Abstract
Patients with decompensated cirrhosis, particularly those with acute‐on‐chronic liver failure (ACLF), show profound alterations in plasma metabolomics. The aim of this study was to investigate the effect of treatment with simvastatin and rifaximin on plasma metabolites of patients with decompensated cirrhosis, specifically on compounds characteristic of the ACLF plasma metabolomic profile. Two cohorts of patients were investigated. The first was a descriptive cohort of patients with decompensated cirrhosis (n = 42), with and without ACLF. The second was an intervention cohort from the LIVERHOPE‐SAFETY randomized, double‐blind, placebo‐controlled trial treated with simvastatin 20 mg/day plus rifaximin 1,200 mg/day (n = 12) or matching placebo (n = 13) for 3 months. Plasma samples were analyzed using ultrahigh performance liquid chromatography–tandem mass spectroscopy for plasma metabolomics characterization. ACLF was characterized by intense proteolysis and lipid alterations, specifically in pathways associated with inflammation and mitochondrial dysfunction, such as the tryptophan–kynurenine and carnitine beta‐oxidation pathways. An ACLF‐specific signature was identified. Treatment with simvastatin and rifaximin was associated with changes in 161 of 985 metabolites in comparison to treatment with placebo. A remarkable reduction in levels of metabolites from the tryptophan–kynurenine and carnitine pathways was found. Notably, 18 of the 32 metabolites of the ACLF signature were affected by the treatment. Conclusion: Treatment with simvastatin and rifaximin modulates some of the pathways that appear to be key in ACLF development. This study unveils some of the mechanisms involved in the effects of treatment with simvastatin and rifaximin in decompensated cirrhosis and sets the stage for the use of metabolomics to investigate new targeted therapies in cirrhosis to prevent ACLF development.
Abbreviations
- ACLF
acute‐on‐chronic liver failure
- AUROC
area under the receiver operating characteristic curve
- PCA
principal component analysis
Patients with cirrhosis may transition from compensated to a decompensated stage. Decompensated cirrhosis is defined by the occurrence of a number of complications that tend to recur, leading to repeated hospital admissions, impaired quality of life, and increased mortality.( 1 , 2 , 3 ) Eventually, during the natural course of the disease, patients may develop acute‐on‐chronic liver failure (ACLF), a syndrome characterized by development of acute decompensation of cirrhosis associated with single or multiple organ failure(s), including not only the liver but also the kidney, lungs, brain, circulation, and coagulation. ACLF is associated with very poor prognosis and represents the most common cause of death in patients with cirrhosis.( 4 , 5 )
It is currently believed that mechanisms leading to cirrhosis progression include abnormalities of the gut–liver axis that induce systemic inflammation triggered by the passage of bacteria or bacterial products from the intestinal lumen to mesenteric lymph nodes and then the systemic circulation.( 6 ) There is a large body of evidence indicating that systemic inflammation increases progressively during the transition from the compensated to decompensated stage and is markedly elevated in patients with ACLF.( 7 ) Therefore, it is currently hypothesized that systemic inflammation is a key player in the pathophysiology of ACLF.( 4 ) Nonetheless, the intimate mechanisms leading to development of ACLF are not completely understood. In addition, to date there are no drugs or interventions that have been shown to act as disease modifiers preventing the progression of decompensated cirrhosis and development of its most severe complication, the ACLF syndrome. With these uncertainties, elucidating the mechanisms that lead to development of ACLF is an important goal that would provide the rationale to investigate new therapeutic targets.
Several lines of evidence show that statins are promising drugs with several potential beneficial effects in patients with cirrhosis.( 8 ) Treatment with statins has been associated with a decrease in the risk of death or hepatic decompensation in these patients.( 9 , 10 ) Mechanisms of action of statins in liver cirrhosis have been investigated in experimental studies, and effects decreasing hepatic inflammation, fibrogenesis, and portal pressure have been described.( 11 ) However, to date there are no studies that have investigated the mechanism of action of statins in human cirrhosis.
Rifaximin is a poorly absorbed antibiotic widely used for prevention of recurrent hepatic encephalopathy in decompensated cirrhosis.( 12 ) There is some evidence suggesting that rifaximin may also exert some disease‐modifying effects in decompensated cirrhosis. However, the mechanisms by which rifaximin may induce these effects are not known. Some studies demonstrated beneficial effects of rifaximin on microbiome composition and preventing bacterial translocation, while others described a modulating effect of rifaximin in the gut independent of its antibacterial action.( 13 , 14 )
In this regard, the LIVERHOPE project is aimed at investigating the effect of the combination of simvastatin and rifaximin in the prevention of disease progression and development of ACLF in patients with cirrhosis.( 15 ) The first study in this project was aimed at investigating the safety of simvastatin in combination with rifaximin in patients with decompensated cirrhosis.( 16 ) Apart from their potential clinical beneficial effects, the mechanism by which these two drugs may affect the pathophysiology of decompensated cirrhosis and ACLF is unknown.
Metabolomics have emerged as a promising tool to explore the pathophysiology of different diseases.( 17 ) Among other omics, metabolomics is considered a very useful technique as the measurement of metabolites and their concentrations directly reflect the underlying biochemical activity of cells and tissues. Therefore, metabolomics may give the best insight into the molecular phenotype of the patients. As for ACLF, recent reports have revealed a profile of serum metabolites that is characteristic of ACLF and distinguishes it from the metabolomic profile of decompensated cirrhosis.( 18 , 19 ) Moreover, the description of the metabolomic profile in serum of patients with ACLF has also provided some clues about the pathophysiology of this condition.
The aim of the current study was to use metabolomics to unveil the potential effects that treatment with simvastatin and rifaximin has in patients with decompensated cirrhosis. Toward this aim, we performed a study that includes a descriptive cohort to assess the metabolomic profile of patients with decompensated cirrhosis and ACLF and an interventional cohort of patients treated with simvastatin plus rifaximin or placebo to investigate the effects of these drugs in the metabolomic profile of patients with decompensated cirrhosis.
Patients and Methods
Study Population
We analyzed two different cohorts of patients. The first cohort was included to perform a descriptive analysis of the metabolomic profile of patients with decompensated cirrhosis with and without ACLF to determine whether patients with ACLF show a specific metabolomic signature. The second cohort was an intervention cohort aimed at assessing the effects of simvastatin plus rifaximin, a pathophysiological‐based treatment, on the metabolomic profile of patients with decompensated cirrhosis.
The first cohort included 42 patients with cirrhosis randomly selected from a prospectively collected database of consecutive patients admitted to the Liver Unit of the Hospital Clinic of Barcelona for acute decompensation of cirrhosis. Demographic, clinical, and analytical data were collected prospectively at admission and during a 3‐month follow‐up period. Blood samples were collected at hospital admission and stored at −80°C. All patients signed a written informed consent for use of blood samples in research studies.
