Abstract
Objective
Non-alcoholic fatty liver disease (NAFLD) is linked to impaired lipid metabolism and systemic insulin resistance, which is partly mediated by altered secretion of liver proteins known as hepatokines. Regular physical activity can resolve NAFLD and improve its metabolic comorbidities, however, the effects of exercise training on hepatokine secretion and the metabolic impact of exercise-regulated hepatokines in NAFLD remain unresolved. Herein, we examined the effect of endurance exercise training on hepatocyte secreted proteins with the aim of identifying proteins that regulate metabolism and reduce NAFLD severity.
Methods
C57BL/6 mice were fed a high-fat diet for six weeks to induce NAFLD. Mice were exercise trained for a further six weeks, while the control group remained sedentary. Hepatocytes were isolated two days after the last exercise bout, and intracellular and secreted proteins were detected using label-free mass spectrometry. Hepatocyte secreted factors were applied to skeletal muscle and liver ex vivo and insulin action and fatty acid metabolism were assessed. Syndecan-4 (SDC4), identified as an exercise-responsive hepatokine, was overexpressed in the livers of mice using adeno-associated virus. Whole-body energy homeostasis was assessed by indirect calorimetry and skeletal muscle and liver metabolism was assessed using radiometric techniques.
Results
Proteomics analysis detected 2657 intracellular and 1593 secreted proteins from mouse hepatocytes. Exercise training remodelled the hepatocyte proteome, with differences in 137 intracellular and 35 secreted proteins. Bioinformatic analysis of hepatocyte secreted proteins revealed enrichment of tumour suppressive proteins and proteins involved in lipid metabolism and mitochondrial function, and suppression of oncogenes and regulators of oxidative stress. Hepatocyte secreted factors from exercise trained mice improved insulin action in skeletal muscle and increased hepatic fatty acid oxidation. Hepatocyte-specific overexpression of SDC4 reduced hepatic steatosis, which was associated with reduced hepatic fatty acid uptake, and blunted pro-inflammatory and pro-fibrotic gene expression. Treating hepatocytes with recombinant ectodomain of SDC4 (secreted form) recapitulated these effects with reduced fatty acid uptake, lipid storage and lipid droplet accumulation.
Conclusions
Remodelling of hepatokine secretion is an adaptation to regular exercise training that induces changes in metabolism in the liver and skeletal muscle. SDC4 is a novel exercise-responsive hepatokine that decreases fatty acid uptake and reduces steatosis in the liver. By understanding the proteomic changes in hepatocytes with exercise, these findings have potential for the discovery of new therapeutic targets for NAFLD.
Keywords: Exercise, Hepatokine, Syndecan-4, Non-alcoholic fatty liver disease, Hepatic steatosis, Lipid metabolism
Abbreviations: AAV, Adeno-associated virus; HFD, High-fat diet; NAFLD, Non-alcoholic fatty liver disease; NAFL, Non-alcoholic fatty liver; NASH, Non-alcoholic steatohepatitis; SDC4, Syndecan-4
Highlights
-
•
Exercise training remodels hepatokine secretion.
-
•
Exercise regulated secreted factors improve insulin action in skeletal muscle.
-
•
Syndecan-4 (SDC4) is a novel exercise-induced hepatokine.
-
•
SDC4 reduces hepatic fatty acid uptake and hepatic steatosis.
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) is the most prevalent chronic liver disease, affecting an estimated 25% of the world's population [1]. It is defined by excessive accumulation of lipids in the liver and encompasses a range of pathologies ranging from hepatic steatosis to the more severe form of the disease, non-alcoholic steatohepatitis (NASH), which is accompanied by inflammation and hepatocyte damage, with or without fibrosis [[2], [3], [4]]. Given the dramatic rise in NAFLD prevalence [5,6], there is an urgent requirement for the development of effective and safe interventions to reduce disease severity and progression to worsening pathologies, such as hepatic cirrhosis and hepatocellular carcinoma (HCC) [5].
Exercise training is effective for the treatment of NAFLD [7], and in the absence of approved pharmacotherapies, remains the current frontline treatment option for patients [2]. Exercise training reduces the severity of hepatic steatosis [8] and causes regression of NASH, with or without accompanying weight loss [9,10]. This is thought to occur by the actions of inter-connected metabolic processes, including increased hepatic fatty acid oxidation, reduced de novo lipogenesis, and enhanced insulin sensitivity [11]. However, adherence to exercise training programs is not consistently achievable, particularly in obesity where NAFLD incidence is highest [12], necessitating the development of pharmacotherapies for NAFLD. We posit that understanding the processes by which exercise reduces NAFLD severity may inform new therapeutic opportunities for the treatment of this disease.
It is well documented that tissues actively secrete hormones (notably proteins), cytokines, metabolites, and nucleic acids, which can induce autocrine, paracrine and endocrine actions to regulate nutrient metabolism and systemic energy homeostasis [13]. Liver secreted proteins, also termed ‘hepatokines’, are important regulators of inter-tissue communication [14]. It is estimated that up to 25% of circulating proteins are liver-derived and a growing body evidence has shown that hepatokines act on numerous organs to induce diverse signalling events that regulate many biological processes [15]. Notably, many hepatokines are important regulators of lipid and glucose metabolism, insulin action, energy expenditure and food intake, and dysregulated hepatokine secretion is a recognised feature of NAFLD [14]. Studies in mice indicate that the liver is highly susceptible to exercise-induced cellular stress [16], resulting in changes in the mRNAs encoding secreted proteins [17], thereby suggesting the likelihood of exercise-induced adaptations in liver protein secretion. However, very few bone fide hepatokines have been identified as exercise responsive and it is unknown whether chronic exercise training alters hepatokine secretion [13,18], and whether exercise-regulated hepatokines impact metabolism in the liver or other tissues. We tested the hypothesis that chronic exercise training induces stable alterations in hepatokine secretion and that such changes can alter processes to alleviate the metabolic burden of NAFLD.
Herein, we demonstrate that exercise training induces marked remodelling of the hepatocyte proteome and secretome, and that hepatocyte-secreted factors from exercise-trained mice improves fatty acid metabolism and insulin action in glucoregulatory tissues. We further identify syndecan-4 (SDC4) as an exercise-responsive hepatokine, which regulates hepatic lipid metabolism.
2. Methods
2.1. Animal housing and experimental design
All procedures complied with the guidelines of the National Health and Medical Research Council of Australia concerning the use and care of experimental animals and were approved by the University of Melbourne Anatomy & Neuroscience, Pathology, Pharmacology, and Physiology animal ethics committee. Eight-week-old male C57BL/6 mice (sourced from Animal Resources Centre, Canning Vale, Australia) were used for the identification of exercise training responsive hepatokines (see 2.1.1) and to investigate the role of SDC4 overexpression on metabolism (see 2.1.2). All mice were maintained at 22 °C ± 1 °C with 12-h light/dark cycle and had ad libitum access to food and water.
2.1.1. Exercise training studies
Mice were fed a high fat diet (HFD, High Fat Rodent Diet SF04-001, 43% energy from fat, Specialty Feeds, Australia) for 6 weeks, and were then assigned to the Sedentary or Exercise group. Mice were maintained on the HFD for a further six weeks. Mice were familiarised to the rodent treadmill once-daily for three days prior to an initial maximal running velocity test [19]. For this, mice commenced running at 10 m/min and the treadmill speed was increased by 2 m/min every 2 min until volitional exhaustion as previously described [19]. Mice in the Exercise group then underwent a progressive endurance exercise training program, which consisted of five days treadmill running per week with progressive increases in exercise intensity and duration, as described previously [20]. Mice in the Sedentary group were placed on a stationary treadmill for the same duration as the exercise trained mice. Following 6 weeks of exercise training, all mice undertook another maximal velocity test. Two days later, mice were anesthetised using 4% isoflurane (SomnoSuite, Kent Scientific, USA), the mixed quadriceps muscle was dissected for later analysis (see 2.8) and hepatocytes were isolated and cultured for the proteomic evaluation of the intracellular proteome and hepatokine secretion (see sections 2.2-2.3).
2.1.2. SDC4 overexpression studies
For Sdc4 overexpression studies, mice were fed a HFD for 6 weeks and then assigned to experimental groups to match pre-intervention adiposity and glucose tolerance. An adeno-associated virus (AAV) serotype 8 (Vector Biolabs, Malvern, PA, USA) driven by a thyroxine-binding globulin promoter containing Sdc4 cDNA was injected into the tail vein at 1 × 1012 genome copies/mouse (Sdc4-AAV). Control mice were injected with the same vector without Sdc4 cDNA (Control-AAV). Mice were maintained on the HFD for a further eight weeks and body mass was recorded weekly. Five weeks after AAV administration mice underwent in vivo metabolic assessments, including evaluation of body composition and whole-body energy metabolism, an oral glucose tolerance test and an insulin tolerance test, each separated by one week (see 2.4). Eight weeks after AAV administration, blood and tissue samples were collected from animals following a 4 h fast (0700–1100 h). Liver slices were immediately prepared for ex vivo metabolic assessments (see 2.3).
2.2. Hepatocyte isolation and cell culture
2.2.1. Hepatocyte isolation and culture
Hepatocytes were isolated from Sedentary, Exercise trained and donor mice (see 2.1.1) by collagenase perfusion as previously described [21]. Briefly, a catheter (BD vacutainer Safety-Lok) was inserted into the inferior vena cava and Hank's Buffered Salt Solution (HBSS + 0.5 mmol/L EGTA, pH 7.4) was infused into the liver, followed by collagenase digestion (HBSS + 0.5 mmol/L CaCl2, containing 50 μg Collagenase H (Roche, cat: 11 074 032 001)). Hepatocytes were firstly settled in adherence media (Gibco M199 media +100 U Penicillin/Streptomycin, 0.1% BSA, 2% FBS, 100 nmoL/l Dexamethasone, 100 nmoL/l Insulin) for 4 h. Following this, hepatocytes from Exercise or Sedentary mice were processed as described in 2.2.2. and hepatocytes from donor mice were assessed as per 2.2.3 and 2.2.4.
2.2.2. Hepatocyte secretion studies
To assess hepatocyte protein secretion, hepatocytes were washed with PBS and incubated in EX-CELL® 325 PF CHO Serum-Free medium (Sigma–Aldrich, Australia) for 16 h. The culture medium containing hepatocyte secreted factors was centrifuged at 300×g for 10 min and stored at −80 °C. Hepatocytes were lysed in 100 mmol/L Tris–HCl + 1% sodium deoxycholate (w/v) (pH 8.1) and snap frozen in liquid nitrogen until proteomic assessment.
2.2.3. Recombinant SDC4 (rSDC4) studies
2.2.3.1. Studies in primary hepatocytes with rSDC4
One million hepatocytes from a donor mouse were plated in a 6 well plate and cultured for 16 h in basal media (Gibco M199 media, 100 U penicillin/streptomycin, 100 nmoL/l dexamethasone, 1 nmoL/l insulin). Hepatocytes were washed 3 × with PBS and cultured for 2 h in 1 g/L glucose DMEM (Gibco, USA) with or without 1 μg/mL of recombinant SDC4 ectodomain (rSDC4) with the Fc region of human IgG1 at the C-terminus (Cat: 50726-M02H, JOMAR Life Research, Australia). Fatty acid metabolism was assessed (see 2.5.1.) as previously described [22].
2.2.3.2. Lipid droplet morphology in primary hepatocytes with chronic rSDC4 exposure
Hepatocytes isolated from high fat fed mice were maintained as per 2.2.3.1. Hepatocytes were plated and cultured in chambered coverslips (Sarstedt, Germany) and treated with or without rSDC4 (1 μg/mL) for 48 h. Culture medium was then incubated with 100 nmoL/l MitoTracker Deep Red (Molecular Probes, USA) for 20 min and 2 μmoL/l BODIPY™ FL C16 (ThermoFisher Scientific, USA) for 15 min. Cells were fixed in 4% paraformaldehyde for 15 min at room temperature, washed with DPBS and stained with 4′,6-diamidino-2-phenylindole (DAPI) (ThermoFisher Scientific, USA) (1 μg/mL) to visualise nuclei. Microscopy was performed as described in section 2.9.
2.3. Hepatocyte proteome processing and mass spectrometry
2.3.1. Intracellular proteome sample preparation
Hepatocytes were boiled at 95 °C for 5 min, followed by three rounds of sonication for 10 s. The protein concentration of the lysate was determined using Pierce™ BCA protein assay kit (Thermo Scientific, #23225, USA). 100 μg of protein was reduced with 10 mmol/L tris(2-carboxyethyl)phosphine (TCEP) for 20 min at 65 °C and alkylated with 40 mmol/L chloroacetamide for 20 min in the dark. Proteins were digested with 1 μg trypsin at 37 °C in a shaking water bath overnight, the peptides acidified with formic acid to a pH of 3, diluted 1:1 (v:v) with ethyl acetate, centrifuged at 15,000×g for 5 min, and peptides purified with Millipore® C18 Ziptips (Sigma Aldrich, USA). Peptides were dried using a Speedvac concentrator (Eppendorf Concentrator Plus, Germany), reconstituted in 0.1% formic acid and 2% acetonitrile (ACN), then sonicated in a water bath for 10 min. Samples were centrifuged at 16,000×g for 5 min and, the supernatant was transferred into mass spectrometry vials and stored at 4 °C.
