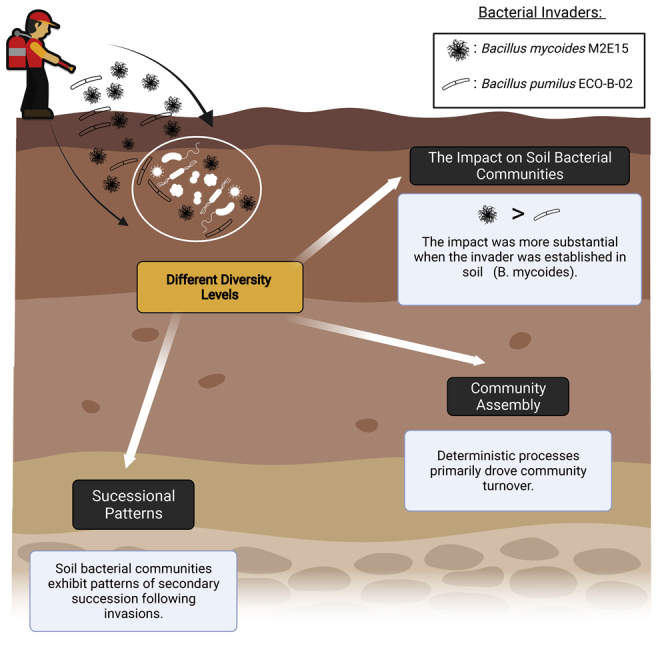
Summary
Microbial inoculants are constantly introduced into the soil as the deployment of sustainable agricultural practices increases. These introductions might induce soil native communities’ dynamics, influencing their assembly process. We followed the impact and successional trajectories of native soil communities of different diversity levels to the invasion by Bacillus mycoides M2E15 (BM) and B. pumilus ECOB02 (BP). Whereas the impact was more substantial when the invader survived (BM), the transient presence of BP also exerted tangible effects on soil bacterial diversity. Community assembly analyses revealed that deterministic processes primarily drove community turnover. This selection acted stronger in highly diverse communities invaded by BM than in those invaded by BP. We showed that resident bacterial communities exhibit patterns of secondary succession following invasions, even if the latter are unsuccessful. Furthermore, the intensification of biotic interactions in more diverse communities might strengthen the deterministic selection upon invasion in communities with higher diversity.
Subject areas: Soil science, Biogeochemistry, Microbiology, Soil biology
Graphical abstract
Highlights
-
•
Microbial invaders altered soil bacterial diversity regardless of their survival
-
•
The impact was more pronounced when the invader was established
-
•
Deterministic selection primarily drove community turnover following invasion
-
•
The dynamic of invaded community showed pattern of secondary succession
Soil science; Biogeochemistry; Microbiology; Soil biology
Introduction
Anthropogenic activities such as agricultural land use intensification alter soil (Emmerson et al., 2016; Tsiafouli et al., 2015; De Vries et al., 2013; Yeates et al., 1999) and plant diversity (Owen et al., 2013; Wagg et al., 2014), threatening various ecosystem functions such as carbon and nutrient cycling (De Graaff et al., 2015; Philippot et al., 2013) and decomposition (Handa et al., 2014; Valentín et al., 2014). To counteract these negative impacts, driven mainly by high fertilizer and pesticide inputs (Cao et al., 2011; Johnsen et al., 2001; Kleijn et al., 2012; Prashar and Shah, 2016), the use of beneficial microorganisms has emerged as an environmentally friendly substitute. The use of microbial inoculants could bypass the environmental problems associated with the excessive and indiscriminate use of those chemicals, such as soil contamination, disease resistance, and food contamination which significantly affect human health (Alori and Babalola, 2018; Baweja et al., 2020). However, releasing these agents into the soil also affects the native soil microbiome, potentially leading to changes in diversity, composition, and functionality in response to invasion (Karpouzas et al., 2011; Ma et al., 2018; Mar Vázquez et al., 2000; Mawarda et al., 2020; Nassal et al., 2018; Xing et al., 2020; Xiong et al., 2017). Considering the impact of biological disturbance caused by the introduction of alien soil microorganisms is thus a crucial aspect in developing sustainable agricultural practices, to ensure that such inoculation is not detrimental for soil diversity and functioning.
Microbial invasions can induce various dynamics in the resident soil microbiome. Susceptible communities can either return to their pre-invasion structure and composition, indicating resilience (Yin et al., 2013; Wang et al., 2018), or progress toward an alternative stable state, defined as the non-transitory change of the community structure away from its initial state after disturbance (Amor et al., 2020; Fu et al., 2017; Mawarda et al., 2020). The soil microbiome might also be resistant to invasions, as indicated by a lack of response (Chowdhury et al., 2013; Lerner et al., 2006). Previous research showed that the response of disturbed microbial communities might follow the “diversity-stability hypothesis”, where more diverse communities are better able to resist disturbance and recover their structure faster than less diverse ones (Girvan et al., 2005; Mallon et al., 2018; Riah-Anglet et al., 2015). Indeed, a recent study by (Xing et al., 2020) showed that the resilience of bacterial communities invaded by E. coli was weaker when communities were less diverse. However, other studies revealed a lack of correlation between community stability and diversity since neutral and negative associations also occur (Pennekamp et al., 2018; Wertz et al., 2007; Zhang and Zhang, 2006). These contrasting results may link to endpoint measurements (single measurement at the end of the experiment) rather than temporal dynamics assessments (multiple points throughout the experiment) of microbiome recovery. Disentangling the temporal variation and the mechanisms controlling community dynamics and assembly might indicate whether an invaded community structure is still recovering, has already recovered, or has switched to an alternative stable state.
Understanding the microbial community dynamics requires examination on their successional pattern, which is conceptually divided into primary (succession occurring in a non-colonized environment) and secondary (succession occurring in a previously colonized environment following a disturbance) (Dini-Andreote et al., 2019). Focusing on the latter, examining community assembly processes, specifically the deterministic and stochastic process, are thus crucial to provide the key to identify the mechanisms driving soil microbiome response to invasion over time (Dini-Andreote et al., 2015; Dumbrell et al., 2010; Nemergut et al., 2013). Deterministic processes, driven by abiotic (environmental) and biotic (competition, inter-taxon interactions) factors, structure microbial diversity and composition, which follow a secondary successional pattern. For instance, this can be described by direct or indirect competition for resources and niche space between invaders and resident communities, coupled with environmental changes resulting from the colonization (Pearson et al., 2018; Vellend, 2010). Meanwhile, stochasticity may also be a major driver of community response, highlighting the importance of randomly occurring events, such as ecological drift and dispersal (Vellend, 2010). Studies of assembly dynamics associated with bacterial community succession show that both deterministic (Jurburg et al., 2017; Lin et al., 2017; Nemergut et al., 2016) and stochastic (Chase, 2007; Zhou and Ning, 2017) processes can be relevant for community structure. Identifying the assembly processes triggered by microbial inoculations provides a dynamic perspective of the soil microbiome response, thus contributing to our ability to predict the consequences of microbial invasions.
