Abstract
Gene therapy has emerged as a potential platform for treating several dreaded and rare diseases that would otherwise not be possible with traditional therapies. Due to their ability to transport genomes to cells, Viral vectors have been a platform of choice in gene delivery applications. However, since their delivery is not precision based, the application has led to off-target toxicities. As such, various strategies in the form of non-viral gene delivery vehicles have been explored and are being developed. In this review, we discuss the opportunities lipid nanoparticles (LNPs) present for gene delivery, efficiently and precisely. We also discuss synthesis strategies via microfluidics used for high throughput fabrication of such non-viral gene delivery vehicles. Finally, the application of these vehicles for the delivery of different genetic materials such as peptides and RNA for different diseases ranging from more common diseases to rare diseases are explored.
Keywords: Non-viral vectors, lipid nanoparticles, microfluidics, gene delivery, CRISPR-delivery
Graphical Abstract
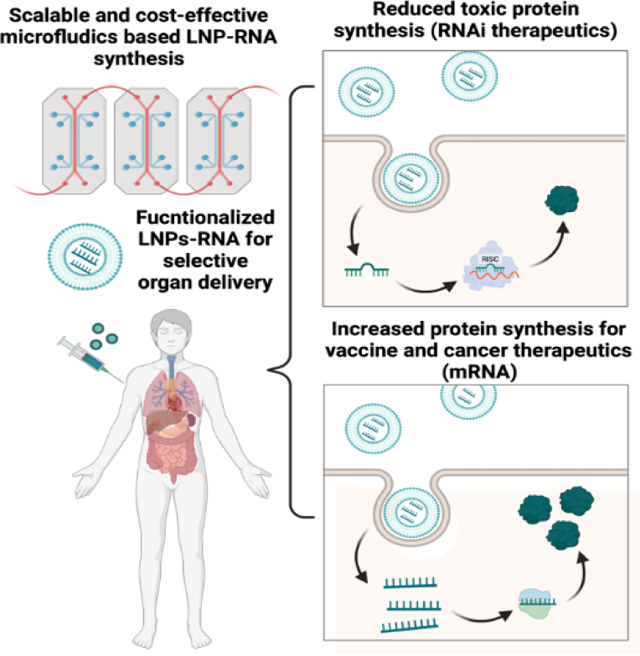
Introduction
Current progress in understanding the genetic causes of diseases and the culmination of the human genome project has paved the way for discovering novel gene editing therapeutics, specifically, perturbation of the gene expression of altered disease-relevant genes1–3. However, treatment of genetic disorders relies on a continuous therapeutic regime, which leads to off-target treatment-related toxicity while reducing the patient’s quality of life4. Gene editing therapies have shown the ability to target the underlying genetic alterations, overcome prolonged treatments and their adverse side effects, and improve treatment efficacy. Cystic fibrosis, sickle cell disease, and Duchene muscular dystrophy (DMD) are among several genetic diseases that have shown significant promises with gene therapies5–7. Furthermore, in vivo delivery of messenger RNA (mRNA) based vaccines have been impactful against Zika virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and several other immune-related diseases8–10. These gene therapies are heavily dependent on the carriers for the delivery of payloads that include clustered regularly interspaced short palindromic repeats (CRISPR) antisense oligonucleotides (ASO), short interfering RNA (siRNA), microRNA (miRNA), and mRNA11.
Based on the delivery routes, gene-editing therapies are often differentiated as viral or non-viral formulations. As the name suggests, viral formulations are centered on the use of viruses to deliver genetic material into the cells12. Several such vectors have been clinically tested, including retroviruses, lentiviruses, and adeno-associated viruses (AAV). Among the first approved gene-editing therapies by FDA are AAV2 and AAV9 vectors for retinitis pigmentosa and spinal muscular atrophy, respectively12. AAV-mediated delivery is efficient for genetic diseases that require long-term gene expression, or genome integration but is less preferred for genetic diseases caused due to point mutations. However, it has been shown that AAVs can lead to off-target effects, robust immune responses, liver damages and fatalities in certain clinical trials11,13–15. Indeed, AAVs have limited transgene carrying capacity (~4.8kb), thus limiting their use with larger CRISPR base editor and other genetic payloads16.
Non-viral therapies are based on engineered lipid nanoparticles for gene delivery that reduces the off-target toxicity related risk compared to viral formulations (Fig 1). This can be attributed to the feasibility and tenability of non-viral therapies that enable efficient nucleic acid encapsulation, cellular delivery, and endosomal release. Non-viral formulations can be efficiently engineered to have prolonged blood circulation and lower renal clearance while having mitigated immune-response17. In some recent studies, LNPs have been conjugated with antibodies for targeted delivery of the payload to cancer and normal tissues18,19. To achieve organ selectivity, alterations in lipid composition of the LNPs have been explored such that they can direct LNP-mRNA vaccines to spleen and targeted genome editing capable LNPs to liver and lungs20–23. Thus, non-viral therapies have emerged as a versatile platform for efficient gene delivery while mitigating the risk and side effects of viral gene delivery and are extensively utilized in several clinical trials to deliver RNA-based therapeutics24–26. However, non-viral platforms have been shown to have the limited capability to deliver CRISPR/Cas9 components, resulting in lower gene editing efficiencies which paves way for novel engineering strategies for the delivery of large payloads while enhancing the efficacy. In this review, we will discuss the current state of non-viral routes, focusing on the synthesis and application of lipid nanoparticles for the delivery of gene editing therapeutics and their limitations and future directions to enhance their targeting and efficacy.15–18
Figure 1. RNA encapsulated lipid nanoparticle delivery for genome editing.
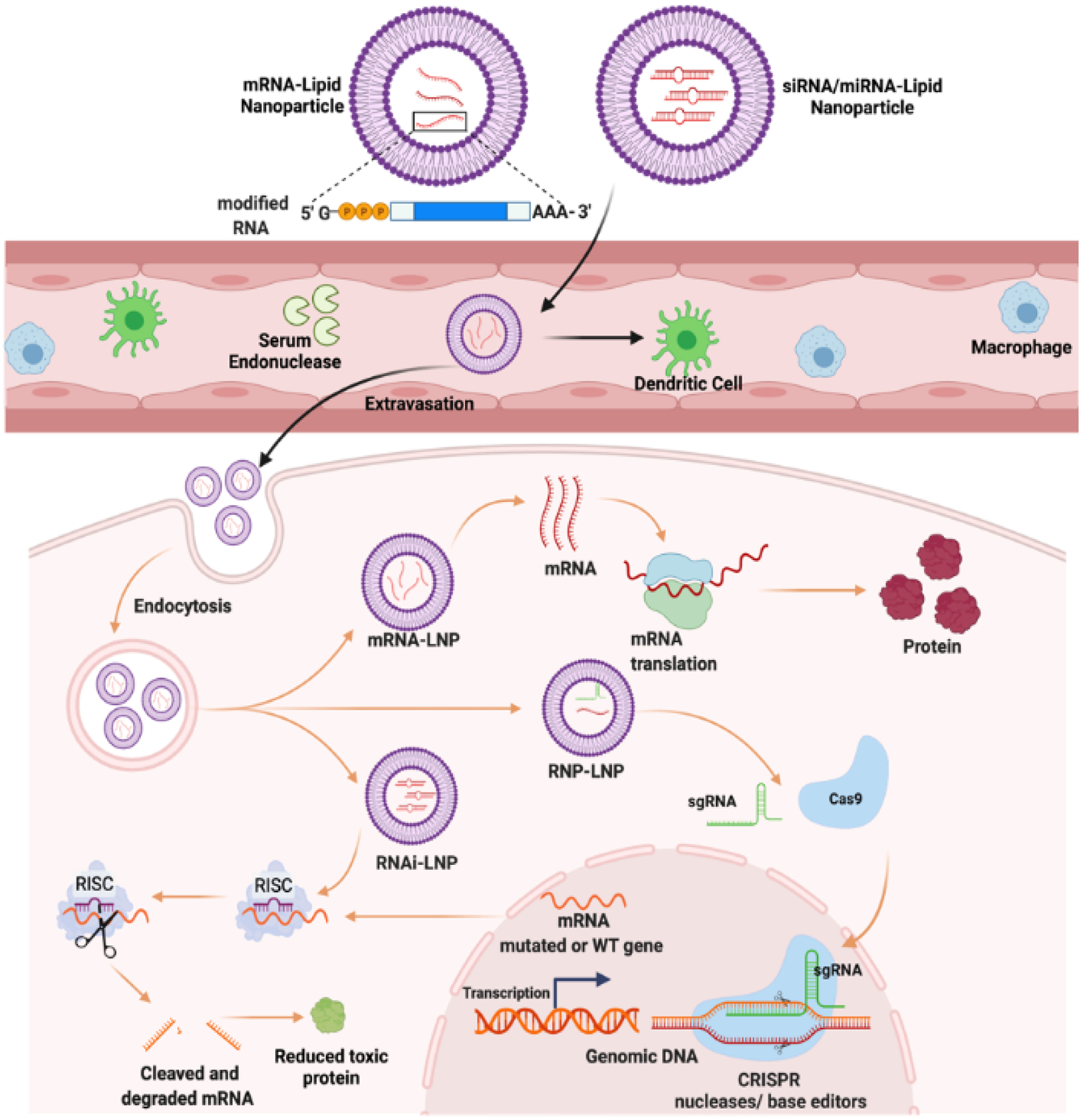
RNAs unstable and prone to degradation by serum endonucleases. LNP encapsulation of RNA have increased circulation time and can be further modified for targeted delivery to specific tissues. LNPs can be used to encapsulated spectrum of RNAs including siRNA and mRNA to modulate protein synthesis.
