Abstract
Inorganic nanocrystals such as quantum dots (QDs) and upconverting nanoparticles (UCNPs) are uniquely suited for quantitative live-cell imaging and are typically functionalized with ligands to study specific receptors or cellular targets. Antibodies (Ab) are among the most useful targeting reagents owing to their high affinities and specificities, but common nanocrystal labeling methods may orient Ab incorrectly, be reversible or denaturing, or lead to Ab-NP complexes too large for some applications. Here, we show that SpyCatcher proteins, which bind and spontaneously form covalent isopeptide bonds with cognate SpyTag peptides, can conjugate engineered Ab to nanoparticle surfaces with control over stability, orientation, and stoichiometry. Compact SpyCatcher-functionalized QDs and UCNPs may be labeled with short-chain variable fragment Ab (scFv) engineered to bind urokinase-type plasminogen activator receptors (uPAR) that are overexpressed in many human cancers. Confocal imaging of anti-uPAR scFv-QD conjugates shows the Ab mediates specific binding and internalization by breast cancer cells expressing uPAR. Time-lapse imaging of photostable scFv-UCNP conjugates show that Ab binding causes uPAR internalization with a ~20-minute half-life on the cell surface, and uPAR is internalized to endolysosomal compartments distinct from general membrane stains and without significant recycling to the cell surface. The controlled and stable conjugation of engineered Ab to NPs enables targeting of diverse receptors for live-cell study of their distribution, trafficking, and physiology.
Keywords: bioconjugation, upconverting nanoparticles, quantum dots, antibodies, uPAR, cancer cells, receptor trafficking
Graphical Abstract
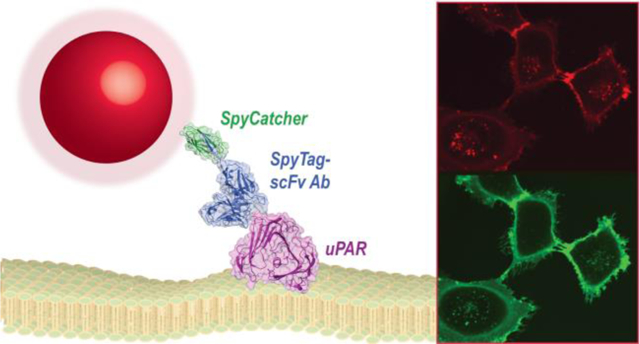
The specific and sensitive detection of cell-surface markers is essential for understanding molecular signaling in both healthy and diseased cells. Cell surface receptors or other biomarkers are typically detected by antibody (Ab) conjugates designed for fluorescence imaging or for targeting therapeutics.1,2 Ab selection and engineering technologies have enabled design of these targeting proteins with high affinity and selectivity for a broad range of biomolecules,3 and Ab miniaturization and labeling strategies have found use in a range of imaging and biotechnology applications.4,5
Ab typically used for immunotargeting are large (150 kDa) and highly polymorphic IgG with 2 identical antigen binding sites, which are spaced up to 17 nm apart in X-ray structures (Fig. 1A).6 Protein engineering has produced monovalent Ab constructs that are significantly smaller and retain antigen affinity and selectivity, including single chain variable fragments (scFv) that are 25–30 kDa and are often more amenable to expression and modification.3 Numerous sidechain and carbohydrate bioconjugation reactions have been developed for synthesizing Ab conjugates, although some of these reactions have been shown to be reversible under physiological conditions7–9 or to reduce Ab stability or affinity for ligand.10–12
Figure 1. Design of nanoparticle-SpyCatcher-scFv antibody conjugates.
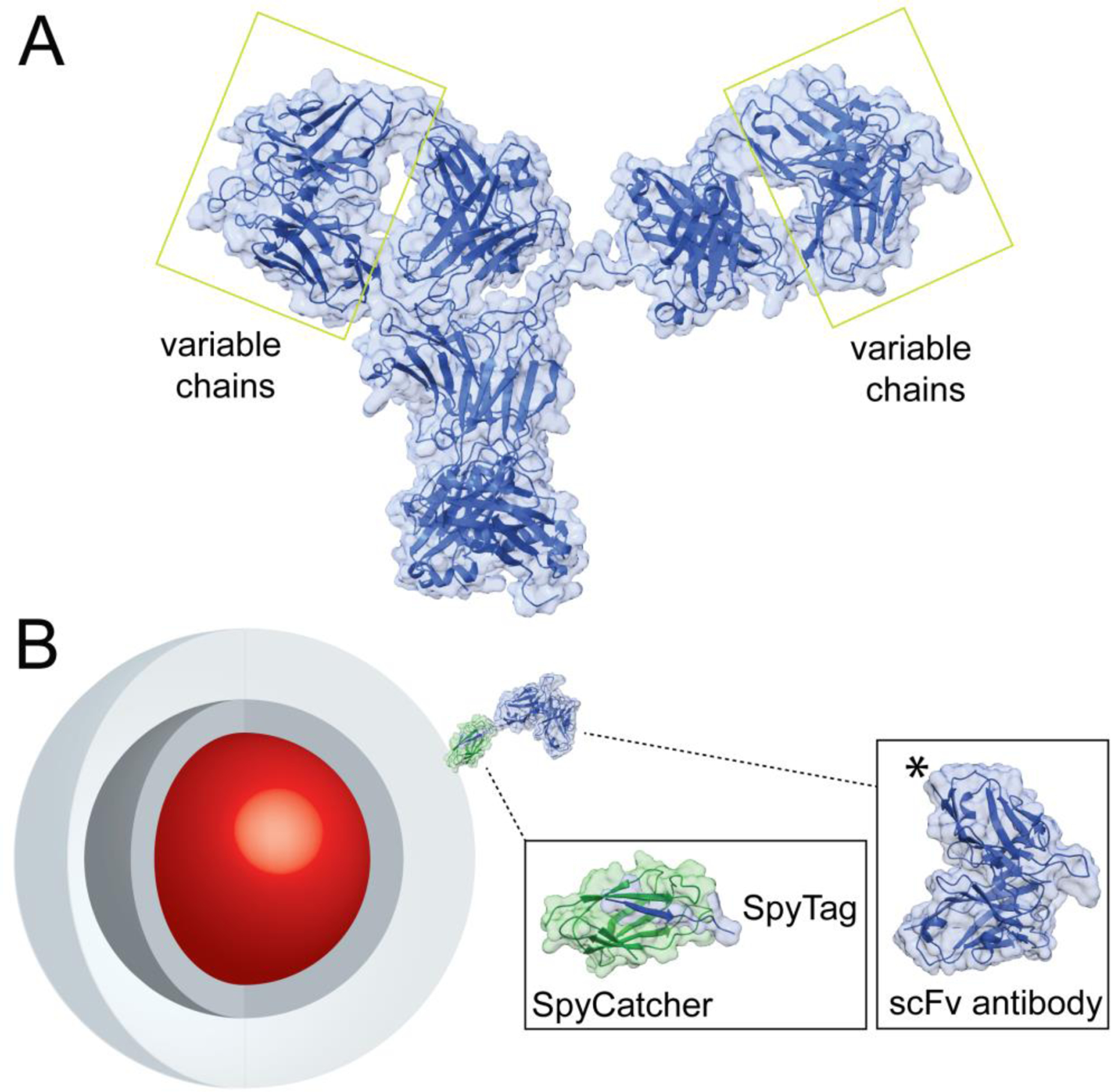
A) Structure of full IgG Ab (PDB file 1HZH).6 Variable chains that bind antigens and are used to engineer miniaturized scFv Ab are highlighted. B) Schematic of NP-SpyCatcher-scFv Ab conjugates. Core/shell nanocrystals (red and dark gray), encapsulated in amphiphilic polymers (light gray), conjugated to a single-Cys SpyCatcher (PDB file 4MLI)23 and SpyTag-scFv (PDB file 5YD5)24 chimera. The SpyTag peptide is added at the scFv C terminus, marked (*) in the structure.
Inorganic nanoparticles (NP) possess optical properties that make them particularly well-suited for live cell imaging. Compared to organic or protein-based fluorophores, semiconductor quantum dots (QDs) have larger optical cross-sections, increased photostability, and broadband excitation spectra.13–16 Similarly, upconverting nanoparticles (UCNPs), which emit at visible wavelengths after absorption of multiple near infrared (NIR) photons, show no overlap with cellular autofluorescence, no measurable blinking or photobleaching at single-molecule powers, and can be imaged millimeters into tissue with exceptionally low laser fluences.17–20 For Ab-based imaging applications, inorganic NPs have adapted organic fluorophore bioconjugation reactions,14,21,22 but difficulties in controlling immunoconjugate size, stoichiometry, Ab orientation, and stability have limited the broad utility of Ab-NP conjugates.25–31 Bioorthogonal covalent reactions that address these issues may be useful in expanding the scope of protein-NP conjugates in bioimaging,32 although these bimolecular reactions typically require higher reactant concentration than is possible for relatively large proteins and colloidal nanoparticles. One exception is the engineered split protein SpyCatcher/SpyTag,33,34 in which the components bind to one another with nanomolar affinity before forming stable isopeptide bonds, and which is emerging as a versatile system for the controlled and stable conjugation of proteins to NPs.35,36
The urokinase-type plasminogen activator receptor (uPAR) is a glycolipid-anchored cell membrane receptor critical to regulation of multiple cellular processes and has been found to have increased plasma membrane expression in many human cancer types.37–40 uPAR activation leads to its internalization in a complex multi-step process that is critical for its activity and which is not fully understood.37,41,42 Here, we describe the SpyCatcher-mediated synthesis of compact nanoparticle conjugates to anti-uPAR scFv Ab for controlled and stable display of engineered Ab on nanoparticle surfaces. scFv-QD conjugates show specific internalization by uPAR-expressing cells, and live-cell imaging of scFv-UCNPs enables extended tracking of uPAR complexes. We observe that scFv-UCNP binding causes receptor internalization with a ~20-minute half-life on the cell surface, and the complex is internalized to endolysosomal compartments distinct from general membrane stains without significant recycling of scFv-UCNP conjugates to the cell surface. SpyCatcher-mediated synthesis of nanoparticle immunoconjugates is applicable to multiple inorganic nanocrystals and to Ab that can be engineered to a scFv, enabling a labeling strategy broadly applicable to receptors and other cellular targets.
RESULTS AND DISCUSSION
Synthesis of SpyCatcher-functionalized nanoparticles
To synthesize stable and well-defined Ab-NP conjugates, we leveraged the capacity of SpyCatcher proteins on nanoparticle surfaces to conjugate SpyTag-bearing Ab under dilute, neutral conditions,36 without chemical modification of the Ab. Inorganic nanocrystals, either 9-nm CdSe/CdS QDs15,16 or 19-nm core/shell NaYb0.8Er0.2F4 alloyed UCNPs (aUCNPs),19,43 were synthesized and characterized by TEM, XRD, dynamic light scattering (DLS), and optical methods (Figs. 2 and S1–S3).15,19 To impart biocompatibility and resistance to aggregation in buffer without degrading optical properties, nanocrystals were encapsulated in amphiphilic copolymers (PAOA44 or PMAO45; see Methods) bearing short PEGs and surface bioconjugation handles. DLS of aqueous aUCNPs shows no apparent aggregation and an increase in hydrodynamic diameter from 19 to 21 nm (Fig. 2). Conjugation to proteins was achieved through introduction of thiol-reactive groups at the NP surfaces, either by PAOA reaction with the amine-thiol crosslinker SM(PEG)2, or by PMAO reaction with pyridyldisulfide-ethylamine (PDEA) to display activated disulfides (Fig. S4). For conjugation to NP surfaces, a single Cys at Ser35 was introduced in an exposed SpyCatcher loop on the face opposing the SpyTag reaction site, and the protein was overexpressed in E. Coli.36 Conjugation to nanoparticle surfaces was analyzed by DLS, with hydrodynamic diameters increasing from 21 to 24 nm (Fig. 2D). The conjugation of S35C SpyCatcher was monitored by absorbance of the leaving group pyridine-2-thione,44,46 showing a stoichiometry of ~20 surface SpyCatcher proteins per aUCNP (Fig. S5).
