Abstract
Background
Early pathogen identification in pulmonary infection is crucial to guide antibacterial therapy and decrease length of hospital stay. We hypothesise that compared to conventional diagnostic methods, a multiplex bacterial polymerase chain reaction assay has a higher diagnostic yield in bronchoalveolar lavage (BAL) fluid and improved clinical outcomes in patients with suspicion of pulmonary infection.
Methods
A prospective, monocentric, quasi-experimental, observational study was carried out. Unselected patients with suspected pulmonary infection who underwent bronchoscopy with BAL were included in the study over a period of 1 year. In addition to conventional diagnostic methods, a multiplex PCR bacterial assay was performed in BAL on a 2 week on: 1 week off pre-determined schedule. No therapeutic recommendations were provided to the treating physician.
Results
605 cases were included, 54% of whom were immunosuppressed. Conventional diagnostic methods detected 56% of the bacteria evidenced by PCR. PCR failed to detect bacteria in 4% of the cases with a positive conventional diagnostic result. After bronchoscopy, 42% of the patients received antibacterial therapy for pulmonary infection for a median of 12 antibiotic days. There was no statistically significant difference in length of hospital stay (median 8 versus 8; p=0.839), antibiotic exposure (median 11 versus 14; p=0.362) or number of antibiotics prescribed (median 2 versus 2; p=0.595) between the two groups.
Conclusions
A multiplex bacterial PCR detected more bacteria in BAL fluid than conventional diagnostic methods. However, without a specific antibiotic stewardship approach and a clear understanding of the clinical implications of a positive or negative PCR result, the PCR results did not influence clinical outcomes.
Short abstract
A multiplex bacterial PCR detects more bacteria in BAL fluid than conventional diagnostic methods; however, without a specific antibiotic stewardship approach, PCR results do not influence clinical outcomes https://bit.ly/3L0nd0U
Introduction
Pulmonary infections are a leading cause of morbidity and mortality worldwide [1]. More than half of deaths from pulmonary infection are attributable to bacterial infections [2]. To reduce the burden and mortality from bacterial pulmonary infection, a timely initiation of adequate antibacterial therapy is required [3–5]. Moreover, early microorganism identification may support antibacterial stewardship and thus reduce costs, reduce complications associated with antibiotic usage and reduce antibacterial resistance [3, 4].
The fast and accurate diagnosis of a pathogen in patients with suspected pulmonary infection remains challenging. Current diagnostic methods fail to establish an etiological diagnosis for >50% of patients [6]. In addition, culture-based methods that are considered the “reference standard” for identifying bacterial microorganisms require 24 to 72 h for a diagnosis and antimicrobial susceptibility testing [7].
The reported range of microbial detection by PCR compared to conventional methods varies from 43% to 89% [8–10]. While some studies have proven a higher identification rate with molecular diagnostics than culture-based microbiological methods, others have shown less promising results [11].
We hypothesise that a multiplex bacterial PCR assay (Curetis Unyvero P50 assay) in the bronchoalveolar lavage (BAL) fluid of patients with suspicion of pulmonary infection has a higher diagnostic yield and can improve clinical outcomes compared to conventional diagnostic methods.
Methods
This was a prospective, monocentric, quasi-experimental, observational study conducted in accordance with the amended Declaration of Helsinki at the University Hospital Basel, Switzerland. The Ethikkommission Beider Basel approved the study (EKBB 120/10) and the subjects provided written informed consent. The primary outcomes of interest were: 1) the diagnostic yield of multiplex bacterial PCR in the BAL compared to conventional microbiology; and 2) clinical outcomes (length of hospital stay, antibiotic exposure and number of antibiotics prescribed) in the group of patients with PCR results compared to the group in which treatment decisions were taken without PCR results. Secondary outcomes were differences across subgroups (immunocompromised versus immunocompetent, with and without consolidation) and the diagnostic performance according to different standard references (clinical diagnosis of bacterial infection, use of antibiotics).
Patients
All patients older than 18 years of age who underwent bronchoscopy with BAL because of suspected pulmonary infection were considered for inclusion in the study over a period of 12 months (figure 1). Patients readmitted with a new episode of suspected pulmonary infection within 14 days of the original infection or patients with an unstable psychiatric or psychological condition were excluded. A randomised list was generated for the use of the PCR device for routine diagnostic on a 2 week on: 1 week off pre-determined schedule.
FIGURE 1.
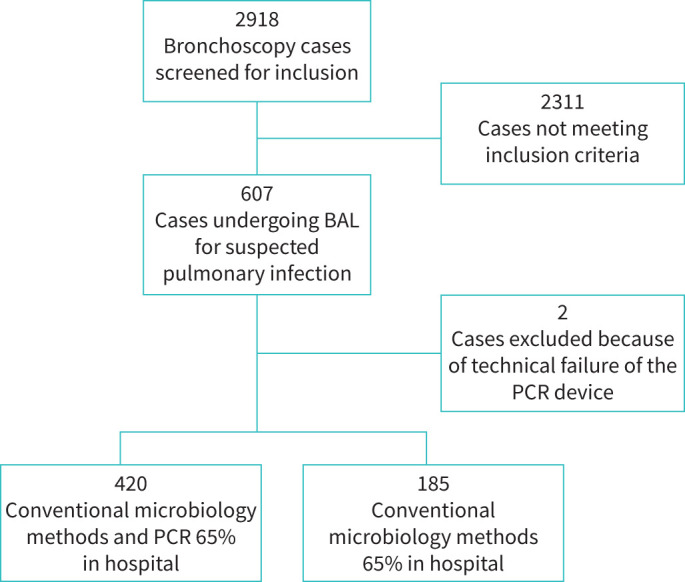
Flow chart depicting study design including 605 cases with suspected pulmonary infection. BAL: bronchoalveolar lavage.
