Summary
Ovarian cancer (OC) is a heterogeneous disease with the highest mortality rate and the poorest prognosis among gynecological malignancies. Because of the absence of specific early symptoms, most OC patients are often diagnosed at late stages. Thus, improved biomarkers of OC for use in research and clinical practice are urgently needed. The last decade has seen increasingly rapid advances in sequencing and biotechnological methodologies. Consequently, multiple omics technologies, including genomic/transcriptomic sequencings and proteomic/metabolomic mass spectra, have been widely applied to analyze tissue- and liquid-derived samples from OC patients. The integration of multi-omics data has increased our knowledge of the disease and identified valuable OC biomarkers. In this review, we summarize the recent advances and perspectives in the use of multi-omics technologies in OC research and highlight potential applications of multi-omics for identifying novel biomarkers and improving clinical assessments.
Keywords: Ovarian cancer, Biomarker, Multi-omics, Translational medicine
Abbreviations: OC, ovarian cancer; EOC, epithelial ovarian cancer; HGSOC, high-grade serous ovarian cancer; TVUS, transvaginal ultrasonography; CA125, Cancer antigen 125; HE4, Human epididymis protein 4; ROCA, risk for ovarian cancer algorithm; SNV, single-nucleotide variation; CNA, copy number alteration; ctDNA, circulating tumor DNA; NGS, next-generation sequencing; WES, whole exome sequencing; WGS, whole genome sequencing; MS, mass spectrometry; UPLC, ultra-performance liquid chromatography; AI, artificial intelligence; ML, machine learning
Introduction
Ovarian cancer (OC) is the eighth most common cause of cancer mortality in women worldwide, accounting for over 310,000 new cases and around 200,000 deaths in 2020.1 Epithelial ovarian cancer (EOC), a heterogeneous disease characterized by great molecular and histological diversity, represents approximately 90% of OC, with high-grade serous ovarian cancer (HGSOC) being the most frequent and lethal subtype. Because of the lack of specific early-stage clinical symptoms, more than 75% of OC patients are diagnosed at an advanced stage. The standard treatments for OC include either primary debulking surgery followed by platinum-based chemotherapy or neoadjuvant chemotherapy followed by interval debulking surgery and additional chemotherapy post-surgery. However, the therapeutic approach is effective for only a small number of patients, and the prognosis of OC remains poor, with an overall 5-year survival rate ranging from 30% to 50%.2, 3, 4
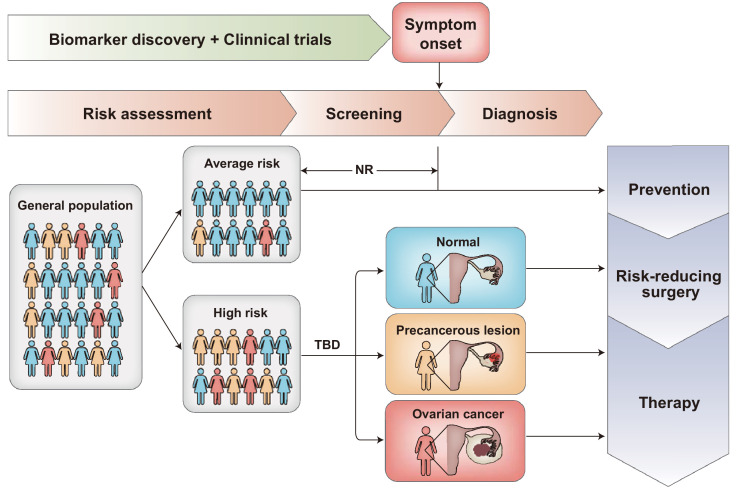
An ideal strategy to improve the low survival rate of OC is early diagnosis. If the disease is detected at low tumor volume or a localized stage (stages IA and IB), approximately 93% of patients can live longer than 5 years after diagnosis.2,3 In patients with indicative symptoms, diagnostic work-up includes physical examination of the patient and radiographic imaging, such as transvaginal ultrasonography (TVUS). For women who are asymptomatic, screening strategy for earlier detection of OC is still not available. Currently, Cancer antigen 125 (CA125) blood test and TVUS have been the most promising screening tools for OC detection.5 Human epididymis protein 4 (HE4) has also been tested as a potential biomarker for use in OC screening, but further studies are required.6 Based on the previous results from the United Kingdom Collaborative Trial of OC screening (UKCTOCS), the largest OC screening trial to date, multimodal screening (MMS) led to an absolute 13% increase of early-stage (stage I or II) cancer detection compared with the no screening group. In the MMS group, serum CA125 concentration was measured and the longitudinal CA125 was interpreted using the risk for ovarian cancer algorithm (ROCA).7 However, the latest results from the UKCTOCS extended follow-up study revealed that neither MMS nor transvaginal ultrasound screening approaches used in the trial significantly reduced deaths from OC.8 There are several possible reasons why detecting OC earlier did not result in fewer deaths in the study. One explanation may be that the 10% increased proportion of detected early-stage OC patients is not sufficient to change the prognosis of OC and translate into saving more lives. Thus, based on the evidence to date, screening in the general or average-risk population for OC cannot be recommended. To achieve mortality reduction for the general population, future screening strategies should be able to detect OC a great deal earlier and in a larger proportion of women than we currently can. As for women at high risk, ROCA-based multimodal screening exhibited high sensitivity and significant stage shift in the UK Familial Ovarian Cancer Screening Study.9 However, the effect of this screening strategy on mortality in this population will not be available because it is unethical and unfeasible to randomly assign women at high risk to a control group. It is believed that sensitive biomarkers for identifying the population at risk and detecting OC at the earliest possible stage are urgently needed, that would help this population get timely prevention and treatment (Figure 1).
Figure 1.
Opportunities for reducing ovarian cancer mortality through early detection.
Many research studies and clinical trials have been conducted to develop sensitive screening tests that could allow for earlier detection of OC in women who are asymptomatic. Recommendations for OC screening need to be dependent on the risk level of the population. Women with genetic mutations known to increase susceptibility to OC or a strong family history of the disease may be at increased risk of developing OC. For women at average risk, there are no recommended screening tests for them to date. For women who have a high risk of developing OC, screening tests may be offered to help this population get timely prevention and treatment. NR = not recommended. TBD = to be determined.
