Abstract
The RNA-binding proteins (RBPs) are critical trans factors that associate with specific cis elements present in mRNAs whose stability and translation are subject to regulation. The RBP Hu antigen R (HuR) is overexpressed in a wide variety of human cancers and serves as a prognostic factor of poor clinical outcome. HuR promotes tumorigenesis by interacting with a subset of oncogenic mRNAs implicated in different cancer hallmarks, and resistance to therapy. Reduction of HuR levels in cancer cells leads to tumor regression in mouse xenograft models. These findings prompt a working model whereby cancer cells use HuR, a master switch of multiple oncogenic mRNAs, to drive drug resistance and promote cell survival and metastasis, thus rendering the tumor cells with high cytoplasmic HuR more progressive and resistant to therapy. This review summarizes the roles of HuR in cancer and other diseases, therapeutic potential of HuR inhibition, and the current status of drug discovery on HuR.
Keywords: RNA-binding protein, HuR, cancer, small molecules, drug discovery, drug development, companion assay, molecular therapy, drug resistance
Graphical Abstract
1. Introduction
Post-transcriptional gene regulation shapes the fate of each transcript, starting from pre-mRNA splicing and maturation, to mRNA nuclear export, and finally mRNA stability and translation [1-3]. This level of gene regulation is essential for normal development, but when dysregulated, has many implications in disease conditions, including cancer. Hu antigen R (HuR) is one of the best-studied RNA-binding post-transcriptional regulators [2, 4]. It belongs to mammalian embryonic abnormal lethal vision-like (ELAVL) protein family with other three members (i.e. HuB, HuC, and HuD), so it is also known as HuA or ELAVL1 [5]. HuR contains three RNA recognition motifs (RRMs) with high degree of sequence homology and structural similarity to other Hu proteins (Figure 1) [3]. The first two tandemly arrayed RRMs (RRM1 and RRM2) near the N-terminal directly interact with target transcripts bearing adenine- and uridine-rich elements (ARE), or uridine-rich sequences, typically located in the 3′-untranslated region (UTR) [6, 7]. The RRM3 domain at C-terminal was thought to contribute to HuR multimerization and assembly of HuR oligomers on target mRNAs [8, 9]. However, recent studies demonstrate that RRM3 is also involved in RNA recognition and binding [10, 11]. The unconserved hinge region between RRM2 and RRM3 contains a HuR nucleocytoplasmic shuttling (HNS) sequence, which is mainly responsible for HuR translocation (Figure 1) [12]. Different from other three family members, which are mainly cytoplasmic localized neuronal proteins, HuR is ubiquitously expressed in human tissues and predominantly localized in nucleus of resting cells [13]. Studies using PAR-CLIP (photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation) method underscore HuR’s nuclear role of associating with target pre-mRNA introns and affecting pre-mRNA processing [14]. Notably, HuR can be shuttled to the cytoplasmic compartment in response to various stimulations, such as stress signals [15], which, in turn, transports mature target mRNAs to the cytoplasm since HuR dimers/oligomers form complex with ARE-containing mRNAs in the nucleus. This translocation is an important aspect of HuR function of stabilizing and promoting translation of target mRNAs.
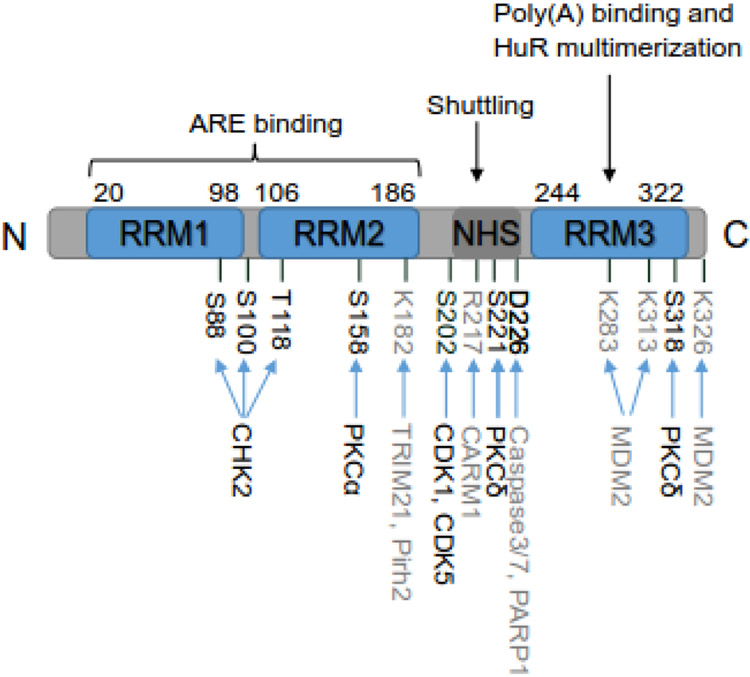
Figure 1:
Schematic structure of HuR. HuR protein consists of 326 amino acids (aa). It has three conserved RNA recognition motifs (RRMs) and an unconserved hinge region containing HuR nucleocytoplasmic shuttling (HNS) sequence. The amino acid positions of these regions are indicated. RRM1 and RRM2 are responsible for recognizing and binding to AREs of mRNA targets. RRM3 is involved in binding to poly(A) tail of mRNA targets and HuR multimerization on mRNA targets. The hinge domain mainly controls the nuclear and cytoplasmic shuttling of HuR. The major post-translational modified amino acids along with the corresponding enzymes are indicated as well, with phosphorylated amino acids and corresponding kinases in black and other modified amino acids and enzymes in grey.
2. Regulation of HuR expression and function
HuR is a ubiquitous protein and it regulates thousands of transcripts. It is not surprising that HuR is found to play essential role in the development biology. Loss of HuR in murine embryos leads to midgestational lethality due to placental insufficiency, while embryos with epiblast-specific HuR deletion have defects in skeletal and splenic development [16]. Postnatal global deletion of HuR in mice is also lethal with impaired hematopoietic and intestinal systems due to the loss of progenitor cells [17]. HuR is also essential for spermatogenesis, as targeted deletion of HuR specifically in germ cells leads to male but not female sterility [18]. Interestingly, HuR-overexpressing in transgenic testis causes compromised fertility as well [19]. Aberrant expression of HuR can lead to many pathological conditions. Indeed, HuR abundance has been founded in malignancies throughout human body and other diseases such as inflammatory diseases. Therefore, tight regulation of HuR expression is critical for maintaining the normal development and preventing pathological processes.
The regulation of HuR expression is reported to be controlled at transcriptional, post-transcriptional, and translational levels (Figure 2). Kang et al. reported that HuR is a direct transcription target of nuclear factor kappaB (NF-κB) and NF-κB activates transcription of HuR to promote gastric tumorigenesis depending on phosphatidylinositol 3-kinase/AKT signaling [20]. Jeyaraj et al. reported that there are multiple Smad 1/5/8-binding motifs in the 5′-UTR of HuR in renal proximal tubule cells indicating the regulation of HuR expression by this Smad family of proteins [21]. However, the full range transcriptional regulation of HuR has not been well defined. HuR gene encodes alternative polyadenylation variants containing functional AREs, which enables HuR protein autoregulates its mRNA by binding to the ARE sites [22, 23]. This post-transcriptional regulation is competed with another RNA-binding protein (RBP) tristetraprolin (TTP), which can also bind to the AREs of HuR mRNA and promote its degradation [22]. HuR was reported to be a target of RBP RNPC1 (also known as RBM38), which stabilizes HuR transcript via binding to the 3′-UTR of HuR [24]. Since the first report of miR-519 repressing HuR translation in 2008 [25], several microRNAs have been identified to regulate HuR biosynthesis. Most of them are tumor-suppressor miRNAs (e.g. miR-16 [26], miR-22 [27], miR-125a [28], miR-125b [29]), which negatively regulates HuR translation.
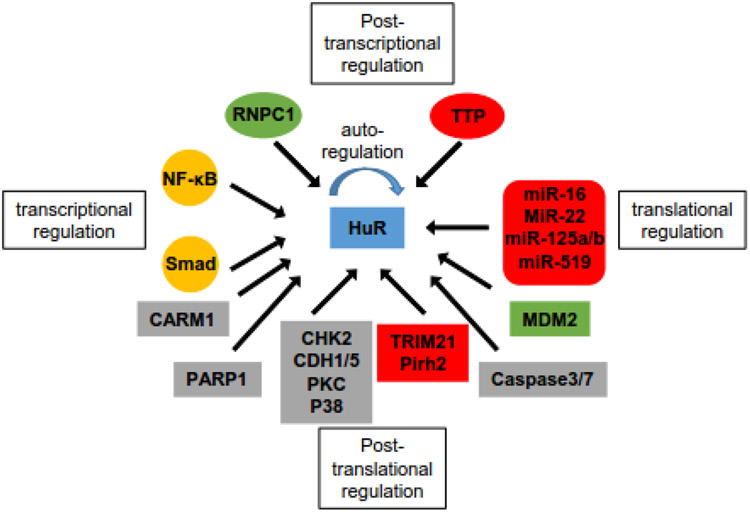
Figure 2:
Map of the regulators of HuR at different levels. Transcriptional factor NF-κB and Smad family proteins regulate the transcription of HuR. RNA-binding protein RNPC1 (green, up-regulation) and TTP (red, down-regulation) oppositely regulate the stability of HuR mRNA, while HuR also autoregulates its own mRNA. MicroRNAs that bind to HuR mRNA and suppress the translation of HuR are listed in red rounded rectangle. Post-translational regulators of HuR are listed in rectangles, with enzymes modulating HuR localization and binding affinity in grey rectangles, MDM2 upregulating HuR expression in green rectangle, and E3 ligases promoting HuR degradation in red rectangle.
