Abstract
Herein, a polymeric nanofiber scaffold loaded with Quercetin (Quer)–gold nanorods (GNR) was developed and characterized. Several parameters related to loading Quer into GNR, incorporating the GNR-Quer into polymeric solutions, and fabricating the nanofibers by electrospinning were optimized. GNR-Quer loaded into a polymeric mixture of poly(lactic-co-glycolic acid) (PLGA) (21%) and poloxamer 407 (23%) has produced intact GNR-Quer-nanofibers with enhanced physical and mechanical properties. GNR-Quer-nanofibers demonstrated a slow pattern of Quer release over time compared to nanofibers free of GNR-Quer. Dynamic mechanical thermal analysis (DMTA) revealed enhanced uniformity and homogeneity of the GNR-Quer-nanofibers. GNR-Quer-nanofibers demonstrated a high ability to retain water upon incubation in phosphate buffer saline (PBS) for 24 h compared to nanofibers free of GNR-Quer. A cellular toxicity study indicated that the average cellular viability of human dermal fibroblasts was 76% after 24 h of exposure to the nanofibers containing a low concentration of GNR-Quer.
Incorporating GNR-Quer into a mixture of 21% PLGA LMWT and 23% poloxamer 407 produced smooth, intact and uniform electrospun nanofibers with enhanced mechanical properties and hydration potential.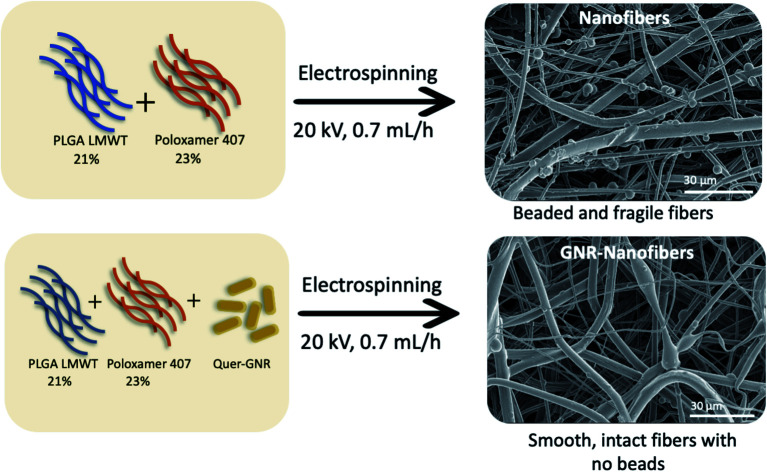
1. Introduction
Nanotechnology is now incorporated into a wide range of applications in numerous fields such as biology, drug delivery, diagnosis and detection, catalysis, optical engineering, electronics, environmental protection, energy storage, and industry.1–9
The various merits that gold nanoparticles (GNP) have in terms of their ability to bind molecules and the possibility to tune their physical properties by changing size and shape make them attractive in many biomedical applications, such as drug delivery, optical diagnosis, photothermal treatment, energy storage, and others.10–22 Gold nanorods (GNR) have gained special interest in biomedical applications because of their unique longitudinal plasmon resonance peaks and their ability to act as photothermal inducing-materials;23 besides, GNP have been reported to promote tissue healing in many previous studies.24,25 GNP bind a wide variety of ligands and biofunctional groups such as polymers, nucleic acids, organic molecules, sugars, and thiol ligands.26–33 Further, GNP-Quercetin (Quer) conjugates were developed for various functions, particularly in cancer and wound healing fields and other applications.34–42
A variety of electrospun nanofiber-based scaffolds have been developed and fabricated for several biomedical applications. By controlling different properties of the nanofibers such as diameter, porosity, surface chemistry, biodegradability, and mechanical properties, they can be used to control and regulate cell behaviors such as cell migration and stem cell differentiation.43–45 Furthermore, electrospun nanofibers could be utilized in drug delivery, tissue engineering, wound healing and biosensing.46–49 A wide range of molecules and medications can be incorporated within the nanofiber systems, ensuring immediate or slow release.50 Researchers have incorporated many molecules within the nanofiber systems: antibiotics, extracellular matrix (ECM) proteins like collagen, antioxidants, metallic nanoparticles, and others.51–53 Incorporating nanoparticles with spun fibers has emerged as an exciting research topic where the functional and mechanical properties of the fibers are improved upon conjugation with nanoparticles.54
This work aims to develop and characterize polymeric nanofibers containing Quer-GNR and evaluate their morphology, physical properties, and biocompatibility against human dermal fibroblasts.
2. Materials and methods
2.1. Materials
Chloroauric acid 99.9% (HAuCl4·3H2O), cetyltrimethylammonium bromide 99% (CTAB), silver nitrate 99% (AgNO3), methoxy-polyethylene glycol-thiol (m-PEG-SH, MW ∼ 5000 g mol−1), sodium borohydride 99% (NaBH4), sodium oleate (NaOL), PEG 4000, l-ascorbic acid (99.9%), poloxamer 407, poloxamer 188 and quercetin (99%) were obtained from Sigma-Aldrich Chemicals, USA. Dimethyl sulfoxide (DMSO) and hydrochloric acid 37% (HCl) were obtained from Alpha Chemika, India. Cholesterol-PEG-SH (MW ∼ 2000 g mol−1) was obtained from Nanosoft Polymers, USA. Chloroform HPLC grade was obtained from Carlo Erba Reagents, Spain, and Ethanol (99.9%) was obtained from Scharlau Chemie s.a, Spain. Nitric acid (HNO3) was obtained from Biosolve–Chimie, France. Resomer® RG 750 S, poly(d,l-lactide-co-glycolide) (PLGA), high molecular weight (HMWT), viscosity of 0.8–1.2 g dL−1, and Resomer® RG 755 S, PLGA, low molecular weight (LMWT), viscosity of 0.5–0.7 g dL−1 were obtained from Evonik, Germany. Tween® 20 from Tedia, USA. Human dermal fibroblast CCD-1064Sk cell line was obtained from American Type Culture Collection (ATCC), USA. Iscove's Modified Dulbecco's Medium (IMDM) was obtained from Biowest, France. Phosphate buffer saline (PBS) was obtained from Lonza, Switzerland. Trypan blue 0.5%, Trypsin–EDTA 0.2% in PBS, and DMSO cell culture grade were obtained from Euro-Clone™, Italy. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was obtained from Bioworld, USA. Potassium bromide (KBr) for Fourier-transform infrared spectroscopy (FTIR) was obtained from AppliChem GmbH, Germany.
The following instruments were used in the study: UV-1800 spectrophotometer (Shimadzu, Japan), Nicomp Nano Z3000 size/zeta potential analyzer (Entegris, USA), Hettich EBA 21 Centrifuge (Germany), formvar-coated copper TEM (Ted Pella Inc., Canada), pH meter (Hanna Instruments, Italy), FT-IR spectroscopy (Shimadzu, Japan), Versa 3D transmission electron microscope (TEM) (FEI, Netherlands), Electrospinning system (EC-DIG. IME Technologies, Netherlands), Laminar airflow cabinet (ESCO Micro, Singapore), Incubator (Avantgarde Binder, Germany), 96-well microplate (Thermo Fisher Scientific, USA), multi-mode microplate reader (BioTek UQuant, USA), Plate shaker (Boekel Scientific 130 000, USA), EZ-LX Long-Stroke Model (Shimadzu, Japan), Hemocytometer (Witeg®, Germany), EVOS™ XL Core Configured Microscope AMEX1200 (Thermo Fischer Scientific, USA), and Q800 dynamic mechanical thermal analysis (DMTA) (Instruments Inc., USA).
