Abstract
Background
Esophageal squamous cell carcinoma (ESCC) is a typical Gastro-Intestinal (GI) tract neoplasm. This study was conducted to know the Differential Expressed Genes (DEGs) profile of ESCC along with hub gene screening, lncRNA identification, and drug-genes interactions.
Methods
GSE161533, GSE20347, GSE45670 microarray datasets were retrieved from the NCBI Gene Expression Omnibus (GEO) database. GEO2R was used for the DEGs identification, whereas GO (Gene Ontology) and KEGG enrichment analysis were performed in DAVID. PPI network constructed using STRING and visualized with Cytoscape app with the help of MCODE. The top ten connectivity genes were selected as hub genes—further survival analysis was performed in the Kaplan-Meier plotter. Moreover, Boxplot, pathological stage plots were constructed using GEPIA (Gene Expression Profiling Interactive Analysis). The methylation heatmap assembled in the DiseaseMeth version 2.0. lncRNA (Long non-coding RNA) was identified comparing the list of genes in HUGO, and Gene-drug interactions were accumulated from the DgiDB platform.
Results
This experiment showed 16 upregulated, and 59 downregulated DEGs shared among the three datasets. Biological process analysis showed significant terms such as extracellular matrix disassembly and collagen catabolism. The extracellular region was detected as the most crucial cellular compartment. Notably, metalloen dopeptidease and serine-type endopeptidase activity showed significant molecular functions term. In contrast, transcriptional misregulation was a highly substantial KEGG pathway. Kaplan-Meier plotter showed higher expression of CXCL8, SPP1, MMP13, CXCL1, and TOP2A have a significant impact on the overall survival of the patients. Nine out of ten hub genes have significantly different expression levels than normal and cancer tissues. HYMAI was the only lncRNA commonly expressed upregulated among the three datasets. Drug-gene interaction showed multiple genes have no drug options exist till now.
Keywords: ESCC, Hub genes, DEGs, lncRNA
Highlights
-
•
GSE161533, GSE20347, and GSE45670 microarray datasets were analyzed.
-
•
16 upregulated and 59 downregulated DEGs shared among the three datasets.
-
•
CXCL8, SPP1, MMP13, CXCL1, and TOP2A have a significant impact on survival.
-
•
HYMAI was the only lncRNA commonly expressed.
-
•
Multiple genes have no drug options that exist.
1. Introduction
Cancer is the cause of a significant number of mortality worldwide. Cancer can be defined as uncontrolled cell growth, and almost every tissue can be affected by this disease [2]. Esophageal Squamous Cell Carcinoma (ESCC) is considered one of the most common gastrointestinal (GI) neoplasms worldwide [1]. In most cases of ESCC, symptoms did not show early, resulting in a higher death rate due to limited treatment regimens in the late stages [11]. Several risk factors were identified for ESCC, including alcohol consumption, tobacco products consumption, smoking, lower fiber intake, etc. Upper Body Mass Index (BMI) and micronutrient deficiency were also hazardous [4].
Due to the present day's advancement of genomic techniques such as microarray analysis and high throughput analysis, genes associated with ESCC are now a topic of interest to discover the specific genes with their expression correlation to the tumor [14]. Differential gene expression and their related activity are the fundamental way to understand the mechanism of disease advancement. Many genes and their co-expression were regularly identified for the progression of ESCC worldwide. Some of the genes showed significant expression results in corresponding survival analyses. CDK1 and TOP2A were analyzed as the critical genes for ESCC neoplasm by Yang and his group (W. [35]. Whereas CDCA5 was considered the crucial gene for the prognosis of ESCC by another author [33]. CFLAR, LAMA5, ITGA6, ITGB4, and SDC4 genes were also validated for ESCC progression (L. [36]. Whereas, there were also few Long Non-coding RNA (lncRNA) identified, which might impact ESCC pathogenesis, and the BANCR gene was identified by another author [26]. HCG22 was also detected as a lncRNA by a previous author (X. [15].
Many publicly available microarray and high throughput genomic data are available, but there is a lack of bioinformatics analysis and correlation of the disease occurrence. That analysis can quickly identify the potential genes associated with cancer or tumor. The following study used three microarray datasets to place the common differential gene expression with functional identification of Esophageal Squamous Cell Carcinoma (ESCC) hub genes.
2. Materials and Method
Data source: Three microarray datasets (GSE161533, GSE20347, and GSE45670) were collected from Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). 28 normal tissue and 28 tumor tissue of esophageal squamous cell carcinoma data were from the GSE161533 dataset. Besides, the GSE20347 dataset contributed 17 samples for both the normal and tumor tissue. At the same time, the GSE45670 dataset has ten normal samples with 28 ESCC samples.
Differential gene expression: GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) was used for the differential gene expression analysis (J. [16]. All the statistical values were auto-selected by the GEO. For the selection of differentially expressed, the genes should qualify the cutoff criteria of Log Fold Change (|logFC|) ≥ 1.5 and the adj. p-value ≤ 0.05. All datasets were analyzed with identical selection criteria. HemI 2.0 - Heatmap Illustrator 2.0 software (http://hemi.biocuckoo.cn:81/) was used to compare the three datasets' expression values [7].
Annotation of DEGs: DAVID platform (https://david.ncifcrf.gov/) was used to annotate the 75 common, regulated DEGs [18]. Upregulated and downregulated genes GO analysis were taken into account, with the p-value less than 0.01 considered significant. Biological process (BP), Cellular Compartment (CC), and Molecular Function (MF) were analyzed. At the same time, KEGG pathway analysis was selected with similar p-values.
Protein-protein interaction (PPI) network of DEGs: STRING (https://string-db.org/) is the online bioinformatics tool to ascertain the hub gene and examine the interactions between the genes [37]. The interaction score and the maximum number of interactions were >0.4 and 10, respectively.
Selection of the hub genes: The highest connectivity of the correlated genes was calculated from the Cytoscape software (https://cytoscape.org/), an available bioinformatic analytical tool. MCODE app used for the selection of hub gene with following criteria- MCODE score >5, degree cut off = 2, node score cut off = 0.2, Max depth = 100, k-score = 2 [41]. The top ten connected genes were considered as hub genes for this study.
Survival analysis: Kaplan-Meier survival analysis was performed in the online analytical tool (https://kmplot.com/analysis/) [28]. Overall survival analysis was performed according to the collected gastric cancer database of the website.
Box plot and pathological stage plot expression comparison: The GEPIA (http://gepia.cancer-pku.cn/index.html) tool was used to construct and compare the boxplots. Overall survival method was taken into consideration along with median group cutoff (50%), Hazards Ratio (HR), and 95% confidence interval [17]. |LogFC| Cutoff value was 1.5. Hub gene pathological stage plot was also constructed to compare the stages of the diseases.
Disease methylation: Normal and diseased methylation data compared in the DiseaseMeth Version 2 (http://bio-bigdata.hrbmu.edu.cn/diseasemeth/index.html) [32]. All default criteria were selected for the analysis.
lncRNA identification: Differentially expressed lncRNA identified compared to the approved lncRNA list from the HUGO database (https://www.genenames.org/) [25]. |LogFC| > 0.5 and adj p-value less than 0.05 were considered as significant during the identification.
