Abstract
Objective
Fibrotic organ responses have recently been identified as long-term complications in diabetes. Indeed, insulin resistance and aberrant hepatic lipid accumulation represent driving features of progressive non-alcoholic fatty liver disease (NAFLD), ranging from simple steatosis and non-alcoholic steatohepatitis (NASH) to fibrosis. Effective pharmacological regimens to stop progressive liver disease are still lacking to-date.
Methods
Based on our previous discovery of transforming growth factor beta-like stimulated clone (TSC)22D4 as a key driver of insulin resistance and glucose intolerance in obesity and type 2 diabetes, we generated a TSC22D4-hepatocyte specific knockout line (TSC22D4-HepaKO) and exposed mice to control or NASH diet models. Mechanistic insights were generated by metabolic phenotyping and single-nuclei RNA sequencing.
Results
Hepatic TSC22D4 expression was significantly correlated with markers of liver disease progression and fibrosis in both murine and human livers. Indeed, hepatic TSC22D4 levels were elevated in human NASH patients as well as in several murine NASH models. Specific genetic deletion of TSC22D4 in hepatocytes led to reduced liver lipid accumulation, improvements in steatosis and inflammation scores and decreased apoptosis in mice fed a lipogenic MCD diet. Single-nuclei RNA sequencing revealed a distinct TSC22D4-dependent gene signature identifying an upregulation of mitochondrial-related processes in hepatocytes upon loss of TSC22D4. An enrichment of genes involved in the TCA cycle, mitochondrial organization, and triglyceride metabolism underscored the hepatocyte-protective phenotype and overall decreased liver damage as seen in mouse models of hepatocyte-selective TSC22D4 loss-of-function.
Conclusions
Together, our data uncover a new connection between targeted depletion of TSC22D4 and intrinsic metabolic processes in progressive liver disease. Hepatocyte-specific reduction of TSC22D4 improves hepatic steatosis and promotes hepatocyte survival via mitochondrial-related mechanisms thus paving the way for targeted therapies.
Keywords: TSC22D4, Fibrosis, NASH, NAFLD, Hepatocyte-specific
Abbreviations: tsc22d4, Transforming growth factor beta-like stimulated clone 22 domain 4; snRNA-seq, single-nuclei RNA-sequencing; TG, triglycerides; MCD, methionine-choline deficient; Met, metformin; LNP, lipid nanoparticle; CCl4, carbon tetrachloride; ETC, electric transport chain
Highlights
-
•
TSC22D4 is significantly upregulated in both patients with NASH and liver fibrosis and in murine models of NAFLD.
-
•
Hepatocyte-specific knockdown of TSC22D4 results in reduced steatosis and hepatocyte damage in MCD diet fed mice.
-
•
Single-nuclei RNA sequencing from TSC22D4-HepaKO mice reveals enrichment in lipid metabolism and OXPHOS pathways.
-
•
Loss of TSC22D4 increases the oxidative consumption rate and upregulates mitochondrial maintenance genes in hepatocytes.
-
•
TSC22D4 expression negatively correlates with mitochondrial targets in NASH patients.
1. Introduction
With more than 1.5 billion overweight subjects worldwide, the prevalence of obesity-related metabolic complications is steadily increasing and imposes continuous pressure on global health care systems. In this regard, non-alcoholic fatty liver disease (NAFLD) is commonly regarded as the hepatic manifestation of the obesity-driven metabolic syndrome. Indeed, fatty liver disease characterizes more than 60% of obese patients, and NAFLD has recently been identified as a main predictor for diabetes risk in humans with prediabetes [1]. Importantly, progression of steatosis to non-alcoholic steatohepatitis (NASH) and eventually fibrosis, represents the key etiology for the development of liver cancer, starting to outpace today's classical, virus-related causes for liver cancer [2]. Despite its clinical importance, effective therapeutic modalities against NAFLD have not been established to date. This can at least partially be explained by the involvement of different hepatic cell types in its pathogenesis and the resulting signaling complexity. In fact, progressive liver disease results from the intricate interplay between distinct hepatic cell populations, including inflammatory cells, hepatic stellate cells and hepatocytes [3]. Indeed, a number of secreted mediators that drive the mutual changes in cellular phenotypes upon NASH progression have been identified, e.g., TGFβ [2,4].
Transforming growth factor stimulated clone (TSC)22D4 is a globally expressed protein that belongs to the TSC protein family, which share a TSC box containing a leucine zipper motif mediating homodimerization or heterodimerization with other family members [5]. Members of the TSC family play diverse roles such as in glucocorticoid and stress signaling, cell proliferation, and apoptosis, yet the exact mechanism of action remains unknown [[6], [7], [8], [9]]. However, we have recently shown that livers from mice with cancer cachexia have elevated TSC22D4 expression levels, positively correlating with the degree of tissue wasting and dysregulated lipid metabolism [10]. Follow-up studies demonstrated elevated hepatic levels of TSC22D4 also in obese patients with type 2 diabetes, particularly linked to insulin resistance [11]. Indeed, initial proof-of-principle studies highlighted the utility of AAV-driven continuous inhibition of TSC22D4 to counteract diabetes-related metabolic phenotypes in mouse models of type 2 diabetes. Hepatic knockdown of TSC22D4 not only ameliorated insulin resistance but also improved blood glucose levels without any adverse changes in body mass [11]. Overall, these studies positioned hepatic TSC22D4 as a potentially druggable upstream regulator of both lipid and glucose metabolism in type 2 diabetes.
However, whether TSC22D4 beyond its acute impact on key metabolic parameters may also exert control in long-term diabetic complications, particularly liver fibrosis, remained unclear but would underscore its pharmacological attractiveness.
Here, we show that TSC22D4 correlated with fibrosis in both murine and human liver samples. Depletion of TSC22D4 in hepatocytes improved liver lipid accumulation in mice fed NASH-promoting diets and reduced liver injury. Furthermore, single-nuclei RNA sequencing revealed in hepatocytes an upregulation of genes involved in enhanced mitochondrial efficiency, lipid metabolism, oxidative phosphorylation under conditions of progressive fatty liver disease. Intriguingly, these TSC22D4 target genes were reciprocally decreased in the livers of patients with progressive NAFLD and NASH.
2. Materials and methods
2.1. Animals
Mice were housed at room temperature with 12-h light–dark cycle on control diet (Research diets, New Brunswick, NJ, USA, D12450B), 60% high fat diet (Research diets, D12492i), control for MCD (Research diets, A02082003BY), MCD (Research diets, A02082002BR) or chow for weeks as indicated in particular study. Mice had ad libitum access to food and water and were weighed weekly and inspected daily for general health. At the conclusion of each study, tissues were flash frozen for analysis or placed in histofix (4% formaldehyde, Carl Roth GmbH, Karlsruhe, Germany) for immunohistochemistry.
Carbon tetrachloride (CCl4,) experiment, 9-week-old, male Balb/c mice were purchased from Janvier Labs (Le Genest-Saint-Isle, France). CCl4 was diluted 1:5 in olive oil (Carl Roth 8873.1) and injected intraperitoneally (i.p.) at a concentration of 0.5 μL/g body weight, (VWR International SIAL289116) 3× weekly for two weeks. Tissues were collected as described above. STAM™ 10 week old liver tissues in Figure 2 were purchased from SMC Laboratories, Inc. (Tokyo, Japan).
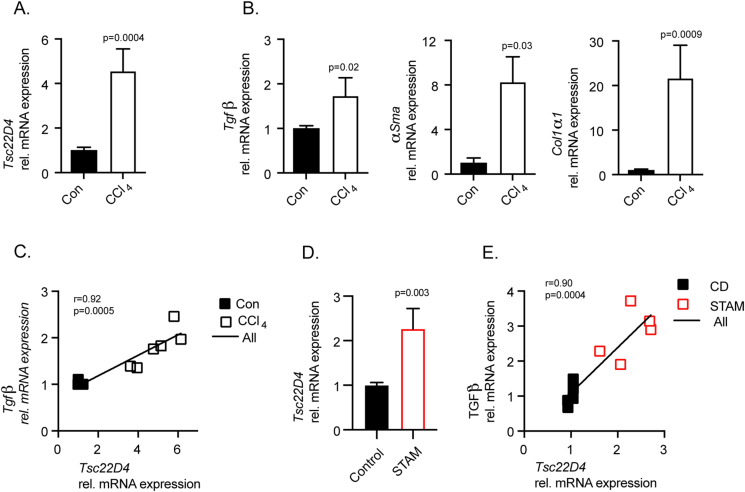
Figure 2.