Diagnostic workup
Flexible bronchoscopy with BAL was performed under conscious sedation using hydrocodone and disoprivan. Pyrogen-free, sterile, 0.9% NaCl solution was inserted three times through the working channel of the bronchoscope into the area corresponding to the infiltrate observed in radiological imaging, according to standard guidelines [12]. A radiologist, a board-certified pulmonologist and a non-certified pulmonology fellow reviewed the radiological images before the bronchoscopy. If no abnormality was detected, BAL was performed in the middle lobe or lingula. BAL fluid was recovered by suction.
Conventional methods
Appropriate stains and cultures for bacteria, mycobacteria, fungi and Pneumocystis jirovecii were performed. Positive bacterial cultures were counted as the number of colony-forming units per millilitre, and identification and susceptibility tests were performed according to standard methods [13]. One microlitre of vortexed BAL fluid was added to culture plates including Columbia sheep blood agar, colistin nalidixic acid blood agar for Gram-positive bacteria, Haemophilus chocolate agar and MacConkey for Gram-negatives. These plates were incubated for 2 days at 36°C. The microbiological diagnostic workup also included a search for Legionella pneumophila, Chlamydia pneumoniae and Mycoplasma pneumoniae, by PCR, culture or immunofluorescence [14, 15], and 18 viruses, i.e. adenovirus, influenza A/B H1–H3, parainfluenza 1–4, RSV A/B, rhinovirus/enterovirus, coronavirus NL63/OC43/229E/HKU1, bocavirus and metapneumovirus (respiratory pathogen panel NxTAG-RPP; Luminex, MV's-Hertogenbosch, The Netherlands). Bacterial infection was diagnosed in cases with positive bacteriology results in the BAL fluid, i.e. a culture yielding a single pathogenic bacterial microorganism at the minimum concentration of 103 cfu·mL−1 or any microorganism excluding mouth flora above the minimum concentration of 104 cfu·mL−1; or the identification of Legionella spp., C. pneumoniae or M. pneumoniae regardless of colony counts.
Curetis unyvero P50 assay
The molecular diagnostic assay, a multiplex PCR-based assay (Curetis Unyvero – P50 pneumonia cartridge; Curetis AG, Holzgerlingen, Germany – obtained commercially) for rapid detection of bacteria and antibiotic resistance genes, was performed on un-centrifuged BAL fluid specimens according to the manufacturer's specifications on a pre-determined 2 weeks on: 1 week off schedule. The assay detects 16 bacteria and one fungus and simultaneously targets different antimicrobial classes by analysing 23 genetic resistance markers (table 1). The process from sample preparation, DNA extraction, purification and amplification to specific detection to obtain diagnostic information is fully automated and requires ∼4 h. Operators of the assay and all other microbiological methods were unaware of the results and did not participate in any data analysis.
TABLE 1.
In-panel microorganisms and resistance genes detected by the Curetis Unyvero - P50 assay
| Gram-positive bacteria | Gram-negative bacteria | Fungi | Resistance genes | Resistance against |
| Staphylococcus aureus | Acinetobacter baumannii | Pneumocystis jirovecii | ctx-M | 3rd generation cephalosporins |
| Streptococcus mitis group | Chlamydophila pneumoniae | ermA | Macrolides/lincosamides | |
| Escherichia coli | ermB | Macrolides/lincosamides | ||
| Enterobacter spp. | ermC | Macrolides/lincosamides | ||
| Haemophilus influenzae | Int1 | MDR | ||
| Klebsiella pneumoniae | kpc | Carbapenems | ||
| Klebsiella oxytoca | mecA | Oxacillin | ||
| Legionella pneumophila | msrA | Macrolide | ||
| Moraxella catarrhalis | mefA | Macrolide | ||
| Morganella morganii | oxa-51 | Carbapenems | ||
| Pseudomonas aeruginosa | shv | Penicillin | ||
| Proteus spp. | sul1 | Sulfonamide | ||
| Serratia marcescens | dha | 3rd generation cephalosporins | ||
| Stenotrophomonas maltophilia | tem-shv | 3rd generation cephalosporins | ||
| shv | Penicillin | |||
| tem1 | Penicillin | |||
| gyrA832_Pseu | Fluoroquinolones | |||
| gyrA833_Pseu | Fluoroquinolones | |||
| gyrA872_Pseu | Fluoroquinolones | |||
| gyrA873_Pseu | Fluoroquinolones | |||
| gyrA83_Ecoli | Fluoroquinolones | |||
| gyrA87_Ecoli | Fluoroquinolones | |||
| parCPseu | Fluoroquinolones |
MDR: multidrug resistant.
