Abstract
Progress in development of prognostic and therapeutic options for the rare cholestatic liver diseases, primary sclerosing cholangitis (PSC) and primary biliary cholangitis (PBC), is hampered by limited knowledge of their pathogeneses. In particular, the potential role of hepatotoxic and/or metabolism‐altering environmental chemicals in the pathogenesis of these diseases remains relatively unstudied. Moreover, the extent to which metabolic pathways are altered due to ongoing cholestasis and subsequent liver damage or possibly influenced by hepatotoxic chemicals is poorly understood. In this study, we applied a comprehensive exposomics‐metabolomics approach to uncover potential pathogenic contributors to PSC and PBC. We used untargeted high‐resolution mass spectrometry to characterize a wide range of exogenous chemicals and endogenous metabolites in plasma and tested them for association with disease. Exposome‐wide association studies (EWAS) identified environmental chemicals, including pesticides, additives and persistent pollutants, that were associated with PSC and/or PBC, suggesting potential roles for these compounds in disease pathogenesis. Metabolome‐wide association studies (MWAS) found disease‐associated alterations to amino acid, eicosanoid, lipid, co‐factor, nucleotide, mitochondrial and microbial metabolic pathways, many of which were shared between PSC and PBC. Notably, this analysis implicates a potential role of the 5‐lipoxygenase pathway in the pathogenesis of these diseases. Finally, EWAS × MWAS network analysis uncovered linkages between environmental agents and disrupted metabolic pathways that provide insight into potential mechanisms for PSC and PBC. Conclusion: This study establishes combined exposomics‐metabolomics as a generalizable approach to identify potentially pathogenic environmental agents and enumerate metabolic alterations that may impact PSC and PBC, providing a foundation for diagnostic and therapeutic strategies.
Abbreviations
- EWAS
exposome‐wide association study
- FDR
false discovery rate
- GC
gas chromatography
- HILIC
hydrophilic interaction liquid chromatography
- HRMS
high‐resolution mass spectrometry
- IBD
inflammatory bowel disease
- LC
liquid chromatography
- MWAS
metabolome‐wide association study
- OR
odds ratio
- PBC
primary biliary cholangitis
- PCA
principal component analysis
- PSC
primary sclerosing cholangitis
- RPC
reverse phase chromatography
- UDCA
ursodeoxycholic acid
Advances in exposome research are emerging to complement genetics for understanding how environmental factors affect disease onset and progression. With knowledge of such factors, development of diagnostic tools and remediation therapies could launch capabilities for exposome medicine to prevent, manage, or cure disease. Because the liver is a primary organ for metabolic homeostasis and xenobiotic metabolism, we designed a study to apply state‐of‐the‐art exposomics and metabolomics methods to test the transformative potential of exposomics in liver disease.
We focused on two rare cholestatic liver diseases, primary sclerosing cholangitis (PSC) and primary biliary cholangitis (PBC), which affect the large and small to medium‐sized bile ducts of the liver, respectively.( 1 , 2 ) PSC and PBC share a characteristic accumulation of toxic bile acids, which drive an inflammatory and tissue remodeling cascade that impairs hepatic function and often progresses to end‐stage liver disease.( 3 ) The pathogeneses of PSC and PBC are complex, involve hereditary and environmental factors, and are poorly understood. Both diseases are associated with comorbid autoimmunity, and approximately 70% of patients with PSC have concurrent inflammatory bowel disease (IBD).( 1 ) Genome‐wide association studies have advanced our understanding of mechanisms underlying PSC and PBC pathogenesis, highlighting the involvement of the immune system in disease development.( 4 ) Although genome‐wide association studies provide important insight into factors contributing to development of these diseases, they do not fully account for the observed risk, nor are they able to identify features of disease pathogenesis driven by persistent cholestasis.
Current and emerging therapeutic strategies are focused primarily on altering the bile acid pool, and two such treatments, ursodeoxycholic acid (UDCA) and obeticholic acid, have shown to be beneficial in PBC, although many patients fail treatment.( 5 ) Neither of these therapies has demonstrated efficacy in PSC; however, many patients with PSC take UDCA off‐label because it improves liver function tests. Lack of effective therapies partially stems from the rarity of these diseases and low fidelity of current markers of disease severity. There have been recent improvements in disease prognostication tools such as the PREsTo score for PSC,( 6 ) and the GLOBE( 7 ) and UK‐PBC scores( 8 ) for PBC. However, there remains a significant need for novel biomarkers and therapies to improve outcomes of these patients.
We apply a clinical exposomics‐metabolomics framework using high‐resolution mass spectrometry (HRMS) capable of detecting >100,000 compounds in biological samples. The method uses gas chromatography (GC) HRMS for detection of semivolatile compounds, and environmental chemicals and liquid chromatography (LC) HRMS to measure endogenous metabolites. We performed separate exposome‐wide association studies (EWAS) and metabolome‐wide association studies (MWAS) of PSC and PBC. These analyses identified disease‐associated chemicals and pathways that were integrated using an exposome‐metabolome correlation matrix to identify exposure–response network relationships in these diseases. The results demonstrate the potential utility of our approach to discover novel aspects of disease pathogenesis, justifying larger future studies.
Experimental Procedures
Study Design and Participant Selection
Patients and controls were selected from the PSC Scientific Community Resource, which integrates the PSC Resource of Genetic Risk, Environment and Synergy Studies (PROGRESS)( 9 ) and the Mayo Clinic PBC Genetic Epidemiology Registry( 10 ) and actively collects biospecimens and clinical data for use in cholestatic disease research. Patients were diagnosed according to the American Association for the Study of Liver Diseases practice guidelines for PSC( 11 ) or PBC.( 12 )
Plasma samples selected for analysis included 40 individuals each of the following groups: (1) PSC with IBD, (2) PSC without IBD, (3) PSC controls, (4) PBC, and (5) PBC controls. Patients meeting predetermined eligibility criteria were randomly selected for inclusion to eliminate selection bias and then were individually matched to a control based on sex and age (closest available). For PBC, the eligibility criteria included sample collection before liver transplantation, antimitochondrial antibody positivity by prospective test, female sex, and negativity for overlap with autoimmune hepatitis and hepatobiliary cancer. For PSC, criteria included sample collection before liver transplantation and/or colectomy and negativity for overlap with autoimmune hepatitis, small duct disease, and hepatobiliary cancer. Patient selection for PSC was driven by the available population of IBD‐negative patients with PSC, as this was the smaller group, and then individually matched to both a patient with PSC and IBD as well as a healthy control. All participants provided written informed consent. The study was approved by the Mayo Clinic Institutional Review Board (protocol 17‐001851) and conforms to standards laid out in the Declaration of Helsinki.
