Abstract
The successful use of artificial intelligence (AI) for diagnostic purposes has prompted the application of AI-based cancer imaging analysis to address other, more complex, clinical needs. In this Perspective, we discuss the next generation of challenges in clinical decision-making that AI tools can solve using radiology images, such as prognostication of outcome across multiple cancers, prediction of response to various treatment modalities, discrimination of benign treatment confounders from true progression, identification of unusual response patterns and prediction of the mutational and molecular profile of tumours. We describe the evolution of and opportunities for AI in oncology imaging, focusing on hand-crafted radiomic approaches and deep learning-derived representations, with examples of their application for decision support. We also address the challenges faced on the path to clinical adoption, including data curation and annotation, interpretability, and regulatory and reimbursement issues. We hope to demystify AI in radiology for clinicians by helping them to understand its limitations and challenges, as well as the opportunities it provides as a decision-support tool in cancer management.
In the past decade, drastic increases in computational power and memory have enabled the development and implementation of state-of-the-art artificial intelligence (AI) techniques for handling radiology images. We are currently witnessing increasing enthusiasm in this field, especially in oncology imaging, although computerized methods have been used in radiology since the 1960s1. Early initiatives did not gain much traction because they relied on analogue image acquisition and limited computational resources. In the 1980s, the advent of digital imaging methods and improvements in computational architecture and storage renewed interest in these computer-aided detection (CAD) techniques2–4. The initial success with AI in breast cancer detection5 paved the way for AI approaches to be used more broadly in diagnostic tasks such as tumour classification and cancer detection. Over the past decade, AI-based diagnostic tools have been continuously refined, and in many cases their diagnostic performance has been shown to match or even surpass that of human experts in multiple different cancer types6,7. This success has led to AI approaches now being evaluated to aid more complex decision-making tasks, such as disease prognostication, prediction of response to different treatment modalities, recognition of treatment-related changes and discovery of imaging representations of phenotypic (for example, sex, age or ethnicity) and genotypic features associated with prognosis.
In this Perspective, we exclusively focus on radiology AI-enabled biomarkers to predict disease outcome and response to treatment, with the ultimate goal of providing individualized management. We aim to equip clinicians interested in state-of-the-art AI approaches for decision-making in oncology with knowledge on the current novel tools being applied to outcome prediction, how these approaches are developed and, specifically, the types of image representation (radiomics or deep learning (DL)) that can be used in AI applications. We discuss the clinical implications of AI in radiology with regard to stratifying patients by disease severity and prognosis, predicting treatment response and benefit, identifying unfavourable treatment outcomes (for example, hyperprogression)8, distinguishing confounding responses (such as pseudoprogression)9,10 from true disease progression, and non-invasively predicting salient molecular and genotypic traits. First, we define AI-enabled imaging biomarkers and their use, contrasting them with existing biomarkers in oncology. We then focus on the general framework of AI-enabled imaging biomarkers, discussing the technical underpinnings of commonly used methods. We describe AI tools used in complex decision-making tasks, providing examples of how these AI indications have been used for the management of common cancer types (further summarized in Supplementary Table 1). Finally, we conclude by summarizing some of the challenges and obstacles along the path towards clinical adoption of these approaches and by discussing future implications for oncology practice.
AI-enabled imaging cancer biomarkers
A biomarker is “a defined characteristic that is measured as an indicator of normal biological processes, pathogenic processes or biological responses to an exposure or intervention, including therapeutic interventions”11. On the basis of the type of clinical decisions they can inform on, biomarkers can be grouped into several categories12. In oncology, biomarkers have applications ranging from prevention, as is the case for biomarkers of cancer susceptibility or risk, to guiding high-level decision-making, among which prognostic and predictive biomarkers are the most clinically relevant.
A prognostic biomarker conveys information pertaining to the risk of a disease-related end point. In oncology, prognostic biomarkers are used to determine the risk profile of a patient with cancer on the basis of tumour characteristics. This knowledge enables the clinician to identify patients with poor prognosis who might be candidates for escalation of therapy and/or clinical trials11. Conversely, if pre-emptively identified, patients with a good prognosis might have favourable outcomes with de-escalated therapy and could thus be spared the physiological and financial toxicities of cancer treatment.
Most prognostic biomarkers currently used in oncology are molecular assays that rely on complex multigene signatures, such as Oncotype DX and MammaPrint in breast cancer13 and Decipher in prostate cancer14. These genomic assays are included in the National Comprehensive Cancer Network (NCCN) guidelines and are routinely used in clinical practice; however, they are prohibitively expensive and require tumour tissue obtained through an invasive procedure, thus limiting their availability and applicability in serial monitoring throughout treatment.
A predictive biomarker enables clinicians to make an informed management choice by identifying patients who would benefit from a particular therapeutic agent. In oncology, a biomarker is considered to be predictive if the treatment effect is statistically different in patients with biomarker-positive versus negative status. For example, in breast, gastric and gastro-oesophageal cancers, among others, HER2 status serves as a biomarker for predicting the effectiveness of HER2-targeted therapies, such as trastuzumab and pertuzumab15. In non-small-cell lung cancer (NSCLC), the presence of EGFR exon 19 deletions or exon 21 mutations serves as a biomarker of eligibility for treatment with EGFR tyrosine kinase inhibitors, such as osimertinib or erlotinib16. Besides being prognostic, Oncotype DX is also a predictive biomarker validated in a prospective clinical trial to determine benefit from chemotherapy in women with early-stage breast cancer17.
Rapid AI-driven advancements in computer vision and pattern recognition tasks have led to the emergence of AI-enabled imaging biomarkers. These biomarkers rely on the extraction of discriminating quantitative representations from radiology that capture properties of the tumour phenotype that correlate with clinical outcomes. Two main categories of AI-enabled biomarker in radiology exist: hand-crafted radiomic and DL approaches18 (TABLE 1). With hand-crafted radiomics, a set of representations are predefined by the AI development team (involving computer scientists, radiologists and oncologists) that are composed of feature measurements with specific algorithmic derivations. These feature representations are then fed into a machine learning (ML) model, which in turn predicts an outcome. Some commonly used radiomic approaches focus on the various attributes of the area inside the tumour (such as shape or texture) as well as the tumour microenvironment (TME; such as texture or tumour vasculature) (TABLE 2). Publicly available radiomics toolkits12,19 enable researchers to apply hand-crafted radiomic features in their work without having to develop the feature pipeline themselves. In DL approaches, the development team defines a DL neural network that can be trained using a large data set to discover new representations that can be synthesized to predict a particular outcome. These approaches have unique strengths and weaknesses, and require distinct development workflows (FIG. 1).
Table 1 |.
Comparison of hand-crafted radiomic and DL strategies for prediction and prognosis
| Characteristic | Radiomics | DL |
|---|---|---|
| Data requirements | Typically require a lesser amount of annotated training data | Typically requires large image data sets for training; this requirement can be reduced with techniques such as transfer learning and augmentation |
| Image representations | Typically predefined; can be chosen from a list of domain- agnostic features or by engineering new features targeted around domain knowledge | Involves learning novel feature representations through trainable convolutional operations based on discriminating patterns in training data |
| Prediction strategy | ML models incorporating radiomic features are trained to predict cancer outcomes and treatment response | Prediction model can be trained simultaneously with learning feature representations |
| Annotation | Usually involves the need for accurate delineation of tumour boundaries and other tissues of interest for feature extraction | Model can be trained with course localization or even without any positional information, if given sufficient training data |
| Interpretability | Predictions can be attributed to values of individual measurements included in the ML model | Challenges in determining factors contributing to predictions, hence these approaches tend to be considered ‘black-box’ approaches; can be coupled with explainability approaches (such as class activation maps) post hoc to provide insight into model decision-making |
| Development resources | Typically, model training and inference is not computationally expensive given the smaller number of model parameters and training data set sizes | Tends to be more expensive computationally than radiomics; usually requires one or more graphical processing units |
DL, deep learning; ML, machine learning.
Table 2 |.
Overview of hand-crafted radiomic features used in oncology
| Class of feature | Features | Common examples |
|---|---|---|
| Intensity-based measures and fi rst-order statistics | Direct physical or functional measures from fully quantitative modalities, and basic statistical measures characterizing the distribution of intensity values within a region | Mean, median, standard deviation, skewness and kurtosis of image intensity values, attenuation values on CT162, 163, maximum standardized uptake value on FDG-PET38 |
| Heterogeneity and texture | Features of spatial arrangement and local heterogeneity of image intensity values | Grey-level co-occurrence matrix44, grey-level run length matrix164, local binary patterns165, Gabor wavelets166, Laws' energy measures45 |
| Shape and volume | Measure of 2D or 3D tumour morphology | Volume, surface-to-volume ratio, sphericity, compactness, fractal dimensionality167 |
| Peritumoural radiomics | Characterization of TME through application of radiomic features (such as texture) within the surrounding TME | Textural heterogeneity of the peritumoural radius surrounding a tumour58, stroma168, lymph nodes169, potential metastatic sites109 |
| Radiomics of tumour vascularity | Measurements of the function or shape of the tumour-associated vasculature | Vessel tortuosity and structural organization78,170,171, kinetic and textural measures of tumour vessels172 |
FDG, 2-deoxy-2-18F-fluoro-D-glucose; TME, tumour microenvironment.
Fig. 1 |. Workflow for AI-enabled biomarkers in radiology.
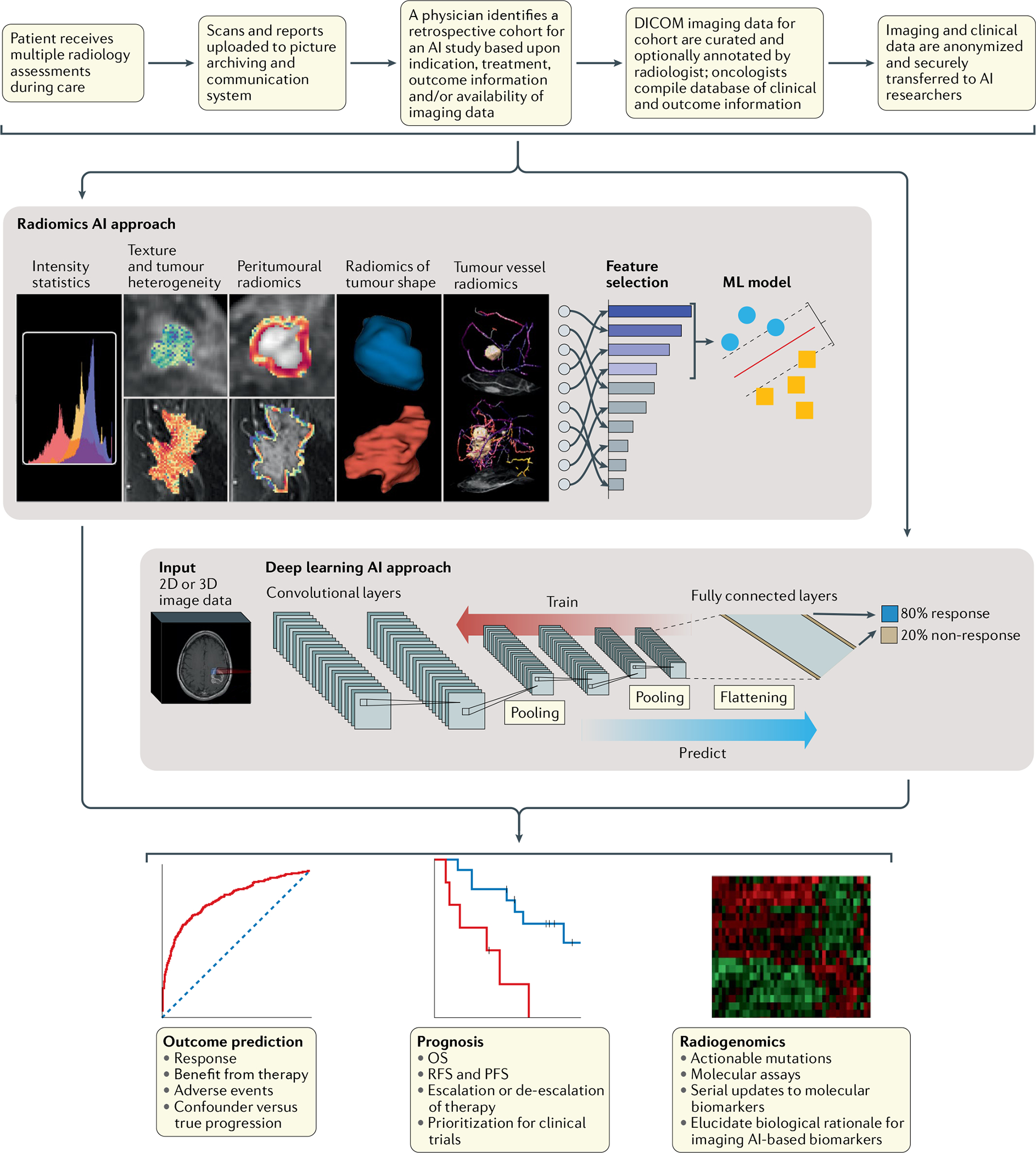
Typical protocol for developing artificial intelligence (AI) radiology biomarkers using radiomic and deep learning approaches, and their clinical applications. Both approaches can be applied in the context of cancer outcome prediction and biomarker discovery for assessment of response to treatment, prognostication and radiogenomics. DICOM, Digital Imaging and Communications in Medicine; ML, machine learning; OS, overall survival; PFS, progression-free survival; RFS, recurrence-free survival.
