Abstract
As a process of cellular uptake, endocytosis, with gradient acidity in different endocytic vesicles, is vital for the homeostasis of intracellular nutrients and other functions. To study the dynamics of endocytic pathway, a membrane-anchored pH probe, ECGreen was synthesized to visualize endocytic vesicles under structured illumination microscopy (SIM), a super-resolution technology. Being sensitive to acidity with increasing fluorescence at low pH, ECGreen, can differentiate early and late endosomes as well as endolysosomes. Meanwhile, membrane-anchoring not only improves the durability of ECGreen, but also provides an excellent anti-photobleaching property for long-time imaging with SIM. Moreover, by taking these advantages of ECGreen, a multi-dimensional analysis model containing spatial, temporal, and pH information was successfully developed for elucidating the dynamics of endocytic vesicles and their interactions with mitochondria during autophagy, and revealed a fast conversion of endosomes near the plasma membrane.
Keywords: endocytosis, super-resolution imaging, pH probes, multi-dimensional models, membrane anchor
Graphical Abstract
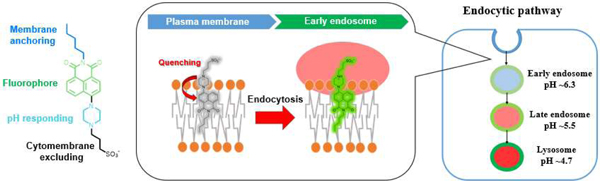
A membrane-anchored pH probe, with excellent durability and photostability in living cells, was developed to study the endocytic dynamics and its interaction with other organelles under nanoscopic visualization.
1. Introduction
As one of the most important species, hydronium, also called proton, plays a critical role in many biological functions, including muscle contraction, ion transport, endocytosis, multidrug resistance, proliferation and apoptosis.[1] The change of proton concentration can make the intracellular pH (pHi) fluctuation, and abnormal pHi values are closely related to illogical cellular processes which lead to diseases such as Alzheimer’s and cancer.[2] The pHi plays important roles in the organelles’ functions, and low pH in lysosomes helps to activate enzyme and protein functions for degradation and the alkalescence makes mitochondria to work regularly.[3] Cellular dysfunction is often associated with abnormal pH values in organelles and hence monitoring pH dynamics of the organelles can help to study the pathological processes.
Fluorescence imaging can help to measure the pH values of organelles using organelles-targeting pH probes.[4] Usually, pH probes are designed based on the special features of the organelles in living cells, such as the acidic environment of lysosomes and the high negative membrane potential of mitochondria.[5] Once the organelles are in an abnormal status, most pH probes will lose their targets and leave the organelles. Therefore, the design of new pH probes that are not affected by the targeting features has attracted great attention.
Endocytosis is a cellular process in which macromolecules are absorbed by plasma membrane-derived vesicles called endosomes.[6] Endocytosis refers to pinocytosis and phagocytosis, and pinocytosis can be divided into macropinocytosis, caveolin- or clathrin-mediated endocytosis, and caveolin- and clathrin-independent endocytosis. Vesicles for the endocytic path, including endosomes (i.e., early endosomes and late endosomes) and endolysosomes, have low pH values from 6.3 to 4.7.[7] The acidic microenvironment can help to maintain the endocytic function of intracellular homeostasis by introducing various nutrients into cells and transporting unwanted components to lysosomes, which collectively act as a waste disposal system.[8] Recent findings also have found that the disruption of endocytosis is related to certain neurodegenerative disorders and immune diseases.[9] Studying the acidic endocytic vesicles at the high spatial resolution can help not only to investigate the dynamics of endocytic pathway but also to understand the relationship between the acidic endocytic vesicles and diseases.
Here, we designed and synthesized a membrane-anchored pH probe, ECGreen, to visualize endocytic vesicles with structured illumination microscopy (SIM) that can overcome the diffraction limit of conventional fluorescence microscopes (~200 nm).[10] ECGreen is a small-molecule membrane-anchored fluorescent probe entering cells via endocytosis. Its fluorescence intensity is enhanced as the acidity of endocytic vesicles increases and thus overcomes the limitations posed by commercial probes for acidic vesicles. Because of membrane anchoring, the probe can stay on the vesicle inner surface stably regardless of pH change and thus effectively avoid the destruction by any reactive species inside the endocytic vesicle, which significantly improves both durability and photostability of ECGreen for long time tracking endocytic vesicles. Moreover, based on ECGreen, we also developed a new multi-dimensional analysis model by incorporating spatial, temporal, and pH information, which can be applied for systematical study of endocytic dynamics. Through this multi-dimensional analysis with ECGreen, we studied the conversation, migration and distribution of endocytic vesicles in live cells and their interaction with mitochondria during autophagy, and discovered a fast conversion of endosomes near the plasma membrane. Our new probe and multi-dimensional analysis pave the way for digitalization of sub-cellular dynamics.
2. Results and Discussion
2.1. Design, Synthesis, and Photophysical Properties
The chemical structure of ECGreen is shown in Figure 1a, it contains a 1,8-naphthalimide moiety as a highly sensitive fluorescent group with “Head” for membrane anchoring and “Tail” group moieties for pH responding and cytomembrane excluding. The “Head” pentyl group, with the dicarboximide moiety, has been reported to possess the strongest signal in liposomal or autophagosomal membranes by us.[11] The pH responding moiety, a piperazine group, is well known for making a fluorophore pH-dependent through a photo induced electron transfer (PeT) mechanism or twisted intramolecular charge transfer (TICT) mechanism.[12] In addition, as the sulfonate group displays a pKa < 0, compounds containing sulfonate moieties are permanently negatively charged in physiological buffers, thereby unable to cross the lipid bilayer membrane.[13] Anchored on the outer membrane, ECGreen can enter live cells via endocytosis and locate on the inner leaflet of endocytic vesicles (Figure 1b). As the acidity of endocytic vesicles increases, the fluorescence intensity of ECGreen is enhanced, that makes it a good probe for tracking the dynamics of endocytic vesicles. The synthetic path of this probe is referred to our reported papers.[11] The identity and purity of the probe were successfully confirmed by the results of ESI-MS, 1H NMR, and HPLC spectra (Figures S1–S3).
Figure 1.
