Abstract
Infections caused by Klebsiella pneumoniae are often difficult to manage due to the high frequency of multidrug resistance, often conferred by efflux pumps. In this study, we analyzed sequence variations of the major RND family multidrug efflux pump AcrB from 387 assembled K. pneumoniae genomes. We confirm that AcrB is a highly-conserved efflux pump in K. pneumoniae, and identified several variants that were prevalent in clinical isolates. Molecular dynamics analyses on two of these variants (L118M and S966A) suggested conformational changes that may correlate with increased drug efflux capabilities. The L118M change resulted in enhanced protein rigidity while the flexibility of drug binding pockets was stable or increased, and the interactions between the proximal pockets and water molecules were stronger. For S966A, the significantly enlarged proximal pocket suggested higher drug accommodation ability. Consistent with these predictions, the L118M and S966A variants conferred a slightly increased ability to grow in the presence of tetracycline and to survive cefoxitin exposure when overexpressed. In summary, our results suggest that the emergence of enhanced-function AcrB variants may be a potential risk for increased antibiotic resistance in clinical K. pneumoniae isolates.
Keywords: AcrAB, efflux pump, resistance-nodulation-cell division, antibiotic resistance
1. Introduction
Klebsiella pneumoniae is one of the most notorious nosocomial pathogens, often causing severe pneumonia with high morbidity [1, 2]. Antibiotic treatment of K. pneumoniae infections has become increasingly difficult due to the emergence of carbapenemase-producing isolates (often concomitant with a reduction in porin levels [3, 4]), and due to multidrug resistance caused by efflux pumps [5, 6]. Indeed, some previously effective antibiotics used for treating K. pneumoniae infections, such as azithromycin, streptomycin, cefoxitin and tetracycline, have become less effective because of efflux pumps [7–9].
The AcrAB-TolC efflux complex is the archetypical representative of the resistance-nodulation-cell division (RND) family, which is one of the most important groups of multidrug efflux pumps in K. pneumoniae and other bacteria [9]. AcrAB-TolC is composed of the integral membrane protein AcrB, the periplasmic RND transporter AcrA and the multifunctional outer membrane channel TolC [10]. The AcrAB system transports a variety of intracellular toxic chemicals directly into the surrounding medium, bypassing the periplasm [11]. In the AcrAB-TolC tripartite complex, AcrB determines substrate specificity and export efficiency [12]. Toxic compounds are actively exported, using the proton motive force to power an antiport mechanism [13].
The structures of AcrB from different bacteria have been extensively characterized [14, 15]. The nearly 1040 amino-acid residues form twelve putative transmembrane α-helices and two large hydrophilic loops on the periplasmic surface [13]. AcrB is a trimeric protein (formed by assembly of three protomers) with a ‘jellyfish’ appearance, where a three-fold symmetry axis is oriented perpendicular to the membrane plane (Fig. S1) [15]. Commonly, the structure of AcrB can be divided into three domains, namely the transmembrane domain, drug binding domain and TolC docking domain. Among these domains, the drug binding domain (which is composed of four subdomains PC1, PC2, PN1 and PN2) is a key structural feature for drug export. All of these subdomains contain a characteristic structure motif, i.e. two β-strand-α-helix-β-strand motifs that are directly repeated, forming a sandwich structure. These motifs form the drug binding site, and a cleft exists between PC1 and PC2 at the periphery of each protomer. Drugs are taken into the pocket via the cleft, then bound by aromatic amino acid residues such as Phe 615 and Phe 178, followed by extrusion into the medium via TolC [16]. Through continuous access, binding and extrusion activities, the three protomers continuously pump toxic compounds out of the cell [16].
Molecular dynamics (MD) simulations on known structures are a powerful tool to predict dynamic conformational changes in proteins. While some simulations have been performed to study the function of the transmembrane domain and the drug binding properties of AcrB in various mutant backgrounds [17, 18], there are few reports of natural mutations of AcrB that may contribute to increased antibiotic resistance[19, 20]. Such an analysis could help predict the emergence of variants with enhanced drug export capabilities.
Here, we have performed sequence analysis of AcrB variants from almost 400 sequenced K. pneumoniae genomes. Mutation analyses identified 6 variants with a frequency of occurrence of > 2 %. We demonstrate that two of the variants, the L118M and S966A variants, exhibited increased resistance to both tetracycline and cefoxitin. By using molecular dynamics simulations, we illustrated the different mechanisms of the increased drug resistance of the variants. These data demonstrate the power of MD simulation analyses in uncovering novel drug resistance determinants and suggest that enhanced multidrug resistance due to natural evolution of more effective efflux pumps might be an emergent clinical threat in K. pneumoniae.
2. Materials and Methods
2.1. Chemicals, media, and growth conditions
Tetracycline (RPI) was formulated as a 10 mg/ml stock solution in 70 % ethanol and stored at 4 °C. Rifampicin (RPI), azithromycin (Sigma) and cefoxitin (RPI) were all formulated as a 1 mg/mL stock solution in distilled water for liquid growth experiments, while more highly-concentrated stock solutions of azithromycin (8 mg/ml in ethanol) and cefoxitin (8 mg/ml in DMSO) were formulated for the killing assays. Ciprofloxacin (RPI, Wilmington, NC) was dissolved in 0.15 M sodium hydroxide at 1 mg/ml. LB medium (per liter: 10 g NaCl, 10 g peptone, 5 g yeast extract, 15 g agar if necessary) was purchased from RPI (Wilmington, NC) and prepared as broth according to the instruction. Chloramphenicol (50 μg/ml) and hygromycin B (100 μg/ml) were added into the medium to sustain the pBAD plasmid, as described below. Arabinose (0.2%) was supplied to induce expression of AcrB. All strains were grown overnight in a 37 °C shaking incubator (200 rpm) prior to initiating the experiment.
2.2. Plasmid and strain construction
Bacterial strains used in this study are listed in Table S1. All genes were PCR amplified from K. pneumoniae KPNIH1 genomic DNA. Plasmids were built using isothermal assembly [21] using primers listed in Table S2. A modified version of plasmid pBAD33 [22] (with the addition of a hygromycin B resistance gene) was used for overexpression. The KPNIH1 acrB∷Tn strain was obtained from the Manoil Lab transposon mutant library [23].
2.3. Sequence variation analysis
AcrB variations were analyzed using the MATLAB bioinformatics toolbox [24]. For each complete K. pneumoniae genome, the ‘getgenbank’ command was used to download the sequence. Next, the nwalign program was used for the alignment between AcrB from KPNIH1 and the target genome. Scores over zero were recorded and those proteins with the highest score in each genome were used for AcrB variation analysis. Next, proteins with the highest score in each genome were collected together, and ‘multiseqalign’ command was used for sequence alignment. Finally the mutation rates were counted in ‘seqalignviewer’ program. The script used in this work is provided in GitHub (https://github.com/liying0128/efflux-pump-finder).
