Abstract
The ability to detect and quantify HIV RNA in blood is essential to sensitive detection of infections and monitoring viremia throughout treatment. Current options for point-of-care HIV diagnosis (i.e. lateral flow rapid tests) lack sensitivity for early detection and are unable to quantify viral load. HIV RNA diagnostics typically require extensive pre-processing of blood to isolate plasma and extract nucleic acids, in addition to expensive equipment for conducting nucleic acid amplification and fluorescence detection. Therefore, molecular HIV diagnostics is still mainly limited to clinical laboratories and there is an unmet need for high sensitivity point-of-care screening and at-home HIV viral load quantification. In this work, we outline a streamlined workflow for extraction of plasma from whole blood coupled with HIV RNA extraction and quantitative polymerase chain reaction (qPCR) in a portable magnetofluidic cartridge platform for use at the point-of-care. Viral particles were isolated from blood using manual filtration through a 3D-printed filter module in seconds followed by automated nucleic acid capture, purification, and transfer to qPCR using magnetic beads. Both nucleic acid extraction and qPCR were integrated within cartridges using compact instrumentation consisting of a motorized magnet arm, miniaturized thermocycler, and image-based fluorescence detection. We demonstrated detection down to 1000 copies of HIV viral particles from whole blood in <30 minutes.
Graphical Abstract
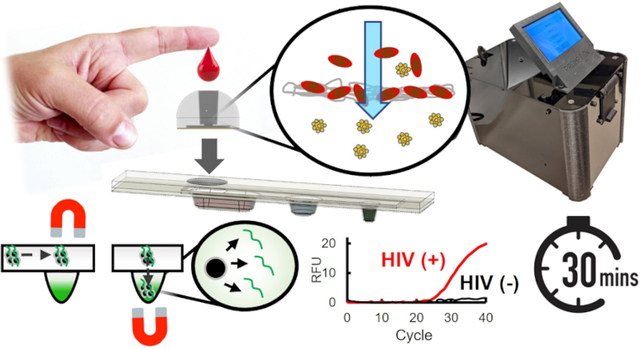
Blood plasma extraction using a 3D-printed filtration module coupled with magnetofluidic nucleic acid purification and quantitative PCR in a plastic cartridge enables a rapid, portable solution to screening and assessing HIV viral load.
1. Introduction
Recent estimates of people living with HIV/AIDS (PLWHA) indicate there are 38 million infected people worldwide of which around 6 million do not know their infection status1. As a comprehensive strategy for improving the clinical management of HIV, the World Health Organization (WHO) specifically calls for improved accessibility of not only anti-retroviral therapy, but also HIV testing and viral load monitoring2. For global eradication of HIV, the tools for treatment and testing must be made universally accessible and easy-to-use at the point-of-care3–5.
Current options for point-of-care testing include lateral flow strip rapid tests that detect viral antigens and host antibodies, which are affordable and fast, but not sensitive for detecting early infections and cannot quantify viral load6,7. Nucleic acid amplification tests (NAATs) such as polymerase chain reaction (PCR) are the standard-of-care for viral load detection and quantification from blood serum or plasma. However, the sample preparation required to separate plasma from blood is typically laborious and instrumentation for NAATs is expensive and not suitable for use at the point-of-care and low and middle-income countries (LMICs) due to bulky and expensive equipment. Both the Abbott m-PIMA and Cepheid GeneXpert platforms for testing HIV are marketed for use at the point-of-care, however each instrument costs $25,000 and $17,500 respectively and each test for assessing viral load (>$25 each) requires trained operators to conduct plasma separation by centrifugation8.
Although the advent of new polymerase enzymes has made direct PCR from blood possible without sample preparation, these assays are limited in the volume of blood that can be processed and require large volumes of reagents to compensate for assay inhibitors9. Detection of HIV nucleic acids directly from lysed whole blood has been investigated in a point-of-care format, though viral RNA was undetectable and the assay was limited to proviral DNA10.
To address the need for simplified extraction of plasma from whole blood, several microfluidic and filtration-based mechanisms have been proposed. Microfluidic techniques have been demonstrated based on sedimentation, microchannel filtration, and inertial forces to separate blood cells from plasma, though the micron-scale channels limit the speed and volumes at which the blood can be processed (0.1–100 μL/min)11. In contrast, membrane-based filtration methods have been developed that can quickly process larger sample volumes (>100 μL/min) with filter membranes that exclude contents of blood based on the size of red blood cells (RBCs) and white blood cells (WBCs) ranging from 6–8 μm and 6–20 μm, respectively, while letting viral particles (~100 nm) pass through for analysis12–15. Other attempts to bring plasma separation to the point-of-care have proposed miniaturization or simplification of centrifugation typically used for laboratory sample preparation with implementation of hand-held separation devices16–18.
Despite some of these techniques achieving a high efficiency of plasma separation, subsequent handling of the plasma for analysis was still required by a trained professional to conduct testing or interpretation of results. For use at the point-of-care and in LMICs, plasma separation should be integrated with nucleic acid purification and amplification with rapid automated results readout for lay-users. We have previously demonstrated use of droplet magnetofluidics to enable automation of nucleic acid purification coupled to nucleic acid amplification directly from clinical samples in a point-of-care format19–24. In contrast to traditional microfluidic techniques that require precise fluidic controls and microchannel fabrication, magnetofluidics leverages transfer of magnetic beads through discrete droplets of reagents for fully integrated sample-to-answer bioassays using simple low-power actuation of permanent magnets25,26. Due to the simplicity of our cartridge construction, each cartridge can be fabricated for around $2 to provide an affordable alternative to currently available commercial products21. However, the complex composition of whole blood does not allow for high recovery of viral nucleic acids on beads and previous work for integrated viral RNA analysis used already separated blood serum samples20.
In this study, we demonstrate filtration-assisted plasma extraction coupled with magnetofluidic cartridges (Fig. 1A) for streamlined HIV viral RNA purification and delivery to PCR assays from fingerprick volumes of whole blood. Viral particles in filtered plasma were lysed and released RNA was captured on silica-coated magnetic beads for purification and transfer to PCR for amplification and detection (Fig. 1B,C). To accommodate silica-based nucleic acid purification within the cartridges, we used a nucleic acid purification strategy composed entirely of aqueous buffers for compatibility with downstream PCR. All magnetic processing steps, PCR, and fluorescence readout were automated within a portable toaster-sized instrument. By combining membrane-based plasma filtration with magnetofluidic cartridges, we introduce a method for on-site plasma extraction with minimal manual steps that is integrated with rapid nucleic acid analysis. Operation with blood volumes easily acquired with self-collected fingerpricks27,28 opens potential for future use as a rapid point-of-care screening device or an at-home solution to viral load monitoring.