Cirrhosis was diagnosed according to standard clinical, analytical, and/or histologic criteria. Exclusion criteria were (1) age <18 or >85 years; (2) hepatocellular carcinoma beyond Milan criteria; (3) severe comorbidities, including extrahepatic malignancies; (4) previous liver or kidney transplant; (5) admission for elective procedures; (6) human immunodeficiency virus (HIV) infection; and (7) lack of written informed consent. Patients were divided into two groups according to presence or absence of ACLF at admission to hospital. The first group consisted of 22 patients with decompensated cirrhosis without ACLF, and the second group included 20 patients with ACLF. ACLF was defined according to the Canonic Study criteria.( 20 ) Baseline characteristics of patients from both groups are shown in Supporting Table S1.
The intervention cohort consisted of patients included in the LIVERHOPE‐SAFETY Trial,( 16 ) a randomized double‐blind placebo‐controlled clinical trial aimed at investigating the safety of the combination of two different doses of simvastatin (20 and 40 mg/day) with rifaximin 400 mg/8 hours versus a placebo of the two medications for 12 weeks in 44 patients with decompensated cirrhosis. Detailed information about inclusion and exclusion criteria is shown in Supporting Table S2. Plasma samples were collected at different time points during the study period and stored at −80°C. Results of this study showed that the safest combination was simvastatin 20 mg/day plus rifaximin because the dose of simvastatin 40 mg/day plus rifaximin was associated with a high rate of liver and muscular toxicity.( 16 ) Therefore, for the purpose of the current study, patients selected were those treated with simvastatin 20 mg/day plus rifaximin or placebo.
Following methodological recommendations for statistical analysis of randomized placebo‐controlled clinical trials, comparisons were made between simvastatin 20 mg/day and rifaximin and the placebo arms, using samples taken at the end of the study period. No intragroup comparisons were done.( 21 , 22 ) Among the 14 patients included in the simvastatin 20 mg/day plus rifaximin arm, we excluded from this analysis 2 patients who received only 4 weeks of treatment. Among the remaining 12 patients, 1 patient received 8 weeks of treatment and 11 patients completed 12 weeks of treatment. Among 13 patients included in the placebo arm, 2 patients received 8 weeks of placebo and 11 patients competed 12 weeks, and all of them were included in this analysis. The baseline demographic, clinical, and analytical data were similar in the two groups, except for a younger age in the simvastatin 20 mg/day plus rifaximin group and a slightly higher frequency of hepatic encephalopathy and variceal bleeding in the placebo group (Supporting Table S3).
Untargeted Metabolomics by Ultrahigh Performance Liquid Chromatography–Tandem Mass Spectroscopy
Analyses were performed by Metabolon, Inc. (Morrisville, NC) using ultrahigh performance liquid chromatography–tandem mass spectroscopy (UPLC/MS‐MS) (Durham, NC). Metabolic extracts were obtained by dry and reconstitution procedures in different solvents. All methods used a Waters ACQUITY UPLC and a Thermo Scientific Q‐Exactive high‐resolution/accurate MS interfaced with a heated electrospray ionization (HESI‐II) source. Four methodological procedures were used to analyze different compounds depending on their hydrophilic/hydrophobic and acid/alkali properties. Raw data were extracted, peak identified, and processed by Metabolon. Peaks were quantified using area under the receiver operator characteristic curve (AUROC) analysis. Metabolites investigated included amino acids, carbohydrates, nucleotides, lipids, cofactors and vitamins, energy molecules, and xenobiotics.
Statistical and Data Analysis
Metabolome values were transformed to log2, adding 0.01 to avoid indetermination. A total of 940 metabolites were identified in the plasma samples of patients from the descriptive cohort, and 1,041 metabolites were detected in the plasma samples of patients included in the intervention cohort. Metabolites corresponding to drug compounds or derived metabolites were excluded from the analysis; therefore, the final analysis included 837 metabolites in the descriptive cohort and 985 metabolites in the intervention cohort.
A systematic analysis of the annotated metabolites was performed by manually querying different databases, including the Human Metabolome Database (HMDB) (http://www.hmdb.ca) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.genome.jp/kegg).
Principal component analysis (PCA) is a dimensionality‐reduction method that transforms a set of variables into a reduced set of noncorrelated variables. PCA was performed with the metabolomics data of the descriptive and the intervention cohort using the open‐source statistical computing environment R (http://www.r‐project.org/) and the prcomp package.
To select metabolites that differed between patients with ACLF versus decompensated cirrhosis and patients treated with simvastatin and rifaximin versus placebo, we applied the unpaired Student t test for each pairwise comparison between groups followed by false discovery rate control using the Benjamini‐Hochberg approach.( 23 )
Pathway analysis was conducted using MetPA (through https://www.metaboanalyst.ca/),( 24 ) which combines several advanced pathway enrichment analyses along with the analysis of pathway topological characteristics across more than 800 metabolic pathways.
We used the nearest shrunken centroid classifier implemented in the open‐source statistical computing environment R (http://www.r‐project.org/) and the PAM package( 25 ) to identify within the metabolites of the descriptive cohort the minimal set of compounds capable of predicting the ACLF state with an overall error rate of less than 10%. This method incorporates an internal cross‐validation step during feature selection in which the model is fit on 90% of the samples and then the class of the remaining 10% is predicted. This procedure is repeated 10 times to compute the overall error (10‐fold cross‐validation). AUROC was computed using posterior probabilities and ACLF category. For the ACLF signature, criteria based on the unpaired Student t test and also fold‐change computation were used.
The effect of the treatment with simvastatin and rifaximin on the ACLF signature was calculated by matching those metabolites from the ACLF signature of the descriptive cohort with significant differences between decompensated cirrhosis with and without ACLF and those with significant differences between the treatment and the placebo groups from the intervention cohort. A chord diagram, using the circlize R package,( 26 ) representing flows or connections between selected metabolites and their HMDB superpathway term, was created.
Results
Plasma Metabolomic Profile of Patients With Decompensated Cirrhosis With or Without ACLF
Patients With ACLF Exhibit a Different Plasma Metabolomic Profile Compared to That of Patients With Decompensated Cirrhosis Without ACLF
A total of 837 metabolites were analyzed in this cohort. PCA including the 837 metabolites showed differences between patients with and without ACLF (Fig. 1). Out of the 837 compounds, we identified 61 metabolites that were significantly increased in patients with ACLF compared to those in patients without ACLF and only eight metabolites that were significantly decreased in ACLF (Supporting Table S4). Metabolites that were significantly increased in patients with ACLF were mainly amino acids and carbohydrates, followed by nucleotides and lipids. The increase in these families of metabolites suggests that ACLF is associated with increased proteolysis and glycolysis that likely reflects an increased catabolic status. In a pathway enrichment analysis, pathways related to histidine metabolism, pyrimidine metabolism, cysteine and methionine metabolism, and pentose and glucuronate interconversions were increased in patients with ACLF (Fig. 2).
