Summary
ATP-dependent chromatin-remodeling complexes (remodelers) modulate gene transcription by regulating the accessibility of highly packaged genomic DNA. However, the molecular mechanisms involved at the nucleosomal level in this process remain controversial. Here, we monitor the real-time activity of single ySWI/SNF or RSC complexes on single, stretched nucleosomal templates under tensions above 1 pN forces. We find that these remodelers can translocate along DNA at rates of ~13 bp/s and generate forces up to ~12 pN, producing DNA loops of a broad range of sizes (20–1200 bp, average ~100 bp) in a nucleosome-dependent manner. This nucleosome-specific activity differs significantly from that on bare DNA observed under low tensions and suggests a nucleosome-remodeling mechanism through intranucleosomal DNA loop formation. Such loop formation may provide a molecular basis for the biological functions of remodelers.
Introduction
The wrapping of DNA around histones and its further packaging into higher-order chromatin structures represent significant barriers for protein binding to DNA and generally inhibit DNA-related transactions in eukaryotes. Chromatin-remodeling factors, along with histone-modifying enzymes, dynamically alter chromatin structures and regulate the accessibility of genomic DNA (Workman and Kingston, 1998; Saha et al., 2006). Although they play diverse roles in chromatin metabolism and are involved in a variety of human diseases (Bochar et al., 2000), over 30 remodelers identified from different organisms share highly conserved SWI2/SNF2 ATPase domains (Workman and Kingston, 1998). Yeast SWI/SNF and RSC (Cairns et al., 1996; Smith et al., 2003a), with molecular weights around 1 million Da and containing 11 and 15 different subunits, respectively, are prototypes of such complexes.
Remodeling enzymes alone can make nucleosomal DNA more accessible in two ways: dynamic formation of large intranucleosomal DNA loops (e.g., ≥20 bp) and nucleosome sliding. Although nucleosome sliding has been shown to be a general activity of remodelers (Hamiche et al., 1999; Whitehouse et al., 1999; Schnitzler et al., 2001; Saha et al., 2005; Shundrovsky et al., 2006), it remains controversial whether large DNA loops are generated on histone surfaces during nucleosome remodeling. Early energetic consideration of nucleosome sliding suggests that no DNA segment needs to be detached from the histone surface in this process (van Holde and Yager, 2003). This view is supported by structural studies of a mononucleosome that contains 147 bp of DNA wrapped around the histone octamer, rather than the canonical 146 bp (Richmond and Davey, 2003). It has been suggested that the extra base pair accommodated by the nucleosome may reflect the structure of an intermediate in nucleosome sliding. Additional experimental evidence comes from nuclease digestion assays, which demonstrate that the predominant, if not the sole, pathway to expose nucleosomal DNA is through the sliding mechanism without significant DNA looping (Saha et al., 2005). However, other observations based on the same assay for the same SWI/SNF-family remodelers have been interpreted as resulting from loop generation (Fan et al., 2003; Lorch et al., 2005). Yet, most of these assays were performed using short DNA templates (<210 bp) and it has been demonstrated that the DNA ends have a strong effect on nucleosome structure upon remodeling of short templates (Lorch et al., 1998; Kassabov et al., 2003). Therefore, the existence of such DNA loops in the natural long nucleosomal array substrate remains to be established (Logie and Peterson, 1997; Shundrovsky et al., 2006). Moreover, even if DNA loops are generated, their mechanism of generation remains unclear. A looping (or loop recapture) model (Langst and Becker, 2001; Lorch et al., 2005; Strohner et al., 2005; Zofall et al., 2006) suggests that DNA loops are produced by DNA bending induced by and coupled to ATP-dependent large remodeler configurational changes (Fitzgerald et al., 2004). This model predicts a single loop size whose maximum is limited by the dimensions of the remodeler that are less than 27 nm, or ~80 bp (Smith et al., 2003a). However, a recent experiment performed on bare DNA showed that larger DNA loops can be formed (Lia et al., 2006). This observation is consistent with an alternative model (Saha et al., 2002, 2005) suggesting that remodeler ATPases are DNA translocases, molecular motors capable of moving along DNA using the energy of ATP hydrolysis. Interestingly, the same experiment also revealed that remodeler motors only generate DNA loops at opposing forces below ~1 pN, i.e., at forces significantly smaller than those generated by most molecular motors (Bustamante et al., 2004). How such bare DNA-associated remodeler translocation relates to the disruption of DNA-histone interactions and exposure of nucleosomal DNA is controversial (Saha et al., 2002, 2005; Lusser and Kadonaga, 2003; Strohner et al., 2005; Zofall et al., 2006). Although a model that connects translocation to possible intranucleosomal DNA loop formation has recently been proposed (Saha et al., 2006), it remains to be tested, and critical parameters, such as loop size, translocation velocity, and processivity have not been measured.
Results
Experimental Setup
To test for the occurrence of DNA looping and its possible molecular mechanism during nucleosome remodeling, we stretched a single nucleosomal template using optical tweezers (Figure 1A). Experiments were designed to monitor DNA end-to-end length changes in real time in response to the remodeling action of individual yeast SWI/SNF or RSC complexes on the template at a given time. The nucleosomal template was made by a salt dialysis method after mixing purified chicken erythrocyte histone octamer and DNA template with proper ratio (see Supplemental Experimental Procedures in the Supplemental Data available with this article online). The DNA template contains either four or six tandem repeats of a 258 bp nucleosome positioning sequence (‘‘601’’) (Lowary and Widom, 1998) in the middle of engineered pUC19 plasmid DNA (Supplemental Experimental Procedures). The total DNA length is 3750 bp for the template with four repeats, and 5040 bp for the one with six repeats. Three nucleosomal templates were mainly used that differ in average number of nucleosomes per template. The first one contains 2.1 (±1.0, SD) nucleosomes on the 3750 bp DNA molecule, and the second contains 4.3 (±2.3) nucleosomes on the 5040 bp DNA molecule. Because the number of nucleosomes varies from template to template even from the same reconstitution solution, a third type of nucleosomal template was selected from the above two templates that contain exactly one nucleosome per template. The average spacing between nucleosomes on these templates is initially >370 bp on the tandem repeats before adding SWI/SNF or RSC. It presumably increases by spreading to the whole DNA template due to remodeler-catalyzed nucleosome sliding when remodeler is introduced. The use of subsaturated nucleosomal arrays with large internucleosomal spacing minimizes the possible collision between nucleosomes during chromatin remodeling while maintaining sufficient signal occurrence frequency. Such collision may complicate our data interpretation and is therefore avoided in the experiments.