Figure 2. Synthesis of scFv Ab-conjugated aUCNPs.
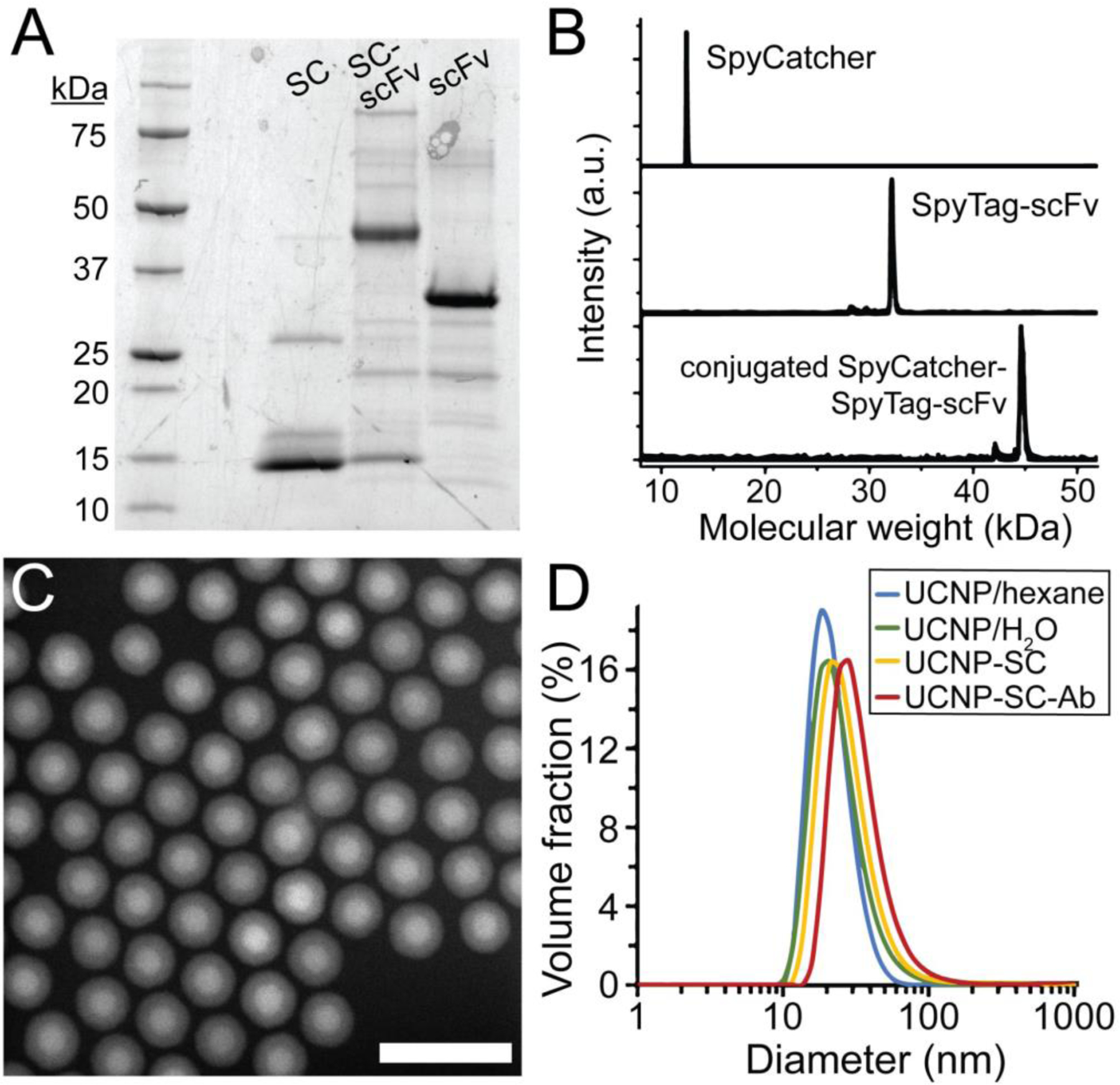
(A) Expression and reaction of SpyCatcher (SC) and SpyTag-scFv Ab (ST-scFv), monitored by SDS-PAGE. Partially purified His-tagged scFv were incubated with an excess of SC. (B) MALDI mass spectra of purified SC and ST-scFv before and after conjugation. (C) High-angle annular dark field transmission electron micrograph of core/shell NaEr0.8Yb0.2F4 @ NaY0.8Gd0.2F4 aUCNPs. Scale bar is 50 nm. (D) Diameters of nanoparticles determined by dynamic light scattering (DLS), of hydrophobic (blue), PMAO-encapsulated (teal), SpyCatcher-functionalized (yellow), and scFv-conjugated (red) UCNPs. Mean diameters (n = 5) are 19, 21, 24, and 28 nm, respectively.
Conjugation of engineered Ab to nanoparticles
The high-affinity 2G10 Ab against human uPAR was previously selected by phage screening of a naive human Fab library against native, folded uPAR.38,39,41 This 2G10 heavy chain sequence has demonstrated monovalent dissociation constants in the nM range, selective binding to uPAR overexpressed HEK-293 cells, and inhibition of uPAR interaction with its uPA ligand. A single chain fragment (scFv) connecting both variable regions, VH and VL, was constructed by standard cloning techniques,47 and a 3mer spacer followed by an 13mer SpyTag sequence33 were added to the C terminus away from the uPAR binding site (Fig. 1). Expression in E. Coli produces a small 2G10 scFv-SpyTag chimera (32 kDa), whose reaction with free SpyCatcher is complete overnight at 4 °C, as confirmed by gel electrophoresis and mass spectrometry (Fig. 2A–B). Conjugation of SpyTag scFv to SpyCatcher-coated NPs is then carried out in neutral buffer at room temperature over 16 h, and monitored by DLS, showing a diameter increase from 24 to 28 nm for aUCNPs (Figure 2D). Because the SpyTag-SpyCatcher reaction goes almost to completion under these conditions,33,34 the average number of surface scFv was controlled by reaction stoichiometry (~2 Ab per QD and ~10 per aUCNP), and the immunoconjugates were used without further purification. The stoichiometry of scFv-aUCNP conjugation was measured using Trp fluorescence, and the average number of scFv per aUCNP was varied from 1 to 20 simply by control of concentrations of each (Fig. S6).
Live-cell imaging of scFv-QD conjugates
To determine whether Ab conjugated through NP surface-bound SpyCatcher proteins can target their antigens in live-cell imaging, we examined whether MDA-MB-231 cells, a breast cancer cell line that express high levels of uPAR receptors,41 would selectively internalize 2G10 scFv-NP immunoconjugates. Cells were incubated with scFv-functionalized QDs for 1 hour at 37 °C, followed by counterstain with membrane lectin-binding wheat germ agglutinin (WGA), and then characterized by live-cell confocal microscopy (Fig 3). The 2G10 scFv-QDs are rapidly internalized, and cells continue to show well-defined filopodia, actin-rich protrusions from the plasma membrane;48 the persistence of these structures even in the absence of extracellular matrix (e.g., fibronectin or collagen) is suggestive that the QD immunoconjugates are not detrimental to cell health.49 Internalized uPAR QDs and WGA show little or no co-localization (Fig. 3C), possibly owing to the sequential timing of labeling. QDs displaying SpyCatcher but without scFv show little attachment to the cell surface or internalization after incubation (Fig. 3D–F). Taken together, these results indicate that the 2G10 scFv remains active and selective for uPAR once conjugated to a nanoparticle surface, and that scFv-QD triggers a uPAR internalization sequence similar to native ligands.50,51
Figure 3. Specificity of anti-uPAR-QD internalization by uPAR-expressing breast cancer cells.
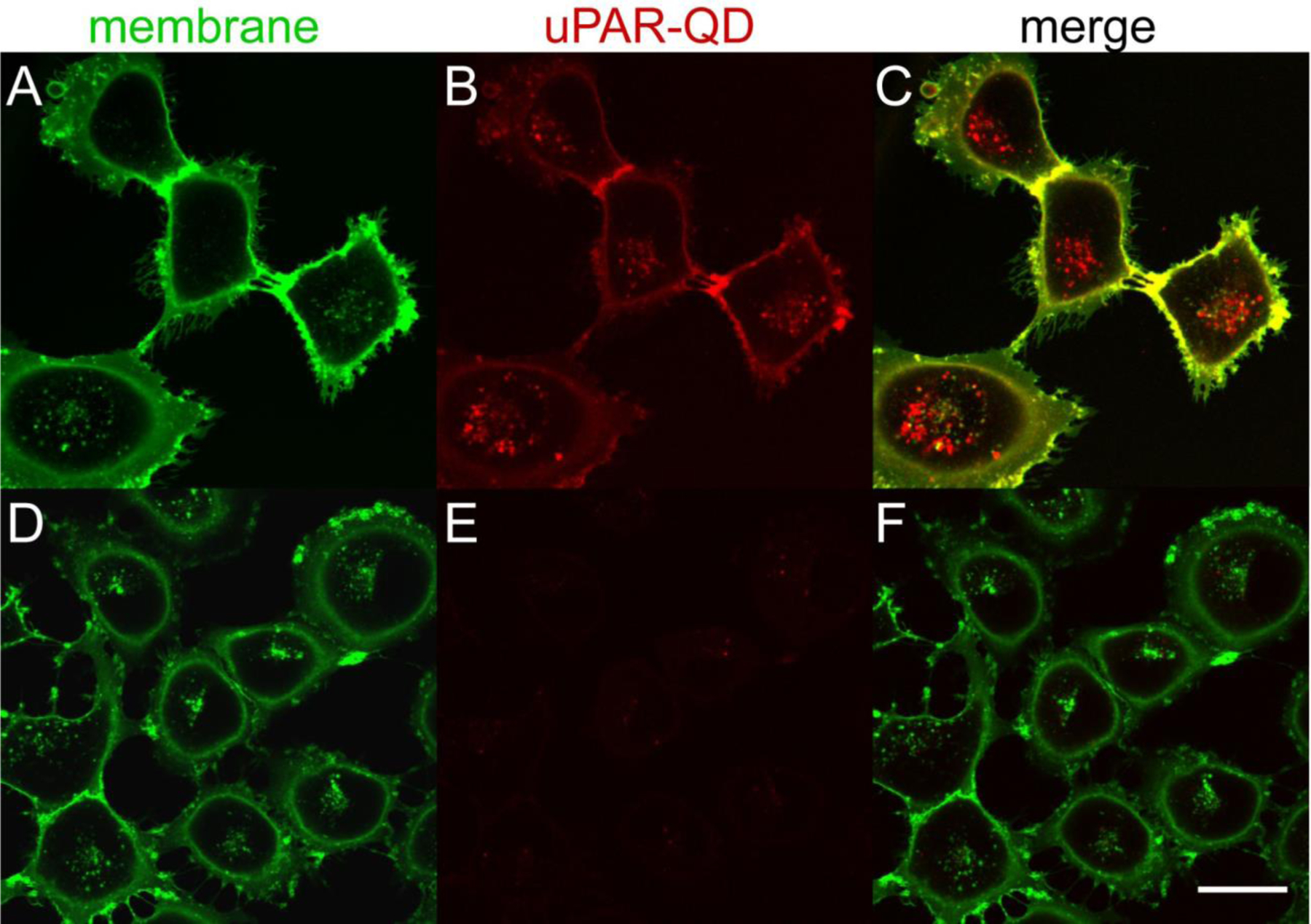
Confocal images of live MDA-MB-231 cells incubated with (A-C) scFv-QD conjugates (610 nm emission peak) for 60 min at 37 °C or (D-F) SpyCatcher-QD conjugates without scFv Ab. (A,D) AlexaFluor 488-WGA fluorescence of the plasma membrane and endolysosomal pathway; (B,E) QD emission with or without surface anti-uPAR scFv. (C,F) Overlay of WGA and anti-uPAR-QD emission. QDs were excited at 405 nm and emission collected from 605 – 690 nm. AlexaFluor488 was excited at 488 nm and emission collected from 495 – 575 nm. Scale bar is 30 μm.
