Abstract
The COrona VIrus Disease 2019 (COVID-19) pandemic is an ongoing global pandemic that has claimed millions of lives till date. Detecting COVID-19 and isolating affected patients at an early stage is crucial to contain its rapid spread. Although accurate, the primary viral test ‘Reverse Transcription Polymerase Chain Reaction’ (RT-PCR) for COVID-19 diagnosis has an elaborate test kit, and the turnaround time is high. This has motivated the research community to develop CXR based automated COVID-19 diagnostic methodologies. However, COVID-19 being a novel disease, there is no annotated large-scale CXR dataset for this particular disease. To address the issue of limited data, we propose to exploit a large-scale CXR dataset collected in the pre-COVID era and train a deep neural network in a self-supervised fashion to extract CXR specific features. Further, we compute attention maps between the global and the local features of the backbone convolutional network while finetuning using a limited COVID-19 CXR dataset. We empirically demonstrate the effectiveness of the proposed method. We provide a thorough ablation study to understand the effect of each proposed component. Finally, we provide visualizations highlighting the critical patches instrumental to the predictive decision made by our model. These saliency maps are not only a stepping stone towards explainable AI but also aids radiologists in localizing the infected area.
Keywords: COVID-19 detection from limited data, AI for COVID-19, Deep learning, Self-supervised learning, Chest radiography, COVID-19 detection using CXR
Graphical abstract
1. Introduction
The COrona VIrus Disease 2019 (COVID-19), epi-centered in Hubei Province of the People’s Republic of China, spread so rapidly across the globe that the World Health Organization (WHO) declared COVID-19 a Public Health Emergency of International Concern on 30 January 2020, and finally a pandemic on 11 March 2020 [1]. It has caused a massive threat to global health with 174,918,667 cases of confirmed coronavirus and 3,782,490 deaths as of 12 June 2021. Once infected with COVID-19, one may experience fever, cough, and respiratory illness. Some may also experience shortness of breath, muscle or body aches, headache, loss of taste or smell, sore throat, and diarrhea [2], [3]. The virus can cause pneumonia or breathing problems in severe cases, leading to multi-organ failure and death [4]. Due to the exponential growth of COVID-19 patients, there is a shortage in supply of diagnostic kits, a limited number of beds in the hospitals to care for critical patients, a dearth of ventilators, scarcity in personal protective equipment (PPE) for healthcare personnel. Despite various preventive measures (such as complete lockdown) adopted by the government of different countries to contain the disease and delay the spread, several developed countries have faced a critical care crisis, and the health system has come to the verge of collapse. It is, therefore, of utmost importance to screen the positive COVID-19 patients accurately for efficient utilization of limited resources. Reverse Transcription Polymerase Chain Reaction (RT-PCR) [5], [6] is the most preferred viral test for COVID-19 detection due to its high sensitivity and specificity. However, the turn-around time of RT-PCR is high.
Consequently, chest radiography such as computerized tomography (CT) scan and X-ray imaging-based detection techniques have emerged as an alternative modality for screening COVID-19 patients. With these modalities, researchers have observed that COVID-19 patients’ lungs exhibit ground-glass opacity and/or mixed ground-glass opacity and mixed consolidation that can separate COVID-19-positive cases from COVID-19-negative cases [7], [8]. In contrast to conventional diagnostic methods, X-ray offers several advantages as it is fast, can simultaneously analyze numerous cases, inexpensive and widely available. It can be very useful in hospitals with limited testing kits and resources.
Deep Machine Learning has revolutionized the field of health care by accurately analyzing, identifying, and classifying patterns in medical images [9]. Artificial neural networks are able to diagnose a variety of illnesses with a high degree of accuracy. The reason for such success is that deep learning techniques do not rely on manual handcrafted features but rather learn features automatically from data itself. This allows the algorithm to be applicable on a broader variety of use cases than traditional machine learning methods and is also faster and more accurate in many cases. Motivated by the remarkable performance of CheXNet in Pneumonia detection from chest X-ray images, artificial intelligence (AI) researchers have put a lot of effort into designing machine learning (ML) algorithms for automated detection of COVID-19 using chest X-rays. However, the biggest challenge lies in the fact that COVID-19 being a novel disease, a limited number of sample images are available for training deep neural networks. Motivated by this, in this work, we propose a novel framework that can be trained using limited labeled data for COVID-19 detection using chest X-rays. In this work, our contributions are as follows.
-
1.
We adopt a self-supervised training methodology to train a CXR feature extractor (a convolutional backbone network) on a large-scale chest X-ray dataset.
-
2.
We design a local–global-attention-based classification network consisting of the pre-trained feature extractor, an attention block, and a classification head.
-
3.
We empirically demonstrate the effectiveness of the proposed framework in the low data regime through extensive experimentation and ablation studies.
-
4.
We present clinically interpretable saliency maps, which are helpful for disease localization and patient triage.
The remainder of this paper is structured as follows: Section 2 provides an overview of related work; Section 3 describes the procedural and methodological stages of the development of this solution; Section 4 evaluates the proposed method and assesses the predictions; finally, Section 5 critically discusses the advantages and the limitations of the proposed framework.
2. Related work
Several deep neural frameworks [10], [11], [12], [13], [14], [15] have been proposed in the past to identify different thoracic diseases such as Pneumonia using chest X-ray (CXR) images and surpassed average radiologist performance. ChestX-ray8 [11] (later extended to constitute ChestX-ray14 dataset), and CheXpert [16] are two large-scale datasets of chest X-rays (CXR) that facilitate the training of deep neural networks (DNN) for automating the interpretation of a wide variety of thoracic diseases. ChexNet [16] is a deep neural network, built using DenseNet-121 [17], for Pneumonia detection using chest X-ray images and it achieved excellent results surpassing average radiologist performance. ChestNet [12] is another deep neural network for thoracic diseases diagnosis using chest radiography images. The authors in [14] propose to learn channel-wise, element-wise, and scale-wise attention (triple attention) simultaneously to classify 14 thoracic diseases using chest radiography. Thorax-Net [15] is an attention regularized deep neural network for the classification of thoracic diseases on chest radiography.
Motivated by this, the research community has examined the possibility of COVID-19 prognosis using CXR.
