Abstract
Understanding racial and ethnic disparities in diagnostic rates of genetic testing is critical for health equity. We sought to understand the extent and cause of racial and ethnic disparities in diagnostic efficacy of comprehensive genetic testing (CGT) for sensorineural hearing loss (SNHL). We performed a retrospective cohort study at two tertiary children’s hospitals on a diverse cohort of 240 consecutive pediatric patients (76% publicly insured, 82% non-White) with SNHL of unknown etiology who underwent CGT. Definite and possible genetic diagnoses were assigned for each patient, representing the likelihood of a genetic cause of hearing loss. Associations between diagnostic rates were examined. 3.8 ± 2.1 variants were detected per patient; this frequency did not vary between White/Asian and Hispanic/Black cohorts. Overall, 82% of variants were variants of uncertain significance (VUS). Compared with White and Asian subjects, variants identified among Hispanic and Black children were less likely to be classified as pathogenic/likely pathogenic (15% vs. 24%, p < 0.001), and Hispanic and Black children were less likely to have a definite genetic diagnosis (10% vs. 37%, p < 0.001). The adjusted odds ratio for definite genetic diagnosis in Black and Hispanic children compared with White and Asian children was 0.19. Expanding genetic diagnostic criteria to include predicted deleterious VUSs reduced these disparities between White/Asian and Hispanic/Black children, with comparable molecular diagnostic rates (41% vs. 38%, p = 0.72). However, in silico predictions are insufficiently valid for clinical use. Increased inclusion of underrepresented groups in genetic hearing-loss studies to clinically validate these variants is necessary to reduce racial and ethnic disparities in diagnostic efficacy of comprehensive genetic testing.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00439-021-02338-4.
Introduction
Hearing loss is the most common congenital sensory deficit, affecting one in 500 newborns (Morton and Nance 2006; Fortnum et al. 2001). 50% of bilateral sensorineural hearing loss (SNHL) is estimated to be caused by genetic factors (Marazita et al. 1993; Smith et al. 2005). Identifying an etiology for childhood SNHL can assist in prognosis and guide management in deaf and hard-of-hearing (D/HH) children (Kimberling et al. 2010; Shearer et al. 2019). Additionally, early identification of syndromic forms of SNHL, prior to the development of overt syndromic phenotypes, can significantly affect management and counseling (Brodie et al. 2020). Consensus statement from the International Pediatric Otolaryngology Group recommended comprehensive genetic testing (CGT) in etiologic testing for children with bilateral SNHL (Liming et al. 2016).
Genetic testing is valuable in the clinical management and understanding of pediatric hearing loss; however, the diagnostic rate has been reported to vary widely, from 10 to 83% (Shearer and Smith 2015). This is in part due to the discrepancy between the vast increase in the amount of genetic information readily available with the advancement and increasing availability of next-generation sequencing (NGS) and our ability to interpret the clinical significance of identified variants. With over 150 genes implicated in SNHL, testing routinely yields a large number of novel variants, most of which are single-nucleotide changes with a small number of indels and copy-number variants (Smith et al. 2005; Hilgert et al. 2009; Shearer et al. 2011). The interpretation of sequence variants is a crucial element of accurate genetic diagnosis, and discrepancies in variant interpretation can have serious implications for patient care (Amendola et al. 2016; Booth 2018; Harrison et al. 2016). The American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP) have published recommendations for the interpretation of sequence variants (Richards et al. 2015). These guidelines require substantial evidence to categorize a variant as disease-causing (Oza et al. 2018), and the paucity of available evidence constrains diagnostic power for populations that are historically underrepresented in genetic studies. Understanding how the efficacy and constraints of genetic testing are affected by demographic disparities is a critical concern when considering health equity.
SNHL is in many ways an ideal human genetic disease model in which to explore complexities of genetic testing—it is a common, narrowly defined, quantifiable clinical entity with a precisely delineated group of highly penetrant genes associated with a clear phenotype. Lessons learned in the study of SNHL may be applicable to the treatment of a range of genetic disorders. Racial and ethnic disparities in genetic testing in hearing loss have been observed. The rate of diagnosis as well as spectrum of genes and variants varies widely by ethnic groups, but the underlying cause of this disparity is not well described (Sloan-Heggen et al. 2016; Yan et al. 2016). The majority of studies have focused on GJB2, yet causative variants in these genes are mostly found in people of European and Asian descent (Chan and Chang 2014). In contrast, GJB2 variants are rarely the cause of SNHL in Black populations (Lebeko et al. 2015).
In recent years, CGT has become more accessible due to decrease in expense and broadening of insurance coverage, increasing the opportunity to examine genetic testing outcomes in historically underrepresented populations. In this study, we report on the molecular diagnostic efficacy of CGT for SNHL in a diverse pediatric population, with a focus on examining the extent and cause of disparities in diagnostic rate and variant distribution among children from different racial and ethnic populations.