Diagnostic criteria and data collection
Demographic data were collected at the time of the bronchoscopy. Suspected pulmonary infection was defined by the presence of respiratory symptoms, fever and/or new or progressive radiology findings, as judged by the attending physician. Chest radiographs were performed in most cases, and computer tomography (CT) scans were obtained, as indicated by the treating physicians. Clinical diagnoses were determined from physician notes, hospital discharge summaries, laboratory studies and radiological examinations, as well as pathology reports and clinical course by two pulmonary physicians. Clinically defined bacterial infection was diagnosed in cases with positive bacteriology results in the BAL fluid or histological specimens and/or a clinical, laboratory and radiological presentation highly suggestive of bacterial infection (i.e. fever, new infiltrate on the chest radiograph accompanied by respiratory symptoms, abnormal breath sounds on auscultation, leukocytosis or leukopenia; increased C-reactive protein and/or procalcitonin). Possible bacterial infection was characterised as a non-suggestive clinical presentation with a proven bacterial pathogen. No bacterial infection was considered to be present when an alternative cause for pulmonary symptoms and/or infiltrates was established (i.e. nonspecific interstitial lung disease, organising pneumonia, bronchiolitis obliterans, graft-versus-host disease, respiratory bronchiolitis, diffuse alveolar haemorrhage, diffuse alveolar damage, invasive pulmonary aspergillosis, pulmonary oedema, acute bronchitis, granuloma, Wegener granulomatosis, malignant pulmonary involvement, or fungal or viral infections). Individual definitions for each of these diagnostic entities were standardised as previously described [13]. The use of antimicrobial therapy was followed for up to 30 days after bronchoscopy, or until the patient was discharged from the hospital. For patients treated in outpatient clinics and after discharge from hospital, antimicrobial therapy was recorded according to the recommendations given at discharge. Multiplex bacterial PCR assay results were provided to the physician who referred the patient for bronchoscopy (oncologist, haematologist, etc.) on the day of the bronchoscopy, between 4 and 24 h after the procedure, through the computerised information system used in the clinical routine of the institution. This information system includes an automated notification indicator for added examination results and is the same one used to provide results of the conventional diagnostic examinations. All members of the attending team, including the pulmonary consultants, had access to the PCR results. However, no specific recommendations regarding antibacterial stewardship, i.e. tailored advice to discontinue or de-escalate antibiotics, were provided with the conventional microbiology or multiplex PCR results.
Statistical analysis
Descriptive analyses performed to elucidate the basic characteristics of the patients were presented as follows: numerical results were expressed as mean±SD or median (interquartile range) unless otherwise stated. When looking at the association between previous antimicrobial therapy, and radiographic findings such as consolidation and cavities, the Chi-square test or Fisher's exact test, as appropriate, was used.
The multiplex PCR diagnostic performance was ascertained by counting the number of pathogens detected and the number of positive cases and comparing to conventional methods. When comparing the multiplex PCR assay with conventional methods, we focused on in-panel pathogens, i.e. pathogens represented on the multiplex bacterial PCR assay. The sensitivity and specificity of the multiplex PCR were assessed and compared to the conventional methods (reference standard). In addition, its diagnostic performance was also assessed by considering the usage of antibacterial therapy and clinically defined bacterial infection as reference standards. We explored the diagnostic performance of the multiplex assay in specific sub-populations, e.g. in immunocompromised patients and in cases with radiological findings such as consolidations and cavities. In these sub-populations, a Chi-square test was performed and using conventional methods as a reference standard, the specificity and sensitivity of the multiplex PCR to identify a bacterial pathogen were calculated.
Length of hospital stay, number of antibiotics, length of antibiotic use and the comparison of all other continuously non-normally distributed variables describing the basic characteristics were evaluated using the non-parametric Mann–Whitney U-test or Kruskal–Wallis test, as appropriate.
All tests were two-tailed; a p-value of <0.05 was considered significant. Data analysis was conducted according to the Statistical Package for Social Sciences (SPSS, version 22 for Windows) program.
Results
Patient characteristics
Of the 2918 bronchoscopy cases screened for inclusion, 2311 did not meet the inclusion criteria, as the bronchoscopies were not for suspicion of lung infection. A total of 605 BAL fluid samples from 470 patients were included in the study. The mean age of the study population was 62±15.5 years and 58% (272 out of 470) were male (table 2). Radiography was performed in 80% (481 out of 605) of the cases within 30 days of admission and/or bronchoscopy, but the radiographs captured within 48 h of the bronchoscopy were analysed. In 58% (348 out of 605) of the cases a radiological examination (CT scan/chest radiograph/both) was done within 48 h of the bronchoscopy. Pathological radiological findings were observed in 82% (284 out of 348) of the cases and were more often seen bilaterally (58%; 164 out of 284). Consolidation was detected in 79% (223 out of 284) of the cases (table 2).
TABLE 2.