2.1. Traditional machine learning for COVID-19 detection using CXR
The proposal in [18] leverages an enhanced cuckoo search algorithm to determine the most significant CXR features and train a -nearest neighbor (KNN) classifier to distinguish between COVID-19 positive and negative cases. In this work, features were extracted from X-ray images using standard feature extraction techniques such as Fractional Zernike Moments (FrZMs), Wavelet Transform (WT), Gabor Wavelet Transform (GW), and Gray Level Co-Occurrence Matrix (GLCM), followed by a fractional order cuckoo search method where the levy flight distribution was replaced with better suited heavy tailed distributions for selecting the most relevant features. Following feature selection, a KNN was used for classification. The work in [19] employs a new set of descriptors, Fractional Multichannel Exponent Moments (FrMEMs) to extract orthogonal moment features. Next, Manta Ray Foraging Optimization (MRFO) using Differential evolution (DE) is utilized to select the most relevant features. Finally, a -nearest neighbor (KNN) classifier is used for prediction. A novel shape-dependent Fibonacci-p patterns-based feature descriptor is proposed in [20] for CXR features extraction, which are classified using conventional ML algorithms such as support vector machine (SVM), -nearest neighbor (KNN), Random Forest, AdaBoost, Gradient Tree Boosting, and Decision Trees. In [21], the author uses Histogram of Oriented Gradients (HOG), Gray-Level Co-Occurrence Matrix (GLCM), Scale-Invariant Feature Transform (SIFT), and Local Binary Pattern (LBP) methods in the feature extraction phase. Next, Principle Component Analysis (PCA) is applied for feature selection. Finally, k-NN, SVM, Bag of Tree, and Kernel Extreme Learning Machine (K-ELM) are used for final classification.
2.2. Deep learning for COVID-19 detection using CXR
Many of the existing deep learning methods [22], [23], [24], [25], [26], [27], [28], [29] use the transfer learning approach by finetuning pre-trained networks such as ResNet-18 [30] or ResNet-50 [30], DenseNet-121 [17], InceptionV3 [31], Xception [32], etc., on COVID- 19 CXR datasets. COVID-SDNet [28] combines segmentation, data-augmentation and data transformations together with a ResNet-50 [30] for inference. The authors in [28] define a novel three-stage segmentation-classification pipeline to solve a binary classification task between COVID-19 and non-COVID-19 CXR. First, the lung region is cropped from CXR using bounding box segmentation. Next, a GAN based class-inherent transformation network is employed to generate two class inherent transformations and from each input image . Finally, the transformed images are used to solve a four-class classification problem using CNN with a Resnet-50 [30] backbone and an aggregation strategy is designed in order to obtain the final class. As the number of classes increase, so will the number of generators to be trained in stage two, which makes scaling inefficient for multi-class classification. In [24] an ensemble of off the shelf pretrained CNNs – InceptionV3 [31], MobileNetV2 [33], ResNet101 [30], NASNet [34] and Xception [32] – is first fine tuned on the chest xray dataset. Their final layer representations are then stacked and then passed through a MLP for COVID-19 diagnosis. The Xception [32] backbone is used in CoroNet [22] for extracting CXR features which are classified using the MLP classification head. In [35] proposed DeepCoroNet where the CXR images are pre-processed using a sobel filter followed by marker-controlled watershed segmentation and then a deep LSTM network is used for classification. The work in [36] uses Google’s Big Transfer models with DenseNet, InceptionV3 and Inception-ResNetV4 models for COVID-19 classification using chest X-rays. COVID-Net [37] proposes a custom architecture for CXR-based COVID-19 detection using a human–machine collaborative design strategy. However, limited COVID-19 samples restrict the generalizability of such large-capacity models. To address this issue, MAG-SD [38] employs a multi-scale attention-guided deep network to augment the data and formulates a new regularization term utilizing soft distance between predictions, to regularize the classifier from producing contradicted output for one target. An attention-based teacher–student framework is proposed in [39]. The teacher network extracts global features and focuses on the infected regions to generate attention maps. An image fusion module transfers the attention knowledge to the student network. CHP-Net [40] involves a discrimination network for lung feature extraction to discriminate COVID-19 cases and a localization network to localize and assign the recognized X-ray images into the left lung, right lung or bipulmonary. In [41] a federated learning model is developed while keeping in mind the privacy of the patients. Individual hospitals or care centers are considered as nodes which have their own datasets and share a common diagnosis model provided by a central server. The individual nodes update the model according to the dataset that they have and their updated weights are averaged and the common server model is updated. In [42] a multimodal system is developed based on data consisting of breathing sounds and chest X-ray images. Sound data is converted to spectrograms and convolutional neural networks are used for analysis for both sound data and chest xray images. An InceptionV3 network is used followed by an MLP for COVID-19 diagnosis. The authors in [43] propose a convolutional CapsNet for COVID-19 detection from chest X-ray images in binary as well as multi-class classification settings. xViTCOS [44] propose a vision transformer based deep neural classifier for COVID-19 prognosis.
3. Proposed method
Supervised learning usually demands a large amount of labeled data. However, collecting quality annotated data is expensive, especially for medical applications. Moreover, COVID-19 being a novel disease, there is a scarcity of well-curated high volume datasets. Therefore, we propose to utilize a self-supervised training methodology to address this issue of data scarcity. In the first stage, we train a convolutional neural network on a large-scale CXR dataset, CheXpert [16] for extracting robust CXR features with self-supervision. Next, we utilize limited COVID-19 CXR images to train a classification network that uses the pretrained backbone to extract local and global features, computes attention maps, and predicts the class label.
3.1. Self-supervised pretraining for representation learning
The fundamental concept behind self-supervised learning is to design some auxiliary pre-text tasks such that the model discovers the underlying structure of the data while solving those tasks. Several state-of-the-art self-supervised methods [45], [46], [47], [48] rely on contrastive strategy to induce similarity between positive pairs (different augmented views of the same image) and dissimilarity between negative pairs (augmented views from different images). These methods, however, require either large batch size, memory bank, or custom mining strategies while selecting negative pairs. Bootstrap Your Own Latent (BYOL) [49] mitigates this issue associated with negative pair selection. In this work, we propose to use BYOL for representation learning.