FIG. 1.
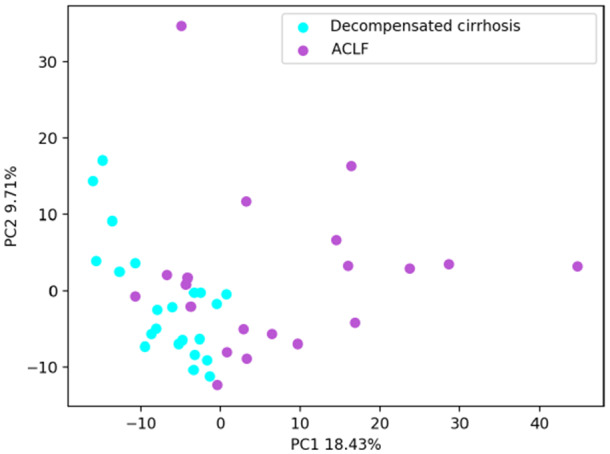
PCA of all patients included in the descriptive cohort and the 837 metabolites analyzed. Each circle corresponds to 1 patient. Light blue circles indicate decompensated cirrhosis without ACLF; purple circles indicate ACLF. PC1 explains 18.43% of the total variation. PC2 explains 9.71% of the total variation. Abbreviations: PC1, principal component 1; PC2, principal component 2.
FIG. 2.
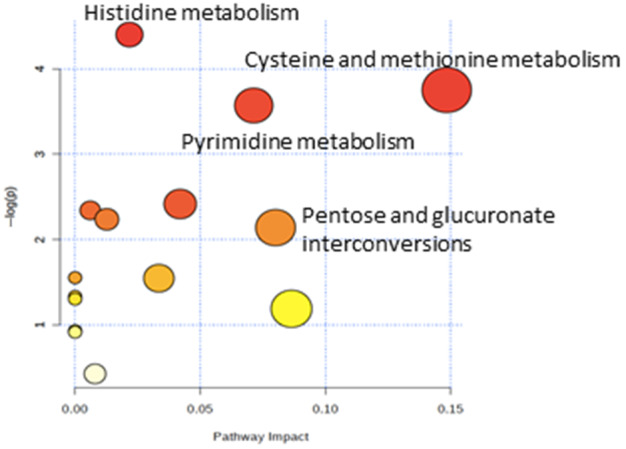
Pathway enrichment analysis, including the metabolites differentially expressed in patients with ACLF compared to patients with decompensated cirrhosis without ACLF. A significant enrichment for several metabolic pathways was identified for these metabolites (P < 0.05). The y axis represents the P value; the x axis represents the pathway impact (pathways more likely to be modified in ACLF compared with decompensated cirrhosis without ACLF). Node color is based on the P value (red indicates a higher level of significance), and node radius is based on the pathway impact value. Metabolites increased in ACLF versus decompensated cirrhosis without ACLF.
We next focused on the top metabolites that were markedly different between patients with ACLF and those without ACLF (fold change >2 or <−2 and P < 0.001), resulting in 21 top increased metabolites. Notably, three out of these 21 metabolites were compounds from the same pathway, the tryptophan–kynurenine pathway (namely, N‐acetyltryptophan, kynurenate, and N‐acetylkinurenine).
Fatty acids, such as adipoyl carnitine and hydroxy adipate, were also included among the top most increased metabolites in patients with ACLF compared to patients without ACLF. Long‐chain fatty acids are transformed into carnitines and carnitine esters to be transported through the mitochondrial membrane and metabolized to generate energy.( 27 ) Increased levels of carnitines and carnitine esters have been reported in the context of impaired fatty acid beta‐oxidation, likely due to mitochondrial dysfunction, in inflammatory conditions and sepsis.( 28 , 29 , 30 )
A Metabolite‐Based Signature That Predicts the Presence of ACLF
A predictive analysis showed that a combination of 32 metabolites was able to predict the presence of ACLF with high accuracy (AUROC, 0.901; 95% confidence interval, 0.820‐1.000) (Fig. 3). Thirty‐one out of the 32 metabolites included in this signature corresponded to compounds that were increased in ACLF compared to patients without ACLF, and the remaining metabolite was decreased in ACLF. The signature included mainly amino acids (19 out of the 32 metabolites, 59%) followed by lipids, carbohydrates, and nucleotides.
FIG. 3.
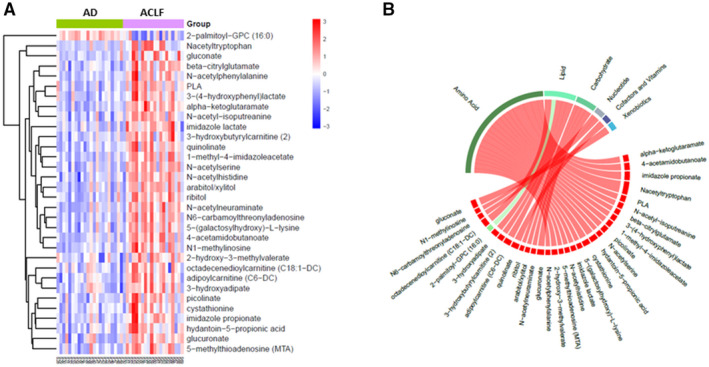
Metabolite‐based signature that predicts the presence of ACLF. (A) Heatmap showing the signature of 32 metabolites that predicts the presence of ACLF (AUROC, 0.91). Each column represents 1 patient, and each row represents one of the 32 metabolites differentially expressed in patients with ACLF compared to patients with decompensated cirrhosis without ACLF. Darker red color represents higher blood levels (fold change), and darker blue color indicates lower levels. (B) Circular chord diagram showing the relationship between the 32 metabolites included in the ACLF signature and their families. Chords connect each metabolite with its respective family. Chords are colored by signal direction (increased metabolites in red and decreased metabolites in green). Chord widths are proportional to magnitude of fold change in ACLF compared to decompensated cirrhosis without ACLF. Abbreviations: AD, acute decompensation of cirrhosis; GPC, glycero‐3‐phosphocholine; PLA, phenyllactate.
Plasma Metabolomic Profile After Treatment With Simvastatin and Rifaximin
Treatment With Simvastatin Plus Rifaximin Induces Relevant Changes in the Plasma Metabolomic Profile of Patients With Decompensated Cirrhosis
A total of 985 metabolites were analyzed in this cohort. PCA obtained from the intervention cohort showed significant differences when comparing patients from the simvastatin plus rifaximin group versus patients from the placebo group at the end of treatment, which suggests the existence of relevant differences in plasma metabolites between the treated group and the placebo group (Fig. 4).