Methods
Study population
We performed a retrospective chart review of 240 consecutive pediatric patients with an unknown etiology of SNHL at two tertiary children's hospitals (UCSF Benioff Children’s Hospital Oakland, UCSF Benioff Children’s Hospital San Francisco) who underwent comprehensive hearing-loss gene-panel testing from 2018 to 2020. Samples were obtained by blood draw or cheek swab. Patients were not excluded based on physical exam, imaging, or other findings. This study was approved by the Institutional Review Board at UCSF.
Demographics
Gender and insurance were extrapolated from the patients’ electronic medical record. Ethnicity, race, and primary home language were based on parents’ self-report, with children categorized as Non-Hispanic White (White), Non-Hispanic Black (Black), Non-Hispanic Asian (Asian), Hispanic, any race (Hispanic), and Other or Unknown (which included Pacific Islander, Native American, Mixed/Multi-race, or Declined to State).
Clinical history
Clinical data were collected from otolaryngology and audiology reports. This included newborn hearing screening (NHS) results as well as earliest and most recent audiogram results. Hearing-loss onset was considered congenital if the patient referred on their NHS and post-natal if the patient passed their NHS. In cases where an NHS result was unavailable, hearing-loss onset was categorized as unknown. Baseline audiogram results are reported from the earliest audiogram report available. Hearing-loss laterality and baseline hearing-loss level were determined using pure-tone average (PTA) for thresholds between 0.5 and 4 kHz. Hearing loss was defined as follows: unilateral (PTA > 15 dB in one ear only); bilateral (PTA > 15 dB in both ears); hearing level (based on worse-ear level): normal (PTA < 15 dB HL), slight-mild (15–40 dB), moderate (41–55 dB), moderately severe (56–70 dB), severe (71–90 dB), and profound (> 90 dB). Comorbidities were defined as developmental or medical comorbid conditions that might indicate a syndromic hearing loss.
Genetic data
Hearing-loss gene-panel testing (GeneDx) was conducted by targeted gene capture followed by massively parallel sequencing. The panel encompassed 146 nuclear genes and 6 variants in 4 mitochondrial genes accounting for non-syndromic and syndromic SNHL. Sequencing of coding regions and splice junctions with selected deletion/duplication analysis and copy-number variant detection was performed (GeneDx 2015). Each variant was classified on the clinical genetic testing report as benign (B), likely benign (LB), variant of uncertain significance (VUS), likely pathogenic (LP), or pathogenic (P) based on the ACMG 2015 Guidelines, and “definite genetic diagnosis” made based on P/LP variants and inheritance pattern (ACMG 2015; Richards et al. 2015).
To examine the distribution of variants classified based on an entirely in silico scheme, we classified VUSs as predicted benign or deleterious, based on PROVEAN (Protein Variation Effect Analyzer) score [< = − 2.5: predicted deleterious (VUS-D); > − 2.5: predicted benign (VUS-B)]. Frameshift VUSs leading to premature truncation and large deletions were classified as VUS-D. Splice-site VUSs and large duplications were classified as unknown (VUS-U). For VUSs, inheritance was defined to accord with the most common mode of inheritance for P/LP variants of that gene. In some cases where a gene exhibits both patterns, inheritance pattern was designated for the VUSs based on clinical history (Supplemental Fig. 1). Several genes with a digenic inheritance pattern were additionally identified (Supplemental Fig. 2). In cases of uncertain inheritance pattern, conflicts were resolved on discussion between the primary and senior authors (MMF, SLR, and DKC). Based on the in silico VUS predictions and these inheritance patterns, in silico “possible genetic diagnoses” were made: subjects with two VUSs in the same AR gene or a single VUS-D in an AD gene were defined as having a “possible genetic diagnosis;” for these individuals, clinically validated P/LP classification of their VUSs in the future would allow a definite genetic diagnosis to be made.
Statistical analyses
Descriptive statistics are presented as percentages or means ± standard deviations. ANOVA was used to identify predictors of our outcomes of interest. Binomial logistic regression was used to assess for predictors and confounding variables.
Results
Sample population and hearing-loss gene-panel testing outcomes
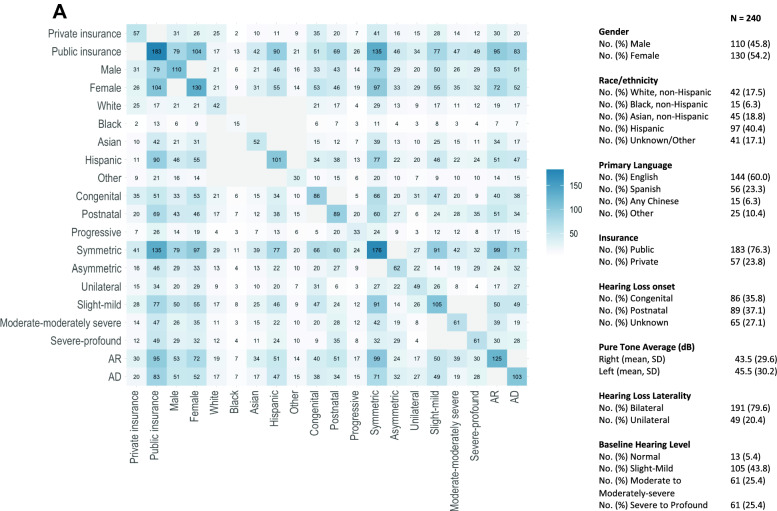
Data were collected from 240 children, 0–22 years old, with SNHL (Fig. 1). The study population was racially, ethnically, and linguistically diverse, with a majority publicly insured (76%). 36% had congenital hearing loss. Children represented a wide range of hearing levels and were predominantly bilaterally affected. Universal public insurance access made possible equitable utilization of hearing-loss gene-panel testing, such that children in different racial and ethnic groups had comparable clinical characteristics; specifically, hearing-loss laterality, hearing level, and comorbidities were not statistically significantly different across Asian, Black, Hispanic, and White subjects. White subjects were more likely to have congenital hearing loss (Table 1).
