Abstract
Get3/4/5 chaperone complex is responsible for targeting C-terminal tail-anchored membrane proteins to the endoplasmic reticulum. Despite the availability of several crystal structures of independent proteins and partial structures of subcomplexes, different models of oligomeric states and structural organization have been proposed for the protein complexes involved. Here, using native mass spectrometry (Native-MS), coupled with intact dissociation, we show that Get4/5 exclusively forms a tetramer using both Get5/5 and a novel Get4/4 dimerization interface. Addition of Get3 to this leads to a hexameric (Get3)2-(Get4)2-(Get5)2 complex with closed-ring cyclic architecture. We further validate our claims through molecular modeling and mutational abrogation of the proposed interfaces. Native-MS has become a principal tool to determine the state of oligomeric organization of proteins. The work demonstrates that for multiprotein complexes, native-MS, coupled with molecular modeling and mutational perturbation, can provide an alternative route to render a detailed view of both the oligomeric states as well as the molecular interfaces involved. This is especially useful for large multiprotein complexes with large unstructured domains that make it recalcitrant to conventional structure determination approaches.
Significance
Despite years of research, molecular architecture and the oligomeric organization of the multiprotein GET chaperone complexes remained elusive and controversial. This is owing to the large unstructured region in Get5, which has proven prohibitively difficult toward structural studies involving the full-length proteins. Addressing this, we have used native mass spectrometry, combined with intact dissociation study of the full-length intact multimeric complexes. Our data unambiguously show that both Get4/5 and Get3/4/5 form cyclic ring-like molecular complexes with heterotetrameric and heterohexameric stoichiometries, respectively. Our work also demonstrates that for multiprotein complexes where inherent conformational dynamics preclude structure determination, native-MS and intact dissociation, in combination with integrative molecular modeling, provide a fantastic alternate avenue to obtain structural and mechanistic insights.
Introduction
From regulating membrane trafficking to the release of neurotransmitters at the neuronal synapses, C-terminal tail-anchored membrane proteins (TAPs) play a central role in eukaryotic biology (1, 2, 3, 4). Trafficking of TAPs to their target physiological membrane follows the usual anterograde trafficking route from the endoplasmic reticulum (ER), where they are first inserted. Nevertheless, unlike other membrane proteins, their insertion into the ER membrane is not mediated via the signal recognition particle pathway (5,6). Instead, a significant portion of TAPs, with hydrophobic C-terminal helix, get inserted into the ER membrane via the GET pathway (TRC pathway in mammals) (7, 8, 9, 10, 11, 12). Here, the nascent TAP gets transferred from the ribosome to an upstream chaperone Sgt2 via cytosolic Hsp70 with the help of J-domain proteins (13). Subsequently, the substrate-bound Sgt2 transfers the TAP to the downstream chaperone Get3, with the help of two other proteins, Get4 and Get5 (14, 15, 16, 17, 18). Finally, TAP-bound Get3 goes to the ER membrane and delivers the substrate with the help of ER receptor Get1 and Get2 (11,19) (19,20). Impairment in the trafficking of the TAPs has been directly implicated in various pathological conditions, such as pediatric cardiomyopathy (21) and Emery-Dreifuss muscular dystrophy (22). Together, this places paramount significance in understanding the mechanism of TAP trafficking by the Get pathway chaperones. Despite the full-length structure of Get3 (23, 24, 25), and partial structures of Get4-Get5(N-term) (26) and Get3-Get4-Get5(N-term) complexes (14,27), the hierarchical structural organization of Get3-Get4-Get5 and how that controls the fidelity of the substrate transfer to Get3 has remained unanswered. Both the oligomeric state of the Get3/4/5 complex, as well as its structural organization has remained controversial (for different proposed models, see Refs. 3,4,14,27,28). This is mostly owing to the large unstructured region in Get5, which resulted in structural studies performed on truncated Get5 that does not contain the C-term dimerization domain (28). This has resulted in different possible oligomeric architecture being proposed for the GET pathway protein assemblies (3,14,27, 28, 29).
Over the last decade, native mass spectrometry (native-MS) has emerged as a flagship technique to determine the oligomeric states of protein assemblies, with success in detecting protein complexes as large as the entire ribosome and as dynamic as the heat shock protein (HSP) chaperone systems (30, 31, 32, 33, 34, 35, 36, 37, 38). Further, using intact dissociation (often referred to as complex-up) (39), subunit stoichiometry of several multiprotein complexes has been unambiguously determined. (40, 41, 42, 43, 44, 45). Here, we show that using native-MS and intact dissociation, beyond stoichiometry, we can further determine interfaces and correct structural organization of multiprotein complexes. The subcomplexes generated through intact MS/MS analysis can report on the assembly interfaces. These interfaces can be further validated through integrative modeling and mutational analysis of key interface residues. Native-MS can be further used to confirm the mutational abrogation of the oligomeric entities, confirming the proposed structural assembly. Here, we used trapping of protein complexes at the front end of the MS, between bent and inject flatapole, to effectively desolvate the multiprotein complexes (46). Subsequently, we used quadrupole isolation and HCD energy to dissociate the target protein complex in subcomplexes.
Applying such approaches on the GET pathway proteins, we show that Get4/5 exists as a tetrameric complex and has a novel Get4/4 interface. We subsequently discovered that Get3/4/5 exists exclusively as a hexameric complex with cyclic architecture. We further substantiate our findings through molecular dynamic simulation and mutational abrogation of the proposed assemblies and identified interfaces. Together, our data reveal the structural organizations of Get4/5 and Get3/4/5 complex and indicate how Get4/5 maintains the fidelity of access of the transmembrane binding hydrophobic cavity of Get3 by forming a closed-ring cyclic structure around it.
Although the use of native-MS and subsequent MS activation is slowly becoming routine in determining subunit stoichiometry of macromolecular protein assemblies, our integrative approach shows that native-MS can also be used to unambiguously determine the three-dimensional structural organization of complexes, as well as identify possible novel interfaces.
Materials and methods
Genetic manipulation
The Get5∗ truncated variant (Δ138–212) was generated with PCR-based site-directed mutagenesis. Plasmid Get5-pCDF1b (23) was used as a template. The primers (F: TAATCTCGAGTCTGGTAAAGAAACCGC and R: AGTGAGTTCCTGTGGAGGAGCAGG) were designed to create Get5 gene coding truncated Get5 protein without the last 36 amino acids, which formed Get5/5 dimerization domain in full-length protein (29).