TSC22D4 is upregulated in mouse models of fibrosis. Carbon tetrachloride (CCl4) treated mice (n = 3 control; n = 6 CCl4) demonstrated significant upregulation of Tsc22d4 mRNA (A) in conjunction with markers of fibrosis collagen 1α1, α-smooth muscle actin, and TGFβ (B). Tsc22d4 correlated with marker of liver disease progression, TGFβ (C). STAM mice (n = 5 control; n = 5 STAM), streptozotocin-induced plus 9w of HFD, exhibited increased Tsc22d4 mRNA expression (D) that correlated with TGFβ (E) in the liver. Data are mean ± SEM. Abbreviations: HFD, high fat diet.
NASH and fibrosis studies were performed with Albumin-cre mice crossed to the Tsc22d4 floxed/floxed line (C57Bl/6N background) (Supp Figure 1). The targeting strategy was carried out by Taconic Biosciences GmbH (Cologne, Germany) based on NCBI transcript NM_023910.6. A targeting vector containing genomic fragments of the mouse Tsc22d4 locus as well as the loxP flanked region was generated. LoxP sites flanked the genomic region containing exons 1 and 2 and 1.5 kb upstream of exon 1 (promoter region).
Genotyping. The presence of the floxed allele was confirmed by PCR performed using DNA extracted from mouse earclip biopsies. Expected PCR products were 266bp for the Tsc22d4 wild-type allele and 385 for the floxed allele. For routine genotyping, primers flanking the loxP site located to the right of exon 2 were used (forward CTTCCCTGATTTCAGGTGATG and reverse ATCCCACACTCCAGGAGAAG). For Albumin-cre crossed mice, a PCR 320bp fragment identifying the presence of cre was observed (forward primer GAACCTGATGGACATGTTCAGG and reverse primer AGTGCGTTCGAACGCTAGAGCCTGT).
STAM experiments. Male mice with hepatocyte-specific deletion of TSC22D4 or cre− littermates (TSC22D4-HepaKO and WT) were treated as previously published [12]. Briefly, 2-day old pups were injected with a low dose of streptozotocin (Sigma) 200 μg i.p., randomized at 3 weeks of age and placed on control (CD) or high fat diet (HFD). Mice continued on diet for 12 weeks until tissue collection as described above.
For MCD diet studies, male, hepatocyte-specific TSC22D4 Ko mice and Wt littermates at 10 weeks of age were fed MCD or matched MCD control diet for 3 weeks and tissues collected as described above.
2.2. Lipid nanoparticle formulation
Preparation of LUNAR-siRNA Nanoparticles. Tsc22d4 siRNA (sense: 5′-UNA-G/mGrAmCrGmUrGmUrGrUrGmGrAmUrGmUrUmUrA/UNA-U/mU-3’; antisense: 5′-mUrAmArAmCrAmUrCmCrAmCmAmCrAmCrGmUrCmC/UNA-U/mU-3′, wherein rN is RNA, mN is 2′-O-methyl RNA, and UNA is unlocked nucleic acid), and non-targeting control siRNA (sense: 5′-UNA-U/mArGmCrGmArCmUrArArAmCrAmCrAmUrCmGrC/UNA-U/mU-3′, antisense: 5′-UNA-G/rCmGrAmUrGmUrGmUrUmUmAmGrUmCrGmCrUmA/UNA-U/mU-3′, wherein rN is RNA, mN is 2′-O-methyl RNA, and UNA is unlocked nucleic acid) were synthesized by Integrated DNA Technologies (Coralville, IA). ATX lipid was synthesized as described previously [13]. LNPs were prepared as described previously [14] by mixing appropriate volumes of lipids in ethanol with an aqueous phase containing siRNA duplexes using a Nanoassemblr microfluidic device, followed by downstream processing. For the encapsulation of siRNA, desired amount of siRNA (Tsc22d4 or non-targeting control siRNA) was dissolved in 5 mM citrate buffer (pH 3.5). Lipids at a molar ratio of 58% ionizable ATX lipid, 7% DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine) (Avanti Polar Lipids), 33.5% cholesterol (Avanti Polar Lipids), and 1.5% DMG-PEG (1,2-dimyristoyl-sn-glycerol, methoxypolyethylene glycol; PEG chain molecular weight: 2000) (NOF America Corporation) were dissolved in ethanol. At a flow ratio of 1:3 ethanol/aqueous phases, the solutions were combined in the microfluidic device using Nanoassemblr (Precision NanoSystems). The total combined flow rate was 12 mL/min per microfluidics chip and the total lipid-to-RNA weight ratio was ∼30:1. The mixed material was then diluted three times with Tris buffer (pH 7.4) containing 50 mM sodium chloride and 9% sucrose after leaving the micromixer outlet, reducing the ethanol content to 6.25%. The diluted LNP formulation was concentrated and diafiltered by tangential flow filtration using spectrum hollow fiber membranes (mPES Kros membranes, Repligen) and Tris buffer (pH 7.4) containing 50 mM sodium chloride and 9% sucrose. A total of 10 diavolumes were exchanged, effectively removing the ethanol. The particle size and polydispersity index (PDI) were characterized using a Zen3600 (Malvern Instruments, with Zetasizer 7.1 software, Malvern, U.K.). Encapsulation efficiency was calculated by determining the unencapsulated siRNA content by measuring the fluorescence intensity (Fi) upon the addition of RiboGreen (Molecular Probes) to the LNP and comparing this value to the total fluorescence intensity (Ft) of the RNA content that is obtained upon lysis of the LNPs by 1% Triton X-100, where % encapsulation = (Ft − Fi)/Ft × 100). Encapsulation efficiencies of these formulations were >90%.
2.3. Gene expression
RNA was prepared using the RNeasy micro kit (QIAGEN) including DNase treatment following the manufacturer's protocol. Complementary DNA (cDNA) synthesis was performed with 200–1,000 ng total RNA using QuantiTect_ Reverse Transcription Kit (Qiagen). Quantitative real time polymerase chain reaction (qRT–PCR) was performed using TaqMan_ Gene Expression Assays (all Life Technologies, Darmstadt, Germany). TaqMan_Gene Expression Master Mix and the StepOnePlusTM Real-Time PCR System (Life Technologies) for Tsc22D4 (Mm00470231_m1), Tfgβ (Mm01178820_m1), αSma (Mm01546133_m1), Col1α1 (Mm00801666_g1), Ndufa1 (Mm00444593_m1), Ndufa12 (Mm01240336_m1), Ndufa8 (Mm00503351_m1), Ndufb2 (Mm00471671_m1), F4/80 (Mm00802529_m1), Tnfα (Mm00443258_m1), Timp1 (Mm01341361_m1), Fasn (Mm00662319_m1), Acly (Mm00652520_m1), Scd1 (Mm00772290_m1), and Lipin1 (Mm00550511_m1). Expression levels were normalized to Tbp (TATA box binding protein, Mm01277042_m1). For human samples, TGFβ (Hs00998133_m1), αSMA (Hs00426835_g1), COL1α1 (Hs00164004_m1), NDUFA1 (Hs00244980_m1), NDUFA12 (Hs00984333_m1), NDUFB2 (Hs00190006_m1), NDUFA8 (Hs00204417_m1), TSC22D4 (Hs00229526_m1) were analyzed and normalized to the expression of 18s rRNA (Hs99999901_s1). Calculation of relative expression values were performed with the ΔΔCT method. DNA was isolated using the DNeasy Blood and Tissue kit (Qiagen) and nuclear DNA marker B2m (beta-2-microglobulin, Mm01269327_g1) with mitochondrial DNA marker Cytb (cytochrome b, Mm04225271_g1) (Life Technologies).