Fig. 1.
Study population demographic and clinical characteristics. A. Blue shading and numbers reported in individual boxes indicate the number of patients with the paired criteria delineated by row and column. B. Categorical variables were reported as both a number and percentage. Descriptive analysis of continuous variables is reported as a mean and standard deviation for normally distributed variables
Table 1.
Comparison of clinical characteristics across racial and ethnic groups with reported p value of one-way ANOVA
| White | Black | Asian | Hispanic | Other/unknown | p value | |
|---|---|---|---|---|---|---|
| % Bilateral | 78.6 | 80.0 | 80.8 | 80.2 | 76.7 | 0.99 |
| % Severe/profound | 28.6 | 26.7 | 21.2 | 23.8 | 33.3 | 0.40 |
| % Congenital | 50.0 | 40.0 | 28.9 | 33.7 | 33.3 | < 0.001 |
| % With comorbidities | 23.8 | 26.7 | 5.8 | 19.8 | 20.0 | 0.12 |
All 240 children underwent comprehensive hearing-loss gene-panel testing. 944 variants were identified in our study population (Supplemental Tables 1 and 2), with an average of 3.8 ± 2.1 variants identified per patient. The majority of variants were Variants of Uncertain Significance (VUSs; 82%). Of the remainder, 14% were identified as Pathogenic (P) and 5% Likely Pathogenic (LP); we analyzed these together as P/LP variants. 22% of subjects overall received a definite genetic diagnosis based on the presence of P/LP variants with the appropriate inheritance pattern.
Racial and ethnic disparities in genetic testing outcomes
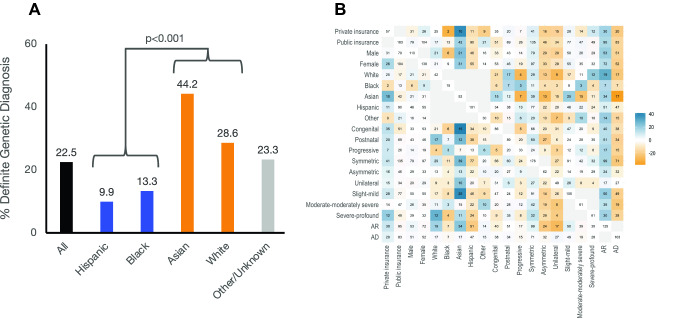
We sought to assess for differences in definite genetic diagnostic rates across racial and ethnic groups. We found that Asian and White children had a higher rate of definite genetic diagnoses (26% and 46%, respectively) when compared to Black and Hispanic children (10% and 13%, respectively) (Fig. 2). The majority of these diagnoses (17/24 Asian, 6/10 Hispanic, and 5/11 White) were attributable to variants in GJB2, particularly the c.109G>A and c.35delG variants common in Asian and European populations, respectively. However, even when these variants are excluded, the disparity remains, with diagnostic rates still lower in Black and Hispanic subjects (7% for these groups together, compared to 20% of White and Asian subjects).
Fig. 2.
Definite genetic diagnostic rate. A. Distribution of definite genetic diagnoses (Patient MD = 4) across race/ethnicity groups. Definite genetic diagnostic rate was compared by ANOVA between dichotomized race/ethnicity groups (White/Asian and Black/Hispanic). B. Distribution of definite genetic diagnosis across insurance, sex, racial-ethnic group, onset, severity, laterality, and inheritance characteristics. Coloring/shading is indicative of difference of diagnostic rate from the average diagnostic rate adjusted by row: orange indicates below the average diagnostic rate for patients with the characteristic defined by the row, white indicates a diagnostic rate average for patients with the characteristic defined by the row, and blue indicates higher diagnostic rate for patients with the characteristic defined by the row. Numbers in boxes indicate the number of patients who received a definite genetic diagnosis, defined as Patient MD of 4, with paired characteristics of the column and row
To reflect this disparity for subsequent analyses, we dichotomized race and ethnicity into a group of Black and Hispanic children and a group of Asian and White children. This grouping is further justified by our finding that published literature on hearing-loss genetics underrepresents African and Latino American subjects by 20–30-fold compared with Asian and European subjects (Rouse et al. 2021). Thus, Black and Hispanic children in our study constitute an underrepresented minority (URM) group in hearing-loss genetics compared with control (Asian and White) subjects. On one-way ANOVA, there was a significant association between definite genetic diagnosis and race/ethnicity, with only 10% of URM children receiving a definite molecular diagnosis compared with 37% of White and Asian controls (p < 0.001).