2.3.2. Secreted protein sample preparation
Proteins within the liver culture medium media were concentrated and purified using Amicon Ultra 4 centrifugal filter devices (Merck, USA) and protein content determined (Pierce™ BCA protein assay, Thermo Scientific, USA). Proteins were reduced with 10 mmol/L TCEP for 20 min at 65 °C, then mixed with 6.6 × volume of 8 mol/L urea. Samples were then spun at 14,000×g for 15 min and the flow through was discarded. 200 μL of 8 mol/L urea was added to the spin filters, samples centrifuged at 14,000×g for 15 min, and the flow through was again discarded. Subsequently, proteins were alkylated through addition of 10 μL 100 mmol/L chloroacetamide and 90 μL 8 mol/L urea in the dark for 20 min. Samples were centrifuged at 14,000×g for 15 min and the flow through was discarded, followed by two wash steps with 100 μL of 8 mol/L urea and three wash steps with 100 μL of 50 mmol/L ammonia bicarbonate (pH = 8.5). Proteins were digested with trypsin (1 μg trypsin/100 μg of protein) overnight at 37 °C. The following day, 40 μL of 50 mmol/L of ammonia bicarbonate was added, samples centrifuged at 14,000×g for 15 min, and this step repeated twice more. Lastly, 50 μL of 0.5 mol/L NaCl was added and the samples centrifuged at 14,000×g for 15 min. Peptides were acidified to pH = 3.0 with formic acid, purified with Millipore® C18 Ziptips (Sigma Aldrich, USA), and dried using a Speedvac concentrator (Eppendorf Concentrator Plus, Germany). The purified peptides were reconstituted in 0.1% formic acid, sonicated for 10 min, centrifuged at 16,000×g and transferred to liquid chromatography vials for analysis. Mass spectrometric analyses and file generation, processing and interpretation was performed as described in 2.3.3.
2.3.3. Mass spectrometry
Peptides were separated and identified by nano electrospray ionisation liquid chromatography coupled with tandem mass spectrometry (Nano-ESI-LC-MS/MS) on an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, USA). Full details are provided in the supplementary methods. All generated files were processed with MaxQuant (v1.6.2.10) [23] to identify (at least 2 razor + unique peptide) as well as to report their label free quantitation intensities. Database searching was performed with settings followed here: methionine oxidation and protein N-terminal acetylation as variable modification, cysteine carbamidomethylation as fixed modification, trypsin as enzyme, maximum missed cleavage as 2, UniProt mouse database with 17,082 entries, LFQ enabled and a minimum of 2 peptides for pair-wise comparison in each protein, false discovery rate at 1% at peptide and protein level, match between run enabled. The MaxQuant output was processed further through Perseus (v1.6.6.0) [24]. The values were Log2 transformed and analysed by two sample t-test (correcting for multiple hypothesis testing using Benjamini-Hochberg set at FDR <5%) to obtain a list of significantly regulated proteins based on q < 0.05. Bioinformatic assessment of the hepatocyte intracellular and secreted proteome data sets was performed using Ingenuity Pathway Analysis (IPA, QIAGEN) [25], and functions assessed included canonical pathways, upstream regulators and disease and biological functions.
2.3.4. Identification of secreted SDC4 by targeted mass spectrometry
Liver slices from Sdc4-AAV mice were collected, settled for 1 h in oxygenated M199 media, then cultured in EX-CELL 325 medium (Sigma–Aldrich, Australia) for 16 h. The incubation medium was collected and frozen at −80 °C until further use. Proteins were processed as per the S-Trap™ method as per manufacturer's instructions (Protifi, USA). The Nano-ESI-LC-MS/MS was used to analyse the peptide samples as described above (see 2.3.3 and Supplementary Methods).
A positive mode, Nano-ESI and Eclipse Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) was employed to execute the MS experiments with settings of spray voltages, S-lens RF, and capillary temperature level of 1.9 kV, 30%, 275 °C, respectively. The mass spectrometry data was acquired with a 3-second cycle time for one full scan MS spectra and as many data-dependent higher-energy C-trap dissociation (HCD)-MS/MS spectra as possible. Full scan MS spectra features ions at m/z of 300–1500, a maximum ion trapping time of 50 msec, an auto gain control target value of 4e5, and a resolution of 120,000 at m/z 200. An m/z isolation window of 1.6, an auto gain control target value of 5e4, a 30% normalized collision energy, a maximum ion trapping time of 22 ms, and a resolution of 15,000 at m/z 200 were used to perform data-dependent HCD-MS/MS of precursor ions having charge states from 2 to 7. A dynamic exclusion of 30 s was applied. After each data-dependent acquisition cycle, targeted HCD-MS/MS using identical parameters as that for data-dependent HCD-MS/MS described above was performed for selected SDC4 peptides based on experimentally observed peptide of mouse SDC4 from Peptide Atlas (see Supplementary Table 1 for peptide sequence, m/z and charge state). The extracted ion chromatogram (XIC) peak area of the detected SDC4 peptides precursors from the secreted medium was extracted from MaxQuant (Supplementary Table 2). Skyline (20.2.0.343) [26] was used to extract the MS2 scan intensity and the XIC peak area of fragment ions on the liquid chromatography run for each peptide detected (Supplementary Table 2). The peptide sequence that was detected in all samples and is unique to SDC4 (ETEVIDPQDLLEGR) was used to determine liver SDC4 secretion.
2.4. In vivo metabolic assessments
2.4.1. Body composition
Body composition (fat and lean mass) was assessed in conscious mice using Time-Domain Nuclear Magnetic Resonance (TD-NMR) relaxometry (MiniSpec LF50, Bruker) as per the manufacturer's instructions.
2.4.2. Metabolic cage studies
Locomotor activity, energy expenditure, food intake and respiratory exchange ratio were evaluated using a 16-chamber Promethion Metabolic cage system (Sable Systems International, USA). Studies were commenced following 12 h of acclimatisation to the metabolic chamber and the parameters were evaluated at 5 or 30 min intervals for 24 h.
2.4.3. Glucose tolerance test
Mice were fasted for 4 h (0700–1100 h). Mice received an oral glucose load (2 g/kg body weight) and blood was obtained from a tail nick before and at 15, 30, 45, 60 and 90 min for glucose assessment (Accu-Check, Roche, USA). Additional blood was collected at 0, 15 and 30 min for later assessment of plasma insulin.
2.4.4. Insulin tolerance test
Mice were fasted for 4 h (0700–1100 h) then injected i.p. with insulin (Actrapid, 1 U/kg body weight). Blood was obtained from a tail nick before and at 20, 30, 45 and 60 min for glucose assessment (Accu-Check, Roche, USA).
2.4.5. Plasma hormones and metabolites
Plasma was isolated by centrifugation of whole blood at 3,000 g at 4 °C for 10 min. Plasma insulin levels were determined using an ultrasensitive mouse insulin ELISA kit (Catalog #90080; Crystal Chem, USA). Plasma free fatty acids (FFAs) and total plasma cholesterol were assessed by colorimetric assay as per manufacturer's instructions (Wako Diagnostics, Japan). Plasma triglyceride levels were evaluated by colorimetric assay as per the manufacturer's instructions (Triglycerides GPOPAP; Roche Diagnostics, USA). Circulating alanine aminotransferase (ALT) was assessed by colorimetric enzymatic assay as previously described [27]. For each assay, absorbance was measured using a spectrophotometer (SPECTROstar Nano, BMG LABTECH, Germany).
2.5. Ex vivo metabolic assessments
2.5.1. Lipid metabolism
Precision-cut liver slices were generated as previously described [22]. Following 1 h of settling in oxygenated M199 media, the liver slices were washed in PBS and transferred to glass vials containing low-glucose DMEM, 0.5 mmol/L palmitic acid and 1 μCi/mL [1–14C] palmitic acid (NEC075H250UC; PerkinElmer) conjugated to 2% BSA for 2 h. Hepatocytes were cultured in the same medium and incubated at 37 °C for 2 h. Fatty acid oxidation was calculated as the sum of 14CO2 production and 14C-palmitate degradation to acid soluble metabolites. Lipids were extracted, separated and incorporation of 14C-palmitate into individual lipid species was determine by thin layer chromatography as previously described [22,28]. Fatty acid uptake was calculated by adding fatty acid oxidation to total lipid storage. Lipogenesis was measured for 2 h in DMEM containing 2 μCi/mL D-[U–14C] glucose (NEC042A001MC; PerkinElmer) and calculated as incorporation of glucose into lipids.
For ‘conditioned medium’ experiments, medium containing the secreted products from the hepatocytes of Exercise or Sedentary mice was applied 1:1 with low-glucose DMEM. Fatty acid metabolism was assessed as described above. Fatty acid metabolism in hepatocytes was normalised to protein content and in liver slices to tissue mass.
2.5.2. Hepatic glucose output
Liver slices were prepared as described in section 2.5.1, then incubated at 37 °C in high-glucose DMEM containing 1 mmol/L sodium pyruvate for 1 h. Liver slices were washed 3 × in PBS and incubated with medium containing liver secreted factors (Exercise or Sedentary) diluted 1:4 with glucose-free DMEM (Gibco, USA; Cat: 11966-025) (v:v) and supplemented with 10 mmol/L sodium pyruvate and 10 mmol/L sodium lactate for 10 min. Slices were washed in PBS then incubated for 1 h in the same pyruvate/lactate-supplemented media containing hepatocyte secreted factors. Glucose content in the medium was assessed using Infinity™ glucose hexokinase reagent (Thermo Fisher, USA), as per the manufacturer's instructions.
2.5.3. 2-Deoxyglucose uptake
2-Deoxyglucose uptake was assessed as previously described [29]. Briefly, intact tendon-to-tendon soleus muscle or epididymal adipose tissue pieces (∼15 mg) from a healthy donor mouse were pre-treated with medium containing secreted factors from hepatocytes of Exercise or Sedentary mice in a 1:1 (v:v) dilution with Krebs buffer or with 1 μg/mL of recombinant SDC4 ectodomain for 30 min.
2.5.4. Lipolysis
Epididymal adipose tissue explants from a healthy donor mouse were cultured in medium containing secreted factors from livers of Exercise or Sedentary mice diluted 1:4 (v:v) with oxygenated phenol red-free low-glucose DMEM and 2% BSA (shaking water bath at 37 °C for 4 h). Glycerol content in the media was assessed by colorimetric assay as per manufacturer's instructions (Free glycerol reagent, Sigma–Aldrich, USA).
2.6. Hepatic triglyceride content
Hepatic triglyceride levels wesre evaluated as described in 2.4.5 or determined by mass spectrometry as previously described [30]. Briefly ∼5 mg of liver was homogenized in solvent containing 10 μL of SPLASH® II LIPIDOMIX® Mass Spec Standard (330709W, Avanti Polar Lipids Inc, USA), dried (Eppendorf Concentrator Plus, Germany), resuspended in 100 μL of chloroform:methanol mixture (1:9, v:v), and transferred into glass vials for analysis by ultrahigh performance liquid chromatography (UHPLC) coupled to tandem mass spectrometry (MS/MS) employing a Vanquish UHPLC linked to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). MS data were processed using LipidSearch software v.4.2.23 (Thermo Fisher Scientific). Relative quantification of lipid species was achieved by comparison of the LC peak areas of identified lipids against those of the corresponding internal lipid standard and the resultant ratio of peak area was then normalized to weight of tissue.
2.7. RNA isolation and quantitative polymerase chain reaction
RNA was isolated from the liver using TRI-Reagent (Sigma–Aldrich, Australia), treated with DNAse (Ambion DNA free kit, Thermo Fisher, Australia) and reversed transcribed into cDNA with iSCRIPT Reverse Transcriptase (Invitrogen, USA) as per the manufacturer's instructions. Real-time PCR was assessed using the SYBR Green PCR master mix (Quantinova® SYBR Green PCR kit, QIAGEN, Germany) and expression was determined using a CFX Connect™ Real-Time PCR Detection System (Biorad, USA). All samples were normalised using the housekeeping gene HPRT, and all primer sequences are provided in Table S3. The mRNA levels were analysed by the 2−ΔΔCT method.
2.8. Immunoblotting
Quadriceps muscle was homogenised in radioimmunoprecipitation assay (RIPA) buffer, separated by SDS-PAGE and immunoblot analysis conducted as previously described [22,28]. Stain-free images were collected after transfer for loading control (ChemiDoc MP and ImageLab software version 4.1; Bio-Rad Laboratories, New South Wales, Australia). Membranes were probed with antibodies raised against PGC1α (cat: ab54481, Cell Signalling, USA) and total OXPHOS (cat: ab110413, Abcam, UK).