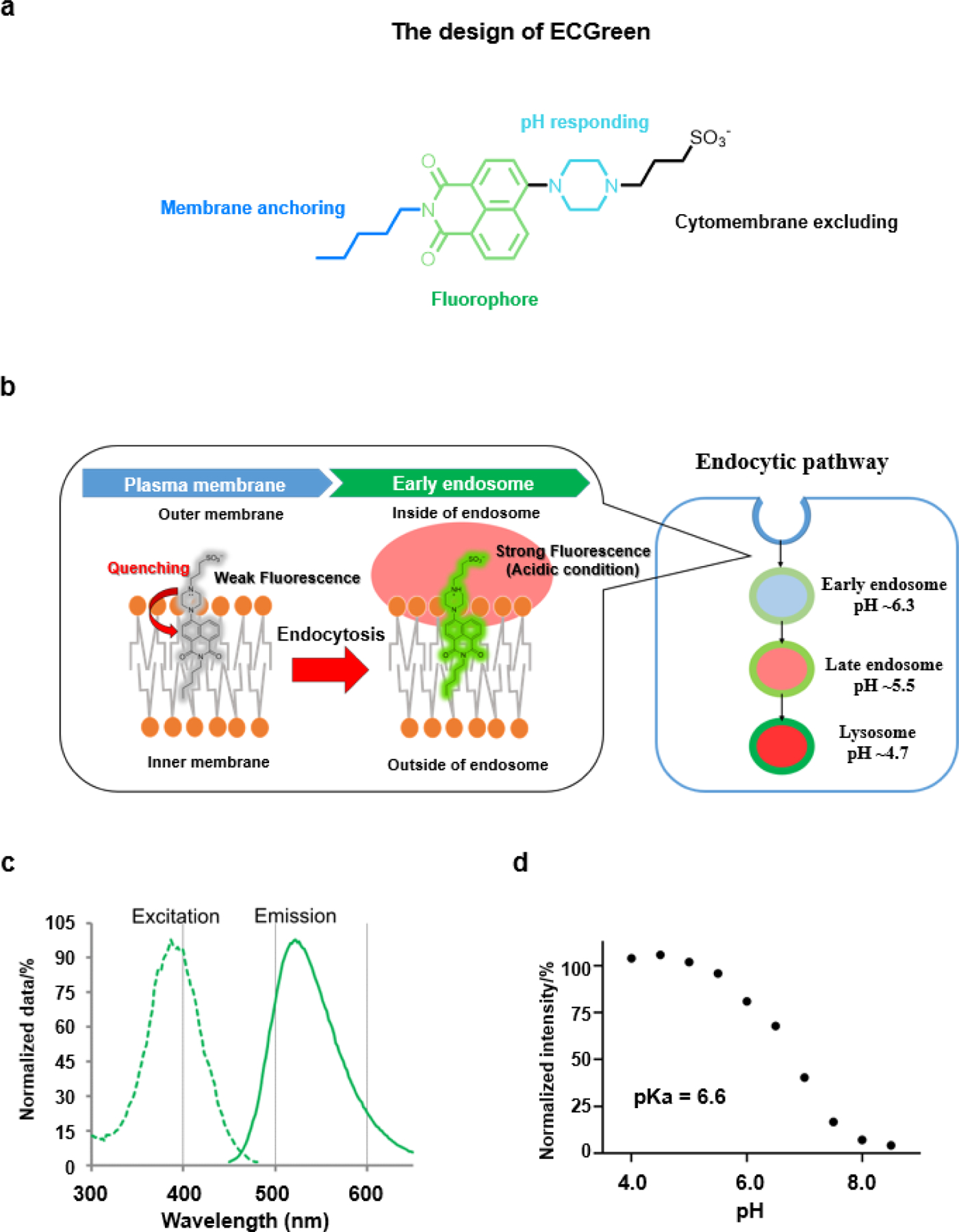
a) The design of the molecular structure for ECGreen. b) The mechanism for visualizing the endocytic pathway. The ECGreen probe is a pH- dependent fluorescence dye without membrane permeability. Anchored on the outer membrane, ECGreen is internalized via endocytosis and on the inner leaflet of endocytic vesicles. As the acidity of endocytic vesicles increases, the fluorescence intensity of ECGreen rises, which makes it a highly specific probe for endocytic vesicles. c) The excitation and emission spectra of ECGreen. d) pH-dependent changes of the fluorescence intensity of ECGreen (1.0 μM) in buffer solutions (pH 4.0–8.5), excited at 405 nm (emission at 520 nm).
To study the photophysical properties, the excitation and emission spectra of ECGreen were first collected. As shown in Figure 1c, ECGreen has an absorption band between 300 and 500 nm, and a band of 450–650 nm for the emission spectrum. Its fluorescence response to different pH is shown in Figure 1d. Lowering the pH value from 8.5 to 4.0, the fluorescence intensity of ECGreen increases and the pKa is calculated to 6.6 (Figure S4). The quantum yields of the probe are 4% at pH 8.0 and 83% at pH 4.0 (refer to fluorescein isothiocyanate, FITC),[14] with a −21-time enhancement (Figure S5). Given the complex intracellular environment, it is important to ensure the probe’s high specificity towards pH change. Control experiments using other essential ions and biologically chemicals confirmed that only proton can make ECGreen light up (Figure S6). In addition, the pH reversibility study showed the fluorescence of ECGreen has no change at the same pH value in different cycle (Figure S7), indicating the probe can be used for illustrating pH changes.
2.2. Calculation of Electronic Structure
Density functional theory (DFT) computation was conducted to aid in the understanding on the electronic structures of ECGreen before and after protonation under acidic conditions. As shown in Figure 2a, its highest occupied molecular orbital (HOMO) is primarily located on the electron-donating piperazine and naphthalene side with an energy of −5.74 eV, while its lowest unoccupied molecular orbital (LUMO) centers over the entire naphthalimide structure with an energy of −2.28 eV. Upon protonation at the terminal N of the piperazine unit, no substantial change in the HOMO/LUMO distribution was observed. However, the energies of HOMO and LUMO of the protonated ECGreen (ECGreen-H) were decreased to −5.83 and −2.35 eV, respectively. More frontier orbitals and energies of ECGreen and ECGreen-H can be found in Tables S1 and S2. The similar HOMO-LUMO gaps of ECGreen before (3.46 eV) and after protonation (3.48 eV) indicate that its UV/visible absorption spectra would not be alternated significantly from neutral to acidic pH, in agreement with reported results of a similar fluorescence probe.[15] Indeed, as shown in Figure 2b, the lowest singlet excited states (S1, primarily from HOMO → LUMO transition) of ECGreen and ECGreen-H were calculated to be located at 411.2 and 405.6 nm, respectively (Tables S3 and S4), well matching the experimentally observed excitation spectra (Figure 1c). Furthermore, the calculated electron density difference maps (EDDMs) of their S1 states are consistent with the distribution of frontier orbitals and plotted in Figure 2b as well.
Figure 2.
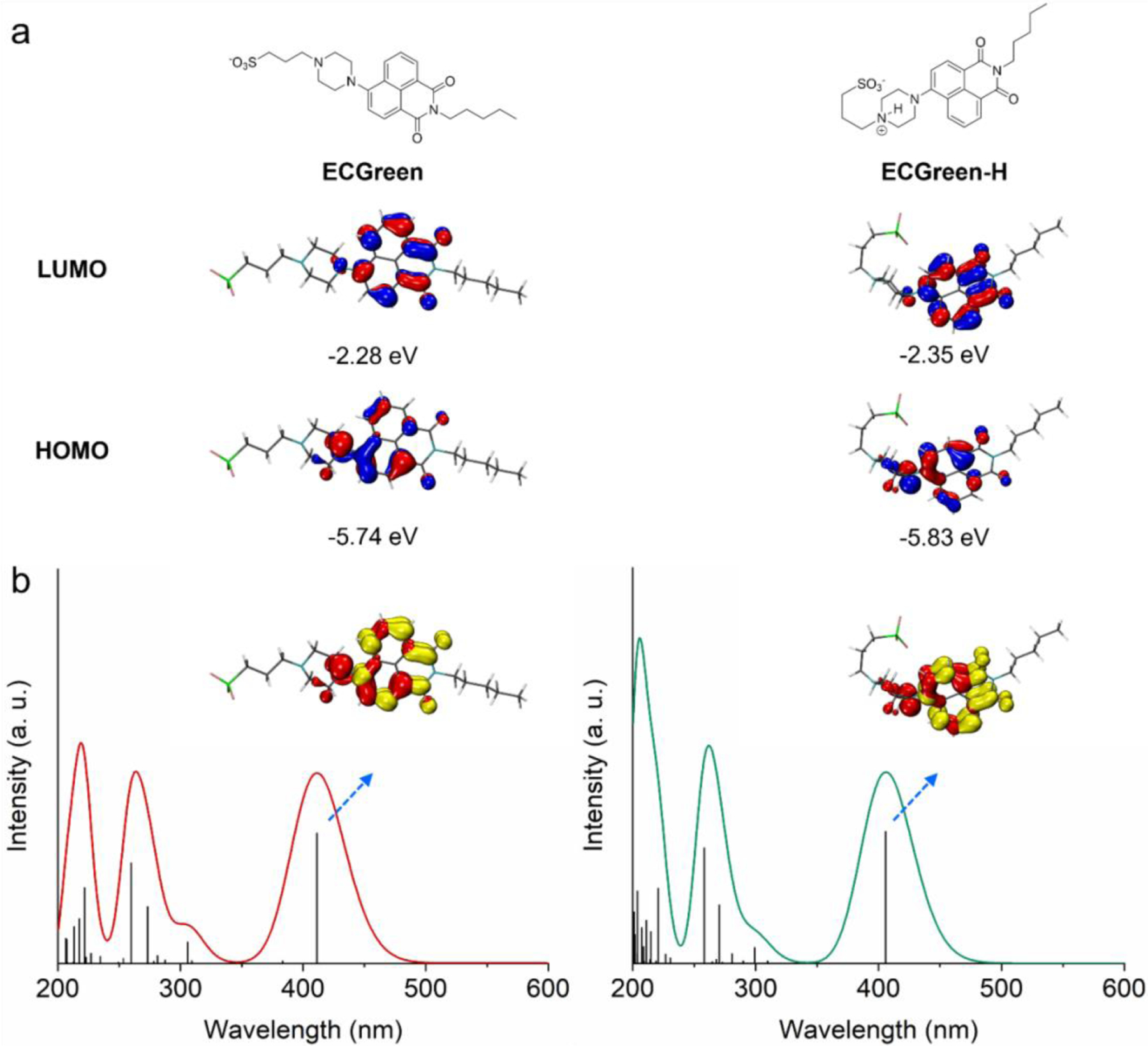
a) Calculated HOMOs and LUMOs and b) simulated UV-vis absorption spectra and EDDMs of the 1st singlet excited states of ECGreen and protonated ECGreen (ECGreen-H) (red and yellow indicate decrease and increase in electron density, Isovalue = 0.04 for plotting HOMOs, LUMOs, and EDDMs).