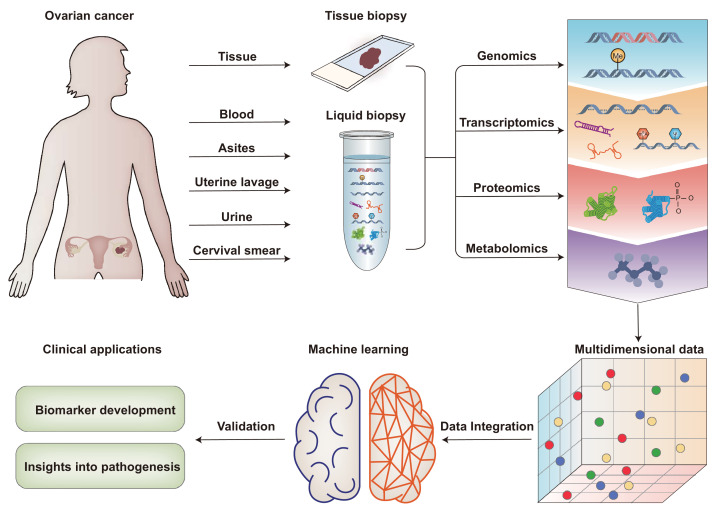
Over the past decade, researchers have made considerable efforts to gain deep molecular profiling of OC that can help guide more precise and individualized clinical decisions. More recently, the developments and availability of multi-omics technologies, including genomics, transcriptomics, proteomics, and metabolomics, are making it possible for more informative biomarkers to be discovered and possibly developed for use in clinical practice.10 In this review, we present an overview of how multi-omics technologies contribute to biomarker discovery for early diagnosis in OC (Figure 2) and discuss future approaches for improving biomarker performance in OC.
Figure 2.
Schematic representation of multi-omics approaches towards biomarker discovery for early diagnosis in ovarian cancer.
Tissue and body fluids, such as blood, ascites, uterine lavage, cervical smear, and urine, can be analyzed by multi-platform omics technologies, including genomic/transcriptomic sequencings and proteomic/metabolomic mass spectra, etc. These multidimensional data could be integrated based on machine learning techniques. The multi-omics approach promotes a comprehensive understanding of OC and biomarker discovery in early diagnosis.
Multi-omics for biomarker discovery for early diagnosis of OC
Sample sources
Tumor tissue biopsy testing has been the standard for evaluating molecular features of a tumor. Since the ovaries are completely intraperitoneal organs, it is impossible to obtain samples of OC tissues without surgical resection. In particular, needle biopsies for early-stage ovarian tumors should be avoided because cancer cells easily disseminate into the peritoneal cavity, and puncture would promote peritoneal metastasis.11 Historically, many researchers chose ovarian surface epithelium as normal control tissue for comparative experiments. With the increasing understanding that fallopian tube is the primary tissue of origin for many OCs,12,13 current studies tend to use normal fallopian tube controls compared with tumor samples to identify molecular biomarkers for OC, especially HGSOCs.
Since tumor tissue biopsies for OC are difficult to obtain serially, informative and less-invasive biomarkers that could be used for OC early detection are urgently needed. Over the past twenty years, advances in instrumentation, sample preparation, and data analysis have enabled the availability of high-quality, reproducible, and comprehensive data from various clinical samples. Liquid biopsy has emerged as a promising alternative for cancer diagnosis. In contrast to conventional tumor biopsies, liquid biopsy has several unique advantages. First, it is minimally invasive and safe, avoiding the potential complications caused by tissue biopsies. Second, it provides an opportunity to identify heterogeneous tumor-specific alterations that may be missed by tissue biopsies. More importantly, liquid biopsies enable serial sampling over time, which provides important information for guiding clinical decisions.14 For occult OC patients, body fluids, such as blood, ascites, uterine lavage, cervical smear, and urine, are characteristically enriched with nucleic acids, proteins, or metabolites that can be a potential source of diagnostic biomarkers.
Genomics
Genomic sequences were the first widespread omics data available for understanding human cancer biology and pathogenesis. As a result of technological innovation and rapid decline in sequencing costs, multigene next-generation sequencing-based tumor genomic profiling has been widely utilized in cancer subtype classification and predictive biomarkers identification. Various genomic assays, such as targeted sequencing, whole-exome sequencing, and whole-genome sequencing (WGS), are commonly used for identifying single-nucleotide variations (SNVs), copy number alterations (CNAs), chromosomal rearrangements, and DNA methylation.15, 16, 17
The Pan-Cancer Analysis of Whole Genomes (PCAWG) consortium aggregated whole-genome sequencing data from thousands of tumors across over 30 cancer types, generated by The Cancer Genome Atlas (TCGA) and The International Cancer Genome Consortium projects.18,19 By analyzing DNA alterations in 489 HGSOC tumors and the DNA sequences of exons from coding genes in 316 of the tumors, TCGA has compiled a catalog of molecular abnormalities in OC, including TP53 mutations, which are found in 96% of the tumors, somatic or germline BRCA1/2 mutations, which are found in ∼25% of the tumors, and CCNE1 aberrations. Other frequently altered pathways in OC include RB1, PI3K/RAS, NOTCH, and FOXM1.20 To decode the genomic complexity of CNAs in OC, Macintyre et al. performed WGS of 117 OC cases. They identified seven copy number signatures that could represent distinct mutational processes and provided a rational framework for the diagnosis and assessment in OC.21
Notably, approximately 50% of HGSOCs are defective in the homologous recombination (HR) DNA repair pathway.20,22 Among these DNA repair genes, BRCA1/2 are best known for their crucial role in HR-mediated DNA double-strand break repair.23 Pathogenic variants in the BRCA1/2 genes are associated with cancer susceptibility and can markedly increase the risk of breast and ovarian cancers.24 Scientists have developed different functional assays and computational prediction methods to assess the effect of missense variants of uncertain significance in BRCA1 and BRCA2 on protein function.25 In 2018, researchers used saturation genome editing to assay 96.5% of all possible SNVs in crucial domains of BRCA1, identifying over 4,000 SNVs associated with pathogenicity. These results could be immediately useful for the clinical interpretation of BRCA1 variants. Also, the approach of saturation genome editing can be extended to overcome the challenge of determining the effects of variants of uncertain significance in additional clinically actionable genes.26 In our recent study, we showed that a single non-pathogenic variant of BARD1, when combined with another variant (R378S) in cis, yields a pathogenic allele. This type of synergetic effect would be more likely to occur in a gene that has two or more functional domains or regions than in those with just one domain. Synergetic effects in these genes may be involved in a significant fraction of all tumor cases and thus have important clinical implications.27 Increased awareness of associations between BRCA1/2 mutations and OC has led to an increased demand for genetic counseling and testing, aiming to identify the individuals at high risk of developing OC. The National Comprehensive Cancer Network genetics guidelines, as well as several European organizations, have recommended universal germline BRCA mutation screening for all women diagnosed with OC, that can help identify family members at high risk. In addition to BRCA1/2, other genes from the Fanconi anemia pathway, such as BRIP1, RAD51C, and RAD51D, have been implicated in hereditary OC.28 For women at high risk of developing OC, risk-reducing surgery, such as bilateral salpingo-oophorectomy (removal of the ovaries and the fallopian tubes) may be an option.5
Apart from genetic changes, epigenetic alterations are also relatively common in all forms of cancer, including OC.29 DNA methylation (DNAme), one of the most common epigenetic modifications, is an early event in cancer, causing chromatin changes or interference with transcription factor binding sites. Aberrant DNAme may ultimately lead to gene transcription silencing.30 Therefore, numerous studies have investigated the use of aberrant DNAme in OC diagnosis. One important study found that 168 genes were epigenetically silenced by increased DNAme in OC samples compared with fallopian tube controls.20 In 2017, Widschwendter et al. analyzed tissue and serum samples using a methylation array or reduced representation bisulfite sequencing. They identified cancer-specific DNAme patterns that could potentially be used to detect OCs up to two years earlier than current diagnosis methods.31 In addition, a subsequent study found that methylation within the promoters of three genes (c17orf64, IRX2, and TUBB6) could accurately distinguish early precursor serous tubal intraepithelial carcinoma lesions from normal or benign gynecologic tissues.32 Overall, these findings highlight the advantages and applications of DNAme analysis in detecting OC at an early stage.