The purpose of this study was to examine the successional trajectory and community assembly processes of soil bacterial communities in response to bacterial invasion. In particular, we questioned whether the diversity of the indigenous soil bacterial communities governs these dynamics. We hypothesized that soil bacterial communities with higher diversity would be more resistant (resist the community) or resilient (return to the original state) to invasion by Bacillus than ones with lower diversity. Moreover, we predicted that deterministic processes would play a major role in high-diversity communities due to greater biotic interactions. In contrast, in lower-diversity ones, the mechanism driving assembly processes would be more stochastic. We tested these hypotheses by inoculating soil microcosms consisting of a gradient of microbial diversity with two soil-borne Bacillus species, B. mycoides M2E15 (from now on refer to BM) and B. pumilus ECO-B-02 (from now on refer to BP), known to promote the growth of potato and grass, potentially via phosphate utilization, iron acquisition, and bacteriocin production (Yi et al., 2016). We further evaluated the impact of invasion by following the changes in soil bacterial community composition for 60 days using amplicon sequencing (16S rRNA gene-based).
Results
Survival of microbial inoculants
The enumeration of total culturable bacteria before soil inoculation by the invaders showed statistical similarity between all treatments (Kruskal, p = 0.2212). The mean value of total culturable bacteria was 8.98 log CFU/g soil. Thus, soil microcosms had similar community sizes across the treatments (Table S1). At this stage, we introduced the Bacillus strains, each in a separate microcosm, with three replicates per diversity treatment.
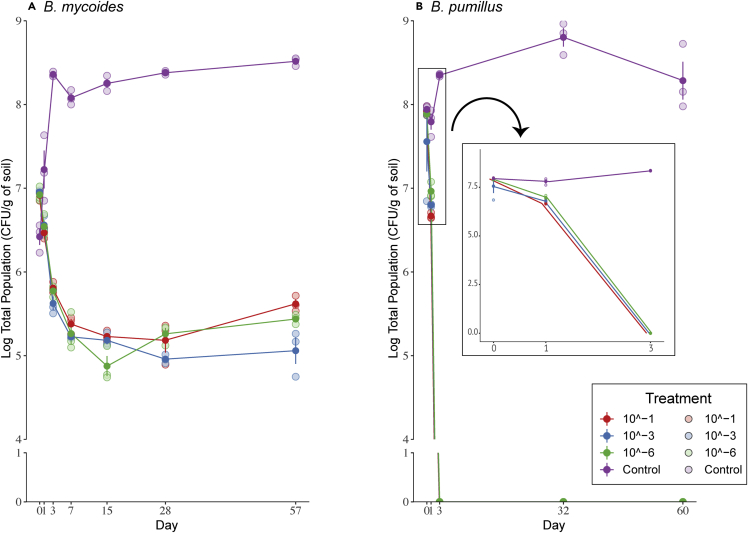
Next, we followed the survival of BM and BP for up to 60 days. The population sizes of both Bacillus spp. were expressed as total and spore populations, in log CFU/g of soil. We observed similar dynamics for the two Bacillus strains, as they both declined over time in all (10−1, 10−3, and 10−6) treatments (Figure 1). However, the decline was much faster for BP than for BM, as its population fell below the detection limit of 1 log CFU/g soil just three days post introduction (p.i). In the (sterile soil) control treatment, both inoculants remained at around 8 and 9 log CFU/g soil (BM and BP, respectively) for the duration of the experiment (Figure 1). Both resident bacterial community diversity and time had a significant effect on the dynamics of BP (Kruskal, P(time) = 0.0072, P(treatment) < 0.0001) and BM (Kruskal, P(time) = 0.0043, P(treatment) < 0.0001). For both inoculants, the survival of the total population was highest in the control treatment (Wilcox's post hoc, p < 0.05), while in the 10−1, 10−3, and 10−6 treatments, the survival was statistically similar (Wilcox's post hoc, p > 0.05).
Figure 1.
Survival of the Bacillus inoculants
(A) Survival of B. mycoides M2E15 and (B) B. pumilus ECO-B-02 across diversity treatments (10−1, 10−3, 10−6, and control) over time. Values represent the log CFU of the total population per gram of soil. Bars represent the standard error of the mean. Full symbols represent the average, whereas the light symbols indicate the values per replicate.
Concerning the spore populations, different trends were observed for both Bacillus inoculants (Figure S2). For BM, the spore population sizes initially increased around 1–2 log CFU/g soil and tended to stabilize around 4–5 log CFU/g soil, at day 3 p.i in 10−1, 10−3, and 10−6 treatments. In the control treatments, the spore increased and stabilized at day 3 p.i, at 8 log CFU/g soil. For BP, spores were only detected around 6–7 log CFU/g soil in the control microcosms (up to day 32; Figure S2), remaining below the detection limit throughout the experiment in the other treatments. Similar to the total population, there were significant effects of time and resident microbiome diversity on the BP (Kruskal, P(time) = 0.043, P(treatment) < 0.001) and BM spore population sizes (Kruskal, P(time) < 0.0001, P(treatment) = 0.0033). The spore numbers were significantly higher in the control treatment (Wilcox's post hoc, p < 0.05) than in the 10−1, 10−3, and 10−6 treatments. In the latter, the spore numbers were not statistically different between each other for both inoculants (Wilcox's post hoc, p > 0.05).
