Abstract
Respiratory Complex I is the site of a large fraction of the mutations that appear to cause mitochondrial disease. Seven of its subunits are mitochondrially encoded, and therefore, such mutants are particularly difficult to construct in cell-culture model systems. We have selected 13 human clinical mutations found in ND2, ND3, ND4, ND4L, ND5 and ND6 that are generally found at subunit interfaces, and not in critical residues. These mutations have been modeled in E. coli subunits of Complex I, nuoN, nuoA, nuoM, nuoK, nuoL, and nuoJ, respectively. All mutants were expressed from a plasmid encoding the entire nuo operon, and membrane vesicles were analyzed for deamino-NADH oxidase activity, and proton translocation activity. ND5 mutants were also analyzed using a time-delayed expression system, recently described by this lab. Other mutants were analyzed for the ability to associate in subcomplexes, after expression of subsets of the genes. For most mutants there was a positive correlation between those that were previously determined to be pathogenic, or likely to be pathogenic, and those that we found with compromised Complex I activity or subunit interactions in E. coli. In conclusion, this approach provides another way to explore the deleterious effects of human mitochondrial mutations, and it can contribute to molecular understanding of such mutations.
Keywords: Complex I, Mitochondria, LHON, Bioenergetics, NADH dehydrogenase, Mutations
1. Introduction
The genes that encode Complex I are found in both nuclear DNA and mitochondrial DNA. Mutations in these genes are responsible for a large fraction of all mitochondrial diseases (Mayr et al., 2015; Scheffler, 2015). Seven of the thirteen protein-coding genes of mammalian mitochondria encode membrane subunits of Complex I (Chomyn et al., 1986; Chomyn et al., 1985). Mutations in these genes have commonly been detected in individuals experiencing various mitochondrial diseases, including Leber’s Hereditary Optic Neuropathy (LHON) (Yu-Wai-Man and Chinnery, 2021) and Leigh syndrome (Bakare et al., 2021), since the 1990s, due to the relative ease of sequencing mitochondrial DNA (Howell et al., 1991; Wallace et al., 1988). It can be difficult to carry out biochemical analyses of mitochondrial function from tissue samples. Moreover, control values of enzyme activities are typically uncertain, and must be specified by a broad range. Due to the high copy number of mitochondrial DNA in most cells, heteroplasmy of a mutation can vary from 0 to 100%, and this value can also vary from one type of tissue to another. The mitochondrial haplotype can also influence the effect of a particular mutation, but generally, in unknown ways (Klink et al., 2021). For all these reasons, the pathogenicity of an identified mitochondrial mutation can be difficult to ascertain (Ratnaike et al., 2021; Wei et al., 2017; Wong et al., 2020).
Complex I is an L-shaped multi-subunit enzyme embedded in the inner mitochondrial membrane (for reviews see (Agip et al., 2019; Hirst, 2013; Parey et al., 2020; Sazanov, 2015)). It is the first member of the electron transport chain and oxidizes NADH while reducing ubiquinone and translocating protons across the membrane. One arm of the “L” extends into the matrix space, and the other is embedded in the mitochondrial inner membrane. The matrix arm contains a flavin mononucleotide (FMN) and all iron–sulfur (FeS) clusters necessary for electron transfer to ubiquinone. The membrane arm contains the subunits that translocate protons from the matrix to the inner membrane space. There are fourteen “core” subunits that have been found in Complex I from all species. The membrane arm contains seven core subunits, and all are encoded by mtDNA: ND1, ND2, ND3, ND4, ND4L, ND5, and ND6. The remaining core subunits are in the matrix arm. In addition to the core subunits, mammalian Complex I contains 31 so-called supernumerary (or accessory) subunits (Elurbe and Huynen, 2016; Vinothkumar et al., 2014). The structure and pathogenicity of human mutations in the supernumerary subunits has been recently reviewed (Dang et al., 2020; Padavannil et al., 2021). The structures of mitochondrial Complex I from several species, including human (Gu et al., 2016), ovine (Fiedorczuk et al., 2016), bovine (Blaza et al., 2018), mouse (Agip et al., 2018) and yeast (Grba and Hirst, 2020; Parey et al., 2018) have been determined.
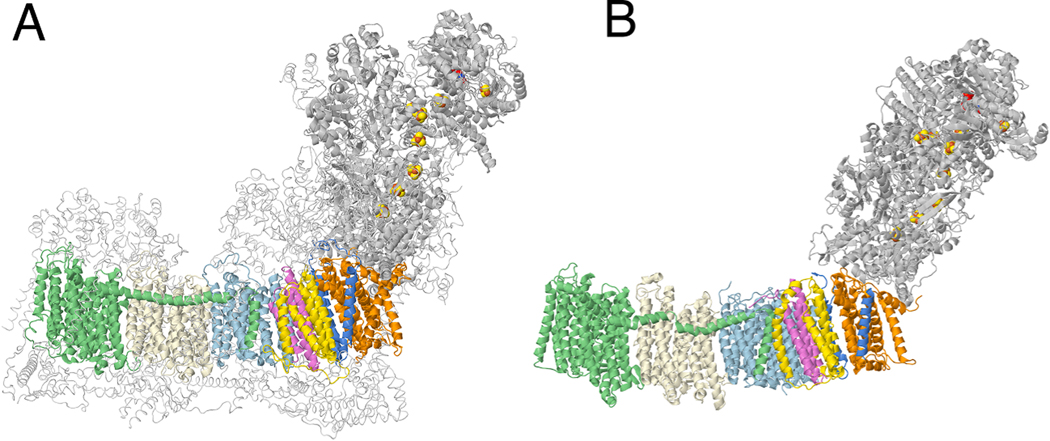
Complex I from E. coli is a useful model system for the mitochondrial enzyme. A partial structure has been determined by crystallography (Efremov and Sazanov, 2011), and full structures by cryo-EM (Kolata and Efremov, 2021; Schimpf et al., 2021), and it appears very similar to the core subunits of the mitochondrial enzyme. The structures of Complex I from ovine and from E. coli are compared in Fig. 1. The enzyme from E. coli contains seven membrane subunits that are homologous to the seven mitochondrially encoded ones in human Complex I. All its subunits are encoded by the nuo operon (Weidner et al., 1993). These genes can be expressed from an inducible plasmid in a strain deleted for the nuo operon, allowing for simple comparison of mutants (Zhang and Vik, 2021).
Fig. 1.
A comparison of the structures of Complex I from ovine and from E. coli. (A) The core membrane subunits are colored: ND1 (orange), ND3 (blue), ND6 (yellow), ND4L (violet), ND2 (light blue), ND4 (light yellow), ND5 (green). Other core subunits from the peripheral arm are shown in gray ribbons. FMN and FeS clusters are in space filling mode. Supernumerary subunits are shown in backbone trace colored light gray. The PDB file is 5lnk, a high-resolution mammalian structure (Fiedorczuk et al., 2016). (B) The same coloring scheme is used for the E. coli enzyme (PDB file 7nyr (Kolata and Efremov, 2021)). nuoH (orange), nuoA (blue), nuoJ (yellow), nuoK (violet), nuoN (light blue), nuoM (light yellow), nuoL (green). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In this project, thirteen human clinical mutations were selected that were found in patients with various conditions thought to be associated with mitochondrial dysfunction. They occur at subunit interfaces between core subunits, and therefore, seemed likely to disrupt assembly. The predicted or observed pathogenicity of these mutations ranged from “confirmed” to “benign” to “uncertain”. All were modeled at corresponding positions in the E. coli proteins and analyzed. In general, a second mutation was constructed, in which the residue was changed to alanine. This helped determine whether loss of function is associated with the loss of the original amino acid, or by the introduction of the mutant amino acid.
2. Materials and methods
2.1. Materials
Restriction endonucleases, T4 DNA ligase, Q5 Site-Directed Mutagenesis Kit, Q5 High-Fidelity PCR Kit, and Monarch PCR Cleanup Kit were from New England BioLabs (Beverly, MA). QuikChange Mutagenesis kit was from Agilent Technologies (Santa Clara, CA). DNA Miniprep kits were purchased from Qiagen (Valencia, CA) and ThermoFisher Scientific (Pittsburgh, PA). The QIAquick and QiaIIX Gel Extraction Kits were from Qiagen (Germantown, MD). Immunoblot PVDF (polyvinylidene difluoride), Mini-PROTEAN TGX gels (12%; 4–15%), Precision Plus Protein ™ Dual Color standards, Precision Plus Protein ™ unstained standards and the DC protein assay kit were from Bio-Rad (Hercules, CA). 9-Amino-6-chloro-2-methoxyacridine (ACMA) and NativeMark™ Unstained Protein Standards were from Invitrogen. The polyclonal antibodies against E. coli Complex I subunits were described and validated in a recent publication (Zhang and Vik, 2021). Subunit N was detected by the monoclonal mouse HA-probe (F7), from Santa Cruz Biotechnology (Dallas, TX), as were Goat anti-rabbit and Goat anti-mouse IgG-HRP, IgG (mouse monoclonal IgG1), and Protein A/G PLUS-Agarose. SuperSignal West Dura Extended Duration Substrate and p-nitro blue tetrazolium chloride (NBT) were from ThermoFisher Scientific (Pittsburgh, PA). n-dodecyl-β-D-maltopyranoside was from Anatrace (Maumee, OH). Other chemicals, including NADH, 6-aminocaproic acid, L-arabinose, D-fucose, glucose, Coomassie Brilliant Blue G, Protease Inhibitor Cocktail (P8465), Bis-Tris, deamino-NADH, FCCP, and IPTG were from MilliporeSigma (Burlington, MA). Oligonucleotides for plasmid construction, mutagenesis, and sequencing were synthesized by Eurofins Genomics (Louisville, KY). DNA sequencing was performed by Lone Star Labs. (Houston, TX).
2.2. Mutagenesis
Single amino acid substitutions were constructed by Q5 Site-Directed Mutagenesis to introduce an additional restriction site, for rapid identification of mutants. Two mutants nuoM_L183A, P, were constructed similarly by QuikChange mutagenesis. Primers used for construction of mutations are found in the supplement (Supplementary Table S1). Mutations constructed in subunits L, M, N, J, K or A were transferred to the expression vector, pBAD33(A-N) that contains a full size nuo operon (Zhang and Vik, 2021) or plasmids with a subset of Complex I genes, using two unique restriction sites. Two strategies were used for generating double mutants. Typically, the mutations were constructed independently, plasmids containing single substitutions were digested with two endonucleases, and the insert was ligated in the vector containing the second mutation. Alternatively, the plasmids containing the first mutations were used as the templates in Q5 Site-Directed mutagenesis for creating the second mutations. The double mutants were transferred to plasmid pBAD33(A-N) and transformed into strain BA14 that carries a chromosomal deletion for the genes of all complex I subunits (Amarneh et al., 2006).
2.3. Growth and membrane preparation
For analysis of mutations expressed by pBAD33(A-N), the cells were grown in a rich chemical medium (0.5% yeast extract, 1.0% peptone, 15 mM Na3PO4, 10 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, 5 mM Na2SO4, 2 mM MgSO4, 50 mg/L riboflavin, 30 mg/L ferric ammonium citrate, and 0.5 mM L-cysteine) at 37 °C (Bungert et al., 1999). For analysis of mutations expressed in a subset of Complex I genes, cells were grown in a rich medium (3% tryptone, 1.5% yeast extract and 0.15% NaCl and 1% (v/v) glycerol) at 37 °C (Amarneh and Vik, 2003). Harvested cells were suspended in 9 ml of 50 mM MES, 25% glycerol, and 10 mM MgSO4 (pH 6.0), and passed through the French press at 8000 psi. Membrane vesicles were prepared as described previously (Zhang and Vik, 2021). These vesicles are considered to be highly oriented, and sealed, as indicated by surface labeling studies (Wada et al., 1999) and ACMA fluorescence quenching assays.
2.4. Time-delayed expression systems
The time-delayed, in vivo assembly procedures developed originally by Brockmann et al. (Brockmann et al., 2013), were used as adapted in this lab, and described recently (Zhang and Vik, 2021). In brief, pBAD33 (A-M) and pET22b(L) bearing nuoL mutations were simultaneously transformed into BA14 (Δnuo) competent cells that already harbored a third plasmid, pT7Pol26. The cells were grown in 500 ml of rich chemical medium (described above) at 37 °C with 100 mg/l ampicillin, 40 mg/l chloramphenicol, and 50 mg/l kanamycin as selection markers. The cultures were induced with 0.03% (w/v) arabinose when A600 = 0.05, to express the genes cloned into pBAD33, and grown to A600 0.3. At this time, 0.5% (w/v) glucose and 0.045% (w/v) D-fucose were added to the medium, to repress further expression of the pBAD-controlled genes. After thirty minutes, induction of genes cloned into pET22b, was started with addition of 0.1 mM IPTG, and the cultures were harvested at A600 = 1.0.