Currently, analysis of circulating tumor DNA (ctDNA) has provided a complementary approach to tissue-based genomic testing for OC. ctDNA is released from tumor cells into the circulation and has been detected in patients with early- and late-stage OCs.33 Thus, there is growing interest in utilizing ctDNA as a biomarker to improve early OC detection. The fraction of ctDNA can be distinguished by the presence of cancer-specific genetic and epigenetic aberrations. Due to the low abundance of ctDNA in samples, highly sensitive techniques, such as digital polymerase chain reaction (PCR) and targeted next-generation sequencing (NGS), are used to detect cancer-specific modifications.34 Sequencing sensitivity can be improved by using random oligonucleotide barcodes called unique molecular identifiers (UMIs). The unique tags facilitate bioinformatic alignment of sequences derived from the same DNA fragment and help with identification of sequencing errors.35,36 Additionally, identifying the presence of aneuploidy in clinical samples also has a broad range of diagnostic applications. Some PCR-based assays, such as Repetitive Element AneupLoidy Sequencing System (RealSeqS), might be an alternative method to WGS for the assessment of aneuploidy.37 Moreover, many studies suggested that multiparameter analyses could lead to an increase in sensitivity.38 For example, the sensitivity of ctDNA detection could be increased from 43% to 63% by combining the analyses of gene mutations and aneuploidy in OC.39 Key studies on ctDNA for early detecting OC are listed in Table 1.
Table 1.
Key studies on ctDNA in ovarian cancer.
ddPCR: droplet digital PCR; NR: Not reported.
| Author (year) | Number of OC Patients | Source samples | Detection method | Genetic Marker | Detection Rate | Sensitivity / Specificity | Refs |
|---|---|---|---|---|---|---|---|
| Paracchini et al. (2021) | 46 HGSOC (III-IV) | plasma | shallow WGS | CNA profiling | 87.8%, 78.05% | NR | 40 |
| Lin et al. (2019) | 112 germline or somatic BRCA-mutant HGSOC | Plasma | Targted NGS | BRCA1, BRCA2, TP53 | 96% for TP53 | NR | 41 |
| Oikkonen et al. (2019) | 12 HGSOCs (II-IV) | Plasma | Targeted NGS | 500 genes+CNA | 100% for TP53 | NR | 42 |
| Wang et al. (2018) | 83 OCs (I-IV) | Plasma /Plasma+Pap Brush samples | multiplex PCR-based test Safe-SeqS |
18 genes+assay for aneuploidy | 43% /63% | NR 100% |
39 |
| Cohen et al. (2018) | 54 OCs (I-III) | Plasma | CancerSEEK multiplex PCR |
16 genes | 98% | Sn: 98% Sp: >99% |
43 |
| Nakabayashi et al. (2018) | 36 OCs (I-IV) | Plasma | WGS | CNA profiling | 16.7%% | NR | 44 |
| Arend et al. (2018) | 14 HGSOCs (III-IV) | Plasma | NGS | 50 genes | 100% | NR | 45 |
| Vanderstichele et al. (2017) | 54 HGSOCs (I-IV) | Plasma | WGS | CNA profiling | 67% | NR 99.6% |
46 |
| Widschwendter et al. (2017) | 151 OCs (I-IV) | Serum | bisulfite sequencing | three-DNA-methylation marker panel | 41% | Sn: 41.4% Sp: 90.7% |
31 |
| Christie et al. (2017) | 30 HGSOCs (I-IV) | Plasma | Targeted NGS | BRCA1/2 | 60% | NR | 47 |
| Phallen et al. (2017) | 42 OCs (I-IV) | Plasma | Targeted NGS (TEC-seq) and ddPCR | 55 gene panel | 71% | Sn:97.4% Sp: 100% |
48 |
Transcriptomics
Unlike the genome, which gives a static view of the genetic information defining a phenotype, the transcriptome varies in different tissues, developmental stages, and disease states. Therefore, knowledge of transcriptomic variation is critical for understanding how genes are regulated in response to internal and external conditions. Modern transcriptomic techniques, such as RNA sequencing (RNA-seq) and full transcript microarrays, have been applied to explore a complete and accurate view of the transcriptome.49
Analysis of the messenger RNA (mRNA) expression data in TCGA classified OC into four transcriptional subtypes: immunoreactive, differentiated, proliferative, and mesenchymal.20 Subsequently, researchers replicated this study using external independent data sets to validate the TCGA transcriptional subtypes.50,51 The findings of these studies suggest that these molecular subtypes are associated with distinct prognoses of OC. The immunoreactive subtype was associated with improved survival outcomes, whereas the mesenchymal and proliferative subtypes were associated with the worst overall survival rates. Another group identified a 39 differentially expressed gene signature that can help further biologically characterize the molecular subtypes and develop targeted clinical trials.52 Until recently, our understanding of the molecular basis of human cancers has mainly relied on bulk sequencing. However, since a homogenized tumor sample can contain millions of cells, bulk RNA-seq data alone are unable to capture the spatial histopathological information and cellular heterogeneity within tumors. The emergence of single-cell RNA sequencing (scRNA-seq) advanced our knowledge of cellular heterogeneity by enabling the characterization of the transcriptomes of individual cells and identification of cell subpopulations in a given tissue.53 A notable example is the study carried out by Izar et al. using scRNA-seq to comprehensively study the ascites and primary tumor samples from OC patients and patient-derived xenograft models. They found that different functional sub-populations of cancer cells contribute to shaping the OC ecosystem. Furthermore, their results indicated that the highly expressed JAK/STAT pathway in both cancer cells and cancer-associated fibroblasts could be an ideal candidate for the diagnosis and treatment of OC.54 Subsequently, a series of spatial transcriptomic methods have been developed and have accelerated the capacity to obtain gene expression profiles for tissues or cell cultures while retaining spatial localization information, resulting in new discoveries in many areas of biology. However, sequencing depth is still a limiting factor for current spatial barcoding techniques, including fluorescence in situ hybridization-based and sequencing-based methods.55 During this couple of years, data integration with spatial transcriptomics and scRNA-seq has developed as a high-resolution approach to map diverse cell subpopulations in tissue. In this approach, reference cells defined by scRNA-seq can be used to infer cell type composition in spatial data using different techniques, providing a great strategy to study the roles of specific cell types and their interactions in development, homeostasis, and disease.56,57 Considering the significant cellular heterogeneity of OC, integrating scRNA-seq and spatial transcriptomics data will undoubtedly help understand the etiology of the disease, and it will be an important future direction for uncovering new diagnostics and treatments in OCs.