Fig. 1.
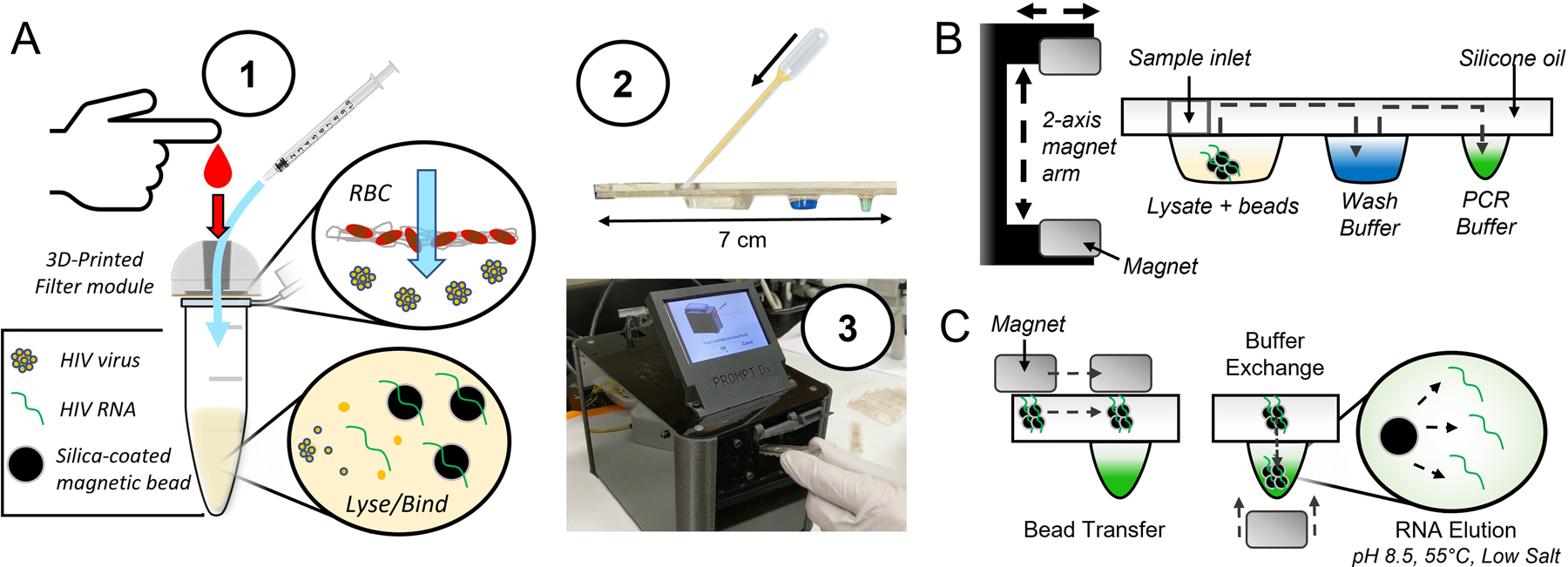
Filtration-assisted magnetofluidic blood-to-PCR workflow. A – To begin the test, (1) a droplet of blood is first deposited into the filter module and viral particles are rinsed through the filter membrane with phosphate buffered saline (PBS) using a syringe. The filter traps red blood cells (RBCs) while viral particles are small enough to pass through into a lysis and binding solution containing magnetic beads for capture of viral RNA. This entire filtered plasma mixture is (2) loaded into the assay cartridge, which is then (3) inserted into the instrument. B – The magnetic beads are transferred through an immiscible silicone oil layer into the cartridge’s extruded wells containing preloaded reagent buffers using a 2-axis motorized magnet arm. C – Bead transfer between wells is conducted by lateral movement of the top permanent magnet with bead exchange into buffers using vertical translation of the magnet arm to attract beads into the well with the bottom magnet. The final transfer of beads into the PCR buffer allows direct elution of RNA due to the relatively alkaline pH, elevated temperature, and low salt conditions.
2. Materials and Methods
Filtration module design
The filtration module (Fig. 2A) was composed of a 3D-printed syringe interface with a filtration membrane (Vivid Plasma separation membrane, GX grade) adhered to the outlet with a pressure-sensitive adhesive (PSA) ring (3M 9472LE). Parts were 3D-printed using clear resin on a Form 2 printer (Formlabs). To conduct plasma filtration, the filtration module could support up to 20 μL whole blood loaded into the syringe interface (Fig. S1 in ESI). All blood samples were acquired through BioIVT (normal human whole blood, K2EDTA Gender Unspecified) and handled in Biological Safety Level 2 (BSL-2) facilities. The blood plasma was then rinsed through the filter with a syringe preloaded with 1X phosphate buffered saline (PBS) press-fit into the 3D-printed body. The resulting filtrate was mixed with magnetic beads and custom buffers for nucleic acid purification and PCR. To assess the improvement in magnetic bead capture after blood filtration (Fig. 2C), 1 μL 104 TCID/mL HIV viral particles (ARP-7689, ATCC) was spiked into 10 μL whole blood, which was either mixed with 100 μL PBS directly or filtered through the membrane. The blood mixtures and filtrate were bound with 4 μL Chargeswitch magnetic beads (Invitrogen) with 10 μL Chargeswitch binding buffer and 1 μL 10% v/v Tween-20 for 1 minute, washed with 50 μL Chargeswitch W12 buffer, and eluted into 10 μL elution buffer, which was assessed with quantitative PCR (qPCR) (see ESI for PCR details).
Fig. 2.

Blood filtration module. A – The filter is integrated into a cavity within a 3D-printed form using a pressure-sensitive adhesive (PSA) ring that secures the filter and allows temporary attachment of the filter module to tubes or cartridges. The tip of a 1 mL syringe press-fit seals into the module for expression of the rinsing PBS through the filter. B – Filtration of blood mixed with PBS shows significant reduction of red blood cells and hemoglobin measured by the resulting reduced optical density of 540 nm wavelength light. C – Real-time PCR curves reveal filtration of whole blood permits higher recovery of RNA using magnetic bead purification.