Alpha diversity levels of invaded soil microbiomes
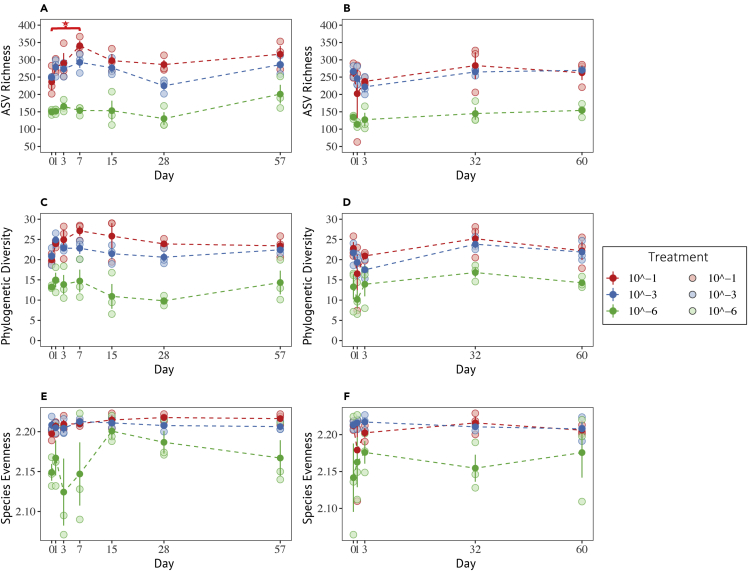
After quality control and processing, 6,969 and 5,013 ASVs (defined at 99% sequence identity) were detected in the soils invaded by BM and BP, respectively. The observed ASV richness, evenness, and phylogenetic diversity levels in the BM experiment were significantly different between the treatments (Kruskal, p < 0.0001) (Figures 2A, 2C, and 2E). These metrics became progressively lower with increasing dilution treatment (Wilcox's post hoc, p < 0.05), indicating that the diversity treatment effect remained constant throughout the experiment. In respect of species evenness, the soil bacterial communities in the 10−1 and 10−3 treatments were statistically similar (Wilcox's post hoc, p > 0.05), whereas the 10−6 treatment was significantly lower from the former treatments (Wilcox's post hoc, p < 0.05).
Figure 2.
Temporal patterns of alpha diversity in response to the Bacillus invasions
(A, C, and E) invaded by B. mycoides M2E15 and (B, D, and F) invaded by B. pumilus ECO-B-02 across diversity treatments (10−1, 10−3, 10−6). The changes in diversity were observed for 3 metrics: Species richness (A and B), represented by the number of ASVs; Faith Phylogenetic diversity (C and D); and species evenness (E and F), based on the Shannon index. Bars represent the standard error of the mean. Full symbols represent the average, whereas the light symbols indicate the values per replicate.
Invasion by BM led to significant changes in ASV evenness and richness in the 10−1 treatment, whereas 10−3 and 10−6 treatments remained similar. Specifically, ASV richness significantly increased from day 0 to day 7 p.i (Tukey's post hoc, p < 0.05) (Figure 2A) while ASV evenness increased progressively and significantly over time (R2 = 0.27; p = 0.017) (Figure S3). In contrast, the phylogenetic diversity levels remained statistically similar during the experiment in each treatment (ANOVA and Kruskal, p > 0.05) (Figure 2C).
We observed significant differences among treatments for ASV richness, evenness, and phylogenetic diversity in the systems invaded by BP (Kruskal, p < 0.0001) (Figures 2B, 2D, and 2F). These diversity metrics were largely similar in the 10−1 and 10−3 treatments (Wilcox's post hoc, p > 0.05), whereas those of the 10−6 treatment were significantly lower than the former (Wilcox's post hoc, p < 0.05). The metrics remained constant over time for each treatment (ANOVA and Kruskal, p > 0.05).
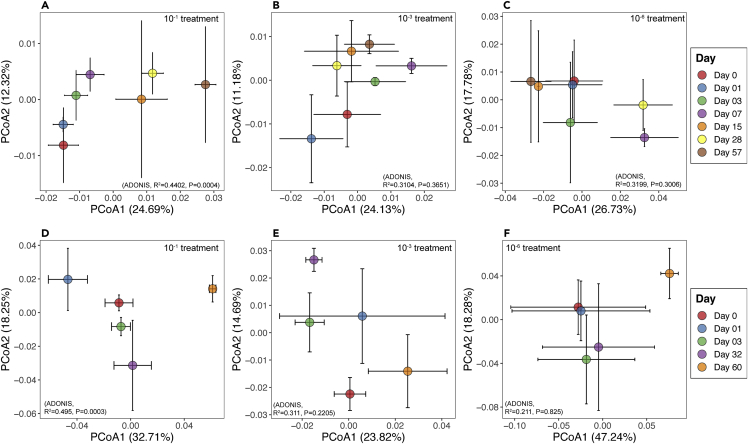
Beta diversity
To examine the impact of Bacillus invasions on the soil bacterial community structure, we calculated Bray-Curtis dissimilarities and weighted Unifrac distances across diversity levels on each sampling day. According to these three metrics, the treatment imposed by dilutions led to the establishment of distinct bacterial community compositions at the onset of the inoculations (Figure S4, p = 0.0001). Effects of time (represented by weighted Unifrac) varied according to diversity treatment (Figures 3A–3F), with a significant effect being visible only in the 10−1 treatment for both Bacillus invaders (Figure 3A, BM - 10−1: ADONIS time, R2 = 04,402, p = 0.0004; Figure 3D, BP - 10−1: ADONIS time, R2 = 0.495, p = 0.0003). Temporal effects based on Bray-Curtis showed the same patterns (Figure S5). For the BM experiment, the community structures in the 10−1 treatment revealed three main clusters (pairwiseAdonis, p < 0.05) along the first axis, according to time p.i: (1) bacterial communities until 7 days p.i, (2) those in the intermediate period (as from 15 to 28 days p.i), and (3) those at day 57. In the BP experiment, the soil bacterial communities in 10−1 treatment also clustered into three groups (pairwiseAdonis, p < 0.05). The first group consisted of the bacterial communities at day 0 and day 1 p.i, while the second group (shifted along the first axis 18.25%) consisted of bacterial communities at days 3 and 32 p.i. The bacterial communities at day 60 p.i moved away from the first and second groups along the second axis (32.71% of variation). For both Bacillus experiments, 10−3 and 10−6 treatments were not significantly different (pairwiseAdonis, p > 0.05)
Figure 3.
Temporal variation of bacterial community structure following the Bacillus invasions
These variations are depicted by PCoA plot of weighted Unifrac distances in 10−1 (A and D), 10−3 (B and E), and 10−6 (C and F) treatment following B. mycoides (A–C) and B. pumilus (D–F) invasions. PCoA in 10−1 treatment (A and D) shows the effect of invasion on bacterial community structure, determined by PERMANOVA, p < 0.001. Centroids for each sampling time are shown along with their standard errors (error bars).
Identification of ASVs that showed temporal responses to invasion
The temporal patterns of bacterial community structures following invasion by the two Bacillus spp. were observed at the ASV level. In both experiments, the most abundant ASVs across all diversity treatments were affiliated to five phyla, i.e., Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Planctomycetes. However, their relative abundance varied according to diversity level (Figure S6). There were 13.21% (921 ASVs) and 12% (602 ASVs) unidentified ASVs in the dataset of 10−1, 10−3, and 10−6 treatments invaded by BM and BP, respectively, suggesting that there were still many unidentified bacterial community members in the system.