2.2. Methods
2.2.1. Synthesis of GNR
GNR were synthesized according to a technique described previously.55 Briefly, seed solution was prepared by mixing 5 mL of CTAB solution (0.20 M) with 5.0 mL of HAuCl4 (0.005 M), then 0.60 mL of ice-cold NaBH4 (0.010 M) was added to the mixture until a honey-colored solution was observed. The growth solution was prepared by dissolving NaOL (1.234 g) and CTAB (7.0 g) in 250 mL of hot water (∼50 °C), which was allowed to cool to 30 °C before adding 18 mL of AgNO3 (4 mM). The mixture was stored in the oven at 30 °C for 15 minutes (min). After that, 250 mL of HAuCl4 (1 mM) was added and stirred for 90 min until it turned into a colorless solution. Then, 2.1 mL of HCl 37 wt%, 0.25 mL of ascorbic acid (64 mM), and 0.8 mL of the seed solution were added to the final solution. The resultant mixture was then left for 48 hour (h) at 30 °C, and the colorless solution turned into a dark orange-brown solution after 12–48 h. Double-round centrifugation of suspensions of GNR was performed for purification, and the pellets were dispersed in Milli-Q water. The concentration of GNR was measured by a validated method of inductively coupled plasma-optical emission spectroscopy (ICP-OES) at a wavelength of 242.795 nm and using a calibration curve of gold standard (0.2–10.0 ppm, r2 = 0.9999).
2.2.2. PEGylation of GNR; GNR-PEG
In order to functionalize the surface of GNR with thiolated PEG, m-PEG-thiol was used by adding 0.1 mL of a 10 mg mL−1 PEG-thiol solution to each 1.0 mL of twice-centrifuged GNR and left for overnight with continuous stirring, followed by centrifugation at 10 000 rpm for 10 min.56
2.2.3. Functionalization of GNR with a cholesterol-PEG-SH moiety; GNR-Chol
One mL of aqueous Chol-PEG-SH solution with a concentration of 20 mg mL−1 was added to 10 mL of twice-centrifuged GNR suspension then mixed overnight. The GNR suspension was centrifuged at 10 000 rpm for 10 min.55
2.2.4. Conjugation of Quer to GNR-PEG and GNR-Chol; GNR-PEG-Quer and GNR-Chol-Quer
A stock solution of Quer in DMSO was prepared. Different parameters, such as type of GNR, time of conjugation reaction, the temperature of the reaction, and concentration of Quer (1–0.25 mg mL−1), were optimized to obtain the most stable conjugated nanoparticles. Then, the solutions were centrifuged twice at 8000 rpm for 8 minutes. The pellet was obtained, then re-suspended in Milli-Q water.
2.3. Characterization of GNR, GNR-PEG, GNR-Chol, GNR-Chol-Quer, and GNR-PEG-Quer
UV-vis absorbance was used to characterize the prepared GNR suspensions at 200–1100 nm. The hydrodynamic radius and zeta potential were measured using a zeta potential/particle size analyzer. Samples of GNR with appropriate dilution (0.1–0.25 nM) were filled into DLS cuvettes for hydrodynamic radius measurement or folded capillary cells for zeta potential measurement at 25 °C. Mean values and standard deviations were calculated from at least three measurements.
FT-IR spectroscopy was used to confirm the surface functionalization of GNR with Quer. GNR before and after loading with Quer were freeze-dried and prepared as potassium bromide (KBr) disks for FT-IR measurements.
The amount of the conjugated Quer to GNR was measured using a validated UV-vis absorption spectroscopy method. A standard calibration curve of Quer was obtained by measuring the UV-vis absorbance of known concentrations of Quer (0.20–0.0312 mg mL−1) in DMSO:PBS; pH 7.4 at 367 nm.
The loading efficiency percentage and the loading content percentage of Quer were calculated as follows:
| Drug loading efficiency (%) = (amount of the drug in the GNR/amount of the added drug) × 100. | 1 |
| Drug loading content (%) = (amount of the drug in the GNR/amount of GNR) × 100. | 2 |
2.4. Formation of the base polymeric nanofibers by electrospinning technique
Different kinds of synthetic polymers and solvents were used to obtain nanofibers without beads while changing the voltage and flow rate of the electrospinning machine. The synthesis of nanofibers started by weighing the desired polymers (poloxamer 188, poloxamer 407, PEG-4000, LMWT PLGA or HMWT PLGA), mixing them with the selected solvent or solvent mixture (ethanol 70%, concentrated ethanol or chloroform) on a vortex or sonicator, depending on the polymers' solubility. The whole formula was drawn by the syringe and fetched to the electrospinning machine, the voltage and flow rate were selected as 20 kV and 0.7 mL h−1, respectively, then the spinning process was performed. The fibers (if any) were collected on the rotating drum, removed and left to dry under the fume hood overnight to assure the remaining solvents' evaporation.
2.5. Incorporation of GNR-PEG-Quer into the polymeric nanofibers; GNR-Quer-nanofibers
The polymeric solution was prepared as described in the previous Section (2.4). The addition of GNR-PEG-Quer of different concentrations (Table 1) was performed drop by drop with the addition of 0.1% (w/v) Tween® 20, letting them mix overnight on a stirrer. On the day after, the whole formula was drawn by the syringe and fetched to the electrospinning machine, the voltage and flow rate were selected as 20 kV and 0.7 mL h−1, respectively, then the spinning process was performed, and the fibers (if any) were collected on the rotating drum. The sheet was then removed, left to dry under the fume hood overnight to assure the remaining solvents' evaporation.
Introducing GNR-PEG-Quer into polymeric solutions and producing GNR-Quer-nanofibers.
| No. | Polymer 1 | Polymer 2 | GNR | Solvent | Flow rate and voltage | Results |
|---|---|---|---|---|---|---|
| 1 | 21% PLGA LMWT | 23% poloxamer 407 | GNR (∼190 μg mL−1)-PEG-Quer | Chloroform | 0.7 mL h−1; 20 kV | Thick fibers, cannot form an intact sheet |
| 2 | 21% PLGA LMWT | 23% poloxamer 407 | GNR (∼95 μg mL−1)-PEG-Quer | Chloroform | 0.7 mL h−1; 20 kV | Intact fibers, no beads |
| 3 | 21% PLGA LMWT | 23% poloxamer 407 | GNR (∼48 μg mL−1)-PEG-Quer | Chloroform | 0.7 mL h−1; 20 kV | Intact fibers, no beads |
2.6. Characterization of the nanofibers
2.6.1. Visual assessment of the nanofibers
Visual examination of nanofiber sheets aimed to investigate the integrity of the produced sheet, its ability to maintain its structure during removal from the drum, and the sheet's texture.
2.6.2. Light microscope imaging of the nanofibers
Fabricated nanofibers sheets (control and GNR-Quer-nanofibers) were investigated under the light microscope to get a general insight into the sheets' structure. Different magnification powers were applied.
2.6.3. Scanning electron microscope (SEM) imaging of the nanofibers
Before starting SEM imaging, sputter coating with gold was applied to increase the signal-to-noise ratio during the imaging process. Ten nm-layer of gold-coated the square-shaped control nanofibers and GNR-Quer-nanofibers were used.
2.6.4. Hydration test of the nanofibers
The nanofiber sheets' swelling was measured by immersing a small sample of the nanofiber sheets in PBS (pH 7.4), then leaving them in the incubator at 37 °C for different periods. Two types of the nanofiber sheets were tested; control sheet and GNR-Quer-nanofibers sheet. Samples were weighed before and after the immersion at different time intervals (1, 4, and 24 h) after placing them on filter paper to remove the excess liquid. The percentage of weight gained was estimated. The experiment was done in triplicate.
2.6.5. Release of Quer from GNR-Quer-nanofibers
One cm2 GNR (∼95 μg mL−1)-Quer-nanofiber was placed in a dialysis bag then immersed in 23 mL of PBS + 1% Tween® 20 in incubator shaker at 37 °C. Aliquots of 0.5 mL were withdrawn from the sample and replaced with fresh 0.5 mL PBS at different time intervals; 2, 4, 6, 24, 48, and 72 h. The same experiment was performed for Quer-nanofibers free of GNR. The released amount of Quer was then calculated based on the concentration equation resulted from the calibration curve (as described in Section 2.3). The release profile of Quer was presented as a percentage of cumulative amount vs. time. The experiment was done in triplicate.