2.4. Lipid and collagen measurements in mice
Triglycerides (TR0100, Sigma Aldrich) and total cholesterol (CH200, Randox, Crumlin, UK) were measured in serum and liver lipid lysates using the Mithras Microplate Reader (Berthold Technologies GmbH & Co, Bad Wildbad, Germany) at 550 nm. Liver lipids were extracted as described previously [15]. Serum levels of HDL-cholesterol were analyzed with an AdviaXPT® 2400 clinical chemistry automation (Siemens Healthineers) according to the standard operating protocol. Sircol soluble collagen assay kit (Biocolor life science assays) was used to quantify collagen in mouse liver tissue according to the manufacturer's instructions. Briefly, approximately 25–50 mg of liver material was digested with pepsin overnight at 4 °C. Neutralized samples were concentrated and mixed with Sirius red dye reagent for 30 min before washing and centrifugation. Alkali reagent was added to release collagen bound dye into solution. Samples were measured on the Mithras Microplate Reader at 555 nm.
2.5. Quantitative nash assessment in mice
Liver samples were fixed with 4% Histofix (Carl Roth) at room temperature for 24 h then embedded in paraffin wax. Blocks were cut into sections 4 μm thick sections each and placed on glass slides. Sections were de-paraffinized with xylene and ethanol dilutions to rehydrate. Following rehydration and rinsing, slides were stained with Picro-Sirius red (ref# 1422.00500, Morphisto GmbH, Vienna, Austria) for 60 min and dehydrated followed by mounting. For pathological assessment hematoxylin and eosin stained mouse liver sections (Leica ST5020 auto-stainer) were evaluated according to Kleiner DE, et al. Hepatology (2005), modified by Liang et al., PLOS (2014) and adopted for the present study consisting of a scoring strategy with features grouped into five broad categories: steatosis, inflammation, hepatocellular injury, fibrosis, and miscellaneous features [16,17].
2.6. Human cohorts
Human NAFLD cohort. The prospective BARIA_DDZ study examines and follows obese individuals undergoing bariatric surgery and lean, healthy humans (controls) undergoing elective surgery such as cholecystectomy or herniotomy (registered clinical trial no. NCT01477957) [18,19]. The cohort evaluated here consisted of 33 females and 12 males with mean age of 42.5 years and mean BMI of 45.8 kg/m2. All participants were stratified based on histological assessment in groups without steatosis (NAFL−) or with steatosis (NAFL+) steatosis and inflammation (NASH) as described previously [18,19]. All participants were examined using hyperinsulinaemic-euglycaemic clamps to determine peripheral and hepatic insulin sensitivity, and blood was sampled for routine lab parameters [18]. Participants maintained stable body weight for at least 2 weeks before the surgery. The study was approved by the Heinrich Heine University Dusseldorf Institutional Review Board and informed consent obtained in writing from all participants.
Human fibrosis cohort. Human liver samples were provided by the Tissue Bank of the National Center for Tumor Diseases (NCT) Heidelberg, Germany in accordance with the regulations of the tissue bank and the approval of the ethics committee of Heidelberg University [20]. The cohort evaluated here comprised 15 females and 24 males with and average age of 60.8 years and BMI 25.6 kg/m2.
Human NAFLD and NASH cohort. A second, independent NAFLD and NASH cohort was included from 26 females and 12 males of the Leipzig Obesity BioBank [21] and represent a mean age of 54.5 years and mean BMI of 45.1 kg/m2. Histological examination by a pathologist classified 9 individuals with no fibrosis, 24 individuals with mild or low fibrosis, and 5 individuals with moderate fibrosis. All individuals were classified as either grade 1 (slight portal inflammation, no or minimal acinar parenchymal cell deaths, no boarder zone hepatitis) or grade 2 (moderate portal inflammatory cell infiltration, slight, focal border zone hepatitis, single parenchymal single cell necrosis, no group necrosis). The study was approved by the Ethics Committee of the University of Leipzig (approval numbers: 363-10-13122010 159-12-21052012), and performed in accordance to the declaration of Helsinki. All subjects gave written informed consent to use their data in anonymized form for research purposes before taking part in this study.
2.7. Rna sequencing
Single nuclei isolation All mouse liver tissues were flash frozen in liquid nitrogen upon collection and then pulverized on dry ice without thawing using a tissue lyser, (Qiagen). Briefly, pulverized liver was placed into a pre-chilled Dounce homogenizer (Lab Logistics, #9651632) with 1 mL of homogenization buffer (HB) [(250 mM sucrose, 25 mM KCl, 5 mM MgCl2, 10 mM Tris buffer, 1 μM DTT), 1 × protease inhibitor tablet (Sigma Aldrich Chemie), 0.4 U/μL RNaseIn (Thermo Fisher Scientific), 0.2 U/μL Superasin (Thermo Fisher Scientific), 0.1% Triton X-100 (v/v) and 10 μg/mL Hoechst 33342 (Thermo Fisher Scientific) in RNase-free water], as described by Krishnaswami et al. [22]. After 5 min incubation in HB, we performed 5 slow strokes with the loose pestle, followed by 10 strokes with the tight tolerance pestle. The suspension was passed through a 50 μm sterilized filter (CellTrics, Symtex, #04-004-2327) into a 2 mL pre-chilled Eppendorf pre-chilled and an additional 1 mL of cold HB was used to wash the Dounce homogenizer and the filter. The sample was then centrifuged for 8 min at 1000×g in a swinging bucket centrifuge. The pellet was resuspended in 250 μL of pre-chilled HB and a density gradient centrifugation clean-up was performed for 20 min, using Iodixanol gradient (Optiprep, D1556, Sigma Aldrich Chemie). The final pellet was resuspended gently in 200 μL of nuclei storage buffer (NSB) (166.5 mM sucrose, 5 mM MgCl2, 10 mM Tris buffer pH 8.0) containing additional RNase inhibitors 0.2 U/μL Superasin (Thermo Fisher Scientific, #AM2696) and 0.4 U/μL Recombinant RNase Inhibitor (Takara Clontech #2313A). The final suspension was filtered through a 35 μm cell strainer cap into a pre-chilled FACS tube prior to FACS sorting.
Flow cytometry Prior to sorting, 384-well PCR plates (thin-walled, BioRad, HSP3901) were freshly prepared with 810 nL of initial Lysis Buffer (LB1) using the Mosquito HV liquid handling robot (STP Labtech) as published previously [23]. Upon sorting, Hoechst stained diploid and tetraploid nuclei were singly sorted using a FACS sorter (BD FACSAria 3 sorter), 1 nucleus per well, with a 100 μm nozzle. Drop delay and cut-off point were optimized prior to every sorting, and the plate holder was maintained at 4 °C. Sorting accuracy was assessed using a colorimetric method with tetramethyl benzidine substrate (TMB, BioLegend, #421501) and Horseradish Peroxidase (HRP, Life Technologies, #31490) as described at Rodriguez et al. [24]. The gating strategy used FSC-A/SSC-A to select intact nuclei stained with Hoechst, in which events above 10ˆ4 are considered Hoechst positive. Subsequently, selection of singlets was performed by FSC-A/FSC-H followed by exclusion of doublets by FSC-A/FSC-W as previously described [25]. After sorting, every plate was firmly sealed (MicroAmp Thermo Seal lid, #AB0558), shortly vortexed for 10 s, centrifuged at 4 °C, 2000×g for 1 min, frozen on dry ice, and stored at −80 °C until cDNA synthesis.
snRNA-seq2 The single-nucleus-RNA-seq2 methodology was used to capture a high number of transcripts from frozen tissues, allowing for the generation of double-stranded full-length cDNA as described by Richter et al. 2021. The SMART-Seq Single Cell Kit (SSsc; Cat. # 634472, Takara) was used and reaction volumes were miniaturized 6 times with the aid of the Mosquito HV robot as per the kit provider User Guide (cDNA synthesis using the mosquito HV genomics with the SMART-Seq® Single Cell Kit) addition of the second lysis buffer (LB2), containing NP40 2%, Triton-X100 1%, 1/400,000 diluted ERCC RNA spike-in, 3′ SMART-seq CDS Primer II A and RNAse-free water, results to a volume of 1.7 uL. Plates were immediately sealed, vortexed 20 s at 2000 rpm, centrifuged at 2000×g for 30 s at 4 °C and incubated at 72 °C for 6 min. ERCC spike-ins (Thermo Fischer Scientific, Ref. 4456740) were diluted 1 in 10 with RNAse-free water, supplemented with 0.4 U/μL Recombinant RNase Inhibitor (Takara Clontech, Ref. 2313A). Fresh dilution of 1 in 400,000 was prepared immediately before the first strand synthesis. Reverse transcription and Pre-PCR amplification steps were followed as described by the manufacturer and the PCR program for the cDNA amplification was performed in a total of 20 cycles of: 1 min at 95 °C, [20 s at 95 °C, 4 min at 58 °C, 6 min at 68 °C] × 5, [20 s at 95 °C, 30 s at 64 °C, 6 min at 68 °C] × 9, [30 s at 95 °C, 30 s at 64 °C, 7 min at 68 °C] × 6, 10 min at 72 °C.
