Abstract
Background/aims
Psoriasis is a chronic immune‐mediated inflammatory skin disease characterized by excessive proliferation of keratinocytes. It has a strong genetic predisposition; gene‐gene interactions are important genetic models for common diseases. In this study, we explore pair‐wise interactions among SNPs contributing to psoriasis susceptibility.
Methods
We first performed gene interactions with exome‐sequencing, next, we analyzed gene interactions combining the exome sequencing data with the targeted sequencing data. After we sequenced HLA region, we analyzed gene interactions including HLA regions and non‐HLA regions.
Results
We found interactions between HLA regions were significant. We observed significant interactions between HLA‐C*06:02 and rs118179173 (snp31443520; p = 8.21 × 10−20, OR = 0.22) and between HLA‐C*06:02 and HLA‐B:AA67 (p = 1.22 × 10−12, OR = 0.45).
Conclusion
This study provides evidence that HLA is the most important susceptibility region on the risk of psoriasis and interactions that occur in this region are still significant.
Keywords: gene‐interactions, HLA, psoriasis
In this study, we explored the roles of interactions among susceptibility genes of psoriasis based on GWAS study.
1. INTRODUCTION
Psoriasis is a complex skin disease characterized by epidermal hyper‐proliferation, vascular remodeling, and inflammation, the joints are frequently involved in pain and stiffness. Approximately 125 million people are affected worldwide (Armstrong & Read, 2020), the prevalence of psoriasis in China is about 0.47% (Ding et al., 2012). It is more common in adults than children, and there is no difference between male and female. Psoriasis is multifactorial in most cases, the exact etiology is unknown. Earlier family linkage studies and twin studies confirm the genetic susceptibility of psoriasis (Brandrup, 1978; Myers et al., 2005), the risk of psoriasis in monozygotic twin pairs are 2–3.5 times than dizygotic twin pairs (Lønnberg et al., 2016), subsequently, the development of genome‐wide association studies (GWAS) laid the genetic foundation of psoriasis. Abnormal proliferation of keratinocytes and infiltration of immune cell means it is a disorder of the immune system, some pro‐inflammatory cytokines have been demonstrated to be related to psoriasis, including interleukin‐23 (IL‐23; Li et al., 2018), IL‐17 as well as tumor necrosis factor α (TNF‐α; Cordiali‐Fei et al., 2014; Girolomoni et al., 2012). Besides, environmental play an important role in the pathogenesis of psoriasis, like drugs, trauma, infection, stress, smoking, cold, and humidity (Gudjonsson et al., 2003; Patel & Weinberg, 2008; Raychaudhuri & Gross, 2000).
It is now well established that heredity is an important part of the etiology of psoriasis. Major histocompatibility complex (MHC) region more precisely the HLA‐C, it accounts for about 35%–50% of disease heritability, is regarded as the most important genetic component of psoriasis, subsequently GWAS identified 86 susceptibility genes associated with psoriasis in total (Hwang et al., 2017). Most association studies detect SNPs independently rather than considering interactions between them, it ignores the complex interactions that often occur in biological systems. There are some gene interaction studies in autoimmune diseases, including rheumatoid arthritis (Liu et al., 2011), systemic lupus erythematosus (Zhang et al., 2016), ankylosing spondylitis (Evans et al., 2011). For psoriasis, researches on the interaction between some susceptibility genes are also being carried out in different populations (Strange et al., 2010; Vasilopoulos et al., 2011). Various algorithms have been proposed to detect the interactions between genes (Niel et al., 2015), In our study, we performed gene interactions using multifactor dimensionality reduction (MDR).
In the last decade, our group has identified eight independent susceptibility loci of psoriasis in HLA region (Zhou et al., 2016) and more than 30 susceptibility genes in non‐HLA regions in Chinese population using GWAS (Cheng et al., 2014; Tang et al., 2014; Zhang et al., 2009; Zuo et al., 2015), based on the previous GWAS data, we performed gene‐gene interactions for psoriasis. The aim of this study is to further explain the genetic mechanism of psoriasis by identifying interacting SNPs and genes.
2. MATERIALS AND METHODS
2.1. Samples
Firstly, 781 psoriasis cases and 676 controls were used to discover gene‐gene interactions among SNPs in exome‐sequencing, and a second cohort was 9946 cases and 9906 controls in targeted sequencing (Tang et al., 2014), thus we analyzed the gene interactions with the two combined datasets, next, we further identified interactions in HLA regions and those susceptibility genes in non‐HLA regions in 10,689 controls and 9946 patients selected from above cohorts (Zhou et al., 2016). All samples were Chinese Han people. Diagnoses of disease, inclusion and exclusion criteria, collection of specimen, and information have been described previously. The study was approved by the institutional ethics committee of each hospital and was conducted according to the principles of the Declaration of Helsinki.
