Abstract
Enzymes within the de novo purine biosynthetic pathway spatially organize into dynamic intracellular assemblies called purinosomes. The formation of purinosomes has been correlated with growth conditions resulting in high purine demand, and therefore, the cellular advantage of complexation has been hypothesized to enhance metabolite flux through the pathway. However, the properties of this cellular structure are unclear. Here, we define the purinosome in a transient expression system as a biomolecular condensate using fluorescence microscopy. We show that purinosomes, as denoted by formylglycinamidine ribonucleotide synthase granules in purine-depleted HeLa cells, are spherical and appear to coalesce when two come into contact, all liquid-like characteristics that are consistent with previously reported condensates. We further explored the biophysical and biochemical means that drive the liquid–liquid phase separation of these structures. We found that the process of enzyme condensation into purinosomes is likely driven by the oligomeric state of the pathway enzymes and not a result of intrinsic disorder, the presence of low-complexity domains, the assistance of RNA scaffolds, or changes in intracellular pH. Finally, we demonstrate that the heat shock protein 90 KDa helps to regulate the physical properties of the condensate and maintain their liquid-like state inside HeLa cells. We show that disruption of heat shock protein 90 KDa activity induced the transformation of formylglycinamidine ribonucleotide synthase clusters into more irregularly shaped condensates, suggesting that its chaperone activity is essential for purinosomes to retain their liquid-like properties. This refined view of the purinosome offers new insight into how metabolic enzymes spatially organize into dynamic condensates within human cells.
Keywords: metabolism, purine biosynthesis, protein complex, liquid condensate, liquid–liquid phase separation
Abbreviations: ADSL, adenylosuccinate lyase; ATIC, aminoimidazole carboxamide ribonucleotide transformylase; DNPB, de novo purine biosynthetic; EYFP, enhanced YFP; FGAMS, formylglycinamidine ribonucleotide synthase; GART, glycinamide ribonucleotide transformylase; HeTOFLI, HeLa Tet-Off cell line; HSP, heat shock protein; IDR, intrinsically disordered region; LLPS, liquid–liquid phase separation; PAICS, phosphoribosylaminoimidazole succinocarboxamide synthetase; PPAT, phosphoribosyl pyrophosphate amidotransferase
Purine production is met through the coordinated actions of two complementary pathways in order to maintain cellular homeostasis and promote cell growth. The purine salvage pathway recycles degraded bases back into their corresponding nucleotide and is commonly employed during normal physiological growth conditions (1). However, when demand is elevated, the de novo purine biosynthetic (DNPB) pathway becomes activated and converts phosphoribosyl pyrophosphate into inosine 5′-monophosphate (2, 3, 4). This process consists of ten concerted reactions catalyzed by six enzymes in humans.
Using fluorescence microscopy, the state of the DNPB pathway has largely been reflected in the behavior of formylglycinamidine ribonucleotide synthase (FGAMS). FGAMS transitions from a diffuse state into multienzyme assemblies called purinosomes upon changes in growth conditions that increase purine demand (5, 6). The purinosome is hypothesized to exist as a metabolon, a complex of sequential metabolic enzymes to enhance metabolite flux through a given pathway (7), and has been further substantiated by identifying intracellular hotspots of purine biosynthesis through mass spectral imaging (8). Components of the purinosome exist beyond the six enzymes in the biosynthetic pathway and include downstream enzymes and regulatory proteins such as molecular chaperones and kinases (6, 9, 10). The collective actions of these regulatory proteins are likely what governs the formation and, in part, the function of the complex. The function of the complex is also reliant on the availability of substrates and cofactors derived from mitochondrial processes (11), further supporting our conclusions that purinosomes and mitochondria have a symbiotic relationship (12, 13).
The compartmentalization of purine biosynthetic enzymes into these intracellular organizations has hinted that these bodies might be an example in the growing list of biomolecular condensates (e.g., nucleoli, P-bodies, stress granules, RNA transport granules). Biomolecular condensates are highly heterogeneous complexes that undergo a liquid–liquid phase separation (LLPS) driven by weak and transient interactions among its constitutive components, commonly referred to as multivalency (14, 15, 16). The multivalency can be achieved as a product of interactions between folded protein domains, such as highly ordered oligomers, and/or self-associating intrinsically disordered regions (IDRs). With the advancements in fluorescence microscopy, we have greatly advanced our understanding of the biophysical properties of these membrane-less organelles. The process of LLPS has been observed within DNA repair (17, 18, 19), transmembrane signaling (20, 21), cytoskeleton formation (22, 23), ribonucleoprotein transport (24, 25), and more recently, metabolism (26). Changes in a cell’s microenvironment and intracellular needs often modify the attractive forces within condensates and impact their formation and stability. Therefore, identifying the mechanism that triggers and regulates the dynamic process might provide insights into their function and the overall cellular advantage of forming condensates over more traditional macromolecular complexes.
In this report, we build on our previous biophysical and biochemical characterizations of the purinosome in purine-depleted HeLa cells to demonstrate its ability to behave similar to other reported biomolecular condensates. Purine depletion of HeLa cells results in activation of the DNPB and has been correlated with purinosome formation (8, 10). We show that purinosomes exist as uniformly spherical intracellular assemblies and readily fuse with one another when they come in close contact. The mechanism that drives the LLPS is likely because of the oligomerization of pathway enzymes into higher ordered structures and not a consequence of intrinsic disorder. Last, we propose a mechanism by which regulation of the condensate’s liquid-like properties is achieved by the assistance of molecular chaperones. Together, these results help us further understand the nature of this assembly and provide further clues into how the purinosome is organized and regulated in cells.