Drugs and genes interactions: The selected 75 genes were analyzed according to their interactions with the currently available and approved drugs. DGIdb (https://www.dgidb.org/) is an open, accessible public repository for the identification of drugs and genes interactions [9]. The exchanges were visualized through Cytoscape software.
3. Results
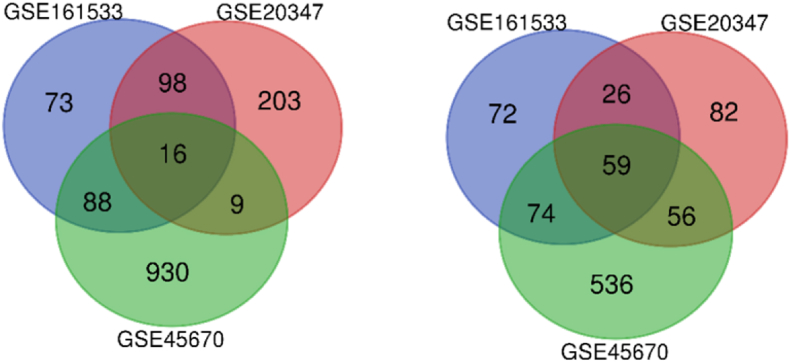
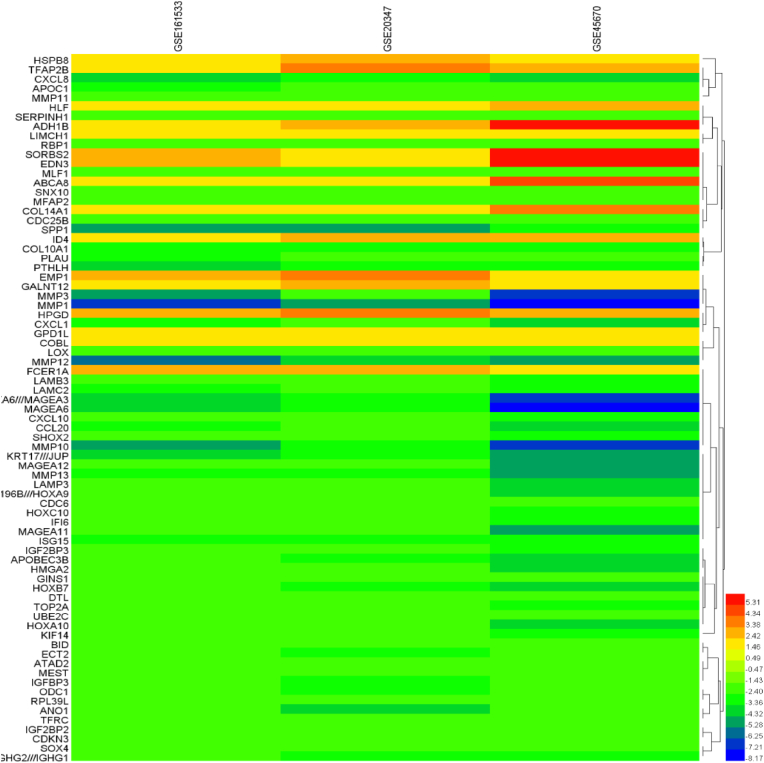
Identification of the DEGs: The number of upregulated and downregulated genes among the three datasets were shown in the Venn diagram (Fig. 1A and B). There were 506, 549, and 1768 significant DEGs for the GSE161533, GSE20347, and GSE45670 datasets with 16 upregulated and 59 downregulated genes in common. Fig. 2 depicts the comparative values of the three datasets. All the gene names are given in Table 1.
Fig. 1.
DEGs among the three datasets (Left - Upregulated; Right - Downregulated).
Fig. 2.
Gene expression values among the three datasets.
Table 1.
Names of the common DEGs from the datasets.
| Upregulated: HSPB8, TFAP2B, HLF, SORBS2, EMP1, ABCA8, EDN3, FCER1A, GALNT12, HPGD, GPD1L, ID4, LIMCH1, COL14A1, COBL, ADH1B |
|---|
|
Downregulated: BID, PLAU, KRT17///JUP, MMP13, IGF2BP3, IGF2BP2, MMP3 SERPINH1, SOX4, HOXA10-HOXA9///MIR196B///HOXA9, CDC25B, GINS1, IFI6, ATAD2, MAGEA6///MAGEA3, UBE2C, LAMP3, PTHLH, HOXC10, MLF1, TOP2A, MAGEA6, CXCL1, CDC6, MFAP2, MIR8071-2///MIR8071-1///IGHV4-31///IGHM///IGHG2///IGHG1, APOBEC3B, SNX10, CXCL8, KIF14, ODC1, CXCL10, LAMB3, DTL, ANO1, LOX, MEST, LAMC2, MAGEA11, HOXA10, MAGEA12, CCL20, SPP1, COL10A1, SHOX2, IGFBP3, TFRC, HOXB7, RBP1, MMP10, CDKN3, MMP1, APOC1, MMP12, HMGA2, ISG15, ECT2, RPL39L, MMP11 |
GO, and KEGG enrichment analysis: Common shared differentially expressed genes; DAVID analysis showed significant terms in Table 2.
Table 2.
GO and KEGG analysis for the DEGs.
