Abstract
Neural precursor cells expressed developmentally downregulated protein 4-2 (Nedd4-2), a homologous to the E6-AP carboxyl terminus (HECT) ubiquitin ligase, triggers the endocytosis and degradation of its downstream target molecules by regulating signal transduction through interactions with other targets, including 14-3-3 proteins. In our previous study, we found that 14-3-3 binding induces a structural rearrangement of Nedd4-2 by inhibiting interactions between its structured domains. Here, we used time-resolved fluorescence intensity and anisotropy decay measurements, together with fluorescence quenching and mass spectrometry, to further characterize interactions between Nedd4-2 and 14-3-3 proteins. The results showed that 14-3-3 binding affects the emission properties of AEDANS-labeled WW3, WW4, and, to a lesser extent, WW2 domains, and reduces their mobility, but not those of the WW1 domain, which remains mobile. In contrast, 14-3-3 binding has the opposite effect on the active site of the HECT domain, which is more solvent exposed and mobile in the complexed form than in the apo form of Nedd4-2. Overall, our results suggest that steric hindrance of the WW3 and WW4 domains combined with conformational changes in the catalytic domain may account for the 14-3-3 binding-mediated regulation of Nedd4-2.
Significance
Ubiquitin ligase Nedd4-2 phosphorylation at residues S342 and S448 triggers its association with 14-3-3 proteins, thus modulating Nedd4-2 function and the ubiquitination of various ion channels. Here, we labeled the individual structured Nedd4-2 domains with the environmentally sensitive extrinsic fluorophore 1,5-IAEDANS and performed fluorescence measurements. This allowed us to monitor the mobility and accessibility of the individual domains of Nedd4-2 upon 14-3-3 protein binding. Our data reveal that the steric hindrance of the WW3 and WW4 domains together with the conformational change in the catalytic domain may be responsible for the 14-3-3 binding-mediated regulation of Nedd4-2 functions. Therefore, 14-3-3-mediated changes in the accessibility and/or mobility of individual WW domains of Nedd4-2 may modulate different dynamic processes of membrane proteins ubiquitination.
Introduction
The Nedd4 family of mammalian homologous to the E6-AP carboxyl terminus (HECT) E3 ubiquitin ligases encompasses nine members, including neural precursor cell-expressed developmentally downregulated protein 4-2 (Nedd4-2). Nedd4-2 originated from the most conserved and ancestral member Nedd4 by gene duplication (1) and is only found in vertebrates, which primarily express this ubiquitin ligase in the lungs, kidneys, brain, and heart (2). In these organs, Nedd4-2 has been shown to modulate epithelial sodium channel ENaC in the lungs and kidneys (3), the renal sodium-chloride symporter NCC (4), several voltage-gated sodium and potassium channels (5,6), and neural receptors (7), among other targets. Some of these targets include membrane proteins, whose activation is controlled by ubiquitination mediated by Nedd4-2. Accordingly, its malfunction has been linked to lung inflammation, Liddle syndrome, salt-sensitive hypertension, epilepsy, and developmental and end-stage renal disorders, highlighting its importance for human physiology (8, 9, 10, 11).
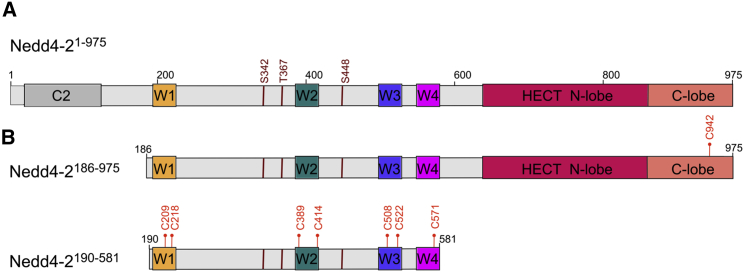
The Nedd4-2 molecule consists of three spatially and functionally distinct domains, the N-terminal calcium/lipid binding (C2) domain, four repeats of 34 amino acids known as WW domains, and the HECT domain (Fig. 1 A). The C2 domain regulates protein function by mediating its translocation to phospholipid membranes in a response to enhanced cytosolic Ca2+ (12). Human Nedd4-2 contains four WW domains located between the C2 and the HECT domains. The WW domains (also known as WWP) are named after their two highly conserved tryptophan residues and proline residue. These domains preferentially bind to proline-rich motifs (PPxY) of regulated substrates because they contain a hydrophobic core bordered by beta sheets with several charged residues (13,14). The catalytic HECT domain catalyzes polyubiquitin chain assembly through a conserved, two-step mechanism, similarly to other HECT family members (15). This substrate ubiquitination by Nedd4-2 through substrate-linked ubiquitin chains promotes degradation and cell signaling (16). WW domains, in the same protein, show different substrate specify in vitro. As such, Nedd4-2 may interact with various proteins through its WW domains in vivo.
Figure 1.
Domain structure of human Nedd4-2 and the protein constructs used in this study. (A) Schematic representation of the Nedd4-2 domain structure showing the positions of 14-3-3 binding motifs S342, T367, and S448; the Ca2+ lipid binding domain is shown in gray (denoted as C2), and the WW1-4 domains are shown in yellow, teal, blue, and magenta (denoted as W1-4). The N- and C-lobes of the HECT domain are shown in raspberry and salmon, respectively. (B) The constructs used in this study are Nedd4-2186−975 (C942) and Nedd4-2190−581 (C209, C218, C389, C414, C508, C522, and C571). The positions of the cysteine residues used for 1,5-IAEDANS labeling are shown in red. To see this figure in color, go online.
In line with the above, the WW3 and WW4 domains of Nedd4-2 directly interact with all epithelial sodium channel (ENaC) subunits and play a key role in the feedback regulation of ENaC in mammalian cells (2,17). In turn, mouse Nedd4 uses all of its three WW domains to interact with ENaC (18), whereas WW2-4 of human Nedd4, which has four WW domains, are required for this interaction (19). Moreover, the WW domains of Nedd4-2 recognize the PY motif in its own HECT domain (20), and WW1 and WW2 interact with TRPV6 and decrease its ubiquitination rate (21). Nedd4-2 function and its substrate specificity are also modulated by phosphorylation and binding to adaptor proteins, which can have both inhibitory and activatory effects. Several protein kinases, such as PKA, Akt, Sgk1, JNK1, and IKKβ, phosphorylate Nedd4-2 at S342, T367, and S448 (22, 23, 24, 25, 26, 27), thus triggering 14-3-3 protein binding (28, 29, 30).
14-3-3 proteins are a family of highly conserved dimeric proteins, which interact with hundreds of other proteins, thereby regulating their functions (31, 32, 33, 34). 14-3-3 proteins recognize pSer/pThr-containing motifs, which are frequently located within disordered regions of their binding partners and far from structured functional domains (35, 36, 37). Nedd4-2 has three 14-3-3 binding motifs, but phosphorylated S342 and S448 are the crucial residues that facilitate high-affinity 14-3-3 binding (30). Both motifs are located within disordered regions bordering the WW2 domain (Fig. 1 A). However, the exact role of 14-3-3 binding in regulating Nedd4-2 remains unclear. For example, Nedd4-2 interaction with 14-3-3 is known to both prevent the ubiquitination of ENaC, increasing ENaC activity (38), and promote the ubiquitination of the GluA1 subunit of the AMPA receptor (7). Nevertheless, we have recently performed a biophysical characterization of the complex between the phosphorylated Nedd4-2 and 14-3-3η, showing that 14-3-3 binding induces a structural rearrangement of Nedd4-2 by altering interactions between its structured domains (30). Our structural analysis also suggested that the formation of this complex may affect the accessibility and/or the mobility of individual WW domains of Nedd4-2.
To test this hypothesis, in this study, we aimed to further characterize interactions between phosphorylated Nedd4-2 and 14-3-3 by investigating changes in the solvent accessibility and mobility of Nedd4-2 domains upon complex formation. For this purpose, we used various fluorescence spectroscopy methods, including time-resolved fluorescence intensity and anisotropy decay measurements and fluorescence quenching. The results clearly indicate that 14-3-3η directly interacts with WW3 and WW4 domains and that the 14-3-3η-protein-induced conformational change of Nedd4-2 involves not only the WW3 and WW4 domains but also its catalytic domain.
Materials and methods
Heterologous expression and purification of 14-3-3 protein
14-3-3η was expressed and purified as described previously (30,39,40). In brief, 14-3-3η was expressed in E. coli BL21(DE3) cells using three purification steps: affinity chromatography, followed by His6-tag cleavage, anion-exchange chromatography (HiTrap Q column, GE Healthcare, Chicago, Illinois, USA), and size-exclusion chromatography (HiLoad Superdex 75, GE Healthcare) in a buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM TCEP, and 10% (w/v) glycerol. The final protein was concentrated to 30 mg∙mL−1, frozen in liquid nitrogen, and stored in aliquots at −80°C (193.15 K).
Heterologous expression, purification, and phosphorylation of Nedd4-2
The coding sequence of Nedd4-2 (residues 190–581) was PCR amplified from the plasmid hNedd4-2 (residues 186–975) previously prepared in our laboratory (30). The PCR product containing residues 190–581 was ligated into the polycistronic pST39 vector SacI/KpnI restriction sites. The entire cloned region was confirmed by sequencing. Thirteen different Nedd4-2 mutants in both Nedd4-2 variants containing a single cysteine residue (Nedd4-2190−581 variant: T209C, S218C, S389C, T414C, A508C, T522C, S571C; Nedd4-2186−975 variant: C702S, C776S, C853S, C874S, C942S) were generated using the QuikChange kit (Stratagene, Santa Clara, California, USA), confirming all mutations by sequencing.