Using the previously published criteria regarding significant temporal variation—i.e., abundance ≥0.5% and presence in ≥3 samples (Jurburg et al., 2017; Shade et al., 2013)—we identified key ASVs that showed temporal responses to the invasion in 21 and 15 samples in the BM and BP experiments, respectively. For BM, we found 45 out of 2,867 ASVs (1.57%), 19 out of 2,624 ASVs (0.72%), and 25 out of 1,463 ASVs (1.71%), in the 10−1, 10−3, and 10−6 treatments, respectively (Figure S7). In the BP experiment, we identified 16 out of 2015 ASVs (0.79%), 25 out of 1943 ASVs (1.28%), and 10 out of 1065 ASVs (0.94%), to represent the community in the 10−1, 10−3, and 10−6 treatments, respectively (Figure S8). Significant correlation of symmetric procrustes rotation based on Bray-Curtis dissimilarity was shown for each treatment in both Bacillus experiments (BM: 9999 permutations 10−1: 0.7655, p = 0.0001; 10−3: 0.6551, p = 0.0014; 10−6: 0.7282, p = 0.0001 || BP: 9999 permutations 10−1: 0.7714, p = 0.0001; 10−3: 0.6874, p = 0.0001; 10−6 treatment: 0.6623, p = 0.0093) confirming that the selected dataset and original dataset were similar, and therefore provide a statistically valid representation of the original community.
Secondary succession patterns in response to invasion
To identify secondary successional patterns, hierarchical cluster analysis (HCA) grouped the previously selected ASVs. In both experiments, some ASV clusters never returned to their initial state. For example, following invasion by BM, ASVs that belong to group 7 (mainly from genus Bacillus) in the 10−1 treatment kept decreasing toward the end of the experiment (see Figure S7A). In the BP experiment, similar decreasing patterns were observed in the first group of the 10−1 treatment (from genus Bacillus, Brevibacillus, and Athrobacter) (Figure S8A).
To uncover possible time-driven trajectories in both experiments, we clustered these groups by time to: (1) Early stage (the groups that peaked at one of 0, 1, 3, and 7 days p.i for BM & day 0, 1, and 3 p.i for BP); (2) Mid-stage (the groups that peaked at day 15 and or 28 p.i for BM & day 32 p.i for BP); and (3) Late-stage (the groups that peaked at day 57 p.i for BM and day 60 p.i for BP). These ASV groups revealed different temporal patterns. For example, in the BM experiment, ASVs that peaked at an early stage kept decreasing toward the end of the experiment (Figure S7A: Group 1, 2, 6, and 7 in 10−1 treatment). There were also ASVs with increasing abundance toward the later stage of succession (Figure S7A: Group 3, 5, and 8 in 10−1 treatment). Other taxa peaked more than once. For instance, the first and second groups in 10−3 treatment peaked at early and mid-stages p.i (Figure S7B). Finally, we observed ASVs having coherent dynamics and similar occurrence, with mild fluctuation throughout the experiment (Figure S7A: Group 4 in 10−1 treatment; Figure S7C: Group 3 in 10−6 treatment).
In the BP experiment, ASVs in the second group of 10−1 treatment (Figure S8A) and the first group of 10−6 treatment (Figure S8C) increased and peaked at the late stage. In addition, we found ASV groups that peaked at multiple stages or maintained their abundance throughout the experiment. The former was observed in the second group of 10−3 treatment, which consisted of ASVs that peaked in abundance at the early and late stages, whereas the latter was observed in the third group of this treatment (Figure S8B). Detailed information about the taxa associated with these successional groups at each diversity level is available in Tables S2 and S3 for BM and BP experiments, respectively.
Processes controlling bacterial community assembly upon invasion
To determine the processes influencing bacterial community composition in response to the invasion, we calculated the stochastic and deterministic turnover rates based on the βNTI. This metric was significantly different across diversity treatments for both inoculants (Figure 4A, BM: Kruskal, p < 0.0001; Figure 4B, BP: Kruskal, p < 0.0001). In both experiments, the effect of selection on community assembly increased as the diversity of the resident community increased (Figure S9: BM: Pearson Correlation, R2 = 0.52, p < 0.0001; BP: Pearson Correlation, R2 = 0.32, p < 0.0001). This indicates that the community assembly in less diverse communities is less deterministic than the more diverse ones, irrespective of the invader.
Figure 4.
Temporal variation of the βNTI index across 10-1, 10-3, 10-6 treatment
(A) Invaded by B. mycoides M2E15 and (B) invaded by B. pumilus ECO-B-02 (B). Values between 2 and −2 indicate stochastic turnover, while βNTI <−2 indicates homogeneous selection. Full symbols represent the average, whereas the light symbols indicate the values per replicate.
BM caused temporal variation of βNTI in the 10−1 and 10−3 treatments (10−1 treatment: ANOVA, p < 0.0001; 10−3 treatment: ANOVA, p < 0.01) but not in the 10−6 one (ANOVA, p > 0.05). Moreover, in the 10−1 treatment, homogeneous selection became stronger after BM invasion, as the mean βNTI significantly dropped toward the end of the experiment (Tukey's post hoc, p < 0.05). Meanwhile, BP did not affect the temporal variation of the community assembly process across diversity gradients, as mean βNTI remains statistically similar within between-replicate variability (ANOVA, p > 0.05). Overall, the effect of invasion on resident community structure was more pronounced in the more diverse communities, invaded by the stronger invader BM than the transient BP.
Discussion
As the deliberate release of microbes into soil becomes a common agricultural practice, ensuring that such practices are not detrimental to soil biodiversity and ecosystem functioning is crucial. Here, we evaluated the temporal dynamics of native soil bacterial communities of several diversity levels to the biological disturbance imposed by the inoculation of soil-borne Bacillus species. We further explored the potential impact of the invasions by placing them into the context of ecological principles associated with community diversity, secondary succession, and community assembly.
The impact of invasion on resident soil microbiomes is related to invader survival
This study confirms previous findings that soil bacterial communities experience temporal shifts induced by Bacillus invasion, in both alpha and beta diversities, irrespective of the invaders' survival (Fu et al., 2017; Wu et al., 2016). We showed that the magnitude of the impact was most pronounced in systems invaded by BM, which established and survived in the soil. Moreover, even though BP went undetectable already 3 days p.i., its release also impacted the resident bacterial communities in a time-dependent manner. Thus, ephemeral and unsuccessful invasion can still leave a footprint in soil microbiomes, in line with previous findings of impacts of ecologically unsuccessful invaders on phylogenetic structures (Mallon et al., 2018) or even transitions to alternative stable states (Amor et al., 2020).