The expression and function of HuR are also regulated by multiple post-translational modifications, including phosphorylation, methylation, ubiquitination, NEDDylation, and proteolytic cleavage (Figure 1 and 2) [30]. The checkpoint kinase 2 (CHK2) [31], cyclin-dependent kinases (CDK1 [32] and CDK5 [33]), and protein kinase C (PKC) family [34, 35] are major kinases that phosphorylate HuR with phosphorylation sites at RRMs, hinge region, or both, respectively. In general, phosphorylation at RRMs affects the binding affinity of HuR to target transcripts while phosphorylation at hinge region affects the subcellular localization of HuR [30]. Coactivator-associated arginine methyltransferase 1 (CARM1) can methylate HuR at R217, which alters both localization and RNA-binding affinity of HuR [36, 37]. Ubiquitination of HuR by E3 ligases (TRIM21, Pirh2) mainly promotes HuR degradation [29, 38, 39], while NEDDylation by murine double minute 2 (MDM2) increases HuR protein stability [40]. Notably, Caspase-mediated cleavage of HuR can shift HuR function from being a prosurvival factor under normal conditions to a promoter of apoptosis in response to a lethal stress [41]. In two recent studies, poly-ADP-ribose polymerase 1 (PARP1) was reported to induce PARylation of HuR at D226, resulting in increased HuR olimerization and binding to pro-inflammatory target mRNAs in response to inflammatory stimuli [42, 43].
3. HuR expression and implication in cancer
The potential function of HuR in carcinogenesis was first revealed by a study in mice demonstrating that differences in HuR levels influence the tumorigenicity of a colon cancer cell line [44]. This study also provided pioneer discovery of broadly cytoplasmic HuR abundance in cancer tissues compared with corresponding normal tissues [44]. The uncoupling of HuR mRNA and protein levels are often observed when comparing normal and malignant tissues, as HuR protein levels are heighted in most cancer tissues compared with corresponding normal tissues, whereas its mRNA levels do not change dramatically between normal and cancer tissues. The underlining mechanisms of this uncoupling are not well elucidated but might be related to the alternative HuR mRNA variants that can be auto-regulated by HuR and other RBPs, and microRNA regulation (discussed above) on HuR translation without altering HuR mRNA levels. Therefore, it is important to measure HuR protein levels, particularly cytoplasmic HuR, to investigate the physiological functions of HuR in cancer. Since the above groundbreaking findings in 2003, there have been numbers of studies that examine the HuR expression and explore the diagnostic and prognostic potential of HuR expression in different cancer types.
3.1. HuR in breast cancer
There are a number of studies, including our own, have examined HuR expression in breast cancer tissues and correlations between HuR expression and the grade and clinical outcomes of breast cancer [45-50]. Cytoplasmic HuR is elevated in atypical ductal hyperplasia (ADH) and in ductal in situ carcinomas (DCIS) when compared to normal controls [48]. More importantly, almost all studies [45-50] consistently report that positive/high cytoplasmic HuR is associated with high tumor grade of DCIS and invasive ductal carcinomas (IDC). Furthermore, positive/high cytoplasmic HuR is an independent prognostic biomarker for reduced overall survival [46, 47, 49, 50], distant disease-free survival [46, 49, 50], recurrence-free survival (RFS) [49], and local recurrence-free survival (LRFS) [49] of breast cancer patients. Several studies also suggest that positive/high cytoplasmic HuR is associated with estrogen receptor (ER) [47, 49] and progesterone receptor (PR) [47-49] negativity. However, a study investigated whether cytoplasmic HuR level could be a predictor for patients’ response to neoadjuvant chemotherapy and found that cytoplasmic HuR level is not predictive for pathologic complete response to neoadjuvant chemotherapy [49]. Those studies suggest cytoplasmic HuR as a diagnostic and prognostic marker for breast cancer but not a predictor for response of breast cancer patients to neoadjuvant chemotherapy.
In estrogen receptor positive breast cancer cells, HuR negatively modulates the sensitivity to tamoxifen treatment by stabilizing the mRNA of ER; in the other hand, acute tamoxifen treatment induces cytoplasmic HuR accumulation, which in turn stabilizes transcripts critical for drug resistance [51]. These finding suggests that cytoplasmic HuR level could be useful in predicting potential response to tamoxifen treatment and HuR inhibition may overcome acquired tamoxifen resistance. Similarly, anticancer agent doxorubicin also induces cytoplasmic HuR accumulation by promoting PKCδ mediated HuR phosphorylation in breast cancer cells. However, this accumulation is necessary for doxorubicin-induced apoptosis as HuR regulates the mRNA of doxorubicin target protein TOP2A, and the selected doxorubicin-resistant cells have a significant HuR and TOP2A downregulation [52, 53]. These findings suggest that cytoplasmic HuR levels may serve as a favorable predictor for response to doxorubicin treatment. Nevertheless, the above studies were conducted in vitro only, the generality of their findings in animal studies and clinical patients’ samples remains to be demonstrated.
3.2. HuR in prostate cancer
The analysis of HuR in prostate cancer tissues and non-tumoral glands reveals similar expression pattern of nuclear HuR but significantly higher cytoplasmic HuR in prostate cancer tissues compared to normal glands [54, 55]. The cytoplasmic HuR expression is even higher in castration-resistant prostate cancer (CRPC) tissues compared to that in hormone-naïve prostate cancer tissues [55], suggesting that high cytoplasmic HuR may contribute to resistance to androgen deprivation therapy. This study also found that cytoplasmic HuR was associated with Gleason score, T stage, and metastasis at diagnosis of hormone-naïve prostate cancer tissues and was a potential predictor for biomedical recurrence after radical prostatectomy [55]. Those findings are consistent with the results from a large-scale tissue microarray (TMA) study with over 12,000 radical prostatectomy specimens. This TMA study demonstrates that high cytoplasmic HuR not only is linked to adverse tumor phenotype (high Gleason score, T stage, and lymph node metastasis) but also correlates with prostate-specific antigen (PSA) recurrence in prostate cancer [56]. Prostate cancer tissues with elevated cytoplasmic HuR also have higher HuR target transcripts COX-2 [54, 55] and VEGF-A/C [55], and harbor higher frequency of PTEN, 5q21, 6q15, and 3p13 deletions [56]. Those studies support the view that cytoplasmic HuR promotes prostate cancer progression and recurrence, and it could be an independent diagnostic and prognostic marker for malignancy and clinical outcome.
3.3. HuR in pancreatic cancer
Cytoplasmic HuR abundance had been reported in pancreatic ductal adenocarcinoma (PDAC) and was associated with poorly differentiated PDAs and higher T stage [57, 58]. In the above two studies, high cytoplasmic HuR was associated with prolonged survival of patients with gemcitabine treatment and proposed to be a favorable predictor for response to gemcitabine-based chemotherapy [57, 58]. Mechanistically, cytoplasmic HuR promotes the stability and translation of its target mRNA deoxycytidine kinase (dCK), which can phosphorylate prodrug gemcitabine to active metabolite [57]. However, in two follow-up studies with larger sample size, cytoplasmic HuR had neither prognostic nor predictive value for gemcitabine adjuvant therapy [59, 60]. A study in pancreatobiliary type periampullary adenocarcinoma reported that high HuR cytoplasmic/nuclear ratio was associated with a significantly reduced five-year overall survival in patients receiving adjuvant gemcitabine, but not in untreated patients [61]. Except this controversial effect on gemcitabine treatment, HuR is mainly considered as a modulator of PDAC drug resistance within the elements of PDAC environment [62]. PDAC-associated stressors (e.g., low glucose and hypoxia) induce cytoplasmic translocation and activation of HuR, which renders PDAC cells resistance to cytotoxic chemotherapeutic agents by post-transcriptional regulation of target mRNAs whose encoded proteins are involved in adaptive survival mechanism, including serine–threonine kinase PIM1, mitotic inhibitor kinase WEE1, and antioxidant enzyme IDH1 [63-65]. Therefore, targeting HuR has the potential to overcome PDAC chemoresistance.
3.4. HuR in other types of cancer
Increased cytoplasmic HuR was found in colon carcinomas compared to adenomas and normal colon mucosa [44, 66]. Furthermore, cytoplasmic HuR correlated with COX-2 expression, lymphatic invasion, the presence of a lymph node metastasis, and tumor grade in colorectal cancer [66, 67]. However, cytoplasmic HuR was not a prognostic factor for overall survival of patients with colorectal adenocarcinomas [66]. Similarly, cytoplasmic HuR accumulation was found in non-small cell lung cancer tissues (NSCLC) compared to benign human lung tissues [68] and was associated with high tumor grade, poor differentiation and lymph node metastasis [68, 69]. Cytoplasmic HuR also positively correlated with lymphatic microvessel density (LVD), or microvessel density (MVD) and negatively correlated with relapse-free survival and overall survival [69]. Total HuR and cytoplasmic HuR levels were increased in glioma tissues as compared to normal brains and positively correlated with tumor grade [70]. Overexpression of HuR could promote chemoresistance to standard glioma therapies in glioma cells, and vice versa [71]. Anti-apoptotic Bcl-2 family members were identified to be regulated by HuR in glioma cells. Silencing HuR led to transcript destabilization and reduced protein translation [71]. In ovarian carcinoma, both nuclear and cytoplasmic HuR were increased as compared with borderline tumors or normal ovaries [72]. The elevated cytoplasmic HuR was associated with increased COX-2 expression, histological grade and mitotic activity, and with reduced progression-free survival as well as overall survival [72]. A later study reported that nuclear HuR in ovarian carcinoma was also associated with tumor grade and an unfavorable indicator for disease-free survival [73]. These findings suggest a more complex model for HuR in ovarian cancer rather than limited to cytoplasmic accumulation.