2.2. Experimental process
Genomic DNAs of the peripheral blood mononuclear cells (PBMCs) was extracted using Flexi Gene DNA Kit (Qiagen), then we used NanoDrop‐2000 to dilute DNA to standard experimental concentration, for the whole genome genotype concentration is 50 ng/μl, the range of A260/A280 is 1.8–2.0, select samples that meet the criteria for the next step of genotyping. Next, SNP genotyping was performed on the Illumina HiSeq 2000 platform to generate 90‐bp paired‐end reads. Locus‐specific polymerase chain reaction (PCR) was performed and PCR reaction products were purified by AgencourtAMpure SPRI XP bead. Genomic DNA for each individual was hybridized with the NimbleGen 2.1M‐probe sequence capture array (Albert et al., 2007) to enrich exonic DNA in each library. Samples were aligned to the NCBI human genome reference assembly (build 36.3) using BWA (Burrows‐Wheeler Aligner; Li & Durbin, 2010), using the Genome Analysis Toolkit (GATK v1.6; McKenna et al., 2010) to perform realignment around known indels. All aligned read data were subjected to CountCovariates (GATK) on the basis of known SNVs (dbSNP135), and base quality was then recalibrated (Table Recalibration in GATK).
2.3. Statistical analysis and quality control
The variants with p < 0.05 in the exome sequencing dataset, the combined datasets of exome sequencing and targeted sequencing as well as the 8 variants reported in the HLA sequencing analysis included in this study. We used PLINK 1.07 to test SNP × SNP interactions for case‐control population‐based samples. Then we could obtain odds ratio for interaction (OR), chi‐square statistic (STAT), asymptotic p‐value (p). The multifactor dimensionality reduction (MDR, V2.0 Beta 2) was used to calculate the interactions between variants according to reported method (Ritchie et al., 2001). Interactions were selected if p < 5 × 10−2. Bonferroni correction was used to define the statistical significance with p < 2.84 × 10−8 (0.05/13262).
3. RESULTS
3.1. The optimal p‐value for psoriasis
The identified variants only explained a small part of genetic risk, GWAS threshold is conventionally using a p‐value of 5 × 10−8 to avoid issues of false‐positive findings due to multiple testing, many genes influence psoriasis but do not reach a significant level (5 × 10−8), here, we used polygenic risk score (PRS) analysis to identify high‐risk loci for psoriasis. PRSice 2.1.11 (https://github.com/choishingwan/PRSice) was used to calculate the p value. Exome sequencing including 1457 participants (859 males and 598 females) was observed. We chose more than 70 psoriasis susceptibility genes reportedly from human genome (HG19), locating upstream and downstream 500K loci, totally 30923 variants. When the p‐value was 0.05, the model was optimal, and the polygenic risk score of the phenotype was 0.77, and the p‐value was 1.1 × 10−87 (Figure 1).
FIGURE 1.
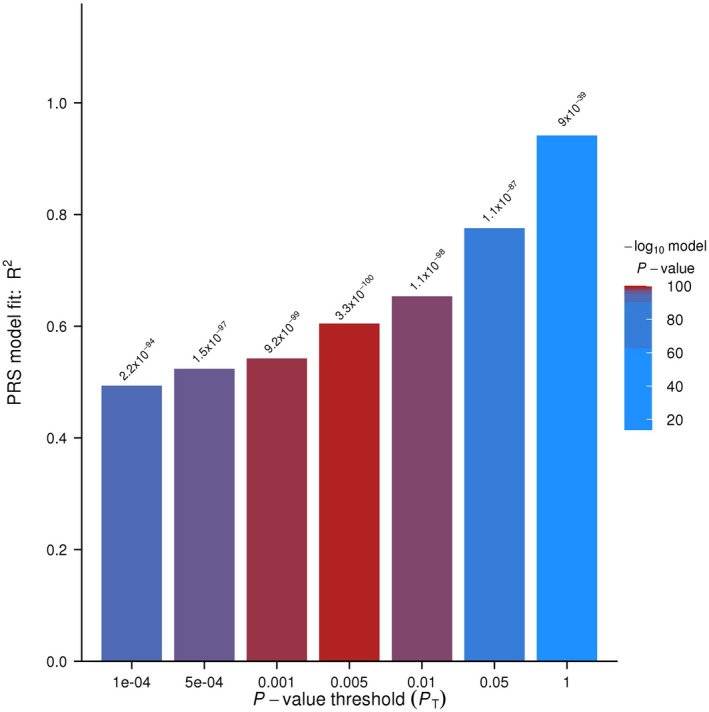
The optimal p‐value for psoriasis using polygenic risk score (PRS) analysis
3.2. Interactions in exome and targeted sequencing data
We performed the interactions of variants with p < 5 × 10−2 in exome sequencing data with 781 psoriasis cases and 676 controls, due to the genetic complexity of HLA region, single nucleotide variants (SNVs) within this region were excluded, 1326 genes was selected (Tang et al., 2014). The interaction between C1orf186 and TNFRSF1B showed the most significance (chr1_12253062 × chr1_206239574, p = 2.77 × 10−5, OR = 7.22). Besides, interaction between COL5A3 and SLC9A4 was also notable; interactions with p < 0.01 were shown in Table S1. Then we enlarged the sample size with the targeted sequencing data of 1326 selected genes in 9946 psoriasis patients and 9906 controls. We analyzed gene‐gene interactions with these two combined datasets. More than 20,000 interactions were identified with p < 0.05 (Table S2). The significant interactions were between MIR548AZ and TYK2 (chr14_64897018 × chr19_10464687, p = 2.34 × 10−5, OR = 0.56; chr14_64897018 × chr19_10472933, p = 2.35 × 10−5, OR = 0.57; chr14_64896783 × chr19_10472933, p = 8.59 × 10−5, OR = 0.59; chr14_64896783 × chr19_10464687, p = 9.56 × 10−5, OR = 0.58); CARD14 and TNP2 (p = 2.34 × 10−5, OR = 3.95); NDST2 and UBLCP1 (p = 2.85 × 10−5, OR = 0.3); TAF1L and TARBP1 (p = 8.06 × 10−5, OR = 7.78); IL12B and SUOX (p = 9.63 × 10−5, OR = 0.2; Table 1).