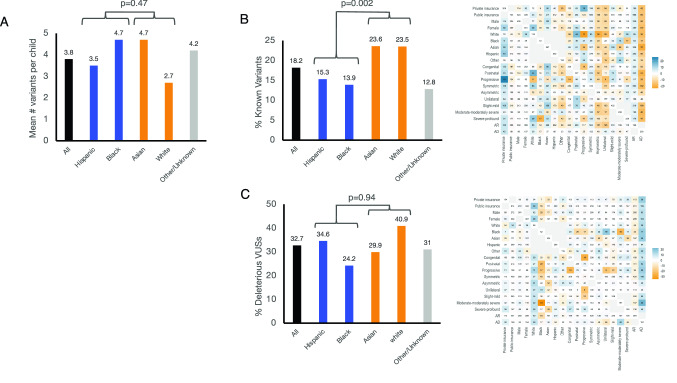
We sought to understand why the definite genetic diagnostic rate varies by race and ethnicity. One possible contributor to this disparity is a difference in the number of rare variants identified between the two groups. After NGS, rare variants are identified by filtering based on racial/ethnic-group-specific allele frequency. One-way ANOVA demonstrated no significant difference between URM and control groups in the number of variants identified per patient (3.8 vs 3.6 variants, p = 0.47; Fig. 3A). Therefore, the number of rare variants detected was unlikely to contribute to the association between race/ethnicity and definite genetic diagnostic rate.
Fig. 3.
Variant distribution. A. Mean number of variants identified per child across race/ethnicity groups with reported p-value of ANOVA for mean number of variants vs dichotomized race/ethnicity (White/Asian and Black/Hispanic). B. Known Variant rate. Left: Distribution of Known Variants across race/ethnicity groups. Known Variant rate was compared by ANOVA between dichotomized race/ethnicity groups (White/Asian and Black/Hispanic). Right: Distribution of Known Variants across characteristics is shown. Color scheme is as described in Fig 2. C. Predicted deleterious VUS rate. Left: Distribution of predicted deleterious VUSs (by PROVEAN prediction) across race/ethnicity groups. Deleterious VUS rate was compared by ANOVA between dichotomized race/ethnicity groups (White/Asian and Black/Hispanic). Right: Distribution of deleterious VUSs across characteristics is shown. Color scheme is as described in Fig 2
We then probed the hypothesis that the variants identified in our control population were better studied than those found in URM subjects and therefore were more likely to lead to a genetic diagnosis. To assess this, we examined the percentage of P/LP variants, many of which are known to be pathogenic because they have been previously described or published. Overall, only 18% of the variants were P/LP variants, with the remainder (82%) classified as VUSs. One-way ANOVA demonstrated that P/LP variants were significantly less common in URM compared with control children (15.0% vs 24.0%, p = 0.002; Fig. 3B).
This disparity in P/LP variants suggests that the body of knowledge contributing to variant classification is inequitable with respect to race and ethnicity. Alternatively, URM children may simply have a lower proportion of deleterious variants in hearing-loss genes. To probe this further, we analyzed variants that were categorized as VUSs and segregated them based on PROVEAN prediction. Because the PROVEAN predictions are based on the in silico predicted effect of the genetic variant on protein function, they should be independent of prior literature, and therefore less subject to prior unequal representation of racial and ethnic groups. We dichotomized VUSs into predicted deleterious VUSs and predicted benign or unknown VUSs, and found no significant difference between the URM and control groups (p = 0.78; Fig. 3C).
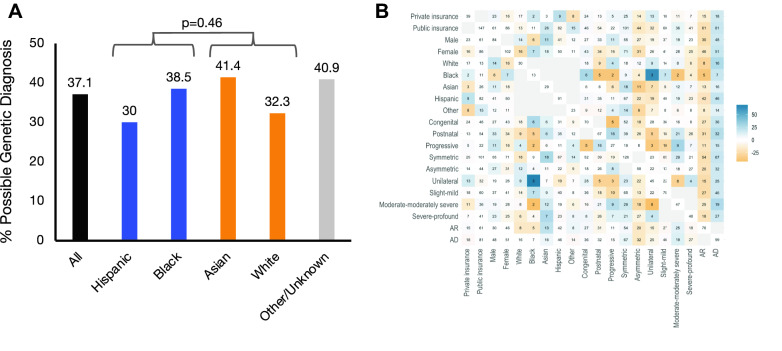
We tested the impact of this racial and ethnically agnostic in silico method of variant prediction on genetic diagnostic rate. Subjects with a single predicted deleterious VUS in an autosomal dominant gene or two VUSs in an autosomal recessive gene were defined as having a “possible genetic diagnosis” based on the supposition that clinical validation of these VUSs as P/LP would yield a genetic diagnosis. Using this in silico classification scheme, URM and control children had comparable possible genetic diagnostic rates (41% vs 38%, p = 0.72; Fig. 4). Thus, when an in silico, racial, and ethnically agnostic variant classification scheme was used, the racial and ethnic disparity in genetic diagnostic rate was eliminated.