Demographics of patients undergoing bronchoalveolar lavage for suspicion of pulmonary infection
| Patient characteristics | n (%), mean±sd | Multiplex PCR | Conventional culture | p-value |
| Age years | 62±15.5 | 60±15.9 | 60±14.6 | 0.82 |
| Female | 198/470 (42) | 140/322 (43) | 58/148 (39) | 0.42 |
| Weight kg | 68±17.96 | 70±18.1 | 69±17.7 | 0.83 |
| Height cm | 169±9.4 | 169±9.6 | 169±9.0 | 0.82 |
| Setting | 0.78 | |||
| Ambulatory | 213/605 (35) | 147/417 (35) | 64/188 (34) | |
| Hospitalised | 392/605 (65) | 270/417 (65) | 124/188 (66) | |
| Smoking status | 0.79 | |||
| Current smokers | 126/605 (21) | 86/403 (21) | 40/183 (22) | |
| Former smokers | 237/605 (39) | 160/403 (40) | 77/183 (42) | |
| Pack-years, median (IQR) | 30 (15–50) | 30 (15–50) | 30 (18.75–50) | 0.78 |
| Respiratory symptoms before bronchoscopy: | ||||
| Cough | 316/399(79) | 215/370 (58) | 101/167 (60) | 0.64 |
| Sputum | 239/537(45) | 167/370 (45) | 72/167 (43) | 0.71 |
| Dyspnoea | 226/399(57) | 156/370 (42) | 70/167 (42) | 1.00 |
| Thoracic pain | 66/399(17) | 50/370 (14) | 16/167 (10) | 0.26 |
| Pathological findings on CT and/or radiograph | 284/348 (82) | 194/235 (83) | 90/113 (80) | 0.56 |
| Consolidation | 223 (79) | 154 (79) | 69 (77) | 0.47 |
| Pleural effusion | 101 (36) | 76 (39) | 25 (28) | 0.06 |
| Interstitial pattern | 63 (22) | 40 (21) | 23 (26) | 0.46 |
| Cavities | 13 (5) | 10 (5) | 3 (3) | 0.56 |
| Laboratory findings values before bronchoscopy | ||||
| C-reactive protein mg·L−1 | 20±87 | 68±93 | 51±70 | 0.27 |
| Procalcitonin ng mL−1 | 0.08±1.22 | 0.4±1.4 | 0.2±0.6 | 0.37 |
| White blood cells (×109) | 7.94±5.8 | 9.0±5.9 | 8.6±5.5 | 0.45 |
| Neutrophil leukocyte count (×109) | 5.28±4.6 | 6.5±4.9 | 6.1±3.7 | 0.95 |
| Haemoglobin g·L−1 | 120±24 | 118±24 | 120±23 | 0.46 |
| Glucose mmol·L−1 | 5.9±2.5 | 6.5±2.2 | 7.0±3.0 | 0.18 |
| BUN mmol·L−1 | 6.3±5.44 | 7.8±5.6 | 7.8±5.2 | 0.46 |
| Creatinine μmol L−1 | 76±63 | 94±62 | 98±68 | 0.24 |
| Final diagnosis | ||||
| Bacterial infection | 188 | 54/144 (37.5) | 45/188 (24) | 0.00 |
| Probably bacterial infection | 183 | 21/116 (18) | 16/183 (9) | |
| No bacterial infection | 234 | 32/160 (20) | 22/234 (9) | |
IQR: interquartile range; CT: computed tomography; BUN: blood urea nitrogen.
Before bronchoscopy, 60% (365 out of 605) of the cases had already received some form of antimicrobial therapy. More than half of the study population were immunosuppressed (54%; 329 out of 605) with solid organ transplantation (116 out of 329; 35%), of which 92 out of 116 (79%) had lung transplantation and three out of 116 (3%) had lung and liver transplantation, being the most prevalent (supplementary table S1).
Comorbidities included congestive heart failure, cardiovascular diseases, solid tumour and COPD (supplementary table S1). There was no significant difference in the patient comorbidities between the groups. Less than half the cases (47%; 284 out of 605) reported having a previous vaccination. Of these, 64% (182 out of 284) were vaccinated against influenza, 1% (3 out of 284) against pneumococcus and 33% (94 out of 284) against both.
Viral infection was analysed in 89% (536 out of 605) of the cases. Diagnostic workup of the BAL yielded a viral pathogen in 152 out of 536 (28%) of the cases. The most common viruses were rhinovirus/enterovirus, cytomegalovirus and human metapneumovirus. In 46 cases, a bacterial (from PCR and/or conventional microbiological methods) and a viral agent were identified in the BAL.
Almost two-thirds of the cases (65%; 392 out of 605) were hospitalised for a median of 8 days (IQR 4–15). Of the hospitalised population, 18% (70 out of 392) were admitted to the intensive care unit for a median of 3.5 days (IQR 1–16). In-hospital 30-day mortality rate was 5% (30 out of 605).
PCR diagnostic performance
Of the 605 cases, 420 BAL fluid samples were examined with both conventional methods and a multiplex bacterial PCR assay, and 185 BAL fluid samples were examined with only conventional methods (figure 1). Pathogen detection rate with the multiplex PCR was 82% (134 out of 163 bacteria detected) versus 56% (91 out of 163 bacteria detected) with conventional methods focusing on in-panel pathogens. Haemophilus influenzae 20% (32 out of 163) followed by the Streptococcus mitis group 19% (31 out of 163) and Staphylococcus aureus 15% (24 out of 163) were detected most often (table 3). The multiplex PCR assay detected H. influenzae, S. mitis group, Moraxella catarrhalis and Acinetobacter baumannii more often than conventional culture (table 3).
TABLE 3.
The number of times the microorganisms present in the Curetis Unyvero P50 assay were detected by conventional methods and the multiplex PCR assay
|
Conventional method +
PCR + |
Conventional method –
PCR + |
Conventional method +
PCR – |
|
| Acinetobacter baumannii | 2 | 5 | 0 |
| Chlamydophila pneumoniae | 0 | 1 | 0 |
| Escherichia coli | 5 | 3 | 5 |
| Haemophilus influenzae | 8 | 20 | 4 |
| Klebsiella oxytoca | 1 | 1 | 0 |
| Klebsiella pneumoniae | 2 | 1 | 2 |
| Legionella pneumophila | 0 | 1 | 0 |
| Moraxella catarrhalis | 2 | 6 | 1 |
| Morganella morganii | 0 | 2 | 0 |
| Proteus spp. | 0 | 1 | 0 |
| Pseudomonas aeruginosa | 13 | 5 | 2 |
| Serratia marcescens | 4 | 1 | 0 |
| Staphylococcus aureus | 11 | 6 | 7 |
| Stenotrophomonas maltophilia | 6 | 3 | 1 |
| Streptococcus mitis/Streptococcus pneumoniae | 8 | 16 | 7 |
| Total | 62 | 72 | 29 |
Conversely, the multiplex PCR assay failed to detect bacteria in 4% (15 out of 420) of the cases where a positive conventional culture result was obtained. Conventional culture detected seven additional pathogens in BAL, which were not available on the multiplex PCR assay (supplementary table S2). Four additional pathogens were detected by culture in the 2-week on group in contrast to the 1-week off group (supplementary table S3). Antibiotic resistance genes and the results of conventional culture antibiogram were evidenced (supplementary table S4).
