Abstract
Cyanovirin-N (CV-N), an 11-kDa protein originally isolated from the cyanobacterium Nostoc ellipsosporum, potently inactivates diverse strains of human immunodeficiency virus type 1 (HIV-1), HIV-2, simian immunodeficiency virus, and feline immunodeficiency virus. It has been well established that the HIV surface envelope glycoprotein gp120 is a molecular target of CV-N. We recently reported that CV-N impaired the binding of virion-associated gp120 to cell-associated CD4 and that CV-N preferentially inhibited binding of the glycosylation-dependent neutralizing monoclonal antibody 2G12 to gp120. However, CV-N did not interfere with the interactions of soluble CD4 (sCD4) with either soluble gp120 (sgp120) or virion-associated gp120. In the present study, we have evaluated the effects of CV-N on the binding of sgp120 to cell-associated CD4 to clarify the experimental basis of the previous binding results, and we further address the detailed mechanism of action of CV-N. Here we present evidence that (i) CV-N impairs both CD4-dependent and CD4-independent binding of sgp120 to the target cells, (ii) CV-N blocks the sCD4-induced binding of sgp120 with cell-associated coreceptor CXCR4, and (iii) CV-N dissociates bound sgp120 from target cells. The results illustrate that the measured effects of CV-N on gp120-CD4 binding interactions depend upon the type of CD4 (soluble or cell associated), but not upon the type of gp120 (soluble or virion associated), employed in the experimental protocol. In addition, this study reinforces that CV-N acts uniquely to prevent essential interactions between the envelope glycoprotein and target cell receptors and further supports the potential broad utility of CV-N as a microbicide to prevent the transmission of HIV and AIDS.
Cyanovirin-N (CV-N) is a potent human immunodeficiency virus (HIV)-inactivating, 11-kDa protein originally isolated from an aqueous extract of the cyanobacterium Nostoc ellipsosporum (6, 14). Recombinant CV-N that is indistinguishable from natural CV-N has subsequently been produced in Escherichia coli (27). In contrast to soluble CD4 (sCD4) and most known neutralizing antibodies against the HIV envelope glycoprotein gp120, CV-N exerts broad virucidal activity, at low nanomolar concentrations, against both primary isolates and laboratory-adapted strains of primate immunodeficiency retroviruses. These include both T-lymphocyte-tropic, macrophage-tropic and dual-tropic primary clinical isolates of HIV type 1 (HIV-1), as well as laboratory-adapted strains of HIV-1, HIV-2, simian immunodeficiency virus, and feline immunodeficiency virus (6, 10). High concentrations (e.g., 9,000 nM) of CV-N are not lethal to cell cultures. CV-N is extremely resistant to physicochemical degradation and can withstand treatment with denaturants, detergents, organic solvents, multiple freeze-thaw cycles, and heat (up to 100°C) with no apparent loss of antiviral activity (6). Both the nuclear magnetic resonance solution structure and the corresponding X-ray crystallographic analysis of recombinant CV-N have been published, revealing a largely β-sheet protein with twofold pseudosymmetry (5, 38). These unique characteristics of CV-N have encouraged ongoing development of this protein as an anti-HIV microbicide, particularly to prevent sexual transmission of HIV (6, 13). CV-N may also have therapeutic applications. For example, to explore one such approach, we have successfully demonstrated the feasibility of using CV-N as a gp120-targeting entity coupled to a cytotoxin (Pseudomonas exotoxin) to produce a conjugate molecule capable of selectively killing HIV-infected gp120-expressing cells (26).
Previous results have indicated that CV-N binds with extremely high avidity to the viral envelope glycoprotein gp120, an interaction essential to its anti-HIV activity (6, 25). Other experiments indicated that CV-N did not visibly disrupt the virion ultrastructure (20). Mapping studies with sCD4 and a series of monoclonal antibodies (MAbs) to defined epitopes on the envelope glycoprotein indicated that the CV-N-binding site(s) on gp120 differed from the primary CD4-binding site, sCD4-induced epitopes, V3 loop, or other domains on gp120 recognized by these reagents (6, 13). However, CV-N apparently bound to gp120 in a manner that occluded or altered the binding site(s) of MAb 2G12 (13), which recognizes a conformational glycosylation-dependent epitope (18, 33, 37). Reciprocal cross-blocking studies showed further that MAb 2G12 pretreatment did not prevent subsequent CV-N binding to sgp120 (13). Thus, CV-N and MAb 2G12 bind to gp120 similarly but not identically.
Previous reports have raised some apparent ambiguities about the experimental effects of CV-N on the binding of gp120 to CD4. Enzyme-linked immunosorbent assay studies showed that prebinding of virus-free, soluble gp120 (sgp120) to cell-free sCD4 did not block the subsequent binding of CV-N with the sgp120; furthermore, pretreatment of sgp120 with CV-N did not inhibit binding of the sgp120 to sCD4 (6). Other studies indicated that CV-N treatment of virion-associated gp120 likewise did not block subsequent binding of the gp120 to sCD4 (13, 20). Moreover, binding of CV-N to virion-associated gp120 did not detectably affect subsequent sCD4-induced conformational changes in the envelope glycoprotein (13). These results initially suggested that CV-N acts primarily at a post-CD4 step but prior to completion of fusion and viral entry (6, 20). However, other experiments described by Esser et al. indicated that CV-N inhibited CD4-dependent binding of HIV-1 virions to target cells (13), reflecting CV-N impairment of virion-associated gp120 binding to cell-associated CD4. Most recently, Dey et al. (10) have presented evidence that CV-N prevents binding of sgp120 to cell-associated CD4, as well as subsequent interaction of sCD4-activated Env with the coreceptor CCR5.
To resolve the aforementioned apparent ambiguities and to further advance the understanding of the effects of CV-N on the interaction between gp120 and its cellular receptors, we present herein further investigations of the effects of CV-N on (i) CD4-dependent and CD4-independent binding of sgp120 to the target cells and (ii) the interaction of sCD4-activated gp120 with the coreceptor CXCR4 (post-CD4 binding steps), using coimmunoprecipitation and flow cytometry analyses. In addition, we have observed a remarkable ability of CV-N to dissociate sgp120 from the target cells. These data clarify that the experimental effects of CV-N on gp120-CD4 binding interactions depend upon the type of CD4 (soluble or membrane associated), but not upon the type of gp120 (soluble or virion associated), employed in the experimental protocol.
MATERIALS AND METHODS
Cell lines.
