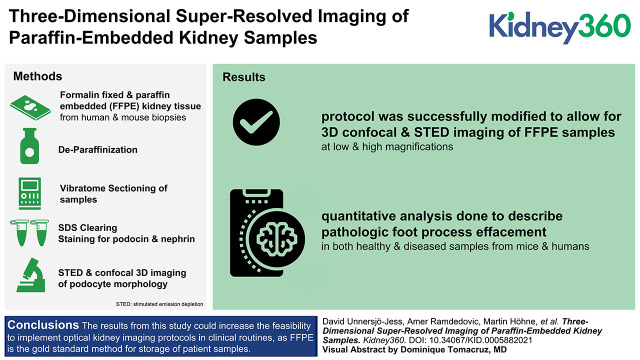
Key Points
Our previous protocols for 3D super-resolution kidney imaging have not been optimized to be compatible with paraffin-embedded samples.
This study overcomes these limitations, allowing 3D super-resolved imaging in FFPE kidney blocks.
This advancement opens up for 3D super-resolution kidney imaging of biobank material and in clinical settings.
Keywords: CKD, diagnostic imaging, foot processes, microscopy, paraffin embedding, renal pathology routine diagnostic tests
Visual Abstract
Abstract
Background
Diseases of the glomeruli, the renal filtration units, are a leading cause of progressive kidney disease. Assessment of the ultrastructure of podocytes at the glomerular filtration barrier is essential for diagnosing diverse disease entities, providing insight into the disease pathogenesis, and monitoring treatment responses.
Methods
Here we apply previously published sample preparation methods together with stimulated emission depletion and confocal microscopy for resolving nanoscale podocyte substructure. The protocols are modified and optimized in order to be applied to formalin-fixed paraffin-embedded (FFPE) samples.
Results
We successfully modified our protocols to allow for deep three-dimensional stimulated emission depletion and confocal imaging of FFPE kidney tissue with similar staining and image quality compared with our previous approaches. We further show that quantitative analysis can be applied to extract morphometrics from healthy and diseased samples from both mice and humans.
Conclusions
The results from this study could increase the feasibility of implementing optical kidney imaging protocols in clinical routines because FFPE is the gold-standard method for storage of patient samples.
Introduction
The three-layered kidney filtration barrier is responsible for filtering 180 L of primary urine each day. It consists of a fenestrated endothelium, a uniquely composed glomerular basement membrane (GBM), and an epithelial layer of podocytes, forming interdigitating structures called foot processes (FP) (1). In particular, the visualization of FP is of high importance to assess glomerular health because effacement (i.e., morphologic broadening and shortening) of FP is seen in almost all types of glomerular disease. Due to the tiny dimensions of these structures (width 200–1000 nm, spacing approximately 50 nm), the standard imaging modality used to visualize FP has previously been scanning or transmission electron microscopy (2). Although powerful in resolution, electron microscopy has drawbacks and limitations when it comes to nondestructive and straightforward three-dimensional (3D) imaging. Further, because electron microscopy produces grayscale contrast images of all constituents of the tissue, segmentation of structures of interest for further analysis can be challenging (3). In optical microscopy, however, tagging specific molecules with fluorescent dyes allows only structures of interest to be visualized, with no signal collected from other regions, thus substantially simplifying automated image segmentation. In the past decade, new optical imaging techniques have been developed for visualizing FP and other parts of the filtration barrier, such as the GBM (4). In 2016, it was shown for the first time that podocyte FP and the slit diaphragm (SD) can be resolved in situ in 3D using a combination of stimulated emission depletion (STED) microscopy and tissue clearing (5). It was also shown in this study that FP pathology (effacement) can be quantified in terms of the SD coverage. Further development, by our group and others, has made it possible to extract the SD coverage (6,7) and a plethora of other quantitative parameters describing effacement semi-automatically (7,8). Our group has recently shown that SD and FP morphometry can be extracted from fluorescence images in a completely automated manner by applying deep-learning segmentation, further establishing the quantitative advantages of optical microscopy for imaging FP and quantifying effacement (Butt L, Unnersjö-Jess D, Höhne M, et al.: Deep learning-based segmentation and quantification of podocyte foot process morphology. bioRxiv. 10.1101/2021.06.14.448284). Before this, moreover, we published a straightforward 3D imaging and quantitative pathologic analysis protocol for both murine and human samples that can be finished for full biopsy diagnostics in only 5 hours (8). All of the protocols previously developed by us are performed in optically cleared and thicker vibratome sections of fresh, fresh-frozen, or paraformaldehyde (PFA) fixed samples/biopsies. Other than the obvious advantage of 3D capacity, which gives access to a much larger volume for imaging and analysis, these protocols have proven highly superior to noncleared samples when it comes to achieving high labeling quality and contrast (5,8). The focus of this study is to investigate how our clearing and swelling protocols perform with paraffin-embedded tissue/biopsies. This will be of importance in many research or pathology laboratories because paraffin embedding is an old standard of storing tissue for later analysis. We show that by slightly modifying our protocols, super-resolved STED and confocal 3D imaging of podocyte morphology can readily be performed in formalin-fixed paraffin-embedded (FFPE) samples. This broadens the use of our fast and simple clearing and swelling protocols for renal research, especially for accelerated use in clinical pathology.