Nedd4-2190−581 WT and single cysteine variants were expressed as a fusion protein with 6× noncleavable His-tag at the C-terminus by isopropyl-1-thioβ-D-galactopyranoside induction for 18– 20 h at 18°C in E. coli BL21 (DE3) cells. The pelleted cells were suspended in lysis buffer (1× PBS, 1 M NaCl, 4 mM β-mercaptoethanol, 2 mM imidazole, and 0.01% (v/v) Tergitol NP-40) and purified using a Chelating Sepharose Fast Flow column (GE Healthcare), according to the standard protocol, followed by gel filtration chromatography on a HiLoad 26/600 Superdex 75 pg column (GE Healthcare) in a buffer containing 20 mM Tris-HCl (pH 7.5), 500 mM NaCl, 1 mM TCEP, 10% glycerol (w/v), and 0.01% (v/v) Tergitol NP-40. Purified Nedd4-2190−581 was phosphorylated with 158 units of PKA (Promega, Santa Clara, California, USA) per milligram of recombinant protein in the presence of 0.75 mM ATP and 20 mM MgCl2 by incubation at 30°C for 2 h and then overnight at 4°C. After phosphorylation, the protein was repurified using a size-exclusion chromatography Superdex 75 Increase 10/300 GL column (GE Healthcare) in a buffer containing 20 mM Tris-HCl (pH 7.5), 500 mM NaCl, 1 mM TCEP, and 10% glycerol (w/v).
Nedd4-2186−975 WT and single cysteine variants were expressed and purified as described previously (30).
Labeling of Nedd4-2 mutants by 1,5-IAEDANS
Nedd4-2190−581 contains only one cysteine residue at position 341. To prepare Nedd4-2190−581 specifically labeled with a fluorescence probe at different positions in the WW domains, we first replaced cysteine 341 by serine and then incorporated a single cysteine into seven different positions (C209, C218, C389, C414, C508, C522, and C571). Nedd4-2186−975 contains six cysteine residues. To prepare Nedd4-2186−975 specifically labeled with a fluorescence probe at different positions of the HECT domain, we mutated C341S and used this mutation as a template for subsequent mutagenesis of five cysteine residues present in the HECT domain. Mutagenesis was performed so that only a single cysteine was present and that all other cysteine residues were mutated to serine residues. Covalent modification of Nedd4-2 containing a single cysteine residue with the thiol-reactive probe 1,5-IAEDANS was performed as described before (41). In brief, the protein (2.0– 8.5 μM) in 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA buffer, and label, were mixed at a molar ratio of 1:40 and incubated at 30°C for 2 h, and then at 4°C overnight in the dark. Subsequent gel filtration chromatography was performed in 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 10% glycerol (w/v) buffer (Sigma-Aldrich, St. Louis, MO) to remove free unreacted label. The incorporation stoichiometry was determined by comparing the peak protein absorbance at 280 nm with the absorbance of bound 1.5-IAEDANS measured at 336 nm using an extinction coefficient of 5700 M−1 cm−1 (Molecular Probes, Eugene, OR).
Mass spectrometry analysis
Proteins were diluted to 50 mM ammonium bicarbonate buffer. Cysteines were reduced with 10 mM DTT for 45 min at 60°C and free cysteines were alkylated with 30 mM iodoacetamide for 30 min at room temperature in the dark. Trypsin digestion proceeded overnight at 37°C with an enzyme/protein ratio of 1:20 (w/w). Peptides were loaded on a trap column (Luna Omega 5 μm Polar C18 100 Å, 20 × 0.3 mm, Phenomenex, Torrance, California, USA) and desalted for 5 min at flow rate 20 μL/min. Peptides were then separated using a reversed-phase C18 column (Luna Omega 3 μm Polar C18 100 Å, 150 × 0.3 mm, Phenomenex) at a flow rate 10 μL/min with a capillary UHPLC system (Agilent Technologies, Santa Clara, California, USA) under the following gradient conditions: 1–10% B in 1 min, 10–45% B in 19 min, 45–95% B in 5 min, where solvent A was 0.1% formic acid and 2.0% acetonitrile in water, and solvent B was 0.1% formic acid in 98% acetonitrile. The column was heated at 50°C and directly connected to a timsTOF Pro mass spectrometer (Bruker Daltonics,Madison, Wisconsin, USA). The instrument was operating in PASEF mode. All data were processed by the PEAKS Studio X software (Bioinformatics Solutions, Waterloo, Ontario, Canada), searching against the database of Nedd4-2190−581 protein. The FDR was set to 1% for the peptides.
Differential scanning fluorimetry
The thermal stability of the Nedd4-2 mutants was checked by measuring the thermally induced protein denaturation, using differential scanning fluorimetry. Nedd4-2190−581 and Nedd4-2186−975 protein variants at concentrations of 0.144–0.37 and 0.185 mg∙mL−1, respectively, were tested in 8× concentrated Sypro Orange (Sigma-Aldrich) in a total reaction volume of 50 μL in buffer containing 100 mM HEPES (pH 7.5) and 150 mM NaCl on a LightCycler 480 Multiwell Plate 96 (Roche Applied Science, Penzberg, Germany). The melting temperature values, Tm, corresponding to the inflection points of the melting curves, were determined as the minima of the negative first derivative using the Roche LightCycler 480 SW 1.5 software (42,43). The final results are expressed as the means from three measurements.
Limited proteolysis
Samples containing 50 pmol of phosphorylated Nedd4-2186−975 WT with or without 100 pmol 14-3-3η were digested by trypsin for 10, 20, and 30 min at 25°C in a buffer containing 50 mM Tris-HCl (pH 8.0), 500 mM NaCl, 1 mM TCEP, 10% (w/v) glycerol, and 0.01% tergitol (protease/protein ratio was 1:1000, w/w). Undigested protein served as the zero-time point. The reactions were terminated by adding SDS-PAGE sample loading buffer and boiling for 5 min. The results were analyzed by SDS-PAGE. The density of the bands resulting from the degradation of Nedd4-2 in respective time points were quantified using Image Lab software and Student's t-test (Bio-Rad, Hercules, California, USA).
Time-resolved fluorescence
Time-resolved fluorescence intensity and anisotropy decay measurements, as well as data analysis, were performed as described previously (44,45). In brief, the apparatus comprised pulsed frequency-tripled Ti:Sapphire laser (Chameleon Ultra II, Coherent, Santa Clara, California, USA) and time-correlated single-photon counting detection (SPC150, Becker & Hickl, Berlin, Germany) with cooled MCP-PMT (R3809U-50, Hamamatsu, Hamamatsu City, Shizuoka, Japan). Dansyl fluorescence was excited at 355 nm and collected at 535 nm on a monochromator (H-20, Horiba, Kyoto, Japan) with a stack of glass OG420 long-pass and dielectric LP520 filters placed in front of the input slit. The repetition frequency of the excitation pulses was lowered to 4 MHZ by the pulse-picker (APE, Berlin, Germany). This corresponds to about 15 dansyl lifetimes (250 ns) allowing dansyl emission to completely decay to zero between the two successive excitation pulses. Intensity decays were accumulated under the magic-angle conditions, typically in 1024 channels with a time resolution of 195 ps/channel, until reaching approximately 1.5 × 105 counts at the decay maximum. The instrument response function was measured by scattered excitation from the diluted ludox solution. FWHM of the response function was about 120 ps, which is well below the decay channel time width. Polarized decays for fluorescence anisotropy were acquired quasi-simultaneously with a switching time between the components of 30 s. The samples were placed in a thermostatic holder.
All experiments were performed at 23°C in a 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, and 10% (w/v) glycerol buffer (Sigma-Aldrich). The concentrations of Nedd4-2190−581 variants (C209, C218, C389, C414, C508, C522, and C571), Nedd4-2186−975 C942, and 14-3-3η ranged from 1.6 to 4 and from 4 to 8 μM, respectively. Both fluorescence intensity and anisotropy decays were analyzed using the model-independent singular value decomposition-maximum entropy method (45).
Fluorescence was assumed to decay multiexponentially
| (1) |
where τi stands for the fluorescence lifetime components, and αi are the corresponding amplitudes. The mean emission lifetime was calculated as follows:
| (2) |
Fluorescence anisotropies r(t) were determined by simultaneous reconvolution of I||(t) and I⊥(t) polarized decays (46,47). The anisotropies were assumed to decay multiexponentially:
| (3) |
where amplitudes βiʹ represent the distribution of the correlation times φiʹ, , and r0 is the initial anisotropy. We typically used 100 correlation times and 100 lifetimes equidistantly spaced in the logarithmic timescale for MEM analysis (45).
Dansyl quenching
Stern-Volmer plots were constructed from changes in mean fluorescence lifetime after adding acrylamide aliquots dissolved in the protein buffer. Stern-Volmer plots were fitted to the modified SV equation (48) transformed to the form of:
| (4) |
where and are the mean fluorescence lifetime with and without quencher, respectively, is acrylamide concentration, is the bimolecular quenching constant, and stands for an inaccessible fraction of the fluorophore.
Results
Construction of the Nedd4-2 protein mutants for dansyl fluorescence measurements
For further insight into interactions between Nedd4-2 and 14-3-3η, we labeled the individual structured Nedd4-2 domains with the environmentally sensitive extrinsic fluorophore 1,5-IAEDANS and performed time-resolved fluorescence intensity and anisotropy decay measurements, in addition to time-resolved collisional quenching of dansyl-labeled proteins. For this purpose, we used two Nedd4-2 variants: Nedd4-2190−581 containing both 14-3-3 binding sites, S342 and S448, and all four WW domains, and Nedd4-2186−975, also containing the HECT domain (Fig. 1 B) (30). Both proteins exhibited sufficient solubility and stability for biophysical studies. A previous study has shown that the Nedd4-2186−975 and 14-3-3η form a complex with a 1:2 stoichiometry and KD < 50 nM (30). Therefore, the concentration used for fluorescence studies enables that ∼99% of proteins are bound in the complex.