Silica bead buffer selection
Binding buffers for viral lysis and binding of RNA to silica beads were made based on GuSCN solutions developed by Scallan et al.29, He et al.,30 or Katevatis et al.31. These buffers comprised a 5M GuSCN 100mM MES solution, a 5M GuSCN magnesium acetate (MgOAC) solution, a 4M GuSCN 55mM Tris-HCl solution, and a 4M GuSCN 55mM Tris-HCl solution with isopropanol. See ESI for full composition and methodology for buffer comparison.
To determine if polyethylene glycol (PEG)-based solutions were a suitable aqueous replacement for the organic solvents typically used as wash buffers for silica-based nucleic acid purification, 104 copies of synthetic RNA (VR-3278SD, ATCC) in 10 μL water was bound to 3 μL silica beads using 30 μL 5M guanidine thiocyanate (GuSCN) in 100mM MES solution (BioPerformance Certified M1317, Sigma-Aldrich). Beads were then washed with 50 μL 10%, 20%, 40% w/v PEG solutions (PEG 6,000, Fluka Analytical), or deionized water followed by elution into 10 μL elution buffer (M-Elution Buffer, Zymo). Amount of recovered RNA was quantified with qPCR run on a Biorad CFX96 thermocycler (see ESI for PCR details).
Comparison of PEG versus ethanol as a wash buffer was conducted by first preparing 12 tubes with 1 μL 103 TCID/mL HIV viral particles (ARP-7689, ATCC) in 50 μL PBS mixed with 150 μL binding buffer (4M GuSCN in 55 mM pH 7.5 Tris-HCl with 25 mM EDTA and 3% v/v Triton-X 100) and 3 μL Zymo Magbinding beads. Tubes were split into two groups of 6 tubes with or without 100 μL silicone oil (100 cSt, Sigma-Aldrich). Beads in three of the tubes of each group were washed with 70% v/v ethanol while the remainder were washed with 20% w/v PEG solution. Beads in tubes without oil were fully resuspended in each solution by vortexing and eluted into elution buffer as previously described, while beads in tubes with oil were moved solely with a permanent magnet and eluted directly into 10 μL PCR buffer to emulate conditions in the magnetofluidic cartridges. Resulting eluates were characterized for quantity of RNA recovered with qPCR.
Magnetofluidic cartridge design
Cartridges were assembled from polyethylene terephthalate glycol (PETG) wells (Welch Fluorocarbon) thermoformed over 3D-printed molds (Black resin, Formlabs Form 2) laminated to a laser-cut acrylic spacer (8560K171, McMaster-Carr) and laser-cut acrylic top section laminated with polytetrafluoroethylene (PTFE) tape (6305A18, McMaster-Carr) as previously described21. Prior to lamination, the thermoform was first filled with 50 μL 20% w/v PEG wash buffer and 7.5 μL PCR buffer in the respective wells. After lamination, the remaining space inside the cartridge was loaded with 100 cSt silicone oil followed by a plug of 40 μL docosane wax to seal off the sample well and keep it empty until loading with plasma filtrate and binding buffer.
Integrated magnetofluidic sample preparation and thermocycler instrumentation
Transfer of the beads with the cartridge was automated with neodymium magnets (K&J Magnetics) attached to a 3D-printed arm on a micro servo motor (TowerPro SG51R) to translate the magnets to the top and bottom of the cartridge wells. Translation of the magnets between cartridge wells was provided by a second servo motor (Hitec HS-485HB). A heat block was custom machined out of 6061 aluminum for heating the sample well to provide viral lysis and melt the wax plug in the cartridge using a power resistor as a heat source (Riedon PF1262-5RF1). A PCR heat block was machined from 145 copper and epoxied onto a thermoelectric element (Peltier Mini Module, Custom Thermoelectric) with temperature monitored on each of the heat blocks with thermistor probes (GA100K6MCD1, TE Connectivity) epoxied to the heat blocks. A 5V fan (Sunon) provided cooling to a heatsink on the side of the thermoelectric module opposite the PCR heat block.
Cartridges were illuminated using the red and blue diodes of a 3-color RGB LED (Vollong) passed through a focusing lens (10356, Carclo) and dual bandpass excitation filter (59003m, Chroma). Fluorescence was captured with a CMOS camera (Pi NoIR Camera V2, Raspberry Pi) through a dual bandpass emission filter (535-700DBEM, Omega Optical). Images of the PCR wells were taken at the end of each PCR annealing step with the blue LED for FAM and red LED for Cy5. Image processing for fluorescence detection and thermocycling was controlled with a Raspberry Pi 3B+ CPU via a motorshield (Dual TB9051FTG Motor Driver, Pololu).
Contrived blood sample testing
HIV viral particles were serially diluted from 104 to 101 TCID/mL in DEPC treated water followed by spiking 1 μL of each dilution into 10 μL whole blood and loading the spiked blood into the filter module. The plasma and viral particles were rinsed through the module with 200 μL PBS injected via syringe and the filtrate was mixed with 150 μL 4M GuSCN 55mM Tris-HCl binding buffer with 3 μL Zymo MagBinding beads and loaded into the first well of the assay cartridge. The cartridge was then loaded into the magnetofluidic instrument for bead transfer and PCR. The sample well was heated at 100°C for 100 seconds to melt the wax plug and encourage viral lysis. Beads were then pelleted and transferred through the oil within the cartridge with the magnet arm to the wash well where the beads were attracted between oil and wash buffer three times to remove PCR inhibitors. Elution of the captured RNA from the beads into PCR was achieved with heating the PCR with beads to 55°C for 1 minute. After elution, beads were extracted from the PCR well followed by 5-minute reverse transcription and 50 cycles of PCR with the heat block set to hold at 100°C for 2 seconds to denature and 55°C for 2 seconds to anneal and extend during each cycle. Cycle threshold (Ct) values from cartridge fluorescence data were determined using the second derivative maximum of a fitted 4-parameter logistic curve following baseline subtraction32. Average and standard error of baseline subtracted fluorescence values were calculated and plotted using the ggplot2 package version 3.3.3 using R software version 3.6.2
To assess the feasibility of our magnetofluidic assay for testing larger sample volumes, HIV viral particles were spiked in triplicate samples of 100 μL blood plasma and processed using either the previously mentioned reagents used in the cartridges to bind and wash the beads in-tube or the Invitrogen MagMAX Blood RNA isolation kit according to manufacturer instructions up until magnetic bead elution. Magnetic beads in both conditions were directly eluted into 7.5 μL PCR followed by qPCR on a Biorad CFX96 thermocycler.