2.5. SDS electrophoresis and immunoblotting
50 μg of membrane protein were mixed with 10 μl 4% SDS and 5 μl of loading dye (60 mM Tris–HCl (pH 6.8), 25% glycerol, 14.4 mM 2-mercaptoethanol, 0.1% bromophenol blue) and then incubated at 37 °C for 15 min. Samples were run on 12% acrylamide gels at 150 V for 1 h and then were transferred to PVDF membrane for immunoblotting as described previously (Zhang and Vik 2021). The PVDF membrane was incubated with rabbit custom antibodies diluted 1:5000 for subunit L or M, 1:10,000 for subunits A, J, K, or mouse anti-HA serum, diluted 1:170 for subunit N at room temperature for at least 2 h. Blots shown are representative of 2 or 3 blots in total.
2.6. Blue-native gel electrophoresis and NADH dehydrogenase activity assay
Blue-native gel electrophoresis was performed according to previous methods (Schägger and von Jagow, 1991; Torres-Bacete et al., 2007). In brief, E. coli membranes equivalent to 800 μg of protein were resuspended in 750 mM aminocaproic acid, 50 mM Bis-Tris-HCl (pH 7.0), 0.1 mg/ml DNase, and 0.8% (w/v) dodecyl maltoside to a total volume of 240 μl. After incubation on ice for 30 min, the samples were centrifuged at 149,000g for 10 min. The supernatants were recovered, and 2% (w/v) of Coomassie Brilliant Blue G in 1 M aminocaproic acid was added at a final concentration of Coomassie Blue of 0.08% (v/v). 50 μg protein from the samples were loaded on a 4–15% gradient gel, and electrophoresis was performed in a 4 °C chamber at 100 V until entry of the protein sample into the stacking gel. After that, the cathode buffer, containing 0.02% of Coomassie Blue, was replaced by the cathode buffer without Coomassie Blue, and the electrophoresis was continued at 150 V until the tracking dye ran out. After the electrophoresis, the gel was washed several times in 2 mM Tris-HCl (pH 7.5) before transfer to PVDF. For the NADH dehydrogenase activity assay, the gel was incubated in 2 mM Tris-HCl (pH 7.5) containing 2.5 mg/ml of p-nitroblue tetrazolium and 150 μM NADH for 40 min at room temperature. The reaction was terminated by 10% acetic acid and 50% methanol.
2.7. Immunoprecipitation
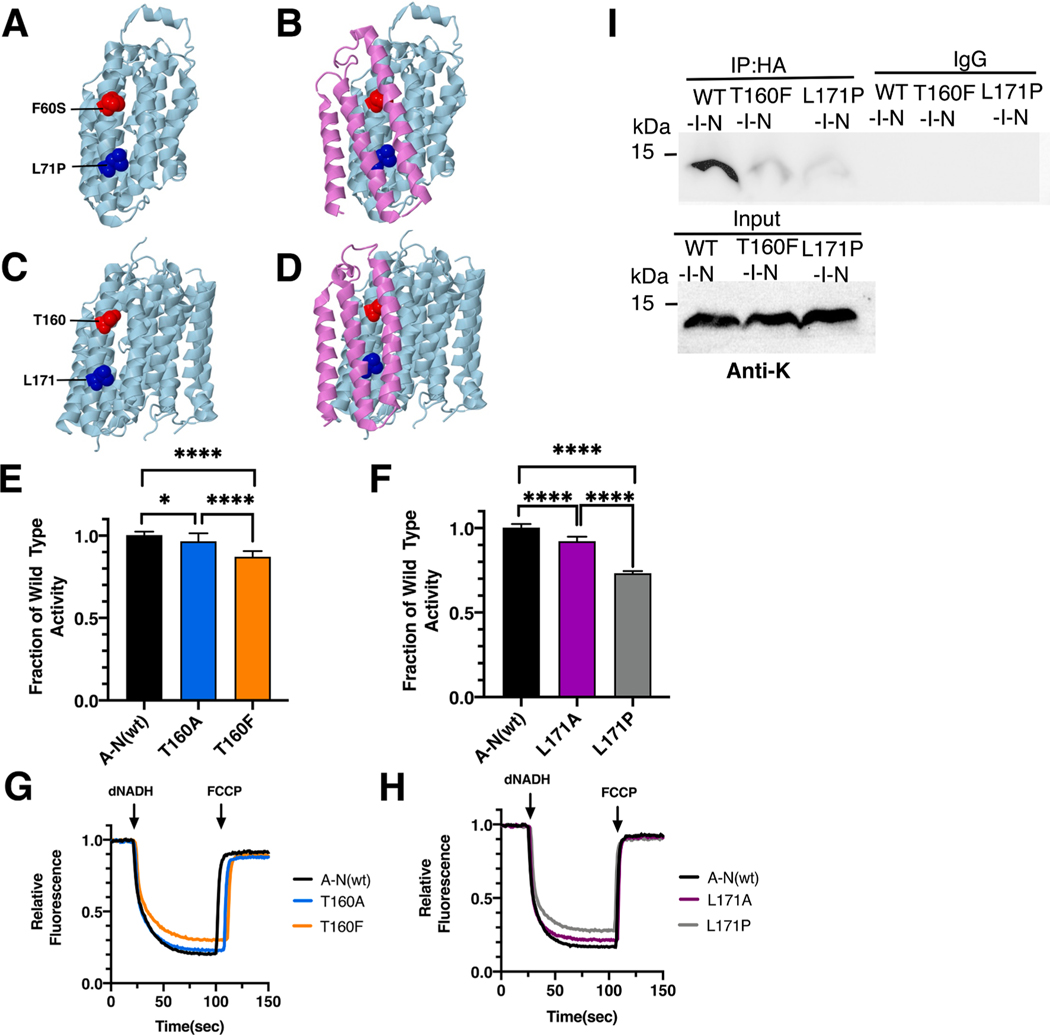
450 μl of protease inhibitor cocktail solution was added to 9 ml cell suspension before the French press procedure. Immunoprecipitation were conducted according to the methods described previously (Zhang and Vik, 2021). In brief, 30 μl of Protein A/G Plus-Agarose beads were incubated with 1 μg of anti-HA in 300 μl of resuspension buffer for 1 h at 4 °C with constant rocking. Beads bound with pull-down antibody were centrifuged at 1470g for 2 min, and the supernatant was discarded. Membranes equivalent to 500 μg of protein were resuspended in 200 μl of the same buffer. After incubation on ice for 30 mins, samples were centrifuged at 149,000g for 10 min at 4 °C and the supernatants were incubated with 25 μl of untreated A/G beads with constant rocking. After 1 h, the samples were centrifuged for 2 min. Supernatants were recovered and incubated overnight with the beads which had bound anti-HA antibodies, at 4 °C with constant rocking, followed by washes with 300 μl of the same resuspension buffer for three times. The immunoprecipitated material was dissolved in 30 μl of SDS sample buffer and incubated at 37 °C for 10 min and then subjected to gel electrophoresis for immunoblot analysis. 50 μg of protein was loaded in “input” lanes.
2.8. Enzyme assay
The activity assays were conducted according to the methods described previously (Amarneh et al., 2006; Michel et al., 2011). In brief, NADH oxidase activity assays were started with 0.25 mM dNADH (extinction coefficient 6.22 mM−1 cm−1) and the absorbance was monitored at 340 nm for 2 min. dNADH oxidase activity assays were conducted by using 50 μg/ml protein membrane in 50 mM MOPS, 10 mM MgCl2, pH 7.3 at room temperature. The uncoupler FCCP was added to 1 μM final concentration from a 1 mM ethanol stock to eliminate the buildup of a proton gradient during NADH oxidase assays. Proton translocation assays were conducted by measuring the fluorescence quenching of the acridine dye ACMA (9-amino, 6-chloro, 2-methoxyacridine) as a ΔpH indicator using excitation and emission wave lengths of 410 and 490 nm, respectively. Proton translocation assays were conducted by using 25 μg/ml protein membrane in 50 mM MOPS, 5 mM MgCl2, 50 mM KCl, at pH 7.3 at 25 °C, with additions of 0.25 mM dNADH, 1 μM valinomycin, and 1 μM ACMA.
3. Results
3.1. Mutations in ND5 (nuoL)
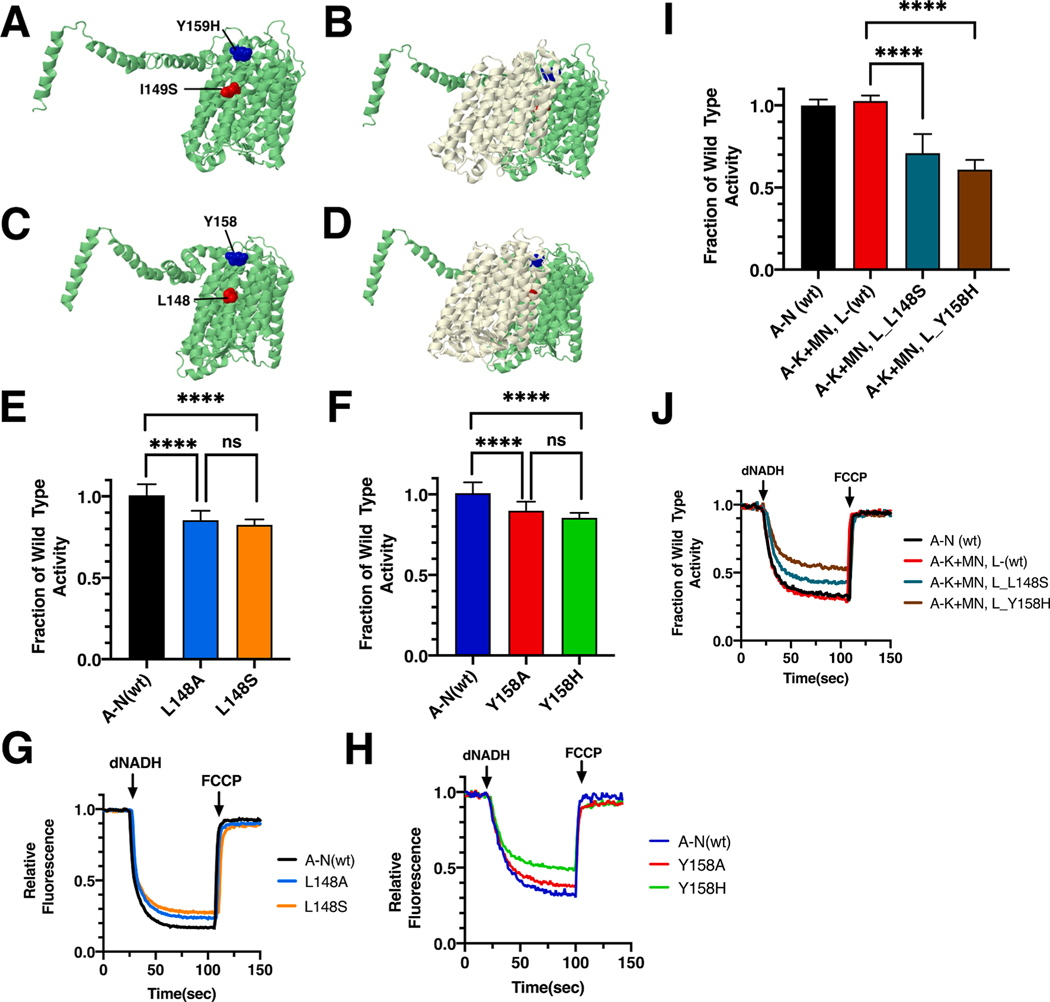
Two mutations found in ND5, I149S (T12782G) and Y159H (T12811C), were modeled in E. coli nuoL. The E. coli residues are L148 and Y158. Both are found at the interface with ND4 (nuoM) in transmembrane helix 5 (TM5) of ND5, which also contains the key residue E145. Both mutations contact the N-terminal half of the “broken-helix” TM12 in ND4. The C-terminal half of this “broken-helix” contains key residue E378. These two key residues constitute the functional interface of ND4 and ND5, which is essential for proton translocation. The locations of the mutations are shown in Fig. 2 for the human and the E. coli proteins.
Fig. 2.