2.3. Improved Durability for Super-Resolution Imaging
We first tested some basic properties of ECGreen in live cells. The highly water-soluble tetrazolium salt WST-8 in Cell Counting Kit-8 (CCK-8), can be reduced by dehydrogenase activities for measuring the dehydrogenase activity with NADH in a live cell. The higher dehydrogenase activity indicates higher cell viability. The CCK-8 assay revealed that ECGreen was safe for the cells at the different concentrations tested (Figure S8). The longer incubation time was used, the more fluorescent signal inside the cells could be observed (Figure S9). Meanwhile, co-localization analysis with LysoTracker™ Red DND-99 (LTR) (Figure S10) confirmed ECGreen localized at the acidic vesicles. The endocytic extracellular vesicles (EVs), labelled with Mem Dye-Deep Red, were used to co-treat with ECGreen. The ECGreen intensity increased and showed a high overlap with EVs (Figure S11), suggesting the intake of ECGreen through endocytosis. To further confirm that ECGreen enters cells via endocytosis, we performed experiments by inhibiting the endocytic process. Knowing that low temperature can inhibit endocytosis,[16] we tested at low temperature and found the decreased fluorescence intensity of ECGreen at 4 °C (Figure S12). ECGreen can also work in Jurkat cells (Figure S12a). Meanwhile, we used wortmannin, a potent inhibitor of class I and III PI3-kinases, to block maturation from early to late endosomes (Figure S13a).[17] Co-localization studies (Figure S13) revealed that ECGreen co-localized with an early endosomal marker Rab5, a late endosomal marker Rab7, and a lysosomal marker LAMP1, each tagged with RFP (Figure S13c–e). In the presence of wortmannin, RFP–Rab5 showed a ring-shaped structure, indicating a swollen endosome phenotype and location at the endosomal membrane, which are co-localized well with the ring structure observed by ECGreen (Figures S13c and S13g). Meanwhiel, RFP–Rab7 and RFP–LAMP1 did not co-localize with the wortmannin-induced ring structure of ECGreen (Figures S13d, S13e, S13h and S13i). On the other hand, transferrin-Alexa Flour 546 conjugate, a well-known tracer for the recycling endosome pathway, was co-localized with ECGreen signal only in the inhibited condition by wortmannin, indicating that ECGreen is mainly transported to the endo-lysosomal pathway, not the recycling pathway (Figures S13b and S13c). These results suggest that ECGreen specifically locates to the early endosomal membrane and then gets transported to the later endo-lysosomal pathway.
SIM offers super-resolution subcellular imaging in living cells with a low background and can overcome the diffraction limit (< 200 nm) of conventional optical microscopes.[18] Figure S14 displays the difference in imaging quality between SIM and widefield microscopy, including that the clearer fluorescent puncta with a lower background were obtained with SIM. The presence or absence of fetal bovine serum (FBS) exerted no obvious effect on the cellular internalization or imaging quality through SIM (Figure S15). By using SIM to acquire images of cells treated with ECGreen for longer treatment times, we observed more and brighter puncta (Figures 3a and S16). At the end of the 180-min treatment, the fluorescence puncta exhibited high overlap (86.6%) with the co-stained acidic vesicle dye LTR (Figure S17), which confirmed that ECGreen is primarily distributed in acidic vesicles.
Figure 3.
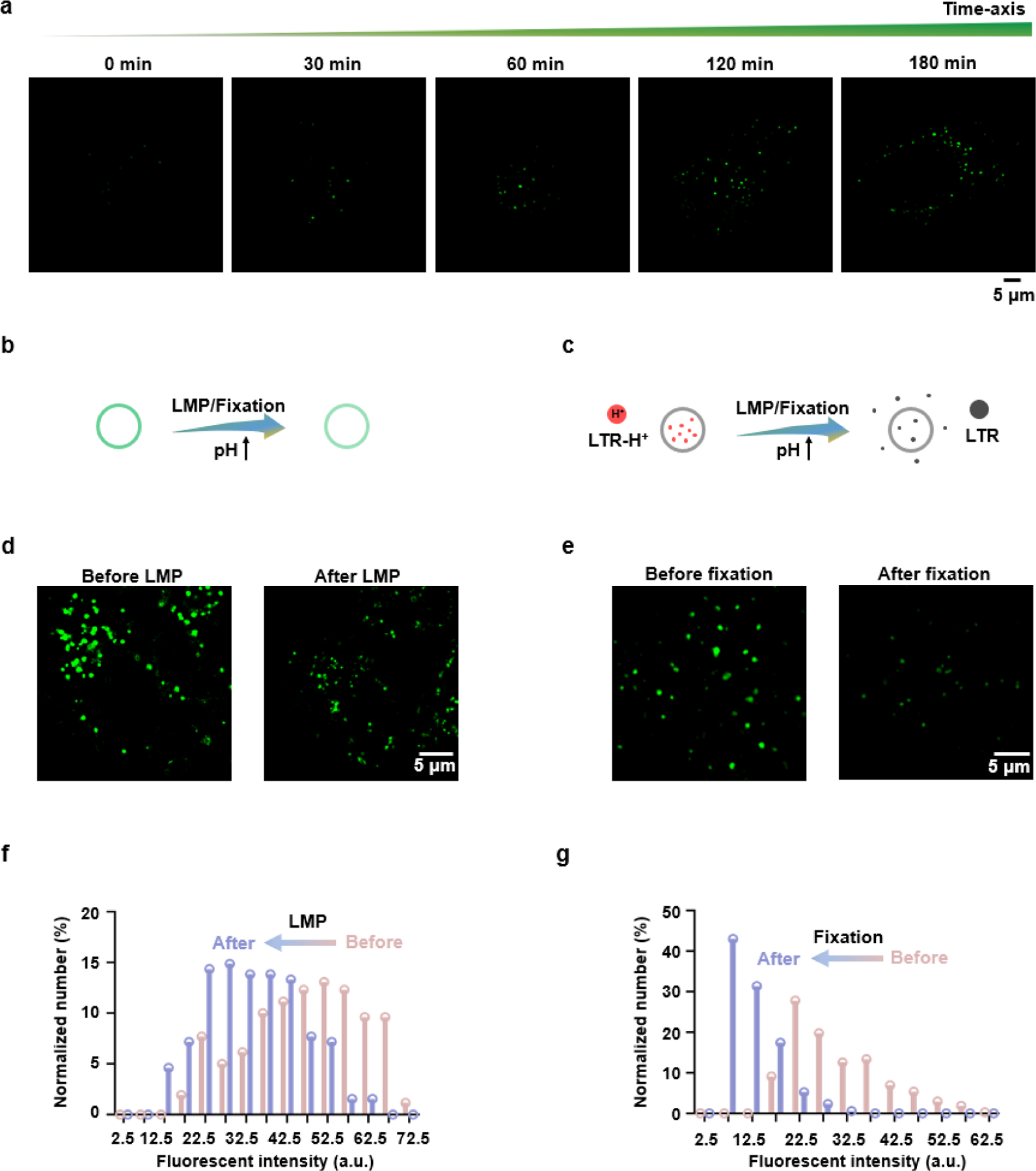
Super-resolution visualization of acidic endocytic vesicles by ECGreen. a) SIM images of HeLa cells labelled with ECGreen for different treatment periods. b, c) Schematic illustration of the acidic vesicles labelled with (b) ECGreen or (c) LTR during treatments with a higher pH. d) SIM images of HeLa cells labelled with ECGreen before and after chloroquine treatment (100 μm, 30 min). e) SIM images of HeLa cells labelled with ECGreen before and after 4% paraformaldehyde treatment (20 min). f) The relationship between the number and fluorescence intensity of ECGreen before and after LMP. g) The relationship between the number and fluorescence intensity of ECGreen before and after fixation.