3. Results and discussion
Blood filtration
Our filter module enables easy manual filtration in a compact format for readily integrating into the magnetofluidic cartridges for downstream analysis (Fig. 2A). Out of 3 candidate filter membranes, the Vivid asymmetric polysulfone filter produced the highest recovery of viral nucleic acids (Fig. S2 in ESI) and has been utilized in previous plasma separation studies13,15,33. Approximately 60 μL of the PBS rinse mixed with blood was recovered in the filtrate with the remainder lost to dead volume in the filter module (Fig. 2B). Despite this volume loss, filtration demonstrated a 16-fold improvement on recovery of viral RNA (Ct = 27.7±0.2) compared to direct bead extraction from unfiltered whole blood (Ct = 31.6±0.5) indicating viral particles were successfully rinsed through the membrane (Fig. 2C). This large shift in the Ct of PCR curves in Fig. 2C indicates components in whole blood (i.e. RBCs) preventing binding of nucleic acids to the magnetic beads is a dominating factor in the reduced sensitivity. If PCR inhibitors carried over from blood were the primary cause of the later Ct values, then we would expect the slope of the curves in Fig. 2C to flatten in the blood condition. The similar shape of curves in both conditions indicates similar amplification efficiency and minor inhibition. Overall recovery using bead processing was 23% for filtered blood compared to between 1–2% when extracting directly from whole blood (Fig. S2).
Optimization of magnetofluidic cartridge buffers for silica beads
Although prior magnetofluidic cartridges used Chargeswitch magnetic beads with polycationic coatings for pH-mediated capture and release of nucleic acids in aqueous buffers19–21, we found silica beads produced less inhibitor carryover into PCR and were more easily extracted out of high-protein blood solutions (Fig. S3 in ESI). Therefore, we sought to adapt silica beads into our cartridges and replace organic solvent washes with a PCR-compatible aqueous wash buffer.
For the first step in the silica bead RNA purification, we assessed several binding buffer conditions for sensitive detection of HIV RNA. All buffers tested used GuSCN as a chaotropic agent to facilitate nucleic acid binding and were based on binding buffers previously described for capture of nucleic acids onto silica surfaces29–31. Recovery was assessed for simultaneous capture of both HIV RNA and human RNA measured by a control assay for the RNAse P (RP) gene34. The best recovery indicated by lowest PCR Ct value was achieved with the buffer based on conditions described by Scallan et al29 using 4M GuSCN in 55mM pH 7.5 Tris-HCl with 3% v/v Triton-X 100 (labelled as Tris-HCl in Fig. 3A). Amplification of the RP gene across all conditions demonstrates its use as a reliable duplexed control to confirm the presence of successfully processed human samples.
Fig. 3.
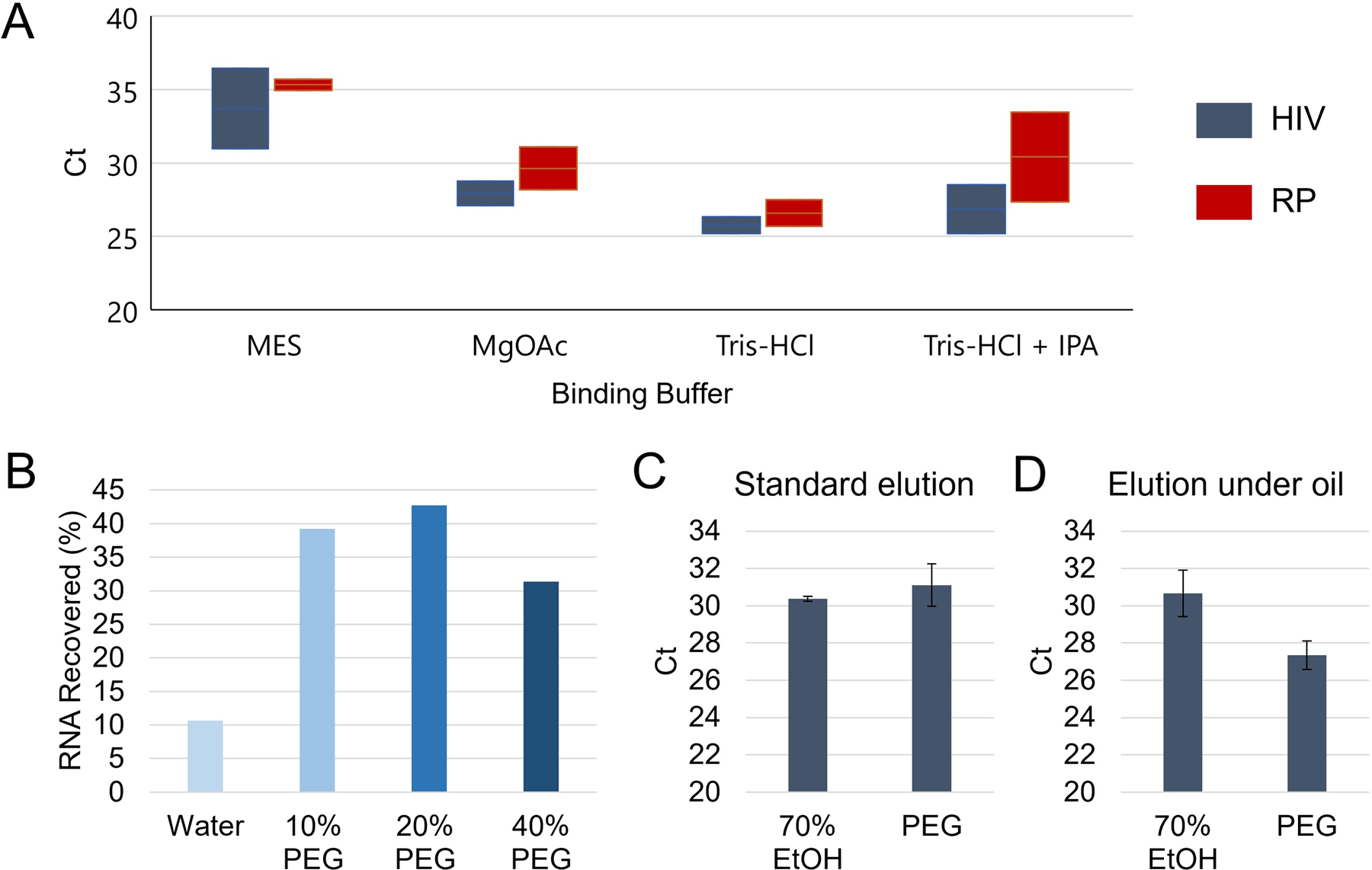
Silica bead buffer characterization. A – Ranges of Ct values for PCR amplification of HIV RNA and human RNAse P (RP) after direct elution of silica beads following blood filtration and varying binding buffer solutions with 20% w/v PEG washes (n=2). B – Aqueous solutions with polyethylene glycol (PEG) showed effective washing of silica beads while improving HIV RNA retention compared to water alone as a wash buffer. C – Compared to traditional ethanol-based washes (n=3), PEG washes (n=3) performed comparably with no significant difference in Ct for captured HIV RNA amplification (p=0.39). D – When using silicone oil to cover the silica beads during binding, wash, and elution, the PEG washes (n=3) exhibited lower Ct values (p=0.03) compared to ethanol washes (n=3) indicating superior RNA recovery.