There was no association between a positive or negative result and the length of time between admission and the bronchoscopy (PCR: p=0.403; culture: p=1.000). In the patients hospitalised for up to 2 days before the bronchoscopy, H. influenza [12], S. mitis group [11] and S. aureus [8] were the most prevalent pathogens. In the patients hospitalised >3 days before the bronchoscopy, Pseudomonas aeruginosa [6], Escherichia coli [5] and Stenotrophomonas maltophilia [5] were the most prevalent pathogens (supplementary table S5).
Immunocompetent versus immunocompromised patients
There was a significant association between immune status and the multiplex PCR positive or negative result (p=0.042) and the culture (all pathogen) positive or negative result (p=0.029). Immunocompetent cases had more positive results (multiplex PCR 56 out of 106, 53%; culture 63 out of 115, 55%) compared to immunocompromised cases (multiplex PCR 50 out of 106, 47%; culture 52 out of 115, 45%). This association did not exist between culture results (in-panel pathogens only) and the immune status (p=0.34). The number of pathogens were more in the immunocompetent group compared to the immunocompromised group (supplementary table S6a and b). More immunocompromised patients (102 out of 310, 33%) had a positive viral infection than immunocompetent patients (50 out of 225, 22%) (p=0.007). Rhinovirus/enterovirus (11%), followed by cytomegalovirus (6%), human metapneumovirus (4%) and herpes simplex virus (4%) were the most prevalent viruses in the immunocompromised patients. In immunocompetent patients, rhinovirus/enterovirus (8.5%), influenza A (4%) and cytomegalovirus (2%) were the most prevalent viruses.
Consolidation and cavities versus no evidence of consolidation or cavities
In cases where no consolidation was evident, PCR identified 24 out of 82 (29%) cases as bacterial positive and 58 out of 82 (71%) as bacterial negative. Culture, focusing on in-panel pathogens, identified 16 out of 125 (13%) cases as bacterial positive and 109 out of 125 (87%) as bacterial negative. Looking at culture with all pathogens, 22 out of 125 (18%) were positive and 103 out of 125 (82%) were bacterial negative.
Using consolidation as reference standard, the multiplex PCR had 26.3% sensitivity and 71% specificity compared to conventional culture (in-panel pathogens), which had a sensitivity of 14% and a specificity of 87%.
In cases where cavities were present, multiplex PCR identified three out of 10 (30%) as bacterial positive. Culture using in-panel pathogens detected two out of 13 (15%) cases as bacterial positive, and culture using all pathogens detected three out of 13 (23%) cases.
Using conventional microbiological methods as reference standard
Overall, multiplex bacterial PCR assay showed a sensitivity of 81.3% and a specificity of 86.9% when using conventional microbiological methods as a reference standard. The positive predictive value was 57.5% and the negative predictive value was 95.2%. The positive likelihood ratio was 6.2 and the negative likelihood ratio 0.22. When using conventional methods including all pathogens detected, i.e. not only in-panel, the multiplex bacterial PCR assay had a sensitivity of 73% and a specificity of 86%. The positive predictive value was 57% and the negative predictive value was 93%. The positive likelihood ratio was 5.2 and the negative likelihood ratio 0.3.
Using clinically defined bacterial infection as a reference standard
Clinically defined bacterial infection was diagnosed in 188 (31%) of the cases whereas in 234 (39%) patients no bacterial infection was considered to be present. In 30% of the patients, a possible bacterial infection was diagnosed on clinical grounds. Multiplex PCR had a sensitivity of 37.5% and specificity of 80% to assess the presence of clinically defined bacterial infection. The positive likelihood ratio was 1.88 and negative likelihood ratio was 0.78. Culture (using in-panel pathogens) had a sensitivity of 31% and a specificity of 86%, respectively. The positive likelihood ratio was 2.21 and negative likelihood ratio 0.80.
PCR impact on antimicrobial therapy
There was no association between previous antimicrobial therapy and either the multiplex bacterial PCR assay (p=0.888) or conventional method positivity (focusing on in-panel pathogens; p=0.100). When considering the relative increase in yield among multiplex PCR-tested patients, we found no association with previous antimicrobial therapy (p=0.991) or with antibiotic therapy at bronchoscopy (p=0.915).
Antibiotic treatment was administered to 313 out of 594 (53%) of the cases at bronchoscopy. Of these, antibiotic therapy was stopped in 106 (34%) cases within 24 h of the bronchoscopy. Of the 281 cases who did not have antibiotics at bronchoscopy, 74 (26%) cases started antibiotic treatment. Of the 74 cases, 57 (77%) started antibiotic treatment within 24 h of the bronchoscopy; five cases started treatment within 48 h and four within 96 h. After bronchoscopy, 42% (254 out of 605) of the patients received antibacterial therapy for suspected pulmonary infection, with a mean of 2.0±1.1 antibacterial agents per case used for a median of 12 (IQR 7–18) antibiotic days. In the cases where antibiotic therapy was started, the main pathogen found was H. influenza, S. mitis group and S. aureus. In the cases where antibiotic therapy was stopped, the main pathogen was S. mitis group, S. maltophilia and P. aeruginosa.
PCR impact on clinical outcomes
There was no statistically significant difference in length of hospital stay (median 8 (IQR 3–14) versus 8 (IQR 4–15); p=0.839), antibiotic days (median 11 (IQR 7–17) versus 14 (IQR 7–20); p=0.362) or number of antibiotics prescribed (median 2 (IQR 1–2) versus 2 (IQR 1–2); p=0.595), between cases analysed using the multiplex PCR assay or those analysed using conventional methods.