Materials and Methods
Murine and Human Kidney Tissue
All mouse experiments were approved by the State Office of North Rhine-Westphalia, Department of Nature, Environment and Consumer Protection (LANUV NRW, Germany), and were performed in accordance with European, national, and institutional guidelines. Mice of 100% C57BL/6N background were used. After anesthesia with ketamine and xylazine, mice were euthanized by cardiac perfusion with HBSS and fixated as described below.
Patient material was obtained from one child suffering from steroid-resistant nephrotic syndrome due to compound-heterozygous point mutations in TRPC6 and NPHS2. Control human tissue was collected from a patient who was nephrectomized due to renal carcinoma. Tissue sample was dissected from the nontumorous pole of the kidney and showed a normal histologic picture in routine histologic examination. All procedures were approved by the Ethics Commission of Cologne (DRKS00024517) and conducted in accordance with the Declaration of Helsinki. The patient or the patient’s parents gave informed consent.
Murine Model for FSGS
Mice with two compound-heterozygous point mutations, PodR231Q/A286V, were generated as previously described (7). Mice were euthanized at 20 weeks of age as stated above.
Fixation and Paraffin Embedding
Mouse kidneys were fixed in 4% PFA in 1× PBS at room temperature for 1–3 hours. Healthy kidney tissue of human control patients was obtained from the healthy contralateral pole of the renal carcinoma and was fixed in 4% PFA in 1× PBS overnight. Murine and human tissue was then paraffin embedded using standard procedures.
Deparaffinization
Pieces of cortex approximately 1–2-mm-thick were cut from the paraffin block using a scalpel. Alternatively, samples were sectioned to a thickness of 10 µm using a microtome and mounted on glass slides. Paraffin was dissolved by incubating the samples in Xylene overnight (three times for 5 minutes for thin sections). After this, the samples were rehydrated by incubation in 100% EtOH three times for 1 hour (three times for 3 minutes for thin sections), 96% EtOH twice for 1 hour (twice for 2 minutes for thin sections), and 70% EtOH for 1 hour (2 minutes for thin sections), and then washed in PBS for 10 minutes (twice for 1 minute for thin sections).
Optical Clearing
Samples were prepared according to a slightly modified version of a previously published fast and simple protocol (8). Samples were sectioned to 200 µm sections using a vibratome and then delipidated in clearing solution for 2 hours at 70°C–80°C (depending on age and species; this has to be optimized for each application). For thin sections, sections were gently removed from glass slides using a razor blade and then cleared for 1 hour at 80°C free-floating in an Eppendorf tube.
Labeling
PBS with Tween 20 (PBST) was used as antibody diluent for murine samples, and HEPES-TCS buffer (10 mM HEPES, pH 7.5, with 200 mM NaCL and 10% TritonX-100) was used for human samples. Samples were incubated in primary antibody at 37°C for 2–24 hours, washed once in antibody diluent for a minimum of 5 minutes, and then incubated in secondary antibody at 37°C for 2–24 hours. Samples were then washed in antibody diluent for a minimum of 5 minutes before mounting. Antibodies used were a rabbit polyclonal antibody to podocin (P0372; Sigma–Aldrich) used at a dilution of 1:100, sheep polyclonal antibody to nephrin (AF4269; R&D systems) used at a dilution of 1:50, and a rabbit polyclonal antibody to collagen IV (ab256353; Abcam). Secondary antibodies used were a donkey anti-rabbit (711–005–152; Jackson ImmunoResearch) conjugated to Atto-594 and a donkey anti-sheep (713–005–147; Jackson ImmunoResearch) conjugated to either Alexa-488 (human samples) or Abberior STAR635P (murine samples). The secondary antibodies were conjugated to dyes in-house as previously described (8). The collagen IV primary antibody above was directly conjugated to Alexa-555 in-house as previously described (8).
For thin sections, PBST supplemented with 3% BSA and 5% normal donkey serum was used as antibody diluent, and PBST was used for washing steps. After deparaffinization, a sheep antinephrin and a rabbit antipodocin (same antibodies as above) antibody was added at dilutions of 1:200 and 1:400, respectively, and incubated at room temperature for 3 hours. After this, sections were washed five times for 2 minutes at room temperature. A donkey anti-rabbit Atto-594 and a donkey anti-sheep Abberior STAR635P antibody was added, both at a 1:400 dilution, and incubated at 3 hours at room temperature.
Mounting
Samples were incubated in 80% wt/wt fructose (1 ml dH2O added to 4 g of fructose, 0.5% 1-thioglycerol was added to inhibit the Maillard reaction) with (human samples) or without (murine samples) 4 M urea at 37°C with shaking at 500 rpm for 15 minutes and then placed in a MatTek or Ibidi dish along with a few drops of fructose solution with a coverslip top (to prevent evaporation) before imaging. Thin sections on slides were mounted in a drop of 80% wt/wt fructose solution (see above).
Imaging
Images were acquired using a Leica SP8 STED 3× system. Objectives and other relevant imaging parameters (such as pinhole diameter) are indicated in the figure legends. For STED imaging, a 300-mW pulsed 775-nm laser was used at 30% power (STAR 635P) and 80% power (Atto594). For excitation, a 0.3-mW white light laser was used at varying intensities between 0.1% and 10%, depending on labeling target and dye.
Image Processing and Analysis
Images were smoothened (unless other stated) by replacing each pixel value by the mean of its 3 × 3 neighborhood using Fiji/ImageJ before inclusion in figures. Details regarding the ImageJ macro and extracted morphometric parameters can be found elsewhere (7) (Butt L, Unnersjö-Jess D, Höhne M, et al.: Deep learning-based segmentation and quantification of podocyte foot process morphology. bioRxiv. 10.1101/2021.06.14.448284).