2.6.6. Dynamic mechanical thermal analysis (DMTA) of the nanofibers
DMTA determined the glass transition temperature (Tg) of the nanofibers. The experiment was conducted in tensile mode at an oscillatory frequency and heating rate of 1 Hz and 3 °C per min, respectively, and at a temperature range −40–80 °C. Control nanofibers and GNR-Quer-nanofibers were dried then cut into rectangular samples (30 mm × 0.5 mm, 0.11 mm thickness, measured using a digital caliper). The Tg of the nanofiber sheets was determined from the tan δ where the fiber sheets were clamped in a loading position inside the furnace. The experiment was done in triplicate.
2.7. Assessment of cellular viability of human dermal fibroblasts upon exposure to the nanofibers
Human dermal fibroblasts were cultured in IMDM. The cells were supplemented with l-glutamine (1.0%, 2.0 mM), FBS (10.0% v/v), penicillin (100 U mL−1), streptomycin (100 μg mL−1), and gentamycin (1% v/v of 200 mM) at 5% CO2 and 99% relative humidity at 37 °C. The cells were stained after confluency with trypan blue dye (0.04%) and counted by a hemocytometer.
The MTT assay was used to measure the cellular viability of the cells upon exposure to the following nanofiber sheets: control nanofibers, GNR (∼95 μg mL−1)-Quer-nanofibers, GNR (∼48 μg mL−1)-Quer nanofibers, and control samples contain only the solution of fibroblast in IMDM. A volume of 100 μL of the cell suspension (5 × 103 cells per well) was seeded in a 96-well plate and incubated for 24 h before the addition of the nanofibers. A small sheet of each of the nanofiber sheet was added to the wells with FBS (10%).
For the MTT assay, the nanofiber sheets and the medium from the wells were removed carefully after incubation, and 100 μL of fresh medium and 10 μL of MTT (5 mg mL−1) were added into each well. The plates were incubated for 4 h in 5% CO2 incubator for cytotoxicity. After incubation, the medium from the wells was removed carefully, and 100 μL of DMSO was added to each well and mixed well by shaking for 10–15 min. The development of purple color measured the viable cells due to the formation of formazan crystals. The absorbance was recorded at 570 nm by an ELISA plate reader, and the cellular viability percentage of the treated cells was calculated relative to the cellular viability of the control untreated cells. The experiment was done in triplicate.
3. Results and discussion
3.1. Synthesis and characterization of GNR, GNR-PEG and GNR-Chol
GNR were synthesized by adopting the seed-mediated surfactant-assisted wet chemical method using CTAB and sodium oleate. A binary surfactant system would provide an extra-fine control over the size uniformity and adjustability.57 Additional functionalization of GNR resulted in GNR-PEG or GNR-Chol to enhance the nanoparticles' colloidal stability and eliminate the CTAB toxicity.
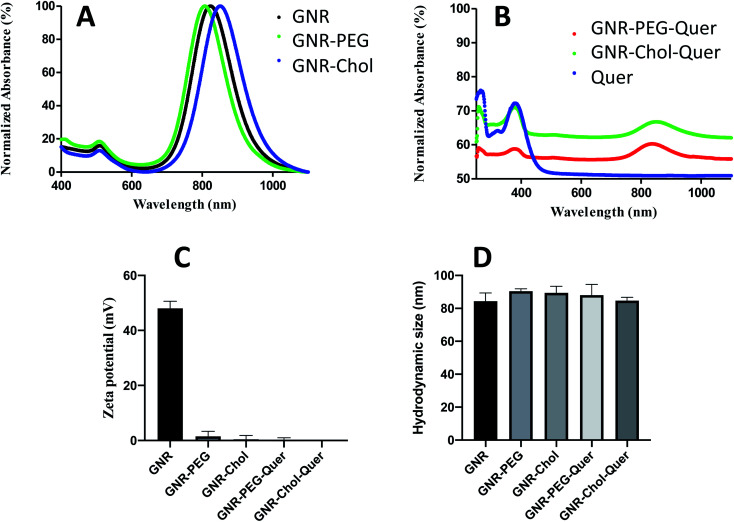
GNR was characterized by UV-vis spectroscopy; they showed transverse and longitudinal peaks at ∼505 nm and ∼817 nm, respectively, without a significant broadening of the peaks (Fig. 1A). The surface-functionalized GNR demonstrated a slight shift of the longitudinal peaks without peak broadening, indicating good stability of the GNR after surface modifications with the polymers (Fig. 1A).
Fig. 1. (A) UV-vis spectra of GNR, GNR-PEG and GNR-Chol. (B) UV-vis of GNR-PEG-Quer, GNR-Chol-Quer and Quer. (C) Zeta potentials of GNR, GNR-PEG, GNR-Chol, GNR-PEG-Quer and GNR-Chol-Quer.
GNR were produced with a CTAB bilayer, exhibiting a positive surface charge. The CTAB bilayer was displaced with PEG-SH or cholesterol-PEG-SH polymers.58 GNR has a zeta potential of +52 mV, and it dropped to +3.3 mV and −0.2 mV after conjugation with PEG-SH and cholesterol-PEG-SH, respectively. The conjugated GNR are stable in the aqueous solution due to PEG or cholesterol-PEG chains' steric repulsion. Fig. 1 demonstrates the zeta potential values and hydrodynamic sizes of the GNR preparations; Fig. 1C and D, respectively.
3.2. Synthesis and characterization of Quer-loaded GNR
Various approaches for loading drugs into nanoparticles are commonly described in the literature. Different methods were utilized to load hydrophobic molecules into nanoparticles such as doxorubicin, rifampicin, cisplatin, antibodies, NSAIDs, and others.21,58 In this study, the nanoparticles were modified with thiolated PEG or thiolated cholesterol-PEG to facilitate loading the hydrophobic drug into the surface of nanoparticles. Quer is a hydrophobic drug soluble in alcohol but insoluble in water (60 mg L−1 in water at 16 °C);59 several nanosystems were utilized to improve the solubility of Quer such as micelles, liposomes, polymeric nanoparticles, nanoemulsions, and inorganic metallic nanoparticles.60,61 In this study, we proposed that Quer was incorporated into the GNR-PEG or GNR-Chol by forming hydrophobic–hydrophobic interactions with the hydrophobic moiety of PEG or cholesterol. A similar conjugation method was used in our previous study of conjugating a hydrophobic chemical compound (PI3Kα inhibitor) to GNR-Chol; the glide docking demonstrated that hydrophobic interactions drive the attachment of the hydrophobic ligand to the cholesterol moiety of the GNR-Chol nanocarrier.55
The amount of loaded Quer to GNR was optimized by controlling defined factors such as the temperature and time of the reaction, coating type of GNR, and concentration of Quer. The best parameters to obtain stable GNR-Quer was achieved upon addition of 0.25 mg mL−1 Quer per every 1 mL of GNR-PEG or GNR-Chol (∼350 μg mL−1) for 24 h. The loaded amount of Quer was similar for GNR-PEG and GNR-Chol (0.17 and 0.19 mg mL−1, respectively). The loading efficiency percentage of Quer is ∼72%, and the loading content percentage is ∼52%.
After conjugating Quer to either GNR-PEG or GNR-Chol, the characteristic bands of Quer at ∼376 nm appeared in the UV-vis spectra leading to the conclusion that binding to GNR has occurred. Fig. 1B shows the UV-vis spectra of GNR-PEG-Quer, GNR-Chol-Quer and Quer. Zeta potential values and hydrodynamic sizes were not significantly changed upon conjugation (Fig. 1C and D, respectively), since Quer is supposed to slide within the cholesterol or PEG pockets. Both GNR-PEG and GNR-Chol were loaded with a similar amount of Quer. GNR-PEG-Quer was selected to perform the rest of the experimental tests.