The sequence of Nedd4-2190−581 contains a single cysteine residue at position C341 preceding the first phosphorylation site, that is, the 14-3-3 binding motif S342. For this reason, we first mutated C341 to serine and subsequently introduced a single cysteine residue at several positions of interest (mutations T209C, S218C, S389C, T414C, A508C, T522C, and S571C) to specifically label the proteins with 1,5-IAEDANS. The longer construct, Nedd4-2186−975, contains five additional cysteine residues in the HECT domain at positions C702, C776, C853, C874, and C942. Thus, we first introduced the C341S mutation and then proceeded with the step-by-step mutagenesis of other Cys residues to prepare single cysteine-containing variants. Unfortunately, of all Nedd4-2186−975 mutant variants, only Nedd4-2186−975 with Cys at position 942 yielded a soluble and stable protein. In total, eight single cysteine-containing Nedd4-2 variants were prepared: seven with the Nedd4-2190−581 construct (C209, C218, C389, C414, C508, C522, and C571) and one with the Nedd4-2186−975 construct (C942). These cysteine residues are located in five different regions of Nedd4-2: C209 and C218 in the WW1 domain, C389 and C414 in the WW2 domain, C508 and C522 in the WW3 domain, C571 in the WW4 domain, and C942 in the active site of the C-lobe of the HECT domain (Fig. 1 B).
The stability of all Nedd4-2 mutants was assessed by measuring the midpoint temperatures of the protein unfolding transition (Tm) by differential scanning fluorimetry. No substantial differences in Tm were observed in all Nedd4-2 variants, except for Nedd4-2190−581C508 (Table 1). The slightly lower Tm of the Nedd4-2190−581C508 variant may reflect a different conformation of the mutated WW3 domain. The binding of all phosphorylated Nedd4-2 mutant variants to 14-3-3η was confirmed by native gel electrophoresis (Fig. S1). The single cysteine-containing variants of Nedd4-2 were labeled with the extrinsic fluorophore 1,5-IAEDANS, confirming the successful modification of all proteins by LC-MS (Figs. S2–S10).
Table 1.
Stability of individual Nedd4-2 mutant variants
| Nedd4-2190−581 variant | Tm (°C) |
|---|---|
| C209 | 54.2 ± 0.1 |
| C218 | 55.3 ± 0.1 |
| C389 | 53.6 ± 0.1 |
| C414 | 55.7 ± 0.2 |
| C508 | 51.06 ± 0.05 |
| C522 | 56.5 ± 0.2 |
| C571 | 54.4 ± 0.3 |
| WT | 56.29 ± 0.05 |
| Nedd4-2186−975 variant | Tm (°C) |
|---|---|
| WT | 41.34 ± 0.06 |
| C942 | 39.5 ± 0.4 |
Midpoint temperatures of the protein unfolding transition (Tm) for Nedd4-2 WT and mutants, as determined by differential scanning fluorimetry. Uncertainties are expressed as the SE values calculated from three experiments.
14-3-3η protein binding affects the conformation of Nedd4-2 WW3, WW4, and the active site of the HECT domain: Time-resolved fluorescence lifetime measurements
We have recently shown that 14-3-3η binding triggers the structural rearrangement of Nedd4-2 (30), which could include conformational changes in the WW and HECT domains. To gather more detailed information about structural changes in individual Nedd4-2 domains induced by their interaction with 14-3-3η, we performed time-resolved fluorescence intensity measurements of all eight AEDANS-labeled cysteine variants of Nedd4-2190−581 and Nedd4-2186−975 in the absence and presence of 14-3-3η. As an environmentally sensitive fluorophore, AEDANS changes its quantum yield and emission lifetime with the polarity of its microenvironment (49). Fluorescence lifetime measurements can therefore sensitively monitor subtle changes in dansyl solvation and/or conformational changes induced by 14-3-3η binding (Fig. 2; Table 2).
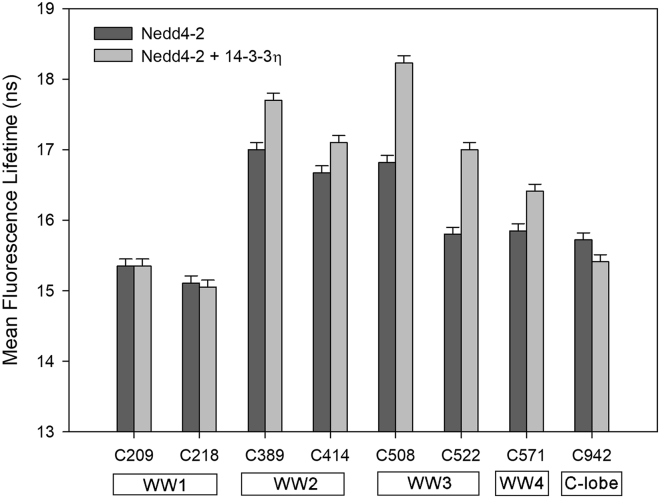
Figure 2.
Mean fluorescence lifetimes of Nedd4-2 variants and their changes upon 14-3-3η binding. The positions of AEDANS-labeled cysteine residues within Nedd4-2 domains are indicated at the bottom. C209 and C218, WW1; C389 and C414, WW2; C414 and C508, WW3; C571, WW4; C943, the C-lobe of HECT domain. Each error bar reflects the standard deviation of a single-curve data analysis.
Table 2.
Summary of the time-resolved AEDANS fluorescence measurements
| Nedd4-2 variant | τmeana,b (ns) |
kq (× 10−8) |
Fb |
φ1c,d |
β1c |
φ2e |
β2 |
φ3e |
β3 |
φ4f |
β4 |
φ5g |
β5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (M−1 s−1)h | (ns) | (ns) | (ns) | (ns) | (ns) | ||||||||
| Nedd4-2190−581 | |||||||||||||
| C209 | 15.4 | 4.3 | 0.27 | 0.1 | 0.19 | 1.4 | 0.05 | 9.5 | 0.05 | 79 | 0.06 | ||
| C209 + 14-3-3η | 15.4 | 4.6 | 0.29 | 0.1 | 0.17 | 0.5 | 0.05 | 3.9 | 0.04 | 15 | 0.02 | >200 | 0.06 |
| C218 | 15.1 | 7.1 | 0.15 | 0.1 | 0.27 | 0.5 | 0.01 | 2.6 | 0.02 | 17 | 0.03 | 170 | 0.01 |
| C218 + 14-3-3η | 15.1 | 6.9 | 0.30 | 0.1 | 0.26 | 0.5 | 0.03 | 3.9 | 0.01 | 15 | 0.02 | >200 | 0.02 |
| C389 | 17.0 | 2.7 | 0.30 | 0.1 | 0.08 | 1.1 | 0.04 | 3.9 | 0.07 | 26 | 0.13 | >200 | 0.04 |
| C389 + 14-3-3η | 17.7 | 1.5 | 0.28 | 0.1 | 0.08 | 1.6 | 0.04 | 6.5 | 0.05 | 50 | 0.11 | >200 | 0.06 |
| C414 | 16.7 | 2.6 | 0.31 | 0.1 | 0.08 | 1.4 | 0.06 | 6.6 | 0.07 | 35 | 0.10 | >200 | 0.03 |
| C414 + 14-3-3η | 17.1 | 2.0 | 0.31 | 0.1 | 0.08 | 1.9 | 0.06 | 9.4 | 0.07 | 97 | 0.04 | >200 | 0.08 |
| C508 | 16.8 | 2.7 | 0.25 | 0.1 | 0.10 | 1.3 | 0.02 | 4 | 0.06 | 12 | 0.08 | 79 | 0.07 |
| C508 + 14-3-3η | 18.2 | 1.5 | 0.22 | 0.1 | 0.07 | 0.6 | 0.03 | 3.8 | 0.05 | 16 | 0.05 | >200 | 0.14 |
| C522 | 15.8 | 3.6 | 0.33 | 0.1 | 0.08 | 1.1 | 0.07 | 6.1 | 0.09 | 39 | 0.07 | >200 | 0.03 |
| C522 + 14-3-3η | 17.0 | 1.7 | 0.31 | 0.1 | 0.09 | 1.4 | 0.05 | 6.2 | 0.05 | 47 | 0.08 | >200 | 0.07 |
| C571 | 15.9 | 3.2 | 0.32 | 0.1 | 0.09 | 1.9 | 0.10 | 14.1 | 0.08 | 67 | 0.03 | >200 | 0.03 |
| C571 + 14-3-3η | 16.4 | 1.8 | 0.26 | 0.1 | 0.09 | 1.1 | 0.06 | 6.0 | 0.07 | 76 | 0.08 | >200 | 0.05 |
| Nedd4-2186−975 | |||||||||||||
| C942 | 15.7 | 5.5 | 0.49 | ||||||||||
| C942 + 14-3-3η | 15.4 | 7.4 | 0.56 | ||||||||||
Mean lifetimes were calculated as τmean = ∑ifiτi, where fi is an intensity fraction of the ith lifetime component τi, see also Eq. 2.
SD = ± 0.1 ns. The SD is a conservative upper estimate derived from the deconvolution analysis of single decay curves. Similary were derived SDs of all other MEM-derived parameters (63).
Anisotropies r(t) were analyzed for a series of exponentials using a model-independent maximum entropy method without setting prior assumptions about the shape of the correlation time distributions (45), r(t) = ∑k’ (βk’ exp(−t/φk’)), where amplitudes βk’ represent the distribution of the correlation times φk’. βn presented in the table are areas of peaks positioned at correlation times φn. SD of βn is ±0.01.
Shortest resolvable correlation time rounded to the first significant digit, SD < ± 0.1 ns.
SD = ± 0.2 ns.
SD = ± 5 ns.
Due to the finite dansyl emission lifetime this correlation time has a larger uncertainty with highly asymmetrical standard deviations. The uncertainty to the + direction (+SD) is much larger then to the − direction (−SD). This is indicated by the “greater then” (>) symbol. The exact value of this correlation time cannot be more accurately determined.
Inaccessible fraction, SD = ± 0.02, apparent bimolecular quenching constant, SD = ± 0.2 (×10−8 M−1 s−1). The standard deviations result from the fitting uncertainty of single quenching curves (64).