The endocytic vesicles including early endosomes, late endosomes, and endolysosomes are all acidic vesicles, for which widely used commercial dyes (e.g., LTR) are membrane-permeable and both lit up and trapped in the acidic microenvironment.[19] However, when the pH changes, the commercial dyes lose their targets and consequently their fluorescence vanishes (Figures 3b,c and S18), which reduces its durability. We then tested the durability of ECGreen during the pH fluctuation. An antimalarial drug chloroquine, a cell-permeable base for endolysosomal membrane permeabilization (LMP),[20] and 4% paraformaldehyde, a solution for cell fixation, were used to stimulate live cells for raising the endolysosomal pH value. After treatment, the fluorescence of ECGreen remained with a reduced intensity corresponding to pH increase, whereas the signal of LTR had completely vanished (Figures 3d–g and S18). These results demonstrate the improved durability of ECGreen for super-resolution imaging.
2.4. Enhanced Photostability for long-time Dynamics Tracking
Anti-photobleaching is an important property of probes that determines whether they can be used for long-time tracking.[21] One of the causes for photobleaching is that reactive species destroy the structure of the fluorophore. Since it’s hard for the reactive species to enter membrane, the membrane insertion should benefit photostability. To confirm this, we did a comparison experiment for the photobleaching time of ECGreen, and LTR. Under 100% laser power, the fluorescence of LTR was quickly bleached, while after long irradiation, the fluorescent intensity of ECGreen still retained one fifth of its original value (Figures 4a and 4b), thereby showing a much better anti-photobleaching property of ECGreen. Next, ECGreen was used to track the dynamics of endocytic vesicles with low laser power. The probe maintained 66.20% fluorescence intensity even after 30 images (Figure S19). Thanks to SIM, the dynamics of endocytic vesicles in those images, including the kiss-and-run process, fusion, and fission, were clearly recorded (Figures 4c–4e, Videos S1–S3). Moreover, live HeLa cells labelled with ECGreen or LTR were cultured for four days, and the cells were washed with phosphate buffered saline (PBS) three times before and after imaging. The fluorescence of inside acidic probe LTR almost completely disappeared at the second day. In contrast, ECGreen can be observed for more than four days (Figures 4d, 4e, and S20). All above results indicate the excellent photostability of ECGreen.
Figure 4.
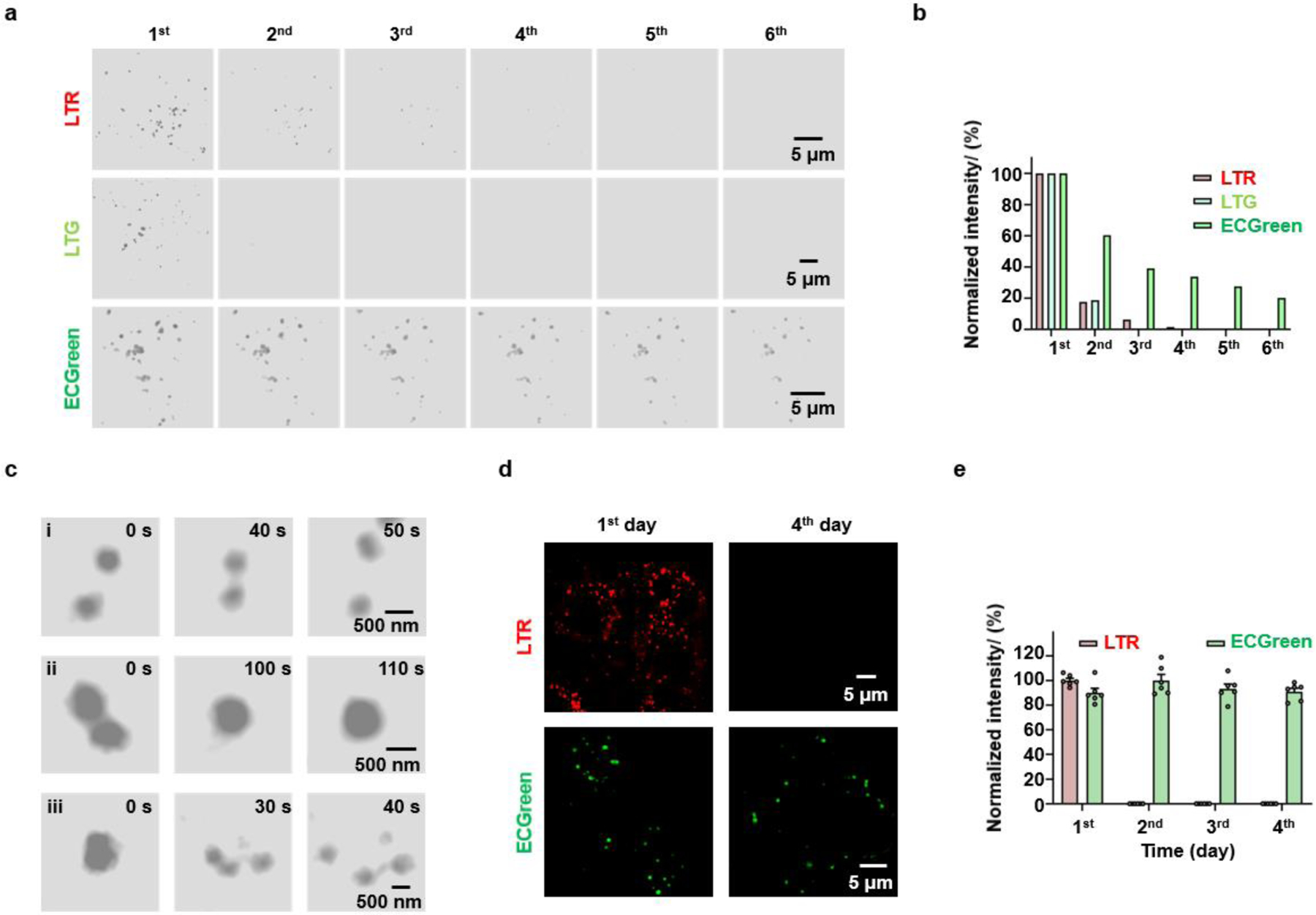
Continuous imaging of endocytic vesicles. a, b) Continuous imaging of HeLa cells labelled with ECGreen, LTR or LTG at the largest laser intensity for photobleaching. The (a) SIM images and (b) normalized fluorescence intensity of ECGreen, LTR and LTG. c) Dynamic SIM images of (i) the kiss-and-run process, (ii) fusion, and (iii) fission process of the endocytic vesicles. d) The images of HeLa cells labelled with LTR or ECGreen for the first day and the fourth day. e) The normalized fluorescence intensity of LTR or ECGreen in HeLa cells at different days.
2.5. Fast Conversion of Endocytic Vesicles Revealed by a Multi-Dimensional Analysis with ECGreen
For the next stage of quantitative cell biology, the digitization of individual cells and sub-cellular organelles has gained its popularity. However, there is still a lack of systematic methods for digitization. Here, we proposed the concept of a multi-dimensional model, containing various parameters including chemical properties (pH, viscosity, polarity, etc.) as well as spatial-temporal information, etc., for highly efficient analysis. With its pH response and excellent durability and photostability for super-resolution imaging, ECGreen was applied to generate a multi-dimensional analysis model for endocytic vesicles. Firstly, without washing, the distance between endocytic vesicles and plasma membrane can be obtained (Figures 5a and S21). Secondly, the fluorescence intensity of ECGreen is correlated with the acidity of the vesicles. Finally, by varying treatment time, spatial, temporal, and pH information can be obtained simultaneously and construct a plot (Figure 5b). As shown in Figure 5c, comparing to evenly distributed fluorescent spots at 3 h, the uneven distribution towards plasma membrane at 0.5 h indicates the fast conversion from early to late endosomes near the plasma membrane. After blocking endocytic pathway by wortmannin treatment (Figure 5d), the distribution of fluorescent spots at 0.5 h became even (Figures 5e, S22, and S23). After 3 h, as the result of endosome conversion, the number and intensity of fluorescent spots both increased (Figures S24, S25, and S26), thus confirming that the high intensity spots were associated with late endosomes or endolysosomes with low pH values. These results demonstrate that the multi-dimensional analysis model with ECGreen can effectively track endocytic vesicles to uncover unknown biological mechanisms.