Fig. 4.
Possible genetic diagnostic rate. A. Distribution of possible genetic diagnoses (Patient MD = 3) across race/ethnicity groups. Possible genetic diagnostic rate was compared by ANOVA between dichotomized race/ethnicity groups (White/Asian and Black/Hispanic). B. Distribution of possible genetic diagnosis across characteristics is shown. Color scheme is as described in Fig. 2
The comparison between genetic testing outcomes and clinical characteristics in URM and control groups is summarized in Table 2. Even though the two groups were indistinguishable clinically and had a comparable number of rare variants detected, URM subjects had significantly fewer P/LP variants, more VUSs, and worse definite genetic diagnostic rate. To determine the effect of racial/ethnic representation on genetic diagnosis adjusting for clinical and demographic confounders, we performed binomial logistic regression (Table 3). The odds of a URM (Black or Hispanic) child receiving a definite genetic diagnosis is 0.19 that of a control (White or Asian) child (95% CI 0.09–0.41, p < 0.001). When the diagnosis is based on race-and-ethnicity agnostic PROVEAN prediction, race and ethnicity no longer affect the overall distribution of genetic diagnoses (p = 0.52).
Table 2.
Comparison of genetic outcomes and clinical characteristics between URM (Black and Hispanic) and control (Asian and White) groups
| White/Asian | Black/Hispanic | p value | |
|---|---|---|---|
| Number of variants per child (mean, SD) | 3.8 (2.1) | 3.6 (2.1) | 0.47 |
| % P/LP variants | 23.6 | 15.1 | 0.002 |
| VUS (%) | 76.4 | 84.9 | < 0.001 |
| % of VUSs that are predicted deleterious | 38.7 | 38.1 | 0.78 |
| Definite genetic diagnosis (%) | 37.1 | 10.3 | < 0.001 |
| Possible genetic diagnosis (%) | 41 | 38.1 | 0.72 |
| % Bilateral | 79.6 | 80.1 | 1.00 |
| % Severe/profound | 23.7 | 24.5 | 0.55 |
| % Congenital | 37.3 | 31.6 | 0.60 |
| % With comorbidities | 15.5 | 20.5 | 0.34 |
| % Publicly insured | 68.1 | 88.4 | < 0.001 |
| % Non-English-speaking | 42.0 | 46.2 | 0.26 |
p values from one-way ANOVA
Table 3.
Logistic regression analysis of definite and possible genetic diagnosis with clinical and demographic covariates, including URM status (URM (Black or Hispanic) vs. control (Asian or White), primary home language, hearing-loss laterality, hearing-loss onset, baseline hearing-loss level, and presence of comorbidities
| Definite genetic diagnosis | |||
|---|---|---|---|
| Odds ratio | p value | 95% CI | |
| Race/ethnicity (URM re: control) | 0.19 | < 0.001 | (0.09–0.41) |
| Language (non-English re: English) | 0.62 | 0.25 | (0.27–1.40) |
| Laterality (unilateral re: bilateral) | 0.28 | 0.03 | (0.08–0.89) |
| Onset (post-natal re: congenital) | 1.67 | 0.26 | (0.68–1.50) |
| Hearing level (severe–profound re: mild–moderate) | 0.63 | 0.30 | (0.26–1.50) |
| Comorbidities (present re: absent) | 0.31 | 0.05 | (0.10–1.00) |
| Possible genetic diagnosis | |||
|---|---|---|---|
| Odds ratio | p value | 95% CI | |
| Race/ethnicity (URM re: control) | 0.86 | 0.66 | (0.43–1.71) |
| Language (non-English re: English) | 1.17 | 0.67 | (0.58–2.37) |
| Laterality (unilateral re: bilateral) | 0.52 | 0.13 | (0.23–1.20) |
| Onset (post-natal re: congenital) | 0.75 | 0.48 | (0.34–1.67) |
| Hearing level (severe–profound re: mild–moderate) | 0.58 | 0.19 | (0.26–1.31) |
| Comorbidities (present re: absent) | 0.83 | 0.64 | (0.37–1.83) |
The reference level “re:” is indicated for each variable. Results include the odds ratio, 95% confidence interval, and p values
Bold values indicate statistically significant associations (p < 0.05)
Discussion
Identifying genetic etiology of hearing loss can improve clinical care. With the advent of NGS, genetic testing is readily available and has become a recommended test for etiologic workup of bilateral SNHL (Shearer et al. 2019), but this approach has its limitations. Access to CGT is often limited by economic factors, and variants are difficult to interpret. The development of massively parallel sequencing technologies has made genetic data more readily available than ever before, shifting the bottleneck in identifying molecular etiology from acquisition of data to meaningful variant interpretation that is accurate, disease-specific, and equitably representative.