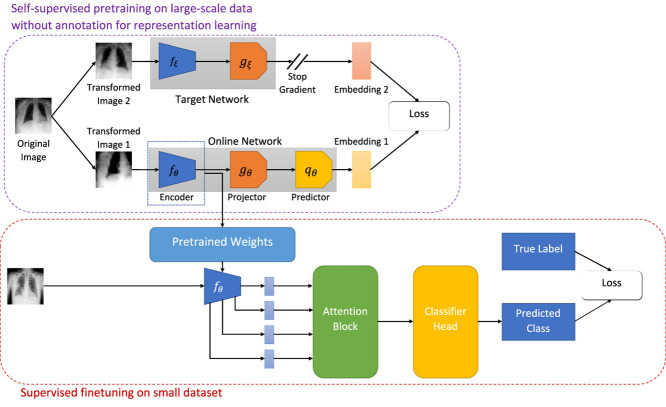
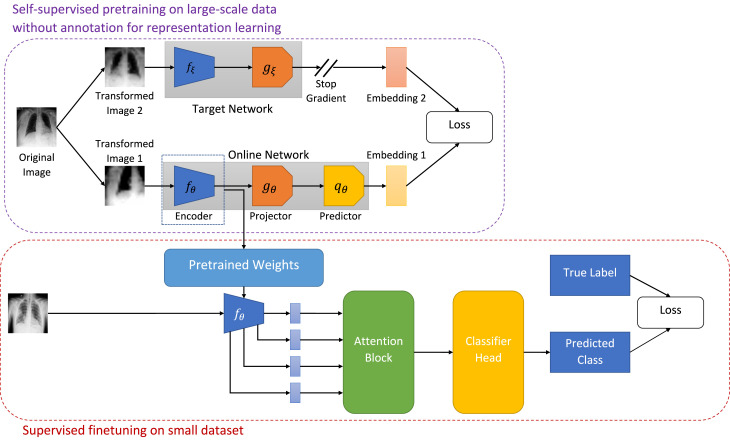
As illustrated in Fig. 1, BYOL consists of two neural networks, viz., online and target networks. These two networks interact and learn together. The online network consists of three sub-networks: an encoder , a projector , and a predictor . denotes the set of trainable parameters of the online network. To break the symmetry between the online and target pipeline, the target network is comprised of two sub-networks: an encoder and a projector . The parameters, of the target network are slow moving average of the online network parameters, i.e.,
| (1) |
where denotes the target decay rate.
Fig. 1.
Illustration of our proposed framework for COVID-19 detection using limited chest X-ray images.
At the beginning of each training step, an original image, is drawn uniformly from the CheXpert [16] dataset. Next, two sets of randomly chosen transformations, are applied on the original image, to obtain two distinct augmented views, of the underlying true image. During training, is fed into the online network, and is fed into the target network. The online network generates a representation, , a projection, , and a prediction, . The target network produces a target representation, and a target projection, . Since the target network is derived from the online network, the online representations should be predictive of the target representations. Consequently, BYOL is trained to maximize the similarity between these two representations. Mathematically, the online network is trained to minimize the mean squared error between the normalized online prediction, and the normalized target projection, :
| (2) |
To make the loss symmetric, next, is passed through the target network and is passed through the online network and loss is computed according to Eq. (2). The total loss is now given as,
| (3) |
The sub-script, in implies that, only the online network is updated to minimize , and the target network is updated as the exponential moving average as indicated in Eq. (1).
3.2. Multi-scale spatial attention based classifier
In the second stage of our proposed method, we utilize the pretrained backbone from the previous step and design a spatial attention network based on the local and the global features. Attention mechanism are widely adapted to enhance the performance of deep neural networks on various downstream tasks such as machine translation, text generation in natural language processing and object classification, image captioning, inpainting, etc., in computer vision. Attention in computer vision tasks can broadly be categorized into spatial attention [50], [51] that captures the local context and channel attention [52] that captures the global semantics. Several works [53], [54] consider a combination of both channel-wise and spatial attention.
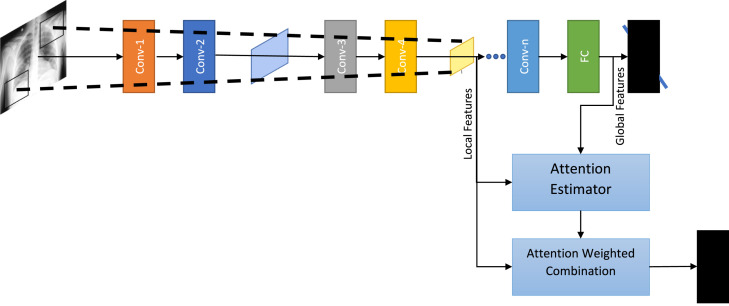
In this work, we adopt the soft trainable visual attention proposed in [55]. Fig. 2 presents an overview of the attention mechanism. We extract local and global features using pretrained feature extractor backbone. ‘Local features’ refer to features extracted by some convolutional layer of the backbone network that have a limited receptive field. In other words the receptive field is a contiguous proper subset of the image (local). The contents of the ‘local features’ can be more specific to a certain region on the image, while ‘global features’ use the entire image as their information source. We insert three attention estimators after ‘layer2’, ‘layer3’, and ‘layer4’ (layer names as per PyTorch implementation) to capture coarse-to-fine attention maps at multiple levels. The local features extracted at these three layers together with the global feature at the penultimate ‘avgpool’ layer produce three attended encodings, which are concatenated and fed into a final classification head.
Fig. 2.
Illustration of the attention mechanism.
Let, denote the set of feature vectors extracted at a given convolutional layer . Where is the vector of output activations at the spatial location of total spatial locations in the layer. The global feature vector, , has the entire input image as the receptive field. Let be the compatibility function that computes a scalar compatibility score between two vectors of equal dimension. Since the dimensionality of the local features and the global features do not match, we first project the low-dimensional local features to the high-dimensional space of . Next, the compatibility score function is employed to compute the compatibility scores as follows:
| (4) |
where and is a learnable vector.
The compatibility scores are then normalized using the softmax function to compute the attention maps.
| (5) |
The attended representations are finally computed as follows:
| (6) |
In this work, we concatenate the three representations obtained from three intermediate layers into a single vector, and feed to a linear classification head.
4. Experiments
In this section, we describe the dataset used in this work and discuss the experimental results.
4.1. Dataset
While some of the works [56] evaluate their proposed algorithm using private datasets, many other works [22], [37], [57] resort to publicly available datasets. In this work, we combine data from several publicly available repositories to create a custom dataset with four classes: Normal, Bacterial Pneumonia, Viral Pneumonia (non-COVID-19), COVID-19. As in [22], we collected Normal, Bacterial Pneumonia, and non COVID-19 Viral Pneumonia chest X-ray images from the Kaggle repository ‘Chest X-ray Images (Pneumonia)’ [58], which is derived from [59]. Chest X-ray images of COVID-19 patients were obtained from the Kaggle repository ‘COVIDx CXR-2’ [60], which is a combination of several publicly available resources [61], [62], [63], [64], [65], [66].