Results
The purinosome demonstrates liquid-like properties in cells
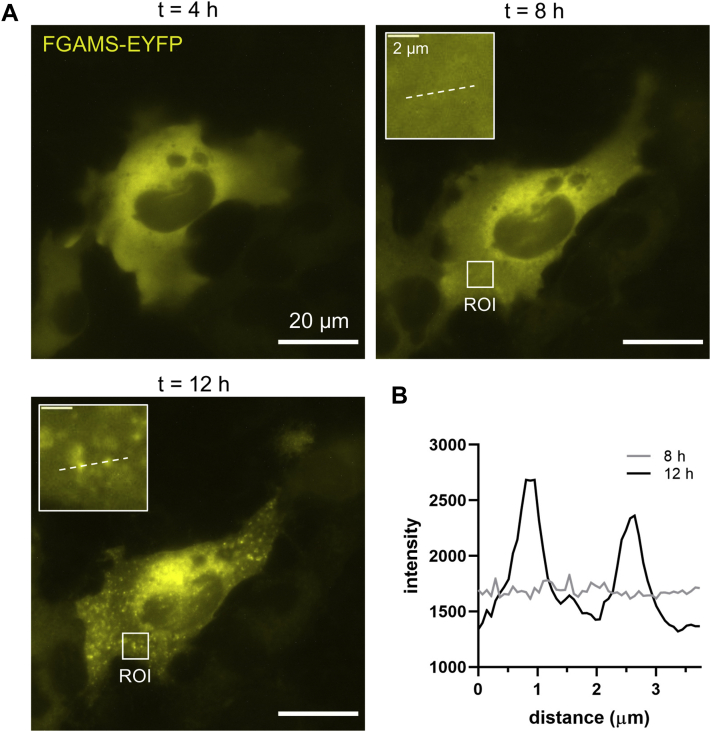
To probe whether the purinosome exhibits liquid-like properties, we first sought out whether an enzyme concentration–dependent threshold exists that might promote an LLPS. For this, we used a modified HeLa Tet-Off cell line (HeTOFLI) that stably expresses LacI-NLS to allow for the IPTG induction of protein expression (27). These cells were transfected with an inducible construct encoding FGAMS-enhanced YFP (EYFP), and the induced expression was monitored over time by fluorescence microscopy (Figs. 1 and S1). Previously, we have used FGAMS as a marker to study the intracellular dynamics of the purinosome, and we infer that most assemblies of FGAMS under purine-depleted growth conditions likely also contain other enzymes in the DNPB pathway (5, 12, 13). At early time points, purine-depleted HeTOFLI cells expressing FGAMS-EYFP displayed a diffuse phenotype that transitioned into discrete granules as time progressed. Figure 1 shows the condensation of FGAMS-EYFP in a representative cell at 12 h post-IPTG induction. Intensity values across a defined region within the same cell before and after triggering condensation verified that the FGAMS-EYFP within the cell was being concentrated into these assemblies (Fig. 1B) further implying that the purinosome is capable of undergoing an LLPS.
Figure 1.
FGAMS condenses with increased protein expression.A, the expression of FGAMS-EYFP was monitored over time post-IPTG induction in a purine-depleted HeTOFLI cell. FGAMS-EYFP transitioned from a diffuse (8 h) to a granular (12 h) phenotype as exemplified in the region of interest (ROI, inset). B, granular structures showed enriched levels of FGAMS-EYFP as noted by profiling their intensities across the ROI (dashed line) from 8 h (gray) to 12 h (black).
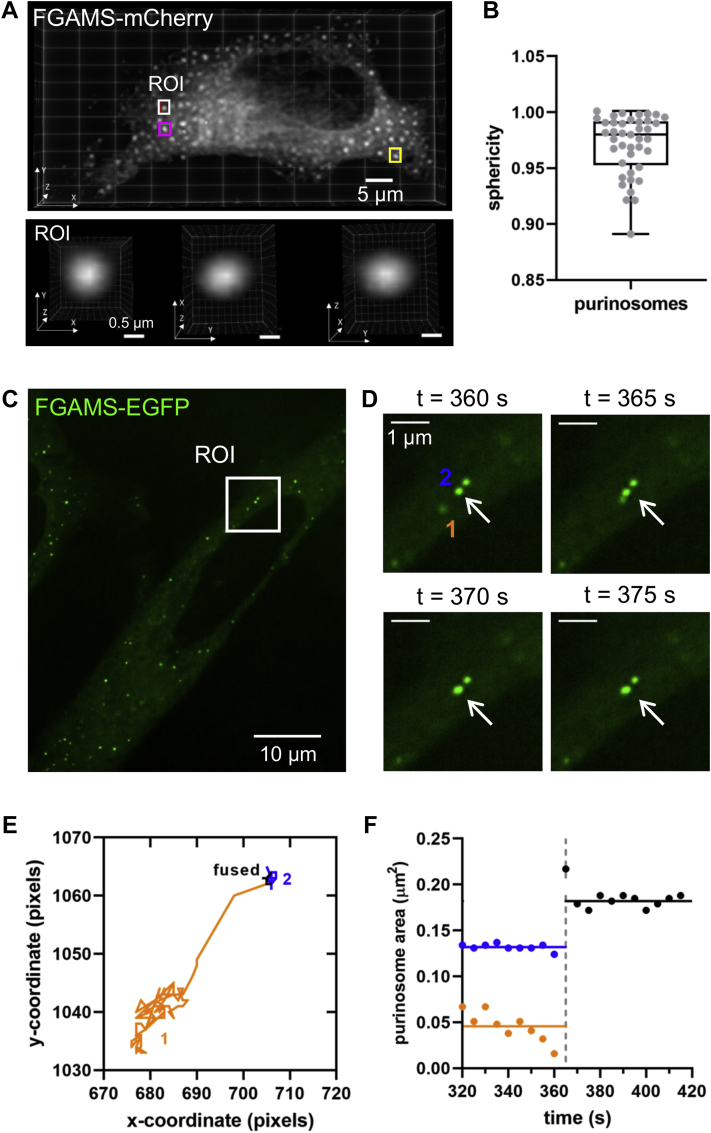
Three properties are widely shared amongst commonly reported liquid-like condensates: the free exchange of molecules in and out of the condensate, the spherical nature of the protein assembly, and the ability of condensates to fuse when in close contact (15). We asked whether the purinosome exhibits these characteristics. Previously, enzymes within the DNPB pathway have been shown to freely exchange between the bulk solvent and the purinosome by fluorescence recovery after photobleaching (5, 28). The calculated diffusion coefficients for the various enzymes as they assemble into purinosomes suggest that the complex is formed through several multivalent interactions in a step-wise manner (28). We continued our investigation by exploring the spherical nature of purinosomes in purine-depleted HeLa cells by lattice light sheet microscopy (29). A characterization of 41 purinosomes demonstrated that those segmented FGAMS-mCherry assemblies were spherical with an average sphericity of 0.97 ± 0.03 (Figs. 2, A and B and S2). Last, we monitored the movement of purinosomes over a 5 min time course (one image every 5 s) using instantaneous structured illumination high-resolution fluorescence microscopy. In several instances, purinosomes, as denoted by FGAMS-enhanced GFP puncta, appear to merge with one another to form larger punctate structures. Figure 2 shows one such event in which purinosome 1 fuses with purinosome 2 at 365 s (Figs. 2, C–E and S3A, Movie S1). The area of the coalesced purinosome was roughly the sum of the two purinosomes combined (Fig. 2F). Together, these data provide overwhelming support that the purinosome is an example of a liquid condensate, and the process of enzyme condensation is likely a regulatory mechanism leveraged in purine metabolism.