Figure 5.
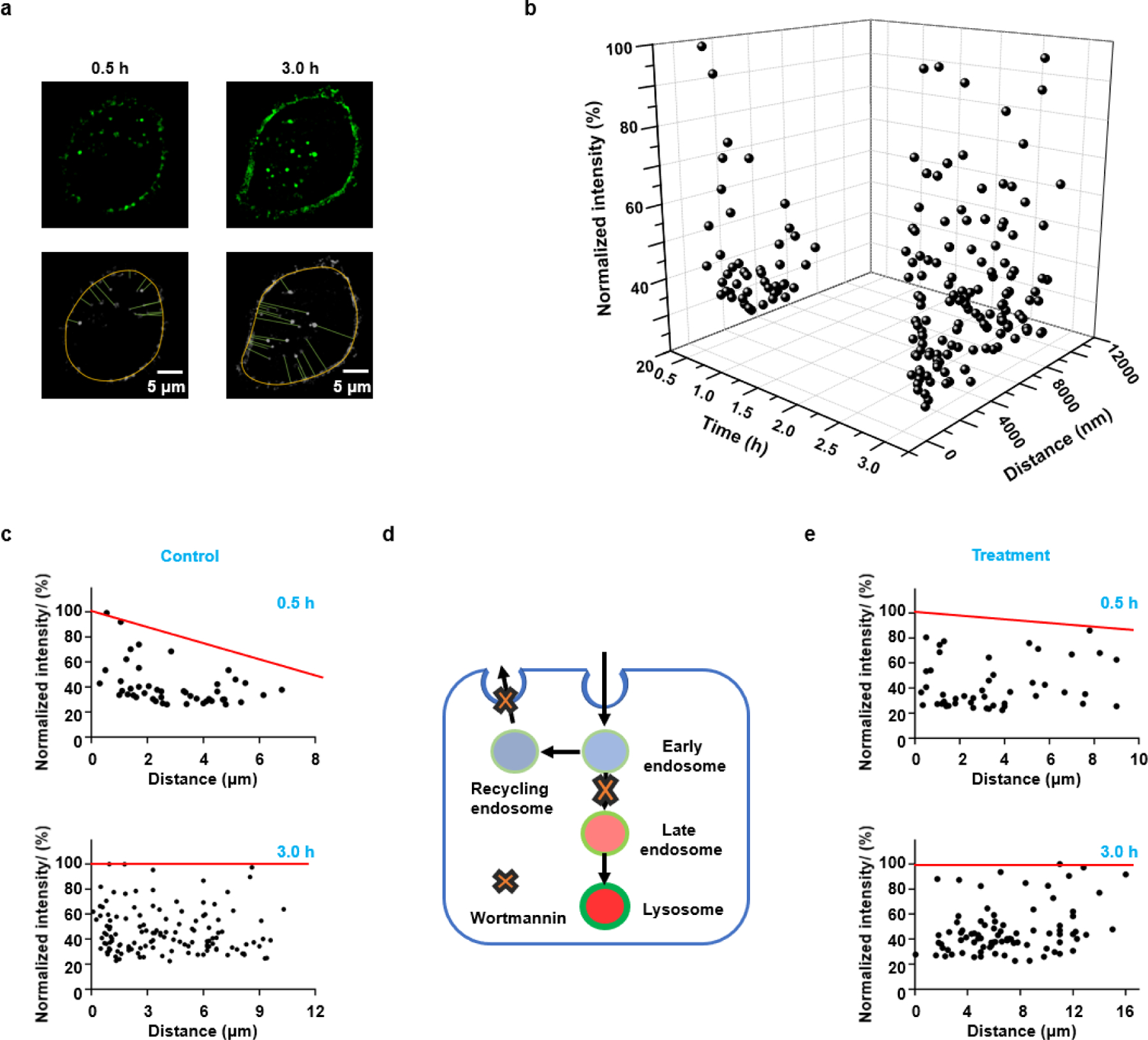
Constructing a multi-dimensional model for endocytic vesicles with ECGreen (5 μM). a) The SIM images of HeLa cells treated with ECGreen for 0.5 h and 3 h, and the quantification of distance between endocytic vesicles and cytomembrane. b) A 3-dimensional space for endocytic vesicles. The parameters were time, distance (between endocytic vesicles and plasma membrane), and fluorescence intensity (pH). c) The relationship between distance and fluorescence intensity for the Control group incubated with ECGreen for 0.5 h and 3 h. d) Schematic illustration of the effect of wortmannin that inhibits the recycling of endosomes and their conversion to endolysosomes. e) The relationship between distance and fluorescence intensity for the group with the treatment of wortmannin (100 nM, with ECGreen) for 0.5 h and 3 h. Red line stands for the tendency of maximum fluorescence intensity for the endocytic vesicles.
2.6. Tracking the Interaction between Endocytic Vesicles and Mitochondria in Autophagy
The interactions of endocytic vesicles with other organelles, such as mitochondria, have been reported to be important for cellular functions.[22] Meanwhile, autophagy is a fundamental process in cellular homeostasis that involves degrading and recycling damaged biomacromolecules or organelles. After autophagosomes engulf damaged biomacromolecules or organelles, they fuse with endolysosomes to degrade their contents.[23] We then used ECGreen and MTDR (MitoTracker™ Deep Red FM) to study the interaction of endocytic vesicles and mitochondria in autophagy. A mitophagy inducer, carbonyl cyanide m‐chlorophenylhydrazone (CCCP) was used to damage mitochondria, by which the morphology of mitochondria became fragmented (Figure 6a). Compared to the control group, the number of ECGreen fluorescent puncta in the CCCP-treated group increased significantly (Figures S27 and S28). Meanwhile, the Pearson’s co-localization coefficient (PCC) value of the interaction increased from 0.96% to 17.12% (Figure 6b), thereby demonstrating the involvement of endocytic vesicles in mitophagy.
Figure 6.