High‐Resolution Exposomics
Environmental chemicals were analyzed using a high‐resolution exposomics framework that included GC interfaced to an ultrahigh‐resolution mass spectrometer. Plasma samples were prepared in batches of 20 study samples and four quality assurance/quality control samples using a modified version of the extraction method described by Lu et al.( 13 ) and detailed in the Supporting Methods. Plasma extracts were then analyzed in duplicate using a Thermo Scientific 1310 gas chromatograph connected to a Q Exactive GC Orbitrap GC tandem mass spectrometry ultrahigh‐resolution mass spectrometer and Triplus RSH autosampler, as described in the Supporting Methods. The ultrahigh‐resolution mass spectrometer was operated in full‐scan mode over mass‐to‐charge (m/z) range of 85‐850 and 60,000 resolution. Features were extracted using XCMS( 14 ); duplicates were averaged; and those detected with intensities greater than two‐fold the levels in a method blank were retained for analysis, which included 56,246 m/z features. To remove redundant m/z features, we applied the data‐driven clustering algorithm available in RamClustR,( 15 ) which groups feature intensities based on correlation and retention‐time grouping, and provides a weight‐averaged intensity for each group of features corresponding to an individual compound. The combination of feature filtering and ion clustering identified 10,121 unique compounds that were used for untargeted EWAS.
High‐Resolution Metabolomics
Untargeted, high‐resolution metabolomic profiling of plasma was completed in batches of 40 study samples using established methods that leverage use of hydrophilic interaction liquid chromatography (HILIC) and reverse phase chromatography (RPC) with Fourier transform HRMS as previously described( 16 ) and detailed in the Supporting Methods. The HRMS was operated in full scan mode at 120,000 resolution and mass‐to‐charge ratio (m/z) range 85‐1,275. Raw data files were extracted and aligned using apLCMS( 17 ) with modifications by xMSanalyzer.( 18 ) Uniquely detected peaks consisted of m/z, retention time and ion abundance, referred to as metabolite features. Before data analysis, metabolite features were batch‐corrected using ComBat( 19 ) and averaged, followed by filtering to remove those with coefficient of variation ≥100% and more than 50% nondetected intensities for study subject samples.
Statistical Analysis
EWAS
PSC and PBC associations were identified using logistic regression in a parallel approach that included untargeted analysis considering all detected compounds and targeted analysis of environmental chemicals with known confirmed identities. For untargeted EWAS, the log2‐transformed averaged intensity for each compound detected using GC‐HRMS was normalized by the interquartile range and used to test for association with PSC or PBC. The statistical model included adjustments for age and sex for PSC, and age only for PBC, as all participants were female. To account for multiple comparisons, we applied a false discovery rate (FDR) threshold of 20%.( 20 ) For targeted EWAS, we limited analysis to a set of 544 environmental chemicals previously confirmed on the same GC‐HRMS or LC‐HRMS platforms using authentic standards. As the sample size was small, we considered P < 0.05 to be significant in the targeted analyses.
MWAS
Disease‐associated metabolites were identified using LC‐HRMS results and logistical regression with adjustments as described for EWAS, considering HILIC and RPC data sets separately.( 21 ) Metabolites meeting the FDR threshold of 20% were selected for additional characterization and pathway enrichment analysis. Metabolite annotations were assigned using the HumanMFN metabolic model and evidence scoring provided in Mummichog.( 22 ) These annotations provide low confidence matches to metabolites by predicting chemical formulas and do not inform on compound structures. Combining low‐confidence annotations with functional activity patterns within the untargeted data improves the ability to uncover insight into metabolite response profiles to disease and other outcomes.( 23 , 24 ) Thus, we combined annotation of features with pathway‐based functional enrichment analysis using Mummichog to characterize metabolic activity patterns associated with each exposure.( 22 ) Enriched metabolic pathways were determined using a Mummichog scoring threshold of 0.05.
Evaluation of IBD Status and UDCA Treatment
To evaluate how IBD status influences PSC metabolite associations, we performed principal components analysis (PCA) of untargeted results from EWAS and MWAS meeting the FDR threshold. A similar approach was used to test how UDCA uses influenced pathways associated with PSC. For all data sets, PCA was completed using mixOmics( 25 ) and PC1/PC2 scores were evaluated using an unpaired t test. Scores with P < 0.05 were considered significant.
Integrated EWAS × MWAS
We used network analysis to test for correlation between environmental chemicals identified in EWAS and enriched pathways, represented by PC1 calculated separately for each enriched pathway using disease‐associated, annotated metabolites. Network analysis was completed separately for patients with PSC and PBC by first calculating the pairwise Pearson correlation, and edges with |r| ≥ 0.3 and P < 0.05 were retained for network analysis. The igraph package in R was then used to generate a multidata integrative network, and community detection was performed using the multilevel community detection method,( 26 ) and visualized in Cytoscape.( 27 )
Results
Study Population
A total of 200 participants were included in the study, equally representing five participant groups. As IBD is a common comorbidity in PSC, this included 40 patients with PSC with concurrent IBD (PSC‐IBD) and 40 patients with PSC without concurrent IBD (PSC‐nIBD) who were age‐matched and sex‐matched to each other as well as to a group of 40 controls. The PBC arm included 40 patients with PBC who were age‐matched and sex‐matched to a separate group of controls due to demographic differences between PBC and PSC. Notably, we restricted the PBC arm of the study to include only females, as greater than 90% of patients with PBC are women. Patient characteristics are summarized in Table 1 (PBC) and Table 2 (PSC).
TABLE 1.
Demographics of Patients With PBC and Matched Controls
| PBC | Control | P Value | |
|---|---|---|---|
| n | 40 | 40 | — |
| Age, median (IQR) | 59.6 (48.0‐64.5) | 59.6 (48.4‐64.4) | 0.9599* |
| Sex, n (%) | >0.9999 † | ||
| Female | 40 (100) | 40 (100) | |
| Male | 0 (0) | 0 (0) | |
| AMA positive, n (%) | 40 (100) | 0 (0) | >0.9999 † |
| Age at diagnosis (years), median (IQR) | 50.0 (40.0‐55.0) | na | |
| Disease duration (years), median (IQR) | 6.0 (3.0‐12.0) | na | |
| UDCA treatment, n (%) | |||
| Yes | 35 (87.5) | na | |
| No | 4 (10.0) | na | |
| Unknown | 1 (2.5) | na |
Mann‐Whitney Test.
Fischer’s exact test.
Abbreviations: AMA, anti‐mitochondrial antibody; IQR, interquartile range; na, not applicable.
TABLE 2.