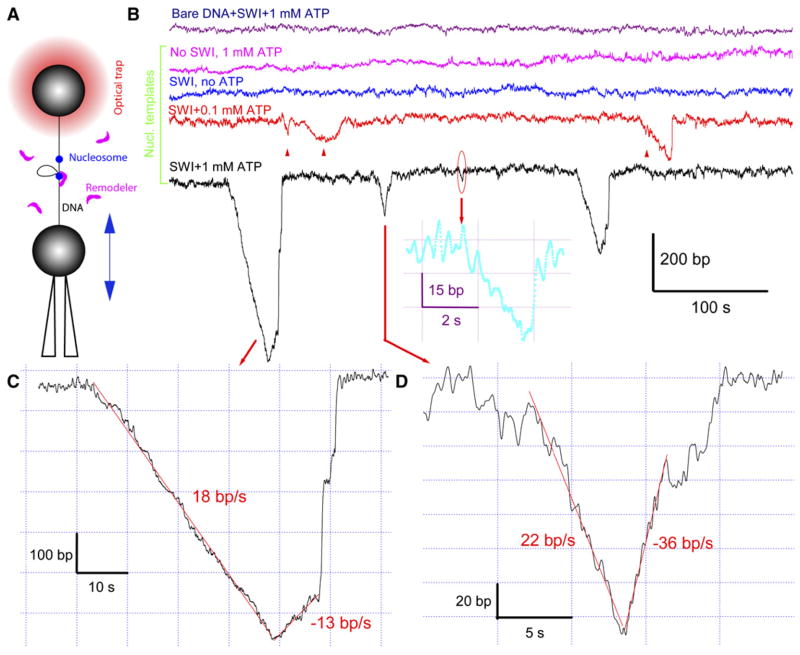
Figure 1.
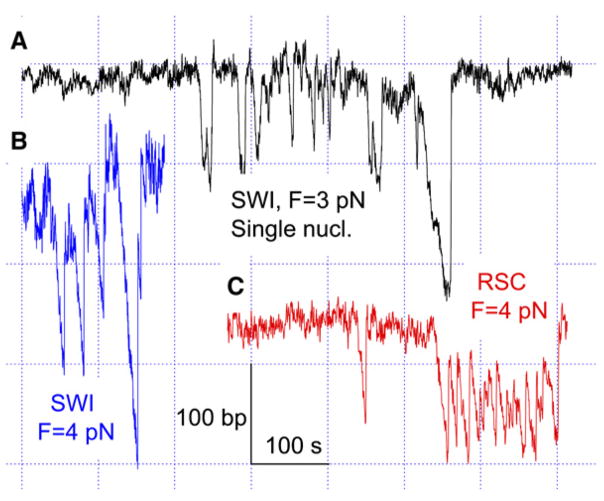
ATP- and Nucleosome-Dependent Translocation and Loop Formation by SWI/SNF at 3 pN Constant Tension
(A) Illustration of the setup used for the remodeling assay (not in scale). DNA ends were attached to two polystyrene beads, one kept in a force-measuring optical trap and another fixed on the tip of a micropipette by suction. The pipette bead can move relative to the trap center, thus maintaining a constant stretching force through a feedback mechanism.
(B) Time-dependent extension of either bare DNA in the presence of 4 nM SWI/SNF (SWI) and 1 mM ATP (purple trace) or nucleosomal template with no SWI/SNF (magenta) or with 4 nMSWI/SNF in the presence of 0mM(blue), 0.1mM(red), or 1mM(black) ATP. In comparison, the Michaelis constant (KM) for ATP is ~0.1 mM (Cairns et al., 1996). Loop formation signals in the red trace are marked with triangles (see Figure S1 for their identification). An enlarged view of a small signal in the black trace is shown as an insert (cyan curve). The ATP- and remodeler-dependent DNA looping on the nucleosomal template was confirmed by more extensive observations. In a total of 8.6 hr accumulated observation time, 12 signals with an average size of 25 (±5) bp were found in the presence of 4 nMSWI/SNF but no ATP. This occurrence rate (0.023 min−1) is close to that in the absence of remodeler (0.019 min−1). In 0.1mMATP, SWI/SNF translocates at lower speed and shorter distance than in 1mMATP (compare red and black traces). An analysis of 92 looping events found in 0.1mM ATP yields 78 bp average loop size and average velocities of 6.2 bp/s for loop generation and of 6.6 bp/s for reverse translocation.
(C and D) Enlarged views of the individual translocation signals from the black trace in (B). Several translocation regions are fitted with straight lines (red) to calculate translocation velocities (Experimental Procedures). The 3750 bp DNA molecule was used in these experiments.
After a DNA or a nucleosomal template was tethered between two polystyrene beads (Figure 1A and Supplemental Experimental Procedures), the free DNA molecules, histones, or nucleosomes in solution were flowed away, so that only a single template was studied at a time. The tethered template was torsionally unconstrained and stretched to facilitate detection of its end-to-end distance change using optical tweezers (Smith et al., 2003b). The optical tweezer instrument can be operated in two modes. In constant-force mode (Figure 1A), a prespecified constant tension was applied to the template through a feedback mechanism. In passive mode (Figure 5, insert), the tension in the template is allowed to increase as the template shortens, and vice versa. In a typical experiment, we first pulled the relaxed template at a constant velocity of 50 nm/s to a certain tension and observed the changes in the template end-to-end distance in either of the above modes in the presence or absence of SWI/SNF or RSC. At the end of an experiment, we stretched the template at the same speed to ~65 pN. Nucleosomes are mechanically disrupted at high forces, yielding characteristic ripping signals in the force-extension curve (Figure 4B) and providing a convenient method to determine the number of nucleosomes on each template (Brower-Toland et al., 2002, 2005). All the single-molecule experiments were performed at room temperature (~21°C).
