by gene modification of primary human stroma. Blood, The Journal of the American Society of Hematology. 1997;90(5):1787–1798. [PubMed] [Google Scholar]
- 123.Marandin A, Canque B, Coulombel L, Gluckman JC, Vainchenker W, Louache F. In vitro infection of bone marrow-adherent cells by human immunodeficiency virus type 1 (HIV-1) does not alter their ability to support hematopoiesis. Virology. 1995;213(1):245–248. doi: 10.1006/viro.1995.1565. [DOI] [PubMed] [Google Scholar]
- 124.Hiener, B., Horsburgh, B. A., Eden, J. S., Barton, K., Schlub, T. E., Lee, E., ... Palmer, S. (2017). Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Reports, 21(3), 813-822. 10.1016/j.celrep.2017.09.081 [DOI] [PMC free article] [PubMed]
- 125.Crooks, A. M., Bateson, R., Cope, A. B., Dahl, N. P., Griggs, M. K., Kuruc, J. D., ... Archin, N. M. (2015). Precise quantitation of the latent HIV-1 reservoir: Implications for eradication strategies. Journal of Infectious Diseases, 212(9), 1361-1365. 10.1093/infdis/jiv218 [DOI] [PMC free article] [PubMed]
- 126.Laird, G. M., Rosenbloom, D. I., Lai, J., Siliciano, R. F., & Siliciano, J. D. (2016). Measuring the frequency of latent HIV-1 in resting CD4+ T cells using a limiting dilution coculture assay. In: HIV protocols (pp. 239-253). Springer. [DOI] [PubMed]
- 127.Archin N, Stuelke E, Katherine S, Kirchherr J, Margolis D. A modified viral outgrowth assay incorporating ultra-sensitive P24 measurements. Journal of Virus Eradication. 2017;3:4. doi: 10.1016/S2055-6640(20)30578-1. [DOI] [Google Scholar]
- 128.Siliciano JD, Siliciano RF. Enhanced Culture Assay for Detection and Quantitation of Latently Infected, Resting CD4+ T-Cells Carrying Replication-Competent Virus in HIV-1-Infected Individuals. In: Zhu T, editor. Human Retrovirus Protocols: Virology and Molecular Biology. Humana Press; 2005. pp. 3–15. [DOI] [PubMed] [Google Scholar]
- 129.Pennings S, Liu KJ, Qian H. The Stem Cell Niche: Interactions between Stem Cells and Their Environment. Stem Cells International. 2018;2018:4879379. doi: 10.1155/2018/4879379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Horwitz EM, Keating A. Nonhematopoietic mesenchymal stem cells: what are they? Cytotherapy. 2000;2(5):387–388. doi: 10.1080/146532400539305. [DOI] [PubMed] [Google Scholar]
- 131.Horwitz, E. M., Le Blanc, K., Dominici, M., Mueller, I., Slaper-Cortenbach, I., Marini, F. C., ... International Society for Cellular, T. (2005). Clarification of the nomenclature for MSC: The International Society for Cellular Therapy position statement. Cytotherapy, 7(5), 393-395. 10.1080/14653240500319234 [DOI] [PubMed]
- 132.Pittenger, M. F., Mackay, A. M., Beck, S. C., Jaiswal, R. K., Douglas, R., Mosca, J. D., ... Marshak, D. R. (1999). Multilineage potential of adult human mesenchymal stem cells. Science, 284(5411), 143-147. 10.1126/science.284.5411.143 [DOI] [PubMed]
- 133.Zuk, P. A., Zhu, M., Mizuno, H., Huang, J., Futrell, J. W., Katz, A. J., ... Hedrick, M. H. (2001). Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Engineering, 7(2), 211-228. 10.1089/107632701300062859 [DOI] [PubMed]
- 134.Bunnell BA, Flaat M, Gagliardi C, Patel B, Ripoll C. Adipose-derived stem cells: isolation, expansion and differentiation. Methods. 2008;45(2):115–120. doi: 10.1016/j.ymeth.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bieback K, Kern S, Kluter H, Eichler H. Critical parameters for the isolation of mesenchymal stem cells from umbilical cord blood. Stem Cells. 2004;22(4):625–634. doi: 10.1634/stemcells.22-4-625. [DOI] [PubMed] [Google Scholar]
- 136.Corrao S, La Rocca G, Lo Iacono M, Corsello T, Farina F, Anzalone R. Umbilical cord revisited: from Wharton's jelly myofibroblasts to mesenchymal stem cells. Histology and Histopathology. 2013;28(10):1235–1244. doi: 10.14670/HH-28.1235. [DOI] [PubMed] [Google Scholar]
- 137.Pereira WC, Khushnooma I, Madkaikar M, Ghosh K. Reproducible methodology for the isolation of mesenchymal stem cells from human umbilical cord and its potential for cardiomyocyte generation. Journal of Tissue Engineering and Regenerative Medicine. 2008;2(7):394–399. doi: 10.1002/term.107. [DOI] [PubMed] [Google Scholar]