| Category | Term | P-Value | Genes |
|---|---|---|---|
| GOTERM_BP_DIRECT | GO:0022617∼extracellular matrix disassembly | 6.35E-10 | MMP12, MMP11, MMP13, LAMB3, MMP1, MMP3, SPP1, LAMC2, MMP10 |
| GOTERM_BP_DIRECT | GO:0030574∼collagen catabolic process | 2.01E-07 | MMP12, MMP11, MMP13, MMP1, MMP3, COL10A1, MMP10 |
| GOTERM_BP_DIRECT | GO:0008284∼positive regulation of cell proliferation | 1.48E-05 | CXCL10, TFAP2B, EDN3, ODC1, ID4, KIF14, LAMC2, PTHLH, SOX4, HOXC10, CDC25B |
| GOTERM_BP_DIRECT | GO:0030198∼extracellular matrix organization | 1.29E-04 | LAMB3, LOX, COL14A1, MFAP2, SPP1, COL10A1, LAMC2 |
| GOTERM_BP_DIRECT | GO:0001501∼skeletal system development | 2.16E-04 | HOXA10, SHOX2, COL10A1, PTHLH, SOX4, HOXC10 |
| GOTERM_BP_DIRECT | GO:0032467∼positive regulation of cytokinesis | 4.28E-04 | KIF14, CDC6, ECT2, CDC25B |
| GOTERM_BP_DIRECT | GO:0030199∼collagen fibril organization | 5.01E-04 | MMP11, LOX, COL14A1, SERPINH1 |
| GOTERM_BP_DIRECT | GO:0008285∼negative regulation of cell proliferation | 9.90E-04 | TFAP2B, CXCL8, IGFBP3, CXCL1, CDC6, PTHLH, SOX4, CDKN3 |
| GOTERM_BP_DIRECT | GO:0032461∼positive regulation of protein oligomerization | 0.001384 | MMP1, MMP3, BID |
| GOTERM_BP_DIRECT | GO:0006935∼chemotaxis | 0.001428 | CXCL10, CXCL8, PLAU, CCL20, CXCL1 |
| GOTERM_BP_DIRECT | GO:0070098∼chemokine-mediated signaling pathway | 0.002859 | CXCL10, CXCL8, CCL20, CXCL1 |
| GOTERM_BP_DIRECT | GO:0007275∼multicellular organism development | 0.004632 | HOXA10, MMP11, ANO1, HLF, EDN3, HMGA2, EMP1, HOXB7 |
| GOTERM_BP_DIRECT | GO:0008544∼epidermis development | 0.004749 | LAMB3, EMP1, LAMC2, PTHLH |
| GOTERM_BP_DIRECT | GO:0043065∼positive regulation of apoptotic process | 0.006892 | TOP2A, IGFBP3, HMGA2, ECT2, BID, SOX4 |
| GOTERM_BP_DIRECT | GO:0042769∼DNA damage response, detection of DNA damage | 0.009053 | HMGA2, DTL, SOX4 |
| GOTERM_BP_DIRECT | GO:0006508∼proteolysis | 0.014588 | MMP12, MMP11, MMP13, PLAU, MMP1, MMP3, MMP10 |
| GOTERM_BP_DIRECT | GO:0045944∼positive regulation of transcription from RNA polymerase II promoter | 0.015501 | TOP2A, HOXA10, CXCL10, TFAP2B, HLF, ATAD2, SHOX2, ID4, HMGA2, SOX4 |
| GOTERM_BP_DIRECT | GO:0097070∼ductus arteriosus closure | 0.019794 | TFAP2B, HPGD |
| GOTERM_BP_DIRECT | GO:0042035∼regulation of cytokine biosynthetic process | 0.027603 | IGF2BP3, IGF2BP2 |
| GOTERM_BP_DIRECT | GO:0060326∼cell chemotaxis | 0.02776 | CXCL10, CCL20, CXCL1 |
| GOTERM_BP_DIRECT | GO:0030593∼neutrophil chemotaxis | 0.028555 | CXCL8, EDN3, CCL20 |
| GOTERM_BP_DIRECT | GO:0043154∼negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | 0.030994 | TFAP2B, LAMP3, IFI6 |
| GOTERM_BP_DIRECT | GO:0045444∼fat cell differentiation | 0.03437 | TFAP2B, ID4, HMGA2 |
| GOTERM_BP_DIRECT | GO:0032330∼regulation of chondrocyte differentiation | 0.03535 | SHOX2, PTHLH |
| GOTERM_BP_DIRECT | GO:0030071∼regulation of mitotic metaphase/anaphase transition | 0.03535 | UBE2C, CDC6 |
| GOTERM_BP_DIRECT | GO:0042127∼regulation of cell proliferation | 0.037708 | CXCL10, TFRC, PLAU, BID |
| GOTERM_BP_DIRECT | GO:2000406∼positive regulation of T cell migration | 0.039201 | CXCL10, CCL20 |
| GOTERM_BP_DIRECT | GO:0001558∼regulation of cell growth | 0.040602 | TFRC, IGFBP3, KIF14 |
| GOTERM_BP_DIRECT | GO:0009952∼anterior/posterior pattern specification | 0.040602 | HOXA10, HOXB7, HOXC10 |
| GOTERM_BP_DIRECT | GO:0043085∼positive regulation of catalytic activity | 0.041525 | IGFBP3, APOC1, CXCL1 |
| GOTERM_BP_DIRECT | GO:0031581∼hemidesmosome assembly | 0.046858 | LAMB3, LAMC2 |
| GOTERM_BP_DIRECT | GO:0021846∼cell proliferation in forebrain | 0.046858 | KIF14, HMGA2 |