‘COVIDx CXR-2’ [60] specifies only train-test split of the dataset. We hold out 20% training examples for automatic model selection based on its performance over the validation set. The validation set in the standard split of ‘Chest X-ray Images (Pneumonia)’ [58] dataset contains only 8 images per class. To avoid a huge class imbalance in the validation set, we combine the training and validation examples and split them into an 80:20 ratio. Table 1 tabulates a summarized description of split-wise image distribution. Note that the test split in the standard data division is left untouched to ensure there is no patient-wise information leakage as multiple CXR images of a patient might be present in the dataset.
Table 1.
Summarized description of CXR dataset.
| Split | Normal | non-COVID Pneumonia |
COVID-19 | Total | |
|---|---|---|---|---|---|
| Bacterial | Viral | ||||
| Train | 1079 | 2030 | 1076 | 1726 | 5911 |
| Validation | 270 | 508 | 269 | 432 | 1479 |
| Test | 234 | 242 | 148 | 200 | 824 |
4.2. Implementation details
4.2.1. Image preprocessing and augmentation
In our compiled dataset and the CheXpert [16] dataset, the images are of variable sizes. To address this issue, we resize all the images to a fixed size of 256 × 256.
For training BYOL, we randomly choose an image from the CheXpert dataset, select a random patch and resize it to 224 ×224. Next, the image is flipped randomly horizontally with 0.5 probability. Apart from these spatial/geometric transformations of data, we apply appearance transformations on the image. Specifically, we apply a random color distortion transformation consisting of a random sequence of brightness, contrast, saturation, and hue adjustments [67], [68]. As noted in previous work [45], stronger color jittering helps self-supervised algorithms learn better representation. We utilize PyTorch’s standard implementation (torchvision.transforms.ColorJitter) for performing color distortion. Following [45], we set brightness, contrast and saturation jitter factor uniformly from . The hue jitter factor is chosen uniformly from . Color distortion is applied randomly 80% of the time. Finally, random Gaussian blur is applied to the patches, and the patches are normalized. We blur the image with 0.5 probability using a Gaussian kernel. We randomly sample , and the kernel size is set to be .
In the second stage of training, we randomly choose an image from the compiled CXR dataset, select a random patch and resize it to 224 × 224 with a random horizontal flip. Finally, the patches are normalized before feeding to the classifier.
We center crop the image to 224 × 224 and normalize it before passing it to the classification network during inference.
4.2.2. Model architecture
We use ResNet-50 [30] pretrained on ImageNet [69] as the online encoder, and the target encoder, . The projector networks, are multi-layer perceptrons with a hidden layer consisting of 4096 neurons followed by batch normalization, ReLU activation, and an output layer of dimension 256. The predictor network, is architecturally the same as the projector.
In our second stage of training, we modify the encoder block architecture to accommodate attention computation and initialize it with the pretrained weights from self-supervised training. We attach three attention estimators after ‘layer2’, ‘layer3’, and ‘layer4’ (layer names as per PyTorch implementation). The local features extracted at these three layers have dimensions (512, 28, 28), (1024, 14, 14) and (2048, 7, 7) respectively using ‘channel first’ representation. These three local features together with the global feature at the ‘avgpool’ layer produce three attended encodings. However, the global feature has a shape of (2048, 1), which causes shape incompatibility. To alleviate this issue, we use projector blocks consisting of 1 × 1 2-D convolution operations, which ensures that the channel dimension of local features matches the channel dimension of the global feature. Next, attention maps are computed using a linear combination of a local feature and the global features, 1 × 1 2-D convolution operations and softmax normalization. Finally, these attended embeddings are concatenated and classified using a linear classifier.
4.2.3. Hyperparameters
For self-supervised training, we use a batch size of , Adam optimizer with a learning rate of , and the model is trained for epochs.
To train the classifier, we use a batch size of , Adam optimizer with an initial learning rate of with a cosine decay learning rate scheduler. Further, we use a global weight decay parameter of .
4.2.4. Computation complexity
For the self-supervised training we use 2 NVIDIA V100 GPU cards with 32 GB memory and 5120 CUDA cores in parallel. One epoch approximately takes 1.5 h to complete execution. For the finetuning stage, we use 1 NVIDIA V100 (32 GB 5120 CUDA cores) GPU card and one epoch takes approximately 8 min to complete execution.
4.3. Quantitative results
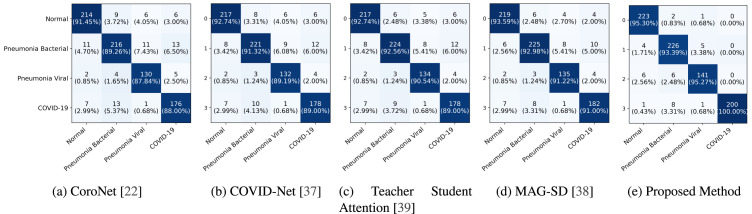
To benchmark the proposed method against other state-of-the-art methodologies, we compute and report class-wise Precision (Positive Prediction Value), Recall (Sensitivity), F1 score, Specificity, Negative Prediction Value (NPV), and overall accuracy along with 95% confidence interval. Table 2 presents our findings. As can be seen from Table 2, the proposed method achieves the best overall accuracy with best 95% confidence interval. Further the proposed method achieves the best precision for COVID-19 cases meaning the proposed classifier rarely label a COVID-19 negative sample as a positive sample. Moreover, the proposed method achieves the best recall score implying the classifier is able to find most of the positive samples belonging to the COVID-19 class. The highest F1 score achieved by the proposed method indicates that the proposed method is the most balanced in terms of both precision and recall as compared to the baseline methods. Similarly, the proposed method achieves high specificity and NPV indicating that the false positive rate is low as well. Finally, from Fig. 3, it can be seen that the proposed method achieves best class-wise accuracy.
Table 2.