- 138.Klein D, Weisshardt P, Kleff V, Jastrow H, Jakob HG, Ergun S. Vascular wall-resident CD44+ multipotent stem cells give rise to pericytes and smooth muscle cells and contribute to new vessel maturation. PLoS One. 2011;6(5):e20540. doi: 10.1371/journal.pone.0020540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Murray, I. R., West, C. C., Hardy, W. R., James, A. W., Park, T. S., Nguyen, A., ... Peault, B. (2014). Natural history of mesenchymal stem cells, from vessel walls to culture vessels. Cellular and Molecular Life Sciences, 71(8), 1353-1374. 10.1007/s00018-013-1462-6 [DOI] [PMC free article] [PubMed]
- 140.Bourin, P., Bunnell, B. A., Casteilla, L., Dominici, M., Katz, A. J., March, K. L., ... Gimble, J. M. (2013). Stromal cells from the adipose tissue-derived stromal vascular fraction and culture expanded adipose tissue-derived stromal/stem cells: a joint statement of the International Federation for Adipose Therapeutics and Science (IFATS) and the International Society for Cellular Therapy (ISCT). Cytotherapy, 15(6), 641-648. 10.1016/j.jcyt.2013.02.006 [DOI] [PMC free article] [PubMed]
- 141.Dominici, M., Le Blanc, K., Mueller, I., Slaper-Cortenbach, I., Marini, F., Krause, D., ... Horwitz, E. (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy, 8(4), 315-317. 10.1080/14653240600855905 [DOI] [PubMed]
- 142.Corselli M, Chen CW, Crisan M, Lazzari L, Peault B. Perivascular ancestors of adult multipotent stem cells. Arteriosclerosis, Thrombosis, and Vascular Biology. 2010;30(6):1104–1109. doi: 10.1161/ATVBAHA.109.191643. [DOI] [PubMed] [Google Scholar]
- 143.da Silva Meirelles L, Caplan AI, Nardi NB. In search of the in vivo identity of mesenchyal stem cells. Stem Cells. 2008;26:2287–2299. doi: 10.1634/stemcells.2007-1122. [DOI] [PubMed] [Google Scholar]
- 144.Crippa, S., Santi, L., Bosotti, R., Porro, G., & Bernardo, M. E. (2019). Bone Marrow-Derived Mesenchymal Stromal Cells: A Novel Target to Optimize Hematopoietic Stem Cell Transplantation Protocols in Hematological Malignancies and Rare Genetic Disorders. Journal of Clinical Medicine, 9(1). 10.3390/jcm9010002 [DOI] [PMC free article] [PubMed]
- 145.Crippa S, Bernardo ME. Mesenchymal Stromal Cells: Role in the BM Niche and in the Support of Hematopoietic Stem Cell Transplantation. Hemasphere. 2018;2(6):e151. doi: 10.1097/HS9.0000000000000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kfoury Y, Scadden DT. Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell. 2015;16(3):239–253. doi: 10.1016/j.stem.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 147.Bourgeois, C., Gorwood, J., Barrail-Tran, A., Lagathu, C., Capeau, J., Desjardins, D., ... Lambotte, O. (2019). Specific Biological Features of Adipose Tissue, and Their Impact on HIV Persistence. Frontiers in Microbiology, 10(2837), 2837. 10.3389/fmicb.2019.02837 [DOI] [PMC free article] [PubMed]
- 148.Eckel-Mahan K, Ribas Latre A, Kolonin MG. Adipose Stromal Cell Expansion and Exhaustion: Mechanisms and Consequences. Cells. 2020;9(4):863. doi: 10.3390/cells9040863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Elliott S, Sinclair A, Collins H, Rice L, Jelkmann W. Progress in Detecting Cell-Surface Protein Receptors: The Erythropoietin Receptor Example. Annals of Hematology. 2014;93(2):181–192. doi: 10.1007/s00277-013-1947-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gao B, Roux PP. Translational Control by Oncogenic Signaling Pathways. Biochimica et Biophysica Acta. 2015;1849(7):753–765. doi: 10.1016/j.bbagrm.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 151.Ghazalpour, A., Bennett, B., Petyuk, V. A., Orozco, L., Hagopian, R., Mungrue, I. N., ... Lusis, A. J. (2011). Comparative Analysis of Proteome and Transcriptome Variation in Mouse. Plos Genetics, 7(6), e1001393. 10.1371/journal.pgen.1001393 [DOI] [PMC free article] [PubMed]
- 152.Richman DD, Bozzette SA. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. Journal of Infectious Diseases. 1994;169(5):968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 153.Honczarenko M, Le Y, Swierkowski M, Ghiran I, Glodek AM, Silberstein LE. Human bone marrow stromal cells express a distinct set of biologically functional chemokine receptors. Stem Cells. 2006;24(4):1030–1041. doi: 10.1634/stemcells.2005-0319. [DOI] [PubMed] [Google Scholar]