| GOTERM_CC_DIRECT | GO:0005576∼extracellular region | 5.78E-09 | CXCL8, LAMB3, EDN3, TFRC, COL14A1, MMP1, CCL20, IGFBP3, MMP3, ISG15, CXCL1, LAMC2, PTHLH, MMP10, MMP12, MMP11, CXCL10, MMP13, LOX, PLAU, MFAP2, APOC1, SPP1, COL10A1 |
| GOTERM_CC_DIRECT | GO:0005578∼proteinaceous extracellular matrix | 6.07E-06 | MMP12, MMP11, MMP13, LOX, MMP1, COL14A1, MMP3, COL10A1, MMP10 |
| GOTERM_CC_DIRECT | GO:0005615∼extracellular space | 2.21E-05 | CXCL8, EDN3, TFRC, COL14A1, CCL20, IGFBP3, MMP3, CXCL1, LAMC2, PTHLH, MMP10, CXCL10, MMP13, LOX, PLAU, SERPINH1, SPP1 |
| GOTERM_CC_DIRECT | GO:0005581∼collagen trimer | 2.22E-05 | MMP13, LOX, MMP1, COL14A1, SERPINH1, COL10A1 |
| GOTERM_CC_DIRECT | GO:0048471∼perinuclear region of cytoplasm | 0.007649 | TFRC, LAMP3, ODC1, SPP1, COBL, SORBS2, LAMC2, CDKN3 |
| GOTERM_CC_DIRECT | GO:0031012∼extracellular matrix | 0.023505 | MMP11, MMP13, MMP1, COL14A1, MMP10 |
| GOTERM_CC_DIRECT | GO:0005654∼nucleoplasm | 0.042373 | GINS1, TOP2A, HPGD, UBE2C, HSPB8, ATAD2, HMGA2, ISG15, CDC6, PTHLH, HOXC10, CDC25B, RBP1, MAGEA11, HOXB7, DTL, SOX4 |
| GOTERM_MF_DIRECT | GO:0004222∼metalloendopeptidase activity | 8.53E-05 | MMP12, MMP11, MMP13, MMP1, MMP3, MMP10 |
| GOTERM_MF_DIRECT | GO:0004252∼serine-type endopeptidase activity | 5.17E-04 | MMP12, MMP11, MMP13, PLAU, MMP1, MMP3, MMP10 |
| GOTERM_MF_DIRECT | GO:0008009∼chemokine activity | 9.66E-04 | CXCL10, CXCL8, CCL20, CXCL1 |
| GOTERM_MF_DIRECT | GO:0001077∼transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | 0.014362 | HOXA10, TFAP2B, HLF, HMGA2, SOX4 |
| GOTERM_MF_DIRECT | GO:0004175∼endopeptidase activity | 0.019447 | MMP12, MMP1, MMP3 |
| GOTERM_MF_DIRECT | GO:0045182∼translation regulator activity | 0.023582 | IGF2BP3, IGF2BP2 |
| GOTERM_MF_DIRECT | GO:0005518∼collagen binding | 0.023691 | MMP13, COL14A1, SERPINH1 |
| GOTERM_MF_DIRECT | GO:0048027∼mRNA 5'-UTR binding | 0.035167 | IGF2BP3, IGF2BP2 |
| KEGG_PATHWAY | hsa05202: Transcriptional misregulation in cancer | 5.57E-06 | HOXA10, CXCL8, HPGD, PLAU, IGFBP3, MMP3, HMGA2, MLF1 |
| KEGG_PATHWAY | hsa05323: Rheumatoid arthritis | 0.0064 | CXCL8, MMP1, CCL20, MMP3 |
| KEGG_PATHWAY | hsa04668: TNF signaling pathway | 0.01095 | CXCL10, CCL20, MMP3, CXCL1 |
| KEGG_PATHWAY | hsa04622: RIG-I-like receptor signaling pathway | 0.036968 | CXCL10, CXCL8, ISG15 |
| KEGG_PATHWAY | hsa04062: Chemokine signaling pathway | 0.046265 | CXCL10, CXCL8, CCL20, CXCL1 |
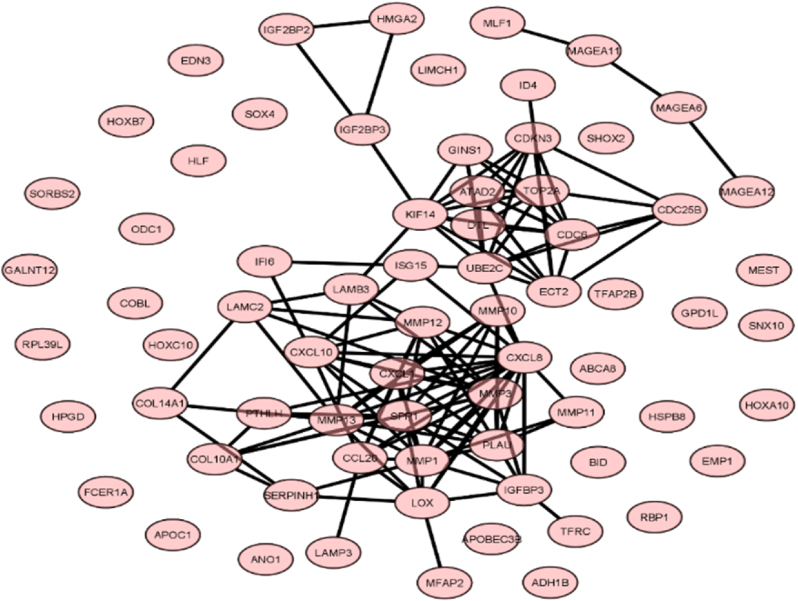
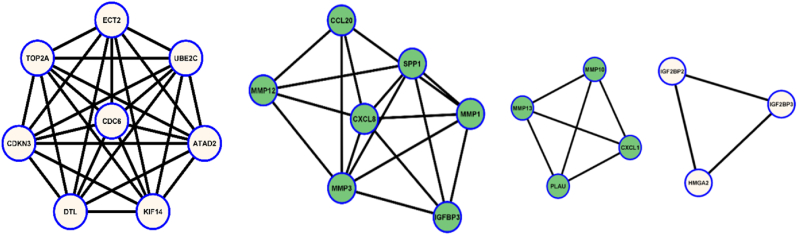
PPI network analysis and hub gene screening: 91 nodes and 120 edges were analyzed during the investigation. Genes having more than or equal ten connectivity were considered as hub genes. The selected gene list is given in Table 3. Fig. 3A, Fig. 3BA-3E showed the entire gene network and subsequent clusters.
Table 3.
Hub genes list with corresponding connectivity.
| Gene Name | Connectivity |
|---|---|
| CXCL8 | 16 |
| MMP3 | 14 |
| MMP1 | 13 |
| SPP1 | 13 |
| MMP13 | 12 |
| UBE2C | 11 |
| PLAU | 10 |
| CXCL1 | 10 |
| KIF14 | 10 |
| TOP2A | 10 |
Fig. 3A.
Overall PPI network of the common upregulated and downregulated genes.
Fig. 3B.
3D: Four cluster of genes from the overall networks after analysis (3B- Left; 3C- Middle left; 3D- Middle right; 3D- Right).
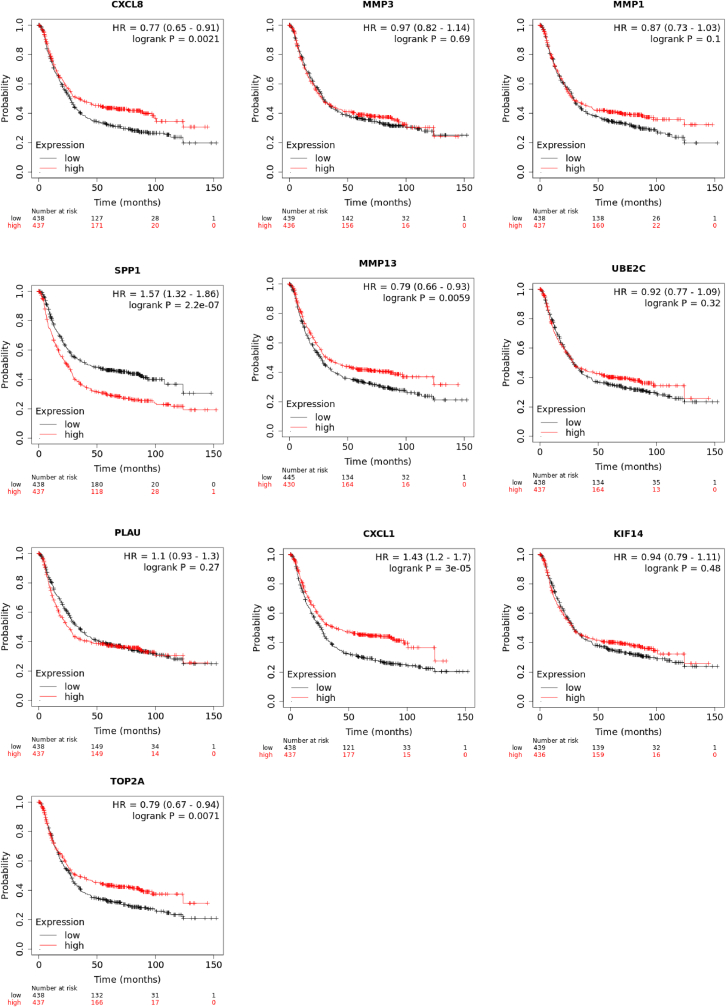
Survival analysis of the hub genes: Kaplan-Meier survival plot analysis showed the hub genes significantly affect the Overall Survival (OS) of the patient affected with gastric cancer (https://kmplot.com/analysis/). CXCL8, SPP1, MMP13, CXCL1, TOP2A were genes that significantly impacted the survivability of the patients (Fig. 4).
Fig. 4.
Overall survival analysis of the hub genes.