We purified the amplified cDNA using Agencourt XP beads at 0,7× (Catalog No. A63881, Beckman) on the Mosquito HV, and then performed quantification of cDNA in an Agilent Bioanalyzer with a High Sensitivity DNA chip.
Library preparation Sequencing libraries were prepared using the Illumina Nextera XT DNA Sample Preparation kit (Illumina, Ref. FC-131-1096) and the combination of 384 Unique Dual Indexes (Illumina- Set A to D, Ref. 20027213–20027216). Using the Mosquito HV robot, the reaction volumes of the Nextera XT chemistry were miniaturized by 10 times, following steps as previously described [26] In brief, 500 nL of undiluted cDNA (∼200pg/uL) were transferred to a new 384 well-plate containing 1500 nL of Tagmentation Mix (TD and ATM reagents). Accordingly, all Nextera XT reagents (NT, NPM and i5/i7 indexes) were added stepwise to a final library volume of 5 μL per well. The final PCR amplification was performed through 12 cycles. Once the libraries were prepared, 500 nL from each well were pooled together into a 1.5 mL tube to perform two consecutive clean-up steps with a ratio of sample to AMPure XP beads of 0,9× (Beckman Coulter, Ref. A63882). The final library sizes ranged between 200 and 1000 bp as assessed using a HS DNA kit in the Agilent Bioanalyzer. Prior to sequencing, the libraries were quantified using a Collibri library quantification kit (Thermo Fischer Scientific, Ref. A38524100) in a QuantStudio 6 Flex (Life Technologies).
Sequencing Each of the pooled libraries from the 384 well plates were sequenced using an Illumina NovaSeq 6000 NGS sequencer in one lane of an SP300 Xp flowcell, in a paired-end 150 bases length resulting on an average of 1Million reads per nuclei.
Analysis STAR aligner (v2.7.1a) was used to align raw sequencing reads against Mouse Genome Assembly GRCm39-mm10 and ERCC92. FastQC (v0.11.9) was used to do the quality control checks for raw and mapped reads. Quantification of the number of reads mapping was done by using featureCounts (v2.0.3) with -t gene (mouse genome) and -t exon (ERCC92). Any nuclei with ERCC spike-in transcripts <5% and >90% were excluded. Any nuclei with no genes detected were omitted in further analysis. Genes that were not detected in any nuclei were skipped. Differential gene expression analysis was performed using the R Bioconductor package DESeq2 (v3.14). For the KEGG pathway and GO analysis, the enrichKEGG and enrichGO function of R clusterProfiler package was used respectively. AnnData object of the count matrix was generated for python (scanpy) to represent the log2 fold changes in gene expression between conditions. The interaction network was constructed and visualized using Cytoscape, and nodes represented enriched terms colored by fold change and edges are colored by interaction type. Resampling was done by sample name switching and repeating the differential expression analysis (Supp. Figure 6D). Gene expression datasets GSE48452 (Ahrens M et al.), GSE49541 (Moylan C et al.), and GSE63067 (Frades I et al.) were obtained from Gene Expression Omnibus (GEO) database and the differentially expressed genes were analyzed by using the NCBI GEO2R tool.
Figure 6.
LNP-siRNA mediated knock down of TSC22D4 in primary hepatocytes increases oxygen consumption rate. Tsc22d4 mRNA levels after 2 days of siControl-LNP or siTSC22D4-LNP knockdown in primary hepatocytes in vitro, n = 6 per group (A). Seahorse measurements of OCR in primary hepatocytes (n = 10 wells per condition) n = 3 independent experiments (B). Metabolic rate calculations from the data in B (C). Violin plots of mitochondrial DNA (Cytb) to nuclear DNA (B2m) relative ratios in primary hepatocytes isolated from AlbCre(−) or AlbCre(+) TSC22D4floxed mice, n = 3 per group (D). RT-qPCR for mitochondrial complex I genes identified by snRNAseq, n = 5 per group (E). Data are mean ± SEM. In (B) all data points are relative to the initial respiration rate of siCtrl (100%), within each experiment. In (C) all data points are relative to basal respiration calculation of siC (100%), within each independent experiment. Abbreviations: OCR, oxygen consumption rate.
2.8. Hepatocyte isolation and transfection
Murine primary hepatocytes were isolated as described previously [27,28]. Briefly, under anesthesia the visceral cavity was opened and the liver perfused via the vena cava first with EGTA-containing HEPES/KH buffer followed by a collagenase-containing HEPES/KH buffer until liver digestion was visible. The liver was excised and placed in suspension buffer where hepatocytes were gently washed out. Cells were filtered through a 100-nm pore mesh, centrifuged, and washed twice in suspension buffer.
Seahorse assay Hepatocytes were seeded at 30′000/well in 24-well seahorse plates that had been coated with a thin layer of collagen and maintained in William's media E (PAN Biotech) containing 10% FBS (Sera Plus, PAN Biotech), 1% P/S, and 100 nM Dexamethasone (Sigma Aldrich, Germany). Two hours after seeding, cells were washed twice with PBS and then incubated with maintenance media in the presence of LNP-siRNAs (40 nM) and 2.4 μg/mL ApoE (Fitzgerald, Acton, MA, USA). Six hours later media was replaced with fresh media and Seahorse assay performed 48 later. A mitostress test was performed on a XFe24 Seahorse analyzer (Agilent Technologies) according to the manufacturer's instructions, with timed sequential injection of oligomycin 2 μM, FCCP 1 μM, and lastly rotenone + antimycin (1 μM each). Oxygen consumption rate (OCR) data were normalized to cell number, as quantified using the CellQUANT cell proliferation Assay Kit (Invitrogen).
2.9. Statistical analysis
Data were graphed and analyzed using GraphPad Prism 9 software (La Jolla, CA, USA).
2.10. Study approval
All experiments were performed in accordance with the European Union directives and the German animal welfare act (Tierschutzgesetz) and were approved by local authorities (Regierungspräsidium Karlsruhe, license #G117/18).
3. Results
3.1. Hepatic TSC22D4 expression correlates with circulating lipids and markers of NASH and fibrosis in patients
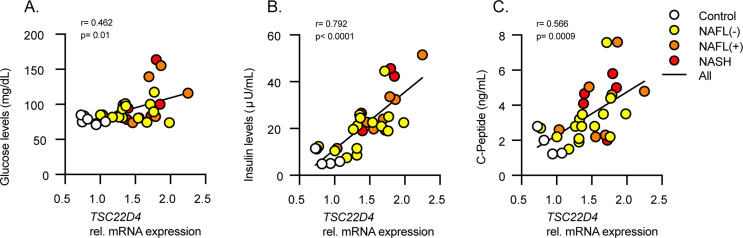
To initially understand the relevance of TSC22D4 in patients with progressive liver disease, we investigated two human cohorts. The first consisted of patients with steatosis or steatohepatitis (NASH), while the second had NASH and liver fibrosis. TSC22D4 mRNA levels were significantly increased in livers from obese persons both without (NAFL-) or with steatosis (NAFL+) and with histologically confirmed NASH (Figure 1A). The levels of TSC22D4 both positively correlated with serum triglycerides and negatively with HDL-cholesterol (Figure 1B) suggesting that increased hepatic levels of TSC22D4 associated with dyslipidemia in NAFLD. Additionally, consistent with our previous report [11], TSC22D4 mRNA positively correlated with increased plasma glucose, insulin, and C-peptide measures (Supp. Figure 2A–C). In patients with liver fibrosis TSC22D4 mRNA expression positively correlated with canonical fibrosis markers COL1α1, αSMA, and TGFβ (Figure 1. C-E). Together these data suggested a potential role for TSC22D4 in different stages of human NAFLD.
Figure 1.