We cannot easily explain these findings, although transient resource competition may have played a role (Mallon et al., 2015b, 2018). Limited niche breadth may constrain the ability of an invader to compete with functionally similar resident taxa, influencing its spread and growth. The resident taxa with similar niche preferences as the invader may “suffer” through direct competition, whereas those that exploit niches not used by the invader are released from competition with the indigenous microbiome, raising in abundance (Mallon et al., 2018). Resource-related mechanisms can thus apply to the impact of unsuccessful invasion where the invaders fail to establish. It is also noteworthy that invader cell death prompts a nutrient pulse in the soil, affecting the taxonomic structure of the resident communities. Transient invaders might also induce community shifts to alternative states by inciting changes in parameters such as pH (Amor et al., 2020). From these examples, it is thus clear that transient invaders contributed to community dynamics and that a successful invasion yields a higher magnitude of an impact than an unsuccessful one (Xing et al., 2020). Although we did not quantify potential shifts in the composition and structure of non-invaded soil microcosms at different diversity levels over time, several other studies have shown that incubation time had no impact on the shift of diversity (Mallon et al., 2018; Xing et al., 2020). Moreover, the difference in successional patterns across diversity levels, in both BM (Figure S7) and BP (Figure S8) experiments, sustains that our results are driven by inoculation and not by incubation biases of soil microcosms. We, therefore, expect that the observed responses of community structure upon inoculation are due to invasion and not lab incubation.
The invasion impact links to the resident soil bacterial diversity
Biodiversity-stability theories suggest that the capacity of a resident community to buffer their structure and function increases with diversity due to the increased presence of functionally redundant taxa (McCann, 2000) and other mechanisms such as complementarity and sampling effect (Tilman et al., 2006). In the context of microbial invasions, several studies have demonstrated that diversity hinders invader survival through resource competition (Mallon et al., 2015a, 2015b; Wei et al., 2015; Xing et al., 2020). In sum, less diverse communities are expected to be less resistant, due to their reduced capacity to explore all available niches, thus leaving unconsumed resources that can support invader establishment (Mallon et al., 2018). However, other studies of this relationship have yielded mixed results, and different underlying principles were invoked (Glasl et al., 2018; Pennekamp et al., 2018; Wertz et al., 2007). Interestingly, our results revealed that the invasion by BM and BP impacted the community structure of the native communities only in the most diverse treatments. Hence, despite the statistical differences in alpha diversity measurements between dilution treatments, the magnitude of the invasion impact on community composition (beta diversity) was not linked to the (positive) biodiversity-stability relationship. One possible explanation is that the species present in the most diverse communities are more sensitive to Bacillus's arrival than the less diverse ones (see discussion in secondary succession and the magnitude of selection and stochastic effects for explanation). Moreover, this response might be associated with the production of spores, which might escape (negative) interactions with the resident microbiota by entering a latent stage. Indeed, in the case of BM, sporulation was triggered across all treatments leading to a similar survival trend (Figures 1 and S2) regardless of differences in resource availability.
The magnitude of selection and stochastic effects link to native soil bacterial diversity
We observed stronger deterministic turnover and (homogeneous) selection in the communities with higher diversity. In contrast, stochasticity was more relevant in less diverse ones, following both Bacillus invasions. The increasing stochastic effect with decreasing bacterial diversity might be due to the potential lower functional redundancy (Delgado-Baquerizo et al., 2016) and higher resource availability for invaders (Mallon et al., 2015b). For instance, weaker resource competition between invaders and resident communities occurred in communities of reduced diversity due to the reduction of functionally different taxa (Eisenhauer et al., 2013). Therefore, community assembly processes tend to be more stochastic as selective forces in less diverse communities wane off. This finding confirms previous studies that observed increasing susceptibility to drift as indicated by higher βNTI values in less diverse communities (Evans et al., 2017; Vellend, 2010). In addition, we found that stochasticity was stronger when less diverse communities got invaded by the transient invader (BP). We hypothesize that the fast elimination of transient invaders contributes to less selective force as resource competition was probably short-lived.
In this study, the community assembly process was significantly more deterministic when the invader was established in more diverse communities. This might be due to higher densities of competing neighbors resulting from more intense resource depletion, strengthening resource competition between invaders and the resident community. Our findings are consistent with studies indicating that at high bacterial diversity, complex biological interactions play a role in community assembly (Brophy et al., 2017; Coyte et al., 2015). Specifically, limited nutrient availability exacerbates competition and intensifies biotic interactions within this community (Fontaine et al., 2003), thus contributing to deterministic processes. Conversely, less diverse communities (10−6 treatment) showed higher variability of taxonomic composition, leading to an increase in the stochastic signal, which was especially evident in response to BP invasion. It is important to note that the high variation between the replicates of the less diverse communities contributes to the detected stochastic signal. However, the differences in stochastic signal between BP and BM observed for the 10−6 treatment suggest that the taxonomic composition of the native bacterial community also plays a role in the community assembly process and temporal dynamics upon invasion. Importantly, the higher level of stochasticity suggests that the impact of Bacillus invasion on native bacterial structure and composition in the less diverse community might be less predictable.
Secondary succession of invaded bacterial communities
Although bacterial community structure in some treatments showed resistance or resilience toward Bacillus invasion, some ASVs within those treatments still experienced temporal dynamics. This finding conforms to previous studies showing that not all components of bacterial communities are resilient, as some ASVs did not return to their initial state (Jurburg et al., 2017). The non-resilient bacterial taxa—especially those that kept decreasing and went undetectable toward the end of succession—could have a detrimental impact if they are key species that hold a role in maintaining soil ecosystem process and functioning. Their elimination could reduce some vital services of terrestrial ecosystems if their functions are irreplaceable by other taxa or replaced by less efficient ones. Hence, evaluating the impact of microbial release in the agricultural sector is critical to ensure that such practices do not harm biodiversity and ecosystem functioning.
Throughout our study, we found three groups of ASVs with different temporal patterns following both Bacillus invasions: early responder, mid responder, late responder, next to combinations of them. According to resource-driven mechanisms, those ASVs that decreased after the early invasion stage might compete for a similar niche as the invader. Although in the BP experiment we cannot verify traits of the responsive groups—as these taxa were mostly from unidentified genera—in BM, these ASVs belong to bacteria with the ability to grow at richer substrate concentrations (r-strategists; e.g., Bacillus, Rhodanobacter, Sphingomonas, Streptomyces, and Burkholderia) (Ho et al., 2017). In line with previous studies, these taxa usually appear in the early stage of bacterial succession (Ditterich et al., 2016; Nemergut et al., 2016). Meanwhile, those taxa that increased at the later stage might have benefited from a release in resource competition with previously dominant taxa that declined after the Bacillus invasion. In BM experiment, these taxa mostly belong to bacteria featured by their relatively slow growth and less reactivity toward abrupt resource availability (k-strategists; e.g., Gemmata, Planctomycetes, Phenylobacterium, Cryocola, and Rhodoplanes) (Moghaddas et al., 2018; Moorhead and Sinsabaugh, 2006). They reach maximal growth efficiency at the later stage of succession, owing to their ability to scavenge and grow when resource availability becomes scarce. Thus, late responder bacteria likely succeed early responders as depletion of resources becomes higher due to stronger competition upon BM invasion.