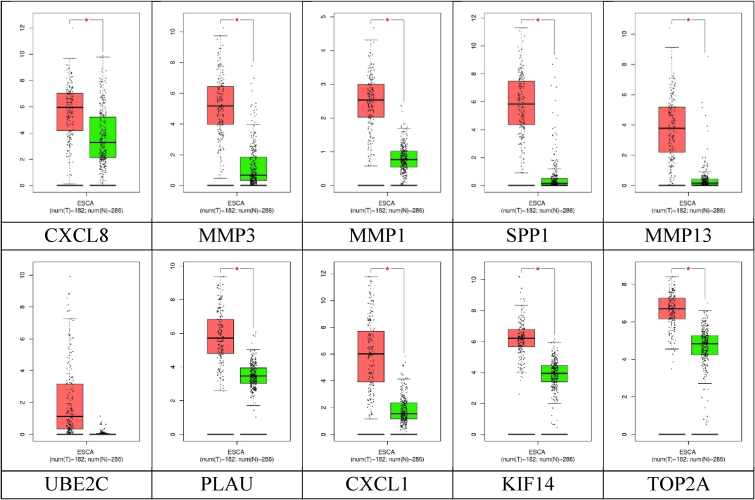
Expression plots of the hub genes: Box plots showed all the genes had significant expression levels compared to the standard and cancer tissues, except UBE2C (Fig. 5).
Fig. 5.
Boxplots of the hub genes (* mark showed significant difference).
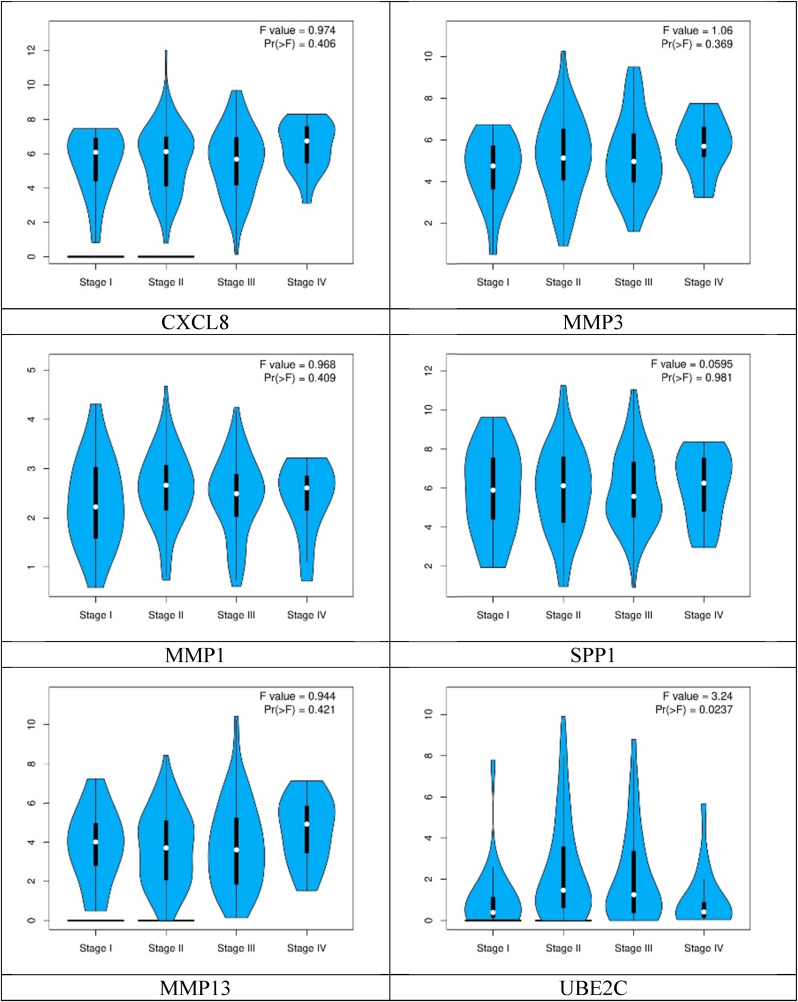
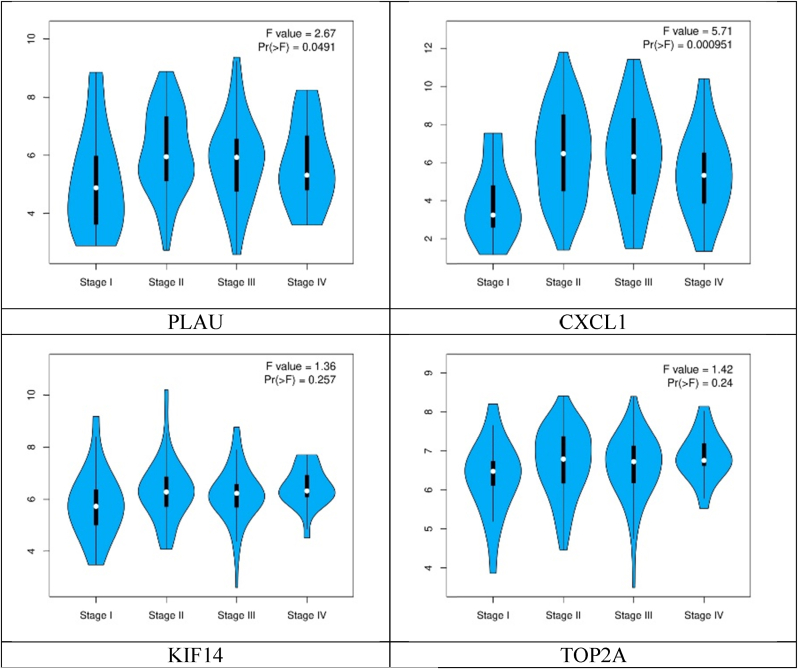
Whereas pathological stage plot analysis of the hub genes showed different expression levels according to their stages (Fig. 6).
Fig. 6.
Pathological stage plots of the hub genes.
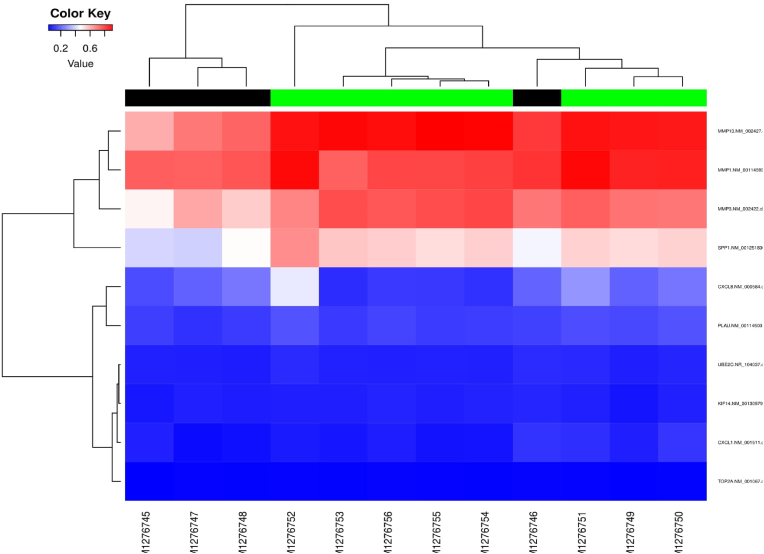
DNA methylation: The resulting heatmap contains methylation data of 8 transcripts from 324 samples of 450k. In the heatmap, rows represent transcripts, and columns represent samples (green color represents standard profiles, black represents disease profiles) (Fig. 7).