FIG. 4.
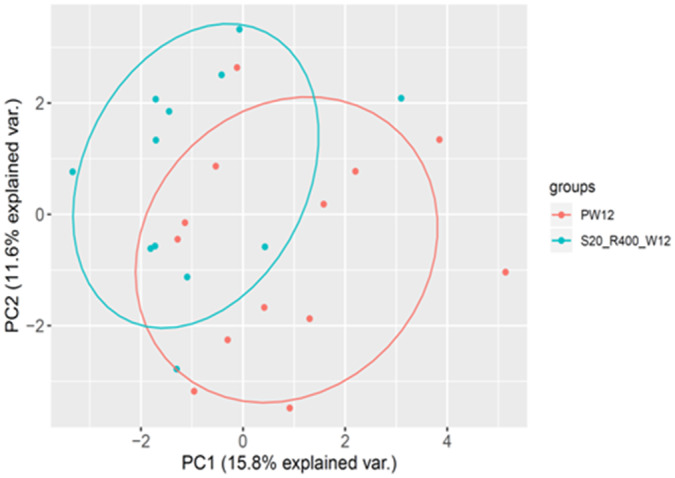
PCA of the intervention cohort. Patients treated with placebo in red, patients treated with simvastatin plus rifaximin in blue. PCA including the 985 metabolites analyzed at the end of treatment versus placebo. PC1 explains 15.8% of the total variation, and PC2 explains 11.6% of the total variation. Abbreviations: PC1, principal component 1; PC2, principal component 2; var., variation.
We then included the list of metabolites that were significantly modified by the treatment with simvastatin plus rifaximin in a pathway analysis. We found that pathways associated with the catabolic state and energy consumption, such as pyrimidine metabolism and pentose and glucuronate interconversions pathways that were increased in the descriptive cohort, were modified by treatment with simvastatin and rifaximin (Fig. 5).
FIG. 5.
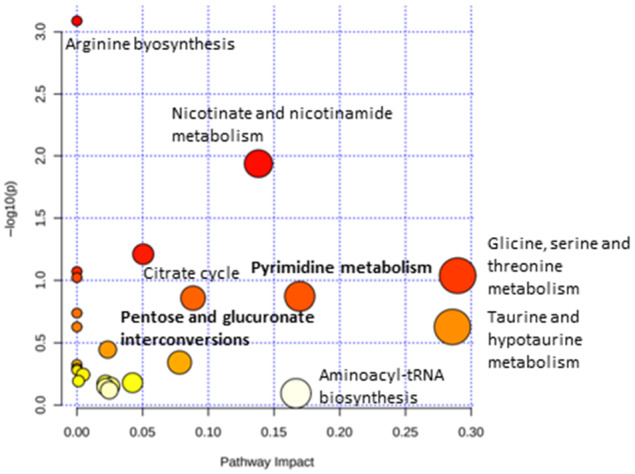
Pathway enrichment analysis, including the metabolites differentially expressed in ACLF in patients treated with simvastatin plus rifaximin compared to those in patients treated with placebo in the intervention cohort. The y axis represents the P value; the x axis represents pathway impact (pathways more likely to be modified in ACLF compared with decompensated cirrhosis without ACLF). Node color is based on the P value (red indicates higher level of significance), and node radius is based on the pathway impact value. A significant enrichment for several metabolic pathways was identified for these metabolites (P < 0.05).
Treatment With Simvastatin and Rifaximin Is Associated With Reduction in Amino Acid and Peptide Levels
Amino acids and peptides metabolism was the metabolic family that was more markedly affected by the treatment with simvastatin plus rifaximin (Supporting Table S5); 54 compounds of this family showed a significantly reduced concentration in the treatment group with respect to placebo, while only five compounds showed a higher concentration in the treatment group with respect to placebo. This suggests a reduction in protein catabolism and degradation in patients receiving treatment with simvastatin plus rifaximin compared to placebo, consistent with a lower catabolic status.
We then investigated tryptophan degradation through the kynurenine pathway because different metabolites of this pathway were found in increased concentrations in patients with ACLF from the first cohort. We found three metabolites, kynurenate, xanthurenate, and 8‐methoxykynurenate, that were at lower concentrations in the treatment arm with respect to placebo; two of these are essential metabolites of this metabolic pathway (Fig. 6). Tryptophan degradation through the kynurenine pathway is induced by proinflammatory molecules and cytokines, and metabolites of this pathway have been involved in the pathogenesis of inflammatory diseases and cancer.( 31 ) The reduction of two of the final metabolites of this pathway in the plasma metabolomic profile of patients treated with simvastatin plus rifaximin strongly suggests an inhibiting effect of treatment on this metabolic pathway.
FIG. 6.
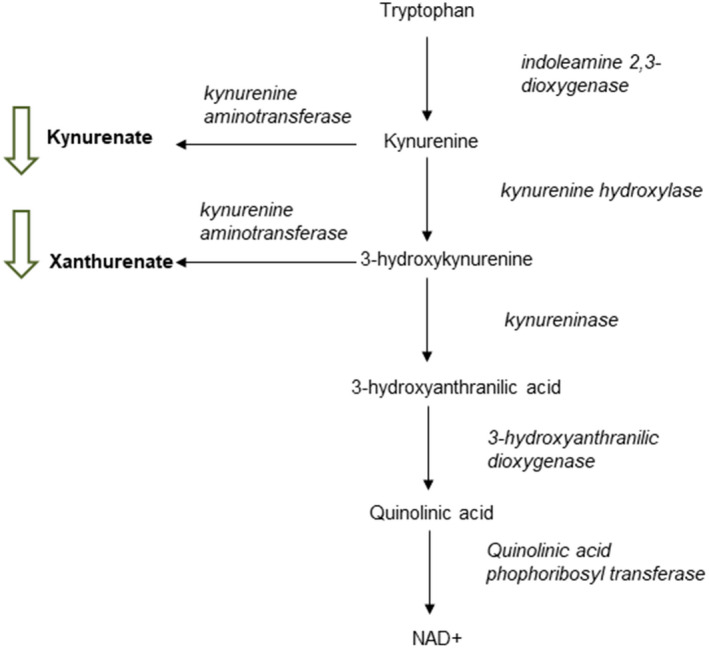
Graphic representation of the most important intermediate metabolites and enzymes involved in the functional pathway of tryptophan degradation through the kynurenine pathway. Green arrows represent metabolites reduced in patients treated with simvastatin plus rifaximin compared to placebo. Abbreviation: NAD, nicotinamide adenine dinucleotide.