For washing the beads after binding, organic solvents such as ethanol or isopropanol are typically used. These solvents serve to dehydrate the silica beads to preserve nucleic acid binding to the silica while allowing diffusion of inhibitory salts, proteins, and other compounds off the beads to purify the nucleic acids35. These solvents themselves are inhibitory to PCR and are allowed to evaporate from the beads in standard procedures prior to bead elution. However, in the enclosed system of our magnetofluidic cartridges, reagents are covered in immiscible oil, which prevents the possibility of evaporation, so direct elution of the beads risks assay inhibition from wash buffer carryover.
A wash buffer should permit diffusion of binding buffer molecules and other PCR inhibitors away from the beads while preserving the binding of nucleic acids, ideally using components that are inert or beneficial to PCR. We chose PEG as a wash buffer additive to act as a crowding agent that maintains RNA in a precipitated state on the beads while permitting diffusion of small molecules like proteins and GuSCN. PEG is a highly soluble polymer previously used for nucleic acid precipitation36,37 and as an additive in PCR to improve specificity38. We compared PEG wash buffers to water alone and found a 3 to 4-fold increase in RNA recovered when using PEG (Fig. 3B). For all subsequent experiments we used 20% w/v PEG washes, which exhibited the highest recovery. When compared side-by-side with a more traditional organic solvent wash buffer (70% ethanol) using standard in-tube conditions where beads were fully resuspended with vortexing and dried after wash, no significant difference (p = 0.39) in recovery was exhibited compared to PEG wash (Fig. 3C). In contrast, when cartridge conditions were mimicked with a layer of silicone oil and beads manipulated solely through magnetic attraction (Fig. 3D), the PEG wash illustrated significantly better recovery (p = 0.03). This inhibited performance for ethanol in oil could be attributed to the carryover of ethanol into the final PCR due to lack of evaporation.
Magnetofluidic PCR instrumentation
After manual transfer of filtered blood with magnetic particles into the cartridge, the instrument allowed for seamless integration of nucleic acid purification and PCR. A motorized magnetic arm (Fig. 4A) transferred the magnetic beads along the internal PTFE coating of the cartridge to move beads between wells using the top magnet. Rotating the arm to bring an opposing magnet to the bottom of the wells coordinated bead transfer into the buffer of choice. By automating sample transport with magnetic attraction instead of fluidic transfer, sample volumes are more flexible compared to traditional microfluidics that require control flowrates on the order of μL per minute. Sample heating mediated by a resistive heater followed by magnetofluidic purification through wash buffer and elution into PCR required <8 minutes. With nucleic acid eluted into the PCR buffer, viral RNA was converted to cDNA with a 5-minute reverse transcription step followed by thermocycling for amplification. Our miniaturized heat block enabled rapid temperature transitions with 40 cycles in less than 13 minutes (Fig. 4B).
Fig. 4.
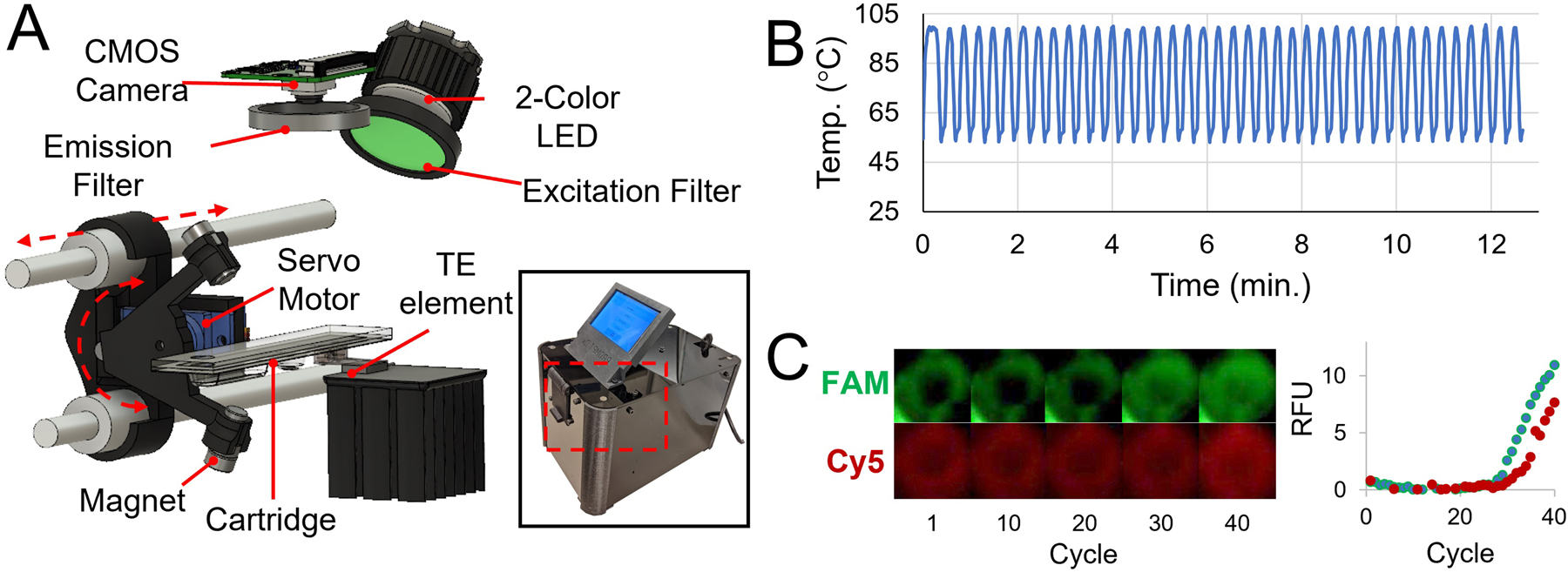
Magnetofluidic instrumentation and PCR. A – Schematic of internal instrumentation with servo-actuated magnet arm, thermoelectric (TE) PCR heat block, and CMOS camera-based fluorescence imaging with LED illumination. B – Temperature readout from thermistor attached to PCR heat block during thermocycling. 40 cycles complete in under 13 minutes. C – Isolated images of PCR well in both FAM (HIV) and Cy5 (RP) channels with corresponding baseline-subtracted real-time fluorescence curve reporting average pixel intensities at each cycle.