4. Targeting HuR in cancer
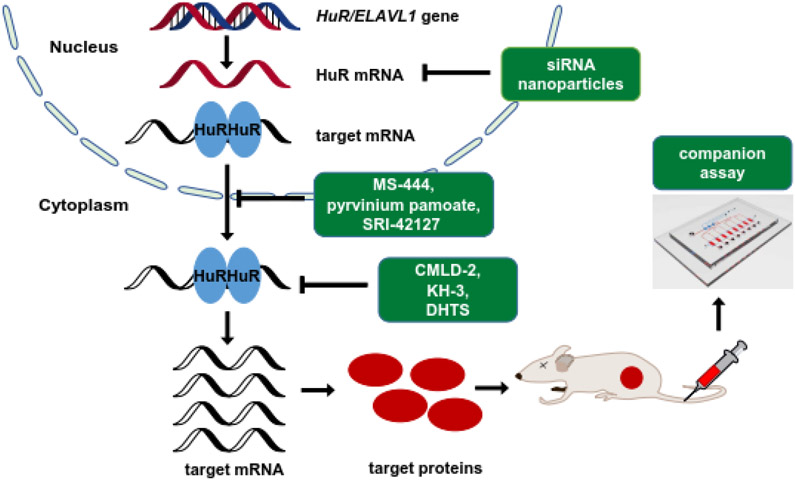
In early reports, upregulated HuR in brain and colon cancer was linked to the enhanced expression of COX-2, VEGF, TGF-β, IL-8, and other cancer-associated proteins [74, 75]. Subsequent studies identified numerous HuR-related mRNAs that regulate and contribute to almost every hallmark of cancer [76-78], supporting a profound impact of HuR on tumorigenesis and progression. In parallel, HuR, especially cytoplasmic HuR, is broadly overexpressed in many types of human cancer and the elevated cytoplasmic HuR is associated with advanced tumor grade and poor clinical outcome as reviewed in Section 3. Therefore, HuR has drawn considerable attention as a cancer therapeutic target and hundreds of studies have explored the therapeutic potential of HuR inhibition, which will be reviewed below. In principle, there are three major strategies used for inhibition of HuR: inhibition of HuR expression, inhibition of HuR cytoplasmic translocation, and inhibition of HuR–RNA interaction (Figure 3).
Figure 3:
Schematic diagram of strategies for targeting HuR. There are three major strategies used for inhibition of HuR: inhibition of HuR expression by RNA interference, inhibition of HuR cytoplasmic translocation and inhibition of HuR–RNA interaction by small molecular compounds. Company assay can be used in parallel to monitor the outcomes of targeting HuR.
4.1. Genetic inhibition of HuR expression
Silencing HuR expression using RNA interference (e.g., siRNA and shRNA) and CRISPR technologies has demonstrated the feasibility of inhibiting tumor growth in vivo in several types of cancer. Our own work in breast cancer cells indicate that knocking down HuR delays tumor formation and inhibits tumor growth while knocking out HuR results in no tumor formation in MDA-MB-231 cells and reduced tumor formation in SUM159 cells [50]. Similarly, HuR-deficient pancreatic cancer cells are unable to engraft tumors in vivo while HuR overexpression in those HuR-null cells restores the tumor formation [79]. Both knocking down and knocking out HuR in colorectal cancer cells inhibit tumor growth [44, 79]. Knocking down HuR in primary glioma cells attenuates tumor initiation, growth and invasiveness in a brain orthotopic model [71]. Ovarian cancer cells with HuR knockdown form much smaller tumors [80]. These findings in broad cancer types fully support the pivotal function of HuR in tumorigenesis and set the fundamentals for genetic inhibition of HuR as a potential cancer therapeutic strategy.
Therapeutic efficacy of genetic HuR inhibition has been examined in several tumor models by delivering HuR siRNA intratumorally or systemically. Intratumor injection of HuR siRNA resulted in ~60% tumor growth inhibition in a renal cell carcinoma xenograft model [81]. In an ovarian cancer xenograft model, treatment with folic acid (FA)-derivatized DNA dendrimer nanocarriers containing HuR siRNA via intraperitoneal injection suppressed tumor growth and ascites formation along with increased survival time of mice [80]. Muralidharan et al. reported to use a transferrin receptor-targeted liposomal nanoparticle system to deliver HuR siRNA in lung cancer models [82]. Both intratumoral and intravenous administration of HuR siRNA encapsulated liposomes inhibited tumor growth in subcutaneous models. Mice treated with HuR siRNA encapsulated liposomes intravenously had fewer tumor modules in the experimental lung metastasis model [82]. These animal studies provide a proof-of-concept for genetic inhibition of HuR expression as an effective means of cancer therapy. Even so, targeted delivery of HuR siRNA to specific tumor tissue will be essential to avoid systemic toxicity considering the ubiquitous expression and multi-facet roles of HuR in normal tissues. This would be the most challenging issue of moving forward such strategy for clinical development.
An attempt to deliver both HuR siRNA and cisplatin using folate receptor-targeted dendrimer nanoparticles as a means of in combination of HuR inhibition with chemotherapy has been reported [83]. Co-delivery had a greater therapeutic effect than individual agent did in lung cancer cells in vitro. However, the feasibility has not yet been established in vivo.
4.2. Inhibition of HuR cytoplasmic translocation
As reviewed above, several post-translational modifications can regulate HuR subcellular translocation. Several transport machinery components, including chromosome region maintenance 1 (CRM1, also known as exportin 1), transportin 1 and 2, and importin α1, are involved in HuR nuclear cytoplasmic shuttling as well [76, 84]. In addition, HuR RRM3 involved dimerization/multimerization is also necessary for HuR subcellular trafficking. Since the pioneer study of identification of small molecule MS-444 as a HuR inhibitor via inhibiting HuR dimerization and disrupting HuR cytoplasmic trafficking [85], several inhibitors of HuR cytoplasmic translocation have been reported by affecting one or more mediators of HuR translocation [32, 86-89]. Below are a few examples whose therapeutic efficacy has been demonstrated in cancer models by targeting HuR translocation.
MS-444, a microbial extract, was identified as an interior of HuR-ARE interaction through HTS, and then found to inhibit HuR dimerization and trafficking as well [85]. Treatment with MS-444 inhibited tumor growth together with decreased COX-2 expression in colorectal cancer xenograft models [90] and diminished the number of small intestinal tumors generated in APCMin mice, a model for familial adenomatosis polyposis and colon cancer [91]. In contract, HuR inhibition by MS-444 in AOM/DSS mice, a model of inflammatory bowel disease and inflammatory colon cancer, enhanced DSS-induced weight loss and increased tumor multiplicity, size, and invasiveness [91]. A recent study reported that MS-444 could attenuate ectopic HuR accelerated Renca allograft tumor progression and improve the survival of syngeneic mice [92]. These differential therapeutic effects of MS-444 may be due to the opposite aspects of HuR in different disease models, suggesting with caution to choose diseases for HuR inhibition.
Pyrvinium pamoate (PP) is an FDA-approved drug to treat pinworms. It was found to inhibit cytoplasmic HuR accumulation by activating AMPK/importin α1 cascade and activation of CDK1, which phosphorylates HuR at Ser202 and leads to nuclear retention of HuR. Pyrvinium pamoate has been shown to potentiate the efficacy of cisplatin in primary bladder cancer xenograft models [88]. A phase I trial was initiated last July to determine the safety and tolerability of pyrvinium pamoate, dosed orally, in patients with PDAC that are surgical candidates. This clinical study is still ongoing. One possible issue of pyrvinium pamoate as a HuR inhibitor is the poor target specificity and significant off-target effects, since it hits many other targets besides the HuR.
A new class of HuR dimerization inhibitors were reported recently, which were identified through high throughput screening (HTS) followed by structural optimization [89]. The lead compound SRI-42127 was established after several round of optimization of hit compound A-92, an inhibitor of the general control nonderepressible-2 kinase, with improved inhibitory potential of HuR dimerization and reduced kinase inhibition. It had favorable attributes to central nervous system penetration and inhibited tumor growth in a primary patient-derived glioblastoma xenograft model [89]. This is the first study to show therapeutic efficacy of a HuR inhibitor in brain tumor.
4.3. Inhibition of HuR–RNA interaction
HuR binds to its target mRNAs at ARE sites and regulates the stability and translation of those mRNAs. Therefore, inhibition of HuR-RNA interaction by competitively binding to HuR blocks HuR function on target mRNAs. So far, most small molecules interfering with HuR-RNA interaction were originally identified through HTS of chemical libraries utilizing biochemical assays, such as RNA electrophoretic mobility gel shift assay (EMSA) [93], fluorescent polarization assay (FPA) [94], amplified luminescent proximity homogenous assay (ALPHA) [95], which use recombinant HuR protein and labeled ARE oligos derived from target mRNAs. The small molecules reported to affect the binding of HuR to RNA are summarized in Table 1.
Table 1.
Published small molecules that interfere with HuR–RNA interaction.