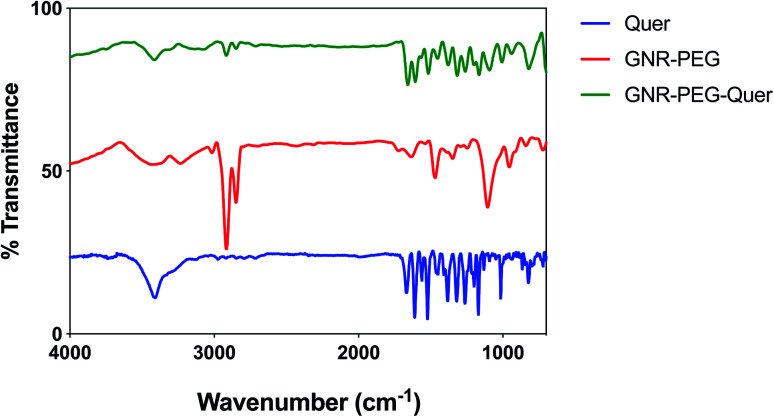
FT-IR spectra for Quer, GNR-PEG, and GNR-PEG-Quer were performed. The characteristic peaks of Quer are indicative for the alcohol group at the 3307–3420 cm−1 range, carbonyl groups at ∼1602 cm−1, and aromatic rings at ∼1512 cm−1. GNR-PEG spectrum represents CH bond stretching at ∼2918 and CO bending at ∼1485 cm−1.62 As conjugating Quer to GNR-PEG, the FT-IR spectra of Quer and GNR-PEG-Quer demonstrated marked similarities; however, the intensities of some characteristic bands such as carbonyl and aromatic bending were slightly decreased upon conjugation. These spectral changes strongly indicate the presence of intermolecular interactions between Quer and GNR-PEG (Fig. 2).
Fig. 2. FT-IR spectra for Quer, GNR-PEG and GNR-PEG-Quer.
3.3. Development, optimization and characterization of the base polymeric nanofibers
Nanofibers represent an exquisite base for several medical applications such as tissue engineering and wound healing. Electrospinning is a technique utilized to generate nanofibers from different materials, particularly organic polymers.45
During the development and optimization process of nanofibers formation, multiple polymers and solvents were first prepared as a base to receive the GNR-Quer later. The base mixture should be clear and of a suitable viscosity to ensure the success of the electrospinning. Nanofibers of concern should be acceptable in terms of continuity, absence of defects, diameter, porosity, interlacing, degradation rate, and biocompatibility. Various factors affecting the electrospinning process have been optimized to produce nanofibers of satisfactory physical and mechanical properties. These, in part, depend on the molecular weight of the polymer, and its concentration and electrical conductivity, the solvent type and the processing parameters such as applied voltage, the flow rate of the electrospinning solution, and distance from the collecting surface.63
Selection of the suitable voltage value is crucial; the relationship between voltage and nanofiber diameter is apparent knowing that a large amount of the solution would be forced out from the injector tip so that the nanofiber diameter would be larger. Thousands of volts are needed to initiate the electrospinning process; 15 kV was tried as a start because of the lower viscosity the initial samples had, which were mainly formed by dissolving poloxamer 188, poloxamer 407, or both in 70% ethanol. Then, as the process of optimization progressed, solutions of different polymers were prepared and utilized. The following polymer solutions were more viscous than the previous solutions, so the need for higher voltage emerged to keep the nanofiber elongation process going. PLGA of high and low molecular weights with or without a hydrophilic polymer (like poloxamer 407 or PEG 4000) or a hydrophobic polymer (poloxamer 188) produced much more viscous solutions; thus, 20 kV was set as a standard and resulted in agreeable results.
In terms of the rotation speed of the collecting drum, the speed must be identical or very close to the solvent's evaporation rate; so that the accumulation produces the best alignment and guarantees the continuous nanofibers that do not break. The rotation speed and the flow rate of the electrospinning liquid were one thousand rounds per min and 0.7 mL h−1, respectively.
Beads are a major problem in nanofibers production; it was considered one of the most important parameters to evaluate the properties of the nanofibers. The formation of beaded fibers depends on several factors, such as the concentration of the polymer and surface tension, viscosity, and density of the surface charges of the electrospinning liquid.45
The ability of several polymers to form fibers was evaluated; poloxamer 188 and poloxamer 407 (20–70% w/v) were tested individually and in combination, with increasing the solution's viscosity using different types of solvents. Individual polymers did not produce fibers, and mixing two polymers slightly improved the mixture's spinnability; however, tiny fibers formed with many beads, particularly at low polymers' concentration. In general, low viscosity of the electrospinning liquid favors beaded nanofibers.45 Further, the polymer's concentration plays a critical role in forming beads; a minimum concentration is required for chain entanglement and electrospinning. The Rayleigh instability could not be avoided at low concentration, and droplets will be formed; thus, fine particles or beads will be obtained.45
Different concentrations of PEG 4000 were investigated; 50–100% w/v in 70% ethanol, and no or little fibers formed with many beads, particularly at low polymers' concentration. Mixing the hydrophilic PEG 4000 with the hydrophobic poloxamer 188 did not enhance the formation of fibers. Consequently, the process was directed towards PLGA; both high and low molecular weight of PLGA were tested. Mixing different ratios of HMWT PLGA with PEG-4000 or poloxamer 407 did not stop bead formation. High concentrations of HMWT PLGA were avoided to ensure a proper release behavior of the sheet's bioactive components; so, LMWT PLGA was used. A low concentration of LMWT PLGA (15%) resulted in beaded fibers, while increasing the concentration (∼20%) of the polymer improved the formation of fibers. Mixing LMWT PLGA (21%) with poloxamer 407 (23%) was the best combination that produced a successful intact electrospun nanofiber sheet with few beads.
The structure and morphology of the obtained polymer nanofibers is determined also by the solvent used. The choosing of a solvent is dependent on the polymer solubility; further, the volatility of the solvent has a crucial role in the success of the spinning; very high volatility may result in immediate jet solidification, and the fibers will still be wet in case of too low volatility.45 In our work, ethanol 70% was selected as a suitable solvent for the poloxamers and PEG 4000; however, it could not evaporate rapidly during the traveling of charged droplets of the electrospinning liquid. Consequently, the solvent was replaced once by 90% ethanol and once by concentrated ethanol; however, the electrospinning process improved slightly. Chloroform was the suitable solvent for PLGA polymers, and its evaporation rate was suitable for producing well-formed fibers. Besides, the surface tension of the electrospinning liquid is affected mainly by the solvent type and to a lesser extent by the polymer concentration; thus, a delicate balance should be achieved considering the polymers and solvents used.64
3.4. Incorporating the GNR-PEG-Quer into the nanofibers; GNR-Quer-nanofibers
The incorporation of functional nanoparticles with electrospun fibers enhances the performance of the nanofibers and preserves their stability. GNP-spun fibers demonstrated enhanced conductivity than fibers free of GNP and could be considered promising scaffold for tissue engineering, surface-enhanced Raman spectroscopy (SERS) and photothermal applications.65–67
GNR-PEG-Quer was introduced into the previously optimized polymeric solution. GNR-PEG-Quer of three different concentrations of GNR (190, 95, and 48 μg mL−1) were added to the polymeric mixtures, then fabricated into nanofibers by electrospinning (Table 1).