Figure 2.
Purinosomes display liquid-like properties.A, a purinosome-positive HeLa cell was imaged by LLSM, and the sphericity of purinosomes was observed (white ROI) and (B) calculated across 41 purinosomes. Additional ROIs (magenta and yellow) are presented in Fig. S2A. C, time-lapse imaging of a purinosome-positive HeLa cell showing (D) the coalescing of a purinosome (1) with another (2, arrow) between 360 and 375 s. E, trajectory of both purinosomes over the entire time-lapse experiment (500 s). F, the area of the individual purinosome 1 (0.045 μm2, orange line) and 2 (0.132 μm2, blue line) before (320–360 s) and after merging (370–415 s). Area of the coalesced purinosome was 0.182 μm2 (black line). Solid lines represent average purinosome area across the time points observed. LLSM, lattice light sheet microscopy; ROI, region of interest.
The oligomeric state of the purinosome contributes to its condensation
The multivalency observed in biomolecular condensates can be induced through a variety of mechanisms including protein oligomerization and self-assembly via IDRs or nucleic acid scaffolds. Physicochemical features, as identified by amino acid sequence, within proteins shown to phase transition have given insight into other proteins within the human proteome that might also drive biomolecular condensate formation by self-assembly. A classical characteristic that differentiates a protein that phase transitions from one that does not is the presence of long IDRs and/or low-complexity domains (15, 30, 31, 32). Therefore, bioinformatic analyses were conducted to predict the likelihood that any of the enzymes in the DNPB pathway have such propensity to drive the observed LLPS. Using the IUPred2A predictor (33, 34) and the Predictor of Natural Disordered Regions (PONDR-VSL2) (35) algorithms, all enzymes did not display long stretches (>30 amino acids) of intrinsic disorder (Fig. S4). In addition, we also queried whether any of these enzymes had stretches of low complexity using the SEG algorithm (36). Low-complexity domains were identified in all enzymes except for phosphoribosylaminoimidazole succinocarboxamide synthetase (PAICS) and aminoimidazole carboxamide ribonucleotide transformylase (ATIC) (Fig. S4); however, the length in any of these regions is relatively very short compared with reported phase-separated proteins (37, 38).
We also investigated the tendency for the enzymes in the DNPB pathway to aggregate using the TANGO (39) and Waltz algorithms (40). The TANGO algorithm accounts for the general physicochemical determinants of protein β-aggregation, whereas Waltz algorithm identifies sequences that promote amyloid structures over amorphous aggregates. Results from the TANGO algorithm predicted that all the pathway enzymes have some degree of aggregation as noted by more than five consecutive amino acids with an aggregation tendency greater than 5% (Fig. S5). The purinosome core proteins (phosphoribosyl pyrophosphate amidotransferase [PPAT], glycinamide ribonucleotide transformylase [GART], and FGAMS) were more aggregation prone, and several of these stretches also predicted to contribute to the formation of amyloid-like assemblies by the Waltz algorithm (Fig. S5). These results are consistent with the overall tendency of globular proteins to have a high degree of β-aggregation nucleating regions when compared with intrinsically disordered proteins (41); however, this does necessarily suggest that these proteins form aggregates. These identified stretches likely are critical for the overall proper folding, stability, and activity of the protein.
Extended nutrient starvation of human cells has been known to induce a stress response to inhibit protein translation and trigger the phase separation of proteins into stress granules (42). We have long prescribed that this is not the case with the purinosome because of its lack of association with the stress granule–nucleating protein G3BP1 (Ras-GAP SH3 binding protein 1) and to differences in their defining characteristics with the purinosome (6, 9, 11, 43). Recent work has suggested that a stress granule might not be comprised of all the traditional protein markers and that the composition is dependent on the type and duration of the stress (44). Therefore, we asked whether poly(A)+ mRNA is present within the purinosome suggesting that these condensates are part of the RNA granule family void of G3BP1. Using fluorescence in situ hybridization, the colocalization of poly(A)+ mRNAs with purinosomes was assessed. Consistent with this body being distinct from stress granules and P-bodies, no colocalization was observed (Fig. S6A). We also investigated whether the purinosome might be a result of nutrient-deprived stalled translational initiation complexes, a known phenomenon giving rise to early stage stress granule formation (42). Purine-depleted HeLa cells were grown in the presence or the absence of purines, and the repression of translation analyzed by the induction of Ser51 phosphorylation on eurkaryotic translation initiation factor 2α. Using a 1 h arsenite treatment as a positive control, we show that purine-depleted HeLa cells do not have repressed translation and are consistent with HeLa cells grown in the presence of purines (Fig. S6B). We also investigated whether overexpression of FGAMS by transient transfection might repress translation. Similar to the untransfected HeLa cells, no increase in the phosphorylation of eurkaryotic translation initiation factor 2α was observed (Fig. S6B). Together, all these data would suggest that despite the chronic starvation of HeLa cells for purines, the purinosome is not an example of a stress granule and that condensate formation is not driven by the presence of an RNA scaffold.