- 154.Li L, Lim RZL, Lee LSU, Chew NSY. HIV glycoprotein gp120 enhances mesenchymal stem cell migration by upregulating CXCR4 expression. Biochimica et Biophysica Acta (BBA) - General Subjects. 2018;1862(8):1790–1800. doi: 10.1016/j.bbagen.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 155.Wynn, R. F., Hart, C. A., Corradi-Perini, C., O'Neill, L., Evans, C. A., Wraith, J. E., ... Bellantuono, I. (2004). A small proportion of mesenchymal stem cells strongly expresses functionally active CXCR4 receptor capable of promoting migration to bone marrow. Blood, 104(9), 2643-2645. 10.1182/blood-2004-02-0526 [DOI] [PubMed]
- 156.Sordi, V., Malosio, M. L., Marchesi, F., Mercalli, A., Melzi, R., Giordano, T., ... Piemonti, L. (2005). Bone marrow mesenchymal stem cells express a restricted set of functionally active chemokine receptors capable of promoting migration to pancreatic islets. Blood, 106(2), 419-427. 10.1182/blood-2004-09-3507 [DOI] [PubMed]
- 157.Pelekanos RA, Ting MJ, Sardesai VS, Ryan JM, Lim YC, Chan JK, Fisk NM. Intracellular trafficking and endocytosis of CXCR4 in fetal mesenchymal stem/stromal cells. BMC Cell Biology. 2014;15(1):15. doi: 10.1186/1471-2121-15-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jones, G. N., Moschidou, D., Lay, K., Abdulrazzak, H., Vanleene, M., Shefelbine, S. J., ... Guillot, P. V. (2012). Upregulating CXCR4 in human fetal mesenchymal stem cells enhances engraftment and bone mechanics in a mouse model of osteogenesis imperfecta. STEM CELLS Translational Medicine, 1(1), 70-78. 10.5966/sctm.2011-0007 [DOI] [PMC free article] [PubMed]
- 159.Alexeev V, Donahue A, Uitto J, Igoucheva O. Analysis of chemotactic molecules in bone marrow-derived mesenchymal stem cells and the skin: Ccl27-Ccr10 axis as a basis for targeting to cutaneous tissues. Cytotherapy. 2013;15(2):171–184.e171. doi: 10.1016/j.jcyt.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ringe, J., Strassburg, S., Neumann, K., Endres, M., Notter, M., Burmester, G. R., ... Sittinger, M. (2007). Towards in situ tissue repair: human mesenchymal stem cells express chemokine receptors CXCR1, CXCR2 and CCR2, and migrate upon stimulation with CXCL8 but not CCL2. Journal of Cellular Biochemistry, 101(1), 135-146. [DOI] [PubMed]
- 161.Fox JM, Chamberlain G, Ashton BA, Middleton J. Recent advances into the understanding of mesenchymal stem cell trafficking. British Journal of Haematology. 2007;137(6):491–502. doi: 10.1111/j.1365-2141.2007.06610.x. [DOI] [PubMed] [Google Scholar]
- 162.Karnoub, A. E., Dash, A. B., Vo, A. P., Sullivan, A., Brooks, M. W., Bell, G. W., ... Weinberg, R. A. (2007). Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature, 449(7162), 557-563. [DOI] [PubMed]
- 163.Novak, M., Koprivnikar Krajnc, M., Hrastar, B., Breznik, B., Majc, B., Mlinar, M., ... Lah Turnšek, T. (2020). CCR5-mediated signaling is involved in invasion of glioblastoma cells in its microenvironment. International Journal of Molecular Sciences, 21(12), 4199. 10.3390/ijms21124199 [DOI] [PMC free article] [PubMed]
- 164.Chamberlain G, Wright K, Rot A, Ashton B, Middleton J. Murine mesenchymal stem cells exhibit a restricted repertoire of functional chemokine receptors: Comparison with human. PLoS One. 2008;3(8):e2934. doi: 10.1371/journal.pone.0002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pesaresi M, Bonilla-Pons SA, Sebastian-Perez R, Di Vicino U, Alcoverro-Bertran M, Michael R, Cosma MP. The chemokine receptors Ccr5 and Cxcr6 enhance migration of mesenchymal stem cells into the degenerating retina. Molecular Therapy. 2021;29(2):804–821. doi: 10.1016/j.ymthe.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ji JF, He BP, Dheen ST, Tay SS. Interactions of chemokines and chemokine receptors mediate the migration of mesenchymal stem cells to the impaired site in the brain after hypoglossal nerve injury. Stem Cells. 2004;22(3):415–427. doi: 10.1634/stemcells.22-3-415. [DOI] [PubMed] [Google Scholar]
- 167.Kroeze KL, Jurgens WJ, Doulabi BZ, van Milligen FJ, Scheper RJ, Gibbs S. Chemokine-mediated migration of skin-derived stem cells: Predominant role for Ccl5/Rantes. Journal of Investigative Dermatology. 2009;129(6):1569–1581. doi: 10.1038/jid.2008.405. [DOI] [PubMed] [Google Scholar]