The CEM-SS cell line was obtained from the American Type Culture Collection, Manassas, Va. The A2.01 and A3.01 cell lines were provided by M. Esser (Science Applications International Corporation, NCI-Frederick). All cell lines were mycoplasma negative (PCR Mycoplasma Detection Kit, American Type Culture Collection) and were cultured in complete medium (RPMI 1640 with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, and 10 μg of gentamicin per ml).
Proteins and antibodies.
Purified CV-N, recombinantly expressed in E. coli, was prepared as described elsewhere (27). Full-length recombinant sgp120 glycoprotein of HIV-1IIIB produced in baculovirus was obtained from Intracel (Issaquah, Wash.). Recombinant sCD4 (amino acids 1 to 369) (1) was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health (contributor; R. Sweet). Fluorescein isothiocyanate (FITC)-conjugated mouse anti-gp120 MAb, raised against the recombinant gp120, and phycoerythrin (PE)-conjugated anti-OKT4 MAb were obtained from Intracel and Ortho Diagnostics (Raritan, N.J.), respectively. Unconjugated and peridinin chlorophyll protein (PerCP)-conjugated anti-Leu3a MAbs were obtained from Becton Dickinson Immunocytometry Systems (San Jose, Calif.). The following anti-gp120 MAbs were obtained through the AIDS Research and Reference Reagent Program: HIV-1 gp120 MAb 2G12 (7, 33), immunoglobulin G1 (IgG1) b12 (3, 8, 9, 30), and 4.8D (23, 31) from (H. Katinger, D. Burton and C. Barbas, and J. Robinson, respectively).
Coimmunoprecipitation assay.
A coimmunoprecipitation assay was performed using modifications to a previously reported assay format (19). CEM-SS cells were washed three times with ice-cold phosphate-buffered saline (PBS) to remove any contaminating proteins by centrifugation at 400 × g for 5 min. The washed cells were suspended in complete medium (2.5 × 107 cells/0.5 ml/condition). A 1.2 μM concentration of sgp120 was preincubated with PBS (mock treatment) or different concentration of CV-N (0.12 to 60 μM) in a total volume of 35 μl of PBS for 1 h at room temperature. Free CV-N was removed by ultrafiltration, using a centrifugal filtration device with a 50-kDa-cutoff membrane (Microcon 50, Amicon, Beverly, Mass.). Cells were then incubated with the protein mixture at 37°C for 2 h in a CO2 incubator with shaking every 20 to 30 min. The cells were washed twice with ice-cold PBS and lysed in buffer containing 1% Brij 97, 150 mM NaCl, 20 mM Tris-HCl (pH 8.0), 5 mM iodoacetamide, 1 mM phenylmethylsulfonyl fluoride, and 4 μM leupeptin. After 20 min on ice, nuclei were pelleted by centrifugation at 13,000 × g for 5 min. The lysates were immunoprecipitated with anti-OKT4 MAb and 50 μl of protein G-Sepharose beads (diluted 1:2 in PBS) at 4°C overnight. The beads were washed five times with lysis buffer and boiled for 5 min with 25 μl of 2× sodium dodecyl sulfate sample buffer. Samples (from 2.5 × 107 cells per lane) were run on a 10% Tris-glycine gel with sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were electrophoretically transferred to nitrocellulose membranes. Membranes were blocked with Tris-buffered saline (pH 7.4) containing 0.1% Tween 20 and 5% skim milk and were incubated with anti-gp120 polyclonal rabbit serum (Intracel) or anti-OKT4 MAb. After three washes with buffer (Tris-buffered saline containing 0.1% Tween 20), the membranes were incubated with anti-rabbit or anti-mouse antibody-conjugated horseradish peroxidase in blocking buffer for 30 min. After being washed, the blots were incubated with SuperSignal substrate (Pierce Chemical Co., Rockford, Ill.) for 1 min and exposed to film.
For the experiments represented in Fig. 5A, CEM-SS cells (2.5 × 107 cells/condition) were incubated with untreated sgp120 (final concentration, 84 nM) in a total of 500 μl of complete medium at 37°C for 1.5 h in a CO2 incubator. After being washed twice, the treated cells were subsequently incubated with CV-N (final concentration 840 nM) or PBS (mock treatment) in a total of 500 μl of complete medium at 37°C for 1.5 h in a CO2 incubator and washed twice with ice-cold PBS. The lysis, coprecipitation, and detection were performed as described above.
FIG. 5.
CV-N induces sgp120 dissociation from cell-associated CD4. (A) Effects when CEM-SS cells pretreated with sgp120 were either mock-treated with PBS (lane 1) or treated with CV-N (lane 2) for 1.5 h at 37°C in a CO2 incubator and washed twice before coimmunoprecipitation using anti-OKT4 MAb as described in Materials and Methods (molar ratio of sgp120 to CV-N, 1 to 100). Anti-gp120 polyclonal serum and anti-OKT4 MAb were used for the detection of sgp120 and CD4, respectively. Numbers at right indicate positions of molecular mass markers. (B) Effect of CV-N on sgp120 dissociation from cell-associated CD4. A flow cytometric analysis was utilized as described in Materials and Methods. The MFI values of untreated cells (black trace), sgp120-bound untreated cells (blue trace), CV-N-treated-sgp120-bound untreated cells (red trace), sgp120-bound unlabeled anti-Leu3a MAb-pretreated cells (green trace), and CV-N-treated sgp120-bound unlabeled anti-Leu3a MAb-pretreated cells (orange trace) are 4.5, 80.7, 17.1, 32.0, and 12.2, respectively. Each data point represents at least 10,000 acquired events.
Flow cytometric assay of binding of sgp120 and cell-associated CD4.
For the experiments represented in Fig. 2 and 3, an immunofluorescence flow cytometry-based, sgp120-binding assay was performed, using modifications to a previously reported assay format (13, 34). The CEM-SS cell line expressing CD4, CXCR4, and CCR5 or A3.01 cells expressing CD4 and CXCR4 (2 × 105 cells per condition) were preincubated at 4°C for 30 min with either blocking buffer (PBS with 1% bovine serum albumin, 1% fetal bovine serum, and 0.02% sodium azide) or 33 nM unlabeled anti-Leu3a MAb in blocking buffer and then washed twice with blocking buffer. A sample containing 167 nM sgp120 was preincubated with PBS (mock treatment) or different concentration of CV-N (66.8 to 534.4 nM) in a total volume of 50 μl of PBS for 30 min at room temperature. Free CV-N was removed as described above. Cells were then incubated with the protein mixture in a total volume of 50 μl of washing buffer (PBS with 1% fetal bovine serum and 0.02% sodium azide) at 37°C for 30 min and washed twice with blocking buffer. Immunofluorescent staining was performed (4°C for 30 min) using FITC-conjugated anti-gp120 MAb, PE-conjugated anti-OKT4 MAb, and PerCP-conjugated anti-Leu3a MAb. A competitive enzyme-linked immunosorbent assay study confirmed that CV-N did not occlude the epitope recognized by the anti-gp120 MAb (data not shown). Following antibody staining, cells were washed twice with washing buffer prior to analysis on a FACScan flow cytometer, using CellQuest software (Becton Dickinson Immunocytometry Systems). Cells were gated by forward and 90° light scatter, and at least 10,000 events were acquired for each sample.