Treatment With Simvastatin and Rifaximin Is Associated With Reduction in Secondary Bile Acids and Dicarboxyl Fatty Acids Concentration
Comparison between patients treated with simvastatin and rifaximin and those treated with placebo showed marked differences in plasma concentrations of bile acids. In general, treatment reduced the concentration of primary and secondary bile acids, but effects were more marked in secondary bile acids (Supporting Table S6). Secondary bile acids have been associated with disease progression in patients with cirrhosis( 32 , 33 ); therefore, the reduction in secondary bile acids levels may account, at least in part, for the potential beneficial effects of rifaximin in the gut of patients with cirrhosis.
Finally, changes in fatty acids metabolism were also explored. We found 21 fatty acids at lower concentrations in the treatment group compared to placebo. Eleven of these were dicarboxyl fatty acids, which are compounds that were found to be increased in ACLF in the descriptive cohort and the ACLF signature. This type of fatty acids is produced when omega oxidation is activated to supply the activity of beta oxidation by the mitochondria, which has been associated with inflammatory states in previous reports.( 29 , 30 , 34 ) The consistent decrease in the concentrations of dicarboxyl fatty acids caused by the treatment medication may reflect a lower inflammatory status associated with treatment and possibly an improved mitochondrial function. Interestingly, mitochondrial dysfunction has been reported as a key component of the metabolomic profile of patients with ACLF.( 18 )
Relationship Between Changes in The Metabolomic Profile and Clinical Outcomes
We next sought to determine the potential relationship between the effects of treatment on the metabolomic profile and the development of complications. Overall, 4 of the 12 patients from the treatment group developed complications of cirrhosis during follow‐up (acute kidney injury, n = 1; hepatic encephalopathy, n = 2; bacterial infections, n = 1). At the end of treatment, 114 out of the 985 metabolites were different between patients who did and did not develop complications within the treatment group. Despite the low number of events, a PCA showed differences between the two subgroups (Supporting Fig. S1). Among the 114 differential metabolites, most of them were amino acids (49, 43%), followed by lipids, carbohydrates, and nucleotides. In patients who developed complications, 42 amino acids were significantly increased compared to patients who did not develop complications and seven amino acids significantly decreased. Interestingly, amino acids were the metabolite family that was found to be more markedly affected by treatment with simvastatin and rifaximin. Therefore, these results suggest that the effect of treatment on the metabolomic profile was reduced in patients who developed complications.
We next compared the metabolomic profile of patients who did and did not develop complications in the treatment group to that of the placebo group. The metabolomic profile of patients who developed complications during follow‐up was more similar to that of the placebo group than to that of patients who did not develop complications. At the end of treatment, only 47 out of the 985 metabolites were significantly different between patients from the treatment group who developed complications and those from the placebo group. Most of these metabolites were amino acids and lipids. In contrast, up to 117 metabolites were significantly different between patients from the treatment group who did not develop complications and those from the placebo group. Overall, these results suggest that treatment with simvastatin and rifaximin had a more marked effect on the metabolomic profile in patients who did not develop complications, and this may have contributed to the prevention of complications. In contrast, the effect of treatment on patients who developed complications may have been lower as their metabolomic profile at the end of treatment was more similar to that of patients from the placebo group.
Effect of Simvastatin and Rifaximin Treatment on the ACLF Plasma Metabolomic Signature
Finally, we investigated whether there was concordance between the ACLF signature derived from the descriptive cohort and the 161 metabolites that were found to be modified by treatment with simvastatin plus rifaximin. Eighteen metabolites that were present in the ACLF signature of the descriptive cohort were decreased by treatment with simvastatin and rifaximin (Fig. 7). Plasma levels of these 18 metabolites were significantly lower in patients treated with simvastatin and rifaximin compared to those of patients treated with placebo and also in patients with ACLF. The 18 metabolites corresponded mainly to amino acids (n = 12), followed by carbohydrates (n = 3), lipids (n = 2), and nucleotide (n = 1). This group of metabolites from the ACLF signature that is targeted by treatment with simvastatin plus rifaximin provides new insights on possible mechanisms of action of this treatment combination to prevent ACLF development.
FIG. 7.
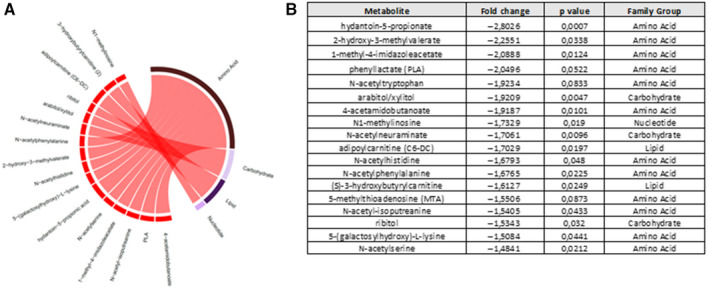
Metabolites from the ACLF signature that are significantly affected by treatment with simvastatin and rifaximin. (A) Circular chord diagram showing the relationship between the 18 metabolites of the ACLF signature affected by the treatment and their families. Chords connect each metabolite with its respective family. Chords are colored by signal direction (increased metabolites in red and decreased metabolites in green). Chord widths are proportional to magnitude of fold change in ACLF compared to decompensated cirrhosis without ACLF. (B) List with the 18 metabolites and the fold change in the treatment group compared to placebo. Abbreviations: PLA, phenyllactic acid.
Discussion
In the current study, we report the effects of treatment with simvastatin plus rifaximin on plasma metabolomic profile of patients with decompensated cirrhosis, using high‐throughput untargeted metabolomic analysis. First, we defined the metabolites and functional pathways associated with ACLF and generated a metabolomic‐based signature characteristic of ACLF. Second, we reported the plasma metabolites that are modified by treatment with simvastatin plus rifaximin in patients with decompensated cirrhosis. Finally, the effects of treatment with simvastatin plus rifaximin on metabolites involved in ACLF pathogenesis were described. To our knowledge, this is the first study in which high‐throughput untargeted metabolomics methodology was used to unveil the effects of a therapeutic intervention aimed at preventing progression of cirrhosis and ACLF development.