We present here an analysis of the clinical efficacy of CGT in a diverse pediatric population. From 240 pediatric patients with SNHL of unknown etiology, 944 variants were identified in 132 genes. Overall, 22% of patients were diagnosed by CGT, with significant variability seen across racial and ethnic groups. Asian (46%) and White (26%) groups had significantly higher diagnostic rates than Black (13%) and Hispanic (10%) children. Though diagnostic rates were overall lower than previous reports, the trend in differences in between racial and ethnic group rates was similar (Sloan-Heggen et al. 2016). Our finding that URM subjects (Blacks and Hispanics) were less likely to receive a definite genetic diagnosis than controls (Asians and Whites) was neither attributable to clinical covariates nor the number of variants identified per child.
Instead, we found that the disparity was related to a significantly lower rate of P/LP variants (and, conversely, higher rate of VUSs) among URM subjects. This inequity in variant classification is likely due to a higher rate of prior genetic studies performed on White and Asian populations compared to Black and Hispanic ones, an inequity that has been demonstrated repeatedly (Edwards et al. 2020). Specific to hearing loss, most SNHL genes have been identified in families from consanguinity belts in the Middle East and India (Bademci et al. 2016). On the other hand, few studies have been done in people of African or indigenous American descent, due to a combination of decreased access to care as well as cultural differences, historic stigmatization, and discrimination that have contributed to avoidance of genetic testing and research (Hall and Olopade 2005, 2006).
We tested to see whether using a method of variant analysis that is agnostic to prior history of testing in different racial and ethnic groups—in silico prediction of deleterious versus benign VUSs—would eliminate this disparity in genetic diagnostic rate. Indeed, these in silico predictions were equitable between URM and control groups, and when we compared the “possible genetic diagnosis” rate, based on the in silico variant classification, URM and control subjects had comparable diagnostic rates. These findings highlight the gap in understanding of variants across these populations and the critical need for increased inclusion of underrepresented groups in genetic hearing-loss studies. Accumulating genetic data on a more diverse group of individuals with and without hearing loss will allow us to better classify VUSs as pathogenic or benign, decreasing the rate of false positives and negatives and increasing the CGT diagnostic value.
Such a classification scheme based on in silico predictions alone without additional clinical validation is clearly not sufficient to make actionable genetic diagnoses. This need for significant evidence to classify variants, however, skews definitive classifications toward populations that are better represented in the literature and variant databases (Gerhard et al. 2018; Manrai et al. 2016). Thus, the clinical value of genetic testing is higher among these groups, exacerbating disparities in treatment and understanding of disease. This is well documented in the study of genetic testing in breast cancer, cardiomyopathy, and chronic kidney disorders, in which racial and ethnic health disparities persist despite the rapid increase in genetic information (Shan et al. 2012; Landry and Rehm 2018; Gasmelseed et al. 2004).
Our study is unique in that 76% of our study population is publicly insured. While Black and Hispanic patients are less likely to have private insurance coverage and therefore less access to genetic testing, access to genetic testing was not a barrier for the Black and Hispanic patients in our study. Additionally, the clinical indications for testing and clinical features themselves were comparable across all groups. The only exception was that White subjects were more likely to have congenital hearing loss (Table 1); however, multiple regression analysis demonstrated that onset of hearing loss (congenital vs post-natal) was not significantly associated with diagnostic rate, whereas race/ethnicity was (Table 3). Because access to testing and clinical indications for obtaining testing was consistent across racial and ethnic groups, differences in diagnostic rate are likely reflective of the disparity in P/LP variants, rather than differences in patients’ clinical features.
There are several limitations to our study. Family history was not available for the majority of patients, so designation of inheritance was not based on family pedigree but instead on clinical report or OMIM classification of affected genes. Thus, the VUS reclassification we designed and tested may disproportionately inflate the role of dominant variants. We mitigated this effect by defaulting to a recessive pattern when inheritance was in question for a VUS. Due to limited family history and parental testing, we cannot confirm biallelic (in trans) variants for most subjects. Pre-clinical syndromic associations may have been missed in clinical evaluations. Though we found equivalent numbers of variants and rate of deleterious VUSs between URM and control groups, individual racial/ethnic groups did show some differences in these rates that did not match the URM/control dichotomization (Fig. 3A, C). Further study is required to understand why these differences may have occurred.
Finally, while our cohort includes many patients underrepresented in studies of genetic HL, only 15 of 240 of our patients were Black, showing the limits to inclusivity and representation even in this relatively large study. Overall, the categorization of race and ethnicity in studies of genetics is complex. Health disparities between vulnerable social groups such as racial and ethnic minorities are frequently based in nonbiologic characteristics such as socio-economic status. Race and ethnicity are sociocultural constructs and are treated as such in this study. It is important to consider disparities within the context of these socially defined categories, as these are the same classifications that lead to disparate treatment. As such, race and ethnicity were self-reported in our study. However, this classification can lead to imprecise designations and limitations in interpretation. Genetic ancestral analysis can provide more precision into classifying the ancestral background of individuals in genetic studies. However, conflating genetic ancestry determined by biological markers and the social construct of race and ethnicity is problematic. Understanding the difference between these constructs is critical in reporting and interpreting genetic studies.