2.9. Liver histology and hepatocyte microscopy
A piece of liver from Sdc4-AAV or Control-AAV mice was fixed in 10% neutral buffered formalin, paraffin embedded, and sections stained with hematoxylin and eosin (H&E) (University of Melbourne Histology Platform, Melbourne, Australia). Lipid droplets were identified by ilastik™ machine learning image analysis [31], converted to an 8-bit image and quantitation of lipid droplet number and area was assessed with ImageJ version 1.8.0_112 (National Institutes of Health, USA).
Hepatocytes were stained as described in 2.2.3.2 and viewed using confocal microscopy (ZEISS LSM 900 with Airyscan 2, Germany) at 63X objective (oil-immersion). Lipid droplet number and area was quantified using Fiji (ImageJ) software version 1.8.0_112 (National Institutes of Health, USA).
2.10. Statistical analysis
Data was assessed for normal distribution using D'Agostino-Pearson test. Statistical analysis was performed using paired and unpaired Student's t-test or two-way analysis of variance (ANOVA) for normally distributed data, where appropriate. Mann–Whitney test was performed for data that was not normally distributed. Means were compared using Bonferroni post hoc analysis where appropriate. All data are shown as mean ± SEM, with statistical significance set as p < 0.05.
3. Results
3.1. Validation of the endurance exercise training program
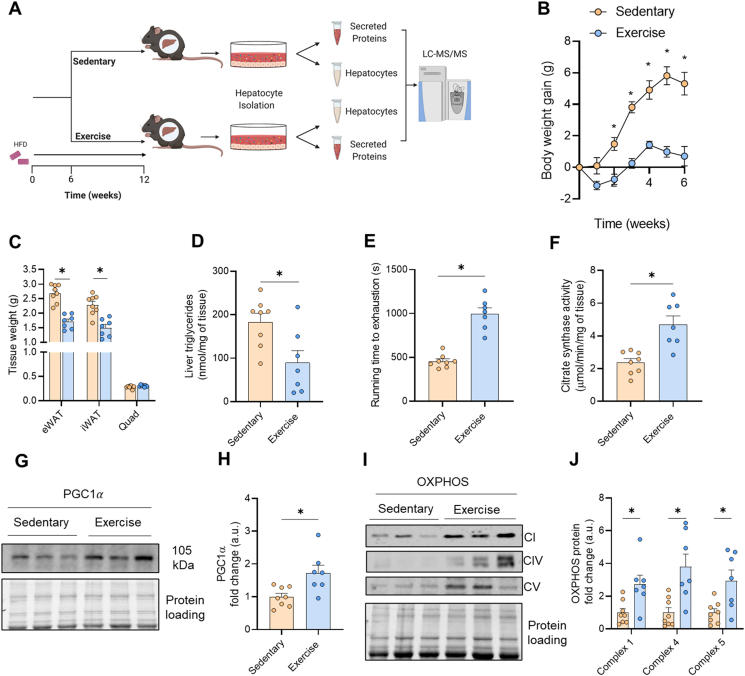
NAFLD was established by feeding mice a high-fat diet (HFD) for six weeks. Mice were matched for body weight and glucose tolerance (Figs. S1A–B) and allocated to either the sedentary group (Sedentary) or the endurance exercise training group (Exercise). Exercise training was performed for a further six weeks and all mice were maintained on the HFD (Figure 1A). Sedentary mice gained ∼5.3 g body weight during this intervention period while exercise trained mice were refractory to HFD-induced body weight gain (Figure 1B). The difference in body mass gain was associated with less adipose tissue mass in exercise-trained compared with sedentary mice (Figure 1C). Skeletal muscle mass (mixed quadriceps) was not different between groups (Figure 1C). Exercise training reduced liver triglyceride content by ∼52% compared with Sedentary (Figure 1D). The efficacy of the exercise program was further demonstrated at the whole-body level by marked improvements in maximal running capacity (Figure 1E), and at the molecular level by increased skeletal muscle citrate synthase activity (Figure 1F), increased protein content of peroxisome proliferator-activated receptor-γ coactivator-1α (PGC1α), a master regulator of mitochondrial biogenesis (Figure 1G–H), as well as increased protein expression of various subunits of the electron transport chain in skeletal muscle (Figure 1I–J). Furthermore, exercise training reduced fasting blood glucose and mildly improved glucose tolerance in mice (Supplementary Figs. 1C–D).
Figure 1.
Validation of exercise adaptations in exercise trained mice. (A) Schematic of the study design. C57Bl/6 mice were fed a high fat diet for 6 weeks and were endurance exercise trained (Exercise) or remained sedentary (Sedentary) for a further 6 weeks, after which their hepatocytes were isolated and cultured for mass spectrometry proteomic evaluation of the intracellular and secreted proteins. (B) Body weight, (C) epididymal (eWAT), inguinal adipose tissue (iWAT) and mixed quadriceps (Quad) mass, (D) maximal running velocity during an incremental test to exhaustion, (E) citrate synthase activity in quadriceps muscle, (F) hepatic triglyceride content, (G–H) representative immunoblot of PGC1α in quadriceps muscle and quantification, and (I–J) representative immunoblot of complex I, IV and V of the electron transport chain in quadriceps muscle and quantification. Data are presented as mean ± SEM, with n = 7–8 per group. ∗P < 0.05 vs. Sedentary. Significance was assessed by two-way ANOVA followed by Bonferroni's post hoc test (panel B) or students two tailed unpaired t-test (panels C–I). Figure 1A was created with BioRender.com.
3.2. Exercise training remodels the hepatocyte proteome and hepatokine secretion
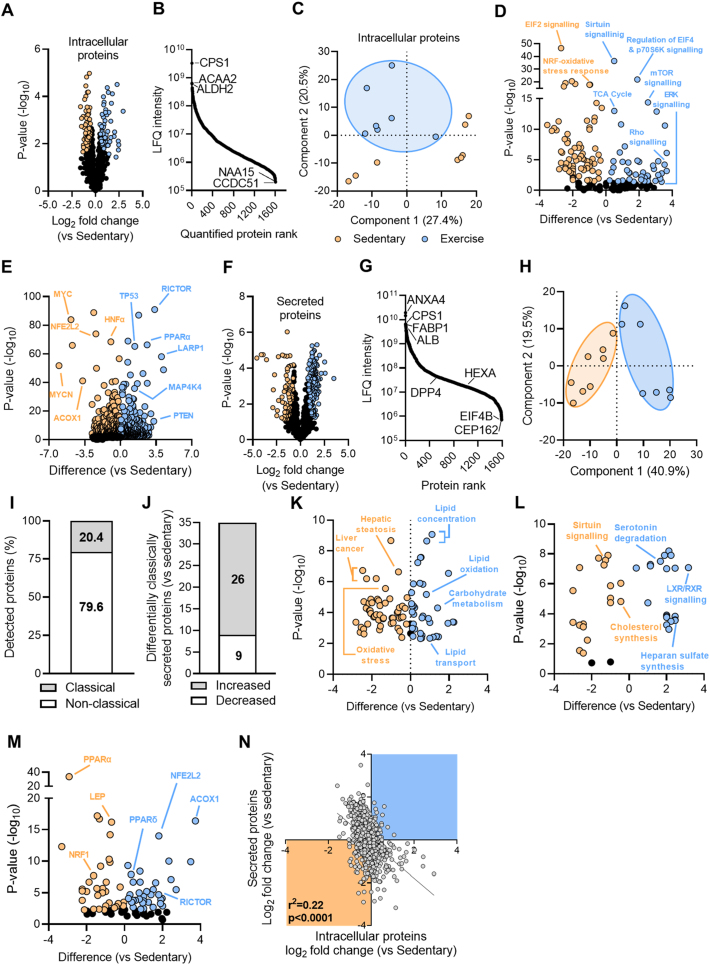
Hepatocytes were isolated from mice 72 h after the last exercise bout to assess adaptive changes to exercise training rather than the effects of an acute exercise bout. The hepatocyte intracellular proteome was assessed using liquid chromatography coupled with tandem mass spectrometry and label-free quantification revealed pronounced differences between the sedentary and exercise trained mice. The analysis identified 2657 proteins in hepatocytes, of which 57 proteins were significantly increased, and 80 proteins were significantly decreased with exercise training compared with sedentary conditions (Figure 2A, Supplementary File 1). Protein LFQ intensity spanned five orders of magnitude and proteins previously shown to be highly enriched in liver [32] were shown to be highly abundant in this analysis, including Carbamoyl-phosphate synthase 1 (CPS), Acetyl-CoA transferase 2 (ACAA2) and Aldehyde dehydrogenase 2 (ALHD2) (Figure 2B). Proteins detected at lower intensities included Coiled-coil domain containing 51 (CCDC51) and N-alpha-acetyltransferase 15 (NAA15). Principle component analysis of the hepatocyte proteomes highlighted that, in general, the Exercise group clustered tightly whereas there was more variability in the Sedentary group (Figure 2C). Qiagen Ingenuity Pathway Analysis (IPA) of canonical pathways highlighted exercise-induced activation of the tricarboxylic acid cycle (TCA) cycle, mTOR and sirtuin signalling, which is indicative of improved mitochondrial function, and suppression of oxidative stress-related pathways (e.g., NRF response, EIF2 signalling) (Figure 2D). Further analysis of potential upstream regulators highlighted enrichment of several tumour suppressive proteins (e.g., PTEN) and proteins involved in lipid metabolism (e.g., RICTOR, PPARα), and suppression of oncogenes (e.g., MYC, MYCN) and regulators of oxidative stress (e.g., NFE2L2) (Figure 2E).
Figure 2.
Identification of the hepatocyte intracellular proteome and hepatocyte secreted proteins. (A) Volcano plot showing hepatocyte intracellular proteins with those increased by exercise training denoted in blue and those decreased denoted in orange. (B) The dynamic range of quantified intracellular proteins. (C) Principal component analysis of the intracellular proteins. (D) Ingenuity Pathway Analysis showing enrichment of upstream factors in exercise-trained mice. (E) Canonical pathways within the hepatocyte. (F) Volcano plot showing hepatocyte secreted proteins with those increased by exercise training denoted in blue and those decreased denoted in orange. (G) The dynamic range of the quantified secreted proteins. (H) Principal component analysis of the hepatocyte secreted proteins. (I) Percentage of proteins that are classically or non-classically secreted. (J) Number of classically secreted proteins that are significantly increased or decreased with exercise training. (K–M) Ingenuity Pathway Analysis showing enrichment of exercise mediated changes in (K) diseases and biological functions, (L) canonical pathways and (M) upstream factors. (N) Correlation between exercise-training induced changes in intracellular proteome and corresponding secreted protein. Shown are means ± SEM, N = 7–8 per group. ∗P < 0.05 vs. Sedentary. Significance was assessed by two-way ANOVA followed by Bonferroni's post hoc test (panel B) or students two tailed unpaired t-test (panels C–I). Proteins identified by corresponding gene name.
We next assessed exercise-induced adaptations in proteins secreted from hepatocytes and identified a total of 1593 proteins across both treatment groups (Figure 2F, Supplementary File 2) spanning six orders of magnitude (Figure 2G). Proteins detected in the secreted medium at higher intensities included Annexin 4 (ANXA4), CPS1, Fatty acid binding protein 1 (FABP1) and Albumin. Hepatocytes secreted other plasma proteins known to be of hepatic origin including Hemopexin [32], Mannose binding lectin [32], Fibrinogen alpha chain [32], Hydroxyacid oxidase 1 (HAO1) [32], Thrombin [32], Complement C9 [32], Apolipoprotein H [32], Dipeptidyl peptidase 4 (DPP4) [33,34] and Hexosaminidase A [30]. In contrast to the intracellular proteome, there was clear clustering of hepatocyte secreted proteins by group as determined by PCA (Figure 2H). Of all secreted proteins, 20.5% were denoted as classically secreted by the presence of a N-terminal signal sequence, while 79.5% were predicted to be secreted via unconventional processes (i.e., in extracellular vesicles) (Figure 2I), which is consistent with previous reports [21,35]. Of the classically secreted proteins, 26 were increased and 9 were decreased following exercise training (Figure 2J). IPA identified overrepresentation of secreted proteins involved in lipid oxidation and transport and downregulation of proteins associated with hepatic steatosis, oxidative stress and liver cancer (Figure 2K). Furthermore, canonical pathways related to heparan sulphate synthesis and degradation of serotonin were overrepresented while cholesterol synthesis was downregulated with exercise training (Figure 2L). Bioinformatic prediction of upstream regulators identified an increase in Acyl-CoA oxidase 1 (ACOX1) and Rapamycin-insensitive companion of mammalian target of rapamycin (RICTOR), and reductions in Nuclear factor-erythroid factor 2-related factor 2 (NRF2) and Peroxisome proliferator activated receptor α (PPARα) (Figure 2M). Together, the results indicate that exercise training regulates the intracellular proteome and protein secretion from hepatocytes and that these adaptive changes are predicted to modulate various aspects of lipid metabolism.