Among the different families of metabolites involved in the ACLF signature, specifically the amino acids and peptides family was significantly modified by treatment. A number of peptides and amino acids were found at lower concentrations in the treatment group with respect to placebo. High levels of amino acids and peptides likely reflect increased proteolysis and have been associated with a high catabolic status characteristic of inflammatory conditions and sepsis.( 35 , 36 , 37 ) Notably, in our descriptive cohort and also in previous reports on the ACLF metabolomic profile, amino acids were the most commonly increased metabolites in patients with ACLF.( 18 ) Our findings show that treatment with simvastatin and rifaximin significantly reduces the concentrations of these metabolites, which suggests that this treatment combination could reduce the catabolic status characteristic of decompensated cirrhosis and ACLF. Considering the mechanistic effects of simvastatin and rifaximin, we can speculate that the decrease in catabolic status may be the result of a decrease in systemic inflammation induced either by the immunomodulatory effects of simvastatin( 38 , 39 ) and/or by the potential effects of rifaximin in decreasing endotoxemia and modulating gut microbiome.( 40 , 41 )
Among the amino acids and peptides family modified by treatment, we found the tryptophan–kynurenine pathway to be of special interest. This pathway is involved in tryptophan degradation by gut microbiome as well as several cell types, and its downstream metabolites are involved in several important physiological metabolic processes.( 42 , 43 ) The kynurenine pathway generates a number of metabolites that are involved in inflammation and immune response. The kynurenine pathway is mainly initiated by indoleamine 2,3‐dioxygenase, which is up‐regulated in the context of increased proinflammatory cytokine levels, such as interferon‐γ or tumor necrosis factor α in inflammatory or infectious conditions.( 42 ) Kynurenine metabolites have been shown to be involved in mechanisms leading to immune suppression by effects on cells from both the innate and the adaptive immune system. In addition, there are data showing that low‐grade chronic inflammation can also lead to increased circulating kynurenine levels.( 44 ) Abnormalities in the kynurenine pathway have been described in patients with cirrhosis and ACLF( 45 ) and were also found in our descriptive cohort. Importantly, treatment with simvastatin plus rifaximin reduced the concentrations of three key compounds of this metabolic pathway, kynurenate, xanthurenate, and 8‐methoxykynurenate, which suggests that this could be a potential mechanism by which the combined treatment with simvastatin plus rifaximin could exert its potential beneficial effects in cirrhosis.
The exact molecular mechanism by which simvastatin and rifaximin treatment can modify the tryptophan–kynurenine pathway cannot be elucidated from the results of our study. Nonetheless, previous studies reported a decrease in kynurenines in patients treated with simvastatin in chronic inflammatory conditions, such as patients with cardiovascular or chronic kidney diseases.( 46 ) Results from these studies suggest that the effect of simvastatin on the kynurenine pathway may be driven by a decrease in systemic inflammation.
In addition, treatment with simvastatin plus rifaximin was associated with reduction in plasma levels of a number of lipid metabolites. Among them, a reduction in the levels of fatty acids, probably associated with simvastatin lipid‐lowering effects, was found. On the other hand, an increase in acyl carnitines has also been reported in the context of impaired fatty acid beta oxidation in inflammatory conditions and sepsis.( 29 , 30 , 34 ) Therefore, the immunomodulatory effects of the combination of simvastatin and rifaximin could also explain the decrease of these fatty acids after treatment.
Secondary bile acids were also found at reduced concentrations after simvastatin plus rifaximin treatment. Secondary bile acids are produced as a result of bacterial metabolism in the gut, and increased plasma concentrations of these metabolites have been described in patients with cirrhosis.( 47 ) A proof‐of‐concept study on the effect of rifaximin on the metabolomic profile of patients with cirrhosis showed an effect of rifaximin treatment reducing plasma concentrations of secondary bile acids; this was associated with changes on the microbiome composition.( 48 ) In the current study, we found similar effects of combined treatment with simvastatin and rifaximin on plasma secondary bile acids concentration that could account, at least in part, for the positive effects of rifaximin in patients with cirrhosis.
The current study has some strengths and limitations that should be mentioned. First, for the first time we provide evidence of the existence of dynamic changes in disease pathophysiology during administration of a pharmacological treatment aimed at targeting some of the pathogenic events of cirrhosis. Although the clinical effects of treatment with the combination of simvastatin and rifaximin have not been investigated, there is strong evidence of the beneficial effect of either drug in reducing mortality and preventing hepatic encephalopathy recurrence, respectively, when given individually.( 10 , 49 ) The study was performed in the context of a double‐blind, placebo‐controlled, randomized trial. Therefore, results in patients treated with simvastatin and rifaximin could be compared to those in patients treated with placebo, thus allowing a precise evaluation of the effects of treatment and avoiding selection bias.
The study has, however, some limitations. The number of patients studied was relatively small, and results will need to be reproduced in a larger series of patients. In addition, treatment was given for a relatively short period of time, 8 or 12 weeks. Therefore, it is not known whether the effects persist during a longer period of treatment or, alternatively, whether a longer duration of treatment could have more marked beneficial effects on metabolic alterations of cirrhosis. The intervention group was treated with the combination of simvastatin and rifaximin, but there were no intervention groups treated with either simvastatin or rifaximin alone. Therefore, the design of our study does not allow us to address the question as to whether the effects on the metabolomic profile are due to either simvastatin, rifaximin, or the combination of both. However, based on existing literature we can speculate which of the drugs may be responsible for the different findings. Finally, the intervention trial was aimed at evaluating the safety of two doses of simvastatin, and the follow‐up period is short; therefore, the number of clinical outcomes developed during follow‐up is low. In this regard, the analysis of the effects of changes on the metabolomic profile on clinical outcomes is limited and should be taken with caution. A large multicenter international study with longer treatment duration and evaluation of clinical outcomes is underway (LIVERHOPE Efficacy Trial, clinical trials.gov NCT03780673).
In conclusion, our results show that treatment with simvastatin and rifaximin modulates the metabolomic profile of patients with decompensated cirrhosis and decreases and deactivates a number of metabolites and metabolomic pathways associated with cirrhosis progression and development of ACLF. In particular, evidence from the present study suggests that the simvastatin and rifaximin combination may decrease protein catabolism, inhibit the tryptophan–kynurenine pathway, reduce lipids and fatty acid plasma levels (possibly reflecting an improvement in mitochondrial function), and decrease plasmatic concentrations of secondary bile acids. Our study shows that some of the pathways that may be key for ACLF development can be targeted and modulated by a specific therapeutic intervention, setting the stage for the use of metabolomics in the development of new targeted therapies in cirrhosis to prevent disease progression and ACLF development.
Supporting information
Fig S1
Table S1‐S6
Acknowledgment
We acknowledge the support of Nicki van Berckel in the overall management of the LIVERHOPE project. We also acknowledge the support of Roser Poblet and Beatriz Márquez in administrative issues related to the current work and manuscript handling. We are very thankful to the physicians and nurses of the different liver units participating in the study. Finally, we thank the patients and their families for their participation in the LIVERHOPE project.
Appendix 1.
LIVERHOPE Consortium investigators: M. Schulz, P. Ferstl, I. Giovo, O. Roux, M. Simon‐Talero, M. Pérez‐Guasch, A.B. Rubio, M. Cervera, S. Martínez, N. Fabrellas, J. Pich, A. Vives, E. Avitabile, I. Graupera, C. Solé, O. Bassegoda, J. Gratacós‐Ginès, M. Joyera, E. Palacio, M. Aban, T. Lanzillotti, G. Nicolao, M.T. Chiappa, V. Esnault, R. Andrade, M. Pavesi, M. Korenjak, J. Farrés, M. Serra‐Burriel, P. Angeli.