- 168.Ge J, Hu Y, Gui Y, Hou R, Yang M, Zeng Q, Xia R. Chemotherapy-induced alteration of Sdf-1/CXCR4 expression in bone marrow-derived mesenchymal stem cells from adolescents and young adults with acute lymphoblastic leukemia. Journal of Pediatric Hematology/Oncology. 2014;36(8):617–623. doi: 10.1097/mph.0000000000000220. [DOI] [PubMed] [Google Scholar]
- 169.Shi, M., Li, J., Liao, L., Chen, B., Li, B., Chen, L., ... Zhao, R. C. (2007). Regulation of CXCR4 expression in human mesenchymal stem cells by cytokine treatment: Role in homing efficiency in NOD/SCID Mice. Haematologica, 92(7), 897-904. 10.3324/haematol.10669 [DOI] [PubMed]
- 170.Yang JX, Zhang N, Wang HW, Gao P, Yang QP, Wen QP. CXCR4 receptor overexpression in mesenchymal stem cells facilitates treatment of acute lung injury in rats. Journal of Biological Chemistry. 2015;290(4):1994–2006. doi: 10.1074/jbc.M114.605063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Schmitt, N., Chêne, L., Boutolleau, D., Nugeyre, M.-T., Guillemard, E., Versmisse, P., ... Israël, N. (2003). Positive regulation of CXCR4 expression and signaling by interleukin-7 in CD4+ mature thymocytes correlates with their capacity to favor human immunodeficiency X4 virus replication. Journal of Virology, 77(10), 5784-5793. [DOI] [PMC free article] [PubMed]
- 172.Busillo JM, Benovic JL. Regulation of CXCR4 signaling. Biochimica et Biophysica Acta. 2007;1768(4):952–963. doi: 10.1016/j.bbamem.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wu, Q., Shao, H., Darwin, E. D., Li, J., Li, J., Yang, B., ... Yu, H. (2009). Extracellular calcium increases CXCR4 expression on bone marrow-derived cells and enhances pro-angiogenesis therapy. Journal of Cellular and Molecular Medicine, 13(9B), 3764-3773. 10.1111/j.1582-4934.2009.00691.x [DOI] [PMC free article] [PubMed]
- 174.Rankin SM. Chemokines and adult bone marrow stem cells. Immunology Letters. 2012;145(1-2):47–54. doi: 10.1016/j.imlet.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 175.Cepeda EB, Dediulia T, Fernando J, Bertran E, Egea G, Navarro E, Fabregat I. Mechanisms regulating cell membrane localization of the chemokine receptor CXCR4 in human hepatocarcinoma cells. Biochimica et Biophysica Acta. 2015;1853(5):1205–1218. doi: 10.1016/j.bbamcr.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 176.Neel NF, Schutyser E, Sai J, Fan GH, Richmond A. Chemokine receptor internalization and intracellular trafficking. Cytokine & Growth Factor Reviews. 2005;16(6):637–658. doi: 10.1016/j.cytogfr.2005.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Brelot A, Heveker N, Pleskoff O, Sol N, Alizon M. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. Journal of Virology. 1997;71(6):4744–4751. doi: 10.1128/JVI.71.6.4744-4751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chabot DJ, Zhang PF, Quinnan GV, Broder CC. Mutagenesis of CXCR4 identifies important domains for human immunodeficiency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates. Journal of Virology. 1999;73(8):6598–6609. doi: 10.1128/JVI.73.8.6598-6609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Doranz, B. J., Orsini, M. J., Turner, J. D., Hoffman, T. L., Berson, J. F., Hoxie, J. A., ... Doms, R. W. (1999). Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. Journal of Virology, 73(4), 2752-2761. [DOI] [PMC free article] [PubMed]
- 180.Kajumo F, Thompson DA, Guo Y, Dragic T. Entry of R5X4 and X4 human immunodeficiency virus type 1 strains is mediated by negatively charged and tyrosine residues in the amino-terminal domain and the second extracellular loop of CXCR4. Virology. 2000;271(2):240–247. doi: 10.1006/viro.2000.0308. [DOI] [PubMed] [Google Scholar]
- 181.Zhou N, Luo Z, Luo J, Liu D, Hall JW, Pomerantz RJ, Huang Z. Structural and functional characterization of human CXCR4 as a chemokine receptor and HIV-1 co-receptor by mutagenesis and molecular modeling studies. Journal of Biological Chemistry. 2001;276(46):42826–42833. doi: 10.1074/jbc.M106582200. [DOI] [PubMed] [Google Scholar]