| ID | Structure | Binding affinity (IC50) |
Source | Ref |
|---|---|---|---|---|
| Quercetin |
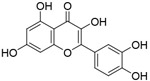
|
1.40μM | RNA EMSA screening of 179 chemicals | [93] |
| b-40 |
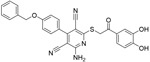
|
0.38μM | ||
| b-41 |
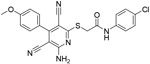
|
6.21μM | ||
| Mitoxantro ne |
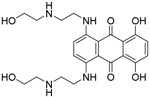
|
NA | AlphaScreen-based HTS of a chemical library of ~2000 small molecules | [95] |
| CMLD-1 |
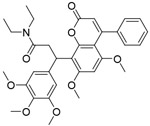
|
4.0μM | FP-based HTS of ~6000 compounds from CMLD library and a library of FDA-approved drugs | [94] |
| CMLD-2 |
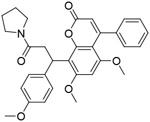
|
2.4μM | ||
| Dihydrotanshinone-l |
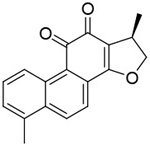
|
68nM | AlphaScreen-based HTS of 107 anti-nflammatory compounds | [101] |
| C10 |
|
NA | FP-based HTS of 1597 compounds from NCI diversity set V library | [125] |
| AZA-9 |
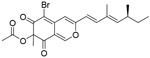
|
1.2μM | FP-based HTS of ~2000 compounds from the NCI library and inhouse compounds | [126] |
| 6a |
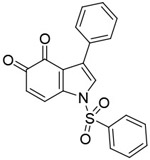
|
12.8nM (Ki) | rationally designed new chemicals | [102] |
| 6n |
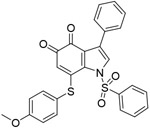
|
15nM (Ki) | ||
| Compoun d 4 |
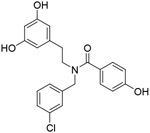
|
NA | rationally designed new chemical | [127] |
| KH-3 |
|
3.54μM | FP-based HTS of ~2000 compounds from the NCI library and inhouse compounds | [50] |
| VP12/14 |
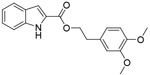
|
NA | rationally designed new chemicals | [128] |
| VP12/110 |
|
NA | ||
| Compound 2 |
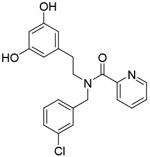
|
105μM | rationally designed new chemicals | [129] |
| Compound 3 |
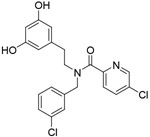
|
92μM |
NA: not available
Taken our own work as an example, we established an FPA using full-length HuR protein and a 16-nt ARE-containing fluorescein-labeled RNA oligo from HuR target Msi1 mRNA. We then applied HTS of ~6,000 compounds using this FPA and identified a class of coumarin-derived compounds as inhibitors of HuR-RNA interaction [94]. Among them, the most potent inhibitor CMLD-2 disrupted HuR interaction with Bcl-2, Msi1, and XIAP mRNA and induced apoptosis in colorectal cancer cells [94]. The follow up studies show that CMLD-2 exhibits cytotoxicity towards lung cancer cells and thyroid cancer cells [96, 97]. A recent study reported that CMLD-2 suppressed the expression of HuR target ARID1A, which plays an important role in HuR-driven resistance to radiation in breast cancer cells [98]. We carried out another HTS of ~2,000 compounds using the same FPA and identified benzothiophene hydroxamate KH-3 as a potent inhibitor of HuR-RNA interaction [50]. KH-3 inhibited breast cancer cell proliferation and invasion in vitro, mimicking genetic HuR knockout by CRISPR. KH-3 suppressed tumor growth in an orthotopic breast cancer xenograft model as well as reduced lung metastasis and improved survival of mice in experimental breast cancer metastasis models [50]. In the study of pancreatic cancer, KH-3 attenuated pancreatic cancer cell viability, epithelial-to-mesenchymal transition (EMT), migration/invasion in vitro and suppressed tumor growth and metastasis in an orthotopic pancreatic cancer xenograft model [99]. Mechanistically, KH-3 interfered with HuR–FOXQ1 interaction in breast cancer cells and HuR–Snail interaction in pancreatic cancer cells. KH-3 demonstrated HuR specificity, as cancer cells with HuR knockout were much less sensitive to KH-3 compared to the parental cells, and xenografts with HuR knockout were not responsive to the KH-3 treatment.
Natural product dihydrotanshinone-I (DHTS) is another disruptor of HuR-RNA interaction that has been evaluated in vivo, which inhibited tumor growth in a HuR-dependent colorectal cancer xenograft model [100]. DHTS was identified to interfere with the RNA binding of HuR via AlphaScreen-based HTS of a set of anti-inflammatory compounds and the following study indicate that inhibition of RNA loading on HuR by DHTS confines HuR into the nuclear compartment at early time points [101]. Inspired by DHTS structure, a series of ortho-quinones (tanshinone mimics) were designed and synthesized. Among them, two compounds turned out to be more effective than DHTS in disrupting HuR binding to RNA in cells [102]. However, the anti-tumor activity of these two compounds in cancer models has yet to be determined.
4.4. Monitoring HuR functional inhibition – a potential companion assay
In modern drug discovery/development for molecularly targeted therapies, molecular target validation is critical for the success of any precision medicine. It is essential to be able to monitor whether a lead compound hits the supposed molecular target in the relevant disease model in vivo. It is increasingly recognized that such real-time target validation assays would significantly facilitate the drug discovery and development process, and help recruit patients that will respond to the molecular therapy and to monitor tumor response and patient outcome: a companion assay.
Liquid biopsy is emerging as an alternative to invasive tissue biopsy and promises to revolutionize disease diagnosis, surveillance, and prognosis of treatment [103-105]. Extracellular vesicles (EVs), including exosomes, present a promising paradigm of liquid biopsy [106, 107]. Exosomes (~40-150 nm in diameter) are formed within cytoplasmic multivesicular bodies (MVBs) and released into the extracellular space when MVBs fuse with cell membrane [108, 109]. Exosomes have been identified in most bodily fluids [110-112] and they consolidate certain biomolecules (e.g., proteins and RNAs) from parent cells, serving as nature’s way of enriching cancer biomarkers [107, 113-115]. Thus, there is a recent surge of interest in exploring exosomes as liquid biopsy biomarkers of tumors.
We recently reported that human breast cancer cells with HuR knockout showed significantly reduced exosome secretion, as well as the exosomal matrix metalloprotease-14 (MMP-14), together with the diminished metastatic potential [116]. We used a microfluid 3D-printed NanoChip to measure the exosomal MMP14 in blood/plasma samples, was able to detect in vitro cell invasiveness and monitor the in vivo tumor metastasis in animal tumor models [116]. This pioneering study provides the feasibility to develop the NanoChip as a Companion Assay in Liquid Biopsy for cancer patient screening, diagnosis and treatment response prognosis, especially to the HuR-targeted therapies.
4.5. Challenges for HuR inhibition in cancer
As discussed above, all three strategies of HuR inhibition consistently show efficacy in terms of tumor growth inhibition in a variety types of cancer, supporting HuR as a promising therapeutic target of cancer. Nevertheless, many challenges need to be overcome before moving these strategies forward for clinical application. Although genetic inhibition is effective and specific, the siRNA-based therapy is mainly hindered from the low stability, low efficiency, and poor site-specific delivery. Optimization of delivery formulations is a direction to overcome these limitations. Furthermore, nanoparticle delivery of HuR siRNA has recently been described as mentioned above. Further studies are needed to confirm the mechanisms of action besides the therapeutic efficiency. In contract, target specificity is the key for small molecule inhibitors of either HuR cytoplasmic translocation or HuR-RNA interaction. The potential off-targets of these reported HuR inhibitors are not well studied to date. Besides that, the physicochemical properties of an inhibitor influence its efficacy in vivo greatly. Therefore, evaluation of the specificity, drug-likeness, and safety of small molecule inhibitors of HuR is critical for moving this strategy to clinical studies.
5. HuR in other diseases
Many studies have demonstrated that HuR is implicated in several other diseases besides cancer. Cytoplasmic HuR accumulation was found in failing human myocardium as compared with health tissues [117]. Here cytoplasmic HuR was elevated similarly in a mouse model of transverse aortic constriction (TAC) to induce left ventricular pressure overload. Cardiomyocyte-specific HuR-deletion could reduce left ventricular hypertrophy, dilation, and fibrosis while preserve cardiac function in this model. Treatment with HuR inhibitor KH-3 phenocopied the HuR deletion and improved the survival of mice [117]. Another study found that cardiomyocyte-specific HuR-deletion aggravated isoproterenol-induced cardiac remodeling [118]. These findings suggest a controversy role of HuR in pathological cardiac hypertrophy.
Muscle wasting, a syndrome also known as cachexia, is often seen in patients with cancer, AIDS, and chronic obstructive pulmonary diseases. Under normal condition, HuR promotes muscle fiber formation. However, HuR switches to an inducer of muscle loss under these disease conditions. A recent study show that muscle-specific HuR knockout mice had high exercise endurance with increasing in the proportion of oxidative type I fibers, which prevented cancer cachexia-induced muscle atrophy [119].
The positive regulation of a number of proinflammatory cytokines and chemokines by HuR has been linked to many chronic disease such as chronic kidney disease. In an experimental nephritis model, total and cytoplasmic HuR levels were increased in glomerular cells of disease rat along with increased profibrotic markers [120]. Disease rat treated with KH-3 showed reduction in glomerular HuR levels and profibrotic markers in our recent study [120]. The syndromes of proteinuria, podocyte injury, and glomerulosclerosis were also ameliorated by KH-3 treatment [120]. The involvement of HuR in various liver diseases has been demonstrated but mechanisms of HuR in different liver diseases is controversy. Contrary to previous evidences that targeting HuR may present beneficial effects against non-alcoholic fatty liver disease (NAFLD) development [121], a recent study using a hepatocyte-specific HuR-deficient mouse model indicates that HuR is a gatekeeper of liver homeostasis and prevents NAFLD-related fibrosis and hepatocellular carcinoma [122].
Obesity and its complications (type 2 diabetes, cardiovascular diseases) are associated with remodeling of adipose tissue. Two recent studies, which investigated the role of adipose HuR, found that adipose-specific HuR knockout mice showed diet induced obesity along with insulin resistance due to decreased expression of adipose triglyceride lipase (ATGL) [123], and adipose-specific HuR knockout mice had spontaneous cardiac hypertrophy and fibrosis [124]. Both studies suggest a protective role of HuR in adipose.
6. Conclusion and perspectives
In summary, differing from the more complicated findings in other diseases, the published studies have demonstrated that the HuR is an attractive target for developing novel molecularly targeted cancer therapy. Recent studies already show some exciting leads in HuR inhibitors and the proof-of-concept of HuR-targeted therapy in various animal tumor models and other disease models. However, so far there are no HuR inhibitors approved by FDA, nor in any clinical trials, HuR remains “undruggable”. More coordinated efforts and investments are needed to achieve a better and deeper understanding of the HuR biology as well as HuR drug discovery / development. Molecularly targeted drugs are often used in combination with chemotherapy and/or radiation therapy in the clinic. Considering some controversy findings of HuR’s impact on some chemotherapeutic agents (e.g. gemcitabine, doxorubicin), the combination of HuR inhibitors with these agents should be designed rationally based on the molecular profiling of the cancer. The latter is consistent with the current effort towards the evidence-based precision medicine.