Analysis of ND5 mutations in nuoL of E. coli. (A) The locations of human mutations I149S (red) and Y159H (blue) are shown in ND5 (green), using PDB file 5xtd (Gu et al., 2016). (B) ND4 (light yellow) is shown. (C) The locations of E. coli residues L148 (red) and Y158 are shown in nuoL (green), using PDB file 3rko (Efremov and Sazanov, 2011). (D) nuoM (light yellow) is shown. (E, F) dNADH-oxidase activities of membrane vesicles prepared from the E. coli mutants are shown compared to a wild type sample prepared the same day. (G, H) Proton translocation rates from the same samples shown in panels E and F are indicated by fluorescence quenching of the acridine dye ACMA. (I) Membrane vesicles from E. coli cells carrying the modeled human mutations, nuoL_L148S and nuoL_Y158H, were prepared using a two-plasmid, time-delayed expression of nuoL. dNADH-oxidase rates are shown compared to that from cells expressing all nuo genes from a single plasmid. (J) Proton translocation rates from the same samples shown in panel I are indicated by fluorescence quenching of ACMA. (E and F) ns, not significant; ****P < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The ND5_I149S mutant was discovered among a group of patients with LHON-like optic neuropathies (Abu-Amero and Bosley, 2006). The individual with this mutation also carried 3 other mitochondrial mutations in ND1 (4216, Y304H), ND5 (13708, A458T), and CYTB (15257, D171N). All are known to be associated with LHON (Leber’s Hereditary Optic Neuropathy), indicating the possibility of an accumulation of negative effects on function. In E. coli the mutations were constructed and moved to the expression vector pBAD33(A-N) for the preparation of membrane vesicles. Both mutants had similarly high levels of dNADH oxidase activity (Fig. 2E), with values compared to wild type of 83 ± 2% for L148S and 85 ± 6% for L148A, suggesting that loss of leucine was the key effect. In proton translocation assays, using the quenching of ACMA fluorescence, both mutants showed a corresponding rate compared to the wild type samples (Fig. 2G). The presence of subunit L was confirmed by immunoblotting, (Supplementary Fig. S1). In BN gels, a complex corresponding to the size of Complex I (540 kDa) was detected by immunoblotting and an in-gel assay showed bands of activity corresponding to full-size Complex I (Supplementary Fig. S1). Finally, the L148S mutant was tested for its ability to function in a time-delayed expression experiment (Fig. 2I and J). When wild type L is expressed from pET22b(L) after a 30 min time delay after all other genes were expressed from pBAD33(A-K + MN), 100% of dNADH oxidase activity is achieved, as compared to the wild type operon, pBAD33(A-N) (Zhang and Vik, 2021). In the case of L148S, only 70 ± 12% activity was found, suggesting a decreased efficiency in assembly due to the mutation.
In a study of a family with LHON (Cai et al., 2008), an individual was identified with an ND5_Y159H mutation, and also carried the common LHON mutation in ND4 (G11778A, R340H) (Wallace et al., 1988). In addition, mutations in CO1 (G6480A) and CYB (A15395G) were identified. A second individual was found with the ND5_Y159H mutation (Bi et al., 2012), and also carried a second mitochondrial mutation, ND1 (S110N, G3635A) that was considered to be a primary LHON mutation (Brown et al., 2001; Yang et al., 2009), and two additional mutations in ND5 (I576T, T14063C) and in CYB (I164T, T15237C). Therefore, it was thought that the ND5_Y159H mutation might contribute to the pathology, without being causative. This was supported by a recent report, in which an individual with the same allele was treated for vision problems as an adult, but recovered with an improved diet, and cessation of alcohol and cigarette consumption (Zhou et al., 2021).
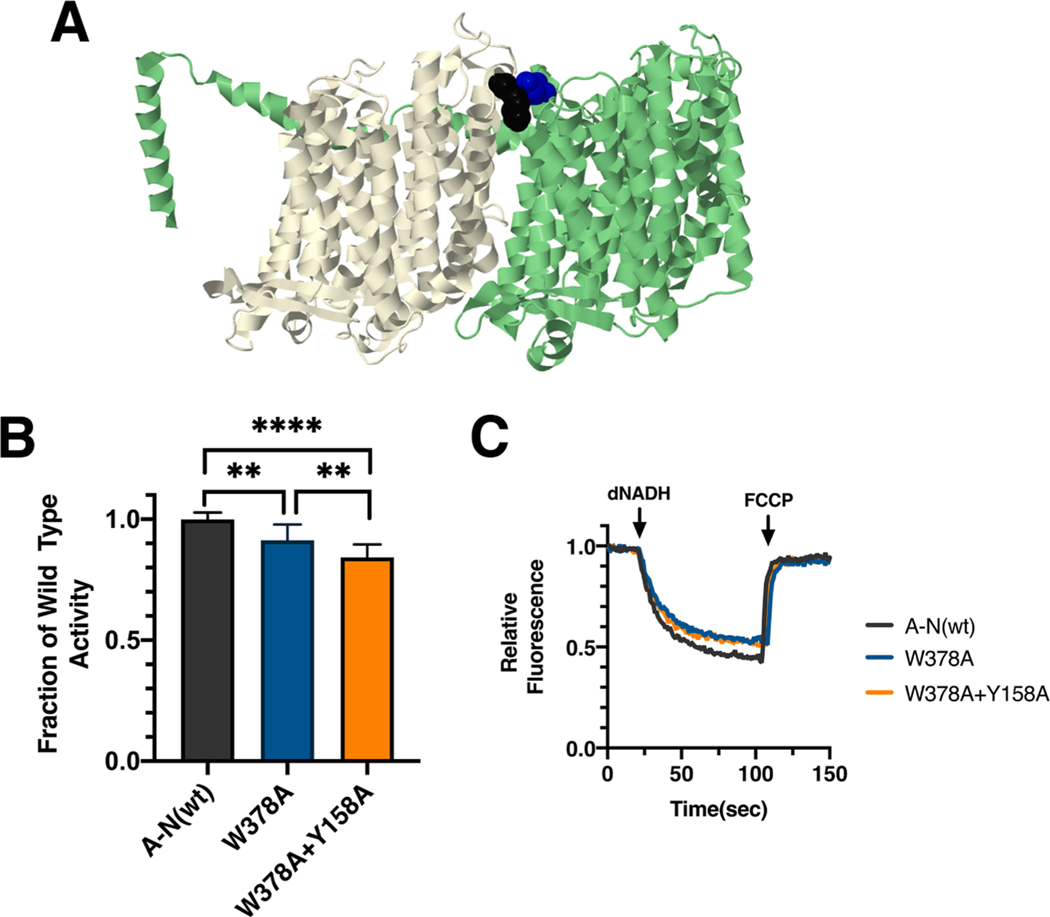
In E. coli two mutations were constructed in subunit L, Y158H and Y158A, and analyzed in membrane vesicles. Both mutants had similarly high levels of dNADH oxidase activity (Fig. 2F), with values compared to wild type of 86 ± 3% for nuoL_Y158H and 90 ± 6% for nuoL_Y158A. In proton translocation assays, both mutants showed a corresponding rate compared to the wild type samples (Fig. 2H). The presence of subunit L was confirmed by immunoblotting (Supplementary Fig. S1). In BN gels, a complex corresponding to the size of Complex I (540 kDa) was detected by immunoblotting, and an in-gel assay showed bands of activity corresponding to full-size Complex I (Supplementary Fig. S1). The nuoL_Y158H mutant was also tested for its ability to function in the time-delayed expression experiment (Fig. 2I and J). When expressed from pET22b(L) after a 30 min time delay after all other genes were expressed from pBAD33(A-K + MN), 60 ± 6% of dNADH oxidase activity was achieved, as compared to the wild type operon, pBAD33(A-N). This indicates a decreased efficiency in assembly due to the mutation. Finally, the Y158A mutation was constructed with a second mutation in an interacting residue of subunit M, W378A, which reduces the possible interaction surface. Activity measurements showed only a slightly reduced level of activity, 84 ± 7%, as compared to the single mutant in L, Y158A, 90% of wild type, suggesting that loss of interactions of Y158A with W378 were not a significant cause of loss of activity (Fig. 3).
Fig. 3.
Analysis of interacting mutations in nuoL of E. coli. (A) The locations of mutations M_W378 (black) and Y158A (blue) are shown in nuoL (green) and nuoM (light yellow), using PDB file 3rko (Efremov and Sazanov, 2011). (B) dNADH-oxidase activities of membrane vesicles prepared from the E. coli mutants nuoM_W378A and double mutant nuoM_W378A/nuoL_Y158A are shown compared to a wild type sample prepared the same day. (C) Proton translocation rates from the same samples shown in panel B are indicated by fluorescence quenching of the acridine dye ACMA. **P < 0.01; ****P < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Mutations in ND4 (nuoM)
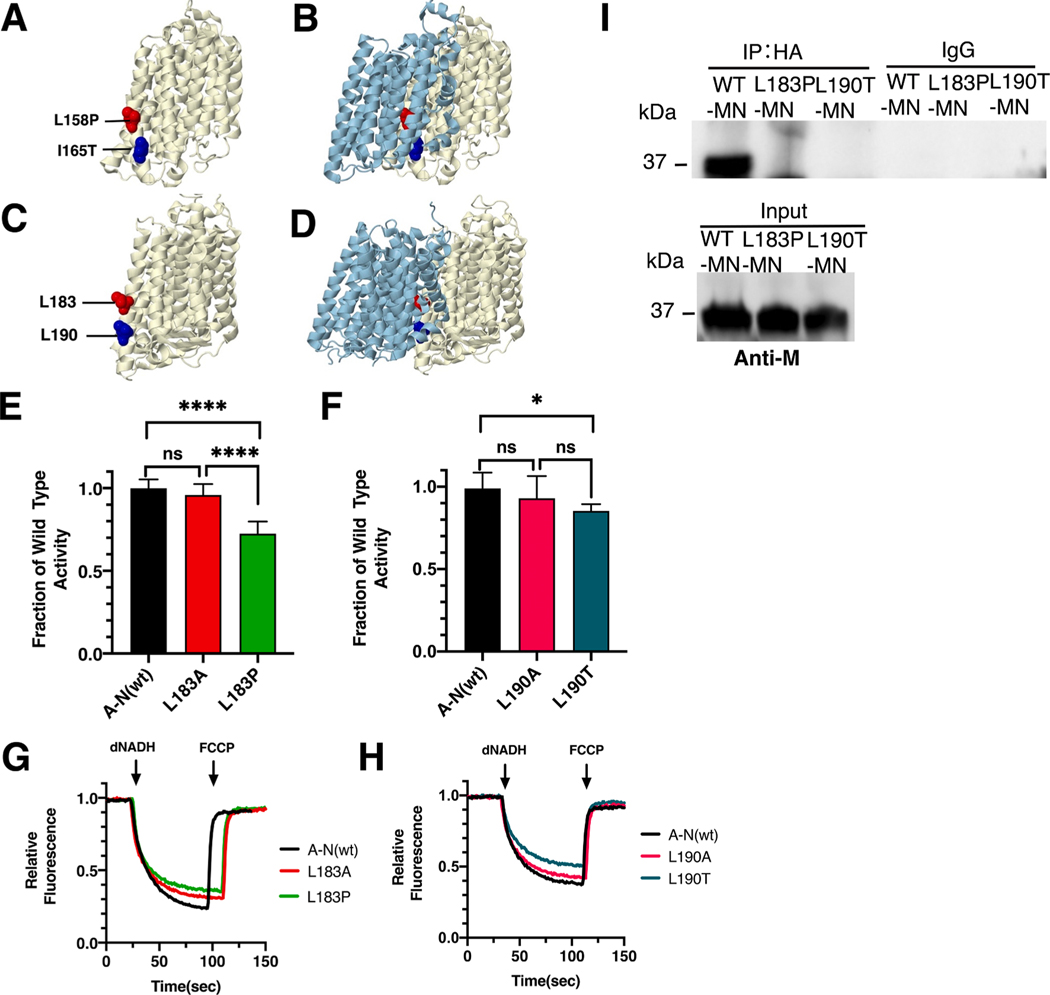
Two clinical mutations in ND4, L158P (red) and I165T (blue), were modeled in the nuoM gene of E. coli. The locations of the mutations for the human and the E. coli proteins are shown in Fig. 4. The E. coli residues are L183 (red) and L190 (blue). Both are found in TM6 of subunit M, at the interface with subunit N (ND2) and in proximity to key residue E144 found in TM5 of M. Both mutations contact the C-terminal half of the “broken-helix” TM12 of subunit N (TM8 in ND2), which contains key residue K395 (K263 in ND2). The interactions of M_E144 and N_K263 are part of the functional interface of these subunits and are essential for proton translocation.
Fig. 4.