Tracking uPAR internalization kinetics with scFv-UCNP conjugates
To better understand the kinetics of ligand-mediated internalization of uPAR, we synthesized and imaged scFv-UCNP conjugates for time-lapse live-cell imaging. Because UCNPs do not photobleach,52,53 are unaffected by cellular components,18 and have no measurable overlap with the excitation/emission of cellular autofluorescence,17 they offer a more quantitative method for tracking receptor trafficking than conventional fluorophores. Core/shell Yb3+/Er3+ aUCNPs were synthesized for optimized brightness under 980 nm scanning continuous-wave laser excitation,19 and microscopy conditions were optimized by varying laser intensity, pinhole size, image resolution, pixel size, scan speed and area, and detector gain. The 980-nm excitation intensity, 105 W/cm2, shows no apparent phototoxicity during confocal imaging, consistent with previous single- UCNP tracking studies.18 The scan rate proved to be a critical parameter, with bidirectional scan mode speeds of 180 μsec/pixel slow enough to afford a strong signal-to-noise ratio (SNR) given the μsec decay times of aUCNP phosphoresence,54 but fast enough to capture physiological events without significant blurring. This optimization of upconverting fast scanning confocal parameters affords roughly 2–3 orders of magnitude increase in SNR compared to standard fluorescence confocal imaging parameters (compare Fig. 4 with Fig. S7).
Figure 4. Kinetics of anti-uPAR-UCNP internalization in breast cancer cells.
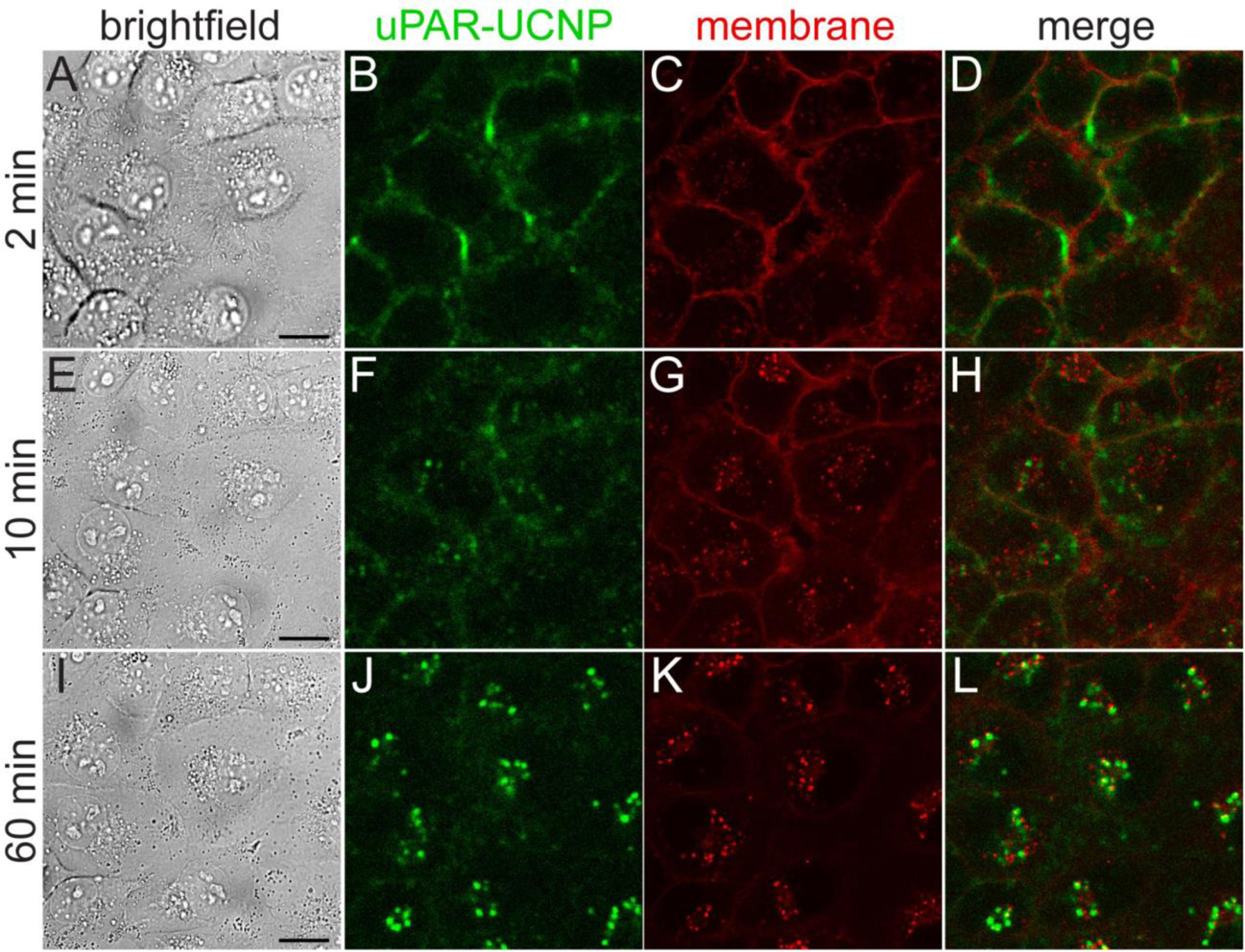
Live MDA-MB-231 cells treated with 2G10 scFv-UCNP immunoconjugates at 37 °C, imaged by: (A,E,I) brightfield; (B,F,J) upconverted confocal microscopy for anti-uPAR-UCNP imaging; and (C,G,K) standard confocal microscopy for lipophilic fluorophore Cellmask Orange. UCNPs were excited at 980 nm and emission collected from 380 – 750 nm. Cellmask Orange was excited at 561 nm and emission collected from 495 – 575 nm. (D,H,L) Overlay of anti-uPAR-UCNP and membrane emission. Frames 2 min (A-D), 10 min (E-H), and 60 min (I-L) of full 70-min time lapse (Supplementary movie 1) are shown here. Scale bars are 20 μm.
MDA-MB-231 cells were pre-incubated with 2G10 scFv-aUCNPs at 13 °C to minimize endocytosis, followed by imaging at 37 °C to track internalization under physiological conditions (Fig. 4 and Supplementary movie 1, green channel). Initial surface staining is followed by the appearance of small puncta at 10 min, which then increase in number and intensity and are accompanied by a near-complete loss of plasma membrane emission. A lipophilic organic fluorophore that non-specifically intercalates in the plasma membrane (Fig. 4, red channel) is similarly internalized,55 but without significant overlap with the scFv-aUCNPs. SpyCatcher-aUCNPs conjugates without 2G10 scFv show modest cell membrane attachment after pre-incubation, with ~25% the emission intensity of Ab conjugates (Fig. S8), and fewer, dimmer puncta are observed (Supplementary movie 2).
To quantify these differences in internalization, we used ImageJ software56 to analyze images for scFv-UCNP and membrane stain distribution, calculating internalized and surface fractions (Figs. 5 and S9). Over half of the scFv-UCNPs are internalized after 20 min at 37 °C, and this reaches almost 75% internalization, with no apparent recycling to the cell surface. The lipophilic membrane stain is internalized more slowly and to a lesser extent than the scFv-UCNP. Limitations of this image analysis may include difficulties in distinguishing plasma membrane emission from that of endosomes residing just below the surface, low precision in identifying the plasma membrane in ImageJ, out-of-plane emission, and organic fluorophore photobleaching.55
Figure 5.
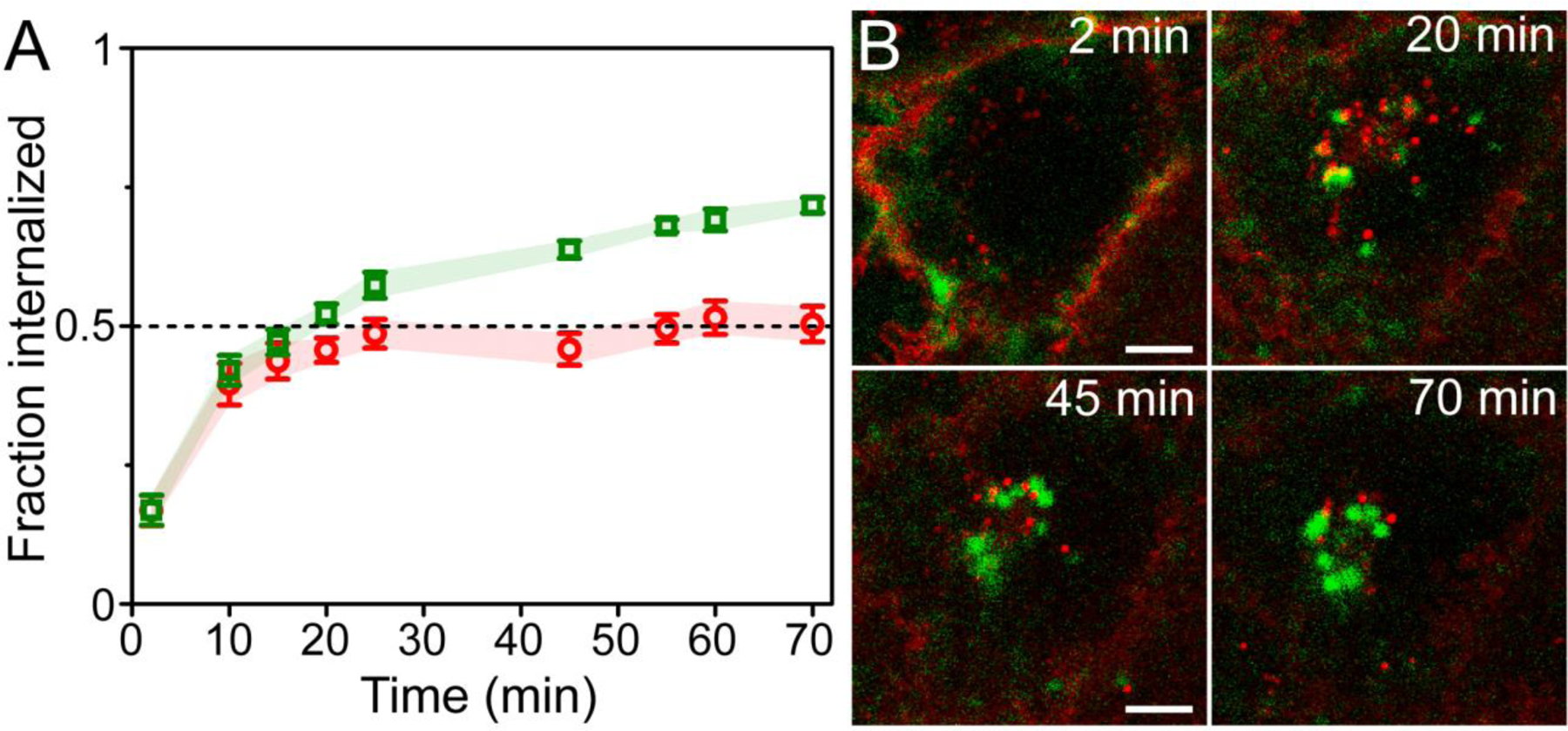
Differences of internalization and recycling between anti-uPAR scFv-UCNPs and a lipophilic membrane stain in breast cancer cells. (A) Fraction internalized of 2G10 scFv-UCNP immunoconjugates in live MDA-MB-231 cells during incubation at 37°C (mean ± SEM; n = 6 cells). (B) Confocal images of anti-uPAR-UCNPs (green squares) and lipophilic membrane fluorophore (red circles) in live MDA-MB-231 cells showing differences in intracellular localization between probes. Details from frames at 2, 20, 45 and 70 min of full 70-min time-lapse (Supplementary movie 1) are shown. Scale bars are 10 μm.