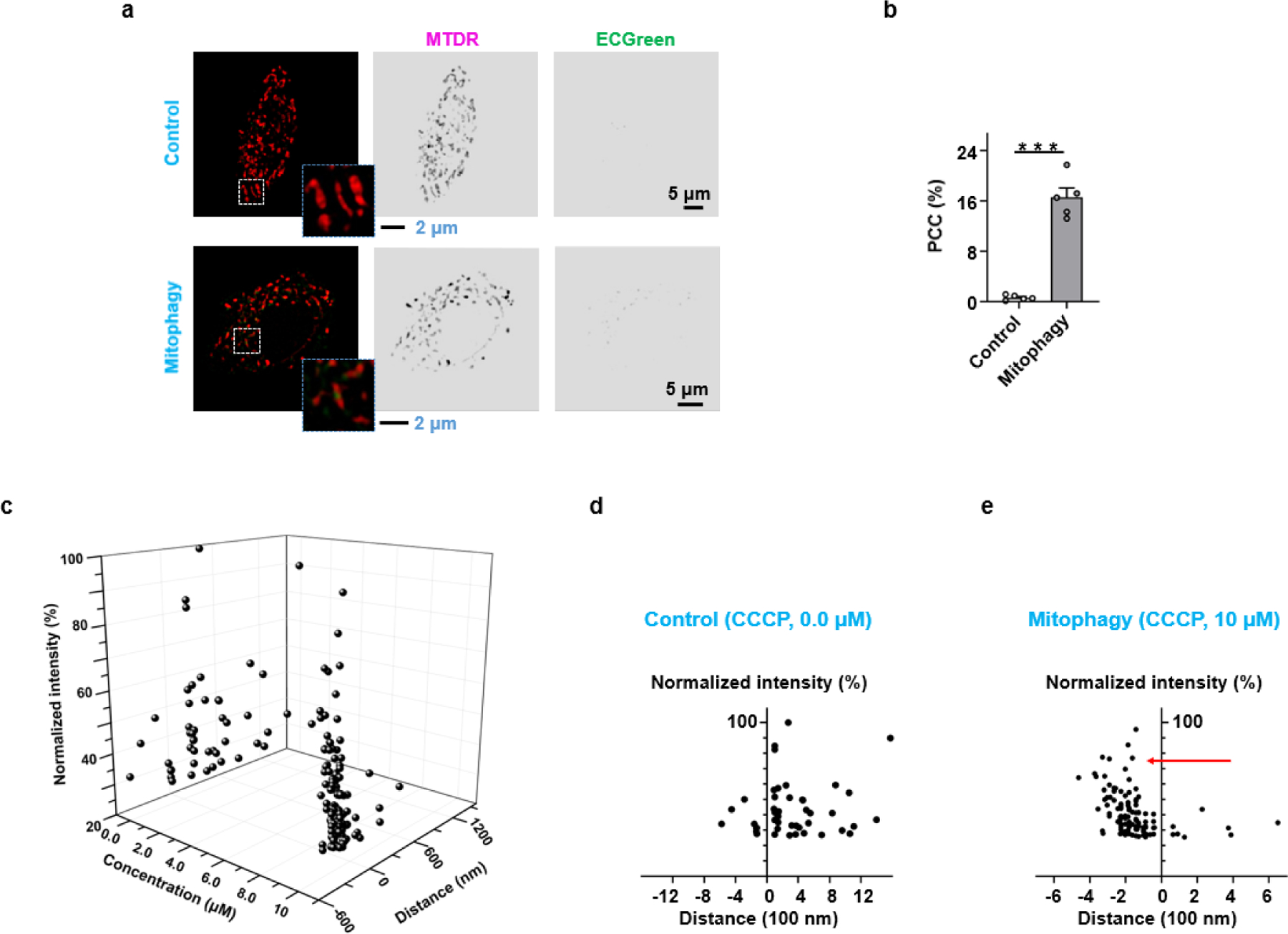
Endocytic vesicles interacted with mitochondria in live HeLa cells labelled with ECGreen and MTDR. The colour of MTDR was false. a) Endocytic vesicles interacting with mitochondria before and after mitophagy. b) PCC values of ECGreen and MTDR before and after mitophagy. c) A 3-dimensional space for endocytic vesicles. The parameters were concentration of CCCP, distance (between endocytic vesicles and mitochondria), and fluorescence intensity (pH). d) The relationship between distance and fluorescence intensity for the Control group without CCCP treatment. e) The relationship between distance and fluorescence intensity for the Mitophagy group with CCCP treatment. Data are given as M ± SEM (n = 6); ***p < 0.001 compared to control cells.
In order to track the dynamics of individual endocytic vesicles in autophagy, we then used the above multi-dimension model to analyze the interaction between endolysosomes and mitochondria in mitophagy. Accordingly, we modified the plotting parameters to the distance between endocytic vesicles and mitochondria, and plotted the fluorescent intensity of individual endocytic vesicles with or without CCCP treatment (Figures 6c and S29). As shown in Figure 6d, most endolysosomes were away from mitochondria in the absence of CCCP. In the mitophagy induced by CCCP treatment, almost all endolysosomes moved close to or into mitochondria (Figures 6e), indicating the fusion between endolysosomes and autophagosomes containing damaged mitochondria. Meanwhile, CCCP might induce a pH alteration in the endolysosomes and affect the distribution of ECGreen in biomembranes. The above results validate that the multi-dimensional analysis with ECGreen is able to catch ongoing cellular process involving endocytic vesicles and other organelles.
3. Conclusion
In sum, we developed ECGreen, a pH probe for endocytic vesicles with membrane anchoring and negligible cytotoxicity. Since the fluorescence intensity of ECGreen correlates with the acidity, it could distinguish different endocytic vesicles (i.e., early endosomes, late endosomes, and endolysosomes). Because of its excellent durability and photostability induced by membrane anchoring, ECGreen is an ideal probe for investigating the dynamics of endocytic vesicles under super-resolution imaging, such as the conversion of endocytic vesicles and the interplay of mitochondria and endocytic vesicles.
In addition, by taking above advantages of ECGreen, we also constructed a multi-dimensional analysis for the dynamics of endocytic vesicles and their interactions with other organelles under various conditions. Digitization of biological system is a trend for qualitative cell biology. With the help of a multi-dimensional analysis system, the correlation of various factors could be easily discovered. As a demonstration, through this multi-dimensional analysis with ECGreen, we found that early endosomes can quickly convert to late endosomes or even endolysosomes right around the plasma membrane. In the future, combined with big data analysis, this method will significantly improve disease diagnosis and promote drug development.
4. Experimental Section
Materials
All reagents and buffers were purchased from Fujifilm Wako Pure Chemical Corporation, unless otherwise noted. Carbonyl cyanide m-chlorophenylhydrazone (CCCP, #C2759) was purchased from Sigma, and MitoTracker™ Deep Red FM (MTDR, #M22426), LysoTracker™ Green DND-26 (LTG, #L7526) and LysoTracker™ Red DND–99 (LTR, #L7528) were purchased from Invitrogen (Thermo Fisher Scientific, USA). Cell Light-Red Fluorescent Protein (RFP) reagents (Thermo Fisher Scientific) were used for staining the early endosome (#C10587), late endosome (#C10589), and lysosome (#C10589). Mem Dye-Deep Red (Dojindo laboratories, #EX03) and PlasMem Bright Red (Dojindo laboratories, #P505) were used for labeling the Extracellular Vesicles (EVs) and for staining the cellular membrane, respectively. All fluorescent dyes were used following the product manuals. Penicillin–streptomycin (#15140163, 10,000 units/mL), fetal bovine serum (FBS, #26140079), and Dulbecco’s modified Eagle’s medium (DMEM, #11965092) were all purchased from Gibco (Thermo Fisher Scientific, USA). Phosphate-buffered saline (PBS, #SH30256.01) was purchased from Hyclone (GE Healthcare Life Sciences).
Instruments
1H-NMR spectra were recorded on a Bruker AVANCE III HD 400 MHz spectroscopy. Mass spectra were measured with a JMS-T100CS (JEOL), Waters SQD2 (Waters). Fluorescence spectra were measured on an FP-6300 fluorescence spectrophotometer (JASCO). Fluorescence images were obtained by LSM 800 confocal laser scanning microscopy (Zeiss), with excitation at 405 nm (for ECGreen), 561 nm (for LTR, RFP and PlasMem Bright Red), and 640 nm (for Mem Dye-Deep Red), using a 500–550-nm filter for ECGreen, a 550–650-nm filter for LTR, RFP and PasMem Bright Red, or a 640–760-nm filter for Mem Dye-Deep Red. Theoretical calculations were carried out using ChemOffice Professional 16 (PerkinElmer). HPLC analysis was performed under an isocratic condition (A: H2O containing 0.1% TFA, B: acetonitrile containing 0.1% TFA; A/B = 60/40) at a flow rate of 1 mL/min using an Inertsil ODS-3 column (250 × 4.6 mm, 5 μm) on a LC-20A (Shimadzu). Absorbance at 254 nm was monitored.
Chemical characteristics of ECGreen
ECGreen (Sodium 3-(4-(1,3-dioxo-2-pentyl-1,3-dihydro-1H-benzo[de]isoquinolin-6-yl)piperazin-1-yl) propansulfonate): 1H-NMR (400 MHz, MeOD) δ: 8.52 (dd, 2H, J = 8.0 Hz), 8.47 (d, 1H, J = 8.0 Hz), 7.80 (t, 1H, J = 8.0 Hz), 7.40 (d, 1H, J = 8.0 Hz), 4.11 (t, 2H, J = 8.0 Hz), 3.47 (s, 4H), 3.38 (s, 4H), 3.21 (s, 2H), 3.02 (t, 2H, J = 8.0 Hz), 3.29–3.22 (m, 2H), 1.74–1.67 (m, 2H), 1.44–1.40 (m, 4H), 0.96–0.93 (m, 3H); ESI-MS: calcd for [M]−, 472.19; found, 472.32.
Spectrophotometric Measurement
The pH-dependent fluorescence intensity of ECGreen was measured in 50% DMSO in MES at pH 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0 and 8.5, using a fluorescence spectrophotometer (excitation 405 nm, emission 520 nm).