Protein expression and purification
6xHIS-TEV site-Get3-pET28 (24) was transformed into Escherichia coli Rosetta DE3 pLys. 6xHIS-Tev site-Get5-pCDF1b (23) or 6xHIS-Tev site-Get5∗-pCDF1b was co-transformed with Get4-pET28 (23) into E. coli Rosetta DE3 pLys. The bacteria with mentioned constructs were grown overnight at 37°C, then the culture was refreshed to OD 0.1 and grew till OD reached between 0.5 and 0.7. The protein expression was induced with 0.2 mM IPTG. After the induction, the bacteria were grown for 6 h at 25°C. The frozen pellets of the bacteria were thawed in the His-tag wash buffer containing 50 mM Tris-Cl, 500 mM NaCl, 50 mM imidazole, 10% glycerol, and Pierce Protease Inhibitors Mini Tablets (pH 8.0). After thawing the cells were lysed with Emulsiflex C3 (Avestin). The protein extract was incubated with HisPur Ni-NTA Superflow (Thermo-Fisher Scientific) on a shaker for 2 h at 4°C. Then the beads were washed four times with His-tag wash buffer (20x beads volume) (washing: mix, spin 2 min 2000 g, discharge supernatant) and one time with TEV protease buffer (20x beads volume): 20 mM Tris-Cl, 150 mM Na Cl, 1mM DTT (pH 8.0). Next, the sample was incubated with TEV protease overnight at 4°C. After the incubation, the sample was loaded on the gravity column and the flow-through with Get4/5 was collected. The proteins were stored at −80°C in TEV protease buffer with 20% glycerol. Get3 was purified in the same way till the washing step. After that, the beads were loaded on the gravity column and Get3 was eluted with the His-tag elution buffer: 50 mM Tris-Cl, 500 mM NaCl, 500 mM imidazole, 10% glycerol (pH 8.0). Then the PD-10 desalting columns (Bio-Rad) were used to transfer the protein to the storage buffer: 20 mM Tris-Cl, 150 mM Na Cl, 1mM DTT, 20 %glycerol (pH 8.0). In this buffer, Get3 was stored at −80°C.
To express the human TRC40-TEV-HA, HEK293 cells on one 10-cm2 plate were transfected with 12 μg plasmid and 30 μg Lipofectamine 2000. 24 h after transfection, the cells were scraped in 10 mL of PBS buffer and centrifuged for 5 min at 2,000 g. To avoid the release of free lipids from membranes, the pellet was resuspended in 1 mL of hypotonic buffer: 10 mM Hepes (pH 7.4), 2 mM MgCl2, 10% glycerol, and 1x protease inhibitor (Pierce Protease Inhibitor Tablets) and passing 15 times via 21-gauge needle. Next, the buffer was adjusted to 50 mM Hepes (pH 7.4), 300 mM NaCl, 2 mM MgCl2, 10% glycerol, and 1x protease inhibitor. After centrifugation at 70,000 rpm for 30 min in TLA 100.3, the supernatant was incubated for 2 h with mouse anti-HA magnetic beads (Thermo Scientific). After washing the beads with 50 mM Hepes (pH 7.4), 300 mM NaCl, 2 mM MgCl2, 10% glycerol, they were incubated overnight at 4°C with TEV protease in buffer: 20 mM Tris-Cl (pH 8.0), 100 mM NaCl, 2 mM MgCl2, 1 mM DTT. The supernatant containing TRC40 was then separated from the beads with the usage of a magnetic rack.
In vitro reconstitution of Get3/4/5 complex
The Get3/4/5 complex was reconstituted in vitro (according to Ref. (27)) with some modification. Zeba Spin Desalting Columns (Thermo-Fisher Scientific) were used to transfer Get3 and Get4/5 into the buffer: 20 mM Tris-Cl, 10 mM NaCl, 2 mm DTT (pH 8.0). Next 50 μL of Get3 at concentration 50 μM was incubated with 1 mM ATP and 1 mM MgCl2 for 5 min at room temperature. Then the mixture was mixed with 50 μL of Get4/5 at a concentration of 25 μM. The sample was incubated on ice for 1 h to allow the complex formation to happen.
Mass spectrometry analysis
All the proteins or protein complexes were buffer exchanged to 200 mM ammonium acetate (MP Biomedicals), 2 mM DTT with Zeba Spin Desalting Columns (Thermo-Fisher Scientific). The protein concentration in the analyzed sample was in the range between 5 μM and 10 μM. Native-MS was performed on Q Exactive UHMR (Thermo-Fisher Scientific) using in-house nano-emitter capillaries. The tips in the capillaries were formed by pulling borosilicate glass capillaries (OD: 1.2 mm, ID: 0.69 mm, length: 10 cm, Sutter Instruments) using a Flaming/Brown micropipette puller (ModelP-1000, Sutter Instruments). Then the nano-emitters were coated with gold using rotary pumped coater Q150R Plus (Quorum Technologies). To perform the measurement the emitter filled with the sample was installed into Nanospray Flex Ion Source (Thermo-Fisher Scientific). MS parameters for the analysis of the proteins or protein complexes include spray voltage 1.3–1.5 kV, capillary temperature 275°C, resolving power 6250 (for Get3 dimer) and 3125 (for the rest of the MS analysis) at m/z of 400, ultrahigh vacuum pressure 4.6e-10–8.18e-10, in-source trapping between −100 V and −300 V. For the MS/MS analysis the main charge state of Get4/5 and Get4/5∗ was isolated in quadrupole and fragmented in HCD cell. During MS/MS, the HCD energy was set to 100 V or 200 V. To perform the HCD dissociation of the GET3/4/5 complex, first the spectra were obtained with the in-source trapping energy of −300 V and with the HCD cell set at –1 V. Next, the 29+ charge state of the complex was quadrupole isolated. Finally, this isolated charge state was dissociated by increasing the HCD to 250 V. MS/MS analysis of Get4/5, Get4/5∗, and GET3/4/5 was performed at resolving power 1563 at m/z 400. Nitrogen was used as the collisional gas. Data were visualized with the Xcalibur software and analyzed using UniDec (47) and assembled into figures using Adobe Illustrator.
Structural modeling of the Get3/4/5 complex
Structural information on the Get3/4/5 complex with full-length Get5 is not available. However, crystal structures for Get3/Get4/Get51-54 with N-terminal part of Get5 (residues 1–54) exist. We stitched together the structure of Get3/Get4/Get51-54 with the model of full-length Get5 to build a structural model for the full-length GET3/4/5 complex using homology modeling (MODELLER (48)) and manual modeling (Coot (49)). The structure of Get3/4 in complex with an N-terminal fragment of Get5 (residue number 1–54 in Q12285) is available (PDB: 4PWX). This structure reveals a Get3 dimer, with the one subunit of Get4 attached to each Get3 subunit (Get32Get42). The C-terminal domain of Get5 (residue number 177–212 in Q12285; PDB: 3VEJ) forms a homodimer in the crystal structure. In addition, the structure of the ubiquitin-like domain (Ubl) of Get5 (residue number 70–152 in Q12285; PDB: 4A20) is also available. We built the full-length Get5 structure by homology modeling using MODELER (48) and the N-terminal fragment from 4PWX, the Ubl domain from 4A20, and the C-terminal domain from 3VEJ as templates. The unstructured linker regions between these three domains were built and refined using MODELLER. These full-length Get5 models were then superposed on 4PWX using the two N-terminal fragments of Get5. The dimeric interface between the C-terminal domain of Get5 is modeled based on the crystal structure 3VEJ. The closed-ring hexameric Get3/4/5 model was obtained by manually refining the unstructured linker connecting the Get5 Ubl and C-terminal domains in using Coot (49). This model was used as a starting structure for successive rounds of energy minimization and molecular dynamics simulations.