Demographics of Patients With PSC and Matched Controls
| PSC (With IBD) | PSC (No IBD) | Control | P Value | |
|---|---|---|---|---|
| n | 40 | 40 | 40 | |
| Age, median (IQR) | 52.0 (43.1‐61.5) | 55.5 (43.5‐64.6) | 52.0 (42.9‐61.5) | 0.8195* |
| Sex, n (%) | >0.9999 † | |||
| Female | 23 (57.5) | 23 (57.5) | 23 (57.5) | |
| Male | 17 (42.5) | 17 (42.5) | 17 (42.5) | |
| Age at diagnosis (years), median (IQR) | 47.0 (31.3‐53.8) | 48.5 (33.5‐57.8) | na | 0.5445 ‡ |
| Disease duration (years), median (IQR) | 5.5 (2.0‐8.0) | 4.0 (2.0‐9.0) | na | 0.9100 ‡ |
| IBD, n (%) | — | |||
| Ulcerative colitis | 30 (75.0) | 0 (0) | na | |
| Crohn’s disease | 4 (10.0) | 0 (0) | na | |
| Indeterminate IBD | 6 (15.0) | 0 (0) | na | |
| UDCA treatment, n (%) | 0.8759 † | |||
| Yes | 17 (42.5) | 19 (47.5) | na | |
| No | 14 (35.0) | 12 (30.0) | na | |
| Unknown | 9 (22.5) | 9 (22.5) | na |
Kruskal‐Wallis test.
Chi‐square test.
Mann‐Whitney test.
Abbreviations: IQR, interquartile range; na, not applicable.
EWAS
Detection of environmental chemicals associated with PSC or PBC was conducted using untargeted and targeted approaches to facilitate inclusion of both unidentified and identity‐confirmed chemicals. We leveraged GC‐HRMS to detect a much larger range of chemicals originating from environmental sources than LC‐HRMS, including many chemicals previously identified as having potential hepatotoxic and steroidogenic effects.
Untargeted EWAS of PSC identified 54 disease‐associated compounds using an FDR threshold of 20% (Fig. 1A). Regression results and spectra for chemicals associated with PSC are provided in Supporting Tables S1 and S2. Notably, no compounds showed an association with PBC at the 20% FDR threshold in the untargeted EWAS (Fig. 1B). Intensity distributions for the top six PSC‐associated compounds with increased levels in patients with PSC relative to controls are shown in Fig. 1C. Annotation of the PSC‐associated compounds in the untargeted EWAS using the NIST 2017 library showed C‐256 (retention time = 4.3 minutes, odds ratio [OR] = 4.3, P = 2.3−05) was a high confidence match to the carbamate pesticide terbucarb (Fig. 1D), with a match score of 840/1,000.
FIG. 1.
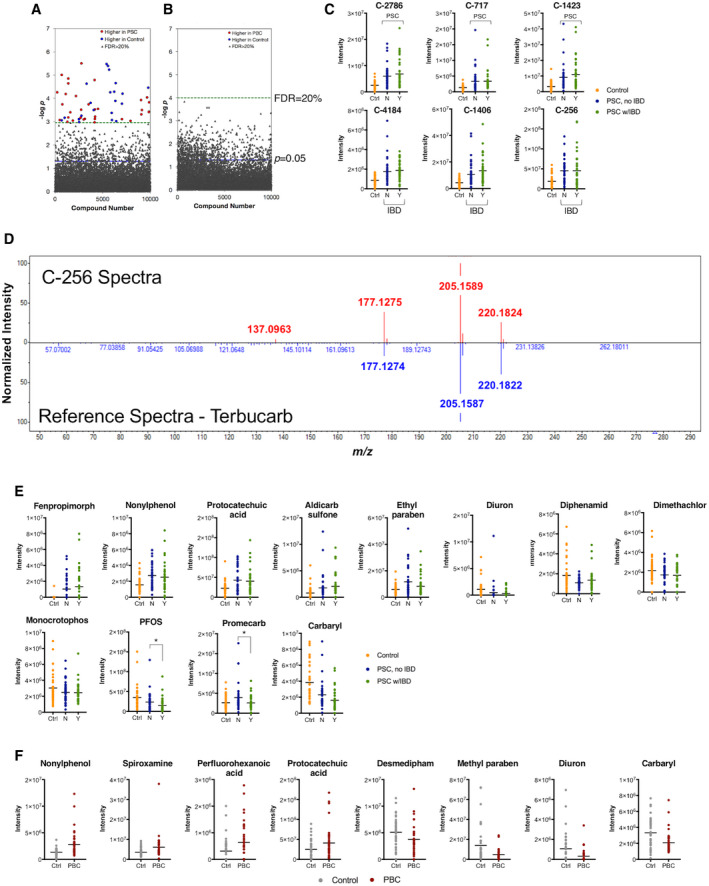
EWAS of PSC and PBC. (A) Untargeted EWAS of 80 patients with PSC and 40 matched controls using >10,000 environmental chemicals detected by GC‐HRMS identified 33 chemicals increased in PSC and 21 elevated in controls at FDR < 20%. (B) Untargeted EWAS of 40 patients with PBC and 40 matched controls found no environmental chemicals to be associated with PBC at FDR < 20%. (C) Intensity values for the top six PSC‐associated environmental chemicals with increased levels in patients with PSC. Patients with PSC are shown separated by IBD status; 40 patients were included in each group. Data for all significant features are provided in Supporting Table S1. (D) Comparison of signal spectra of 54 PSC‐associated environmental chemicals from untargeted EWAS to the NIST database identified terbucarb as a likely match to chemical signal C‐256. (E) Intensity values for PSC‐associated environmental chemicals identified by targeted EWAS of 80 patients with PSC and 40 matched controls at P < 0.05. Levels of perfluorooctanesulfonic acid and promecarb differed between patients with PSC with and without concurrent IBD. (F) Intensity values for PBC‐associated environmental chemicals identified by targeted EWAS of 40 patients with PBC and 40 matched controls at P < 0.05. Complete results of the targeted EWAS are provided in Supporting Tables S3 and S4 for PSC and PBC, respectively. Abbreviations: Ctrl, control; N, no; Y, yes.
For targeted EWAS analysis, we limited evaluation of disease associations to environmental chemicals with confirmed identities by comparison of mass m/z and retention time to a database of standards previously analyzed and confirmed on each platform.( 28 , 29 ) We detected a total of 126 identity‐confirmed environmental chemicals across the two HRMS platforms: 55 by GC‐HRMS and 71 by LC‐HRMS. We found 12 of the 126 environmental chemicals to be associated with PSC at P < 0.05 (Fig. 1E). Environmental chemicals found to be elevated in PSC included the fungicide fenpropimorph (OR = 7.8, P = 0.01), the surfactant nonylphenol (OR = 2.8, P = 0.002), the dietary phenol metabolite protocatechuic acid (OR = 2.75, P = 0.003), the preservative ethyl paraben (OR = 1.9, P = 0.01), and the carbamate pesticide aldicarb sulfone (OR = 2.1, P = 0.01). Of the 12 chemicals associated with PSC, two also differed between patients with PSC‐nIBD and PSC‐IBD, including the perfluoroalkyl surfactant perfluorooctanesulfonic acid (P = 0.049) and the carbamate pesticide promecarb (P = 0.04). Targeted EWAS results for PSC are provided in Supporting Table S3.