TABLE 1.
Interaction analysis in exome and targeted sequencing data
| SNP1 | SNP2 | OR | p | Gene 1 | Gene 2 |
|---|---|---|---|---|---|
| chr14_64897018 | chr19_10464687 | 0.5653 | 2.34E‐05 | MIR548AZ | TYK2 |
| chr16_11363025 | chr17_78171944 | 3.954 | 2.34E‐05 | CARD14 | TNP2 |
| chr14_64897018 | chr19_10472933 | 0.5768 | 2.35E‐05 | MIR548AZ | TYK2 |
| chr5_158705065 | chr10_75564624 | 0.302 | 2.85E‐05 | NDST2 | UBLCP1 |
| chr1_234565787 | chr9_32633719 | 7.783 | 8.06E‐05 | TAF1L | TARBP1 |
| chr14_64896783 | chr19_10472933 | 0.594 | 8.95E‐05 | MIR548AZ | TYK2 |
| chr14_64896783 | chr19_10464687 | 0.5831 | 9.56E‐05 | MIR548AZ | TYK2 |
| chr5_158750013 | chr12_56398287 | 0.2036 | 9.63E‐05 | IL12B | SUOX |
3.3. Interactions in HLA and non‐HLA regions
Then we explored interactions adding HLA region. Datasets are obtained from the entire 5‐Mb MHC region in 20,635 individuals of Chinese Han population (including 10,689 controls and 9946 patients with psoriasis selected from the exome and targeted sequencing). MHC sequencing identified eight independent variants, including HLA‐C*07:04, HLA‐C*06:02, rs118179173 (snp31443520), HLA‐B amino acid9 (HLA‐B:AA9), HLA‐B:AA67, HLA‐B:AA116, HLA‐DPB1*05:01, and BTNL2:AA281 (snp32472030; Zhou et al., 2016). The strong interactions with these 3 sequencing datasets were located in HLA regions between HLA‐C*06:02 and snp31443520 (rs118179173; p = 8.21 × 10−20, OR = 0.22) and between HLA‐C*06:02 and HLA‐B:AA67 (p = 1.22 × 10−12, OR = 0.45; Table 2, Table S3).
TABLE 2.
Interaction analysis in exome, targeted and HLA sequencing data
| Vatiant 1 | Vatiant 2 | OR | p | Gene 1 | Gene 2 |
|---|---|---|---|---|---|
| HLA‐C*06:02 | rs118179173 | 0.22 | 8.21E‐20 | HLA‐C | HLA‐B |
| HLA‐C*06:02 | HLA‐B:AA67 | 0.45 | 1.22E‐12 | HLA‐C | HLA‐B |
| chr5_96139250 | HLA‐C*06:02 | 0.74 | 3.61E‐07 | ERAP1 | HLA‐C |
| chr5_96117300 | HLA‐C*06:02 | 0.75 | 1.68E‐06 | ERAP1 | HLA‐C |
| HLA‐C*06:02 | HLA‐C*07:04 | 0.28 | 6.86E‐06 | HLA‐C | HLA‐C |
| chr5_96118852 | HLA‐C*06:02 | 0.78 | 3.43E‐05 | ERAP1 | HLA‐C |
| chr5_96125910 | HLA‐C*06:02 | 0.78 | 3.81E‐05 | ERAP1 | HLA‐C |
| snp31443520 | HLA‐DPB1*05:01 | 1.47 | 6.07E‐05 | HLA‐B | HLA‐DPB1 |
| chr5_96124447 | HLA‐C*06:02 | 0.79 | 6.44E‐05 | ERAP1 | HLA‐C |
| chr5_96121994 | HLA‐C*06:02 | 0.79 | 6.50E‐05 | ERAP1 | HLA‐C |
| chr5_96121715 | HLA‐C*06:02 | 0.79 | 6.96E‐05 | ERAP1 | HLA‐C |
| chr5_96129512 | HLA‐C*06:02 | 0.80 | 0.0002089 | ERAP1 | HLA‐C |
| chr5_96222183 | HLA‐C*06:02 | 0.80 | 0.0002627 | ERAP2 | HLA‐C |
| HLA‐C*06:02 | HLA‐DPB1*05:01 | 1.26 | 0.0002793 | HLA‐C | HLA‐DPB1 |
| chr5_96222185 | HLA‐C*06:02 | 0.81 | 0.0005506 | ERAP2 | HLA‐C |
| chr5_96232222 | HLA‐C*06:02 | 0.82 | 0.0009334 | ERAP2 | HLA‐C |
| chr5_96237114 | HLA‐C*06:02 | 0.82 | 0.0009459 | ERAP2 | HLA‐C |
| chr5_96132795 | HLA‐C*06:02 | 0.77 | 0.0009922 | ERAP1 | HLA‐C |
Previously reported interactions have also been confirmed, interactions between the ERAP1 and HLA‐C was obvious (chr5_96139250 × HLA‐C*06:02, p = 3.61 × 10−7, OR = 0.74;chr5_96117300 × HLA‐C*06:02, p = 1.68 × 10−6, OR = 0.75; chr5_96118852 × HLA‐C*06:02, p = 3.43 × 10−5, OR = 0.78; chr5_96125910 × HLA‐C*06:02, p = 3.81 × 10−5, OR = 0.78; chr5_96124447 × HLA‐C*06:02, p = 6.44 × 10−5, OR = 0.79; chr5_96121994 × HLA‐C*06:02, p = 6.50 × 10−5, OR = 0.79; chr5_96121715 ×HLA‐C*06:02, p = 6.96 × 10−5, OR = 0.79; Interactions between them were shown in Table 3). Besides, We also observed significant interactions between HLA and IL12B (chr5_158750013 × HLA‐C*06:02, p = 0.009494).