Challenges of synthesizing nanocrystal immunoconjugates
The ubiquity of antibodies in cell imaging arises from the ability to select specific, high-affinity probes against a wide range of receptors and other cellular components.1,3 Advances in Ab selection methods and engineering have both expanded the range of possible targets and enabled the development of miniaturized Ab designs with specific conjugation handles that avoid some of the pitfalls of traditional Ab labeling techniques, such as breakdown under physiological conditions, either for non-covalent28–30,57 or equilibrium covalent7,9,32 interactions. Common labeling techniques that require chemical modification of the Ab, such as oxidation or reduction, also run the risk of diminishing Ab-ligand affinity or Ab stability and need to be evaluated on a case-by-case basis.9–12,21 For synthesis of NP-Ab complexes, most reactions adapted from organic fluorophore chemistry, such as lysine sidechain modification, are far more difficult to control with the multivalent reactivity of both Ab11,58 and NP surfaces,14,44 which presents the risk of significant aggregation. Similarly, nonspecific adsorption of full Ab to NP surfaces exposes their Fc regions, which have high affinity for off-target Fc receptors.59 Common methods that avoid these problems include streptavidin coating of NP surfaces, which are then recognized with a biotinylated secondary antibody to a primary antigen-specific antibody, although this results in complexes >40 nm in diameter that dwarf the size of the typical protein target and leave the large complexes prone to aggregation or unable to reach their targets.15,25
Reactions designed for site-specific conjugation of drugs or fluorophores can overcome these issues,12,32,51 but those that rely on diffusion for reactants to interact are constrained by inherent limits on NP and Ab concentrations (i.e., low μM, well below the mM concentrations typical for bimolecular reactions), as well as slower diffusion rates compared to small organics. Our initial attempts to use aldehyde-reactive Pictet-Spengler reagents8 conjugated to UCNP surfaces showed no apparent conjugation with aldehyde-tagged antibodies51 (not shown). In contrast, SpyTag peptides bind to SpyCatcher proteins with low nM affinities before isopeptide bond formation,23 enabling these irreversible reactions to proceed nearly to completion at low concentrations of both Ab and NPs.36 The stability of the SpyCatcher-SpyTag isopeptide bond is critical for extended imaging experiments, as some common methods of Ab attachment have been shown to dissociate or reverse under live-cell imaging conditions, including those based on electrostatic,28–30,60 biotin-avidin,57 or protein-protein interactions,61 as well as aldehyde-based oxime or hydrazide bonds.7,8 Unlike many methods developed specifically for protein functionalization of NPs,13,14,27,31,60,61 SpyCatcher-mediated bioconjugation does not depend on a specific nanocrystal surface chemistry, making it compatible with any hydrophobic nanocrystal passivated with amphiphilic polymers, which have found use for improving both colloidal stability and optoelectronic properties of diverse hydrophobic nanocrystals.15,16,44,45
Antibody engineering enables improved nanoparticle conjugation
Ab engineering efforts over the last 2 decades have demonstrated that selective and high-affinity binding sequences against a variety of antigens can be selected and then subcloned into Ab scaffolds to create stable Ab with non-natural properties, and that this requires only basic recombinant DNA technology.3 Unlike naturally occurring IgG Ab, which are >15 nm along their longest axes,6 scFv Ab are 2–3 nm in diameter,24 can readily be expressed in E. coli, and do not have Fc domains that bind to macrophages, lymphocytes, and other cells expressing Fc receptors.59 A key advantage of recombinant Ab technology is the ease of introducing C-terminal modifications for various applications, ranging from purification (e.g., hexahistidine, myc tags), to valency engineering, to biotechnology applications such as drug conjugation and enzyme fusion.62–64 Here, the C-terminal SpyTag enables stable, oriented, and stoichiometric self-assembly and -reaction to form scFv-UCNP conjugates, which allows long-term tracking of the fate of uPAR without concern that the Ab may dissociate from the UCNP in the complex cellular environment. The generality of SpyCatcher-mediated conjugation of scFv Ab to NP surfaces, unlimited by either Ab target or NP chemistry, suggests potential in a variety immunotargeting applications, ranging from deeply subwavelength NIR imaging65 to whole animal imaging.19,66
Quantitative bioimaging with upconverting nanoparticles
UCNPs differ from organic and protein fluorophores in significant ways that address some of the confounding factors in quantitative image analysis, such as loss of probe signal to photobleaching, uneven distribution of cellular autofluorescence, bleed-through between channels in multicolor imaging, and changes in cell health during live cell imaging. UCNPs have shown complete resistance to photobleaching, even under extreme conditions,52,53 they have no measurable NIR-excited emission overlap with fluorophores or cellular autofluorescence,17,18 and are excited at NIR wavelengths that are significantly less phototoxic to cells than visible wavelengths.18,19 However, imaging UCNPs by scanning microscopy presents unique challenges owing to the relative inefficiency and longer lifetimes of multiphoton Ln3+ upconversion.19,54,67 The aUCNPs in this study have been optimized for both higher quantum yields and shorter luminescence lifetimes,19 and confocal parameters also needed to be optimized for fast scanning under non-phototoxic 980-nm excitation intensities (ca. 105 W/cm2)18 to account for the trade-off between SNR and the kinetic resolution needed for live-cell processes. uPAR localization is limited primarily by the diffraction limit (~300 nm for UCNP wavelengths65), which makes it difficult to pinpoint the exact plasma membrane boundary or to distinguish between receptors on the plasma membrane from those sitting in endosomes just inside the surface. Previous live-cell UCNP imaging has relied on exceptionally high intracellular UCNP concentrations, by microinjection or extended incubations,68,69 or on using relatively large (>50 nm) UCNPs.70 For receptor tracking experiments, UCNP concentration is limited by receptor surface expression levels, and large UCNPs run the risk of altering trafficking patterns or may be too large to reach their targets.25
Live-cell imaging of uPAR trafficking
The primary function of the uPAR system is to degrade the proximal extracellular matrix, making the density of active receptors on the cell surface critical to uPAR function.40 uPAR surface density is controlled by the endolysosomal pathway, which internalizes and then sorts activated uPAR complexes, either for recycling to the cell surface or degradation in lysosomes. A ligand-induced clathrin-dependent endocytic pathway internalizes a complex of uPAR with its soluble ligand uPA and other membrane-associated proteins.71 These are subject to proteolysis or glycolipid cleavage to dissociate the components and produce soluble forms of uPAR.50 Alternatively, a constitutive clathrin- and ligand-independent internalization has been found to contribute to rapid uPAR endocytosis and recycling.72 In both of these processes, uPAR is returned to the cell surface without its ligands, which are dissociated and sent to lysosomes.71 In this study, the intracellular localization of uPAR-specific UCNPs is distinct from a lipophilic dye that intercalates non-specifically in the plasma membrane and is subject to constitutive membrane recycling (Figs. 4, 5),73 suggesting that uPAR internalization is induced by ligand binding rather than constitutive in MDA-MB-231 cells. The internalization kinetics of the scFv-UCNP complex with uPAR are similar to those observed for uPA-uPAR complexes,50,74 which also suggests a ligand-induced mechanism, and that the 2G10 anti-uPAR scFv evokes similar internalization responses as the endogenous ligand uPA. There appears to be no recycling to the plasma membrane of scFv-UCNP during the 70-min experiment (Fig. 5), which may reflect its dissociation from uPAR and targeting to lysosomes, where the UCNPs remain luminescent, or differences in trafficking between uPA-uPAR and Ab-uPAR complexes. The photostability and chemical stability of scFv-UCNP conjugates throughout the endolysosomal pathway, from plasma membrane to lysosomes, makes them particularly useful for quantitative live-cell trafficking studies.
CONCLUSION
We have demonstrated that SpyCatcher proteins can attach engineered SpyTag-Ab fusions to different nanoparticle surfaces with control over stability, orientation, and stoichiometry. scFv-conjugated QDs and UCNPs are specific in binding target uPAR, a key protein in the development of many human cancers, and are selectively internalized by cultured cancer cells overexpressing uPAR. Time course confocal imaging of scFv-UCNP shows fast uptake where at least half of the available immunoconjugates are internalized after 20 min incubation and accumulation over time with no apparent recycling to the cell surface. This strategy for specific and stable conjugation of antibodies to NPs as compact probes may be extended to many other engineered Ab fragments quantitative live-cell imaging of a range of biological targets.
MATERIALS AND METHODS
Chemicals and materials.
Erbium chloride hydrate (ErCl3·xH2O, 99.999% Er) was purchased from Strem Chemicals; poly(maleic anhydride-alt-1-octadecene) (PMAO) was purchased from Polysciences; 2-(2-(2-methoxyethoxy)ethoxy)ethanamine (m-PEG3-amine) was purchased from Aurum Pharmatech; and SM(PEG)2 was purchased from ThermoFisher. Mutagenesis kits and competent cells were purchased from New England Biolabs. Custom DNA oligos were purchased from Genscript. Plasmids were procured from Addgene, and cell lines were purchased from ATCC. All other reagents and chemicals were purchased from MilliporeSigma and used without further purification.
Aqueous passivation of CdSe/CdS QDs.
CdSe/CdS QDs with emission maxima of 610 nm were synthesized as described15,44 and dispersed in hexane with 1% (v/v) oleic acid to 8 μM, as determined by first exciton absorbance. PAOA polymer (20 mg, 6.25 μmol, 3000-fold molar excess over QDs) was dissolved in 1 mL of MeOH and 19 mL of CHCl3. QDs in hexane (250 μL of 8 μM, 2.0 nmol) were added with stirring, and the solvents were removed under a gentle stream of N2 overnight. The dry QD/polymer residue was then resuspended in 15 mL of 200 mM sodium bicarbonate buffer, pH 8.0. This suspension was sonicated for 30 min, heated in an 80 °C water bath for 60 min, slowly cooled in the bath to room temperature, and then sonicated for 30 min. Excess polymer was removed by spin dialysis (Amicon Ultra15, 50 kDa MWCO), washing with 3 × 15 mL of 100 mM HEPES, pH 7.8. The retentate was diluted to 1 mL with HEPES buffer and centrifuged at 16100 × g for 5 min to remove residual polymer and insoluble aggregates. Aqueous QD dispersions were stored under ambient conditions.