Schemes
The scheme in Figure 1A was produced by D. Unnersjö-Jess using BioRender.
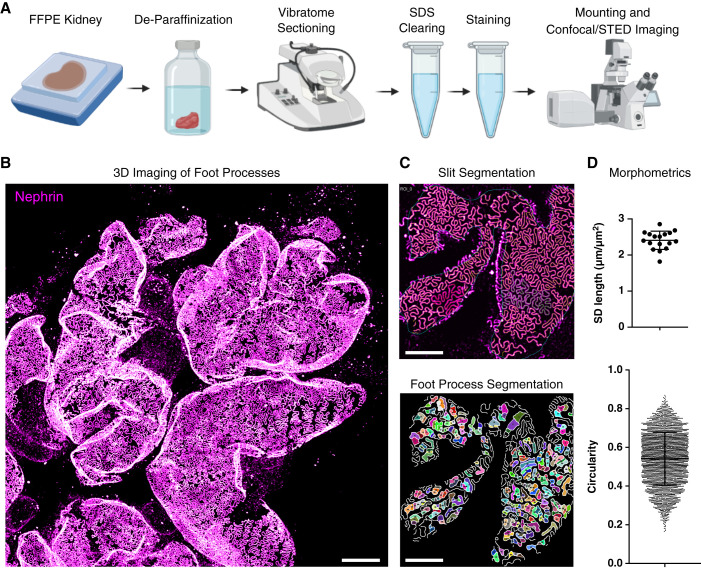
Figure 1.
Fast sample preparation and imaging protocol for the visualization of foot processes (FP) in formalin-fixed paraffin-embedded (FFPE) kidney tissue. (A) Overview of the sample preparation and imaging protocol. (B–D) This protocol allows for fast, high-throughput confocal imaging of FP on glomerular capillaries (B), with semi-automatic segmentation of the slit diaphragm (SD) and FP (C) and quantification of morphometry (D). The images and plots are illustrative examples; see Figures 2–4 for detailed results. Scale bars: 10 µm (B), 5 µm (C).
Results
Optical Clearing and Mild Swelling of Kidney Tissue
The general workflow is shown schematically in Figure 1A. We used our recently published fast clearing and swelling protocol (8) as a starting point, adding a deparaffinization step before vibratome sectioning. Samples were treated according to a slightly modified simple clearing protocol, and the podocyte substructure could be imaged using either confocal or STED microscopy with subsequent segmentation and morphologic analysis (Figure 1B–D). One of the most important advantages of our previously published protocols is the access to the depth dimension, and we therefore wanted to explore if the depth dimension would also be accessible in FFPE tissue. To this end, pieces of kidney tissue were cut out of the paraffin block using a razor blade and were then deparaffinized using standard deparaffinization protocols, utilizing Xylene for dissolving paraffin followed by rehydration in decreasing concentrations of ethanol in water. After this, samples were cut into sections 200-µm-thick using a vibratome. Samples were then delipidated using SDS, and different clearing times and temperatures were tried (Supplemental Figure 1). Compared with nonparaffinized PFA or formalin-fixed samples, slightly longer clearing times and higher temperatures were needed for sufficient clearing (Supplemental Figure 1, A and B). Of note is that the nephrin staining, the basis for later quantitative analysis, showed sufficient signal after only 15 minutes of clearing, whereas podocin needed a longer clearing time for sufficient antigenicity (Supplemental Figure 1A). Thus, if only nephrin staining is used, the protocol can be shortened. Standard antigen retrieval by boiling in Tris-EDTA was tried, resulting in slightly lower staining quality and less optical transparency, and we thus decided to keep the SDS clearing step (Supplemental Figure 1C). On the basis of the results in Supplemental Figure 1, we established an SDS clearing step of 2 hours at 80°C as a starting point. However, as with all clearing protocols (and also with standard antigen retrieval on thin sections), temperature and time need to be optimized on the basis of tissue type, fixation time and conditions, different epitopes/antibodies, etc. Our final protocol resulted in a linear swelling of around 25%, slightly lower than earlier reported in non-FFPE samples (8) (Supplemental Figure 1D).