All-atom molecular dynamics simulations
The molecular dynamics (MD) trajectories were generated using Amber18 (50). The MD simulations were performed on Extreme Science and Engineering Discovery Environment (XSEDE) using the Comet-GPU nodes (51). The ff14SB (52) force field was used for all the calculations. MD simulations were carried out in water using the TIP3P water model, and counter ions (Na+ in this case) were added to neutralize the net charges on the system. Energy minimization was performed in three steps. The water molecules were minimized first with the protein held fixed. The entire system was then minimized without any restraints. After initial energy minimization, the system was subjected to two rounds of 500 ps constant number, volume, and temperature equilibration, during which the system was heated to 300 K. This was followed by two short equilibrations at a constant number, pressure, and temperature (NPT) for 1 ns each. Finally, the production simulations were performed under constant NPT conditions, with the Langevin thermostat in Amber18, in the absence of any positional restraints. Periodic boundary conditions were imposed, and particle-mesh Ewald summations were used for long-range electrostatics. A time step of 2 fs was employed and the structures were stored every 10 ps.
Results
The architecture of the Get4-Get5 complex
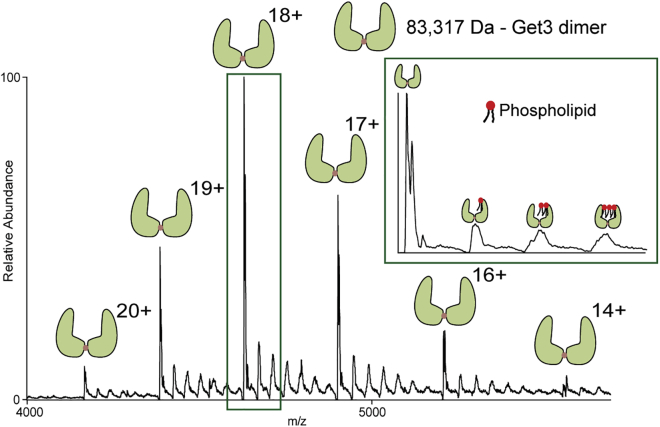
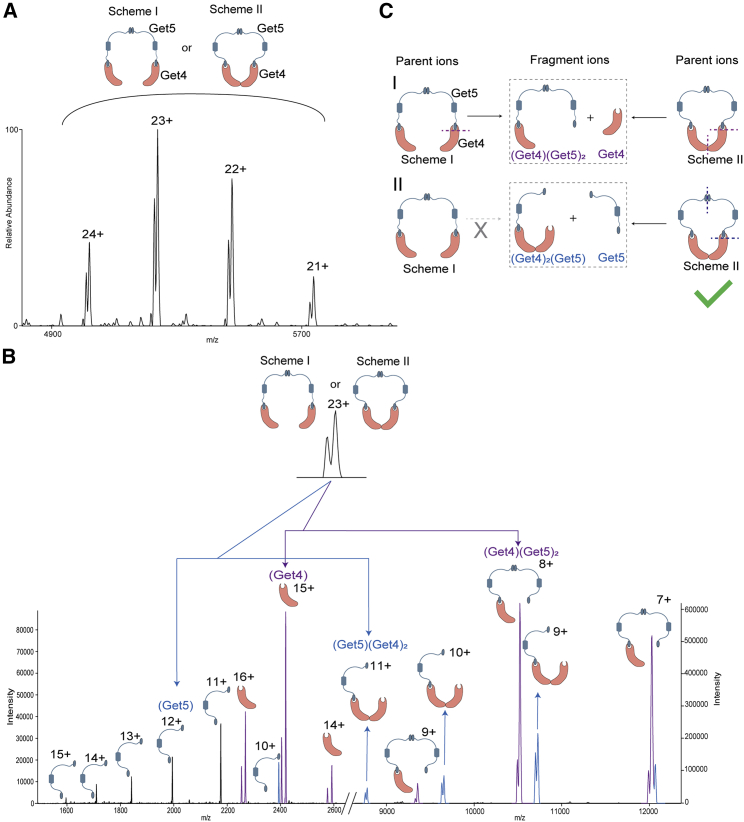
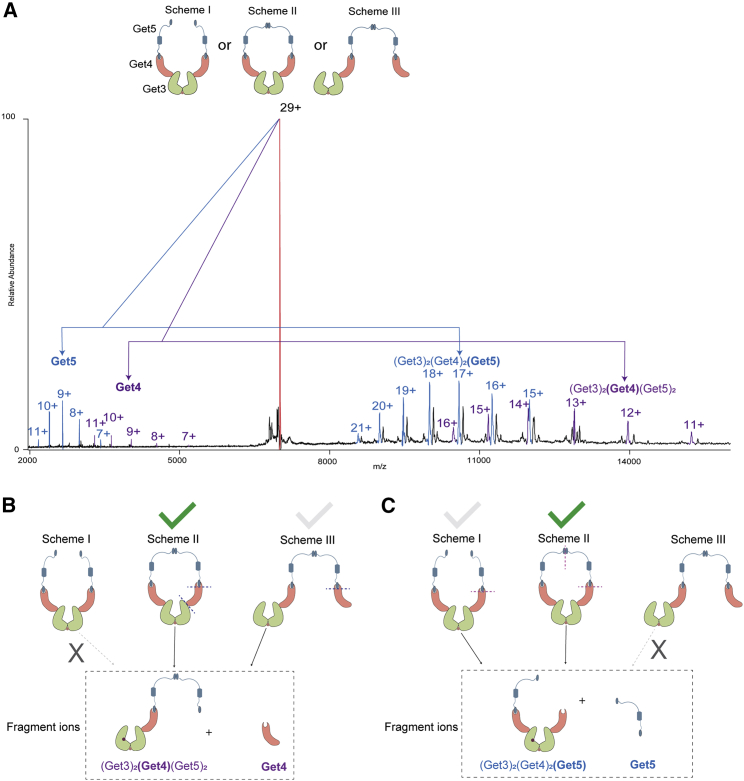
Our initial experiments on Get3 show the presence of dimer (Fig. 1), with the mass matching with that of the zinc-bound dimer. Subsequently, we co-expressed full-length Get4 and Get5 and purified them as a complex. Native-MS analysis of the complex revealed an exclusively tetrameric species with (Get4)2-(Get5)2 stoichiometry (Fig. 2 A). Based on the previous studies using the C-terminal dimerization domain of Get5 (29), it was presumed that any tetramerization in Get4/5 complex must be through dimerization of the Get4/5 heterodimer through the Get5/5 interface (Fig. 2 A, Scheme I). But our MS/MS dissociation of the complex led to the discovery of a new interface, indicating different structural organization (Fig. 2 A, Scheme II).
Figure 1.
Mass spectrum of Get3 dimer. Get3 dimer stabilized by the zinc ion: beige dot between Get3 monomers. Nonspecific bindings of endogenous lipids to Get3 dimers are visible as the six small peaks between main peaks with assigned charge states. Since Get3 harbors a hydrophobic transmembrane (TM) binding groove, it is perceivable that overexpressing it in E. coli can lead to nonspecific capture of lipids. Inset shows the expansion of the m/z region showing Get3 dimer bound to phospholipids. Similar spectra were obtained from three independent protein preparations. To see this figure in color, go online.