Cases with a conventional method positive result had the same length of hospital stay (median 9.5 (IQR 6–21.5) versus 8 (IQR 4–15); p=0.168), antibiotic days (median 14 (IQR 8–20) versus 8 (IQR 7–18); p=0.362) or number of antibiotics (median 2 (IQR 1–3) versus 2 (IQR 1–2); p=0.081) as patients with a negative conventional method result. The same was seen in multiplex PCR assay positive samples compared to negative samples.
Discussion
In this study, we evaluated the diagnostic performance and clinical impact of a multiplex PCR bacterial assay compared to conventional diagnostic methods in BAL fluid from patients with suspected pulmonary infection. The study took advantage of the availability of the PCR device for use in the clinical routine on a 2 week on: 1 week off pre-determined schedule. Attending physicians were provided with the results of the molecular testing of the BAL fluid in unselected cases undergoing bronchoscopy in a quasi-experimental design. Multiplex PCR detected more bacteria than conventional culture, but failed to influence antibiotic usage and clinical outcomes, thus suggesting that a specific antibiotic stewardship action plan as well as more in-depth understanding of the PCR results may be necessary to support antibiotic de-escalation based on fast molecular diagnostic results.
Previous studies suggest that multiplex PCR bacterial assays are faster than conventional culture and have a higher pathogen detection rate [16, 17]. Our study confirmed the better performance of molecular diagnostics in patients undergoing BAL for suspicion of pulmonary infection. The most commonly detected microorganisms in our study were H. influenzae and the S. mitis group, which included Streptococcus pneumoniae. These results are in line with former reports [18, 19].
Immunocompetent patients had more positive results and more bacterial pathogens than the immunocompromised patients. Conversely, more immunocompromised patients were positive for viral infection than immunocompetent patients. This could be because immunocompromised patients are often treated with antibiotics as soon as an infection is suspected. Indeed, of the patients who had received previous antimicrobial treatment, 58% were immunocompromised. Of the immunocompromised patients, 59% were receiving antimicrobial treatment at the time of the bronchoscopy. However, in the immunocompromised population, we found no association between either previous antibiotic therapy or antibiotic therapy on the day of the bronchoscopy and whether the conventional culture or multiplex PCR were positive or negative.
Conventional diagnostic methods detected seven additional pathogens (6 Gram-negative and 1 Gram-positive) in BAL fluid, which were not available on the multiplex PCR assay. While it is obvious that only pathogens included in a panel can be detected by a certain test, this finding stresses the fact that, so far, molecular diagnostic tests may have to be seen as complementary rather than as a stand-alone tool.
The evaluation of the diagnostic performance of a test is usually based on a reference standard [8–10, 20, 21]. Consensus is that the reference standard for the etiological diagnosis of respiratory infections is far from reliable. Indeed, even in an experimental setting, less than half of the patients with pneumonia have a pathogenic agent identified in the course of their disease [22]. Nevertheless, using radiographic evidence of a consolidation as a reference standard for bacterial infection, we found that the multiplex PCR had a better sensitivity, but lower specificity than conventional diagnostic methods. This observation is in alignment with other studies [8–10, 20, 21]. The observed lower specificity of the bacterial multiplex PCR assay could be attributed to potential amplification of dead, clinically non-relevant pathogens [23]. In clinical practice a pathological finding in the chest radiograph could suggest more severe bacterial pneumonia and could be used to help guide clinical decisions for the therapy [24] and hospitalisation. A negative chest radiograph is, however, not indicative of no infection [25].
Diagnosing a bacterial infection involves many factors, not least of which is the microbiological result. It is, therefore, unsurprising that the multiplex PCR and conventional culture had similar sensitivities and specificities when using the bacterial infection diagnosis as a reference standard.
In our cohort, in 60% of the cases, antimicrobial therapy was already administered before sampling. However, we found no association between previous antimicrobial therapy and findings in either the multiplex bacterial PCR or conventional culture. Data in this regard are contradictory. Some studies show similar findings [16, 26], whereas others report that previous antibiotic therapy decreases diagnostic yield in PCR and especially in conventional culture [17, 23, 26–28]. Characteristics of the patient population receiving antibacterial therapy before bronchoscopy vary across studies. They are governed by the previous duration of antibacterial therapy before sampling, and the indications for bronchoscopic sampling itself, such as worsening of disease, treatment failure, or analysing BAL fluid after the start of effective empiric antibacterial therapy or prophylactic antimicrobial treatment in immunocompromised patients. It is therefore important to exercise caution in drawing conclusions about the diagnostic effectiveness of diverse diagnostic methods.
We were particularly keen on evaluating whether PCR results, available as soon as 4 h after the bronchoscopy, would significantly impact clinical outcomes. However, we found a similar duration of antibacterial therapy, antibiotic exposure and hospital stay after bronchoscopy for cases where multiplex PCR assay results were available and those in whom only conventional methods were applied. This result remained true when comparing immunocompromised and immunocompetent patients, and patients with or without consolidation. Taking into account that the PCR results were available to the treating physician with no recommendations about management, our observation strengthens previously reported inertia in antibacterial stewardship in clinical practice [29]. It is also striking that antibacterial therapy was still administered in up to 40% of cases even though both multiplex PCR assay and the conventional method results were negative. These findings are in line with previously reported data from randomised controlled trials, which show that treatment decision-making in clinical practice is based on several factors including possible infiltrate on the chest radiograph, severity of illness, culture, abnormal inflammatory markers, fever, age, patient expectations and others [24].