Hepatic TSC22D4 expression correlates with circulating lipids and markers of NASH and fibrosis in patients. Liver samples from patients classified as control, NAFLD negative, NAFLD positive, and NASH, (33 females, 12 males; mean age 42.5 years; mean BMI 45.8 kg/m2). TSC22D4 mRNA expression (A) and correlation with plasma triglycerides and HDL (B). NASH and fibrosis patient liver samples (15 females, 24 males; mean age 60.8 years; mean BMI 25.6 kg/m2) TSC22D4 mRNA expression correlated with profibrotic markers collagen 1α1 (C) α-smooth muscle actin (D) and TGFβ (E). Data are mean ± SEM. Abbreviations: Nafl (−), patients without NAFLD diagnosis; Nafl (+), patients with NAFLD diagnosis; NASH, nonalcoholic steatohepatitis.
3.2. Tsc22d4 is upregulated in mouse models of fibrosis
CCl4 is a well-known potent chemical inducer of fibrosis in mice [29]. In agreement with results from patients presenting with clinical fibrosis, mice with a CCl4-induced fibrotic liver displayed increased Tsc22D4 mRNA expression (Figure 2A). Indeed, mice treated with CCl4 had a significant induction of several profibrotic markers, Tgfβ, αSma and Col1α1 (Figure 2B), and hepatic Tgfβ significantly correlated with Tsc22D4 mRNA levels (Figure 2C). A more recent model used to recapitulate the NASH-fibrosis-hepatocellular carcinoma transition in mice is the STAM™ model [12]. STAM mouse livers also demonstrated a clear induction of Tsc22D4 expression, which correlated with the induction of the profibrotic marker Tgfβ (Figure 2D–E). Overall, these results supported the hypothesis that the aberrant activation of TSC22D4 expression represents a common feature of both murine and human progressive liver disease.
3.3. Tsc22d4 knockout in hepatocytes decreases liver lipid accumulation in models of NASH and liver fibrosis in mice
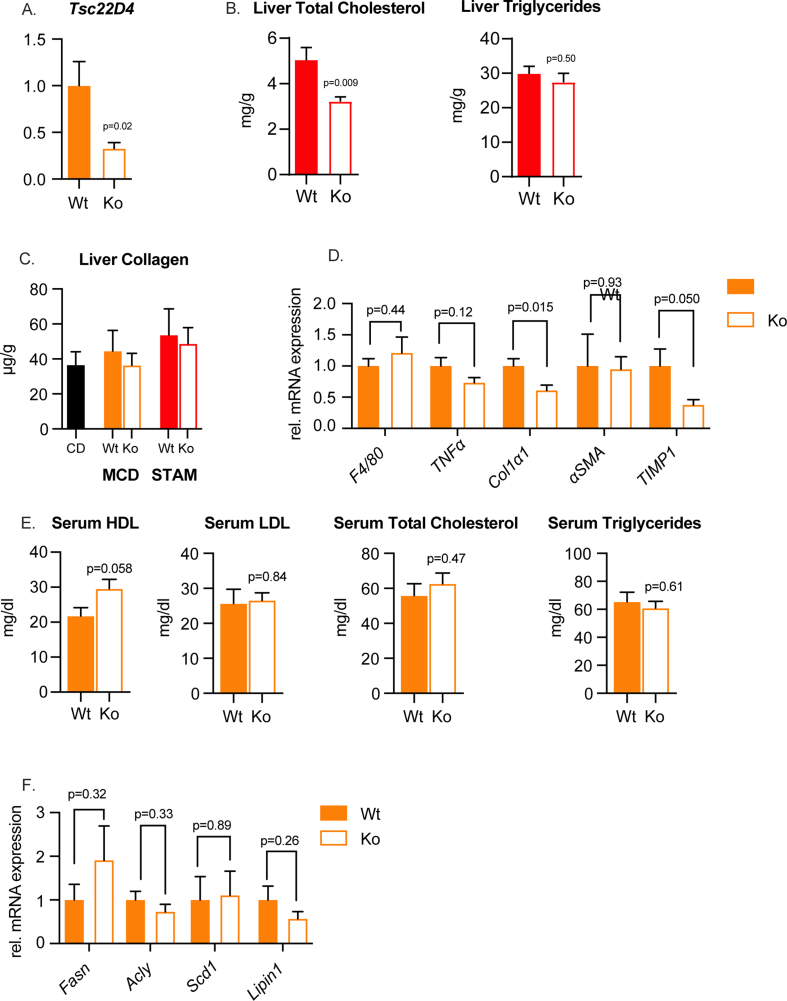
Triglyceride accumulation within hepatocytes is a hallmark of NAFLD, and chronic steatosis leads to inflammation, apoptosis, and fibrosis [30]. The MCD diet model is a robust inducer of progressive NAFLD in mice [31]. In agreement with our earlier observation in human progressive NAFLD and STAM mice, animals fed the MCD diet had an increased expression of hepatic Tsc22D4 (Figure 3A). When mice lacking TSC22D4 only in hepatocytes (TSC22D4-HepaKO) (Supp. Fig. 3A) were given the same diet, pathological examination revealed a decrease in liver lipid droplets and an improved sum of scores [16] relating to classic NASH characteristics (Figure 3B–D). The improvement in liver injury was mainly the result of fewer apoptotic hepatocytes, reduced steatosis and inflammation scoring (Figure 3E–F). In conjunction with the improved NASH phenotype seen in TSC22D4-HepaKO MCD fed mice we measured decreased levels of liver triglycerides and total cholesterol (Figure 3G–H). TSC22D4-HepaKO mice exposed to the STAM model also showed a decrease in liver total cholesterol but not liver triglycerides (Supp. Fig. 3B). Neither NASH-inducing model significantly increased collagen content, a surrogate for fibrosis (Supp. Fig. 3C). In TSC22D4-HepaKO MCD fed mice mRNA expression of neither inflammatory nor pro-fibrotic markers were altered (Supp. Fig. 3D), serum lipids were also unchanged (Supp. Fig. 3E), and lipogenic gene expression was similar to Wt (Supp. Fig. 3F). Tsc22d4 expression both positively correlated with the amount of liver triglycerides and negatively with serum HDL-cholesterol (Supp. Figure 4A) as we observed in patient cohorts (Fig. 1B). Furthermore, Tsc22D4 mRNA expression significantly correlated with TGFβ in both models of NAFLD (Supp. Figure 4B–C) suggesting a robust connection between TSC22D4 and progressive liver disease. These experiments demonstrated that TSC22D4 contributed to key parameters of NAFLD progression, including dyslipidemia, inflammation, and hepatocyte apoptosis.
Figure 3.
Hepatocyte specific loss of TSC22D4 decreases liver lipids in mice. Mice carrying the Tsc22d4 floxed allele with (Ko) or without (Wt) albumin cre-recombinase ensuring hepatocyte targeted deletion, were fed control or methionine-choline deficient diet for 3 weeks. Hepatic levels of Tsc22d4 in Wt mice fed CD or MCD (A). Representative images of Picro- Sirius red-stained liver tissue from CD and MCD fed mice (B) and H&E (C). Hepatocyte-specific deletion of TSC22D4 reduced liver injury sum of scores (D) number of apoptotic hepatocytes (E) steatosis and inflammatory scores (F) on MCD diet. Liver triglycerides (G) and total cholesterol (H) were also reduced in Ko mice on MCD diet. Data are mean ± SEM. n = 6 CD, n = 6 MCD (A); n = 11 Wt, n = 11 Ko (B–G) Abbreviations: MCD, methionine-choline deficient; CD, control diet.
Figure 4.
Gene signature in single-nuclei RNAseq from TSC22D4-HepaKO and WT mice. T-distributed stochastic neighborhood embedding (tSNE) analysis of hepatocyte fraction from single-nuclei RNAseq of TSC22D4-HepaKO and WT mice on control or MCD diet (A). Ten of top 100 pathways identified by gene ontology (GO) analysis (B). A dot plot of the genes significantly enriched in TSC22D4-HepaKO cells under MCD diet (C). Abbreviations: MCD, methionine-choline deficient.