Multiplexed fluorescence detection was accomplished with CMOS camera imaging coupled with LED illumination using dual bandpass excitation and emission filters for FAM and Cy5 fluorescence spectra (Fig. 4A). Sequential illumination with blue then red LEDs at the end of each PCR annealing step was synchronized with imaging to capture fluorescence in each channel (Fig.4C). Averaged pixel intensity was extracted in real-time to calculate fluorescence signal and analyze curves for amplification.
PCR assay
The PCR assay was designed to detect both HIV RNA and human RNAse P (RP) as an endogenous control in a single reaction. Prior to incorporating the assay into our cartridges, we validated sensitivity of the HIV primer design down to 10 copies per reaction with rapid thermocycling conditions using just one second hold times for denaturation at 95°C and annealing at 60°C (Fig. S6 in ESI). RNA copy number conversion of the viral particles stock was determined to be 1 TCID50/mL = 20 copies/μL. Using rapid thermocycling PCR with our cartridge, the assay sensitivity maintained similar sensitivity with detection of directly spiked HIV particles (Fig. S7 in ESI). Our assay used glycerol free reagents to allow potential future lyophilization for stable dry storage of reagents on cartridge for point-of-care use in low-resource settings.
Testing with contrived blood samples
Full processing of whole blood samples spiked with HIV viral particles through filtration, magnetic bead processing, and integrated PCR enabled successful amplification and automated detection of HIV in our magnetofluidic instrument (Fig. 5A). Cartridges demonstrated a limit of detection with detectable amplification in all 3 replicates down to 1000 copies of HIV RNA per blood sample with assay completion in <30 minutes after insertion of the cartridge into the instrument. Serial dilutions of HIV viral particles resulted in a strong linear trend in the Ct values that could be leveraged for quantification of viral load (Fig. 5B).
Fig. 5.
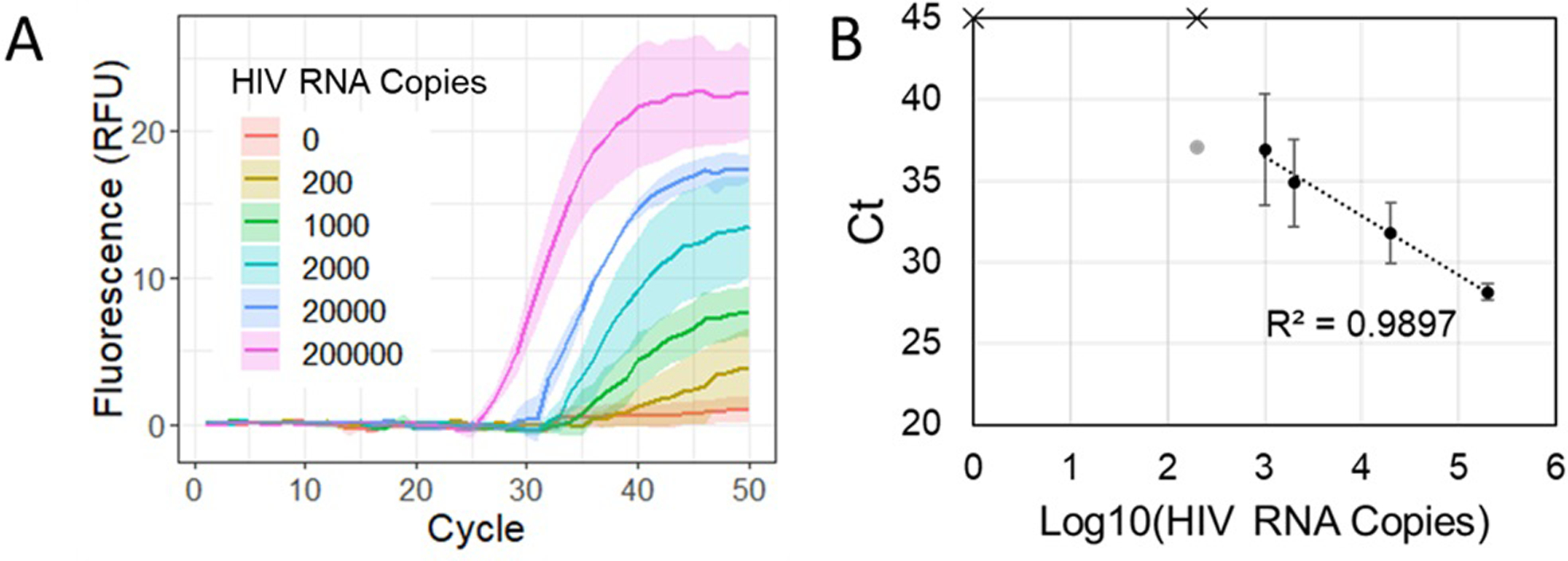
Cartridge tests with blood samples. A – Real-time fluorescence for cartridge amplification of HIV RNA processed from HIV viral particles spiked into whole blood (n = 3 cartridges for each concentration). Solid lines indicate average fluorescence and bands show standard error. B – Standard curve with Ct values generated from cartridge amplification data. Solid circles indicate successful amplification in all 3 replicates while faded circles indicate a lone positive result at the given concentration. Data points with an ‘x’ indicate no amplification detected.
Given the 10 μL starting sample volume, the current cartridge results yielded a limit of detection of 105 copies/mL. To demonstrate the feasibility of processing larger sample volumes, we used 100 μL of blood plasma and the same binding and wash buffer reagents used in the cartridge. We showed consistent detection of 1000 copies per sample (Fig. S8 in ESI) for a potential improvement to 104 copies/mL assuming equivalent scale-up of the filtration module. Additional testing with the commercial MagMAX kit using 100 μL of blood plasma demonstrated recovery as low as 103 copies/mL, illustrating further optimization of our magnetic bead chemistry and larger sample volumes may enable cartridges sensitive enough to detect the threshold of low-level viremia and virological failure39.