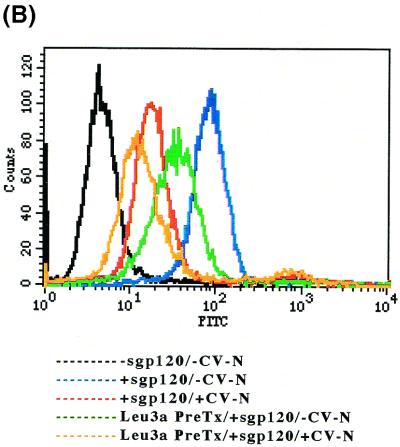
FIG. 2.
Binding of sgp120 to CEM-SS cells demonstrated by flow cytometry. CEM-SS (CD4-, CXCR4-, and CCR5-positive) cells were either mock treated or treated with unlabeled anti-Leu3a MAb for 30 min at 4°C and washed twice before addition of sgp120 that was either mock treated or treated with CV-N (molar ratio of sgp120 to CV-N, 1 to 2.4). The sgp120 was added to CEM-SS cells, and the binding was determined from the sgp120 signal using flow cytometry, where anti-gp120 MAb–FITC acquisition indicates overall sgp120 binding, and by anti-Leu3a MAb–PerCP staining, where loss of availability of the Leu3a epitope is an indirect indication of CD4-dependent sgp120 binding. (A) sgp120 signal. CEM-SS cells are sgp120 negative (black trace). Addition of untreated sgp120 results in acquisition of anti-gp120 MAb–FITC staining (compare black and blue traces). CV-N treatment of sgp120 inhibits overall sgp120 binding (compare blue and red traces). Approximately 74% of overall sgp120 binding is CD4 dependent (compare green and blue traces). MFI values for untreated sgp120: binding to untreated cells, 100.3, binding to unlabeled anti-Leu3a MAb-pretreated cells 30.1. CV-N inhibits CD4-dependent and CD4-independent binding of sgp120; note the increased inhibition of sgp120 binding for CV-N-treated sgp120 on unlabeled anti-Leu3a MAb-pretreated cells (compare green and orange traces). (B) Leu3a signal (gp120-binding epitope on CD4). CEM-SS cells express the Leu3a epitope on CD4 (black trace), and saturating unlabeled anti-Leu3a MAb pretreatment blocks binding of anti-Leu3a MAb–PerCP (compare green and black traces). Binding of sgp120 blocks the Leu3a epitope (compare black and blue traces). Cells incubated with CV-N-treated sgp120 have availability of Leu3a epitopes comparable to that of those incubated with untreated sgp120 (compare red and black traces). (C) OKT4 signal (non-gp120-binding epitope on CD4). Neither sgp120 nor CV-N interferes with the level of CD4 or the availability of the OKT4 epitope. At least 10,000 events were acquired for each sample.
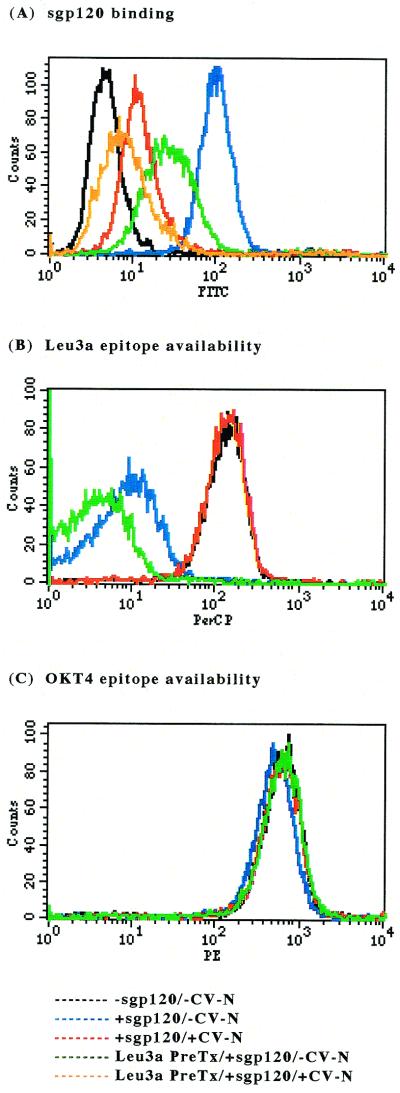
FIG. 3.
CV-N concentration-dependent effects on sgp120 binding. CEM-SS cells were either mock treated with PBS or treated with unlabeled anti-Leu3a MAb for 30 min at 4°C and washed twice before addition of sgp120 that was either mock-treated or treated with CV-N at various concentrations. The indicated ratio represents the molar ratio of sgp120 to CV-N when those proteins were preincubated. Each data point represents at least 10,000 acquired events. (A) Percentage of overall and CD4-independent sgp120 binding. The MFI value for untreated cells with sgp120 (100.3) was considered 100%, that for untreated cells without sgp120 (5.0) was considered 0%. (B) Percentage of CD4-dependent sgp120 binding, calculated by subtracting the MFI for CD4-independent sgp120 binding (sgp120 binding in the presence of unlabeled anti-Leu3a MAb) from that for overall sgp120 binding, with 100% binding equal to sgp120 binding in the absence of CV-N. (C) Percentage of Leu3a epitope availability. The MFI value for untreated cells with anti-Leu3a MAb–perCP (137.4) was considered 100%, that for untreated cells without anti-Leu3a MAb–PerCP (5.2) was considered 0%. (D) percentage of OKT4 epitope availability. The MFI value for untreated cells with anti-OKT4 MAb–PE (630.3) was considered 100%, that for untreated cells without anti-OKT4 MAb–PE (3.7) was considered 0%. In panels A, C, and D, closed circles and open circles represent untreated samples and unlabeled anti-Leu3a MAb-pretreated samples, respectively.