It was an utmost priority to assess the polymer mixture's stability after incorporating GNR-PEG-Quer as GNR-PEG-Quer were suspended in water, and the solvents used in the preparation of the polymer mixture are organic. Visual examination showed good stability of the GNR–polymer mixture as the solution color remained pink-brown even after two weeks of preparation. Tween® 20 was added to the GNR-Quer-polymer mixture to make the solution homogenous and avoid separation of the overall mixture; further, Tween® 20 reduced the surface tension of the electrospinning liquid since as it increases, more beads are likely to be formed.68
3.5. Characterization of nanofibers
3.5.1. Visual assessment and imaging of nanofibers (light microscope and SEM imaging)
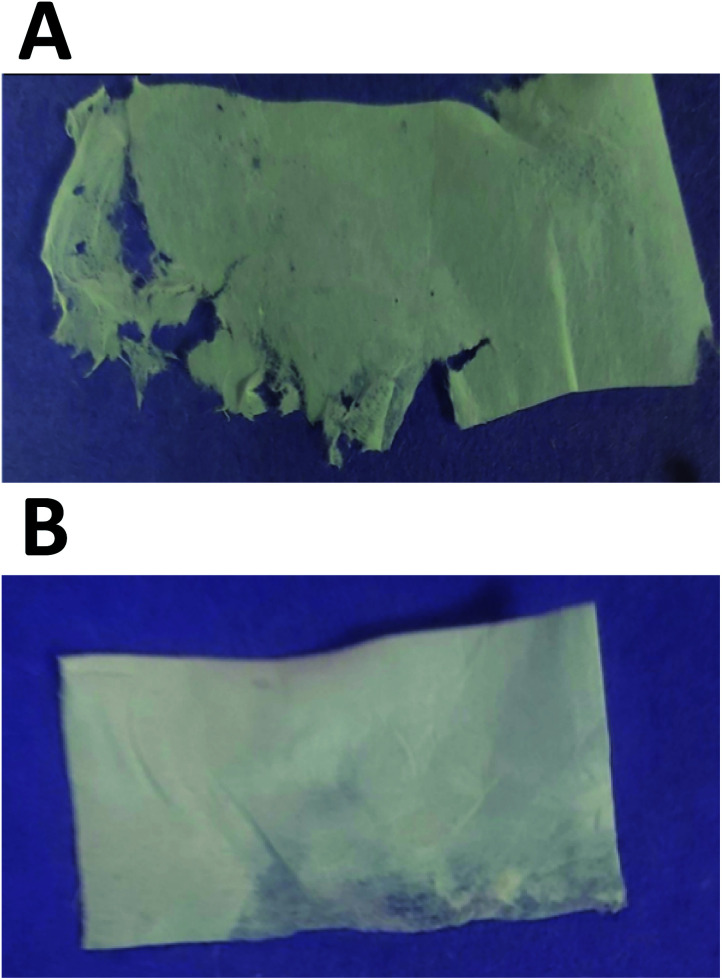
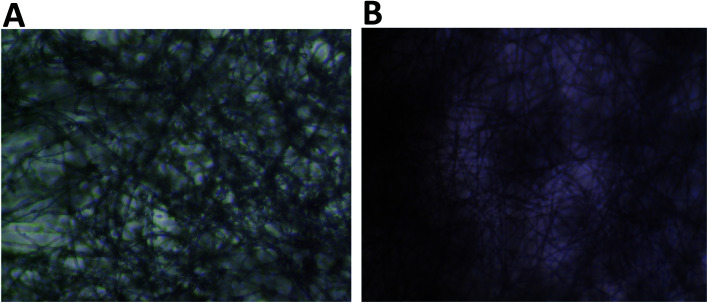
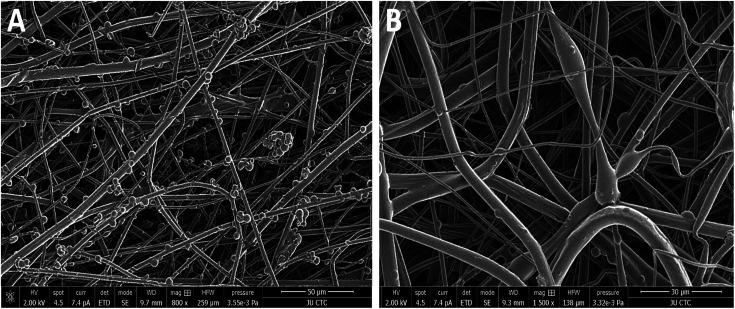
Visual examination of the nanofiber sheets revealed the basic structural intactness of the sheets. The control nanofibers sheet was slightly fragile compared to the sheet that had GNR-PEG-Quer within. GNR (∼95–48 μg mL−1)-Quer-nanofiber sheets were more intact than the control sheet, which were tared upon removing them from the collecting drum (Fig. 3A and B). Further, the control sheet was made of many layers over each other that are separated and noticed upon inspection by the naked eye. However, the nanofibers sheet with a high concentration of GNR (∼190 μg mL−1)-PEG-Quer was fragile and demonstrated very poor consistency. The light microscope showed clumps of the GNR-free nanofibers that were not of a consistent diameter, while GNR-containing nanofibers sheet has a better quality in terms of texture, homogeneity, diameter, and morphology (Fig. 4A and B).
Fig. 3. Electrospun nanofiber sheets of (A) mixture of 21% (w/v) LMWT PLGA and 23% (w/v) poloxamer 407 in chloroform (control), and (B) GNRs-PEG-Quer (∼48 μg gold per mL) mixed with 21% (w/v) LMWT PLGA; 23% (w/v) poloxamer 407 in chloroform. Visual examination reveals better integrity of the whole sheet containing GNR-PEG-Quer compared to the control.
Fig. 4. Light microscope images of nanofibers showing the gross overview of the nanofiber morphology. (A) Nanofibers of a mixture of 21% (w/v) LMWT PLGA and 23% (w/v) poloxamer 407 in chloroform; clumping of the polymer mixture is obvious on the nanofibers. (B) Nanofibers of GNRs-PEG-Quer incorporated into 21% (w/v) LMWT PLGA and 23% (w/v) poloxamer 407 in chloroform. Few beads and clumps are observed in the GNR-containing nanofibers compared to the control nanofibers sheet.
SEM images represent that GNR-Quer-nanofibers are smoother, more uniform than GNR-free nanofibers and contained no beads (Fig. 5A and B). The decreased number of beads improves the nanofibers sheet's quality as the sheet becomes more filamentous and more surface area is available to receive the incorporated materials. GNR (conductive particles) in the polymeric mixture may hold a charge during the electrospinning process, elevating each nanofiber's surface's smoothness to affect the morphology and decrease the diameter because the flowing and elongation process becomes much more manageable. It should be pointed out that the compatibility and homogeneity between the polymers in the electrospinning liquid might be improved upon the inclusion of GNR.
Fig. 5. SEM images of electrospun nanofibers. (A) Control nanofibers clearly show the beads and clusters of aggregation of polymer mixture as well as the rough surface morphology. (B) GNR-Quer-nanofibers have a smooth surface morphology and the beads are scarcely present.
Another possible explanation of improving the nanofibers' physical properties upon addition of GNR is the ability of PEG that was conjugated to the surface of GNR to form intermolecular interactions with the polymeric fibers that may improve their structure. A recent study showed that a smooth surface was observed for the electrospun polyacrylonitrile nanofibers upon incorporating GNP without aggregation of nanoparticles on the nanofibers' surface.69
3.5.2. Hydration test of the nanofibers
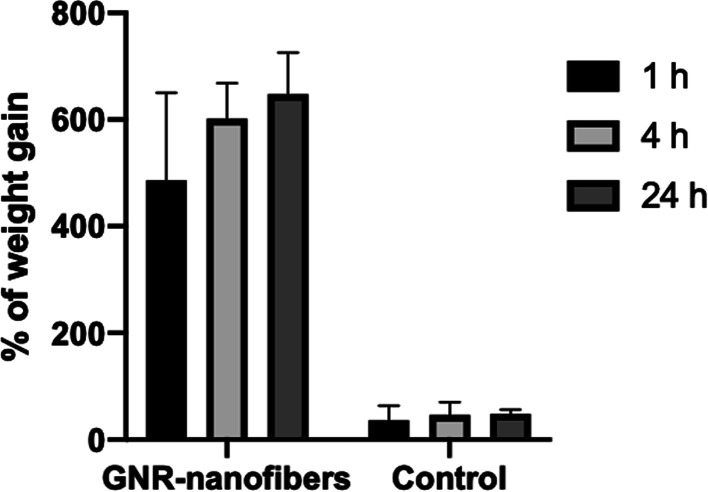
The hydration test is performed to describe the ability of the sheets to swell in physiological conditions. Our results indicate that GNR-containing nanofibers are more able to absorb water (an average of 647% of weight gain) than the control nanofibers (an average of ∼48%) after 24 h of incubation in PBS (Fig. 6). It is proposed that the well-formed nanofibers in GNR addition may provide more channels for water absorption than fragile control nanofibers. Further, GNR's coating, i.e., polyethylene glycol, which has a hydrophilic nature that recruits water molecules, contributed to the drastic ability of GNR-containing nanofibers to absorb water. These results indicate that such nanofibers could be considered promising dressing material for wound healing due to ability to contain the produced exudate at the injury site and reducing the risk of infections.70,71 Recently, research has focused on developing nanofiber-based dressings with high porosity and hydration potential to reduce infection, inflammation and promote wound healing.72
Fig. 6. Weight gain percentages of GNR-containing nanofibers and control nanofibers upon incubation in PBS for 1, 4, and 24 h.