Analysis of ND4 mutations in nuoM of E. coli. (A) The locations of human mutations L158P (red) and I165T (blue) are shown in ND4 (light yellow), using PDB file 5xtd (Gu et al., 2016). (B) ND2 (light blue) is shown. (C) The locations of E. coli residues L183 (red) and L190 are shown in nuoM (light yellow), using PDB file 3rko (Efremov and Sazanov, 2011). (D) nuoN (light blue) is shown. (E, F) dNADH-oxidase activities of membrane vesicles prepared from the E. coli mutants are shown compared to a wild type sample prepared the same day. (G, H) Proton translocation rates from the same samples shown in panels E and F are indicated by fluorescence quenching of the acridine dye ACMA. (I) Membrane vesicles from E. coli cells carrying the modeled human mutations, M_L183P and M_L190T, in pBAD33-(MN), were prepared and solubilized with dodecyl maltoside. The samples, including pBAD33-(A-N), a control expressing the entire nuo operon, were immunoprecipitated with an HA antibody to bring down the HA-tagged subunit N. The input panel shows the presence of subunit M in all samples, while the IgG panel shows that in the absence of HA-antibody, no M subunits were precipitated. (E and F) ns, not significant; *P < 0.05; ****P < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The ND4_L158P mutation (T11232C) was first identified in a 48 year old male with symptoms of chronic progressive external ophthalmoplegia (CPEO) (Pulkes et al., 2003). In muscle cells the mutant load was about 40%, but the rates of Complex I activity were well within the normal range, suggesting a mild effect. In E. coli two mutations, nuoM_L183P and nuoM_L183A, were constructed and analyzed in membrane vesicles. The L183P mutant had a diminished level of dNADH oxidase activity of 72 ± 7% as compared to wild type (Fig. 4E), while the L183A had higher activity of 87 ± 6%, suggesting that the proline was deleterious. In proton translocation assays, both mutants showed a corresponding level compared to the wild type (Fig. 4G). The presence of subunit M was confirmed by immunoblotting (Supplementary Fig. S2). A band corresponding to the size of Complex I (540 kDa) was detected in BN gels, and an in-gel assay showed bands of activity corresponding to full-size Complex I (Supplementary Fig. S2). Finally, the L183P mutant was tested for its ability to disrupt a complex of subunits M and N. This mutant was expressed from pBAD33(MN), and after preparation of membrane vesicles and solubilization by dodecyl maltoside, subunit M was tested for co-immunoprecipitation by an anti-HA antibody, which precipitates subunit N. The results (Fig, 4I), show that nuoM_L183P is not co-immunoprecipitated with N, unlike the wild type M. This shows a potential disruption of assembly due to the mutation.
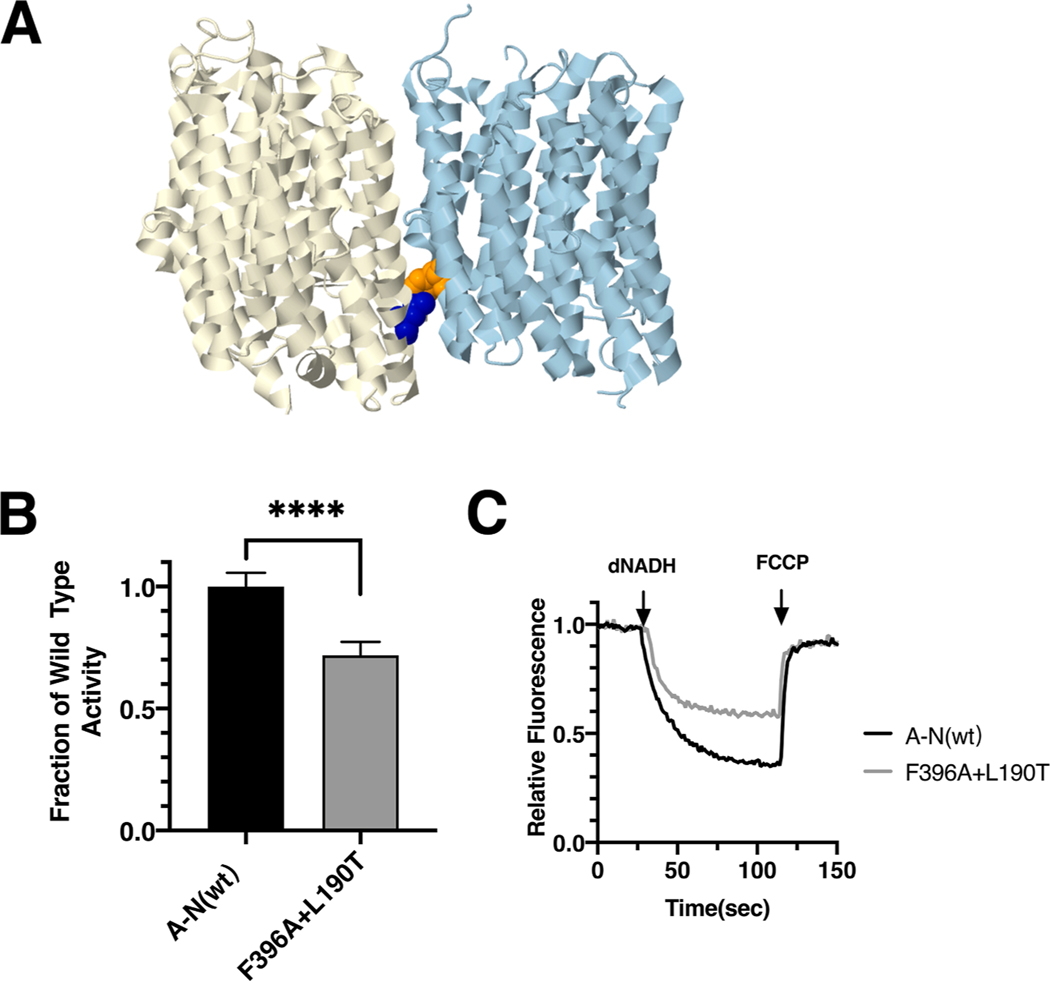
The ND4_I165T mutation was discovered in a 29 year old male with LHON symptoms (Leo-Kottler et al., 2002) and in a patient with idiopathic Parkinson disease by PCR analysis of the mitochondrial DNA from the substantia nigra (Kösel et al., 1998). Vision of the individual with LHON declined for several months after initial diagnosis, but the following year began to improve to moderate levels. In E. coli two mutations, nuoM_L190T and nuoM_L190A, were constructed and analyzed in membrane vesicles. The nuoM_L190T mutant had a slightly diminished level of dNADH oxidase activity of 85 ± 3% as compared to wild type (Fig. 4F), while nuoM_L190A had higher activity of 93 ± 14%. In proton translocation assays, both mutants showed a corresponding rate compared to the wild type (Fig. 4H). The presence of subunit M was confirmed by immunoblotting (Supplementary Fig. S2). A complex corresponding to the size of Complex I (540 kDa) was detected in BN gels by immunoblotting, and an in-gel assay showed bands of activity corresponding to full-size Complex I (Supplementary Fig. S2). The nuoM_L190T mutant was also tested for its ability to disrupt the formation of a complex of subunits M and N. The results showed that nuoM_L190T is not co-immunoprecipitated with N, unlike the wild type M (Fig. 4I). This suggests a potential disruption of assembly like that of the nuoM_L183P mutant. Finally, the nuoM_L190T mutation was constructed with a second mutation in an interacting residue of subunit N, F396A, which reduces the possible interaction surface. The results indicate a further reduced level of activity, 72 ± 5%, as compared to the single mutant in M, L190T, 85% of wild type (Fig. 5). These results suggest that interactions of subunit M with nuoN_F396 contribute to enzyme stability or function.
Fig. 5.
Analysis of interacting mutations in nuoM of E. coli. (A) The locations of mutations nuoN_F396A (orange) and nuoM_L190T (blue) are shown in nuoN (light blue) and nuoM (light yellow), using PDB file 3rko (Efremov and Sazanov, 2011). (B) dNADH-oxidase activity of membrane vesicles prepared from the E. coli double mutant nuoN_F396A/nuoM_L190T is shown compared to a wild type sample prepared the same day. (C) Proton translocation rates from the same samples shown in panel B are indicated by fluorescence quenching of the acridine dye ACMA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Mutations in ND2 (nuoN)
Two clinical mutations in ND2, F60S (red) and L71P (blue), were modeled in the nuoN gene of E. coli. The E. coli residues are T160 (red) and L171 (blue). Both are found at the interface with ND4L (nuoK) in TM2 (TM6 in E. coli). The mammalian ND2 lacks the first four transmembrane helices, and about 100 amino acids, that are found in bacterial species. Mutated residue F60 contacts TM3 of subunit K (ND4L), near the key residue E70, while mutated residue L71 contacts TM2 of subunit K (ND4L) near key residue E34. These interactions are part of the functional interface of subunits K and N (ND4L and ND2) and are essential for proton translocation. The locations of the mutations in the human and the E. coli proteins are shown in Fig. 6.
Fig. 6.
Analysis of ND2 mutations in nuoN of E. coli. (A) The locations of human mutations F60S (red) and L71P (blue) are shown in ND5 (light blue), using PDB file 5xtd (Gu et al., 2016). (B) ND4L (violet) is shown. (C) The locations of E. coli residues T160 (red) and L171 are shown in nuoN (light blue), using PDB file 3rko (Efremov and Sazanov, 2011). (D) nuoK (violet) is shown. (E, F) dNADH-oxidase activities of membrane vesicles prepared from the E. coli mutants are shown compared to a wild type sample prepared the same day. (G, H) Proton translocation rates from the same samples shown in panels E and F are indicated by fluorescence quenching of the acridine dye ACMA. (I) Membrane vesicles from E. coli cells carrying the modeled human mutations, nuoN_T160F and nuoN_L171P, in pBAD33-(I-N), were prepared and solubilized with dodecyl maltoside. The samples, including a control expressing the entire nuo operon, pBAD33-(A-N), were immunoprecipitated with an HA antibody to bring down the HA-tagged subunit N. The input panel shows the presence of subunit K in all samples, while the IgG panel shows that in the absence of HA-antibody, no K subunits were precipitated. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The ND2_F60S mutation (T4648C) was first identified in a 65 year old male with pseudoexfoliation syndrome (PES), a less severe form of pseudoexfoliation glaucoma (PEG) (Abu-Amero et al., 2008). Limited biochemical analysis indicated no difference in respiration rates relative to controls. In E. coli two mutations, nuoN_T160A and nuoN_T160F, were constructed and analyzed in membrane vesicles. Due to lack of sequence conservation, T160F was chosen to be disruptive. Both mutants had high levels of dNADH oxidase activity (Fig. 6E), with values compared to wild type of 97 ± 5% for nuoN_T160A and 87 ± 3% for nuoN_T160F. In proton translocation assays, both mutants showed a corresponding level compared to the wild type (Fig. 6G). The presence of subunit N was confirmed by immunoblotting (Supplementary Fig. S3). A complex corresponding to the size of Complex I (540 kDa) was detected in BN gels by immunoblotting, and an in-gel assay showed bands of activity corresponding to full-size Complex I (Supplementary Fig. S3).
The ND2_L71P (T4681C) mutation was discovered in an infant boy with Leigh Syndrome, who died at age 12 (Ugalde et al., 2007). In blood, muscle, and fibroblast cells the mutant load was about 95%, and the rates of Complex I activity were well below the normal range. Analysis by BN gel electrophoresis found decreased levels of fully assembled Complex I and increased levels of sub-complexes. In E. coli two mutations, L171P and L171A, were constructed in nuoN and analyzed in membrane vesicles. The nuoN_L171P mutant had a diminished level of dNADH oxidase activity of 73 ± 1% as compared to wild type, while the nuoN_L171A had higher activity of 92 ± 3% (Fig. 6F). In proton trans- location assays, both mutants showed a corresponding level compared to the wild type (Fig. 6H). The presence of subunit N was confirmed by immunoblotting (Supplementary Fig. S3). A band corresponding to the size of Complex I (540 kDa) was detected in BN gels, and an in-gel assay showed bands of activity corresponding to full-size Complex I (Supplementary Fig. S3).