Comparison of performance of the proposed method on chest X-ray dataset against state-of-the-art methods.
| Method | Class label | Precision | Recall | F1-Score | Specificity | NPV | Overall accuracy (95% CI) |
|---|---|---|---|---|---|---|---|
| CoroNet [22] | Normal | 0.9106 | 0.9145 | 0.9126 | 0.9644 | 0.9660 | 0.8932 (0.8701, 0.9135) |
| Pneumonia Bacterial | 0.8606 | 0.8926 | 0.8763 | 0.9399 | 0.9546 | ||
| Pneumonia Viral | 0.9220 | 0.8784 | 0.8997 | 0.9837 | 0.9736 | ||
| COVID-19 | 0.8934 | 0.8800 | 0.8866 | 0.9663 | 0.9617 | ||
| COVIDNet [37] | Normal | 0.9156 | 0.9274 | 0.9214 | 0.9661 | 0.9710 | 0.9078 (0.8859, 0.9266) |
| Pneumonia Bacterial | 0.8840 | 0.9132 | 0.8984 | 0.9502 | 0.9634 | ||
| Pneumonia Viral | 0.9362 | 0.8919 | 0.9135 | 0.9867 | 0.9766 | ||
| COVID-19 | 0.9082 | 0.8900 | 0.8990 | 0.9712 | 0.9650 | ||
| Teacher Student Attention [39] | Normal | 0.9274 | 0.9134 | 0.9203 | 0.9712 | 0.9711 | 0.9138 (0.8926, 0.9321) |
| Pneumonia Bacterial | 0.8889 | 0.9256 | 0.9069 | 0.9519 | 0.9685 | ||
| Pneumonia Viral | 0.9371 | 0.9054 | 0.9210 | 0.9867 | 0.9794 | ||
| COVID-19 | 0.9128 | 0.8900 | 0.9013 | 0.9728 | 0.9650 | ||
| MAG-SD [38] | Normal | 0.9399 | 0.9359 | 0.9379 | 0.9763 | 0.9746 | 0.9235 (0.9032, 0.9408) |
| Pneumonia Bacterial | 0.9036 | 0.9298 | 0.9165 | 0.9588 | 0.9704 | ||
| Pneumonia Viral | 0.9375 | 0.9122 | 0.9247 | 0.9867 | 0.9809 | ||
| COVID-19 | 0.9192 | 0.9100 | 0.9146 | 0.9744 | 0.9712 | ||
| Proposed Method | Normal | 0.9867 | 0.9530 | 0.9696 | 0.9949 | 0.9816 | 0.9587 (0.9428, 0.9713) |
| Pneumonia Bacterial | 0.9617 | 0.9339 | 0.9476 | 0.9845 | 0.9728 | ||
| Pneumonia Viral | 0.9216 | 0.9527 | 0.9369 | 0.9822 | 0.9896 | ||
| COVID-19 | 0.9524 | 1.0000 | 0.9756 | 0.9840 | 1.0000 | ||
Fig. 3.
Confusion Matrix: The horizontal axis and the vertical axis correspond to the ground truth labels and the predicted classes respectively.
4.4. Ablation studies
In this section, we examine the impact of different training components proposed in this work. Specifically, we study the effect of pretraining on ImageNet [69], self-supervised pretraining on CheXpert [16] and the attention map. Table 3 presents the findings. When a ResNet-50 architecture is trained on the COVID-19 CXR dataset from scratch, its performance is the worst. Transfer learning (ResNet-50 [30] pretrained using ImageNet [69]) improves the model’s classification performance. Attention mechanism provides a further boost in performance. Finally, self-supervised pretraining using ChexPert [16] helps the model extract useful CXR specific features and enhance the model’s classification accuracy.
Table 3.
Ablation studies to understand the impact of each training component.
| Training components |
Class label | Performance metrics |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Pretrained on ImageNet [69] |
Self-supervised learning on CheXpert [16] | Attention | Precision | Recall | F1 | Specificity | NPV | Overall accuracy | |
| No | No | No | Normal | 0.8628 | 0.8333 | 0.8478 | 0.9475 | 0.9348 | 0.8483 |
| Pneumonia Bacterial | 0.8772 | 0.8264 | 0.8511 | 0.9519 | 0.9295 | ||||
| Pneumonia Viral | 0.8217 | 0.8716 | 0.8459 | 0.9586 | 0.9715 | ||||
| COVID-19 | 0.8216 | 0.875 | 0.8474 | 0.9391 | 0.9591 | ||||
| Yes | No | No | Normal | 0.8811 | 0.8547 | 0.8676 | 0.9542 | 0.943 | 0.8786 |
| Pneumonia Bacterial | 0.897 | 0.8636 | 0.88 | 0.9588 | 0.9442 | ||||
| Pneumonia Viral | 0.8571 | 0.8919 | 0.8742 | 0.9675 | 0.9761 | ||||
| COVID-19 | 0.8714 | 0.915 | 0.8927 | 0.9567 | 0.9723 | ||||
| Yes | No | Yes | Normal | 0.8991 | 0.8761 | 0.8874 | 0.961 | 0.9513 | 0.8956 |
| Pneumonia Bacterial | 0.9056 | 0.8719 | 0.8884 | 0.9622 | 0.9475 | ||||
| Pneumonia Viral | 0.8671 | 0.9256 | 0.8954 | 0.9689 | 0.9835 | ||||
| COVID-19 | 0.9024 | 0.925 | 0.9136 | 0.9679 | 0.9758 | ||||
| No | Yes | No | Normal | 0.908 | 0.9274 | 0.9175 | 0.9627 | 0.9709 | 0.915 |
| Pneumonia Bacterial | 0.9177 | 0.9215 | 0.9196 | 0.9656 | 0.9673 | ||||
| Pneumonia Viral | 0.9241 | 0.9054 | 0.9147 | 0.9837 | 0.9794 | ||||
| COVID-19 | 0.9137 | 0.9 | 0.9068 | 0.9728 | 0.9681 | ||||
| No | Yes | Yes | Normal | 0.9163 | 0.9359 | 0.926 | 0.9661 | 0.9744 | 0.9345 |
| Pneumonia Bacterial | 0.9574 | 0.9298 | 0.9434 | 0.9828 | 0.9711 | ||||
| Pneumonia Viral | 0.9388 | 0.9324 | 0.9356 | 0.9867 | 0.9852 | ||||
| COVID-19 | 0.9261 | 0.94 | 0.9330 | 0.976 | 0.9807 | ||||
| Yes | Yes | No | Normal | 0.9212 | 0.9487 | 0.9347 | 0.9678 | 0.9794 | 0.9454 |
| Pneumonia Bacterial | 0.9664 | 0.9504 | 0.9583 | 0.9863 | 0.9795 | ||||
| Pneumonia Viral | 0.9456 | 0.9392 | 0.9424 | 0.9882 | 0.9867 | ||||
| COVID-19 | 0.9495 | 0.9400 | 0.9447 | 0.9840 | 0.9808 | ||||
| Yes | Yes | Yes | Normal | 0.9867 | 0.9530 | 0.9696 | 0.9949 | 0.9816 | 0.9587 |
| Pneumonia Bacterial | 0.9617 | 0.9339 | 0.9476 | 0.9845 | 0.9728 | ||||
| Pneumonia Viral | 0.9216 | 0.9527 | 0.9369 | 0.9822 | 0.9896 | ||||
| COVID-19 | 0.9524 | 1.0000 | 0.9756 | 0.9840 | 1.0000 | ||||
4.5. Qualitative results
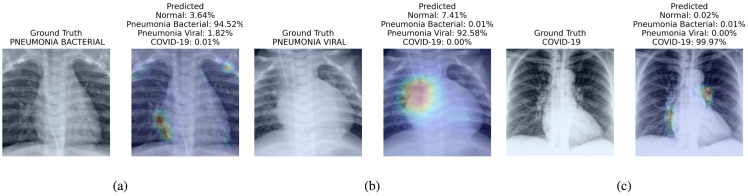
Fig. 4 presents the attention map instrumental for the prognosis made by the proposed method. We present three visualizations one for each of bacterial pneumonia (Fig. 4(a)), viral pneumonia (Fig. 4(b)) and COVID-19 (Fig. 4(c)).