The mRNA expression or intracellular protein abundance is often used as a proxy to predict protein secretion from cells, despite the requirement for extensive posttranslational glycosylation and organellar shuttling to enable protein secretion [36]. We identified a significant negative correlation (r2 = 0.22, p < 0.0001) between the abundance of intracellular proteins and the corresponding secreted proteins (Figure 2N). This data suggests that the change in the intracellular abundance of a protein with an exercise intervention is a poor predictor of the likely change in protein secretion from hepatocytes.
3.3. Hepatocyte-secreted factors of exercise trained mice improve lipid metabolism and insulin action in isolated tissues
Hepatocyte secreted factors derived from mice with NAFL cause inflammation and insulin resistance [37], and numerous hepatokines contribute to dysregulated glucose and lipid metabolism in NAFLD [14,36]. We tested whether hepatocyte-derived factors from exercise trained mice would improve metabolic processes that are often dysfunctional in NAFLD. To do this, we bathed isolated epididymal adipose tissue, intact soleus muscles and precision-cut liver slices from healthy donor mice in the medium containing hepatocyte-secreted factors from sedentary or exercise trained mice and assessed metabolism.
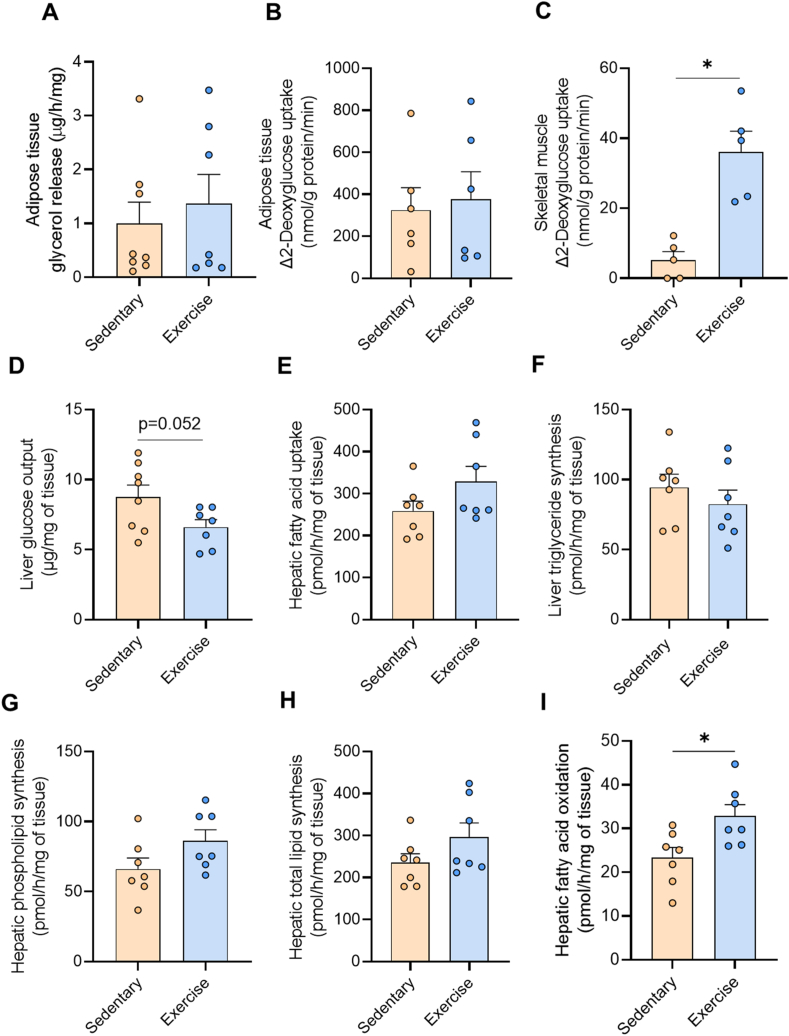
Exercise-secreted factors did not alter lipolysis or insulin-stimulated 2-deoxyglucose uptake in epidydimal adipose tissue (Figure 3A–B). In contrast, hepatocyte secreted factors from exercise trained mice significantly increased insulin-stimulated 2-deoxyglucose uptake in skeletal muscle (Figure 3C) when compared to effects induced by hepatocyte secreted factors from sedentary mice. Liver glucose output was also reduced by hepatocyte secreted factors from exercised compared with sedentary mice (Figure 3D). Fatty acid uptake (Figure 3E), triglyceride synthesis (Figure 3F), phospholipid synthesis (Figure 3G), and total lipid synthesis in the liver (Figure 3H) were not significantly different between treatment conditions, whereas hepatocyte secreted factors from exercise trained mice increased fatty acid oxidation by 39% when compared to hepatocyte factors secreted from sedentary mice (Figure 3I). Collectively, these data provide evidence that regular endurance exercise training alters the complement of secreted factors from hepatocytes, which in turn improves metabolic functions that are often dysregulated in NAFLD.
Figure 3.
Impact of secreted factors derived from hepatocytes of sedentary and exercise-trained mice on metabolism. Hepatocyte-secreted factors from Sedentary or Exercise trained mice were applied to tissues followed by metabolic assessment. (A) Lipolysis and (B) insulin stimulated 2-deoxyglucose uptake in epididymal adipose tissue. (C) Insulin stimulated glucose uptake in soleus muscle. (D) Glucose output from precision cut liver slices. (E–I) Radiometric tracing of lipid metabolism in liver slices showing (E) fatty acid uptake, (F) triglyceride synthesis, (G) phospholipid synthesis, (H) total lipid synthesis, and (I) fatty acid oxidation. Data are presented as mean ± SEM, with n = 5–8 per group. ∗P < 0.05 vs. Sedentary. Significance was assessed by students two tailed unpaired t-test.
3.4. Identification of syndecan-4 as an exercise-responsive hepatokine
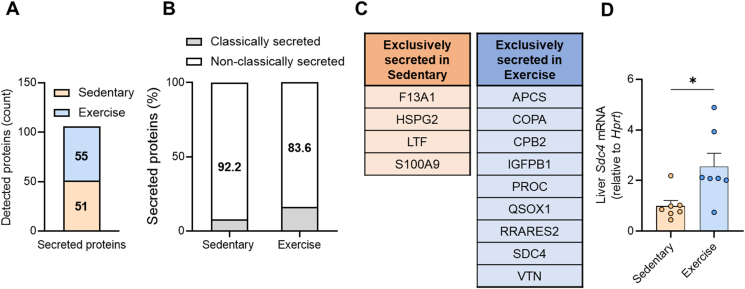
We next sought to identify the secreted proteins that may be mediating these metabolic effects and detected 55 proteins that were exclusively secreted from hepatocytes of exercise trained mice and 51 proteins secreted only from hepatocytes of sedentary mice (Figure 4A, Supplementary File 3). A small proportion of these proteins were denoted as classically secreted (Figure 4B) and these are shown in Figure 4C. Classically secreted proteins exclusively secreted by hepatocytes in Sedentary mice included Coagulation factor XIII a chain (F13A1), Lactotransferrin (LTF), S100 calcium-binding protein A9 (S100A9) and Perlecan (HSPG2), which is associated with glucose intolerance [38] and adiposity [39]. Classically secreted proteins only secreted from hepatocytes of exercise trained mice (Figure 4C) included Insulin like growth factor binding protein (IGFBP1), which was previously shown to protect against cardiovascular pathologies [40], and Retinoic acid receptor responder 2 (RARRES2), also known as Chemerin, which was shown to improve glycaemic control [41].
Figure 4.
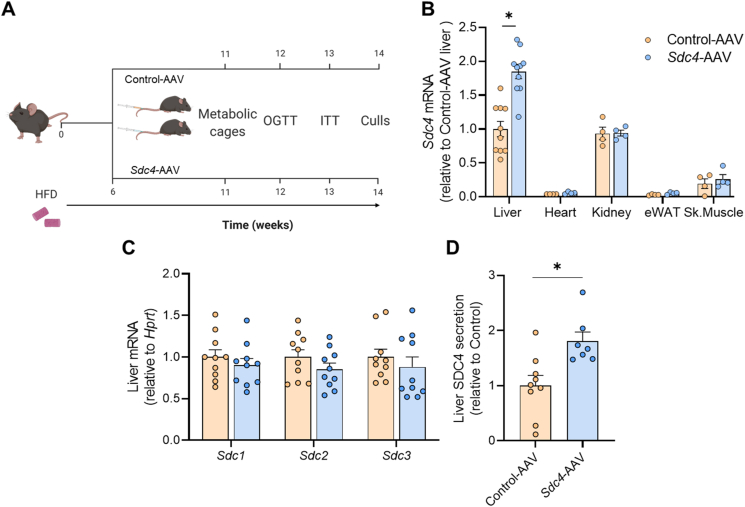
Identification of SDC4 as an exercisetraining-responsivehepatokine. (A) Identification of proteins only secreted from hepatocytes derived from Sedentary and Exercise-trained mice. (B) The percentage of classically and non-classically secreted proteins secreted from hepatocytes derived from Sedentary and Exercise-trained mice. (C) Classically secreted hepatokines derived from hepatocytes of Sedentary and Exercise-trained mice. (D)Sdc4 mRNA in the livers of Sedentary and Exercise-trained mice. Data are presented as mean ± SEM, with n = 7–8 per group. ∗P < 0.05 vs. Sedentary. Significance was assessed by students two tailed unpaired t-test (panel D).
One protein that sparked our interest was Syndecan-4 (SDC4) (Figure 4C). SDC4 is a transmembrane heparan sulfate proteoglycan localized to the plasma membrane, which is cleaved by a range of extracellular proteases [42,43] to release the ectodomain for secretion. We detected three peptides of SDC4 in our mass spectrometry analysis and all are contained in the ectodomain, which spans amino acids 28–41, 106–116, and 100–116 (peptides: 28-ETEVIDPQDLLEGR-41, 106-ELEENEVIPK-116, 100-VPSEPKELEENEVIPK-116). We did not detect peptides of the transmembrane or cytosolic domains, indicating detection of the secreted form of SDC4. Circulating SDC4 levels have been reported to be ∼10–20 ng/mL in humans [44]. Plasma SDC4 is increased during exercise in humans [44] and polymorphisms in SDC4 are associated with abdominal fat [45]. Moreover, ablation of the SDC4 orthologue in Drosophila (known as Sdc) reduced Spargel (i.e., PGC1α) expression and decreased metabolic rate [46], whereas, seemingly paradoxically, plasma SDC4 levels are higher in men with dysregulated glycemic control compared to normoglycemic men [44]. Notably, Sdc4 expression was significantly increased in the livers of exercise trained compared with sedentary mice (Figure 4D).
3.5. Hepatocyte-specific Sdc4 overexpression in mice with NAFLD
To investigate the metabolic roles of hepatic SDC4 and its potential to reduce the burden of NAFLD and its associated co-morbidities, mice were fed a high-fat diet for six weeks to induce mild obesity and NAFLD (Figure 5A). Mice were segregated into experimental groups after matching for body weight, fat mass and glucose tolerance (Figs. S2A–C). Mice were then injected i.v with an AAV8 vector containing a hepatocyte-specific promotor (TBG) and Sdc4 (Sdc4-AAV) or the same AAV8 vector containing scrambled DNA (Control-AAV) (Figure 5A). Sdc4 expression was increased 2-fold in the liver following Sdc4-AAV administration, in the absence of changes in Sdc4 expression in other tissues (Figure 5B), and this occurred independent of changes in the expression of other SDC family members, including Sdc1, Sdc2 and Sdc3 (Figure 5C). SDC4 secretion from precision-cut liver slices was increased by 80% in Sdc4-AAV compared with Control-AAV (Figure 5D), noting that the same three ectodomain spanning peptides in the exercise secretome were detected in this preparation. We were unable to detect SDC4 in the plasma of mice when using a commercially available ELISA or targeted mass spectrometry, which has been reported previously [47] and is thought to be due to excessive glycosylation [48].
Figure 5.
Validation of Sdc4 overexpression in mice. (A) Schematic representation of the experimental design. C57BL/6 mice were fed a high fat diet (HFD) for 6 weeks then injected with Sdc4-AAV or Control-AAV. Following AAV administration, mice were maintained on a HFD for a further 6 weeks. (B)Sdc4 expression in liver, heart, kidney, skeletal muscle and epididymal adipose tissue. (C)Sdc1, Sdc2 and Sdc3 mRNA expression in the liver. (D) Targeted mass spectrometry analysis of SDC4 secretion from precision cut liver slices. Data are presented as mean ± SEM, with n = 4–10 per group. ∗P < 0.05 vs. Control-AAV. Significance was assessed by students two tailed unpaired t-test. Figure 5A was created with Biorender.com.