All AEDANS-labeled Nedd4-2 proteins exhibited multiexponential decays, typically containing two major and up to two minor components. Examples of decays and lifetime distributions are shown in Figs. S16 and S17. Since the protein is expected to scan a conformational space and the individual components are difficult to unequivocally assign, we used the mean fluorescence lifetime (τmean) as a qualitative indicator of changes in the local environment of AEDANS-labeled cysteines. AEDANS-labeled Nedd4-2190−581C209 and Nedd4-2190−581C218 (both Cys are located in the WW1 domain) exhibited relatively short emission lifetimes, 15.4 and 15.1 ns, respectively, unresponsive to 14-3-3η binding. Conversely, AEDANS-labeled Nedd4-2190−581C389, Nedd4-2190−581C414, and Nedd4-2190−581C508 (Cys located in WW2 and WW3 domains) exhibited the highest τmean of 17.0, 16.7 and 16.8 ns, respectively, in the absence of 14-3-3η, indicating a less polar fluorophore microenvironment (49). Lower τmean values were also detected for Nedd4-2190−581C522 (the WW3 domain), Nedd4-2190−581C571 (the WW4 domain), and Nedd4-2186−975C942 (the C-lobe of the HECT domain).
The strongest effect of 14-3-3η binding was observed in AEDANS moieties attached within the WW3 domain (Nedd4-2190−581C508 and Nedd4-2190−581C522), whose τmean values increased by ∼1.4 ns. AEDANS-labeled Nedd4-2190−581C389, C414, and C571 mutants (Cys in WW2 and WW4 domains) also exhibited significantly increased τmean in the presence of 14-3-3η, particularly the Nedd4-2190−581C389 variant (the WW2 domain), whose increase in τmean was close to 0.7 ns. As the dansyl fluorophore is known to exhibit an increased τmean in a less polar environment (49), these changes in τmean may result from 14-3-3η-induced conformational changes in Nedd4-2, which affect interactions between dansyl moieties and amino acid residues in their vicinity. Alternatively, the increase in τmean may reflect reduced polar contacts and modulated dansyl solvation due to a direct interaction of the fluorophore with bound 14-3-3η.
In contrast to C389, the AEDANS-labeled catalytic C942 in the active site of the HECT domain revealed a decrease in τmean by 0.3 ns, which can be interpreted as increased contacts with the polar environment resulting from increased solvation and/or quenching interactions near this residue. This suggests that 14-3-3η binding induces conformational changes within the active site of Nedd4-2 HECT domain (Fig. 2; Table 2).
14-3-3η protein binding reduces the mobility of Nedd4-2 WW domains: Time-resolved fluorescence anisotropy decay measurements
To investigate how 14-3-3η binding affects the mobility of Nedd4-2 AEDANS-labeled domains, we performed time-resolved emission polarization anisotropy measurements. For all seven variants of Nedd4-2190−581, the results are summarized in Table 2. Unfortunately, we were unable to prepare Nedd4-2186−975C942 in a concentration comparable with the other mutants. Low emission intensity called for longer measurement times, stronger excitation, and larger light exposure of the sample. Consequently, resulting progressive photobleaching prevented reliable collection of sequentially measured polarized decays and evaluation of the Nedd4-2186−975C942 emission anisotropy.
The AEDANS fluorescence anisotropy decays were complex, usually with five classes of correlational times. Typical data are shown in Fig. S18. The first two correlation times likely reflect the fast hindered motion of the AEDANS fluorophore itself (φ1 ∼ 0.1 and φ2 ∼ 0.5–1.9 ns). The slower decay components φ3 and φ4, with correlation times ranging from 2.6 to 14 ns and from 12 to 97 ns, respectively, reflect slower internal movements and components arising from the free rotation of the generally asymmetric rotor (46). The longest φ5, poorly resolved due to the finite dansyl lifetime, reflects the slow rotational movement of the Nedd4-2:14-3-3η complex (Mw = 150 kDa).). The φ5 component is generally modulated by the asymmetricity of the complex (47) and it may also reflect minor aggregation.
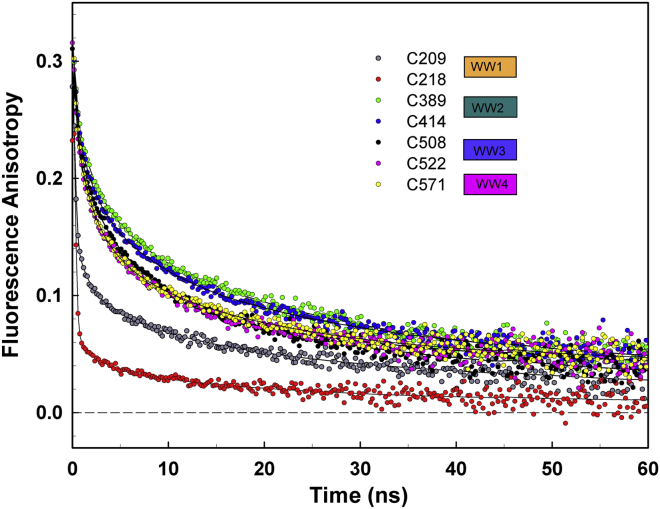
Based on the position of the label, the time-resolved fluorescence anisotropy decays of AEDANS-labeled Nedd4-2190−581 mutants can be separated into three distinct groups with different extent of the dansyl mobilities (Fig. 3) reflected by the depolarization amplitudes βi. The extent of segmental motion of the AEDANS-labeled cysteines of Nedd4-2190−581 and/or their 14-3-3η binding-induced changes can be assessed by visual comparison of the initial part of the anisotropy decay of each mutant variant and from the amplitude β1. The higher the β1 is, the lower the steric hindrance of the fast-depolarizing motion of the fluorophore and the higher the internal protein mobility will be. According to these correlations, C209 and C218 from the WW1 domain formed the group with the highest internal mobility (β1 = 0.19 and 0.27, respectively). C218 was the AEDANS-labeled cysteine with the highest angular mobility, which suggest its high accessibility to the solvent and quencher molecules. The large extent of the fast initial fluorescence depolarization is clearly shown in Fig. 4 A). The proposed high solvent accessibility of AEDANS-labeled C209 and C218 is supported by their relatively short emission lifetimes, 15.4 and 15.1 ns, respectively (Table 2). The group with the lowest internal mobility contains the C389 and C414 from the WW2 domain (β1 = 0.08). As expected, the high protein rigidity near C389 and C414 residues is accompanied by longer emission lifetimes of 17.0 and 16.7 ns, respectively, thus suggesting a less polar dansyl microenvironment (Table 2). In the third group, Nedd4-2190−581 mutants with cysteines inserted into the WW3 and WW4 domains (C508, C522, and C571) showed intermediate internal flexibility. In the apo form, the mutants with cysteines in the WW2-4 domains exhibited almost identical fluorescence anisotropy decays, indicating the similar internal mobilities of dansyl-labeled protein segments and the similar overall hydrodynamic properties of the whole protein (Fig. 3; Table 2).
Figure 3.
WW domains of Nedd4-2 exhibit different mobilities. Fluorescence anisotropy decays of AEDANS-labeled Nedd4-2190−581 variants. To see this figure in color, go online.
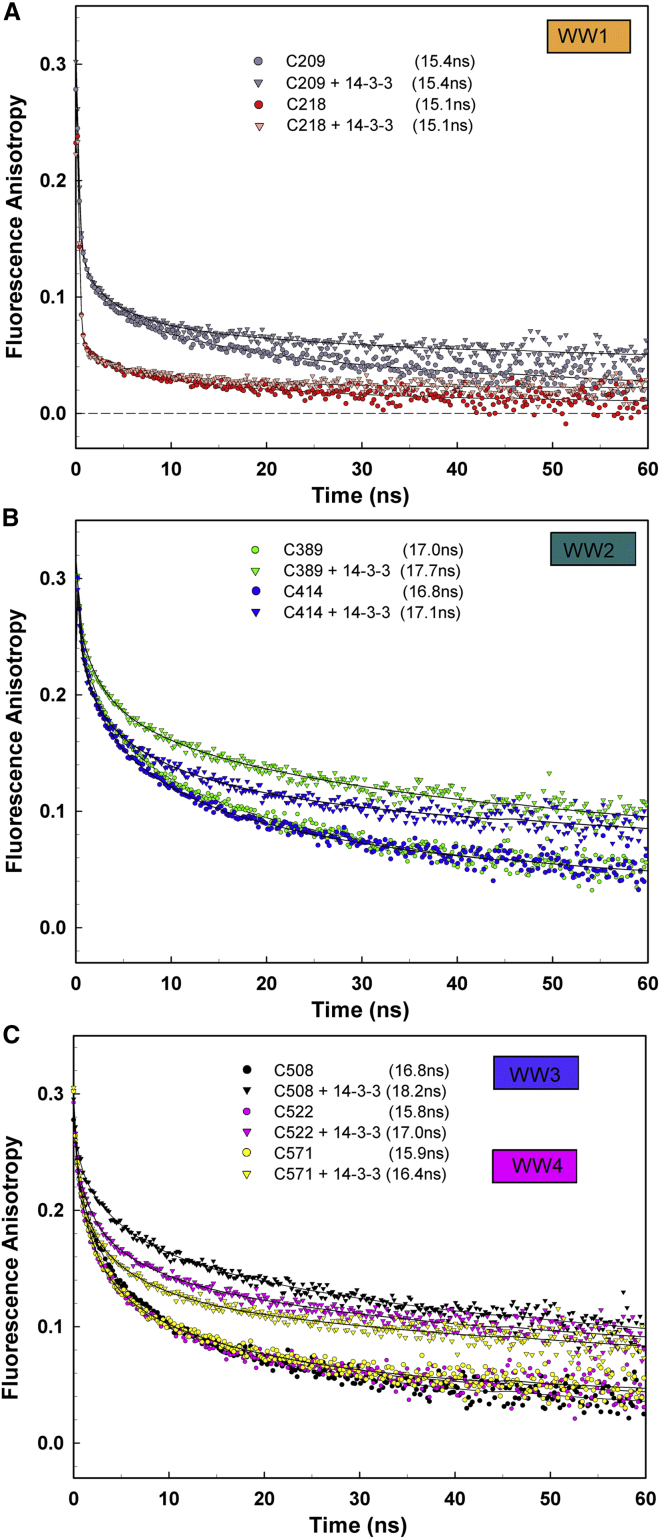
Figure 4.