Fig. 4.
Visualization of different cases (Bacterial Pneumonia, Viral Pneumonia, and COVID-19) considered in this study and their associated critical factors in decision making by our proposed method. In each subfigure, the left figure presents the input to the model and its ground truth label; the right figure presents the predicted probabilities for each class and highlight the factors critical corresponding to the top predicted class. We have used jet colormap to colorize heatmap.
5. Discussion and conclusion
This work introduces a method for automated COVID-19 prognosis using a limited amount of labeled COVID-19 CXR data. We have empirically demonstrated the effectiveness of the proposed method over existing SOTA methods as measured using various metrics such as precision, recall, F1 score, specificity, and NPV. While the proposed methodology is highly performant, it is not error-free as the CXR findings due to COVID-19 are not exclusive and overlap with other thoracic infections [70]. Therefore, to improve the efficiency of diagnosis and efficient resource utilization, we suggest the proposed method to be used in conjunction with RT-PCR, and first-line treatment may be initiated based on CXR findings while the RT-PCR test report is awaited.
Despite, the great success achieved by deep learning models in different machine learning tasks, they are prone to various biases such as selection bias (distribution of training examples is not reflective of their real-world distribution), group attribution bias (tendency to generalize what is true of individuals to an entire group to which they belong) and so on. Therefor, to deploy the proposed method clinically, it is imperative to thoroughly evaluate the model through clinical trials to examine its generalization capabilities and stability. Although, the method proposed in this work is highly performant on multinational dataset (since the datasets used in this study were compiled from several repositories), to further improve the generalization ability of the proposed method, the model needs to be trained on a large, diverse, high-quality dataset.
To conclude, preventing the spread of COVID-19 requires early diagnosis. While RT-PCR is highly accurate when the test is conducted appropriately, its turn-around time is high. Therefore, our proposed deep neural framework might be useful to initiate the first line treatment. Further, the proposed method, when used in conjunction with RT-PCR can be thought of as a complimentary diagnosis or a second opinion to ensure efficient utilization of limited resources. In our future work, we intend to extend this work to automate the analysis of infection severity.
CRediT authorship contribution statement
Arnab Kumar Mondal: Conceptualization, Data curation, Methodology, Formal analysis, Validation, Software, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We thank IIT Delhi HPC facility1 for computational resources.
Biography

Arnab Kumar Mondal received his Bachelor of Engineering in Electronics and Telecommunication from Jadavpur University, India in 2013. Right after his graduation, he joined Centre for Development of Telematics (C-DOT), Delhi, and served as a research engineer there until July 2018. In C-DOT he had the opportunity to participate in cutting-edge projects such as the Dense Wavelength Division Multiplexing (DWDM) and Packet Optical Transport Platform (P-OTP). He joined IIT Delhi as a Ph.D. scholar in July, 2018 under the guidance of Prof. Prathosh AP and Prof. Parag Singla. His research interests lie primarily within the field of deep generative models and applied deep learning. He has published a couple of top-tier peer reviewed conference papers and journal papers so far. His Ph.D. is supported by the Prime Minister’s Research Fellows (PMRF) Scheme by Govt. of India.
Footnotes
References
- 1.W.H. Organization, WHO updates on COVID-19, URL https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen.
- 2.W.H. Organization, COVID-19 symptoms, URL https://www.who.int/health-topics/coronavirus#tab=ta_3.
- 3.Carfì A., Bernabei R., Landi F., et al. Persistent symptoms in patients after acute COVID-19. JAMA. 2020;324(6):603–605. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahase E. 2020. Coronavirus: covid-19 has killed more people than SARS and MERS combined, despite lower case fatality rate. [DOI] [PubMed] [Google Scholar]
- 5.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296(2):E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen D., Wu G., Suk H.-I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017;19:221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajpurkar P., Irvin J., Zhu K., Yang B., Mehta H., Duan T., Ding D.Y., Bagul A., Langlotz C., Shpanskaya K.S., Lungren M.P., Ng A.Y. 2017. CheXNet: RAdiologist-level pneumonia detection on chest X-Rays with deep learning. CoRR abs/1711.05225, arXiv:1711.05225. [Google Scholar]
- 11.X. Wang, Y. Peng, L. Lu, Z. Lu, M. Bagheri, R.M. Summers, ChestX-Ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases, in: Proc. of CVPR, 2017.
- 12.Wang H., Xia Y. 2018. Chestnet: A deep neural network for classification of thoracic diseases on chest radiography. arXiv preprint arXiv:1807.03058. [DOI] [PubMed] [Google Scholar]
- 13.Ranjan E., Paul S., Kapoor S., Kar A., Sethuraman R., Sheet D. Proceedings of the 11th Indian Conference on Computer Vision, Graphics and Image Processing. 2018. Jointly learning convolutional representations to compress radiological images and classify thoracic diseases in the compressed domain. [DOI] [Google Scholar]
- 14.Wang H., Wang S., Qin Z., Zhang Y., Li R., Xia Y. Triple attention learning for classification of 14 thoracic diseases using chest radiography. Med. Image Anal. 2021;67 doi: 10.1016/j.media.2020.101846. [DOI] [PubMed] [Google Scholar]
- 15.Wang H., Jia H., Lu L., Xia Y. Thorax-Net: An attention regularized deep neural network for classification of thoracic diseases on chest radiography. IEEE J. Biomed. Health Inf. 2020;24(2):475–485. doi: 10.1109/JBHI.2019.2928369. [DOI] [PubMed] [Google Scholar]
- 16.J. Irvin, P. Rajpurkar, M. Ko, Y. Yu, S. Ciurea-Ilcus, C. Chute, H. Marklund, B. Haghgoo, R.L. Ball, K. Shpanskaya, J. Seekins, D.A. Mong, S.S. Halabi, J.K. Sandberg, R. Jones, D.B. Larson, C.P. Langlotz, B.N. Patel, M.P. Lungren, A.Y. Ng, CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison, in: Proc. of AAAI, 2019.