Figure 2.
Deciphering the molecular organization of Get4-Get5 complex. (A) Mass spectrum of the Get4-Get5 heterotetramer: (Get4)2(Get5)2. Scheme I and Scheme II show the possible organization of the complex. Scheme I: the complex is formed only via the Get5/5 interface, Scheme II: complex is formed via Get5/5 and Get4/4 interface. The shorter peaks visible in the mass spectrum were formed by the complex with one truncated form of the Get4 lacking the first two residues (Met-Val), and the higher peaks were formed by complex with two full-length Get4. Inset shows the two peaks for the representative 23+ charge state. (B) MS/MS spectra of (Get4)2(Get5)2. CID of the (Get4)2(Get5)2 produced two sets of subcomplexes. Peaks in purple highlight the subcomplexes generated by the loss of a monomeric Get4 unit from the overall complex. The remainder of the trimer Get4-(Get5)2 species was observed at the higher m/z range (purple peaks in the right panel). The lost, monomeric Get4 molecule was detected in the lower m/z region (purple peaks in the left panel). Peaks in blue highlight the subcomplexes generated by the loss of a monomeric Get5 unit from the overall complex. The remainder of the trimer (Get4)2-Get5 species was observed at the higher m/z range (purple peaks in the right panel). The lost, monomeric Get5 molecule was detected in the lower m/z region (blue peaks in the left panel). Similar to panel A, subcomplexes containing Get4 showed a small amount of truncated Get4 containing population. (C) Deciphering the molecular organization of (Get4)2(Get5)2. Ions owing to the subcomplex (Get4) (Get5)2 and Get4 can be produced by CID of parent ions with molecular organization presented in both Schemes I and II. In contrast, ions owing to dissociated subcomplex (Get4)2(Get5) and Get5 can be produced by CID of parent ions with molecular organization presented only in Scheme II. Dotted lines on the structure indicate interaction interfaces dissociated by CID during the generation of daughter ions. Similar spectra were obtained from three independent protein preparations.
Fig. 2B shows the MS activation of the 23+ charge state of the intact Get4/5 tetramer under collision-induced dissociation (CID) conditions. The spectra show two pairs of subcomplexes derived through the dissociation of the intact tetramer. The pair of ions highlighted in purple originates through the dissociation of the Get4/5 interface that yields a stripped subunit of Get4 and the remainder of the trimeric complex Get4-(Get5)2 (Fig. 2 C (I)). But interestingly, we also observed another pair of subcomplexes (highlighted in blue) that stem from the loss of a single subunit of Get5 from the overall complex and the remainder of the trimeric complex (Get4)2-Get5. This striking observation negates the existing structural model of the tetramer (Scheme I), as from this structure it is not possible to excise out a single subunit of Get5, keeping the remainder of the trimer intact (Fig. 2 C). This is because in Scheme I, the loss of Get5 requires the dissociation of both the 5/5 and 4/5 interface. This in turn should also lead to the loss of the Get4 subunit. Hence, observation of the loss of the single Get5 subunit indicates the presence of an additional Get4/4 interface, as shown in Scheme II (Fig. 2 C (II)). This cyclic organization of the tetramer can explain both loss of a single subunit of Get4, as well as Get5.
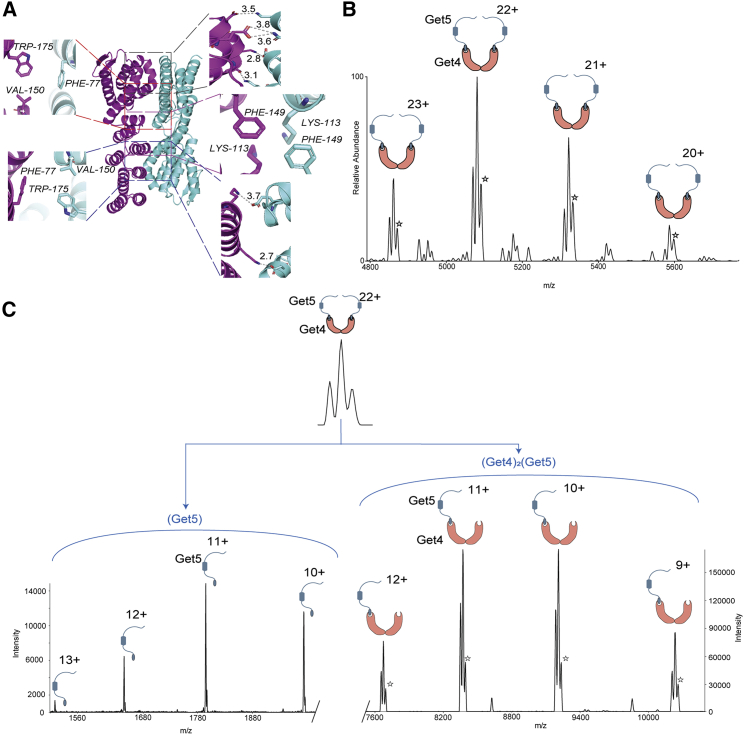
We subsequently inspected the previously determined Get4-Get5(N-term) structure. Careful analysis revealed a putative homodimeric interface in Get4 (PDB: 2WPV) that is formed by a series of intersubunit hydrogen bonds and ionic interactions (Fig. 3 A). This indicates a Get4/4 homodimerization interface that could be used to cyclize the Get4/5 tetrameric complex.
Figure 3.
(Get4)2(Get5)2 tetrameric complex is stabilized via Get4/4 interface. (A) Structural representation highlighting a putative homodimeric interface in Get4 (PDB: 2WPV), formed by intersubunit hydrogen bonds, and hydrophobic and ionic interactions. (B) Mass spectrum of the Get4-Get5∗ heterotetramer. Like before, we observe a population of Get4 without the first two residues. This population is present in all the complexes containing Get4, as shown in the inset. (C) MS/MS spectra of (Get4)2(Get5∗)2. CID MS/MS of the (Get4)2(Get5∗)2 produced led to the loss of monomeric Get5 (left panel), leaving behind the remainder of the trimeric (Get4)2Get5 (right panel). Similar spectra were obtained for two independent protein preparations. To see this figure in color, go online.
To further confirm the presence of the Get4/4 interface, we co-expressed Get4 with a truncated Get5 (Get5∗) that does not have the C-terminal dimerization motif (29). If the Get5/5 interface is the only tetramerization interface (Scheme I, Fig. 2 C (I)), then this truncated Get4/5∗ must yield an exclusive Get4-Get5∗ dimer. Nevertheless, native-MS analysis of the purified complex yet again yielded a tetrameric species (Fig. 3 B), explicitly establishing the presence of a 4/4 dimerization motif. We also performed the CID dissociation of this truncated (Get4)2-(Get5∗)2 tetrameric complex. This yielded dissociated monomeric Get5 and the remainder of the trimer (Get4)2-Get5∗ (Fig. 3 C), confirming the presence of the Get4-4 interface. Finally, to further prove this, we expressed and purified Get4 alone. Native-MS analysis of Get4 decidedly showed the presence of a dimeric species, proving the existence of a novel dimeric interface (Fig. S2). Together, this unambiguously establishes the presence of a previously unknown Get4/4 interface and cyclic organization of the (Get4/5)2 tetramer.
Architecture of the Get3/4/5 complex
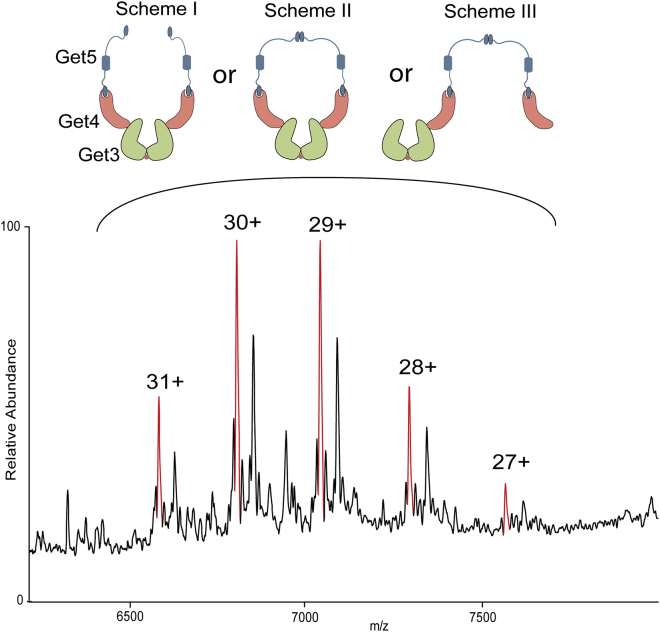
Next, we sought to determine the oligomeric organization of the Get3/4/5 complex. We formed the complex by mixing purified Get3 with the purified full-length Get4/5 complex using previously established protocols (27). Native-MS analysis of the formed complex revealed an exclusive presence of a heterohexameric complex with (Get3)2-(Get4)2-(Get5)2 stoichiometry (Fig. 4).
Figure 4.
Oligomeric organization of the Get3/4/5 complex. Mass spectrum of Get3/4/5 complex. The red peaks correspond to the macromolecule with a mass of 204,228 Da. This mass matches with the complex consisting of two copies of Get3, Get4, and Get5 each. The theoretical mass of the complex with mentioned stoichiometry is 204,228 Da. Scheme I, II, and III show possible molecular organization of the (Get3)2(Get4)2(Get5)2 complex. The zoom of the peak with 29 charges and analysis of its satellite peaks is shown in Fig. S1. To see this figure in color, go online.
Like Get4/5 complex, here again, we sought to use intact dissociation to unambiguously determine the molecular organization of this hetero-hexameric complex. The inset to Fig. 4 shows the possible structural schemes for the hexameric organization. We subsequently used CID MS/MS of the overall hexameric complex to further identify the correct connectivity (Fig. 5 A).
Figure 5.
Decoding the molecular organization of Get3/4/5complex. (A) MS/MS spectra of (Get3)2(Get4)2(Get5)2. CID of the (Get3)2(Get4)2(Get5)2 produced two sets of subcomplexes. Peaks in purple highlight the subcomplexes generated by the loss of a monomeric Get4 unit from the overall complex. The remainder of the trimer (Get3)2-Get4-(Get5)2 species was observed at the higher m/z range (purple peaks in the right panel). The lost, monomeric Get4 molecule was detected in the lower m/z region (purple peaks in the left panel). Peaks in blue highlight the subcomplexes generated by the loss of a monomeric Get5 unit from the overall complex. The remainder of the trimer (Get3)2-(Get4)2-Get5 species was observed at the higher m/z range (blue peaks on the right-hand side). The lost, monomeric Get5 molecule was detected in the lower m/z region (blue peaks in the left panel). (B) Subcomplexes (Get3)2(Get4) (Get5)2 and Get4 can be produced by CID of parent ions with molecular organization presented in both Scheme II and III but not in Scheme I. (C) In contrast, subcomplexes (Get3)2(Get4)2(Get5) and Get5 can be produced by CID of parent ions with molecular organization presented in the Scheme I and II but not in Scheme III. Altogether the data showed that only fragmentation of (Get3)2(Get4)2(Get5)2 with molecular organization presented in Scheme II can give the fragmentation pattern shown in panel A. Dotted lines on parent ions indicate interaction interfaces destroyed in CID during generation of fragment ions. Similar spectra were obtained for three independent protein preparations.
We isolated the 29+ charge state of the hetero-hexamer and subjected it to CID MS/MS. Two pairs of subcomplexes were observed upon dissociating the hexameric complex. The peaks highlighted in blue correspond to a dissociated Get5 monomer and remainder of the hetero-pentameric complex of (Get3)2-(Get4)2-Get5. Among the three possible structures, Scheme I cannot yield these subcomplexes (Fig. 5 B). This is because from Scheme I, the loss of a single monomeric Get5 is not feasible. In Scheme I, the loss of monomeric Get5 requires dissociation of the Get5/5 dimer interface, which also leads to the loss of the entire Get4/5 heterodimer. Similarly, the peaks highlighted in purple correspond to the dissociated Get4 monomer and remainder of the hetero-pentameric complex of (Get3)2-Get4-(Get5)2. Among the three possible structures, Scheme III cannot lead to these subcomplexes. This is because from Scheme III, loss of a single monomeric Get4 is not feasible. In Scheme III, loss of monomeric Get4 requires dissociation of the Get4/3 interface, which also leads to the loss of the entire Get4/5 heterodimer. As shown in Fig. 5 B, it is only Scheme II that can justify both of these dissociation pathways, assigning a cyclic heterohexameric structure for the Get3/4/5 complex.
We also considered the hypothetical scenario that the Get3/4/5 complex exists as a mixture of both Scheme I and III, which together can yield the observed dissociation pathways. To negate this possibility, we used the truncated Get5 that lacks the C-term dimerization motif (Get5∗). Mixing Get4/5∗ with Get3 did not yield any detectable amount of the Get3/4/5∗ complex, or the potential products of dissociation of the hexamer, for example, Get3/Get4/Get5∗, under identical MS conditions (Fig. S3). This also confirms that the observed hexameric molecular organization of the complex was not created as a result of gas-phase rearrangement of the subunits present in the protein complex (53). Further, it proves that the Get5/5 interface is essential for the stability of the complexes. Hence, Scheme I type arrangements, lacking the Get5/5 interface are not a viable structural organization. Interestingly, this also rationalizes the previous observation that mutation of the Get5 dimer interface is deleterious to Get3/4/5 complex formation. Our data clearly show that the wild-type Get3/4/5 complex has a cyclic architecture, and abrogation of the cyclization leads to lack of stability. Together, this negates any possibility of Get3/4/5 hexamer coexisting in two different structural organizations. Hence, it is only Scheme II that can rationalize all generated subcomplexes and dissociation pathways.
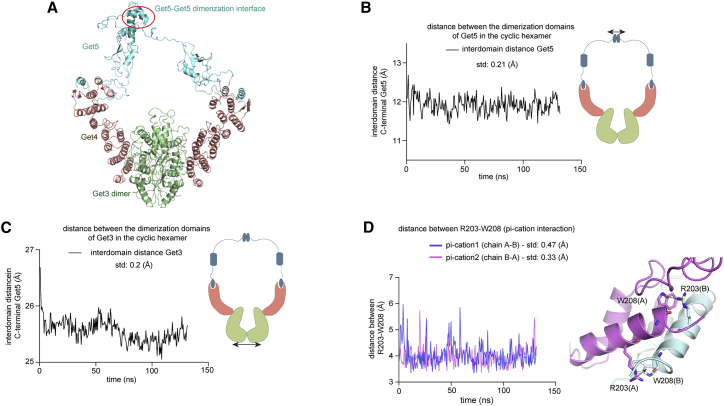
We next sought to use computational modeling and MD simulations to show that the MS-derived closed-ring hexameric Get3/4/5 complex (Scheme II) is conformationally feasible. The available crystal structures of the Get3/GetGet51-54 and fragments of Get5 are used to build a closed-ring, hexameric model (see Materials and Methods section for details). This modeled Get3/4/5 complex (Scheme II) is energy minimized and subjected to MD simulations for about 120 ns (three replicates). Analysis of the MD simulation trajectories confirms the stability of this closed-ring, hexameric Get3/4/5 complex. The dimeric interfaces between Get3 and the C-terminal domains of Get5 remained intact during the course of the simulations (Video S1). To quantitatively show this, we calculated the distance between the center-of-mass of the dimerization domains in Get3 (Fig. 6 B) and the C-terminal domains of Get5 (Fig. 6 C) along the MD simulation trajectory. Variations in these distances are low, with a maximum deviation of about ±1 Å from that seen in the corresponding crystal structures. In addition, the dimeric interface between the C-terminal domains of Get5 in the crystal structure is largely mediated by hydrophobic and pi-cation interactions. We chose a residue pair of Trp 208 and Arg 203 that participate in pi-cation interactions at the dimeric interface of Get5. The residue-pairwise distance between Trp 208 and Arg 203 along the MD simulation trajectory also showed minimal change (Fig. 6 D; deviation of about ±1 Å from that seen in the crystal structures). These results suggest that the dimeric interfaces between Get3 and Get5 in our modeled hexameric, closed-ring cyclic Get3/4/5/complex remain stable throughout the course of the MD simulations. Next, we checked if the long unstructured linkers between the structured domains of Get5 may provide the required conformational flexibility to form the closed-ring hexameric complex. We computed the distances between two pairs of points: the linkers 1 connecting the N-terminal and Ubl domains of Get5 (residues 31–64, Uniprot: Q12285) and the linkers 2 connecting the Ubl and C-terminal domains of Get5 (residues 152–156, Uniprot: Q12285) for the complex. As shown in Fig. S6 variation in the distances for linker 1 and linker 2 (standard deviation: ±6.78 Å and ±4.05 Å respectively) are much higher in comparison to the rigid domains in the complex (standard deviation: ±0.2 Å). All together the MD simulations show that individual structured “rigid” domains in the Get3/4/5 complex are held together using large variations in the unstructured linker regions. This allows immense flexibility in the relative positioning of structured GET domains.
Figure 6.
Analysis of the Get3/4/5 MD simulation trajectories. (A) Structural model of the full-length GET3/4/5 complex. (B) Plot of the distance between the center of mass of the C-terminal domains of Get5 during the course of MD simulation. (C) Plot of the distance between the center of mass of the dimerization domains in Get3 during the course of MD simulation. Variations in the distances in (B) and (C) are low, with a maximum deviation of about ±1 Å from that seen in the corresponding crystal structures. (D) Plot of the distances between residue pairs Trp 208 and Arg 203 that participate in pi-cation interactions at the dimeric interface of Get5 during the course of the MD simulation. Residue-pairwise distance between Trp 208 and Arg 203 shows a small change, with a deviation of about ±1 Å from that seen in the crystal structures. To see this figure in color, go online.
Taken together, although multiple models have been previously proposed for the Get3/4/5 heteromer, our data unambiguously establishes the hexameric, symmetric, cyclic architecture of the complex. Further addition of a large excess of membrane protein substrate (Vamp2) to the wild-type Get3/4/5 complex led to the dissociation of Get4/5 tetramer from the substrate-bound Get3 (Fig. S4). This is consistent with the view that substrate transfer to Get3 leads to the dissociation of the Get3/4 interface, leading to the dissolution of the complex.
Discussions
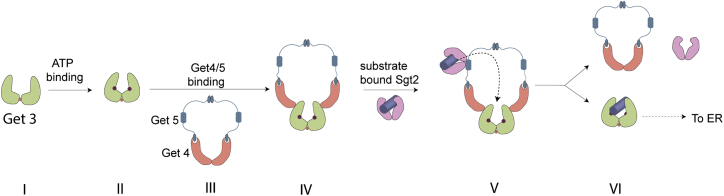
Native-MS is routinely used to determine subunit compositions and oligomeric stoichiometry of protein complexes. Here, we employed intact dissociation of multiprotein complexes to further discern the spatial organization of different subunits and identify the interfaces involved. Coupled with molecular modeling, this can provide a detailed structural model of macromolecular complexes. Applying this strategy, combined with the key mutational analysis, we have unambiguously determined the molecular architecture of the chaperone complexes present in the GET pathway. Our results show that Get4/5 forms a closed-ring tetrameric complex. This Get4/5 tetramer subsequently interacts with the nucleotide bound, conformationally primed Get3 dimer to form a hexameric ring complex of Get32-Get42-Get52 stoichiometry, as shown in Fig. 7. We have further validated our findings through molecular modeling and mutational abrogation of key interfaces. Previous results have shown that only one copy of the substrate-bound Sgt2 dimer can bind to the Get5 dimer (54). Integrating that with our findings on Get3/4/5 renders a detailed molecular view of the pathway where substrate-bound Sgt2 dimer associates with the hexameric Get3/4/5 ring via its interaction with the Get5. Subsequent substrate transfer from Sgt2 to Get3 leads to the dissociation of substrate-bound Get3 from the complex.
Figure 7.
Overall scheme of Get3 chaperone pathway. Get3 dimer (I) after binding to ATP (II) can interact with the Get4/5 heterotetrameric complex stabilized by Ge4/4 and Get5/5 binding interfaces (III). The ATP binding increases the affinity of Get3 dimer to Get4, and Get3 can disrupt the Get4/4 binding interface and create the hexameric complex with Get4/5 via two Get3/4 interaction interfaces (IV). Sgt2 dimer with TA protein cargo binds to Get3/4/5complex via Sgt2/Get5 interface. After the binding, the cargo is transferred to Get3 dimer (V). The binding of the cargo to Get3 triggers dissociation of the complex to Sgt2 dimer, Get4/5 heterotetramer, and Get3 dimer bound to TA protein (VI). In the next steps, the Get3 with the cargo is transferred to the ER. To see this figure in color, go online.
It is worth emphasizing that previous structural studies on the complexes present in the GET pathway were performed on truncated proteins. For example, the structures of the Get4/5 and Get3/4/5 complexes were obtained using an N-terminal truncated peptide of Get5 that only has the Get4 binding interface. Similarly, the Get5 dimer interface was obtained by using a C-terminal polypeptide stretch alone. This is owing to the large unstructured domain in Get5 that can pose a significant challenge in structure determination. Our ability to detect and selectively dissociate the full-length complex highlights the role of native-MS in providing critical structural information in such cases.
An intriguing question here is why Get3/4/5 need to form a closed-ring cyclic complex. One obvious clue lies in the apparent thermodynamic stability of closed-ring complexes. This is indeed evident in markedly less stability of the Get3/4/5∗ complex (Fig. S2) where the Get5 dimer interface has been deleted. Such structural stability upon ring closure is a common theme in many higher-order oligomeric complexes (55). Another interesting justification could be maintaining the fidelity of access of the Get3 transmembrane binding grove. Indeed, we showed that overexpressed Get3 from E. coli co-purifies with endogenous lipids bound to its transmembrane binding grove (Fig. 1). Intrigued by this observation, we further expressed and purified the human homolog of Get3, TRC40, from HEK293 cells. Native-MS analysis of TRC40 also shows the presence of similar binding of endogenous glycerophospholipids (Fig. S5). This indicates that the hydrophobic groove of Get3 is relatively exposed and can pick up nonspecific hydrophobic moieties. In fact, when Get3 was separately expressed and purified in E. coli and incubated with Get4/5, these bound endogenous lipids were even carried forward to the Get3/4/5 complex (Figs. 4 and S1). Hence, keeping it in a closed-ring form may hinder such nonspecific access and ensure that the hydrophobic cavity can only be accessed via the upstream chaperons that bind to the Get3/4/5 complex.
Together, our data provide a detailed molecular architecture of the protein complexes in the GET chaperone pathway and trace out a molecular mechanism of substrate transfer. Further, we show that for complexes where inherent conformational dynamics preclude structure determination, intact dissociation of complexes in native-MS, in combination with integrative molecular modeling, provides a fantastic alternate avenue to obtain structural and mechanistic insights.
Author contributions
F.G. expressed and purified all the proteins and did the mass spectrometric experiments. M.B. performed the MD simulations. K.G. and F.G. designed all the experiments, with the help and critical insights from M.M. K.G. and F.G. wrote the manuscript with the help from all the authors.
Acknowledgments
The authors acknowledge Vicky Wysocki and Sophie Harvey for insightful discussions. K.G. and M.M. acknowledge NIH (USA) for respective fundings: R01GM141192 and R01GM117386. M.B. acknowledges the use of the Extreme Science and Engineering Discovery Environment (XSEDE, which is supported by National Science Foundation (USA) grant number ACI-1548562) SDSC (USA) Comet-GPU nodes through allocation MCB200152. We thank Robert Keenan for Get3, Get4, and Get5 plasmids.
All the raw data can be downloaded from Figshare: https://doi.org/10.6084/m9.figshare.17207996.
Editor: Jill Trewhella.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.02.026.
Supporting material
References
- 1.Borgese N., Fasana E. Targeting pathways of C-tail-anchored proteins. Biochim. Biophys. Acta. 2011;1808:937–946. doi: 10.1016/j.bbamem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Chio U.S., Cho H., Shan S.-O. Mechanisms of tail-anchored membrane protein targeting and insertion. Annu. Rev. Cell Dev. Biol. 2017;33:417–438. doi: 10.1146/annurev-cellbio-100616-060839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shan S.-O. Guiding tail-anchored membrane proteins to the endoplasmic reticulum in a chaperone cascade. J. Biol. Chem. 2019;294:16577–16586. doi: 10.1074/jbc.REV119.006197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chartron J.W., Clemons W.M., Suloway C.J.M. The complex process of GETting tail-anchored membrane proteins to the ER. Curr. Opin. Struct. Biol. 2012;22:217–224. doi: 10.1016/j.sbi.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kutay U., Hartmann E., Rapoport T.A. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- 6.Kutay U., Ahnert-Hilger G., et al. Rapoport T.A. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuldiner M., Metz J., et al. Weissman J.S. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chio U.S., Chung S., et al. Shan S.-O. A chaperone lid ensures efficient and privileged client transfer during tail-anchored protein targeting. Cell Rep. 2019;26:37–44.e7. doi: 10.1016/j.celrep.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guna A., Hegde R.S. Transmembrane domain recognition during membrane protein biogenesis and quality control. Curr. Biol. 2018;28:R498–R511. doi: 10.1016/j.cub.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Denic V., Dötsch V., Sinning I. Endoplasmic reticulum targeting and insertion of tail-anchored membrane proteins by the GET pathway. Cold Spring Harb. Perspect. Biol. 2013;5:a013334. doi: 10.1101/cshperspect.a013334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mariappan M., Mateja A., et al. Keenan R.J. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature. 2011;477:61–66. doi: 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suloway C.J.M., Chartron J.W., et al. Clemons W.M. Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc. Natl. Acad. Sci. U S A. 2009;106:14849–14854. doi: 10.1073/pnas.0907522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho H., Shim W.J., et al. Shan S.-O. J-domain proteins promote client relay from Hsp70 during tail-anchored membrane protein targeting. J. Biol. Chem. 2021;296:100546. doi: 10.1016/j.jbc.2021.100546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gristick H.B., Rao M., et al. Clemons W.M. Crystal structure of ATP-bound Get3-Get4-Get5 complex reveals regulation of Get3 by Get4. Nat. Struct. Mol. Biol. 2014;21:437–442. doi: 10.1038/nsmb.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mateja A., Keenan R.J. A structural perspective on tail-anchored protein biogenesis by the GET pathway. Curr. Opin. Struct. Biol. 2018;51:195–202. doi: 10.1016/j.sbi.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chartron J.W., Gonzalez G.M., Clemons W.M. A structural model of the Sgt2 protein and its interactions with chaperones and the Get4/Get5 complex. J. Biol. Chem. 2011;286:34325–34334. doi: 10.1074/jbc.M111.277798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F., Brown E.C., et al. Denic V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol. Cell. 2010;40:159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao S., Rodrigo-Brenni M.C., et al. Hegde R.S. Mechanistic basis for a molecular triage reaction. Science. 2017;355:298–302. doi: 10.1126/science.aah6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F., Chan C., et al. Denic V. The Get1/2 transmembrane complex is an endoplasmic-reticulum membrane protein insertase. Nature. 2014;512:441–444. doi: 10.1038/nature13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDowell M.A., Heimes M., et al. Sinning I. Structural basis of tail-anchored membrane protein biogenesis by the GET insertase complex. Mol. Cell. 2020;80:72–86.e7. doi: 10.1016/j.molcel.2020.08.012. [DOI] [PubMed] [Google Scholar]
- 21.Verhagen J.M.A., van den Born M., et al. van de Laar I.M.B.H. Biallelic variants in ASNA1, encoding a cytosolic targeting factor of tail-anchored proteins, cause rapidly progressive pediatric cardiomyopathy. Circ. Genom. Precis. Med. 2019;12:397–406. doi: 10.1161/CIRCGEN.119.002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaff J., Rivera Monroy J., et al. Kehlenbach R.H. Emery-Dreifuss muscular dystrophy mutations impair TRC40-mediated targeting of emerin to the inner nuclear membrane. J. Cell Sci. 2016;129:502–516. doi: 10.1242/jcs.179333. [DOI] [PubMed] [Google Scholar]
- 23.Mateja A., Paduch M., et al. Keenan R.J. Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science. 2015;347:1152–1155. doi: 10.1126/science.1261671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mateja A., Szlachcic A., et al. Keenan R.J. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461:361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J., Li J., et al. Sha B. The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion. PLoS One. 2009;4:e8061. doi: 10.1371/journal.pone.0008061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang Y.-W., Chuang Y.-C., et al. Wang C. Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J. Biol. Chem. 2010;285:9962–9970. doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gristick H.B., Rome M.E., et al. Clemons W.M. Mechanism of assembly of a substrate transfer complex during tail-anchored protein targeting. J. Biol. Chem. 2015;290:30006–30017. doi: 10.1074/jbc.M115.677328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chartron J.W., Suloway C.J.M., et al. Clemons W.M. Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc. Natl. Acad. Sci. U S A. 2010;107:12127–12132. doi: 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chartron J.W., VanderVelde D.G., et al. Clemons W.M. Get5 carboxyl-terminal domain is a novel dimerization motif that tethers an extended Get4/Get5 complex. J. Biol. Chem. 2012;287:8310–8317. doi: 10.1074/jbc.M111.333252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamin D.R., Robinson C.V., et al. Dobson C.M. Mass spectrometry of ribosomes and ribosomal subunits. Proc. Natl. Acad. Sci. U S A. 1998;95:7391–7395. doi: 10.1073/pnas.95.13.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt C., Beilsten-Edmands V., Robinson C.V. The joining of the Hsp90 and Hsp70 chaperone cycles yields transient interactions and stable intermediates: insights from mass spectrometry. Oncotarget. 2015;6:18276–18281. doi: 10.18632/oncotarget.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Berkel W.J., van den Heuvel R.H., et al. Heck A.J. Detection of intact megaDalton protein assemblies of vanillyl-alcohol oxidase by mass spectrometry. Protein Sci. 2000;9:435–439. doi: 10.1110/ps.9.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharyya M., Stratton M.M., et al. Kuriyan J. Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II. Elife. 2016;5:e13405. doi: 10.7554/eLife.13405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahdash Z., Lau A.M., et al. Politis A. Mechanistic insight into the assembly of the HerA-NurA helicase-nuclease DNA end resection complex. Nucleic Acids Res. 2017;45:12025–12038. doi: 10.1093/nar/gkx890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hochberg G.K.A., Shepherd D.A., et al. Benesch J.L.P. Structural principles that enable oligomeric small heat-shock protein paralogs to evolve distinct functions. Science. 2018;359:930–935. doi: 10.1126/science.aam7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olinares P.D.B., Chait B.T. Native mass spectrometry analysis of affinity-captured endogenous yeast RNA exosome complexes. Methods Mol. Biol. 2020;2062:357–382. doi: 10.1007/978-1-4939-9822-7_17. [DOI] [PubMed] [Google Scholar]
- 37.Marcoux J., Robinson C.V. Twenty years of gas phase structural biology. Structure. 2013;21:1541–1550. doi: 10.1016/j.str.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Ruotolo B.T., Benesch J.L.P., et al. Robinson C.V. Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 39.Lermyte F., Tsybin Y.O., et al. Loo J.A. Top or middle? Up or down? Toward a standard lexicon for protein top-down and allied mass spectrometry approaches. J. Am. Soc. Mass Spectrom. 2019;30:1149–1157. doi: 10.1007/s13361-019-02201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harvey S.R., Seffernick J.T., et al. Wysocki V.H. Relative interfacial cleavage energetics of protein complexes revealed by surface collisions. Proc. Natl. Acad. Sci. U S A. 2019;116:8143–8148. doi: 10.1073/pnas.1817632116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H., Nguyen H.H., et al. Loo J.A. An integrated native mass spectrometry and top-down proteomics method that connects sequence to structure and function of macromolecular complexes. Nat. Chem. 2018;10:139–148. doi: 10.1038/nchem.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehaffey M.R., Xia Q., Brodbelt J.S. Uniting native capillary electrophoresis and multistage ultraviolet photodissociation mass spectrometry for online separation and characterization of Escherichia coli ribosomal proteins and protein complexes. Anal. Chem. 2020;92:15202–15211. doi: 10.1021/acs.analchem.0c03784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker L.R., Marzluff E.M., et al. Marty M.T. Native mass spectrometry of antimicrobial peptides in lipid nanodiscs elucidates complex assembly. Anal. Chem. 2019;91:9284–9291. doi: 10.1021/acs.analchem.9b02261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ebong I.-O., Beilsten-Edmands V., et al. Robinson C.V. The interchange of immunophilins leads to parallel pathways and different intermediates in the assembly of Hsp90 glucocorticoid receptor complexes. Cell Discov. 2016;2:16002. doi: 10.1038/celldisc.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sipe S.N., Patrick J.W., et al. Brodbelt J.S. Enhanced characterization of membrane protein complexes by ultraviolet photodissociation mass spectrometry. Anal. Chem. 2020;92:899–907. doi: 10.1021/acs.analchem.9b03689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fort KL K.L., van de Waterbeemd M., Heck A.J.R. Expanding the structural analysis capabilities on an Orbitrap-based mass spectrometer for large macromolecular complexes. Analyst. 2017;143:100–105. doi: 10.1039/c7an01629h. [DOI] [PubMed] [Google Scholar]
- 47.Reid D.J., Diesing J.M., et al. Marty M.T. MetaUniDec: high-throughput deconvolution of native mass spectra. J. Am. Soc. Mass Spectrom. 2019;30:118–127. doi: 10.1007/s13361-018-1951-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D, Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Case D.A., Walker R.C., et al. Kollman P.A. University of California; 2018. Amber 2018. [Google Scholar]
- 51.Towns J., Cockerill T., et al. Wilkens-Diehr N. XSEDE: accelerating scientific discovery. Comput. Sci. Eng. 2014;16:62–74. [Google Scholar]
- 52.Maier J.A., Martinez C., et al. Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou M., Dagan S., Wysocki V.H. Impact of charge state on gas-phase behaviors of noncovalent protein complexes in collision induced dissociation and surface induced dissociation. Analyst. 2013;138:1353–1362. doi: 10.1039/c2an36525a. [DOI] [PubMed] [Google Scholar]
- 54.Lin K.-F., Fry M.Y., et al. Clemons W.M. Molecular basis of tail-anchored integral membrane protein recognition by the cochaperone Sgt2. J. Biol. Chem. 2021;296:100441. doi: 10.1016/j.jbc.2021.100441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deeds E.J., Bachman J.A., Fontana W. Optimizing ring assembly reveals the strength of weak interactions. Proc. Natl. Acad. Sci. U S A. 2012;109:2348–2353. doi: 10.1073/pnas.1113095109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.