Visualizing FP and the SD using STED and Confocal Microscopy
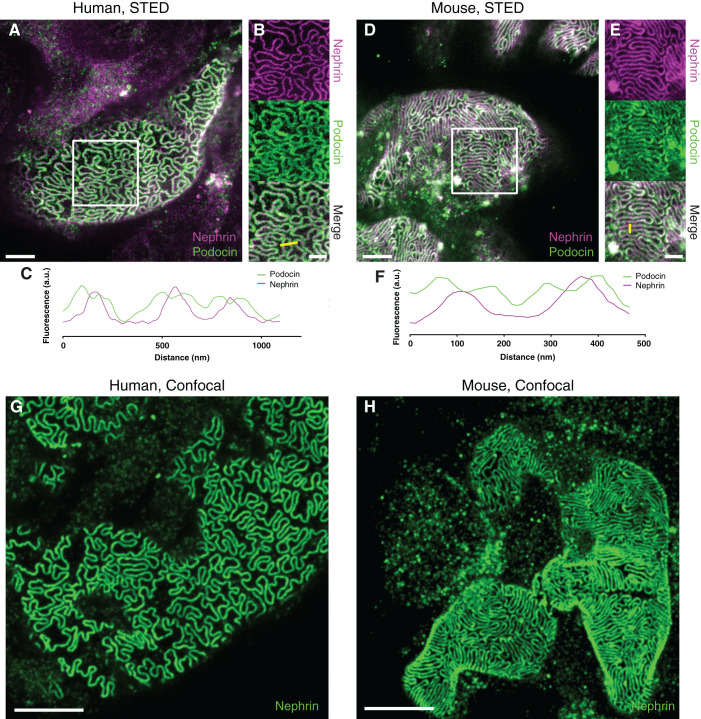
Next, by staining for podocin and nephrin, we validated that FP in both murine and human tissue could be clearly resolved using STED microscopy (Figure 2, A and D). Importantly, the expression of podocin on both sides of the slit could be resolved (Figure 2, B, C, E, and F), which validates that the resolution achieved with this FFPE-optimized protocol is similar compared with our previous, more tedious, STED-imaging protocol (5). In our latest fast and simple protocol, we show that the podocyte substructure can be resolved using confocal microscopy if samples are embedded in a fructose solution containing 4 M urea in order to induce slight swelling in the sample (8). We show that this approach can also be applied to FFPE tissue by demonstrating that FP can clearly be resolved in both human and murine samples (Figure 2, G and H).
Figure 2.
Validation of the protocol for imaging podocyte ultrastructure in human and murine FFPE tissue samples. All samples were imaged using a Leica SP8 3× STED microscope with a 100×/1.4NA oil objective. All images are maximum intensity projections of z-stacks approximately 2-µm-thick acquired at depths of 0–20 µm. Samples were stained for nephrin using either Abberior STAR-635P (A–E) or Alexa-488 (G and H) and for podocin using Atto-594 (A–E). Scale bars: 2 µm (A and D), 1 µm (B and E), 5 µm (G and H). (A) A human control sample treated according to the protocol shows that FP are clearly outlined by staining for the SD. (B) Single-channel views of the area outlined by a white rectangle in (A) shows that the two-sided expression of podocin on both sides of the SD can be resolved. (C) A line profile along the yellow line in (B) shows that podocin is flanking the nephrin signal in three SDs with a separation of the podocin signal of around 80–100 nm. (D) In addition, in a wild-type mouse sample, FP are clearly outlined by staining for the SD. (E) Single-channel views of the area outlined by a white rectangle in (D) shows that the two-sided expression of podocin on both sides of the SD can also be resolved in the murine sample. (F) A line profile along the yellow line in (E) shows that podocin is flanking the nephrin signal in two SDs, also here with a separation of the podocin signal of around 80–100 nm. (G and H) Confocal images of human (G) and murine (H) tissue, showing that FP morphology can be resolved with diffraction-limited microscopy by using a 1.4 NA 100× objective with the green-emitting dye Alexa-488 and a confocal pinhole size of 0.3 airy units.
We next applied our validated clearing and swelling protocol to clinical samples. Occasionally, such as when retrieving samples from a biobank or a pathology laboratory, the only option is to obtain thin (<10 µm) paraffin sections on glass slides. Thus, we investigated if our protocol could also be applied to such samples. Applying a standard Tris-EDTA antigen retrieval step before staining with the simplified staining protocol used for cleared sections (8) did not result in sufficient staining quality to resolve FP (Supplemental Figure 2A). Interestingly, we found that removal of the section from the glass slide using a razor blade and performing the staining free-floating substantially increased staining quality (Supplemental Figure 2B). However, it was only after adding a 1-hour delipidation step at 80°C that sufficient staining quality could be achieved to resolve the SD by staining for podocin (Supplemental Figure 2, C and D). Importantly, we did manage to achieve sufficient staining quality in sections mounted on a glass slide by extending the length of the Tris-EDTA boil to 40 minutes and by applying a more laborious standard immunohistochemistry staining protocol, using blocking with BSA/NDS and by adding several washing steps between and after antibody incubations (Supplemental Figure 2F). However, we were not able to resolve the SD when staining for podocin in sections still mounted on a glass slide (Supplemental Figure 2G). The results from Supplemental Figure 2 further establish what has been shown previously—that SDS delipidation of thick sections results in higher staining quality, and that this staining quality can be achieved also by using a drastically simplified staining protocol with no blocking and very little washing (5,8).