Supported by The European Commission Horizon 2020 (LIVERHOPE project number 731875), ISCIII‐Subdirección General de Evaluación and European Regional Development Fund for the Plan Nacional I+D+I (grant number PI20/00579 to P.G. and PI18/00727 to E.S.), and the Agency for Administration of University and Research (grant number 2017SGR‐01281 to E.P., E.S., P.G., and other members of the LIVERHOPE Consortium). The H2020 LIVERHOPE GRANT supports the research of the following authors: E.P., E.S., A.J., G.Z., K.dW., F.U., M.T., K.K., C.J., D.C., M.C., M.M.‐R., C.A., U.B., P.C., F.D., R.P.M., J.T., V.V., S.P., and P.G.
Clinical Trials Register, 2016‐004499‐23
Registration on ClinicalTrials.gov, NCT03150459
Part of the work reported here is an ancillary study of previously reported work published in Lancet Gastroenterology and Hepatology (Lancet Gastroenterol Hepatol 2020;5:31‐41).
Potential conflict of Interest: Dr. Durand consults for Biotest. Dr. Vargas consults for Promethera and is on the speakers’ bureau for Intercept. Dr. Piano advises Mallinckrodt. Dr. Watson is employed by Evotech and owns stock in Sanofi. Dr. Gines consults for and received grants from Gilead, Grifols, and Mallinckrodt; he consults for Novartis, Martin, and Ferring. The other authors have nothing to report.
Contributor Information
Elsa Solà, Email: esola@clinic.cat.
for the investigators of the LIVERHOPE Consortium:
M. Schulz, P. Ferstl, I. Giovo, O. Roux, M. Simon‐Talero, M. Pérez‐Guasch, A.B. Rubio, M. Cervera, S. Martínez, N. Fabrellas, J. Pich, A. Vives, E. Avitabile, I. Graupera, C. Solé, O. Bassegoda, J. Gratacós‐Ginès, M. Joyera, E. Palacio, M. Aban, T. Lanzillotti, G. Nicolao, M.T. Chiappa, V. Esnault, R. Andrade, M. Pavesi, M. Korenjak, J. Farrés, M. Serra‐Burriel, and P. Angeli
References
Authors names in bold designate shared co‐first authorship.
- 1. Ginés P, Quintero E, Arroyo V, Terés J, Bruguera M, Rimola A, et al. Compensated cirrhosis: natural history and prognostic factors. Hepatology 1987;7:122‐128. 10.1002/hep.1840070124. [DOI] [PubMed] [Google Scholar]
- 2. D’Amico G, Garcia‐Tsao G, Pagliaro L. Natural history and prognostic indicators of survival in cirrhosis: a systematic review of 118 studies. J Hepatol 2006;44:217‐231. [DOI] [PubMed] [Google Scholar]
- 3. Gines P, Krag A, Abraldes JG, Solà E, Fabrellas N, Kamath PS. Liver cirrhosis. Lancet 2021;398:1359‐1376. [DOI] [PubMed] [Google Scholar]
- 4. Hernaez R, Solà E, Moreau R, Ginès P. Acute‐on‐chronic liver failure: an update. Gut 2017;66:541‐553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mezzano G, Juanola A, Cardenas A, Mezey E, Hamilton JP, Pose E, et al. Global burden of disease: acute‐on‐chronic liver failure, a systematic review and meta‐analysis. Gut 2021; 10.1136/gutjnl-2020-322161. [DOI] [PubMed] [Google Scholar]
- 6. Wiest R, Lawson M, Geuking M. Pathological bacterial translocation in liver cirrhosis. J Hepatol 2014;60:197‐209. 10.1016/j.jhep.2013.07.044. [DOI] [PubMed] [Google Scholar]
- 7. Solé C, Solà E, Morales‐Ruiz M, Fernàndez G, Huelin P, Graupera I, et al. Characterization of inflammatory response in acute‐on‐chronic liver failure and relationship with prognosis. Sci Rep 2016;6:32341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pose E, Trebicka J, Mookerjee RP, Angeli P, Ginès P. Statins: old drugs as new therapy for liver diseases? J Hepatol 2019;70:194‐202. [DOI] [PubMed] [Google Scholar]
- 9. Simon TG, Bonilla H, Yan P, Chung RT, Butt AA. Atorvastatin and fluvastatin are associated with dose‐dependent reductions in cirrhosis and hepatocellular carcinoma, among patients with hepatitis C virus: results from ERCHIVES. Hepatology 2016;64:47‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abraldes JG, Villanueva C, Aracil C, Turnes J, Hernandez‐Guerra M, Genesca J, et al.; BLEPS Study Group . Addition of simvastatin to standard therapy for the prevention of variceal rebleeding does not reduce rebleeding but increases survival in patients with cirrhosis. Gastroenterology 2016;150:1160‐1170.e3. [DOI] [PubMed] [Google Scholar]
- 11. Trebicka J, Hennenberg M, Odenthal M, Shir K, Klein S, Granzow M, et al. Atorvastatin attenuates hepatic fibrosis in rats after bile duct ligation via decreased turnover of hepatic stellate cells. J Hepatol 2010;53:702‐712. [DOI] [PubMed] [Google Scholar]
- 12. Caraceni P, Vargas V, Solà E, Alessandria C, Wit K, Trebicka J, et al.; Liverhope Consortium . The use of rifaximin in patients with cirrhosis. Hepatology 2021;74:1660‐1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng J, Shah YM, Ma X, Pang X, Tanaka T, Kodama T, et al. Therapeutic role of rifaximin in inflammatory bowel disease: clinical implication of human pregnane X receptor activation. J Pharmacol Exp Ther 2010;335:32‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Solé C, Guilly S, Da Silva K, Llopis M, Le‐Chatelier E, Huelin P, et al. Alterations in gut microbiome in cirrhosis as assessed by quantitative metagenomics. Relationship with acute‐on‐chronic liver failure and prognosis. Gastroenterology 2021;160:206‐218.e13. [DOI] [PubMed] [Google Scholar]
- 15. LIVERHOPE Consortium . LIVERHOPE project. https://www.liverhope‐h2020.eu. Published January 1, 2017. Accessed October 2020.