Synthesis of LNPs using microfluidics
The non-viral therapies provide tunable formulations and enhance the delivery of larger payloads, including plasmid DNA, RNA, and CRISPR based genetic materials. Lipid nanoparticles (LNPs) are FDA approved non-viral nucleic acid delivery vehicles capable of delivering a broad spectrum of payloads. LNPs were initially designed for the delivery of small molecule therapeutics and are now being adapted for nucleic acid delivery29–31. The essential design parameters for nucleic acid delivery include proper (nano)particle size for an efficient terminal filtration, long-term stability for preservation, enhanced payload release rates, scalable manufacturing capacity, and efficient entrapments32. The first nucleic acid formulations, containing only phosphatidylcholine and cholesterol, demonstrated that nucleic acid entrapment within a particle suffered from poor entrapment efficiency32,33. In addition, ionic interactions between the lipids and payload were shown to increase entrapment efficiencies and negatively affect intracellular delivery dramatically. However, cationic lipids’ positive charge and non-biodegradable nature have led to initial lipoplex-like formulations with significant toxicity that limited their use in gene delivery applications34,35. Four LNPs including ionizable amino-lipid (e.g., dilinoleylmethyl 4 dimethylaminobutyrate, DLin-MC3-DMA), a helper lipid (e.g., 1,2-distearoyl-sn-glycero-3-phosphocholine, DSPC), cholesterol, and a polyethylene glycol (PEG)-lipid (e.g., 1,2-dimyristoyl-sn-glycerol, methoxypolyethylene glycol, PEG-DMG) have been introduced based on the evolution of the composition, and the manufacturing processes. Among these four groups, PEG-lipids shield the LNP surface and protects against opsonins and uptake by the mononuclear phagocyte system, preventing their aggregation in the circulation36. While PEG-lipids prevent aggregation during production and storage, their incorporation affects the LNP size37.
The properties of LNPs are greatly influenced by the particle size and size distribution38–40. Smaller LNPs (±100–200 nm) with narrow size distribution are ideal for biodistribution, also enhancing their ability to cross biological barriers (e.g. endothelial border)40–44. Moreover, the drug loading efficiency of particles increases with decreasing particle size45. As such, the stress on fabricating of LNPs with homogenous size distribution and smaller sizes is of priority vis-à-vis delivery of genetic material, in vivo. Current top-down strategies, such as ultrasonication, high pressure homogenization, and emulsification can produce high quantities of lipid particles, but cannot produce particles in the nanoscale range and lack reproducibility46–48. Moreover, these methods can have a major destructive impact on the particles due to high thermal energy and mechanical abrasive shear stresses that are produced during the syntheses processes, deeming them inefficient for nucleic acid encapsulation49. Although these processes have evolved, however they still lack accurate control over large scale mixing, leading to high polydispersity with low encapsulation efficiency causing batch-to-batch variability and hampering scalability50,51.
Microfluidic devices enable a continuous, controllable, and reproducible production of small sized LNPs with a narrow size distribution in a single-step process52–55. Lipids are generally dissolved in an aqueous-miscible solvent, together with an active ingredient (e.g., RNA, proteins or drugs), and a surfactant. Subsequent mixing with an aqueous phase causes the solution to become supersaturated with lipids, leading to LNP precipitation56,57. Microfluidic LNP production is highly versatile with a range in channel dimensions, multiple fabrication materials (e.g., polymers or glass), and a wide range of lipid formulations that eventually lead to the production of homogenously sized particles. However, due to the laminar flow (Reynolds number <1), the mixing of liquids is limited to molecular diffusion, which is relatively slow58. Fast mixing is essential to create uniform supersaturation of LNPs throughout the microfluidic system59. The inclusion of micromixers enables a higher contact area between the liquids and reduces the diffusion length, significantly increasing the mixing efficiency. Table 1 outlines major micromixers for LNP production with their advantages and limitation.
Table 1.
Mechanism, advantages, and disadvantages of various micromixers and fabrication platforms for the synthesis of LNPs.
| References | ||||
|---|---|---|---|---|
| Staggered herringbone micromixer | PDMS/glass | Large size range (20–250 nm) Low PDI Widely used (and easy to use) | Relatively low throughput Potential clogging in micromixer by clustering of LNPs | 52–55 |
| Segmented flow micromixer | Introduces a gas phase to generate a gas-liquid flow | Shortened mixing time and length | More complex design | 56–58 |
| High-pressure micromixer | Introduces pressure intensifiers to increase pressure at the inflow channels. | Small size and low PDI Higher flow rates | Limited use of materials to withstand high pressures Need for specialized equipment | 59,60,63 |
| Flow-focusing micromixer | Mixing based on droplet formation by flowing a stream of lipid phase into a channel containing the continuous phase | Short mixing time Low sample amounts Easy design | High concentration of lipids Low flow rates | 61, 62, 63 |
| Toroidal micromixer | Circular structures are introduced in the flow path which increases centrifugal forces for increased chaotic mixing. | High throughput Small size and low PDI | Potental clogging in micromixer by clustering of LNPs | 45 |
| iLiNP device | A novel chaotic mixer that uses a baffle structure for more efficient mixing | Large size range and low PDI No clogging of LNPs | Relatively low throughput | 66 |
The staggered herringbone micromixer (SHM) is the most widely used microfluidic platform to produce LNPs53,60–62. Due to the herringbone structure, the fluids inside the microchannel are rapidly mixed. Therefore, the mixing efficiency and subsequent LNP properties (i.e., size and polydispersity index (PDI)) are highly influenced by the design of the micromixer, e.g., dimensions of the micromixer channels. SHMs are low-throughput (<100ml/hr) microfluidic devices but provides rapid and controlled mixing within a narrow size range of ~20–50nm. Cheung et al reported the production of PEGylated LNPs using SHM. They studied the effect of multiple formulation parameters, including aqueous media, lipid components and composition, lipid ratio, and processing parameters. The investigators also characterized different lipid formulations based on their fluidity, showing that DOPC5 (fluid lipid) had smaller size than more rigid lipids DPPC5 and DSPC5. Furthermore, it was also observed that using higher concentrations of 2–5% PEG decreased the LNP size and increased the polydispersity. By optimizing these parameters, it was possible to formulate three PEGylated LNPs based on 2.5% PEG with ±100 nm size and a PDI of <0.2 that was lower than microfluidic LNP production without mixing63.
Several microfluidic designs have been devised over the last decade for LNP production, including the segmented flow micromixer64–66, high-pressure micromixer67,68, and a flow-focusing micromixer69,70. Recently, Riewe et al studied LNP production (e.g., reproducibility, size and size distribution) via SHM, high-pressure, and a segmented-flow micromixer with different types of lipid carriers (Fig 2A–D). Castor oil and glycerol monooleate were chosen due to their high solubility in ethanol. Furthermore, they performed LNP production on systems with different channel diameters (58 and 29 μm diameter, respectively). All the systems were shown to produce LNPs of smaller sizes compared to their reference batch systems. Particle size and PDI could be controlled by changing flow rates and process pressure, especially in the SHM (Fig 2E). As expected, the high-pressure micromixer resulted in the highest throughput (approximately 10 −100 times as much as SHM and segmented flow micromixers, respectively), owing to its relatively high flow rates and pressures. Moreover, LNPs of different sizes could be synthesized within the same micromixer depending on the lipid formulation, showing that different microsystems are preferred depending on the choice of lipid formulation71.
Figure 2. Micromixers for comparative study of lipid nanoparticle synthesis.
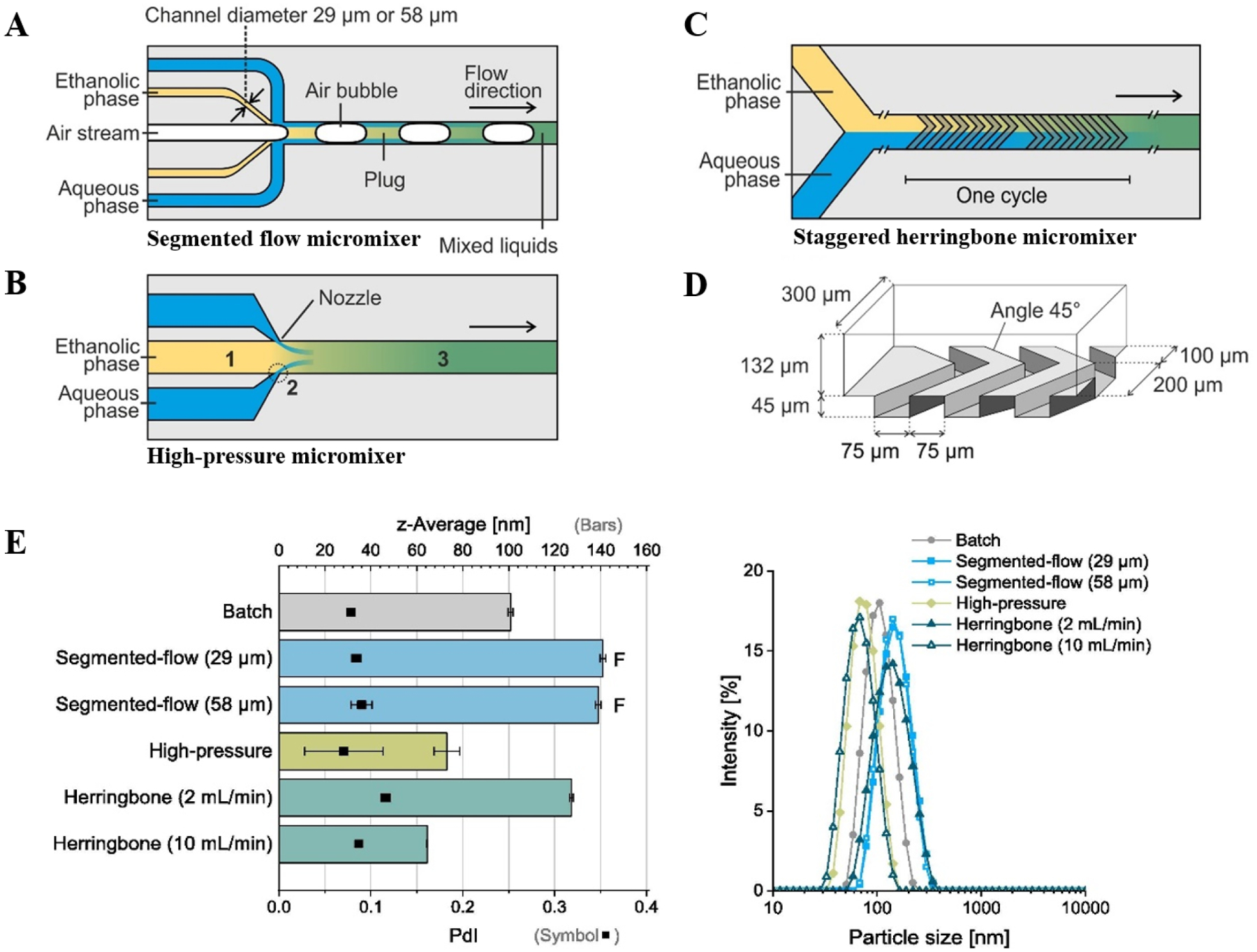
(A) Segmented-flow micromixer. Two designs were used in the study with varying diameter of ethanol channel. (B) High-pressure micromixer (C) Staggered herringbone micromixer. The micromixer was arranged in nine cycles each containing twelve grooves. (D) Perspective view on channel structures in the herringbone micromixer. (E) Mixture of 10 mg/mL glycerol monooleate in ethanol with 0.222 mg/mL poloxamer 407 in water; Segmented-flow micromixer showed fouling in microchannels (“F”). A selection of corresponding representative intensity weighted particle size distributions (DLS), shown on right. Adapted from Reiwe et. at.71. All the figures are adapted with permission from Elsevier.