2.4. Modeling and molecular dynamics (MD)
The structures of AcrB and its variants were generated based on the characterized structures of AcrB from PDB Bank (PDB: 4U8Y, 4U95, 5JMN, 6Q4O and 6Q4P). Homology modeling was performed in YASARA structure with the macro ‘hm_built’. All molecular dynamics simulations were also conducted in YASARA Structure using AMBER14 force field. AcrB was embedded in a membrane in a simulation cell containing 99679 water molecules. Several Na+ cations were added into the simulation cell to neutralize the system. The solvent molecules left a 10 Å space around the protein, and the final volume of the simulation cell was set at 150 Å *150 Å *150 Å. An energy minimization was performed to eliminate improper contacts in each system. The steepest descent method was used in the first 5000 steps and the conjugate gradient method was applied in the last 5000 steps. After the energy minimization, each system was heated gradually from 0 K to 310 K. Each system was then equilibrated for 500 ps at constant temperature (310K) and pressure (1 bar) conditions via the Langevin dynamics (the collision frequency is 1.0 ps−1). Each simulation was performed for 100 ns at 310 K and 1 bar using a time step of 0.1 ns.
2.5. MD data analysis
MD data were analyzed by the marco ‘MD_analyze’ in YASARA. Root mean square deviation (RMSD) between structures following least-squares fitting to a referenced initial structure was computed. Root mean square fluctuations (RMSF) of residues were computed to check the flexibility of each residue. The key residues of proximal pocket, distal pocket, external cleft and entrance cleft were used to calculate the volume and the distance as in [17]. The surface volume formed by these key residues was calculated in YASARA’s ‘Volume’ command, and group distance was calculated by measuring the geometric center of relative groups. Group dihedral angle was defined as the angle between the line (from 101D to 107V) and the bottom surface. Triangle area formed by Phe 178, Phe 613 and Phe 615 was calculated by Heron’s formula
| (1) |
| (2) |
where a, b, and c represent the respective distance between the three residues. The data were plotted using the Python “matplotlib” module.
2.6. Growth curve, MIC and killing assay
Strains were grown overnight in 5 mL LB medium containing chloramphenicol (50 μg/ml), hygromycin B (100 μg/ml) and inducer (1 % arabinose) in a 10 mL borosilicate culture tube. The tubes were incubated at 37 °C at 200 rpm. The harvested cells were separated by centrifugation at 12000 rpm for 10 min, and the supernatant was discarded. After washing with fresh LB medium, cells were diluted 100-fold into fresh medium containing 0.4 % arabinose and then transferred into a 100-well honeycomb plate (200 μL volume/well) for growth curve determination at 37 °C in a bioscreen growth curve analyzer without shaking. Increasing concentrations of antibiotic were added to the medium. The growth of each 200 μL culture in the plate was monitored by optical density at 600 nm (OD600) every 20 minutes for 24 h.
Killing assays were used to determine antibiotic resistance of strains. Overnight cultures of each strain were grown in LB medium at 37 °C and diluted 1:10 the following day in fresh, prewarmed LB liquid medium containing a final concentration of each antibiotic. Then cell cultures were incubated at 37 °C without agitation. After 24 h, cells were centrifugally separated (8000 g, 5 min) from the medium containing antibiotics, and viable cell counts were determined by 10-fold serial dilution of cells in LB broth and spot-plating 10 μL of each on plates. Colonies were counted after 12 h of incubation at 37 °C. The experiment was independently carried out for three times.
Minimum inhibitory concentrations (MICs) were determined by use of a microplate assay to test antibiotic activity against strains used in this work. First, OD600 of the overnight cultures was measured to maintain the same amount of cells in each group, and cells were diluted 10,000 fold into a 96 well plate. Then twofold dilutions of each working solution were prepared, and the last cell in each row was set as control. Plates were incubated at 37 °C for 12 h. The MIC determinations were repeated independently 3 times.
3. Results
3.1. Global analysis of AcrB across K. pneumoniae isolates reveals variants with increased antibiotic resistance
To assess amino acid variations in AcrB among K. pneumoniae, sequence analysis was conducted on 387 NCBI assembled K. pneumoniae genomes (Table S3), using the AcrB sequence of the well-characterized clinical isolate KPNIH1 [25] as a baseline. This analysis revealed that AcrB is highly conserved among sequenced K. pneumoniae isolates. Significantly, 374 of the sequences showed higher than 95% similarity and 121 of them were identical to AcrB in KPNIH1. Within the 374 high similarity sequences, we focused on amino acids variations occurring with a frequency of over 2 % (Table 1). These sequence variations were evenly distributed in the binding pocket domain, transmembrane domain and TolC docking domain.
Table 1.
Variant distribution of AcrB among K. pneumoniae isolates
| Mutations | Amount | Frequency | Location |
|---|---|---|---|
| E639K | 128 | 34.2% | PC1 |
| H558P | 250 | 66.8% | TM7 |
| S966A | 18 | 4.8% | TM4 |
| L118M | 18 | 4.8% | PN1 |
| R806H | 15 | 4.0% | DC |
| D242E | 10 | 2.7% | DN |
3.2. The L118M mutation enhances AcrB stability in molecular dynamics simulations
Since these variations were observed in clinical isolates, we hypothesized that at least some of them might have evolved to enhance AcrB drug efflux capabilities. A preliminary screen for growth in 5 μg/mL tetracycline (0.5 x MIC) indeed suggested that overexpression of any of the six variants conferred a moderate increase in growth yield during drug treatment (Fig. S2). We chose to focus on two variants (L118M and S966A) for further molecular characterization. To predict how these variants affected AcrB protein dynamics, we conducted MD of AcrB (from KPNIH1, henceforth “wild type”), AcrBS966A and AcrBL118M (100 ns timeframe). The backbone RMSD value exhibited in each MD run reached a plateau after 5 ns (Fig. 1A). From 10 ns to the end of the simulation, AcrBL118M exhibited lower RMSD values than wild-type AcrB, suggesting this variant enhanced the stability of AcrB, while RMSD of S966A showed no significant change compared to wild type AcrB. Total relative RMSF values (RMSF of AcrB minus RMSF of variants, Fig. 1B) revealed significantly increased rigidity of the DN (near Leu 230), TN7-Iα (near Ser 530) and DC (near Ser 800) subdomains for both variants. Since the DN and DC subdomains mediate docking with the periplasmic domain of AcrAB-TolC, the increased rigidity may indicate a more stable connection with other components of the assembled pump. In contrast, the RMSFs of residues that formed the binding pocket (e.g. Ser 128 to Ser 134, Gln 176 to Ala 180, Glu 273 to Val 277 and Met 660 to Thr 674) of both L118M and S966A were increased compared to the control. Since the flexible conformation of the binding pocket helps AcrB in accommodating substrates of very different properties [17], the increased RMSF of these residues may indicate their enhanced ability to bind and export substrates.