Quantifying Glomerular Nanoscale Pathology in FFPE Samples from Mice and Humans
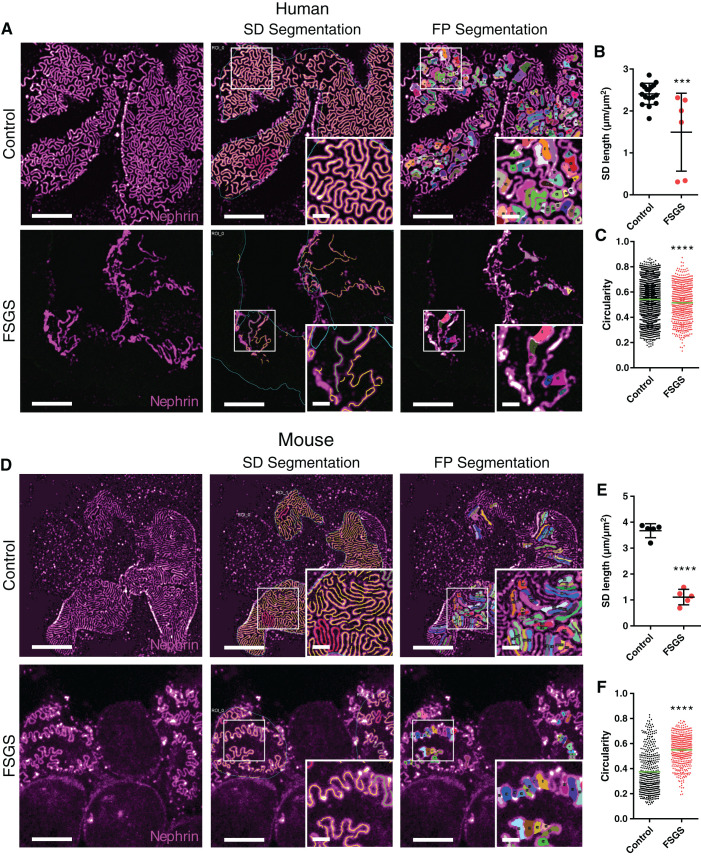
We further went on to validate that by applying our previously published analysis approaches, we could quantitatively describe pathologic FP effacement in both humans and mice. To do this, we applied the protocol to FFPE tissue from a patient with trans-associated mutations in TRPC6 and NPHS2, and to mice with compound-heterozygous mutations in the nphs2 gene (R231Q/A286V), both leading to an FSGS phenotype (Figure 3). Using a previously published ImageJ macro (7), we show that both the filtration slit pattern and individual FP could be segmented from images acquired using confocal microscopy (Figure 3, A and D). From these segmentations, the SD length per area (Figure 3, B and E) and FP circularity (Figure 3, C and F) could be extracted. The results for the murine FSGS model are in line with what has previously been published (7), with a significantly lower SD length and a significantly increased FP circularity compared with controls (Figure 3, E and F). A decrease in SD length was also observed in the patient (Figure 3B), whereas the increased circularity seen in the murine FSGS model was not observed. Instead, a decrease in circularity was observed for the patient with genetic FSGS (Figure 3C). In addition, for other morphometric parameters, except for FP area, results for FSGS mice and the FSGS patient showed opposite significant differences (Supplemental Figure 3). One reason for this variability could be that the control human sample was taken from a very old patient (>70 years old), whereas the FSGS sample was taken from a young child (<1 year old), and mouse samples were age matched. Moreover, we have recently shown that different disease types can present with different morphologic effacement “fingerprints,” and thus effacement patterns might differ between subtypes of FSGS (Butt L, Unnersjö-Jess D, Höhne M, et al.: Deep learning-based segmentation and quantification of podocyte foot process morphology. bioRxiv. 10.1101/2021.06.14.448284). Even though the segmentation and analysis work well in images acquired with confocal microscopy, we further demonstrate that as a result of the higher spatial resolution, the ImageJ macros perform slightly better in images acquired with STED microscopy (Supplemental Figure 4). As part of the SD density macro, the user must define the capillary surface area in which the analysis is to be carried out. This is not always straightforward, especially in capillary regions with severe FP effacement, and might lead to erroneous results. We found that staining the GBM using a collagen IV antibody enabled us to define the region of interest automatically and accurately without user intervention (Supplemental Figure 5).
Figure 3.
Imaging and segmentation/morphometry of the SD/FP pattern in healthy and diseased human and murine FFPE tissue imaged with confocal microscopy. All samples were stained for nephrin using Alexa-488 and imaged using a 100× NA 1.4 oil objective with a confocal pinhole setting of 0.3 AU. Scale bars: 5 µm and 1 µm (zoomed insets). (A) Maximum intensity projections of z-stacks approximately 2-µm-thick, showing the global slit pattern in a control human sample and a sample from a patient diagnosed with FSGS. Insets are zooms of the area marked with a white square, showing effacement in higher detail. (B) SD length per capillary area in control and FSGS patients, showing a significant decrease for the FSGS patient. Each dot represents one image. Black line represents the mean, and error bars represent SD. Two-tailed t test, P=0.001. (C) FP circularity score for control and FSGS patients, showing an unexpected but significant decrease in circularity score for the FSGS patient. Each dot represents one FP, and the green line represents the mean. Two-tailed t test, P<0.0001. (D) Maximum intensity projections of z-stacks approximately 2-µm-thick, showing the global SD pattern in a wild-type (WT) murine sample and a sample from a mouse with genetic FSGS. Insets show zooms of the area marked with a white square, showing effacement of FP in the FSGS mouse in higher detail. (E) SD length per capillary area in WT and FSGS mice, showing a significant decrease for the mutated mouse. Each dot represents one image. Black line represents the mean, and error bars represent SD. Two-tailed t test, P<0.0001. (F) FP circularity score for WT and FSGS mice, showing a significant increase in circularity score for the FSGS mouse. Each dot represents one FP, and the green line represents the mean. Two-tailed t test, P<0.0001.