3.4. Loss of TSC22D4 in hepatocytes upregulates a mitochondrial-protective gene network in murine NAFLD
To gain mechanistic insight into the protective effect of hepatocyte-specific loss of TSC22D4 in NAFLD, single-nuclei RNA sequencing (snRNAseq-2) [23] was performed on liver samples from TSC22D4-HepaKO and WT mice on MCD and control diet. The 4n-cell fraction is tetraploid and highly enriched for hepatocytes. T-distributed stochastic neighborhood embedding (tSNE) of 4n-hepatocytes revealed clustering based on control or MCD diet (Fig. 4A) with no detectable batch effect (Supp. Figure 5A). Tsc22d4 was not significantly enriched in other liver cell types (Supp. Fig. 5B). Within the MCD diet treatment 4n-nuclei from TSC22D4-HepaKO and WT tended to overlap (Figure 4A, green vs purple) whereas 4n-nuclei from mice on control diet tended to cluster within genotype (Figure 4A, red vs blue dots).
Figure 5.
Gene networking reveals mitochondrial maintenance and lipid metabolism. KEGG gene interaction network created from the genes identified in the top ten GO biological pathways.
Subsequent gene ontology (GO) analysis of the MCD diet groups identified gene signatures related to mitochondrial maintenance, the TCA cycle and electron transport chain as well as cellular respiration, triglyceride metabolism, and lipid catabolic processes among the top 100 significantly enriched pathways (Fig. 4B). The top 10 GO biological pathways within control diet groups also included metabolic and catabolic processes as well as DNA repair and autophagy (Supp. Fig. 5C). However, the metabolic parameters, liver triglycerides, total cholesterol and liver injury as measured by sum of score were not significantly altered by loss of hepatic TSC22D4 (Supp. Fig. 5D).
Intriguingly TSC22D4-HepaKO mice on MCD diet did not show significant down-regulation of any genes but rather increased the expression of genes related to mitochondrial complex I and cellular respiration (Ndufa1,7,8,12, Ndufs6, and Ndufb9), lipid metabolism (Apoe, Apoa4 and Fgf21) and mitochondrial organization and transport (Idh3a and g, Timm13 and Tomm5) (Figure 4C., Supp. Figure 6A–B). Genes contributing to the top 10 pathways identified by GO analysis (Supp. Fig. 6C) were applied to the KEGG pathway database to generate a gene network signature (Fig. 5). KEGG annotated gene clusters identified gene clusters related to NAFLD and oxidative phosphorylation as well as cholesterol metabolism.
To evaluate the functional impact of TSC22D4 knock down on mitochondrial respiration in hepatocytes, isolated primary cells transfected with lipid nanoparticles (LNP) carrying specific TSC22D4-directed siRNA were subjected to mitochondrial stress tests. In line with our single-nuclei RNAseq-2 analysis, primary hepatocytes lacking Tsc22d4 (Fig. 6A) had increased oxygen consumption rate when compared with siRNA-Ctrl (Fig. 6B). This stemmed from enhanced basal and maximal respiration as well as increased ATP-linked respiration and proton-leak (Fig. 6B). Non-mitochondrial respiration was not significantly altered by Tsc22d4 knock down. Measurement of mitochondrial DNA to nuclear DNA ratios in primary hepatocytes taken from TSC22D4-HepaKO and WT mice revealed no significant change in mitochondrial content (Fig. 6D) however, the mitochondrial genes idendified by snRNAseq were upregulated (Fig. 6E). Together these data supported a link between TSC22D4 and mitochondrial function in hepatocytes.
Research suggests that mitochondrial dysfunction is an early event in the progression of NAFLD [32] and that alongside hepatic insulin resistance leads to deficits in maximal respiration (30–40% reduced) in NASH patients compared to lean controls [18]. Evaluation of liver material from NAFLD and NASH patients revealed an upregulation of TSC22D4 (Figure 7A) as shown above, but also significant down-regulation of selected mitochondrial genes, including electron transport complex I (NDUFA1, 8, 12 and NDUFB2) (Fig. 7B). TSC22D4 mRNA expression was also significantly correlated with these, further confirming the importance of hepatic TSC22D4 in mitochondrial health in progressive stages of fatty liver disease also in humans.
Figure 7.
TSC22D4 correlates with mitochondrial markers in human NAFLD and NASH. TSC22D4 mRNA levels were increased in NAFLD and NASH livers (A). Gene expression of electron transport complex I components identified in snRNAseq-2 were significantly decreased in liver tissue from NAFLD and NASH patients (26 females, 12 males; mean age 54.5 years; mean BMI 45.1 kg/m2). (B) TSC22D4 expression positively correlated with genes in A (C). Data are mean ± SEM.
4. Discussion
NAFLD is the most common cause of chronic liver disease worldwide [33,34]. Stages of NAFLD range from early steatosis (NAFL) to non-alcoholic steatohepatitis (NASH) which can lead to irreversible conditions such as cirrhosis or hepatocellular carcinoma. Distinct biological pathways and molecular mechanisms converge to drive NAFLD progression including oxidative stress, autophagy, apoptosis, and inflammation, ultimately resulting in fibrosis and eventually liver cancer [35,36]. A key event in the progression of NAFLD is lipotoxicity resulting from disproportionate fat storage in hepatocytes.
Excess liver fat can result from several sources in the development of NAFLD from overnutrition to de novo lipogenesis. In fact, the prevalence of liver steatosis worldwide has sparked discussion over altering terminology to include ‘metabolic-associated fatty liver disease’ (MAFLD) [37,38]. While both diseases emphasize the presence of excess fat storage in the liver, NAFLD requires liver imaging or biopsy and the exclusion of other liver disease (e.g. alcoholism or viral hepatitis) while MAFLD is diagnosed concurrently with obesity, diabetes or other metabolic disorders [37].
In the current study mice were fed the NAFLD-promoting MCD diet. Deficiencies in dietary methionine and choline promote fatty liver disease through impairing the mechanisms by which the liver catabolizes fatty acids and generates VLDL for secretion [30]. In contrast to the high fat diet promoting obesity, insulin resistance, adipose overload, and de novo lipogenesis, the MCD diet induces a program of lipotoxicity by impairing central functions of lipid handling in the liver. In this context TSC22D4 depletion was able to reduce liver fat accumulation through modifying mitochondrial function in the absence of changes in serum LDL, total cholesterol, or triglycerides (Supp. Fig 3E).
NAFLD represents the hepatic manifestation of the metabolic syndrome, and indeed the majority of patients affected by obesity-driven type 2 diabetes display signs of liver dysfunction. Of note, NAFLD not only impairs liver function but is also a key driver of systemic insulin resistance as well as macro-vascular complications in long-term diabetes [39]. This suggests the existence of hepatic molecular checkpoints serving as connecting hubs between the intra-hepatic and systemic consequences of liver metabolic dysfunction. In this respect, our previous work has identified hepatic TSC22D4 as a key regulator of systemic insulin sensitivity and hepatic lipogenesis. Hepatocyte-selective knock down of TSC22D4 in type 2 diabetic mice improved glucose metabolism by reducing circulating glucose levels, increasing insulin sensitivity and Akt phosphorylation in the liver [11]. As shown in the current study, hepatic TSC22D4 expression also positively correlated with circulating glucose, insulin, and C-peptide levels in patients with steatosis or steatohepatitis (Supp. Figure 2A–C), which may highlight a broader link between TSC22D4 and systemic glucose control in situations of perturbed liver health. Furthermore, in the context of tumor-mediated metabolic wasting, hepatic TSC22D4 was linked to systemic lipid metabolism [10], while hepatic knockdown improved dyslipidemia in murine type 2 diabetes [11]. In mice and humans, we could show that higher hepatic TSC22D4 is linked to liver injury and fatty liver, while lower levels correspond to decreased serum triglycerides and increased serum HDL. TSC22D4 was increased at the mRNA level in patients with progressive NAFLD as well as histologically confirmed NASH (Figure 1A and Figure 7A), however, evaluation of additional bulk RNAseq datasets revealed mild changes in TSC22D4 expression in NASH or liver fibrosis (Supp. Figure 6E). Potential explanations for the inconsistency between these datasets and ours are the heterogeneity in diseased liver as well as differences in control material. Furthermore, while we did measure a consistent increase in TSC22D4 mRNA levels, that increase was modest, and the fact that TSC22D4 is expressed in other liver niche cells could account for variation that led to the differences in these datasets. Due to the influence of other liver cell types, our data from hepatocyte-specific TSC22D4-null mice allow us to investigate a direct role for this factor in systemic lipid metabolism, as these mice displayed attenuated hepatic inflammation, steatosis and hepatocyte injury following MCD-feeding.