Fig. 1. MD analysis reveals that L118M confers increased stability of AcrB, while both variants exhibit increased flexibility of binding pockets.
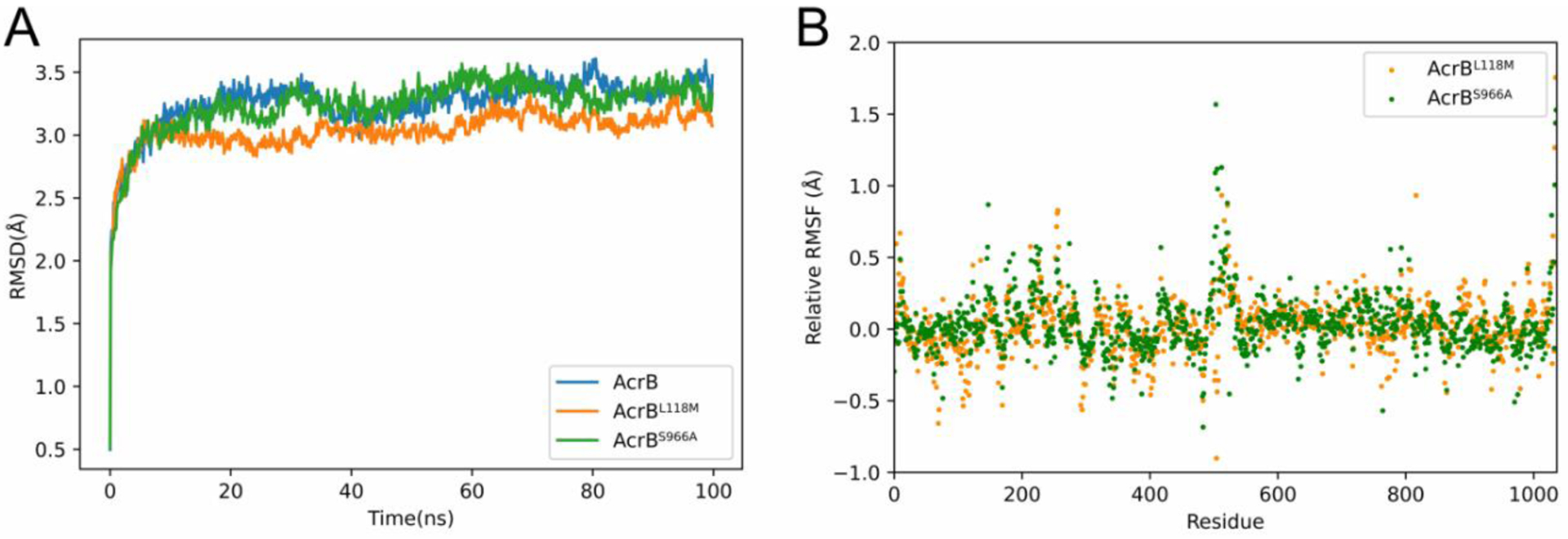
(A) The AcrBL118M variant exhibits decreased RMSD values at extended simulation times. Values were derived from 100 ns molecular dynamics simulations. (B) AcrBL118M and AcrBS966A exhibit enhanced rigidity of periplasm docking domains, while the flexibilities of drug binding pockets were increased. RMSF values were calculated from the average RMSF of their constituting atoms. Relative RMSF value means RMSF of AcrB minus RMSF of variants at each residue.
3.3. Enlarged proximal pocket of S966A
Except for flexibility, the volume of the drug binding pocket and its interaction with water are also important for drug export efficiency [17]. Therefore, the size of the drug binding pocket in each molecule was calculated (Fig. 2). On average, no significant changes of postulated gate and external cleft were observed in AcrBL118M and AcrBS966A compared to the control (Fig. 2A and 2B). Strikingly, however, AcrBS966A displayed enlarged volumes of the proximal pockets of all the three protomers (Fig. 2C and 2D), while AcrBL118M was similar to wild-type AcrB. We also calculated the number of water molecules that formed hydrogen bonds with key residues (Table 2). The results indicated that the proximal pockets of both AcrBL118M and AcrBS966A interacted more strongly with water than that of wild-type AcrB, while the distal pockets of the variants showed slightly reduced interactions with water even though the volume of one protomer of AcrBS966A was enlarged. The proximal pockets are suggested for accommodation of small molecule drugs while the distal pockets are responsible for larger compounds [26, 27]. Thus, the enlarged proximal volume of AcrBS966A and the stronger interaction with water for both of AcrBL118M and AcrBS966A may indicate their higher export efficiency.
Fig. 2. AcrBS966A exhibits enlarged proximal pockets and distal pockets.