Some early responder taxa became predominant again at a later stage of succession, possibly due to the resurgence of their previously seized niche due to invader exclusion or dormancy. In addition, some ASVs showed significant fluctuations that persisted over time, which could be explained by their ability to explore a niche different from the invader, being excluded from direct competition with the invader. However, we cannot exclude the fact that the death of Bacillus cells might have provided small pulses of nutrients in the soil, giving an initial flush of energy resources which may contribute to the dynamics of invaded communities. Finally, the observed changes in composition are not necessarily driven only by the resources, as biotic interaction with native bacterial communities might also cause the temporal dynamics we observed on each diversity gradients, especially at the late stage of succession. Together, our data provide evidence for successional patterns that mimic secondary succession in macro-organisms. Further work is needed to disentangle whether this pattern is driven by resource competition or other mechanisms such as antagonism, predation, and parasitism (Mallon et al., 2015a, 2015b).
Our findings show that invasion of soil systems can impact the resident community regardless of whether the invasion is successful or not, with a stronger impact in the community where the invader gets established. The impact is also more pronounced in more diverse communities, suggesting that the invasion outcome may not obey a positive biodiversity-stability relationship. Furthermore, we showed the dominance of deterministic turnover with homogeneous selection as a general process driving bacterial community assembly. Together with the pattern of secondary succession, it suggests that biotic interactions may be the main mechanisms contributing to bacterial assembly in response to invasion. Future work should focus on (i) identifying the functional consequences of the invasion on soil processes; (ii) on whether the impact can be attenuated if competition is relieved, either through resource pulse or plant exudation; and (iii) to what extent the germination of the spores from the invasive species will worsen the shifts in soil native bacterial community composition. Regardless of the mechanisms, our data revealed that the introduction of bacterial inoculants generates patterns of secondary succession and that even brief and unsuccessful invasion can still leave a footprint in soil microbiomes.
Limitations of the study
There are several limitations of our study. First, the response of soil bacterial communities following disturbance depends on the type, duration, strength, and frequency of disturbance events (Shade et al., 2012). Thus, the fundamental knowledge on the impact of microbial invasion generated in this work needs to be further investigated in the experiment leveraging different type of microbial invaders, longer experimental time-span, different concentration of inoculants, and single vs multiple inoculations. Furthermore, there is a need to test microbial communities from different type of soil since different microbial community structure and composition may influence the resistance and resilience capacity toward microbial invasion. Finally, in order to see the impact of soil microbial invasion holistically, we cannot rely solely on the changes of native community structure, as the function has to be concomitantly assessed. Thus, future experiment aiming at soil microbial community function is required to foster this field. In spite of these limitations, we expect our work to set the stage for research on the impact of microbial invasion to the diversity of soil bacterial communities. This reinforces the idea that despite the growing practice of deliberately releasing microbes to open environment, research needs to better evaluate its consequences to the invaded biodiversity and ecosystem.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| B. mycoides M2E15 | This paper | N/A |
| B. pumilus ECO-B-02 | This paper | DSM 32048 |
| Critical commercial assays | ||
| Dneasy Powersoil Kit | Qiagen, Hilden (Germany) | #Cat. No. / ID: 47016 |
| Deposited data | ||
| Raw reads of DNA-based sequences | Sequence Read Archive of the National Center for Biotechnology Information | PRJNA721368 |
| Original code | This Paper | https://doi.org/10.5281/zenodo.6382466 |
| Software and algorithms | ||
| Quantitative Insights Into Microbial Ecology (QIIME 2) | (Bolyen et al., 2019) | https://qiime2.org/ |
| DADA2 | (Callahan et al., 2016) | https://doi.org/10.1038/nmeth.3869 |
| MAFFT (ver. 7) | (Katoh and Standley, 2013) | https://doi.org/10.1093/molbev/mst010 |
| FastTree (ver. 2.1) | (Price et al., 2010) | https://doi.org/10.1371/journal.pone.0009490 |
| Ribosomal Database Project classifier | (Wang et al., 2007) | https://doi.org/10.1128/AEM.00062-07 |
| Bacterial Succession Algorithm | (Shade et al., 2013) | https://doi.org/10.1128/mBio.00602-12 |
| Vegan (R package) | (Oksanen et al., 2020) | https://CRAN.R-project.org/package=vegan |
| Phyloseq (R package) | (McMurdie and Holmes, 2013) | https://joey711.github.io/phyloseq/index.html |
| pairwise-Adonis (R package) | (Martinez Arbizu, 2020) | https://github.com/pmartinezarbizu/pairwiseAdonis |
| Phylogenetic Turnover | (Stegen et al., 2013) | https://doi.org/10.1038/ismej.2013.93 |
Resource availability
Lead contact
Further information and requests for resources, data, and codes should be directed to and will be fulfilled by the lead contact, Panji Cahya Mawarda (p.c.mawarda@rug.nl/panji.cahya@gmail.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Soil and microcosm preparation
The used soil was sampled from a potato field (sandy loam, pH 4.75) in Friesland, the Netherlands, and sieved through a 2 mm mesh, homogenized, and adjusted to pH 7.0 by adding 2.33 g Ca(OH)2/kg soil. The soil was divided into two parts; the first part was non-sterile soil (natural soil) used as the source of the indigenous communities, whereas the second part was sterilized via gamma irradiation (50 kGy). The sterilized soil was aseptically divided into 60-g portions in sterile glass flasks capped by sterile aluminum foil. Soil moisture was adjusted at 50% of the water holding capacity (WHC) and kept constant by replenishing the water until the setup of diversity gradients.