Three-Dimensional Confocal and STED Imaging of Thick Samples
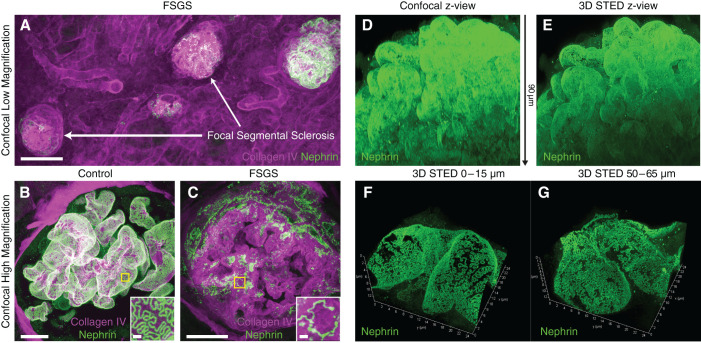
We have previously shown that our optical clearing protocols allow for both confocal and STED imaging deep inside intact kidney tissue (5,8,9). In addition, with our FFPE-optimized protocol, we validated that thick samples can be imaged in 3D at both low and high magnifications (Figure 4, A–C). By mixing the fructose-embedding medium at a lower concentration, the refractive index can be freely adjusted to 1.45, perfectly matching a longer working distance glycerol objective lens. With this approach, confocal imaging is possible at depths of at least 120 µm (Supplemental Figure 6) in cleared FFPE samples, although the lower NA of the glycerol objective lens of 1.3 results in slightly lower lateral and axial resolution compared with the 1.4 NA oil objective otherwise used. Although sufficient lateral resolution for resolving FP can be achieved with a confocal microscope, the z-resolution is still not sufficient to resolve FP when projecting x-z or y-z directions on capillaries (Figure 4D). However, by applying 3D STED, which increases the axial resolving power by a factor of 5, sufficient isotropic resolution is reached for resolving FP nanoscale morphology in any spatial orientation (Figure 4, E–G, and Supplemental Movie 1). We have thus demonstrated that our in situ 3D imaging capacity shown with previous protocols is transferable to clinically stored FFPE tissue samples as well.
Figure 4.
Three-dimensional confocal and STED imaging in human samples (same samples as in Figure 3). All images acquired using a Leica SP8 confocal system using either a 20× NA 0.75 oil (A), a 100× NA 1.4 oil objective, or a 93× NA 1.3 glycerol motCorr objective (B-C). Samples were stained for nephrin with either Alexa-488 (A–C) or Abberior STAR635P (D-G). (A) Maximum intensity projection of a z-stack approximately 200-µm-thick in the sample from the FSGS patient, showing clear focal and segmental sclerosis of glomeruli (high collagen expression and low nephrin expression). Scale bar: 100 µm. (B and C) Maximum intensity projections of confocal z-stacks approximately 100-µm-thick of control (B) and FSGS (C) patients, showing apparent effacement of FP in a sclerosed glomerulus of the FSGS patient. Insets show zooms of areas marked with yellow squares. Scale bars: 10 µm and 1 µm (zoomed insets). (D and E) Maximum intensity projections (x-z views) of confocal (D) and 3D STED (E) stacks of the same glomerulus. The z-depth is indicated with the arrow between the images. See Supplemental Video 1 for better visualization of the data. (F and G) Maximum intensity projections of 3D STED stacks of glomerular capillaries in the control patient, showing that FP can be clearly resolved at depths of at least 65 µm. Stacks are tilted to show the high z-resolution provided by the STED depletion in the z-dimension. Dimensions are indicated on the axes.
Discussion
In the last years, we have shown that by applying novel sample preparation technologies, superb staining quality can be achieved when imaging the podocyte substructure in intact kidney tissue. Apart from higher signal-to-noise ratio and higher labeling density, the protocols also allow for 3D visualization of FP morphology in health and disease. The protocols have proven to be superior for later semi-automatic and automatic segmentation to extract quantitative parameters describing podocyte morphology (7) (Butt L, Unnersjö-Jess D, Höhne M, et al.: Deep learning-based segmentation and quantification of podocyte foot process morphology. bioRxiv. 10.1101/2021.06.14.448284). By modifying these protocols, here we show that FFPE tissue can also be treated according to our protocol to achieve similar imaging results as with samples not embedded in paraffin. We also validated that our previously developed image analysis/quantification strategies can be applied to FFPE samples, with correlating results. In FFPE tissue, we found that SDS delipidation acts not only to increase tissue transparency, but also as an antigen retrieval step, with increasing epitope availability upon increased clearing time. We further show that the increase in staining quality can be partially attributed to the fact that samples are stained in a free-floating manner because an identical staining protocol applied to an FFPE section mounted on a glass slide resulted in substantially lower staining quality. However, as has also been demonstrated previously (6,10,11), we show that it is also possible to achieve a decent staining quality in sections mounted on glass slides by applying a longer antigen retrieval step and a more tedious labeling protocol with extensive washing and blocking. For maximal staining quality and effective resolution, FFPE tissue samples had to be cleared and stained in a free-floating manner. We thus show that apart from the obvious disadvantage of having access to less in situ 3D volume for imaging, thin FFPE sections mounted on slides are suboptimal for visualizing and segmenting podocyte substructure.