Nutrient starvation can also lead to changes in intracellular pH previously shown to promote an LLPS in eukaryotes (45, 46, 47). To assess whether the pH is different in HeLa cells under purinosome-forming (purine-depleted) growth conditions, we used the pH-sensitive fluorescent dye, cSNARF-1 (48). HeLa cells costained with cSNARF-1 and Hoechst 33342 were imaged by confocal microscopy. Background-subtracted ratios of non-nuclear cSNARF-1 emissions between 561 to 598 nm and 620 to 753 nm were converted to pH values after establishment of a calibration curve (Fig. S7, A and B) using the nigericin method (49, 50). Intracellular pH values for normal and purine-depleted HeLa cells were not significantly different at 7.61 ± 0.10 and 7.66 ± 0.07, respectively (Fig. S7, C and D). Therefore, we conclude that pH is likely not a factor in the formation of the purinosome condensate.
Our investigations to date do not point to a self-assembly mechanism that drives purinosome formation. Therefore, we hypothesize that the multivalency could be achieved through the oligomerization of the given pathway enzymes. Within the DNPB pathway, PAICS is an octamer, PPAT and adenylosuccinate lyase (ADSL) exist between dimeric and tetrameric states, and GART and ATIC are functional dimers (6). A recent study showing the purinosome in neurons suggested that PAICS might act as a scaffold for the purinosome (51). Other studies have suggested that perturbing the oligomeric structure and/or composition of the purinosome impacts its formation. First, purine-depleted HeLa cells deficient in the expression of one pathway enzyme showed a reduction (FGAMS and PAICS) or a complete loss in purinosome content compared with wildtype cells (52). Second, disruption of the active ATIC homodimer with a cyclic peptide-derived inhibitor, compound 14, resulted in a loss of hypoxia-induced purinosome formation (11). Therefore, the increased weak and transient interactions between enzymes in the pathway could drive a thermodynamically favorable process of an LLPS and promote a unique composition. Given these findings, it is plausible that higher order macromolecular structures, such as metabolons, can emerge solely between pathway enzymes that induce the phase transition.
Molecular chaperones help regulate the purinosomes’ biophysical properties
The process of condensation proceeds along a continuum ranging from LLPS driven by weak, transient, and reversible interactions to stronger and irreversible interactions such as those observed in insoluble aggregates and polymeric structures. The regulation of condensate maturation has been proposed to be controlled through the inclusion of modifying enzymes (53), such as kinases (54, 55) and heat shock proteins (HSPs) (56, 57, 58, 59).
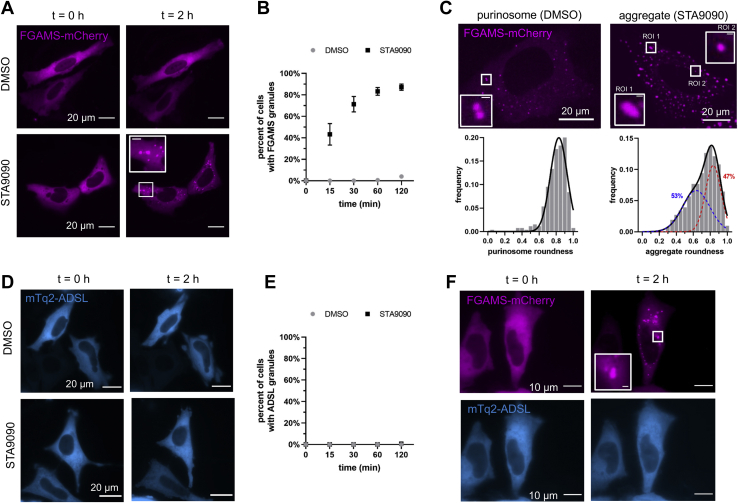
Previously, we have demonstrated a role for molecular chaperones in the formation of the purinosome (9, 60). Molecular chaperones, HSP 90 KDa (HSP90) and HSP70, colocalized with purinosomes in purine-depleted HeLa cells (9), and the biochemical interactions between HSP90 and pathway enzymes, PPAT and FGAMS, were no longer detected upon the inhibition of HSP90 activity (60). Given this, we explored whether HSP90 might help preserve the liquid-like state of purinosomes by acting as a modifying enzyme for the condensate. We first asked whether HSP90 inhibition can artificially induce the formation of intracellular FGAMS granules in the absence of purinosome-forming growth conditions. For these studies, we used STA9090 (ganetesib), an inhibitor of HSP90 activity, at a concentration not shown to significantly perturb HeLa cell viability (Fig. S8). HeLa cells transiently expressing FGAMS-mCherry were treated with STA9090, and the number of cells displaying intracellular FGAMS granules was monitored over a 2 h period. After 2 h of exposure, a majority of all transfected cells showed the emergence of FGAMS granules compared with transfected cells treated with dimethyl sulfoxide (Fig. 3, A and B). The emergence of granular FGAMS structures was also observed in purine-depleted HeLa cells suggesting that the impact that HSP90 inhibition has on FGAMS is independent of purine levels (Fig. S9). Using instantaneous structured illumination high-resolution fluorescence microscopy, we were able to show that roughly half of the FGAMS clusters in purine-depleted HeLa cells became less circular after a 2 h treatment with STA9090. This emergent subpopulation of FGAMS containing assemblies was distinct from the highly circular purinosomes (Fig. 3C).
Figure 3.
Inhibition of HSP90 results in FGAMS aggregation.A, HSP90 inhibition by STA9090 resulted in the aggregation of FGAMS-mCherry over 2 h. The scale bar for inset represents 1 μm. B, the percentage of cells showing FGAMS granules as a function of time. Data represent mean ± standard deviation, N = 3. C, roughly half of these FGAMS-mCherry aggregates displayed a more irregular morphology (inset, ROI 1) when compared with the purinosome morphology in purine-depleted HeLa cells (DMSO, inset ROI). The scale bar for insets represents 1 μm. Distributions were generated from roundness calculations of 262 purinosomes and 1146 STA9090-induced granules. D, HeLa cells transiently expressing mTq2-ADSL did not aggregate upon HSP90 inhibition. E, the percentage of cells showing ADSL granules as a function of time. Data represent mean ± standard deviation, N = 3. F, purine-depleted HeLa cells coexpressing FGAMS-mCherry and mTq2-ADSL did not cocluster upon STA9090 treatment. The scale bar for inset represents 1 μm. DMSO, dimethyl sulfoxide; FGAMS, formylglycinamidine ribonucleotide synthase; HSP90, heat shock protein 90 kDa; mTq2-ADSL, mTurquoise2-ADSL; ROI, region of interest.