Synthesis of core UCNPs.
he β-phase NaYb0.8Er0.2F4 nanocrystals were synthesized with minor modifications of the methodology previously described.19,52 YbCl3·6H2O (0.24 mmol, 93 mg), ErCl3·xH2O (0.06 mmol, 23 mg), oleic acid (OA, 3.25 g) and 1-octadececene (ODE, 4 mL) were added into a 50-mL 3-neck flask. The precursors were then heated to 110 °C under vacuum and stirred for 45 min, yielding Ln oleates in a clear, homogeneous solution. The flask was filled with N2 and cooled to room temperature. Sodium oleate (1.25 mmol, 382 mg), NH4F (2.0 mmol, 74 mg), and ODE (3 mL) were added, the mixture was stirred under vacuum at room temperature for 20 min and then heated at 305 °C under N2 for 45 min. The heating mantle was removed and a strong stream of air was used to cool the reaction flask to room temperature. To purify the UCNPs, the suspension was transferred to a 50 mL centrifuge tube, and 15 mL of EtOH was added to precipitate the nanocrystals. The tube was centrifuged at 3000 × g for 5 min forming a white pellet. The supernatant was disposed and the solid was sonicated in n-hexane (5 mL) to disperse the nanocrystals. The dispersion was centrifuged at 3000 × g for 3 min and transferred to a new tube leaving behind insoluble impurities. The purification cycle was repeated one more time, and then the nanocrystals were stored in 10 mL of n-hexane with 0.2% (v/v) oleic acid under ambient conditions.
Synthesis of core/shell UCNPs.
Epitaxial NaY0.8Gd0.2F4 shells were grown on β-NaYb0.8Er0.2F4 cores using a layer-by-layer method19 in WANDA, a nanocrystal synthesis robot at the Molecular Foundry.75 A precursor solution of 0.10 M oleates was prepared by heating YCl3 (0.8 mmol, 156 mg) and GdCl3 (0.2 mmol, 53 mg) in 10 mL 2:3 (v:v) OA/ODE to 110 °C for 30 min under vacuum and 160 °C for about 30 min, until the solution became clear. Then, the solution was cooled to 110 °C and purged under vacuum for 30 min. A 0.20 M sodium trifluoroacetate (NaTFA) solution with OA was prepared in a separate flask by stirring NaTFA (2 mmol, 272 mg), 5 mL OA, and 5 mL ODE under vacuum at room temperature for 2 h ensuring that no solid remained. Purified core UCNPs in n-hexane (15 nmol) were stirred under N2 stream to evaporate the solvent and redispersed in 4 mL of OA and 6 mL of ODE. Then, the reaction was carried out using a robotic WANDA protocol under air-free conditions. The core UCNP dispersion was stirred and heated to 280 °C at 20 °C/min. Then, 0.10 M Ln-OA and 0.20 M NaTFA-OA solutions were injected in alternating cycles into the reaction at 17 μL/s. Every cycle began with the Ln-OA addition, followed by the Na-TFA-OA addition, waiting 20 min between each injection to form a single 0.5-nm unit cell layer. After the last injection, the dispersion was stirred for an additional 30 min at 280 °C and cooled rapidly to room temperature. Core/shell UCNPs were purified and stored using the procedure described for core UCNPs.
Crystalline structure determination by X-ray powder diffraction.
All oleate-capped nanocrystals were characterized by X-ray powder diffraction on a Bruker AXS D8 Discover GADDS X-ray diffractometer operating with 35 kV/40 mA Co Kα (λ = 1.78897 Å) radiation. For XRD samples, 200 μL of a UCNP stock solution in n-hexane was spotted onto a glass coverslip and allowed to air dry completely until an opaque white film formed. All NaLnF4 nanocrystals were found to be pure hexagonal β phase.
Synthesis of 2-(2-pyridyldisulfide)ethylamine (PDEA).
Aldrithiol-2 (2,2’-Dipyridyldisulfide, 20.0 mmol, 4.41 g) was dissolved in 20 mL of MeOH with 0.8 mL of glacial AcOH in a 300 mL round-bottom flask. Cysteamine-HCl (10.0 mmol, 1.14 g) dissolved in 20 mL of MeOH was added dropwise over 1 h to the vigorously mixing flask and stirred for 16 h under N2 atmosphere. MeOH was removed by rotary evaporation, and the yellow oil product was washed twice with 50 mL of dry, ice-cooled Et2O. The oil was dissolved in 10 mL of MeOH and precipitated by addition of 200 mL of cold Et2O. The flask was stored at −20 °C for 36 h to ensure full precipitation. The pale-yellow solid was vacuum filtered, rinsing with 3 × 50 mL of dry, cold Et2O, and left under vacuum for 20 min to remove residual solvent, giving 2.02 g (93% yield) of PDEA. 1H NMR (500 MHz, CD3OD): δ 9.39 (broad s, 3H), 9.29 (d, 1H), 8.45 (t, 1H), 8.32 (d, 1H), 7.97 (t, 1H), 3.92 (m, 4H). Mass spectrum, C7H11N2S2 (MH+) calculated: 187.0; found: 187.2.
Aqueous passivation of core/shell UCNPs.
PMAO polymer45 (2 mg, 0.8 monomer units per nm2 of UCNP surface area) was dissolved in 1 mL of acetone, 14 mL of CHCl3 was added, and the mixture was quickly sonicated to obtain a homogeneous solution. Approximately 200 μL of 0.5 μM core/shell UCNPs (0.1 nmol) in n-hexane was added to the polymer dispersion. The solution was stirred overnight under a gentle stream of N2 until the complete evaporation of the solvents. The UCNP/polymer residue was then resuspended in 15 mL of 100 mM sodium borate buffer, pH 8.6 with 1:99 ratio of PDEA (1.27 mg, 5.7 μmol) to 2-(2-(2-methoxyethoxy)ethoxy)-ethylamine (92 μL, 564 μmol). The reaction flask was sonicated during 4 h at a water bath. The excess polymer was removed with 100 mM HEPES, pH 7.4, by five rounds of centrifugal dialysis (Amicon Ultra-15, 100 kDa MWCO). Aqueous dispersions of UCNPs were further purified to remove excess polymer by dialysis (Spectra-Por Float-A-Lyzer G2, 10 mL, 100 kDa MWCO) and concentrated by spin dialysis (Amicon Ultra-15, 100 kDa MWCO) to a final 1 mL dispersion in 20 mM HEPES buffer, pH 7.4. The final concentration was determined as 100 nM by an emission versus concentration curve measured for the parent hydrophobic UCNPs.
Emission of UCNP dispersions.
Low-power upconverted emission spectra were measured from aqueous UCNPs diluted at 25 nM in 20 mM HEPES, pH 7.4. The spectra were performed in FLS980 Spectrometer (Edinburg Instruments) equipped with a 980-nm laser (CrystaLaser) from 495 to 800 nm, using a 2-nm slit bandwidth and 1 s integration times. Spectra were corrected for the sensitivity of the detector using a calibrated light source.
scFv antibody cloning.
The anti-uPAR antibody 2G10 was previously discovered from a highly diverse and naïve human Fab phage display library.38 Expression plasmids were cloned by standard PCR methods and assembled by Gibson assembly. To generate the 2G10 scFv-SpyTag construct, VH and VL domains of 2G10 were cloned into the expression vector pSYN1 (James D. Marks, UCSF) using cutting sites NcoI and NheI for VH and BssHII and NotI for VL. SpyTag (AHIVMVDAYKPTK)33 was added to the C-terminus of the VL domain with a GSG linker sequence creating a -GSG-SpyTag-Myc-His6 carboxy terminus.
SpyCatcher and scFv antibody expression.
Plasmids encoding SpyCatcher and 2G10-SpyTag scFv were transformed into the E. coli expression cell line BL21(DE3) (MilliporeSigma) for heterologous protein expression. Single colonies from agar plates were used to inoculate cultures of LB broth (25 mL) containing ampicillin (100 μg/mL) and grown at 37 °C overnight (250 rpm). This starter culture was then used to inoculate 2 × 500 mL Terrific Broth cultures containing ampicillin (100 μg/mL) at 0.05 OD600 and grown at 37 °C with shaking (250 rpm). Once cell density reached 0.4 – 0.6 (2 – 4 h post-induction), protein expression was induced with 1 mM IPTG. Cultures were then grown 18 h at 20 °C with shaking (250 rpm) to avoid the formation of inclusion bodies. Cells were centrifuged (6000 × g, 20 min, 4 °C) and pellets either resuspended in 30 mL loading buffer (20 mM Tris pH 8.0, 300 mM NaCl, 10 mM imidazole) or stored at −80 °C.
Recombinant His6-tagged proteins were isolated using a prepacked 1 mL Ni2+-NTA column (HisTrap Excel, GE Healthcare). To remove loosely bound proteins, the column was washed with 50 mM imidazole until 280 nm absorbance stabilized. His6-tagged protein was eluted utilizing a linear imidazole gradient up to 250 mM. Chromatography steps were conducted at 1 mL/min with an ÄKTA Pure FPLC system (GE Healthcare). Successful protein expression was confirmed by a band shift in whole cell lysate in Laemmli buffer and 0.05% 2-mercaptoethanol on a BioRad Criterion Stain-free 4−20% gradient SDS-PAGE. The gel was UV-activated for 2 min before imaging on a ProteinSimple FluorChem E system.
All protein structure graphics were created by us from Protein Data Bank (PDB) files using ChimeraX.76
Mass spectrometry.
SpyCatcher S35C, 2G10-SpyTag scFv, and SpyCatcher-SpyTag scFv conjugate masses were determined by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). The matrix solution was prepared by dissolving 10 mg of sinapic acid in 1 mL of 50% CH3CN and 0.1% TFA in distilled water. Each protein stock solution (10 to 40 μM in 50 mM NH4HCO3) was mixed with the matrix solution in a 1:1 (v/v) ratio. Then, 2 μL of each of these mixtures was loaded onto the MALDI plate and dried at atmospheric pressure. Mass spectrometric analyses of crystallized mixtures were conducted on a MALDI TOF/TOF TF4800 (AB SCIEX) mass spectrometer.
Nanoparticle-SpyCatcher conjugation.
To generate SpyCatcher-QD conjugates, PAOA-encapsulated QDs (2 μM, 200 μL) in 100 mM HEPES, pH 7.8, were combined with SM(PEG)2 (100 mM, 50 μL) in DMSO. The reaction was mixed at room temperature for 30 min. The mixture was diluted to 4 mL with 100 mM HEPES, pH 7.0, and excess reagents were removed by centrifugal dialysis (Amicon Ultra-4, 100 kDa MWCO), washing with 3 × 4 mL of HEPES buffer. An aliquot of SpyCatcher-S35C protein (200 μM, 100 μL) was added to a 1.5 mL low protein-binding centrifuge tube, the QDs were added, and the reaction was mixed overnight at 4 °C on a rotary mixer. The reaction mixture was diluted to 4 mL with 100 mM HEPES, pH 7.5, and excess SpyCatcher protein was removed by centrifugal dialysis (Amicon Ultra-4, 100 kDa MWCO), washing with 4 × 4 mL of buffer. The retentate was diluted to 400 μL in HEPES buffer, and the SpyCatcher-QD conjugate was stored at 4 °C.
For disulfide conjugated UCNP-SpyCatcher, 50 μL of 100 nM PMAO-encapsulated UCNPs were diluted to 400 μL with 100 mM HEPES, pH 7.2. Separately, an aliquot of S35C SpyCatcher (200 μM, 100 μL) was desalted on a Biospin-6 Desalting Column (Bio-Rad) and further washed by centrifugal dialysis (Amicon Ultra-4, 3 kDa MWCO), washing with 3 × 4 mL of HEPES buffer to remove all TCEP storage buffer. The retentate was diluted to 100 μL in a 1.5 mL low protein-binding centrifuge tube, the UCNPs were added, and the reaction was mixed overnight at 4 °C on a rotary mixer. The reaction mixture was diluted to 4 mL with 100 mM HEPES, pH 7.5, and excess SpyCatcher was removed by spin dialysis (Amicon Ultra-4, 100 kDa MWCO), washing with 4 × 4 mL of buffer. The retentate was diluted to 400 μL in HEPES buffer, and the UCNP-SpyCatcher conjugate was stored at 4 °C.