Taken together, findings in this study show that our fast and simple clearing protocols can also be applied to FFPE tissue blocks, which has its main effect when it comes to the study of human clinical samples. In most clinical pathology laboratories, it is standard procedure to embed biopsies in paraffin before thin sectioning and histologic evaluation. Thus, the threshold for implementing our clearing protocols in a clinical pathology workflow is drastically lowered. It should be pointed out that the long deparaffinization protocol applied here to tissue chunks several millimeters thick can be drastically shortened when deparaffinizing smaller samples, such as human biopsies. It should also be apparent that paraffinization and deparaffinization of tissue is still laborious and time-consuming, and there is thus no benefit in applying it for the purpose of imaging FP using optical microscopy if one has the choice. Moreover, our recently shown automatic multiparametric morphologic analysis of filtration structure morphology, on the basis of deep learning, gives additional information regarding differential patterns of effacement across different diagnoses (Butt L, Unnersjö-Jess D, Höhne M, et al.: Deep learning-based segmentation and quantification of podocyte foot process morphology. bioRxiv. 10.1101/2021.06.14.448284). By applying this analysis strategy to images provided by the protocol in this study with just classic confocal imaging, a faster and completely automated and bias-free evaluation of effacement in biopsies could possibly be implemented in clinical pathology in the future. Finally, it should be mentioned that other protocols exist for imaging FP in FFPE samples, but all of these super-resolution approaches are applied to thin sections, with their aforementioned disadvantages (6,12). In addition, we have previously shown that all of these published protocols, including FFPE protocols presently used clinically, require more reagents and user interactions compared with the protocol applied in this study. We therefore believe that our fast and simple protocol, which can be carried out with a conventional confocal microscope, could prove beneficial for use in routine kidney pathology in the future.
Disclosures
T. Benzing reports advisory activity for Otsuka in the field of cystic kidney disease and hyponatremia; support for ADPKD registry by Otsuka; speaker honoraria and travel support from Amgen, Hexal, Novartis, Otsuka, Roche, and Sanofi-Genzyme. He is also on the editorial board of the Journal of the American Society of Nephrology, Nephrology Dialysis Transplantation, and Science Signaling. H. Blom reports other interests/relationships with MedTechLabs, BioClinicum, Karolinska University Hospital, Solna, Sweden. P. Hoyer reports consultancy agreements with Boehringer Ingelheim and is a scientific advisor for Archives of Disease in Childhood. F. Koehler reports other interests/relationships with Else Kröner-FreseniusStiftung, German Research Foundation under Germany’s Excellence Strategy EXC 2030: CECAD—Excellent in Aging Research and Koeln Fortune program/Faculty of Medicine, University of Cologne, Germany. R. Mueller reports consultancy agreements with Alnylam and Sanofi; research funding from Otsuka Pharmaceuticals and Thermo Fisher Scientific, which was paid to the employer (Department II of Internal Medicine); and honoraria from Alnylam and Sanofi. He is also a board member of the WGIKD (ERA-EDTA), Scientific Advisory Board Santa Barbara Nutrients, Editorial Board “Kidney and Dialysis.” All remaining authors have nothing to disclose.
Funding
This study was supported by the Clinical Research Unit (CRU) 329 of the Deutsche Forschungsgemeinschaft ([DFG]; Project Nos.: SCHE 1562/7-1 to B.S. and BE 2212/23-1 + BE 2212/24-1 to T.B.) and partly by FOR 2743 of DFG (Project No.: BE 2212/25-1 + BE 2212/25-2), Else Kröner-Fresenius-Stiftung, and the Eva Luise und Horst Köhler Stiftung (Project No.: 2019_KollegSE.04); and by the German Research Foundation under Germany’s Excellence Strategy EXC 2030: CECAD—Excellent in Aging Research (Project No.: 390661388).
Acknowledgments
We thank the CECAD Imaging Facility for their support in the acquisition of microscopy data.
Portions of this article previously appeared as a preprint in bioRxiv as https://doi.org/10.1101/2021.09.02.458679.
Additional funding was received by the Koeln Fortune Program/Faculty of Medicine, University of Cologne, outside the conduct of this project.
Data Sharing Statement
Data cannot be shared. The data sets in this study are large image files. Specific data sets can be provided upon request.
Author Contributions
T. Benzing, H. Blom, and B. Schermer conceptualized the study; L. Butt, M. Hoehne, P. Hoyer, F. Koehler, R.-U. Mueller, A. Ramdedovic, and D. Unnersjoe-Jess curated the data; M. Hoehne, A. Ramdedovic, and D. Unnersjoe-Jess undertook the formal analysis; D. Unnersjoe-Jess was responsible for the investigation, methodology, and visualization, and wrote the original draft of the manuscript; and all authors were responsible for reviewing and editing the manuscript.
Supplemental Material
This article contains supplemental material online at http://kidney360.asnjournals.org/lookup/suppl/doi:10.34067/KID.0005882021/-/DCSupplemental.