- 182.Tarasova NI, Stauber RH, Michejda CJ. Spontaneous and ligand-induced trafficking of CXC-chemokine receptor 4. Journal of Biological Chemistry. 1998;273(26):15883–15886. doi: 10.1074/jbc.273.26.15883. [DOI] [PubMed] [Google Scholar]
- 183.Badr G, Borhis G, Treton D, Moog C, Garraud O, Richard Y. HIV type 1 glycoprotein 120 inhibits human B cell chemotaxis to CXC chemokine ligand (CXCL) 12, CC chemokine ligand (CCL)20, and CCL21. Journal of Immunology. 2005;175(1):302–310. doi: 10.4049/jimmunol.175.1.302. [DOI] [PubMed] [Google Scholar]
- 184.Deng, X., Ueda, H., Su, S. B., Gong, W., Dunlop, N. M., Gao, J. L., ... Wang, J. M. (1999). A synthetic peptide derived from human immunodeficiency virus type 1 gp120 downregulates the expression and function of chemokine receptors CCR5 and CXCR4 in monocytes by activating the 7-transmembrane G-protein-coupled receptor FPRL1/LXA4R. Blood, 94(4), 1165-1173. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/10438703 [PubMed]
- 185.Su, S. B., Gong, W., Grimm, M., Utsunomiya, I., Sargeant, R., Oppenheim, J. J., & Ming Wang, J. (1999). Inhibition of tyrosine kinase activation blocks the down-regulation of CXC chemokine receptor 4 by HIV-1 gp120 in CD4+ T cells. Journal of Immunology, 162(12), 7128-7132. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/10358157 [PubMed]
- 186.Chen, L., Li, Y., Chen, W., Han, N., Li, K., Guo, R., ... Xiao, Y. (2020). Enhanced Recruitment and Hematopoietic Reconstitution of Bone Marrow-Derived Mesenchymal Stem Cells in Bone Marrow Failure by the Sdf-1/CXCR4. Journal of Tissue Engineering and Regenerative Medicine, 14(9), 1250-1260. 10.1002/term.3096 [DOI] [PubMed]
- 187.Soundararajan M, Kannan S. Fibroblasts and Mesenchymal Stem Cells: Two Sides of the Same Coin? Journal of Cellular Physiology. 2018;233(12):9099–9109. doi: 10.1002/jcp.26860. [DOI] [PubMed] [Google Scholar]
- 188.Persaud D, Siberry GK, Ahonkhai A, Kajdas J, Monie D, Hutton N, et al. Continued Production of Drug-Sensitive Human Immunodeficiency Virus Type 1 in Children on Combination Antiretroviral Therapy Who Have Undetectable Viral Loads. Journal of Virology. 2004;78(2):968–979. doi: 10.1128/jvi.78.2.968-979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Weissman D, Li Y, Orenstein JM, Fauci AS. Both a precursor and a mature population of dendritic cells can bind HIV. However, only the mature population that expresses CD80 can pass infection to unstimulated CD4+ T cells. The Journal of Immunology. 1995;155(8):4111–4117. [PubMed] [Google Scholar]
- 190.Fazeley F, Hu J, Thirkill TL, Douglas GC. Infection of Primary Human Placental Fibroblasts with HIV-1, HIV-2, and SIV. Archives of Virology. 1997;142(11):2237–2248. doi: 10.1007/s007050050238. [DOI] [PubMed] [Google Scholar]
- 191.Wu, M., Zhang, R., Zou, Q., Chen, Y., Zhou, M., Li, X., ... Chen, Q. (2018). Comparison of the Biological Characteristics of Mesenchymal Stem Cells Derived from the Human Placenta and Umbilical Cord. Scientific Reports, 8(1), 5014. 10.1038/s41598-018-23396-1 [DOI] [PMC free article] [PubMed]
- 192.Robert AW, Marcon BH, Dallagiovanna B, Shigunov P. Adipogenesis, Osteogenesis, and Chondrogenesis of Human Mesenchymal Stem/Stromal Cells: A Comparative Transcriptome Approach. Frontiers in Cell and Development Biology. 2020;8:561. doi: 10.3389/fcell.2020.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Beresford JN, Bennett JH, Devlin C, Leboy PS, Owen ME. Evidence for an inverse relationship between the differentiation of adipocytic and osteogenic cells in rat marrow stromal cell cultures. Journal of Cell Science. 1992;102(Pt 2):341–351. doi: 10.1242/jcs.102.2.341. [DOI] [PubMed] [Google Scholar]
- 194.Chen, Q., Shou, P., Zheng, C., Jiang, M., Cao, G., Yang, Q., ... Shi, Y. (2016). Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death & Differentiation, 23(7), 1128-1139. 10.1038/cdd.2015.168 [DOI] [PMC free article] [PubMed]
- 195.Stechschulte LA, Lecka-Czernik B. Reciprocal regulation of PPARgamma and RUNX2 activities in marrow mesenchymal stem cells: Fine balance between p38 MAPK and Protein Phosphatase 5. Current Molecular Biology Reports. 2017;3(2):107–113. doi: 10.1007/s40610-017-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Gibellini, D., Miserocchi, A., Tazzari, P. L., Ricci, F., Clo, A., Morini, S., ... Re, M. C. (2012). Analysis of the effects of HIV-1 Tat on the survival and differentiation of vessel wall-derived mesenchymal stem cells. Journal of Cellular Biochemistry, 113(4), 1132-1141. 10.1002/jcb.23446 [DOI] [PubMed]