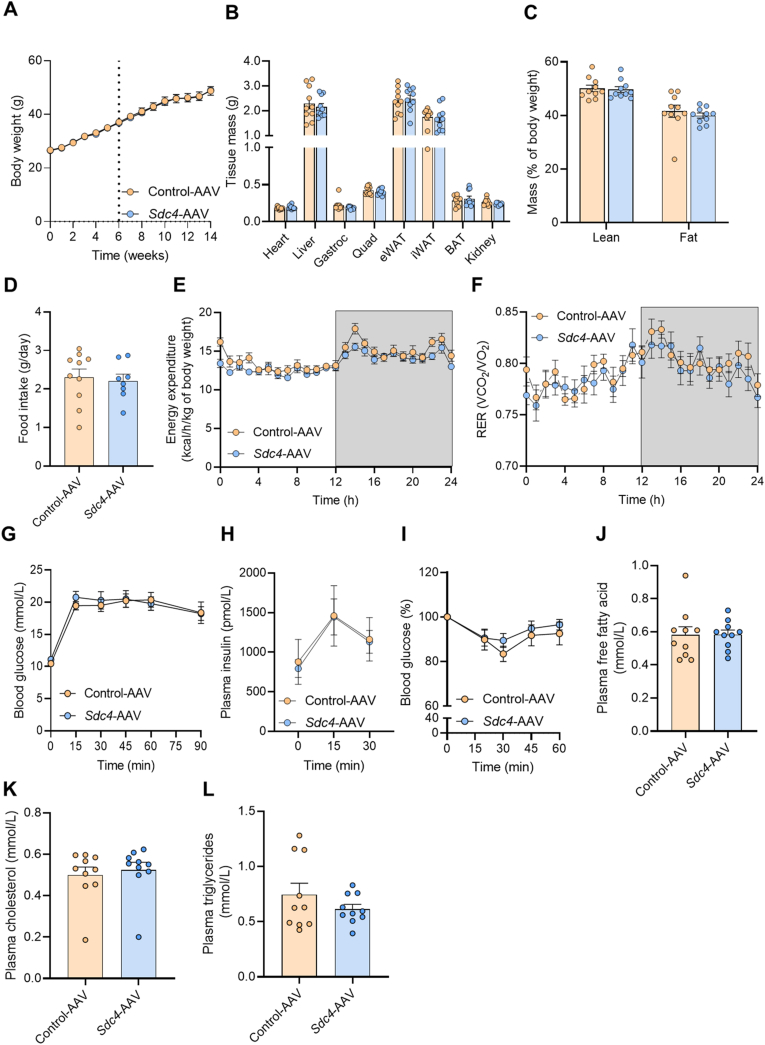
3.6. Hepatic overexpression of Sdc4 does not alter energy homeostasis or glycaemic control
Given the known links between SDC4, metabolism and energy expenditure in other species [45,49,50], we first assessed the impact of Sdc4 overexpression on body mass regulation and energy homeostasis. Body weight, individual tissue weights, adiposity, food intake, daily energy expenditure and substrate partitioning were not different between Sdc4-AAV and Control-AAV (Figure 6A–F). Similarly, increasing Sdc4 expression in the liver did not alter glucose tolerance (Figure 6G), fasting or glucose-stimulated insulin secretion (Figure 6H), or whole-body insulin action (Figure 6I). Plasma lipids including free fatty acids, total cholesterol and triglycerides were not different between groups (Figure 6J-L).
Figure 6.
Hepatic Sdc4 overexpression does not alter whole body energy homeostasis or glycemic control. (A) Body weight (dotted line indicates Sdc4-AAV or Control-AAV injection), (B) tissue weights, (C) lean and fat mass, (D) daily food intake, (E) energy expenditure, (F) respiratory exchange ratio (RER), (G) glucose tolerance following oral glucose administration (2 g/kg), (H) fasting and glucose-stimulated plasma insulin levels, and (I) blood glucose in response to i.p insulin administration (1 U/kg). (J) Plasma free fatty acids, (K) cholesterol and (L) triglycerides. Data are presented as mean ± SEM, with n = 7–10 per group. ∗P < 0.05 vs. Control-AAV. Significance was assessed by students two tailed unpaired t-test (panels B, C, D, J, K, L) or two-way ANOVA followed by Bonferroni's post hoc test (panels A, E, F, G, H, I).
3.7. SDC4 reduces fatty acid uptake and markers of inflammation and fibrosis in the liver
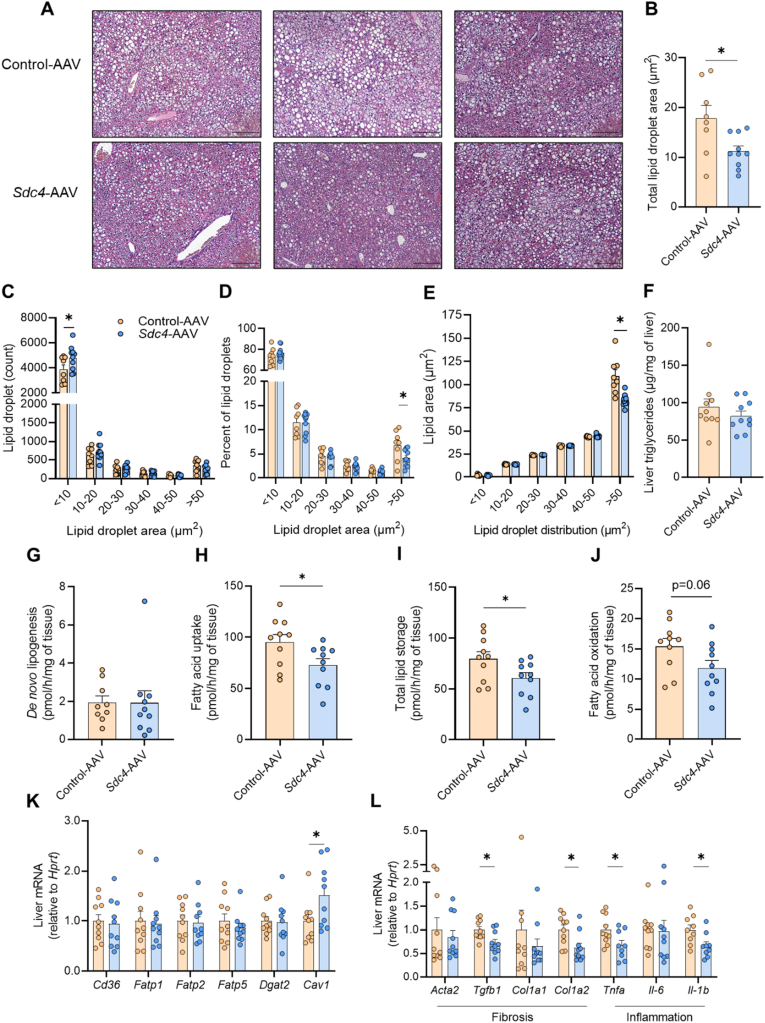
Histological assessment of H&E-stained liver sections revealed a reduction in macrosteatosis in Sdc4-AAV compared with Control-AAV livers (Figure 7A). Unbiased assessment of lipid droplet number and size demonstrated an overall reduction of total lipid droplet area with Sdc4-AAV (Figure 7B), which was associated with a change in the size distribution of lipid droplets. SDC4 overexpression increased the number of ‘smaller’ lipid droplets (<10 μm2) without altering the number of larger lipid droplets (10–50 μm2) (Figure 7C). However, relative to the total number of lipid droplets, there was a 40% reduction in lipid droplets with an area >50 μm2 in mice overexpressing Sdc4 in the liver and, importantly, the average area of ‘larger’ lipid droplets (>50 μm2) was reduced in Sdc4-AAV compared with Control-AAV (Figure 7E). These findings are consistent with the histological finding of reduced macrosteatosis in Sdc4-AAV mice (Figure 7A). Despite the significant lipid droplet remodelling, we did not observe any differences in total liver triglyceride levels as determined by biochemical assessment (Figure 7F). Taken together, this remodelling of lipid droplet size and number indicated a plausible role for SDC4 in regulating hepatic lipid metabolism.
Figure 7.
SDC4 regulates hepatic lipid metabolism in mice. (A) Representative liver histology stained with hematoxylin and eosin. Scale bar denotes 200 μm. Sections from n = 3 individual mice per group. (B) Quantification of the average lipid droplet area in liver sections. (C) Lipid droplet number in livers presented as a function of lipid droplet size. (D) Proportion of different sized lipid droplets in livers of mice. (E) Average area of lipid droplets highlighting a reduction in the area of larger (>50 μm2) lipid droplets in Sdc4-AAV vs. Control-AAV mice. (F) Liver triglyceride content. (G) De novo lipogenesis, (H) fatty acid uptake, (I) total lipid storage and (J) fatty acid oxidation in liver. (K) mRNA expression of lipid transporter and esterification enzymes. (L) mRNA expression pro-fibrotic and pro-inflammatory genes in the liver. Data are presented as mean ± SEM, with n = 8–10 per group. ∗P < 0.05 vs. Control-AAV. Significance was assessed by students two tailed unpaired t-test.
In light of these alterations in liver lipid droplets, and given that SDC4 is localized to the plasma membrane [51] and is linked to triglyceride storage in Drosophila [52], we surmised that SDC4 might induce autocrine/paracrine regulation of lipid metabolism. Accordingly, we investigated the effect of SDC4 overexpression on lipid metabolism in precision-cut liver slices. De novo lipogenesis was assessed by tracing 14C-glucose into cellular lipid pools and was not different between groups (Figure 7G). In contrast, fatty acid uptake was significantly reduced in Sdc4-AAV compared with Control-AAV (Figure 7H), and this coincided with a 25% reduction in fatty acid incorporation into cellular lipids (Figure 7I). Fatty acid oxidation was also reduced (P = 0.06) in livers overexpressing Sdc4 (Figure 7J). Given the marked reduction in fatty acid uptake, we assessed the expression of genes encoding proteins involved in lipid transport and intracellular trafficking. There was no difference in the expression of fatty acid transporters including fatty acid translocase (Cd36), fatty acid transport protein −1 (Fatp1), −2 (Fatp2) and −5 (Fatp5). Diacylglycerol O-acyltransferase 2 (Dgat2), a key enzyme involved in triglyceride synthesis, was also similar between groups. In contrast, caveolin-1 (Cav-1) expression was increased with SDC4 overexpression compared with control (Figure 7K). This latter finding is of interest because Cav-1 is required to create membrane invaginations termed caveolae and form a complex with CD36 and other proteins to regulate fatty acid uptake and intracellular trafficking [53,54].
Inflammation and fibrosis are common features of NAFLD. While this short-term HFD dietary intervention does not induce significant adverse histopathology such as lobular inflammation, hepatocellular ballooning, or fibrosis (data not shown), Sdc4-AAV reduced the expression of pro-inflammatory genes including tumour necrosis factor α (Tnfα) and interleukin-1β (Il-1b) and several pro-fibrotic genes including transforming growth factor β1 (Tgfb1) and interstitial collagen type I α2 chain (Col1a2) (Figure 7L). Thus, increasing SDC4 in the liver may reduce the rate of fatty acid uptake, and may prevent the eventual induction of inflammation and fibrosis in NAFLD.