It is known that most of the human genome encoding RNAs do not code for proteins. RNA species beyond mRNA include microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs, which are collectively known as non-coding RNAs (ncRNAs).58 Although ncRNAs are not translated into proteins, compelling evidence shows that they affect the expression and biological functions of other genes through various mechanisms and could serve as potential biomarkers for human cancer diagnosis.59 MiRNAs are endogenous, small ncRNAs (19-25 nucleotides) that form a major class of functional ncRNAs. It is well established that miRNAs are critical regulators of post-transcriptional gene expression.60 The TCGA project showed that miRNAs have a widespread impact on gene expression and molecular heterogeneity in OC.20 In addition, Todeschini et al. performed microarrays to profile serum miRNA expression and validated the differentially expressed miRNAs from two independent cohorts. Their results revealed circulating miR-1246 as a potential diagnostic biomarker for HGSOC.61 Similarly, another group constructed a diagnostic model based on ten miRNAs after obtaining comprehensive miRNA profiles from 4046 serum samples from 428 OCs, 2759 non-cancer controls, and 859 other solid cancers.62 These studies proved that profiling serum miRNA is a promising strategy for OC diagnosis. In addition to miRNAs, lncRNAs, which are ncRNAs longer than 200 nucleotides, also play important roles at the transcriptomic level, and they are involved in a wide range of biological processes.63 Wang et al. performed lncRNA and mRNA microarray profiling in the normal ovary, benign cysts, and malignant EOC and identified 18 lncRNAs differentially expressed between these groups.64 With the rapid developments of liquid biopsy technology, circulating miRNAs and lncRNAs may become more reliable biomarkers for OC prediction in the future.
Thus far, more than 100 types of RNA modifications have been identified in various RNA molecules from all domains of life.65 However, their critical roles in gene expression regulation at the post-transcriptional level were not uncovered until several years ago, when sufficiently sensitive tools and high-resolution genome-wide techniques were developed.66 The most abundant and well-characterized RNA modification on mRNA, the m6A (N6-methyladenosine), has attracted considerable attention. M6A modifications have the ability to alter mRNA splicing, cause mRNA decay, and affect translation; thus, they can govern major cellular processes.67 Many high-throughput methods (e.g., m6A-seq, MeRIP-Seq), that rely on immunoprecipitation of methylated RNAs using m6A-recognizing antibodies, have been widely used to globally profile m6A in various cell lines and tissue types. In 2012, Dominissini et al. profiled the transcriptome-wide m6A modification landscape using m6A-seq and identified over 12,000 m6A sites in the transcripts of more than 7000 human genes.68 A recent study revealed that YTHDF1, one of m6A “reader” proteins, regulated translation of EIF3C in an m6A-dependent manner in OC tumorigenesis. These findings suggest the potential role of transcriptome and m6A methylome analysis in OC diagnosis.69
Another of the most abundant and widespread types of RNA epigenetic modifications in living organisms is pseudouridine (Ψ). Pseudouridines are present in all major types of cellular RNAs, including mRNA, rRNA, and tRNA. Recently, high-throughput pseudouridine-seq studies revealed hundreds of pseudouridines sites in 509 different mRNAs. Most of this mRNA pseudouridylation activity is catalyzed by members of the stand-alone tRNA pseudouridine synthase (PUS) family.70 Guzzi et al. revealed that PUS7-mediated pseudouridylation activates a new class of tRNA-derived small RNAs to control protein synthesis and stem cell fate determination, revealing a critical function of Ψ in controlling translation in stem cells with important implications for development and disease.71 By developing a small RNA Ψ sequencing method, we and our collaborators revealed that PUS10 plays a role in both miRNA biogenesis and tRNA pseudouridylation. These findings provided initial evidence that PUS10-catalyzed pseudouridylation may be essential for cell fate determination.72 Consistent with this, Cui et al. reported that the expression and catalytic activity of PUS7 are critical for tumorigenesis.73 Taken together, the above results indicate the potential importance of RNA modification activity for cancer screening and diagnosis.
Proteomics
Since proteins are the primary functional elements of most biological processes, proteins and their post-translational modifications (PTMs) are being studied to provide deeper insights into disease.74 Mass spectrometry (MS)-based proteomics is a sensitive and accurate method for large-scale, unbiased proteomic analysis that enables characterization of nearly complete proteomes. In addition, multiple quantitative proteomics methods have been developed to isolate and quantify proteins, including stable isotope labeling with amino acids in cell culture (SILAC), isotope-coded affinity tag (ICAT), isobaric tags for relative and absolute quantitation (iTRAQ), tandem mass tags (TMT), and label-free methods.75,76 Besides MS, new technologies, such as proximity ligation and extension assays, that enable large-scale targeted protein detection by using a matched pair of DNA-conjugated antibodies, provide a feasible method for identifying low-abundant proteins from limited clinical samples.77, 78, 79
Advances in MS-based proteomics have identified many novel biomarkers, leading to the development of multivariate index assays for OC, such as OVA1, the Risk of Ovarian Malignancy Algorithm (ROMA) and Overa.80, 81, 82, 83 To study the impact of genomic alterations on OC biology at a functional level, the Clinical Proteomic Tumor Analysis Consortium (CPTAC) performed an extensive MS-based proteomic and phosphoproteomic characterization of 174 OC tumor samples available from the TCGA. The findings from this study provided a detailed analysis of the molecular components and underlying mechanisms associated with OC, as well as views on how the somatic genome drives the cancer proteome and the association between protein levels and clinical outcomes in OC.84 Recently, Lee et al. performed multi-platform omics analysis of differences in molecular and cellular features of HGSOC tissue samples from clinically defined subgroups, including the patients with no gross residual disease (R0) after primary surgery and the patients who received neoadjuvant chemotherapy (NACT). They found that the R0 group had a higher rate of NF1 copy number loss, reduced chromothripsis-like patterns, higher levels of strong-binding neoantigens, and a higher number of infiltrated T cells compared with the NACT group.85
PTMs, such as phosphorylation, SUMOylation, acetylation, and other novel modifications, are becoming more appreciated for their diverse roles in the regulation of gene expression, protein structure, and molecular interactions.86 Increasing evidence shows that changes in the PTMs of proteins that are essential for cell survival and proliferation can cause tumorigenesis.87 In addition to the above mentioned PTMs, which have been well studied for decades, glycosylation and poly(ADP-ribosyl)ation (PARylation) have attracted the most attention in tumor biology. In a systematic proteomic and glycoproteomic analysis of 83 HGSOC and 23 non-tumor tissues collected from fallopian tubes, Hu et al. revealed tumor-specific glycosylation, which facilitates the development of diagnostic strategies for OCs.88 It is becoming more interesting for the PTM of PARylation since the polymer modification is highly dynamic and plays multiple roles in DNA damage response (DDR), chromatin remodeling, transcription, and regulation of cell death. In mammals, PARylation is catalyzed by a class of enzymes called poly-ADP-ribose polymerases (PARPs) from the substate of NAD+. One of the 17 members of the PARP family, PARP1 has been shown to synthesize nearly 90% of cellular poly(ADP-ribose) (PAR) in response to DNA damage.89 Upon genotoxic stress, PARP1 is rapidly activated and recruited to DNA lesions, which leads to PARylation of itself and hundreds of other DNA repair proteins, initiating the DDR. Because of its prominent role in maintaining genome integrity, PARP1 has become an important pharmacologic target for therapeutic interventions. The synthetic lethal interaction between BRCA1/2 mutations and PARP inhibition was first observed in 2005,90 and PARP inhibitors including olaparib, rucaparib, and niraparib have been approved by the Federal Drug Administration (FDA) to treat women with BRCA1/2 mutations.91 Because of the labile nature of the ADP-ribose moiety, it has been challenging to identify PAR-associated proteins and unambiguously map PAR acceptor sites. Recent advances in MS-based proteomics and various PAR enrichment strategies have tremendously broadened our knowledge of PAR-associated proteins and offered insight into the molecular mechanisms by which PAR exerts its many biological functions.92 Consistently, scientists revealed a significant reduction of total PAR adducts of peripheral blood lymphocyte proteins in advanced cancers of head & neck, breast, and cervix compared to healthy controls.93 Therefore, knowledge gained from proteomics studies can extend our understanding of cancer-relevant mechanisms and help identify new biomarkers to improve early detection and diagnosis in OC.