- 16. Pose E, Napoleone L, Amin A, Campion D, Jimenez C, Piano S, et al. Safety of two different doses of simvastatin plus rifaximin in decompensated cirrhosis (LIVERHOPE‐SAFETY): a randomised, double‐blind, placebo‐controlled, phase 2 trial. Lancet Gastroenterol Hepatol 2020;5:31‐41. [DOI] [PubMed] [Google Scholar]
- 17. Tolstikov V, Moser AJ, Sarangarajan R, Narain NR, Kiebish MA. Current status of metabolomic biomarker discovery: Impact of study design and demographic characteristics. Metabolites 2020;10:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moreau R, Clària J, Aguilar F, Fenaille F, Lozano JJ, Junot C, et al.; CANONIC Study Investigators of the EASL Clif Consortium; Grifols Chair; European Foundation for the Study of Chronic Liver Failure . Blood metabolomics uncovers inflammation‐associated mitochondrial dysfunction as a potential mechanism underlying ACLF. J Hepatol 2020;72:688‐701. Erratum in: J Hepatol 2020;72:1218‐1220. [DOI] [PubMed] [Google Scholar]
- 19. Bajaj JS, Reddy KR, O’Leary JG, Vargas HE, Lai JC, Kamath PS, et al. Serum levels of metabolites produced by intestinal microbes and lipid moieties independently associated with acute on chronic liver failure and death in patients with cirrhosis. Gastroenterology 2020;159:1715‐1730.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moreau R, Jalan R, Gines P, Pavesi M, Angeli P, Cordoba J, et al.; CANONIC Study Investigators of the EASL–CLIF Consortium . Acute‐on‐chronic liver failure is a distinct syndrome that develops in patients with acute decompensation of cirrhosis. Gastroenterology 2013;144:1426‐1437.e1‐e9. [DOI] [PubMed] [Google Scholar]
- 21. Bland JM, Altman DG. Comparisons within randomised groups can be very misleading. BMJ 2011;342:d561. [DOI] [PubMed] [Google Scholar]
- 22. Bland JM, Altman DG. Comparisons against baseline within randomised groups are often used and can be highly misleading. Trials 2011;12:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 1995;57:289‐300. [Google Scholar]
- 24. Xia J, Wishart DS. MetPA: a web‐based metabolomics tool for pathway analysis and visualization. Bioinformatics 2010;26:2342‐2344. [DOI] [PubMed] [Google Scholar]
- 25. Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A 2002;99:6567‐6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gu Z, Gu L, Eils R, Schlesner M, Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics 2014;30:2811‐2812. [DOI] [PubMed] [Google Scholar]
- 27. Houten SM, Wanders RJA. A general introduction to the biochemistry of mitochondrial fatty acid β‐oxidation. J Inherit Metab Dis 2010;33:469‐477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langley RJ, Tsalik EL, Velkinburgh JCV, Glickman SW, Rice BJ, Wang C, et al. An integrated clinico‐metabolomic model improves prediction of death in sepsis. Sci Transl Med 2013;5.195ra95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Puskarich MA, Evans CR, Karnovsky A, Das AK, Jones AE, Stringer KA. Septic shock nonsurvivors have persistently elevated acylcarnitines following carnitine supplementation. Shock 2018;49:412‐419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eaton S, Pierro A. Carnitine and fatty acid oxidation in sepsis. Monatsh Chem 2005;136:1483‐1492. [Google Scholar]
- 31. Platten M, Nollen EAA, Röhrig UF, Fallarino F, Opitz CA. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discov 2019;18:379‐401. [DOI] [PubMed] [Google Scholar]
- 32. Kakiyama G, Hylemon PB, Zhou H, Pandak WM, Heuman DM, Kang DJ, et al. Colonic inflammation and secondary bile acids in alcoholic cirrhosis. Am J Physiol Liver Physiol 2014;306:G929‐G937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Albillos A, de Gottardi A, Rescigno M. The gut‐liver axis in liver disease: pathophysiological basis for therapy. J Hepatol 2020;72:558‐577. [DOI] [PubMed] [Google Scholar]
- 34. Van Wyngene L, Vandewalle J, Libert C. Reprogramming of basic metabolic pathways in microbial sepsis: therapeutic targets at last? EMBO Mol Med 2018;10.e8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang A, Luan HH, Medzhitov R. An evolutionary perspective on immunometabolism. Science 2019;363:eaar3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hotamisligil GS. Inflammation, metaflammation and immunometabolic disorders. Nature 2017;542:177‐185. [DOI] [PubMed] [Google Scholar]
- 37. Hotamisligil GS. Foundations of immunometabolism and implications for metabolic health and disease. Immunity 2017;47:406‐420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liao JK, Laufs U. Pleiotropic effects of statins. Annu Rev Pharmacol Toxicol 2005;45:89‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kwak B, Mulhaupt F, Myit S, Mach F. Statins as a newly recognized type of immunomodulator. Nat Med 2000;6:1399‐1402. [DOI] [PubMed] [Google Scholar]
- 40. Kalambokis GN, Mouzaki A, Rodi M, Pappas K, Fotopoulos A, Xourgia X, et al. Rifaximin improves systemic hemodynamics and renal function in patients with alcohol‐related cirrhosis and ascites. Clin Gastroenterol Hepatol 2012;10:815‐818. [DOI] [PubMed] [Google Scholar]
- 41. Kalambokis GN, Mouzaki A, Rodi M, Tsianos EV. Rifaximin improves thrombocytopenia in patients with alcoholic cirrhosis in association with reduction of endotoxaemia. Liver Int 2012;32:467‐475. [DOI] [PubMed] [Google Scholar]
- 42. Cervenka I, Agudelo LZ, Ruas JL. Kynurenines: tryptophan’s metabolites in exercise, inflammation, and mental health. Science 2017;357:eaaf9794. [DOI] [PubMed] [Google Scholar]
- 43. Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018;23:716‐724. [DOI] [PubMed] [Google Scholar]
- 44. Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, et al. An endogenous tumour‐promoting ligand of the human aryl hydrocarbon receptor. Nature 2011;478:197‐203. [DOI] [PubMed] [Google Scholar]
- 45. Clària J, Moreau R, Fenaille F, Amorós A, Junot C, Gronbaek H, et al.; CANONIC Study Investigators of the EASL Clif Consortium, Grifols Chair and the European Foundation for the Study of Chronic Liver Failure . Orchestration of tryptophan‐kynurenine pathway, acute decompensation, and acute‐on‐chronic liver failure in cirrhosis. Hepatology 2019;69:1686‐1701. [DOI] [PubMed] [Google Scholar]
- 46. Zinellu A, Sotgia S, Mangoni AA, Sotgiu E, Ena S, Satta AE, et al. Effect of cholesterol lowering treatment on plasma markers of endothelial dysfunction in chronic kidney disease. J Pharm Biomed Anal 2016;129:383‐388. [DOI] [PubMed] [Google Scholar]
- 47. Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Gut microbiota, cirrhosis, and alcohol regulate bile acid metabolism in the gut. Dig Dis 2015;33:338‐345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol 2013;58:949‐955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bass NM, Mullen KD, Sanyal A, Poordad F, Neff G, Leevy CB, et al. Rifaximin treatment in hepatic encephalopathy. N Engl J Med 2010;362:1071‐1081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Table S1‐S6