AI-enabled predictive or prognostic imaging biomarkers can offer certain advantages over molecular assays. Given that they are assessed using routine clinical radiological scans, AI-enabled imaging biomarkers are non-invasive, non-tissue-destructive, rapidly analysed, easily serialized, fairly inexpensive20 and fully compatible with existing clinical workflows, similar to AI-enabled pathology biomarkers21, with the added advantage of being non-invasive22,23. They additionally offer the ability to characterize a tumour over its full 3D volume, avoiding sampling errors that can occur with biopsy samples from heterogeneous tumours24, as well as enabling the detection of changes in the TME. Owing to these advantages over molecular testing, another category of AI-enabled biomarkers that reflect the genotype of a tumour has been developed using imaging representations, an approach known as radiogenomics. Radiogenomic approaches predictive of tumour mutational status could potentially become surrogate non-invasive biomarkers for established molecular biomarkers and could be applied in routine imaging. This approach would be similar to circulating tumour DNA-based liquid biopsy approaches, which are being developed as minimally invasive tools for cancer surveillance25. Such tests could also be used serially to detect changes in the predominant genotype of a tumour following initiation of treatment, a known cause of acquired resistance to targeted therapy26 that cannot be monitored accurately with invasive molecular testing. At present, however, radiogenomic approaches have several limitations, including difficulty in assembling comprehensive data sets containing imaging, genomics and clinical information as well as being restricted to retrospective studies, and thus currently they are limited to research settings27. Indeed, these techniques need further optimization and prospective validation before clinical deployment28.
A framework for AI-derived biomarkers
Two main AI approaches are currently used to develop AI-enabled biomarkers in radiology: radiomics and DL (TABLE 1). These approaches can be leveraged separately or used in combination29–31.
Hand-crafted radiomic models
Several radiomic representations have proved effective in outcome prediction (FIG. 2; TABLE 2). These representations can be translated into a predictive or prognostic model; typically a ML model is trained using a set of features. A common first step in this process is feature selection, which involves algorithmically narrowing down a large pool of explicit features to a smaller subset of features best suited for a particular task. Features can be chosen to optimize predictive performance32,33, reduce correlation within a feature set34 or maximize robustness and stability35. This reduced feature representation is then fed into a statistical ML model (for example, a random forest classifier) to predict clinical outcomes.
Fig. 2 |. Examples of the types of radiomic feature used in oncology.
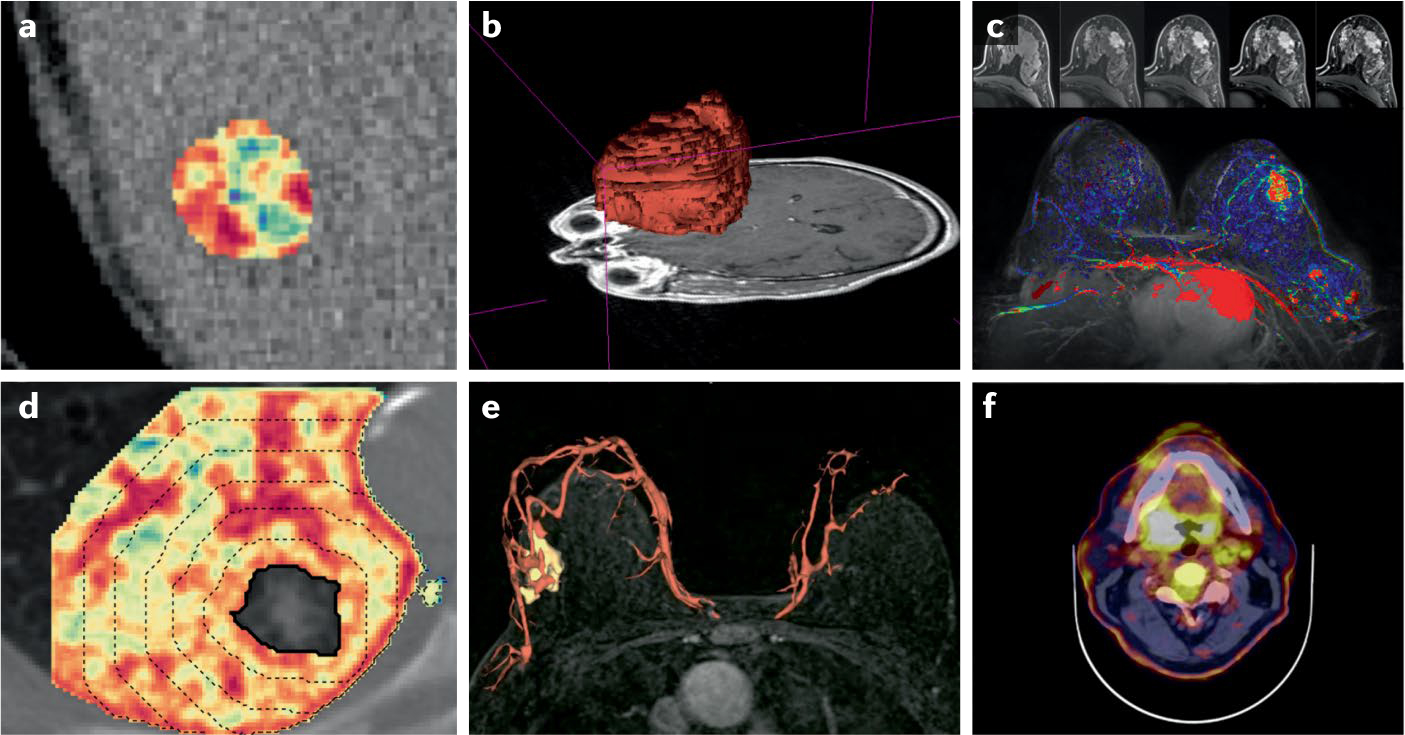
a | Grey-level co-occurrence matrix entropy in a metastatic liver lesion detected by CT. b | Shape of a glioblastoma detected on gadolinium-enhanced T1-weighted MRI. c | Kinetic measure of contrast enhancement over time in breast tissue using contrast-enhanced MRI. d | Peritumoural radiomics measuring textural heterogeneity in the lung stroma surrounding a non-small-cell carcinoma. e | Shape of breast vasculature and tumour-associated vessel network detected using contrast-enhanced MRI. f | Enhanced standardized uptake values on 2-deoxy-2-18F-fiuoro-D-giucose PET-CT scans showing increased metabolic activity in a head and neck carcinoma.
Intensity-based measures.
In many cases, image intensity-based values correspond to some underlying physiological property of the tissue and can be leveraged in radiomic approaches. For example, attenuation values derived from CT scans directly correspond to tissue density, and these values can be used to develop a prognostic biomarker of outcome36 or tumour phenotype37. Similar physiological measures on 2-deoxy-2-18F-fluoro-D-glucose (FDG)-PET scans, which enable quantification of tumour metabolic activity based on positron emissions from a metabolized radiotracer, are highly effective in the early prediction of outcome for patients with several cancer types and across treatment modalities38–42. The distribution of voxel intensity across the tumour or other regions of interest can be further characterized with a broader range of statistical measures (such as standard deviation, skewness or kurtosis), commonly referred to as first-order statistics.
Subvisual heterogeneity and texture.
Tumour heterogeneity can be quantified through radiological imaging using textural heterogeneity features, which involves determination of spatial relationships between image voxel intensities within a region of interest. Statistical measures, such as standard deviation, can provide insights into the variability of an imaging signal, but do so across an entire region of interest (in this case, the whole tumour). By contrast, texture features quantify the relationship between voxels and their surroundings as a function of both distance and intensity. Accordingly, texture features might be better suited to detect tissue architecture heterogeneity on imaging43.
Signal measurements on radiology typically correspond to some physical or biological property of a tissue, and thus a spatial pattern of greater intensity variation on imaging is usually reflective of the underlying anatomical or physiological heterogeneity of the tissue itself. For example, intensity interaction features are commonly used to explore correlative patterns between intensities of adjacent voxels, such as grey-level co-occurrence matrix features44. Other varieties of texture features involve the application of targeted image filters to isolate spatial patterns potentially relevant to patient outcomes. For example, laws’ energy measures use filters that target specific texture patterns, such as speckling and waves45.
Shape and volumetric features.
Measuring tumour size over the course of treatment is standard practice in oncology, and is commonly performed using the Response Evaluation Criteria in Solid Tumors (RECIST)46, an algorithm for monitoring patient response on longitudinal imaging. A limited number of strictly 2D tumour measurements are collected and compared between examinations to assess whether a tumour is stable, progressing or responding. However, RECIST measurements can vary considerably between radiologists47 and the criteria are not well suited for certain therapeutic scenarios, such as identifying pseudoprogression following immunotherapy48 or monitoring response to systemic therapy in patients with metastatic cancers47. Shape radiomics, which refers to any feature characterizing the shape of a tissue of interest, enables more sophisticated analysis of the 3D shape and growth of a tumour, with higher reproducibility than clinical assessment of radiology images. Prospective trials have demonstrated that tumour volume or changes in tumour volume during the course of treatment outperform planar RECIST assessment for longitudinal monitoring in multiple cancer types49,50. More sophisticated morphological measurements, such as surface-to-volume ratio51 and fractal dimensionality52, offer detailed characterization of aberrations of tumour shape and growth patterns. Often, increased tumour shape complexity is associated with poor outcome53–57.
Peritumoural and TME radiomics.
A growing body of work has explored the application of radiomic features beyond the tumour to characterize the surrounding TME. TME radiomic approaches often involve other radiomic feature families to characterize signal properties, such as heterogeneity within non-tumour stroma. Peritumoural radiomics, which involves extraction of texture and statistical features within a radius of tissue surrounding the tumour, has been shown to have predictive and prognostic value across a number of treatment contexts in breast58–60, lung61–66, brain67,68, oesophageal69, gastric70,71 and prostate cancers72, and head and neck squamous cell carcinoma (HNSCC)73. The inclusion of analyses of the peritumoural region increases the predictive power of radiomic signatures over intratumoural radiomics alone29,58,59,68,74–77. Specialized TME radiomics approaches to focus on tumour-associated vasculature have also shown increasing promise and are discussed below.
Radiomics of tumour vascularity.
Shape-based radiomic analysis can also be applied to quantify structural abnormalities in the tumour-associated vasculature and effects of tumour angiogenesis. Vessel tortuosity, a category of features measuring the abnormal shape of the tumour-associated vasculature, has shown promise in the prediction of response to chemotherapy in patients with breast cancer78,79 or malignant gliomas50 and response to targeted agents in patients with breast cancer brain metastases80. Measurements of vessel tortuosity have also shown promise for identifying those patients with NSCLC who are likely to have hyperprogression when receiving immune-checkpoint inhibitors (ICIs)65. This atypical response pattern is characterized by a paradoxical acceleration of tumour growth following ICIs and requires immediate therapy cessation.
DL AI models
DL strategies leverage deep neural networks for pattern recognition, which typically comprise a series of trainable nonlinear operations, known as layers, each of which transforms input data into a representation that facilitates pattern recognition. As more layers apply transformations to the input data, such data become increasingly abstracted into a deep-feature representation. The resulting deep features can eventually be translated by the final layer of a network into a desired output, such as the likelihood of a therapeutic outcome or the molecular subtype of a tumour. DL is a vast, technical and dynamically evolving field. We provide a brief introduction to the most frequently encountered topics in the context of prediction-based radiology AI, with a more detailed supplementary discussion of the types of deep neural network (Supplementary Box 1), popular architectures (Supplementary Box 2; Supplementary Table 2) and strategies for addressing data limitations (Supplementary Box 3). All these aspects have been reviewed elsewhere81,82.
Convolutional neural networks used for outcome prediction.