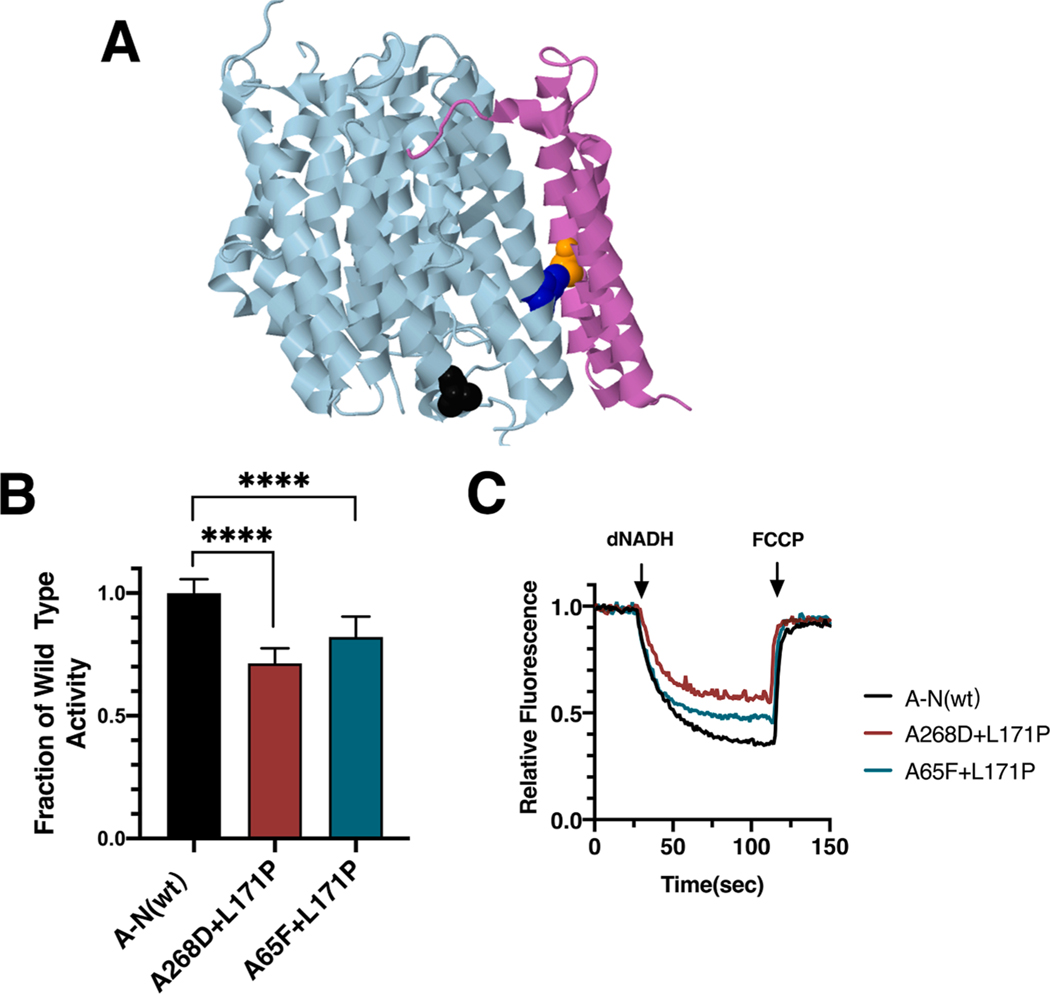
In the original report of the ND2_L71P (T4681C) mutation (Ugalde et al., 2007), 25 polymorphisms in the mtDNA of the individual were reported, including N150D in ND2 (A4917G). N150 is not conserved in E. coli nuoN, but the corresponding residue appears to be A268 (see Fig. 7A). The double mutant in subunit N, A268D/L171P, was constructed to probe whether the polymorphism contributed to the pathology. The double mutant had a level of dNADH oxidase activity of 71 ± 6% (Fig. 7B), while the single mutant, nuoN_L171P, had an indistinguishable activity of 73 ± 1% (Fig. 6F). This suggests that the polymorphism had no impact on the identified mutation. Finally, the nuoN_L171P mutation was constructed with a second mutation in an interacting residue of subunit K, A65F, which increases the possible interaction surface. The results indicate a somewhat increased level of activity, 82 ± 8% (Fig. 7B), as compared to the single mutant nuoN_L171P, 73% of wild type (Fig. 6F). This result suggests that the introduced nuoK_A65F mutation partially suppresses the nuoN_L171P mutation through increased nonbonding interactions.
Fig. 7.
Analysis of interacting mutations in nuoN of E. coli. (A) The locations of mutations K_A65F (orange), N_A268D (black), and N_L171P (blue) are shown in nuoN (light blue) and nuoK (violet), using PDB file 3rko (Efremov and Sazanov, 2011). (B) dNADH-oxidase activities of membrane vesicles prepared from the E. coli double mutants nuoN_A268D/nuoN_L171P and nuoK_A65F/nuoN_L171P are shown compared to a wild type sample prepared the same day. (C) Proton translocation rates from the same samples shown in panel B are indicated by fluorescence quenching of the acridine dye ACMA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Mutations in ND4L and ND6 (nuoK and nuoJ)
One clinical mutation in ND4L, H25R (blue), was modeled in the nuoK gene of E. coli. The E. coli residue is nuoK_N27 (blue). It is found at the interface with ND6 (nuoJ), which is one of the largest subunit interfaces in the E. coli enzyme. The three helices of K are nearly surrounded by the five helices of J, on three sides. In the human enzyme, H25 is contacted by S24 at the N-terminal end of TM2, and by E76 at the C-terminal end of TM3. In E. coli, the primary contact is from E84 of subunit J, found in a similar position in a helical connection between TM2 and TM3. The location of the mutation in the human and the E. coli proteins is shown in Fig. 8.
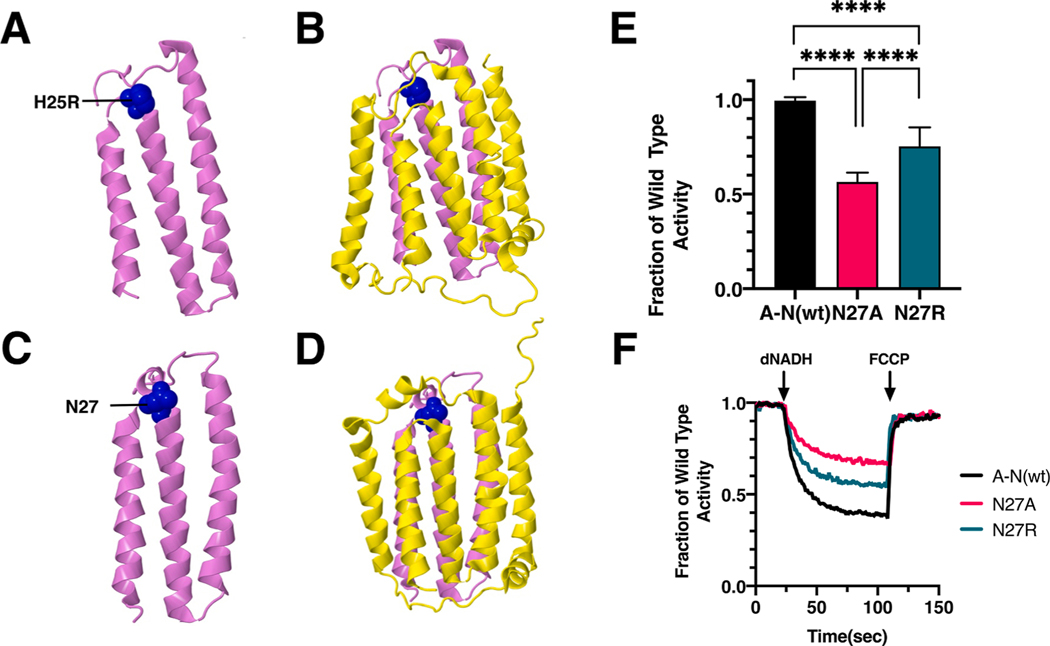
Fig. 8.
Analysis of an ND4L Mutation in nuoK of E. coli. (A) The location of human mutation H25R (blue) is shown in ND4L (violet), using PDB file 5xtd (Gu et al., 2016). (B) Neighboring subunit ND6 (yellow) is shown. (C) The location of E. coli residue N27 is shown in nuoK (violet), using PDB file 3rko (Efremov and Sazanov, 2011). (D) Neighboring subunit nuoJ (yellow) is shown. (E) dNADH-oxidase activities of membrane vesicles prepared from the E. coli mutants are shown compared to a wild type sample prepared the same day. (F) Proton translocation rates from the same samples shown in panel E are indicated by fluorescence quenching of the acridine dye ACMA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The ND4L_H25R mutation (A10543G) was first identified in a 65 year old male with pseudoexfoliation syndrome (PES) (Abu-Amero and Bosley, 2006). Limited analysis of biochemical function indicated no difference in respiration rates relative to controls. In E. coli two mutations, N27R and N27A, were constructed in nuoK and analyzed in membrane vesicles. The nuoK_N27R mutant had a diminished level of dNADH oxidase activity of 74 ± 10% as compared to wild type (Fig. 8E), while nuoK_N27A had a lower activity of 54 ± 5%, suggesting that a polar residue was preferred at this position. In proton translocation assays, corresponding levels were seen, as compared to the wild type (Fig. 8F). The presence of subunit K was confirmed by immunoblotting (Supplementary Fig. S4). A band corresponding to the size of Complex I (540 kDa) was detected in BN gels by immunoblotting, and an in-gel assay showed bands of activity corresponding to full-size Complex I (Supplementary Fig. S4).
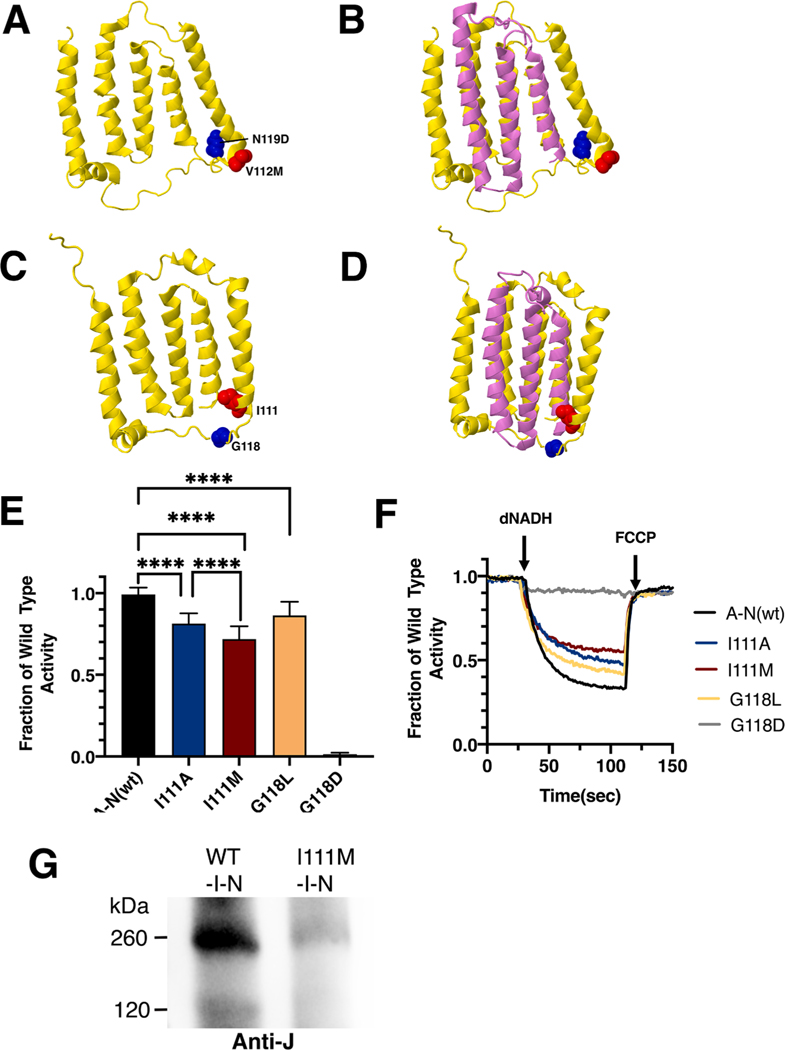
Two clinical mutations in ND6, V112M (red) and N119D (blue), were modeled in the nuoJ gene of E. coli. The E. coli residues are I111 (red) and G118 (blue). The locations of the mutations are shown in Fig. 9 for the human and the E. coli proteins. In all cases, the mutated residues are found near the N-terminus of ND4L (nuoK).
Fig. 9.
Analysis of ND6 mutations in nuoJ of E. coli. (A) The locations of human mutations V112M (red) and N117D (blue) are shown in ND6 (yellow), using PDB file 5xtd (Gu et al., 2016). (B) Neighboring subunit ND4L (violet) is shown. (C) The locations of E. coli residues I111 (red) and G118 (blue) are shown in nuoJ (yellow), using PDB file 3rko (Efremov and Sazanov, 2011). (D) Neighboring subunit nuoK (violet) is shown. (E) dNADH-oxidase activities of membrane vesicles prepared from the E. coli mutants are shown compared to a wild type sample prepared the same day. (F) Proton translocation rates from the same samples shown in panel E are indicated by fluorescence quenching of the acridine dye ACMA. (G) Membrane vesicles were prepared from cells expressing wild type pBAD33-(I-N) and from the same plasmid carrying the I111M mutation. The samples were solubilized with dodecyl maltoside and analyzed by BN gel electrophoresis. After probing with the blot with anti-J antibody, the wild type sample showed the sub-complex of JKLMN (~260 kDa), while the mutant showed only a faint band at that position. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The ND6_V112M mutation (C14340T) was first identified in a 27 year old female with auditory neuropathy along with the 12S rRNA mutation T1095C, previously known to be associated with hearing loss (Wang et al., 2005). No biochemical analysis was done. In E. coli two mutations were constructed in nuoJ, I111M and I111A, and analyzed in membrane vesicles. The nuoJ_I111M mutant had a diminished level of dNADH oxidase activity of 72 ± 8% as compared to wild type (Fig. 9E), while nuoJ_I111A had a slightly higher activity of 81 ± 6%. In proton translocation assays, both mutants showed a corresponding level compared to the wild type (Fig. 9F). The presence of subunit J was confirmed by immunoblotting (Supplementary Fig. S5). A band corresponding to the size of Complex I (540 kDa) was detected in BN gels by immunoblotting, and an in-gel assay showed bands of activity corresponding to full-size Complex I (Supplementary Fig. S5). The mutant I111M was tested for its ability to assemble in the subcomplex of J-N. The mutants were expressed from pBAD33(I-N), and after preparation of membrane vesicles, and solubilization by dodecyl maltoside. Blue native gel electrophoresis was performed, immunoblotting used antibody against subunit J. A very faint band at the size of 260KDa was seen in I-N (I111M), suggesting that mutant I111M greatly decreases the efficiency of assembly of J-N (Fig. 9G).