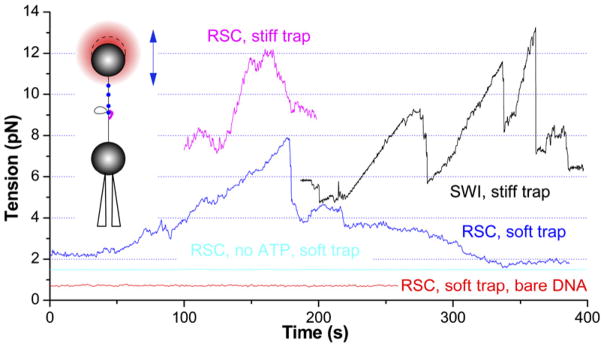
Figure 5.
SWI/SNF and RSC Can Translocate against High Forces
Force is monitored in passive mode by pulling the array template initially to <5 pN here, with the micropipette then retained in a fixed position relative to the trap center (insert). The engagement in translocation sometimes is not disrupted by abrupt loop relaxation (black), which can remain at high force and restart translocation after pauses (magenta) without initiation at low force. Due to the low processivity of remodeler translocase, a stiff trap (~20 bp/pN) is required to measure the stall force, which can be seen in comparison with a soft trap (~150 bp/pN). The remodeler-, nucleosome-, and ATP-dependent (1 mM) DNA translocation and loop formation activity was detected at any tension above 1 pN force (blue, magenta, and black). In contrast, DNA looping was not found on nucleosome templates with remodeler but without ATP (cyan) or on bare DNA at tensions ≥0.7 pN (red) in the presence of both remodeler and ATP under our experimental conditions.
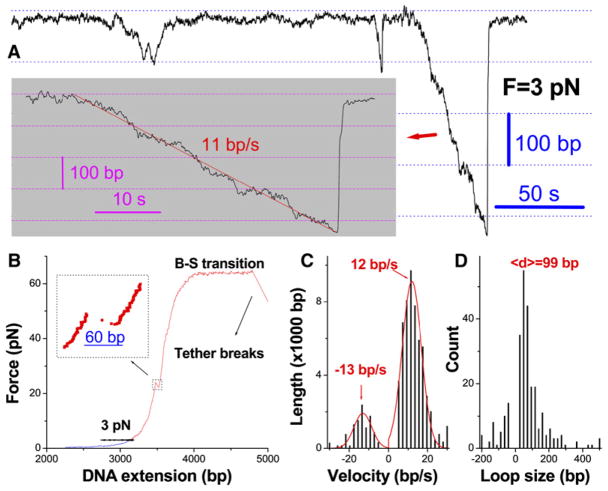
Figure 4.
SWI/SNF-Dependent DNA Translocation and Loop Formation on DNA Templates Containing Single Nucleosomes
(A) A DNA extension-time trace at 3 pN tension in the presence of 4 nM SWI/SNF and 1 mM ATP. A close-up view of an individual loop formation event is shown in insert.
(B) The complete force-extension curve corresponding to the same DNA molecule as in (A). The nucleosomal template was pulled to (blue) and then held at (black) 3 pN, at which point the nucleosome-remodeling activity shown in (A) was detected. After this observation phase, the template was pulled to high forces to disrupt the nucleosome on the template (red) and to confirm the involvement of a single DNA molecule (Smith et al., 1996). Here, the single rip in the force-extension curve (insert) confirms the presence of a single nucleosome on this template (Brower-Toland et al., 2002).
(C and D) Velocity and loop size distributions for SWI/SNF. They were calculated from a total 217 looping events. Here, the looping events on single-nucleosome templates obtained at 3 pN and 4 pN constant tensions were combined to achieve statistical significance, since such translocation and loop formation properties were not altered by the tension within this range (Figure 3A).
Nucleosome- and ATP-Dependent DNA Looping
Because the nucleosomal templates consist primarily of bare DNA, we first tested whether the ATP-dependent remodeler activities cause changes in the length of a bare DNA molecule at 3 pN tension. In a total of 7.5 hr accumulated observation time, only six length changes were picked as signals using our chosen criteria (Experimental Procedures). A typical DNA extension versus time trace is shown as a purple curve in Figure 1B. Furthermore, these signals were ATP independent, as their occurrence frequency (0.013 min−1) and average size (23 ± 2 bp) were approximately the same when ATP or remodeler was omitted (data not shown). We interpret these signals as resulting from Brownian fluctuations and possible instrument drift. Similar results were obtained at stretching forces >1 pN for both SWI/SNF and RSC. Therefore, we conclude that neither remodeler generates appreciable DNA length changes on bare, tethered DNA molecules above 1 pN tension, regardless of the presence of ATP. This conclusion is consistent with the observation by Lia and coworkers in the same force range (Lia et al., 2006). Nevertheless, using a magnetic trap, these authors have described an ATP-dependent DNA-looping activity for RSC on bare DNA at tensions below 1 pN (typically 0.3 pN). This activity sharply decreases with increasing force. To eliminate such bare DNA-dependent looping activity, and to observe nucleosome- and ATP-dependent chromatin-remodeling activity of SWI/SNF and RSC, we applied forces >1 pN to the nucleosomal templates (typically ≥3 pN).
Since nucleosomes have a limited lifetime when subjected to high tension (>16 pN) (Brower-Toland et al., 2002), we characterized their mechanical stability under a constant force of 3 pN. We found that most nucleosomes are stable for more than 20 min without appreciable DNA unwrapping (Figure 1B, magenta), a result that is consistent with previous observations (Brower-Toland et al., 2005). However, more extended stretching and/or application of higher tension enhanced histone dissociation above the spontaneous dissociation induced by dilution effects (Gottesfeld and Luger, 2001). The experiments presented here were therefore first obtained at a constant tension of 3 pN and within 15 min of their initial stretching, using the same 3750 bp nucleosomal template.