Fig. 7.
DNA methylation profile of the hub genes.
lncRNA identification: Three datasets shared only one lncRNA during the screening procedure. Fig. 8 illustrates the lncRNA numbers among the three studies datasets as a Venn diagram. All the names are given in Table 4.
Fig. 8.
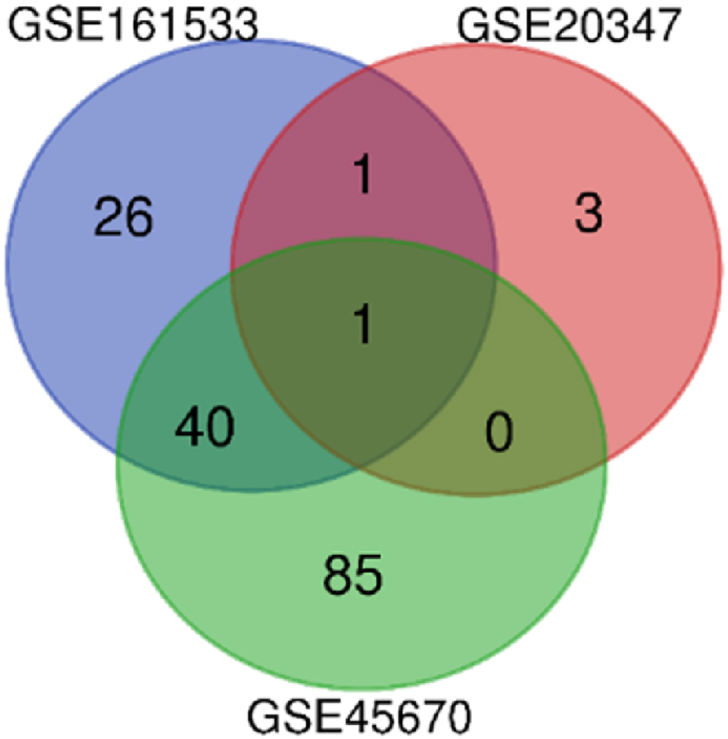
lncRNA among the three sets of GEO data.
Table 4.
lncRNA names of the datasets.
| Gene Sets | Common lncRNA | Names |
|---|---|---|
|
GSE161533 GSE20347 GSE45670 |
1 | HYMAI |
|
GSE161533 GSE20347 |
1 | SNHG17 |
|
GSE161533 GSE45670 |
40 | TMPO-AS1, PGM5-AS1, BBOX1-AS1, PSMD6-AS2, KCNMB2-AS1, ZFAS1, HCG11, LINC00702, LCAL1, LINC00491, NR2F2-AS1, SLC8A1-AS1, CARMN, ADAMTS9-AS1, LINC01315, MIR100HG, MIAT, PWAR6, LINC00467, PCBP1-AS1, ZNF667-AS1, POU6F2-AS2, CADM3-AS1, LINC00240, FENDRR, TP73-AS1, WDFY3-AS2, LINC01082, LINC00472, MAGI2-AS3, ZNF790-AS1, HLA-F-AS1, ADAMTS9-AS2, ATP2A1-AS1, LINC00938, LINC01140, PDZRN3-AS1, KLF3-AS1, HAGLROS, LINC00844 |
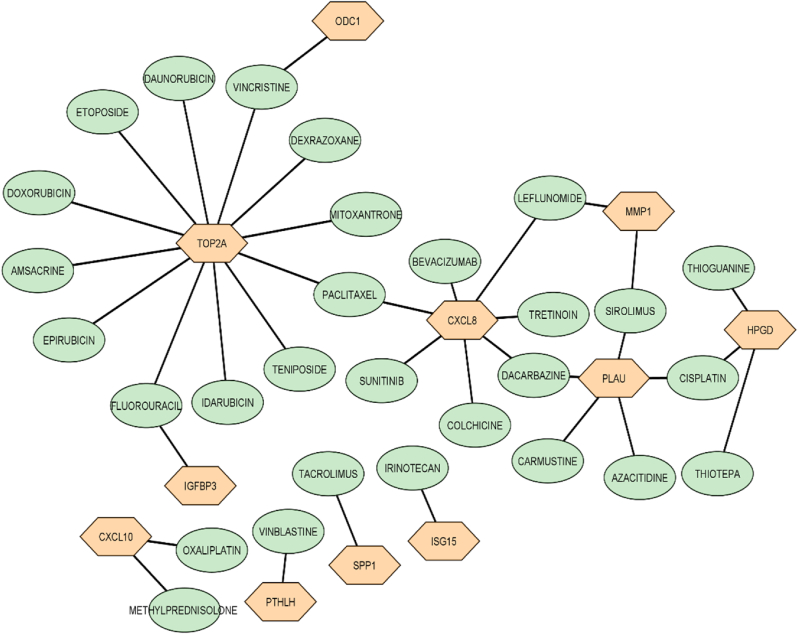
Drug-genes interactions: Interaction showed that not every DEG has specific drugs to choose from in the current therapeutic practice. According to the findings of this study, very few genes have targeted medication (Fig. 9).
Fig. 9.
Interaction between drugs and genes (Oval shapes represent drugs, Hexagonal shapes represent genes).
4. Discussion
GO enrichment analysis of this study found several terms associated with this neoplasm. This study detects extracellular matrix disassembly as the most significant biological process related to ESCC approved by the previously published article [24]. Collagen catabolic process, positive regulation of cell proliferation, extracellular matrix organizations were also detected as important biological process terms reported by previous authors [10,19,20,27].
The most important cellular component identified was the extracellular region aligned with the findings of the previous studies [29,31]. According to published articles, some other terms on the same functional dysregulation were proteinaceous extracellular matrix, extracellular matrix, collagen trimer, etc. that significantly impacted the ESCC oncogenesis [22,40]. Apart from the metallo endopeptidase activity, several other molecular functions, including chemokine activity, transcriptional activator activity, translation regulatory activity, were included in this study. Previous studies reported similar KEGG pathways linked to the ESCC occurrence [3,13,30].
This study identified five pathways for ESCC pathogenesis. Transcriptional misregulation in cancer was the most significant pathway, a similar route acknowledged by previous authors [6,12,23,39].
PPI networks showed four gene clusters in this study. CXCL8 was the highest corresponding gene which might have a crucial effect on ESCC metastasis. Previous authors detected this gene as the primary cause for ESCC occurrence [5,42]. MMP3, MMP1, and MMP13 gene expression related to cancer occurrence is explained by previous studies [8,38]. TOP2A and UBE2C were also detected by the published articles for cancer progression [21,34].
lncRNA identified in this study was not identified by the previous studies for ESCC occurrence. However, no previous reports about the HYMAI involvements, but all the three datasets shared this lncRNA. This might have an impact on the ESCC progression in a novel way. Several lncRNA identified were shared between 2 datasets, which might have a more significant effect on ESCC.
Drug interactions indicated that all the significant genes targeted therapy were unavailable until now. There needed much more research to precisely target the genes that cause the ESCC.