We also investigated whether ADSL, another pathway enzyme not previously shown to be dependent on HSP90 (60), also formed granules upon HSP90 inhibition. During the 2 h study, HeLa cells transiently expressing mTurquoise2-ADSL did not have a tendency to form granular structures upon STA9090 treatment (Figs. 3, D and E and S9). We next asked whether the STA9090-induced FGAMS granules would result in the formation of purinosome-like structures as evident in the coclustering of ADSL granules. In this study, we transiently overexpressed FGAMS-mCherry with mTurquoise2-ADSL in purine-depleted HeLa cells and investigated whether upon HSP90 inhibition, the association of FGAMS and ADSL seen in purinosomes was ablated. Consistent with previous observations, HSP90 inhibition resulted in the formation of FGAMS clusters; however, the diffuse ADSL phenotype persisted (Fig. 3F). These results demonstrate that inhibiting HSP90 activity results in the formation of FGAMS-only assemblies with a morphology distinct from that of purinosome condensates and suggests that HSP90 likely assists in regulating its liquid-like properties.
Discussion
Emerging evidence across many metabolic processes suggests that enzymes within a given pathway condense and undergo a phase transition to maintain cellular homeostasis (26). With this study, we present further evidence supporting the notion that the purinosome, as denoted by discrete FGAMS assemblies under purine-depleted growth conditions, behaves like a classically defined biomolecular condensate with liquid-like properties in a transient expression system.
We showed that the condensation of FGAMS was concentration dependent; however, we do not fully understand whether there are size restrictions to retain the liquid-like properties of the purinosome. We can hypothesize that the size of a functional purinosome is limited based on the expression of pathway enzymes, their stoichiometric makeup, and their subcellular localization. Calculations representing an equimolar distribution of pathway enzymes suggest that purinosome sizes likely will not exceed 300 nm in diameter (61). In addition, the subcellular localization might restrict its overall size. In this study, we observed the directed motion of purinosomes (consistent with their movement along microtubules) until they merge with a stationary purinosome (Fig. S3, B–F). Based on our previous characterizations of purinosome motion and subcellular localization (12), we prescribe those stationary purinosomes are at the mitochondria–microtubule interface. This interface might limit the size a purinosome can be in order to retain its function. Investigations into the size requirements and composition of the purinosome are ongoing.
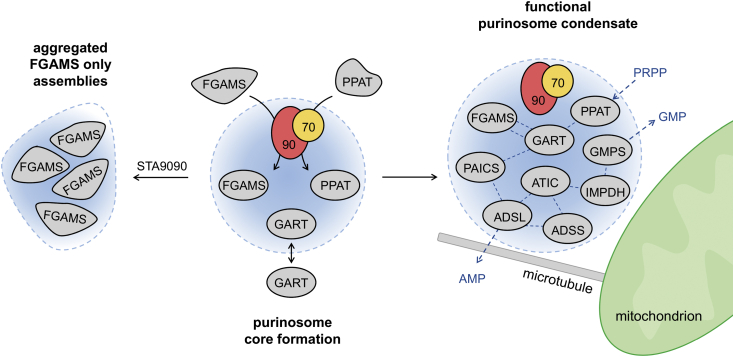
We have only started to understand the biochemical mechanisms that govern purinosome formation and its stability. This study highlighted HSP90 as a way to sustain the properties of the condensate. Inhibition of HSP90 activity altered the properties of the purinosome driven likely by an increase in misfolded FGAMS protein within the condensate. This accumulation of the misfolded protein created complexes that diminished the available contacts needed with other enzymes in the DNPB pathway. Therefore, the inclusion of this regulatory protein in the condensate could limit the number of possible transient interactions and influence the overall stoichiometry of the purinosome simply by modulating the folded state of FGAMS. Recently, a chemical affinity strategy using immobilized PU-H71, an inhibitor of HSP90, was shown to coisolate a majority of the enzymes within the DNPB pathway with HSP90 (62). This study is consistent with our findings and suggests that HSP90 might serve as a core scaffold (Fig. 4).
Figure 4.
Hypothesized model for the purinosome as a liquid condensate regulated by molecular chaperones. Inactive forms of FGAMS and PPAT are recruited to chaperoning complexes, where they are folded into their active state to promote transient interactions with GART to form the purinosome core. These associations promote the recruitment of the other enzymes into purinosome condensates localized at microtubule–mitochondrion interfaces. Complete purinosome condensates are hypothesized to be responsible for the channeled conversion of PRPP into GMP or AMP (blue dashed line). Disruption of HSP90 activity through STA9090 was shown to perturb the properties of purinosome condensates and result in FGAMS-only assemblies. FGAMS, formylglycinamidine ribonucleotide synthase; GART, glycinamide ribonucleotide transformylase; HSP90, heat shock protein 90 kDa; PPAT, phosphoribosyl pyrophosphate amidotransferase; PRPP, phosphoribosyl pyrophosphate.
This study does not rule out the possibility that the purinosome is a multiphasic condensate as shown in ATPase-modulated stress granules (63). We can conceive that chaperoning is imperative and isolated to a substructure within the purinosome (Fig. 4). Based on the diffusion coefficients and protein proximity studies, the first three enzymes in the pathway (PPAT, GART, and FGAMS) have been assigned to act as the core of the purinosome (28, 64). Interestingly, two of these three proteins are also under regulation by HSP90 (60). The proposed stepwise assembly of the purinosome supports this hypothesis as once the HSP90 clients adopt a conformation for purinosome formation, they are able to move to a second compartment where the other enzymes are recruited to adopt a unique stoichiometry and facilitate substrate channeling.
This study begs to ask if the purinosome exists as a condensate outside a transient overexpression system. Recent platforms studying the purinosome with endogenous proteins indicate that the number of functional purinosomes per purine-depleted HeLa cell is approximately 10-fold lower than those inferred from overexpressed systems (8, 11). This discrepancy points to the overexpression of pathway enzymes promoting subcellular organizations void of specific components, and therefore, not contributing fully to the observed substrate channeling and enhanced purine production. Yet, at endogenous levels, not currently amendable to live cell fluorescence imaging, the enzymes still appear to cluster into purinosome metabolons suggesting that a condensate is still likely present (8, 11, 52). The focus for subsequent studies is to characterize how the activity of the purinosome condensate is regulated on the endogenous level, possibly through its post-translational modifications, after various pathway enzymes assembly within the condensate.