When 4 nM SWI/SNF and 1 mM ATP are added to the remodeling reaction, a series of prominent downward spikes appears in a plot of array extension versus time (Figure 1B, black), corresponding to a large reduction in end-to-end DNA tether length. These spikes either jump back suddenly (Figure 1C) or continuously return to the baseline (Figure 1D). The appearance of these spikes is ATP dependent, as they were barely detected without ATP (Figure 1B, blue) and both their average size and rate of formation decrease at 0.1 mM ATP concentration (Figure 1B, red). We observed spikes ranging from the resolution limit of ~20 bp in these experiments (Figure S1) up to ~1200 bp (Figure 2B). A similar ATP-dependent activity is observed for RSC (Figure S2). Three individual spikes are shown in Figure 1B (insert, cyan) and Figures 1C and 1D, (see more in Figure S2). We contend here that each spike is produced by a single remodeler complex because (1) the actual remodeler concentration is relatively low (<4 nM, see Experimental Procedures). Assuming similar Michaelis constants for the binding of ySWI/SNF and human SWI/SNF to nucleosomes, we estimate that the probability of a nucleosome bound by ySWI/SNF is less than ~10% under our experimental conditions (Supplemental Discussion). The probability is even lower that more than one SWI/SNF complex binds these nucleosomes simultaneously to generate the observed spikes (Supplemental Discussion); accordingly, simultaneous appearances of two or more spikes are negligible (~0.2% as estimated from a Poisson distribution) due to a low overall occurrence frequency of these spikes (~0.4 min−1). In addition, (2) further decrease by 15-fold or increase by 2-fold in actual remodeler concentration, estimated from the occurrence frequency of the spikes, does not change the average size of the spikes nor their rate of formation (data not shown), and (3) similar activity is seen on templates containing a single nucleosome (Figure 4). Furthermore, gel shift and footprinting assays with RSC (Lorch et al., 1998; Saha et al., 2005), along with EM and AFM imaging studies with SWI/SNF (Schnitzler et al., 2001; Smith et al., 2003a), strongly suggest that only one remodeler binds tightly to one nucleosome at a time.
Figure 2.
Distributions of the Translocation Velocity and Loop Size for SWI/SNF
Distributions of the translocation velocity (A) and loop size (B) for SWI/SNF. In (A), the histograms for positive and negative velocity values are separately fitted with Gaussian functions (red), yielding the indicated means and standard deviations of 5 bp/s for the positive translocation and of 6 bp/s for the reverse translocation. The accumulative translocation length at the specified velocity is shown. In (B), the loop size for each loop formation event is defined as the corresponding DNA contour-length change from the start point of the first translocation phase to the endpoint of the last translocation phase. The loop size for reverse translocation is the DNA length that is removed from the loop after its maximum size is attained. Note that velocity and loop size values outside the range drawn (±30 bp/s for velocity and −200/+500 bp for loop size) are binned to the corresponding endpoints. The distributions were calculated (Experimental Procedures) from 241 DNA-looping events measured on 67 different DNA molecules.
The great variation in spike sizes, and the existence of large spikes (>80 bp), cannot be rationalized entirely by remodeler-induced DNA-wrapping models. Instead, the continuous DNA shortening suggests the formation of DNA loops by a processive DNA translocation mechanism through the remodeler. Following this processive phase, one or several jumps are observed (Figure 1C and Figure S2), with DNA extension finally returning to baseline. The exact origin of these jumps remains to be identified. For example, the DNA loop may propagate along the histone octamer surface to eventually dissipate at the other edge of the nucleosome in a discrete fashion, leading to a nucleosome position shift. Alternatively, the discrete jumps may simply reflect the relaxation of the DNA loop due to the dissociation of the remodeler from the nucleosome, or the partial detachment of at least one of its points of contact with the DNA.
The DNA loop relaxes by a sudden jump in ~70% of translocation events for SWI/SNF (Figure 2B). The remaining ~30% of events mainly undergo a continuous, controlled release process as shown in Figure 1D and Figure S2E. The velocity of this release decreases with decreasing ATP concentration (Figure 1B), indicating that it also involves an active translocation process. The release process following loop formation suggests that the remodeler may be able to change its direction of translocation (reverse translocation) (Lia et al., 2006), leading to removal of the pumped loop. Either mode of loop relaxation eventually leads to a complete release of the loop, as revealed by the coincidence of the baselines before and after the looping event. Therefore, in contrast to the permanent loop formation model proposed for mononucleosome on short DNA templates (Lorch et al., 1998), the loops found here are short-lived (~15 s average lifetime). This observation is consistent with results from experiments using nucleosomes reconstituted on long DNA templates (Logie and Peterson, 1997; Shundrovsky et al., 2006).
The distributions of translocation velocity and the loop size are shown in Figure 2, where minus signs correspond to the observed reverse translocation. The velocities approximately follow a unimodal, Gaussian distribution, and their means are equal within experimental error for loop generation and loop dissipation by reverse translocation, further supporting the involvement of single remodeler complexes in these events. The step size of remodeler translocation is below our machine resolution, as was demonstrated by no distinct peaks in the pair-wise distance distribution of the translocation phases (data not shown). The loop size varies greatly, with an average of 108 bp (for a comparison of the loop sizes obtained with different approaches, see Supplemental Discussion).
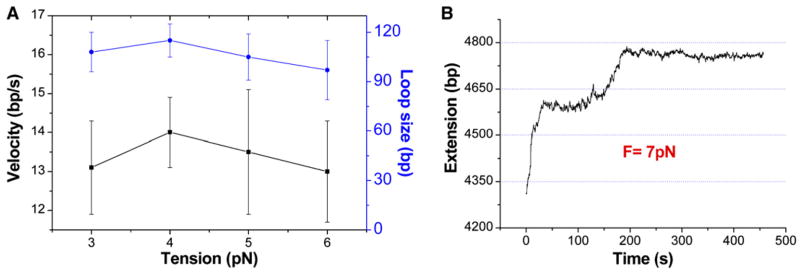
Effect of Constant Tension upon Remodeler-Induced DNA Looping
To characterize the effect of the force on remodeler activity, we repeated the experiments at higher constant tension from 4–7 pN. No stretching forces between 1 pN and 3 pN were tested in the constant-force mode (Supplemental Experimental Procedures). The translocation and loop formation properties of SWI/SNF and RSC at a tension between 4 pN and 6 pN are indistinguishable from those at 3 pN within experimental error (Figures 3A and 2 and Figures S2 and S3). The average velocities and loop sizes determined at 3 pN, 4 pN, 5 pN, and 6 pN are approximately equal, and their mean values are shown in Table S1. This force independence suggests that the remodeling parameters such as loop size and translocation velocity measured in this force range can be interpolated to zero force and represent the intrinsic properties of these remodelers. However, the frequency of loop formation decreases with tension. To obtain sufficient loop formation frequency, nucleosomal arrays containing an average of 4.3 (±2.3) nucleosomes were mainly used for assays performed above 3 pN. On this nucleosomal template, the measured occurrence rates of the loop formation events are 0.52 min−1, 0.37 min−1, and 0.20 min−1 at 4 pN, 5 pN, and 6 pN tensions, respectively. Interestingly, loop formation activity is not observed when a tension of 7 pN is applied to the nucleosomal array (Figure 3B). At this tension, the nucleosomal template becomes much less stable, leading to fast nucleosome disruption and resulting in tether length increases.