14-3-3η binding affects the mobility of Nedd4-2 WW domains. Fluorescence anisotropy decays of Nedd4-2 variants with AEDANS-labeled individual WW domains in the absence and presence of 14-3-3η. (A) WW1 domain variants C209 and C218, (B) WW2 domain variants C389 and C414, and (C) WW3 and WW4 domain variants C508, C522, and C571. To see this figure in color, go online.
Time-resolved dansyl fluorescence anisotropy measurements performed in the presence of 14-3-3η revealed that complex formation does not lower the high mobility of the dansyl moiety at C209 and C218 within the WW1 domain because the anisotropy decays initially overlap (Fig. 4 A). This overlap correlates with the unchanged τmean (Table 2).
The mobilities of all other mutants differed upon complex formation. Fig. 4 B demonstrates that the segmental mobility of the most rigid C389 and C414 mutants (the WW2 domain) further decreased, particularly in the C389 mutant, as shown by the elevation of the corresponding anisotropy curve. Since Nedd4-2190−581C389:14-3-3η and Nedd4-2190−581C414:14-3-3η complexes have the same molecular weight, the slower initial depolarization should be caused by the decrease in dansyl mobility. The increase in fluorescence lifetime by 0.7 and 0.4 ns (Table 2) correlates with the observed decrease in the internal mobility of segments containing AEDANS-labeled C389 and C414, respectively.
A similar behavior was observed in the last group of mutants with cysteine residues in WW3 and WW4 domains (C508, C522, and C571) (Fig. 4 C). Although the anisotropy decays of these Nedd4-2190−581 variants in the absence of 14-3-3η seem to be similar, the presence of 14-3-3η significantly reduced their dansyl mobility in the following order: C571, C522, and C508 (Fig. 4 C). Fluorescence lifetime values increased in the same order upon 14-3-3η binding (Table 2). Among all other mutants, Nedd4-2190−581C508 exhibited the highest rigidization and a considerable lifetime increase of 1.4 ns. This increase likely reflects a conformational transition in the C508-containing WW3 domain combined with an increase in the solvent shielding of its dansyl group.
14-3-3η protein binding decreases the accessibility of Nedd4-2 WW3 and WW4 domains to the quencher, conversely increasing the accessibility of the HECT C-lobe: Time-resolved acrylamide quenching measurements
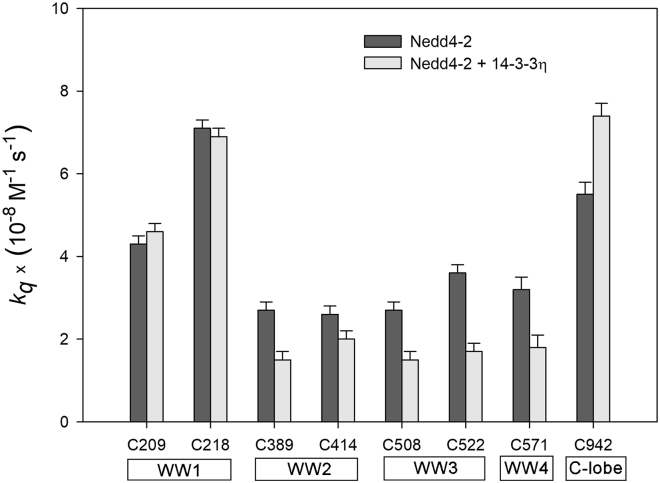
To further explore 14-3-3-induced structural changes in Nedd4-2, we performed time-resolved quenching experiments with all eight variants of Nedd4-2190−581 and Nedd4-2186−975, probing fluorophore accessibility to acrylamide. This time-resolved approach eliminates static quenching effects, which often complicate intensity-based quenching experiments and result in curved Stern-Volmer plots. Fluorescence decays of Nedd4-2 labeled by a single AEDANS fluorophore typically contained two closely spaced major peaks accompanied by up to two minor peaks, as shown in Figs. S16 and S17. Being aware of the same simplification, we represented decays by τmean that allowed as to reduce data, construct a single Stern-Volmer plot for each mutant, and retrieve an apparent collisional quenching constants for the each variant. As shown in Figs. S11–S14, this approach led to curved Stern-Volmer plots that indicate at least two classes of the fluorophore with different accessibilities to the collisional quencher, that is, acrylamide. Although the underlying molecular mechanism remains unknown, a likely explanation is the dynamic conformational heterogeneity of the proteins, fluctuating between conformations with different dansyl accessibilities. Hence, for another simplification, we assumed two classes of dansyl only, i.e., accessible and fully inaccessible forms. The values of the apparent bimolecular quenching constant kq determined by fitting the Stern-Volmer plots are presented in Table 2 and compared in Fig. 5. Generally, the inaccessible fractions of dansyl (Fb) were similar across all mutants ∼0.3, except for Nedd4-2190−581C218, whose Fb was lower.
Figure 5.
14-3-3η binding affects the solvent exposure of Nedd4-2 domains, as shown by the apparent bimolecular quenching constants of AEDANS-labeled Nedd4-2 cysteine variants in the absence and presence of 14-3-3η. Each error bar reflects the standard deviation of a single-curve data analysis.
The quenching experiments revealed that 14-3-3η binding does not significantly affect the quenching efficiency of C209, or C218 in the WW1 domain, with an apparent kq of 4.3 and 7.1 × 10−8 M−1 s−1, respectively (Table 2; Fig. S11). This lack of an effect suggests a high dansyl accessibility to the quencher molecules dissolved in the aqueous environment, regardless of the presence of 14-3-3η. Among the mutants, Nedd4-2190−581C218 exhibited the highest accessibility to the quencher, in line with the highest internal mobility of C218 and the shortest emission lifetime (Table 2). Moreover, only this mutant showed a significant change in Fb upon 14-3-3η binding, which is the only reason for the dramatic change in the shape of the quenching curves, as shown in Fig. S11. Although the molecular mechanism is still unknown, unbound Nedd4-2190−581C218 has approximately two times lower Fb than the other mutants. This difference indicates that the conformational heterogeneity of dansyl at position 218 in the WW1 domain is lower in the unbound protein. The steady-state emission spectra of Nedd4-2190−581C218 are shown in Fig. S19. In contrast to the other mutants, the spectrum in the presence of 14-3-3η displays a blue-shifted shoulder, which is consistent with the quenching experiment and suggests the appearance of new conformers upon 14-3-3η binding.
All mutants with cysteine residues in WW2-4 domains exhibited a significantly decreased kq after14-3-3η protein binding and only minor changes in Fb. Only the Nedd4-2186−975C942 mutant with dansyl positioned in the HECT domain displayed an increased value of kq upon 14-3-3η binding. The relative decrease in kq induced by 14-3-3 binding was approximately 23% for Nedd4-2190−581C414, 44% for Nedd4-2190−581C389, C508, and C571, and 53% for Nedd4-2190−581C522 (Fig. 5). These changes are also in line with the results from our fluorescence anisotropy measurements, because the strongest effect of 14-3-3η binding on dansyl rotational mobility was identified in the Nedd4-2190−581C508 mutant (Fig. 4 C) and the weakest in the rigid Nedd4-2190−581C414 (Figure 2, Figure 4 B). In contrast, the Nedd4-2186−975C942 exhibited the opposite behavior, with a 35% increase in kq in the presence of 14-3-3η, matching the decrease in τmean after 14-3-3η binding (Figs. S14 and 5).
14-3-3 protein binding protects the WW3 and WW4 domains of Nedd4-2 from proteolytic degradation in vitro
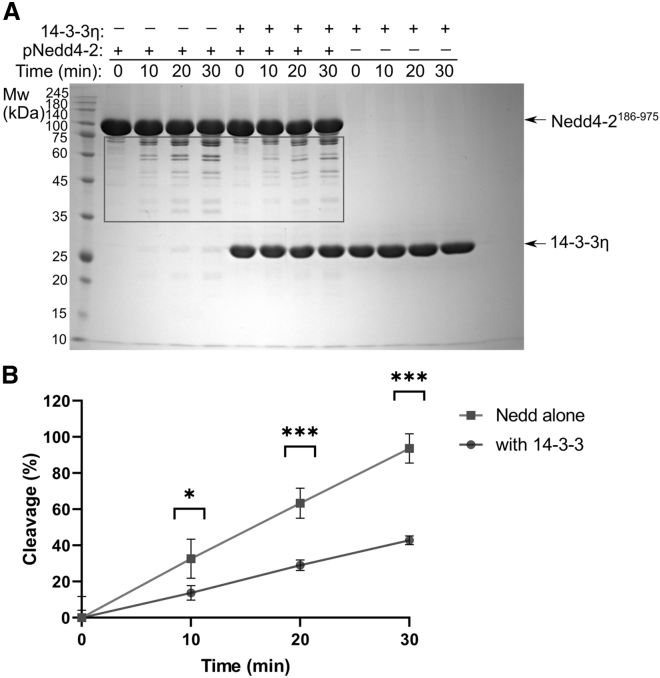
Our previous structural characterization of Nedd4-2186−975 and its complex with 14-3-3η based on SAXS and chemical cross-linking coupled to MS (30), and the fluorescence spectroscopy data gathered in this study, suggest that 14-3-3η protein binding alters interactions between its structured domains, consequently changing their mobility and solvent exposure. To further corroborate these findings, we investigated which Nedd4-2 regions are sensitive to proteolysis in the absence and presence of 14-3-3η. For this purpose, we performed limited proteolysis experiments with trypsin (Figs. 6 A and S15). The results of Nedd4-2 digestion with low trypsin levels revealed that Nedd4-2186−975 alone is highly sensitive to proteolysis, whereas adding 14-3-3η significantly slowed the degradation of Nedd4-2186−975 (Fig. 6 B).