The ND6_N119D mutation (T14325C) was identified in a single Dutch pedigree of individuals with LHON that lack the 3 most common mutations in mitochondrial DNA at positions 3460, 11778, or 14,484 (Howell et al., 2003). In E. coli two mutations, G118D and G118L, were constructed in nuoJ and analyzed in membrane vesicles. The nuoJ_G118D mutant showed no dNADH oxidase activity, while the nuoJ_G118L had a slightly reduced activity of 86 ± 8% as compared to wild type (Fig. 9E). In proton translocation assays, both mutants showed a corresponding level compared to the wild type (Fig. 9F). The presence of G118L was confirmed by immunoblotting, but G118D was absent (Supplementary Fig. S5). A band corresponding to the size of Complex I (540 kDa) was detected for G118L in BN gels, but not for G118D (Supplementary Fig. S5). Similarly, an in-gel assay showed a band of activity corresponding to full-size Complex I for G118L, but only a faint band of activity for G118D, consistent with a very low level of assembled Complex I (Supplementary Fig. S5).
3.5. Mutations in ND3 (nuoA)
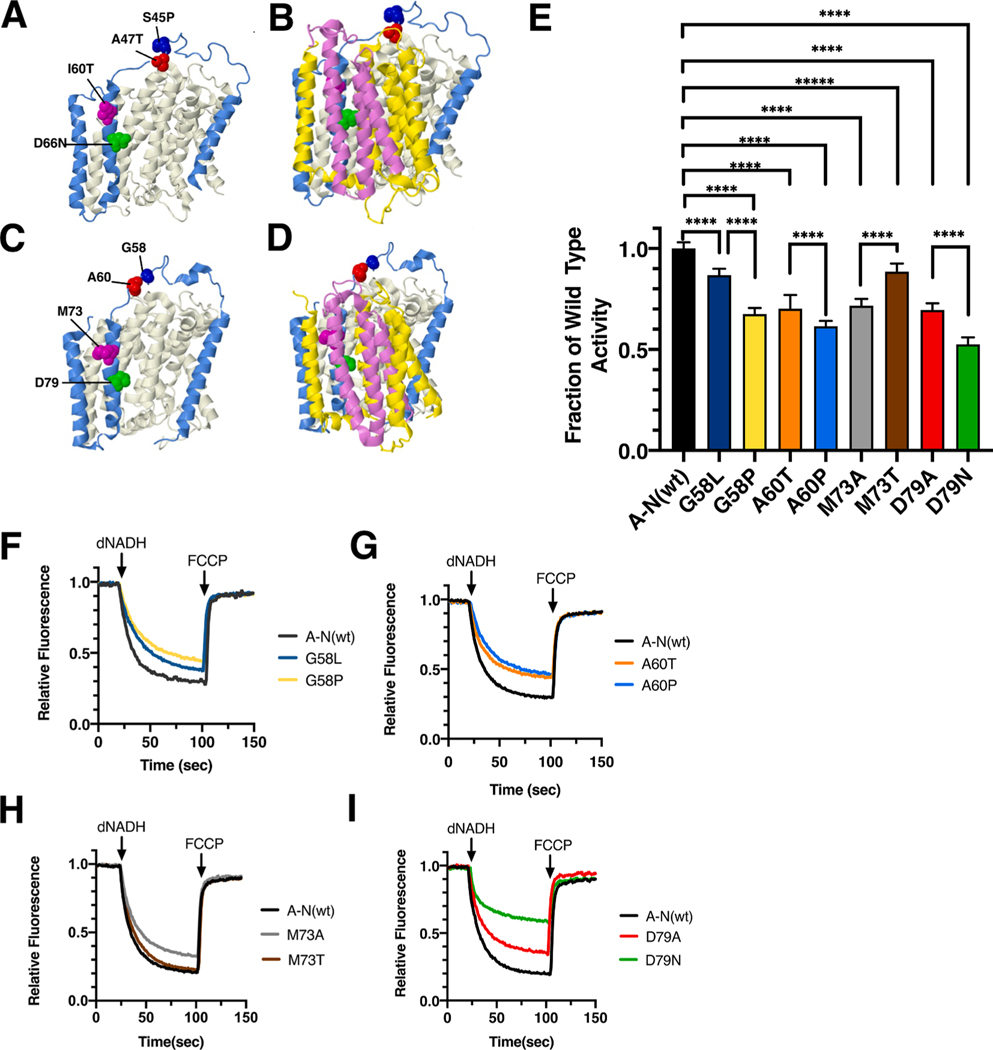
Four clinical mutations in ND3, S45P (blue), A47T (red), I60T (magenta), and D66N (lime), were modeled in the nuoA gene of E. coli. The E. coli residues are G58 (blue), A60 (red), M73 (magenta), and D79 (lime). The locations of the mutations are shown in Fig. 10 for the human and the Thermus thermophilus proteins. In this case, the structure of different bacterial Complex I is shown, because it shows a more complete ND3-ND1 architecture (Baradaran et al., 2013). Both S45P and A47T are found in the long loop (residues 25–50) of ND3 between TM1 and TM2. These two residues are found at the interface between the loop connecting TM3-TM4 in ND1 (nuoH) and the loop connecting TM3-TM4 in ND6 (nuoJ). This region undergoes important conformational changes during catalytic turnover. Residues I60 and D66 are found in TM2 of ND3, which is situated between TM4 of ND1 (nuoH), TM5 of ND6 (nuoJ), and TM3 of ND4L (nuoK). Residue D66 is in a hydrophobic environment in the conformation shown here, but during turnover, it becomes hydrated, and is part of the proton translocation pathway.
Fig. 10.
Analysis of ND3 mutations in nuoA of E. coli. (A) The locations of human mutations S45P (red), A47T (blue), I60T (magenta), and D66N (green) are shown in ND3 (blue). ND1 is shown in beige, using PDB file 5xtd (Gu et al., 2016). (B) Neighboring subunits ND4L (violet) and ND6 (yellow) are shown. (C) The locations of E. coli residues G58 (red), A60 (blue), M73 (magenta), and D79 (green) are shown in nuoA (blue) as modeled in the T. thermophilus structure (PDB file 4hea (Baradaran et al., 2013)). nuoH is shown in beige. (D) Neighboring subunits nuoK (violet) and nuoJ (yellow) are shown. (E) dNADH-oxidase activities of membrane vesicles prepared from the E. coli mutants are shown compared to a wild type sample prepared the same day. (F, G, H, I) Proton translocation rates from the same samples shown in panel E are indicated by fluorescence quenching of the acridine dye ACMA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The ND3_S45P mutation (T10191C) was first identified in a 42 year old male that had developed symptoms including ataxia and seizures beginning at age 24 (Taylor et al., 2001). Initial analysis of Complex I activity indicated a loss of respiration rate, depending upon the mutant load in the tissue assayed. This mutation was found again in 2003 in a second individual that died after 23 days (McFarland et al., 2004). This individual had a high mutant load (>98%) in all tissues that were analyzed and showed 60% level of Complex I assembly by native gel electrophoresis. In E. coli two mutations, G58P and G58L, were constructed in nuoA and analyzed in membrane vesicles. The nuoA_G58P mutant had a diminished level of dNADH oxidase activity of 65 ± 3% as compared to wild type, while nuoA_G58L had a higher activity of 85 ± 3% (Fig. 10E). In proton translocation assays, both mutants showed a corresponding rate compared to the wild type (Fig. 10F). The presence of subunit A was confirmed by immunoblotting (Supplementary Fig. S6). A band corresponding to the size of Complex I (540 kDa) was detected in BN gels by immunoblotting, and an in-gel assay showed bands of activity corresponding to full-size Complex I (Supplementary Fig. S6).
The ND3_A47T (G10197A) mutation was discovered in 3 children with basal ganglia lesions and Complex I deficiencies [Chae 2007], and in 3 other families, where affected individuals had Leigh syndrome or dystonia [Sarzi 2007]. Severity of the disease was associated with high mutant loads (>95%), and rates of Complex I activity well below the normal range. In E. coli two mutations, A60P and A60T, were constructed in nuoA and analyzed in membrane vesicles. The nuoA_A60P mutant had a diminished level of dNADH oxidase activity of 60 ± 3% as compared to wild type, while nuoA_A60T had a slightly higher activity of 70 ± 7% (Fig. 10E). In proton translocation assays, both mutants showed a corresponding rate compared to the wild type (Fig. 10G). The presence of subunit A was confirmed by immunoblotting (Supplementary Fig. S6). A band corresponding to the size of Complex I (540 kDa) was detected in BN gels, and an in-gel assay showed bands of activity corresponding to full-size Complex I (Supplementary Fig. S6).
The ND3_I60T (T10237C) mutation was discovered in an LHON patient, but was not characterized biochemically (Horvath et al., 2002). In E. coli two mutations were constructed in nuoA, M73T and M73A, and analyzed in membrane vesicles. The nuoA_M73T mutant had a somewhat high level of dNADH oxidase activity of 88 ± 4% as compared to wild type, while the nuoA_M73A had a lower activity of 70 ± 3% (Fig. 10E). In proton translocation assays, both mutants showed a corresponding rate compared to the wild type samples (Fig. 10H). The presence of subunit A was confirmed by immunoblotting (Supplementary Fig. S6). A band corresponding to the size of Complex I (540 kDa) was detected in BN gels, and an in-gel assay showed bands of activity corresponding to full-size Complex I (Supplementary Fig. S6).
The ND3_D66N (G10254A) mutation was discovered in a patient with Leigh syndrome that died at the age of 8 months (Leshinsky-Silver et al., 2010). In muscle cells the mutant load was 90%, and the level of Complex I was low, as judged by BN gel electrophoresis. In E. coli two mutations, D79N and D79A, were constructed in nuoA and analyzed in membrane vesicles. The nuoA_D79N mutant had a diminished level of dNADH oxidase activity of 52 ± 4% as compared to wild type, while the nuoA_D79A had a higher activity of 69 ± 3% (Fig. 10E). In proton translocation assays, both mutants showed a corresponding rate compared to the wild type (Fig. 10I). The presence of subunit A was confirmed by immunoblotting (Supplementary Fig. S6). A band corresponding to the size of Complex I (540 kDa) was detected in BN gels, and an in-gel assay showed bands of activity corresponding to full-size Complex I (Supplementary Fig. S6). In all three blots, the level of D79N was reduced relative to the wild type and the D79A mutant.
4. Discussion
The thirteen clinical mutations described in this report are listed in Table 1, along with the activities of the modeled residues in E. coli and whether an assembly defect was observed. The pathogenicity of the human mutations is listed, as described at the Mitomap (Lott et al., 2013) website (www.mitomap.org) and at the NCBI site of Clinical Variations (ClinVar) [Henrie 2018]. Only two of these mutations, ND3_S45P and ND3_A47T, were considered to be pathogenic by both sites. The activities of the corresponding E. coli mutants seem consistent with pathogenicity, at 65% and 70%, respectively. This region of ND3 undergoes large conformational changes as the enzyme transitions between closed and open states (Grba and Hirst, 2020; Gu et al., 2022; Kampjut and Sazanov, 2020; Parey et al., 2018), consistent with the sensitivity to mutation. Two additional mutations are described as pathogenic at ClinVar: ND2_L71P and ND3_D66N. The latter shows an activity of 52% when modeled in E. coli, also consistent with pathogenicity. The same mutation had been constructed and analyzed in 2004 (Kao et al., 2004), and found to have 44% of the wild type activity, similar to our results. This residue in ND3 is found near a chain of water molecules connecting the quinone binding region with the distal membrane arm, and it adopts different positions depending upon whether the enzyme is in a closed or open conformation, during enzyme turnover (Grba and Hirst, 2020; Gu et al., 2022; Kampjut and Sazanov, 2020). When modeled in E. coli, ND2_L71P has a somewhat higher activity of 73%. However, this mutation shows a clear assembly defect when subunits JKLMN are expressed alone, in that the K subunit (ND4L) fails to co-immunoprecipitate with N (ND2). Therefore, it appears that these 4 mutants, which are described as pathogenic at Mitomap and/or ClinVar, are successfully modeled in E. coli.
Table 1.