A major limitation of bottom-up strategies of LNP production is the relatively low throughput that microfluidic systems offer. Webb et. al. recently compared two different micromixers (a staggered herringbone and a toroidal micromixer) on their LNP production and LNP properties (.e.g. size and PDI)51 (Fig 3A–C). A major advantage of the toroidal micromixer is the ability to increase production irrespective of channel size, while maintaining the same parameter set points. The LNPs were prepared with the NanoAssemblr® Benchtop, the Ignite™ or the NxGen Blaze™ devices from Precision NanoSystems Inc. (Vancouver, Canada). Using the toroidal micromixer, the authors were able to scale up the production of LNPs from 12 mL/min to 200 mL/min without changing any of the process parameters. To integrate SHMs in the clinical and benchtop applications, a recent work developed a parallel microfluidic device (PMD) that can incorporate SHMs at 1x, 10x and 128x arrays to operate simultaneously72. Thus, enhancing the device capability to work on a broader volume scale while maintaining the LNPs size distribution and PDI (Fig 3D–E). The in vivo efficacy of PMD produced LNP-siRNA against factor VII was >90% compared to bulk LNP-siRNA which decreased factor VII only by 20%. To test PMD based mRNA-LNPs, luciferase encoded mRNA-LNP was delivered by tail-vein injection in mice to demonstrate 5-fold higher luciferase expression compared to bulk LNPs. In these mice no liver toxicity was apparent. This shows that efficient scaling of microfluidic systems can aid in the translation of LNPs from bench to production of large-scale LNPs, which is the need of the future therapies and to meeting the demand of large scale vaccinations51.
Figure 3. Microfluidics mixers for scalable manufacturing of lipid nanoparticles.
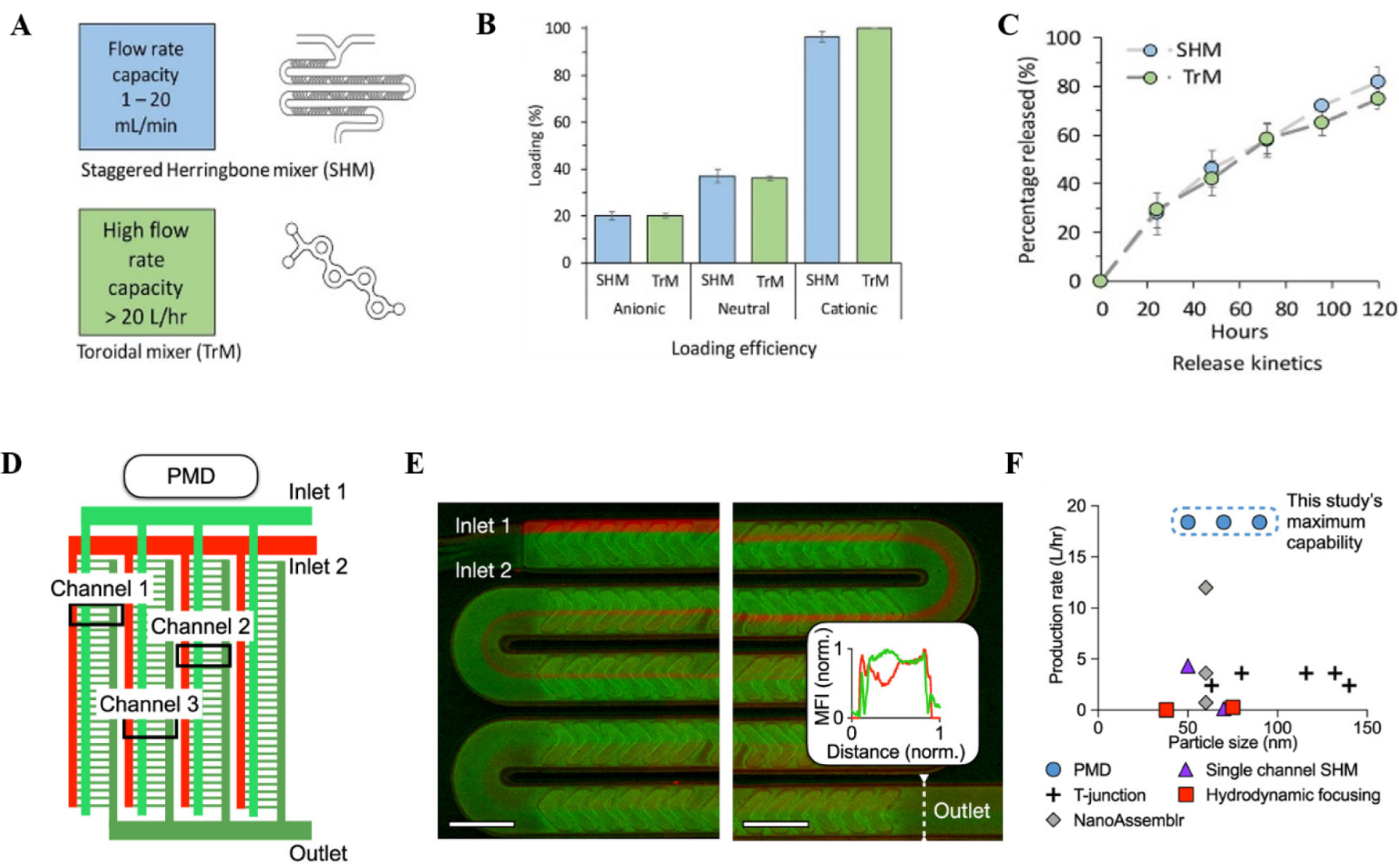
(A) Microfluidic design used for the Webb et. al. study51. Two micromixers were employed, including parallelization of multiple Staggered herringbone micromixer (SHM) using NanoAssemblr® and toroidal mixer(TrM) using NxGen Blaze™. (B) Production of drug loaded liposomes with different zeta potential using SHM and TrM. (C) Drug release profile from LNPs synthesized using SHM and TrM. (D) Outline of parallel SHM microfluidic device (PMD). 128 SHMs are incorporated in parallel. (E) Fluorescent images of mixing in a channel, showing the red and green plot profiles versus channel distance at the outlet. (F) LNP production rate comparison. Panels D-E are adapted from Shepherd et. al.72. All the figures are adapted with permission from Elsevier and American Chemical Society.
CRISPR-Cas9 based nucleases and DNA base editors have gained significant traction in clinical settings2. However, efficient and targeted delivery of the CRISPR-Cas9/sgRNA ribonucleoprotein (RNP) is still a significant challenge due to the large size of Cas9 and stability of the complex in the LNPs73. Non-viral formulations, especially the LNPs have been used for the efficient delivery and enhanced the efficacy of the RNPs. Suzuki et al used an invasive lipid nanoparticle production (iLiNP) microfluidic system to synthesize RNP-loaded LNPs (Fig 4). The lipids, (a pH sensitive cationic lipid, a phospholipid, cholesterol, and PEG-DMG) dissolved in ethanol, and an on-chip S-shaped micromixer rapidly mixed the RNP, suspended in acidic buffer74. The addition of a third inlet for introducing the buffer solution reduced the Cas9 RNP exposure to high concentrations of ethanol, thus decreasing Cas9-RNP aggregation at the junction. This further improved the mixing efficiency and quality of the RNP-loaded LNPs. The delivery of the RNPs was optimized by adding negative charges by complexing the RNPs with single stranded oligonucleotides. PEG-DMG was found to significantly impact zeta-average and knock-out (KO) efficiency, using different % PEG. The study showed that although lower % PEG had higher efficiency, but it reflected a reduced colloidal stability. After multiple rounds of parameter optimization (e.g. mol% of lipids or RNP/lipid molar ratio), the investigators obtained 100–200 nm spherical LNPs with 2% PEG which remained stable over four weeks74. Using this system, it was demonstrated that the efficient delivery of Cas9n-RNP-LNPs lead to >90% reduction in EGFP expression. To test targeted inhibition against hepatitis B virus, LNP-RNPs were delivered in a cell culture model to inhibit HBV DNA and cccDNA by ~60% and ~80%, respectively. Compared to AAV2, LNP-RNPs showed higher inhibitory effect towards HBV DNA (Fig 4C).
Figure 4. Ribonucleoprotein loaded lipid nanoparticle synthesized using microfluidic device.