Figure 6.
Limited proteolysis assay showing the protective effect of 14-3-3 to pNedd4-2186−975. (A) Phosphorylated Nedd4-2186−975 in the absence and presence of 14-3-3η digested with trypsin for 10, 20, and 30 min. The protease/Nedd4-2 ratio was 1:1000 (w/w). The reactions were stopped by boiling the samples with SDS/PAGE loading buffer at the times indicated before SDS-PAGE analysis. The black rectangle marks the quantified region. Unedited gel image is provided in supporting material (Fig. S15). (B) The density of the bands representing degraded Nedd4-2186−975 in the presence and absence of 14-3-3η were quantified in ImageLab (Bio-Rad). Error bars represent the standard deviation of four independent experiments. Asterisks indicate significant differences, according to Student's t-tests comparing relative changes between samples with and without 14-3-3η at selected time points (∗p ≤ 0.01, ∗∗∗p ≤ 0.0001).
Mass spectrometry analysis of the Nedd4-2 fragments after 30 min of digestion with trypsin in the absence and presence of 14-3-3η showed that the most sensitive Nedd4-2 region, in both cases, is the N-terminus of our Nedd4-2186−975 construct containing the WW1 and WW2 domains and the phosphorylation sites S342 and T367, because the bands with an apparent Mw ∼60 kDa correspond to the Nedd4-2 sequence 446–975 (the first peptide identified from the N-terminus was peptide 446–462, phosphorylated at S448, with an m/z signal of 1736.86). Peptides containing the preceding phosphorylation sites S342 and T367 (the peptide 339–362 phosphorylated at S342 with an m/z signal of 2383.06 and the peptide 365–394 phosphorylated at T367 with an m/z signal of 3207.44) were detected only in control samples without trypsin. Combined, these data indicate that 14-3-3η binding protects the 14-3-3 binding motif S448 and the WW3-4 and HECT domains at the C-terminus of Nedd4-2 against proteolytic degradation, and are thus in line with the results from our fluorescence analysis (Fig. 4).
Discussion
Nedd4-2 WW domains function as protein interaction modules and participate in a wide range of eukaryotic signaling processes, as shown in numerous studies (50,51). These WW domains of Nedd4-2 bind PY motifs present in ENaC subunits (17) and in SGK1, ACK1, and WNK1 (52, 53, 54). Nedd4-2 contains four WW domains with different sequences. Therefore, each WW domain likely has a specific function.
In this study, we analyzed the domain specificity of Nedd4-2 to two binding partners, namely ENaC and SGK1. The in vitro characterization of the binding of individual domains of Nedd4-2 by surface plasmon resonance demonstrated that WW2 and WW3 bind to SGK1 both individually and cooperatively, whereas only WW3 and WW4 bind to ENaC (17,55,56). The findings of Wiemuth et al. support a model where SGK binds to WW2 and WW3 and then phosphorylates Nedd4-2 and pulls it back from the ENaC (56).
Nedd4-2 phosphorylation triggers the binding of scaffolding 14-3-3 proteins, which further modulate Nedd4-2 function, albeit through mechanisms not yet fully understood (29). Our recently reported structural characterization of the Nedd4-2:14-3-3η complex based on SAXS, cross-linking, and crystallography data suggested that 14-3-3η binding alters interactions between the structured domain of Nedd4-2, thus indicating that complex formation may affect the accessibility of the catalytic HECT and/or individual WW domains (30). To test this hypothesis, we studied the 14-3-3η-induced conformational changes, mobility, and solvent accessibility of individual WW domains and HECT domains of Nedd4-2 by time-resolved dansyl fluorescence.
The data reported here clearly indicate that 14-3-3η directly interacts with WW3 and WW4 domains, as shown by the large increase in dansyl fluorescence lifetime and by the decrease in solvent accessibility and mobility of AEDANS-labeled Cys residues at positions 508, 522, and 571 (Table 2; Figs. 2, 4 C, and 5). The blue shift of the emission spectra of about 4–5 nm for labeled Cys508 and Cys522 residues and ∼1 nm for Cys571 upon 14-3-3η binding is consistent with this conclusion, Fig. S19. The shift indicates a less polar microenvironment, as shown in Fig. S20 for free AEDANS in water and DMSO. The observed changes can be interpreted as a modulation of the AEDANS microenvironment, mobility, and quencher accessibility due to at least two distinct actions: 1) the 14-3-3η-protein-induced conformational change involving Nedd4-2 WW3 and WW4 domains, and/or 2) the direct contact of the 14-3-3η protein with these labeled cysteine residues and consequent shielding from the polar solvent and restriction of their mobility.
The fluorescence properties of AEDANS-labeled Cys residues at positions 389 and 414 within the WW2 domain were affected by 14-3-3η binding to a lesser extent. The mobility of the WW2 domain, which exhibited the lowest internal mobility of all four WW domains of Nedd4-2 (Fig. 3), was only weakly affected by 14-3-3η binding (Fig. 4 B). Nevertheless, as in the WW3 and WW4 domains, complex formation induced some structural changes near this domain, as indicated by the increase in τmean. The decrease in kq values (Table 2; Figs. 2 and 5) and a slight blue spectral shift (Fig. S19) suggest decreased solvent accessibility of the WW2 domain after 14-3-3η binding.
In contrast, the fluorescent lifetimes and apparent kq of AEDANS-Cys at positions 209 and 218 within the WW1 domain remained unaffected by 14-3-3η binding. This domain exhibits high internal mobility, high collisional quenching of the AEDANS emission, and short emission lifetimes, both in the absence and in the presence of 14-3-3η. Taking together, this indicates that WW1 is relaxed and highly accessible to the solvent (Table 2; Figs. 2, 3, 4 A and 5).
While emission spectrum of AEDANS-C209 is also insensitive to 14-3-3η binding, the appearance of blue-shifted shoulder on the emission spectrum of AEDANS-C218 in the presence of 14-3-3η suggests binding-induced increase of the conformational heterogeneity in this region (Fig. S19). Considering the above, 14-3-3η does not affect considerably the interactions and structure of the WW1 domain, but the 14-3-3η binding-induced conformational change of Nedd4-2 also involves its catalytic domain, as shown by analysis of the fluorescent properties of AEDANS-labeled C942 within the active site of the HECT domain (Table 2; Figs. 2 and 5).Nedd4-2 is key regulator of Na+ transport in mammalian cells (3,57, 58, 59). When cells need to reduce Na+ transport, Nedd4-2 binds to the PY motif of the epithelial sodium channel ENaC and catalyzes its ubiquitination. Conversely, when increased Na+ transport is required, Nedd4-2 is phosphorylated by various kinases, which induces 14-3-3 protein binding and blocks ENaC ubiquitination. As a result, ENaC remains permanently active on the cellular surface (Fig. 7 A) (reviewed in (58)).
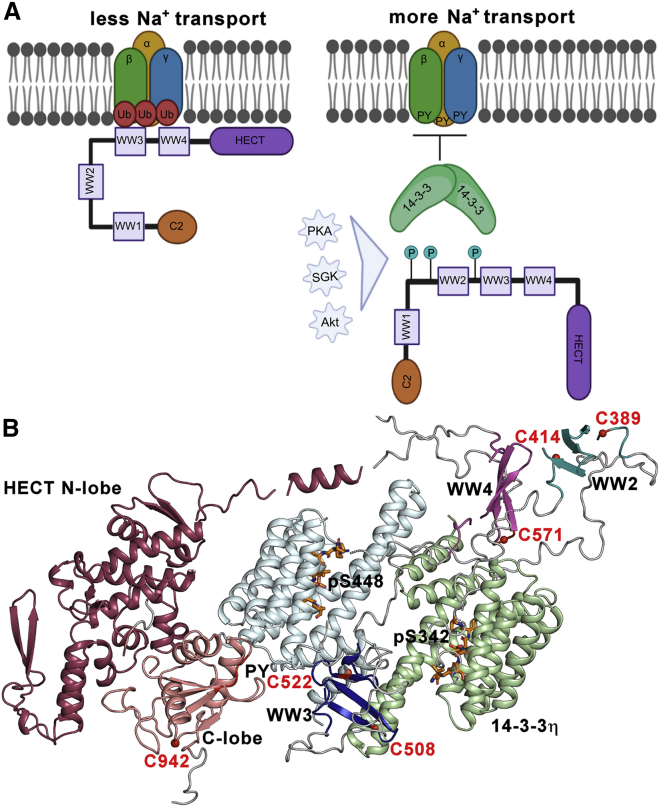
Figure 7.
Phosphorylation followed by 14-3-3 binding regulates Nedd4-2. (A) Nedd4-2 binds to ENaC PY motifs and catalyzes its ubiquitination, reducing the rate of sodium transport and consequently the surface expression of these channels. Phosphorylation by various kinases (PKA, SGK, and Akt) triggers 14-3-3 protein binding, which sterically blocks WW domains and affects the structure of the active site, thereby preventing Nedd4-2 binding to ENaC and its ubiquitination. The 14-3-3 binding motifs S342, T367, and S448 are shown as teal circles with P. (B) Positions of AEDANS-labeled cysteine residues in the SAXS-based model of the pNedd4-2186−975:14-3-3η complex (30). The 14-3-3η protomers are shown in pale green and pale cyan. In the HECT domain of Nedd4-2186−975, the N-lobe is indicated in raspberry and the C-lobe in salmon. WW2, WW3, and WW4 domains are indicated in teal, blue, and magenta, respectively. Phosphorylated 14-3-3 binding motifs of Nedd4-2 are shown as orange sticks (PDB: 6ZBT and 6ZC9 (30)). The positions of the cysteine residues in the WW2, WW3, WW4, and HECT domains labeled by AEDANS are indicated as red balls. The WW1 domain is not shown. The PY motif (L948PPY951) is shown in red. To see this figure in color, go online.