Our results now support the hypothesis that cell specific targeting of hepatic TSC22D4 may provide a therapeutic strategy to diminish the early adverse events before NASH or other progression occurs, thereby not only ameliorating glucose tolerance and insulin sensitivity but also diminishing the degree of hepatic fibrosis as a long-term consequence of diabetes-related metabolic dysfunction.
Single-nuclei transcriptomics analysis of whole livers in the context of MCD-feeding demonstrated a significant improvement of mitochondrial complex I gene expression, upon hepatocyte TSC22D4 loss-of-function. Although systemic improvements in lipid metabolism could be a causal explanation for this observation in vivo, increased oxygen consumption rate following knockdown of TSC22D4 in cultured hepatocytes suggests that a cell-autonomous primary involvement for this factor in mitochondrial function may exist.
Little is known about TSC22D4 or other TSC22-family members in relation to mitochondrial function. One report described four forms of TSC22D4 in developing cerebellar granule neurons (CGN) and identified the 67 kDa modified form to be specifically enriched in the mitochondria of differentiated CGNs [40]. TSC22D4 has been shown to heterodimerize with apoptosis inducing factor (AIF), a key protein in mitochondrial redox metabolism [41].
Of note, mitochondrial impairments represent key feature of NAFLD, varying in different stages of the disease [18,42]. Thus, the hepatocyte-selective activity of TSC22D4 appears to target one of the critical cellular events in the progression of NAFLD.
Furthermore, additional data suggest mitochondrial dysregulation to be an initial step in the pathway to insulin resistance and hepatic steatosis by highlighting the role of mitochondrial complexes in obesity and progression to NAFLD [32,43,44]. Others have demonstrated deficits in specific complex subunit proteins such as Ndufa 9 [45] Ndufa (1, 2, 11), Ndufb (6, 7, 9) in progressive NAFLD and NASH [46]. Our data support these findings that mitochondrial complex I genes are downregulated in patients with NAFLD and NASH while the targeted inhibition of TSC22D4 selectively restores their abundance (Figure 7B and Figure 4C, respectively).
As an outlook, our data suggest that the hepatocyte-selective targeting of TSC22D4 may provide a new path forward to improve systemic insulin sensitivity and glucose handling as well as improving mitochondrial function, thereby preventing, and potentially even reversing diabetes-related liver fibrosis.
Author contributions
GW contributed to the investigation, formal analysis, visualization, and writing the original draft. PW and AMh contributed formal analysis, visualization, reviewing and editing the manuscript. AW, MT, IKD, KY, YK, RS, and JM contributed to the investigation. AZ, MR, MB, NV, TH, FDAR, JVR, AF, RM, PL, KT, PK, JP, CPMJ, and JS contributed resources. MS, AMa, and BEU contributed to writing the original draft. TP and LW contributed resources and investigation. PN and JS contributed to the conceptualization, funding acquisition and supervision. SH contributed to the conceptualization, funding acquisition, supervision and writing the original draft.
Financial Support
This work was supported by the Deutsche Forschungsgemeinschaft (SFB1118 to S.H. and P.N.; SFB1116 and GRK2576 to M.R.; CRC1118 to the Biobank for Diabetes), the Helmholtz Association (to S.H.), and the Helmholtz Pioneer Campus (C.P.M-J., K.Y.,), and Helmholtz future topic Aging and Metabolic Programming (AMPro ZT-0026 to I.K.D).
Acknowledgements
We would like to thank Bettina Walter and the Center for Model System and Comparative Pathology for their technical expertise (human and mouse disease), the IBF and clinical experimental area of Heidelberg University for technical expertise (mouse studies), the Helmholtz Center München (HMGU) Genomics Core facility and Inti Velazquez, Ana Alfaro for scientific discussions and expertise in mouse disease models, Julia Zuber for expertise with Seahorse analysis, Estefania Simoes Fernandez for support with mitochondrial target analysis, Mateusz Strzelecki for FACS sorting support, and Luke Harrison for editorial contributions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101487.
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
figs6.
References
- 1.Wagner R., Heni M., Tabák A.G., Machann J., Schick F., Randrianarisoa E., et al. Pathophysiology-based subphenotyping of individuals at elevated risk for type 2 diabetes. Natura Med. 2021 doi: 10.1038/s41591-020-1116-9. [DOI] [PubMed] [Google Scholar]
- 2.Tilg H., Moschen A.R., Roden M. NAFLD and diabetes mellitus. Nature Reviews Gastroenterology & Hepatology. 2017;14(1):32–42. doi: 10.1038/nrgastro.2016.147. [DOI] [PubMed] [Google Scholar]
- 3.Loft A., Alfaro A.J., Schmidt S.F., Pedersen F.B., Terkelsen M.K., Puglia M., et al. Liver-fibrosis-activated transcriptional networks govern hepatocyte reprogramming and intra-hepatic communication. Cell Metabolism. 2021;33(8):1685–1700. doi: 10.1016/j.cmet.2021.06.005. e1689. [DOI] [PubMed] [Google Scholar]
- 4.Yang L., Roh Y.S., Song J., Zhang B., Liu C., Loomba R., et al. Transforming growth factor beta signaling in hepatocytes participates in steatohepatitis through regulation of cell death and lipid metabolism in mice. Hepatology. 2014;59(2):483–495. doi: 10.1002/hep.26698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohta S., Shimekake Y., Nagata K. Molecular cloning and characterization of a transcription factor for the C-type natriuretic peptide gene promoter. European Journal of Biochemistry. 1996;242(3):460–466. doi: 10.1111/j.1432-1033.1996.460rr.x. [DOI] [PubMed] [Google Scholar]
- 6.Khoury C.M., Yang Z., Li X.Y., Vignali M., Fields S., Greenwood M.T. A TSC22-like motif defines a novel antiapoptotic protein family. FEMS Yeast Research. 2008;8(4):540–563. doi: 10.1111/j.1567-1364.2008.00367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H., Xia L., Chen J., Zhang S., Martin V., Li Q., et al. Stress-glucocorticoid-TSC22D3 axis compromises therapy-induced antitumor immunity. Natura Med. 2019;25(9):1428–1441. doi: 10.1038/s41591-019-0566-4. [DOI] [PubMed] [Google Scholar]
- 8.Fiol D.F., Mak S.K., Kültz D. Specific TSC22 domain transcripts are hypertonically induced and alternatively spliced to protect mouse kidney cells during osmotic stress. FEBS Journal. 2007;274(1):109–124. doi: 10.1111/j.1742-4658.2006.05569.x. [DOI] [PubMed] [Google Scholar]
- 9.Canterini S., Bosco A., Carletti V., Fuso A., Curci A., Mangia F., et al. Subcellular TSC22D4 localization in cerebellum granule neurons of the mouse depends on development and differentiation. The Cerebellum. 2012;11(1):28–40. doi: 10.1007/s12311-010-0211-8. [DOI] [PubMed] [Google Scholar]
- 10.Jones A., Friedrich K., Rohm M., Schäfer M., Algire C., Kulozik P., et al. TSC22D4 is a molecular output of hepatic wasting metabolism. EMBO Molecular Medicine. 2013;5(2):294–308. doi: 10.1002/emmm.201201869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ekim Ustunel B., Friedrich K., Maida A., Wang X., Krones-Herzig A., Seibert O., et al. Control of diabetic hyperglycaemia and insulin resistance through TSC22D4. Nature Communications. 2016;7:13267. doi: 10.1038/ncomms13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii M., Shibazaki Y., Wakamatsu K., Honda Y., Kawauchi Y., Suzuki K., et al. A murine model for non-alcoholic steatohepatitis showing evidence of association between diabetes and hepatocellular carcinoma. Medical Molecular Morphology. 2013;46(3):141–152. doi: 10.1007/s00795-013-0016-1. [DOI] [PubMed] [Google Scholar]
- 13.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., et al. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proceedings of the National Academy of Sciences of the U S A. 2017;114(10):E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajappan K., Tanis S.P., Mukthavaram R., Roberts S., Nguyen M., Tachikawa K., et al. Property-driven design and development of lipids for efficient delivery of siRNA. Journal of Medicinal Chemistry. 2020;63(21):12992–13012. doi: 10.1021/acs.jmedchem.0c01407. [DOI] [PubMed] [Google Scholar]