We found that current CGT for SNHL is five times less effective in Black and Hispanic children compared to White and Asian ones, accounting for covariates (Table 3). This decreased hit rate may lead clinical providers to utilize this resource less often in these populations. Compared to White children, Black children with SNHL in the US are already half as likely to receive genetic testing (Qian et al. 2021). If applications of new technology, such as CGT, continue to be utilized disproportionally, the models generated from newly available data risk perpetuating and exacerbating health disparities (Smith et al. 2016). This has been seen in the genetic diagnosis of cancers, in which disparities in genetic testing of cancer predisposition have increased disparities in clinical management (Cragun et al. 2017; Thompson et al. 2003). Similarly, misclassification of hypertrophic cardiomyopathy has been demonstrated in African Americans due to lack of accessible genetic data from appropriate control populations (Gerhard et al. 2018).
The gap in diagnostic utility between racial and ethnic groups highlights the need for expansion of genetic knowledge among traditionally underrepresented groups of D/HH individuals. Targeted studies of underrepresented groups to understand these hearing-loss genes and variants, as well as acquisition of large-scale sequencing data from diverse populations, are necessary to close this gap. Expanding the scope of testing to involve whole-exome sequencing will allow better identification of novel variants in genes not currently represented in existing panels. While CGT is one of the strongest tools in the clinical evaluation of HL, there is a need to establish that it is equivalently useful and clinically valid across all populations, or else it threatens to exacerbate existing disparities (Smith et al. 2016). As efforts increase to develop gene therapy for hearing loss, ensuring an inclusive basis of genetic diagnosis is critical to avoid propagation of historical inequities from testing to treatment.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
MMF, SLR, and DKC contributed to the study conception and design. All authors contributed to material preparation, data collection, and analysis. The first draft of the manuscript was written by MMF and SLR and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by an award from the Claire Giannini Fund, and by NIDCD R01DC018583 to DKC.
Availability of data and materials
All relevant data are included in this manuscript.
Declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval and consent
This study was approved by the Institutional Review Board at UCSF.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Michelle M. Florentine and Stephanie L. Rouse have contributed equally to this manuscript.
References
- ACMG (2015) Analysis TD, Review D. and variant on process data analysis classification
- Amendola LM, Jarvik GP, Leo MC, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;98(6):1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bademci G, Foster J, Mahdieh N, et al. Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive non-syndromic deafness in a large multiethnic cohort. Genet Med. 2016;18(4):364–371. doi: 10.1038/gim.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth KT (2018) Unraveling the genotypic and phenotypic complexities of genetic hearing loss. 10.17077/etd.h3om-klhg
- Brodie KD, Moore AT, Slavotinek AM, et al. Genetic testing leading to early identification of childhood ocular manifestations of Usher syndrome. Laryngoscope. 2020 doi: 10.1002/lary.29193. [DOI] [PubMed] [Google Scholar]
- Chan DK, Chang KW. GJB2-associated hearing loss: systematic review of worldwide prevalence, genotype, and auditory phenotype. Laryngoscope. 2014;124(2):E34–53. doi: 10.1002/lary.24332. [DOI] [PubMed] [Google Scholar]
- Cragun D, Kinney AY, Pal T. Care delivery considerations for widespread and equitable implementation of inherited cancer predisposition testing. Expert Rev Mol Diagn. 2017;17(1):57–70. doi: 10.1080/14737159.2017.1267567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TL, Breeyear J, Piekos JA, Edwards DRV. Equity in health: consideration of race and ethnicity in precision medicine. Trends Genet. 2020;36(11):807–809. doi: 10.1016/j.tig.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortnum HM, Summerfield AQ, Marshall DH, Davis AC, Bamford JM. Prevalence of permanent childhood hearing impairment in the United Kingdom and implications for universal neonatal hearing screening: questionnaire based ascertainment study. BMJ. 2001;323(7312):536. doi: 10.1136/bmj.323.7312.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasmelseed NMA, Schmidt M, Magzoub MMA, et al. Low frequency of deafness-associated GJB2 variants in Kenya and Sudan and novel GJB2 variants. Hum Mutat. 2004;23(2):206–207. doi: 10.1002/humu.9216. [DOI] [PubMed] [Google Scholar]
- GeneDx (2015) Test Information Sheet—XomeDx, pp 10–13
- Gerhard GS, Fisher SG, Feldman AM. Genetic testing for inherited cardiac diseases in underserved populations of non-European ancestry: double disparity. JAMA Cardiol. 2018;3(4):273. doi: 10.1001/jamacardio.2017.5345. [DOI] [PubMed] [Google Scholar]