Ethical approval
This study has no invasive methods involvement regarding ethical approval requirements.
Declaration of competing interest
There is no conflict of interest about the article.
Acknowledgment
The author would like to acknowledge Zhaojun Ding and COSTA DA SILVA Raniere for the manuscript's critical revising and language editing.
Data availability
No data was used for the research described in the article.
References
- 1.Abnet C.C., Arnold M., Wei W.Q. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2018;154:360–373. doi: 10.1053/J.GASTRO.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous What is cancer? - National cancer Institute [WWW document] 2021. https://www.cancer.gov/about-cancer/understanding/what-is-cancer 1.8.22.
- 3.Bhat A.A., Nisar S., Maacha S., Carneiro-Lobo T.C., Akhtar S., Siveen K.S., Wani N.A., Rizwan A., Bagga P., Singh M., Reddy R., Uddin S., Grivel J.C., Chand G., Frenneaux M.P., Siddiqi M.A., Bedognetti D., El-Rifai W., Macha M.A., Haris M. Cytokine-chemokine network driven metastasis in esophageal cancer; promising avenue for targeted therapy. Mol. Cancer. 2021:1–20. doi: 10.1186/S12943-020-01294-3. 2020 201 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chetwood J.D., Garg P., Finch P., Gordon M. Vol. 13. 2018. pp. 71–88. (Systematic review: the etiology of esophageal squamous cell carcinoma in low-income settings). [DOI] [PubMed] [Google Scholar]
- 5.Cui K., Hu S., Mei X., Cheng M. Innate immune cells in the esophageal tumor microenvironment. Front. Immunol. 2021;12 doi: 10.3389/FIMMU.2021.654731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang T., Chang Z., Meng J., Cui X., Wang P., Chai J. TNF antagonizes CCN1 in apoptosis in esophageal adenocarcinoma. Cytokine. 2022;149:155728. doi: 10.1016/J.CYTO.2021.155728. [DOI] [PubMed] [Google Scholar]
- 7.Deng W., Wang Y., Liu Z., Cheng H., Xue Y. HemI: a toolkit for illustrating heatmaps. PLoS One. 2014;9 doi: 10.1371/JOURNAL.PONE.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di C., Cheung A.L. Roles of microRNAs in tumorigenesis and metastasis of esophageal squamous cell carcinoma. World J. Clin. Oncol. 2021;12:609. doi: 10.5306/WJCO.V12.I8.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freshour S.L., Kiwala S., Cotto K.C., Coffman A.C., McMichael J.F., Song J.J., Griffith M., Griffith O.L., Wagner A.H. Integration of the drug–gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49:D1144–D1151. doi: 10.1093/NAR/GKAA1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goda K., Murao T., Handa Y., Katsumata R., Fukushima S., Nakato R., Osawa M., Ishii M., Fujita M., Handa O., Matsumoto H., Fujita Y., Nishio K., Wallace T.M., Gomez-Esquivel R., Berzosa M., Wolfsen H.C., Wallace M.B., Umegaki E., Shiotani A. Molecular biomarker identification for esophageal adenocarcinoma using endoscopic brushing and magnified endoscopy. Esophagus. 2021;18:306–314. doi: 10.1007/S10388-020-00762-5/FIGURES/5. [DOI] [PubMed] [Google Scholar]
- 11.Hirano H., Kato K. Systemic treatment of advanced esophageal squamous cell carcinoma: chemotherapy, molecular-targeting therapy and immunotherapy. Jpn. J. Clin. Oncol. 2019;49:412–420. doi: 10.1093/JJCO/HYZ034. [DOI] [PubMed] [Google Scholar]
- 12.Hou Q., Jiang Z., Li Z., Jiang M. Identification and functional validation of radioresistance-related genes AHNAK2 and EVPL in esophageal squamous cell carcinoma by exome and transcriptome sequencing analyses. OncoTargets Ther. 2021;14:1131. doi: 10.2147/OTT.S291007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakeji Y., Oshikiri T., Takiguchi G., Kanaji S., Matsuda T., Nakamura T., Suzuki S. Multimodality approaches to control esophageal cancer: development of chemoradiotherapy, chemotherapy, and immunotherapy. Esophagus. 2021;18:25–32. doi: 10.1007/S10388-020-00782-1/TABLES/2. [DOI] [PubMed] [Google Scholar]
- 14.Li Jieling, Wang X., Zheng K., Liu Y., Li Junjun, Wang Shaoqi, Liu K., Song X., Li N., Xie S., Wang Shaoxiang. The clinical significance of collagen family gene expression in esophageal squamous cell carcinoma. PeerJ. 2019 doi: 10.7717/PEERJ.7705/SUPP-14. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J., Xie Y., Wang X., Jiang C.…O X.Y. undefined, 2020. Identification of hub genes associated with esophageal cancer progression using bioinformatics analysis. 2020. spandidos-publications.com 20. [DOI] [PMC free article] [PubMed]
- 16.Li X., Xiao X., Chang R., Zhang C. Comprehensive bioinformatics analysis identifies lncRNA HCG22 as a migration inhibitor in esophageal squamous cell carcinoma. J. Cell. Biochem. 2020;121:468–481. doi: 10.1002/JCB.29218. [DOI] [PubMed] [Google Scholar]
- 17.Lin L., Lei Q., Zhang S., Kong L., Qin B. Screening and identification of key biomarkers in hepatocellular carcinoma: evidence from bioinformatic analysis. Oncol. Rep. 2017;38:2607–2618. doi: 10.3892/OR.2017.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z., Zhang R., Chen X., Yao P., Yan T., Liu W., PeerJ J.Y.- 2019. U., 2019. Identification of Hub Genes and Small-Molecule Compounds Related to Intracerebral Hemorrhage with Bioinformatics Analysis. peerj.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y., Xue J., Zhong M., Wang Z., Li J., Zhu Y. Prognostic prediction, immune microenvironment, and drug resistance value of collagen type I Alpha 1 chain: from gastrointestinal cancers to pan-cancer analysis. Front. Mol. Biosci. 2021;8 doi: 10.3389/FMOLB.2021.692120/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Y., Wang Z., Zhou L., Ma Z., Zhang J., Wu Y., Shao Y., Yang Y. FAT1 and PTPN14 regulate the malignant progression and chemotherapy resistance of esophageal cancer through the hippo signaling pathway. Anal. Cell Pathol. 2021 doi: 10.1155/2021/9290372. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luan S., Yang Y., Zhou Y., Zeng X., Xiao X., Liu B., Yuan Y. The emerging role of long noncoding RNAs in esophageal carcinoma: from underlying mechanisms to clinical implications. Cell. Mol. Life Sci. 2021;78:3403–3422. doi: 10.1007/S00018-020-03751-0/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meiqi W., Liu D., Huang Y., Jiang Z., Wu F., Cen Y., Ma L. Identification of key genes related to the prognosis of esophageal squamous cell carcinoma based on chip Re-annotation. Appl. Sci. 2021;11:3229. doi: 10.3390/APP11073229. 2021. Page 3229 11. [DOI] [Google Scholar]