Our fluorescence spectroscopy measurements, together with a recent structural analysis of apo Nedd4-2 and the Nedd4-2:14-3-3η complex (30), suggest a plausible mechanism for this 14-3-3-mediated inhibition of Nedd4-2 binding to ENaC (Fig. 7). Models of apo Nedd4-2 and its complex with 14-3-3η indicate that the WW2 and WW3 domains, in the apo form, interact with the HECT domain and that these interactions are disrupted in the presence of 14-3-3η, which apparently sequesters WW3 into the central channel of its dimeric molecule, away from the HECT domain (Fig. 7 B). This model of the complex also suggests interactions of 14-3-3η with WW4 and HECT domains.
In line with these models, our time-resolved fluorescence measurements (Figs. 2, 3, 4, and 5), together with the results of limited proteolysis (Fig. 6) reported in this study, confirm the interactions between 14-3-3η and WW3 and WW4 domains and the structural change within the HECT domain upon complex formation. Moreover, our time-resolved fluorescence anisotropy and quenching measurements show that complex formation decrease the mobility and solvent accessibility of WW2-4, most likely because 14-3-3η binding sterically masks these domains. Therefore, this physical obstruction of WW domains may be responsible for the 14-3-3-dependent modulation of Nedd4-2 functions.
In a second modulatory mechanism, 14-3-3 directly interacts with the HECT domain, changes the relative positions of the N- and C-lobes of the HECT domain (30), and thus may also affect the catalytic activity of this domain. Indeed, our experiments revealed that the AEDANS moiety attached to C942, which is a catalytic residue (60), exhibits a somewhat lower τmean and a considerably higher accessibility to the solvent in the complexed form than in the apo form (Figs. 2 and 5), thus indicating a structural change within the catalytic core of the enzyme.
In conclusion, the fluorescence spectroscopy analysis of the Nedd4-2:14-3-3η complex reported in this study indicates that the steric hindrance of the WW3 and WW4 domains, together with the conformational change in the catalytic domain, may be responsible for the 14-3-3 binding-mediated regulation of Nedd4-2 functions. Moreover, our data provide a platform for future studies targeting the 14-3-3:Nedd4-2 interface for potential therapeutic purposes for treating Nedd4-2-related diseases, such as Parkinson and kidney disease, hypertension, etc. Recent advances in the development of small-molecule compounds that stabilize protein-protein interactions in 14-3-3 protein complexes have demonstrated the feasibility of this approach. For example, Ottmann's group recently identified compounds able to stabilize complexes between 14-3-3 and the adaptor protein SLP76 (61) or the p65 subunit of NF-κB (62). Our finding that Nedd4-2 interacts with 14-3-3η not only through phosphorylated motifs, but also other regions, including WW3 and WW4 domains, suggests the presence of a sufficiently large binding interface that could be targeted by small-molecule compounds as an alternative or complimentary strategy in suppressing the activity of Nedd4-2.
Author contributions
V.O. and T.O. conceived the study and provided scientific guidance. R.J. performed mutagenesis, prepared the recombinant proteins, performed protein labeling by 1,5-IAEDANS, the differential scanning fluorimetry measurements, and limited proteolysis, in addition to preparing samples for fluorescence measurements. P.P. prepared the recombinant proteins. D.S. and P.H. performed the fluorescence measurements and analyzed the fluorescence data. V.O. and T.O. wrote the paper. All co-authors revised the manuscript.
Acknowledgments
This study was funded by the Czech Science Foundation (to V.O., grant no. 20-00058S), the Grant Agency of Charles University (to R.J., grant no. 348421), the Czech Academy of Sciences (to R.V.O. grant no. 67985823 of the Institute of Physiology). We thank the Czech Infrastructure for Integrative Structural Biology (CIISB) for access to the CMS facilities at BIOCEV (project LM2018127 by MEYS). P.H. and D.S. acknowledge the institutional support UNCE/SCI/010. We also thank Dr. Petr Pompach for mass spectrometry analysis and Dr. Carlos V. Melo for proofreading the article.
Editor: Samrat Mukhopadhyay.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.02.025.
Contributor Information
Tomas Obsil, Email: obsil@natur.cuni.cz.
Veronika Obsilova, Email: veronika.obsilova@fgu.cas.cz.
Supporting material
References
- 1.Yang B., Kumar S. Nedd4 and Nedd4-2: closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ. 2010;17:68–77. doi: 10.1038/cdd.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itani O.A., Campbell J.R., et al. Thomas C.P. Alternate promoters and variable splicing lead to hNedd4-2 isoforms with a C2 domain and varying number of WW domains. Am. J. Physiol. Ren. Physiol. 2003;285:F916–F929. doi: 10.1152/ajprenal.00203.2003. [DOI] [PubMed] [Google Scholar]
- 3.Fotia A.B., Ekberg J., et al. Kumar S. Regulation of neuronal voltage-gated sodium channels by the ubiquitin-protein ligases Nedd4 and Nedd4-2. J. Biol. Chem. 2004;279:28930–28935. doi: 10.1074/jbc.M402820200. [DOI] [PubMed] [Google Scholar]
- 4.Arroyo J.P., Lagnaz D., et al. Staub O. Nedd4-2 modulates renal Na+-Cl- cotransporter via the aldosterone-SGK1-Nedd4-2 pathway. J. Am. Soc. Nephrol. 2011;22:1707–1719. doi: 10.1681/ASN.2011020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamynina E., Debonneville C., Bens M., Vandewalle A., Staub O. A novel mouse Nedd4 protein suppresses the activity of the epithelial Na+ channel. FASEB J. 2001;15:204–214. doi: 10.1096/fj.00-0191com. [DOI] [PubMed] [Google Scholar]
- 6.Ekberg J., Schuetz F., et al. Adams D.J. Regulation of the voltage-gated K(+) channels KCNQ2/3 and KCNQ3/5 by ubiquitination. Novel role for Nedd4-2. J. Biol. Chem. 2007;282:12135–12142. doi: 10.1074/jbc.M609385200. [DOI] [PubMed] [Google Scholar]
- 7.Zhu J., Lee K.Y., et al. Tsai N.P. Epilepsy-associated gene Nedd4-2 mediates neuronal activity and seizure susceptibility through AMPA receptors. PLoS Genet. 2017;13:e1006634. doi: 10.1371/journal.pgen.1006634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broix L., Jagline H., et al. Chelly J. Mutations in the HECT domain of NEDD4L lead to AKT-mTOR pathway deregulation and cause periventricular nodular heterotopia. Nat. Genet. 2016;48:1349–1358. doi: 10.1038/ng.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popovic D., Vucic D., Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014;20:1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 10.Rizzo F., Staub O. NEDD4-2 and salt-sensitive hypertension. Curr. Opin. Nephrol. Hypertens. 2015;24:111–116. doi: 10.1097/MNH.0000000000000097. [DOI] [PubMed] [Google Scholar]
- 11.Vanli-Yavuz E.N., Ozdemir O., et al. Baykan B. Investigation of the possible association of NEDD4-2 (NEDD4L) gene with idiopathic photosensitive epilepsy. Acta Neurol. Belg. 2015;115:241–245. doi: 10.1007/s13760-014-0412-x. [DOI] [PubMed] [Google Scholar]
- 12.Corbalan-Garcia S., Gomez-Fernandez J.C. Signaling through C2 domains: more than one lipid target. Biochim. Biophys. Acta. 2014;1838:1536–1547. doi: 10.1016/j.bbamem.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Bork P., Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem. Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 14.Chen H.I., Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl. Acad. Sci. U S A. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Todaro D.R., Augustus-Wallace A.C., et al. Haas A.L. The mechanism of neural precursor cell expressed developmentally down-regulated 4-2 (Nedd4-2)/NEDD4L-catalyzed polyubiquitin chain assembly. J. Biol. Chem. 2017;292:19521–19536. doi: 10.1074/jbc.M117.817882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.French M.E., Klosowiak J.L., et al. Hunter T. Mechanism of ubiquitin chain synthesis employed by a HECT domain ubiquitin ligase. J. Biol. Chem. 2017;292:10398–10413. doi: 10.1074/jbc.M117.789479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fotia A.B., Dinudom A., et al. Kumar S. The role of individual Nedd4-2 (KIAA0439) WW domains in binding and regulating epithelial sodium channels. FASEB J. 2003;17:70–72. doi: 10.1096/fj.02-0497fje. [DOI] [PubMed] [Google Scholar]
- 18.Harvey K.F., Dinudom A., et al. Kumar S. All three WW domains of murine Nedd4 are involved in the regulation of epithelial sodium channels by intracellular Na+ J. Biol. Chem. 1999;274:12525–12530. doi: 10.1074/jbc.274.18.12525. [DOI] [PubMed] [Google Scholar]
- 19.Snyder P.M., Olson D.R., et al. Bucher D.B. Multiple WW domains, but not the C2 domain, are required for inhibition of the epithelial Na+ channel by human Nedd4. J. Biol. Chem. 2001;276:28321–28326. doi: 10.1074/jbc.M011487200. [DOI] [PubMed] [Google Scholar]
- 20.Bruce M.C., Kanelis V., et al. Rotin D. Regulation of Nedd4-2 self-ubiquitination and stability by a PY motif located within its HECT-domain. Biochem. J. 2008;415:155–163. doi: 10.1042/BJ20071708. [DOI] [PubMed] [Google Scholar]