3.8. SDC4 ectodomain mediates changes in fatty acid metabolism and lipid droplet remodelling in hepatocytes
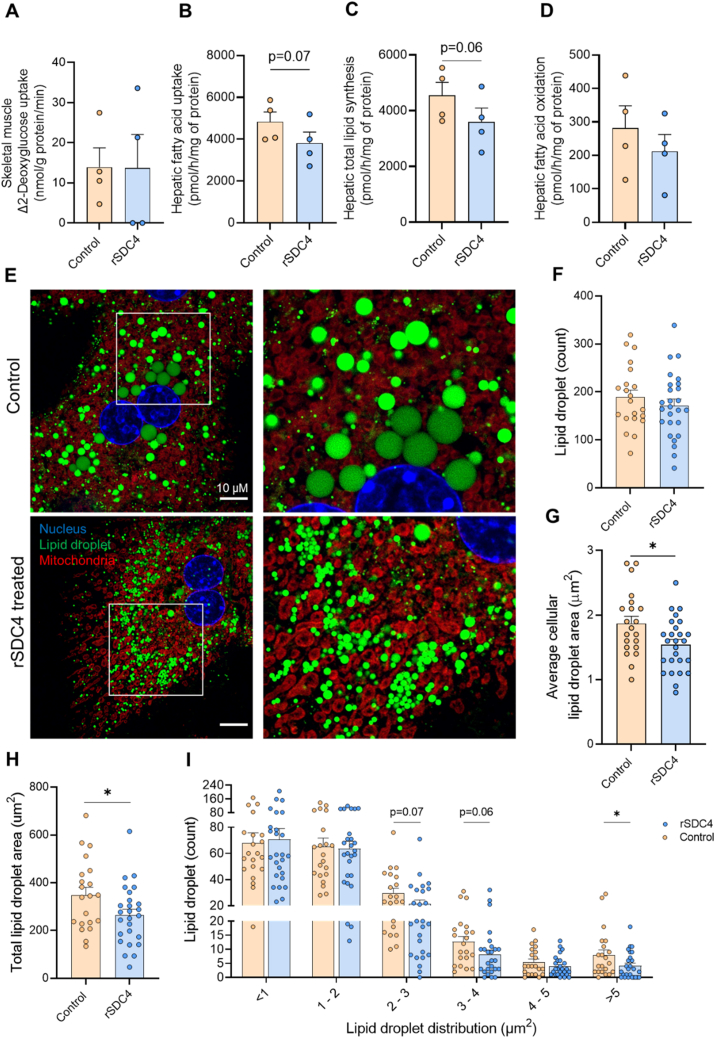
While Sdc4 overexpression in mice induced changes in hepatocyte fatty acid metabolism and lipid droplet morphology, we were unable to definitively ascribe these effects to SDC4's function as a hepatokine. Accordingly, we next assessed the role of the SDC4 ectodomain in regulating these processes. To do this, we cultured intact soleus muscle and isolated hepatocytes with recombinant SDC4 ectodomain (rSDC4, 1 μg/mL) and assessed insulin action and fatty acid metabolism. rSDC4 did not affect insulin-stimulated glucose uptake in intact soleus muscle (Figure 8A) whereas rSDC4 reduced fatty acid uptake (p = 0.07) (Figure 8B) and lipid storage (p = 0.06) (Figure 8C) in hepatocytes without significant changes in fatty acid oxidation (Figure 8D).
Figure 8.
SDC4 ectodomain regulates fatty acid metabolism and lipid droplet distribution.(A) Insulin stimulated glucose uptake with or without recombinant SDC4 in isolated soleus muscle. (B–D) Radiometric tracing of lipid metabolism in isolated hepatocytes with or without acute recombinant SDC4 treatment showing (B) fatty acid uptake, (C) total lipid storage and, (D) fatty acid oxidation. (E) Representative image of lipid droplet morphology stained with Bodipy, Mitotracker and DAPI. Scale bar denotes 10 μm. Hepatocytes were isolated from n = 3 mice, 5–8 hepatocytes were imaged across two technical replicates and quantified per mouse totalling n = 21 Control and n = 26 rSDC4 treated hepatocytes. (F) Quantification of the lipid droplet number within the cell, (G) the total area that lipid droplets occupy in each hepatocyte, and (H) the average lipid droplets area within the hepatocyte. (I) Proportion of different sized lipid droplets in isolated hepatocytes. Data are presented as mean ± SEM, ∗P < 0.05 vs. Control-AAV, with n = 4 per group for paired analyses. Significance was assessed by students two tailed unpaired t-test (panel B–E) or paired t-test (panel G–I).
To ascertain whether the secreted form of SDC4 was responsible for altering lipid droplet morphology, primary hepatocytes were isolated and cultured with or without rSDC4 for 48 h. Visual inspection revealed marked remodelling of lipid droplet size with rSDC4 treatment (Figure 8E). While there was no difference in the number of lipid droplets between conditions (Figure 8F), rSDC4 administration reduced the average cellular lipid droplet area by ∼20% (Figure 8C), and the total area occupied by lipid droplets within the cell by 25% (Figure 8D). Further, these changes were associated with changes in the size distribution of lipid droplets, with reductions in the number of lipid droplets with an area of 2–3, 3–4 and >5 μm2 with rSDC4 treatment (Figure 8E). Thus, the secreted ectodomain of SDC4 impacts fatty acid metabolism and reduces lipid droplet size in hepatocytes.
4. Discussion
Exercise training induces a myriad of health promoting effects in many tissues, including the liver [55]. However, the role of exercise in regulating the liver proteome and secretome is not well described, particularly in the context of NAFLD, which is the world's most common liver disease. In this study, we employed unbiased mass spectrometry-based proteomics analyses to identify substantial remodelling of the intracellular proteome and secreted proteins from hepatocytes and offer insights into changes that may contribute to the metabolic benefits associated with regular exercise training. First, we identified exercise training regulation of the liver proteome. Second, we described the effect of exercise training on liver secreted proteins. Third, we show that the complement of liver secreted factors is altered with exercise training, which in turn alters glucose and lipid metabolism in liver and skeletal muscle. And lastly, we identified SDC4 as a novel hepatokine that is increased with exercise training and its secreted ectodomain regulates hepatic lipid metabolism and lipid droplet morphology in NAFLD.
The effects of exercise on liver metabolism are becoming increasingly well described [56] and include increased mitochondrial content and glycolysis, and reduced expression of lipid synthesis proteins [57,58], which is associated with partial resolution of hepatic steatosis [7]. Furthermore, transcriptomic analysis conducted in the liver of mice immediately after an acute bout of exercise revealed marked increases in the expression of genes involved in mitogen-activated protein kinase (MAPK) and PPAR signalling, and fatty acid metabolism [17]. However, whether such acute changes lead to stable adaptations at the protein level is not well defined [58]. To address this knowledge gap, we performed unbiased assessment of the hepatocyte proteome following chronic exercise training. Aligned with previous transcriptomic analysis [17] and the known protective effects of exercise training on NAFLD and hepatocellular carcinoma [59,60], our bioinformatic investigation of the hepatocyte proteome highlighted upregulation of metabolic processes including the TCA cycle, mTOR signalling and PPARα signalling, downregulation of pathways involved in cancer and inflammation (e.g., Nrf2-oxidative stress response, Myc), and upregulation of tumour suppressor pathways (e.g., PTEN and TP53). Collectively, these findings support the notion that the hepatocyte proteome is remodelled in response to regular exercise training in the context of NAFLD, and that these changes are associated with improved metabolic health and anti-tumorigenic effects.
The liver is an active endocrine organ that produces and secretes a large proportion of the body's circulating proteins [14,15] and these hepatokines influence a myriad of biological functions [13,16,18,61]. Expanding this knowledge base, we identified 1593 hepatocyte secreted proteins, of which 20.5% are classically secreted. This analysis has the advantage of providing cell-specific knowledge of the hepatocyte secretome. However, hepatocyte isolation and prolonged ex vivo incubation/culture of cells may influence protein secretion and future studies are required to compare the hepatocyte secretome with the secretion pattern of liver tissue. Bioinformatic investigation revealed an enrichment of proteins regulating carbohydrate metabolism, lipid transport and lipid oxidation following chronic exercise training. This prediction is supported by our finding that exercise-responsive factors secreted from hepatocytes improved skeletal muscle insulin action, reduced liver glucose output and increased liver fatty acid oxidation, adaptations that are often reported with exercise training [11]. It should be noted that secreted factors other than proteins, including metabolites, lipids and extracellular vesicles (containing lipids, non-coding RNA, proteins) [14,62] are likely to contribute to these metabolic changes. We also identified significant alterations in specific hepatokines with chronic exercise training, including reductions in known mediators of insulin resistance (e.g., dipeptidyl peptidase) [34] pathological angiogenesis (e.g., pantetheinase) [63], adipose tissue lipolysis (e.g., cathepsin B) [64] and fatty acid uptake (e.g., fatty acid binding protein 4). On the other hand we identified increased secretion of proteins involved in the preservation of insulin sensitivity (e.g., insulin degrading enzyme) [65], alleviation of oxidative stress (e.g., n-myc downstream regulated gene 2, heme oxgenase-1) [66,67] and the liver X receptor/retinoid X receptor activation that dampens the inflammatory response (e.g., α-1-microglobulin, transthyretin, Serpina1, α-1-antitrypsin) [68]. Notably, of the 26 hepatokines upregulated with exercise-training, there remains a paucity of evidence describing their role in glucose and lipid metabolism. An intriguing observation from these studies was the clear negative correlation between secreted and intracellular proteins in exercise-trained hepatocytes. It is possible that secreted proteins induce negative feedback loops, whereby increased protein degradation and turnover, and proteolytic ectodomain shedding of proteins explained the negative correlation in secreted versus intracellular proteins. It is also possible that there is a ‘lag’ in protein translation behind transcription of highly secreted proteins. Indeed, others have reported that a group of proteins in the secretome of activated macrophages did not show a concomitant change in transcript or intracellular protein levels [69], and further studies combining isotopic labelling and mass spectrometry to measure the rate of protein synthesis, cellular distribution and secretion are required to test these possibilities in hepatocytes. Together, the present study shows that remodelling of hepatokine secretion is an adaptation to regular exercise training, and that these changes in hepatokine secretion are associated with changes in metabolism in the liver (autocrine/paracrine effects) and in distant tissues that would be predicted to resolve several pathogenic features of NAFLD.
The proteomic analysis identified SDC4 as an exercise-responsive hepatokine. SDC4 is a transmembrane proteoglycan that comprises an extracellular ectodomain, a transmembrane domain, and a cytoplasmic tail that binds to the actin cytoskeleton and intracellular binding partners. Extracellular proteases can cleave the ectodomain of SDC4 [70] and subsequently regulate its secretion. SDC4 has been implicated in controlling triglyceride storage [49] and dyslipidaemia [50], while SNPs in Sdc4 are associated with energy expenditure, fasting blood glucose, insulin sensitivity, and abdominal fat [45], processes known to be impacted in NAFLD and by exercise training. However, previous studies by our group have not detected SDC4 secretion from hepatocytes isolated from mice fed a chow or a high-fat diet [71,72], indicating that SDC4 secretion is unlikely to be regulated by an obesogenic diet per se. In this study, the ectodomain of SDC4 was reproducibly detected in the secretome of hepatocytes derived from exercise trained mice, indicating that SDC4 is an exercise-responsive hepatokine.
Our studies using recombinant protein of the SDC4 ectodomain and hepatocyte-specific AAV-mediated SDC4 overexpression in mice identified a role for SDC4 in regulating hepatic lipid metabolism. Histological assessment revealed a reduction in macrosteatosis and concomitant accumulation of smaller lipid droplets, without altering total triglyceride content with either AAV-mediated overexpression or chronic rSDC4 exposure in primary hepatocytes. Such lipid droplet remodelling is reminiscent of the changes seen in liver and muscle after exercise training [8,73] and aligns with the view that smaller lipid droplets differ from larger lipid droplets with respect to their protein composition, inter-organellular contacts and functions [74]. Smaller lipid droplets are considered ‘healthier’ than larger lipid droplets [73,75], a notion supported by parallel lines of evidence demonstrating that exercise training reduces intramyocellular lipid droplet size without altering lipid droplet quantity or intramyocellular lipid content in patients with type 2 diabetes [73], and that athletes, when compared to individuals with obesity and type 2 diabetes, have a reduction in lipid droplet area and size of ‘larger’ lipid droplets, independent of changes in the total number of lipid droplets and intramyocellular lipid content [75]. Whether this association holds true in the human liver remains to be determined.
SDC4 reduced fatty acid uptake in hepatocytes, which was associated with less incorporation of fatty acids into cellular lipids, factors that might contribute to the reduced prevalence of larger lipid droplets in Sdc4-AAV mice and hepatocytes chronically exposed to the SDC4 ectodomain. Changes in the expression of fatty acid transporters do not appear to be mediating SDC4 effects on fatty acid uptake and storage. Rather, SDC4 overexpression was associated with increased Cav1 expression in the liver. While the role of CAV-1 in mediating fatty acid uptake remains contentious [76], previous studies show that Cav1 overexpression decreases fatty acid uptake in multiple cells types and deletion of Cav1 in the liver reduces hepatic lipid accumulation [77,78]. Further, others report binding of SDC4 to CAV-1 [79], which triggers caveolar dependent endocytosis [47], a process that influences fatty acid transport and lipid droplet growth [80]. Together, these findings provide a possible mechanism to explain the reduction in SDC4-mediated fatty acid uptake and lipid droplet storage reported in the present study.
Loss of SDC4 results in an aggravated inflammatory response and impairments in wound healing [47,[81], [82], [83]]. Here, we showed that increasing Sdc4 content and secretion in the liver reduces the expression of pro-inflammatory and pro-fibrotic genes in NAFLD. Given that reductions in hepatic inflammation and fibrosis are associated with improved health outcomes [84] and progression to NASH [85], further studies examining the anti-inflammatory and anti-fibrotic effects of SDC4 in NASH are warranted.
In summary, we have developed a novel resource detailing the proteomic changes that occur in hepatocytes with regular exercise training in mice with NAFLD. These studies identify marked remodelling of the liver intracellular proteome and altered hepatokine secretion with exercise training, effects that impacted metabolism in the liver and skeletal muscle. SDC4 was identified as a novel exercise-responsive hepatokine that decreases fatty acid uptake, reduces macrosteatosis and dampens the expression of inflammatory and pro-fibrotic genes in the liver. By understanding the proteomic changes in hepatocytes with exercise, we believe that these findings have strong potential for the discovery of new therapeutic targets for NAFLD, which remains an unmet clinical need.