Co-development of HuR inhibitors with a Companion Assay, which provides a convenient and reliable way for target validation in vivo, would facilitate the drug development and approval process. For example, the reported NanoChip may be further developed as a Companion Assay in Liquid Biopsy for cancer patients screening in future clinical trials of HuR inhibitors, as well as prognosis for treatment response. This drug + companion assay approach has been very successful in the past and welcomed by the pharmaceutical industry and FDA.
Acknowledgements
This study was supported in part by the National Institutes of Health grants R01 CA191785 and CA243445, R33 CA252158, Department of Defense Breast Cancer Research Program Breakthrough Level II grant BC151845, Midwest Biomedical Accelerator Consortium (MBArC), an NIH Research Evaluation and Commercialization Hub (REACH), Kansas Bioscience Authority Rising Star Award (to L.X.); the Susan G. Komen Career Catalyst Research grant CCR18548252 and Department of Defense Prostate Cancer Research Program Idea Development Award PC200447 (to X.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
References
- [1].Glisovic T, Bachorik JL, Yong J, Dreyfuss G, RNA-binding proteins and post-transcriptional gene regulation, FEBS Lett., 582 (2008) 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pascale A, Govoni S, The complex world of post-transcriptional mechanisms: is their deregulation a common link for diseases? Focus on ELAV-like RNA-binding proteins, Cell Mol Life Sci, 69 (2012) 501–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Keene JD, RNA regulons: coordination of post-transcriptional events, Nat Rev Genet, 8 (2007) 533–543. [DOI] [PubMed] [Google Scholar]
- [4].Srikantan S, Gorospe M, HuR function in disease, Front Biosci (Landmark Ed), 17 (2012) 189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H, Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein, J Biol Chem, 271 (1996) 8144–8151. [DOI] [PubMed] [Google Scholar]
- [6].Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M, Identification of a target RNA motif for RNA-binding protein HuR, Proceedings of the National Academy of Sciences of the United States of America, 101 (2004) 2987–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Brennan CM, Steitz JA, HuR and mRNA stability, Cell Mol Life Sci, 58 (2001) 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fialcowitz-White EJ, Brewer BY, Ballin JD, Willis CD, Toth EA, Wilson GM, Specific protein domains mediate cooperative assembly of HuR oligomers on AU-rich mRNA-destabilizing sequences, J. Biol. Chem, 282 (2007) 20948–20959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Scheiba RM, de Opakua AI, Diaz-Quintana A, Cruz-Gallardo I, Martinez-Cruz LA, Martinez-Chantar ML, Blanco FJ, Diaz-Moreno I, The C-terminal RNA binding motif of HuR is a multi-functional domain leading to HuR oligomerization and binding to U-rich RNA targets, RNA Biol, 11 (2014) 1250–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ripin N, Boudet J, Duszczyk MM, Hinniger A, Faller M, Krepl M, Gadi A, Schneider RJ, Sponer J, Meisner-Kober NC, Allain FH, Molecular basis for AU-rich element recognition and dimerization by the HuR C-terminal RRM, Proceedings of the National Academy of Sciences of the United States of America, 116 (2019) 2935–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pabis M, Popowicz GM, Stehle R, Fernandez-Ramos D, Asami S, Warner L, Garcia-Maurino SM, Schlundt A, Martinez-Chantar ML, Diaz-Moreno I, Sattler M, HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs, Nucleic Acids Res., 47 (2019) 1011–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fan XC, Steitz JA, HNS, a nuclear-cytoplasmic shuttling sequence in HuR, Proceedings of the National Academy of Sciences of the United States of America, 95 (1998) 15293–15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hinman MN, Lou H, Diverse molecular functions of Hu proteins, Cell Mol Life Sci, 65 (2008) 3168–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Srikantan S, Gorospe M, UneCLIPsing HuR nuclear function, Mol Cell, 43 (2011) 319–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].von Roretz C, Di Marco S, Mazroui R, Gallouzi IE, Turnover of AU-rich-containing mRNAs during stress: a matter of survival, Wiley Interdiscip Rev RNA, 2 (2011) 336–347. [DOI] [PubMed] [Google Scholar]
- [16].Katsanou V, Milatos S, Yiakouvaki A, Sgantzis N, Kotsoni A, Alexiou M, Harokopos V, Aidinis V, Hemberger M, Kontoyiannis DL, The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development, Mol Cell Biol, 29 (2009) 2762–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ghosh M, Aguila HL, Michaud J, Ai Y, Wu MT, Hemmes A, Ristimaki A, Guo C, Furneaux H, Hla T, Essential role of the RNA-binding protein HuR in progenitor cell survival in mice, J Clin Invest, 119 (2009) 3530–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chi MN, Auriol J, Jegou B, Kontoyiannis DL, Turner JM, de Rooij DG, Morello D, The RNA-binding protein ELAVL1/HuR is essential for mouse spermatogenesis, acting both at meiotic and postmeiotic stages, Mol Biol Cell, 22 (2011) 2875–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Levadoux-Martin M, Gouble A, Jegou B, Vallet-Erdtmann V, Auriol J, Mercier P, Morello D, Impaired gametogenesis in mice that overexpress the RNA-binding protein HuR, EMBO Rep, 4 (2003) 394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kang MJ, Ryu BK, Lee MG, Han J, Lee JH, Ha TK, Byun DS, Chae KS, Lee BH, Chun HS, Lee KY, Kim HJ, Chi SG, NF-kappaB activates transcription of the RNA-binding factor HuR, via PI3K-AKT signaling, to promote gastric tumorigenesis, Gastroenterology, 135 (2008) 2030–2042, 2042 e2031-2033. [DOI] [PubMed] [Google Scholar]
- [21].Jeyaraj SC, Singh M, Ayupova DA, Govindaraju S, Lee BS, Transcriptional control of human antigen R by bone morphogenetic protein, J. Biol. Chem, 285 (2010) 4432–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Al-Ahmadi W, Al-Ghamdi M, Al-Haj L, Al-Saif M, Khabar KS, Alternative polyadenylation variants of the RNA binding protein, HuR: abundance, role of AU-rich elements and auto-Regulation, Nucleic Acids Res., 37 (2009) 3612–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dai W, Zhang G, Makeyev EV, RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage, Nucleic Acids Res., 40 (2012) 787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cho SJ, Jung YS, Zhang J, Chen X, The RNA-binding protein RNPC1 stabilizes the mRNA encoding the RNA-binding protein HuR and cooperates with HuR to suppress cell proliferation, J. Biol. Chem, 287 (2012) 14535–14544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M, miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels, Proceedings of the National Academy of Sciences of the United States of America, 105 (2008) 20297–20302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xu F, Zhang X, Lei Y, Liu X, Liu Z, Tong T, Wang W, Loss of repression of HuR translation by miR-16 may be responsible for the elevation of HuR in human breast carcinoma, J. Cell. Biochem, 111 (2010) 727–734. [DOI] [PubMed] [Google Scholar]
- [27].Liu Y, Chen X, Cheng R, Yang F, Yu M, Wang C, Cui S, Hong Y, Liang H, Liu M, Zhao C, Ding M, Sun W, Liu Z, Sun F, Zhang C, Zhou Z, Jiang X, Chen X, The Jun/miR-22/HuR regulatory axis contributes to tumourigenesis in colorectal cancer, Mol Cancer, 17 (2018) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Guo X, Wu Y, Hartley RS, MicroRNA-125a represses cell growth by targeting HuR in breast cancer, RNA Biol, 6 (2009) 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Guha A, Ahuja D, Das Mandal S, Parasar B, Deyasi K, Roy D, Sharma V, Willard B, Ghosh A, Ray PS, Integrated Regulation of HuR by Translation Repression and Protein Degradation Determines Pulsatile Expression of p53 Under DNA Damage, iScience, 15 (2019) 342–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Grammatikakis I, Abdelmohsen K, Gorospe M, Posttranslational control of HuR function, Wiley Interdiscip Rev RNA, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Abdelmohsen K, Pullmann R Jr., Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M, Phosphorylation of HuR by Chk2 regulates SIRT1 expression, Mol Cell, 25 (2007) 543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kim HH, Abdelmohsen K, Lal A, Pullmann R Jr., Yang X, Galban S, Srikantan S, Martindale JL, Blethrow J, Shokat KM, Gorospe M, Nuclear HuR accumulation through phosphorylation by Cdk1, Genes Dev, 22 (2008) 1804–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Filippova N, Yang X, King P, Nabors LB, Phosphoregulation of the RNA-binding protein Hu antigen R (HuR) by Cdk5 affects centrosome function, J. Biol. Chem, 287 (2012) 32277–32287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Doller A, Huwiler A, Muller R, Radeke HH, Pfeilschifter J, Eberhardt W, Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2, Mol Biol Cell, 18 (2007) 2137–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Doller A, Schlepckow K, Schwalbe H, Pfeilschifter J, Eberhardt W, Tandem phosphorylation of serines 221 and 318 by protein kinase Cdelta coordinates mRNA binding and nucleocytoplasmic shuttling of HuR, Mol Cell Biol, 30 (2010) 1397–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Li H, Park S, Kilburn B, Jelinek MA, Henschen-Edman A, Aswad DW, Stallcup MR, Laird-Offringa IA, Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase, J. Biol. Chem, 277 (2002) 44623–44630. [DOI] [PubMed] [Google Scholar]