Nanoparticle characterization.
To determine the size of as-synthesized nanocrystals, a dilute dispersion of nanocrystals in hexane was drop cast onto an ultrathin carbon film on lacey carbon support, 400 mesh copper TEM grid (Ted Pella) and dried in a fume hood. Images were collected on a Gemini Ultra-55 Analytical Field Emission Scanning Electron Microscope (Zeiss) in dark-field transmission mode under 30 kV accelerating voltage or on a JEOL 2100-F in HAADF mode under 200 kV accelerating voltage. Diameters for 100 random nanoparticles were manually designated in ImageJ and a distribution was plotted. To determine the size of aqueous nanoparticles, dispersions were diluted to 20 nM in 100 mM HEPES, pH 7.8 for PAOA-encapsulated nanoparticles, and in 100 mM HEPES, pH 7.0, for PMAO-encapsulated and SpyCatcher-functionalized nanoparticles. The dispersions were sonicated for 30 min prior to measurement. Diameters were measured by dynamic light scattering using a Malvern Zetasizer with typical count rates of 150 kilocounts per second. Data were collected for 60 sec each in 5 separate runs and fits using Malvern Zetasizer software to a volume-weighted size distribution of hydrodynamic diameter.
SpyCatcher quantification on NP surfaces.
Tryptophan fluorescence was used to quantify SpyCatcher on nanoparticle surfaces. Emission spectra of UCNPs were taken under 290 nm excitation prior to and after SpyCatcher conjugation, with difference curves showing a Trp peak at 340 nm. A calibration curve was generated of known concentrations of purified SpyCatcher, to determine SpyCatcher:UCNP stoichiometry.
To follow the disulfide exchange of S35C SpyCatcher onto the nanoparticle surface, the absorption signature of the leaving group (pyridine-2-thione) was measured at 343 nm in a plate reader as a function of time. The concentration of pyridine-2-thione was determined from this absorbance and its extinction coefficient (8,080 M−1 cm−1), and a ratio with the nanoparticle concentration defined the number of SpyCatcher proteins exchanged per nanoparticle.
scFv antibody conjugation and characterization.
In order to generate the final QD-Ab conjugate, SpyCatcher-coated QDs (100 μl, 1 μM, 100 pmol) and SpyTag-2G10 antibody (8.9 μl, 22.6 μM, 201 pmol, ~2x excess) were gently mixed overnight and used without further purification. Ab to nanoparticle stoichiometries were tested by varying the volume of added SpyTag-scFv solution. To verify the presence of Ab on the nanoparticle surface, fluorescence spectra were taken under 270 nm excitation prior to and after conjugation, with the difference curve showing a Trp peak at ~340 nm. A calibration curve was generated of known concentrations of purified SpyTag-scFv, and the difference curve peak was used to determine the concentration of Ab conjugated to the nanoparticle. The final UCNP-Ab conjugate was prepared by mixing SpyCatcher-coated UCNPs (200 μl, 25 nM, 5 pmol) and SpyTag-scFv (10 μl, 5 μM, 50 pmol) overnight without further purification. scFv-UCNP conjugates were sterilized with 0.22-μm filters before addition to cells.
Cell culture.
MDA-MB-231 cells were maintained in tissue culture polystyrene dishes (Corning) at 37 °C in a 5% CO2 atmosphere in Dulbecco’s modified Eagle’s medium (DMEM, Sigma) containing 10% fetal bovine serum (FBS, Gibco) and 1% penicillin–streptomycin solution (Life Technologies).
Cell labeling and microscopy.
Cells were grown in an 8-well chamber slide (μ-Slide 8 Well, IbiTreat, Polymer No. 1.5 and thickness of 180 μm) to >80% confluency. For QD imaging: media was aspirated and the wells were washed with 2 × 500 μL DMEM without phenol red. QD constructs were diluted to 10 nM in DMEM without phenol red and added to the cells. After 1 h in a 37 °C incubator, cells were washed 1 x with 500 μL PBS + 10 mM glucose. The cells were then incubated in a 5 μg/mL solution of AlexaFluor488-WGA (ThermoFisher) in PBS + glucose at room temperature for 10 min, and they were then washed 3 times with 500 μL of PBS + glucose. Fresh PBS + glucose was added, and the cells were imaged by confocal microscopy. Confocal images of the focal plane at the chamber interface were obtained using an inverted Zeiss LSM710 system with a 1.4 NA 63x Apochromat oil immersion objective. QDs were excited by a 405 nm diode laser with an MBS-458/514 main dichroic beam splitter, and emission was collected from 605 – 690 nm. AlexaFluor488 was excited with a 488 nm diode laser with an MBS-488/594 main dichroic beam splitter, and emission was collected from 495 – 575 nm.
For UCNP imaging: media was aspirated and the wells were washed with 3 × 500 μL PBST (PBS + 0.1% (w/w) TWEEN-20). The cells were pre-incubated for 5 min at 13 °C with 400 μL of PBST supplemented with 1% (w/w) BSA and 300 mM glycine. UCNP constructs were added to obtain a final concentration of 5 nM and maintained for 30 min at room temperature. In the last 5 min, 3 μL of CellMask Orange Plasma membrane Stain (ThermoFisher) at 50x in PBS was added to the mixture. The solution was aspirated and the well was washed 5 times with PBST. Opti-MEM without phenol red and supplemented with 10% (v/v) FBS were added and the cells were imaged by time-lapse confocal microscopy at 37 °C. Confocal images of the focal plane at the chamber interface were obtained using an inverted Zeiss LSM710 system with a 40x/1.2NA Apochromat water immersion objective. UCNPs were excited by a 980 nm diode laser (500 mW) at maximum intensity reflected by a dichroic beam splitter MBS-760+. The pinhole was adjusted to 2.7 Airy units and section thickness of 0.7 μm to obtain an optimal compromise of emission detection and image resolution. The UCNP images were recorded from 380 to 750 nm with 1200 Maximum Gain and 15x Digital Gain which provided maximum upconverted emission without detectable autofluorescence under 980 nm excitation. The lipophilic membrane stain (CellMask Orange) was excited with a 561 nm diode laser (20 mW) at 0.2% of maximum intensity and reflected by MBS-488/561 dichroic beam splitter. The dye emission was collected from 575 to 700 nm using 900 Maximum Gain and 1x Digital Gain combined with a pinhole set to 1.0 Airy units and section thickness of 1.0 μm. Each confocal image was collected at scan speed of 177 μsec/pixel. The detection time was slowed to the limit of the Zeiss 710LSM confocal to accommodate Er3+ lifetimes. Other parameters like 512 × 512-pixel resolution, scan area, 16-bit dynamic range, 0.23 μm pixel size, 1 line step, mode line bidirectional and two-frame averaging provided optimal compromise between image resolution and detection time. Bright-field images of MDA-MB-231 cells were captured using the brightfield mode of Zeiss 710 LSM with X-Cite 120Q light source, AxioCam MRm camera, 100-ms acquisition times, and 1388 × 1040-pixel resolution.
Confocal images and data processing.
ImageJ (Fiji)56 was used to analyze the 16-bit images such as adjusting brightness, contrast and image offset. Membranes and intracellular ROIs of individual cells were selected manually. Six cells were selected for quantitative analysis of uPAR-UCNP internalization by OriginPro (OriginLab). Histograms from the ROIs were obtained in the ImageJ (FIJI) and pixel values lower than 3600 counts were identified as background noise. The ratios of intracellular to membrane integrated intensities are average results of 6 cells were calculated as the mean ± standard error of the mean. Time-lapse videos were created from the individual images in RGB format.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Cheryl Tajon for preliminary immunoconjugation experiments and Dr. Behzad Rad for technical support. The scFv expression vector pSYN1 was kindly provided by Dr. James D. Marks (UCSF). This work was supported by National Institutes of Health awards R01NS096317 (BEC) and P50AI150476 (CSC). Work at the Molecular Foundry was supported by the Director, Office of Science, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Funding Sources
US National Institutes of Health
US Department of Energy
Footnotes
Supporting Information. The following files are available free of charge via the internet at http://pubs.acs.org.
Supplementary reaction schemes and figures with nanoparticle characterization, cell imaging, and analysis (PDF); supplementary time-lapse videos of live-cell imaging (MOV).
Competing Financial Interests
The authors declare no competing financial interests.
