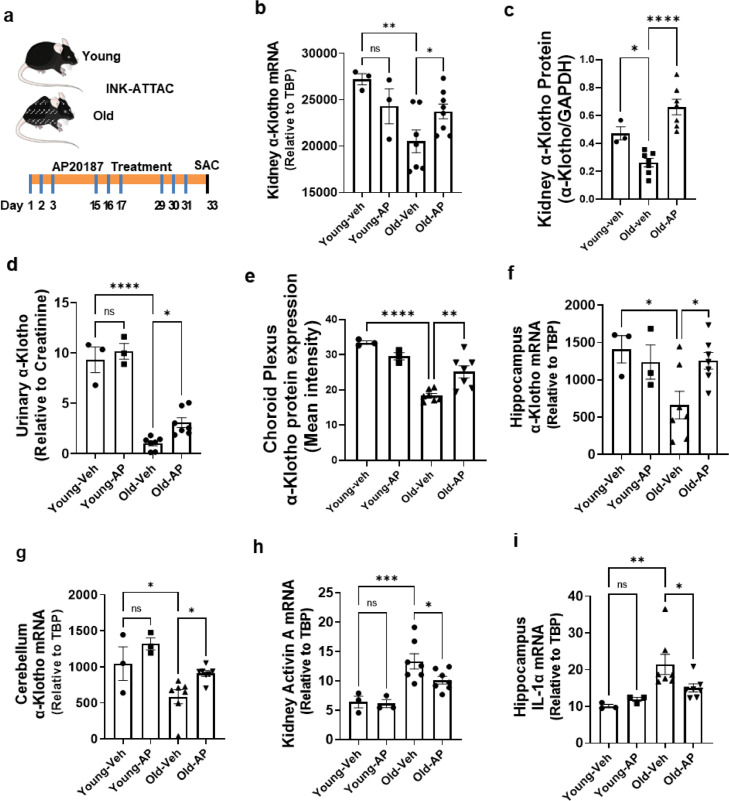
Figure 3.
Genetic clearance of highly p16Ink4a-expressing cells increases α-Klotho. Young (8-month-old) or old (27-29-month-old) INK-ATTAC male mice were treated with vehicle or AP20187 (n = 3 young+ vehicle; n = 3 young+ AP20187; n = 7 old + vehicle; n = 7 old + AP20187) to dimerize the FKBP-caspase-8 fusion protein expressed in highly p16Ink4a-expressing cells to selectively eliminate p16Ink4a+ cells. AP20187 (10 mg/kg) or vehicle was administered i.p. every 2 weeks for 6 weeks. (a) Schematic view of treatments. (b) Kidney α-Klotho mRNA was assayed by qPCR. One-way ANOVA, F = 4.81, p = 0.01, R2 = 0.46; *p = 0.03, **p < 0.01. (c) α-Klotho protein was assayed relative to GAPDH (Western blots in Supplementary Fig. 7). One-way ANOVA: F = 4.81, p = 0.01, R2 = 0.46; *p < 0.05, ****p < 0.0001. (d) Mouse urine was collected 2 days after the last dose of AP20187. Urinary α-Klotho was assayed by ELISA and expressed as a function of creatinine. One-way ANOVA, F = 61.53, p < 0.00001, R2 = 0.90; *p = 0.04, ****p < 0.0001. Choroid Plexus (e) α-Klotho protein was assayed by immunofluorescence. One-way ANOVA, F = 20.98, R2 = 0.80, **p < 0.01, ****p < 0.0001. Hippocampal (f) and cerebellar (g) α-Klotho mRNA was assayed by qPCR. Activin A mRNA in kidney (h) and IL-1α mRNA in hippocampus (i) were assayed by qPCR. Means ± SEM; one-way ANOVA and post hoc Tukey's tests; F = 3.84, p = 0.038, R2 = 0.40; *p = 0.04 in (f); *p = 0.04, F = 7.87, p < 0.01, R2 = 0.60; *p = 0.03 in (g); F = 10.85; p < 0.001, R2 = 0.64; *p = 0.03, ***p < p < 0.0001 in (h); F = 5.49; p = 0.01, R2 = 0.49; *p < 0.05, **p = 0.01 in (i).