Experimental procedures
Full experimental procedures including cell culture, fluorescence microscopy, data analysis (Figs. S1–S9) are provided in the supporting information.
Data availability
The data supporting the findings of this study are available within the article and supporting information files. Additional data files underpinning this study are available upon request.
Supporting information
This article contains supporting information (5, 12, 27, 29, 33, 34, 35, 36, 39, 40, 43, 49, 50, 60, 65, 66, 67, 68).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We acknowledge helpful discussions with Xin Zhang, Songon An, and members of the Benkovic Laboratory. We also thank Richard Morimoto for the HeTOFLI cell line and associated plasmids, Ling-Nan Zou for the mTurquoise2-ADSL expression plasmid, and Missy Hazen and the Penn State Microscopy Facility—University Park, PA for technical expertise.
Author contributions
A. M. P., M. K., and S. J. B. methodology; A. M. P., J. P. B., E. L. K., and M. K. formal analysis; A. M. P., J. P. B., C. Y. C., and E. L. K. investigation; A. M. P., J. P. B., C. Y. C., E. L. K., M. K., and S. J. B. writing–original draft; A. M. P., J. P. B., C. Y. C., E. L. K., M. K., and S. J. B. writing–review & editing.
Funding and additional information
Financial support for this study was provided by the Huck Institutes of the Life Sciences at Penn State (Huck Innovative and Transformative Seed Fund grant to A. M. P.) and The National Institutes of Health (grant nos.: R01GM024129 [to S. J. B.], R01GM134086 [to M. K.], T32GM066706 [to E. L. K.], and R25GM55036 [to E. L. K.]). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Craig Cameron
Footnotes
Present address for Jack P. Boylan: Molecular, Cellular, and Integrative Biosciences Graduate Program, Huck Institutes of the Life Sciences, The Pennsylvania State University, University Park, Pennsylvania 16802, USA.
Present address for Chung Yu Chan: National Institute of Neurological Disorders and Stroke, The National Institutes of Health, Bethesda, Maryland 20892, USA.
Supplorting information
References
- 1.Murray A.W. The biological significance of purine salvage. Annu. Rev. Biochem. 1971;40:811–826. doi: 10.1146/annurev.bi.40.070171.004115. [DOI] [PubMed] [Google Scholar]
- 2.Mayer D., Natsumeda Y., Ikegami T., Faderan M., Lui M., Emrani J., Reardon M., Olah E., Weber G. Expression of key enzymes of purine and pyrimidine metabolism in a hepatocyte-derived cell line at different phases of the growth cycle. J. Cancer Res. Clin. Oncol. 1990;116:251–258. doi: 10.1007/BF01612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Natsumeda Y., Prajda N., Donohue J.P., Glover J.L., Weber G. Enzymic capacities of purine de novo and salvage pathways for nucleotide synthesis in normal and neoplastic tissues. Cancer Res. 1984;44:2475–2479. [PubMed] [Google Scholar]
- 4.Yamaoka T., Kondo M., Honda S., Iwahana H., Moritani M., Ii S., Yoshimoto K., Itakura M. Amidophosphoribosyltransferase limits the rate of cell growth-linked de novo purine biosynthesis in the presence of constant capacity of salvage purine biosynthesis. J. Biol. Chem. 1997;272:17719–17725. doi: 10.1074/jbc.272.28.17719. [DOI] [PubMed] [Google Scholar]
- 5.An S., Kumar R., Sheets E.D., Benkovic S.J. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- 6.Pedley A.M., Benkovic S.J. A new view into the regulation of purine metabolism: The purinosome. Trends Biochem. Sci. 2017;42:141–154. doi: 10.1016/j.tibs.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srere P.A. The metabolon. Trends Biochem. Sci. 1985;10:109–110. [Google Scholar]
- 8.Pareek V., Tian H., Winograd N., Benkovic S.J. Metabolomics and mass spectrometry imaging reveal channeled de novo purine synthesis in cells. Science. 2020;368:283–290. doi: 10.1126/science.aaz6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.French J.B., Zhao H., An S., Niessen S., Deng Y., Cravatt B.F., Benkovic S.J. Hsp70/Hsp90 chaperone machinery is involved in the assembly of the purinosome. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2528–2533. doi: 10.1073/pnas.1300173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao H., Chiaro C.R., Zhang L., Smith P.B., Chan C.Y., Pedley A.M., Pugh R.J., French J.B., Patterson A.D., Benkovic S.J. Quantitative analysis of purine nucleotides indicates that purinosomes increase de novo purine biosynthesis. J. Biol. Chem. 2015;290:6705–6713. doi: 10.1074/jbc.M114.628701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doigneaux C., Pedley A.M., Mistry I.N., Papayova M., Benkovic S.J., Tavassoli A. Hypoxia drives the assembly of the multienzyme purinosome complex. J. Biol. Chem. 2020;295:9551–9566. doi: 10.1074/jbc.RA119.012175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan C.Y., Pedley A.M., Kim D., Xia C., Zhuang X., Benkovic S.J. Microtubule-directed transport of purine metabolons drives their cytosolic transit to mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2018;115:13009–13014. doi: 10.1073/pnas.1814042115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.French J.B., Jones S.A., Deng H., Pedley A.M., Kim D., Chan C.Y., Hu H., Pugh R.J., Zhao H., Zhang Y., Huang T.J., Fang Y., Zhuang X., Benkovic S.J. Spatial colocalization and functional link of purinosomes with mitochondria. Science. 2016;351:733–737. doi: 10.1126/science.aac6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberti S., Gladfelter A., Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bracha D., Walls M.T., Brangwynne C.P. Probing and engineering liquid-phase organelles. Nat. Biotechnol. 2019;37:1435–1445. doi: 10.1038/s41587-019-0341-6. [DOI] [PubMed] [Google Scholar]