In PBC, we found 8 of the 126 environmental chemicals to be associated with disease at P < 0.05 (Fig. 1F). These included two chemicals also elevated in patients with PSC (nonylphenol [OR = 3.5, P = 0.0008] and protocatechuic acid [OR = 1.85, P = 0.03]) and two also decreased in patients with PSC (carbaryl [OR = 0.33, P = 0.006] and diuron [OR = 0.41, P = 0.008]). Additional environmental chemicals found to be elevated in patients with PBC include the fluorinated surfactant perfluorohexanoic acid (OR = 1.89, P = 0.03) and the fungicide spiroxamine (OR = 3.0, P = 0.005). Targeted EWAS results for PBC are provided in Supporting Table S4.
MWAS
Plasma samples were analyzed using HILIC and RPC to maximize detection of polar and nonpolar metabolites.( 21 ) We focused on alterations in endogenous metabolic pathways associated with PSC or PBC to generate insight into systemic biological changes underlying pathogenesis of these diseases. We detected a total of 11,634 (HILIC) and 9,109 (RPC) metabolite features in the 80 patients with PSC and their 40 matched controls. MWAS of these found 1,204 to be associated with PSC at an FDR threshold of 20% (Fig. 2A). To evaluate whether IBD status had a significant impact on metabolic differences, we performed a PCA using the metabolite features associated with PSC. The PCA plot shows that controls form a distinct cluster, albeit overlapping with some of the patients, whereas the patients with PSC do not appear to separate based on IBD status (Fig. 2B). Furthermore, comparison of scores for PC1 and PC2 showed that PC1 significantly differed between patients with PSC and their matched controls (P = 0.0001) and that IBD status did not significantly influence PSC‐associated metabolic differences (PC1: P = 0.55; PC2: P = 0.38) (Fig. 2B). As our approach to the PCA may underestimate the impact of IBD on metabolic profiles in PSC, we performed a second untargeted MWAS that tested for metabolite association with IBD status in patients with PSC only (Fig. 2C). Notably, none of the metabolite features tested in this analysis met the FDR threshold of 20%. A separate MWAS analysis of the 40 patients with PBC and their 40 matched controls detected a comparable number of metabolite features as the PSC MWAS, including 11,729 and 9,294 from HILIC and RPC, respectively. Of these, 703 were associated with PBC at an FDR threshold of 20% (Fig. 2D).
FIG. 2.
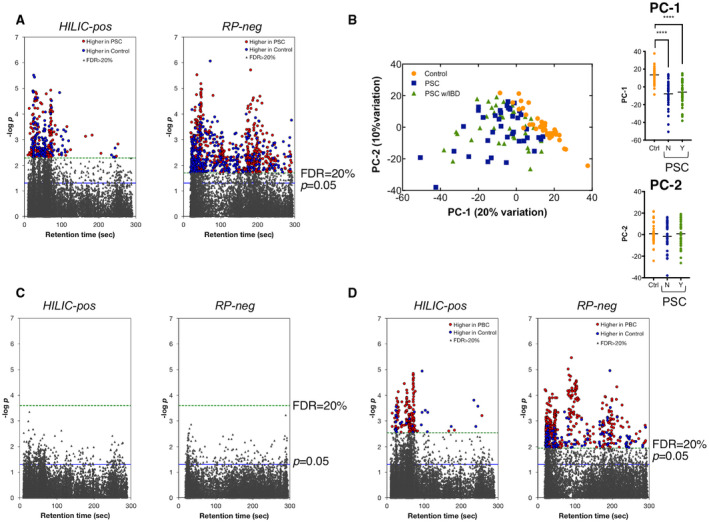
MWAS of PSC and PBC. (A) MWAS of 80 patients with PSC and 40 matched controls identified 1,204 metabolites associated with the disease at FDR < 20%. (B) Comparison of all metabolites associated with PSC at FDR < 20% using PCA showed no significant differences between the 40 patients with PSC with concurrent IBD and 40 without concurrent IBD. (C) MWAS of 40 patients with PSC with concurrent IBD and 40 patients with no IBD detected (no IBD‐associated metabolites at FDR < 20%). (D) MWAS of 40 patients with PBC and 40 matched controls identified 703 metabolites associated with PBC at FDR < 20%. ****P < 0.0001. Abbreviations: PC, principal component; RP, reverse phase.
To determine potential biological mechanisms underlying disease‐associated metabolites from MWAS, we performed pathway enrichment analyses using Mummichog,( 22 ) which was developed specifically for untargeted HRMS data. Mummichog, whose algorithms have been extensively validated,( 30 ) predicts metabolic activity directly from HRMS data without a priori metabolite identification and has been shown to improve biological interpretation while balancing type I and type II error in enrichment of high‐dimensional mass spectral data. For PSC, 27 pathways were significantly enriched for disease‐associated metabolites (Fig. 3 and Supporting Table S5A). For PBC, we identified 10 enriched pathways (Fig. 3 and Supporting Table S5B), nine of which overlap with those associated with PSC. For both diseases, bile acid biosynthesis was the most significant pathway for both HILIC and RPC modes. Pathways relating to metabolism of amino acids, lipids, and eicosanoids were highly represented in both PSC and PBC. Pathways involving metabolism of co‐factors, including vitamins B3 and D, were associated with PSC, while vitamin E metabolism was associated with both diseases. Finally, microbiome‐related metabolic pathways were also present, including butanoate metabolism enriched in both PBC and PSC and propanoate metabolism enriched only in PBC. Complete results are provided in Supporting Tables S6 and S7 for PSC and PBC, respectively.
FIG. 3.