The majority of DL-enabled biomarker applications in radiology use convolutional neural networks (CNNs)83 (FiG. 3a) to derive predictions from imaging data. CNNs are a specialized type of neural network designed to learn spatial patterns in images and they have received substantial attention owing to their performance in diagnostic tasks. In several high-profile studies, CNN-based models have even surpassed the performance of expert human readers in interpreting chest radiography84 and CT85, and digital mammography6,86. Just as CNNs have been shown to be capable of learning image features indicative of malignancy, a growing body of research has shown that they can stratify patients according to subtle differences in tumour properties related to outcome, risk and molecular profiles (FiG. 3a). When trained with patient outcome data, the convolutional layers of a CNN can learn to recognize novel imaging phenotypes reflective of prognosis. CNNs can be applied to 2D or 3D inputs, and can be modified with multiple inputs for learning from a combination of image types, such as multiparametric or dynamic MRI scans87,88. A substantial number of CNN architectures can be chosen from for AI-based biomarker studies (Supplementary Table 2), and their histories and strengths are discussed in further detail in Supplementary Box 2.
Fig. 3 |. Building blocks and types of neural network commonly applied to medical imaging data.
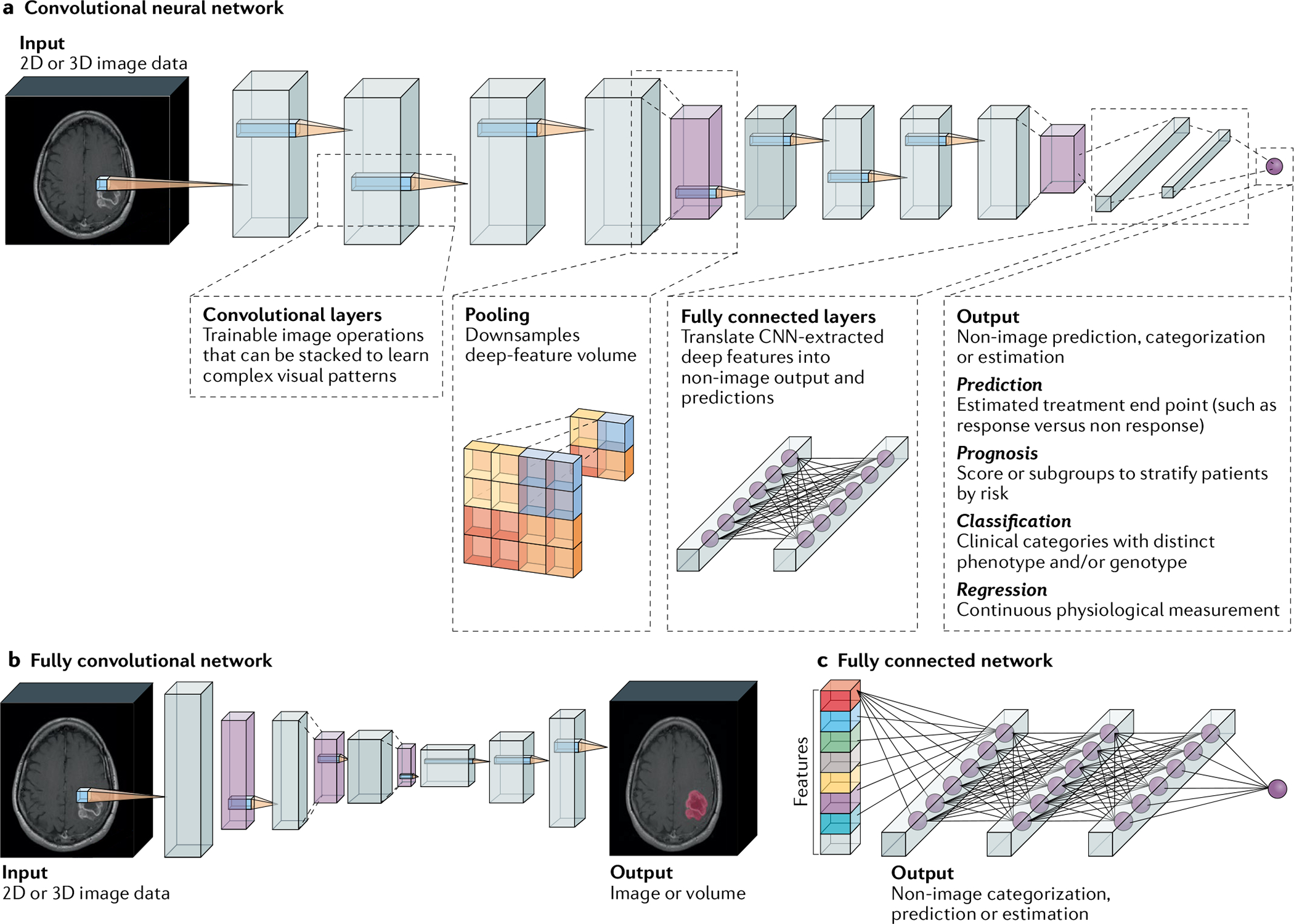
a | Example of a convolutional neural network (CNN) model configured for prediction. Input images or volumes are passed through the CNN layers, which perform operations and translate them into a target output vector. Convolutional layers are sets of operations that transform imaging data into deep-feature representations. Each filter is passed over the image and paired with a nonlinear activation function to emphasize visual patterns of interest for a certain task. As more convolutional layers are stacked, a CNN can learn more complex visual patterns within an image. Throughout a CNN classifier, deep features are periodically aggregated through pooling operations. After processing by convolutional and pooling layers, deep-feature representations are eventually flattened into a vector. Next, fully connected layers translate these CNN-derived image features into a vector that corresponds to a target output. These models can be applied to the prediction of treatment response, prognostication, classification of tumour subtypes and biomarkers, and prediction of physiological values. b | Fully convolutional neural networks are a type of CNN comprising only convolutional layers that yield image-like outputs, such as a map of a tumour’s location. c | Fully connected networks can be trained to make predictions based on non-image data, such as radiomic features and clinical variables.
Other neural networks in radiology.
Fully convolutional neural networks (FCNs)89 (FiG. 3b) are a type of CNN that produces image-like outputs. FCNs can be used to map the boundaries of a tumour within an image for downstream radiomic analyses (a process known as segmentation) or unsupervised feature learning when data are limited (such as by training a convolutional autoencoder). Likewise, fully connected networks (FiG. 3c) are neural networks without convolutional layers that can make predictions from various lists of measurements, such as radiomic features. Other varieties of neural network can be combined with CNNs to process multiple sets of radiological data collected over time, enabling longitudinal analysis of imaging data (for example, for response assessment). These and other variations are discussed in greater depth in Supplementary Box 1.
Training DL models.
To train a DL model, neural networks are updated iteratively with subsets of the training data set known as batches. For each batch, a neural network first generates predictions of patient outcomes based on imaging data. These predictions are then compared with the corresponding real treatment outcomes via a loss function — an equation that measures the correctness of the network outputs. The value obtained from the loss function is then used to update the operations performed by the network layers (FiG. 1), making changes informed most by samples for which the network performed poorly. A second set of patient data, known as the tuning data set, is used to monitor performance while training and optimizing the configuration and learning processes of the model before it is applied to an independent data set, referred to as the test or external validation data set.
Training a neural network typically requires a substantially larger amount of data than that required for development of a radiomic model. All ML models are defined by a set of parameters, which are variables that specify all the possible configurations of the algorithm. Increasing the number of parameters in a model expands the range of possible solutions it can discover, but the quantity of data it requires to learn effectively will also be greater relative to simpler models. State-of-the-art CNN architectures comprise millions of parameters in order to discover novel prognostic representations directly from the original data. By contrast, radiomics restricts prediction problems to a limited pool of prespecified features combined within a statistical model with fewer parameters (typically dozens to hundreds).
The need for a vast quantity of data can be especially constraining when models are trained for outcome prediction, a setting in which viable patient data might be more limited than in diagnostic studies. Fortunately, several strategies exist for leveraging the benefits of neural networks despite sparse training data. For example, transfer learning80, in which a model trained for one pattern recognition task is repurposed to perform a new task, is frequently used to achieve strong CNN performance with substantially less training data. Further strategies are available to handle limited or flawed training data80,90 (Supplementary Box 2; Supplementary Fig. 1).
Risk assessment and response prediction
Prognostic approaches
Lung cancer.
Most radiomic approaches for lung cancer management have focused on NSCLC. Huang et al.91 were among the first groups to use texture-based hand-crafted radiomics to develop a prognostic nomogram to predict disease-free survival (DFS) in patients with stage I–II NSCLC. Interestingly, they showed that first-order statistical measures inside the tumour (for example, kurtosis) were indicative of tumour heterogeneity and 3-year DFS, and their combination with routine clinicopathological data (such as sex and histological grade) outperformed the tumour, node, metastasis (TNM) staging criteria alone (C-index 0.72 (95% CI 0.71–0.73) versus 0.63 (95% CI 0.62–0.64)). Kamran et al.92 developed a radiomic model using CT scans from patients with limited-stage small-cell lung cancer to predict 2-year overall survival (OS), locoregional recurrence and distant metastases. They observed that radiomic tumour elongation on radiomics was strongly associated with locoregional recurrence (HR 1.10; P = 0.003) and 2-year OS (HR 1.10; P = 0.03). Pavic et al.93 developed a radiomic model using FDG-PET images from patients with mesothelioma to stratify them on the basis of progression-free survival (PFS) and OS. The feature with the best discriminative power was long-run high-grey-level emphasis, which reflects the intratumoural heterogeneity of standardized uptake values (SUVs) on PET scans. The C-index for PFS was 0.66 (95% CI 0.57–0.78). However, a radiomic model developed using CT scans from the same patients had no discriminative power for outcome prediction93. This study is worth highlighting because the investigators applied novel radiomic approaches on state-of-the-art FDG-PET and CT scans to prognosticate outcome in mesothelioma, a rare cancer type.
In the DL domain, Hosny et al.94 trained a 3D CNN to predict 2-year OS following radiotherapy using CT data and then adapted the model to predict OS following surgery with an area under the curve (AUC) of 0.71 (95% CI 0.60–0.82) via transfer learning. The study was unique for several reasons: first, the researchers used seven independent data sets involving ~1,200 patients from five different institutions; second, genomic association studies revealed correlations of the DL feature representations with cell cycle and transcriptional processes, providing a biological interpretation; and third, DL features from the area immediately surrounding the tumour had the highest prognostic signal.
Breast cancer.
Park et al.95 trained an elastic net survival model to combine radiomic intensity, texture and morphology features derived from preoperative MRI scans of patients with invasive breast cancer into a radiomics-derived prognostic score; higher scores were significantly associated with worse DFS in the testing data set (P = 0.036). The investigators not only created a radiomic method for breast cancer prognostication but also developed a nomogram combining radiomics and clinicopathological features for integrated DFS estimation that performed better than scores based on each class of feature alone. Wu et al.96 identified subregions of the intratumoural environment corresponding to different levels of perfusion on contrast-enhanced MRI and quantified interactions between these subregions through network analysis. A radiomic signature indicative of the abundance and distribution of poorly perfused areas was predictive of recurrence-free survival (RFS) on multivariable analysis, adjusting for clinical variables such as age, volume, receptor status and pathological response. Interestingly, tumours with unfavourable prognosis had a higher proportion of poorly perfused regions on breast MRI scans than indolent tumours. Another group97 developed radiomic signatures using dynamic contrast-enhanced (DCE) MRI scans from patients with early-stage breast cancer enrolled on a completed clinical trial. The developed signatures independently predicted axillary lymph node metastasis and 3-year DFS. These investigators extracted radiomic features from not only intratumoural and peritumoural regions but also sentinel and non-sentinel axillary lymph nodes. The study revealed that radiomic features of axillary lymph nodes were equivalent in prognostic performance to those from tumour radiomic features alone or combined with those from lymph nodes. Chitalia et al.98 used imaging and outcome data from patients involved in a completed clinical trial to develop an imaging phenotype through clustering of radiomic features on pretreatment DCE-MRI scans. They found three phenotypes with significant variation in image heterogeneity (P < 0.01) that enabled stratification in subgroups with significant differences in 10-year RFS (P < 0.05). The signature was also successfully validated on a publicly available data set. These researchers showed that AI can uncover potential intrinsic imaging phenotypes, corresponding to different degrees of tumour heterogeneity, which in turn might be associated with histologically poorly differentiated tumours and higher mitotic grades. Drukker et al.99 used a long short-term memory DL model developed from radiomic features related to the kinetics of contrast enhancement from dynamic breast MRI scans performed throughout neoadjuvant chemotherapy, which predicted 2-year RFS with a C-index of 0.80. The study was unique in using a recurrent neural network (RNN), a specialized category of DL network, which integrates and learns using features derived from images across different time points.
Brain cancer.