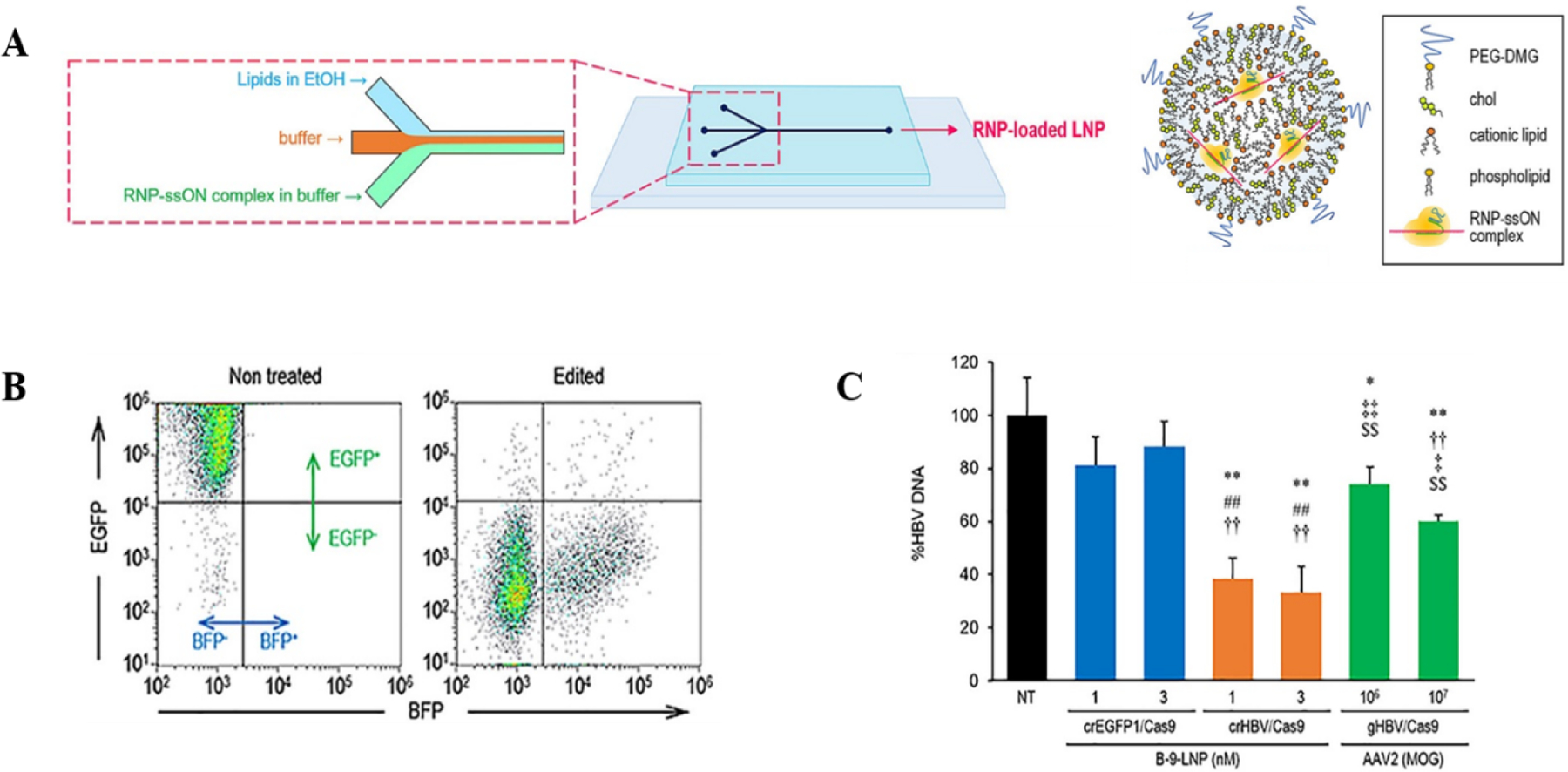
(A) Microfluidic design with solvent inlets for synthesizing RNP-loaded LNPs. The injected buffer helps to mitigate the exposure of RNPs to high concentrations of EtOH. (B) RNP-LNP induces gene knockout tested using FACS in HeLA-GFP cells. (C) Inhibition of HBV by Cas9 RNP-loaded B-9-LNPs. All the figures are adapted from Suzuki et. el.74. All the figures are adapted with permission from Elsevier.
Microfluidics based LNPs synthesis provides scalable and cost-effective technologies, which can be easily modulated for novel payload delivery. These systems are amenable for a broad range of lipids and solvent to fine-tune several parameters in LNPs synthesis75. Furthermore, microfluidic devices provide an advantage to scale-up the system by using multiple micromixers in parallel72. Several micromixers can also be scaled up for production, while toroidal mixtures provide scale independent production systems51. These large-scale production devices have removed the scalability bottleneck in LNP production. However, large payloads such as nucleic acids and RNP often lead to lower efficacy and off-target effect, thus there is a need for lipid-wide library screen with barcoded LNPs, such that their delivery across tissues can be monitored to elucidate the in vivo pharmacodynamics and biodistribution. Additionally, multifunctionality and biodegradability of lipids should be considered while designing microfluidic based LNPs. Multifunctional lipids can act as adjuvants to boost the efficacy of the payloads, while biodegradable lipids will minimize long term genome integration and immunogenicity. Thus, the benefits these systems offer only strengthen the need for a microfluidic route for the synthesis of LNPs playing a critical role in transitioning these platforms to a clinical setting.
Nature and delivery of payload
A broad spectrum of payload can be delivered using LNPs, including small molecules (neuroactive agents), peptides and proteins (recombinant hormone antigen, receptor agonist, and antagonist), and nucleic acids76. Here, we will discuss recently explored mRNA payload developments and outline their evolution for stable delivery while mitigating immune response. Additionally, we discuss the delivery of CRISPR base-editor that have become the cornerstone of gene-editing therapeutics. Finally, we discuss LNP based delivery of mRNAs encoding for chimeric antigen receptor that hold the key to the future clinical trials of CAR T-cell therapies.
Modified RNA:
LNPs have played a significant role in the delivery of mRNA that has aided the clinical translation of genome engineering technologies, resulting in successful FDA approvals and clinical trials25,77. LNP formulations encapsulating chemotherapeutics were first clinically approved in 1990 and ever since several LNPs encapsulating small molecules have been FDA approved78. However, biologics (siRNA) encapsulated by LNPs were clinically approved only in 2017 for treatment of transthyretin-mediated amyloidosis. Thus, paving way for clinical approval of nucleic acid therapeutics, including DNA, RNA, and genome editors that hold significant potential in cancer therapies, genetic diseases, vaccinations, and infectious disease treatments79. However, there are significant impediments related to the stability of the nucleic acids, intracellular delivery, and toxicity80. Unmodified mRNA can undergo rapid degradation by endonucleases and can activate several immune response pathways, such as retinoic acid-inducible gene (RIG-I) and toll-like receptors (TLRs) that can induce toxicity25,81. Several recent works have explored the chemical modification of mRNA as an efficient way to enhance their stability and dampen the immune response82,83.
mRNA modifications such as 2-thiouridine, 5-methylcytidine, and N1-methyl-pseudouridylation (1mΨ-mRNA) have been shown to prevent the activation of immune response sensors. Moreover, these modifications stabilized the mRNA against cleavage and degradation, thus enhancing their editing efficacy80 (Fig 5A,B). LNP containing canonical uridine were found to be immunostimulatory, while 1mΨ-mRNA-LNPs had low immune simulation84. These modifications have been explored in mRNA-based vaccines where the LNP encapsulated with modified 1-methyl- Ψ-mRNA towards Zika prM-E exhibited strong protective immune response in mice and rhesus macaques at low doses85. In addition, another group working on influenza vaccines reported LNP complexed with modified mRNA elicited protective effects upon single low dose intradermal immunization86. Currently, both the vaccines NCT03014089 (Zika) and NCT03076385 (Influenza) are under clinical phase I/II trial. Moreover, mRNA-1273 and BNT162B2 SARS-CoV2 vaccines which have been extremely well tolerated and efficient against COVID-19 are also synthesized by replacing uridine with 1mΨ-mRNA10. Thus, mRNA modification provides an efficient way to stabilize the mRNA while enhancing its translation efficiency and low immunogenicity. Substantial research in understanding mRNA biology, such as 5’cap, 5’ and 3’ untranslated regions of mRNA and the length of poly(A) tail, along with elucidation of efficient position for the modification of mRNA, would provide future directions in the field of mRNA-based therapeutics.
Figure 5. Lipid nanoparticle for efficient delivery of therapeutic RNAs.
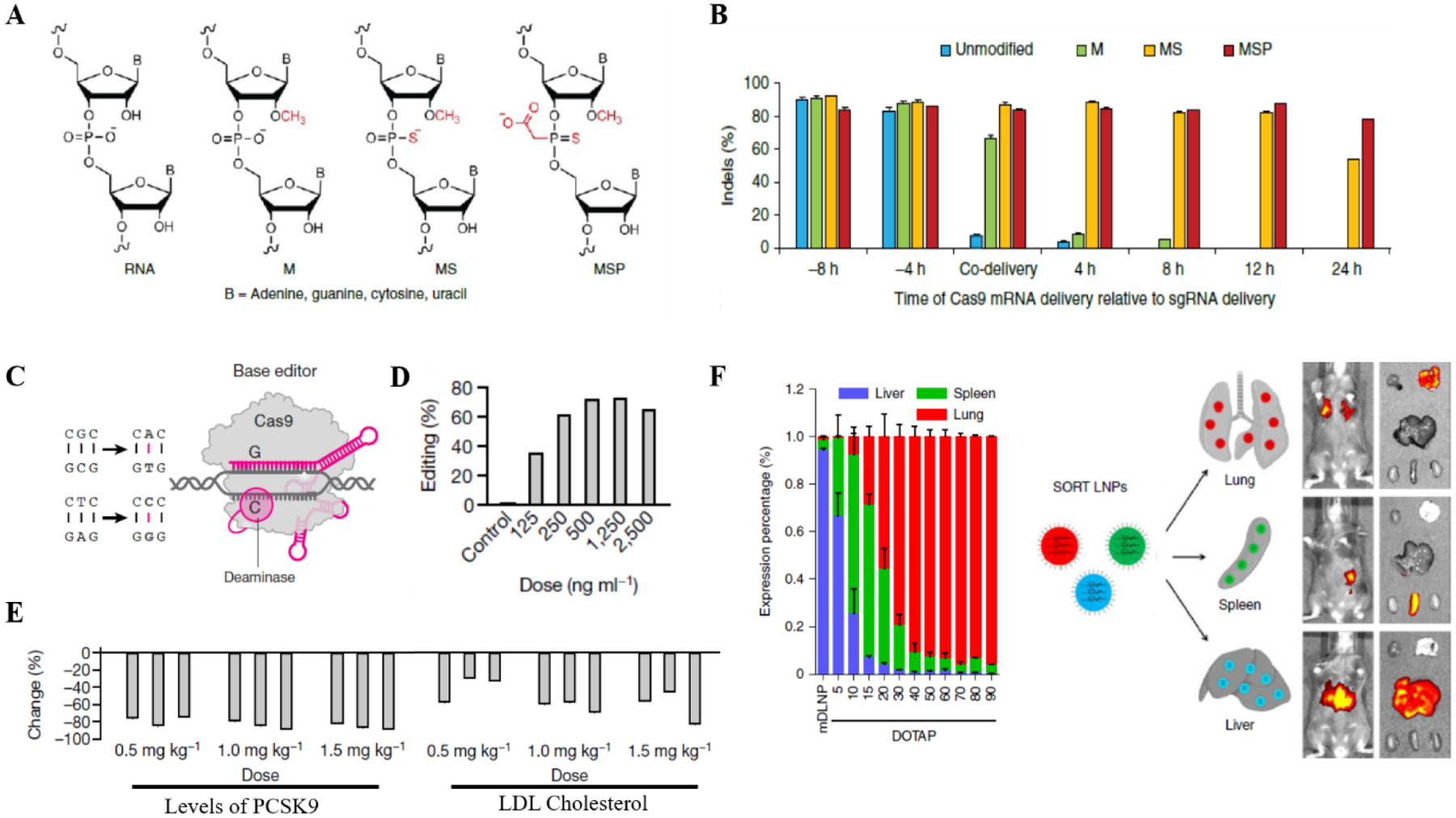
(A) Outline of the chemical modifications on the sgRNAs83. (B) Average indel frequencies as measured by tracking of indel by decomposition analysis upon delivery of modified sgRNAs83. (C) CRISPR base editor depicting a fused domain that replaces a single base through deamination and DNA replication/repair1. (D) Editing of the PCSK9 adenine base in primary human hepatocytes14 (E) Dose–response study, with liver PCSK9 editing showing reduction in the levels of PCSK9 and LDL cholesterol14. (F) Selective organ targeting of lipid nanoparticle22. Engineered LNPs to modulate their charge for accurate control of delivery into specific organs. Luciferase expression in each organ is shown to illustrate specific organ delivery. All the figures are adapted with permission from Elsevier Springer Nature.