TABLE 3.
Interactions between HLA and ERAP1
| SNP1 | SNP2 | OR | p | Gene 1 | Gene 2 |
|---|---|---|---|---|---|
| C*06:02 | 5_96139250 | 0.74 | 3.61E‐07 | HLA‐C | ERAP1 |
| C*06:02 | 5_96117300 | 0.75 | 1.68E‐06 | HLA‐C | ERAP1 |
| C*06:02 | 5_96118852 | 0.78 | 3.43E‐05 | HLA‐C | ERAP1 |
| C*06:02 | 5_96125910 | 0.78 | 3.81E‐05 | HLA‐C | ERAP1 |
| C*06:02 | 5_96124447 | 0.79 | 6.44E‐05 | HLA‐C | ERAP1 |
| C*06:02 | 5_96121994 | 0.79 | 6.50E‐05 | HLA‐C | ERAP1 |
| C*06:02 | 5_96121715 | 0.79 | 6.96E‐05 | HLA‐C | ERAP1 |
| C*06:02 | 5_96129512 | 0.80 | 0.0002089 | HLA‐C | ERAP1 |
| C*06:02 | 5_96132795 | 0.77 | 0.0009922 | HLA‐C | ERAP1 |
4. DISCUSSION
In recent years, the exploring of gene‐gene interaction is an important supplement to the genetic mechanism of psoriasis and enriches the genetic pattern of psoriasis in Chinese Han population. Based on GWAS data, this study further explores interactions between susceptibility genes of psoriasis. With a large sample size, we have identified significant genetic interacting signals within the HLA complex as expected, more than 60% of the interactions SNP are located at 6p21. We observed significant interactions between HLA‐C*06:02 and rs118179173 (snp31443520; p = 8.21 × 10−20, OR = 0.22) and between HLA‐C*06:02 and HLA‐B:AA67 (p = 1.22 × 10−12, OR = 0.45). Previous reported interactions have also been confirmed, including interactions between HLA and ERAP1 (Strange et al., 2010), HLA and IL12B (Zheng et al., 2011).
HLA has strong homology and complex polymorphism, HLA‐C may affect both innate and adaptive immune system, it plays an important role in antigen recognition, presentation, immune response, and regulation, the skin lesions of psoriasis are rich in activated CD8+ T cells (Chang et al., 1995), HLA‐C participates in the immune response by presenting antigens to CD8+ T cells and interacting with natural killer (NK) cell receptors (Prinz, 2017). The frequency of HLA‐Cw6 alleles in Chinese patients with psoriasis is 16.18%–18.6% (Chang et al., 2003; Tsai et al., 2002), it is a major contributor to psoriasis (psoriasis susceptibility 1 [PSORS1]). HLA‐C has been recognized as the acknowledged susceptibility gene in multiple populations (Capon et al., 2004), association studies have defined the HLA‐Cw6 allele as a factor that predisposes to early‐onset disease (Das et al., 2017), moreover, patients with HLA‐Cw6 are usually earlier to develop psoriatic arthritis (Chen & Tsai, 2018), research around HLA accounts for the majority studies regard to gene interactions.
This study also verified previously reported gene interactions, among which the interaction between HLA‐C and ERAP1 is the most significant. Interactions between HLA and ERAP1 also define the importance of ERAP1 in the risk of psoriasis. In a study by Yin et al. (2013), when ERAP1 and HLA‐C co‐exist in an individual, the interaction between them was obvious, which increases the risk of psoriasis by 20 times compared with individuals with HLA‐C alone. ERAP1 is an immune‐functional gene that encodes a multifunctional aminopeptidase and is involved in processing HLA molecular antigen peptides (Kochan et al., 2011). Enzymes encoded by ERAP1 genes can produce pro‐inflammatory cytokines and chemokines, which also inhibit immune cells maturation, then induce psoriasis (Aldhamen et al., 2013). Deficiency in ERAP1 expression was shown to result in unstable and highly immunogenic peptides presented in the context of HLA class I, which elicited potent CD8+ T cell and B cell responses (Hammer et al., 2007).