Most of the radiology AI research in brain cancer focuses on glioblastoma, one of the brain tumour types associated with substantially worse outcomes. Kickingereder et al.100 used a hand-crafted radiomic model incorporating volume, shape and texture features from multiparametric MRI scans and used a supervised principal component analysis to predict PFS (HR 2.43; P = 0.002) and OS (HR 4.33; P < 0.001) in patients with glioblastoma. An interesting finding of this analysis was that all radiomic features selected for the model were exclusively from the fluid-attenuated inversion recovery (FLAIR) sequence, a common MRI modality, and included grey-level features indicative of intratumoural heterogeneity. Beyond intratumoural features, another group67 developed a radiomic risk score using 25 texture and entropy features from both within and outside the tumour, and integrated these features with molecular information that included IDH and MGMT status to predict PFS in the validation data set (C-index 0.84; P = 0.03). Additionally, the radiomic risk score was associated with biological pathways of cell differentiation, adhesion and angiogenesis. This study was one of the first to leverage peritumoural radiomic features for estimating survival in patients with glioblastoma and to comprehensively develop an imaging biomarker by leveraging hand-crafted radiomics, clinical attributes and mutational information.
Lao et al.31 extracted ~98,000 features from multiparametric MRI (T1-weighted (T1w), T1 contrast (T1c), T2w and FLAIR modalities) with a transfer learning approach using a pretrained CNN to predict OS (C-index 0.71, 95% CI 0.588–0.932) in glioblastoma. Following feature selection, a LASSO Cox regression model including six of the top DL features enabled accurate stratification of patients in the validation data set (HR 5.13, 95% CI 2.03–12.96; P < 0.001) on the basis of OS. Kickingereder et al.101 developed and validated an automatic neural network (ANN) for the identification and volumetric segmentation of contrast-enhancing tumours and non-enhancing T2w signal abnormalities on MRI scans. The ANN-based model was trained on a data set of patients from one institution and validated using two data sets: one internal and another from a completed clinical trial (EORTC-26101), in which it had almost a 25% higher performance in survival prediction relative to the Response Assessment in Neuro-Oncology (RANO) criteria (with hazard ratios of 2.59 (95% CI 1.86–3.60) versus 2.07 (95% CI 1.46–2.92) for ANN and RANO, respectively). This study was unique in using a clinical trial data set for validation of the performance, although this validation was retrospective. Zhou et al.87 presented a novel neural network approach incorporating brain multiparametric MRI data (T1w, T1c, T2w and FLAIR) projected along three spatial dimensions to form RGB images for a four-input CNN, which fused data from these images with lesion measurements and patient age. The model was able to stratify patients into subgroups with an expected median OS of 0–10 months, 10–15 months and >15 months with an average accuracy of 0.664 ± 0.061 in tenfold cross-validation.
Prostate cancer.
Both DL and hand-crafted radiomics have been applied to multiparametric MRI scans obtained after definitive therapy to predict the risk of prostate cancer recurrence. Shiradkar et al.102 used texture-based radiomics of pretreatment multiparametric MRI scans to predict biochemical recurrence after radical prostatectomy. These investigators showed that textural heterogeneity and gradient orientation radiomic features derived not only from T2w images, but also from apparent diffusion coefficient maps, were strongly associated with cancer recurrence. Zhang et al.103 developed an AI model using MRI features as well as clinical parameters to predict 3-year biochemical recurrence after radical prostatectomy through cross-validation. A support vector machine-based ML classifier, which integrated several imaging features, PI-RADS score (a structured reporting system for evaluating clinically significant cancer on multiparametric MRI) and clinicopathological features, predicted 3-year biochemical recurrence with an AUC of 0.95 (95% CI 0.92–0.98). This study was unique in integrating parameters from multiple scales and sources to build an accurate prognostic biomarker. Zhong et al.104 used a deep transfer learning-based model to distinguish indolent from clinically significant prostate cancer using multiparametric MRI. In the validation data set, the model outperformed the standard PI-RADS v2 score in identifying clinically significant prostate cancer (AUC of 0.726 versus 0.711).
Other cancer types.
Wang et al.105 trained a prognostic model using a set of 16 deep features obtained via unsupervised feature learning with a convolutional autoencoder (Supplementary Box 1) trained on contrast-enhanced CT images from patients with high-grade serous ovarian cancer. This model accurately predicted 3-year RFS in two different validation data sets (with AUCs of 0.77 and 0.83; P < 0.05). Parmar et al.106 developed a radiomic model using pretreatment CT scans from patients with NSCLC or HNSCC. Consensus clustering was performed to select the top radiomic features for each tumour type, predicting OS with C-indexes of 0.61 and 0.63 in NSCLC and HNSCC, respectively. Interestingly, the NSCLC model had AUCs of 0.56 and 0.61 for predicting tumour histology and stage, respectively. The HNSCC model was even more predictive of histology (AUC 0.80) and moderately predictive of human papillomavirus status (AUC 0.58). Zheng et al.107 showed that a radiomic score that included the top six texture features relating to architectural heterogeneity extracted from the arterial phase of pretreatment abdominal CT scans from patients with solitary hepatocellular carcinoma was associated with RFS (P = 0.004) and OS (P = 0.039). In a radiomic signature108 using first-order statistics of molecular profiling and pretreatment contrast-enhanced CT scans from patients with stage IV colorectal cancer, skewness was associated with 5-year OS (P = 0.025). In addition, the mean value of positive pixels was significantly lower in BRAF-mutated tumours than in BRAF-wild-type tumours (P = 0.007). Creasy et al.109 demonstrated that radiomic analysis of the liver parenchyma on presurgical CT scans could predict the future development of hepatic metastases in patients following resection for colon cancer, with 17% of 254 radiomic features distinguishing between hepatic recurrence, extrahepatic recurrence and non-recurrence (P < 0.05). This finding suggests that heterogeneity measures of healthy organ tissue beyond the site of primary disease might be reflective of biology that might provide a more viable premetastatic niche for invasive tumours110.
In the domain of DL, Peng et al.111 developed an AI model using DL features extracted from four CNNs and hand-crafted radiomic features from PET and CT images of patients with nasopharyngeal carcinoma. This AI model was combined with relevant clinicopathological parameters to develop an integrated nomogram that accurately predicted DFS in an independent validation data set. Zhang et al.112 used an AI model combining features learnt from a CNN pretrained using CT scans from patients with NSCLC and hand-crafted radiomic features on CT scans from patients with pancreatic ductal adenocarcinoma to predict 2-year OS in the latter group, outperforming traditional DL or radiomic methods.
Predicting response to therapy
Chemotherapy and chemoradiotherapy.
In patients with NSCLC, tumour stage usually determines treatment stratification. Patients with stage IA disease generally receive surgery alone, whereas those with stage IB–IIB NSCLC tend to undergo surgical resection followed by adjuvant chemotherapy. Combination chemotherapy with a pemetrexed and platinum doublet is the standard of care for patients with stage III NSCLC without metastases, although some receive radiotherapy or neoadjuvant chemoradiotherapy followed by surgery. In a study involving two different validation data sets of patients with early-stage NSCLC64, a radiomic nomogram incorporating features within and outside the lung nodule on CT scans predicted benefit from adjuvant chemotherapy and was prognostic of 3-year DFS (C-index 0.74, 95% CI 0.72–0.76). The score was used to stratify patients into three groups according to risk (high, intermediate or low). Patients in the high-risk group had a significant DFS benefit with adjuvant chemotherapy (P = 0.003 in the validation data sets), whereas those in the low-risk group had no such benefit. Analysis of radiomic, pathology and genomic data revealed that radiomic score was associated with the spatial arrangement of tumour-infiltrating lymphocytes (TILs) on histology images (P = 0.036) and with biological pathways related to cellular differentiation and angiogenesis64. Our group61 showed that a radiomic model comprising intratumoural and peritumoural texture features could predict response to pemetrexed–platinum chemotherapy (AUC 0.77; P < 0.05) and was strongly associated with OS in patients with locally advanced NSCLC (HR 2.35, 95% CI 1.41–3.94). The above authors also developed a radiomic model62 using non-contrast CT scans from patients with locally advanced NSCLC receiving neoadjuvant chemoradiotherapy followed by surgery to enable stratification by OS (HR 11.18, 95% CI 3.17–44.1) and predict major pathological response. Coroller et al.113 used radiomic features from both primary tumours and lymph nodes from patients with locally advanced NSCLC to predict pathological complete response (pCR) to neoadjuvant chemoradiotherapy before surgery. Three radiomic features that describe tumour sphericity and lymph node homogeneity predicted pCR with an AUC of 0.67 (P < 0.05), while features quantifying lymph node homogeneity could also accurately predict residual disease (AUC 0.72–0.75; P < 0.05). Wei et al.114 developed and validated a radiomic model to predict response to platinum-based chemotherapy using data from patients included in a completed clinical trial, which achieved an AUC of 0.79 (P < 0.05) on cross-validation. Regarding DL approaches, Xu et al.115 combined a pretrained CNN with a RNN to analyse longitudinal CT scans of patients with stage III NSCLC before and after treatment. The AI method had high performance in predicting pathological response (P = 0.016) in a validation data set, and this performance improved as the number of scans analysed was increased.
With regard to breast cancer, radiomics and DL approaches have largely been focused on predicting response to neoadjuvant chemotherapy116. In a large-scale multicentre validation study117, a multiparametric radiomic model incorporating features from contrast-enhanced T1w, T2w MRI and diffusion-weighted imaging accurately predicted pCR (AUC 0.79; P < 0.05) in validation data sets from three institutions. Mazurowski et al.118 found 20 prognostic radiomic features on DCE-MRI in patients with invasive breast cancer that were significantly associated with distant RFS. Descriptors of size (with the highest C-index, 0.77, 95% CI 0.67–0.86), heterogeneity (C-index 0.64, 95% CI 0.52–0.76) and perfusion (C-index 0.70, 95% CI 0.60–0.80) were found to have the most predictive value. Cain et al.119 evaluated a predictive radiomic signature on MRI scans from patients who received neoadjuvant chemotherapy and found it to be highly predictive of pCR (AUC 0.71, 95% CI 0.58–0.83) in patients with breast cancer subtypes associated with poor outcomes (triple-negative breast cancer (TNBC) and HER2+ disease). Interestingly, we were among the first groups to show that adding textural radiomics of the peritumoural region immediately surrounding the tumour to intratumoural features from pretreatment MRI scans improves predictions of response to neoadjuvant chemotherapy (AUC 0.74; P < 0.05). To date, most studies have aimed to predict response to neoadjuvant chemotherapy primarily using dynamic MRI scans, although Tadayyon et al.120 predicted such responses by demonstrating significant survival differences between responders and non-responders at weeks 1 (P = 0.035) and 4 (P = 0.027) using texture features from breast ultrasonography images with a cross-validation strategy. Regarding DL, Ha et al.121 trained a CNN to predict response to neoadjuvant chemotherapy based on pretreatment MRI scans and reported an accuracy of 88% in a testing data set. The pCR rate in the study was higher in patients with TNBC (36%) or HER2+ (50%) breast cancer compared with those with luminal A (18%) subtypes, which is concordant with population studies122. These investigators hence provided a potential way to use non-invasive imaging even before treatment initiation to select those patients most likely to respond to neoadjuvant treatment, in contrast to current standard-of-care imaging methods, which use post-treatment serial MRIs to assess response to therapy.
Nie et al.123 developed a radiomic signature using T2w MRI scans from patients with confirmed locally advanced rectal cancer comprising 30 features from within the tumour, which significantly predicted pCR (AUC 0.84; P < 0.05) following neoadjuvant chemoradiotherapy. Antunes et al.124 built a radiomic model to predict pCR in a similar patient population, showing that it was robust and reproducible across a validation set comprising patients from two different institutions (AUC 0.71; P < 0.05) and was consistent across two different expert tumour annotations (Dice Similarity coefficient 73.7 ± 14.1 for gross tumour volume).
Cha et al.125 compared multiple AI methods, including hand-crafted radiomics and CNN-based DL radiomics, to predict pCR in patients with bladder cancer using CT scans performed before and after neoadjuvant chemotherapy. The hand-crafted model and the DL model achieved AUCs of 0.77 and 0.73, respectively.
Fang et al.126 developed a MRI radiomic signature derived from the TME using sagittal T2w, contrast-enhanced T1w and apparent diffusion coefficient MRI images from patients with locally advanced cervical cancer. This model accurately predicted RECIST response in patients undergoing concurrent chemoradiotherapy (AUC 0.80, 95% CI 0.68–0.92).
Jiang et al.127 developed a novel DL AI biomarker using portal venous phase contrast-enhanced CT scans to predict DFS and OS in a training data set of patients with gastric cancer. The model was then used to build an integrated nomogram with clinicopathological features that not only predicted DFS (C-index 0.85, 95% CI 0.83–0.88) and OS (C-index 0.86, 95% CI 0.84–0.89) but also benefit from adjuvant chemotherapy, in an extensive independent validation data set.