Financial support
WD is supported by The University of Melbourne Research Scholarship. PMM was supported by a Canadian Institutes of Health Research (CIHR) post-doctoral fellowship and a Natural Sciences and Engineering Research Council of Canada (NSERC) post-doctoral fellowship. MKM is supported by a Career Development Fellowship from the National Health and Medical Research Council of Australia (ID: APP1143224). These studies were supported by the NHMRC (APP1162511, APP1156508).
Acknowledgements
We thank the Melbourne Mass Spectrometry and Proteomics Facility of The Bio21 Molecular Science and Biotechnology Institute at The University of Melbourne for the support of mass spectrometry analysis. This work was supported by infrastructure and technical assistance from the Melbourne Mouse Metabolic Phenotyping Platform (MMMPP) at the University of Melbourne. This work was supported by infrastructure and technical assistance from the Melbourne Histology Platform at the University of Melbourne and the biological optical microscopy platform (BOMP), with thanks for the technical assistance of Dr Ellie Cho. Figure 1A & 5A were created with BioRender.com
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101491.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease—meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64(1):73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Chalasani N., Younossi Z., Lavine J.E., Charlton M., Cusi K., Rinella M., et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2017;67(1):328–357. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
- 3.Loomba R., Sanyal A.J. The global NAFLD epidemic. Nature Reviews Gastroenterology and Hepatology. 2013;10:686. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- 4.Kleiner D.E., Berk P.D., Hsu J.Y., Courcoulas A.P., Flum D., Khandelwal S., et al. Hepatic Pathology among patients without known liver disease undergoing bariatric surgery: observations and a perspective from the longitudinal assessment of bariatric surgery (LABS) study. Seminars in Liver Disease. 2014;34(1):98–107. doi: 10.1055/s-0034-1371083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams L.A., Roberts S.K., Strasser S.I., Mahady S.E., Powell E., Estes C., et al. Nonalcoholic fatty liver disease burden: Australia, 2019-2030. Journal of Gastroenterology and Hepatology. 2020;35(9):1628–1635. doi: 10.1111/jgh.15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Estes C., Razavi H., Loomba R., Younossi Z., Sanyal Arun J. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology. 2017;67(1):123–133. doi: 10.1002/hep.29466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashida R., Kawaguchi T., Bekki M., Omoto M., Matsuse H., Nago T., et al. Aerobic vs. resistance exercise in non-alcoholic fatty liver disease: a systematic review. Journal of Hepatology. 2017;66(1):142–152. doi: 10.1016/j.jhep.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 8.la Fuente F.P., Quezada L., Sepúlveda C., Monsalves-Alvarez M., Rodríguez J.M., Sacristán C., et al. Exercise regulates lipid droplet dynamics in normal and fatty liver. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2019;1864(12):158519. doi: 10.1016/j.bbalip.2019.158519. [DOI] [PubMed] [Google Scholar]
- 9.Promrat K., Kleiner D.E., Niemeier H.M., Jackvony E., Kearns M., Wands J.R., et al. Randomized controlled trial testing the effects of weight loss on nonalcoholic steatohepatitis. Hepatology. 2010;51(1):121–129. doi: 10.1002/hep.23276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckard C., Cole R., Lockwood J., Torres D.M., Williams C.D., Shaw J.C., et al. Prospective histopathologic evaluation of lifestyle modification in nonalcoholic fatty liver disease: a randomized trial. Therapeutic Advances in Gastroenterolog. 2013;6(4):249–259. doi: 10.1177/1756283X13484078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rector R.S., Thyfault J.P., Morris R.T., Laye M.J., Borengasser S.J., Booth F.W., et al. Daily exercise increases hepatic fatty acid oxidation and prevents steatosis in Otsuka Long-Evans Tokushima Fatty rats. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2008;294(3):G619–G626. doi: 10.1152/ajpgi.00428.2007. [DOI] [PubMed] [Google Scholar]
- 12.Inelmen E.M., Toffanello E.D., Enzi G., Gasparini G., Miotto F., Sergi G., et al. Predictors of drop-out in overweight and obese outpatients. International Journal of Obesity. 2005;29(1):122–128. doi: 10.1038/sj.ijo.0802846. [DOI] [PubMed] [Google Scholar]
- 13.Murphy R.M., Watt M.J., Febbraio M.A. Metabolic communication during exercise. Nature Metabolism. 2020;2(9):805–816. doi: 10.1038/s42255-020-0258-x. [DOI] [PubMed] [Google Scholar]
- 14.Watt M.J., Miotto P.M., De Nardo W., Montgomery M.K. The liver as an endocrine organ—linking NAFLD and insulin resistance. Endocrine Reviews. 2019;40(5):1367–1393. doi: 10.1210/er.2019-00034. [DOI] [PubMed] [Google Scholar]
- 15.Lai K.K.Y., Kolippakkam D., Beretta L. Comprehensive and quantitative proteome profiling of the mouse liver and plasma. Hepatology. 2008;47(3):1043–1051. doi: 10.1002/hep.22123. [DOI] [PubMed] [Google Scholar]
- 16.Hoene M., Weigert C. The stress response of the liver to physical exercise. Exercise Immunology Review. 2010;16:163–183. [PubMed] [Google Scholar]
- 17.Hoene M., Franken H., Fritsche L., Lehmann R., Pohl A.K., Häring H.U., et al. Activation of the mitogen-activated protein kinase (MAPK) signalling pathway in the liver of mice is related to plasma glucose levels after acute exercise. Diabetologia. 2010;53(6):1131–1141. doi: 10.1007/s00125-010-1666-3. [DOI] [PubMed] [Google Scholar]
- 18.Ennequin G., Sirvent P., Whitham M. Role of exercise-induced hepatokines in metabolic disorders. American Journal of Physiology. Endocrinology and Metabolism. 2019;317(1):e11–e24. doi: 10.1152/ajpendo.00433.2018. [DOI] [PubMed] [Google Scholar]
- 19.Mohktar R.A., Montgomery M.K., Murphy R.M., Watt M.J. Perilipin 5 is dispensable for normal substrate metabolism and in the adaptation of skeletal muscle to exercise training. American Journal of Physiology. Endocrinology and Metabolism. 2016;311(1):E128–E137. doi: 10.1152/ajpendo.00084.2016. [DOI] [PubMed] [Google Scholar]
- 20.Borg M.L., Omran S.F., Weir J., Meikle P.J., Watt M.J. Consumption of a high-fat diet, but not regular endurance exercise training, regulates hypothalamic lipid accumulation in mice. The Journal of Physiology. 2012;590(17):4377–4389. doi: 10.1113/jphysiol.2012.233288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meex Ruth C., Hoy Andrew J., Morris A., Brown Russell D., Lo Jennifer C.Y., Burke M., et al. Fetuin B is a secreted hepatocyte factor linking steatosis to impaired glucose metabolism. Cell Metabolism. 2016;22(6):1078–1089. doi: 10.1016/j.cmet.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 22.Keenan S.N., De Nardo W., Lou J., Schittenhelm R.B., Montgomery M.K., Granneman J.G., et al. Perilipin 5 S155 phosphorylation by PKA is required for the control of hepatic lipid metabolism and glycemic control. Journal of Lipid Research. 2021;62 doi: 10.1194/jlr.RA120001126. 100016-100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26:1367. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 24.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods. 2016;13:731. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 25.Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergmeyer H.U. IFCC methods for the measurement of catalytic concentrations of enzymes: Part 3. IFCC method for alanine aminotransferase (L-alanine: 2-oxoglutarate aminotransferase, EC 2.6.1.2) Clinica Chimica Acta. 1980;105(1):147–154. [PubMed] [Google Scholar]
- 28.Keenan S.N., Meex R.C., Lo J.C.Y., Ryan A., Nie S., Montgomery M.K., et al. Perilipin 5 deletion in hepatocytes remodels lipid metabolism and causes hepatic insulin resistance in mice. Diabetes. 2019;68(3):543–555. doi: 10.2337/db18-0670. [DOI] [PubMed] [Google Scholar]
- 29.Watt M.J., Dzamko N., Thomas W.G., Rose-John S., Ernst M., Carling D., et al. CNTF reverses obesity-induced insulin resistance by activating skeletal muscle AMPK. Nature Medicine. 2006;12(5):541–548. doi: 10.1038/nm1383. [DOI] [PubMed] [Google Scholar]
- 30.Montgomery M.K., Taddese A.Z., Bayliss J., Nie S., Williamson N.A., Watt M.J. Hexosaminidase A (HEXA) regulates hepatic sphingolipid and lipoprotein metabolism in mice. The FASEB Journal. 2021;35(12) doi: 10.1096/fj.202101186R. [DOI] [PubMed] [Google Scholar]
- 31.Berg S., Kutra D., Kroeger T., Straehle C.N., Kausler B.X., Haubold C., et al. ilastik: interactive machine learning for (bio)image analysis. Nature Methods. 2019;16(12):1226–1232. doi: 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 32.Kampf C., Mardinoglu A., Fagerberg L., Hallström B.M., Edlund K., Lundberg E., et al. The human liver-specific proteome defined by transcriptomics and antibody-based profiling. The FASEB Journal. 2014;28(7):2901–2914. doi: 10.1096/fj.14-250555. [DOI] [PubMed] [Google Scholar]
- 33.Baumeier C., Schluter L., Saussenthaler S., Laeger T., Rodiger M., Alaze S.A., et al. Elevated hepatic DPP4 activity promotes insulin resistance and non-alcoholic fatty liver disease. Molecular Metabolism. 2017;6(10):1254–1263. doi: 10.1016/j.molmet.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghorpade D.S., Ozcan L., Zheng Z., Nicoloro S.M., Shen Y., Chen E., et al. Hepatocyte-secreted DPP4 in obesity promotes adipose inflammation and insulin resistance. Nature. 2018;555:673. doi: 10.1038/nature26138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franko A., Hartwig S., Kotzka J., Ruoß M., Nüssler A.K., Königsrainer A., et al. Identification of the secreted proteins originated from primary human hepatocytes and HepG2 cells. Nutrients. 2019;11(8):1795. doi: 10.3390/nu11081795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montgomery M.K., De Nardo W., W M.J. Impact of lipotoxicity on tissue “cross-talk” and metabolic regulation. Physiology. 2019;34 doi: 10.1152/physiol.00037.2018. [DOI] [PubMed] [Google Scholar]
- 37.Meex Ruth C., Hoy Andrew J., Morris A., Brown Russell D., Lo Jennifer C.Y., Burke M., et al. Fetuin B is a secreted hepatocyte factor linking steatosis to impaired glucose metabolism. Cell Metabolism. 2015;22(6):1078–1089. doi: 10.1016/j.cmet.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 38.Templin A.T., Mellati M., Soininen R., Hogan M.F., Esser N., Castillo J.J., et al. Loss of perlecan heparan sulfate glycosaminoglycans lowers body weight and decreases islet amyloid deposition in human islet amyloid polypeptide transgenic mice. Protein Engineering Design and Selection. 2019;32(2):95–102. doi: 10.1093/protein/gzz041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamashita Y., Nakada S., Yoshihara T., Nara T., Furuya N., Miida T., et al. Perlecan, a heparan sulfate proteoglycan, regulates systemic metabolism with dynamic changes in adipose tissue and skeletal muscle. Scientific Reports. 2018;8(1):7766. doi: 10.1038/s41598-018-25635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajwani A., Ezzat V., Smith J., Yuldasheva N.Y., Duncan E.R., Gage M., et al. Increasing circulating IGFBP1 levels improves insulin sensitivity, promotes nitric oxide production, lowers blood pressure, and protects against atherosclerosis. Diabetes. 2012;61(4):915–924. doi: 10.2337/db11-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi M., Takahashi Y., Takahashi K., Zolotaryov F.N., Hong K.S., Kitazawa R., et al. Chemerin enhances insulin signaling and potentiates insulin-stimulated glucose uptake in 3T3-L1 adipocytes. FEBS Letters. 2008;582(5):573–578. doi: 10.1016/j.febslet.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 42.Rodríguez-Manzaneque J.C., Carpizo D., Plaza-Calonge M.d.C., Torres-Collado A.X., Thai S.N.M., Simons M., et al. Cleavage of syndecan-4 by ADAMTS1 provokes defects in adhesion. The International Journal of Biochemistry & Cell Biology. 2009;41(4):800–810. doi: 10.1016/j.biocel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramnath R., Foster R.R., Qiu Y., Cope G., Butler M.J., Salmon A.H., et al. Matrix metalloproteinase 9-mediated shedding of syndecan 4 in response to tumor necrosis factor α: a contributor to endothelial cell glycocalyx dysfunction. The FASEB Journal. 2014;28(11):4686–4699. doi: 10.1096/fj.14-252221. [DOI] [PubMed] [Google Scholar]