- 23.Pal R.R., Rajpal V., Singh P., Saraf S.A. Recent findings on thymoquinone and its Applications as a Nanocarrier for the treatment of cancer and rheumatoid Arthritis. Pharm. Times. 2021;13:775. doi: 10.3390/PHARMACEUTICS13060775. 2021. Page 775 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng H., Wang S., Pang L., Yang L., Chen Y., Cui X. bin. Comprehensive bioinformation analysis of methylated and differentially expressed genes in esophageal squamous cell carcinoma. Mol Omi. 2019;15:88–100. doi: 10.1039/C8MO00218E. [DOI] [PubMed] [Google Scholar]
- 25.Sabaie H., Moghaddam, Mazaheri Madiheh, Moghaddam, Mazaheri Marziyeh, Ahangar N.K., Asadi M.R., Hussen B.M., Taheri M., Rezazadeh M. Bioinformatics analysis of long non-coding RNA-associated competing endogenous RNA network in schizophrenia. Sci. Rep. 2021;111(11):1–13. doi: 10.1038/s41598-021-03993-3. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadeghpour S., Ghorbian S. Evaluation of the potential clinical prognostic value of lncRNA-BANCR gene in esophageal squamous cell carcinoma. Mol. Biol. Rep. 2019;46:991–995. doi: 10.1007/S11033-018-4556-2/TABLES/2. [DOI] [PubMed] [Google Scholar]
- 27.Song W., Dai W.J., Zhang M.H., Wang H., Yang X.Z. Comprehensive analysis of the expression of TGF- β signaling regulators and prognosis in human esophageal cancer. Comput. Math. Methods Med. 2021 doi: 10.1155/2021/1812227. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun C., Yuan Q., Wu D., Meng X., Oncotarget B.W.- 2017. U., 2017. Identification of Core Genes and Outcome in Gastric Cancer Using Bioinformatics Analysis. ncbi.nlm.nih.gov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verma R., Sattar R.S.A., Nimisha Apurva, Kumar A., Sharma A.K., Sumi M.P., Ahmad E., Ali A., Mahajan B., Saluja S.S. Cross-talk between next generation sequencing methodologies to identify genomic signatures of esophageal cancer. Crit. Rev. Oncol. Hematol. 2021;162:103348. doi: 10.1016/J.CRITREVONC.2021.103348. [DOI] [PubMed] [Google Scholar]
- 30.Visaggi P., Barberio B., Ghisa M., Ribolsi M., Savarino V., Fassan M., Valmasoni M., Marchi S., de Bortoli N., Savarino E. Modern diagnosis of early esophageal cancer: from blood biomarkers to advanced endoscopy and Artificial Intelligence. Cancers. 2021;13:3162. doi: 10.3390/CANCERS13133162. 2021. Page 3162 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiaojie Y., Tian M., Zhang W., Chai T., Shen Z., Kang M., Lin J. Identification of potential core genes in esophageal carcinoma using bioinformatics analysis. Medicine (Baltim.) 2021;100 doi: 10.1097/MD.0000000000026428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong Y., Wei Y., Gu Y., Zhang S., Lyu J., Zhang B., Chen C., Zhu J., Wang Y., Liu H., Zhang Y. DiseaseMeth version 2.0: a major expansion and update of the human disease methylation database. Nucleic Acids Res. 2017;45:D888–D895. doi: 10.1093/NAR/GKW1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu J., Zhu C., Yu Y., Wu W., Cao J., Li Z., Dai J., Wang C., Tang Y., Zhu Q., Wang J., Wen W., Xue L., Zhen F., Liu J., Huang C., Zhao F., Zhou Y., He Z., Pan X., Wei H., Zhu Y., He Y., Que J., Luo J., Chen L., Wang W. Systematic cancer-testis gene expression analysis identified CDCA5 as a potential therapeutic target in esophageal squamous cell carcinoma. EBioMedicine. 2019;46:54–65. doi: 10.1016/J.EBIOM.2019.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu W., Xu J., Wang Z., Jiang Y. Weighted gene correlation network analysis identifies specific functional modules and genes in esophageal cancer. JAMA Oncol. 2021:1–13. doi: 10.1155/2021/8223263. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L., Zhang X., Hou Q., Huang M., Zhang H., Jiang Z., Yue J., Wu S. Single-cell RNA-seq of esophageal squamous cell carcinoma cell line with fractionated irradiation reveals radioresistant gene expression patterns. BMC Genom. 2019;20:1–11. doi: 10.1186/S12864-019-5970-0/FIGURES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang W., Zhao X., Han Y., Duan L., Lu X., Wang X., Zhang Y., Zhou W., Liu J., Zhang H., Zhao Q., Hong L., Fan D. Identification of hub genes and therapeutic drugs in esophageal squamous cell carcinoma based on integrated bioinformatics strategy. Cancer Cell Int. 2019;19:1–15. doi: 10.1186/S12935-019-0854-6/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L., Chen T.Y., Yang Z.Y., Fang W., Wu Q., Zhang C. Identification of hub genes in papillary thyroid carcinoma: robust rank aggregation and weighted gene co-expression network analysis. J. Transl. Med. 2020;18 doi: 10.1186/S12967-020-02327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yue L., Liu J., Cai X. wei, Li H. xuan, Cheng Y., Dong, huan X., Yu W., Fu X. long. Biomarkers for the prediction of esophageal cancer neoadjuvant chemoradiotherapy response: a systemic review. Crit. Rev. Oncol. Hematol. 2021;167:103466. doi: 10.1016/J.CRITREVONC.2021.103466. [DOI] [PubMed] [Google Scholar]
- 39.Zheng Z.Y., Yang P.L., Luo W., Yu S.X., Xu H.Y., Huang Y., Li R.Y., Chen Y., Xu X.E., Liao, Di L., Wang S.H., Huang H.C., Li E.M., Xu L.Y. STAT3β enhances sensitivity to concurrent chemoradiotherapy by Inducing cellular Necroptosis in esophageal squamous cell carcinoma. Cancers. 2021;13 doi: 10.3390/CANCERS13040901. 2021. Page 901 13, 901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhicheng W., Chen M., Qiu Y., Yang Y., Huang Y., Li X., Zhang W. Identification of potential biomarkers associated with immune infiltration in the esophageal carcinoma tumor microenvironment. Biosci. Rep. 2021;41 doi: 10.1042/BSR20202439/227787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Zengyuan, Li Y., Hao H., Wang Y., Zhou Zihao, Wang Z., Chu X. Screening hub genes as prognostic biomarkers of hepatocellular carcinoma by bioinformatics analysis. Cell Transplant. 2019;28:76S–86S. doi: 10.1177/0963689719893950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zou T., Huang Y., Hu Y., Wu M., Zhao Y., Du F., Li M., Wu X., Ji H., Kaboli P.J., Wang S., Xiao Z., Wu Z. The Anti-tumor mechanism and target of triptolide based on network pharmacology and molecular docking. Recent Pat. Anti-Cancer Drug Discov. 2021;16:426–435. doi: 10.2174/1574892816666210211143059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.