Targeted therapy.
Our group59 showed that a combination of peritumoural and intratumoural radiomic features from DCE-MRI scans of patients with invasive HER2+ breast cancer could help to identify intrinsic molecular cancer subtypes, providing insights into the immune response within the peritumoural environment as well as predicting response to HER2-targeted therapy. In an exploratory study, Mehta et al.128 demonstrated that pharmacokinetic modelling on baseline breast dynamic MRI could help to identify patients with downregulation of angiogenesis pathways following bevacizumab treatment, which might be indicative of response to therapy. In a preliminary study involving patients with hormone receptor-positive metastatic breast cancer treated with CDK4/6 inhibitors, our group129 showed that a radiomic feature-derived risk score of liver metastases on CT scans indicating intratumoural heterogeneity was prognostic of OS (HR 2.02, 95% CI 1.13–3.61; P = 0.0027) and response to therapy (AUC 0.68; P < 0.05).
Aerts et al.130 analysed data from a completed clinical trial of patients with early-stage NSCLC treated with the EGFR inhibitor gefitinib. They developed a radiomic model using pretreatment CT scans, and found that the Laws’ energy feature was strongly associated with EGFR mutation status (AUC 0.67; P = 0.03) and thus associated with a gefitinib response phenotype.
Immunotherapy.
Sun et al.131 used a radiomic approach based on CT scans to estimate the presence of CD8+ TILs and also to predict response to ICIs across four solid tumour types (HNSCC, NSCLC, hepatocellular carcinoma and urothelial carcinoma). They modelled the radiomic analysis on the completed MOSCATO trial of ICIs, which collected RNA sequencing data and tumour biopsy samples. The radiomic signature was validated using a data set from The Cancer Genome Atlas (TCGA) for correlation with CD8 gene expression, and on two other independent data sets with baseline imaging data available for tumour immune phenotype association and ICI response prediction, respectively. In the response prediction validation set, the radiomic signature was associated with OS (HR 0.52, 95% CI 0.35–0.79) and could also accurately predict response to ICIs (P = 0.025). Our group63 developed a radiomic model using both pretreatment and immediate post-treatment (6–8 weeks) CT scans of patients with NSCLC receiving ICIs. The intratumoural and peritumoural radiomic models predicted RECIST response (with AUCs of 0.85 and 0.81, respectively; P < 0.05) and OS (HR 1.64, 95% CI 1.22–2.21) in two independent data sets. In an exploration of pathological associations of radiomic features, we found that peritumoural texture features were associated with TIL density on tissue biopsy samples (P < 0.05). Trebeschi et al.132 developed a radiomic biomarker using contrast-enhanced CT scans of primary and metastatic lesions in patients with melanoma or NSCLC receiving ICIs; the model predicted response to ICIs with high performance across both tumour types (P < 0.001). Independent gene set enrichment analysis of patients with NSCLC revealed radiogenomic associations with pathways involved in mitosis and proliferation132. A unique study by Yang et al.133 introduced a transformer network able to integrate clinical measurements, previous interventions and radiomic features from imaging scans over a timeline to predict response before treatment with anti-PD-1 antibodies, with an AUC of 0.80 in a cross-validation data set. This approach is innovative owing to its potential for analysing longitudinal, real-world clinical data from multiple modalities that are not available in fixed orders or time intervals. Our group65 developed a radiomic predictor that could classify patients with NSCLC receiving ICIs not only as responders or non-responders but also as hyperprogressors. Tunali et al.134 retrospectively developed clinical radiomic models based on four clinical features together with radiomic textural features of patients receiving single-agent or doublet ICIs in clinical trials. These models successfully identified hyperprogressors on cross-validation (with AUCs 0.81–0.84) using only CT scans performed before ICI treatment initiation.
Radiogenomic approaches.
Wu et al.135 described three imaging subtypes in breast cancer based on the enhancement profile of the tumour and surrounding parenchyma on dynamic MRI and explored the association of these subtypes with prognosis and genotype. The subtype characterized by prominent enhancement in the TME was associated with the poorest 5-year RFS and with increasing dysregulation of certain signalling pathways, including those involved in angiogenesis and protein export. In another study136, these authors developed a radiomic signature to estimate percentage of stromal TILs in pathology samples (ρ= 0.40, 95% CI 0.24–0.54) and evaluated the association of the signature with RFS (P = 0.0008) in an external validation data set. This signature enabled stratification of patients into two subgroups, which were significantly associated with RFS in patients with TNBC (P = 0.04), for whom the presence of TILs is highly prognostic137. Rao et al.138 used an unsupervised hierarchical clustering approach to identify novel phenotypes defined by multiparametric MRI features in samples from a TCGA glioblastoma collection with available microRNA and mRNA expression data. They identified such a phenotype using three features that stratified patients into two subgroups with a statistically significant difference in OS (P = 0.0002) and differential expression of transcripts involved in several immune-related and metabolic pathways. DL models developed using CT139 and PET–CT scans140 efficiently predicted EGFR mutational status in patients with NSCLC with AUCs of 0.81, for both CT and PET–CT. A radiogenomic approach141 predicted KRAF, NRAS and BRAF mutational status in patients with colorectal cancer. Pernicka et al.142 analysed radiomic features in pretreatment CT scans from patients with resected stage II–III colon cancer to predict microsatellite instability (MSI)-positive status, which is associated with a favourable prognosis. They observed increased textural homogeneity in MSI-positive tumours relative to MSI-negative tumours (AUC 0.79; specificity 96.9%; sensitivity 92.5%; P < 0.05). Finally, Liu et al.143 developed a CT-based radiomic signature to predict the expression status of the genes encoding E-cadherin, Ki-67, VEGFR2 and EGFR, in patients with gastric cancer.
Challenges and opportunities
Data curation and annotation
Obtaining sufficient data to develop an AI-based model is always a challenge, which is especially pronounced when developing predictive and prognostic radiology AI tools. Data from retrospectively acquired data sets are often most convenient to aggregate, but raise challenges related to data purity for both model training and validation because predefined inclusion and exclusion criteria might result in unconscious biases in AI algorithms144. For example, a requirement for completion of a treatment regimen might inadvertently exclude patients who discontinued that regimen owing to an exceptionally poor response. Hence, randomized controlled trials (RCTs) are the gold standard for modelling and validating biomarkers. AI-based imaging techniques depend on the signal-to-noise ratio of both imaging and outcome data. RCTs provide unbiased and homogeneous data with well-curated arms for comparative experimental analysis. Nevertheless, unlike retrospective data, accessing these RCT data sets is time consuming and challenging, often requiring extensive and lengthy approvals from pharmaceutical companies or cancer collaborative organizations.
The difficulty in acquiring unbiased and homogeneous data sets has revealed the importance of multi-institutional collaborations in building large data sets for training and validation of these techniques. One of these, The Cancer Imaging Archive145, convened by the National Cancer Institute (NCI), is a publicly available repository of aggregated and prescreened multi-institutional data sets. This initiative has also brought to the forefront the importance of cooperative organizations in oncology, which in the USA involves the NCI National Clinical Trial Network groups (such as SWOG, ECOG and NRG), and worldwide it involves the European Oganization for Research and Treatment of Cancer, the Canadian Cancer Trial Group and the Japan Clinical Oncology group, which are responsible for funding and running RCTs. These organizations already have a crucial role in biomarker development given that data sets from completed cooperative group-led clinical trials can provide enough power to validate some radiomic algorithms, enabling prospective evaluation in RCTs. Additionally, federated learning techniques146, which are DL AI techniques for training models from multi-institutional data sets without actually exchanging data but instead by sharing training parameters and weights, might have a role in large-scale validation of prognostic AI methods.
Once data are acquired, a key preliminary step in many radiology AI studies is annotation, the process of defining the spatial boundaries within which imaging analysis should be performed. The level of detail necessary and intensiveness of the annotation effort depend on the nature of the study (FiG. 4). Radiomics generally requires precise delineation of tumour boundaries or other regions of interest, enabling the computation of measurements specific to the tumour, such as shape and heterogeneity. Annotations can be provided manually by a radiologist or as the outputs of another ML model, such as a FCN. Either way, this step should be handled thoughtfully owing to the high susceptibility of some features to variations in spatial delineation147. Alternatively, DL models can be trained effectively from coarser labels, such as the approximate location of a tumour in a volume, drastically reducing the effort and expertise required for annotation. With sufficient data, the need for spatial localization can even be entirely obviated80.
Fig. 4 |. Different levels of annotation detail in radiomics and deep learning studies.
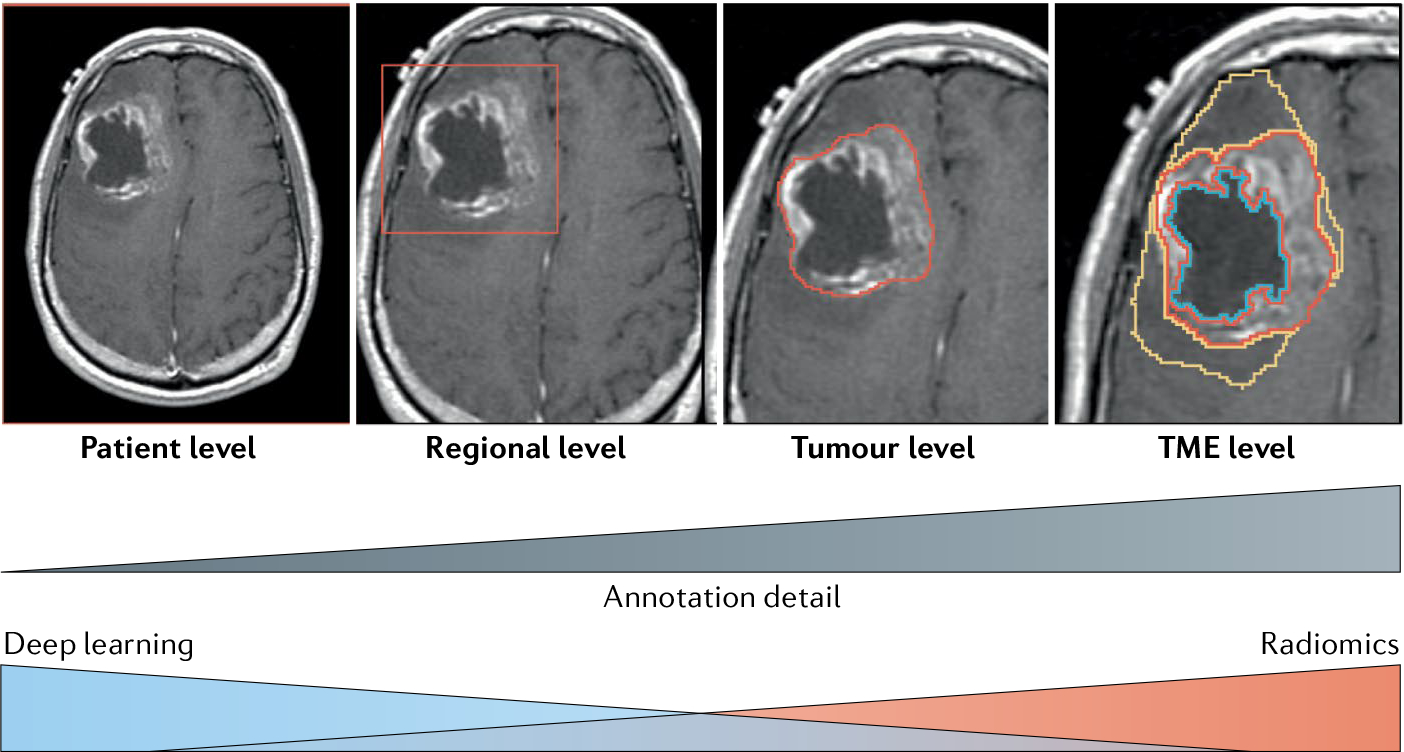
Defining the region of interest and level of annotation detail in radiomics and deep learning studies. TME, tumour microenvironment.