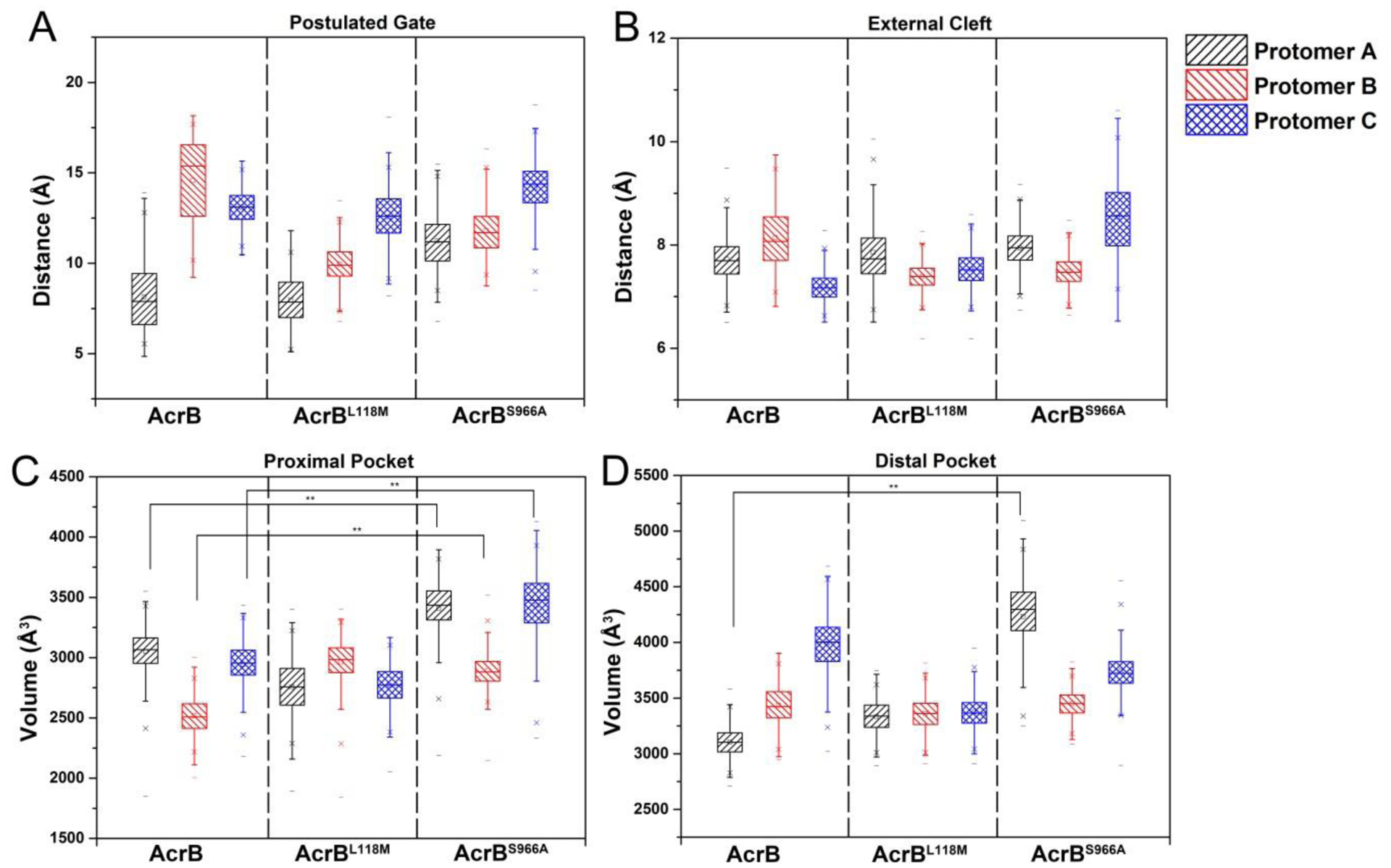
The distributions of distance of postulated gate (A) and external cleft (B) in each protomer during 100s MD simulations displayed no difference for wild-type AcrB, AcrBL118M and AcrBS966A. The volumes of three proximal pockets (C) and one distal pocket (D) of AcrBS966A were significantly enlarged compared to wild-type AcrB, while those of AcrBL118M showed no difference.
Table 2.
Average numbers of water molecule that formed hydrogen bonds with key residues during simulations. P(A): protomer A; P(B): protomer B; P(C): protomer C.
| Proximal Pocket | Distal Pocket | External Cleft | Postulated Gate | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P(A) | P(B) | P(C) | P(A) | P(B) | P(C) | P(A) | P(B) | P(C) | P(A) | P(B) | P(C) | |
| AcrB | 14.9 | 14.8 | 14.6 | 15.7 | 16.2 | 16.5 | 2.0 | 2.0 | 1.7 | 2.0 | 2.0 | 2.0 |
| AcrBL118M | 14.9 | 15.0 | 14.9 | 15.6 | 15.9 | 15.5 | 2.0 | 2.0 | 2.0 | 1.9 | 2.0 | 1.9 |
| AcrBS966A | 14.8 | 15.1 | 14.9 | 15.7 | 15.9 | 15.9 | 2.0 | 2.0 | 2.0 | 1.8 | 1.9 | 1.9 |
3.4. AcrB variants exhibit conformational differences to wild type in the central helix and aromatic-rich residues
In addition to a change in drug binding pocket volume, extrusion is also a crucial process for drug export. Active drug extrusion co-incides with structural re-arrangements, e.g. an inclined central helix. We thus also computed the dihedral angle of the three central helices in our MD simulations (Fig. 3). For wild-type AcrB, most dihedral angles of protomer A and B were less than 80 °, while the central helix of protomer C (blue) was distributed over 80 ° throughout the simulation. In contrast, most of the dihedrals of protomer A and protomer B in AcrBL118M were larger than 80 °, while protomer C displayed an inclined central helix from 60 ns to the end. For AcrBS966A, the dihedrals of protomers were concentrated in over 80°. The upright central helix suggested the protomer has a vacant binding site, which is waiting for the next substrate binding event [16]. Thus, the increased dihedrals of AcrBL118M and AcrBS966A may indicate their increased capacity for accessing antibiotics.
Fig. 3. L118M and S966A confer increased and balanced distributed dihedral angles of the central helix.
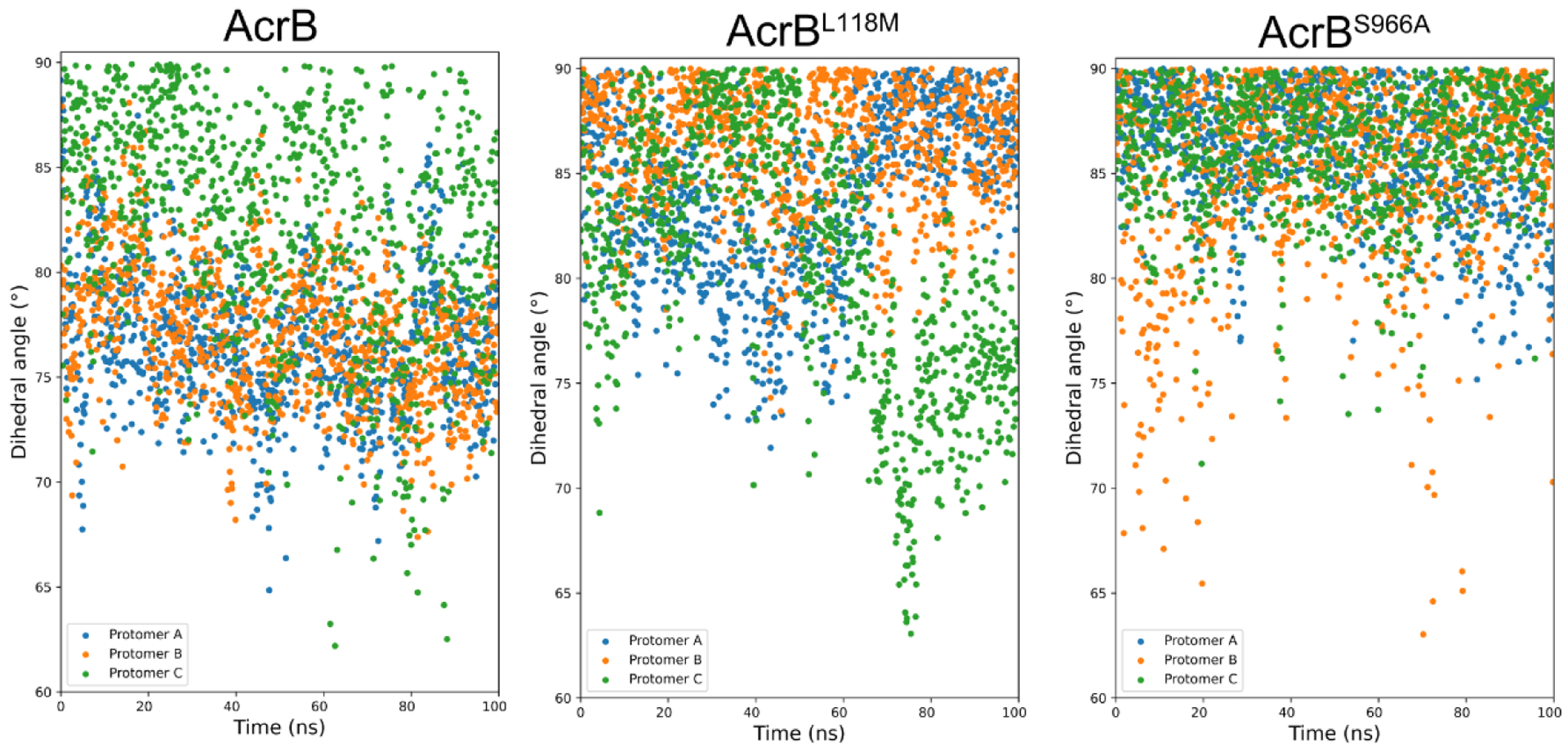
Central helix dihedral angle distributions during 100 ns MD simulations for AcrB, AcrBL118M and AcrBS966A. The angle between the central helix of each protomer and the bottom surface was calculated and plotted as scatter.