- [37].Vigouroux C, Casse JM, Battaglia-Hsu SF, Brochin L, Luc A, Paris C, Lacomme S, Gueant JL, Vignaud JM, Gauchotte G, Methyl(R217)HuR and MCM6 are inversely correlated and are prognostic markers in non small cell lung carcinoma, Lung Cancer, 89 (2015) 189–196. [DOI] [PubMed] [Google Scholar]
- [38].Lucchesi C, Sheikh MS, Huang Y, Negative regulation of RNA-binding protein HuR by tumor-suppressor ECRG2, Oncogene, 35 (2016) 2565–2573. [DOI] [PubMed] [Google Scholar]
- [39].Daks A, Petukhov A, Fedorova O, Shuvalov O, Kizenko A, Tananykina E, Vasileva E, Semenov O, Bottrill A, Barlev N, The RNA-binding protein HuR is a novel target of Pirh2 E3 ubiquitin ligase, Cell Death Dis, 12 (2021) 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Embade N, Fernandez-Ramos D, Varela-Rey M, Beraza N, Sini M, Gutierrez de Juan V, Woodhoo A, Martinez-Lopez N, Rodriguez-Iruretagoyena B, Bustamante FJ, de la Hoz AB, Carracedo A, Xirodimas DP, Rodriguez MS, Lu SC, Mato JM, Martinez-Chantar ML, Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation, Hepatology, 55 (2012) 1237–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].von Roretz C, Lian XJ, Macri AM, Punjani N, Clair E, Drouin O, Dormoy-Raclet V, Ma JF, Gallouzi IE, Apoptotic-induced cleavage shifts HuR from being a promoter of survival to an activator of caspase-mediated apoptosis, Cell Death Differ, 20 (2013) 154–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ke Y, Lv X, Fu X, Zhang J, Bohio AA, Zeng X, Hao W, Wang R, Boldogh I, Ba X, Poly(ADP-ribosyl)ation enhances HuR oligomerization and contributes to pro-inflammatory gene mRNA stabilization, Cell Mol Life Sci, 78 (2021) 1817–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ke Y, Han Y, Guo X, Wen J, Wang K, Jiang X, Tian X, Ba X, Boldogh I, Zeng X, PARP1 promotes gene expression at the post-transcriptiona level by modulating the RNA-binding protein HuR, Nature communications, 8 (2017) 14632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M, Role of the RNA-binding protein HuR in colon carcinogenesis, Oncogene, 22 (2003) 7146–7154. [DOI] [PubMed] [Google Scholar]
- [45].Denkert C, Weichert W, Winzer KJ, Muller BM, Noske A, Niesporek S, Kristiansen G, Guski H, Dietel M, Hauptmann S, Expression of the ELAV-like protein HuR is associated with higher tumor grade and increased cyclooxygenase-2 expression in human breast carcinoma, Clin Cancer Res, 10 (2004) 5580–5586. [DOI] [PubMed] [Google Scholar]
- [46].Heinonen M, Bono P, Narko K, Chang SH, Lundin J, Joensuu H, Furneaux H, Hla T, Haglund C, Ristimaki A, Cytoplasmic HuR expression is a prognostic factor in invasive ductal breast carcinoma, Cancer research, 65 (2005) 2157–2161. [DOI] [PubMed] [Google Scholar]
- [47].Heinonen M, Fagerholm R, Aaltonen K, Kilpivaara O, Aittomaki K, Blomqvist C, Heikkila P, Haglund C, Nevanlinna H, Ristimaki A, Prognostic role of HuR in hereditary breast cancer, Clin Cancer Res, 13 (2007) 6959–6963. [DOI] [PubMed] [Google Scholar]
- [48].Heinonen M, Hemmes A, Salmenkivi K, Abdelmohsen K, Vilen ST, Laakso M, Leidenius M, Salo T, Hautaniemi S, Gorospe M, Heikkila P, Haglund C, Ristimaki A, Role of RNA binding protein HuR in ductal carcinoma in situ of the breast, J Pathol, 224 (2011) 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wang J, Li D, Wang B, Wu Y, Predictive and prognostic significance of cytoplasmic expression of ELAV-like protein HuR in invasive breast cancer treated with neoadjuvant chemotherapy, Breast Cancer Res Treat, 141 (2013) 213–224. [DOI] [PubMed] [Google Scholar]
- [50].Wu X, Gardashova G, Lan L, Han S, Zhong C, Marquez RT, Wei L, Wood S, Roy S, Gowthaman R, Karanicolas J, Gao FP, Dixon DA, Welch DR, Li L, Ji M, Aube J, Xu L, Targeting the interaction between RNA-binding protein HuR and FOXQ1 suppresses breast cancer invasion and metastasis, Commun Biol, 3 (2020) 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hostetter C, Licata LA, Witkiewicz A, Costantino CL, Yeo CJ, Brody JR, Keen JC, Cytoplasmic accumulation of the RNA binding protein HuR is central to tamoxifen resistance in estrogen receptor positive breast cancer cells, Cancer Biol Ther, 7 (2008) 1496–1506. [DOI] [PubMed] [Google Scholar]
- [52].Latorre E, Tebaldi T, Viero G, Sparta AM, Quattrone A, Provenzani A, Downregulation of HuR as a new mechanism of doxorubicin resistance in breast cancer cells, Mol Cancer, 11 (2012) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Latorre E, Castiglioni I, Gatto P, Carelli S, Quattrone A, Provenzani A, Loss of protein kinase Cdelta/HuR interaction is necessary to doxorubicin resistance in breast cancer cell lines, J. Pharmacol. Exp. Ther, 349 (2014) 99–106. [DOI] [PubMed] [Google Scholar]
- [54].Niesporek S, Kristiansen G, Thoma A, Weichert W, Noske A, Buckendahl AC, Jung K, Stephan C, Dietel M, Denkert C, Expression of the ELAV-like protein HuR in human prostate carcinoma is an indicator of disease relapse and linked to COX-2 expression, Int. J. Oncol, 32 (2008) 341–347. [PubMed] [Google Scholar]
- [55].Mitsunari K, Miyata Y, Asai A, Matsuo T, Shida Y, Hakariya T, Sakai H, Human antigen R is positively associated with malignant aggressiveness via upregulation of cell proliferation, migration, and vascular endothelial growth factors and cyclooxygenase-2 in prostate cancer, Transl Res, 175 (2016) 116–128. [DOI] [PubMed] [Google Scholar]
- [56].Melling N, Taskin B, Hube-Magg C, Kluth M, Minner S, Koop C, Grob T, Graefen M, Heinzer H, Tsourlakis MC, Izbicki J, Wittmer C, Huland H, Simon R, Wilczak W, Sauter G, Steurer S, Schlomm T, Krech T, Cytoplasmic accumulation of ELAVL1 is an independent predictor of biochemical recurrence associated with genomic instability in prostate cancer, Prostate, 76 (2016) 259–272. [DOI] [PubMed] [Google Scholar]
- [57].Costantino CL, Witkiewicz AK, Kuwano Y, Cozzitorto JA, Kennedy EP, Dasgupta A, Keen JC, Yeo CJ, Gorospe M, Brody JR, The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR Up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase, Cancer research, 69 (2009) 4567–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Richards NG, Rittenhouse DW, Freydin B, Cozzitorto JA, Grenda D, Rui H, Gonye G, Kennedy EP, Yeo CJ, Brody JR, Witkiewicz AK, HuR status is a powerful marker for prognosis and response to gemcitabine-based chemotherapy for resected pancreatic ductal adenocarcinoma patients, Ann Surg, 252 (2010) 499–505; discussion 505-496. [DOI] [PubMed] [Google Scholar]
- [59].McAllister F, Pineda DM, Jimbo M, Lal S, Burkhart RA, Moughan J, Winter KA, Abdelmohsen K, Gorospe M, Acosta Ade J, Lankapalli RH, Winter JM, Yeo CJ, Witkiewicz AK, Iacobuzio-Donahue CA, Laheru D, Brody JR, dCK expression correlates with 5-fluorouracil efficacy and HuR cytoplasmic expression in pancreatic cancer: a dual-institutional follow-up with the RTOG 9704 trial, Cancer Biol Ther, 15 (2014) 688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Tatarian T, Jiang W, Leiby BE, Grigoli A, Jimbo M, Dabbish N, Neoptolemos JP, Greenhalf W, Costello E, Ghaneh P, Halloran C, Palmer D, Buchler M, Yeo CJ, Winter JM, Brody JR, Cytoplasmic HuR Status Predicts Disease-free Survival in Resected Pancreatic Cancer: A Post-hoc Analysis From the International Phase III ESPAC-3 Clinical Trial, Ann Surg, 267 (2018) 364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Elebro J, Ben Dror L, Heby M, Nodin B, Jirstrom K, Eberhard J, Prognostic effect of hENT1, dCK and HuR expression by morphological type in periampullary adenocarcinoma, including pancreatic cancer, Acta Oncol, 55 (2016) 286–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Brody JR, Dixon DA, Complex HuR function in pancreatic cancer cells, Wiley interdisciplinary reviews. RNA, 9 (2018) e1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Blanco FF, Jimbo M, Wulfkuhle J, Gallagher I, Deng J, Enyenihi L, Meisner-Kober N, Londin E, Rigoutsos I, Sawicki JA, Risbud MV, Witkiewicz AK, McCue PA, Jiang W, Rui H, Yeo CJ, Petricoin E, Winter JM, Brody JR, The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells, Oncogene, 35 (2016) 2529–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lal S, Burkhart RA, Beeharry N, Bhattacharjee V, Londin ER, Cozzitorto JA, Romeo C, Jimbo M, Norris ZA, Yeo CJ, Sawicki JA, Winter JM, Rigoutsos I, Yen TJ, Brody JR, HuR posttranscriptionally regulates WEE1: implications for the DNA damage response in pancreatic cancer cells, Cancer research, 74 (2014) 1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zarei M, Lal S, Parker SJ, Nevler A, Vaziri-Gohar A, Dukleska K, Mambelli-Lisboa NC, Moffat C, Blanco FF, Chand SN, Jimbo M, Cozzitorto JA, Jiang W, Yeo CJ, Londin ER, Seifert EL, Metallo CM, Brody JR, Winter JM, Posttranscriptional Upregulation of IDH1 by HuR Establishes a Powerful Survival Phenotype in Pancreatic Cancer Cells, Cancer research, 77 (2017) 4460–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Denkert C, Koch I, von Keyserlingk N, Noske A, Niesporek S, Dietel M, Weichert W, Expression of the ELAV-like protein HuR in human colon cancer: association with tumor stage and cyclooxygenase-2, Mod Pathol, 19 (2006) 1261–1269. [DOI] [PubMed] [Google Scholar]