REFERENCES
- (1).Freise AC; Wu AM In Vivo Imaging with Antibodies and Engineered Fragments. Mol. Immunol 2015, 67 (2 Pt A), 142–152. 10.1016/j.molimm.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Specht EA; Braselmann E; Palmer AE A Critical and Comparative Review of Fluorescent Tools for Live-Cell Imaging. Annu. Rev. Physiol 2017, 79 (1), 93–117. 10.1146/annurev-physiol-022516-034055. [DOI] [PubMed] [Google Scholar]
- (3).Basu K; Green EM; Cheng Y; Craik CS Why Recombinant Antibodies - Benefits and Applications. Curr. Opin. Biotechnol 2019, 60, 153–158. 10.1016/j.copbio.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wu AM; Senter PD Arming Antibodies: Prospects and Challenges for Immunoconjugates. Nat. Biotechnol 2005, 23 (9), 1137–1146. 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]
- (5).Rathore AS; Sarker A; Gupta RD Recent Developments toward Antibody Engineering and Affinity Maturation. Protein Pept. Lett 2018, 25 (10), 886–896. 10.2174/0929866525666180925142757. [DOI] [PubMed] [Google Scholar]
- (6).Saphire EO; Parren PW; Pantophlet R; Zwick MB; Morris GM; Rudd PM; Dwek RA; Stanfield RL; Burton DR; Wilson IA Crystal Structure of a Neutralizing Human IGG against HIV-1: A Template for Vaccine Design. Science 2001, 293 (5532), 1155–1159. 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- (7).Agarwal P; van der Weijden J; Sletten EM; Rabuka D; Bertozzi CR A Pictet-Spengler Ligation for Protein Chemical Modification. Proc. Natl. Acad. Sci. U. S. A 2013, 110 (1), 46–51. 10.1073/pnas.1213186110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kudirka R; Barfield RM; McFarland J; Albers AE; de Hart GW; Drake PM; Holder PG; Banas S; Jones LC; Garofalo AW; Rabuka D Generating Site-Specifically Modified Proteins via a Versatile and Stable Nucleophilic Carbon Ligation. Chem. Biol 2015, 22 (2), 293–298. 10.1016/j.chembiol.2014.11.019. [DOI] [PubMed] [Google Scholar]
- (9).Shen B-Q; Xu K; Liu L; Raab H; Bhakta S; Kenrick M; Parsons-Reponte KL; Tien J; Yu S-F; Mai E; Li D; Tibbitts J; Baudys J; Saad OM; Scales SJ; McDonald PJ; Hass PE; Eigenbrot C; Nguyen T; Solis WA; et al. Conjugation Site Modulates the in Vivo Stability and Therapeutic Activity of Antibody-Drug Conjugates. Nat. Biotechnol 2012, 30 (2), 184–189. 10.1038/nbt.2108. [DOI] [PubMed] [Google Scholar]
- (10).Shrestha D; Bagosi A; Szöllősi J; Jenei A Comparative Study of the Three Different Fluorophore Antibody Conjugation Strategies. Anal. Bioanal. Chem 2012, 404 (5), 1449–1463. 10.1007/s00216-012-6232-z. [DOI] [PubMed] [Google Scholar]
- (11).Szabó Á; Szendi-Szatmári T; Ujlaky-Nagy L; Rádi I; Vereb G; Szöllősi J; Nagy P The Effect of Fluorophore Conjugation on Antibody Affinity and the Photophysical Properties of Dyes. Biophys. J 2018, 114 (3), 688–700. 10.1016/j.bpj.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Chudasama V; Maruani A; Caddick S Recent Advances in the Construction of Antibody–Drug Conjugates. Nat. Chem 2016, 8 (2), 114–119. 10.1038/nchem.2415. [DOI] [PubMed] [Google Scholar]
- (13).Medintz IL; Uyeda HT; Goldman ER; Mattoussi H Quantum Dot Bioconjugates for Imaging, Labelling and Sensing. Nat. Mater 2005, 4 (6), 435–446. 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- (14).Barat B; Sirk SJ; McCabe KE; Li J; Lepin EJ; Remenyi R; Koh AL; Olafsen T; Gambhir SS; Weiss S; Wu AM Cys-Diabody Quantum Dot Conjugates (ImmunoQdots) for Cancer Marker Detection. Bioconjug. Chem 2009, 20 (8), 1474–1481. 10.1021/bc800421f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Wichner SM; Mann VR; Powers AS; Segal MA; Mir M; Bandaria JN; DeWitt MA; Darzacq X; Yildiz A; Cohen BE Covalent Protein Labeling and Improved Single-Molecule Optical Properties of Aqueous CdSe/CdS Quantum Dots. ACS Nano 2017, 11 (7), 6773–6781. 10.1021/acsnano.7b01470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Thal LB; Mann VR; Sprinzen D; McBride JR; Reid KR; Tomlinson ID; McMahon DG; Cohen BE; Rosenthal SJ Ligand-Conjugated Quantum Dots for Fast Sub-Diffraction Protein Tracking in Acute Brain Slices. Biomaterials Science. 2020, 8, 837–845. 10.1039/c9bm01629e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wu S; Han G; Milliron DJ; Aloni S; Altoe V; Talapin DV; Cohen BE; Schuck PJ Non-Blinking and Photostable Upconverted Luminescence from Single Lanthanide-Doped Nanocrystals. Proc. Natl. Acad. Sci. U. S. A 2009, 106 (27), 10917–10921. 10.1073/pnas.0904792106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Nam SH; Bae YM; Park YI; Kim JH; Kim HM; Choi JS; Lee KT; Hyeon T; Suh YD Long-Term Real-Time Tracking of Lanthanide Ion Doped Upconverting Nanoparticles in Living Cells. Angew. Chem. Int. Ed Engl 2011, 50 (27), 6093–6097. 10.1002/anie.201007979. [DOI] [PubMed] [Google Scholar]
- (19).Tian B; Fernandez-Bravo A; Najafiaghdam H; Torquato NA; Altoe MVP; Teitelboim A; Tajon CA; Tian Y; Borys NJ; Barnard ES; Anwar M; Chan EM; Schuck PJ; Cohen BE Low Irradiance Multiphoton Imaging with Alloyed Lanthanide Nanocrystals. Nat. Commun 2018, 9 (1), 3082. 10.1038/s41467-018-05577-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Liang L; Feng Z; Zhang Q; Cong TD; Wang Y; Qin X; Yi Z; Ang MJY; Zhou L; Feng H; Xing B; Gu M; Li X; Liu X Continuous-Wave Near-Infrared Stimulated-Emission Depletion Microscopy Using Downshifting Lanthanide Nanoparticles. Nat. Nanotechnol 2021, 16, 975–980. 10.1038/s41565-021-00927-y. [DOI] [PubMed] [Google Scholar]
- (21).Algar WR; Prasuhn DE; Stewart MH; Jennings TL; Blanco-Canosa JB; Dawson PE; Medintz IL The Controlled Display of Biomolecules on Nanoparticles: A Challenge Suited to Bioorthogonal Chemistry. Bioconjug. Chem 2011, 22 (5), 825–858. 10.1021/bc200065z. [DOI] [PubMed] [Google Scholar]
- (22).Wegner KD; Jin Z; Lindén S; Jennings TL; Hildebrandt N Quantum-Dot-Based Förster Resonance Energy Transfer Immunoassay for Sensitive Clinical Diagnostics of Low-Volume Serum Samples. ACS Nano 2013, 7 (8), 7411–7419. 10.1021/nn403253y. [DOI] [PubMed] [Google Scholar]
- (23).Li L; Fierer JO; Rapoport TA; Howarth M Structural Analysis and Optimization of the Covalent Association between SpyCatcher and a Peptide Tag. J. Mol. Biol 2014, 426 (2), 309–317. 10.1016/j.jmb.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Miyanabe K; Akiba H; Kuroda D; Nakakido M; Kusano-Arai O; Iwanari H; Hamakubo T; Caaveiro JMM; Tsumoto K Intramolecular H-Bonds Govern the Recognition of a Flexible Peptide by an Antibody. J. Biochem 2018, 164 (1), 65–76. 10.1093/jb/mvy032. [DOI] [PubMed] [Google Scholar]
- (25).Francis JE; Mason D; Lévy R Evaluation of Quantum Dot Conjugated Antibodies for Immunofluorescent Labelling of Cellular Targets. Beilstein J. Nanotechnol 2017, 8, 1238–1249. 10.3762/bjnano.8.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Tajima N; Takai M; Ishihara K Significance of Antibody Orientation Unraveled: Well-Oriented Antibodies Recorded High Binding Affinity. Anal. Chem 2011, 83 (6), 1969–1976. 10.1021/ac1026786. [DOI] [PubMed] [Google Scholar]
- (27).Qiu X; Wegner KD; Wu Y-T; van Bergen en Henegouwen, P. M. P.; Jennings, T. L.; Hildebrandt, N. Nanobodies and Antibodies for Duplexed EGFR/HER2 Immunoassays Using Terbium-to-Quantum Dot FRET. Chem. Mater 2016, 28 (22), 8256–8267. 10.1021/acs.chemmater.6b03198. [DOI] [Google Scholar]
- (28).Bhuckory S; Mattera L; Wegner KD; Qiu X; Wu Y-T; Charbonnière LJ; Reiss P; Hildebrandt N Direct Conjugation of Antibodies to the ZnS Shell of Quantum Dots for FRET Immunoassays with Low Picomolar Detection Limits. Chem. Commun. (Camb.) 2016, 52 (100), 14423–14425. 10.1039/c6cc08835j. [DOI] [PubMed] [Google Scholar]
- (29).Léger C; Yahia-Ammar A; Susumu K; Medintz IL; Urvoas A; Valerio-Lepiniec M; Minard P; Hildebrandt N Picomolar Biosensing and Conformational Analysis Using Artificial Bidomain Proteins and Terbium-to-Quantum Dot Förster Resonance Energy Transfer. ACS Nano 2020, 14 (5), 5956–5967. 10.1021/acsnano.0c01410. [DOI] [PubMed] [Google Scholar]
- (30).Wu Y-T; Qiu X; Lindbo S; Susumu K; Medintz IL; Hober S; Hildebrandt N Quantum Dot-Based FRET Immunoassay for HER2 Using Ultrasmall Affinity Proteins. Small 2018, 14 (35), e1802266. 10.1002/smll.201802266. [DOI] [PubMed] [Google Scholar]
- (31).Annio G; Jennings TL; Tagit O; Hildebrandt N Sensitivity Enhancement of Förster Resonance Energy Transfer Immunoassays by Multiple Antibody Conjugation on Quantum Dots. Bioconjug. Chem 2018, 29 (6), 2082–2089. 10.1021/acs.bioconjchem.8b00296. [DOI] [PubMed] [Google Scholar]
- (32).Agarwal P; Bertozzi CR Site-Specific Antibody-Drug Conjugates: The Nexus of Bioorthogonal Chemistry, Protein Engineering, and Drug Development. Bioconjug. Chem 2015, 26 (2), 176–192. 10.1021/bc5004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zakeri B; Fierer JO; Celik E; Chittock EC; Schwarz-Linek U; Moy VT; Howarth M Peptide Tag Forming a Rapid Covalent Bond to a Protein, through Engineering a Bacterial Adhesin. Proc. Natl. Acad. Sci. U. S. A 2012, 109 (12), E690–E697. 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Keeble AH; Turkki P; Stokes S; Khairil IN; Rahikainen R; Hytönen VP; Howarth M Approaching Infinite Affinity through Engineering of Peptide–Protein Interaction. Proc. Natl. Acad. Sci. U. S. A 2019, 116 (52), 26523–26533. 10.1073/pnas.1909653116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Charrier M; Li D; Mann V; Yun L; Jani S; Rad B; Cohen BE; Ashby PD; Ryan K; Ajo-Franklin CM Engineering the S-Layer of Caulobacter Crescentus as a Foundation for Stable, High-Density, 2D Living Materials. ACS Synth. Biol 2018, 8 (1), 181–190. 10.1021/acssynbio.8b00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Mann VR; Manea F; Borys NJ; Ajo-Franklin CM; Cohen BE Controlled and Stable Patterning of Diverse Inorganic Nanocrystals on Crystalline Two-Dimensional Protein Arrays. Biochemistry 2021, 60 (13), 1063–1074. 10.1021/acs.biochem.1c00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Mauro CD; Pesapane A; Formisano L; Rosa R; D’Amato V; Ciciola P; Servetto A; Marciano R; Orsini RC; Monteleone F; Zambrano N; Fontanini G; Servadio A; Pignataro G; Grumetto L; Lavecchia A; Bruzzese D; Iaccarino A; Troncone G; Veneziani BM; et al. Urokinase-Type Plasminogen Activator Receptor (UPAR) Expression Enhances Invasion and Metastasis in RAS Mutated Tumors. Sci. Rep 2017, 7 (1), 9388. 10.1038/s41598-017-10062-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Duriseti S; Goetz DH; Hostetter DR; LeBeau AM; Wei Y; Craik CS Antagonistic Anti-Urokinase Plasminogen Activator Receptor (UPAR) Antibodies Significantly Inhibit UPAR-Mediated Cellular Signaling and Migration. J. Biol. Chem 2010, 285 (35), 26878–26888. 10.1074/jbc.M109.077677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).LeBeau AM; Duriseti S; Murphy ST; Pepin F; Hann B; Gray JW; VanBrocklin HF; Craik CS Targeting UPAR with Antagonistic Recombinant Human Antibodies in Aggressive Breast Cancer. Cancer Res. 2013, 73 (7), 2070–2081. 10.1158/0008-5472.CAN-12-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Smith HW; Marshall CJ Regulation of Cell Signalling by UPAR. Nat. Rev. Mol. Cell Biol 2010, 11 (1), 23–36. 10.1038/nrm2821. [DOI] [PubMed] [Google Scholar]