Diversity gradient experiment: Setup and design
Diversity gradients were created by inoculating the microcosms with different dilutions of community inocula, next to sterile water as a control. The inoculum was prepared by mixing the natural soil (non-sterile soil) with sterile water in a ratio of 1:10. The mixture was homogenized by shaking for 20 min at 28°C and serially diluted (10-fold) with sterile water until 10−6. Volumes of 16 mL inoculum from the 10−1, 10−3, 10−6 dilutions were transferred to 60 g sterile soil in each microcosm, increasing soil moisture to 65% of the WHC. The control was prepared by transferring 16 mL of sterile water to the remaining microcosms containing sterilized soil, raising the WHC to 65%. Thus, the four diversity treatments were as follows: 10−1, 10−3, 10−6, and control. To allow for microbiomes establishment, all systems were incubated for 60 days at room temperature. The soil moisture was kept constant, at 65% of the WHC, by measuring the weight of each microcosm and replenishing the sterile water. Destructive samplings were initially prepared at days 0, 3, 10, 15, 30, and 90 for microcosms invaded by both Bacillus strains. The destructive samplings were done to ensure that no jar was ever sampled more than once. The microcosms were set up in triplicate for each treatment and each sampling date. This comprised 144 microcosms (4 diversity treatments x 2 bacterial inoculants x 6 sampling dates x 3 replicates).
Method details
Determination of initial bacterial abundance, Bacillus spp invasion, and survival monitoring
The total culturable bacteria were enumerated before the invasion in all diversity treatments via the plate count method. Material from each microcosm was serially diluted, in a ratio of 1:10, plated on TSA medium, and counted after incubation at 28°C for 4 d. All microcosms had roughly the same number of culturable cells per gram of soil before Bacillus spp invasion.
To differentiate our invaders from the resident soil bacteria, we developed antibiotic rifampicin-resistant strains of BM and BP by a spontaneous mutation, which did not influence the fitness of the mutated strains compared to that of the wild-type strains (based on growth rate – ANOVA, p > 0.05, data not shown). The rifampicin-resistant strains were grown overnight, after which they were washed 3× with saline solution (0.85% NaCl) via centrifugation at 6,000 rpm for 3 min. The cell pellets from each culture were resuspended in 1 mL of saline solution (OD = 0.8 at 590 nm), after which the resulting cell suspensions were used for introduction into the soil microcosms. The introductions raised soil moisture in each microcosm to 75% of WHC. After the introduction, the WHC values were kept constant by replenishing the water until the end of the experiments. Each invader was enumerated by 1:10 serial dilution plating on TSA medium containing rifampicin (50 μg/mL) and cycloheximide (400 μg/mL). The plates were incubated at 28°C for 24 h for enumeration of BM and 48 h for BP. In addition, to perform spore counts of each bacterium, each diluted sample was heated for 20 min at 80°C to kill vegetative cells and maintain the spores and plated on antibiotic-containing TSA as described above. Due to the filamentous nature of BM, the enumeration of total cell population and spore number were adjusted as such that spore counts could not be higher than the total population.
Three days after introducing the Bacillus strains, their population densities were below the detection limit of 1 log CFU/g soil, likely caused by a decrease in soil pH (to 4.9) upon inoculation, given that invaded sterile soils did not vary in pH. Thus, 7 days after the first inoculation attempt, we pooled soils from replicate jars together, adjusted the soil pH to 7.0 by adding sterile 1 M Ca(OH)2, and transferred 39 g of soil at 65% WHC from each treatment to a new, sterile jar. Control jars were made anew by adding sterile demineralized water to 39 g of gamma-sterilized soil to reach 65% WHC. The soil depth and soil bulk densities were around 2 cm and 1.2 gr/cm3, respectively. Soil microcosms were incubated at room temperature for at least 2 weeks for stabilization purposes. After verifying pH stability, soils were reinoculated with either BP or BM (3 weeks after the first inoculation attempt) using the procedure described above. The second inoculation was successful, as indicated by the enumeration (destructive samplings) of both vegetative cells and spores of each invader at days 0, 1, 3, 7, 15, 28, and 57 for microcosms invaded by BM and at days 0, 1, 3, 32 and 60 for microcosms invaded by BP. All data provided here refer thus to the time points right after (Day 0) and after the second inoculation (Day 1 onwards). The sampling times were chosen based on the survival pattern for each invader during the pilot experiment (data not published). Hence, this comprised a total of 144 microcosms ([4 diversity treatments x 7 sampling dates x 3 replicates for BM] + [4 diversity treatments x 5 sampling dates x 3 replicates for BP]). Soil moisture levels were kept at 75% WHC until the end of the experiment. The overview of the experimental set up and its adjustment is available in Figure S1.
Soil DNA extraction, 16S rRNA gene sequencing and processing
Five g of soil was taken from each microcosm at each sampling date and stored at −80°C until further use. The DNA was extracted using the DNeasy Powersoil Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The extracted DNA was quantified using NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). The extracted DNA was stored at −20°C and sent for 16S rRNA gene sequencing targeting the V4 region (forward primer 16S-515F: 5′-GTGCCAGCMGCCGCGGTAA-3′; reverse primer 16S-806R: 5′-GGACTACHVGGGTWTCTAAT-3′) to the University of Minnesota Genomic Centre (Minneapolis, MN, USA), using Ilumina Miseq 2 × 300 base paired-end reads (Illumina, San Diego, California) and their dual indexing method (Gohl et al., 2016). Sequence processing and analyses were conducted in Quantitative Insights Into Microbial Ecology (QIIME 2, https://qiime2.org). Demultiplexed sequences were provided directly by UMGC. DADA2 (Callahan et al., 2016) was employed to trim, denoise, dereplicate, and paired end merge these sequences. This tool was also used to filter chimeric sequences and remove primer sequence and singleton. Sequences were trimmed at positions 241 for the forward reads and at 215 bp or 217 bp (reverse reads) for the B. mycoides and B. pumilus experiment, respectively. For both experiments, the expected error for forward and reverse reads was set at 5 and the minimum overlap in pairing end merging was set at 20 bases. Sequences meeting these requirements were binned into Amplicon Sequence Variants (ASVs), which were provided in the form of feature tables and the representative sequences after being processed in DADA2. Representative sequence sets containing these ASVs were each aligned by MAFFT (ver. 7) (Katoh and Standley, 2013) from which a phylogenetic tree was created using FastTree (ver. 2.1) (Price et al., 2010). The Naïve Bayes classifier was trained on the Greengenes 13_8 99% ASV database (McDonald et al., 2012) using forward and reverse primers that were used to amplify V4 region of 16S rRNA gene. Afterwards, the representative sequences were compared and classified using the Greengenes taxonomy via Ribosomal Database Project classifier (Wang et al., 2007) at a nucleotide sequence similarity of 99%.