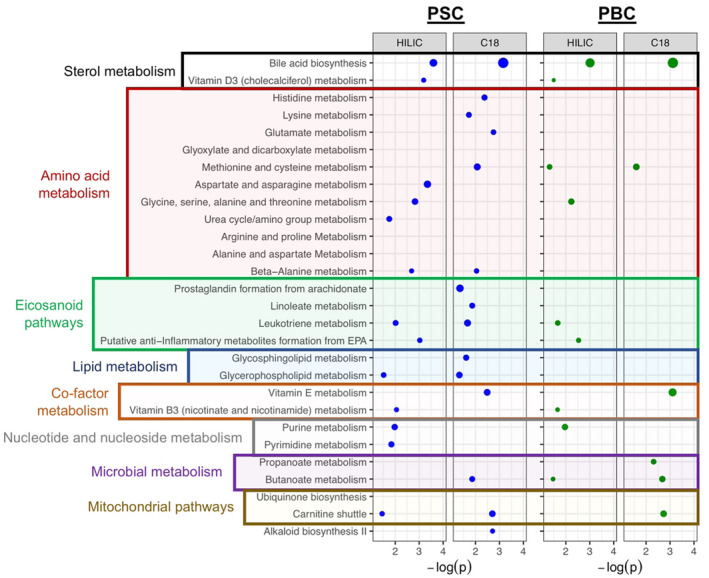
Metabolic pathways associated with PSC and PBC. Pathways enriched in disease‐associated metabolites were identified using Mummichog and grouped based on major biological function. Position of the dot on the x‐axis corresponds to the −log P value calculated for the pathway; and size of the dot corresponds to the number of metabolites meeting the FDR threshold of <20% in the pathway.
As PBC and PSC patients are often treated with UDCA, we performed PCA to assess the potential impact of UDCA treatment on identified disease‐associated metabolic pathways. The high prevalence of UDCA use in PBC (87.5%) precluded useful application of this approach in the patients with PBC. For PSC, patients did not appear to cluster based on UDCA treatment status (Fig. 4A). However, there was a small, yet significant, difference in PC1 (P = 0.034) between patients being treated with UDCA and those who were not (Fig. 4B). To identify which pathways were contributing to this difference, we performed a second PCA analysis considering PC1 score distributions for each individual disease‐associated pathway. Notably, only bile acid biosynthesis (P = 0.01) and vitamin D3 metabolism (P = 0.04) pathways differed between the two groups (Supporting Table S8). This is consistent with the known sterol modulating effect of UDCA and suggests that other pathway associations are largely independent of UDCA treatment.
FIG. 4.
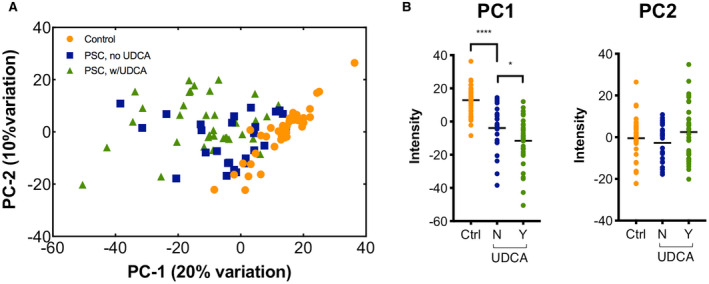
UDCA in treated and untreated patient groups. (A) Comparison of all metabolites associated with PSC at FDR < 20% using PCA showed no apparent clustering of 36 UDCA‐treated and 26 untreated patients with PSC. (B) PC1 differed between controls and untreated patients and between UDCA‐treated and untreated patients with PSC. PC2 did not differ between groups. *P < 0.05, ****P < 0.0001.
EWAS × MWAS
To examine potential environmental impacts on metabolic pathway associations observed in PSC and PBC, we performed an integrated EWAS and MWAS network analysis. All identified disease‐associated environmental chemicals were included. We tested these chemicals for correlation with each disease‐associated pathway in PSC or PBC using a network‐based correlation analysis that enables identification of relationships and characterization of tightly clustered nodes. Results for PSC are shown in Fig. 5A; network correlations and P values for all edges are provided in Supporting Table S9A. For PSC, the resulting network included three interconnected clusters, with the largest (cluster 1) centered on aldicarb sulfone, a carbamate pesticide. This cluster contained metabolic pathways from several biological processes, including most of the amino acid–related pathways, eicosanoid metabolism, and nucleotide pathways. Cluster 2 contained the environmental chemicals fenpropimorph, nonylphenol, and terbucarb in association with pathways related to amino acid metabolism. Finally, cluster 3 grouped pathways from mitochondrial metabolism, bile acid biosynthesis, and eicosanoid metabolism with ethyl paraben and protocatechuic acid. Network correlation results for PBC are shown in Fig. 5B ; correlations and P values for all edges are provided in Supporting Table S9B. For PBC, two connected clusters were identified. Cluster 1 included the fungicide spiroxamine and the carbamate pesticide carbaryl in correlation with five pathways related to metabolism of amino acids, eicosanoids, and nucleotides. Cluster 2 included the chemicals nonylphenol and desmedipham as well as two pathways related to microbiome metabolism.( 31 )
FIG. 5.
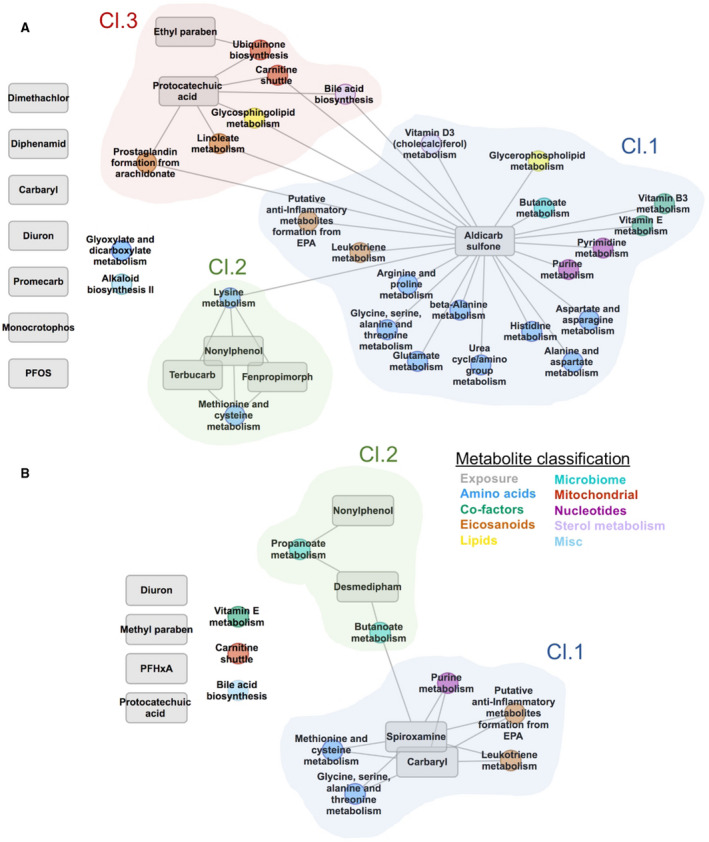
Integrated EWAS × MWAS network analysis of PSC and PBC. The first principal component of each disease‐associated pathway was tested for association with the level of each positively identified disease‐associated environmental chemical, and clusters were identified using a multilevel community detection approach to identify communities of nodes that are tightly connected with each other, but sparsely connected with the rest of the network. (A) Network analysis for PSC. (B) Network analysis for PBC. Abbreviation: PFOS, perfluorooctanesulfonic acid.