- [67].Lim SJ, Lee SH, Joo SH, Song JY, Choi SI, Cytoplasmic expression of HuR is related to cyclooxygenase-2 expression in colon cancer, Cancer Res Treat, 41 (2009) 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Wang J, Zhao W, Guo Y, Zhang B, Xie Q, Xiang D, Gao J, Wang B, Chen Z, The expression of RNA-binding protein HuR in non-small cell lung cancer correlates with vascular endothelial growth factor-C expression and lymph node metastasis, Oncology, 76 (2009) 420–429. [DOI] [PubMed] [Google Scholar]
- [69].Wang J, Wang B, Bi J, Zhang C, Cytoplasmic HuR expression correlates with angiogenesis, lymphangiogenesis, and poor outcome in lung cancer, Med Oncol, 28 Suppl 1 (2011) S577–585. [DOI] [PubMed] [Google Scholar]
- [70].Bolognani F, Gallani AI, Sokol L, Baskin DS, Meisner-Kober N, mRNA stability alterations mediated by HuR are necessary to sustain the fast growth of glioma cells, J Neurooncol, 106 (2012) 531–542. [DOI] [PubMed] [Google Scholar]
- [71].Filippova N, Yang X, Wang Y, Gillespie GY, Langford C, King PH, Wheeler C, Nabors LB, The RNA-binding protein HuR promotes glioma growth and treatment resistance, Mol Cancer Res, 9 (2011) 648–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Denkert C, Weichert W, Pest S, Koch I, Licht D, Kobel M, Reles A, Sehouli J, Dietel M, Hauptmann S, Overexpression of the embryonic-lethal abnormal vision-like protein HuR in ovarian carcinoma is a prognostic factor and is associated with increased cyclooxygenase 2 expression, Cancer Res, 64 (2004) 189–195. [DOI] [PubMed] [Google Scholar]
- [73].Yi X, Zhou Y, Zheng W, Chambers SK, HuR expression in the nucleus correlates with high histological grade and poor disease-free survival in ovarian cancer, Aust N Z J Obstet Gynaecol, 49 (2009) 93–98. [DOI] [PubMed] [Google Scholar]
- [74].Nabors LB, Gillespie GY, Harkins L, King PH, HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3' untranslated regions of cytokine and angiogenic factor mRNAs, Cancer research, 61 (2001) 2154–2161. [PubMed] [Google Scholar]
- [75].Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM, Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells, J Clin Invest, 108 (2001) 1657–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Abdelmohsen K, Gorospe M, Posttranscriptional regulation of cancer traits by HuR, Wiley Interdiscip Rev RNA, 1 (2010) 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Schultz CW, Preet R, Dhir T, Dixon DA, Brody JR, Understanding and targeting the disease-related RNA binding protein human antigen R (HuR), Wiley Interdiscip Rev RNA, (2020) e1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hanahan D, Hallmarks of Cancer: New Dimensions, Cancer Discovery, 12 (2022) 31. [DOI] [PubMed] [Google Scholar]
- [79].Lal S, Cheung EC, Zarei M, Preet R, Chand SN, Mambelli-Lisboa NC, Romeo C, Stout MC, Londin E, Goetz A, Lowder CY, Nevler A, Yeo CJ, Campbell PM, Winter JM, Dixon DA, Brody JR, CRISPR Knockout of the HuR Gene Causes a Xenograft Lethal Phenotype, Mol Cancer Res, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Huang YH, Peng W, Furuuchi N, Gerhart J, Rhodes K, Mukherjee N, Jimbo M, Gonye GE, Brody JR, Getts RC, Sawicki JA, Delivery of Therapeutics Targeting the mRNA-Binding Protein HuR Using 3DNA Nanocarriers Suppresses Ovarian Tumor Growth, Cancer research, 76 (2016) 1549–1559. [DOI] [PubMed] [Google Scholar]
- [81].Danilin S, Sourbier C, Thomas L, Lindner V, Rothhut S, Dormoy V, Helwig JJ, Jacqmin D, Lang H, Massfelder T, Role of the RNA-binding protein HuR in human renal cell carcinoma, Carcinogenesis, 31 (2010) 1018–1026. [DOI] [PubMed] [Google Scholar]
- [82].Muralidharan R, Babu A, Amreddy N, Srivastava A, Chen A, Zhao YD, Kompella UB, Munshi A, Ramesh R, Tumor-targeted Nanoparticle Delivery of HuR siRNA Inhibits Lung Tumor Growth In Vitro and In Vivo By Disrupting the Oncogenic Activity of the RNA-binding Protein HuR, Mol Cancer Ther, 16 (2017) 1470–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Amreddy N, Babu A, Panneerselvam J, Srivastava A, Muralidharan R, Chen A, Zhao YD, Munshi A, Ramesh R, Chemo-biologic combinatorial drug delivery using folate receptor-targeted dendrimer nanoparticles for lung cancer treatment, Nanomedicine, 14 (2018) 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Doller A, Pfeilschifter J, Eberhardt W, Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR, Cell Signal, 20 (2008) 2165–2173. [DOI] [PubMed] [Google Scholar]
- [85].Meisner NC, Hintersteiner M, Mueller K, Bauer R, Seifert JM, Naegeli HU, Ottl J, Oberer L, Guenat C, Moss S, Harrer N, Woisetschlaeger M, Buehler C, Uhl V, Auer M, Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR, Nat Chem Biol, 3 (2007) 508–515. [DOI] [PubMed] [Google Scholar]
- [86].Cheng YC, Liou JP, Kuo CC, Lai WY, Shih KH, Chang CY, Pan WY, Tseng JT, Chang JY, MPT0B098, a novel microtubule inhibitor that destabilizes the hypoxia-inducible factor-1alpha mRNA through decreasing nuclear-cytoplasmic translocation of RNA-binding protein HuR, Mol Cancer Ther, 12 (2013) 1202–1212. [DOI] [PubMed] [Google Scholar]
- [87].Doller A, Badawi A, Schmid T, Brauss T, Pleli T, zu Heringdorf DM, Piiper A, Pfeilschifter J, Eberhardt W, The cytoskeletal inhibitors latrunculin A and blebbistatin exert antitumorigenic properties in human hepatocellular carcinoma cells by interfering with intracellular HuR trafficking, Exp Cell Res, 330 (2015) 66–80. [DOI] [PubMed] [Google Scholar]
- [88].Guo J, Lv J, Chang S, Chen Z, Lu W, Xu C, Liu M, Pang X, Inhibiting cytoplasmic accumulation of HuR synergizes genotoxic agents in urothelial carcinoma of the bladder, Oncotarget, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Filippova N, Yang X, Ananthan S, Calano J, Pathak V, Bratton L, Vekariya RH, Zhang S, Ofori E, Hayward EN, Namkoong D, Crossman DK, Crowley MR, King PH, Mobley J, Nabors LB, Targeting the HuR Oncogenic Role with a New Class of Cytoplasmic Dimerization Inhibitors, Cancer research, 81 (2021) 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Blanco FF, Preet R, Aguado A, Vishwakarma V, Stevens LE, Vyas A, Padhye S, Xu L, Weir SJ, Anant S, Meisner-Kober N, Brody JR, Dixon DA, Impact of HuR inhibition by the small molecule MS-444 on colorectal cancer cell tumorigenesis, Oncotarget, 7 (2016) 74043–74058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Lang M, Berry D, Passecker K, Mesteri I, Bhuju S, Ebner F, Sedlyarov V, Evstatiev R, Dammann K, Loy A, Kuzyk O, Kovarik P, Khare V, Beibel M, Roma G, Meisner-Kober N, Gasche C, HuR Small-Molecule Inhibitor Elicits Differential Effects in Adenomatosis Polyposis and Colorectal Carcinogenesis, Cancer research, 77 (2017) 2424–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Liu Y, Li X, Zhang H, Zhang M, Wei Y, HuR up-regulates cell surface PD-L1 via stabilizing CMTM6 transcript in cancer, Oncogene, 40 (2021) 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Chae MJ, Sung HY, Kim EH, Lee M, Kwak H, Chae CH, Kim S, Park WY, Chemical inhibitors destabilize HuR binding to the AU-rich element of TNF-alpha mRNA, Exp Mol Med, 41 (2009) 824–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Wu X, Lan L, Wilson DM, Marquez RT, Tsao WC, Gao P, Roy A, Turner BA, McDonald P, Tunge JA, Rogers SA, Dixon DA, Aube J, Xu L, Identification and Validation of Novel Small Molecule Disruptors of HuR-mRNA Interaction, ACS chemical biology, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].D'Agostino VG, Adami V, Provenzani A, A novel high throughput biochemical assay to evaluate the HuR protein-RNA complex formation, PLoS One, 8 (2013) e72426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Muralidharan R, Mehta M, Ahmed R, Roy S, Xu L, Aube J, Chen A, Zhao YD, Herman T, Ramesh R, Munshi A, HuR-targeted small molecule inhibitor exhibits cytotoxicity towards human lung cancer cells, Scientific reports, 7 (2017) 9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Allegri L, Baldan F, Roy S, Aube J, Russo D, Filetti S, Damante G, The HuR CMLD-2 inhibitor exhibits antitumor effects via MAD2 downregulation in thyroid cancer cells, Scientific reports, 9 (2019) 7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Andrade