Metabolomics
It has been demonstrated that metabolic reprogramming is a hallmark of cancer, and recently there has been growing interest in characterizing the altered metabolism across many types of cancer.94 By analyzing the endogenous and exogenous small molecules that are the substrates and products of metabolic process, scientists have discovered that metabolomics may provide more information about the subtle alterations occurring during various biological processes and diseases.95 Developments in analytical tools such as nuclear magnetic resonance, MS, and ultra-performance liquid chromatography (UPLC) have improved understanding of the metabolome.
Various studies have assessed the efficacy of metabolites in detecting early-stage OC. In 2015, Ke et al. performed a large-scale metabolic investigation of 448 plasma samples related to OCs through the use of a UPLC/MS platform. They identified OC-related metabolic signatures and potential biomarkers that were able to facilitate early detection and could be used to distinguish early and late stages of OC.96 Similarly, plasma metabolic changes were used to differentiate OC from benign ovarian tumors and help boost the accuracy of CA125 for clinical triage.97 Another study investigated the low-concentration metabolites in OC by performing targeted metabolomics and revealed that serum metabolite changes of phospholipids and essential amino acids are associated with specific characteristics and clinical outcomes in OC.98 Since metabolites are highly abundant and known to alter phenotypes in human cells, it is reasonable to expect that recent developments in metabolomics have the potential to improve OC diagnosis.
Artificial intelligence for multi-omics data integration
As the big “omics” data proliferate, questions remain about how to improve and make the best use of the data from various studies. The large amounts of multi-omics data in cancer research are often biologically and computationally heterogeneous, noisy and lack statistical power, making it extremely difficult to gain biological insights from these high-dimensional datasets using traditional data analytical methods. Analysis of datasets generated by multi-omics sequencing requires the development of computational approaches spanning from data integration, statistical methods, and artificial intelligence (AI) systems.
In recent years, the application of AI in preclinical and translational cancer research has increased rapidly due to advances in computer science. Machine learning (ML), a branch of AI, enables robust interrogation of multiple datasets to identify previously undiscovered patterns and relationships in the data.99 In a study by Kawakami et al., researchers developed an OC-specific prediction approach based on AI using multiple markers in peripheral blood and clinical factors for pretreatment estimation of clinical stages, histotypes, surgical outcomes, and prognosis of patients with EOC. They found that ML approach could predict malignant tumors with appreciably high accuracy compared with early reports.100 Using ML-based multi-omics analylsis, Hu et al. integrated the expression data from global proteomics and glycoproteomics of OC and non-tumor tissues, and identified different glycosylations associated with three tumor clusters.88 The integration and analysis of high-throughput molecular assays based on ML techniques promote the understanding of specific variations for the disease and the discovery of further biomarkers. Biomarker candidates based on integrated analysis of big data would be biologically relevant regardless of the changes at each single omics level. Hence, it is believed that the rapidly evolving AI-based analysis will aid precision medicine in OC significantly.
Outstanding questions
Although multi-omics technologies have extensively promoted the discovery of candidate biomarkers during the past few years, gaps between discovery research and clinical application remain. One major reason for the low translation rate of discovery research into clinical practice is the weaknesses of study design, resulting in low statistical power of many studies. To increase the predictive power of potential diagnostic biomarkers, experimental setups need to be carefully designed. For example, the sample size should be calculated based on the sample type, omics technical characteristics, and statistical analysis methods. Also, standardization of sample collection and storage can help reduce biological variability between different studies. Ultimately, further in-depth validation must be done before implementing the findings in routine clinical care. It is vital to perform large-scale studies with robust quality control and appropriately stringent statistical methods. The recent UKCTOCS study highlighted the importance of having OC mortality as the primary outcome in screening trials. Future large trials are supposed to take a long period of time (e.g., a decade) to monitor survival outcomes and answer whether specific screening methods could reduce mortality. The identified biomarkers for diagnosis in OC that have been validated in independent cohorts are presented in Table 2. The multi-omics studies for identifying potential OC biomarkers are listed in Table 3.
Table 2.
Validated biomarkers for diagnosis in ovarian cancer.
qMSP: quantitative methylation-specific real-time PCR; NR: not reported.
| Biomarker/signature | Technology | Sample | No. of OC patients | No. of controls | Sensitivity | specificity | Refs. |
|---|---|---|---|---|---|---|---|
| Methylation within the promoters of 3 genes (c17orf64, IRX2, and TUBB6) | Genome-wide methylation analysis and qMSP assays | Tissue | 23 (HGSOC) | 36 | 100% | 100% | 32 |
| miR-1246, miR-595, miR-2278 | Microarray, RT-qPCR | Serum, tissue | 168 (HGSOC) | 65 | 87% | 77% | 61 |
| 10-miRNA profile (miR-320a, miR-665, miR-3184-5p, miR-6717-5p, miR-4459, miR-6076, miR-3195, miR-1275, miR-3185, and miR-4640-5p) | Microarray | Serum | 428 (OC) | 2759 | 99% | 100% | 62 |
| 18 lncRNAs | Microarray and qPCR | Tissue | 18 (EOC) | 31 | NR | NR | 64 |
| 53 metabolites | UPLC-MS | Plasma | 140 (EOC) | 308 | NR | NR | 96 |
| Four lipid metabolites | LC-MS | Plasma | 50 (Serous OC) | 50 | 95% | 35% | 97 |
Table 3.
Potential ovarian cancer biomarkers identified in multi-omics studies.