3.5.3. Release of Quer from GNR-Quer-containing nanofibers and Quer-nanofibers sheets
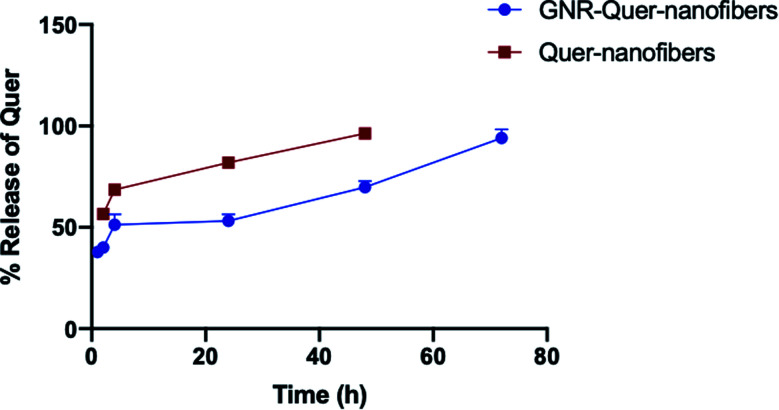
The incorporation of molecules into nanofibers may modulate and target the release of these molecules into the biological systems, thus enhancing their effectiveness and reducing side effects. The release profile of Quer from the nanofibers was illustrated in Fig. 7. Incorporating the GNR-Quer into nanofibers resulted in slow release of Quer over 72 h of incubation. About half the Quer amount was released after 24 h of incubation, and almost full release of Quer (an average of ∼95%) was achieved after 72 h of incubation. This slow-release pattern is most likely attributed to the conjugation of Quer to GNR via the cholesterol moiety and incorporating PLGA in the nanofibers. On the other hand, the release of Quer from GNR-free nanofibers demonstrated a faster release pattern (an average of ∼82% after 24 h) and almost complete release (an average of ∼96%) after 48 h compared to those containing GNR. The slow-release profile of drugs has advantages in certain biomedical applications such as drug delivery and therapy; several studies in the literature demonstrated the modulation effect of nanofibers on the release of various incorporated molecules and drugs. Several changes in environmental parameters such as pH value, temperature and light can modulate the drug release rate at the site of action.46
Fig. 7. Release pattern of Quer from GNR-Quer-nanofibers and Quer-nanofibers over time.
3.5.4. Determination of Tg temperature of GNR-Quer-nanofibers using DMTA
Transition temperatures are useful as indicators of many structural changes correlated with the chemical and physical behaviors of the polymers. The Tg temperature is one such parameter that indicates the glassy to viscous liquid change, and it is correlated to the polymer chain mobility and homogeneity, and mechanical properties.63 In our study, the determination of Tg was utilized to evaluate the dynamic mechanical properties, the uniformity and homogeneity of the nanofiber scaffolds, and the effect of GNR's incorporation into them.
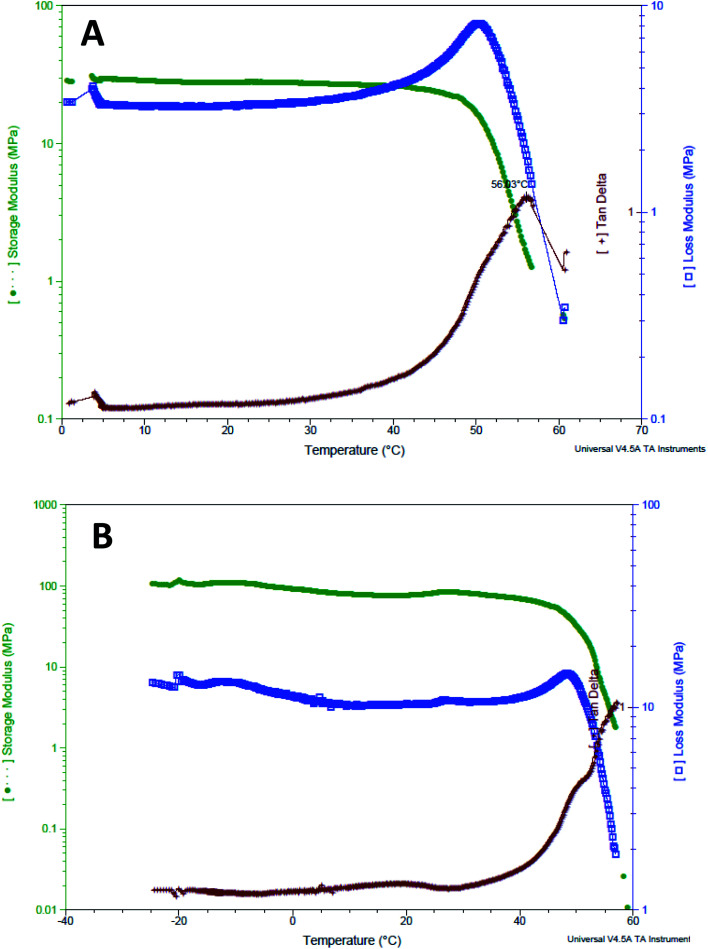
The GNR-Quer-nanofibers showed a typical DMTA thermogram where one apparent glass transition temperature was noticed at 56.03 °C and no observation of minor relaxation events (Fig. 8A). The nanofibers melted at 60.66 °C, and at this temperature, the nanofibers were found split and cut upon removal from the furnace. Interestingly, the DMTA thermogram of control nanofibers showed multiple glass transition events indicating that the nanofibers free of GNR are irregular, where an α relaxation was detected at 19.4 °C, and β relaxation was observed at 30.6 °C (Fig. 8B). The control nanofibers eventually melted at 58.9 °C. The DMTA thermograms suggest that the addition of GNR improved the uniformity, homogeneity, and smoothness of the nanofibers; this is also supported by the SEM images, where irregular clusters of polymer aggregation of the control nanofibers were prominent. These irregularities will dictate uneven absorption of energy; thus, different transition states may be observed. However, the upgraded morphology provided by the addition of the GNR would favor an improved fashion of energy absorption leading to one Tg presence.
Fig. 8. DMTA thermogram of (A) GNR-Quer-nanofibers, and (B) control nanofibers.
3.5.5. Cellular viability of human dermal fibroblasts upon exposure to GNR-Quer-nanofibers
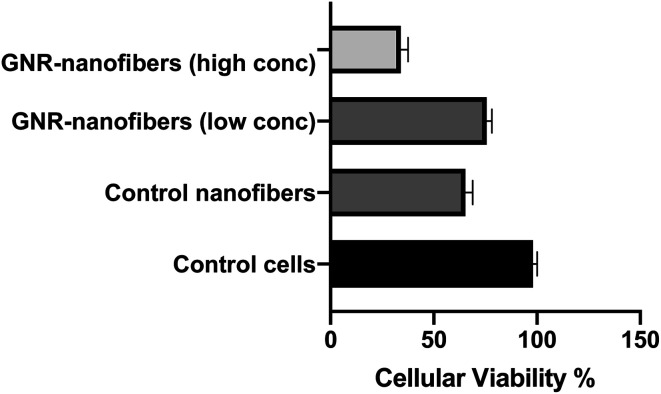
The MTT test was applied to determine the cytotoxicity of both control and GNR-containing nanofiber sheets towards human dermal fibroblasts. GNR-Quer-nanofibers showed concentration-dependent cytotoxicity; the cellular viability of the cells exposed to nanofibers containing 96 μg mL−1 of GNR was high compared to nanofibers containing a low concentration of GNR (∼48 μg mL−1) (celuular viability; 76% vs. 35%, respectively, Fig. 9).
Fig. 9. Cellular viability of human dermal fibroblast after 24 h of exposure to nanofiber sheets containing high and low concentration of GNR-Quer compared to control nanofibers and untreated control cells.
Interestingly, the fibroblasts exhibited similar cellular viability upon exposure to nanofibers containing a low concentration of GNR or the control nanofibers free of GNR (76% vs. 66%, respectively, Fig. 9). We propose that the presence of nanofiber sheets added directly on the top of cells could have compromised the tissue's microenvironment, and thus a reduction in cell viability was seen in even control nanofibers where the components are known to be safe. To minimize the interference of the nanofiber sheets with the absorbance of the MTT dye, the sheets were carefully removed before the dye's addition, which could have been another cause for reducing the number of living cells.