- 197.Cotter EJ, Malizia AP, Chew N, Powderly WG, Doran PP. HIV proteins regulate bone marker secretion and transcription factor activity in cultured human osteoblasts with consequent potential implications for osteoblast function and development. AIDS Research and Human Retroviruses. 2007;23(12):1521–1530. doi: 10.1089/aid.2007.0112. [DOI] [PubMed] [Google Scholar]
- 198.Cotter EJ, Ip HS, Powderly WG, Doran PP. Mechanism of HIV protein induced modulation of mesenchymal stem cell osteogenic differentiation. BMC Musculoskeletal Disorders. 2008;9:33. doi: 10.1186/1471-2474-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Beaupere C, Garcia M, Larghero J, Feve B, Capeau J, Lagathu C. The HIV proteins Tat and Nef promote human bone marrow mesenchymal stem cell senescence and alter osteoblastic differentiation. Aging Cell. 2015;14(4):534–546. doi: 10.1111/acel.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yuan YH, Zhao SS, Wang XL, Teng ZP, Li DS, Zeng Y. HIV-1 p55-gag protein induces senescence of human bone marrow mesenchymal stem cells and reduces their capacity to support expansion of hematopoietic stem cells in vitro. Cell Biology International. 2017;41(9):969–981. doi: 10.1002/cbin.10791. [DOI] [PubMed] [Google Scholar]
- 201.Yuan, Y., Zhou, C., Yang, Q., Ma, S., Wang, X., Guo, X., ... Li, D. (2019). HIV-1 Tat protein inhibits the hematopoietic support function of human bone marrow mesenchymal stem cells. Virus Research, 273, 197756. 10.1016/j.virusres.2019.197756 [DOI] [PubMed]
- 202.Gorwood, J., Bourgeois, C., Mantecon, M., Atlan, M., Pourcher, V., Pourcher, G., ... Lagathu, C. (2019). Impact of HIV/simian immunodeficiency virus infection and viral proteins on adipose tissue fibrosis and adipogenesis. AIDS, 33(6), 953-964. 10.1097/QAD.0000000000002168 [DOI] [PubMed]
- 203.Cheng K, Rai P, Lan X, Plagov A, Malhotra A, Gupta S, Singhal PC. Bone-derived mesenchymal stromal cells from HIV transgenic mice exhibit altered proliferation, differentiation capacity and paracrine functions along with impaired therapeutic potential in kidney injury. Experimental Cell Research. 2013;319(14):2266–2274. doi: 10.1016/j.yexcr.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Couturier J, Lewis DE. HIV Persistence in Adipose Tissue Reservoirs. Current HIV/AIDS Reports. 2018;15(1):60–71. doi: 10.1007/s11904-018-0378-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wong JK, Yukl SA. Tissue reservoirs of HIV. Current Opinion in HIV and AIDS. 2016;11(4):362–370. doi: 10.1097/COH.0000000000000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gomez-Salazar, M., Gonzalez-Galofre, Z. N., Casamitjana, J., Crisan, M., James, A. W., & Péault, B. (2020). Five Decades Later, Are Mesenchymal Stem Cells Still Relevant? Frontiers in Bioengineering and Biotechnology, 8(148). 10.3389/fbioe.2020.00148 [DOI] [PMC free article] [PubMed]
- 207.Zhang, Z., Fu, J., Xu, X., Wang, S., Xu, R., Zhao, M., ... Wang, F.-S. (2013). Safety and Immunological Responses to Human Mesenchymal Stem Cell Therapy in Difficult-to-Treat HIV-1-Infected Patients. AIDS (London, England), 27(8), 1283-1293. 10.1097/QAD.0b013e32835fab77 [DOI] [PMC free article] [PubMed]
- 208.Thanunchai M, Hongeng S, Thitithanyanont A. Mesenchymal Stromal Cells and Viral Infection. Stem Cells International. 2015;2015:860950. doi: 10.1155/2015/860950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bermejo, M., Martín-Serrano, J., Oberlin, E., Pedraza, M. A., Serrano, A., Santiago, B., ... Alcami, J. (1998). Activation of Blood T Lymphocytes Down-Regulates CXCR4 Expression and Interferes with Propagation of X4 HIV Strains. European Journal of Immunology, 28(10), 3192-3204. [DOI] [PubMed]
- 210.Anton, P. A., Elliott, J., Poles, M. A., McGowan, I. M., Matud, J., Hultin, L. E., ... Giorgi, J. V. (2000). Enhanced Levels of Functional HIV-1 Co-Receptors on Human Mucosal T Cells Demonstrated Using Intestinal Biopsy Tissue. AIDS, 14(12), 1761-1765. 10.1097/00002030-200008180-00011 [DOI] [PubMed]