Except for the different conformations of the central helix, the conformations of aromatic amino acid-rich residues in the binding pocket also play crucial roles in drug accessing ability [16]. Thus, the area of the triangle formed by Phe 178, Phe 613 and Phe 615 in each protomer was calculated based on Heron’s formula (Fig. 4 and Fig. S5). The triangle areas of AcrBS966A were significantly enlarged compared to those of wild-type AcrB. However, the areas of AcrBL118M were slightly decreased. Since the expanded distance of aromatic rings is favorable for hydrophobic interaction with substrates [16], the enlarged Phe triangle area of AcrBS966A potentially indicates increased AcrB-antibiotic interactions.
Fig. 4. S966A and L118M have opposing effects on a key Phe triangle that mediates AcrB-antibiotic interactions.
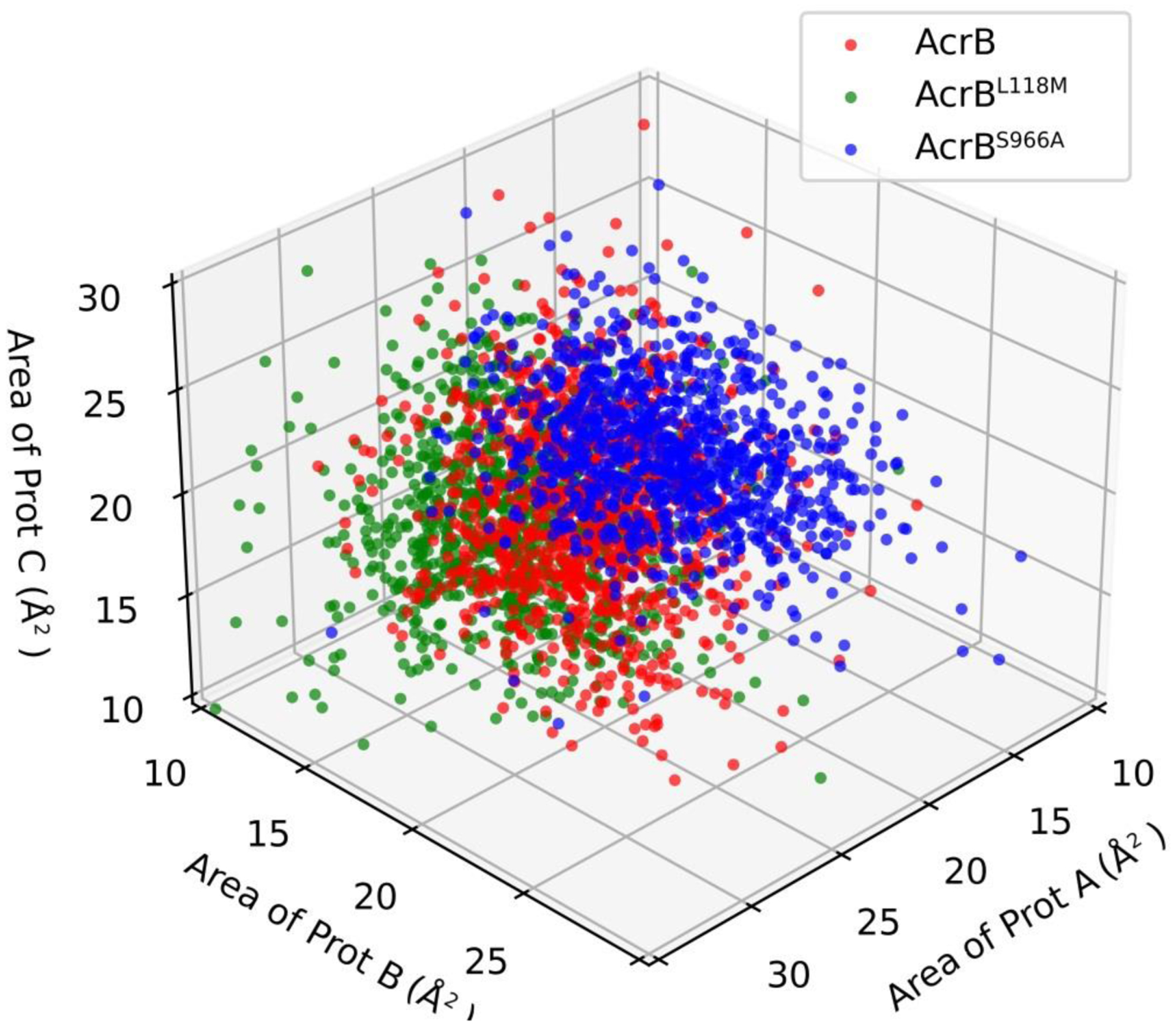
The triangle areas formed by Phe 178, Phe 613 and Phe 615 of each protomer were calculated (Details in Fig. S5) by Heron’s formula during the 100 ns simulations. Prot A, Prot B and Prot C indicate protomer A, protomer B and protomer C in each AcrB molecule.