The biocompatibility of nanofibers was demonstrated in the literature towards several types of normal cells; recently, mouse fibroblast exhibited high viability upon treatment with electrospun carbon nanofibers modified with hydroxyapatite.73
Our results indicate the biocompatibility of GNR-containing nanofibers, particularly those containing a low concentration of GNR.
4. Conclusions
GNR-PEG-Quer incorporated into polymeric nanofibers of poloxamer 407 (23%) and PLGA (21%) has drastically improved the properties of nanofibers in different aspects. The nanofibers' structure and morphology, intactness, flexibility and mechanical properties were dramatically improved upon the incorporation of GNR. GNR-Quer-nanofibers demonstrated enhanced hydration ability compared to control nanofibers free of GNR (weight gain of ∼647% vs. ∼48%, respectively), and their cytotoxicity was dependent on the GNR's concentration. Conjugation of Quer into GNR sustained the release of Quer over time compared to the release of Quer from GNR-free nanofibers. The development of GNR-incorporated nanofibers provides significant advances toward promising nanomaterials for biomedical applications such as drug delivery, tissue and wound healing.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was funded by the Deanship of Scientific Research and Graduate Studies at Al-Zaytoonah University of Jordan (2020-2019/12/28). Authors would like to thank Mrs Maha Rashid and Miss Sabaa Al-Dabash for assistance in conducting the experiments.
References
- Nasrollahzadeh M., Sajadi M. S., Atarod M., Sajjadi M. and Isaabadi Z., An Introduction to Green Nanotechnology, Academic Press, 2019 [Google Scholar]
- Ummi A. S. and Siddiquee S., in Nanotechnology: Applications in Energy, Drug and Food, ed. S. Siddiquee, G. J. H. Melvin and M. M. Rahman, Springer International Publishing, Cham, 2019, pp. 295–308, 10.1007/978-3-319-99602-8_15 [DOI] [Google Scholar]
- Ahmadi M. H. Ghazvini M. Alhuyi Nazari M. Ahmadi M. A. Pourfayaz F. Lorenzini G. Ming T. Int. J. Energy Res. 2019;43:1387–1410. doi: 10.1002/er.4282. [DOI] [Google Scholar]
- Karn B. Kuiken T. Otto M. Environ. Health Perspect. 2009;117:1813–1831. doi: 10.1289/ehp.0900793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q. Shi J. J. Mater. Chem. 2011;21:5845–5855. doi: 10.1039/C0JM03851B. [DOI] [Google Scholar]
- Oberdörster G. Sharp Z. Atudorei V. Elder A. Gelein R. Kreyling W. Cox C. Inhalation Toxicol. 2004;16:437–445. doi: 10.1080/08958370490439597. [DOI] [PubMed] [Google Scholar]
- Conner S. D. Schmid S. L. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- Ghosh P. Han G. De M. Kim C. K. Rotello V. M. Adv. Drug Delivery Rev. 2008;60:1307–1315. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Jain P. K. Huang X. El-Sayed I. H. El-Sayed M. A. Acc. Chem. Res. 2008;41:1578–1586. doi: 10.1021/ar7002804. [DOI] [PubMed] [Google Scholar]
- Kim T. J. Colloid Interface Sci. 2008;318:238–243. doi: 10.1016/j.jcis.2007.10.029. [DOI] [PubMed] [Google Scholar]
- Pourhashem S. Vaezi M. R. Rashidi A. Surf. Coat. Technol. 2017;311:282–294. doi: 10.1016/j.surfcoat.2017.01.013. [DOI] [Google Scholar]
- Hassan S. Singh A. V. J. Nanosci. Nanotechnol. 2014;14:402–414. doi: 10.1166/jnn.2014.8747. [DOI] [PubMed] [Google Scholar]
- Cai J. Ding L. Gong P. Huang J. J. Nanotechnol. 2019;31:095501. doi: 10.1088/1361-6528/ab55b7. [DOI] [PubMed] [Google Scholar]
- Chang C.-C. Chen C.-P. Wu T.-H. Yang C.-H. Lin C.-W. Chen C.-Y. Nanomaterials. 2019;9:861. doi: 10.3390/nano9060861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghaie T. Jazayeri M. H. Manian M. Khani l. Erfani M. Rezayi M. Ferns G. A. Avan A. J. Cell. Biochem. 2019;120:2749–2755. doi: 10.1002/jcb.27415. [DOI] [PubMed] [Google Scholar]
- Yang W. Liang H. Ma S. Wang D. Huang J. Sustainable Mater. Technol. 2019:e00109. doi: 10.1016/j.susmat.2019.e00109. [DOI] [Google Scholar]
- Mahato K. Nagpal S. Shah M. A. Srivastava A. Maurya P. K. Roy S. Jaiswal A. Singh R. Chandra P. 3 Biotech. 2019;9:57. doi: 10.1007/s13205-019-1577-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bakri A. G. Mahmoud N. N. Molecules. 2019;24:2661. doi: 10.3390/molecules24142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud N. N. Hikmat S. Abu Ghith D. Hajeer M. Hamadneh L. Qattan D. Khalil E. A. Int. J. Pharm. 2019;565:174–186. doi: 10.1016/j.ijpharm.2019.04.079. [DOI] [PubMed] [Google Scholar]
- Mahmoud N. N. Alkilany A. M. Khalil E. A. Al-Bakri A. G. Sci. Rep. 2018;8:6881. doi: 10.1038/s41598-018-24837-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud N. N. Sabbah D. A. Abu-Dahab R. Abuarqoub D. Abdallah M. Khalil E. A. RSC Adv. 2019;9:12718–12731. doi: 10.1039/C9RA01041F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjana P. M. Bindhu M. R. Rakhi R. B. Mater. Sci. Energy Technol. 2019;2:389–395. [Google Scholar]
- Alkilany A. M. Thompson L. B. Boulos S. P. Sisco P. N. Murphy C. J. Adv. Drug Delivery Rev. 2012;64:190–199. doi: 10.1016/j.addr.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Lau P. Bidin N. Islam S. Shukri W. N. B. W. M. Zakaria N. Musa N. Krishnan G. Lasers Surg. Med. 2017;49:380–386. doi: 10.1002/lsm.22614. [DOI] [PubMed] [Google Scholar]
- Mahmoud N. N. Hikmat S. Ghith D. A. Hajeer M. Hamadneh L. Qattan D. Khalil E. A. Int. J. Pharm. 2019;565:174–186. doi: 10.1016/j.ijpharm.2019.04.079. [DOI] [PubMed] [Google Scholar]
- Niemeyer C. M. Angew. Chem. 2001;40:4128–4158. doi: 10.1002/1521-3773(20011119)40:22<4128::AID-ANIE4128>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Shenhar R. Rotello V. M. Acc. Chem. Res. 2003;36:549–561. doi: 10.1021/ar020083j. [DOI] [PubMed] [Google Scholar]
- Daniel M.-C. Astruc D. Chem. Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- Rosi N. L. Mirkin C. A. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- Ghadiali J. E. Stevens M. M. Adv. Mater. 2008;20:4359–4363. doi: 10.1002/adma.200703158. [DOI] [Google Scholar]
- Stewart M. E. Anderton C. R. Thompson L. B. Maria J. Gray S. K. Rogers J. A. Nuzzo R. G. Chem. Rev. 2008;108:494–521. doi: 10.1021/cr068126n. [DOI] [PubMed] [Google Scholar]