In addition, interactions between MIR548AZ and TYK2 were references to explore more pathogenesis of psoriasis. In a study by Zhao et al. (2018), miR‐548a‐3p was unregulated after treating human keratinocytes with IL‐22, miR‐548a‐3p may promote keratinocytes proliferative disorder. IL‐22 is a member of the IL‐10 cytokine family, Th17 cells could produce both IL‐17 and IL‐22 in psoriatic skin (Liang et al., 2006). IL‐22 signaling is mediated by TYK2 and JAK1 (Works et al., 2014). Tyrosine kinase 2 (TYK2) belongs to the JAK family protein tyrosine kinase, by catalyzing TYK2, the downstream signaling pathway of IL‐12 and IL‐23 can be activated (Sohn et al., 2013). IL‐23 enhances IL‐22 expression during Th17 differentiation.
In a word, our study analyzed the susceptibility gene interactions of psoriasis in Chinese population and confirmed the important roles of HLA region, interactions between HLA and ERAP1, MIR548AZ, and TYK2. The results just refer to the Chinese Han population and it is worth repeating for other populations. Gene interactions provide a new perspective for understanding the genetic mechanism of psoriasis, which puts forward the ways to reveal the complex mechanisms of interactions for other common diseases.
CONFLICT OF INTEREST
The authors have declared no conflict of interest.
ETHICS STATEMENT
Written informed consent was obtained from the patient. The study was approved by Ethics Committee of Anhui Medical University and was conducted according to the Principles of the Declaration of Helsinki.
AUTHOR CONTRIBUTIONS
Liangdan Sun, Wenjun Wang, and Hui Cheng conceived and designed the research. Xiaodong Zheng designed the structure of the manuscript. Qiongqiong Xu and Yiwen Mao wrote the manuscript. Weiwei Chen,Shirui Chen,Hui Zhang, Huiyao Ge, Ruixue Zhang, and Lu Cao were responsible for selecting samples. Qi Zhen, Bao Li, Liang Yong, and Yafen Yu analyzed the data. All authors contributed literature search, analysis of literature, and drafting of the manuscript.
Supporting information
Table S1 Interaction analysis in exome sequencing data with p<0.01
Table S2 Interaction analysis in exome and targeted sequencing data with p<0.05
Table S3 Interaction analysis in exome, targeted and HLA sequencing data with p<0.05
ACKNOWLEDGMENTS
We thank all the individuals who participated in this study. We also thank the participants for contributing to the collection of samples and phenotype data from the Genetic Resources Collection Collaboration, China.
Xu, Q. , Zheng, X. , Mao, Y. , Chen, W. , Chen, S. , Zhang, H. , Zhen, Q. , Li, B. , Yong, L. , Ge, H. , Yu, Y. , Zhang, R. , Cao, L. , Cheng, H. , Wang, W. , & Sun, L. (2022). Gene interaction analysis of psoriasis in Chinese Han population. Molecular Genetics & Genomic Medicine, 10, e1858. 10.1002/mgg3.1858
Qiongqiong Xu, Xiaodong Zheng, Yiwen Mao equally contributed.
Funding information
This study was supported by the National Natural Science Foundation of China (81972927 and 81773313) and the General Program of National Natural Science Foundation of China (31671307).
REFERENCES
- Albert, T. J. , Molla, M. N. , Muzny, D. M. , Nazareth, L. , Wheeler, D. , Song, X. , Richmond, T. A. , Middle, C. M. , Rodesch, M. J. , Packard, C. J. , Weinstock, G. M. , & Gibbs, R. A. (2007). Direct selection of human genomic loci by microarray hybridization. Nature Methods, 4(11), 903–905. 10.1038/nmeth1111 [DOI] [PubMed] [Google Scholar]
- Aldhamen, Y. A. , Seregin, S. S. , Rastall, D. P. W. , Aylsworth, C. F. , Pepelyayeva, Y. , Busuito, C. J. , Godbehere‐Roosa, S. , Kim, S. , & Amalfitano, A. (2013). Endoplasmic reticulum aminopeptidase‐1 functions regulate key aspects of the innate immune response. PLoS One, 8(7), e69539. 10.1371/journal.pone.0069539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, A. W. , & Read, C. (2020). Pathophysiology, clinical presentation, and treatment of psoriasis. JAMA, 323(19), 1945. 10.1001/jama.2020.4006 [DOI] [PubMed] [Google Scholar]
- Brandrup, F. (1978). Psoriasis in an unselected series of twins. Archives of Dermatology, 114(6), 874–878. 10.1001/archderm.1978.01640180008002 [DOI] [PubMed] [Google Scholar]