Despite the well-characterised patient population including BAL and the quasi-experimental design, our study had several limitations. Firstly, this was a monocentric study performed in a tertiary hospital, and our cohort included a high proportion of patients requiring hospitalisation (65%) or heavily immunocompromised patients (54%). Therefore, the results might not be generalisable to patients presenting with uncomplicated community-acquired pneumonia or other centres with a different or less vulnerable patient population. Secondly, with the lack of a robust reference standard to diagnose bacterial infection, we have used several proxies to calculate the test performance of the molecular diagnostic assay. In addition to the comparison to the conventional microbiological methods, we used immune status and the presence or absence of radiographic consolidation.
Finally, in this study, we evaluated the effect of providing results of a multiplex bacterial PCR in the BAL to the treating physician without additional advice regarding antibiotic discontinuation or de-escalation, mimicking a real-life approach. Thus, the design explored the effect of a specific component of the antibiotic stewardship strategy. Antibiotic stewardship remains a challenging chapter, particularly in a vulnerable population such as immunocompromised patients. We believe that it is fair to assume that the impact generated by a molecular assay in the BAL combined with a targeted recommendation regarding antibiotic therapy and/or the presence of antibiotic resistance genes might be more pronounced. Thus, a further study focusing on the combined impact of the molecular assay and a tailored recommendation for antibiotic stewardship should be undertaken. Nevertheless, our study fills a gap in the literature in informing about the potential role of molecular diagnostics in analysing the BAL of patients with suspected pulmonary infection – and the potential need for its implementation.
In summary, our study corroborates the notion that a multiplex PCR panel detects more microorganisms in BAL fluid than conventional diagnostic methods, therefore potentially allowing faster and targeted antibiotic therapy. However, it also strongly emphasises the requirement for a broader approach to antibiotic stewardship than a single test in informing clinicians to endorse confident and timely decisions in a hospitalised setting of high-risk patients. A proper randomised interventional study would be required to explore the potential role of a fast diagnostic PCR panel in analysing BAL fluid and in treatment decision-making.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00595-2021.SUPPLEMENT (359.3KB, pdf)
Footnotes
Provenance: Submitted article, peer reviewed.
Author contributions: Data collection, accuracy of data, contribution to discussion of results, statistical analysis, writing of the manuscript, finalisation of the manuscript and approval of the submitted article: A. Salina, D.M. Schumann, L. Franchetti, K. Jahn, K. Purkabiri, R. Müller, W. Strobel and N. Khanna. Conception of the research project, contribution to clinical work, integrity and accuracy of data, preparation and approval of the submitted article: M. Tamm and D. Stolz.
Ethics approval and consent to participate: The Ethikkommission Beider Basel approved the study (EKBB 120/10) and the subjects provided written informed consent.
Consent for publication: All subjects provided written informed consent.
Availability of data and materials: The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Conflict of interest: D. Stolz has received grants or contracts from AstraZeneca AG, Curetis AG and Boston Scientific. D. Stolz and M. Tamm have received consulting fees from AstraZeneca AG, Sanofi, MSD and Novartis. D. Stolz and M. Tamm have received payment or honoraria for lectures, presentations, speakers bureaus or educational events from AstraZeneca AG, Novartis AG, GSK AG, Roche AG, Schwabe Pharma AG, Vifor AG, Chiesi AG, MSD and Sanofi. D. Stolz has participated on a data safety monitoring board or advisory board of CSL Behring. The remaining authors have no conflicts of interest.
References
- 1.Gibson J, Loddenkemper R, Sibille Y, et al. The Acute Respiratory Infections Atlas. New York, World Lung Foundation, 2019. [Google Scholar]
- 2.GBD2016 Lower Respiratory Infections Collaborators . Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis 2018; 18: 1191–1210. doi: 10.1016/S1473-3099(18)30310-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houck PM, Bratzler DW, Nsa W, et al. Timing of antibiotic administration and outcomes for Medicare patients hospitalized with community-acquired pneumonia. Arch Intern Med 2004; 164: 637–644. doi: 10.1001/archinte.164.6.637 [DOI] [PubMed] [Google Scholar]
- 4.Houck PM, Bratzler DW, Nsa W, et al. Antibiotic administration in community-acquired pneumonia. Chest 2004; 126: 320–321. doi: 10.1378/chest.126.1.320 [DOI] [PubMed] [Google Scholar]
- 5.Claeys KC, Zasowski EJ, Trinh TD, et al. Antimicrobial stewardship opportunities in critically ill patients with gram-negative lower respiratory tract infections: a multicenter cross-sectional analysis. Infect Dis Ther 2018; 7: 135–146. doi: 10.1007/s40121-017-0179-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Musher DM, Abers MS, Bartlett JG. Evolving understanding of the causes of pneumonia in adults, with special attention to the role of pneumococcus. Clin Infect Dis 2017; 65: 1736–1744. doi: 10.1093/cid/cix549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caliendo AM, Gilbert DN, Ginocchio CC, et al. Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis 2013; 57: Suppl. 3, S139–S170. doi: 10.1093/cid/cit578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mustafa MI, Al-Marzooq F, How SH, et al. The use of multiplex real-time PCR improves the detection of the bacterial etiology of community acquired pneumonia. Trop Biomed 2011; 28: 531–544. [PubMed] [Google Scholar]