- 17.Altmeyer M., Neelsen K.J., Teloni F., Pozdnyakova I., Pellegrino S., Grofte M., Rask M.D., Streicher W., Jungmichel S., Nielsen M.L., Lukas J. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose) Nat. Commun. 2015;6:8088. doi: 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilic S., Lezaja A., Gatti M., Bianco E., Michelena J., Imhof R., Altmeyer M. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 2019;38 doi: 10.15252/embj.2018101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oshidari R., Huang R., Medghalchi M., Tse E.Y.W., Ashgriz N., Lee H.O., Wyatt H., Mekhail K. DNA repair by Rad52 liquid droplets. Nat. Commun. 2020;11:695. doi: 10.1038/s41467-020-14546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banjade S., Rosen M.K. Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife. 2014;3 doi: 10.7554/eLife.04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Case L.B., Ditlev J.A., Rosen M.K. Regulation of transmembrane signaling by phase separation. Annu. Rev. Biophys. 2019;48:465–494. doi: 10.1146/annurev-biophys-052118-115534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang H., Wang S., Huang Y., He X., Cui H., Zhu X., Zheng Y. Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell. 2015;163:108–122. doi: 10.1016/j.cell.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weirich K.L., Banerjee S., Dasbiswas K., Witten T.A., Vaikuntanathan S., Gardel M.L. Liquid behavior of cross-linked actin bundles. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2131–2136. doi: 10.1073/pnas.1616133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alami N.H., Smith R.B., Carrasco M.A., Williams L.A., Winborn C.S., Han S.S.W., Kiskinis E., Winborn B., Freibaum B.D., Kanagaraj A., Clare A.J., Badders N.M., Bilican B., Chaum E., Chandran S., et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao Y.C., Fernandopulle M.S., Wang G., Choi H., Hao L., Drerup C.M., Patel R., Qamar S., Nixon-Abell J., Shen Y., Meadows W., Vendruscolo M., Knowles T.P.J., Nelson M., Czekalska M.A., et al. RNA granules Hitchhike on lysosomes for long-distance transport, using annexin A11 as a molecular tether. Cell. 2019;179:147–164.e20. doi: 10.1016/j.cell.2019.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prouteau M., Loewith R. Regulation of cellular metabolism through phase separation of enzymes. Biomolecules. 2018;8:160. doi: 10.3390/biom8040160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto G., Kim S., Morimoto R.I. Huntingtin and mutant SOD1 form aggregate structures with distinct molecular properties in human cells. J. Biol. Chem. 2006;281:4477–4485. doi: 10.1074/jbc.M509201200. [DOI] [PubMed] [Google Scholar]
- 28.Kyoung M., Russell S.J., Kohnhorst C.L., Esemoto N.N., An S. Dynamic architecture of the purinosome involved in human de novo purine biosynthesis. Biochemistry. 2015;54:870–880. doi: 10.1021/bi501480d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.An S., Jeon M., Kennedy E.L., Kyoung M. Phase-separated condensates of metabolic complexes in living cells: Purinosome and glucosome. Methods Enzymol. 2019;628:1–17. doi: 10.1016/bs.mie.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darling A.L., Zaslavsky B.Y., Uversky V.N. Intrinsic disorder-based emergence in cellular biology: Physiological and pathological liquid-liquid phase transitions in cells. Polymers (Basel) 2019;11:990. doi: 10.3390/polym11060990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin E.W., Mittag T. Relationship of sequence and phase separation in protein low-complexity regions. Biochemistry. 2018;57:2478–2487. doi: 10.1021/acs.biochem.8b00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uversky V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017;44:18–30. doi: 10.1016/j.sbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 33.Erdos G., Dosztanyi Z. Analyzing protein disorder with IUPred2A. Curr. Protoc. Bioinformatics. 2020;70 doi: 10.1002/cpbi.99. [DOI] [PubMed] [Google Scholar]
- 34.Meszaros B., Erdos G., Dosztanyi Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018;46:W329–W337. doi: 10.1093/nar/gky384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng K., Radivojac P., Vucetic S., Dunker A.K., Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wootton J.C. Non-globular domains in protein sequences: Automated segmentation using complexity measures. Comput. Chem. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- 37.Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., Grishin N.V., Frantz D.E., Schneider J.W., Chen S., Li L., et al. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue S., Gong R., He F., Li Y., Wang Y., Tan T., Luo S.Z. Low-complexity domain of U1-70K modulates phase separation and aggregation through distinctive basic-acidic motifs. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aax5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez-Escamilla A.M., Rousseau F., Schymkowitz J., Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 40.Maurer-Stroh S., Debulpaep M., Kuemmerer N., Lopez de la Paz M., Martins I.C., Reumers J., Morris K.L., Copland A., Serpell L., Serrano L., Schymkowitz J.W., Rousseau F. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat. Methods. 2010;7:237–242. doi: 10.1038/nmeth.1432. [DOI] [PubMed] [Google Scholar]
- 41.Linding R., Schymkowitz J., Rousseau F., Diella F., Serrano L. A comparative study of the relationship between protein structure and beta-aggregation in globular and intrinsically disordered proteins. J. Mol. Biol. 2004;342:345–353. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- 42.Reineke L.C., Cheema S.A., Dubrulle J., Neilson J.R. Chronic starvation induces noncanonical pro-death stress granules. J. Cell Sci. 2018;131 doi: 10.1242/jcs.220244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan C.Y., Zhao H., Pugh R.J., Pedley A.M., French J., Jones S.A., Zhuang X., Jinnah H., Huang T.J., Benkovic S.J. Purinosome formation as a function of the cell cycle. Proc. Natl. Acad. Sci. U. S. A. 2015;112:1368–1373. doi: 10.1073/pnas.1423009112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aulas A., Fay M.M., Lyons S.M., Achorn C.A., Kedersha N., Anderson P., Ivanov P. Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 2017;130:927–937. doi: 10.1242/jcs.199240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kroschwald S., Munder M.C., Maharana S., Franzmann T.M., Richter D., Ruer M., Hyman A.A., Alberti S. Different material states of Pub1 condensates define distinct modes of stress adaptation and recovery. Cell Rep. 2018;23:3327–3339. doi: 10.1016/j.celrep.2018.05.041. [DOI] [PubMed] [Google Scholar]