Discussion
We applied a multiplatform exposomic and metabolomic analytical framework in PSC and PBC that allows characterization of the blood exposome and metabolome, including many environmental chemicals with known hepatotoxic properties and endogenous metabolic pathways potentially underlying liver dysfunction. Taken together, our results support a role for environmental chemicals in PSC and PBC pathogenesis and represent an innovative step in adapting exposomic methodologies for precision medicine approaches to study liver disease.
We applied untargeted GC‐HRMS profiling to identify potential environmental chemicals contributing to pathogenesis of PSC and PBC, which expands beyond what is traditionally measured using LC‐HRMS and enables detection and characterization of volatile and semivolatile organic pollutants, many of which have previously been linked to liver toxicity. Using this approach, we identified 54 compounds associated with PSC, while none were associated with PBC, highlighting the importance of power in studies of the exposome. Attempted annotation of the 54 PSC‐associated compounds using data available in the NIST 2017 library identified only one high‐confidence match. Although current efforts are focused on generating spectral libraries of unknown metabolites, these methods have not been extended to GC‐HRMS.( 29 , 32 ) The limited annotation of environmental chemicals associated with PSC underscores a major challenge in gaining biological insight using untargeted GC‐HRMS and the need for improved annotation tools for exposome research. This chemical, terbucarb, is a carbamate pesticide, a class of insecticide widely used in household, agricultural, and industrial applications.( 33 ) Carbamate pesticides are acetylcholinesterase inhibitors that have been linked to hepatotoxicity and liver cancer( 34 ) and inhibit activity of key xenobiotic detoxification enzymes in the liver and gut, potentially resulting in accumulation of hepatoxic compounds.( 35 )
We also performed a targeted analysis limited to environmental chemicals with known identities. Using this approach, we found that two environmental chemicals, nonylphenol and protocatechuic acid, were elevated in both patients with PSC and PBC relative to their control groups. Nonylphenol is a suspected endocrine‐disrupting nonionic surfactant used in many consumer products, which contributes to liver damage in animal models.( 36 ) Notably, nonylphenol stimulates transcription of the Pregnane X nuclear receptor, a critical contributor to bile acid homeostasis.( 37 ) Protocatechuic acid is formed from metabolism of dietary polyphenols found in numerous foods, including coffee, green tea, and fruits.( 38 ) Protocatechuic acid has antioxidant and anti‐inflammatory effects and has been shown to be protective against exposure‐induced toxicity in the liver.( 39 ) It is possible that the increased level of protocatechuic acid observed in patients is due to postdiagnostic changes in diet, such as increased coffee consumption, which has been shown to be beneficial in liver disease.( 40 , 41 ) The results from this targeted EWAS identify associations between disease and environmental chemicals with suspected hepatotoxic effects, supporting the possible role of such chemicals in the pathogenesis of PSC and PBC.
Evaluation of metabolites by MWAS identified extensive differences in metabolite levels of patients with PSC and PBC relative to controls. Pathway enrichment analysis found that the bile acid biosynthesis pathway included the greatest number of disease‐associated metabolites in both PSC and PBC, consistent with known mechanisms of cholestatic liver disease.( 42 ) Other metabolomic alterations identified in the MWAS suggest liver dysfunction associated with cholestatic liver disease results in extensive shifts in systemic metabolism. These changes included alterations in pathways related to amino acid, eicosanoid, lipid, cofactor, nucleotide, mitochondrial, and microbial metabolism. The liver is the primary site of amino acid metabolism, and our findings of alterations to many of these pathways in patients is expected and consistent with previous studies.( 43 , 44 ) Furthermore, we found significantly elevated levels of plasma glutamate in patients with PSC and PBC; this is consistent with urea cycle deficiency known to occur with advancing liver disease, which can lead to cognitive dysfunction and hepatic encephalopathy.( 45 ) Although bile acid alterations and the urea cycle are known to influence pathogenesis in PSC and PBC, it remains unclear to what extent broader alterations in amino acid metabolism influence or exacerbate disease. Considering that many of these pathways and their constituent molecules play important roles in regulating oxidative stress and immunity, they could indeed have significant effects. Larger studies capable of stratifying patients based on disease severity will be of interest and could determine whether the observed alterations in various amino acid metabolic pathways are actively pathogenic or a relatively benign consequence of ongoing liver disease.
Eicosanoids are important signaling molecules resulting from metabolism of polyunsaturated fatty acids that have numerous and diverse roles, including modulation of inflammation and immune responses.( 46 ) We found two eicosanoid pathways, leukotriene metabolism and anti‐inflammatory metabolites from eicosapentaenoic acid, to be altered in patients with PSC and PBC. In both groups, changes in leukotriene metabolism were characterized by increased levels of leukotriene B4 and its various metabolites. Leukotriene B4 is formed through the 5‐lipoxygenase pathway and stimulates the production of proinflammatory cytokines.( 47 , 48 ) Notably, alteration of the anti‐inflammatory metabolites from the eicosapentaenoic acid pathway was also characterized by increased levels of products of 5‐lipoxygenase metabolism, including leukotriene B5 and related compounds. Of interest, leukotriene B5 is much less biologically active than leukotriene B4 and can somewhat counteract its inflammatory effects.( 49 ) Taken together, alterations in these pathways suggest that the 5‐lipoxygenase pathway may be involved with the pathogenesis of PSC and PBC, highlighting the ability of our approach to uncover previously unappreciated aspects of cholestatic liver disease.
Cholestasis is known to result in disrupted homeostasis of xenobiotic and internal metabolism; thus, observed associations likely arise due to presence of PSC or PBC. Although these are important to better understanding disease pathogenesis, the combined exposomics‐metabolomics approach provides the added strength of assessing the metabolic response to disease‐associated chemical exposures. To identify such potential relationships between environmental chemicals and metabolic changes, we performed an integrative network analysis that evaluated the correlation between disease‐associated chemicals identified in EWAS and the enriched metabolic pathways from MWAS. The largest cluster in PSC centered on aldicarb sulfone, a commercial‐use carbamate pesticide that is classified as an extremely hazardous substance in the United States and is no longer being distributed. This cluster contains 17 of the 27 PSC‐associated pathways. An additional six pathways were correlated with aldicarb sulfone but were in other clusters, suggesting that this chemical may have broad metabolic effects. This finding is consistent with a recent study of aldicarb exposure in a mouse model that showed chronic exposure resulting in systemic alterations to multiple ‐omic levels including the microbiome, metabolome, and lipidome.( 50 ) Of interest, we noted some inconsistencies with clustering of chemicals that were associated with both PSC and PBC. For instance, in PSC, protocatechuic acid is correlated with changes to carnitine shuttle and bile acid biosynthesis pathways, whereas in PBC it is not. These observations suggest that our integrative network analysis may be underpowered, especially in PBC, and findings should be considered more as a proof‐of‐principle rather than definitive. Indeed, larger studies capable of detecting more robust chemical and metabolic associations and incorporating alternative strategies for integration will provide improved insight into the relationship between environmental chemicals and altered metabolic pathways in the context of PSC and PBC.