Summary of the Pathogenicity of Clinical Mutants.
| Clinical Mutation | E. coli mutation | dNADH activity of E. coli mutanta | Defective Assembly observedb | Pathogenicity by Mitomapc (Lott et al., 2013) | Pathogenicity by ClinVard (Landrum et al., 2016) |
|---|---|---|---|---|---|
|
| |||||
| ND5-I149S | nuoL_L148S | 83 ± 2% | yes | Reported | |
| ND5-Y159H | nuoL_Y158H | 86 ± 3% | yes | Reported | conflict |
| ND4-L158P | nuoM_L183P | 72 ± 7% | yes | Reported | uncertain |
| ND4-I165T | nuoM_L190T | 85 ± 3% | yes | Reported | benign |
| ND2-F60S | nuoN_T160F | 87 ± 3% | yes | Reported | |
| ND2-L71P | nuoN_L171P | 73 ± 1% | yes | Reported | pathogenic |
| ND4L-H25R | nuoK_N27R | 74 ± 10% | no | Reported | |
| ND6-V112M | nuoJ_I111M | 72 ± 8% | yes | Reported | likely benign |
| ND6-N117D | nuoJ_G118D | 2% | yes | Reported | benign |
| ND3-S45P | nuoA_G58P | 65 ± 3% | no | Confirmed | pathogenic |
| ND3-A47T | nuoA_A60T | 70 ± 7% | no | Confirmed | pathogenic |
| ND3-I60T | nuoA_M73T | 88 ± 4% | no | Reported | benign |
| ND3-D66N | nuoA_D79N | 52 ± 4% | yes | Reported | pathogenic |
Activities are calculated relative to the wild type plasmid, and are reported as mean ± standard deviation with 2–3 biological replicates and at least 3 technical replicates each.
Defective assembly includes a low yield of Complex I as seen in native gel electrophoresis, low yield in co-immunoprecipitation, or low yield in sub-complex formation.
Mitomap is found at https://www.mitomap.org.
ClinVar is found at https://www.ncbi.nlm.nih.gov/clinvar/.
Two of the clinical mutations were considered uncertain in pathogenicity: ND5_Y159H and ND4-L158P. The former residue is found near a chain of water molecules that connect to the matrix space (Grba and Hirst, 2020; Gu et al., 2022; Kampjut and Sazanov, 2020), and showed a relatively high activity of 86% when modeled in E. coli as Y158H, although a deficiency in assembly was discovered. When it was tested in the time-delayed expression system, where subunit L (ND5) is expressed last, it showed only 60% of the activity of wild type L, indicating a likely assembly defect. Similarly, ND4_L158P showed an activity of 73% when expressed in E. coli as nuoM_L183P, but in an assembly test when only subunits M and N were expressed, subunit M (ND4) failed to associate with subunit N (ND2). These results support the hypothesis that these mutations could cause assembly defects in humans. This residue was found near the chain of water molecules in the central axis of the membrane arm in mitochondrial Complex I, and so might also impact activity by disturbing the connected water molecules (Grba and Hirst, 2020; Gu et al., 2022; Kampjut and Sazanov, 2020).
Four of the clinical mutations are described as benign, or likely benign, at the ClinVar site. ND4_I165T is associated with Parkinson’s Disease and with a mild case of LHON that exhibited partial recovery. In E. coli it showed 85% of the activity of the wild type, but also an inability to associate with the N subunit (ND2) when expressed together. Formation of the MN dimer is likely to be very sensitive to mutations at the interface. The ND3_I60T mutation, when modeled in E. coli, gave 88% of the activity, which seems consistent with a benign mutation. Complex I appeared to assemble normally, when it was expressed in the while operon plasmid, pBAD33(A-N), and no further analysis of assembly was carried out. The two ND6 mutations, V112M and N117D, are described as likely benign, or benign at ClinVar. The former mutation was found with a second mutation in a tRNA gene that was considered more likely to be pathogenic (Wang et al., 2005). The latter was found in Dutch pedigree of LHON patients, and its effects are likely to depend upon the mitochondrial haplotype (Howell et al., 2003). In general, it has been difficult to model ND6 mutations in the E. coli nuoJ subunit (Kao et al., 2005; Pätsi et al., 2008), especially the loop containing these two residues (see Fig. 5), due to differences in sequence, and in the resulting tertiary structure. That likely explains the very low activity of the G118D mutation in E. coli, which probably does not reflect the ND6_N117D phenotype. The E. coli protein has two additional aspartate residues in that region, 116 and 120, which might contribute to the phenotype we observed. In contrast, the human protein has no charged amino acids in the corresponding region, residues 117–121.
The remaining three mutations were not characterized at ClinVar: ND5_I149S, ND2_F60S, and ND4L_H25R. When modeled in E. coli, the first two both showed deficiencies in assembly. The ND5 mutation was tested by the time-delayed expression of L, where it showed only 70% of the wild type activity, as compared to its 83% when expressed from the whole operon plasmid. The ND2 mutation also showed a deficiency in assembly, when only M and N subunits were expressed, So, although the activity of this mutant when expressed from the whole operon plasmid was 87% of wild type, it is possible that it could be defective in assembly. In a previous report from this lab (Amarneh and Vik, 2003), the nuoN_T160I mutant was shown to have about 80% of wild type activity. Later, using molecular dynamics simulations (Kaila et al., 2014) it was found that this mutation caused a blockage of the chain of water mol- ecules in the central axis. In the case of ND4L_H25R no further analysis of assembly was done, but the activity was only 74% of the wild type in E. coli, consistent with pathogenicity. This residue is found in a polar environment, in both human and E. coli Complex I provided by E77 (ND6) or E84 (nuoJ), and in our analysis the alanine mutant had lower activity than the arginine mutant. This would be consistent with His25 being a sensitive site for mutation. This residue was previously mutated in the E. coli subunit, with activities of 82 ± 8% for N27C and 59 ± 8% for N27S as compared to wild type (Torres-Bacete et al., 2012). In that study the mutations were incorporated into the chromosome, and so were expressed from a single copy gene.
In summary, the clinical mutations under consideration in this project were all from membrane arm subunits. They ranged from those that are confirmed or highly likely to be pathogenic, to those that are considered unlikely to be pathogenic. Assessment from reading the clinical literature is difficult because not all relevant information is available, such as biochemical analysis of tissue samples, measurement of the heteroplasmy of the mutation in various tissues, analysis of family members, and knowledge and interpretation of mitochondrial haplotypes. As modeled in E. coli most mutants in this group showed modest effects on Complex I activity, from 60 to 85% of the wild type activity. In general, there was a correlation between the loss of activity seen in the E. coli enzyme and the reported pathogenicity of the clinical mutation, especially when the amino acid sequence and structure was conserved. A limitation of a high expression system, as was used here, might be that a very large amount of Complex I is produced, and this might obscure poor assembly that would have occurred at normal levels of expression. The level of Complex I activity in this system is about 2-fold higher than the normal level as expressed from the chromosome (Amarneh et al., 2006). The use of several assembly assays, expressing individual genes, or genes in small subsets revealed more extensive assembly defects with many of the mutations. We conclude that the E. coli model system can be effective in identifying deleterious human mutations, especially if appropriate assembly assays are carried out.
Supplementary Material
Acknowledgement
We thank Hind Alkhaldi for construction of plasmids, including pBAD33(A-N), and for other assistance. Q.-C.L.D. acknowledges support of the Engaged Learning and Hamilton Scholars programs at Southern Methodist University.
Funding
This research was funded by the National Institutes of Health, USA, grant number 1R15GM126507. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional funding came from the American Heart Association, grant number 17AIREA33661165.
Abbreviations:
- ACMA
9-amino, 6-chloro, 2-methoxyacridine
- BA14
nuoA-Ndeletion strain in E. coli
- BN
Blue native
- dodecyl maltoside
n-Dodecyl-03B2-D-maltopyranoside
- FCCP
carbonyl cyanidep-(trifluoromethoxy) phenylhydrazone
- dNADH
deamino-NADH
- LHON
Leber’s Hereditary Optic Neuropathy
- PES
pseudoexfoliation syndrome
- TM5
transmembrane helix 5 (for example)
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mito.2022.03.001.
References
- Abu-Amero KK, Bosley TM, 2006. Mitochondrial abnormalities in patients with LHON-like optic neuropathies. Invest. Ophthalmol. Vis. Sci 47, 4211–4220. 10.1167/iovs.06-0295. [DOI] [PubMed] [Google Scholar]
- Abu-Amero KK, Bosley TM, Morales J, 2008. Analysis of nuclear and mitochondrial genes in patients with pseudoexfoliation glaucoma. Mol Vis 14, 29–36. [PMC free article] [PubMed] [Google Scholar]
- Agip AA, Blaza JN, Bridges HR, Viscomi C, Rawson S, Muench SP, Hirst J, 2018. Cryo-EM structures of Complex I from mouse heart mitochondria in two biochemically defined states. Nat. Struct. Mol. Biol 25, 548–556. 10.1038/s41594-018-0073-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agip AA, Blaza JN, Fedor JG, Hirst J, 2019. Mammalian respiratory Complex I through the lens of cryo-EM. Annu. Rev. Biophys 48, 165–184. 10.1146/annurev-biophys-052118-115704. [DOI] [PubMed] [Google Scholar]
- Amarneh B, De Leon-Rangel J, Vik SB, 2006. Construction of a deletion strain and expression vector for the Escherichia coli NADH:ubiquinone oxidoreductase (Complex I). BBA 1757, 1557–1560. 10.1016/j.bbabio.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Amarneh B, Vik SB, 2003. Mutagenesis of subunit N of the Escherichia coli Complex I. Identification of the initiation codon and the sensitivity of mutants to decylubiquinone. Biochemistry 42, 4800–4808. 10.1021/bi0340346. [DOI] [PubMed] [Google Scholar]
- Bakare AB, Lesnefsky EJ, Iyer S, 2021. Leigh Syndrome: A Tale of Two Genomes. Front. Physiol 12, 693734 10.3389/fphys.2021.693734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baradaran R, Berrisford JM, Minhas GS, Sazanov LA, 2013. Crystal structure of the entire respiratory Complex I. Nature 494, 443–448. 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi R, Zhang AM, Jia X, Zhang Q, Yao YG, 2012. Complete mitochondrial DNA genome sequence variation of Chinese families with mutation m.3635G>A and Leber hereditary optic neuropathy. Mol Vis 18, 3087–3094. [PMC free article] [PubMed] [Google Scholar]
- Blaza JN, Vinothkumar KR, Hirst J, 2018. Structure of the deactive state of mammalian respiratory Complex I. Structure 26 (312–319), e313. 10.1016/j.str.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann B, Koop Genannt Hoppmann KD, Strahl H, Deckers-Hebestreit G, 2013. Time-delayed in vivo assembly of subunit a into preformed Escherichia coli FoF1 ATP synthase. J. Bacteriol 195, 4074–4084. 10.1128/JB.00468-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MD, Zhadanov S, Allen JC, Hosseini S, Newman NJ, Atamonov VV, Mikhailovskaya IE, Sukernik RI, Wallace DC, 2001. Novel mtDNA mutations and oxidative phosphorylation dysfunction in Russian LHON families. Hum. Genet 109, 33–39. [DOI] [PubMed] [Google Scholar]
- Bungert S, Krafft B, Schlesinger R, Friedrich T, 1999. One-step purification of the NADH dehydrogenase fragment of the Escherichia coli Complex I by means of Strep-tag affinity chromatography. FEBS Lett. 460, 207–211. [DOI] [PubMed] [Google Scholar]
- Cai W, Fu Q, Zhou X, Qu J, Tong Y, Guan MX, 2008. Mitochondrial variants may influence the phenotypic manifestation of Leber’s hereditary optic neuropathy-associated ND4 G11778A mutation. J Genet Genomics 35, 649–655. 10.1016/S1673-8527(08)60086-7. [DOI] [PubMed] [Google Scholar]
- Chomyn A, Cleeter MW, Ragan CI, Riley M, Doolittle RF, Attardi G, 1986. URF6, last unidentified reading frame of human mtDNA, codes for an NADH dehydrogenase subunit. Science 234, 614–618. [DOI] [PubMed] [Google Scholar]
- Chomyn A, Mariottini P, Cleeter MW, Ragan CI, Matsuno-Yagi A, Hatefi Y, Doolittle RF, Attardi G, 1985. Six unidentified reading frames of human mitochondrial DNA encode components of the respiratory-chain NADH dehydrogenase. Nature 314, 592–597. [DOI] [PubMed] [Google Scholar]