- 46.Munder M.C., Midtvedt D., Franzmann T., Nuske E., Otto O., Herbig M., Ulbricht E., Muller P., Taubenberger A., Maharana S., Malinovska L., Richter D., Guck J., Zaburdaev V., Alberti S. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. Elife. 2016;5 doi: 10.7554/eLife.09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Werley C.A., Boccardo S., Rigamonti A., Hansson E.M., Cohen A.E. Multiplexed optical sensors in arrayed islands of cells for multimodal recordings of cellular physiology. Nat. Commun. 2020;11:3881. doi: 10.1038/s41467-020-17607-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buckler K.J., Vaughan-Jones R.D. Application of a new pH-sensitive fluoroprobe (carboxy-SNARF-1) for intracellular pH measurement in small, isolated cells. Pflugers Arch. 1990;417:234–239. doi: 10.1007/BF00370705. [DOI] [PubMed] [Google Scholar]
- 49.Michl J., Park K.C., Swietach P. Evidence-based guidelines for controlling pH in mammalian live-cell culture systems. Commun. Biol. 2019;2:144. doi: 10.1038/s42003-019-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas J.A., Buchsbaum R.N., Zimniak A., Racker E. Intracellular pH measurements in Ehrlich ascites tumor cells utilizing spectroscopic probes generated in situ. Biochemistry. 1979;18:2210–2218. doi: 10.1021/bi00578a012. [DOI] [PubMed] [Google Scholar]
- 51.Yamada S., Sato A., Sakakibara S.I. Nwd1 regulates neuronal differentiation and migration through purinosome formation in the developing cerebral cortex. iScience. 2020;23:101058. doi: 10.1016/j.isci.2020.101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baresova V., Krijt M., Skopova V., Souckova O., Kmoch S., Zikanova M. CRISPR-Cas9 induced mutations along de novo purine synthesis in HeLa cells result in accumulation of individual enzyme substrates and affect purinosome formation. Mol. Genet. Metab. 2016;119:270–277. doi: 10.1016/j.ymgme.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 53.Soding J., Zwicker D., Sohrabi-Jahromi S., Boehning M., Kirschbaum J. Mechanisms for active regulation of biomolecular condensates. Trends Cell Biol. 2020;30:4–14. doi: 10.1016/j.tcb.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 54.Rai A.K., Chen J.X., Selbach M., Pelkmans L. Kinase-controlled phase transition of membraneless organelles in mitosis. Nature. 2018;559:211–216. doi: 10.1038/s41586-018-0279-8. [DOI] [PubMed] [Google Scholar]
- 55.Wippich F., Bodenmiller B., Trajkovska M.G., Wanka S., Aebersold R., Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 56.Mateju D., Franzmann T.M., Patel A., Kopach A., Boczek E.E., Maharana S., Lee H.O., Carra S., Hyman A.A., Alberti S. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 2017;36:1669–1687. doi: 10.15252/embj.201695957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Z., Zhang S., Gu J., Tong Y., Li Y., Gui X., Long H., Wang C., Zhao C., Lu J., He L., Li Y., Liu Z., Li D., Liu C. Hsp27 chaperones FUS phase separation under the modulation of stress-induced phosphorylation. Nat. Struct. Mol. Biol. 2020;27:363–372. doi: 10.1038/s41594-020-0399-3. [DOI] [PubMed] [Google Scholar]
- 58.Qamar S., Wang G., Randle S.J., Ruggeri F.S., Varela J.A., Lin J.Q., Phillips E.C., Miyashita A., Williams D., Strohl F., Meadows W., Ferry R., Dardov V.J., Tartaglia G.G., Farrer L.A., et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-pi interactions. Cell. 2018;173:720–734.e715. doi: 10.1016/j.cell.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang K., Liu J.Q., Zhong T., Liu X.L., Zeng Y., Qiao X., Xie T., Chen Y., Gao Y.Y., Tang B., Li J., Zhou J., Pang D.W., Chen J., Chen C., et al. Phase separation and cytotoxicity of tau are modulated by protein disulfide isomerase and S-nitrosylation of this molecular chaperone. J. Mol. Biol. 2020;432:2141–2163. doi: 10.1016/j.jmb.2020.02.013. [DOI] [PubMed] [Google Scholar]
- 60.Pedley A.M., Karras G.I., Zhang X., Lindquist S., Benkovic S.J. Role of HSP90 in the regulation of de novo purine biosynthesis. Biochemistry. 2018;57:3217–3221. doi: 10.1021/acs.biochem.8b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pareek V., Sha Z., He J., Wingreen N.S., Benkovic S.J. Metabolic channeling: Predictions, deductions, and evidence. Mol. Cell. 2021;81:3775–3785. doi: 10.1016/j.molcel.2021.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Calvo-Vidal M.N., Zamponi N., Krumsiek J., Stockslager M.A., Revuelta M.V., Phillip J.M., Marullo R., Tikhonova E., Kotlov N., Patel J., Yang S.N., Yang L., Taldone T., Thieblemont C., Leonard J.P., et al. Oncogenic HSP90 facilitates metabolic alterations in aggressive B-cell lymphomas. Cancer Res. 2021;81:5202–5216. doi: 10.1158/0008-5472.CAN-21-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deng Y., Gam J., French J.B., Zhao H., An S., Benkovic S.J. Mapping protein-protein proximity in the purinosome. J. Biol. Chem. 2012;287:36201–36207. doi: 10.1074/jbc.M112.407056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang F., Watson E., Dempsey C., Suh J. Real-time particle tracking for studying intracellular trafficking of pharmaceutical nanocarriers. Methods Mol. Biol. 2013;991:211–223. doi: 10.1007/978-1-62703-336-7_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Royer L.A., Weigert M., Gunther U., Maghelli N., Jug F., Sbalzarini I.F., Myers E.W. ClearVolume: Open-source live 3D visualization for light-sheet microscopy. Nat. Methods. 2015;12:480–481. doi: 10.1038/nmeth.3372. [DOI] [PubMed] [Google Scholar]
- 67.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wadell H. Volume, shape, and roundness of quartz particles. J. Geol. 1935;43:250–280. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the article and supporting information files. Additional data files underpinning this study are available upon request.