CRISPR base-editors: Recent genetic screens have highlighted that several pathogenic alleles arise due to single nucleotide variations87,88. Thus, targeted and ‘hit-and-run’ CRISPR base editors have shown significant promises in several genetic disorders89. Cytosine base editors and adenine base editors are the critical genome engineering technologies that have enabled the precise installation of point mutations (Fig 5C). LNPs are preferred as delivery vehicles for the base editors due to their longer mRNA as compared to CRISPR nucleases. Furthermore, in tissues with slow turnover rate, LNPs based delivery leads to transient RNA expression and a lower probability of genome integration90. High tissue tropism and specificity of the LNP-RNPs are essential criteria for the safety of CRISPR base editing therapies. In recent work, on-target editing rates in nine different organs from macaques treated with 1.5mg/kg LNP-RNA complex were evaluated as less than 1%90. Only in the spleen, editing rates were in the range of 10 – 15 % in single- and repeat-dose treated animals, while hepatocytes exhibited 70–90% editing efficiency90 (Fig 5D,E). These efficiencies have been shown to be sufficient for therapeutic application in several genetic liver diseases, including urea cycle disorders, phenylketonuria and tyrosinemia. Moreover, it is conceivable that adjustment of dose levels and schedules could further increase editing rates. However, these approaches still need to be further optimized for targeting different tissues with higher efficacy. Some recent works have modified LNPs with antibodies to increase their tissue specificity, while a recent work, selective organ targeting (SORT) optimized the constituents of LNPs for targeted organ delivery22. The authors demonstrated that SORT is adaptable with various cargos, including mRNA, Cas9 mRNA/gRNA and RNPs for efficient editing in the lung, liver and spleen following intravenous (i.v.) administration (Fig 5F). Organ selective SORT LNPs resulted in 40% transfection efficiency in the epithelial cells and 65% in the endothelial cells. Additionally, 10–15% efficiency was observed for B-cells and T-cells, while over 90% transfection efficiency was achieved in the hepatocytes.
CRISPR based cytosine-base editors, adenine-base editors and prime editors have significantly advanced the treatment of diseases with single-nucleotide mutations. LNP-RNP (CRISPR-base editor) complex has been crucial in the delivery of base-editing RNPs. However, targeted organ delivery and tissue tropism remain a challenge that outweigh the advantages in the treatment of hereditary cardiovascular and neurological disorders. Rapidly evolving CRISPR-based platforms with minimized off-target efficacy such as CRISPRi, CRPSPRa, and prime-editing may offer key avenues for clinical trials once their specificity and delivery issues are addressed.
Chimeric antigen receptors:
Immunotherapy has emerged as a successful platform for personalized cancer treatment. Chimeric antigen receptor (CAR) is a synthetic construct that is expressed in T cells to mimic the T cell activation and to target them towards a specific antigen79,91. In 2017, FDA approved CD19 CAR-T cell therapy to treat relapsed acute lymphoblastic leukemia and large B cell Lymphoma. However, virally engineered CAR-T cells have major side effects, including cytokine storm and neurotoxicity caused by cerebral edemas that has resulted in several fatalities92. In this regard, mRNAs encoding CAR encapsulated by LNPs have been proposed for delivery into the human T cells, which ensures transient CAR expression and lower peripheral toxicity. In a recent study, researchers screened a library of lipids which were formulated into LNPs for mRNA delivery into T cells, showing efficient cytotoxicity of CAR-T cells engineered with mRNA-LNPs while exhibiting lower cytotoxicity to the T cells compared to electroporation93. Another study used mRNA to engineer CAR-T cells against chondroitin sulfate proteoglycan 4 (CSPG4) to treat melanoma94. Albeit with low in vivo efficacy, the investigators were able to demonstrate high levels of CAR-positive cells having high potency against melanoma.
In a recent work, mRNA that encodes fibroblast activation protein-CAR was encapsulated in LNPs conjugated with CD5-tageting antibodies to treat hypertensive cardiac injury95. The study demonstrated that mice with cardiac injury showed 17.5–24.7% FAPCAR+-T cell populations upon RNA-LNP injection. Three weeks post RNA-LNP injections, the animals had decreased interstitial fibrosis and improved cardiac function. This pioneering work paves way for further investigations of LNPs based delivery of mRNA/RNPs for in vivo human CAR therapeutic towards cardiovascular diseases and cancer. In patients with solid tumor, CAR-T cells have been shown to decline over time in circulation due to a lack of targets, affecting their therapeutic efficacy. Finding new targets such as Claudin6 and CAR-T ligands, and optimizing their in vivo efficacy and delivery are likely to initiate new clinical trails96,97.
Disease Outlook
Microfluidic fabrication of LNPs has found utility in diverse medical sciences, particularly in targeted nucleic acid delivery. We briefly highlight recent clinical advancements in cancer therapy, rare genetic diseases, and vaccine development.
Cancer therapeutics and vaccines:
LNPs have been extensively explored for cancer drug delivery applications. Doxorubicin was the first LNP encapsulated chemotherapeutic drug, which is currently being used to treat HIV, AIDS-related Kaposi’s sarcoma, and multiple myeloma78,98. Since then LNPs have been used for development of several chemotherapeutics, including DepoCyt to treat neoplastic meningitis and Abraxane for the treatment of cancer, while several other clinical trials are underway98. LNP based preparations were equivalent or more effective than conventional techniques used for the delivery of nucleic acid therapeutics99. Microfluidic based LNPs are also widely used as carriers for RNA interference (RNAi) based cancer therapeutics, that rely on the siRNA and miRNA pathway25,100. RNAi therapeutic targets to selectively reduce the levels of the protein of interest by either degrading their mRNA (siRNA) or by mitigating the translation of mRNA (miRNA)100,101. LNP-siRNA based therapeutic has been used against broad cancers for their effectiveness due to its long circulatory half-life of over 12 hours102.
Indeed, synergistic activity in retardation of tumor growth was observed in enzalutamide-resistant tumors when LNP-siRNA against clusterin was used with antisense oligonucleotides103. In addition, TKM080301, lipid encapsulated siRNA targeting polo-like kinase 1(PLK1), has shown anti-tumor activity in the mouse xenograft model and is currently under phase II trial for patients with advanced hepatocellular carcinoma (NCT02191878)104. Presently there are several clinical trials on LNP-RNAi complex, targeting metastatic pancreatic cancer (NCT01808638)105, lymphoma (NCT03323398)106, breast cancer (NCT02316457)107, recurrent glioblastoma (NCT02340156)108 and liver cancer (NCT02716012)109. Furthermore, studies have reported therapeutic activity of LNPs equipped with tumor-suppressive miRNA payloads, which are surface embedded with monoclonal antibodies against different subsets of leukocytes and cellular receptors that provide a potential use of this novel procedure for targeted drug delivery to a particular subset of cells110.
Cancer vaccines benefit from the feasibility in modulating the constituents of LNPs to target specific organs and prolong mRNA translation resulting in increased protein synthesis111–113. This helps in the efficient presentation of neoantigen and anti-tumor antigen expression in the immune cells. Thus, the LNP-mRNA complex favors sustained antigen availability during vaccination, which drives high antibody titer and immune cell response114,115. In recent work, intranasal LNP-mRNA delivery delayed tumor onset and increased survival in mouse prophylactic and therapeutic immunization models116. Interestingly, the authors demonstrated that the observed tumor immunity is limited to mice when mRNA is delivered in LNPs and correlates with splenic antigen-specific CD8+ T cells116. For efficient priming anti-tumor T-cells, systemic delivery of LNP-mRNA to the dendritic cell plays a critical role. In another work, engineered lipid-to-mRNA showed exclusive delivery of negatively charged LNP-mRNA complex to dendritic cells in lymphoid tissues20. Currently, several clinical trials have been initiated using this approach to treat advanced melanoma (Clinical trial number: NCT 02410733)117 and triple-negative breast cancer (Clinical trial number: NCT02316457)107. A deeper understanding of immune cell diversity and the ability to specifically deliver mRNA-LNPs to a subset of immune cells is expected to improve therapeutic efficacy while lowering immunogenicity related issues. Adoptive T cell therapies can be engineered with mRNA-LNPs, where subset of immunosuppressive T cell can be targeted and likely enhance the combination clinical efficacy of immune-checkpoint blockade therapies. These methods will evolve based on a deeper understanding of tissue immune architecture using scRNA sequencing and cytometry-time-of-flight to target novel immune cell population and receptors.
Rare genetic diseases:
In the case of rare genetic diseases, protein replacement therapeutic intervention based on in vitro transcribed (IVT) mRNAs delivered by LNPs fabricated by microfluidics has yielded successful results118,119. The rationale relies on the synthesis of deficient/down-regulated proteins from delivered mRNA. For instance, 26 genes were successfully encoded in animal models for William-Beuren syndrome (WBS), which results from the microdeletion of chromosomal 7q11.23 fragment120,121. Recently some researchers have proposed that modification of mRNA can determine the subcellular localization of encoded proteins, and hence distal organs like the liver can be used as repository/production depot for therapeutically active proteins122,123. LNP based siRNA formulation (Onpattro) has been FDA approved to treat transthyretin-mediated amyloidosis (ATTR), a fatal genetic condition characterized by accumulation of amyloid fibrils124. Recently, LNPs based CRISPR formulation (NTLA-2001) targeting ATTR in clinical phase I trial demonstrated up to 87% reduction in toxic TTR protein levels at 0.3mg/kg125. Several RNAi-based drugs are under trial to treat hypercholesterolemia (Proprotein convertase subtilisin/kexin type 9 serine protease; PCSK9) and autosomal dominant familial disease. CRISPR base editor has shown significant potential towards gene editing and recently advanced the treatment of familial hypercholesterolemia with LOF mutations in LDLR or GOF mutations in PCSK9. Using LNP based CRISPR base editor, authors demonstrated editing rates of PCSK9 up to 80% in mouse liver hepatocytes14,90. These emerging genome engineering technologies have shown tremendous results in the treatment of genetic disorders. CRISPR interference, CRISPR activation and CRISPR base and prime editing have been the cornerstone of these technologies and their efficient organ delivery and potency remains to be fully addressed before they can be employed for human trials. mRNA-LNPs can also be tailored for protein replacement therapy for treatment of hereditary metabolic disorders such as methylmalonic acidemia and propionic acidemia. Moreover, broad spectrum diseases, such as glycogen storage disease and hematological diseases provide ample opportunities of LNP-mRNA/RNP based therapeutics.