3.5. L118M and S996A modulate susceptibility to tetracycline and cefoxitin
Lastly, we sought to characterize in more detail the impact of L118M and S966A on K. pneumoniae drug resistance. To this end, we measured the susceptibility of a K. pneumoniae acrB∷Tn mutant overexpressing AcrB, AcrBL118M and AcrBS966A to multiple antibiotics (Fig. 5). Intriguingly, MICs of the first line antibiotics tetracycline, rifampicin, ciprofloxacin, azithromycin and cefoxitin against different recombinant strains were unchanged in the variants, suggesting they do not confer full clinical resistance under these conditions (Table S4). However, we also turned to a growth-based assay, similar to our preliminary screen (Fig. S3). Strikingly, in the presence of a tetracycline concentration (10 μ g/ml) around the MIC, cells overexpressing acrBS966A and acrBL118M displayed substantially higher cell growth than cells carrying empty plasmid (negative control) or those overexpressing wild-type acrB (Fig. 5A). At 0.5 x MIC, both the S966A and L118M mutant conferred a moderate, but reproducible growth advantage compared to the other strains, while growth rate was the same for all in the absence of antibiotic (Fig. 5A).
Fig. 5. AcrBL118M and AcrBS966A confer enhanced tetracycline and cefoxitin resistance.
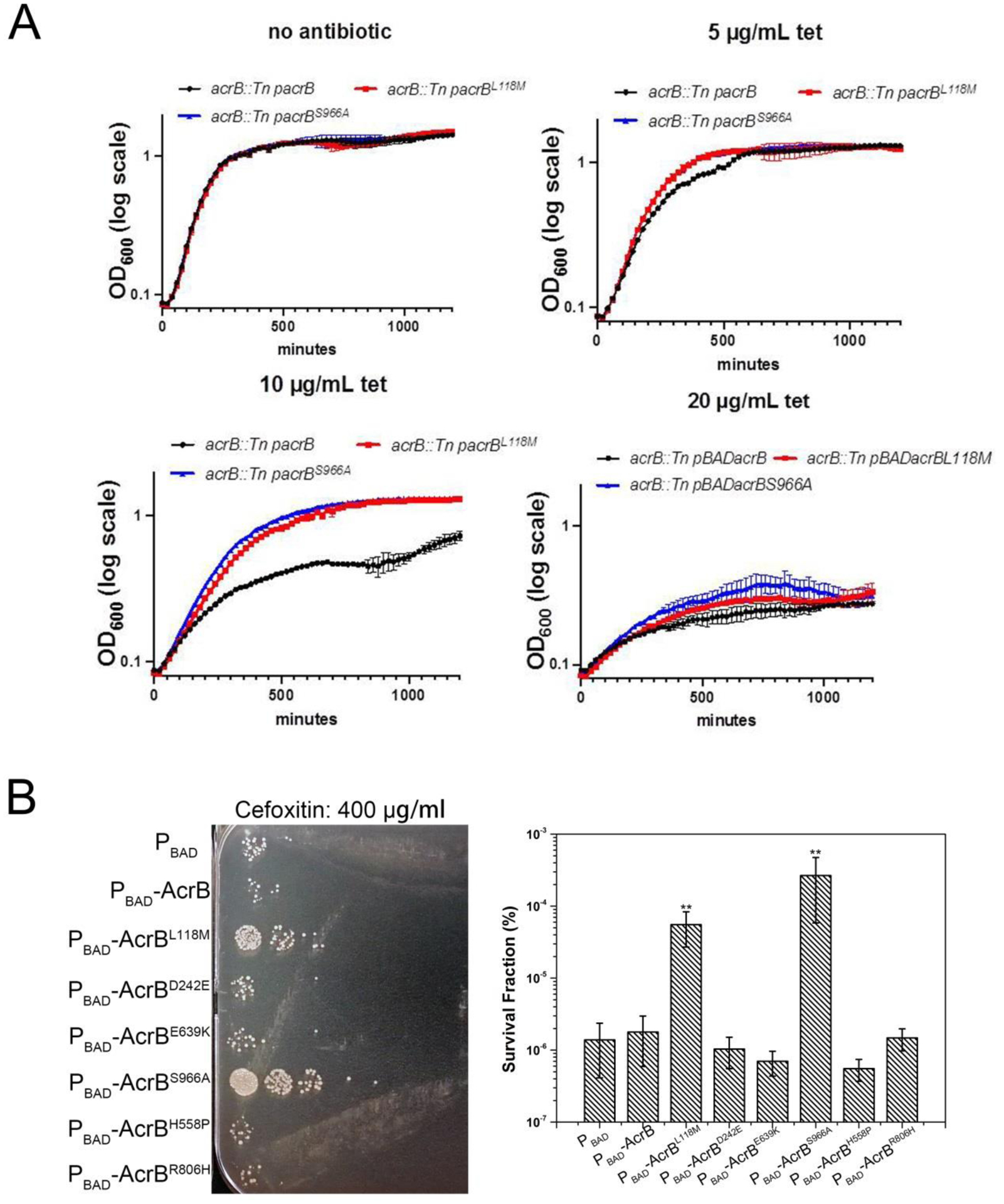
(A) Overnight cultures of KPNIH1 acrB∷Tn carrying either empty plasmid or inducible copies of wild type AcrB or variants were diluted 100-fold into fresh growth medium containing inducer and increasing concentrations of tetracycline. Growth was monitored via OD600 readings in 200 μL volume in a bioscreen plate reader. Shown are mean values of 3 technical replicates (representative of two biological replicates with similar results), error bars represent standard error. (B) Overnight cultures of KPNIH1 acrB∷Tn carrying either empty plasmid or inducible copies of wild type AcrB, AcrBL118M and AcrBS966A were diluted 10-fold into fresh growth medium containing inducer and antibiotics. After incubation at 30 °C for 24 h, viable cell counts were determined by 10-fold serial dilution of cells in LB agar and spot-plating 10 μL of each on plates. Shown are plates from one representative experiment and mean survival data from 3 independent replicates (**, P<0.01). Error bars represent standard error.
Next, we turned to killing experiments to test whether increased efflux pump activity may result in reduced killing. Indeed, efflux pumps have previously been implicated in antibiotic tolerance and persistence, i.e. the ability to survive for extended time periods in the presence of ordinarily bactericidal antibiotics [28, 29]. Strikingly, overexpressing AcrBS966A and AcrBL118M resulted in a 10- to 100fold increase in survival after 24 h of exposure to high concentration of cefoxitin (400 μg/ml, 2 x MIC), but not azithromycin, tetracycline, rifampicin and ciprofloxacin, compared to a strain overexpressing either no efflux pump or wild-type AcrB (Fig. 5B and Fig. S4). In total, our data indicate that the L118M and S966A variants can confer enhanced resistance against two antibiotics, suggesting that their prevalence in clinical isolates may pose a potential risk for the emergence of increased antibiotic resistance in K. pneumoniae.