- Capon, F. , Trembath, R. , & Barker, J. (2004). An update on the genetics of psoriasis. Dermatologic Clinics, 22(4), 339–347. 10.1016/S0733-8635(03)00125-6 [DOI] [PubMed] [Google Scholar]
- Chang, J. , Smith, L. R. , Froning, K. J. , Schwabe, B. J. , Laxer, J. A. , Caralli, L. L. , Kurkland, H. H. , Karasek, M. A. , Wilkinson, D. I. , & Carlo, D. J. (1995). CD8+ T‐cells in psoriatic lesions preferentially use T‐cell receptors V beta 3 and/or V beta 13.1 genes. Annals of the New York Academy of Sciences, 756, 370–381. [DOI] [PubMed] [Google Scholar]
- Chang, Y. T. , Tsai, S. F. , Lee, D. D. , Shiao, Y. M. , Huang, C. Y. , Liu, H. N. , Wang, W. J. , & Wong, C. K. (2003). A study of candidate genes for psoriasis near HLA‐C in Chinese patients with psoriasis. British Journal of Dermatology, 148(3), 418–423. 10.1046/j.1365-2133.2003.05166.x [DOI] [PubMed] [Google Scholar]
- Chen, L. , & Tsai, T. (2018). HLA‐Cw6 and psoriasis. The British Journal of Dermatology, 178(4), 854–862. [DOI] [PubMed] [Google Scholar]
- Cheng, H. , Li, Y. , Zuo, X.‐B. , Tang, H.‐Y. , Tang, X.‐F. , Gao, J.‐P. , Sheng, Y.‐J. , Yin, X.‐Y. , Zhou, F.‐S. , Zhang, C. , Chen, G. , Zhu, J. , Pan, Q. , Liang, B. O. , Zheng, X.‐D. , Li, P. , Ding, Y.‐T. , Cheng, F. , Luo, J. , … Zhang, X.‐J. (2014). Identification of a missense variant in LNPEP that confers psoriasis risk. Journal of Investigative Dermatology, 134(2), 359–365. 10.1038/jid.2013.317 [DOI] [PubMed] [Google Scholar]
- Cordiali‐Fei, P. , Bianchi, L. , Bonifati, C. , Trento, E. , Ruzzetti, M. , Francesconi, F. , Bultrini, S. , D’Agosto, G. , Bordignon, V. , Francavilla, V. , & Tripiciano, A. (2014). Immunologic biomarkers for clinical and therapeutic management of psoriasis. Mediators of Inflammation, 2014, 236060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A. , Chandra, A. , Chakraborty, J. , Chattopadhyay, A. , Senapati, S. , Chatterjee, G. , & Chatterjee, R. (2017). Associations of ERAP1 coding variants and domain specific interaction with HLA‐C∗06 in the early onset psoriasis patients of India. Human Immunology, 78(11‐12), 724–730. 10.1016/j.humimm.2017.08.006 [DOI] [PubMed] [Google Scholar]
- Ding, X. , Wang, T. , Shen, Y. , Wang, X. , Zhou, C. , Tian, S. , Liu, Y. , Peng, G. , Zhou, J. , Xue, S. , & Wang, R. (2012). Prevalence of psoriasis in China: A population‐based study in six cities. European Journal of Dermatology, 22(5), 663–667. [DOI] [PubMed] [Google Scholar]
- Evans, D. M. , Spencer, C. C. A. , Pointon, J. J. , Su, Z. , Harvey, D. , Kochan, G. , Oppermann, U. , Dilthey, A. , Pirinen, M. , Stone, M. A. , Appleton, L. , Moutsianas, L. , Leslie, S. , Wordsworth, T. , Kenna, T. J. , Karaderi, T. , Thomas, G. P. , Ward, M. M. , Weisman, M. H. , … Donnelly, P. (2011). Interaction between ERAP1 and HLA‐B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA‐B27 in disease susceptibility. Nature Genetics, 43(8), 761–767. 10.1038/ng.873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girolomoni, G. , Mrowietz, U. , & Paul, C. (2012). Psoriasis: Rationale for targeting interleukin‐17. The British Journal of Dermatology, 167(4), 717–724. 10.1111/j.1365-2133.2012.11099.x [DOI] [PubMed] [Google Scholar]
- Gudjonsson, J. E. , Thorarinsson, A. M. , Sigurgeirsson, B. , Kristinsson, K. G. , & Valdimarsson, H. (2003). Streptococcal throat infections and exacerbation of chronic plaque psoriasis: A prospective study. British Journal of Dermatology, 149(3), 530–534. 10.1046/j.1365-2133.2003.05552.x [DOI] [PubMed] [Google Scholar]
- Hammer, G. E. , Gonzalez, F. , James, E. , Nolla, H. , & Shastri, N. (2007). In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nature Immunology, 8(1), 101–108. 10.1038/ni1409 [DOI] [PubMed] [Google Scholar]
- Hwang, S. , Nijsten, T. , & Elder, J. (2017). Recent Highlights in Psoriasis Research. The Journal of Investigative Dermatology, 137(3), 550–556. 10.1016/j.jid.2016.11.007 [DOI] [PubMed] [Google Scholar]