- 15.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. Journal of Biological Chemistry. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 16.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 17.Liang W., Menke A.L., Driessen A., Koek G.H., Lindeman J.H., Stoop R., et al. Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0115922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koliaki C., Szendroedi J., Kaul K., Jelenik T., Nowotny P., Jankowiak F., et al. Adaptation of hepatic mitochondrial function in humans with non-alcoholic fatty liver is lost in steatohepatitis. Cell Metabolism. 2015;21(5):739–746. doi: 10.1016/j.cmet.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Apostolopoulou M., Gordillo R., Koliaki C., Gancheva S., Jelenik T., De Filippo E., et al. Specific hepatic sphingolipids relate to insulin resistance, oxidative stress, and inflammation in nonalcoholic steatohepatitis. Diabetes Care. 2018;41(6):1235–1243. doi: 10.2337/dc17-1318. [DOI] [PubMed] [Google Scholar]
- 20.Lydia Kynast K., Volk N., Fleming T., Herpel E. Diabetes-associated biobanking: more topical than ever? Experimental and Clinical Endocrinology & Diabetes. 2017;125(9):603–609. doi: 10.1055/s-0042-123039. [DOI] [PubMed] [Google Scholar]
- 21.Xia Y., Caputo M., Cansby E., Anand S.K., Sutt S., Henricsson M., et al. STE20-type kinase TAOK3 regulates hepatic lipid partitioning. Molecular Metabolism. 2021;54:101353. doi: 10.1016/j.molmet.2021.101353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnaswami S.R., Grindberg R.V., Novotny M., Venepally P., Lacar B., Bhutani K., et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nature Protocols. 2016;11(3):499–524. doi: 10.1038/nprot.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richter M.L., Deligiannis I.K., Yin K., Danese A., Lleshi E., Coupland P., et al. Single-nucleus RNA-seq2 reveals functional crosstalk between liver zonation and ploidy. Nature Communications. 2021;12(1):4264. doi: 10.1038/s41467-021-24543-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodrigues O.R., Monard S. A rapid method to verify single-cell deposition setup for cell sorters. Cytometry, Part A. 2016;89(6):594–600. doi: 10.1002/cyto.a.22865. [DOI] [PubMed] [Google Scholar]
- 25.Duncan A.W., Taylor M.H., Hickey R.D., Hanlon Newell A.E., Lenzi M.L., Olson S.B., et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467(7316):707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mora-Castilla S., To C., Vaezeslami S., Morey R., Srinivasan S., Dumdie J.N., et al. Miniaturization Technologies for efficient single-cell library preparation for next-generation sequencing. Journal of Laboratory Automation. 2016;21(4):557–567. doi: 10.1177/2211068216630741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seitz S., Kwon Y., Hartleben G., Jülg J., Sekar R., Krahmer N., et al. Hepatic Rab24 controls blood glucose homeostasis via improving mitochondrial plasticity. Nat Metab. 2019;1(10):1009–1026. doi: 10.1038/s42255-019-0124-x. [DOI] [PubMed] [Google Scholar]
- 28.Godoy P., Hewitt N.J., Albrecht U., Andersen M.E., Ansari N., Bhattacharya S., et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Archiv für Toxikologie. 2013;87(8):1315–1530. doi: 10.1007/s00204-013-1078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholten D., Trebicka J., Liedtke C., Weiskirchen R. The carbon tetrachloride model in mice. Labaratory Animals. 2015;49(1 Suppl):4–11. doi: 10.1177/0023677215571192. [DOI] [PubMed] [Google Scholar]
- 30.Ibrahim S.H., Hirsova P., Malhi H., Gores G.J. Animal models of nonalcoholic steatohepatitis: eat, delete, and inflame. Digestive Diseases and Sciences. 2016;61(5):1325–1336. doi: 10.1007/s10620-015-3977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machado M.V., Michelotti G.A., Xie G., Almeida Pereira T., Boursier J., Bohnic B., et al. Mouse models of diet-induced nonalcoholic steatohepatitis reproduce the heterogeneity of the human disease. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0127991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rector R.S., Thyfault J.P., Uptergrove G.M., Morris E.M., Naples S.P., Borengasser S.J., et al. Mitochondrial dysfunction precedes insulin resistance and hepatic steatosis and contributes to the natural history of non-alcoholic fatty liver disease in an obese rodent model. Journal of Hepatology. 2010;52(5):727–736. doi: 10.1016/j.jhep.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wree A., Broderick L., Canbay A., Hoffman H.M., Feldstein A.E. From NAFLD to NASH to cirrhosis-new insights into disease mechanisms. Nature Reviews Gastroenterology & Hepatology. 2013;10(11):627–636. doi: 10.1038/nrgastro.2013.149. [DOI] [PubMed] [Google Scholar]
- 34.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64(1):73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 35.Bence K.K., Birnbaum M.J. Metabolic drivers of non-alcoholic fatty liver disease. Molecular Metabolism. 2021;50:101143. doi: 10.1016/j.molmet.2020.101143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rada P., González-Rodríguez Á., García-Monzón C., Valverde Á M. Understanding lipotoxicity in NAFLD pathogenesis: is CD36 a key driver? Cell Death & Disease. 2020;11(9):802. doi: 10.1038/s41419-020-03003-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Q., Zou X., Wen X., Zhou X., Ji L. NAFLD or MAFLD: which has closer association with all-cause and cause-specific mortality?-results from NHANES III. Frontiers of Medicine. 2021;8:693507. doi: 10.3389/fmed.2021.693507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heeren J., Scheja L. Metabolic-associated fatty liver disease and lipoprotein metabolism. Molecular Metabolism. 2021;50:101238. doi: 10.1016/j.molmet.2021.101238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radaelli M.G., Martucci F., Perra S., Accornero S., Castoldi G., Lattuada G., et al. NAFLD/NASH in patients with type 2 diabetes and related treatment options. Journal of Endocrinological Investigation. 2018;41(5):509–521. doi: 10.1007/s40618-017-0799-3. [DOI] [PubMed] [Google Scholar]
- 40.Canterini S., Carletti V., Nusca S., Mangia F., Fiorenza M.T. Multiple TSC22D4 iso-/phospho-glycoforms display idiosyncratic subcellular localizations and interacting protein partners. FEBS Journal. 2013;280(5):1320–1329. doi: 10.1111/febs.12123. [DOI] [PubMed] [Google Scholar]
- 41.Lim J., Hao T., Shaw C., Patel A.J., Szabo G., Rual J.F., et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125(4):801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 42.Gancheva S., Jelenik T., Alvarez-Hernandez E., Roden M. Interorgan metabolic crosstalk in human insulin resistance. Physiological Reviews. 2018;98(3):1371–1415. doi: 10.1152/physrev.00015.2017. [DOI] [PubMed] [Google Scholar]
- 43.Eccleston H.B., Andringa K.K., Betancourt A.M., King A.L., Mantena S.K., Swain T.M., et al. Chronic exposure to a high-fat diet induces hepatic steatosis, impairs nitric oxide bioavailability, and modifies the mitochondrial proteome in mice. Antioxidants and Redox Signaling. 2011;15(2):447–459. doi: 10.1089/ars.2010.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simoes I.C.M., Fontes A., Pinton P., Zischka H., Wieckowski M.R. Mitochondria in non-alcoholic fatty liver disease. The International Journal of Biochemistry & Cell Biology. 2018;95:93–99. doi: 10.1016/j.biocel.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 45.Aharoni-Simon M., Hann-Obercyger M., Pen S., Madar Z., Tirosh O. Fatty liver is associated with impaired activity of PPARgamma-coactivator 1alpha (PGC1alpha) and mitochondrial biogenesis in mice. Laboratory Investigation. 2011;91(7):1018–1028. doi: 10.1038/labinvest.2011.55. [DOI] [PubMed] [Google Scholar]
- 46.Bellanti F., Villani R., Tamborra R., Blonda M., Iannelli G., di Bello G., et al. Synergistic interaction of fatty acids and oxysterols impairs mitochondrial function and limits liver adaptation during nafld progression. Redox Biology. 2018;15:86–96. doi: 10.1016/j.redox.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]