We acknowledge limitations in this work. This is a small, cross‐sectional study that is not able to evaluate clinical disease features such as differences in disease presentation, progression, and outcome. Moreover, results of the EWAS focus on identifiable environmental chemicals that could be annotated using available chemical databases, because the limited power precluded challenging and costly in‐depth structural characterization of nonidentified metabolites. Indeed, even in larger studies, such characterization would likely be limited to signals demonstrating an effect on disease progression or outcome rather than those simply associated with disease status. In addition, the results of this study are correlative in nature, and should not be assumed to represent causal associations. In fact, the rarity of PSC and PBC prohibit predisease observational studies required to directly implicate environmental chemicals with disease risk. Finally, we could not account for unknown confounders such as potential differences in lifestyle and nutrition between patients and controls, nor could we identify the exact mechanism through which the associations occurred. Indeed, validation in larger independent cohorts, particularly across geographically and genetically diverse patient groups, as well as functional studies using in vitro and in vivo model systems will be key to gaining actionable insights into the pathogeneses of PSC and PBC. Despite these limitations, the results from this study provide a characterization of PSC and PBC using advanced strategies to simultaneously evaluate the exposome and metabolome. Although the chemicals and metabolic pathways identified in the present study are not necessarily causal factors in PSC or PBC, elevated levels of toxic chemicals in patients could contribute to accelerated liver damage and disease progression, and alterations to metabolic pathways could provide important insight into disease mechanisms.
In conclusion, we used an HRMS‐based approach to evaluate the plasma exposome and metabolome in PSC and PBC. We identified associations between disease and environmental chemicals with suspected hepatotoxic effects. We found alterations in metabolic pathways known to be disrupted in PSC and PBC and identified alterations in pathways and metabolite levels, suggesting the potential involvement of the 5‐lipoxygenase pathway in PSC and PBC. Finally, we applied an innovative network‐based approach to integrate exposome and metabolome data, which identified significant correlations between the environmental toxin aldicarb sulfone and numerous metabolic pathways altered in patients with PSC. This effort provided important insights that will impact the design and execution of future studies. Information obtained from such studies will lead to new lines of research and eventually improved prognostic tools and treatment options for clinicians and their patients that are currently limited.
Supporting information
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Table S8
Table S9
Supplementary Material
Supported by the National Institutes of Health (P30 ES023515, RC2 DK118619, and U2C ES030859), European Commission (874627), the Chris M. Carlos and Catharine N. Nicole Jockisch Carlos Foundation for Endowment Fund in Primary Sclerosing Cholangitis, and the Mayo Clinic.
Potential conflict of interest: Nothing to report.
References
Author names in bold designate shared co‐first authorship.
- 1. Lazaridis KN, LaRusso NF. Primary sclerosing cholangitis. N Engl J Med 2016;375:1161‐1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lleo A, Marzorati S, Anaya JM, Gershwin ME. Primary biliary cholangitis: a comprehensive overview. Hepatol Int 2017;11:485‐499. [DOI] [PubMed] [Google Scholar]
- 3. Lazaridis KN, LaRusso NF. The cholangiopathies. Mayo Clin Proc 2015;90:791‐800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Webb GJ, Hirschfield GM. Using GWAS to identify genetic predisposition in hepatic autoimmunity. J Autoimmun 2016;66:25‐39. [DOI] [PubMed] [Google Scholar]
- 5. Goldstein J, Levy C. Novel and emerging therapies for cholestatic liver diseases. Liver Int 2018;38:1520‐1535. [DOI] [PubMed] [Google Scholar]
- 6. Eaton JE, Vesterhus M, McCauley BM, Atkinson EJ, Schlicht EM, Juran BD, et al. Primary Sclerosing Cholangitis Risk Estimate Tool (PREsTo) predicts outcomes of the disease: a derivation and validation study using machine learning. Hepatology 2020;71:214‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lammers WJ, Hirschfield GM, Corpechot C, Nevens F, Lindor KD, Janssen HLA, et al. Development and validation of a scoring system to predict outcomes of patients with primary biliary cirrhosis receiving ursodeoxycholic acid therapy. Gastroenterology 2015;149:1804‐1812.e1804. [DOI] [PubMed] [Google Scholar]
- 8. Carbone M, Sharp SJ, Flack S, Paximadas D, Spiess K, Adgey C, et al. The UK‐PBC risk scores: derivation and validation of a scoring system for long‐term prediction of end‐stage liver disease in primary biliary cholangitis. Hepatology 2016;63:930‐950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ji S‐G, Juran BD, Mucha S, Folseraas T, Jostins L, Melum E, et al. Genome‐wide association study of primary sclerosing cholangitis identifies new risk loci and quantifies the genetic relationship with inflammatory bowel disease. Nat Genet 2017;49:269‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cordell HJ, Han Y, Mells GF, Li Y, Hirschfield GM, Greene CS, et al. International genome‐wide meta‐analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nat Commun 2015;6:8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chapman R, Fevery J, Kalloo A, Nagorney DM, Boberg KM, Shneider B, et al. Diagnosis and management of primary sclerosing cholangitis. Hepatology 2010;51:660‐678. [DOI] [PubMed] [Google Scholar]
- 12. Lindor KD, Bowlus CL, Boyer J, Levy C, Mayo M. Primary biliary cholangitis: 2018 Practice Guidance from the American Association for the Study of Liver Diseases. Hepatology 2019;69:394‐419. [DOI] [PubMed] [Google Scholar]
- 13. Lu D, Wang D, Ip HS, Barley F, Ramage R, She J. Measurements of polybrominated diphenyl ethers and polychlorinated biphenyls in a single drop of blood. J Chromatogr B Analyt Technol Biomed Life Sci 2012;891‐892:36‐43. [DOI] [PubMed] [Google Scholar]
- 14. Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 2006;78:779‐787. [DOI] [PubMed] [Google Scholar]
- 15. Broeckling CD, Afsar FA, Neumann S, Ben‐Hur A, Prenni JE. RAMClust: a novel feature clustering method enables spectral‐matching‐based annotation for metabolomics data. Anal Chem 2014;86:6812‐6817. [DOI] [PubMed] [Google Scholar]