- Dang QL, Phan DH, Johnson AN, Pasapuleti M, Alkhaldi HA, Zhang F, Vik SB, 2020. Analysis of Human Mutations in the Supernumerary Subunits of Complex I. Life (Basel) 10. 10.3390/life10110296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov RG, Sazanov LA, 2011. Structure of the membrane domain of respiratory Complex I. Nature 476, 414–420. 10.1038/nature10330. [DOI] [PubMed] [Google Scholar]
- Elurbe DM, Huynen MA, 2016. The origin of the supernumerary subunits and assembly factors of Complex I: A treasure trove of pathway evolution. BBA 1857, 971–979. 10.1016/j.bbabio.2016.03.027. [DOI] [PubMed] [Google Scholar]
- Fiedorczuk K, Letts JA, Degliesposti G, Kaszuba K, Skehel M, Sazanov LA, 2016. Atomic structure of the entire mammalian mitochondrial Complex I. Nature 538, 406–410. 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grba DN, Hirst J, 2020. Mitochondrial complex I structure reveals ordered water molecules for catalysis and proton translocation. Nat. Struct. Mol. Biol 27, 892–900. 10.1038/s41594-020-0473-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Liu T, Guo R, Zhang L, Yang M, 2022. The coupling mechanism of mammalian mitochondrial complex I. Nat. Struct. Mol. Biol 10.1038/s41594-022-00722-w. [DOI] [PubMed] [Google Scholar]
- Gu J, Wu M, Guo R, Yan K, Lei J, Gao N, Yang M, 2016. The architecture of the mammalian respirasome. Nature 537, 639–643. 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- Hirst J, 2013. Mitochondrial complex I. Annu. Rev. Biochem 82, 551–575. 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- Horvath J, Horvath R, Karcagi V, Komoly S, Johns DR, 2002. Sequence analysis of Hungarian LHON patients not carrying the common primary mutations. J. Inherit. Metab. Dis 25, 323–324. [DOI] [PubMed] [Google Scholar]
- Howell N, Bindoff LA, McCullough DA, Kubacka I, Poulton J, Mackey D, Taylor L, Turnbull DM, 1991. Leber hereditary optic neuropathy: identification of the same mitochondrial ND1 mutation in six pedigrees. Am. J. Hum. Genet 49, 939–950. [PMC free article] [PubMed] [Google Scholar]
- Howell N, Oostra RJ, Bolhuis PA, Spruijt L, Clarke LA, Mackey DA, Preston G, Herrnstadt C, 2003. Sequence analysis of the mitochondrial genomes from Dutch pedigrees with Leber hereditary optic neuropathy. Am. J. Hum. Genet 72, 1460–1469. 10.1086/375537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaila VR, Wikstrom M, Hummer G, 2014. Electrostatics, hydration, and proton transfer dynamics in the membrane domain of respiratory complex I. Proc. Natl. Acad. Sci. U. S. A 111, 6988–6993. 10.1073/pnas.1319156111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampjut D, Sazanov LA, 2020. The coupling mechanism of mammalian respiratory complex I. Science 370, eabc4209. 10.1126/science.abc4209. [DOI] [PubMed] [Google Scholar]
- Kao MC, Di Bernardo S, Nakamaru-Ogiso E, Miyoshi H, Matsuno-Yagi A, Yagi T, 2005. Characterization of the membrane domain subunit NuoJ (ND6) of the NADH-quinone oxidoreductase from Escherichia coli by chromosomal DNA manipulation. Biochemistry 44, 3562–3571. 10.1021/bi0476477. [DOI] [PubMed] [Google Scholar]
- Kao MC, Di Bernardo S, Perego M, Nakamaru-Ogiso E, Matsuno-Yagi A, Yagi T, 2004. Functional roles of four conserved charged residues in the membrane domain subunit NuoA of the proton-translocating NADH-quinone oxidoreductase from Escherichia coli. J. Biol. Chem 279, 32360–32366. 10.1074/jbc.M403885200. [DOI] [PubMed] [Google Scholar]
- Klink GV, O’Keefe H, Gogna A, Bazykin GA, Elson JL, 2021. A broad comparative genomics approach to understanding the pathogenicity of Complex I mutations. Sci. Rep 11, 19578. 10.1038/s41598-021-98360-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolata P, Efremov RG, 2021. Structure of Escherichia coli respiratory complex I reconstituted into lipid nanodiscs reveals an uncoupled conformation. eLife 10. 10.7554/eLife.68710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kösel S, Grasbon-Frodl EM, Mautsch U, Egensperger R, von Eitzen U, Frishman D, Hofmann S, Gerbitz KD, Mehraein P, Graeber MB, 1998. Novel mutations of mitochondrial complex I in pathologically proven Parkinson disease. Neurogenetics 1, 197–204. [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. , 2016. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44 (D1), D862–D868. 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo-Kottler B, Luberichs J, Besch D, Christ-Adler M, Fauser S, 2002. Leber’s hereditary optic neuropathy: clinical and molecular genetic results in a patient with a point mutation at np T11253C (isoleucine to threonine) in the ND4 gene and spontaneous recovery. Graefes Arch. Clin. Exp. Ophthalmol 240, 758–764. 10.1007/s00417-002-0494-7. [DOI] [PubMed] [Google Scholar]
- Leshinsky-Silver E, Lev D, Malinger G, Shapira D, Cohen S, Lerman-Sagie T, Saada A, 2010. Leigh disease presenting in utero due to a novel missense mutation in the mitochondrial DNA-ND3. Mol. Genet. Metab 100, 65–70. 10.1016/j.ymgme.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, Procaccio V, Wallace DC, 2013. mtDNA Variation and Analysis Using Mitomap and Mitomaster. Curr Protoc Bioinformatics 44, 1 23 21–26. 10.1002/0471250953.bi0123s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr JA, Haack TB, Freisinger P, Karall D, Makowski C, Koch J, Feichtinger RG, Zimmermann FA, Rolinski B, Ahting U, Meitinger T, Prokisch H, Sperl W, 2015. Spectrum of combined respiratory chain defects. J. Inherit. Metab. Dis 38, 629–640. 10.1007/s10545-015-9831-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland R, Kirby DM, Fowler KJ, Ohtake A, Ryan MT, Amor DJ, Fletcher JM, Dixon JW, Collins FA, Turnbull DM, Taylor RW, Thorburn DR, 2004. De novo mutations in the mitochondrial ND3 gene as a cause of infantile mitochondrial encephalopathy and complex I deficiency. Ann. Neurol 55, 58–64. 10.1002/ana.10787. [DOI] [PubMed] [Google Scholar]
- Michel J, DeLeon-Rangel J, Zhu S, Van Ree K, Vik SB, 2011. Mutagenesis of the L, M, and N subunits of Complex I from Escherichia coli indicates a common role in function. PLoS ONE 6, e17420. 10.1371/journal.pone.0017420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padavannil A, Ayala-Hernandez MG, Castellanos-Silva EA, Letts JA, 2021. The Mysterious Multitude: Structural Perspective on the Accessory Subunits of Respiratory Complex I. Front Mol Biosci 8, 798353. 10.3389/fmolb.2021.798353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parey K, Brandt U, Xie H, Mills DJ, Siegmund K, Vonck J, Kuhlbrandt W, Zickermann V, 2018. Cryo-EM structure of respiratory Complex I at work. eLife 7, e39213 10.7554/eLife.39213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parey K, Wirth C, Vonck J, Zickermann V, 2020. Respiratory complex I - structure, mechanism and evolution. Curr. Opin. Struct. Biol 63, 1–9. 10.1016/j.sbi.2020.01.004. [DOI] [PubMed] [Google Scholar]
- Pätsi J, Kervinen M, Finel M, Hassinen IE, 2008. Leber hereditary optic neuropathy mutations in the ND6 subunit of mitochondrial complex I affect ubiquinone reduction kinetics in a bacterial model of the enzyme. Biochem. J 409, 129–137. 10.1042/BJ20070866. [DOI] [PubMed] [Google Scholar]
- Pulkes T, Liolitsa D, Nelson IP, Hanna MG, 2003. Classical mitochondrial phenotypes without mtDNA mutations: the possible role of nuclear genes. Neurology 61, 1144–1147. 10.1212/01.wnl.0000090465.27024.3d. [DOI] [PubMed] [Google Scholar]
- Ratnaike TE, Greene D, Wei W, Sanchis-Juan A, Schon KR, van den Ameele J, Raymond L, Horvath R, Turro E, Chinnery PF, 2021. MitoPhen database: a human phenotype ontology-based approach to identify mitochondrial DNA diseases. Nucleic Acids Res. 49, 9686–9695. 10.1093/nar/gkab726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazanov LA, 2015. A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat. Rev. Mol. Cell Biol 16, 375–388. 10.1038/nrm3997. [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G, 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem 199, 223–231. [DOI] [PubMed] [Google Scholar]
- Scheffler IE, 2015. Mitochondrial disease associated with complex I (NADH-CoQ oxidoreductase) deficiency. J. Inherit. Metab. Dis 38, 405–415. 10.1007/s10545-014-9768-6. [DOI] [PubMed] [Google Scholar]
- Schimpf J, Oppermann S, Gerasimova T, Santos Seica AF, Hellwig P, Grishkovskaya I, Wohlwend D, Haselbach D, Friedrich T, 2021. Structure of the peripheral arm of a minimalistic respiratory complex I. Structure. 10.1016/j.str.2021.09.005. [DOI] [PubMed] [Google Scholar]
- Taylor RW, Singh-Kler R, Hayes CM, Smith PE, Turnbull DM, 2001. Progressive mitochondrial disease resulting from a novel missense mutation in the mitochondrial DNA ND3 gene. Ann. Neurol 50, 104–107. [DOI] [PubMed] [Google Scholar]
- Torres-Bacete J, Nakamaru-Ogiso E, Matsuno-Yagi A, Yagi T, 2007. Characterization of the NuoM (ND4) subunit in Escherichia coli NDH-1: conserved charged residues essential for energy-coupled activities. J. Biol. Chem 282, 36914–36922. 10.1074/jbc.M707855200. [DOI] [PubMed] [Google Scholar]
- Torres-Bacete J, Sinha PK, Sato M, Patki G, Kao MC, Matsuno-Yagi A, Yagi T, 2012. Roles of subunit NuoK (ND4L) in the energy-transducing mechanism of Escherichia coli NDH-1 (NADH:quinone oxidoreductase). J. Biol. Chem 287, 42763–42772. 10.1074/jbc.M112.422824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugalde C, Hinttala R, Timal S, Smeets R, Rodenburg RJ, Uusimaa J, van Heuvel LP, Nijtmans LG, Majamaa K, Smeitink JA, 2007. Mutated ND2 impairs mitochondrial complex I assembly and leads to Leigh syndrome. Mol. Genet. Metab 90, 10–14. 10.1016/j.ymgme.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Vinothkumar KR, Zhu J, Hirst J, 2014. Architecture of mammalian respiratory complex I. Nature 515, 80–84. 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Long JC, Zhang D, Vik SB, 1999. A novel labeling approach supports the five-transmembrane model of subunit a of the Escherichia coli ATP synthase. J. Biol. Chem 274, 17353–17357. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ 2nd, Nikoskelainen EK, 1988. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science 242, 1427–1430. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li R, Zhao H, Peters JL, Liu Q, Yang L, Han D, Greinwald JH Jr., Young WY, Guan MX, 2005. Clinical and molecular characterization of a Chinese patient with auditory neuropathy associated with mitochondrial 12S rRNA T1095C mutation. Am J Med Genet A 133A, 27–30. 10.1002/ajmg.a.30424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Gomez-Duran A, Hudson G, Chinnery PF, 2017. Background sequence characteristics influence the occurrence and severity of disease-causing mtDNA mutations. PLoS Genet. 13, e1007126 10.1371/journal.pgen.1007126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H, 1993. The gene locus of the proton-translocating NADH: ubiquinone oxidoreductase in Escherichia coli. Organization of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. J. Mol. Biol 233, 109–122. 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- Wong LC, Chen T, Schmitt ES, Wang J, Tang S, Landsverk M, Li F, Zhang S, Wang Y, Zhang VW, Craigen WJ, 2020. Clinical and laboratory interpretation of mitochondrial mRNA variants. Hum. Mutat 41, 1783–1796. 10.1002/humu.24082. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhu Y, Tong Y, Chen L, Liu L, Zhang Z, Wang X, Huang D, Qiu W, Zhuang S, Ma X, 2009. Confirmation of the mitochondrial ND1 gene mutation G3635A as a primary LHON mutation. Biochem. Biophys. Res. Commun 386, 50–54. 10.1016/j.bbrc.2009.05.127. [DOI] [PubMed] [Google Scholar]
- Yu-Wai-Man P, Chinnery PF, 2021. Leber Hereditary Optic Neuropathy, GeneReviews; ®. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Vik SB, 2021. Analysis of the assembly pathway for membrane subunits of Complex I reveals that subunit L (ND5) can assemble last in E. coli. BBA Advances 1, 100027. doi. 10.1016/j.bbadva.2021.100027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HP, Ishikawa H, Yasumoto R, Sakurai K, Sawamura H, 2021. Leber hereditary optic neuropathy harboring a rare m.12811 T>C mitochondrial DNA mutation. Can. J. Ophthalmol 56, e82–e84. 10.1016/j.jcjo.2020.12.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.