- 17.G. Huang, Z. Liu, L. Van Der Maaten, K.Q. Weinberger, Densely connected convolutional networks, in: Proc. of CVPR, 2017.
- 18.Yousri D., Abd Elaziz M., Abualigah L., Oliva D., Al-qaness M.A., Ewees A.A. COVID-19 X-ray images classification based on enhanced fractional-order cuckoo search optimizer using heavy-tailed distributions. Appl. Soft Comput. 2021;101 doi: 10.1016/j.asoc.2020.107052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elaziz M.A., Hosny K.M., Salah A., Darwish M.M., Lu S., Sahlol A.T. New machine learning method for image-based diagnosis of COVID-19. PLoS One. 2020;15(6) doi: 10.1371/journal.pone.0235187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panetta K., Sanghavi F., Agaian S., Madan N. Automated detection of COVID-19 cases on radiographs using shape-dependent fibonacci-p patterns. IEEE J. Biomed. Health Inf. 2021;25(6):1852–1863. doi: 10.1109/JBHI.2021.3069798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saygılı A. A new approach for computer-aided detection of coronavirus (COVID-19) from CT and X-ray images using machine learning methods. Appl. Soft Comput. 2021;105 doi: 10.1016/j.asoc.2021.107323. URL https://www.sciencedirect.com/science/article/pii/S1568494621002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan A.I., Shah J.L., Bhat M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsiknakis N., Trivizakis E., Vassalou E.E., Papadakis G.Z., Spandidos D.A., Tsatsakis A., Sánchez-García J., López-González R., Papanikolaou N., Karantanas A.H., et al. Interpretable artificial intelligence framework for COVID-19 screening on chest X-rays. Exp. Ther. Med. 2020;20(2):727–735. doi: 10.3892/etm.2020.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta A., Anjum E., Gupta S., Katarya R. InstaCovNet-19: A deep learning classification model for the detection of COVID-19 patients using chest X-ray. Appl. Soft Comput. 2021;99 doi: 10.1016/j.asoc.2020.106859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heidari M., Mirniaharikandehei S., Khuzani A.Z., Danala G., Qiu Y., Zheng B. Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. Int. J. Med. Inf. 2020;144 doi: 10.1016/j.ijmedinf.2020.104284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ezzat D., Hassanien A.E., Ella H.A. An optimized deep learning architecture for the diagnosis of COVID-19 disease based on gravitational search optimization. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang R., Tie X., Qi Z., Bevins N.B., Zhang C., Griner D., Song T.K., Nadig J.D., Schiebler M.L., Garrett J.W., Li K., Reeder S.B., Chen G.-H. Diagnosis of coronavirus disease 2019 pneumonia by using chest radiography: Value of artificial intelligence. Radiology. 2021;298(2):E88–E97. doi: 10.1148/radiol.2020202944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabik S., Gómez-Ríos A., Martín-Rodríguez J.L., Sevillano-García I., Rey-Area M., Charte D., Guirado E., Suárez J.L., Luengo J., Valero-González M.A., García-Villanova P., Olmedo-Sánchez E., Herrera F. COVIDGR dataset and COVID-sdnet methodology for predicting COVID-19 based on chest X-Ray images. IEEE J. Biomed. Health Inf. 2020;24(12):3595–3605. doi: 10.1109/JBHI.2020.3037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain G., Mittal D., Thakur D., Mittal M.K. A deep learning approach to detect Covid-19 coronavirus with X-ray images. Biocybern. Biomed. Eng. 2020;40(4):1391–1405. doi: 10.1016/j.bbe.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.K. He, X. Zhang, S. Ren, J. Sun, Deep residual learning for image recognition, in: Proc. of CVPR, 2016.
- 31.C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens, Z. Wojna, Rethinking the inception architecture for computer vision, in: Proc. of CVPR, 2016, pp. 2818–2826.
- 32.F. Chollet, Xception: Deep learning with depthwise separable convolutions, in: Proc. of CVPR, 2017.
- 33.M. Sandler, A. Howard, M. Zhu, A. Zhmoginov, L.-C. Chen, Mobilenetv2: Inverted residuals and linear bottlenecks, in: Proc. of CVPR, 2018, pp. 4510–4520.
- 34.B. Zoph, V. Vasudevan, J. Shlens, Q.V. Le, Learning transferable architectures for scalable image recognition, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2018, pp. 8697–8710.
- 35.Demir F. DeepCoroNet: A deep LSTM approach for automated detection of COVID-19 cases from chest X-ray images. Appl. Soft Comput. 2021;103 doi: 10.1016/j.asoc.2021.107160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albahli S., Ayub N., Shiraz M. Coronavirus disease (COVID-19) detection using X-ray images and enhanced DenseNet. Appl. Soft Comput. 2021;110 doi: 10.1016/j.asoc.2021.107645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L., Lin Z.Q., Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020;10(1):19549. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J., Wang Y., Wang S., Wang J., Liu J., Jin Q., Sun L. Multiscale attention guided network for COVID-19 diagnosis using chest X-Ray images. IEEE J. Biomed. Health Inf. 2021;25(5):1336–1346. doi: 10.1109/JBHI.2021.3058293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi W., Tong L., Zhu Y., Wang M.D. COVID-19 automatic diagnosis with radiographic imaging: Explainable attention transfer deep neural networks. IEEE J. Biomed. Health Inf. 2021:1. doi: 10.1109/JBHI.2021.3074893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z., Xiao Y., Li Y., Zhang J., Lu F., Hou M., Liu X. Automatically discriminating and localizing COVID-19 from community-acquired pneumonia on chest X-rays. Pattern Recognit. 2021;110 doi: 10.1016/j.patcog.2020.107613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feki I., Ammar S., Kessentini Y., Muhammad K. Federated learning for COVID-19 screening from chest X-ray images. Appl. Soft Comput. 2021;106 doi: 10.1016/j.asoc.2021.107330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sait U., K.V. G.L., Shivakumar S., Kumar T., Bhaumik R., Prajapati S., Bhalla K., Chakrapani A. A deep-learning based multimodal system for Covid-19 diagnosis using breathing sounds and chest X-ray images. Appl. Soft Comput. 2021;109 doi: 10.1016/j.asoc.2021.107522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toraman S., Alakus T.B., Turkoglu I. Convolutional capsnet: A novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mondal A.K., Bhattacharjee A., Singla P., Prathosh A. xViTCOS: Explainable vision transformer based COVID-19 screening using radiography. IEEE J. Transl. Eng. Health Med. 2021;10:1–10. doi: 10.1109/JTEHM.2021.3134096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.T. Chen, S. Kornblith, M. Norouzi, G. Hinton, A Simple Framework for Contrastive Learning of Visual Representations, in: Proc. of ICML, 2020.