Figure 3.
Effect of Constant Nucleosomal Template Tension upon SWI/SNF Translocation
(A) SWI/SNF translocation velocity and loop size are not affected by tensions between 3 pN and 6 pN. The error bars shown are standard deviations of three independent calculations performed on the same set of extension-time data (Experimental Procedures).
(B) The SWI/SNF-dependent DNA translocation and loop formation activity is inhibited at 7 pN tension. Instead, nucleosomal array length quickly increases due to DNA unwrapping from histones. This DNA-unwrapping process contains continuous and discontinuous DNA releases from histones under tension (Brower-Toland et al., 2002). Note that at least two nucleosomes still remain on the 5040 bp DNA molecule between 200 s and 500 s.
Single-Nucleosome-Dependent DNA Looping
Under our experimental conditions, with template tensions ≥3 pN, DNA translocation and loop formation activity is observed only on nucleosomal templates, not on bare DNA. Thus, SWI/SNF and RSC have to target a nucleosome and undergo the formation of an intermediate remodeler-nucleosome complex to generate ATP-dependent DNA loops. Such complex could contain either a single nucleosome or more than one nucleosome. Previous experiments showed that a dinucleosome structure can be formed during chromatin remodeling by SWI/SNF-like remodelers (Schnitzler et al., 2001). To investigate whether a similar dinucleosome or multinucleosome structure is required for the DNA loop formation activity observed here, we investigated remodeling activity on single-nucleosome-containing templates.
Two kinds of nucleosomal templates were used to ensure that single nucleosomes were indeed studied in these experiments. The first type of single-nucleosome templates was selected from the nucleosomal array described above. The second type (3.4 kbp long) was obtained by ligating a mononucleosome (reconstituted on a 254 bp DNA molecule) with two DNA handles. In both cases, the presence of a single rip associated with nucleosomal disruption in the force-extension curve, was used as evidence of the existence of a single nucleosome (Figure 4B). DNA loop formation activity can be detected on both types of single-nucleosome-containing templates (Figure 4A). Detailed analyses of such activity yielded the same velocity and loop size distributions (Figures 4C and 4D) as those obtained from nucleosomal array templates (Figure 2). These observations indicate that remodelers only require a single nucleosome to generate the DNA loops observed here, and that formation of a multinucleosome-remodeler complex is not essential in our assays. Moreover, these results also indicate that the interaction between adjacent nucleosomes in nucleosomal arrays is probably small, at least at the level of resolution of the current experiments. This conclusion is to be expected, given the large internucleosomal spacing (>370 bp) relative to the average loop size (~100 bp). Taken together, these observations suggest that the measured velocity and loop size distributions observed here represent intrinsic properties of the remodelers.
Remodeler Translocation against Increasing Forces
Remodelers must generate enough mechanical force to efficiently disrupt histone-DNA interactions. Forces >3 pN are required to mechanically disrupt histone-DNA interactions (Figure 1B, magenta) at rates that match those observed occurrence rates for the looping events. The above experiments showed that SWI/SNF and RSC can translocate against at least 6 pN external tension to produce DNA loops. To monitor the maximum force that a remodeler can generate, the optical tweezers were operated in passive mode (Figure 5, insert). Here, template shortening caused by remodeler-mediated loop formation leads to a proportional increase in the force opposing the remodeler (Figure 5). Interestingly, RSC and SWI/SNF translocate at nearly uniform speeds against increasing force up to ~12 pN (see distributions of stall force in Figure S4) until jumping (black) and/or continuously returning (magenta) to low forces, indicating that the translocation phase is indeed force insensitive, even in this larger force regime. Closer analysis of the data shows, however, that the translocation always initiates at a force lower than 7 pN (143 and 97 events for SWI/SNF and RSC, respectively). These observations suggest that translocation initiation is force sensitive and probably involves a DNA-wrapping or bulging step that decreases DNA end-to-end distance, resulting in its force sensitivity; alternatively, it is also possible that translocation initiation requires a transient nucleosome or nucleosome-remodeler structure that is disrupted by forces ≥7 pN. However, once translocation is initiated and the actual translocation is under way, remodelers can operate against forces up to 12 pN. Precedent for the existence of this distinct initiation event is found in studies of Rad54 and type I restriction enzyme EcoR124I. Both enzymes belong to the same SF2 helicase superfamily as remodelers and translocate along DNA (Seidel et al., 2004; Amitani et al., 2006). Rad54 shows a lag phase before the start of translocation, and EcoR124I displays an initial complex containing an 8 nm DNA bulge trapped by ATPγS (van Noort et al., 2004; Amitani et al., 2006). The initiation step characterized in the present studies must occur after remodeler binding, since its force sensitivity is not attenuated by high remodeler concentration (data not shown).
Persistent DNA Loop Generation and Dissipation
Although most of the DNA loop formation events seem to appear independently in time on the same nucleosomal template, sometimes series of events of loop formation followed by dissipation occur in a row (Figure 6). In the example shown in Figure 6A, 14 events were observed within 330 s, a number far greater than the value of 2.2 events predicted from the average signal occurrence rate (0.4 min−1). The probability that these events were generated independently by different remodelers acting on the same or on different nucleosomes during the same time period would be 8 × 10−8, assuming a Poisson distributed process. In contrast, 26 similar events were found out of 1045 loop formation events for SWI/SNF, and 17 out of 836 events for RSC, at forces between 3 pN and 6 pN, with actual occurrence probability >2% for both remodelers. Therefore, significant correlation was present for at least a fraction of the loop formation/dissipation events. Moreover, such persistent events could be found on templates containing single nucleosomes (Figure 6A). We thus propose that these correlated loop formation events are caused by single remodeler molecules acting on the same nucleosomes. Thus, nucleosome-remodeler complexes may conduct a series of translocation and loop formation events before complete remodeler dissociation from the nucleosome.
Figure 6.
Processive Nucleosome Remodeling Caused by a Single Remodeler Complex
(A) SWI/SNF generates a burst of looping events on a single-nucleosome-containing DNA template under a constant tension of 3 pN. (B and C) (B) SWI/SNF or (C) RSC generates correlated looping events on nucleosomal array templates at 4 pN tension.