Standardization and reproducibility
Reproducibility across heterogeneous acquisition protocols, multiple institutions and patient populations is one of the primary challenges that AI imaging techniques must overcome for clinical deployment. Most radiomic methods have a sharp drop in performance metrics from training to independent validation. Lambin et al.148 proposed a quality score indicative of the robustness of radiomics studies based on 16 components of the radiomics workflow. Park et al.149 performed a meta-analysis of 77 studies, finding a mean radiomic quality score of only 26.1% of the maximum and identifying some key areas for improvement. In addition to metrics to quantify robustness, several approaches incorporate stability measures to build more reproducible radiomic models. For example, our group150 developed a radiomic method accounting for both stability and discriminability, and applied it to predict disease recurrence in patients with early-stage NSCLC. In three multi-institutional data sets, the radiomic model incorporating stability was substantially stronger in predicting recurrence than the conventional radiomic model, despite both models having similar performance in the training data set. Researchers have also used statistical approaches (such as ComBat harmonization)151 to correct for batch effects in reconstruction methods (for example, radiomic feature differences caused by the use of multiple different image protocols). Orlhac et al.152 used ComBat on ‘phantom images’ on CT scans and found that it enabled realignment of radiomic feature distributions from multi-institutional data sets using different CT protocols. Only models that are robust and reproducible as well as discriminative will find use in clinical practice. For this purpose, multicentre initiatives, such as the Quantitative Imaging Network153 and the Image Biomarker Standardization Initiative154, have developed standardized and optimized sets of radiomic features for use in research.
Interpretability
Interpretability is one of the challenges that AI-enabled biomarkers must overcome to be broadly adopted. Hand-crafted radiomic tools can offer some intuitiveness into how an AI algorithm makes its decision; for example, vessel tortuosity metrics are attributable to the physical and biological properties of the vasculature resulting from tumour angiogenesis. Additionally, several of the studies previously discussed herein have focused on explaining the biological rationale behind radiomic features through correlation with computational pathology features63, radiology–pathology coregistration58 and analysis of biological pathways or genomic correlations64,94,131. Nevertheless, major gaps in knowledge regarding the biological cause of disease outcomes and treatment responses are areas that clearly need additional research.
This problem is further compounded in the context of CNNs or DL networks, which even lack the limited interpretability offered by hand-crafted methods and instead, focus solely on maximizing performance155. Many of these so-called ‘black-box’ approaches might be perfectly viable in the diagnostic setting (for example, AI tools deployed primarily for triaging time-sensitive scans); however, when it comes to AI-enabled imaging biomarkers for optimizing treatment, the question of interpretability becomes more paramount because a biomarker-driven treatment decision needs an explanation rooted in pathophysiology.
Although researchers are currently trying to develop models to explain black-box approaches, an essential caveat is why the original model is needed at all if better models are available. For example, these approaches can involve saliency or attention maps integrated into the model itself94, indicating the specific area of the image the signal emanates from. Such models are trained to localize the prognostic and predictive signal within an image; however, the specific information contributing to a prediction within that region cannot be readily ascertained and might require additional post hoc biological correlation. Hence, some researchers have called for the development of interpretable models from the outset155, whereas other investigators contend that performance compared with present gold standard should be the most important metric to determine the usability of the imaging biomarker156, while others feel that there is a need to go even beyond explainable AI157.
Regulatory framework and reimbursement
The pathway for regulatory approval is a key roadblock in the clinical adoption of imaging-based AI-enabled prognostic and predictive tools. One of the principles for regulatory permission includes the necessary explanation of how the software works. In the USA, the FDA is working on simplifying the AI approval mechanisms; in the meantime, AI tools are classified as medical devices. The FDA has a three-class system in place to determine the risk posed by the device, in which class I devices are those that require the least regulatory hurdles before they can be marketed.
AI-based devices tend to be categorized as class II or III. To date, the FDA has not approved any imaging AI-based prognostic or predictive tool. Several genomic assays (such as MammaPrint, a prognostic multigene assay for breast cancer)158 have received FDA approval through the 510(k) pathway for class II devices. These approvals might set a precedent for prognostic and predictive AI-enabled imaging biomarkers in oncology to be pursued via the less rigorous 510(k) approval process instead of the more restrictive premarket approval (PMA) process for class III devices. Akin to the FDA’s tiered device classification, European Union regulations involve a four-tiered risk classification system (A–D) for medical devices, which includes AI decision-support tools. Only A, the lowest tier, does not need oversight from the regulatory body. Similar policies have been adopted worldwide to regulate AI-based medical decision-support tools. In an Action Plan published in January 2021 (REF.159), the FDA proposed a ‘predetermined change control plan’ in premarket submissions for AI tools. This plan will include the types of anticipated modification in such submissions and also how they expect algorithms to change in a controlled manner that manages risk to patients. The FDA thus expects AI device providers to commit to real-world performance monitoring of these tools and to be able to evaluate such tools from premarket development to postmarket performance.
In terms of reimbursement, AI tools do not currently have dedicated common procedural technology (CPT) codes for billing. In the USA, CPT codes are maintained by the American Medical Association to standardize billing practices across the country. For AI tools to be translated into practice, new CPT codes must be created, but the tool needs to be approved by the FDA for clinical use beforehand. Opting out of FDA approval and going through the Clinical Laboratory Improvement Amendments (CLIA) route, a regulatory pathway for lab-based diagnostic tests (including prognostic and predictive genomic assays such as Oncotype DX) might be an interesting option160; however, the FDA has put out a statement indicating that it might also regulate CLIA tests in the future161.
Conclusions
In this Perspective, we provide an overview of the present and future of AI in radiology as a tool to identify new predictive and prognostic biomarkers for use in clinical decision-making. We believe that this article will provide clinicians with a firm foundation on the emerging field of AI-enabled response and outcome prediction. In particular, we hope to facilitate an understanding of the tools and practices common in radiology AI, and in particular of which clinical scenarios they can be used for. We also expect to contribute to a greater interest in the development and adoption of AI-enabled imaging biomarkers. Just as the digitization of radiology in the past 50 years completely revolutionized the field with increased resolution and wider availability, the next decade is poised for an AI-fuelled revolution in radiology — not to replace radiologists, oncologists or clinicians in general, but to provide them with a new arsenal of tools to better guide treatment and, ultimately, improve patient care.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the Clinical and Translational Science Collaborative of Cleveland (UL1TR0002548) from the National Center for Advancing Translational Sciences (NCATS) component of the NIH and NIH roadmap for Medical Research; the Kidney Precision Medicine Project (KPMP) Glue Grant; the National Cancer Institute (award numbers 1F31CA221383–01A1, 1U24CA199374-01, R01CA249992-01A1, R01CA202752-01A1, R01CA208236-01A1, R01CA216579-01A1, R01CA220581-01A1, R01CA257612-01A1, 1U01CA239055-01, 1U01CA248226-01 and 1U54CA254566-01); the National Center for Research Resources (award number 1C06RR12463-01); the National Heart, Lung and Blood Institute (1R01HL15127701A1 and R01HL15807101A1); the National Institute of Biomedical Imaging and Bioengineering (1R43EB028736-01 and T32EB007509); the Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program (W81XWH-19-1-0668), the Lung Cancer Research Program (W81XWH-18-1-0440, W81XWH-20-1-0595), the Peer Reviewed Cancer Research Program (W81XWH-18-1-0404, W81XWH-21-1-0345) and the Prostate Cancer Research Program (W81XWH-15-1-0558, W81XWH-20-1-0851); the Ohio Third Frontier Technology Validation Fund; the VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service; and The Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University; and through sponsored research agreements from AstraZeneca, Boehringer Ingelheim and Bristol Myers Squibb. The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the institutions named.
Glossary
- Cross-validation
A method of analysing model validity without an independent validation set on a limited data sample by dividing the training data into subsets for training and assessing the performance on the complementary subset of data. Several methods of cross-validation include holdout, k-fold or leave-one-out
- Elastic net survival model
Type of Cox proportional hazard model that is used to calculate hazard ratios, which are a way of evaluating the strength of the association of a variable (for example, survival outcomes) with a time point. an elastic net has the added advantage over a standard Cox model of adjusting for high dimensional data and covariates that might be correlated with each other, while making survival estimations
- Grey-level co-occurrence matrix features
Class of commonly used radiomic features, also known as Haralick features, which rely on higher-order statistics to describe the spatial arrangement and apparent position of the different grey levels present throughout the analysed image
- Kurtosis
Statistical measure to indicate the shape of a probability distribution in terms of its ‘tailedness’. High kurtosis means high deviation from the mean
- Laws’ energy measures
Eponymously named after K. I. Laws, this radiomic feature focuses on measuring variations of energy within a fixed window size, to calculate a combined texture energy of the pixels analysed
- Long short-term memory
Type of recurrent neural network that has been supplemented by the addition of recurrent or ‘forget’ gates, which enables the network to learn by looking back at propagated errors
- Skewness
Statistical measure to indicate the apparent distance between the mean and mode of a distribution. Skewness = (mean–mode)/standard deviation
- Support vector machine
Supervised machine learning model used to classify data by constructing hyperplanes and choosing the hyperplane that has the largest separation between the two classes of interest
- Tumour-infiltrating lymphocytes
(TILs). Lymphocytes that have invaded the tumour tissue from the bloodstream. in the past few years, studies have found TILs to be prognostic of survival and predictive of treatment benefit in several solid tumour types, including breast and lung tumours
Footnotes
Competing interests
N.B. is a current employee of Tempus Labs and a former employee of IBM Research, with both of which he is an inventor on several pending patents pertaining to medical image analysis. He additionally holds equity in Tempus Labs. V.V. is a consultant for Alkermes, AstraZeneca, Bristol Myers Squibb, Celgene, Foundation Medicine, Genentech, Merck, Nektar Therapeutics and Takeda, has current or pending grants from Alkermes, AstraZeneca, Bristol Myers Squibb, Genentech and Merck, is on the speakers’ bureaus of Bristol Myers Squibb, Celgene, Foundation Medicine and Novartis, and has received payment for the development of educational presentations from Bristol Myers Squibb and Foundation Medicine. A.M. holds equity in Elucid Bioimaging and Inspirata, has been or is a scientific advisory board member for Aiforia, AstraZeneca, Bristol Myers Squibb, Inspirata and Merck, serves as a consultant for Caris, Inc. and Roche Diagnostics, has sponsored research agreements with AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb and Philips, has developed a technology relating to cardiovascular imaging that has been licensed to Elucid Bioimaging, and is involved in an NIH U24 grant with PathCore and three different NIH R01 grants with Inspirata. The other authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at https://doi.org/10.1038/s41571-021-00560-7.