FFPE: formalin-fixed paraffin-embedded.
| Type of biomarker | Technology | Sample | Evidence | Refs. |
|---|---|---|---|---|
| DNA | ||||
| Mutation | WES | Tissue | TP53, BRCA1, BRCA2, RB1, NF1, FAT3, CSMD3, GABRA6, CDK12 mutations found in HGSOC tumors | 19 |
| Copy number aberrations | WGS | Tissue | Seven copy number signatures represent distinct mutational processes and provide a rational framework for the diagnosis and assessment in HGSOC | 20 |
| RNA | ||||
| mRNA | Microarray | Tissue | Four expression subtypes (immunoreactive, differentiated, proliferative, and mesenchymal) exist in HGSOC | 19,50,51 |
| mRNA | Taqman-based, fluorescent oligonucleotides, targeted RNA sequencing (Illumina) assays | FFPE | A 39 differentially expressed gene signature for classification of four transcriptional subtypes in HGSOC | 52 |
| mRNA | single-cell RNA sequencing | Cells from ascites | Different functional sub-populations of cancer cells contribute to shaping the HGSOC ecosystem and highly expressed JAK/STAT pathway in both cancer cells and cancer-associated fibroblasts could be an ideal candidate for the diagnosis and treatment of HGSOC | 54 |
| Protein | ||||
| NF1 | LC-MS/MS, RPPA | Tissue | NF1 is significantly lower in abundance in HGSOC patients who underwent complete gross resection (R0) versus neoadjuvant chemotherapy (NACT) groups | 85 |
| Glycosylation | LC-MS/MS | Tissue | Different glycosylation associated with three tumor clusters in HGSOC | 88 |
Liquid biopsies are increasingly applied in the clinical setting for patients with OC. The minimally invasive and rapid nature of liquid biopsy fulfills the needs of large screens in healthy individuals. However, current liquid biopsy assays lack consistency and precision. Future efforts are required to standardize the liquid biopsy assay procedures and analysis platforms, which will enable the comparison and combination of results from different studies. Hopefully, the new generation of liquid biopsy-based screening approaches will contribute towards mortality reduction for OC.
Given the evidence that multivariate assays, such as OVA1 and Overa, demonstrated higher sensitivity than CA125 alone for detecting OC, especially for early-stage disease,80,83 future developments in biomarker discovery will be likely to involve multivariate signatures. Fortunately, dimension reduction methods, such as machine learning techniques, have been proposed for analyzing of multi-omics data, thus enabling the identification of multi-omics signatures that are associated with phenotypes of the disease. These different types of molecular profiles will provide a comprehensive view and accelerate the discovery of biomarker candidates for OC screening and diagnosis.
Search strategy and selection criteria
Data for this review were identified by searches of MEDLINE, Pubmed, and references from relevant articles using the search terms “ovarian cancer”, “Epithelial ovarian cancer”, “omics”, “liquid biopsy”, “ctDNA”, “genomics”, “DNA methylation”, “transcriptomics”, “microRNA”, “long non-coding RNA” “proteomics”, “metabolomics”, “artificial intelligence”, “machine learning”, “screening”, “diagnosis” and “biomarker discovery”. Only articles published in English between Jan 1, 2001 and Sep 1, 2021 were included. The final reference list was generated based on originality and relevance to the scope of this Review.
Contributors
ML conceptualized and supervised the writing of the manuscript. YX and MB performed the literature search and wrote the original draft. YX and ML designed and produced the figures and tables. ML and HG reviewed and edited the final manuscript. All authors have read and approved the final version of the manuscript.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (NSFC) (81672610, 81521002), and the “Clinic + X” program of Peking University to ML; the research fund from the China Postdoctoral Science Foundation (2021M700289) and “Boya” postdoctoral program of Peking University to YX. All the funders did not play any role in paper design, interpretation, or writing of the paper.
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Torre L.A., Trabert B., DeSantis C.E., et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68(4):284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 4.Lheureux S., Gourley C., Vergote I., Oza A.M. Epithelial ovarian cancer. Lancet. 2019;393(10177):1240–1253. doi: 10.1016/S0140-6736(18)32552-2. [DOI] [PubMed] [Google Scholar]
- 5.Matulonis U.A., Sood A.K., Fallowfield L., Howitt B.E., Sehouli J., Karlan B.Y. Ovarian cancer. Nat Rev Dis Prim. 2016;2:16061. doi: 10.1038/nrdp.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hellstrom I., Raycraft J., Hayden-Ledbetter M., et al. The HE4 (WFDC2) protein is a biomarker for ovarian carcinoma. Cancer Res. 2003;63(13):3695–3700. [PubMed] [Google Scholar]
- 7.Jacobs I.J., Menon U., Ryan A., et al. Ovarian cancer screening and mortality in the UK collaborative trial of ovarian cancer screening (UKCTOCS): a randomised controlled trial. Lancet. 2016;387(10022):945–956. doi: 10.1016/S0140-6736(15)01224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menon U., Gentry-Maharaj A., Burnell M., et al. Ovarian cancer population screening and mortality after long-term follow-up in the UK collaborative trial of ovarian cancer screening (UKCTOCS): a randomised controlled trial. Lancet. 2021;397(10290):2182–2193. doi: 10.1016/S0140-6736(21)00731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenthal A.N., Fraser L.S.M., Philpott S., et al. Evidence of Stage shift in women diagnosed with ovarian cancer during phase II of the United Kingdom Familial ovarian cancer screening study. J Clin Oncol. 2017;35(13):1411–1420. doi: 10.1200/JCO.2016.69.9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C., Sun Y.D., Yu G.Y., et al. Integrated omics of metastatic colorectal cancer. Cancer Cell. 2020;38(5):734–747.e9. doi: 10.1016/j.ccell.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Miralles R.M., Petit J., Gine L., Balaguero L. Metastatic cancer spread at the laparoscopic puncture site. Report of a case in a patient with carcinoma of the ovary. Case report. Eur J Gynaecol Oncol. 1989;10(6):442–444. [PubMed] [Google Scholar]
- 12.Kurman R.J. Origin and molecular pathogenesis of ovarian high-grade serous carcinoma. Ann Oncol. 2013;24(Suppl 10):x16–x21. doi: 10.1093/annonc/mdt463. [DOI] [PubMed] [Google Scholar]
- 13.Labidi-Galy S.I., Papp E., Hallberg D., et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat Commun. 2017;8(1):1093. doi: 10.1038/s41467-017-00962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan J.C.M., Massie C., Garcia-Corbacho J., et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 15.Chakravarty D., Solit D.B. Clinical cancer genomic profiling. Nat Rev Genet. 2021;22(8):483–501. doi: 10.1038/s41576-021-00338-8. [DOI] [PubMed] [Google Scholar]
- 16.Beane J., Campbell J.D., Lel J., Vick J., Spira A. Genomic approaches to accelerate cancer interception. Lancet Oncol. 2017;18(8):e494–e502. doi: 10.1016/S1470-2045(17)30373-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherman R.M., Salzberg S.L. Pan-genomics in the human genome era. Nat Rev Genet. 2020;21(4):243–254. doi: 10.1038/s41576-020-0210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The International Cancer Genome Consortium. Hudson T.J., Anderson W., Artez A., et al. International network of cancer genome projects. Nature. 2010;464(7291):993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium ITP-CAoWG Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research N Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macintyre G., Goranova T.E., De Silva D., et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat Genet. 2018;50(9):1262–1270. doi: 10.1038/s41588-018-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y.K., Bashashati A., Anglesio M.S., et al. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat Genet. 2017;49(6):856–865. doi: 10.1038/ng.3849. [DOI] [PubMed] [Google Scholar]