- 16. Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High‐performance metabolic profiling with dual chromatography‐Fourier‐transform mass spectrometry (DC‐FTMS) for study of the exposome. Metabolomics 2013;9:S132‐S143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu T, Park Y, Li S, Jones DP. Hybrid feature detection and information accumulation using high‐resolution LC‐MS metabolomics data. J Proteome Res 2013;12:1419‐1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Uppal K, Soltow QA, Strobel FH, Pittard WS, Gernert KM, Yu T, et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large‐scale, non‐targeted metabolomics data. BMC Bioinformatics 2013;14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118‐127. [DOI] [PubMed] [Google Scholar]
- 20. Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Series B‐Methodological 1995;57:289‐300. [Google Scholar]
- 21. Liu KH, Walker DI, Uppal K, Tran V, Rohrbeck P, Mallon TM, et al. High‐resolution metabolomics assessment of military personnel: evaluating analytical strategies for chemical detection. J Occup Environ Med 2016;58:S53‐S61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol 2013;9:e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uppal K, Walker DI, Liu K, Li S, Go YM, Jones DP. Computational metabolomics: a framework for the million metabolome. Chem Res Toxicol 2016;29:1956‐1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang LE, Barrette M, et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 2021;49:W388‐W396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rohart F, Gautier B, Singh A, Le Cao KA. mixOmics: an R package for 'omics feature selection and multiple data integration. PLoS Comput Biol 2017;13:e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech 2008;2008:P10008. [Google Scholar]
- 27. Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics 2014;47:8.13.1‐18.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith MR, Uppal K, Walker DI, Utell MJ, Hopke PK, Mallon TM, et al. Environmental chemicals altered in association with deployment for high risk areas. J Occup Environ Med 2019;61(Suppl 12):S15‐S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu X, Walker DI, Liang Y, Smith MR, Orr ML, Juran BD, et al. A scalable workflow to characterize the human exposome. Nat Commun 2021;12:5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li S, Sullivan NL, Rouphael N, Yu T, Banton S, Maddur MS, et al. Metabolic phenotypes of response to vaccination in humans. Cell 2017;169:862‐877.e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheung AC, Walker DI, Juran BD, Miller GW, Lazaridis KN. Studying the exposome to understand the environmental determinants of complex liver diseases. Hepatology 2020;71:352‐362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu KH, Lee CM, Singer G, Bais P, Castellanos F, Woodworth MH, et al. Large scale enzyme based xenobiotic identification for exposomics. Nat Commun 2021;12:5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Struger J, Grabuski J, Cagampan S, Sverko E, Marvin C. Occurrence and distribution of carbamate pesticides and metalaxyl in Southern Ontario Surface Waters 2007‐2010. Bull Environ Contam Toxicol 2016;96:423‐431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morgan MM, Sohn JH, Liebeskind JC. Stimulation of the periaqueductal gray matter inhibits nociception at the supraspinal as well as spinal level. Brain Res 1989;502:61‐66. [DOI] [PubMed] [Google Scholar]
- 35. Di L. The impact of carboxylesterases in drug metabolism and pharmacokinetics. Curr Drug Metab 2019;20:91‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kazemi S, Mousavi Kani SN, Ghasemi‐Kasman M, Aghapour F, Khorasani H, Moghadamnia AA. Nonylphenol induces liver toxicity and oxidative stress in rat. Biochem Biophys Res Commun 2016;479:17‐21. [DOI] [PubMed] [Google Scholar]
- 37. Masuyama H, Hiramatsu Y, Kunitomi M, Kudo T, MacDonald PN. Endocrine disrupting chemicals, phthalic acid and nonylphenol, activate Pregnane X receptor‐mediated transcription. Mol Endocrinol 2000;14:421‐428. [DOI] [PubMed] [Google Scholar]
- 38. Noh H, Freisling H, Assi N, Zamora‐Ros R, Achaintre D, Affret A, et al. Identification of urinary polyphenol metabolite patterns associated with polyphenol‐rich food intake in adults from four European countries. Nutrients 2017;9:796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adefegha SA, Omojokun OS, Oboh G. Modulatory effect of protocatechuic acid on cadmium induced nephrotoxicity and hepatoxicity in rats in vivo. SpringerPlus 2015;4:619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heath RD, Brahmbhatt M, Tahan AC, Ibdah JA, Tahan V. Coffee: the magical bean for liver diseases. World J Hepatol 2017;9:689‐696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lammert C, Juran BD, Schlicht E, Xie X, Atkinson EJ, de Andrade M, et al. Reduced coffee consumption among individuals with primary sclerosing cholangitis but not primary biliary cirrhosis. Clin Gastroenterol Hepatol 2014;12:1562‐1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mousa OY, Juran BD, McCauley BM, Vesterhus MN, Folseraas T, Turgeon CT, et al. Bile acid profiles in primary sclerosing cholangitis and their ability to predict hepatic decompensation. Hepatology 2021;74:281‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tietz‐Bogert P, Kim M, Cheung A, Tabibian J, Heimbach J, Rosen C, et al. Metabolomic profiling of portal blood and bile reveals metabolic signatures of primary sclerosing cholangitis. Int J Mol Sci 2018;19:3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hao J, Yang T, Zhou Y, Gao G‐Y, Xing F, Peng Y, et al. Serum metabolomics analysis reveals a distinct metabolic profile of patients with primary biliary cholangitis. Sci Rep 2017;7:784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lemberg A, Fernandez MA. Hepatic encephalopathy, ammonia, glutamate, glutamine and oxidative stress. Ann Hepatol 2009;8:95‐102. [PubMed] [Google Scholar]
- 46. Lone AM, Tasken K. Proinflammatory and immunoregulatory roles of eicosanoids in T cells. Front Immunol 2013;4:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Crooks SW, Stockley RA. Leukotriene B4. Int J Biochem Cell Biol 1998;30:173‐178. [DOI] [PubMed] [Google Scholar]
- 48. Fainboim L, Cherñavsky A, Paladino N, Flores AC, Arruvito L. Cytokines and chronic liver disease. Cytokine Growth Factor Rev 2007;18:143‐157. [DOI] [PubMed] [Google Scholar]
- 49. Calder PC. The relationship between the fatty acid composition of immune cells and their function. Prostaglandins Leukot Essent Fatty Acids 2008;79:101‐108. [DOI] [PubMed] [Google Scholar]
- 50. Gao B, Chi L, Tu P, Gao N, Lu K. The carbamate aldicarb altered the gut microbiome, metabolome, and lipidome of C57BL/6J mice. Chem Res Toxicol 2019;32:67‐79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Table S8
Table S9
Supplementary Material