Related links
Genomic Data commons Data Portal: https://portal.gdc.cancer.gov/
The Cancer Imaging Archive: https://www.cancerimagingarchive.net/
References
- 1.Giger ML, Chan H-P & Boone J Anniversary Paper: History and status of CAD and quantitative image analysis: the role of medical physics and AAPM. Med. Phys. 35, 5799–5820 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giger ML, Doi K & MacMahon H Computerized detection of lung nodules in digital chest radiographs. Med. Imaging Proc. 767, 384–387 (1987). [Google Scholar]
- 3.Carmody DP, Nodine CF & Kundel HL An analysis of perceptual and cognitive factors in radiographic interpretation. Perception 9, 339–344 (1980). [DOI] [PubMed] [Google Scholar]
- 4.Kundel HL & Hendee WR The perception of radiologic image information. Report of an NCI workshop on April 15–16, 1985. Invest. Radiol. 20, 874–877 (1985). [DOI] [PubMed] [Google Scholar]
- 5.Rao VM et al. How widely Is computer-aided detection used in screening and diagnostic mammography? J. Am. Coll. Radiol. 7, 802–805 (2010). [DOI] [PubMed] [Google Scholar]
- 6.McKinney SM et al. International evaluation of an AI system for breast cancer screening. Nature 577, 89–94 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Bejnordi BE et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318, 2199–2210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frelaut M, Le Tourneau C & Borcoman E Hyperprogression under immunotherapy. Int. J. Mol. Sci. 20, 2674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frelaut M, du Rusquec P, de Moura A, Le Tourneau C & Borcoman E Pseudoprogression and hyperprogression as new forms of response to immunotherapy. BioDrugs 34, 463–476 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Cruz LCH, da, Rodriguez I, Domingues RC, Gasparetto EL & Sorensen AG Pseudoprogression and pseudoresponse: imaging challenges in the assessment of posttreatment glioma. Am. J. Neuroradiol. 32, 1978–1985 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and other Tools) Resource (FDA, 2016). [PubMed] [Google Scholar]
- 12.Griethuysen JJMV et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77, e104–e107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicolini A, Ferrari P & Duffy MJ Prognostic and predictive biomarkers in breast cancer: past, present and future. Semin. Cancer Biol. 52, 56–73 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Cucchiara V et al. Genomic markers in prostate cancer decision making. Eur. Urol. 73, 572–582 (2018). [DOI] [PubMed] [Google Scholar]
- 15.LI SG & LI L Targeted therapy in HER2-positive breast cancer. Biomed. Rep. 1,499–505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan BA & Hughes BG Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl. Lung Cancer Res. 4, 36 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sparano JA et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N. Engl. J. Med. 379, 111–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeCun Y, Bengio Y & Hinton G Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Pfaehler E, Zwanenburg A, Jong J Rde & Boellaard, R. RaCaT: an open source and easy to use radiomics calculator tool. PLoS ONE 14, e0212223 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verma V et al. The rise of radiomics and implications for oncologic management. J. Natl Cancer Inst. 109, djx055 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Bera K, Schalper KA, Rimm DL, Velcheti V & Madabhushi A Artificial intelligence in digital pathology — new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 16, 703–715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan T et al. A radio-genomics approach for identifying high risk estrogen receptor-positive breast cancers on DCE-MRI: preliminary results in predicting oncotypeDX risk scores. Sci. Rep. 6, 21394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of mammaprint, oncotype DX, and PAM50 gene assays. Radiology 281, 382–391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cyll K et al. Tumour heterogeneity poses a significant challenge to cancer biomarker research. Br. J. Cancer 117, 367–375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crowley E, Di Nicolantonio F, Loupakis F & Bardelli A Liquid biopsy: monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 10, 472–484 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Lim Z-F & Ma PC Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J. Hematol. Oncol. 12, 134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazurowski MA Radiogenomics: what it is and why it is important. J. Am. Coll. Radiol. 12, 862–866 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Bodalal Z, Trebeschi S, Nguyen-Kim TDL, Schats W & Beets-Tan R Radiogenomics: bridging imaging and genomics. Abdom. Radiol. 44, 1960–1984 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Eben J, Braman N & Madabhushi A in Medical Image Computing and Computer Assisted Intervention Vol. 11767 (eds Shen D et al.) 602–610 (Springer, 2019). [Google Scholar]
- 30.Bizzego A et al. Integrating deep and radiomics features in cancer bioimaging. IEEE Conf. Comput. Intell. Bioinform. Comput. Biol. 10.1109/CIBCB.2019.8791473 (2019). [DOI] [Google Scholar]
- 31.Lao J et al. A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci. Rep. 7, 10353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.‘Student’. The probable error of a mean. Biometrika 6, 1–25 (1908). [Google Scholar]
- 33.Wilcoxon F Individual comparisons by ranking methods. Biometrics Bull. 1, 80–83 (1945). [Google Scholar]
- 34.Ding C & Peng H Minimum redundancy feature selection from microarray gene expression data. J. Bioinform. Comput. Biol. 3, 185–205 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Chirra P et al. Multisite evaluation of radiomic feature reproducibility and discriminability for identifying peripheral zone prostate tumors on MRI. J. Med. Imaging 6, 024502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JW & et al. Prognostic significance of CT-attenuation of tumor-adjacent breast adipose tissue in breast cancer patients with surgical resection. Cancers 11, 1135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eguchi T et al. Tumor size and computed tomography attenuation of pulmonary pure ground-glass nodules are useful for predicting pathological invasiveness. PLoS ONE 9, e97867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinahan PE & Fletcher JW PET/CT standardized uptake values (SUVs) in clinical practice and assessing response to therapy. Semin. Ultrasound CT MR 31, 496–505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kubota K From tumor biology to clinical PET: a review of positron emission tomography (PET) in oncology. Ann. Nucl. Med. 15, 471–486 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Sheikhbahaei S et al. The value of FDG PET/CT in treatment response assessment, follow-up, and surveillance of lung cancer. Am. J. Roentgenol. 208, 420–433 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Eckstein JM et al. Primary vs nodal site PET/CT response as a prognostic marker in oropharyngeal squamous cell carcinoma treated with intensity-modulated radiation therapy. Head Neck 42, 2405–2413 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Lin C et al. Early 18F-FDG PET for prediction of prognosis in patients with diffuse large B-cell lymphoma: SUV-based assessment versus visual analysis. J. Nucl. Med. 48, 1626–1632 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Prasanna P, Tiwari P & Madabhushi A Co-occurrence of local anisotropic gradient orientations (CoLlAGe): a new radiomics descriptor. Sci. Rep. 6, 37241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haralick RM, Shanmugam K & Dinstein I Textural features for image classification. IEEE Trans. Syst. Man Cybern. SMC-3, 610–621 (1973). [Google Scholar]
- 45.Laws KI Rapid texture identification. SPIE Proc. 10.1117/12.959169 (1980). [DOI] [Google Scholar]
- 46.Eisenhauer EA et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Kuhl CK et al. Validity of RECIST version 1.1 for response assessment in metastatic cancer: a prospective, multireader study. Radiology 290, 349–356 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Nishino M Tumor response assessment for precision cancer therapy: response evaluation criteria in solid tumors and beyond. Am. Soc. Clin. Oncol. Educ. Book 38, 1019–1029 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Hylton NM et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy–results from ACRIN 6657/I-SPY TRIAL. Radiology 263, 663–672 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao J et al. Tumor volume reduction rate is superior to RECIST for predicting the pathological response of rectal cancer treated with neoadjuvant chemoradiation: results from a prospective study. Oncol. Lett. 9, 2680–2686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Decazes P et al. Tumor fragmentation estimated by the volume surface ratio of tumors measured on FDG PET/CT is an independent prognostic factor of diffuse large B-cell lymphoma. J. Nucl. Med. 59, 1416–1416 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Jang K, Russo C & Di leva A Radiomics in gliomas: clinical implications of computational modeling and fractal-based analysis. Neuroradiology 62, 771–790 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Ismail M et al. Shape features of the lesion habitat to differentiate brain tumor progression from pseudoprogression on routine multiparametric MRI: a multisite study. Am. J. Neuroradiol. 39, 2187–2193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghose S et al. Prostate shapes on pre-treatment MRI between prostate cancer patients who do and do not undergo biochemical recurrence are different: preliminary findings. Sci. Rep. 7, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grove O et al. Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma. PLoS ONE 10, e0118261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prasanna P et al. Mass effect deformation heterogeneity (MEDH) on gadolinium-contrast T1-weighted MRI is associated with decreased survival in patients with right cerebral hemisphere glioblastoma: a feasibility study. Sci. Rep. 9, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Antunes J et al. in Medical Image Computing and Computer Assisted Intervention Vol. 11767 (eds Shen D et al.) 611–619 (Springer, 2019). [Google Scholar]
- 58.Braman NM et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 19, 57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Braman N et al. Association of peritumoral radiomics with tumor biology and pathologic response to preoperative targeted therapy for HER2 (ERBB2)–positive breast cancer. JAMA Netw. Open. 2, e192561 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones EF et al. MRI enhancement in stromal tissue surrounding breast tumors: association with recurrence free survival following neoadjuvant chemotherapy. PLoS ONE 8, e61969 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khorrami M et al. Combination of peri-and intratumoral radiomic features on baseline CT scans predicts response to chemotherapy in lung adenocarcinoma. Radiol. Artif. Intell. 1, 180012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khorrami M et al. Predicting pathologic response to neoadjuvant chemoradiation in resectable stage III non-small cell lung cancer patients using computed tomography radiomic features. Lung Cancer 135, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khorrami M et al. Changes in CT radiomic features associated with lymphocyte distribution predict overall survival and response to immunotherapy in non–small cell lung cancer. Cancer Immunol. Res. 8, 108–119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vaidya P et al. CT derived radiomic score for predicting the added benefit of adjuvant chemotherapy following surgery in stage I, II resectable non-small cell lung cancer: a retrospective multicohort study for outcome prediction. Lancet Digital Health 2, e116–e128 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Vaidya P et al. Novel, non-invasive imaging approach to identify patients with advanced non-small cell lung cancer at risk of hyperprogressive disease with immune checkpoint blockade. J. Immunother. Cancer 8, e001343 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Akinci D’Antonoli T et al. CT radiomics signature of tumor and peritumoral lung parenchyma to predict nonsmall cell lung cancer postsurgical recurrence risk. Acad. Radiol. 27, 497–507 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Beig N et al. Radiogenomic-based survival risk stratification of tumor habitat on Gd-T1w MRI is associated with biological processes in glioblastoma. Clin. Cancer Res. 26, 1866–1876 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prasanna P, Patel J, Partovi S, Madabhushi A & Tiwari P Radiomic features from the peritumoral brain parenchyma on treatment-naive multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: preliminary findings. Eur. Radiol. 27, 4188–4197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu Y et al. Assessment of intratumoral and peritumoral computed tomography radiomics for predicting pathological complete response to neoadjuvant chemoradiation in patients with esophageal squamous cell carcinoma. JAMA Netw. Open 3, e2015927 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li J et al. Intratumoral and peritumoral radiomics of contrast-enhanced CT for prediction of disease-free survival and chemotherapy response in stage II/III gastric cancer. Front. Oncol. 10, 552270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang Y et al. Non-invasive imaging evaluation of tumor immune microenvironment to predict outcomes in gastric cancer. Ann. Oncol. 31, 760–768 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Algohary A et al. Combination of peri-tumoral and intra-tumoral radiomic features on Bi-Parametric MRI accurately stratifies prostate cancer risk: a multi-site study. Cancers 12, 2200 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keek S et al. Computed tomography-derived radiomic signature of head and neck squamous cell carcinoma (peri)tumoral tissue for the prediction of locoregional recurrence and distant metastasis after concurrent chemo-radiotherapy. PLoS ONE 15, e0232639 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shan Q et al. CT-based peritumoral radiomics signatures to predict early recurrence in hepatocellular carcinoma after curative tumor resection or ablation. Cancer Imaging 19, 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ding J et al. Optimizing the peritumoral region size in radiomics analysis for sentinel lymph node status prediction in breast cancer. Acad. Radiol. 10.1016/j.acra.2020.10.015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dou TH, Coroller TP, Griethuysen JJMV, Mak RH & Aerts HJWL Peritumoral radiomics features predict distant metastasis in locally advanced NSCLC. PLoS ONE 13, e0206108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen S et al. Pretreatment prediction of immunoscore in hepatocellular cancer: a radiomics-based clinical model based on Gd-EOB-DTPA-enhanced MRI imaging. Eur. Radiol. 29, 4177–4187 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Braman N, Prasanna P, Alilou M, Beig N & Madabhushi A in Medical Image Computing and Computer Assisted Intervention Vol. 11071 (eds Frangi AF, Schnabel JA, Davatzikos C, Alberola-López C& Fichtinger G) 803–811 (Springer, 2018). [Google Scholar]
- 79.Bullitt E et al. Blood vessel morphologic changes depicted with MR angiography during treatment of brain metastases: a feasibility study. Radiology 245, 824–830 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheplygina V, Bruijne MD & Pluim JPW Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med. Image Anal. 54, 280–296 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Chartrand G et al. Deep learning: a primer for radiologists. RadioGraphics 37, 2113–2131 (2017). [DOI] [PubMed] [Google Scholar]
- 82.Miotto R, Wang F, Wang S, Jiang X & Dudley JT Deep learning for healthcare: review, opportunities and challenges. Brief. Bioinform. 19, 1236–1246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.LeCun Y, Bottou L, Bengio Y & Haffner P Gradient-based learning applied to document recognition. Proc. IEEE 86, 2278–2324 (1998). [Google Scholar]
- 84.Rajpurkar P et al. Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 15, e1002686 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ardila D et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 25, 954–961 (2019). [DOI] [PubMed] [Google Scholar]
- 86.Wu N et al. Deep neural networks improve radiologists’ performance in breast cancer screening. IEEE Trans. Med. Imaging 39, 1184–1194 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou T et al. in Medical Image Computing and Computer Assisted Intervention Vol. 12262 (eds Martel AL et al.) 221–231 (Springer, 2020). [Google Scholar]
- 88.Braman N et al. Deep learning-based prediction of response to HER2-targeted neoadjuvant chemotherapy from pre-treatment dynamic breast MRI: a multi-institutional validation study. Preprint at arXiv https://arxiv.org/abs/2001.08570 (2020).