- Kochan, G. , Krojer, T. , Harvey, D. , Fischer, R. , Chen, L. , Vollmar, M. , von Delft, F. , Kavanagh, K. L. , Brown, M. A. , Bowness, P. , Wordsworth, P. , Kessler, B. M. , & Oppermann, U. (2011). Crystal structures of the endoplasmic reticulum aminopeptidase‐1 (ERAP1) reveal the molecular basis for N‐terminal peptide trimming. Proceedings of the National Academy of Sciences, 108(19), 7745–7750. 10.1073/pnas.1101262108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , & Durbin, R. (2010). Fast and accurate long‐read alignment with Burrows‐Wheeler transform. Bioinformatics (Oxford, England), 26(5), 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Yao, Q. I. , Mariscal, A. G. , Wu, X. , Hülse, J. , Pedersen, E. , Helin, K. , Waisman, A. , Vinkel, C. , Thomsen, S. F. , Avgustinova, A. , Benitah, S. A. , Lovato, P. , Norsgaard, H. , Mortensen, M. S. , Veng, L. , Rozell, B. , & Brakebusch, C. (2018). Epigenetic control of IL‐23 expression in keratinocytes is important for chronic skin inflammation. Nature Communications, 9(1), 1–18. 10.1038/s41467-018-03704-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, S. C. , Tan, X.‐Y. , Luxenberg, D. P. , Karim, R. , Dunussi‐Joannopoulos, K. , Collins, M. , & Fouser, L. A. (2006). Interleukin (IL)‐22 and IL‐17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. Journal of Experimental Medicine, 203(10), 2271–2279. 10.1084/jem.20061308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. , Ackerman, H. H. , & Carulli, J. P. (2011). A genome‐wide screen of gene–gene interactions for rheumatoid arthritis susceptibility. Human Genetics, 129(5), 473–485. 10.1007/s00439-010-0943-z [DOI] [PubMed] [Google Scholar]
- Lønnberg, A. , Skov, L. , Duffy, D. , Skytthe, A. , Kyvik, K. , Pedersen, O. , & Thomsen, S. (2016). Genetic factors explain variation in the age at onset of psoriasis: A population‐based twin study. Acta Dermato Venereologica, 96(1), 35–38. 10.2340/00015555-2171 [DOI] [PubMed] [Google Scholar]
- McKenna, A. , Hanna, M. , Banks, E. , Sivachenko, A. , Cibulskis, K. , Kernytsky, A. , Garimella, K. , Altshuler, D. , Gabriel, S. , Daly, M. , & DePristo, M. A. (2010). The genome analysis toolkit: A MapReduce framework for analyzing next‐generation DNA sequencing data. Genome Research, 20(9), 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, A. , Kay, L. J. , Lynch, S. A. , & Walker, D. J. (2005). Recurrence risk for psoriasis and psoriatic arthritis within sibships. Rheumatology, 44(6), 773–776. 10.1093/rheumatology/keh589 [DOI] [PubMed] [Google Scholar]
- Niel, C. , Sinoquet, C. , Dina, C. , & Rocheleau, G. (2015). A survey about methods dedicated to epistasis detection. Frontiers in Genetics, 6, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, R. , & Weinberg, J. (2008). Psoriasis in the patient with human immunodeficiency virus, part 1: Review of pathogenesis. Cutis, 82(2), 117–122. [PubMed] [Google Scholar]
- Prinz, J. (2017). Autoimmune aspects of psoriasis: Heritability and autoantigens. Autoimmunity Reviews, 16(9), 970–979. 10.1016/j.autrev.2017.07.011 [DOI] [PubMed] [Google Scholar]
- Raychaudhuri, S. , & Gross, J. (2000). Psoriasis risk factors: Role of lifestyle practices. Cutis, 66(5), 348–352. [PubMed] [Google Scholar]
- Ritchie, M. D. , Hahn, L. W. , Roodi, N. , Bailey, L. R. , Dupont, W. D. , Parl, F. F. , & Moore, J. H. (2001). Multifactor‐dimensionality reduction reveals high‐order interactions among estrogen‐metabolism genes in sporadic breast cancer. The American Journal of Human Genetics, 69(1), 138–147. 10.1086/321276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn, S. , Barrett, K. , Van Abbema, A. , Chang, C. , Kohli, P. B. , Kanda, H. , Smith, J. , Lai, Y. , Zhou, A. , Zhang, B. , & Yang, W. (2013). A restricted role for TYK2 catalytic activity in human cytokine responses revealed by novel TYK2‐selective inhibitors. Journal of Immunology, 191(5), 2205–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange, A. , Barrett, K. , Van Abbema, A. , Chang, C. , Kohli, P. B. , Kanda, H. , Smith, J. , Lai, Y. , Zhou, A. , Zhang, B. , & Yang, W. (2010). A genome‐wide association study identifies new psoriasis susceptibility loci and an interaction between HLA‐C and ERAP1. Nature Genetics, 42(11), 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, H. , Jin, X. , Li, Y. , Jiang, H. , Tang, X. , Yang, X. U. , Cheng, H. , Qiu, Y. , Chen, G. , Mei, J. , Zhou, F. , Wu, R. , Zuo, X. , Zhang, Y. , Zheng, X. , Cai, Q. I. , Yin, X. , Quan, C. , Shao, H. , … Zhang, X. (2014). A large‐scale screen for coding variants predisposing to psoriasis. Nature Genetics, 46(1), 45–50. 10.1038/ng.2827 [DOI] [PubMed] [Google Scholar]