Discussion
Comparison with DNA Loop Formation Activity on Bare DNA
DNA translocation and loop formation activities for SWI/SNF-like remodelers have been found on bare DNA by Lia et al. (Lia et al., 2006) and on the nucleosomal templates reported here. The two activities share the property of switching translocation direction, although with lower probability on the nucleosomal template (~30%) than on bare DNA (~60%). This observation is consistent with the recent finding that remodelers track along one of the DNA strands with a 3′ to 5′ directionality (Saha et al., 2005; Zofall et al., 2006). In addition, the average loop sizes, or apparent average translocation distances, measured on both kinds of templates increase with increasing ATP concentration below saturating concentrations as observed for other DNA or RNA translocases (Jankowsky et al., 2000). However, the two activities differ both qualitatively and quantitatively, as shown in Table S2. Specifically, loop formation on bare DNA occurs only at very low DNA tensions (< ~1 pN). In contrast, on the nucleosomal templates, loop formation can be observed at tensions from 1 pN to 6 pN and against forces up to 12 pN after translocation initiation (Figure 5, Figure S4, and data not shown). The ability to generate high forces on nucleosomal arrays is consistent with the role of remodelers for disrupting DNA-histone interactions during chromatin remodeling. Moreover, loop formation activity displays different force sensitivity on the two types of templates: on bare DNA, the loop size attenuates very quickly upon increasing the force even below 1 pN, whereas on nucleosomal templates both translocation velocity and loop size do not change significantly in the force range between 3 pN and 6 pN. The average loop size on bare DNA substrate (~700 bp) is significantly larger than the average loop size (~100 bp) observed on nucleosomal substrates. Furthermore, translocation rates on bare DNA (>500 bp/s) greatly exceed those observed on nucleosomal templates (12 bp/s), whereas bare DNA translocation duration (≤1.0 s) is more than an order of magnitude shorter than those observed on nucleosomal DNA (~10 s, including pause duration). These differences well exceed experimental errors (Gosse and Croquette, 2002; Lia et al., 2006). Finally, the burst of loop formation/dissipation activity found on nucleosome substrates (Figure 6) has not been reported for bare DNA. We speculate that this persistent or processive activity might be a feature of remodelers subjected to regulation, possibly being enhanced by tethering the remodeler to an activator or through remodeler interaction with acetylated histone tails (Workman and Kingston, 1998).
The dramatic difference in remodeler translocation properties between nucleosomal and bare DNA suggests a specific recognition of the nucleosome by remodelers and a strong coupling between nucleosome association and remodeler translocation. A preference of SWI/SNF or RSC for nucleosomes over bare DNA has been described in a DNA-supercoiling or gel shift assay (Lorch et al., 1998; Smith and Peterson, 2005). We have found that SWI/SNF cannot generate DNA loops on templates containing nonspecifically bound histones (data not shown), consistent with the requirement of a specific remodeler/nucleosome interaction. Recent experiments suggest that the ATPase domain of the remodeler engages the DNA at about two helical turns from the dyad of the nucleosome and that the DNA translocation is internally elicited (Saha et al., 2005, 2006; Zofall et al., 2006). Taken together, these observations indicate that SWI/SNF and RSC generate intranucleosomal DNA loops by binding to the nucleosome and by drawing the flanking DNA toward the nucleosome through a processive translocation mechanism.
A DNA Translocation and Loop Formation Model for Chromatin Remodeling
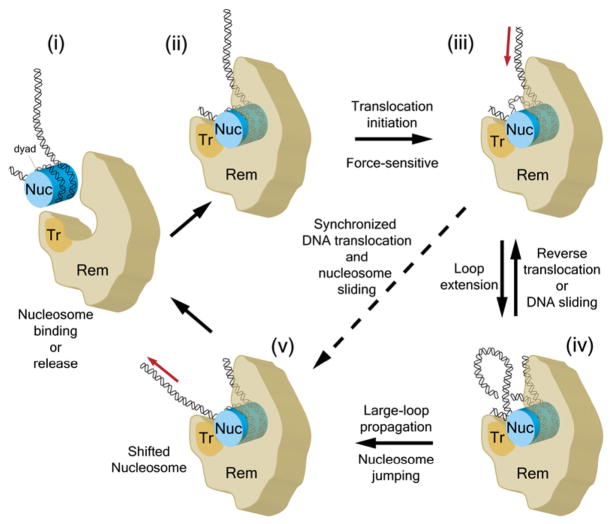
The results presented here, together with previous ensemble observations (Saha et al., 2006), support a DNA translocation and loop formation model of nucleosome remodeling by SWI/SNF-family remodelers, as illustrated in Figure 7. First, the remodeler binds the nucleosome core and its translocation domain engages DNA close to the nucleosomal dyad, either producing a bulge that is presumably the force-sensitive step of initiation (from state [ii] to state [iii]) or requiring a nucleosome-remodeler structure that is undone at forces above 7 pN. The subsequent translocation generates strained DNA that will be temporally accumulated to form an intranucleosomal loop as observed in our assay (state [iv]). This loop may resolve at one of the two DNA entry/exit sites of the nucleosome: from the same edge that the DNA entered the loop, by a disengagement of the translocation domain from the DNA (DNA sliding) or by reverse translocation (from [iv] to [iii]), or from the other edge, thus resulting in nucleosome jumping (from [iv] to [v]). In either case, the relaxation of the loop is not synchronized with DNA translocation. Note that this scenario supports two mechanisms for exposure of nucleosomal DNA, loop formation and nucleosome mobilization. The frequency of DNA translocation and loop formation activities seen in our assay at 3 pN is comparable to those inferred from bulk assays under optimal temperature for SWI/SNF-like remodelers (Logie and Peterson, 1997; Narlikar et al., 2001), suggesting that our observations are basic features of nucleosome remodeling. Loop formation provides also a molecular mechanism for nucleosome mobilization (Langst and Becker, 2001; Kassabov et al., 2003; Owen-Hughes, 2003; Strohner et al., 2005; Zofall et al., 2006; see also Supplemental Discussion). However, we cannot rule out the existence of other mechanisms that support nucleosome mobilization without large-loop generation (Figure 7, from [iii] to [v]), as has been previously suggested (Saha et al., 2005, 2006), because such activity cannot be directly detected in our current assay.