To evaluate whether or not CV-N could dissociate the binding between sgp120 and cell-expressed CD4, CEM-SS cells (2 × 105 per condition) were preincubated with sgp120 (final concentration, 167 nM) in a total volume of 50 μl of washing buffer at 37°C for 30 min; the pretreated cells were washed twice with blocking buffer. Subsequently, the cells were incubated in 668 nM CV-N, 2G12 MAb, or IgG1 b12 MAb in a total volume of 50 μl of washing buffer at 37°C for different durations and washed twice with blocking buffer. Immunofluorescent staining and analysis by flow cytometry were performed as described above.
Flow cytometric assay of sCD4-induced binding of sgp120 and cell-associated CXCR4.
For the experiments represented in Fig. 4, A2.01 cells (2 × 105 per condition), expressing CXCR4 but not CD4, were preincubated at 4°C for 30 min with blocking buffer. For Fig. 4B, C, and D, 278 nM sgp120 was preincubated with PBS (mock treatment), 1112 nM, CV-N, and 1,112 nM sCD4 respectively, in a total volume of 60 μl of PBS for 30 min at room temperature. For Fig. 4E, 278 nM sgp120 was first preincubated with 1,112 nM CV-N for 30 min at room temperature and then activated by incubation with 1,112 nM sCD4 for 30 min at room temperature. For Fig. 4F, 278 nM sgp120 was first activated by 1,112 nM sCD4 and then incubated with 1,112 nM CV-N. Free CV-N was removed as described above. Cells were then incubated with the protein mixture in a total volume of 50 μl of washing buffer at 37°C for 30 min and washed twice with blocking buffer. Immunofluorescent staining was performed (4°C for 30 min) using FITC-conjugated anti-gp120 MAb. Following antibody staining, cells were analyzed as described above.
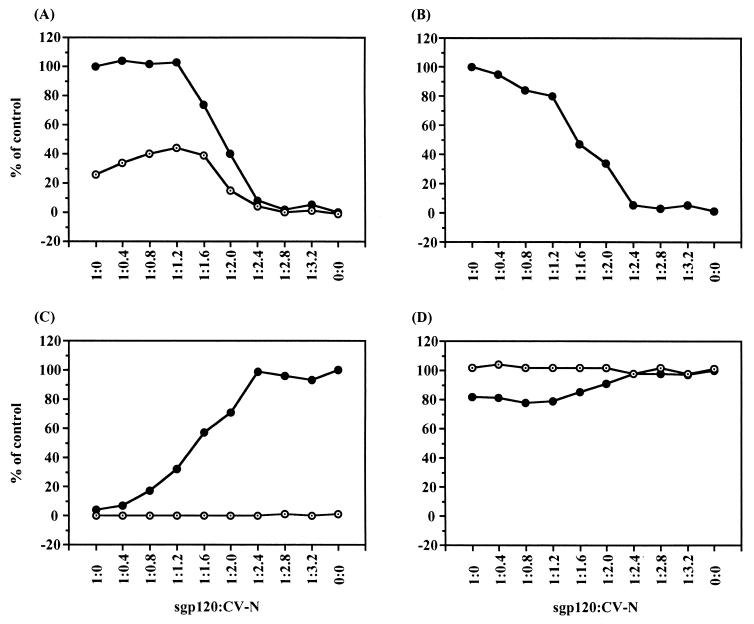
FIG. 4.
Inhibition of sgp120 and sCD4-induced sgp120 binding to the target cell coreceptor CXCR4 by CV-N. Binding of CV-N-treated sgp120 to A2.01 cells (CD4 negative, CXCR4 positive) upon activation by sCD4 was determined by a flow cytometry-based sgp120-binding assay (see Materials and Methods). Incubation of the cells with sgp120 results in acquisition of anti-gp120 MAb–FITC staining. (A) Background (A2.01 cells without sgp120, CV-N, and sCD4). MFI value, 8.0. (B) Effects when sgp120 was used as positive control. MFI value, 16.9. (C and D) Effects when sgp120 was preincubated with sCD4(C) and CV-N (D). MFI values, 29.4 (C) and 7.7 (D). (E) Effects when sgp120 was first preincubated with CV-N and then activated by incubation with sCD4. MFI values, 8.6. (F) sgp120 was first activated by sCD4 and then incubated with CV-N. MFI value, 7.8. The molar ratios of sgp120 to CV-N and sCD4 are 1 to 4 and 4, respectively, in the final mixtures. At least 10,000 events were acquired for each sample.
RESULTS
CV-N impairs sgp120 binding to CD4-expressing cells.
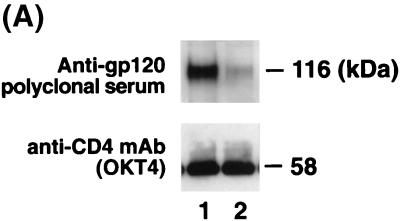
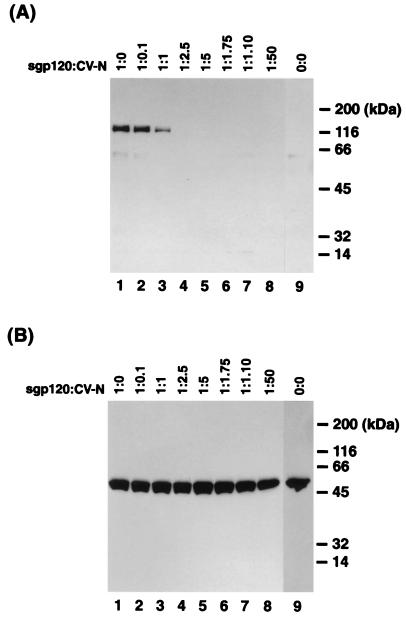
To further understand the mechanism of CV-N inhibition of HIV-1 envelope-mediated binding and infection, we examined the effects of CV-N on sgp120 binding to CD4-expressing cells. The sgp120 of HIV-1IIIB (T-cell-line-tropic, X4, syncytium-inducing phenotype) (see references 4 and 24 for reviews) was utilized. CEM-SS cells expressing CD4, CXCR4, and CCR5 were incubated with sgp120, centrifuged, and washed; cell-associated sgp120 was detected by coimmunoprecipitation assay. It is well established that sgp120 of HIV-1IIIB interacts with CXCR4 but not CCR5 coreceptor (11, 17). As shown in Fig. 1A, cell-associated, CD4-bound sgp120 was coprecipitated (lane 1). When coprecipitated sgp120 was detected by anti-gp120 polyclonal serum, additional background bands were commonly seen after immunoprecipitation with anti-OKT4 MAb. Pretreatment of sgp120 with CV-N clearly blocked binding of the sgp120 to cell-associated CD4 in a concentration-dependent manner (Fig. 1A, lanes 2 to 8). When the pretreatment molar ratio of sgp120 to CV-N was 1 to 2.5, sgp120 binding to cell-associated CD4 was completely inhibited, indicating that more than one molecule of CV-N per sgp120 molecule was necessary to fully block CD4-dependent sgp120 binding to target cells. CD4 was consistently precipitated regardless of treatment with sgp120 and CV-N (Fig. 1B).