- Hall M, Olopade OI. Confronting genetic testing disparities knowledge is power. JAMA. 2005;293(14):1783–1785. doi: 10.1001/jama.293.14.1783. [DOI] [PubMed] [Google Scholar]
- Hall MJ, Olopade OI. Disparities in genetic testing: thinking outside the BRCA box. J Clin Oncol. 2006;24(14):2197–2203. doi: 10.1200/JCO.2006.05.5889. [DOI] [PubMed] [Google Scholar]
- Harrison SM, Riggs ER, Maglott DR, et al. Using ClinVar as a resource to support variant interpretations. Curr Protoc Hum Genet. 2016;89:8–16. doi: 10.1002/0471142905.hg0816s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgert N, Smith RJH, Van Camp G. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res. 2009;681(2–3):189–196. doi: 10.1016/j.mrrev.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberling WJ, Hildebrand MS, Shearer AE, et al. Frequency of Usher syndrome in two pediatric populations: implications for genetic screening of deaf and hard of hearing children. Genet Med. 2010;12(8):512–516. doi: 10.1097/GIM.0b013e3181e5afb8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry LG, Rehm HL. Association of racial/ethnic categories with the ability of genetic tests to detect a cause of cardiomyopathy. JAMA Cardiol. 2018;3(4):341. doi: 10.1001/jamacardio.2017.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeko K, Bosch J, Noubiap JJN, Dandara C, Wonkam A. Genetics of hearing loss in Africans: use of next generation sequencing is the best way forward. Pan Afr Med J. 2015 doi: 10.11604/pamj.2015.20.383.5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liming BJ, Carter J, Cheng A, et al. International Pediatric Otolaryngology Group (IPOG) consensus recommendations: hearing loss in the pediatric patient. Int J Pediatr Otorhinolaryngol. 2016;90:251–258. doi: 10.1016/j.ijporl.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Manrai AK, Funke BH, Rehm HL, et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. 2016;375(7):655–665. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML, Ploughman LM, Rawlings B, Remington E, Arnos KS, Nance WE. Genetic epidemiological studies of early-onset deafness in the U.S. school-age population. Am J Med Genet. 1993;46(5):486–491. doi: 10.1002/ajmg.1320460504. [DOI] [PubMed] [Google Scholar]
- Morton CC, Nance WE. Newborn hearing screening—a silent revolution. N Engl J Med. 2006;354(20):2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- On behalf of the ACMG Laboratory Quality Assurance Committee. Richards S, Aziz N, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza AM, DiStefano MT, Hemphill SE, et al. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum Mutat. 2018;39(11):1593–1613. doi: 10.1002/humu.23630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian ZJ, Chang KW, Ahmad IN, Tribble MS, Cheng AG. Use of diagnostic testing and intervention for sensorineural hearing loss in US children from 2008 to 2018. JAMA Otolaryngol Neck Surg. 2021;147(3):253–260. doi: 10.1001/jamaoto.2020.5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse SL, Florentine MM, Taketa E, Chan DK (2021) Racial and ethnic disparities in genetic testing for hearing loss: a systematic review and synthesis. Rev Hum Genet [DOI] [PMC free article] [PubMed]
- Shan J, Mahfoudh W, Dsouza SP, et al. Genome-wide association studies (GWAS) breast cancer susceptibility loci in Arabs: susceptibility and prognostic implications in Tunisians. Breast Cancer Res Treat. 2012;135(3):715–724. doi: 10.1007/s10549-012-2202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Smith RJH. Massively parallel sequencing for genetic diagnosis of hearing loss: the new standard of care. Otolaryngol Head Neck Surg. 2015;153(2):175–182. doi: 10.1177/0194599815591156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Hildebrand MS, Sloan CM, Smith RJH. Deafness in the genomics era. Hear Res. 2011;282(1–2):1–9. doi: 10.1016/j.heares.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Shen J, Amr S, Morton CC, Smith RJ. A proposal for comprehensive newborn hearing screening to improve identification of deaf and hard-of-hearing children. Genet Med. 2019;21(11):2614–2630. doi: 10.1038/s41436-019-0563-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan-Heggen CM, Bierer AO, Shearer AE, et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet. 2016;135:441–450. doi: 10.1007/s00439-016-1648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJH, Bale JF, White KR. Sensorineural hearing loss in children. Lancet Lond Engl. 2005;365(9462):879–890. doi: 10.1016/S0140-6736(05)71047-3. [DOI] [PubMed] [Google Scholar]
- Smith CE, Fullerton SM, Dookeran KA, et al. Using genetic technologies to reduce, rather than widen, health disparities. Health Aff Proj Hope. 2016;35(8):1367–1373. doi: 10.1377/hlthaff.2015.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson HS, Valdimarsdottir HB, Jandorf L, Redd W. Perceived disadvantages and concerns about abuses of genetic testing for cancer risk: differences across African American, Latina and Caucasian women. Patient Educ Couns. 2003;51(3):217–227. doi: 10.1016/s0738-3991(02)00219-7. [DOI] [PubMed] [Google Scholar]
- Yan D, Tekin D, Bademci G, et al. Spectrum of DNA variants for nonsyndromic deafness in a large cohort from multiple continents. Hum Genet. 2016;135(8):953–961. doi: 10.1007/s00439-016-1697-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in this manuscript.