Calculation
All calculations were performed with the Gaussian 16[24] program package employing the DFT method with Becke’s three-parameter hybrid functional and Lee-Yang-Parr’s gradient corrected correlation functional (B3LYP).[25] 6–31G* basis set was applied for H, C, O, S and N.[26] The geometries of the singlet ground states of compounds were optimized in H2O using the conductive polarizable continuum model (CPCM). The local minimum on each potential energy surface was confirmed by frequency analysis. Time-dependent DFT calculations produced the singlet excited states of each compound starting from the optimized geometry of the corresponding singlet ground state, using the CPCM method with H2O as the solvent. The calculated absorption spectra, electronic transition contributions, and electron density difference maps (EDDMs) were generated by GaussSum 3.0.[27] The molecular orbitals were visualized using VMD 1.9.4a51.[28]
Cell culture
The HeLa cell line (cervical cancer cell, a gift from Dr. Carolyn M. Price, University of Cincinnati) and other cell lines were cultured in DMEM containing 10% FBS, 100 units/mL of penicillin, and 100 units/mL of streptomycin in a 5% CO2 cell incubator (Thermo Fisher Scientific, USA) with 100% humidity at 37 °C.
Isolation of HEK293S-derived EVs and fluorescent labeling
For HEK293S-derived EVs isolation,1.5 × 107 cells were cultured in fresh growth medium for 48h before collecting the supernatant. The resulting conditioned media were centrifuged at 125 × g for 10 min, 10,000 × g for 20 min, and then 100,000 × g for 120 min at 4 °C. Subsequently, the EVs pellets were washed with phosphate-buffered saline (PBS) by ultracentrifugation at 120,000 × g for 120 min at 4 °C. EVs pellets were resuspended in PBS and stored at −80 °C until use. The concentration of the proteins of the EVs were determined using a Micro BCA assay kit. Fluorescent labeling of Evs with Mem Dye-Deep Red was perfomed following the product manuals.
Structured illumination microscopy imaging
All SIM images were performed with a Nikon structured illumination microscopy (N-SIM, version AR5.11.00 64bit, Tokyo, Japan), a 3D-SIM equipped with an Apochromat 100×/1.49 numerical aperture oil-immersion objective lens and solid-state lasers (488 nm, 561 nm, 640 nm, the output powers at the fiber end: 15 mW). Images were captured using Nikon NIS-Elements 512 × 512 using Z-stacks with a step size of 0.2 μm and the raw images were reconstructed and processed with NIS-Elements AR Analysis. Cells were seeded on glass-bottomed culture dishes (MatTek; P35G-1.5–14-C) for 24 h to adhere. ECGreen was treated with cells as indication. Commercial dyes staining with 30 min were performed. Before imaging, cells were washed with PBS for 3 times. The green channel images with emission bandwidth at 500–550 nm were excited by a 488 nm laser for ECGreen. The red channel images with emission bandwidth at 570–640 nm were excited by a 561 nm laser for LTR. The deep red channel images with emission bandwidth at 660–735 nm were excited by a 640 nm laser for MTDR. All of Pearson’s colocalization coefficients (PCC) were analyzed and quantified in the open-source software CellProfiler. The imaging data analysis was performed with ImageJ.
Cell viability test
Cell Counting Kit-8 (CCK-8, Dojindo Molecular Technologies, Inc., Japan) was used to determine the cytotoxicity of ECGreen. HeLa cells were seeded in a 96-well plate with 1 × 104 cells/well. After 24 h to adhere, different concentrations of ECGreen were added to the wells and placed in the incubator for 24 h. Then 10 μL of CCK-8 solution was added to each well, the culture plate was incubated for 1 h. Absorbance at 490 nm was determined with the Synergy Mx microplate reader (BioTek Instruments, Inc., USA).
Statistical Analysis
Statistical significance of data was evaluated using Student’s t test. Data were presented as M ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 were considered statistically significant in analyses. Statistics and graphing were performed using Prism 8 (GraphPad) and Excel (Microsoft).
Supplementary Material
Acknowledgements
We thank Yuki Tatenaka for coordinating this project. J.D. was supported by the National Institutes of Health (NIH R35GM128837). K.Q. was supported by the seed grant (202101) of the Center for Chemical Imaging in Biomedicine at the University of Cincinnati. Y.S. acknowledges the support of the National Science Foundation (CHE1955358).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of interest
Ryo Seino, Munetaka Ishiyama, and Yuichiro Ueno are employees of Dojindo Laboratories, manufacturer of ECGreen.
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request
References
- [1].a) Gottlieb RA, Dosanjh A, Proc. Natl. Acad. Sci. U. S. A 1996, 93, 3587–3591; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chin ER, Allen DG, J. Physiol 1998, 512, 831–840; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Varadi A, Rutter GA, Endocrinology 2004, 145, 4540–4549; [DOI] [PubMed] [Google Scholar]; d) Han J, Burgess K, Chem. Rev 2010, 110, 2709–2728; [DOI] [PubMed] [Google Scholar]; e) Steinegger A, Wolfbeis OS, Borisov SM, Chem. Rev 2020, 120, 12357–12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Izumi H, Torigoe T, Ishiguchi H, Uramoto H, Yoshida Y, Tanabe M, Ise T, Murakami T, Yoshida M. Nomoto, Kohno K, Cancer Treat. Rev 2003, 29, 541–549; [DOI] [PubMed] [Google Scholar]; b) Davies TA, Fine RE, Johnson RJ, Levesque CA, Rathbun WH, Seetoo KF, Smith SJ, Strohmeier G, Volicer L, Delva L, Simons ER, Biochem. Biophys. Res. Commun 1993, 194, 537–543; [DOI] [PubMed] [Google Scholar]; c) Hamilton GRC, Sahoo SK, Kamila S, Singh N, Kaur N, Hyland BW, Callan JF, Chem. Soc. Rev 2015, 44, 4415–4432; [DOI] [PubMed] [Google Scholar]; d) Yin J, Hu Y, Yoon J, Chem. Soc. Rev 2015, 44, 4619–4644 [DOI] [PubMed] [Google Scholar]
- [3].a) Casey JR, Grinstein S, Orlowski J, Nat. Rev. Mol. Cell Biol 2010, 11, 50–61; [DOI] [PubMed] [Google Scholar]; b) Davidson SM, Heiden MGV, Annu. Rev. Pharmacol. Toxicol 2017, 57, 481–507; [DOI] [PubMed] [Google Scholar]; c) Qiu K, Ke L, Zhang X, Liu Y, Rees TW, Ji L, Diao J, Chao H, Chem. Commun 2018, 54, 2421–2424; [DOI] [PubMed] [Google Scholar]; d) Fang H, Yao S, Chen Q, Liu C, Cai Y, Geng S, Bai Y, Tian Z, Zacharias AL, Takebe T, Chen Y, Guo Z, He W, Diao J ACS Nano 2019, 13, 14426–14436; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Qiu K, Du Y, Liu J, Guan J-L, Chao H, Diao J, Theranostics 2020, 10, 6072–6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Kowada T, Maeda H, Kikuchi K, Chem. Soc. Rev 2015, 44, 4953–4972; [DOI] [PubMed] [Google Scholar]; b) Yue Y, Huo F, Lee S, Yin C, Yoon J, Analyst 2017, 142, 30–41. [DOI] [PubMed] [Google Scholar]
- [5].a) Xu W, Zeng Z, Jiang J-H, Chang Y-T, Yuan L, Angew. Chem. Int. Ed 2016, 55, 13658–13699; [DOI] [PubMed] [Google Scholar]; b) Qiu K, Chen Y, Rees TW, Ji L, Chao H, Coord. Chem. Rev 2019, 378, 66–86; [Google Scholar]; c) Qiu K, Zhu H, Rees TW, Ji L, Zhang Q, Chao H, Coord. Chem. Rev 2019, 378, 66–86. [Google Scholar]