- Murphy C. J. Science. 2002;298:2139–2141. doi: 10.1126/science.1080007. [DOI] [PubMed] [Google Scholar]
- Murphy C. J. Gole A. M. Stone J. W. Sisco P. N. Alkilany A. M. Goldsmith E. C. Baxter S. C. Acc. Chem. Res. 2008;41:1721–1730. doi: 10.1021/ar800035u. [DOI] [PubMed] [Google Scholar]
- Madhyastha H. Halder S. Intan N. Madhyastha R. Mohanapriya A. Sudhakaran R. Sajitha L. Banerjee K. Bethasiwi P. Daima H. Navya P. N. Maruyama M. Nakajima Y. RSC Adv. 2020;10:37683–37694. doi: 10.1039/D0RA06690G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan S. Mukherjee S. Das S. Bhat F. A. Raja Singh P. Patra C. R. Arunakaran J. Cell Biochem. Funct. 2017;35:217–231. doi: 10.1002/cbf.3266. [DOI] [PubMed] [Google Scholar]
- Ajmal G. Bonde G. V. Mittal P. Khan G. Pandey V. K. Bakade B. V. Mishra B. Int. J. Pharm. 2019;567:118480. doi: 10.1016/j.ijpharm.2019.118480. [DOI] [PubMed] [Google Scholar]
- Ezzati M. Yousefi B. Velaei K. Safa A. Life Sci. 2020:117463. doi: 10.1016/j.lfs.2020.117463. [DOI] [PubMed] [Google Scholar]
- Marunaka Y. Ann. N. Y. Acad. Sci. 2017;1398:142–151. doi: 10.1111/nyas.13361. [DOI] [PubMed] [Google Scholar]
- Patel R. V. Mistry B. M. Shinde S. K. Syed R. Singh V. Shin H.-S. Eur. J. Med. Chem. 2018;155:889–904. doi: 10.1016/j.ejmech.2018.06.053. [DOI] [PubMed] [Google Scholar]
- Ulusoy H. G. Sanlier N. Crit. Rev. Food Sci. Nutr. 2020;60:3290–3303. doi: 10.1080/10408398.2019.1683810. [DOI] [PubMed] [Google Scholar]
- Wang D. Sun-Waterhouse D. Li F. Xin L. Li D. Crit. Rev. Food Sci. Nutr. 2019;59:2189–2201. doi: 10.1080/10408398.2018.1441123. [DOI] [PubMed] [Google Scholar]
- Xiao L. Luo G. Tang Y. Yao P. Food Chem. Toxicol. 2018;114:190–203. doi: 10.1016/j.fct.2018.02.022. [DOI] [PubMed] [Google Scholar]
- Qu J. Zhou D. Xu X. Zhang F. He L. Ye R. Zhu Z. Zuo B. Zhang H. Appl. Surf. Sci. 2012;261:320–326. doi: 10.1016/j.apsusc.2012.08.008. [DOI] [Google Scholar]
- Wang X. Ding B. Li B. Mater. Today. 2013;16:229–241. doi: 10.1016/j.mattod.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J. Wu T. Dai Y. Xia Y. Chem. Rev. 2019;119:5298–5415. doi: 10.1021/acs.chemrev.8b00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L. Xie J. Curr. Pharm. Des. 2015;21:1944–1959. doi: 10.2174/1381612821666150302151959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj N. Kundu S. C. Biotechnol. Adv. 2010;28:325–347. doi: 10.1016/j.biotechadv.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Kim H. N. Hong Y. Kim M. S. Kim S. M. Suh K.-Y. Biomaterials. 2012;33:8782–8792. doi: 10.1016/j.biomaterials.2012.08.038. [DOI] [PubMed] [Google Scholar]
- Liu Y. Zhou S. Gao Y. Zhai Y. Asian J. Pharm. Sci. 2019;14:130–143. doi: 10.1016/j.ajps.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai R. S. Bachu R. D. Boddu S. H. Bhaduri S. Pharmaceutics. 2019;11:5. doi: 10.3390/pharmaceutics11010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. Duan X.-P. Li Y.-M. Yang D.-P. Long Y.-Z. Mater. Sci. Eng., C. 2017;76:1413–1423. doi: 10.1016/j.msec.2017.03.034. [DOI] [PubMed] [Google Scholar]
- Thakur R. Florek C. Kohn J. Michniak B. Int. J. Pharm. 2008;364:87–93. doi: 10.1016/j.ijpharm.2008.07.033. [DOI] [PubMed] [Google Scholar]
- Fouda M. M. El-Aassar M. Al-Deyab S. S. Carbohydr. Polym. 2013;92:1012–1017. doi: 10.1016/j.carbpol.2012.10.047. [DOI] [PubMed] [Google Scholar]
- Zhang C.-L. Yu S.-H. Chem. Soc. Rev. 2014;43:4423–4448. doi: 10.1039/C3CS60426H. [DOI] [PubMed] [Google Scholar]
- Mahmoud N. N. Sabbah D. A. Abu-Dahab R. Abuarqoub D. Abdallah M. Hasan Ibrahim A. Khalil E. A. RSC Adv. 2019;9:12718–12731. doi: 10.1039/C9RA01041F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud N. N. Harfouche M. Alkilany A. M. Al-Bakri A. G. El-Qirem R. A. Shraim S. A. Khalil E. A. Colloids Surf., B. 2018;165:118–126. doi: 10.1016/j.colsurfb.2018.02.021. [DOI] [PubMed] [Google Scholar]
- Ye X. Zheng C. Chen J. Gao Y. Murray C. B. Nano Lett. 2013;13:765–771. doi: 10.1021/nl304478h. [DOI] [PubMed] [Google Scholar]
- Park H. Yang S. Kang J. Y. Park M.-H. ACS Med. Chem. Lett. 2016;7:1087–1091. doi: 10.1021/acsmedchemlett.6b00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis R. A., Hawley's condensed chemical dictionary, John Wiley & Sons, 2016 [Google Scholar]
- Jeon S. Yoo C. Y. Park S. N. Colloids Surf., B. 2015;129:7–14. doi: 10.1016/j.colsurfb.2015.03.018. [DOI] [PubMed] [Google Scholar]
- Kumar S. Gokhale R. Burgess D. J. Int. J. Pharm. 2014;464:234–242. doi: 10.1016/j.ijpharm.2013.12.039. [DOI] [PubMed] [Google Scholar]
- Heneczkowski M. Kopacz M. Nowak D. Kuźniar A. Acta Pol. Pharm. 2001;58:415. [PubMed] [Google Scholar]
- Jones D. S. Tian Y. Abu-Diak O. Andrews G. P. Adv. Drug Delivery Rev. 2012;64:440–448. doi: 10.1016/j.addr.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Demir M. M. Yilgor I. Yilgor E. Erman B. Polymer. 2002;43:3303–3309. doi: 10.1016/S0032-3861(02)00136-2. [DOI] [Google Scholar]
- McKeon-Fischer K. D. Freeman J. W. J. Tissue Eng. Regener. Med. 2011;5:560–568. doi: 10.1002/term.348. [DOI] [PubMed] [Google Scholar]
- Shi W. Lu W. Jiang L. J. Colloid Interface Sci. 2009;340:291–297. doi: 10.1016/j.jcis.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Maity S. Wu W.-C. Tracy J. B. Clarke L. I. Bochinski J. R. Nanoscale. 2017;9:11605–11618. doi: 10.1039/C7NR04613H. [DOI] [PubMed] [Google Scholar]
- Fong H. Reneker D. H. J. Polym. Sci., Part B: Polym. Phys. 1999;37:3488–3493. doi: 10.1002/(SICI)1099-0488(19991215)37:24<3488::AID-POLB9>3.0.CO;2-M. [DOI] [Google Scholar]
- Shamsabadi A. S. Ranjbar M. Tavanai H. Farnood A. Mater. Res. Express. 2019;6:055051. doi: 10.1088/2053-1591/ab0709. [DOI] [Google Scholar]
- Kim T. G. Park T. G. Tissue Eng. 2006;12:221–233. doi: 10.1089/ten.2006.12.221. [DOI] [PubMed] [Google Scholar]
- Pereira R. F. Bártolo P. J. Adv. Wound Care. 2016;5:208–229. doi: 10.1089/wound.2013.0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine R. Kalarikkal N. Thomas S. Tissue Eng. Regener. Med. 2015;12:12–19. doi: 10.1007/s13770-014-0032-9. [DOI] [Google Scholar]
- Abd El-Aziz A. M. El-Maghraby A. Ewald A. Kandil S. H. Molecules. 2021;26:1552. doi: 10.3390/molecules26061552. [DOI] [PMC free article] [PubMed] [Google Scholar]