D, Mehta M, Griffith J, Oh S, Corbin J, Babu A, De S, Chen A, Zhao YD, Husain S, Roy S, Xu L, Aube J, Janknecht R, Gorospe M, Herman T, Ramesh R, Munshi A, HuR Reduces Radiation-Induced DNA Damage by Enhancing Expression of ARID1A, Cancers (Basel), 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Dong R, Chen P, Polireddy K, Wu X, Wang T, Ramesh R, Dixon DA, Xu L, Aube J, Chen Q, An RNA-Binding Protein, Hu-antigen R, in Pancreatic Cancer Epithelial to Mesenchymal Transition, Metastasis, and Cancer Stem Cells, Mol Cancer Ther, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Lal P, Cerofolini L, D'Agostino VG, Zucal C, Fuccio C, Bonomo I, Dassi E, Giuntini S, Di Maio D, Vishwakarma V, Preet R, Williams SN, Fairlamb MS, Munk R, Lehrmann E, Abdelmohsen K, Elezgarai SR, Luchinat C, Novellino E, Quattrone A, Biasini E, Manzoni L, Gorospe M, Dixon DA, Seneci P, Marinelli L, Fragai M, Provenzani A, Regulation of HuR structure and function by dihydrotanshinone-I, Nucleic Acids Res., 45 (2017) 9514–9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].D'Agostino VG, Lal P, Mantelli B, Tiedje C, Zucal C, Thongon N, Gaestel M, Latorre E, Marinelli L, Seneci P, Amadio M, Provenzani A, Dihydrotanshinone-I interferes with the RNA-binding activity of HuR affecting its post-transcriptional function, Scientific reports, 5 (2015) 16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Manzoni L, Zucal C, Maio DD, D'Agostino VG, Thongon N, Bonomo I, Lal P, Miceli M, Baj V, Brambilla M, Cerofolini L, Elezgarai S, Biasini E, Luchinat C, Novellino E, Fragai M, Marinelli L, Provenzani A, Seneci P, Interfering with HuR-RNA Interaction: Design, Synthesis and Biological Characterization of Tanshinone Mimics as Novel, Effective HuR Inhibitors, J. Med. Chem, 61 (2018) 1483–1498. [DOI] [PubMed] [Google Scholar]
- [103].Goldman JW, Noor ZS, Remon J, Besse B, Rosenfeld N, Are liquid biopsies a surrogate for tissue EGFR testing?, Ann. Oncol, 29 (2018) I38–I46. [DOI] [PubMed] [Google Scholar]
- [104].Pantel K, Alix-Panabieres C, Real-time Liquid Biopsy in Cancer Patients: Fact or Fiction?, Cancer Res., 73 (2013) 6384–6388. [DOI] [PubMed] [Google Scholar]
- [105].Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A, Liquid biopsy: monitoring cancer-genetics in the blood, Nat. Rev. Clin. Oncol, 10 (2013) 472–484. [DOI] [PubMed] [Google Scholar]
- [106].Siravegna G, Marsoni S, Siena S, Bardelli A, Integrating liquid biopsies into the management of cancer, Nature Reviews Clinical Oncology, 14 (2017) 531. [DOI] [PubMed] [Google Scholar]
- [107].Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, Xia H, Man Q, Zhong W, Antelo LF, Wu B, Xiong X, Liu X, Guan L, Li T, Liu S, Yang R, Lu Y, Dong L, McGettigan S, Somasundaram R, Radhakrishnan R, Mills G, Lu Y, Kim J, Chen YH, Dong H, Zhao Y, Karakousis GC, Mitchell TC, Schuchter LM, Herlyn M, Wherry EJ, Xu X, Guo W, Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response, Nature, 560 (2018) 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Gajos-Michniewicz A, Duechler M, Czyz M, MiRNA in melanoma-derived exosomes, Cancer Lett, 347 (2014) 29–37. [DOI] [PubMed] [Google Scholar]
- [109].van Niel G, D'Angelo G, Raposo G, Shedding light on the cell biology of extracellular vesicles, Nature Reviews Molecular Cell Biology, 19 (2018) 213. [DOI] [PubMed] [Google Scholar]
- [110].Kharaziha P, Ceder S, Li Q, Panaretakis T, Tumor cell-derived exosomes: a message in a bottle, Biochim Biophys Acta, 1826 (2012) 103–111. [DOI] [PubMed] [Google Scholar]
- [111].Hannafon BN, Ding WQ, Intercellular Communication by Exosome-Derived microRNAs in Cancer, Int J Mol Sci, 14 (2013) 14240–14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Ge R, Tan E, Sharghi-Namini S, Asada HH, Exosomes in Cancer Microenvironment and Beyond: have we Overlooked these Extracellular Messengers?, Cancer Microenviron, 5 (2012) 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO, Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers, Nature Cell Biology, 10 (2008) 1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Szabo G, Momen-Heravi F, Extracellular vesicles in liver disease and potential as biomarkers and therapeutic targets, Nature Reviews Gastroenterology & Amp; Hepatology, 14 (2017) 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Robbins PD, Morelli AE, Regulation of immune responses by extracellular vesicles, Nature Reviews Immunology, 14 (2014) 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zhang P, Wu X, Gardashova G, Yang Y, Zhang Y, Xu L, Zeng Y, Molecular and functional extracellular vesicle analysis using nanopatterned microchips monitors tumor progression and metastasis, Science Translational Medicine, 12 (2020) eaaz2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Green LC, Anthony SR, Slone S, Lanzillotta L, Nieman ML, Wu X, Robbins N, Jones SM, Roy S, Owens AP 3rd, Aube J, Xu L, Lorenz JN, Blaxall BC, Rubinstein J, Benoit JB, Tranter M, Human antigen R as a therapeutic target in pathological cardiac hypertrophy, JCI Insight, 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Hu H, Jiang M, Cao Y, Zhang Z, Jiang B, Tian F, Feng J, Dou Y, Gorospe M, Zheng M, Zheng L, Yang Z, Wang W, HuR regulates phospholamban expression in isoproterenol-induced cardiac remodelling, Cardiovasc Res, 116 (2020) 944–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Janice Sanchez B, Tremblay AK, Leduc-Gaudet JP, Hall DT, Kovacs E, Ma JF, Mubaid S, Hallauer PL, Phillips BL, Vest KE, Corbett AH, Kontoyiannis DL, Hussain SNA, Hastings KEM, Di Marco S, Gallouzi IE, Depletion of HuR in murine skeletal muscle enhances exercise endurance and prevents cancer-induced muscle atrophy, Nature communications, 10 (2019) 4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Liu S, Huang Z, Tang A, Wu X, Aube J, Xu L, Xing C, Huang Y, Inhibition of RNA-binding protein HuR reduces glomerulosclerosis in experimental nephritis, Clin Sci (Lond), 134 (2020) 1433–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Liu R, Wu K, Li Y, Sun R, Li X, Human antigen R: A potential therapeutic target for liver diseases, Pharmacol. Res, (2020) 104684. [DOI] [PubMed] [Google Scholar]
- [122].Subramanian P, Gargani S, Palladini A, Chatzimike M, Grzybek M, Peitzsch M, Papanastasiou AD, Pyrina I, Ntafis V, Gercken B, Lesche M, Petzold A, Sinha A, Nati M, Thangapandi VR, Kourtzelis I, Andreadou M, Witt A, Dahl A, Burkhardt R, Haase R, Domingues AMJ, Henry I, Zamboni N, Mirtschink P, Chung KJ, Hampe J, Coskun U, Kontoyiannis DL, Chavakis T, The RNA binding protein human antigen R is a gatekeeper of liver homeostasis, Hepatology, (2021). [DOI] [PubMed] [Google Scholar]
- [123].Li J, Gong L, Liu S, Zhang Y, Zhang C, Tian M, Lu H, Bu P, Yang J, Ouyang C, Jiang X, Wu J, Zhang Y, Min Q, Zhang C, Zhang W, Adipose HuR protects against diet-induced obesity and insulin resistance, Nature communications, 10 (2019) 2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Guarnieri AR, Anthony SR, Gozdiff A, Green LC, Fleifil SM, Slone S, Nieman ML, Alam P, Benoit JB, Owens AP, Kanisicak O, Tranter M, Adipocyte-specific deletion of HuR induces spontaneous cardiac hypertrophy and fibrosis, Am J Physiol Heart Circ Physiol, 321 (2021) H228–H241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Wang Z, Bhattacharya A, Ivanov DN, Identification of Small-Molecule Inhibitors of the HuR/RNA Interaction Using a Fluorescence Polarization Screening Assay Followed by NMR Validation, PLoS One, 10 (2015) e0138780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Kaur K, Wu X, Fields JK, Johnson DK, Lan L, Pratt M, Somoza AD, Wang CCC, Karanicolas J, Oakley BR, Xu L, De Guzman RN, The fungal natural product azaphilone-9 binds to HuR and inhibits HuR-RNA interaction in vitro, PLoS One, 12 (2017) e0175471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Della Volpe S, Nasti R, Queirolo M, Unver MY, Jumde VK, Domling A, Vasile F, Potenza D, Ambrosio FA, Costa G, Alcaro S, Zucal C, Provenzani A, Di Giacomo M, Rossi D, Hirsch AKH, Collina S, Novel Compounds Targeting the RNA-Binding Protein HuR. Structure-Based Design, Synthesis, and Interaction Studies, ACS Med Chem Lett, 10 (2019) 615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Platania CBM, Pittala V, Pascale A, Marchesi N, Anfuso CD, Lupo G, Cristaldi M, Olivieri M, Lazzara F, Di Paola L, Drago F, Bucolo C, Novel indole derivatives targeting HuR-mRNA complex to counteract high glucose damage in retinal endothelial cells, Biochem. Pharmacol, 175 (2020) 113908. [DOI] [PubMed] [Google Scholar]
- [129].Della Volpe S, Linciano P, Listro R, Tumminelli E, Amadio M, Bonomo I, Elgaher WAM, Adam S, Hirsch AKH, Boeckler FM, Vasile F, Rossi D, Collina S, Identification of N, N-arylalkylpicolinamide derivatives targeting the RNA-binding protein HuR, by combining biophysical fragment-screening and molecular hybridization, Bioorg. Chem, 116 (2021) 105305. [DOI] [PubMed] [Google Scholar]