- 9.Affolter K, Schumann DM, Tamm M, et al. Multiplex PCR on the bronchoalveolar lavage fluid of immunocompromised patients. Chest 2018; 154: 722–725. doi: 10.1016/j.chest.2018.06.018 [DOI] [PubMed] [Google Scholar]
- 10.Trotter AJ, Aydin A, Strinden MJ, et al. Recent and emerging technologies for the rapid diagnosis of infection and antimicrobial resistance. Curr Opin Microbiol 2019; 51: 39–45. doi: 10.1016/j.mib.2019.03.001 [DOI] [PubMed] [Google Scholar]
- 11.Kunze N, Moerer O, Steinmetz N, et al. Point-of-care multiplex PCR promises short turnaround times for microbial testing in hospital-acquired pneumonia: an observational pilot study in critical ill patients. Ann Clin Microbiol Antimicrob 2015; 14: 33. doi: 10.1186/s12941-015-0091-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stolz D, Kurer G, Meyer A, et al. Propofol versus combined sedation in flexible bronchoscopy: a randomised non-inferiority trial. Eur Respir J 2009; 34: 1024–1030. doi: 10.1183/09031936.00180808 [DOI] [PubMed] [Google Scholar]
- 13.Stolz D, Stulz A, Müller B, et al. BAL neutrophils, serum procalcitonin, and C-reactive protein to predict bacterial infection in the immunocompromised host. Chest 2007; 132: 504–514. doi: 10.1378/chest.07-0175 [DOI] [PubMed] [Google Scholar]
- 14.Stolz D, Christ-Crain M, Bingisser R, et al. Antibiotic treatment of exacerbations of COPD: a randomized, controlled trial comparing procalcitonin-guidance with standard therapy. Chest 2007; 131: 9–19. doi: 10.1378/chest.06-1500 [DOI] [PubMed] [Google Scholar]
- 15.Buess M, Cathomas G, Halter J, et al. Aspergillus-PCR in bronchoalveolar lavage for detection of invasive pulmonary aspergillosis in immunocompromised patients. BMC Infect Dis 2012; 12: 237. doi: 10.1186/1471-2334-12-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baudel JL, Tankovic J, Dahoumane R, et al. Multiplex PCR performed of bronchoalveolar lavage fluid increases pathogen identification rate in critically ill patients with pneumonia: a pilot study. Ann Intensive Care 2014; 4: 35. doi: 10.1186/s13613-014-0035-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tschiedel E, Goralski A, Steinmann J, et al. Multiplex PCR of bronchoalveolar lavage fluid in children enhances the rate of pathogen detection. BMC Pulm Med 2019; 19: 132. doi: 10.1186/s12890-019-0894-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodhead M, Blasi F, Ewig S, et al. Guidelines for the management of adult lower respiratory tract infections: full version. Clin Microbiol Infect 2011; 17: Suppl. 6, E1–59. doi: 10.1111/j.1469-0691.2011.03602.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ieven M, Coenen S, Loens K, et al. Aetiology of lower respiratory tract infection in adults in primary care: a prospective study in 11 European countries. Clin Microbiol Infect 2018; 24: 1158–1163. doi: 10.1016/j.cmi.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozongwu C, Personne Y, Platt G, et al. The Unyvero P55 ‘sample-in, answer-out’ pneumonia assay: a performance evaluation. Biomol Detect Quantif 2017; 13: 1–6. doi: 10.1016/j.bdq.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulte B, Eickmeyer H, Heininger A, et al. Detection of pneumonia associated pathogens using a prototype multiplexed pneumonia test in hospitalized patients with severe pneumonia. PLoS One 2014; 9: e110566. doi: 10.1371/journal.pone.0110566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain S, Self WH, Wunderink RG, et al. Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med 2015; 373: 415–427. doi: 10.1056/NEJMoa1500245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strålin K, Korsgaard J, Olcén P. Evaluation of a multiplex PCR for bacterial pathogens applied to bronchoalveolar lavage. Eur Respir J 2006; 28: 568–575. doi: 10.1183/09031936.06.00006106 [DOI] [PubMed] [Google Scholar]
- 24.Branche AR, Walsh EE, Jadhav N, et al. Provider decisions to treat respiratory illnesses with antibiotics: insights from a randomized controlled trial. PLoS One 2016; 11: e0152986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basi SK, Marrie TJ, Huang JQ, et al. Patients admitted to hospital with suspected pneumonia and normal chest radiographs: epidemiology, microbiology, and outcomes. Am J Med 2004; 117: 305–311. doi: 10.1016/j.amjmed.2004.03.029 [DOI] [PubMed] [Google Scholar]
- 26.Abdeldaim GM, Strålin K, Korsgaard J, et al. Multiplex quantitative PCR for detection of lower respiratory tract infection and meningitis caused by Streptococcus pneumoniae, Haemophilus influenzae and Neisseria meningitidis. BMC Microbiol 2010; 10: 310. doi: 10.1186/1471-2180-10-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rampini SK, Bloemberg GV, Keller PM, et al. Broad-range 16S rRNA gene polymerase chain reaction for diagnosis of culture-negative bacterial infections. Clin Infect Dis 2011; 53: 1245–1251. doi: 10.1093/cid/cir692 [DOI] [PubMed] [Google Scholar]
- 28.Harris AM, Bramley AM, Jain S, et al. Influence of antibiotics on the detection of bacteria by culture-based and culture-independent diagnostic tests in patients hospitalized with community-acquired pneumonia. Open Forum Infect Dis 2017; 4: ofx014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Houten CB, Cohen A, Engelhard D, et al. Antibiotic misuse in respiratory tract infections in children and adults: a prospective, multicentre study (TAILORED Treatment). Eur J Clin Microbiol Infect Dis 2019; 38: 505–514. doi: 10.1007/s10096-018-03454-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00595-2021.SUPPLEMENT (359.3KB, pdf)