Figure 7.
A Model for Nucleosome Remodeling
(i) Unbound state. (ii) The remodeler (Rem) binds the nucleosome (Nuc) in a pocket. (iii) The ATPase/translocase subunit (Tr) engages nucleosomal DNA at a position flanking the dyad, forming a small bulge near the dyad. (iv) Subsequent processive translocation generates large intranucleosomal DNA loops that have three possible fates: forward propagation (resulting in nucleosome jumping [v]), active reverse translocation, or DNA sliding (which may reflect the disengagement of the translocase subunit). Alternatively, the translocation can lead to immediate nucleosome sliding, as indicated by the dashed line, without large loops having been accumulated (from state [iii] to state [v]). (v) Following a remodeling cycle, the remodeler may release the nucleosome (from state [v] to state [i]).
For the DNA loop formation observed here, the loop size is determined by the translocation processivity of the remodeling complex and the balance between the rates of loop generation and dissipation processes. Thus, the ~100 bp average loop sizes constitute a lower bound for the average processivity of the remodeler translocase. A majority of sudden loop release events suggest that the loop dissipation process (at either end of the nucleosomal DNA) is discontinuous (Figure 7, from state [iv] to state [v] by nucleosome jumping or to state [iii] by DNA sliding) and, in contrast to the previous translocation model, it indicates that the DNA pumping and dissipation processes are not necessarily synchronized. The similarity of the translocation and loop formation activities of SWI/SNF and RSC complexes (Table S1) suggests that these are general features of chromatin remodeling by SWI/SNF-family remodelers and raises the possibility that they may also be the mechanism underlying the activity of other remodeler families (Saha et al., 2002; Strohner et al., 2005; Zofall et al., 2006).
Several qualitative and quantitative similarities between chromatin remodelers and RNA polymerases in regard to DNA translocation are worth emphasizing. First, their average translocation velocities are nearly identical (at saturating NTPs) (Adelman et al., 2002). Second, the measured stall forces for the remodelers fall into (Yin et al., 1995) or close to the lower bound (Bustamante et al., 2004) of the range of stall forces obtained for RNA polymerases. Because of the difficulties in measuring the stall forces of remodelers, due to their small apparent translocation distances and the limited stability of nucleosomes under high tensions, the actual stall forces for remodelers might be higher than our current values of ~12 pN. Finally, remodelers display a susceptibility to DNA applied tension similar to that of RNA polymerases to force (Yin et al., 1995). We speculate that these similarities may reflect a functional synergy between remodelers and RNA polymerases during gene transcription (Wilson et al., 1996).
Restriction enzyme accessibility assays (Narlikar et al., 2001; Lorch et al., 2005; Saha et al., 2005) and DNA footprinting assays (Langst and Becker, 2001; Zofall et al., 2006) have been widely used to study chromatin remodeling. These assays detect certain local changes in DNA conformation on the histone surface and only infer from these local changes the corresponding global DNA conformational changes, such as DNA looping and nucleosome sliding. In addition, the interpretations of experimental results from these kinetic assays are critically dependent upon synchronization of the remodeling reaction and the initial positioning of histone octamer relative to DNA. In contrast, the single-molecule assay for nucleosome remodeling presented here directly monitors in real time the global DNA conformational changes caused by DNA looping. Although the two types of assays are complementary, their different focuses may explain why the intranucleosomal DNA loops detected here are much larger than in bulk assays. Moreover, the limited DNA lengths (<210 bp) used in most bulk assays may prevent the formation of large DNA loops during chromatin remodeling. Comparisons with other techniques, such as AFM imaging (Schnitzler et al., 2001) and a recent single-molecule measurement of nucleosome mobilization (Shundrovsky et al., 2006), are given in Supplemental Discussion.
In conclusion, the single-molecule approach presented here to investigate the dynamics of remodelers reveals the formation of large intranucleosomal DNA loops during nucleosome remodeling and has provided nucleosome- and ATP-dependent real-time kinetics of DNA translocation. Such loops may play a role not only as a dynamic product of chromatin remodeling, making nucleosomal DNA accessible, but also as an intermediate state in the process of nucleosome mobilization and histone eviction (Lorch et al., 2006). Future experiments will focus on dissecting the different loop dissipation pathways and their relationship to nucleosome mobilization, and on characterizing the remodeling mechanisms of other remodeler families (Workman and Kingston, 1998; Fan et al., 2003; Zofall et al., 2006).
Experimental Procedures
Enzymes and Buffers
SWI/SNF and RSC were purified as reported (Saha et al., 2002; Smith et al., 2003a). If not specified, enzyme concentrations in the remodeling assays were 4 nM for SWI/SNF and 10 nM for RSC, respectively. Different buffers were used: for SWI/SNF (Logie and Peterson, 1997), 20mMTris-HCl (pH 8.0), 125mMNaCl, 5mMMgCl2, 1%glycerol, 0.2 mM DTT, 0.1 mg/ml BSA, and 0.05% NP-40; and for RSC (Cairns et al., 1996), 20 mM HEPES (pH 7.6), 100 mM potassium acetate, 5 mM MgCl2, 1% glycerol, 0.2 mM DTT, 0.1 mg/ml BSA, and 0.05% NP-40. If not specified, all remodeling assays were done in 1 mM ATP. To minimize enzyme adsorption, all tubing and chamber surfaces that touch the enzyme were soaked in a 0.05% powdered milk solution overnight and then rinsed thoroughly with the buffer before use. In addition, enzyme was injected using a computer-controlled syringe directly to the microchamber ~1 mm away from the tip of the micropipette through a glass tube sandwiched in the microchamber. However, some enzyme adsorption cannot be completely ruled out.
Data Analysis
The time-dependent extension data were filtered using a moving Gaussian function as weight with a standard deviation of 0.1 s, and then presented in this paper or further analyzed. To identify the translocation and looping signals from an extension-time trace, its corresponding velocity trace was calculated using a method similar to that previously adopted (Adelman et al., 2002), utilizing a 2 s time window. Only extension changes with their corresponding absolute velocities >4.4 bp/s were considered as signals (Figure S1). This approach identifies looping signals > ~20 bp and smoothes out most of the possible smaller signals due to their poor signal-to-noise ratio and short duration. Once a looping signal was identified, its translocation phase was fitted by a straight line to calculate translocation velocity more accurately (Figures 1C and 1D), and validated against the velocity threshold. Such data processing generates a small gap in its distribution profiles between positive and negative translocation velocities and loop sizes (Figures 2, 4C, and 4D and Figure S3).