4. Discussion
Drug export by efflux pumps is a crucial means for K. pneumoniae to survive antibiotic exposure. Indeed, we found that more than 95% of sequenced K. pneumoniae isolates have ten or more AcrB paralogs (Fig. S6). The widely distributed AcrB homologs make K. pneumoniae a “super bacterium” to resist clinical treatment [30]. While increases in efflux-mediated antibiotic resistance are typically associated with upregulation of efflux pumps as well as with the accumulation of efflux pump paralogs in clinical isolates [31, 32], naturally occurring sequence variations can also result in an increase in the efflux efficiency of specific pumps [19]. Such variants may then be inadvertently selected for during clinical antibiotic therapy. It is thus important to test these variants for their drug export capabilities to aid in efforts to predict the emergence of multidrug-resistant K. pneumoniae. Here, we have characterized two such AcrB variants that are prevalent in clinical isolates of K. pneumoniae.
Most research on AcrB or its paralogs has focused on how engineered mutations in the proton relay network of the TM subdomain inactivate the exporter as a means to understand drug efflux function [20, 33]. In contrast, only a few studies have reported on naturally occurring mutations in AcrB that confer increased antibiotic resistance [19]. In E. coli, the AcrBG288D mutation confers ciprofloxacin resistance by increased efflux, likely via an enlarged distal pocket [19]. Interestingly, however, this amino acid substitution created a trade-off where it resulted in increased susceptibility to other drugs, indicating that G288D altered substrate specificity based on the substrate binding sites. A recent study indicated that this mutation results in a subtle expansion of the distal pocket, which could account for the changes in substrate transport; however, the significance of this substitution requires further investigation [34]. Similarly, a naturally occurring Q733R substitution in AcrB from Salmonella was reported to have little effect on antimicrobial resistance [35]. In contrast, our work reports naturally occurring variants of AcrB from K. pneumoniae that confer increased multidrug resistance (albeit with moderate effectiveness), raising the alarming possibility that enhanced efflux pump mutants may emerge as a clinical threat under selective pressure of antibiotic therapy in the future.
Our MD analyses revealed possible reasons for the variants’ increased drug export capabilities. The proximal and distal pockets are suggested to mediate interactions with different efflux substrates [17]. Tetracycline and cefoxitin likely interact with the proximal pocket due to their low molecular weight and molecular surfaces (Table S5) [27]. Thus, the significantly enlarged proximal pocket might be the reason for tetracycline and cefoxitin resistance of AcrBS966A. Moreover, as the drug binding pocket was enlarged, the increased triangle area formed by three crucial aromatic residues (Phe 178, Phe 613 and Phe 615) suggested a higher ability for accommodation of substrates by aromatic-aromatic interactions [16]. A common method for uncovering specificities of efflux pumps toward antibiotics is molecular docking, in which the efflux pump is set as a rigid conformation for acceptance of ligands [36]. However, the export of antibiotics by efflux pump is a multi-step process, and antibiotics may interact with different regions during different phases. In contrast, MD simulations of efflux pump provided a wealth of information on the detailed trajectory of the important domains such as binding pocket and entrance cleft, and our results of wet-experiments indicated they were in accordance with their efficiencies in drug export. Thus, our data also provided a unique view in predicting the efficiency of this process with MD simulations.
Interestingly, both AcrBL118M and AcrBS966A displayed increased resistance in low concentration of tetracycline, but altered tolerance to a high concentration of cefoxitin. Likely, this reflects different survival mechanisms in the presence of these antibiotics, e.g., a role for persisters (that may rely on AcrAB for their survival [39, 40]) in tolerance to the bactericidal cefoxitin [37]. Drastically increased survival in cefoxitin vs. the narrow range of tetracycline concentrations around the MIC where we observed a fitness advantage of the variants suggests that cefoxitin might be a better substrate for the variants than tetracycline. This is in line with recent research showing that a mutation of AcrB dose not affect E. coli’s survival in tetracycline [38, 39].
In summary, our work demonstrates that the naturally occurring AcrBL118M and AcrBS966A increase resistance/tolerance to tetracycline and cefoxitin, posing a potential risk in clinical settings. While the effects of these variants appear to be limited, and possibly by themselves not broadly clinically relevant (unchanged MIC), such small differences could result in just enough of a growth advantage during clinical therapy that the emergence of more robustly resistant variants might ultimately be favored. More broadly, our analyses support the underappreciated notion that antibiotic selection might create efflux pumps with higher efficiency, as demonstrated previously [41, 42]. Our study thus underscores the value of analyzing variations in sequenced genomes alongside functional characterization to get a more comprehensive understanding of how sequence variations in multidrug efflux pumps can increase resistance to clinically-relevant antibiotics. These data will ultimately promote a more complete prediction of the emergence of novel mechanisms of antibiotic resistance.