Vaccines for Infectious Diseases:
Infectious diseases are one of the significant contributors to global mortality rates and have had a severe impact on healthcare and socio-economic development. Bacterial and viral pathogens are the leading causes of widespread diseases, where the live attenuated and inactivated pathogen-based vaccine approach has shown successful outcome. However, these conventional vaccines face hurdles against rapidly evolving and immune evading pathogens. Furthermore, non-infectious diseases such as cancer may not benefit from the conventional vaccine approach that rely on live attenuated and inactivated pathogens. Tumors have broad range of genomic alterations with only a small fraction shared among patients along with altered immune microenvironment which makes them difficult candidates for conventional vaccine development. Therefore, many efforts have been made to develop mRNA-based vaccines as they have several advantages over the conventional approaches, including ease of mRNA synthesis and modification, controlled toxicity profile, regulated stability and durable expression.
Moreover, it has been reported that the SARS-CoV2 mRNA-based vaccines BNT162b2 and mRNA-1273 are easily scalable and inexpensive.
mRNA vaccines encode for the pathogenic antigens and also induce strong CD8+ and CD4+ T cell responses. These vaccines can generate a higher titer of neutralizing antibody with a lower immunization112,113. Thus, mRNA vaccines have shown to provide immunity against several viral outbreaks in the last decade, including Ebola, Zika and SARS viruses126. However, as previously discussed, mRNAs are unstable in the blood and reflect a poor cellular uptake127. LNPs have evolved as the major carriers for the delivery of mRNA-based vaccines. Currently, mRNA containing LNPs are being clinically tested like the mRNA-1647 encoding 6 CMV protein is in trial for cytomegalovirus treatment, VAL-339851 (mRNA-1851), VAL-506440 (mRNA-1440) coding H7 and H10 region of influenza, VAL-181388 (mRNA-1388) which codes for CHIKV proteins for Chikungunya virus therapy, and mRNA-1325 coding for Zika-virus proteins to cure Zika virus disease31. In addition, the recently approved and widely used SARS-CoV2 vaccines from BioNTech/Pfizer (BNT162b2) and Moderna (mRNA-1273) also rely on the LNPs for mRNA delivery127.
Self-amplifying mRNAs (sa-RNA) are another class of mRNAs that can replicate based on the same template to enable high-expression and translation of mRNA. These sa-RNA-LNP vaccines have been explored in the treatment of non-viral infection, such as bacterial and parasite infections121,128,129. It is expected that engineering mRNA-LNPs in the future to target broad spectrum bacterial diseases and induce memory immune response will prove beneficial in long term in low-income countries.
Conclusion:
Non-viral gene delivery platforms have emerged as a preferred vehicle for therapeutic nucleic acid administration. Among these, LNPs are being explored extensively for their scalability towards GMP synthesis, where microfluidics has been used to efficiently synthesize LNPs with low polydispersity index, resulting in homogenous sizes eventually leading to higher targeting efficiency and enhanced biodistribution. As discussed, recent works have modified the LNPs to bolster tissue specificity via lipid modification or bioconjugation with antibodies for cell specific delivery. Their role in vaccine formulations which garnered FDA approval for SARS-CoV2 vaccine has been a pivotal in extending these LNP-RNA technology to spectrum of the diseases. Several clinical trials are now underway with LNPs as carrier of genome engineering tools for therapeutic purposes in cancer, viral, and rare gene disorders. Since efficacy and safety profiles of LNP-mRNAs have been well documented since the SARS-CoV2 vaccine approvals, they can be readily expanded to address cell engineering bottleneck for therapeutic applications. We opine that the next few years will see LNPs being developed for increased retention of the genetic payload, higher targeting efficiency, and less biodistribution. We predict these LNP-mRNA complexes will prove crucial for in vivo cellular reprogramming which will be beneficial in reprogramming fibroblast and stem cells to diverse cell types depending on the disease phenotype130–132. Although several advances have been made in this regard, however, cell type specific delivery remains a challenge. Understanding of tissue cell composition with single cell RNA sequencing technology is expected to guide vaccine development and therapeutics for diseases such as rheumatoid arthritis, multiple sclerosis and aging related issues. In the age of ‘omics’, several population-wide studies have been conducted, interrogating genomic loci responsible for disease susceptibility. Integrating these omics data with a rational design of LNP-CRISPR gene-editing using machine learning and AI will be crucial in drug development89,133,134. Further, AI based technologies have significantly enhanced the high-throughput and optimized the LNP, while reducing the production error and costs134. Thus, streamlined integration of the AI technologies for LNP synthesis along with deconvolution of disease associated gene therapy will hold the key for future personalized therapeutics.
To conclude, nucleic acid-LNP based therapeutics hold tremendous potential in personalized medicine that can be explored in a broad spectrum of applications. The development and engineering of novel LNPs will further support the accuracy and development of CRISPR and other gene-editing based tools to improve several aspects of biology and healthcare.
Table 2.
Lipid nanoparticle-based therapeutics and vaccines against cancer, rare diseases, and viruses in clinical trial.
| Clinical Trial Identifier | |||
|---|---|---|---|
| LNPs-mRNA based therapeutics in clinical trials against cancer and rare disease | |||
| Ovarian cancer | |||
| Relapsed/Refractory solid tumor or lymphoma | mRNA-2752 - LNPs encapsulating mRNA encoding OX40L, IL23 and IL36Y, alone or in combination with immune checkpoint blockade | Phase I | NCT03739931 |
| Hepatocellular Cancer | NCT01437007 | ||
| Transthyretin amyloidosis with polyneuropathy | NTLA-2001 - CRISPR/Cas9 based gene editing with guide RNA against TTR | Phase I | NCT04601051 |
| Methylmalonic Acidemia | mRNA-3704 - LNPs encapsulating mRNA encoding human methylamalonyl-CoA mutase | Phase I/II | NCT03810690 |
| Propionic Acidemia | mRNA-3927 - LNPs encapsulating mRNA encoding alpha and beta subunits of mitochondrial enzyme propionyl-CoA carboxylase | Phase I/II | NCT04159103 |
| Cystic Fibrosis | MRT5005 - LNPs encapsulating mRNA encoding Cystic fibrosis transmembrane conductance regulator (CFTR) protein. | Phase I/II | NCT03375047 |
| LNPs-mRNA based vaccines in clinical trials against cancer and viruses | |||
| Pancreatic Adenocarcinoma | |||
| Triple Negative Breast Cancer | TNBC-MERIT - Individualized cancer immunotherapy, immunogenic RNA vaccine | Phase I | NCT02316457 |
| Stage III or IV Melanoma | BNT 111 - LNPs encapsulating mRNA encoding fixed set of four cancer specific antigen alone or in combination with cemiplimab | Phase II | NCT04526899 |
| Resected solid tumors | mRNA-4157 - Personalized cancer vaccine targeting twenty tumor-based antigens identified from each patient alone or in combination with pembrolizumab | Phase I | NCT03313778 |
| Locally advanced or metastatic tumors | RO7198457 - Contains up to 20 patients specific neoantigens (alone or in combination with atezolizumab) | Phase I | NCT03289961 |
| Influenza H10N8 | mRNA-1440 - LNPs encapsulating mRNA encoding for the membrane bound hemagglutinin (H10) protein | Phase I | NCT03076385 |
| Influenza H7N9 | mRNA-1851 - LNPs encapsulating mRNA encoding for the membrane bound hemagglutinin (H7) protein | Phase I | NCT03345043 |
| Zika Virus | mRNA-1893 - LNPs encapsulating mRNA encoding for the structural proteins of Zika Virus | Phase I | NCT04064905 |
| Cytomegalovirus | mRNA-1647 - LNPs encapsulating mRNA encoding for six different mRNA. Five mRNAs encode for CMV pentamer complex and one encode for glycoprotein B | Phase III | NCT05085366 |
| Chikungunya Virus | mRNA-1388 - LNPs encapsulating mRNA encoding for viral antigenic proteins associated with CHIKV | Phase I | NCT03325075 |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Doudna JA The promise and challenge of therapeutic genome editing. Nature 578, 229–236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anzalone AV, Koblan LW & Liu DR Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol 38, 824–844 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Knott GJ & Doudna JA CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim D, Luk K, Wolfe SA & Kim J-S Evaluating and Enhancing Target Specificity of Gene-Editing Nucleases and Deaminases. Annu. Rev. Biochem 88, 191–220 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Christopher Boyd A et al. New approaches to genetic therapies for cystic fibrosis. J. Cyst. Fibros 19, S54–S59 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Wu Y et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat. Med 25, 776–783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amoasii L et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 362, 86–91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richner JM et al. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell 169, 176 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Aldosari BN, Alfagih IM & Almurshedi AS Lipid Nanoparticles as Delivery Systems for RNA-Based Vaccines. Pharmaceutics 13, 206 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbett KS et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M, Glass ZA & Xu Q Non-viral delivery of genome-editing nucleases for gene therapy. Gene Ther. 24, 144–150 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Bulcha JT, Wang Y, Ma H, Tai PWL & Gao G Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther 6, 1–24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finn JD et al. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 22, 2227–2235 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Musunuru K et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature 593, 429–434 (2021). [DOI] [PubMed] [Google Scholar]
- 15.High-dose AAV gene therapy deaths. Nat. Biotechnol 38, 910–910 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Song C-Q et al. Adenine base editing in an adult mouse model of tyrosinaemia. Nat. Biomed. Eng 4, 125–130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Eygeris Y, Gupta M & Sahay G Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev 170, 83–112 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenblum D et al. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci. Adv 6, eabc9450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veiga N et al. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun 9, 4493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kranz LM et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Krienke C et al. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science (2021) doi: 10.1126/science.aay3638. [DOI] [PubMed] [Google Scholar]