- 23.Roy R., Chun J., Powell S.N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2011;12(1):68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuchenbaecker K.B., Hopper J.L., Barnes D.R., et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317(23):2402–2416. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 25.Bouwman P., van der Gulden H., van der Heijden I., et al. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 2013;3(10):1142–1155. doi: 10.1158/2159-8290.CD-13-0094. [DOI] [PubMed] [Google Scholar]
- 26.Findlay G.M., Daza R.M., Martin B., et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562(7726):217–222. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W., Gu X., Liu C., et al. A synergetic effect of BARD1 mutations on tumorigenesis. Nat Commun. 2021;12(1):1243. doi: 10.1038/s41467-021-21519-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones M.R., Kamara D., Karlan B.Y., Pharoah P.D.P., Gayther S.A. Genetic epidemiology of ovarian cancer and prospects for polygenic risk prediction. Gynecol Oncol. 2017;147(3):705–713. doi: 10.1016/j.ygyno.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Jones P.A., Baylin S.B. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26(4):577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Widschwendter M., Zikan M., Wahl B., et al. The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer. Genome Med. 2017;9(1):116. doi: 10.1186/s13073-017-0500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pisanic T.R., Cope L.M., Lin S.F., et al. Methylomic analysis of ovarian cancers identifies tumor-specific alterations readily detectable in early precursor lesions. Clin Cancer Res. 2018;24(24):6536–6547. doi: 10.1158/1078-0432.CCR-18-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamat A.A., Sood A.K., Dang D., Gershenson D.M., Simpson J.L., Bischoff F.Z. Quantification of total plasma cell-free DNA in ovarian cancer using real-time PCR. Ann N Y Acad Sci. 2006;1075:230–234. doi: 10.1196/annals.1368.031. [DOI] [PubMed] [Google Scholar]
- 34.Diaz L.A., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman A.M., Bratman S.V., To J., et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman A.M., Lovejoy A.F., Klass D.M., et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34(5):547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Douville C., Cohen J.D., Ptak J., et al. Assessing aneuploidy with repetitive element sequencing. Proc Natl Acad Sci U S A. 2020;117(9):4858–4863. doi: 10.1073/pnas.1910041117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Springer S.U., Chen C.H., Rodriguez Pena M.D.C., et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. Elife. 2018;7 doi: 10.7554/eLife.32143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y., Li L., Douville C., et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci Transl Med. 2018;10(433) doi: 10.1126/scitranslmed.aap8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paracchini L., Beltrame L., Grassi T., et al. Genome-wide copy-number alterations in circulating tumor DNA as a novel biomarker for patients with high-grade serous ovarian cancer. Clin Cancer Res. 2021;27(9):2549–2559. doi: 10.1158/1078-0432.CCR-20-3345. [DOI] [PubMed] [Google Scholar]
- 41.Lin K.K., Harrell M.I., Oza A.M., et al. BRCA reversion mutations in circulating tumor DNA predict primary and acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2019;9(2):210–219. doi: 10.1158/2159-8290.CD-18-0715. [DOI] [PubMed] [Google Scholar]
- 42.Oikkonen J., Zhang K., Salminen L., et al. Prospective longitudinal ctDNA workflow reveals clinically actionable alterations in ovarian cancer. JCO Precis Oncol. 2019;3 doi: 10.1200/PO.18.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen J.D., Li L., Wang Y., et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakabayashi M., Kawashima A., Yasuhara R., et al. Massively parallel sequencing of cell-free DNA in plasma for detecting gynaecological tumour-associated copy number alteration. Sci Rep. 2018;8(1):11205. doi: 10.1038/s41598-018-29381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arend R.C., Londono A.I., Montgomery A.M., et al. Molecular response to neoadjuvant chemotherapy in high-grade serous ovarian carcinoma. Mol Cancer Res. 2018;16(5):813–824. doi: 10.1158/1541-7786.MCR-17-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanderstichele A., Busschaert P., Smeets D., et al. Chromosomal instability in cell-free DNA as a highly specific biomarker for detection of ovarian cancer in women with adnexal masses. Clin Cancer Res. 2017;23(9):2223–2231. doi: 10.1158/1078-0432.CCR-16-1078. [DOI] [PubMed] [Google Scholar]
- 47.Christie E.L., Fereday S., Doig K., Pattnaik S., Dawson S.J., Bowtell D.D.L. Reversion of BRCA1/2 germline mutations detected in circulating tumor DNA from patients with high-grade serous ovarian cancer. J Clin Oncol. 2017;35(12):1274–1280. doi: 10.1200/JCO.2016.70.4627. [DOI] [PubMed] [Google Scholar]
- 48.Phallen J., Sausen M., Adleff V., et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9(403) doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu W., Seok J., Mindrinos M.N., et al. Human transcriptome array for high-throughput clinical studies. Proc Natl Acad Sci U S A. 2011;108(9):3707–3712. doi: 10.1073/pnas.1019753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verhaak R.G., Tamayo P., Yang J.Y., et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. 2013;123(1):517–525. doi: 10.1172/JCI65833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konecny G.E., Wang C., Hamidi H., et al. Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J Natl Cancer Inst. 2014;106(10) doi: 10.1093/jnci/dju249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leong H.S., Galletta L., Etemadmoghadam D., et al. Efficient molecular subtype classification of high-grade serous ovarian cancer. J Pathol. 2015;236(3):272–277. doi: 10.1002/path.4536. [DOI] [PubMed] [Google Scholar]
- 53.Stuart T., Satija R. Integrative single-cell analysis. Nat Rev Genet. 2019;20(5):257–272. doi: 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]
- 54.Izar B., Tirosh I., Stover E.H., et al. A single-cell landscape of high-grade serous ovarian cancer. Nat Med. 2020;26(8):1271–1279. doi: 10.1038/s41591-020-0926-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lewis S.M., Asselin-Labat M.L., Nguyen Q., et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods. 2021;18(9):997–1012. doi: 10.1038/s41592-021-01203-6. [DOI] [PubMed] [Google Scholar]