- 46.T. Chen, S. Kornblith, K. Swersky, M. Norouzi, G. Hinton, Big Self-Supervised Models are Strong Semi-Supervised Learners, in: Proc. of NeurIPS, 2020.
- 47.K. He, H. Fan, Y. Wu, S. Xie, R. Girshick, Momentum Contrast for Unsupervised Visual Representation Learning, in: Proc. of CVPR, 2020.
- 48.Y. Tian, D. Krishnan, P. Isola, Contrastive Multiview Coding, in: Proc. of ECCV, 2020.
- 49.J.-B. Grill, F. Strub, F. Altché, C. Tallec, P. Richemond, E. Buchatskaya, C. Doersch, B. Avila Pires, Z. Guo, M. Gheshlaghi Azar, B. Piot, k. kavukcuoglu, R. Munos, M. Valko, Bootstrap Your Own Latent - A New Approach to Self-Supervised Learning, in: Proc. of NeurIPS, Vol. 33, 2020, pp. 21271–21284.
- 50.K. Xu, J. Ba, R. Kiros, K. Cho, A. Courville, R. Salakhudinov, R. Zemel, Y. Bengio, Show, attend and tell: Neural image caption generation with visual attention, in: Proc. of ICML, 2015, pp. 2048–2057.
- 51.Y. Zhu, O. Groth, M. Bernstein, L. Fei-Fei, Visual7w: Grounded question answering in images, in: Proc. of CVPR, 2016, pp. 4995–5004.
- 52.Hu J., Shen L., Sun G. Proc. of CVPR. 2018. Squeeze-and-excitation networks; pp. 7132–7141. [DOI] [Google Scholar]
- 53.L. Chen, H. Zhang, J. Xiao, L. Nie, J. Shao, W. Liu, T.-S. Chua, SCA-CNN: Spatial and channel-wise attention in convolutional networks for image captioning, in: Proc. of CVPR, 2017, pp. 5659–5667.
- 54.S. Woo, J. Park, J.-Y. Lee, I.S. Kweon, CBAM: Convolutional Block Attention Module, in: Proceedings of the European Conference on Computer Vision, ECCV, 2018, pp. 3–19.
- 55.S. Jetley, N.A. Lord, N. Lee, P.H.S. Torr, Learn To Pay Attention, in: Proc. of ICLR, 2018.
- 56.Yu X., Lu S., Guo L., Wang S.-H., Zhang Y.-D. ResGNet-C: A graph convolutional neural network for detection of COVID-19. Neurocomputing. 2021;452:592–605. doi: 10.1016/j.neucom.2020.07.144. URL https://www.sciencedirect.com/science/article/pii/S0925231220319184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cohen J.P., Morrison P., Dao L. 2020. COVID-19 image data collection. arXiv preprint arXiv:2003.11597. [Google Scholar]
- 58.Mooney P. 2018. Chest X-Ray images (Pneumonia) https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia. [Google Scholar]
- 59.Kermany D., Zhang K., Goldbaum M. 2018. Labeled optical coherence tomography (OCT) and chest X-Ray images for classification. Mendeley Data, V2. [Google Scholar]
- 60.Zhao A., Aboutalebi H., Wong A., Gunraj H., Terhljan N., et al. 2021. COVIDX CXR-2: Chest x-ray images for the detection of COVID-19. https://www.kaggle.com/andyczhao/covidx-cxr2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cohen J.P., Morrison P., Dao L., Roth K., Duong T.Q., Ghassemi M. 2020. COVID-19 image data collection: Prospective predictions are the future. arXiv:2006.11988. [Google Scholar]
- 62.Wang L., Wong A., Lin Z.Q., McInnis P., Chung A., Gunraj H., Lee J., Ross M., VanBerlo B., Ebadi A., Git K.-A., Al-Haimi A. 2020. Figure 1 COVID-19 chest X-ray dataset initiative. https://github.com/agchung/Figure1-COVID-chestxray-dataset. [Google Scholar]
- 63.Wang L., Wong A., Lin Z.Q., McInnis P., Chung A., Gunraj H., Lee J., Ross M., VanBerlo B., Ebadi A., Git K.-A., Al-Haimi A. 2020. Actualmed COVID-19 chest X-ray dataset initiative. https://github.com/agchung/Actualmed-COVID-chestxray-dataset. [Google Scholar]
- 64.Chowdhury M.E.H., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Emadi N.A., Reaz M.B.I., Islam M.T. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- 65.of North America R.S. 2018. RSNA pneumonia detection challenge: Can you build an algorithm that automatically detects potential pneumonia cases? https://www.kaggle.com/c/rsna-pneumonia-detection-challenge. [Google Scholar]
- 66.Tsai E.B., Simpson S., Lungren M., Hershman M., Roshkovan L., Colak E., Erickson B.J., Shih G., Stein A., Kalpathy-Cramer J., et al. 2021. Data from medical imaging data resource center (MIDRC) - RSNA international COVID radiology database (RICORD) release 1c - chest x-ray, Covid+ (MIDRC-RICORD-1c) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Howard A.G. 2013. Some improvements on deep convolutional neural network based image classification. arXiv preprint arXiv:1312.5402. [Google Scholar]
- 68.C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, A. Rabinovich, Going deeper with convolutions, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2015, pp. 1–9.
- 69.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., Berg A.C., Fei-Fei L. ImageNet Large scale visual recognition challenge. Int. J. Comput. Vis. (IJCV) 2015;115(3):211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 70.of Radiology A.C. 2020. ACR recommendations for the use of chest radiography and computed tomography (CT) for suspected COVID-19 infection. URL https://www.acr.org/Advocacy-and-Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CT-for-Suspected-COVID19-Infection. [Google Scholar]