- 22.Cheng Q et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol 15, 313–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR–Cas gene editing. Nat. Mater 20, 701–710 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomeh MA & Zhao X Recent Advances in Microfluidics for the Preparation of Drug and Gene Delivery Systems. Mol. Pharm 17, 4421–4434 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Titze-de-Almeida R, David C & Titze-de-Almeida SS The Race of 10 Synthetic RNAi-Based Drugs to the Pharmaceutical Market. Pharm. Res 34, 1339–1363 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Shim G et al. Therapeutic gene editing: delivery and regulatory perspectives. Acta Pharmacol. Sin 38, 738–753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang K-W et al. Highly efficient and tumor-selective nanoparticles for dual-targeted immunogene therapy against cancer. Sci. Adv 6, eaax5032 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeki M, Kimura N, Sato Y, Harashima H & Tokeshi M Advances in microfluidics for lipid nanoparticles and extracellular vesicles and applications in drug delivery systems. Adv. Drug Deliv. Rev 128, 84–100 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Miao L, Satterlee A & Huang L Delivery of oligonucleotides with lipid nanoparticles. Recent Dev. Oligonucleotide Based Ther 87, 68–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen TM & Cullis PR Liposomal drug delivery systems: From concept to clinical applications. Adv. Drug Deliv. Perspect. Prospects 65, 36–48 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Kulkarni JA, Cullis PR & van der Meel R Lipid Nanoparticles Enabling Gene Therapies: From Concepts to Clinical Utility. Nucleic Acid Ther. 28, 146–157 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Witzigmann D et al. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv. Drug Deliv. Rev 159, 344–363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraley R, Subramani S, Berg P & Papahadjopoulos D Introduction of liposome-encapsulated SV40 DNA into cells. J. Biol. Chem 255, 10431–10435 (1980). [PubMed] [Google Scholar]
- 34.Whitehead KA, Langer R & Anderson DG Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov 8, 129–138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo X et al. Transfection reagent Lipofectamine triggers type I interferon signaling activation in macrophages. Immunol. Cell Biol 97, 92–96 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Allen TM The use of glycolipids and hydrophilic polymers in avoiding rapid uptake of liposomes by the mononuclear phagocyte system. Adv. Drug Deliv. Rev 13, 285–309 (1994). [Google Scholar]
- 37.Chen S et al. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Controlled Release 235, 236–244 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Nakamura T et al. The Effect of Size and Charge of Lipid Nanoparticles Prepared by Microfluidic Mixing on Their Lymph Node Transitivity and Distribution. Mol. Pharm (2020) doi: 10.1021/acs.molpharmaceut.9b01182. [DOI] [PubMed] [Google Scholar]
- 39.Sato Y et al. Elucidation of the physicochemical properties and potency of siRNA-loaded small-sized lipid nanoparticles for siRNA delivery. J. Controlled Release (2016) doi: 10.1016/j.jconrel.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 40.Cabral H et al. Accumulation of sub-100 nm polymeric micelles in poorly permeable tumours depends on size. Nat. Nanotechnol (2011) doi: 10.1038/nnano.2011.166. [DOI] [PubMed] [Google Scholar]
- 41.Gaumet M, Vargas A, Gurny R & Delie F Nanoparticles for drug delivery: The need for precision in reporting particle size parameters. European Journal of Pharmaceutics and Biopharmaceutics (2008) doi: 10.1016/j.ejpb.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Tadros T, Izquierdo P, Esquena J & Solans C Formation and stability of nano-emulsions. Adv. Colloid Interface Sci (2004) doi: 10.1016/j.cis.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 43.Wei Y, Quan L, Zhou C & Zhan Q Factors relating to the biodistribution & clearance of nanoparticles & their effects on in vivo application. Nanomedicine (2018) doi: 10.2217/nnm-2018-0040. [DOI] [PubMed] [Google Scholar]
- 44.Hoshyar N, Gray S, Han H & Bao G The effect of nanoparticle size on in vivo pharmacokinetics and cellular interaction. Nanomedicine (2016) doi: 10.2217/nnm.16.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kupetz E & Bunjes H Lipid nanoparticles: Drug localization is substance-specific and achievable load depends on the size and physical state of the particles. J. Controlled Release (2014) doi: 10.1016/j.jconrel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 46.Mehnert W & Mäder K Solid lipid nanoparticles: Production, characterization and applications. Advanced Drug Delivery Reviews (2012) doi: 10.1016/j.addr.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 47.Hou DZ, Xie CS, Huang KJ & Zhu CH The production and characteristics of solid lipid nanoparticles (SLNs). Biomaterials (2003) doi: 10.1016/S0142-9612(02)00578-1. [DOI] [PubMed] [Google Scholar]
- 48.Joseph S & Bunjes H Solid Lipid Nanoparticles for Drug Delivery. in Drug Delivery Strategies for Poorly Water-Soluble Drugs (2013). doi: 10.1002/9781118444726.ch4. [DOI] [Google Scholar]
- 49.Liu Y, Xie P, Zhang D & Zhang Q A mini review of nanosuspensions development. Journal of Drug Targeting (2012) doi: 10.3109/1061186X.2011.645161. [DOI] [PubMed] [Google Scholar]
- 50.Roces CB et al. Manufacturing Considerations for the Development of Lipid Nanoparticles Using Microfluidics. Pharmaceutics 12, 1095 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webb C et al. Using microfluidics for scalable manufacturing of nanomedicines from bench to GMP: A case study using protein-loaded liposomes. Int. J. Pharm 582, 119266 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Lorenz T, Bojko S, Bunjes H & Dietzel A An inert 3D emulsification device for individual precipitation and concentration of amorphous drug nanoparticles. Lab. Chip (2018) doi: 10.1039/c7lc01313b. [DOI] [PubMed] [Google Scholar]
- 53.Zhigaltsev IV et al. Bottom-up design and synthesis of limit size lipid nanoparticle systems with aqueous and triglyceride cores using millisecond microfluidic mixing. Langmuir (2012) doi: 10.1021/la204833h. [DOI] [PubMed] [Google Scholar]
- 54.Shepherd SJ, Issadore D & Mitchell MJ Microfluidic formulation of nanoparticles for biomedical applications. Biomaterials 274, 120826 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian F, Cai L, Liu C & Sun J Microfluidic Technologies for Nanoparticle Formation. Lab. Chip (2021) doi: 10.1039/D1LC00812A. [DOI] [PubMed] [Google Scholar]
- 56.Schubert MA & Müller-Goymann CC Solvent injection as a new approach for manufacturing lipid nanoparticles - Evaluation of the method and process parameters. Eur. J. Pharm. Biopharm (2003) doi: 10.1016/S0939-6411(02)00130-3. [DOI] [PubMed] [Google Scholar]
- 57.Hu FQ, Yuan H, Zhang HH & Fang M Preparation of solid lipid nanoparticles with clobetasol propionate by a novel solvent diffusion method in aqueous system and physicochemical characterization. Int. J. Pharm (2002) doi: 10.1016/S0378-5173(02)00081-9. [DOI] [PubMed] [Google Scholar]
- 58.Dietzel A A brief introduction to microfluidics. in Microsystems for Pharmatechnology: Manipulation of Fluids, Particles, Droplets, and Cells (2016). doi: 10.1007/978-3-319-26920-7_1. [DOI] [Google Scholar]
- 59.Dong Y, Ng WK, Shen S, Kim S & Tan RBH Solid lipid nanoparticles: Continuous and potential large-scale nanoprecipitation production in static mixers. Colloids Surf. B Biointerfaces (2012) doi: 10.1016/j.colsurfb.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 60.Maeki M et al. A strategy for synthesis of lipid nanoparticles using microfluidic devices with a mixer structure. RSC Adv. (2015) doi: 10.1039/c5ra04690d. [DOI] [Google Scholar]
- 61.Chen D et al. Rapid discovery of potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. J. Am. Chem. Soc (2012) doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 62.Kimura N et al. Development of the iLiNP Device: Fine Tuning the Lipid Nanoparticle Size within 10 nm for Drug Delivery. ACS Omega (2018) doi: 10.1021/acsomega.8b00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheung CCL & Al-Jamal WT Sterically stabilized liposomes production using staggered herringbone micromixer: Effect of lipid composition and PEG-lipid content. Int. J. Pharm (2019) doi: 10.1016/j.ijpharm.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 64.Erfle P, Riewe J, Bunjes H & Dietzel A Optically monitored segmented flow for controlled ultra-fast mixing and nanoparticle precipitation. Microfluid. Nanofluidics (2017) doi: 10.1007/s10404-017-2016-2. [DOI] [Google Scholar]
- 65.Shao N, Gavriilidis A & Angeli P Flow regimes for adiabatic gas-liquid flow in microchannels. Chemical Engineering Science (2009) doi: 10.1016/j.ces.2009.01.067. [DOI] [Google Scholar]
- 66.Xu L et al. Formulation of poorly water-soluble compound loaded solid lipid nanoparticles in a microchannel system fabricated by mechanical microcutting method: Puerarin as a model drug. Ind. Eng. Chem. Res (2012) doi: 10.1021/ie300592u. [DOI] [Google Scholar]
- 67.Richter C, Krah T & Büttgenbach S Novel 3D manufacturing method combining microelectrial discharge machining and electrochemical polishing. in Microsystem Technologies (2012). doi: 10.1007/s00542-012-1452-x. [DOI] [Google Scholar]
- 68.Melzig S et al. Fluid mechanics and process design of high-pressure antisolvent precipitation of fenofibrate nanoparticles using a customized microsystem. Chem. Eng. J (2019) doi: 10.1016/j.cej.2019.04.051. [DOI] [Google Scholar]
- 69.Carugo D, Bottaro E, Owen J, Stride E & Nastruzzi C Liposome production by microfluidics: Potential and limiting factors. Sci. Rep (2016) doi: 10.1038/srep25876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang S. h. et al. Formation of solid lipid nanoparticles in a microchannel system with a cross-shaped junction. Chem. Eng. Sci (2008) doi: 10.1016/j.ces.2008.08.005. [DOI] [Google Scholar]
- 71.Riewe J et al. Antisolvent precipitation of lipid nanoparticles in microfluidic systems – A comparative study. Int. J. Pharm (2020) doi: 10.1016/j.ijpharm.2020.119167. [DOI] [PubMed] [Google Scholar]
- 72.Shepherd SJ et al. Scalable mRNA and siRNA Lipid Nanoparticle Production Using a Parallelized Microfluidic Device. Nano Lett. (2021) doi: 10.1021/acs.nanolett.1c01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moss KH, Popova P, Hadrup SR, Astakhova K & Taskova M Lipid Nanoparticles for Delivery of Therapeutic RNA Oligonucleotides. Mol. Pharm 16, 2265–2277 (2019). [DOI] [PubMed] [Google Scholar]