4. Conclusions
Magnetofluidic technology provides streamlined sample-to-answer nucleic acid purification and amplification for point-of-care screening of infectious diseases. Integration of assays into magnetofluidic cartridges provides a compact format that can be made with scalable manufacturing techniques at low-cost. Improving ease-of-use and access to molecular diagnostics for HIV would enable higher sensitivity screening of infections compared to existing point-of-care assays that require confirmatory tests and open potential for use in self-administered at-home viral load monitoring.
Detection of RNA allows for identification of early infections prior to seroconversion, which would be undetectable by rapid antibody test strips often used for HIV screening6,40. Recently developed tests for HIV-1 p24 antigen targeted early identification, though assessments of commercially available products found laboratory p24 tests had limits of detection corresponding to viral loads of approximately 40,000 copies/mL or higher, while the point-of-care rapid antigen tests failed outright to detect positive samples used in the study41. We have demonstrated feasibility of our assay detecting 10,000 copies/mL. with 100 μL sample volumes, and even with just 10 μL of blood, our current cartridge platform would outperform the point-of-care p24 antigen tests. Furthermore, automated analysis and interpretation of fluorescent results with our platform reduces the risk of error due to operator interpretation, and rapid results provide an opportunity for earlier linkage to care in a clinical context without necessitating additional visits.
Although our platform shows promise for more sensitive screening compared to antigen tests, using fingerprick volumes in its current form would not allow detection at the threshold for identifying low-level viremia (<200 copies/mL) and virological failure (>200 copies/mL)39. Optimizing our magnetic bead assay to match the nucleic acid recovery efficiency of commercial magnetic bead kits (Fig. S8 in ESI) combined with an increase in sample volume would improve assay sensitivity to meet these goals. It has been demonstrated that 10–250 μL27,28,42 or even more (250–500 μL43) capillary blood could be collected from a fingerprick although a maximum volume of 200 μL from fingerprick sampling is recommended44. For larger capillary blood volumes, blood collection devices such as Tasso+ that enable untrained user to painlessly sample up to 600 μL capillary blood45 could be an option. While using larger sample volumes, the captured nucleic acids would still be concentrated into a small magnetic bead volume for transport. Therefore, increasing the sample volume will only require equivalent scaling-up of the filtration module and increasing the cartridge’s sample well volume without significant changes to the remainder of the cartridge, thus keeping PCR thermal mass low for similar rapid thermocycling and fast turnaround times.
While many proposed point-of-care molecular platforms rely on isothermal assays such as LAMP and RPA46,47, these assays lack the quantification capabilities of PCR and multiplexing is made difficult due to large numbers of primer interactions in LAMP and non-specific binding at low temperatures in RPA48. The ability to quantify viral load with PCR enables potential monitoring of patient response to treatment or development of viral resistance to treatment49 and multiplexing enables integration of control assays and opportunities for simultaneous screening of other bloodborne pathogens like Hepatitis C.
To make testing truly amenable to point-of-care and home use, there are a few features that would be necessary for translation. Future designs should incorporate the filtration module onto the cartridge for a fully integrated user workflow. Storage of PCR reagents in a dry format within the cartridge would enable shelf-stability at ambient temperatures to increase testing accessibility and reduce cost where cold-chain transport and storage is difficult. Although the feasibility of HIV viral load quantification has been previously demonstrated from fingerprick blood samples using plasma separation for sensitive detection of viral RNA from blood42, further testing will be necessary with clinical capillary blood samples instead of venous whole blood to evaluate the performance of this platform with varying input volumes and compositions from a variety of patients.
Supplementary Material
Acknowledgements
This work was supported by funding through the National Institutes of Health (R01AI138978, R01AI137272, R61AI154628).
Footnotes
Conflicts of interest
T.-H.W and A.Y.T are co-inventors on patents related to magnetofluidic cartridge technology.
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
References
- 1.UNAIDS, Global HIV & AIDS statistics — Fact sheet, https://www.unaids.org/en/resources/fact-sheet, (accessed 26 June 2021).
- 2.World Health Organization, Viral suppression for HIV treatment success and prevention of sexual transmission of HIV, https://www.who.int/news/item/20-07-2018-viral-suppression-for-hiv-treatment-success-and-prevention-of-sexual-transmission-of-hiv, (accessed 2 July 2021).
- 3.Brook G, Sex. Transm. Infect, 2018, 94, 394–395. [DOI] [PubMed] [Google Scholar]
- 4.Dorward J, Drain PK and Garrett N, Lancet HIV, 2018, 5, e8–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hellard M, Ryan KE and Stoové M, Lancet HIV, 2020, 7, e216–e217. [DOI] [PubMed] [Google Scholar]
- 6.Parker MM, Bennett SB, Sullivan TJ, Fordan S, Wesolowski LG, Wroblewski K and Gaynor AM, J. Clin. Virol, 2018, 104, 89–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Neal JD, Golden MR, Branson BM and Stekler JD, JAIDS J. Acquir. Immune Defic. Syndr, 2012, 60, e119–e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherjee S, Cohn J, Ciaranello AL, Sacks E, Adetunji O, Chadambuka A, Mafaune H, Makayi M, McCann N and Turunga E, J. Acquir. Immune Defic. Syndr, 2020, 84, S63–S69. [DOI] [PubMed] [Google Scholar]