FIG. 1.
Effect of CV-N on binding of sgp120 to cell-associated CD4. Binding of CV-N-treated sgp120 to cell-associated CD4 was determined by coimmunoprecipitation using anti-OKT4 MAb (see Materials and Methods). (A) Detection of sgp120 by anti-gp120 polyclonal serum. (B) Detection of CD4 by anti-OKT4 MAb. Lanes 1, mock-treated sgp120; lanes 2 to 8, sgp120 preincubated with different concentration of CV-N; lanes 9, no sgp120 and no CV-N. The indicated molar ratio represents that of sgp120 to CV-N when those proteins were preincubataed. Numbers at the right indicate positions of molecular mass markers.
To confirm these results, we utilized a flow cytometry-based, sgp120-binding assay using CEM-SS cells. The CEM-SS cell line expresses CD4, as demonstrated by the staining of both anti-Leu3a (Fig. 2B) and anti-OKT4 (Fig. 2C) MAbs. After incubation of CEM-SS cells with sgp120, the cells were stained by anti-gp120 MAb–FITC (Fig. 2 A), with a concomitant decrease in the availability of the Leu3a epitope (gp120-binding site [2]) (Fig. 2B) but with little change in OKT4 (non-gp120-binding epitope) staining (Fig. 2C), all consistent with CD4-dependent sgp120 binding to the target cells. Preincubation of CEM-SS cells with unlabeled anti-Leu3a MAb inhibited acquisition of approximately 74% of the anti-gp120 MAb–FITC signal (Fig. 2A), indicating that approximately 74% of sgp120 binding detected by acquisition of sgp120 staining was CD4 dependent while the remaining 26% of the binding was CD4 independent. This observation was consistent with the evidence previously reported by Hoffman et al., in which sgp120 of HIV-1IIIB could interact with CXCR4 independently of CD4 but this binding was markedly enhanced by the previous interaction of envelope with sCD4 (17). Pretreatment of sgp120 with CV-N (molar ratio of sgp120 to CV-N, 1:2.4) substantially inhibited overall sgp120 binding as assessed by acquisition of anti-gp120 MAb–FITC signal (Fig. 2A). Importantly, there was little or no significant decrease in the anti-Leu3a MAb signal when CV-N-treated sgp120 was added to the cells (Fig. 2B), indicating that CV-N completely blocked CD4-dependent sgp120 binding. Binding of CV-N-treated sgp120 to anti-Leu3a-pretreated cells was much lower than binding of native sgp120 to anti-Leu3a MAb-pretreated cells (Fig. 2B). The additive inhibitory effect suggests that CV-N treatment of sgp120 inhibited CD4-independent binding of sgp120 to cells, as well as CD4-dependent binding. Using the A3.01 cell line expressing CD4 and CXCR4, but not CCR5, we obtained essentially the same results described above (data not shown).
The aforementioned results perhaps are better appreciated in Fig. 3, which graphically summarizes data for sgp120 binding based on measurements of mean fluorescence intensity (MFI) of staining for positive cells in the experiments described above in the absence or presence of unlabeled anti-Leu3a MAb pretreatment of CEM-SS cells. When the molar ratio of sgp120 to CV-N was 1 to ≥2.4, CV-N inhibited acquisition of anti-gp120 MAb–FITC signal, reflective of CV-N blocking both the overall sgp120 binding and CD4-independent sgp120 binding (sgp120 binding in the presence of unlabeled anti-Leu3a MAb) to the target cells (Fig. 3A). CD4-dependent sgp120 binding was calculated by subtracting the MFI for CD4-independent sgp120 binding from that for overall sgp120 binding. As shown in Fig. 3B, CV-N inhibited CD4-dependent sgp120 binding, as well as CD4-independent binding of sgp120 to the target cells. This was paralleled by an increase in the availability of the Leu3a epitope, reflecting a decrease in the blockade of the epitope associated with CD4-dependent binding of sgp120 to target cells (Fig. 3C). Treatment of sgp120 with CV-N thus blocks CD4-dependent binding of sgp120 to target cells. Binding of neither native sgp120 nor CV-N-treated sgp120 interfered with detection of the OKT4 epitope on CEM-SS cells (no concentration-dependent inhibition) (Fig. 3D), revealing that bound sgp120 was not masking this epitope or inducing CD4 degradation or endocytosis. Thus, at a molar ratio of sgp120 to CV-N of 1 to 2.4 or more, CD4-dependent sgp120 binding to the target cells was completely inhibited by CV-N, confirming the results in Fig. 1; likewise, CV-N nearly completely blocked CD4-independent sgp120 binding to the target cells.
CV-N blocks direct sgp120 binding, as well as sCD4-induced binding of sgp120, to the cell-associated CXCR4 coreceptor.
Binding of the HIV-1 envelope protein to CD4 induces structural alterations in the gp120 subunit that enables it to interact with an appropriate coreceptor (15, 19, 32, 36). Since CV-N did not detectably affect the binding of sCD4 to sgp120 or virions, or subsequent sCD4-induced conformational changes in the envelope glycoprotein (6, 13, 20), we investigated the effects of CV-N on the interaction of sCD4-activated sgp120 with the target cells coreceptor, CXCR4. We tested binding of HIV-1IIIB sgp120 to A2.01 cells, expressing CXCR4 but not CD4, in a flow cytometric binding study (Fig. 4). In agreement with previous observations (11, 17), the sgp120 itself very modestly bound to the CXCR4-positive, CD4-negative cells (Fig. 4B) (MFI value of 16.9, compared with blank MFI value of 8.0), but sgp120 preincubated with sCD4 was well bound to the CXCR4-expressing cells (Fig. 4C) (MFI value of 29.4) as a result of sCD4-induced conformational change of sgp120. Preincubation of sgp120 with CV-N apparently inhibited its modest binding to the cells (Fig. 4D) (MFI value of 7.7). The sCD4-induced binding of sgp120 to the CXCR4-expressing cells was also completely blocked by the CV-N treatment of sgp120 before and after sCD4 activation (Fig. 4E and F) (MFI values of 8.6 and 7.8, respectively), indicating that CV-N possesses blocking activity at the level of sCD4-activated sgp120 interaction with coreceptor, in addition to the sgp120 interaction with cell-associated CD4.