- 89.Shelhamer E, Long J & Darrell T Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39, 640–651 (2017). [DOI] [PubMed] [Google Scholar]
- 90.Zhou Z-H A brief introduction to weakly supervised learning. Natl Sci. Rev. 5, 44–53 (2018). [Google Scholar]
- 91.Huang Y et al. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non — small cell lung cancer. Radiology 281, 947–957 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Kamran SC et al. The impact of quantitative CT-based tumor volumetric features on the outcomes of patients with limited stage small cell lung cancer. Radiat. Oncol. 15, 14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pavic M et al. FDG PET versus CT radiomics to predict outcome in malignant pleural mesothelioma patients. EJNMMI Res. 10, 81 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hosny A et al. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med. 15, e1002711 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Park H et al. Radiomics signature on magnetic resonance imaging: association with disease-free survival in patients with invasive breast cancer. Clin. Cancer Res. 24, 4705–4714 (2018). [DOI] [PubMed] [Google Scholar]
- 96.Wu J et al. Intratumoral spatial heterogeneity at perfusion MR imaging predicts recurrence-free survival in locally advanced breast cancer treated with neoadjuvant chemotherapy. Radiology 288, 26–35 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu Y et al. Development and validation of a preoperative magnetic resonance imaging radiomics-based signature to predict axillary lymph node metastasis and disease-free survival in patients with early-stage breast cancer. JAMA Netw. Open 3, e2028086 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chitalia RD et al. Imaging phenotypes of breast cancer heterogeneity in preoperative breast dynamic contrast enhanced magnetic resonance imaging (DCE-MRI) scans predict 10-year recurrence. Clin. Cancer Res. 26, 862–869 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Drukker K, Edwards A, Papaioannou J & Giger M Deep learning predicts breast cancer recurrence in analysis of consecutive MRIs acquired during the course of neoadjuvant chemotherapy. Proc. SPIE 11314, 1131410 (2020). [Google Scholar]
- 100.Kickingereder P et al. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology 280, 880–889 (2016). [DOI] [PubMed] [Google Scholar]
- 101.Kickingereder P et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol. 20, 728–740 (2019). [DOI] [PubMed] [Google Scholar]
- 102.Shiradkar R et al. Radiomic features from pretreatment biparametric MRI predict prostate cancer biochemical recurrence: preliminary findings. J. Magn. Reson. Imaging 48, 1626–1636 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang Y-D et al. An imaging-based approach predicts clinical outcomes in prostate cancer through a novel support vector machine classification. Oncotarget 7, 78140–78151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhong X et al. Deep transfer learning-based prostate cancer classification using 3 Tesla multi-parametric MRI. Abdom. Radiol. 44, 2030–2039 (2019). [DOI] [PubMed] [Google Scholar]
- 105.Wang S et al. Deep learning provides a new computed tomography-based prognostic biomarker for recurrence prediction in high-grade serous ovarian cancer. Radiother. Oncol. 132, 171–177 (2019). [DOI] [PubMed] [Google Scholar]
- 106.Parmar C et al. Radiomic feature clusters and Prognostic Signatures specific for Lung and Head & Neck cancer. Sci. Rep. 5, 11044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zheng B-H et al. Radiomics score: a potential prognostic imaging feature for postoperative survival of solitary HCC patients. BMC Cancer 18, 1148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Negreros-Osuna AA et al. Radiomics texture features in advanced colorectal cancer: correlation with BRAF mutation and 5-year overall survival. Radiology 2, e190084 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Creasy JM et al. Differences in liver parenchyma are measurable with CT radiomics at initial colon resection in patients that develop hepatic metastases from stage II/III colon cancer. Ann. Surg. Oncol. 28, 1982–1989 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Langley RR & Fidler IJ The seed and soil hypothesis revisited–the role of tumor–stroma interactions in metastasis to different organs. Int. J. Cancer 128, 2527–2535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Peng H et al. Prognostic value of deep learning PET/CT-based radiomics: potential role for future individual induction chemotherapy in advanced nasopharyngeal carcinoma. Clin. Cancer Res. 25, 4271–4279 (2019). [DOI] [PubMed] [Google Scholar]
- 112.Zhang Y et al. Improving prognostic performance in resectable pancreatic ductal adenocarcinoma using radiomics and deep learning features fusion in CT images. Sci. Rep. 11, 1378 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Coroller TP et al. Radiomic-based pathological response prediction from primary tumors and lymph nodes in NsCLC. J. Thorac. Oncol. 12, 467–476 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wei H et al. Application of computed tomography-based radiomics signature analysis in the prediction of the response of small cell lung cancer patients to first-line chemotherapy. Exp. Ther. Med. 17, 3621–3629 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu Y et al. Deep learning predicts lung cancer treatment response from serial medical imaging. Clin. Cancer Res. 25, 3266–3275 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Granzier RWY, van Nijnatten TJA, Woodruff HC, Smidt ML & Lobbes MBI Exploring breast cancer response prediction to neoadjuvant systemic therapy using MRI-based radiomics: a systematic review. Eur. J. Radiol 121, 108736 (2019). [DOI] [PubMed] [Google Scholar]
- 117.Liu Z et al. Radiomics of multi-parametric MRI for pretreatment prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clin. Cancer Res. 25, 3538–3547 (2019). [DOI] [PubMed] [Google Scholar]
- 118.Mazurowski MA et al. Association of distant recurrence-free survival with algorithmically extracted MRI characteristics in breast cancer. J. Med. Reson. Imaging 49, e231–e240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cain EH et al. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: a study using an independent validation set. Breast Cancer Res. Treat. 173, 455–463 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tadayyon H et al. A priori prediction of breast tumour response to chemotherapy using quantitative ultrasound imaging and artificial neural networks. Oncotarget 10, 3910–3923 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ha R et al. Prior to initiation of chemotherapy, can we predict breast tumor response? Deep learning convolutional neural networks approach using a breast MRI tumor dataset. J. Digit. Imaging 32, 693–701 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Houssami N, Macaskill P, von Minckwitz G, Marinovich ML & Mamounas E Meta-analysis of the association of breast cancer subtype and pathologic complete response to neoadjuvant chemotherapy. Eur. J. Cancer 48, 3342–3354 (2012). [DOI] [PubMed] [Google Scholar]
- 123.Nie K et al. Rectal cancer: assessment of neoadjuvant chemoradiation outcome based on radiomics of multiparametric MRI. Clin. Cancer Res. 22, 5256–5264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Antunes JT et al. Radiomic features of primary rectal cancers on baseline T2-weighted MRI are associated with pathologic complete response to neoadjuvant chemoradiation: a multisite study. J. Magn. Reson. Imaging 52, 1531–1541 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cha KH et al. Bladder cancer treatment response assessment in CT using radiomics with deep-learning. Sci. Rep. 7, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fang M et al. Multi-habitat based radiomics for the prediction of treatment response to concurrent chemotherapy and radiation therapy in locally advanced cervical cancer. Front. Oncol. 10, 563 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jiang Y et al. Development and validation of a deep learning CT signature to predict survival and chemotherapy benefit in gastric cancer: a multicenter, retrospective study. Ann. Surg. 10.1097/SLA.0000000000003778 (2020). [DOI] [PubMed] [Google Scholar]
- 128.Mehta S et al. Radiogenomics monitoring in breast cancer identifies metabolism and immune checkpoints as early actionable mechanisms of resistance to anti-angiogenic treatment. EBioMedicine 10, 109–116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kunte S et al. Radiomics risk score (RRS) on CT to predict survival and response to CDK 4/6 inhibitors in hormone receptor (HR) positive metastatic breast cancer (MBC). J. Clin. Oncol. 38, e13041–e13041 (2020). [Google Scholar]
- 130.Aerts HJWL et al. Defining a radiomic response phenotype: a pilot study using targeted therapy in NSCLC. Sci. Rep. 6, 33860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sun R et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 19, 1180–1191 (2018). [DOI] [PubMed] [Google Scholar]
- 132.Trebeschi S et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann. Oncol. 30, 998–1004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yang J et al. in Medical Image Computing and Computer Assisted Intervention Vol. 12262 (eds Martel AL et al.) 211–220 (2020). [Google Scholar]
- 134.Tunali I et al. Novel clinical and radiomic predictors of rapid disease progression phenotypes among lung cancer patients treated with immunotherapy: an early report. Lung Cancer 129, 75–79 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wu J et al. Unsupervised Clustering of quantitative image phenotypes reveals breast cancer subtypes with distinct prognoses and molecular pathways. Clin. Cancer Res. 23, 3334–3342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu J et al. Magnetic resonance imaging and molecular features associated with tumor-infiltrating lymphocytes in breast cancer. Breast Cancer Res. 20, 101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Loi S et al. Tumor-infiltrating lymphocytes and prognosis: a pooled individual patient analysis of early-stage triple-negative breast cancers. J. Clin. Oncol. 37, 559–569 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rao A et al. A combinatorial radiographic phenotype may stratify patient survival and be associated with invasion and proliferation characteristics in glioblastoma. J. Neurosurg. 124, 1008–1017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang S et al. Predicting EgFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur. Respir. J. 53, 1800986 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mu W et al. Non-invasive decision support for NSCLC treatment using PET/CT radiomics. Nat. Commun. 11, 5228 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang L et al. Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur. Radiol. 28, 2058–2067 (2018). [DOI] [PubMed] [Google Scholar]
- 142.Golia Pernicka JS et al. Radiomic-based prediction of microsatellite instability in colorectal cancer at initial computed tomography evaluation. Abdom. Radiol. 44, 3755–3763 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu S et al. CT textural analysis of gastric cancer: correlations with immunohistochemical biomarkers. Sci. Rep. 8, 11844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Park SH & Han K Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology 286, 800–809 (2018). [DOI] [PubMed] [Google Scholar]
- 145.Clark K et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26, 1045–1057 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sheller MJ et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci. Rep. 10, 12598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Traverso A, Wee L, Dekker A & Gillies R Repeatability and reproducibility of radiomic features: a systematic review. Int. J. Radiat. Oncol. Biol. Phys. 102, 1143–1158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lambin P et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 14, 749–762 (2017). [DOI] [PubMed] [Google Scholar]
- 149.Park JE et al. Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement. Eur. Radiol. 30, 523–536 (2020). [DOI] [PubMed] [Google Scholar]
- 150.Khorrami M et al. Stable and discriminating radiomic predictor of recurrence in early stage non-small cell lung cancer: multi-site study. Lung Cancer 142, 90–97 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Johnson WE, Li C & Rabinovic A Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 152.Orlhac F, Frouin F, Nioche C, Ayache N & Buvat I Validation of a method to compensate multicenter effects affecting CT radiomics. Radiology 291, 53–59 (2019). [DOI] [PubMed] [Google Scholar]
- 153.Kumar V et al. Radiomics: the process and the challenges. Magn. Reson. Imaging 30, 1234–1248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zwanenburg A et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295, 328–338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rudin C Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 1, 206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.London AJ Artificial intelligence and black-box medical decisions: accuracy versus explainability. Hastings Cent. Rep. 49, 15–21 (2019). [DOI] [PubMed] [Google Scholar]
- 157.Holzinger A, Langs G, Denk H, Zatloukal K & Müller H Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 9, e1312 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.US Food and Drug Administration. MammaPrint 510(k) premarket notification. FDA; https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpmn/pmn.cfm?ID=k070675 (2007). [Google Scholar]
- 159.US Food and Drug Administration. FDA releases artificial intelligence/machine learning action plan. FDA; https://www.fda.gov/news-events/press-announcements/fda-releases-artificial-intelligencemachine-learning-action-plan (2021). [Google Scholar]
- 160.Institute of Medicine. Policy Issues in the Development of Personalized Medicine in Oncology: Workshop Summary (National Academies, 2010). [PubMed] [Google Scholar]
- 161.US Food and Drug Administration. Discussion paper on laboratory developed tests (LDTs) (FDA, 2017). [Google Scholar]
- 162.Nakasu S, Onishi T, Kitahara S, Oowaki H & Matsumura K CT Hounsfield unit is a good predictor of growth in meningiomas. Neurol. Med. Chir. 59, 54–62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Urata M et al. Computed tomography Hounsfield units can predict breast cancer metastasis to axillary lymph nodes. BMC Cancer 14, 54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Galloway MM Texture analysis using gray level run lengths. Comput. Graph. Image Process. 4, 172–179 (1975). [Google Scholar]
- 165.Wang L & He D-C Texture classification using texture spectrum. Pattern Recognit. 23, 905–910 (1990). [Google Scholar]
- 166.Fogel I & Sagi D Gabor filters as texture discriminator. Biol. Cybern. 61, 103–113 (1989). [Google Scholar]
- 167.Chen SS, Keller JM & Crownover RM On the calculation of fractal features from images. IEEE Trans. Pattern Anal. Mach. Intell. 15, 1087–1090 (1993). [Google Scholar]
- 168.Kontos D et al. Radiomic phenotypes of mammographic parenchymal complexity: toward augmenting breast density in breast cancer risk assessment. Radiology 290, 41–49 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yang J et al. Integrating tumor and nodal radiomics to predict lymph node metastasis in gastric cancer. Radiother. Oncol. 150, 89–96 (2020). [DOI] [PubMed] [Google Scholar]
- 170.Bullitt E et al. Abnormal vessel tortuosity as a marker of treatment response of malignant gliomas: preliminary report. Technol. Cancer Res. Treat. 3, 577–584 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Alilou M et al. Quantitative vessel tortuosity: a potential CT imaging biomarker for distinguishing lung granulomas from adenocarcinomas. Sci. Rep. 8, 1–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wu C, Pineda F, Hormuth DA, Karczmar GS & Yankeelov TE Quantitative analysis of vascular properties derived from ultrafast DCE-MRI to discriminate malignant and benign breast tumors. Magn. Reson. Med. 81,2147–2160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