- Tsai, T.‐F. , Hu, C.‐Y. , Tsai, W.‐L. , Chu, C.‐Y. , Lin, S.‐J. , Liaw, S.‐H. , Chiang, B.‐L. , Lin, P.‐J. , & Jee, S.‐H. (2002). HLA‐Cw6 specificity and polymorphic residues are associated with susceptibility among Chinese psoriatics in Taiwan. Archives of Dermatological Research, 294(5), 214–220. 10.1007/s00403-002-0324-0 [DOI] [PubMed] [Google Scholar]
- Vasilopoulos, Y. , Sagoo, G. S. , Cork, M. J. , Walters, K. , & Tazi‐Ahnini, R. (2011). HLA‐C, CSTA and DS12346 susceptibility alleles confer over 100‐fold increased risk of developing psoriasis: Evidence of gene interaction. Journal of Human Genetics, 56(6), 423–427. 10.1038/jhg.2011.33 [DOI] [PubMed] [Google Scholar]
- Works, M. , Yin, F. , Yin, C. C. , Yiu, Y. , Shew, K. , Tran, T. T. , Dunlap, N. , Lam, J. , Mitchell, T. , Reader, J. , & Stein, P. L. (2014) Inhibition of TYK2 and JAK1 ameliorates imiquimod‐induced psoriasis‐like dermatitis by inhibiting IL‐22 and the IL‐23/IL‐17 axis. Journal of Immunology, 193(7), 3278–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, X. , Zhang, R. , Cheng, H. , Pan, Q. , Shen, C. B. , Fan, X. , Wang, Z. X. , Sun, L. D. , Yang, S. , & Zhang, X. J. (2013). Gene‐gene interactions between HLA‐C, ERAP1, TNFAIP3 and TRAF3IP2 and the risk of psoriasis in the Chinese Han population. The British Journal of Dermatology, 169(4), 941–943. [DOI] [PubMed] [Google Scholar]
- Zhang, X.‐J. , Huang, W. , Yang, S. , Sun, L.‐D. , Zhang, F.‐Y. , Zhu, Q.‐X. , Zhang, F.‐R. , Zhang, C. , Du, W.‐H. , Pu, X.‐M. , Li, H. , Xiao, F.‐L. , Wang, Z.‐X. , Cui, Y. , Hao, F. , Zheng, J. , Yang, X.‐Q. , Cheng, H. , He, C.‐D. , … Liu, J.‐J. (2009). Psoriasis genome‐wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nature Genetics, 41(2), 205–210. 10.1038/ng.310 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Yang, J. , Zhang, J. , Sun, L. , Hirankarn, N. , Pan, H. F. , Lau, C. S. , Chan, T. M. , Lee, T. L. , Leung, A. M. , & Mok, C. C. (2016). Genome‐wide search followed by replication reveals genetic interaction of CD80 and ALOX5AP associated with systemic lupus erythematosus in Asian populations. Annals of the Rheumatic Diseases, 75(5), 891–898. [DOI] [PubMed] [Google Scholar]
- Zhao, X. , Li, R. , Qiao, M. , Yan, J. , & Sun, Q. (2018). MiR‐548a‐3p promotes keratinocyte proliferation targeting PPP3R1 after being induced by IL‐22. Inflammation, 41(2), 496–504. 10.1007/s10753-017-0705-3 [DOI] [PubMed] [Google Scholar]
- Zheng, H.‐F. , Zuo, X.‐B. , Lu, W.‐S. , Li, Y. , Cheng, H. , Zhu, K.‐J. , Yin, X.‐Y. , Zhang, C. , Ren, Y.‐Q. , & Wang, W.‐J. (2011). Variants in MHC, LCE and IL12B have epistatic effects on psoriasis risk in Chinese population. Journal of Dermatological Science, 61(2), 124–128. 10.1016/j.jdermsci.2010.12.001 [DOI] [PubMed] [Google Scholar]
- Zhou, F. , Cao, H. , Zuo, X. , Zhang, T. , Zhang, X. , Liu, X. , Xu, R. , Chen, G. , Zhang, Y. , Zheng, X. , Jin, X. , Gao, J. , Mei, J. , Sheng, Y. , Li, Q. , Liang, B. O. , Shen, J. , Shen, C. , Jiang, H. , … Zhang, X. (2016). Deep sequencing of the MHC region in the Chinese population contributes to studies of complex disease. Nature Genetics, 48(7), 740–746. 10.1038/ng.3576 [DOI] [PubMed] [Google Scholar]
- Zuo, X. , Sun, L. , Yin, X. , Gao, J. , Sheng, Y. , Xu, J. , Zhang, J. , He, C. , Qiu, Y. , Wen, G. , & Tian, H. (2015). Whole‐exome SNP array identifies 15 new susceptibility loci for psoriasis. Nature Communications, 6, 6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Interaction analysis in exome sequencing data with p<0.01
Table S2 Interaction analysis in exome and targeted sequencing data with p<0.05
Table S3 Interaction analysis in exome, targeted and HLA sequencing data with p<0.05