CV-N dissociates the interaction between sgp120 and cell-associated CD4.
Finally, we investigated whether CV-N could dissociate sgp120 from the target cells. Following incubation of CEM-SS cells with untreated sgp120, the cells were centrifuged, washed, and subsequently incubated with CV-N and washed again; CD4-bound sgp120 was detected by coimmunoprecipitation analysis as before. As shown in Fig. 5A, CV-N dissociated the interaction between sgp120 and cell-associated CD4 nearly completely. To further characterize this unique dissociative activity of CV-N, we utilized a flow cytometric analysis (Fig. 5B). CEM-SS cells were either mock treated or treated with unlabeled anti-Leu3a MAb for 30 min at 4°C and washed twice before addition of sgp120. After incubation for 30 min at 37°C and washing twice, sgp120-treated cells were either mock treated or treated with CV-N (molar ratio of sgp120 to CV-N, 1 to 4). As shown in Fig. 5B, addition of sgp120 resulted in acquisition of anti-gp120 MAb–FITC staining (compare black and blue traces). CV-N treatment of sgp120 bound cells caused a substantial overall dissociation of sgp120 from cells (compare blue and red traces). The increased dissociation of sgp120 from unlabeled anti-Leu3a MAb-pretreated cells by posttreatment with CV-N (compare green and orange traces) indicated that CV-N detached the gp120, whether CD4-dependently or CD4-independently bound, from the cells. This unique activity of CV-N was manifest very rapidly (less than 2.5 min) after incubation of sgp120-bound cells with CV-N (data not shown). Moreover, neither 2G12 MAb nor IgG1 b12 MAb, which binds to the CD4-binding site on gp120, showed any such dissociative activity (data not shown).
DISCUSSION
Previous studies assessing the mechanism of action of CV-N on gp120-mediated binding to CD4 have yielded some contrasting results. Experiments with cell-free CD4 (sCD4) indicated that CV-N failed to block sCD4 binding to virus-free gp120 (sgp120) as well as virion-associated gp120 (6, 13, 20). However, flow cytometry experiments reported by Esser et al. (13) indicated that CV-N blocked cell-associated CD4-dependent binding of virion-associated gp120. Other results obtained by Dey et al. (10) indicated that CV-N blocked binding of sgp120 to cell-associated CD4. However, they did not undertake detailed quantitative analyses.
In the present study, we sought to further resolve the aforementioned ambiguities by utilizing complementary coimmunoprecipitation analyses and flow cytometry-based sgp120-target cell-binding assays. The present results directly indicate that CV-N blocks both CD4-dependent and CD4-independent binding of sgp120 to target cells in a concentration-dependent manner and that more than one CV-N molecule binding to each sgp120 molecule is required to inhibit both types of sgp120 binding (Fig. 1 to 3). These results are interesting in light of a theoretical surface analysis of the solution structure of CV-N in which two potential binding sites for protein-protein interactions were proposed (5). However, data generated using sgp120 should be interpreted with some caution because several reports have shown that the binding of sgp120 to cells is not fully representative of the binding of virion-associated gp120 (22, 34). Nevertheless, studies with sgp120 allow quantitative analyses of gp120-receptor complex interactions, something that is not yet possible with whole-virus assays.
The present data thus clarify that the experimental effects of CV-N on gp120-CD4 binding interactions depend upon the type of CD4 (soluble or membrane associated) but not upon the type of gp120 (soluble or virion associated) employed in the study protocol. Since the steric context may be quite different for virion-associated gp120 binding to sCD4 compared to binding to membrane-bound CD4 (28), CV-N may sterically hinder gp120 binding to cell-associated CD4 but still allow binding to sCD4. However, additional experiments will be required to determine the molecular basis for the different effects seen with sCD4 versus cell-surface CD4.
Furthermore, we observed that CV-N can dissociate both CD4-dependently and CD4-independently bound sgp120 from target cells (Fig. 5). We are not aware of any antibody capable of dissociating sgp120 from cell-associated CD4. Since MAb 2G12 is known neither to block sgp120 binding to CD4-expressing target cells (34) nor to dissociate bound sgp120 from CD4-expressing cells (data not shown), the modes and/or site(s) of binding of CV-N and MAb 2G12 must differ substantially. These results further indicate that CV-N uniquely acts to prevent essential interactions between gp 120 and target cell receptors, consistent with the extensive inhibitory activity profile of CV-N against numerous isolates of HIV-1, as well as other lentiviruses. However, Esser et al. (13) reported that CV-N did not impair sCD4-triggered conformational changes in virion-associated gp120 required for coreceptor binding (18, 37), and it was reported that sCD4 bound on sgp120 was not dissociated from the sgp120 by either pre- or posttreatment of sgp120 with CV-N (6). These results together indicated that CV-N did not dissociate sCD4 bound to the envelope glycoprotein. Again, the apparent discrepancies can be explained by the type of CD4 (soluble or membrane associated) employed. To further ascertain the implications and relevance of gp120-CD4 dissociative activity of CV-N, additional experiments would be needed to determine whether the infectivity of viruses initially bound to but not yet fused with or entered into cells (e.g., at 4°C) could still be inhibited by the subsequent addition of CV-N, which presumably could displace the virus from the cells. The apparent dissociation observed in the present study occurred after incubation of sgp120 with the cells for 30 min at 37°C, which is long enough to form gp120-CD4-coreceptor tricomplexes. Thus, the results seen in Fig. 5 could be alternatively explained by an enhanced rate of internalization or degradation. However, we have previously shown that a substantial delay of addition of CV-N after exposure of cells to virus could still result in maximum antiviral activity (maximum activity still after a 30-min delay and very high antiviral activity even after a 60-min delay) (6). This may indicate that virions were dissociated by CV-N from CD4-positive cells.