- 44.Lee S., Kolset S.O., Birkeland K.I., Drevon C.A., Reine T.M. Acute exercise increases syndecan-1 and -4 serum concentrations. Glycoconjugate Journal. 2019;36(2):113–125. doi: 10.1007/s10719-019-09869-z. [DOI] [PubMed] [Google Scholar]
- 45.De Luca M., Klimentidis Y.C., Casazza K., Chambers M.M., Cho R., Harbison S.T., et al. A conserved role for syndecan family members in the regulation of whole-body energy metabolism. PLoS One. 2010;5(6) doi: 10.1371/journal.pone.0011286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davenport L., Johari Y., Klejn A., Laurie C., Smith A., Ooi G.J., et al. Improving compliance with very low energy diets (VLEDs) prior to bariatric surgery-a randomised controlled trial of two formulations. Obesity Surgery. 2019;29(9):2750–2757. doi: 10.1007/s11695-019-03916-2. [DOI] [PubMed] [Google Scholar]
- 47.Bass M.D., Williamson R.C., Nunan R.D., Humphries J.D., Byron A., Morgan M.R., et al. A syndecan-4 hair trigger initiates wound healing through caveolin- and RhoG-regulated integrin endocytosis. Developmental Cell. 2011;21(4):681–693. doi: 10.1016/j.devcel.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karamanos N.K., Piperigkou Z., Theocharis A.D., Watanabe H., Franchi M., Baud S., et al. Proteoglycan chemical diversity drives multifunctional cell regulation and therapeutics. Chemistry Review. 2018;118(18):9152–9232. doi: 10.1021/acs.chemrev.8b00354. [DOI] [PubMed] [Google Scholar]
- 49.De Luca M., Yi N., Allison D.B., Leips J., Ruden D.M. Mapping quantitative trait loci affecting variation in Drosophila triacylglycerol storage. Obesity Research. 2005;13(9):1596–1605. doi: 10.1038/oby.2005.196. [DOI] [PubMed] [Google Scholar]
- 50.Rose G., Crocco P., De Rango F., Corsonello A., Lattanzio F., De Luca M., et al. Metabolism and successful aging: polymorphic variation of syndecan-4 (SDC4) gene associate with longevity and lipid profile in healthy elderly Italian subjects. Mechanism of Ageing and Development. 2015;150:27–33. doi: 10.1016/j.mad.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Agere S.A., Kim E.Y., Akhtar N., Ahmed S. Syndecans in chronic inflammatory and autoimmune diseases: pathological insights and therapeutic opportunities. Journal of Cellular Physiology. 2018;233(9):6346–6358. doi: 10.1002/jcp.26388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eveland M., Brokamp G.A., Lue C.-H., Harbison S.T., Leips J., De Luca M. Knockdown expression of Syndecan in the fat body impacts nutrient metabolism and the organismal response to environmental stresses in Drosophila melanogaster. Biochemical and Biophysical Research Communications. 2016;477(1):103–108. doi: 10.1016/j.bbrc.2016.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pohl J., Ring A., Stremmel W. Uptake of long-chain fatty acids in HepG2 cells involves caveolae: analysis of a novel pathway. The Journal of Lipid Research. 2002;43(9):1390–1399. doi: 10.1194/jlr.m100404-jlr200. [DOI] [PubMed] [Google Scholar]
- 54.Stremmel W., Staffer S., Wannhoff A., Pathil A., Chamulitrat W. Plasma membrane phospholipase A2 controls hepatocellular fatty acid uptake and is responsive to pharmacological modulation: implications for nonalcoholic steatohepatitis. The FASEB Journal. 2014;28(7):3159–3170. doi: 10.1096/fj.14-249763. [DOI] [PubMed] [Google Scholar]
- 55.Hawley John A., Hargreaves M., Joyner Michael J., Zierath Juleen R. Integrative biology of exercise. Cell. 2014;159(4):738–749. doi: 10.1016/j.cell.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 56.Thyfault J.P., Rector R.S. Exercise combats hepatic steatosis: potential mechanisms and clinical implications. Diabetes. 2020;69(4):517. doi: 10.2337/dbi18-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trefts E., Williams A.S., Wasserman D.H. Progress in molecular biology and translational science. Vol. 135. 2015. Exercise and the regulation of hepatic metabolism; pp. 203–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li F., Li T., Liu Y. Proteomics-based identification of the molecular signatures of liver tissues from aged rats following eight weeks of medium-intensity exercise. Oxidative Medicine and Cellular Longevity 2016. 2016 doi: 10.1155/2016/3269405. 3269405-3269405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arfianti A., Pok S., Barn V., Haigh W.G., Yeh M.M., Ioannou G.N., et al. Exercise retards hepatocarcinogenesis in obese mice independently of weight control. Journal of Hepatology. 2020;73(1):140–148. doi: 10.1016/j.jhep.2020.02.006. [DOI] [PubMed] [Google Scholar]
- 60.Saran U., Guarino M., Rodríguez S., Simillion C., Montani M., Foti M., et al. Anti-tumoral effects of exercise on hepatocellular carcinoma growth. Hepatology communications. 2018;2(5):607–620. doi: 10.1002/hep4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seo D.Y., Park S.H., Marquez J., Kwak H.-B., Kim T.N., Bae J.H., et al. Hepatokines as a molecular transducer of exercise. Journal of Clinical Medicine. 2021;10(3) doi: 10.3390/jcm10030385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji Y., Luo Z., Gao H., Dos Reis F.C.G., Bandyopadhyay G., Jin Z., et al. Hepatocyte-derived exosomes from early onset obese mice promote insulin sensitivity through miR-3075. Nature Metabolism. 2021;3(9):1163–1174. doi: 10.1038/s42255-021-00444-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Povero D., Eguchi A., Niesman I.R., Andronikou N., de Mollerat du Jeu X., Mulya A., et al. Lipid-induced toxicity stimulates hepatocytes to release angiogenic microparticles that require Vanin-1 for uptake by endothelial cells. Science Signaling. 2013;6(296) doi: 10.1126/scisignal.2004512. ra88-ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mizunoe Y., Kobayashi M., Hoshino S., Tagawa R., Itagawa R., Hoshino A., et al. Cathepsin B overexpression induces degradation of perilipin 1 to cause lipid metabolism dysfunction in adipocytes. Scientific Reports. 2020;10(1):634. doi: 10.1038/s41598-020-57428-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Villa-Pérez P., Merino B., Fernández-Díaz C.M., Cidad P., Lobatón C.D., Moreno A., et al. Liver-specific ablation of insulin-degrading enzyme causes hepatic insulin resistance and glucose intolerance, without affecting insulin clearance in mice. Metabolism - Clinical and Experimental. 2018;88:1–11. doi: 10.1016/j.metabol.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anderson K.J., Russell A.P., Foletta V.C. NDRG2 promotes myoblast proliferation and caspase 3/7 activities during differentiation, and attenuates hydrogen peroxide - but not palmitate-induced toxicity. FEBS Open Bio. 2015;5:668–681. doi: 10.1016/j.fob.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malaguarnera L., Madeddu R., Palio E., Arena N., Malaguarnera M. Heme oxygenase-1 levels and oxidative stress-related parameters in non-alcoholic fatty liver disease patients. Journal of Hepatology. 2005;42(4):585–591. doi: 10.1016/j.jhep.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 68.Zelcer N., Tontonoz P. Liver X receptors as integrators of metabolic and inflammatory signaling. The Journal of Clinical Investigation. 2006;116(3):607–614. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eichelbaum K., Krijgsveld J. Rapid temporal dynamics of transcription, protein synthesis, and secretion during macrophage activation. Molecular & Cellular Proteomics : MCP. 2014;13(3):792–810. doi: 10.1074/mcp.M113.030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rodríguez-Manzaneque J.C., Carpizo D., Plaza-Calonge Mdel C., Torres-Collado A.X., Thai S.N., Simons M., et al. Cleavage of syndecan-4 by ADAMTS1 provokes defects in adhesion. The International Journal of Biochemistry & Cell Biology. 2009;41(4):800–810. doi: 10.1016/j.biocel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meex R.C., Hoy A.J., Morris A., Brown R.D., Lo J.C., Burke M., et al. Fetuin B is a secreted hepatocyte factor linking steatosis to impaired glucose metabolism. Cell Metabolism. 2015;22(6):1078–1089. doi: 10.1016/j.cmet.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 72.Montgomery M.K., Bayliss J., Nie S., De Nardo W., Keenan S.N., Miotto P.M., et al. Deep proteomic profiling unveils arylsulfatase A as a non-alcoholic steatohepatitis inducible hepatokine and regulator of glycemic control. Nature Communications. 2022;13(1):1259. doi: 10.1038/s41467-022-28889-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daemen S., Gemmink A., Brouwers B., Meex R.C.R., Huntjens P.R., Schaart G., et al. Distinct lipid droplet characteristics and distribution unmask the apparent contradiction of the athlete's paradox. Molecular Metabolism. 2018;17:71–81. doi: 10.1016/j.molmet.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang S., Wang Y., Cui L., Deng Y., Xu S., Yu J., et al. Morphologically and functionally distinct lipid droplet subpopulations. Scientific Reports. 2016;6:29539. doi: 10.1038/srep29539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gemmink A., Daemen S., Brouwers B., Hoeks J., Schaart G., Knoops K., et al. Decoration of myocellular lipid droplets with perilipins as a marker for in vivo lipid droplet dynamics: a super-resolution microscopy study in trained athletes and insulin resistant individuals. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2021;1866(2):158852. doi: 10.1016/j.bbalip.2020.158852. [DOI] [PubMed] [Google Scholar]
- 76.Fernandez-Rojo M.A., Ramm G.A. Caveolin-1 function in liver Physiology and disease. Trends in Molecular Medicine. 2016;22(10):889–904. doi: 10.1016/j.molmed.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 77.Mattern H.M., Raikar L.S., Hardin C.D. The effect of caveolin-1 (Cav-1) on fatty acid uptake and CD36 localization and lipotoxicity in vascular smooth muscle (VSM) cells. International Journal of Physiology, Pathophysiology and Pharmacology. 2009;1(1):1–14. [PMC free article] [PubMed] [Google Scholar]
- 78.Li M., Chen D., Huang H., Wang J., Wan X., Xu C., et al. Caveolin1 protects against diet induced hepatic lipid accumulation in mice. PLoS One. 2017;12(6) doi: 10.1371/journal.pone.0178748. e0178748-e0178748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mathiesen S.B., Lunde M., Aronsen J.M., Romaine A., Kaupang A., Martinsen M., et al. The cardiac syndecan-4 interactome reveals a role for syndecan-4 in nuclear translocation of muscle LIM protein (MLP) Journal of Biological Chemistry. 2019;294(22):8717–8731. doi: 10.1074/jbc.RA118.006423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hao J.-W., Wang J., Guo H., Zhao Y.-Y., Sun H.-H., Li Y.-F., et al. CD36 facilitates fatty acid uptake by dynamic palmitoylation-regulated endocytosis. Nature Communications. 2020;11(1):4765. doi: 10.1038/s41467-020-18565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scarpellini A., Huang L., Burhan I., Schroeder N., Funck M., Johnson T.S., et al. Syndecan-4 knockout leads to reduced extracellular transglutaminase-2 and protects against tubulointerstitial fibrosis. Journal of the American Society of Nephrology. 2014;25(5):1013. doi: 10.1681/ASN.2013050563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Echtermeyer F., Streit M., Wilcox-Adelman S., Saoncella S., Denhez F., Detmar M., et al. Delayed wound repair and impaired angiogenesis in mice lacking syndecan-4. Journal of Clinical Investigation. 2001;107(2):R9–R14. doi: 10.1172/JCI10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tanino Y., Chang M.Y., Wang X., Gill S.E., Skerrett S., McGuire J.K., et al. Syndecan-4 regulates early neutrophil migration and pulmonary inflammation in response to lipopolysaccharide. American Journal of Respiratory Cell and Molecular Biology. 2012;47(2):196–202. doi: 10.1165/rcmb.2011-0294OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dong J., Viswanathan S., Adami E., Singh B.K., Chothani S.P., Ng B., et al. Hepatocyte-specific IL11 cis-signaling drives lipotoxicity and underlies the transition from NAFLD to NASH. Nature Communications. 2021;12(1):66. doi: 10.1038/s41467-020-20303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kamari Y., Shaish A., Vax E., Shemesh S., Kandel-Kfir M., Arbel Y., et al. Lack of interleukin-1α or interleukin-1β inhibits transformation of steatosis to steatohepatitis and liver fibrosis in hypercholesterolemic mice. Journal of Hepatology. 2011;55(5):1086–1094. doi: 10.1016/j.jhep.2011.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.