- 74.Suzuki Y et al. Lipid nanoparticles loaded with ribonucleoprotein–oligonucleotide complexes synthesized using a microfluidic device exhibit robust genome editing and hepatitis B virus inhibition. J. Controlled Release (2021) doi: 10.1016/j.jconrel.2020.12.013. [DOI] [PubMed] [Google Scholar]
- 75.Shrimal P, Jadeja G & Patel S A review on novel methodologies for drug nanoparticle preparation: Microfluidic approach. Chem. Eng. Res. Des 153, 728–756 (2020). [Google Scholar]
- 76.Cao S et al. Nanoparticles: Oral Delivery for Protein and Peptide Drugs. AAPS PharmSciTech 20, 190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hou X, Zaks T, Langer R & Dong Y Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater 6, 1078–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hassan S et al. Evolution and clinical translation of drug delivery nanomaterials. Nano Today 15, 91–106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mukalel AJ, Riley RS, Zhang R & Mitchell MJ Nanoparticles for nucleic acid delivery: Applications in cancer immunotherapy. Cancer Lett. 458, 102–112 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin H et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol 35, 1179–1187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Deleavey GF & Damha MJ Designing Chemically Modified Oligonucleotides for Targeted Gene Silencing. Chem. Biol 19, 937–954 (2012). [DOI] [PubMed] [Google Scholar]
- 82.Behlke MA Chemical Modification of siRNAs for In Vivo Use. Oligonucleotides 18, 305–320 (2008). [DOI] [PubMed] [Google Scholar]
- 83.Hendel A et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol 33, 985–989 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nelson J et al. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv (2020) doi: 10.1126/sciadv.aaz6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pardi N et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 543, 248–251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bahl K et al. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol. Ther 25, 1316–1327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Landrum MJ et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44, D862–D868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stenson PD et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet 136, 665–677 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arbab M et al. Determinants of Base Editing Outcomes from Target Library Analysis and Machine Learning. Cell 182, 463–480.e30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rothgangl T et al. In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels. Nat. Biotechnol 1–9 (2021) doi: 10.1038/s41587-021-00933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guevara ML, Persano F & Persano S Advances in Lipid Nanoparticles for mRNA-Based Cancer Immunotherapy. Front. Chem 8, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Clinical development of CAR T cells—challenges and opportunities in translating innovative treatment concepts. EMBO Mol. Med 9, 1183–1197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Billingsley MM et al. Ionizable Lipid Nanoparticle-Mediated mRNA Delivery for Human CAR T Cell Engineering. Nano Lett. 20, 1578–1589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wiesinger M et al. Clinical-Scale Production of CAR-T Cells for the Treatment of Melanoma Patients by mRNA Transfection of a CSPG4-Specific CAR under Full GMP Compliance. Cancers 11, 1198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rurik JG et al. CAR T cells produced in vivo to treat cardiac injury. Science (2022) doi: 10.1126/science.abm0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma L et al. Enhanced CAR-T cell activity against solid tumors by vaccine boosting through the chimeric receptor. Science 365, 162–168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cully M Driving CARs to last. Nat. Rev. Drug Discov 19, 91–91 (2020). [DOI] [PubMed] [Google Scholar]
- 98.Beltrán-Gracia E, López-Camacho A, Higuera-Ciapara I, Velázquez-Fernández JB & Vallejo-Cardona AA Nanomedicine review: clinical developments in liposomal applications. Cancer Nanotechnol. 10, 11 (2019). [Google Scholar]
- 99.Allen TM & Cullis PR Drug Delivery Systems: Entering the Mainstream. Science 303, 1818–1822 (2004). [DOI] [PubMed] [Google Scholar]
- 100.Setten RL, Rossi JJ & Han S The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov 18, 421–446 (2019). [DOI] [PubMed] [Google Scholar]
- 101.Tian Z et al. Insight Into the Prospects for RNAi Therapy of Cancer. Front. Pharmacol 0, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee JB et al. A Glu-urea-Lys Ligand-conjugated Lipid Nanoparticle/siRNA System Inhibits Androgen Receptor Expression In Vivo. Mol. Ther. - Nucleic Acids 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yamamoto Y et al. siRNA Lipid Nanoparticle Potently Silences Clusterin and Delays Progression When Combined with Androgen Receptor Cotargeting in Enzalutamide-Resistant Prostate Cancer. Clin. Cancer Res 21, 4845–4855 (2015). [DOI] [PubMed] [Google Scholar]
- 104.Semple SC et al. Abstract 2829: Preclinical characterization of TKM-080301, a lipid nanoparticle formulation of a small interfering RNA directed against polo-like kinase 1. Cancer Res. 71, 2829 (2011). [Google Scholar]
- 105.Schultheis B et al. First-in-Human Phase I Study of the Liposomal RNA Interference Therapeutic Atu027 in Patients With Advanced Solid Tumors. J. Clin. Oncol 32, 4141–4148 (2014). [DOI] [PubMed] [Google Scholar]
- 106.Jimeno A et al. Abstract CT032: A phase 1/2, open-label, multicenter, dose escalation and efficacy study of mRNA-2416, a lipid nanoparticle encapsulated mRNA encoding human OX40L, for intratumoral injection alone or in combination with durvalumab for patients with advanced malignancies. Cancer Res. 80, CT032–CT032 (2020). [Google Scholar]
- 107.Schmidt M et al. 88MO T-cell responses induced by an individualized neoantigen specific immune therapy in post (neo)adjuvant patients with triple negative breast cancer. Ann. Oncol 31, S276 (2020). [Google Scholar]
- 108.51. Results of a Phase I Trial of SGT-53: A Systemically Administered, Tumor-Targeting Immunoliposome Nanocomplex Incorporating a Plasmid Encoding wtp53. Mol. Ther 20, S21 (2012). [Google Scholar]
- 109.Sarker D et al. MTL-CEBPA, a Small Activating RNA Therapeutic Upregulating C/EBP-α, in Patients with Advanced Liver Cancer: A First-in-Human, Multicenter, Open-Label, Phase I Trial. Clin. Cancer Res 26, 3936–3946 (2020). [DOI] [PubMed] [Google Scholar]
- 110.Van Hoecke L & Roose K How mRNA therapeutics are entering the monoclonal antibody field. J. Transl. Med 17, 54 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fiedler K, Lazzaro S, Lutz J, Rauch S & Heidenreich R mRNA Cancer Vaccines. in Current Strategies in Cancer Gene Therapy (ed. Walther W) 61–85 (Springer International Publishing, 2016). doi: 10.1007/978-3-319-42934-2_5. [DOI] [PubMed] [Google Scholar]
- 112.Aldosari BN, Alfagih IM & Almurshedi AS Lipid Nanoparticles as Delivery Systems for RNA-Based Vaccines. Pharmaceutics 13, 206 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pardi N, Hogan MJ, Porter FW & Weissman D mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pardi N et al. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Controlled Release 217, 345–351 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tam HH et al. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proc. Natl. Acad. Sci 113, E6639–E6648 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Phua KKL, Staats HF, Leong KW & Nair SK Intranasal mRNA nanoparticle vaccination induces prophylactic and therapeutic anti-tumor immunity. Sci. Rep 4, 5128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Loquai C et al. A shared tumor-antigen RNA-lipoplex vaccine with/without anti-PD1 in patients with checkpoint-inhibition experienced melanoma. J. Clin. Oncol 38, 3136–3136 (2020). [Google Scholar]
- 118.Sahu I, Haque AKMA, Weidensee B, Weinmann P & Kormann MSD Recent Developments in mRNA-Based Protein Supplementation Therapy to Target Lung Diseases. Mol. Ther 27, 803–823 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Magadum A, Kaur K & Zangi L mRNA-Based Protein Replacement Therapy for the Heart. Mol. Ther 27, 785–793 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.DeRosa F et al. Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther. 23, 699–707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Berraondo P, Martini PGV, Avila MA & Fontanellas A Messenger RNA therapy for rare genetic metabolic diseases. Gut 68, 1323 (2019). [DOI] [PubMed] [Google Scholar]
- 122.Svitkin YV et al. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 45, 6023–6036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Anderson BR et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 38, 5884–5892 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Akinc A et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol 14, 1084–1087 (2019). [DOI] [PubMed] [Google Scholar]
- 125.Gillmore JD et al. CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. N. Engl. J. Med 0, null. [DOI] [PubMed] [Google Scholar]
- 126.Zhang C, Maruggi G, Shan H & Li J Advances in mRNA Vaccines for Infectious Diseases. Front. Immunol 10, 594 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Let’s talk about lipid nanoparticles. Nat. Rev. Mater 6, 99–99 (2021). [Google Scholar]
- 128.Versteeg L, Almutairi MM, Hotez PJ & Pollet J Enlisting the mRNA Vaccine Platform to Combat Parasitic Infections. Vaccines 7, 122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Baeza Garcia A et al. Neutralization of the Plasmodium-encoded MIF ortholog confers protective immunity against malaria infection. Nat. Commun 9, 2714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gallego-Perez D et al. Topical tissue nano-transfection mediates non-viral stroma reprogramming and rescue. Nat. Nanotechnol 12, 974–979 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jin Y et al. Triboelectric Nanogenerator Accelerates Highly Efficient Nonviral Direct Conversion and In Vivo Reprogramming of Fibroblasts to Functional Neuronal Cells. Adv. Mater 28, 7365–7374 (2016). [DOI] [PubMed] [Google Scholar]
- 132.Wang S, Hashemi S, Stratton S & Arinzeh TL The Effect of Physical Cues of Biomaterial Scaffolds on Stem Cell Behavior. Adv. Healthc. Mater 10, 2001244 (2021). [DOI] [PubMed] [Google Scholar]
- 133.Egorov E, Pieters C, Korach-Rechtman H, Shklover J & Schroeder A Robotics, microfluidics, nanotechnology and AI in the synthesis and evaluation of liposomes and polymeric drug delivery systems. Drug Deliv. Transl. Res 11, 345–352 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu L et al. Artificial intelligence-powered microfluidics for nanomedicine and materials synthesis. Nanoscale 13, 19352–19366 (2021). [DOI] [PubMed] [Google Scholar]