We tested whether CV-N could block the sCD4-induced sgp120 binding to CXCR4-expressing cells. The results showed that sCD4-activated sgp120 treated with CV-N either before or after sCD4 activation failed to bind to CXCR4-expressing cells (Fig. 4), indicating a direct effect on activated sgp120 binding to the coreceptor. Therefore, it is suggested that, regardless of an sCD4-activated conformational change of gp120, CV-N binding to sgp120 leads to steric blockage and/or conformational changes of sgp120, resulting in the inaccessibility of the coreceptor to its binding site on the glycoprotein. Since none of the anti-gp120 MAbs directed to the third hypervariable domain of gp120 (the V3 loop), the major determinant of CXCR4 interaction with gp120 (16, 17) blocked subsequent CV-N binding to sgp120, or vice versa (6), it seems unlikely that CV-N directly binds to the coreceptor binding site(s) on gp120, but rather that it induces conformational changes of sgp120, leading to CXCR4 inaccessibility to its binding site. This interpretation also suggested that CV-N's inhibition of Env-coreceptor interactions would not be limited to CXCR4. This view was confirmed by recent results of Dey et al. (10), using a different experimental protocol than that employed here, indicating that CV-N blocked interaction of sCD4-activated Env with CCR5.
The inhibitory effect of CV-N on the interaction between sCD4-activated sgp120 and CXCR4 may help explain the potent inhibitory effects of CV-N on feline immunodeficiency virus (10), a virus which infects feline cells in a CD4-independent (29), but strictly CXCR4-dependent (35), manner. This model would also predict that CD4-independent strains of HIV and simian immunodeficiency virus (12, 21), in which the chemokine receptor-binding domain of gp120 is already sufficiently exposed to obviate the need for initial gp120-CD4-binding induced conformational changes, should be susceptible to inhibition by CV-N.
Mapping studies with sCD4 and a series of MAbs to defined epitopes on the envelope glycoprotein indicated that the CV-N-binding site(s) on gp120 differed from the primary CD4-binding site, sCD4-induced epitopes, V3 loop, or other domains on gp120 recognized by these reagents (6, 13). However, Esser et al. (13) described reciprocal cross-blocking studies in which CV-N pretreatment prevented subsequent MAb 2G12 binding to gp120 but MAb 2G12 pretreatment did not prevent subsequent CV-N binding to gp120. Moreover, our recent studies indicated that rather than CV-N binding to essentially the same site(s) on gp120 as MAb 2G12 with even higher affinity, CV-N binds to unique sites on gp120 in a manner that alters the affinity of, or renders inaccessible, the 2G12 epitope for the antibody (T. Mori, unpublished data).
The 2G12 MAb is known to be directed against a conserved conformational epitope that is highly dependent on glycosylation (33). From several indirect lines of evidence, it is apparent that CV-N may interact with carbohydrate moieties on gp120; however, a nonspecific carbohydrate interaction model would not easily explain the distinctions between the potent effects of CV-N seen against HIV-1, HIV-2, simian immunodeficiency virus, and feline immunodeficiency virus versus the reportedly negligible effects of CV-N on certain other enveloped viruses having glycosylated envelope proteins, such as human cytomegalovirus and human herpesvirus 1 (6), murine leukemia virus (M. T. Esser, personal communication), and vaccinia virus (10). Interestingly, Dey et al. (10) have observed potent inhibitory effects of CV-N against certain other enveloped, but nonretroviral, viruses, specifically human herpesvirus 6 and measles virus. The possibility that CV-N's antiviral effects are mediated through specific carbohydrate interactions available only on susceptible viruses remains to be explored.
Overall, the present studies demonstrate that CV-N binds to sgp120 in a manner that alters and/or masks the 2G12 epitope, prevents both CD4-dependent and CD4-independent gp120 binding to target cells, and blocks sCD4-induced gp120 binding to cell-associated coreceptor CXCR4. Furthermore, CV-N induces sgp120 dissociation from target cells. These data indicate that the mechanism(s) of action of CV-N involves interference with essential interactions between the viral envelope glycoprotein and target cell receptors. CV-N should be a valuable reagent to further examine the early steps of virion binding and fusion. Furthermore, since CV-N has shown minimal or no toxicity to cultured cells (6, 13), benign behavior in a rabbit vaginal toxicity model (National Institute of Allergy and Infectious Diseases, unpublished data), and potent activity against multiple pathogens, including macrophage-tropic HIV-1, CV-N appears promising as a candidate microbicide to prevent the sexual transmission of HIV and AIDS.
ACKNOWLEDGMENTS
We thank C. K. Lapham for technical input on coimmunoprecipitation assays, M. T. Esser for providing A2.01 and A3.01 cells for flow cytometry studies, M. L. Hursey for flow cytometry support, and M. T. Esser, J. B. McMahon, and B. R. O'Keefe for critical review of the manuscript prior to submission.
Footnotes
Paper 67 in the NCI Laboratory of Drug Discovery Research and Development series “HIV-Inhibitory Natural Products.”