- 211.Bergamini A, Bolacchi F, Pesce CD, Carbone M, Cepparulo M, Demin F, Rocchi G. Increased CD4 and CCR5 Expression and Human Immunodeficiency Virus Type 1 Entry in CD40 Ligand-Stimulated Macrophages. The Journal of Infectious Diseases. 2002;185(11):1567–1577. doi: 10.1086/340413. [DOI] [PubMed] [Google Scholar]
- 212.Lv FJ, Tuan RS, Cheung KM, Leung VY. Concise review: the surface markers and identity of human mesenchymal stem cells. Stem Cells. 2014;32(6):1408–1419. doi: 10.1002/stem.1681. [DOI] [PubMed] [Google Scholar]
- 213.Ostrowski, M. A., Justement, S. J., Catanzaro, A., Hallahan, C. A., Ehler, L. A., Mizell, S. B., ... Fauci, A. S. (1998). Expression of Chemokine Receptors CXCR4 and CCR5 in HIV-1-Infected and Uninfected Individuals. The Journal of Immunology, 161(6), 3195-3201. Retrieved from https://www.jimmunol.org/content/jimmunol/161/6/3195.full.pdf [PubMed]
- 214.Lama J, Mangasarian A, Trono D. Cell-Surface Expression of CD4 Reduces HIV-1 Infectivity by Blocking Env Incorporation in a Nef- and Vpu-Inhibitable Manner. Current Biology. 1999;9(12):622–631. doi: 10.1016/S0960-9822(99)80284-X. [DOI] [PubMed] [Google Scholar]
- 215.Cortés MAJ, Wong-Staal F, Lama J. Cell Surface CD4 Interferes with the Infectivity of HIV-1 Particles Released from T Cells *. Journal of Biological Chemistry. 2002;277(3):1770–1779. doi: 10.1074/jbc.M109807200. [DOI] [PubMed] [Google Scholar]
- 216.Prévost, J., Richard, J., Medjahed, H., Alexander, A., Jones, J., Kappes, J. C., ... Finzi, A. (2018). Incomplete downregulation of CD4 expression affects HIV-1 Env conformation and antibody-dependent cellular cytotoxicity responses. Journal of Virology, 92(13). 10.1128/jvi.00484-18 [DOI] [PMC free article] [PubMed]
- 217.Heigele A, Joas S, Regensburger K, Kirchhoff F. Increased Susceptibility of CD4+ T Cells from Elderly Individuals to HIV-1 Infection and Apoptosis Is Associated with Reduced CD4 and Enhanced CXCR4 and Fas Surface Expression Levels. Retrovirology. 2015;12(1):86. doi: 10.1186/s12977-015-0213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Carl S, Greenough TC, Krumbiegel M, Greenberg M, Skowronski J, Sullivan JL, Kirchhoff F. Modulation of Different Human Immunodeficiency Virus Type 1 Nef Functions During Progression to AIDS. Journal of Virology. 2001;75(8):3657–3665. doi: 10.1128/JVI.75.8.3657-3665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Santosuosso M, Righi E, Lindstrom V, Leblanc PR, Poznansky MC. HIV-1 envelope protein gp120 is present at high concentrations in secondary lymphoid organs of individuals with chronic HIV-1 infection. Journal of Infectious Diseases. 2009;200(7):1050–1053. doi: 10.1086/605695. [DOI] [PubMed] [Google Scholar]
- 220.Banda NK, Bernier J, Kurahara DK, Kurrle R, Haigwood N, Sekaly RP, Finkel TH. Crosslinking CD4 by human immunodeficiency virus gp120 primes T cells for activation-induced apoptosis. Journal of Experimental Medicine. 1992;176(4):1099–1106. doi: 10.1084/jem.176.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Re, M. C., Zauli, G., Gibellini, D., Furlini, G., Ramazzotti, E., Monari, P., ... La Placa, M. (1993). Uninfected haematopoietic progenitor (CD34+) cells purified from the bone marrow of AIDS patients are committed to apoptotic cell death in culture. AIDS, 7(8), 1049-1055. 10.1097/00002030-199308000-00004 [DOI] [PubMed]
- 222.Dessels C, Ambele MA, Pepper MS. The effect of medium supplementation and serial passaging on the transcriptome of human adipose-derived stromal cells expanded in vitro. Stem Cell Research & Therapy. 2019;10(1):253. doi: 10.1186/s13287-019-1370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Gundry RL, Boheler KR, Van Eyk JE, Wollscheid B. A novel role for proteomics in the discovery of cell-surface markers on stem cells: Scratching the surface. Proteomics Clinical Applications. 2008;2(6):892–903. doi: 10.1002/prca.200780122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lin C-S, Xin Z-C, Dai J, Lue TF. Commonly used mesenchymal stem cell markers and tracking labels: Limitations and challenges. Histology and Histopathology. 2013;28(9):1109. doi: 10.14670/hh-28.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Rolandsson, S., Andersson Sjoland, A., Brune, J. C., Li, H., Kassem, M., Mertens, F., ... Westergren-Thorsson, G. (2014). Primary mesenchymal stem cells in human transplanted lungs are CD90/CD105 perivascularly located tissue-resident cells. BMJ Open Respiratory Research, 1(1), e000027. 10.1136/bmjresp-2014-000027 [DOI] [PMC free article] [PubMed]