Supplementary Material
Supplemental Data include four figures, two tables, Supplemental Discussion, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://www.molecule.org/cgi/content/full/24/4/559/DC1/.
Acknowledgments
We thank J. Widom for providing histone octamer for initial experiments and plasmid containing the 601 sequence; G. Bowman, J. Core, and J. Quinlan for critical reading of the manuscript; and J. Wang and E. Watson for technical assistance. This research was supported by the Jane Coffin Childs Memorial Fund (Y.Z.), NIH grants (GM32543 to C.B., GM49650 to C.L.P., and GM60415 to B.R.C.), and DOE grants to C.B.
References
- Adelman K, La Porta A, Santangelo TJ, Lis JT, Roberts JW, Wang MD. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proc Natl Acad Sci USA. 2002;99:13538–13543. doi: 10.1073/pnas.212358999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitani I, Baskin RJ, Kowalczykowski SC. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Bochar DA, Wang L, Beniya H, Kinev A, Xue YT, Lane WS, Wang WD, Kashanchi F, Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci USA. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Wacker DA, Fulbright RM, Lis JT, Kraus WL, Wang MD. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J Mol Biol. 2005;346:135–146. doi: 10.1016/j.jmb.2004.11.056. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu Rev Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang MC, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Fan HY, He X, Kingston RE, Narlikar GJ. Distinct strategies to make nucleosomal DNA accessible. Mol Cell. 2003;11:1311–1322. doi: 10.1016/s1097-2765(03)00192-8. [DOI] [PubMed] [Google Scholar]
- Fitzgerald DJ, DeLuca C, Berger I, Gaillard H, Sigrist R, Schimmele K, Richmond TJ. Reaction cycle of the yeast Isw2 chromatin remodeling complex. EMBO J. 2004;23:3836–3843. doi: 10.1038/sj.emboj.7600364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosse C, Croquette V. Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophys J. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld JM, Luger K. Energetics and affinity of the histone octamer for defined DNA sequences. Biochemistry. 2001;40:10927–10933. doi: 10.1021/bi0109966. [DOI] [PubMed] [Google Scholar]
- Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- Kassabov SR, Zhang B, Persinger J, Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- Langst G, Becker PB. ISWI induces nucleosome sliding on nicked DNA. Mol Cell. 2001;8:1085–1092. doi: 10.1016/s1097-2765(01)00397-5. [DOI] [PubMed] [Google Scholar]
- Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, Croquette V, Bensimon D, Owen-Hughes T. Direct observation of DNA distortion by the RSC complex. Mol Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logie C, Peterson CL. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 1997;16:6772–6782. doi: 10.1093/emboj/16.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Cairns BR, Zhang MC, Kornberg RD. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Davis B, Kornberg RD. Chromatin remodeling by DNA bending, not twisting. Proc Natl Acad Sci USA. 2005;102:1329–1332. doi: 10.1073/pnas.0409413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci USA. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25:1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Phelan ML, Kingston RE. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol Cell. 2001;8:1219–1230. doi: 10.1016/s1097-2765(01)00412-9. [DOI] [PubMed] [Google Scholar]
- Owen-Hughes T. Pathways for remodelling chromatin. Biochem Soc Trans. 2003;31:893–905. doi: 10.1042/bst0310893. [DOI] [PubMed] [Google Scholar]
- Richmond RK, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- Schnitzler GR, Cheung CL, Hafner JH, Saurin AJ, Kingston RE, Lieber CM. Direct imaging of human SWI/SNF-remodeled mono- and polynucleosomes by atomic force microscopy employing carbon nanotube tips. Mol Cell Biol. 2001;21:8504–8511. doi: 10.1128/MCB.21.24.8504-8511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel R, van Noort J, van der Scheer C, Bloom JGP, Dekker NH, Dutta CF, Blundell A, Robinson T, Firman K, Dekker C. Real-time observation of DNA translocation by the type I restriction modification enzyme EcoR124I. Nat Struct Mol Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- Shundrovsky A, Smith CL, Lis JT, Peterson CL, Wang MD. Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nat Struct Mol Biol. 2006;13:549–554. doi: 10.1038/nsmb1102. [DOI] [PubMed] [Google Scholar]
- Smith CL, Peterson CL. A conserved Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin remodeling. Mol Cell Biol. 2005;25:5880–5892. doi: 10.1128/MCB.25.14.5880-5892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SB, Cui YJ, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- Smith CL, Horowitz-Scherer R, Flanagan JF, Woodcock CL, Peterson CL. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat Struct Biol. 2003a;10:141–145. doi: 10.1038/nsb888. [DOI] [PubMed] [Google Scholar]
- Smith SB, Cui YJ, Bustamante C. Optical-trap force transducer that operates by direct measurement of light momentum. Methods Enzymol. 2003b;361:134–162. doi: 10.1016/s0076-6879(03)61009-8. [DOI] [PubMed] [Google Scholar]
- Strohner R, Wachsmuth M, Dachauer K, Mazurkiewicz J, Hochstatter J, Rippe K, Langst G. A ‘loop recapture’ mechanismfor ACF-dependent nucleosome remodeling. Nat Struct Mol Biol. 2005;12:683–690. doi: 10.1038/nsmb966. [DOI] [PubMed] [Google Scholar]
- van Holde K, Yager T. Models for chromatin remodeling: a critical comparison. Biochem Cell Biol. 2003;81:169–172. doi: 10.1139/o03-038. [DOI] [PubMed] [Google Scholar]
- van Noort J, van der Heijden T, Dutta CF, Firman K, Dekker C. Initiation of translocation by Type I restriction-modification enzymes is associated with a short DNA extrusion. Nucleic Acids Res. 2004;32:6540–6547. doi: 10.1093/nar/gkh999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- Wilson CJ, Chao DM, Imbalzano AN, Schnitzler GR, Kingston RE, Young RA. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- Yin H, Wang MD, Svoboda K, Landick R, Block SM, Gelles J. Transcription against an applied force. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data include four figures, two tables, Supplemental Discussion, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://www.molecule.org/cgi/content/full/24/4/559/DC1/.