REFERENCES
- 1.Arthos J, Deen K C, Chaikin M A, Fornwald J A, Sathe G, Sattentau Q J, Clapham P R, Weiss R A, McDougal J S, Pietropaolo C, Axel R, Truneh A, Maddon P J, Sweet R W. Identification of the residues in human CD4 critical for the binding of HIV. Cell. 1989;57:469–481. doi: 10.1016/0092-8674(89)90922-7. [DOI] [PubMed] [Google Scholar]
- 2.Attanasio R, Dilley D, Buck D, Maino V C, Lohman K L, Kanda P, Kennedy R C. Structural characterization of a cross-reactive idiotype shared by monoclonal antibodies specific for the human CD4 molecule. J Biol Chem. 1991;266:14611–14619. [PubMed] [Google Scholar]
- 3.Barbas C F, III, Bjorling E, Chiodi F, Dunlop N, Cababa D, Jones T M, Zebedee S L, Persson M A, Nara P L, Norrby E, Burton D R. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc Natl Acad Sci USA. 1992;89:9339–9343. doi: 10.1073/pnas.89.19.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger E A. HIV entry and tropism. When one receptor is not enough. Adv Exp Med Biol. 1998;542:151–157. [PubMed] [Google Scholar]
- 5.Bewley C A, Gustafson K R, Boyd M R, Covell D G, Bax A, Clore G M, Gronenborn A M. Solution structure of cyanovirin-N, a potent HIV-inactivating protein. Nat Struct Biol. 1998;5:571–578. doi: 10.1038/828. [DOI] [PubMed] [Google Scholar]
- 6.Boyd M R, Gustafson K R, McMahon J B, Shoemaker R H, O'Keefe B R, Mori T, Gulakowski R J, Wu L, Rivera M I, Laurencot C M, Currens M J, Cardellina II J H, Buckheit R W, Jr, Nara P L, Pannell L K, Sowder II R C, Henderson L E. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrob Agents Chemother. 1997;41:1521–1530. doi: 10.1128/aac.41.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, Katinger H. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 8.Burton D R, Barbas III C F, Persson M A, Koenig S, Chanock R M, Lerner R A. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burton D R, Pyati J, Koduri R, Sharp S J, Thornton G B, Parren P W, Sawyer L S, Hendry R M, Dunlop N, Nara P L, Lamacchia M, Garratty E, Stiehm E R, Bryson Y J, Cao Y, Moore J P, Ho D D, Barbas C F., III Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 10.Dey B, Lerner D L, Lusso P, Boyd M R, Elder J H, Berger E A. Multiple antiviral activities of cyanovirin-N: blocking of human immunodeficiency virus type 1 gp120 interaction with CD4 and coreceptor and inhibition of diverse enveloped viruses. J Virol. 2000;74:4562–4569. doi: 10.1128/jvi.74.10.4562-4569.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doranz B J, Orsini M J, Turner J D, Hoffman T L, Berson J F, Hoxie J A, Peiper S C, Brass L F, Doms R W. Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. J Virol. 1999;73:2752–2761. doi: 10.1128/jvi.73.4.2752-2761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 13.Esser M T, Mori T, Mondor I, Sattentau Q J, Dey B, Berger E A, Boyd M R, Lifson J D. Cyanovirin-N binds to gp120 to interfere with CD4-dependent human immunodeficiency virus type 1 virion binding, fusion, and infectivity but does not affect the CD4 binding site on gp120 or soluble CD4-induced conformational changes in gp120. J Virol. 1999;73:4360–4371. doi: 10.1128/jvi.73.5.4360-4371.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gustafson K R, Sowder II R C, Henderson L E, Cardellina II J H, McMahon J B, Rajamani U, Pannell L K, Boyd M R. Isolation, primary sequence determination, and disulfide bond structure of cyanovirin-N, an anti-HIV (human immunodeficiency virus) protein from the cyanobacterium Nostoc ellipsosporum. Biochem Biophys Res Commun. 1997;238:223–228. doi: 10.1006/bbrc.1997.7203. [DOI] [PubMed] [Google Scholar]
- 15.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffman T L, Stephens E B, Narayan O, Doms R W. HIV type I envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors. Proc Natl Acad Sci USA. 1998;95:11360–11365. doi: 10.1073/pnas.95.19.11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman T L, LaBranche C C, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie J A, Doms R W. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 20.Mariner J M, McMahon J B, O'Keefe B R, Nagashima K, Boyd M R. The HIV-inactivating protein, cyanovirin-N, does not block gp120-mediated virus-to-cell binding. Biochem Biophys Res Commun. 1998;248:841–845. doi: 10.1006/bbrc.1998.9060. [DOI] [PubMed] [Google Scholar]
- 21.Martin K A, Wyatt R, Farzan M, Choe H, Marcon L, Desjardins E, Robinson J, Sodroski J, Gerard C, Gerard N P. CD4-independent binding of SIV gp120 to rhesus CCR5. Science. 1997;278:1470–1473. doi: 10.1126/science.278.5342.1470. [DOI] [PubMed] [Google Scholar]
- 22.Mondor I, Ugolini S, Sattentau Q J. Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J Virol. 1998;72:3623–3634. doi: 10.1128/jvi.72.5.3623-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore J P, Yoshiyama H, Ho D D, Robinson J E, Sodroski J. Antigenic variation in gp120s from molecular clones of HIV-1 LAI. AIDS Res Hum Retroviruses. 1993;9:1185–1193. doi: 10.1089/aid.1993.9.1185. [DOI] [PubMed] [Google Scholar]
- 24.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 25.Mori T, Shoemaker R H, Gulakowski R J, Krepps B L, McMahon J B, Gustafson K R, Pannell L K, Boyd M R. Analysis of sequence requirements for biological activity of cyanovirin-N, a potent HIV (human immunodeficiency virus)-inactivating protein. Biochem Biophys Res Commun. 1997;238:218–222. doi: 10.1006/bbrc.1997.7202. [DOI] [PubMed] [Google Scholar]
- 26.Mori T, Shoemaker R H, McMahon J B, Gulakowski R J, Gustafson K G, Boyd M R. Construction and enhanced cytotoxicity of a [cyanovirin-N]-[Pseudomonas exotoxin] conjugate against human immunodeficiency virus-infected cells. Biochem Biophys Res Commun. 1997;239:884–888. doi: 10.1006/bbrc.1997.7505. [DOI] [PubMed] [Google Scholar]
- 27.Mori T, Gustafson K R, Pannell L K, Shoemaker R H, Wu L, McMahon J B, Boyd M R. Recombinant production of cyanovirin-N, a potent human immunodeficiency virus-inactivating protein derived from a cultured cyanobacterium. Protein Expr Purif. 1998;12:151–158. doi: 10.1006/prep.1997.0838. [DOI] [PubMed] [Google Scholar]
- 28.Parren P W, Mondor I, Naniche D, Ditzel H J, Klasse P J, Burton D R, Sattentau Q J. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J Virol. 1998;72:3512–3519. doi: 10.1128/jvi.72.5.3512-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poeschla E M, Looney D J. CXCR4 is required by a nonprimate lentivirus: heterologous expression of feline immunodeficiency virus in human, rodent, and feline cells. J Virol. 1998;72:6858–6866. doi: 10.1128/jvi.72.8.6858-6866.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roben P, Moore J P, Thali M, Sodroski J, Barbas III C F, Burton D R. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 33.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ugolini S, Mondor I, Parren P W, Burton D R, Tilley S A, Klasse P J, Sattentau Q J. Inhibition of virus attachment to CD4+ target cells is a major mechanism of T cell line-adapted HIV-1 neutralization. J Exp Med. 1997;186:1287–1298. doi: 10.1084/jem.186.8.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willett B J, Picard L, Hosie M J, Turner J D, Adema K, Clapham P R. Shared usage of the chemokine receptor CXCR4 by the feline and human immunodeficiency viruses. J Virol. 1997;71:6407–6415. doi: 10.1128/jvi.71.9.6407-6415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 37.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 38.Yang F, Bewley C A, Louis J M, Gustafson K R, Boyd M R, Gronenborn A M, Clore G M, Wlodawer A. Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping. J Mol Biol. 1999;288:403–412. doi: 10.1006/jmbi.1999.2693. [DOI] [PubMed] [Google Scholar]