- 9.Cai D, Behrmann O, Hufert F, Dame G and Urban G, Sci. Rep, 2018, 8, 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McFall SM, Wagner RL, Jangam SR, Yamada DH, Hardie D and Kelso DM, J. Virol. Methods, 2015, 214, 37–42. [DOI] [PubMed] [Google Scholar]
- 11.Kersaudy-Kerhoas M and Sollier E, Lab Chip, 2013, 13, 3323–3346. [DOI] [PubMed] [Google Scholar]
- 12.Wang SQ, Sarenac D, Chen MH, Huang SH, Giguel FF, Kuritzkes DR and Demirci U, Int. J. Nanomedicine, 2012, 7, 5019–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Homsy A, van der Wal PD, Doll W, Schaller R, Korsatko S, Ratzer M, Ellmerer M, Pieber TR, Nicol A and de Rooij NF, Biomicrofluidics, 2012, 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gan W, Zhuang B, Zhang P, Han J, Li CX and Liu P, Lab Chip, 2014, 14, 3719–3728. [DOI] [PubMed] [Google Scholar]
- 15.Liu C, Liao S-C, Song J, Mauk MG, Li X, Wu G, Ge D, Greenberg RM, Yang S and Bau HH, Lab Chip, 2016, 16, 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhamla MS, Benson B, Chai C, Katsikis G, Johri A and Prakash M, Nat. Biomed. Eng, 2017, 1, 0009. [Google Scholar]
- 17.Vemulapati S and Erickson D, Anal. Chem, 2019, 91, 14824–14828. [DOI] [PubMed] [Google Scholar]
- 18.Liu CH, Chen CFCACF, Chen SJ, Tsai TT, Chu CC, Chang CC and Chen CFCACF, Anal. Chem, 2019, 91, 1247–1253. [DOI] [PubMed] [Google Scholar]
- 19.Shin DJ, Athamanolap P, Chen L, Hardick J, Lewis M, Hsieh YH, Rothman RE, Gaydos CA and Wang TH, Sci. Rep, 2017, 7, 4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin DJ, Trick AY, Hsieh Y-H, Thomas DL and Wang T-H, Sci. Rep, 2018, 8, 9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trick AY, Melendez JH, Chen F-E, Chen L, Onzia A, Zawedde A, Nakku-Joloba E, Kyambadde P, Mande E, Matovu J, Atuheirwe M, Kwizera R, Gilliams EA, Hsieh Y-H, Gaydos CA, Manabe YC, Hamill MM and Wang T-H, Sci. Transl. Med, 2021, 13, eabf6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trick AY, Chen F, Chen L, Lee P, Hasnain AC, Mostafa HH, Carroll KC and Wang T, Adv. Mater. Technol, DOI: 10.1002/admt.202101013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen F-E, Lee P-W, Trick AY, Park JS, Chen L, Shah K, Mostafa H, Carroll KC, Hsieh K and Wang T-H, Biosens. Bioelectron, 2021, 190, 113390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Wen K, Chen F-E, Trick AY, Liu H, Shao S, Yu W, Hsieh K, Wang Z, Shen J and Wang T-H, Anal. Chem, 2021, 93, 10940–10946. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y and Wang TH, Adv. Mater, 2013, 25, 2903–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y and Nguyen N-T, Lab Chip, 2017, 17, 994–1008. [DOI] [PubMed] [Google Scholar]
- 27.Serafin A, Malinowski M and Prażmowska-Wilanowska A, Postgrad. Med, 2020, 132, 288–295. [DOI] [PubMed] [Google Scholar]
- 28.Tang R, Yang H, Choi JR, Gong Y, You M, Wen T, Li A, Li X, Xu B, Zhang S, Mei Q and Xu F, Crit. Rev. Clin. Lab. Sci, 2017, 54, 294–308. [DOI] [PubMed] [Google Scholar]
- 29.Scallan M, Dempsey C, MacSharry J, O’Callaghan I, O’Connor P, Horgan C, Durack E, Cotter P, Hudson S, Moynihan H and Lucey B, bioRxiv, 2020, 2–7. [Google Scholar]
- 30.He H, Li R, Chen YY, Pan P, Tong W, Dong X, Chen YY and Yu D, Sci. Rep, 2017, 7, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katevatis C, Fan A and Klapperich CM, PLoS One, 2017, 12, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tichopad A, Dilger M, Schwarz G and Pfaffl MW, Nucleic Acids Res, 2003, 31, 122e–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C, Mauk M, Gross R, Bushman FD, Edelstein PH, Collman RG and Bau HH, Anal. Chem, 2013, 85, 10463–10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Centers for Disease Control and Prevention (CDC), CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel, Revision: 06, https://www.fda.gov/media/134922/download, (accessed 21 July 2021).
- 35.Jue E, Witters D and Ismagilov RF, Sci. Rep, 2020, 10, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lis JT and Schleif R, Nucleic Acids Res, 1975, 2, 383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paithankar KR and Prasad KSN, Nucleic Acids Res, 1991, 19, 1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim HR, Baek A, Lee IJ and Kim DE, ACS Appl. Mater. Interfaces, 2016, 8, 33521–33528. [DOI] [PubMed] [Google Scholar]
- 39.Department of Health and Human Services, Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV, https://clinicalinfo.hiv.gov/sites/default/files/guidelines/documents/AdultandAdolescentGL.pdf, (accessed 3 November 2021).
- 40.Hurt CB, Nelson JAE, Hightow-Weidman LB and Miller WC, Sex. Transm. Dis, 2017, 44, 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stone M, Bainbridge J, Sanchez AM, Keating SM, Pappas A, Rountree W, Todd C, Bakkour S, Manak M, Peel SA, Coombs RW, Ramos EM, Shriver MK, Contestable P, Nair SV, Wilson DH, Stengelin M, Murphy G, Hewlett I, Denny TN and Busch MP, J. Clin. Microbiol, 2018, 56, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fidler S, Lewis H, Meyerowitz J, Kuldanek K, Thornhill J, Muir D, Bonnissent A, Timson G and Frater J, Sci. Rep, 2017, 7, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kattenberg JH, Tahita CM, Versteeg IAJ, Tinto H, Traoré-Coulibaly M, Schallig HDFH and Mens PF, Trop. Med. Int. Heal, 2012, 17, 550–557. [DOI] [PubMed] [Google Scholar]
- 44.Drain PK, Dorward J, Bender A, Lillis L, Marinucci F, Sacks J, Bershteyn A, Boyle DS, Posner JD and Garrett N, Clin. Microbiol. Rev, DOI: 10.1128/CMR.00097-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tasso+, https://www.tassoinc.com/tasso-plus, (accessed 16 January 2022).
- 46.Moehling TJ, Choi G, Dugan LC, Salit M and Meagher RJ, Expert Rev. Mol. Diagn, 2021, 21, 43–61. [DOI] [PubMed] [Google Scholar]
- 47.Lillis L, Lehman DA, Siverson JB, Weis J, Cantera J, Parker M, Piepenburg O, Overbaugh J and Boyle DS, J. Virol. Methods, 2016, 230, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Macdonald J and Von Stetten F, Analyst, 2019, 144, 31–67. [DOI] [PubMed] [Google Scholar]
- 49.Stevens WS, Scott LE and Crowe SM, J. Infect. Dis, DOI: 10.1086/650392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