- 226.Pinho S, Lacombe J, Hanoun M, Mizoguchi T, Bruns I, Kunisaki Y, Frenette PS. PDGFRalpha and CD51 mark human nestin+ sphere-forming mesenchymal stem cells capable of hematopoietic progenitor cell expansion. Journal of Experimental Medicine. 2013;210(7):1351–1367. doi: 10.1084/jem.20122252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Svensson A, Ramos-Moreno T, Eberstal S, Scheding S, Bengzon J. Identification of two distinct mesenchymal stromal cell populations in human malignant glioma. Journal of Neuro-Oncology. 2017;131(2):245–254. doi: 10.1007/s11060-016-2302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Affan A, Al-Jezani N, Railton P, Powell J, Krawetz R. Hip derived synovial mesenchymal progenitor cell surface markers as indicators for differentiation potential. Osteoarthritis and Cartilage. 2016;24:S459–S460. doi: 10.1016/j.joca.2016.01.839. [DOI] [Google Scholar]
- 229.Gray ER, Bain R, Varsaneux O, Peeling RW, Stevens MM, McKendry RA. p24 revisited: a landscape review of antigen detection for early HIV diagnosis. AIDS. 2018;32(15):2089–2102. doi: 10.1097/QAD.0000000000001982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Massanella M, Richman DD. Measuring the latent reservoir in vivo. Journal of Clinical Investigation. 2016;126(2):464–472. doi: 10.1172/JCI80567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Boulle, M., Muller, T. G., Dahling, S., Ganga, Y., Jackson, L., Mahamed, D., ... Sigal, A. (2016). HIV Cell-to-Cell Spread Results in Earlier Onset of Viral Gene Expression by Multiple Infections per Cell. PLoS Pathogens, 12(11), e1005964. 10.1371/journal.ppat.1005964 [DOI] [PMC free article] [PubMed]
- 232.Iyengar SS. Computer Modeling and Simulations of Complex Biological Systems. 2. Taylor & Francis; 1997. [Google Scholar]
- 233.Trible RP, Emert-Sedlak L, Wales TE, Ayyavoo V, Engen JR, Smithgall TE. Allosteric loss-of-function mutations in HIV-1 Nef from a long-term non-progressor. Journal of Molecular Biology. 2007;374(1):121–129. doi: 10.1016/j.jmb.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Cuevas JM, Geller R, Garijo R, Lopez-Aldeguer J, Sanjuan R. Extremely High Mutation Rate of HIV-1 In Vivo. PLoS Biology. 2015;13(9):e1002251. doi: 10.1371/journal.pbio.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Chun, T.-W., Carruth, L., Finzi, D., Shen, X., DiGiuseppe, J. A., Taylor, H., ... Quinn, T. C. (1997). Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature, 387(6629), 183-188. [DOI] [PubMed]
- 236.Seibert SA, Howell CY, Hughes MK, Hughes AL. Natural Selection on the Gag, Pol, and Env Genes of Human Immunodeficiency Virus 1 (HIV-1) Molecular Biology and Evolution. 1995;12(5):803–813. doi: 10.1093/oxfordjournals.molbev.a040257. [DOI] [PubMed] [Google Scholar]
- 237.Chandra, P. K., Gerlach, S. L., Wu, C., Khurana, N., Swientoniewski, L. T., Abdel-Mageed, A. B., ... Mondal, D. (2018). Mesenchymal stem cells are attracted to Latent HIV-1-infected cells and enable virus reactivation via a non-canonical PI3K-NFκb signaling pathway. Scientific Reports, 8(1), 14702. 10.1038/s41598-018-32657-y [DOI] [PMC free article] [PubMed]
- 238.Brandenberg OF, Magnus C, Rusert P, Regoes RR, Trkola A. Different Infectivity of HIV-1 Strains Is Linked to Number of Envelope Trimers Required for Entry. PLoS Pathogens. 2015;11(1):e1004595–e1004595. doi: 10.1371/journal.ppat.1004595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Berger, E. A., Doms, R. W., Fenyö, E. M., Korber, B. T. M., Littman, D. R., Moore, J. P., ... Weiss, R. A. (1998). A New Classification for HIV-1. Nature, 391(6664), 240-240. 10.1038/34571 [DOI] [PubMed]
- 240.Coakley E, Petropoulos CJ, Whitcomb JM. Assessing Chemokine Co-Receptor Usage in HIV. Current Opinion in Infectious Diseases. 2005;18(1):9–15. doi: 10.1097/00001432-200502000-00003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable