Abstract
Changes in intracellular pH (pHi) reflect metabolic states of cancer cells during tumor growth and dissemination. Therefore, monitoring of pHi is essential for understanding the metabolic mechanisms that support cancer progression. Genetically encoded fluorescent pH sensors have become irreplaceable tools for real-time tracking pH in particular subcellular compartments of living cells. However, ratiometric readout of most of the pH probes is poorly suitable to measure pH in thick samples ex vivo or tissues in vivo including solid tumors. Fluorescence lifetime imaging (FLIM) is a promising alternative to the conventional fluorescent microscopy. Here, we present a quantitative approach to map pHi in cancer cells and tumors in vivo, relying on fluorescence lifetime of a genetically encoded pH sensor SypHerRed. We demonstrate the utility of SypHerRed in visualizing pHi in cancer cell culture and in mouse tumor xenografts using fluorescence lifetime imaging microscopy and macroscopy. For the first time to our knowledge, the absolute pHi value is obtained for tumors in vivo by an optical technique. In addition, we demonstrate the possibility of simultaneous detection of pHi and endogenous fluorescence of metabolic cofactor NADH, which provides a complementary insight into metabolic aspects of cancer. Fluorescence lifetime-based readout and red-shifted spectra make pH sensor SypHerRed a promising instrument for multiparameter in vivo imaging applications.
Significance
This work is important because intracellular pH is a key homeostasis parameter involved in regulation of numerous biological processes. In cancer cells, pH controls molecular processes leading to cell cycle initiation, proliferation, and adaptation. Many techniques for measuring pH in vivo have low spatial and temporal resolution, require the exogenous contrast agents, and complex methods for data processing. The combination of FLIM with genetically encoded sensor SypHerRed allowed both the spatially resolved quantitative imaging of intracellular pH at the micro- and macroscopic scales and the dynamic measurements in real time.
Introduction
The intracellular pH (pHi) is a key homeostasis parameter, important for a variety of cellular functions. In regard to cancer progression, pH controls molecular processes leading to cell cycle initiation, proliferation, and adaptation. It is known that the cytosolic pH (pHc) of tumor cells is more alkaline than that of normal cells (7.12–7.65 vs. 6.99–7.2), and the extracellular pH (pHe) of a tumor is more acidic (6.2–6.9 vs. 7.3–7.4) (1, 2, 3). As a result, a reverse pH gradient is formed in the tumor (pHe < pHc), compared with normal physiological states (pHe > pHс), which creates favorable conditions for malignant cell transformation, metastasis, and angiogenesis, increases proliferative activity, and promotes evasion from apoptosis and tumor drug resistance (3, 4, 5). In addition, the higher pHc is supposed to drive a metabolic switch from oxidative phosphorylation to aerobic glycolysis, a preferable metabolic pathway for rapidly growing tumors (5), although this link between pHc and metabolism is still debatable (6). Acidic pHc favors apoptosis, as it is optimal for activation of caspases and endonucleases (7,8). To prevent cytosol acidification due to the high glycolytic rate, tumor cells have an excessive number of mechanisms for maintaining pHc, such as H+-ATPase, alkaline cation-H+ exchangers, lactate-H+ cotransporters, bicarbonate transporters, and acid transporters (4). Despite the great importance of pH in regulation of biological behavior of tumor cells, there is a lack of noninvasive methods for its measuring in the tumor.
There are several in vivo methods of pH registration: positron emission tomography, techniques based on nuclear magnetic resonance (magnetic resonance spectroscopy and magnetic resonance imaging), and fluorescence imaging (9). However, the first three techniques have low spatial and temporal resolution, require the exogenous contrast agents, some of which are toxic and radioactive, use complex methods of data processing, and require high-cost equipment and materials.
Fluorescence imaging of pH-sensitive synthetic or genetically encoded indicators is a versatile and sensitive instrument for mapping pH in living systems at different scales, from subcellular organelles to a whole body of small animals. Although some of the small synthetic molecules are able to penetrate cultured cells, their intracellular delivery in tumors remains challenging (10). Genetically encoded probes require incorporation of plasmid DNA into cells, which, on the one hand, might complicate in vivo studies, but on the other hand give an advantage of targeted localization of the probe in the cytosol or in any targeted cellular organelles and a stable expression in cells during long time periods (11,12). Therefore, they provide an opportunity to monitor intracellular pH in vitro in transduced cells and in vivo in a tumor generated from these cells.
Currently existing genetically encoded pH sensors are based on the proteins of the GFP (green fluorescent protein) family, in which fluorescence intensity or lifetime depend on pH (12,13). The utility of these sensors has been widely demonstrated on cultured cells, although their in vivo applications are still limited. Although fluorescence intensity is a simpler metric than fluorescence lifetime, in terms of data acquisition, the use of single-color fluorescence intensity as a readout can report only relative changes of pH in individual “matched” cells. Calibration and determination of absolute pH value using the intensity is difficult because it is affected by a number of factors such as concentration of the fluorophore, photobleaching, excitation and detection efficiency, absorption, and scattering events (11). Ratiometric (dual-excitation or dual-emission) pH sensors partially address these artifacts and enable pH quantification in cultured cells under standardized settings. Yet, they do not allow recording absolute pH values in tissue due to the impossibility of calibrating the signal, while providing only a snapshot of spatial distribution of the intensity ratio. An alternative approach to improved quantification of pH is to measure fluorescence lifetime, which is an intrinsic characteristic of a fluorophore. Fluorescence lifetime imaging (FLIM) offers a number of advantages over the conventional intensity-based techniques. First, it is quantitative, and the values of lifetime depend only on the properties of the fluorophore and its microenvironment. Consequently, FLIM can avoid many artifacts that contaminate the intensity measurements. Second, sample thickness, light scattering, and other parameters of the specimen affect resolution of the image, but not the measured parameter, fluorescence lifetime. Therefore, availability of a pH-responsive fluorescence lifetime-based probe could potentially solve the problems associated with heterogeneity and large size of a sample (14, 15, 16, 17, 18).
The present work was aimed at the development of methodology for probing intracellular pH in living cancer cells in vitro and in tumor xenografts in vivo, using a genetically encoded sensor and FLIM, both at the micro- and macroscopic scales. At first, we tested the fluorescence lifetime response to pH for three genetically encoded pH sensors, SypHer, SypHer-2, and SypHerRed. The sensor SypHerRed with the substantial lifetime difference was then applied to measure intracellular pH in cultured HeLa cells and in mouse tumors in vivo using two-photon excited FLIM microscopy and one-photon excited macro-FLIM. Since SypHerRed is a single wavelength red-color sensor, the possibility of multiparameter imaging was demonstrated. Simultaneous probing of pHc by SypHerRed fluorescence lifetime and metabolic state by endogenous fluorescence of the reduced nicotinamide adenine dinucleotide NADH was performed in vitro on cancer cells upon metabolic perturbations.
Materials and methods
pH sensors
The genetically encoded pH sensors SypHer and SypHer-2 are based on the cpYFP protein and created from the sensors for hydrogen peroxide HyPer and HyPer-2, respectively, by directional mutagenesis, the replacement of the first cysteine with serine (C199S mutation) (19). SypHer-2 has twice higher brightness than SypHer (20). The key role in this is in Ala406Val substitution corresponding to the Ala233Val mutation in wtOxyR domain. SypHer and SypHer-2 have two peaks of the absorption with maxima at 420 nm and 500 nm, which correspond to protonated and anionic forms of the chromophore, respectively. The maximum emission of fluorescence is at 516 nm. Acidification of the environment leads to an increase in the protonated form (excitation maximum 420 nm) and a decrease in the deprotonated form (excitation maximum 500 nm). Therefore, measurements of pH can be performed using ratiometric recording of the fluorescence with the excitation at these two peaks (I488/I405).
SypHerRed is based on the circularly permuted red fluorescent protein mApple and created from the hydrogen peroxide biosensor HyPerRed via C199S mutation (21). SypHerRed has excitation maximum at 575 nm and emission maximum at 605 nm. The intensiometric pH response of purified SypHerRed was shown earlier by Ermakova et al. (21). Fluorescence intensity of SypHerRed increases with increase of pH.
Cell cultures and transfection
A CT26 (mouse colon carcinoma) cell line, stably expressing SypHer (22), and HeLa Kyoto (human cervical cancer) cell line, stably expressing SypHer-2 (23), were used.
To obtain a HeLa Kyoto cell line, stably expressing the SypHerRed sensor, lentiviral transduction was used. Vector particles were generated by calcium phosphate transient transfection of HEK293T cells with the two packaging plasmids pR 8.91 and pMD.G, kindly provided by Didier Trono (Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland), and the transfer vector plasmid pLCMV-PL4-Puro-SypHerRed. Transfection of HEK293T cells was performed using the FuGene transfection reagent (Sigma-Aldrich, Burlington, MA, USA). Supernatant containing the lentiviral particles was collected in 48 h and 72 h after transfection, filtered (0.45-μm filter) and used for transduction of HeLa Kyoto cells. Note, that the plasmid encoding SypHerRed is already available from AddGene (24).
To create a stable cell line, 3 × 106 lentiviral particles were added to 1 × 105 HeLa Kyoto cells in the presence of 8 μg/mL Polybrene (Sigma-Aldrich). To select the cells with a stable expression, 0.5–1 μg/mL puromycin was added in culture medium 48–72 h after transduction. Cell populations with the brightest fluorescence were sorted using BD FACS Aria (Biosciences, San Jose, CA, USA). In the obtained HeLa-SypHerRed cell line, the pH sensor was localized to both the cytosol and the nucleus.
All cell lines were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA), 2 mM glutamine (PanEco, Moscow, Russia), 10 U/ml penicillin, and 10 mg/mL streptomycin in CO2 incubator at 37°C, 5% CO2, and 80% relative humidity.
pH calibration
To translate the fluorescence readings into pH, calibration curves were generated. Calibrations were performed in the range 6.0–8.0 pH units on live cells, stably expressing the pH sensors, as described in Ref. (25). Briefly, the cells were incubated with buffer solutions with the specified pH in the presence of two ionophores nigericin (5 μM) and monensin (5 μM), for 3–4 min to equilibrate the extracellular and intracellular pH. Buffer composition was 130 mM potassium gluconate, 20 mM sodium gluconate, 0.5 mM MgSO4, 0.2 mM EDTA, and 30 mM HEPES (for pH 6.9–8.0) or MES (for pH 6.0–6.8). pH of the buffer solutions was adjusted to the required value using 1 mM HCl or 1 mM KOH.
To carry out pH calibration on a confocal microscope, cells were seeded on glass-bottom 35-mm FluoroDishes (World Precision Instruments, Sarasota, FL, USA) in an amount of 105 cells in 200 μL FluoroBright DMEM (Life Technologies, Carlsbad, CA, USA) 24 h before calibration. A separate dish was used for each pH value. The microscopic images were recorded from five fields of view, with further measurements of fluorescence intensity and lifetime done for the same 20–30 cells. The obtained values from 20 to 30 cells were then averaged and plotted against pH. To construct the curve fit for the pH calibration data the least-squares method was used in GraphPad Prism 9.
For the macro-FLIM system, a separate pH calibration was done to account for signal integration effects on a macroscale. For this, single-cell suspension of HeLa-SypHerRed cells was prepared (5 x 105 cells/mL FluoroBright DMEM) and 500 μL of the suspension was placed in the Eppendorf tube and incubated with the buffer solutions. The macroscopic images of fluorescence intensity and lifetime at the specific pH were obtained from the Eppendorf tubes.
Mice with tumors
All animal protocols were approved by the Ethics Committee of the Privolzhsky Research Medical University. Experiments were performed on female nu/nu mice purchased from the Pushchino animal nursery (Pushchino, Russia). Mice of 20–22 g body weight were inoculated subcutaneously in the left flank with HeLa-SypHerRed cells (2 x 106 cells) in 150 μL phosphate buffered saline (PBS). Imaging was performed on days 7, 11, 16, 22, and 25 after the cell injection. The mice were anesthetized intramuscularly with a mixture of Zoletil (40 mg/kg, Virbac, Carros, France) and 2% Rometar (10 mg/kg, Spofa, Jičín, Czech Republic) before the imaging experiments. To increase signal/noise ratio, a skin flap over the tumor was surgically opened for the time of image acquisition and closed immediately afterward in sterile conditions. On day 25 the animals were sacrificed by cervical dislocation and the tumors were excised for histological analysis.
Fluorescence intensity and lifetime imaging of pH sensors
Microscopic fluorescence intensity and lifetime images were collected using laser-scanning microscope LSM 880 (Carl Zeiss, Jena, Germany) equipped with FLIM module Simple Tau 152 TCSPC (Becker & Hickl GmbH) and a femtosecond Ti:Sa laser (Spectra Physics, Milpitas, CA, USA; 80 MHz, 100 fs). A water-immersion objective С-Apochromat 40×/1.2 NA W Korr was used for image acquisition.
For fluorescence microscopy, the cells were seeded (1 x 105 in 2 mL) into glass-bottom 35-mm FluoroDishes and incubated overnight. Then the cells were washed with PBS and placed in DMEM life medium without phenol red (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), for imaging. In the animal study, a mouse was placed in the holder so that the tumor was in a tight contact with the coverslip above the objective.
In one-photon mode on the LSM 880 microscope, SypHer and SypHer-2 were excited at a wavelength of 405 nm with a diode laser and at 488 nm with an argon laser. Emission was detected in the ranges 435–689 nm and 509–689 nm for excitation at 405 nm (I405) and 488 nm (I488), respectively. For one-photon excitation of SypHerRed fluorescence, a wavelength of 543 nm was used; the signal was recorded in the range of 570–754 nm.
For two-photon FLIM, a wavelength of 980 nm was used for excitation of SypHer and SypHer-2; the signal was registered in the range of 500–550 nm. A wavelength of 1040 nm was used for two-photon excitation of SypHerRed; the signal was recorded in the range of 570–640 nm. A hybrid detector HPM-100-40 (Becker & Hickl GmbH) was used for all two-photon experiments for fluorescence detection. The average power applied to the samples was ∼6 mW. Image collection time was 60 s to provide pixel intensities ≥3000 photons per decay curve at binning 1 in most cells on the image. The cells with lower pixel intensities were excluded from the analysis.
Fluorescence lifetime macro-images were obtained using confocal FLIM macroscanner DCS-120 MACRO (Becker & Hickl GmbH) (26). The macro-FLIM setup provides a field of view up to 18 × 18 mm and a lateral spatial resolution of 15 μm. A picosecond diode laser with a wavelength of 594 nm was used as an excitation source. Fluorescence was detected using hybrid detector HPM-100-40 in the range 610–690 nm. Image collection time was 60 s.
The fluorescence intensity images were processed with ZEN 3.0 (Carl Zeiss) and ImageJ 1.39p software (NIH, Bethesda, MD, USA). The background signal, taken from an empty region of an image, was subtracted from the measurements. For SypHer and SypHer-2, the ratio of emission intensities resulting from excitation at the two wavelengths was calculated (I488/I405). FLIM data were processed using the SPCImage software (Becker & Hickl GmbH). The fluorescence decay curves of the pH sensors were fitted with a biexponential decay model. For adequate fitting, pixel intensities were adjusted to ∼5000 photons using the binning option. The goodness of the fit, the χ2 value, was in the range from 0.8 to 1.2. The short and long lifetime components (τ1 and τ2, respectively), the relative amplitudes of the lifetime components (a1 and a2; a1+a2 = 100%), and the amplitude-weighted (also denoted as weighted average) fluorescence lifetime (τm = (a1 · τ1 + a2 · τ2)/(a1+ a2)), were estimated. Fluorescence intensity and lifetime of SypHerRed were assessed in the individual cells, where the regions of interest (ROIs) were selected manually from the cell morphology.
For the macro-FLIM data, the average fluorescence lifetime (τm) was calculated for each Eppendorf tube with the cell suspension or mouse tumor.
Fluorescence imaging of SypHerRed-expressing tumors in vivo
A molecular imaging system IVIS-Spectrum (Caliper Life Sciences, Hopkinton, MA, USA) was used for fluorescence whole-body imaging. Fluorescence of SypHerRed was excited at 540 nm and detected using a 600–620 nm band filter.
Cell treatments
HeLa-SypHerRed cells were seeded in gridded 35-mm glass-bottom dishes (Ibidi, Gräfelfing, Germany) and incubated with either 15 μM 3-bromopyruvate (Sigma-Aldrich) for 24 h or with 1 μM rotenone (Sigma-Aldrich) for 1 h to induce metabolic alterations. Simultaneous FLIM microscopy of NADH and pHc was performed before and after incubation with the agents in the same 5–7 randomly selected fields of view.
FLIM of NADH
FLIM microscopy of NADH in SypHerRed-expressing HeLa cells was performed using laser-scanning microscope LSM 880 (Carl Zeiss). Two-photon fluorescence of NADH was excited at the wavelength of 750 nm and registered in the range 455–500 nm. The average power applied to the samples was ∼6 mW, and the approximate photon count rate was 100–200 kHz. Image collection time was 60 s.
FLIM images were processed in the SPCImage software (Becker & Hickl GmbH) following the protocol described in Ref. (27). Fluorescence decay curves of NADH were fitted with a biexponential model, with the goodness of the fit, χ2 value 0.8–1.2. Pixel intensities were adjusted to ≥5000 photons per decay curve with binning factor 1–2. In each cell of interest the maximal cytoplasmic region with the appropriate χ2 was manually selected as ROI. The weighted average fluorescence lifetime was estimated: τm=(a1 · τ1 + a2 · τ2)/(a1+ a2), where τ1 and τ2 are the short and long lifetime components, and a1 and a2 are the relative amplitudes of the lifetime components.
Histopathology
For histological analysis, the tumors were surgically removed and fixed in 10% neutral buffered formalin, dehydrated, and embedded in paraffin according to the standard protocol. Ten μm tumor tissue paraffin sections were stained with hematoxylin and eosin (H&E) and examined with a Leica DM 2500 microscope (Leica, Wetzlar, Germany). The histological structures were matched with the SypHerRed fluorescence signals registered in vivo for the corresponding plane.
Statistics
The results are expressed, below, as mean ± standard deviation (SD) for monolayer cells and as mean ± standard error of the mean (SEM) for tumors in vivo. Student's t-test was used for data comparison, with p ≤ 0.05 being considered statistically significant.
Results
Testing pH sensitivity of fluorescence lifetime
For the ratiometric sensors SypHer and SypHer-2 the fluorescence lifetime did not show the pH response, whereas the fluorescence intensities at two excitation wavelengths, and consequently the I488/I405 ratio, did (Figs. S1 and S2). Both sensors displayed biexponential fluorescence decay with a short component τ1 ∼0.6 ns (a1 ∼70%) and a long component τ2 ∼2 ns (a2 ∼30%) without any significant variations in the range of pH from 6.9 to 8.1.
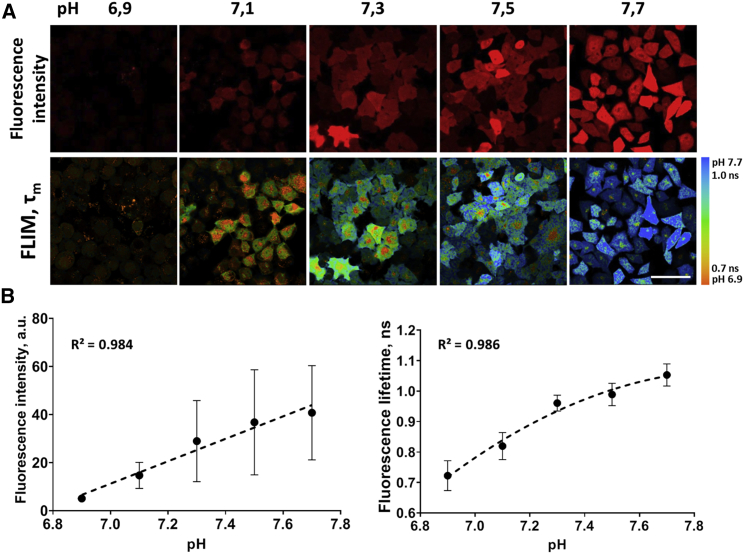
In the case of SypHerRed the fluorescence intensity and lifetime increased with an increase in pH in cells (Fig. 1 A). For the fluorescence intensity, a linear dependence on pHi was obtained for pH range 6.9–7.7 (Fig. 1 B). However, due to different levels of expression of the sensor, large variations of the intensity between cells were observed independently on pH.
Figure 1.
Fluorescence lifetime dependence of SypHerRed on pH. (A) Representative images of fluorescence intensity and FLIM images of HeLa-SypHerRed cells at different intracellular pH. Bar: 40 μm. (B) The fluorescence intensity and τm plotted against pH. Mean ± SD, n = 20–30 cells for each pH value. Black dots are the experimental measurements, and dashed lines are the approximation curves (y = 39.113x – 263.44 for the intensity, y = −0.4177x2 + 6.5475x – 24.503 for τm). To see this figure in color, go online.
Time-resolved measurements showed that the fluorescence decay of SypHerRed fits to a biexponential model (χ2 ∼ 1.05). Among the parameters of the decay, weighted average fluorescence lifetime τm demonstrated the strongest (nonlinear) dependence on pH in the physiological range 6.9–7.7 with increase of the value from 0.72 to 1.05 ns (Figs. S3 and 1 B). The relationship between τm and pH was then employed as a standard curve to determine pH from microscopy data. The intercellular variability in the fluorescence intensity (i.e., concentration) of SypHerRed did not affect the resulting fluorescence lifetimes, at least in the range of the protein concentrations produced by the cells (Fig. S4).
Thus, we concluded that, out of the three tested genetically encoded pH sensors, only SypHerRed can operate in FLIM mode as it has a pH-responsive fluorescence lifetime.
Intracellular pH imaging in cultured HeLa cells
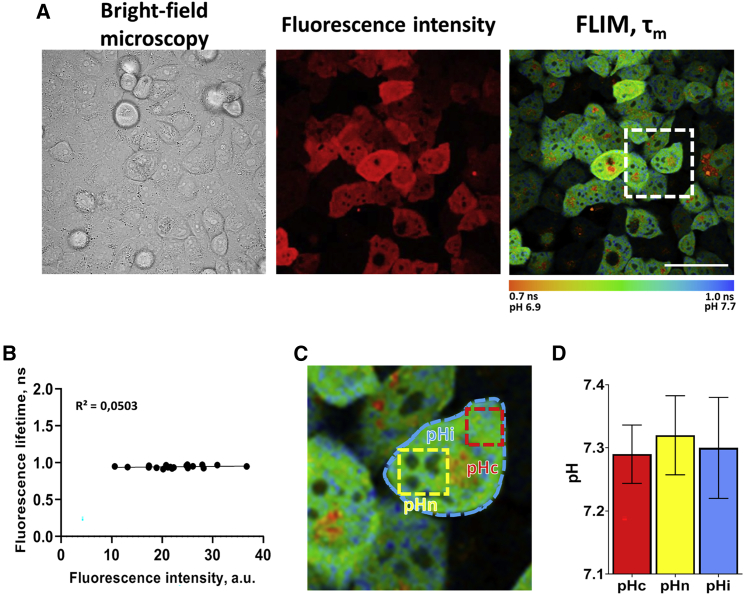
Next, using FLIM microscopy and SypHerRed, we assessed pH in HeLa cells under conventional cultivation conditions (Fig. 2).
Figure 2.
Measurements of pH in HeLa-SypHerRed cells under standard cultivation conditions. (A) Representative bright-field, fluorescence intensity and FLIM images of cells. Bar: 40 μm. (B) Scatter plot of fluorescence lifetimes versus intensity in the cytoplasm. Dots are the measurements in individual cells. (С) Enlarged area shown by the white square in (A). The dashed red, yellow, and blue lines indicate the areas selected for analysis. Note, that the small areas with short lifetime (∼0.7 ns, not pH-responsive) in the perinuclear zone were avoided upon ROI selection in the cytoplasm. (D) Quantification of pH in the cytoplasm (pHc), in the nucleus (pHn), and in the whole cell (pHi). Mean ± SD. N = 20–30 cells. To see this figure in color, go online.
Again, fluorescence intensity significantly varied between cells. Since it is not possible to reliably determine whether observed differences in fluorescence are due to variations in pH or sensor concentration, or both, single-cell pH measurements using SypHerRed in the intensiometric mode are problematic. Plotting pH-dependent fluorescence lifetime τm against intensity showed no correlation between these estimates, indicating that intercellular variability of the intensity resulted from different expression levels of the sensor but not from different pH (Fig. 2 B).
Since the sensor was localized both to the cytosol and the nucleus, it enabled pH measurements in both compartments as well as across the whole cells (Fig. 2 C and D). To account for the local variations of the fluorescence lifetime across the cell, separate calibration curves were used for the different areas (Fig. 1 B, right: for the cytosol and nucleus, Fig. S5: for the whole cell). We found that the fluorescence lifetime of SypHerRed in the cell cytoplasm was 0.93 ± 0.02 ns, which corresponded to 7.29 ± 0.05 pH units. The fluorescence lifetime and pH in the nuclei were nearly the same as in the cytoplasm, 0.97 ± 0.03 ns, 7.32 ± 0.06 pH units. In a whole cell, lifetime was slightly shorter than that measured precisely in the cytoplasm or in the nucleus, 0.95 ± 0.08 ns, because of the small inclusions with short lifetime present in the perinuclear area, which could presumably be the immature protein. pH evaluated in a whole cell was 7.31 ± 0.07 pH units.
pH mapping in HeLa tumor xenografts in vivo
The next step was to demonstrate the applicability of SypHerRed for pHi assessment in an animal tumor model in vivo. FLIM was performed both on the macro- and microscopic scales, which allowed us to measure pHi in a whole tumor and at the cellular level.
Since macro-imaging collects the signal integrally, from the whole cells, a separate calibration on the cell suspensions was performed (Fig. 3). As expected, the fluorescence lifetime and pH for the suspension were nearly the same as for cells in a monolayer when the calculations were done for the whole cells (pH 7.30 and 7.31 ± 0.07, correspondingly). Therefore, any of two standard curves could be used to translate lifetime values into pH in vivo, but we prefer to use that from macro-FLIM to simulate the situation when FLIM microscopy is not available in a lab.
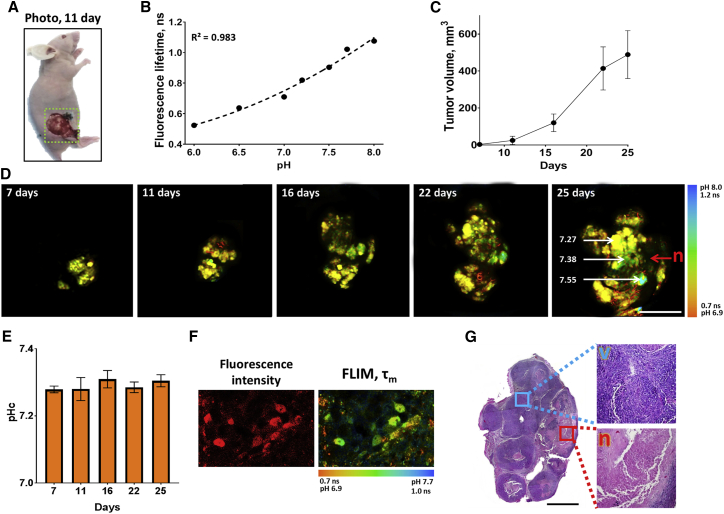
Figure 3.
In vivo mapping of pHi in HeLa tumor xenografts stably expressing genetically encoded pH sensor SypHerRed. (A) Photograph of HeLa tumor on 11th day of growth. Yellow dashed square corresponds to the area imaged in (D). (B) The fluorescence lifetime τm plotted against pH on a macro scale (y = 0.062x2 – 0.5818x + 17838). (C) Dynamics of tumor growth. Mean ± SEM, n = 3 tumors. (D) Macro-FLIM during tumor growth. Bar: 2 mm. pH values in the specific spots of the image on day 25 are shown. n, necrosis. (E) Quantification of pH in the tumors. Mean ± SEM, three tumors. (F) Fluorescence intensity and FLIM microscopy of tumors on the 25th day of growth. Bar: 40 μm. (G) Histopathology of HeLa tumor shown in (D) on 25th day of growth. H&E staining. Bar: 100 μm. Viable tumor tissue (v) is indicated by the blue square, necrosis (n) by the red square. To see this figure in color, go online.
Monitoring of pHi on a macro scale was implemented in dynamics from day 7–25 of the tumor growth (Fig. 3 C). As one can see from the macro-FLIM images, HeLa tumors displayed some degree of intra-tumor heterogeneity of pH at all stages of growth. The local pHi within a tumor amounted from 7.21 to 7.55 in different areas.
On day 7 after tumor inoculation, the average fluorescence lifetime in the tumors was 0.81 ± 0.01 ns, which corresponded to a pHi value of 7.27 ± 0.03. As tumors grew, only insignificant variations in pHi were detected with a tendency to increase maximum to 7.31 ± 0.06 pH units on day 16.
Using FLIM microscopy, in vivo measurements of pHi in the same tumors were performed on day 25 (Fig. 3 F). The fluorescence lifetime calculated for individual tumor cells was 0.95 ± 0.05 ns, which corresponds to pHi 7.30 ± 0.12.
Histological analysis on day 25 showed that the tumors had a dense, multinodular structure and consisted mainly of vital tumor cells (∼70%–80% of tumor area) (Fig. 3 G). Connective tissue fibers and spontaneous necrosis composed the remaining 20%–30% of the tumors. Matching of the histological slices with macro-FLIM images revealed the loss of SypHerRed fluorescence in necrotic zones. Accordingly, the in vivo measurements of pHi with SypHerRed are related to the vital tumor tissue.
Therefore, these results demonstrate the possibility of spatiotemporal imaging and quantification of pHi in tumors in vivo using SypHerRed and FLIM and elaborate on previous findings that suggest heterogeneity of pHi throughout the tumor.
Simultaneous imaging of pHc and metabolism using FLIM
SypHerRed is a single-color variant of pH sensor, which opens the opportunity to combine the sensor with fluorophores of other colors and, thus, to visualize pH simultaneously with other cellular parameters. To demonstrate this advantage of SypHerRed, we performed imaging of pHc in conjunction with endogenous fluorescence of metabolic cofactor NADH. It is known that fluorescence lifetime of NADH is a sensitive metric of cellular metabolic state, as NADH is involved in different biochemical reactions in the forms having different fluorescence decay times. The free form of NADH with short lifetime (∼0.4 ns, ∼85%) is attributed to glycolysis, and the protein-bound form with long lifetime (∼2.2–4.5 ns, ∼15%) is attributed to mitochondrial oxidative phosphorylation (28,29). Therefore, the shift to a more glycolytic state results in a decrease of the mean fluorescence lifetime τm of NADH.
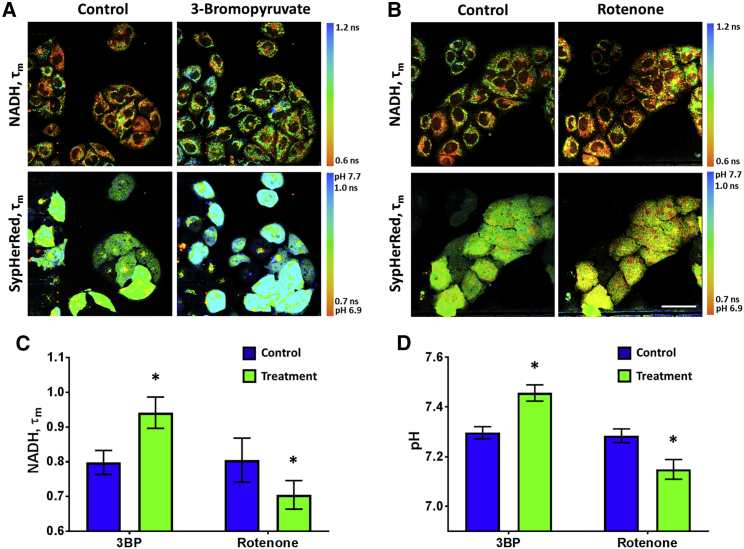
We measured simultaneously pHc using SypHerRed and the mean lifetime of NADH in living HeLa cells exposed to metabolic perturbations (Fig. 4). 3-bromopyruvate and rotenone have been used to inhibit glycolysis and complex I of the mitochondrial respiratory chain, respectively. Upon incubation with 3-bromopyruvate, an increase in τm NADH from 0.79 ± 0.03 ns to 0.94 ± 0.04 ns (p = 0.00014) was detected. The pHc also increased from 7.29 ± 0.02 to 7.45 ± 0.03 (p = 0.00011), which is rational as inhibition of glycolysis leads to decrease in the production of lactate. The opposite effects were observed when cells were treated with rotenone (Fig. 4 B). The mean lifetime of NADH τm decreased from 0.80 ± 0.06 ns to 0.70 ± 0.04 ns (p = 0.00011), and the cytosol became more acidic (7.28 ± 0.03 versus 7.15 ± 0.04 pH units, p = 0.00048). This result demonstrates that simultaneous assessments of pHc and NADH pool can be implemented using FLIM, which provide complementary insights into cancer metabolism.
Figure 4.
Simultaneous visualization of pHc and NADH in HeLa cells exposed to metabolic perturbations. Representative FLIM images of NADH and genetically encoded pH sensor SypHerRed upon treatment with 3-bromopyruvate (A) and rotenone (B). Images before (control) and after treatment were captured from the same fields of view. In the case of 3-bromopyruvate, the differences with control in the cell number and position are associated with a long (24 h) time lapse between image acquisition. Bar: 40 μm, applicable to all images. Quantification of τm NADH (C) and pHc (D) in control and treated cells. Mean ± SD, n = 25–35 cells. ∗, p ≤ 0.0005 with control. To see this figure in color, go online.
Discussion
In this work, we have developed a method for assessment of the intracellular pH of living tumor cells using genetically encoded pH sensor SypHerRed and FLIM at the cellular level and at the level of the whole tumor. It is important that the use of FLIM enabled us to obtain not only a picture of the spatial distribution of the sensor signal, but also the absolute pH values in tumor cells in vivo, which was done for the first time to our knowledge.
FLIM is an attractive technique for pH measurements in tissues because it has no limitations associated with variations in fluorophore concentrations, fluctuations in the intensity of the excitation light and emission light paths, and focusing. Previously, attempts to measure pH in live cells and tissues using fluorescence lifetime of a fluorescent sensor have been made in several studies. For example, in the study by Sanders et al. the intracellular pH was imaged in Chinese hamster ovary (CHO) cells using FLIM microscopy and the chemical fluorescein-based ratiometric probe BCECF since this probe is sensitive in the physiological pH range (30). Hanson et al. also identified that the fluorescence lifetime of BCECF is pH dependent and assessed pH gradient in the skin samples with two-photon FLIM microscopy (31). Hille et al. tested the suitability of different commercial fluorescence dyes for lifetime-based pH sensing. They showed that, out of four dyes (BCECF-AM, HPTS, LysoSensor Green DND-153, and SNARF-5F), only BCECF provided reliable intracellular pH recordings in living cells, and they used it to measure pHi in situ in isolated salivary glands (32). Lin et al. used C-SNAFL2 as a lifetime-based fluorescent pH indicator to image the cytosolic pH of 3T3 fibroblasts, CHO cells, and MCF-7 breast cancer cells (33). In the same study, the dextran-conjugated acidic pH indicators DM-NERF and OG-514 were applied for imaging of lysosomal pH in 3T3 fibroblasts. Recently, a time-domain macroscopic FLIM of upper gastrointestinal pH in mice models in vivo was performed with a lanthanide-based near-infrared probe (34). Intracellular pH measurements by FLIM microscopy were also performed with functionalized CdSe/ZnS quantum dots in stem cells C3H10T1/2 and melanoma cells SK-MEL-2 (35). Although the use of time-resolved measurements instead of intensity (or the ratio of intensities) helped to overcome the issues associated with inhomogeneous distribution of the chemical sensors in cells and tissue samples, in vivo imaging of the intracellular pH remained unrealizable, which is, in part, due to nonspecific distribution of the dye in the body without entering in the target cells.
The possibility to measure intracellular pH using FLIM was demonstrated not only with synthetic probes, mentioned earlier, but also with pH-sensitive proteins, genetically targeted to specific cell organelles or diffusely distributed in the cytosol. Schmitt et al. reported on cytoplasmic and mitochondrial pH measurements in CHO-K1 cells using FLIM microscopy with eGFP-pHsens, a mutant of eGFP (14). pH-associated lifetime changes were also identified for E2GFP; pH analysis in 3T3 fibroblasts and HeLa cells expressing E2GFP in the cytoplasm or mitochondria was performed in Ref. (15). Another example of genetically encoded pH sensor, the fluorescence lifetime of which depends on pH, is pHRed, a mutant of mKeima. Its performance with FLIM microscopy was illustrated in mouse neuroblasts Neuro2A cells to image cytosolic and mitochondrial pH (16). The pH sensitivity of the fluorescence lifetime of ECFP was used by Poëa-Guyon et al. to measure pH in secretory granules of rat pheochromocytoma PC12 cells (17). Novel pH sensor, pH-Lemon, showed pH-dependent fluorescence lifetime change, suitable for the study of pH in acidic cellular compartments (18). However, in all these studies, imaging of pHi was carried out only in the in vitro experiments. The advantages of SypHerRed as a lifetime-based probe allowed us to measure absolute pH in a mouse tumor in vivo, which was impossible to do previously with a ratiometric sensor SypHer-2 due to calibration issues (23).
The average pHi registered in HeLa tumors during growth was ∼7.29 pH units. However, at all stages of growth local heterogeneities of pHi were seen with variations throughout a tumor from 7.21 to 7.55. Previous works have suggested spatial heterogeneity of pHi in tumors as cancer cells exist in a heterogeneous microenvironment and exhibit a variety of phenotypes (36). Unlike extracellular pH gradients, pHi heterogeneity is much less explored (first of all, because of the lack of reliable methods to measure it). Further studies are needed to understand the origin of pHi variations in cancer.
A serious limitation of most of the fluorescent pH-sensitive proteins for imaging pH in thick samples ex vivo or tissues in vivo is their blue to green absorption-emission bands that significantly limit the penetration of light into tissue. In addition, some of them exhibit pH sensitivity only in acidic to near-neutral environments, which restricts the measurements to acidic organelles. Fluorescent lifetime-based pH sensor SypHerRed, described here, has an absorption and fluorescence in the yellow-red range (excitation maximum 575 nm; emission maximum 605 nm), which falls into the optical transparency window of tissues, and a lifetime sensitivity for pH up to 8.0, therefore covering an entire physiological range.
The sensor SypHerRed is originally intensiometric (fluorescence intensity increases with increasing pH). On a cellular level, the intensity-based assessment of pH requires taking into account the levels of protein expression in cells. Indeed, individual cells in a cell culture exhibited different fluorescence intensities, even if intracellular pH was equal (Fig. 1). In contrast, fluorescence lifetime showed only minor variations in the cell population, as it does not depend on the concentration of a fluorophore, within reasonable limits. On the macro level, multiple factors influence fluorescence intensity, besides pH response of the sensor, e.g., local concentration of the fluorophore, sample shape and thickness, proximity to the detector, tissue density. Tumor xenografts generated from SypHerRed-expressing cells showed a high intra-tumor heterogeneity of fluorescence signal, typically with a brighter area in the center (Fig. S7). A single emission nature of SypHerRed does not allow to eliminate the interference of pH-independent factors, unlike ratiometric approaches, so it is problematic to interpret the steady-state fluorescence images. But in principle, the intensiometric response of SypHerRed could be used to visualize real-time dynamic changes of pH in biological systems when quantification is not critical.
In the present study, we compared the sensitivity of fluorescence lifetime to pH for three genetically encoded sensors- SypHer, SypHer-2, and SypHerRed. Among three sensors, only the latter one displayed a large dynamic range of lifetimes suitable for pH measurements with FLIM. These results are in line with previous works that show that not all fluorescence dyes that are suitable for intensity-based or ratiometric pH sensing are useful for FLIM.
Although SypHerRed, used in this study, was designed to localize to the cytosol, it inevitably penetrated into the cell nucleus. The ability of GFP-like fluorescent proteins to diffuse in the nucleus was previously confirmed in several studies (37,38). As a result, fluorescence of SypHerRed was observed in the cytoplasm and in the nucleus and, therefore, resolved pH measurement in both compartments using FLIM microscopy. The obtained values of the cytosolic and nuclear pH appeared identical, which is consistent with some other reports and supports the assumption that the nuclear envelope is highly permeable to H+ (or acid equivalents) and readily equilibrate its pH to the cytoplasmic value (1,38). At the same time, there are the papers that demonstrate cytosol-nucleus pH gradients (39,40). It cannot be excluded that the presence of the gradients is specific to the cell types or cell lines.
Because of the quantitative nature of FLIM, one expects that the sensor calibration should be equipment independent, and therefore, translatable across different FLIM systems. In practice, however, this is only partly true. Typically, due to specifics of the fitting algorithms and type of the equipment for data acquisition used, the results that can be easily reproduced and compared can be obtained only among the systems of one supplier. Even so, this is a substantial practical advantage of FLIM over conventional intensity-based techniques that the probe does not need to be recalibrated whenever image acquisition settings (e.g., laser power, exposure time, detection range) are changed. Consequently, one calibration can be used for different datasets or experiments. In our study, pH imaging with SypHerRed was performed using two setups, two-photon FLIM microscopy and one-photon macro-FLIM, both being based on the time-correlated single photon counting technique from Becker & Hickl. Test measurements on solution of Rhodamin 110 in water verified that the fluorescence lifetimes measured using both systems are 4 ns and 4.05 ns, which basically are the same within the error of the measurements. Therefore, we can exclude any artifacts related to the measurements on different systems. However, since a macrosystem probes whole cells, to account for intracellular heterogeneity of the sensor fluorescence lifetime, quantification of pH for the tumors on a macro scale has required a different calibration. As a result of integral signal collection, the lifetimes recorded from the cultured cells on the macro-FLIM system were slightly shorter (by ∼ 0.1 ns) than those obtained on the microscope upon accurate selection of ROIs in the cytoplasm or in the nucleus, but the resulting pH in the whole cells was identical.
One more advantage of SypHerRed as a lifetime-based sensor is its single-color operation. Unlike ratiometric variants, it occupies only one spectral channel, releasing other channels for possible combinations with fluorophores of a different color for multiparameter imaging. Using combined imaging of SypHerRed (in red range) and autofluorescence of the metabolic redox cofactor NADH (in blue range), we were able to monitor pHc and metabolic shifts simultaneously in the same live cells. Since pHc is intrinsically coupled to cellular metabolism, we observed correlative changes in fluorescence lifetime of NADH and pHc upon inhibition of glycolysis and complex I of the mitochondrial electron transport chain. Such multiparameter measurements are not restricted to in vitro studies but also can be performed in vivo on animal tumor models.
Conclusion
The intracellular pH measurements are important for the understanding of biological roles of pH in disease progression and the development of pH-based therapeutic approaches. In this paper, we report a method that allows quantitative measurement of intracellular pH in live cultured cells and tumors in vivo using FLIM, the latter being done for the first time to our knowledge. We demonstrate the approach to simultaneous visualization of pH and NADH in cells and believe that, due to fluorescence lifetime-based readout and red-shifted emission spectrum, pH sensor SypHerRed will be a valuable instrument for multiparameter imaging in vivo. An ability to determine absolute pH value in a tissue using SypHerRed might be applied not only for cancer studies, but also for various pathological states (e.g., ischemic stroke, neurotrauma, wounds, inflammation) associated with the alterations of pH homeostasis.
Author contributions
L.S., M.L., I.D., and V.D. collected the data. L.S., V.S., M.L., and V.S. analyzed and interpreted FLIM data. E.P. and V.B. prepared the cell lines expressing the sensors. M.S. and E.Z. designed the study. L.S., V.S., V.B., and M.S. wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
The study of pH was supported by the Russian Foundation for Basic Research (Project No. 19-32-90139). The study of metabolism was supported by the Russian Science Foundation (Project No. 20-65-46018). The development of the probes for tumor imaging was supported by the Ministry of Science and Higher Education of the Russian Federation, grant 075-15-2019-1933.
Editor: Amy E. Palmer.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.02.036.
Contributor Information
Vladislav Shcheslavskiy, Email: vis@becker-hickl.de.
Marina Shirmanova, Email: shirmanovam@gmail.com.
Supporting material
References
- 1.Casey J.R., Grinstein S., Orlowski J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 2.Song C.W., Griffin R., Park H.J. Cancer Drug Discovery and Development: Cancer Drug Resistance. Springer Nature; 2006. Influence of tumor pH on therapeutic response; pp. 21–42. [DOI] [Google Scholar]
- 3.Webb B.A., Chimenti M., et al. Barber D.L. Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer. 2011;11:671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- 4.Damaghi M., Wojtkowiak J.W., Gillies R.J. pH sensing and regulation in cancer. Front. Physiol. 2013;4:370. doi: 10.3389/fphys.2013.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderon-Montano J.M., Burgos-Moron E., et al. Lopez-Lazaro M. Role of the intracellular ph in the metabolic switch between oxidative phosphorylation and aerobic glycolysis - relevance to cancer. WebmedCentral Cancer. 2011;3:WMC001716. doi: 10.9754/journal.wmc.2011.001716. [DOI] [Google Scholar]
- 6.Manoli S.S., Kisor K., et al. Barber D.L. Ethyl isopropyl amiloride decreases oxidative phosphorylation and increases mitochondrial fusion in clonal untransformed and cancer cells. Am. J. Physiol. Cell Physiol. 2021;321:C147–C157. doi: 10.1152/ajpcell.00001.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagadic-Gossmann D., Huc L., Lecureur V. Alterations of intracellular pH homeostasis in apoptosis: origins and roles. Cell Death Differ. 2004;11:953–961. doi: 10.1038/sj.cdd.4401466. [DOI] [PubMed] [Google Scholar]
- 8.Zanke B.W., Lee C., et al. Tannock I.F. Death of tumor cells after intracellular acidification is dependent on stress-activated protein kinases (SAPK/JNK) pathway activation and cannot be inhibited by Bcl-2 expression or interleukin 1/3-converting enzyme inhibition. Cancer Res. 1998;58:2801–2808. [PubMed] [Google Scholar]
- 9.Anemone A., Consolino L., et al. Longo D.L. Imaging tumor acidosis: a survey of the available techniques for mapping in vivo tumor pH. Cancer Metastasis Rev. 2019;38:25–49. doi: 10.1007/s10555-019-09782-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang R., Yu C., et al. Yu C. Molecular fluorescent probes for monitoring pH changes in living cells. TrAC Trends Anal. Chem. 2010;29:1004–1013. doi: 10.1016/j.trac.2010.05.005. [DOI] [Google Scholar]
- 11.Benčina M. Illumination of the spatial order of intracellular pH by genetically encoded pH-sensitive sensors. Sensors (Basel) 2013;13:16736–16758. doi: 10.3390/s131216736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martynov V.I., Pakhomov A.A., et al. Petrenko A.G. Genetically encoded fluorescent indicators for live cell pH imaging. Biochim. Biophys. Acta Gen. Subj. 2018;1862:2924–2939. doi: 10.1016/j.bbagen.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Frommer W.B., Davidson M.W., Campbell R.E. Genetically encoded biosensors based on engineered fluorescent proteins. Chem. Soc. Rev. 2009;38:2833. doi: 10.1039/b907749a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitt F.-J., Thaa B., et al. Friedrich T. eGFP-pHsens as a highly sensitive fluorophore for cellular pH determination by fluorescence lifetime imaging microscopy (FLIM) Biochim. Biophys. Acta. 2014;1837:1581–1593. doi: 10.1016/j.bbabio.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Battisti A., Digman M.A., et al. Bizzarri R. Intracellular pH measurements made simple by fluorescent protein probes and the phasor approach to fluorescence lifetime imaging. Chem. Commun. 2012;48:5127. doi: 10.1039/c2cc30373f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tantama M., Hung Y.P., Yellen G. Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor. J. Am. Chem. Soc. 2011;133:10034–10037. doi: 10.1021/ja202902d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poëa-Guyon S., Pasquier H., et al. Erard M. The enhanced cyan fluorescent protein: a sensitive pH sensor for fluorescence lifetime imaging. Anal. Bioanal. Chem. 2013;405:3983–3987. doi: 10.1007/s00216-013-6860-y. [DOI] [PubMed] [Google Scholar]
- 18.Burgstaller S., Bischof H., et al. Malli R. pH-Lemon, a fluorescent protein-based pH reporter for acidic compartments. ACS Sensors. 2019;4:883–891. doi: 10.1021/acssensors.8b01599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poburko D., Santo-Domingo J., Demaurex N. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J. Biol. Chem. 2011;286:11672–11684. doi: 10.1074/jbc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matlashov M.E., Bogdanova Y.A., et al. Belousov V.V. Fluorescent ratiometric pH indicator SypHer2: applications in neuroscience and regenerative biology. Biochim. Biophys. Acta. 2015;1850:2318–2328. doi: 10.1016/j.bbagen.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ermakova Y.G., Bilan D.S., et al. Belousov V.V. Red fluorescent genetically encoded indicator for intracellular hydrogen peroxide. Nat. Commun. 2014;5:5222. doi: 10.1038/ncomms6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sergeeva T.F., Shirmanova M.V., et al. Zagaynova E.V. Relationship between intracellular pH, metabolic co-factors and caspase-3 activation in cancer cells during apoptosis. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:604–611. doi: 10.1016/j.bbamcr.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 23.Shirmanova M.V., Druzhkova I.N., et al. Snopova L.B. Intracellular pH imaging in cancer cells in vitro and tumors in vivo using the new genetically encoded sensor SypHer2. Biochim. Biophys. Acta. 2015;1850:1905–1911. doi: 10.1016/j.bbagen.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Addgene Vsevolod Belousov lab plasmids. 2014. http://www.addgene.org/browse/article/9267/
- 25.Zagaynova E.V., Druzhkova I.N., et al. Shirmanova M.V. Imaging of intracellular pH in tumor spheroids using genetically encoded sensor SypHer2. Adv. Exp. Med. Biol. 2017;1035:105–119. doi: 10.1007/978-3-319-67358-5_7. [DOI] [PubMed] [Google Scholar]
- 26.Shcheslavskiy V.I., Shirmanova M.V., et al. Becker W. Fluorescence time-resolved macroimaging. Opt. Lett. 2018;43:3152. doi: 10.1364/ol.43.003152. [DOI] [PubMed] [Google Scholar]
- 27.Shimolina L., Lukina M., et al. Shirmanova M. Probing metabolism and viscosity of cancer cells using fluorescence lifetime imaging microscopy. J. Vis. Exp. 2021 doi: 10.3791/62708. [DOI] [PubMed] [Google Scholar]
- 28.Kolenc O.I., Quinn K.P. Evaluating cell metabolism through autofluorescence imaging of NAD(P)H and FAD. Antioxid. Redox Signal. 2018;20:875–889. doi: 10.1089/ars.2017.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shirmanova M.V., Shcheslavskiy V.I., et al. Zagaynova E.V. Multimodal Optical Diagnostics of Cancer. Springer Nature; 2020. Exploring tumor metabolism with time-resolved fluorescence methods: from single cells to a whole tumor; pp. 133–155. [DOI] [Google Scholar]
- 30.Sanders R., Gerritsen H.C., et al. Levine Y.K. Confocal fluorescence lifetime imaging of pH in single cells. SPIE Proc. 1994;2137:56–62. doi: 10.1117/12.182761. [DOI] [PubMed] [Google Scholar]
- 31.Hanson K.M., Behne M.J., et al. Clegg R.M. Two-photon fluorescence lifetime imaging of the skin stratum corneum pH gradient. Biophys. J. 2002;83:1682–1690. doi: 10.1016/s0006-3495(02)73936-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill C., Berg M., et al. Dosche C. Time-domain fluorescence lifetime imaging for intracellular pH sensing in living tissues. Anal. Bioanal. Chem. 2008;391:1871–1879. doi: 10.1007/s00216-008-2147-0. [DOI] [PubMed] [Google Scholar]
- 33.Lin H.-J., Herman P., Lakowicz J.R. Fluorescence lifetime-resolved pH imaging of living cells. Cytometry. 2003;52A:77–89. doi: 10.1002/cyto.a.10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ning Y., Cheng S., et al. Zhang J.-L. Fluorescence lifetime imaging of upper gastrointestinal pH in vivo with a lanthanide based near-infrared τ probe. Chem. Sci. 2019;10:4227–4235. doi: 10.1039/c9sc00220k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pacheco-Liñán P.J., Bravo I., et al. Garzón-Ruiz A. Functionalized CdSe/ZnS quantum dots for intracellular pH measurements by fluorescence lifetime imaging microscopy. ACS Sensors. 2020;5:2106–2117. doi: 10.1021/acssensors.0c00719. [DOI] [PubMed] [Google Scholar]
- 36.Korenchan D.E., Flavell1 R.R. Spatiotemporal pH heterogeneity as a promoter of cancer progression and therapeutic resistance. Cancers (Basel) 2019;11:1026. doi: 10.3390/cancers11071026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dross N., Spriet C., et al. Langowski J. Mapping eGFP oligomer mobility in living cell nuclei. PLoS One. 2009;4:e5041. doi: 10.1371/journal.pone.0005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Llopis J., McCaffery J.M., et al. Tsien R.Y. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. U S A. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santos J.M., Martínez-Zaguilán R., et al. Sennoune S.R. Vacuolar H+-ATPase in the nuclear membranes regulates nucleo-cytosolic proton gradients. Am. J. Physiol. Cell Physiol. 2016;311:C547–C558. doi: 10.1152/ajpcell.00019.2016. [DOI] [PubMed] [Google Scholar]
- 40.Seksek O., Bolard J. Nuclear pH gradient in mammalian cells revealed by laser microspectrofluorimetry. J. Cell Sci. 1996;109:257–262. doi: 10.1242/jcs.109.1.257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.