Effect of different times and temperatures of the SDS delipidation step as evaluated by stimulated emission depletion (STED) imaging of the two slit diaphragm proteins nephrin and podocin. Download Supplemental Figure S1, PDF file, 4.5 MB (4.5MB, pdf)
Performance of our protocol on 10-µm paraffin sections. Download Supplemental Figure S2, PDF file, 4.5 MB (4.5MB, pdf)
Further morphometric parameters extracted from the patients/mice in Figure 3. Download Supplemental Figure S3, PDF file, 4.5 MB (4.5MB, pdf)
Representative comparison of performance of the ImageJ macro-based analysis on images acquired with confocal and STED microscopy. Download Supplemental Figure S4, PDF file, 4.5 MB (4.5MB, pdf)
Automatic assignment of region of interest for analyzing slit diaphragm length with the ImageJ macro based on a collagen IV staining. Download Supplemental Figure S5, PDF file, 4.5 MB (4.5MB, pdf)
Three-dimensional confocal imaging of a whole human glomerulus at imaging depths >100 µm. Download Supplemental Figure S6, PDF file, 4.5 MB (4.5MB, pdf)
3D renderings of STED and Confocal z-stacks of the same human glomerulus as in Figure 4, D–E. Download Supplemental Movie 1, AVI file, 62 MB (60.7MB, avi)
References
- 1.Pavenstädt H, Kriz W, Kretzler M: Cell biology of the glomerular podocyte. Physiol Rev 83: 253–307, 2003. 10.1152/physrev.00020.2002 [DOI] [PubMed] [Google Scholar]
- 2.Amann K, Haas CS: What you should know about the work-up of a renal biopsy. Nephrol Dial Transplant 21: 1157–1161, 2006. 10.1093/ndt/gfk037 [DOI] [PubMed] [Google Scholar]
- 3.Wanner AA, Kirschmann MA, Genoud C: Challenges of microtome-based serial block-face scanning electron microscopy in neuroscience. J Microsc 259: 137–142, 2015. 10.1111/jmi.12244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pullman JM: New views of the glomerulus: Advanced microscopy for advanced diagnosis. Front Med (Lausanne) 6: 37, 2019. 10.3389/fmed.2019.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unnersjö-Jess D, Scott L, Blom H, Brismar H: Super-resolution stimulated emission depletion imaging of slit diaphragm proteins in optically cleared kidney tissue. Kidney Int 89: 243–247, 2016. 10.1038/ki.2015.308 [DOI] [PubMed] [Google Scholar]
- 6.Siegerist F, Ribback S, Dombrowski F, Amann K, Zimmermann U, Endlich K, Endlich N: Structured illumination microscopy and automatized image processing as a rapid diagnostic tool for podocyte effacement. Sci Rep 7: 11473, 2017. 10.1038/s41598-017-11553-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butt L, Unnersjö-Jess D, Höhne M, Edwards A, Binz-Lotter J, Reilly D, Hahnfeldt R, Ziegler V, Fremter K, Rinschen MM, Helmstädter M, Ebert LK, Castrop H, Hackl MJ, Walz G, Brinkkoetter PT, Liebau MC, Tory K, Hoyer PF, Beck BB, Brismar H, Blom H, Schermer B, Benzing T: A molecular mechanism explaining albuminuria in kidney disease. Nat Metab 2: 461–474, 2020. 10.1038/s42255-020-0204-y [DOI] [PubMed] [Google Scholar]
- 8.Unnersjö-Jess D, Butt L, Höhne M, Witasp A, Kühne L, Hoyer PF, Patrakka J, Brinkkötter PT, Wernerson A, Schermer B, Benzing T, Scott L, Brismar H, Blom H: A fast and simple clearing and swelling protocol for 3D in-situ imaging of the kidney across scales. Kidney Int 99: 1010–1020, 2021. 10.1016/j.kint.2020.10.039 [DOI] [PubMed] [Google Scholar]
- 9.Unnersjö-Jess D, Scott L, Sevilla SZ, Patrakka J, Blom H, Brismar H: Confocal super-resolution imaging of the glomerular filtration barrier enabled by tissue expansion. Kidney Int 93: 1008–1013, 2018. 10.1016/j.kint.2017.09.019 [DOI] [PubMed] [Google Scholar]
- 10.Wunderlich LCS, Ströhl F, Ströhl S, Vanderpoorten O, Mascheroni L, Kaminski CF: Superresolving the kidney—A practical comparison of fluorescence nanoscopy of the glomerular filtration barrier. Anal Bioanal Chem 413: 1203–1214, 2021. 10.1007/s00216-020-03084-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ilgen P, Stoldt S, Conradi L-C, Wurm CA, Rüschoff J, Ghadimi BM, Liersch T, Jakobs S: STED super-resolution microscopy of clinical paraffin-embedded human rectal cancer tissue. PLoS One 9: e101563, 2014. 10.1371/journal.pone.0101563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bucur O, Fu F, Calderon M, Mylvaganam GH, Ly NL, Day J, Watkin S, Walker BD, Boyden ES, Zhao Y: Nanoscale imaging of clinical specimens using conventional and rapid-expansion pathology. Nat Protoc 15: 1649–1672, 2020. 10.1038/s41596-020-0300-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of different times and temperatures of the SDS delipidation step as evaluated by stimulated emission depletion (STED) imaging of the two slit diaphragm proteins nephrin and podocin. Download Supplemental Figure S1, PDF file, 4.5 MB (4.5MB, pdf)
Performance of our protocol on 10-µm paraffin sections. Download Supplemental Figure S2, PDF file, 4.5 MB (4.5MB, pdf)
Further morphometric parameters extracted from the patients/mice in Figure 3. Download Supplemental Figure S3, PDF file, 4.5 MB (4.5MB, pdf)
Representative comparison of performance of the ImageJ macro-based analysis on images acquired with confocal and STED microscopy. Download Supplemental Figure S4, PDF file, 4.5 MB (4.5MB, pdf)
Automatic assignment of region of interest for analyzing slit diaphragm length with the ImageJ macro based on a collagen IV staining. Download Supplemental Figure S5, PDF file, 4.5 MB (4.5MB, pdf)
Three-dimensional confocal imaging of a whole human glomerulus at imaging depths >100 µm. Download Supplemental Figure S6, PDF file, 4.5 MB (4.5MB, pdf)
3D renderings of STED and Confocal z-stacks of the same human glomerulus as in Figure 4, D–E. Download Supplemental Movie 1, AVI file, 62 MB (60.7MB, avi)