- [6].Huotari J, Helenius A, EMBO J 2011, 30, 3481–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Raiborg C, Wenzel EM, Stenmark H, EMBO J 2015, 34, 1848–1858; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Eisenberg-Bord M, Shai N, Schuldiner M, Bohnert M, Dev. Cell 2016, 39, 395–409. [DOI] [PubMed] [Google Scholar]
- [8].a) Brunt L, Scholpp S, Cell Mol. Life Sci 2018, 75, 785–795; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sorkin A, Zastrow MV, Nat. Rev. Mol. Cell Biol 2009, 10, 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Vidyadhara DJ, Lee JE, Chandra SS, J. Neurochem 2019, 150, 487–506; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Heckmann BL, Teubner BJW, Tummers B, Boada-Romero E, Harris L, Yang M, Guy CS, Zakharenko SS, Green DR, Cell 2019, 178, 536–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) Li D, Shao L, Chen BC, Zhang X, Zhang M, Moses B, Milkie DE, Beach JR, Hammer JA 3rd, Pasham M, Kirchhausen T, Baird MA, Davidson MW, Xu P, Betzig E, Science 2015, 349, aab3500; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Heintzmann R, Huser T, Chem. Rev 2017, 117, 13890–13908; [DOI] [PubMed] [Google Scholar]; c) Huang X, Fan J, Li L, Liu H, Wu R, Wu Y, Wei L, Mao H, Lal A, Xi P, Tang L, Zhang Y, Liu Y, Tan S, Chen L, Nat. Biotechnol 2018, 36, 451–459; [DOI] [PubMed] [Google Scholar]; d) Wang L, Frei MS, Salim A, Johnsson K, J. Am. Chem. Soc 2019, 141, 2770–2781; [DOI] [PubMed] [Google Scholar]; e) Chen Q, Shao X, Hao M, Guan R, Tian Z, Li M, Wang C, Ji L, Chao H, Guan J-L, Diao J Biomaterials 2020, 250, 120059; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Shao X, Chen Q, Hu L, Tian Z, Liu L, Liu F, Wang F, Ling P, Mao Z-W, Diao J Nano Res 2020, 13, 2149–2155; [Google Scholar]; g) Chen Q, Fang H, Shao X, Tian Z, Geng S, Zhang Y, Fan H, Xiang P, Zhang J, Tian X, Zhang K, He W, Guo Z, Diao J Nat. Commun 2020, 11, 6290; [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Fang H, Geng S, Hao M, Chen Q, Liu M, Liu C, Tian Z, Wang C, Takebe T, Guan J-L, Chen Y, Guo Z, He W, Diao J Nat. Commun 2021, 12, 109; [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Chen Q, Jin C, Shao X, Guan R, Tian Z, Wang C, Liu F, Ling P, Guan J, Ji L, Wang F, Chao H, Diao J, Small 2018, 14, 1802166; [DOI] [PubMed] [Google Scholar]; k) Chen Q, Hao M, Wang L, Li L, Chen Y, Shao X, Tian Z, Pfuetzner RA, Zhong Q, Brunger AT, Guan J-L, Diao J, Cell Death Dis 2021, 12, 939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Iwashita H, Sakurai HT, Nagahora N, Ishiyama M, Shioji K, Sasamoto K, Okuma K, Shimizu S, Ueno Y FEBS Lett 2018, 592, 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Poc P, Gutzeit VA, Ast J, Lee J, Jones BJ, D’Este E, Mathes B, Lehmann M, Hodson DJ, Levitz J, Broichhagen J, Chem. Sci 2020, 11, 7871–7883; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chi W, Chen J, Qiao Q, Gao Y, Xu Z, Liu X, Phys. Chem. Chem. Phys 2019, 21, 16798–16803. [DOI] [PubMed] [Google Scholar]
- [13].Kucherak OA, Oncul S, Darwich Z, Yushchenko DA, Arntz Y, Didier P, Mély Y, Klymchenko AS, J. Am. Chem. Soc 2010, 132, 4907–4916. [DOI] [PubMed] [Google Scholar]
- [14].Liu B, Fletcher S, Avadisian M, Gunning PT, Gradinaru CC, J. Fluoresc 2009, 19, 915–920. [DOI] [PubMed] [Google Scholar]
- [15].Gan J, Chen K, Chang C-P, Tian H, Dyes. Pigm 2003, 57, 21–28. [Google Scholar]
- [16].Punnonen E, Ryhänen K, Marjomi VS, Eur. J. Cell Biol 1998, 75, 344–352. [DOI] [PubMed] [Google Scholar]
- [17].a) Shpetner H, Joly M, Hartley D, Corvera S, J. Cell Biol 1996, 132, 595–605; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Spiro DJ, Boll W, Kirchhausen T, Wessling-Resnick M, Mol. Biol. Cell 1996, 7, 355–367; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Liu K, Jian Y, Sun X, Yang C, Gao Z, Zhang Z, Liu X, Li Y, Xu J, Jing Y, Mitani S, He S, Yang C, J. Cell Biol 2016, 212, 181–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].a) Chen Q, Shao X, Tian Z, Chen Y, Mondal P, Liu F, Wang F, Ling P, He W, Zhang K, Guo Z, Diao J, Nano Res 2019, 12, 1009–1015; [Google Scholar]; b) Liu L-Y, Fang H, Chen Q, Chan MH-Y, Ng M, Wang K-N, Liu W, Tian Z, Diao J, Mao Z-W, Yam VW-W, Angew. Chem. Int. Ed 2020, 59, 19229–19236. [DOI] [PubMed] [Google Scholar]
- [19].Pierzynska-Mach A, Janowski PA, Dobrucki JW, Cytometry A 2014, 85, 729–737. [DOI] [PubMed] [Google Scholar]
- [20].Liu X, Su Y, Tian H, Yang L, Zhang H, Song X, Foley JW, Anal. Chem 2017, 89, 7038–7045. [DOI] [PubMed] [Google Scholar]
- [21].Yang Z, Li L, Ling J, Liu T, Huang X, Ying Y, Zhao Y, Zhao Y, Lei K, Chen L, Chen Z, Chem. Sci 2020, 11, 8506–8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].a) Han Y, Li M, Qiu F, Zhang M, Zhang Y, Nat. Commun 2017, 8, 1307; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wong YC, Ysselstein D, Krainc D, Nature 2018, 554, 382–386; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Nixon RA, Nat. Med 2013, 19, 983–997; [DOI] [PubMed] [Google Scholar]; d) Levine B, Nature 2007, 446, 745–747. [DOI] [PubMed] [Google Scholar]
- [23].a) Hurley JH, Schulman BA, Cell 2014, 157, 300–311; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang Y, Li Y, Wei F, Duan Y, Trends Biotechnol 2017, 35, 1181–1193; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Dikic I, Elazar Z, Nat. Rev. Mol. Cell Biol 2018, 19, 349–364; [DOI] [PubMed] [Google Scholar]; d) Nakatogawa H, Nat. Rev. Mol. Cell Biol 2020, 21, 439–458. [DOI] [PubMed] [Google Scholar]
- [24].Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Petersson GA, Nakatsuji H, Li X, Caricato M, Marenich AV, Bloino J, Janesko BG, Gomperts R, Mennucci B, Hratchian HP, Ortiz JV, Izmaylov AF, Sonnenberg JL, Williams-Young D, Ding F, Lipparini F, Egidi F, Goings J, Peng B, Petrone A, Henderson T, Ranasinghe D, Zakrzewski VG, Gao J, Rega N, Zheng G, Liang W, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Throssell K, Montgomery JA Jr., Peralta JE, Ogliaro F, Bearpark MJ, Heyd JJ, Brothers EN, Kudin KN, Staroverov VN, Keith TA, Kobayashi R, Normand J, Raghavachari K, Rendell AP, Burant JC, Iyengar SS, Tomasi J, Cossi M, Millam JM, Klene M, Adamo C, Cammi R, Ochterski JW, Martin RL, Morokuma K, Farkas O, Foresman JB, Fox DJ, Gaussian 16, Revision C.01, Gaussian, Inc., Wallingford CT, 2019. [Google Scholar]
- [25].a) Becke AD, Phys. ReV. A: Gen. Phys 1988, 38, 3098–3100; [DOI] [PubMed] [Google Scholar]; b) Becke AD, J. Chem. Phys 1993, 98, 5648–5652; [Google Scholar]; c) Lee C, Yang W, Parr RG, Phys. ReV. B: Condens. Matter Phys 1988, 37, 785–789. [DOI] [PubMed] [Google Scholar]
- [26].Hehre WJ, Radom L, Schleyer P. v. R., Pople JA, Ab Initio Molecular Orbital Theory, John Wiley & Sons: New York, 1986. [Google Scholar]
- [27].O’Boyle NM, Tenderholt AL, Langner KM, J. Comp. Chem 2008, 29, 839–845. [DOI] [PubMed] [Google Scholar]
- [28].Humphrey W, Dalke A, Schulten K, J. Mol. Graph 1996, 14, 33–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request