- (41).LeBeau AM; Sevillano N; King ML; Duriseti S; Murphy ST; Craik CS; Murphy LL; VanBrocklin HF Imaging the Urokinase Plasminongen Activator Receptor in Preclinical Breast Cancer Models of Acquired Drug Resistance. Theranostics 2014, 4 (3), 267–279. 10.7150/thno.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Jo M; Eastman BM; Webb DL; Stoletov K; Klemke R; Gonias SL Cell Signaling by Urokinase-Type Plasminogen Activator Receptor Induces Stem Cell–Like Properties in Breast Cancer Cells. Cancer Res. 2010, 70 (21), 8948–8958. 10.1158/0008-5472.can-10-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Tajon CA; Yang H; Tian B; Tian Y; Ercius P; Schuck PJ; Chan EM; Cohen BE Photostable and Efficient Upconverting Nanocrystal-Based Chemical Sensors. Opt. Mater 2018, 84, 345–353. 10.1016/j.optmat.2018.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Mann VR; Powers AS; Tilley DC; Sack JT; Cohen BE Azide–Alkyne Click Conjugation on Quantum Dots by Selective Copper Coordination. ACS Nano 2018, 12 (5), 4469–4477. 10.1021/acsnano.8b00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Di Corato R; Quarta A; Piacenza P; Ragusa A; Figuerola A; Buonsanti R; Cingolani R; Manna L; Pellegrino T Water Solubilization of Hydrophobic Nanocrystals by Means of Poly(maleic Anhydride-Alt-1-Octadecene). J. Mater. Chem 2008, 18 (17), 1991–1996. 10.1039/b717801h. [DOI] [Google Scholar]
- (46).Myers DA; Murdoch WJ; Villemez CL Protein-Peptide Conjugation by a Two-Phase Reaction. Biochem. J 1985, 227 (1), 343. 10.1042/bj2270343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Sgier D; Zuberbuehler K; Pfaffen S; Neri D Isolation and Characterization of an Inhibitory Human Monoclonal Antibody Specific to the Urokinase-Type Plasminogen Activator, UPA. Protein Eng. Des. Sel 2010, 23 (4), 261–269. 10.1093/protein/gzp089. [DOI] [PubMed] [Google Scholar]
- (48).Jacquemet G; Baghirov H; Georgiadou M; Sihto H; Peuhu E; Cettour-Janet P; He T; Perälä M; Kronqvist P; Joensuu H; Ivaska J L-Type Calcium Channels Regulate Filopodia Stability and Cancer Cell Invasion Downstream of Integrin Signalling. Nat. Commun 2016, 7, 13297. 10.1038/ncomms13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Franchi M; Piperigkou Z; Riti E; Masola V; Onisto M; Karamanos NK Long Filopodia and Tunneling Nanotubes Define New Phenotypes of Breast Cancer Cells in 3D Cultures. Matrix Biol Plus 2020, 6–7, 100026. 10.1016/j.mbplus.2020.100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Czekay R-P; Kuemmel TA; Orlando RA; Farquhar MG Direct Binding of Occupied Urokinase Receptor (UPAR) to LDL Receptor-Related Protein Is Required for Endocytosis of UPAR and Regulation of Cell Surface Urokinase Activity. Mol. Biol. Cell 2001, 12 (5), 1467–1479. 10.1091/mbc.12.5.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Harel ET; Drake PM; Barfield RM; Lui I; Farr-Jones S; Van’t Veer L; Gartner ZJ; Green EM; Lourenço AL; Cheng Y; Hann BC; Rabuka D; Craik CS Antibody-Drug Conjugates Targeting the Urokinase Receptor (UPAR) as a Possible Treatment of Aggressive Breast Cancer. Antibodies (Basel) 2019, 8 (4). 10.3390/antib8040054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Ostrowski AD; Chan EM; Gargas DJ; Katz EM; Han G; Schuck PJ; Milliron DJ; Cohen BE Controlled Synthesis and Single-Particle Imaging of Bright, Sub-10 Nm Lanthanide-Doped Upconverting Nanocrystals. ACS Nano 2012, 6 (3), 2686–2692. 10.1021/nn3000737. [DOI] [PubMed] [Google Scholar]
- (53).Gargas DJ; Chan EM; Ostrowski AD; Aloni S; Altoe MVP; Barnard ES; Sanii B; Urban JJ; Milliron DJ; Cohen BE; Schuck PJ Engineering Bright Sub-10-nm Upconverting Nanocrystals for Single-Molecule Imaging. Nat. Nanotechnol 2014, 9 (4), 300–305. 10.1038/nnano.2014.29. [DOI] [PubMed] [Google Scholar]
- (54).Teitelboim A; Tian B; Garfield DJ Energy Transfer Networks within Upconverting Nanoparticles Are Complex Systems with Collective, Robust, and History-Dependent Dynamics. J. Phys. Chem. C 2019, 123 (4), 2678–2689. 10.1021/acs.jpcc.9b00161. [DOI] [Google Scholar]
- (55).Jia H-R; Wang H-Y; Yu Z-W; Chen Z; Wu F-G Long-Time Plasma Membrane Imaging Based on a Two-Step Synergistic Cell Surface Modification Strategy. Bioconjug. Chem 2016, 27 (3), 782–789. 10.1021/acs.bioconjchem.6b00003. [DOI] [PubMed] [Google Scholar]
- (56).Schindelin J; Arganda-Carreras I; Frise E; Kaynig V; Longair M; Pietzsch T; Preibisch S; Rueden C; Saalfeld S; Schmid B; Tinevez J-Y; White DJ; Hartenstein V; Eliceiri K; Tomancak P; Cardona A Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9 (7), 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Chivers CE; Crozat E; Chu C; Moy VT; Sherratt DJ; Howarth M A Streptavidin Variant with Slower Biotin Dissociation and Increased Mechanostability. Nat. Methods 2010, 7 (5), 391–393. 10.1038/nmeth.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Dennler P; Fischer E; Schibli R Antibody Conjugates: From Heterogeneous Populations to Defined Reagents. Antibodies (Basel) 2015, 4 (3), 197–224. 10.3390/antib4030197. [DOI] [Google Scholar]
- (59).Aragnol D; Leserman LD Immune Clearance of Liposomes Inhibited by an Anti-Fc Receptor Antibody in Vivo. Proc. Natl. Acad. Sci. U. S. A 1986, 83 (8), 2699–2703. 10.1073/pnas.83.8.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Goldman ER; Anderson GP; Tran PT; Mattoussi H; Charles PT; Mauro JM Conjugation of Luminescent Quantum Dots with Antibodies Using an Engineered Adaptor Protein to Provide New Reagents for Fluoroimmunoassays. Anal. Chem 2002, 74 (4), 841–847. 10.1021/ac010662m. [DOI] [PubMed] [Google Scholar]
- (61).Ma L; Geng J; Kolossov VL; Han Z; Pei Y; Lim SJ; Kilian KA; Smith AM Antibody Self-Assembly Maximizes Cytoplasmic Immunostaining Accuracy of Compact Quantum Dots. Chem. Mater 2021, 33 (13), 4877–4889. 10.1021/acs.chemmater.1c00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Albrecht H; Burke PA; Natarajan A; Xiong C-Y; Kalicinsky M; DeNardo GL; DeNardo SJ Production of Soluble ScFvs with C-Terminal-Free Thiol for Site-Specific Conjugation or Stable Dimeric ScFvs on Demand. Bioconjug. Chem 2004, 15 (1), 16–26. 10.1021/bc030018+. [DOI] [PubMed] [Google Scholar]
- (63).Nuñez-Prado N; Compte M; Harwood S; Álvarez-Méndez A; Lykkemark S; Sanz L; Álvarez-Vallina L The Coming of Age of Engineered Multivalent Antibodies. Drug Discov. Today 2015, 20 (5), 588–594. 10.1016/j.drudis.2015.02.013. [DOI] [PubMed] [Google Scholar]
- (64).Hashad RA; Lange JL; Tan NCW; Alt K; Hagemeyer CE Engineering Antibodies with C-Terminal Sortase-Mediated Modification for Targeted Nanomedicine. Methods Mol. Biol 2019, 2033, 67–80. 10.1007/978-1-4939-9654-4_6. [DOI] [PubMed] [Google Scholar]
- (65).Lee C; Xu EZ; Liu Y; Teitelboim A; Yao K; Fernandez-Bravo A; Kotulska AM; Nam SH; Suh YD; Bednarkiewicz A; Cohen BE; Chan EM; Schuck PJ Giant Nonlinear Optical Responses from Photon-Avalanching Nanoparticles. Nature 2021, 589 (7841), 230–235. 10.1038/s41586-020-03092-9. [DOI] [PubMed] [Google Scholar]
- (66).Najafiaghdam H; Papageorgiou E; Torquato NA; Tian B; Cohen BE; Anwar MA 25 Micron-Thin Microscope for Imaging Upconverting Nanoparticles with NIR-I and NIR-II Illumination. Theranostics 2019, 9 (26), 8239–8252. 10.7150/thno.37672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Del Rosal B; Jaque D Upconversion Nanoparticles for in Vivo Applications: Limitations and Future Perspectives. Methods Appl. Fluoresc 2019, 7 (2), 022001. 10.1088/2050-6120/ab029f. [DOI] [PubMed] [Google Scholar]
- (68).Li X; Zhao H; Ji Y; Yin C; Li J; Yang Z; Tang Y; Zhang Q; Fan Q; Huang W Lysosome-Assisted Mitochondrial Targeting Nanoprobe Based on Dye-Modified Upconversion Nanophosphors for Ratiometric Imaging of Mitochondrial Hydrogen Sulfide. ACS Appl. Mater. Interfaces 2018, 10 (46), 39544–39556. 10.1021/acsami.8b16818. [DOI] [PubMed] [Google Scholar]
- (69).Wang H; Li Y; Yang M; Wang P; Gu Y FRET-Based Upconversion Nanoprobe Sensitized by Nd3+ for the Ratiometric Detection of Hydrogen Peroxide in Vivo. ACS Appl. Mater. Interfaces 2019, 11 (7), 7441–7449. 10.1021/acsami.8b21549. [DOI] [PubMed] [Google Scholar]
- (70).Liang T; Li Z; Wang P; Zhao F; Liu J; Liu Z Breaking through the Signal-to-Background Limit of Upconversion Nanoprobes Using a Target-Modulated Sensitizing Switch. J. Am. Chem. Soc 2018, 140 (44), 14696–14703. 10.1021/jacs.8b07329. [DOI] [PubMed] [Google Scholar]
- (71).Pasupuleti N; Grodzki AC; Gorin F Mis-Trafficking of Endosomal Urokinase Proteins Triggers Drug-Induced Glioma Nonapoptotic Cell Death. Mol. Pharmacol 2015, 87 (4), 683–696. 10.1124/mol.114.096602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Cortese K; Sahores M; Madsen CD; Tacchetti C; Blasi F Clathrin and LRP-1-Independent Constitutive Endocytosis and Recycling of UPAR. PLoS One 2008, 3 (11), e3730. 10.1371/journal.pone.0003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Rennick JJ; Johnston APR; Parton RG Key Principles and Methods for Studying the Endocytosis of Biological and Nanoparticle Therapeutics. Nat. Nanotechnol 2021, 16 (3), 266–276. 10.1038/s41565-021-00858-8. [DOI] [PubMed] [Google Scholar]
- (74).Nykjaer A; Conese M; Christensen EI; Olson D; Cremona O; Gliemann J; Blasi F Recycling of the Urokinase Receptor upon Internalization of the UPA:Serpin Complexes. EMBO J 1997, 16 (10), 2610–2620. 10.1093/emboj/16.10.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Chan EM; Xu C; Mao AW; Han G; Owen JS; Cohen BE; Milliron DJ Reproducible, High-Throughput Synthesis of Colloidal Nanocrystals for Optimization in Multidimensional Parameter Space. Nano Lett. 2010, 10 (5), 1874–1885. 10.1021/nl100669s. [DOI] [PubMed] [Google Scholar]
- (76).Pettersen EF; Goddard TD; Huang CC; Meng EC; Couch GS; Croll TI; Morris JH; Ferrin TE UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. 2021, 30 (1), 70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