- 21.Zhang W., Na T., et al. Peng J.B. Down-regulation of intestinal apical calcium entry channel TRPV6 by ubiquitin E3 ligase Nedd4-2. J. Biol. Chem. 2010;285:36586–36596. doi: 10.1074/jbc.M110.175968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichimura T., Yamamura H., et al. Isobe T. 14-3-3 proteins modulate the expression of epithelial Na+ channels by phosphorylation-dependent interaction with Nedd4-2 ubiquitin ligase. J. Biol. Chem. 2005;280:13187–13194. doi: 10.1074/jbc.M412884200. [DOI] [PubMed] [Google Scholar]
- 23.Snyder P.M., Olson D.R., et al. Steines J.C. cAMP and serum and glucocorticoid-inducible kinase (SGK) regulate the epithelial Na(+) channel through convergent phosphorylation of Nedd4-2. J. Biol. Chem. 2004;279:45753–45758. doi: 10.1074/jbc.M407858200. [DOI] [PubMed] [Google Scholar]
- 24.Bhalla V., Daidie D., et al. Pearce D. Serum- and glucocorticoid-regulated kinase 1 regulates ubiquitin ligase neural precursor cell-expressed, developmentally down-regulated protein 4-2 by inducing interaction with 14-3-3. Mol. Endocrinol. 2005;19:3073–3084. doi: 10.1210/me.2005-0193. [DOI] [PubMed] [Google Scholar]
- 25.Lee I.H., Dinudom A., et al. Cook D.I. Akt mediates the effect of insulin on epithelial sodium channels by inhibiting Nedd4-2. J. Biol. Chem. 2007;282:29866–29873. doi: 10.1074/jbc.M701923200. [DOI] [PubMed] [Google Scholar]
- 26.Hallows K.R., Bhalla V., et al. Pearce D. Phosphopeptide screen uncovers novel phosphorylation sites of Nedd4-2 that potentiate its inhibition of the epithelial Na+ channel. J. Biol. Chem. 2010;285:21671–21678. doi: 10.1074/jbc.M109.084731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edinger R.S., Lebowitz J., et al. Hallows K.R. Functional regulation of the epithelial Na+ channel by IkappaB kinase-beta occurs via phosphorylation of the ubiquitin ligase Nedd4-2. J. Biol. Chem. 2009;284:150–157. doi: 10.1074/jbc.M807358200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagaki K., Yamamura H., et al. Ichimura T. 14-3-3 Mediates phosphorylation-dependent inhibition of the interaction between the ubiquitin E3 ligase Nedd4-2 and epithelial Na+ channels. Biochemistry. 2006;45:6733–6740. doi: 10.1021/bi052640q. [DOI] [PubMed] [Google Scholar]
- 29.Chandran S., Li H., et al. Bhalla V. Neural precursor cell-expressed developmentally down-regulated protein 4-2 (Nedd4-2) regulation by 14-3-3 protein binding at canonical serum and glucocorticoid kinase 1 (SGK1) phosphorylation sites. J. Biol. Chem. 2011;286:37830–37840. doi: 10.1074/jbc.M111.293233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pohl P., Joshi R., et al. Obsilova V. 14-3-3-protein regulates Nedd4-2 by modulating interactions between HECT and WW domains. Commun. Biol. 2021;4:899. doi: 10.1038/s42003-021-02419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 2004;381(Pt 2):329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Obsilova V., Obsil T. The 14-3-3 proteins as important allosteric regulators of protein kinases. Int. J. Mol. Sci. 2020;21:8824. doi: 10.3390/ijms21228824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sluchanko N.N. Association of multiple phosphorylated proteins with the 14-3-3 regulatory hubs: problems and perspectives. J. Mol. Biol. 2018;430:20–26. doi: 10.1016/j.jmb.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Gogl G., Tugaeva K.V., et al. Sluchanko N.N. Hierarchized phosphotarget binding by the seven human 14-3-3 isoforms. Nat. Commun. 2021;12:1677. doi: 10.1038/s41467-021-21908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sluchanko N.N., Bustos D.M. Intrinsic disorder associated with 14-3-3 proteins and their partners. Prog. Mol. Biol. Transl Sci. 2019;166:19–61. doi: 10.1016/bs.pmbts.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Bustos D.M. The role of protein disorder in the 14-3-3 interaction network. Mol. Biosyst. 2012;8:178–184. doi: 10.1039/c1mb05216k. [DOI] [PubMed] [Google Scholar]
- 37.Horvath M., Petrvalska O., et al. Obsil T. 14-3-3 proteins inactivate DAPK2 by promoting its dimerization and protecting key regulatory phosphosites. Commun. Biol. 2021;4:986. doi: 10.1038/s42003-021-02518-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rotin D., Staub O. Nedd4-2 and the regulation of epithelial sodium transport. Front Physiol. 2012;3:212. doi: 10.3389/fphys.2012.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obsil T., Ghirlando R., et al. Dyda F. Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation. Cell. 2001;105:257–267. doi: 10.1016/s0092-8674(01)00316-6. [DOI] [PubMed] [Google Scholar]
- 40.Obsilova V., Herman P., et al. Obsil T. 14-3-3zeta C-terminal stretch changes its conformation upon ligand binding and phosphorylation at Thr232. J. Biol. Chem. 2004;279:4531–4540. doi: 10.1074/jbc.M306939200. [DOI] [PubMed] [Google Scholar]
- 41.Boura E., Silhan J., et al. Obsil T. Both the N-terminal loop and wing W2 of the forkhead domain of transcription factor Foxo4 are important for DNA binding. J. Biol. Chem. 2007;282:8265–8275. doi: 10.1074/jbc.M605682200. [DOI] [PubMed] [Google Scholar]
- 42.Niesen F.H., Berglund H., Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 43.Ballone A., Centorrino F., et al. Ottmann C. Protein X-ray crystallography of the 14-3-3zeta/SOS1 complex. Data Brief. 2018;19:1683–1687. doi: 10.1016/j.dib.2018.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kacirova M., Kosek D., et al. Obsil T. Structural characterization of phosducin and its complex with the 14-3-3 protein. J. Biol. Chem. 2015;290:16246–16260. doi: 10.1074/jbc.M115.636563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vecer J., Herman P. Maximum entropy analysis of analytically simulated complex fluorescence decays. J. Fluorescence. 2011;21:873–881. doi: 10.1007/s10895-009-0589-1. [DOI] [PubMed] [Google Scholar]
- 46.Lakowicz J.R. Springer; 2006. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 47.Lakowicz J.R. Plenum Press; 1983. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 48.Lehrer S.S. Solute perturbation of protein fluorescence. The quenching of the tryptophyl fluorescence of model compounds and of lysozyme by iodide ion. Biochemistry. 1971;10:3254–3263. doi: 10.1021/bi00793a015. [DOI] [PubMed] [Google Scholar]
- 49.Yong W., Tsukasa I., et al. Fujio T. Dansyl-β-cyclodextrins as fluorescent sensors responsive to organic compounds. Bull. Chem. Soc. Jpn. 1994;67:1598–1607. [Google Scholar]
- 50.Sudol M., Chen H.I., et al. Bork P. Characterization of a novel protein-binding module--the WW domain. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 51.Staub O., Rotin D. WW domains. Structure. 1996;4:495–499. doi: 10.1016/s0969-2126(96)00054-8. [DOI] [PubMed] [Google Scholar]
- 52.Snyder P.M., Olson D.R., Thomas B.C. Serum and glucocorticoid-regulated kinase modulates Nedd4-2-mediated inhibition of the epithelial Na+ channel. J. Biol. Chem. 2002;277:5–8. doi: 10.1074/jbc.C100623200. [DOI] [PubMed] [Google Scholar]
- 53.Chan W., Tian R., et al. Manser E. Down-regulation of active ACK1 is mediated by association with the E3 ubiquitin ligase Nedd4-2. J. Biol. Chem. 2009;284:8185–8194. doi: 10.1074/jbc.M806877200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roy A., Al-Qusairi L., et al. Subramanya A.R. Alternatively spliced proline-rich cassettes link WNK1 to aldosterone action. J. Clin. Invest. 2015;125:3433–3448. doi: 10.1172/JCI75245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Asher C., Sinha I., Garty H. Characterization of the interactions between Nedd4-2, ENaC, and sgk-1 using surface plasmon resonance. Biochim. Biophys. Acta. 2003;1612:59–64. doi: 10.1016/s0005-2736(03)00083-x. [DOI] [PubMed] [Google Scholar]
- 56.Wiemuth D., Lott J.S., et al. McDonald F.J. Interaction of serum- and glucocorticoid regulated kinase 1 (SGK1) with the WW-domains of Nedd4-2 is required for epithelial sodium channel regulation. PLoS One. 2010;5:e12163. doi: 10.1371/journal.pone.0012163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhalla V., Hallows K.R. Mechanisms of ENaC regulation and clinical implications. J. Am. Soc. Nephrol. 2008;19:1845–1854. doi: 10.1681/ASN.2008020225. [DOI] [PubMed] [Google Scholar]
- 58.Snyder P.M. Down-regulating destruction: phosphorylation regulates the E3 ubiquitin ligase Nedd4-2. Sci. Signal. 2009;2:pe41. doi: 10.1126/scisignal.279pe41. [DOI] [PubMed] [Google Scholar]
- 59.Manning J.A., Kumar S. Physiological functions of Nedd4-2: lessons from knockout mouse models. Trends Biochem. Sci. 2018;43:635–647. doi: 10.1016/j.tibs.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 60.Wu P.Y., Hanlon M., et al. Pickart C.M. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003;22:5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soini L., Redhead M., et al. Ottmann C. Identification of molecular glues of the SLP76/14-3-3 protein-protein interaction. RSC Med. Chem. 2021;12:1555–1564. doi: 10.1039/d1md00172h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolter M., Valenti D., et al. Ottmann C. An exploration of chemical properties required for cooperative stabilization of the 14-3-3 interaction with NF-kappaB-Utilizing a reversible covalent tethering approach. J. Med. Chem. 2021;64:8423–8436. doi: 10.1021/acs.jmedchem.1c00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bryan R.K. Maximum entropy analysis of oversampled data problems. Eur. Biophys. J. 1990;18:165–174. [Google Scholar]
- 64.Marquardt D.W. An algorithm for least-squares estimation of Nonlinear parameters. J. Soc. Ind. Appl. Math. 1963;11:431–441. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.