- 56.Longo S.K., Guo M.G., Ji A.L., Khavari P.A. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat Rev Genet. 2021;22(10):627–644. doi: 10.1038/s41576-021-00370-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu S.Z., Al-Eryani G., Roden D.L., et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021;53(9):1334–1347. doi: 10.1038/s41588-021-00911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Djebali S., Davis C.A., Merkel A., et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 61.Todeschini P., Salviato E., Paracchini L., et al. Circulating miRNA landscape identifies miR-1246 as promising diagnostic biomarker in high-grade serous ovarian carcinoma: a validation across two independent cohorts. Cancer Lett. 2017;388:320–327. doi: 10.1016/j.canlet.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 62.Yokoi A., Matsuzaki J., Yamamoto Y., et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat Commun. 2018;9(1):4319. doi: 10.1038/s41467-018-06434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 64.Wang H., Fu Z., Dai C., et al. LncRNAs expression profiling in normal ovary, benign ovarian cyst and malignant epithelial ovarian cancer. Sci Rep. 2016;6:38983. doi: 10.1038/srep38983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Machnicka M.A., Milanowska K., Osman Oglou O., et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013;41(Database issue):D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X., Zhao B.S., Roundtree I.A., et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 69.Liu T., Wei Q., Jin J., et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48(7):3816–3831. doi: 10.1093/nar/gkaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwartz S., Bernstein D.A., Mumbach M.R., et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159(1):148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guzzi N., Ciesla M., Ngoc P.C.T., et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell. 2018;173(5):1204–1216. doi: 10.1016/j.cell.2018.03.008. e26. [DOI] [PubMed] [Google Scholar]
- 72.Song J., Zhuang Y., Zhu C., et al. Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat Chem Biol. 2020;16(2):160–169. doi: 10.1038/s41589-019-0420-5. [DOI] [PubMed] [Google Scholar]
- 73.Cui Q.Y.K., Zhang X., Ye P., et al. Targeting PUS7 suppresses tRNA pseudouridylation and glioblastoma tumorigenesis. Nat Cancer. 2021;2:932–949. doi: 10.1038/s43018-021-00238-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uhlen M., Fagerberg L., Hallstrom B.M., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220) doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 75.Angel T.E., Aryal U.K., Hengel S.M., et al. Mass spectrometry-based proteomics: existing capabilities and future directions. Chem Soc Rev. 2012;41(10):3912–3928. doi: 10.1039/c2cs15331a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang E.H., Combe P.C., Schug K.A. Multiple reaction monitoring for direct quantitation of intact proteins using a triple quadrupole mass spectrometer. J Am Soc Mass Spectrom. 2016;27(5):886–896. doi: 10.1007/s13361-016-1368-2. [DOI] [PubMed] [Google Scholar]
- 77.Fredriksson S., Gullberg M., Jarvius J., et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20(5):473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 78.Oliveira F.M.S., Mereiter S., Lonn P., et al. Detection of post-translational modifications using solid-phase proximity ligation assay. N Biotechnol. 2018;45:51–59. doi: 10.1016/j.nbt.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 79.Shen Q., Polom K., Williams C., et al. A targeted proteomics approach reveals a serum protein signature as diagnostic biomarker for resectable gastric cancer. EBioMedicine. 2019;44:322–333. doi: 10.1016/j.ebiom.2019.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ueland F.R., Desimone C.P., Seamon L.G., et al. Effectiveness of a multivariate index assay in the preoperative assessment of ovarian tumors. Obstet Gynecol. 2011;117(6):1289–1297. doi: 10.1097/AOG.0b013e31821b5118. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Z., Chan D.W. The road from discovery to clinical diagnostics: lessons learned from the first FDA-cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol Biomarkers Prev. 2010;19(12):2995–2999. doi: 10.1158/1055-9965.EPI-10-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moore R.G., Jabre-Raughley M., Brown A.K., et al. Comparison of a novel multiple marker assay vs the risk of malignancy index for the prediction of epithelial ovarian cancer in patients with a pelvic mass. Am J Obstet Gynecol. 2010;203(3):228. doi: 10.1016/j.ajog.2010.03.043. e1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Coleman R.L., Herzog T.J., Chan D.W., et al. Validation of a second-generation multivariate index assay for malignancy risk of adnexal masses. Am J Obstet Gynecol. 2016;215(1):82. doi: 10.1016/j.ajog.2016.03.003. e1- e11. [DOI] [PubMed] [Google Scholar]
- 84.Zhang H., Liu T., Zhang Z., et al. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell. 2016;166(3):755–765. doi: 10.1016/j.cell.2016.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee S., Zhao L., Rojas C., et al. Molecular analysis of clinically defined subsets of high-grade serous ovarian cancer. Cell Rep. 2020;31(2) doi: 10.1016/j.celrep.2020.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Deribe Y.L., Pawson T., Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17(6):666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 87.Chen L., Liu S., Tao Y. Regulating tumor suppressor genes: post-translational modifications. Signal Transduct Target Ther. 2020;5(1):90. doi: 10.1038/s41392-020-0196-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hu Y., Pan J., Shah P., et al. Integrated Proteomic and Glycoproteomic characterization of human high-grade serous ovarian carcinoma. Cell Rep. 2020;33(3) doi: 10.1016/j.celrep.2020.108276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vivelo C.A., Wat R., Agrawal C., Tee H.Y., Leung A.K. ADPriboDB: the database of ADP-ribosylated proteins. Nucleic Acids Res. 2017;45(D1):D204–D2D9. doi: 10.1093/nar/gkw706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Farmer H., McCabe N., Lord C.J., et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 91.Curtin N.J., Szabo C. Poly(ADP-ribose) polymerase inhibition: past, present and future. Nat Rev Drug Discov. 2020;19(10):711–736. doi: 10.1038/s41573-020-0076-6. [DOI] [PubMed] [Google Scholar]
- 92.Daniels C.M., Ong S.E., Leung A.K. The promise of proteomics for the study of ADP-ribosylation. Mol Cell. 2015;58(6):911–924. doi: 10.1016/j.molcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lakadong R.O., Kataki A.C., Sharan R.N. ADP-ribose polymer–a novel and general biomarker of human cancers of head & neck, breast, and cervix. Mol Cancer. 2010;9:286. doi: 10.1186/1476-4598-9-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 95.Vander Heiden M.G., DeBerardinis R.J. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168(4):657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ke C., Hou Y., Zhang H., et al. Large-scale profiling of metabolic dysregulation in ovarian cancer. Int J Cancer. 2015;136(3):516–526. doi: 10.1002/ijc.29010. [DOI] [PubMed] [Google Scholar]
- 97.Buas M.F., Gu H., Djukovic D., et al. Identification of novel candidate plasma metabolite biomarkers for distinguishing serous ovarian carcinoma and benign serous ovarian tumors. Gynecol Oncol. 2016;140(1):138–144. doi: 10.1016/j.ygyno.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bachmayr-Heyda A., Aust S., Auer K., et al. Integrative systemic and local metabolomics with impact on survival in high-grade serous ovarian cancer. Clin Cancer Res. 2017;23(8):2081–2092. doi: 10.1158/1078-0432.CCR-16-1647. [DOI] [PubMed] [Google Scholar]
- 99.Yu K.H., Beam A.L., Kohane I.S. Artificial intelligence in healthcare. Nat Biomed Eng. 2018;2(10):719–731. doi: 10.1038/s41551-018-0305-z. [DOI] [PubMed] [Google Scholar]
- 100.Kawakami E., Tabata J., Yanaihara N., et al. Application of artificial intelligence for preoperative diagnostic and prognostic prediction in epithelial ovarian cancer based on blood biomarkers. Clin Cancer Res. 2019;25(10):3006–3015. doi: 10.1158/1078-0432.CCR-18-3378. [DOI] [PubMed] [Google Scholar]