Supplementary Material
Acknowledgements
Research on antibiotic tolerance in the Dörr laboratory is supported by NIH grant R01AI143704; Dr. Ying Li was supported by China Scholarship Council fellowship 201908110070.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- [1].Troeger C, Blacker B, Khalil IA, Rao PC, Cao J, Zimsen SR, et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis 2018;18:1191–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lin Y-T, Wang Y-P, Wang F-D, Fung C-P. Community-onset Klebsiella pneumoniae pneumonia in Taiwan: clinical features of the disease and associated microbiological characteristics of isolates from pneumonia and nasopharynx. Front Microbiol 2015;6:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 2009;9:228–36. [DOI] [PubMed] [Google Scholar]
- [4].Brunson DN, Maldosevic E, Velez A, Figgins E, Ellis TN. Porin loss in Klebsiella pneumoniae clinical isolates impacts production of virulence factors and survival within macrophages. Int J Med Microbiol 2019;309:213–24. [DOI] [PubMed] [Google Scholar]
- [5].Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Albertí S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother 2010;54:177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pages J-M, Lavigne J-P, Leflon-Guibout V, Marcon E, Bert F, Noussair L, et al. Efflux pump, the masked side of ß-lactam resistance in Klebsiella pneumoniae clinical isolates. PLoS One 2009;4:e4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bialek-Davenet S, Marcon E, Leflon-Guibout V, Lavigne J-P, Bert F, Moreau R, et al. In vitro selection of ramR and soxR mutants overexpressing efflux systems by fluoroquinolones as well as cefoxitin in Klebsiella pneumoniae. Antimicrob Agents Chemother 2011;55:2795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Srinivasan VB, Singh BB, Priyadarshi N, Chauhan NK, Rajamohan G. Role of novel multidrug efflux pump involved in drug resistance in Klebsiella pneumoniae. PloS One 2014;9:e96288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ni RT, Onishi M, Mizusawa M, Kitagawa R, Kishino T, Matsubara F, et al. The role of RND-type efflux pumps in multidrug-resistant mutants of Klebsiella pneumoniae. Sci Rep 2020;10:10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, et al. Structure of the AcrAB–TolC multidrug efflux pump. Nature 2014;509:512–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Reens AL, Crooks AL, Su C-C, Nagy TA, Reens DL, Podoll JD, et al. A cell-based infection assay identifies efflux pump modulators that reduce bacterial intracellular load. PLoS Pathog 2018;14:e1007115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zwama M, Yamaguchi A. Molecular mechanisms of AcrB-mediated multidrug export. Res Microbiol 2018;169:372–83. [DOI] [PubMed] [Google Scholar]
- [13].Matsunaga Y, Yamane T, Terada T, Moritsugu K, Fujisaki H, Murakami S, et al. Energetics and conformational pathways of functional rotation in the multidrug transporter AcrB. eLife 2018;7:e31715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nakashima R, Sakurai K, Yamasaki S, Nishino K, Yamaguchi A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 2011;480:565–9. [DOI] [PubMed] [Google Scholar]
- [15].Reading E, Ahdash Z, Fais C, Ricci V, Wang-Kan X, Grimsey E, et al. Perturbed structural dynamics underlie inhibition and altered efflux of the multidrug resistance pump AcrB. Nat Commun 2020;11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 2006;443:173–9. [DOI] [PubMed] [Google Scholar]
- [17].Vargiu AV, Nikaido H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc Natl Acad Sci USA 2012;109:20637–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Silva L Jr, Carrion LL, von Groll A, Costa SS, Junqueira E, Ramos DF, et al. In vitro and in silico analysis of the efficiency of tetrahydropyridines as drug efflux inhibitors in Escherichia coli. Int J Antimicrob Agents 2017;49:308–14. [DOI] [PubMed] [Google Scholar]
- [19].Blair JM, Bavro VN, Ricci V, Modi N, Cacciotto P, Kleinekathӧfer U, et al. AcrB drug-binding pocket substitution confers clinically relevant resistance and altered substrate specificity. Proc Natl Acad Sci USA 2015;112:3511–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Takatsuka Y, Nikaido H. Threonine-978 in the transmembrane segment of the multidrug efflux pump AcrB of Escherichia coli is crucial for drug transport as a probable component of the proton relay network. J Bacteriol 2006;188:7284–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 2009;6:343–5. [DOI] [PubMed] [Google Scholar]
- [22].Guzman L-M, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 1995;177:4121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ramage B, Erolin R, Held K, Gasper J, Weiss E, Brittnacher M, et al. Comprehensive arrayed transposon mutant library of Klebsiella pneumoniae outbreak strain KPNIH1. J Bacteriol 2017;199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Singh GB. Fundamental of bioinformatics and computational biology. Springer, 2015. [Google Scholar]
- [25].Snitkin ES, Zelazny AM, Thomas PJ, Stock F, Henderson DK, Palmore TN, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 2012;4:148ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 2006;313:1295–8. [DOI] [PubMed] [Google Scholar]
- [27].Zwama M, Yamasaki S, Nakashima R, Sakurai K, Nishino K, Yamaguchi A. Multiple entry pathways within the efflux transporter AcrB contribute to multidrug recognition. Nat Commun 2018;9:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pu Y, Zhao Z, Li Y, Zou J, Ma Q, Zhao Y, et al. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol Cell 2016;62:284–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Westblade LF, Errington J, Dörr T. Antibiotic tolerance. PLoS Pathog 2020;16:e1008892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shon AS, Russo TA. Hypervirulent Klebsiella pneumoniae: the next superbug? Future Microbiol 2012;7:669–71. [DOI] [PubMed] [Google Scholar]
- [31].Frimodt-Møller J, Løbner-Olesen A. Efflux-pump upregulation: from tolerance to high-level antibiotic resistance? Trends Microbiol 2019;27:291–3. [DOI] [PubMed] [Google Scholar]
- [32].Duan W, Li X, Ge Y, Yu Z, Li P, Li J, et al. Mycobacterium tuberculosis Rv1473 is a novel macrolides ABC Efflux Pump regulated by WhiB7. Future Microbiol 2019;14:47–59. [DOI] [PubMed] [Google Scholar]
- [33].Su C-C, Li M, Gu R, Takatsuka Y, McDermott G, Nikaido H, et al. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J Bacteriol 2006;188:7290–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Johnson RM, Fais C, Parmar M, Cheruvara H, Marshall RL, Hesketh SJ, et al. Cryo-EM structure and molecular dynamics analysis of the fluoroquinolone resistant mutant of the AcrB transporter from Salmonella. Microorganisms 2020;8:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Elkins CA, Mullis LB, Lacher DW, Jung CM. Single nucleotide polymorphism analysis of the major tripartite multidrug efflux pump of Escherichia coli: functional conservation in disparate animal reservoirs despite exposure to antimicrobial chemotherapy. Antimicrob Agents Chemother 2010;54:1007–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Byrd BA, Zenick B, Rocha-Granados MC, Englander HE, Hare PJ, LaGree TJ, et al. The AcrAB-TolC Efflux Pump Impacts Persistence and Resistance Development in Stationary-Phase Escherichia coli following Delafloxacin Treatment. Antimicrob Agents Chemother 2021;65:e00281–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cui P, Niu H, Shi W, Zhang S, Zhang W, Zhang Y. Identification of genes involved in bacteriostatic antibiotic-induced persister formation. Front Microbiol 2018;9:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nolivos S, Cayron J, Dedieu A, Page A, Delolme F, Lesterlin C. Role of AcrAB-TolC multidrug efflux pump in drug-resistance acquisition by plasmid transfer. Science 2019;364:778–82. [DOI] [PubMed] [Google Scholar]
- [40].Wu Y, Vulić M, Keren I, Lewis K. Role of oxidative stress in persister tolerance. Antimicrob Agents Chemother 2012;56:4922–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Fernandes P, Ferreira BS, Cabral J. Solvent tolerance in bacteria: role of efflux pumps and cross-resistance with antibiotics. Int J Antimicrob Agents 2003;22:211–6. [DOI] [PubMed] [Google Scholar]
- [42].Sandoval MS, Philippe C, Maximino A, Choong-Min R. Adaptive Resistance in Bacteria Requires Epigenetic Inheritance, Genetic Noise, and Cost of Efflux Pumps. PLoS One 2015;10:e0118464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