Quantification and statistical analysis
Bacterial community dynamics
Amplicon sequences, taxonomy data, and phylogenetic tree were used for community analyses using Vegan and Phyloseq packages (R 1.2.5019). Feature tables were rarefied at 5954 sequences and 5057 sequences for the BM and BP experiments, respectively. These data were used to calculate alpha diversity metrics such as species richness, phylogenetic diversity, and evenness using Pielou's evenness index (Figure 2). Shapiro Wilk’s Test and Bartlett’s test were used to examine the data normality and their homogeneity of variance, respectively. The results were used to categorize parametric and non-parametric data. To evaluate changes in alpha diversity over time on each diversity treatment, Kruskal-Wallis and Wilcox's post hoc were used for non-parametric data, while ANOVA and post-hoc Tukey tests were carried out for parametric data (Figure 2). Variation in community structure and composition (beta diversity) was assessed by calculating Bray Curtis dissimilarity and visualized it via Principle Coordinates Analysis (PCoA). Temporal patterns of beta diversity on each diversity treatment were assessed with PERMANOVA. Differences between time points were determined by pairwiseAdonis (Martinez Arbizu, 2020) (Figure 3). The bacterial community dynamics after invasion was also identified through their successional patterns (Figures S7 and S8), according to the method proposed by Jurburg et al. (2017) and Shade et al. (2013). The ASVs across each treatment in both Bacillus experiments were clustered using taxon relative abundances z-scored across the time series. ASVs were selected based on three criteria: ASVs that (1) significantly varied with time after each Bacillus invasion (ANOVA, p < 0.05), (2) made up at least 0.5% of the community at least once throughout the experiment, and (3) appeared in three or more samples. The validity of the selected set of ASVs was verified by comparing it to the original dataset using procrustes analysis on Bray-Curtis dissimilarity matrices using “protest”. Each data subset was then clustered by time using hierarchical cluster analysis based on Bray-Curtis dissimilarity matrices. The number of clusters formed per treatment was determined by cutting the dendrogram at specific height, using the function rect.hclust (Figures S7 and S8).
Phylogenetic turnover
Phylogenetic turnover was quantified by calculating the abundance weighted β-mean nearest taxon distance (βMNTDobs) using the R function comdistnt in the picante package (Figure 4). For the null modeling approach, the tip labels of the phylogeny were randomized 999 times, generating null βMNTD distributions. The deviation magnitude and direction between an observed βMNTD value and the null βMNTD distribution was quantified using the β-nearest taxon index (βNTI) (Stegen et al., 2013):
If βNTI was significantly lower than −2 or greater than +2, the observed βMNTD deviates from stochastic phylogenetic turnover (null βMNTD), and changes in community composition are deterministic. βNTI < −2 suggests phylogenetic turnover was driven by homogenous selection, while βNTI > +2 means variable selection. βNTI values between −2 and +2 indicate that the observed βMNTD does not deviate from stochastic phylogenetic turnover (null βMNTD), and therefore communities are assembled by stochastic processes (Dini-Andreote et al., 2015).
Acknowledgments
We thank EcoStyle for providing B. pumilus ECO-B-02, Jan Spoelder and HLB for providing B. mycoides M2E15, and Moniek Bloemsma for helping with the experiment. This work was supported by the Indonesia Endowment Fund for Education (LPDP, Departemen Keuangan, Republik Indonesia) scholarship to P.C.M. J.F.S. was financed by ERA-NET Cofund SusCrop project potatoMETAbiome (Grant No 771134), supported by EU Horizon 2020 research and innovation program and NWO, and part of the Joint Programming Initiative on Agriculture, Food Security and Climate Change (FACCE-JPI).
Author contributions
P.C.M. as the main author conceived the research question, designed the experiment, conducted the experiment, and analyzed the data. S.L.L. helped the experiments. J.F.S. and J.V.E. helped analyzing the data. All authors discussed the results. P.C.M. wrote the manuscript, J.F.S. and J.V.E. finalized it with inputs from all authors.
Declaration of interests
The authors declare no competing interests.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104185.
Contributor Information
Panji Cahya Mawarda, Email: p.c.mawarda@rug.nl, panji.cahya.mawarda@brin.go.id.
Joana Falcao Salles, Email: j.falcao.salles@rug.nl.
Supplemental information
Data and code availability
-
•
Raw reads of DNA-based sequences used in this study are available in the Sequence Read Archive of the National Center for Biotechnology Information under the accession number PRJNA721368.
-
•
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alori E.T., Babalola O.O. Microbial inoculants for improving crop quality and human health in Africa. Front. Microbiol. 2018;9:2213. doi: 10.3389/FMICB.2018.02213/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor D.R., Ratzke C., Gore J. Transient invaders can induce shifts between alternative stable states of microbial communities. Sci Adv. 2020;6:7–8. doi: 10.1126/sciadv.aay8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baweja P., Kumar S., Kumar G. Fertilizers and pesticides: their impact on soil health and environment. Soil Health. 2020;59:265–285. doi: 10.1007/978-3-030-44364-1_15. [DOI] [Google Scholar]
- Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy C., Dooley Á., Kirwan L., Finn J.A., McDonnell J., Bell T., Cadotte M.W., Connolly J. Biodiversity and ecosystem function: making sense of numerous species interactions in multi-species communities. Ecology. 2017;98:1771–1778. doi: 10.1002/ecy.1872. [DOI] [PubMed] [Google Scholar]
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Han X., Hu C., Chen J., Zhang D., Steinberger Y. Changes in the abundance and structure of a soil mite (Acari) community under long-term organic and chemical fertilizer treatments. Appl. Soil Ecol. 2011;49:131–138. [Google Scholar]
- Chase J.M. Drought mediates the importance of stochastic community assembly. Proc. Natl. Acad. Sci. U S A. 2007;44:17430–17434. doi: 10.1073/pnas.0704350104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S.P., Dietel K., Rändler M., Schmid M., Junge H., Borriss R., Hartmann A., Grosch R. Effects of Bacillus amyloliquefaciens FZB42 on lettuce growth and health under pathogen pressure and its impact on the rhizosphere bacterial community. PLoS One. 2013;8:1–10. doi: 10.1371/journal.pone.0068818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyte K.Z., Schluter J., Foster K.R. The ecology of the microbiome: networks, competition, and stability. Science. 2015;350:663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- De Graaff M.A., Adkins J., Kardol P., Throop H.L. A meta-analysis of soil biodiversity impacts on the carbon cycle. Soil Discuss. 2015;1:907–945. [Google Scholar]
- De Vries F.T., Thébault E., Liiri M., Birkhofer K., Tsiafouli M.A., Bjørnlund L., Jørgensen H.B., Brady M.V., Christensen S., De Ruiter P.C., et al. Soil food web properties explain ecosystem services across European land use systems. Proc. Natl. Acad. Sci. U S A. 2013;110:14296–14301. doi: 10.1073/pnas.1305198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Baquerizo M., Giaramida L., Reich P.B., Khachane A.N., Hamonts K., Edwards C., Lawton L.A., Singh B.K. Lack of functional redundancy in the relationship between microbial diversity and ecosystem functioning. J. Ecol. 2016;104:936–946. [Google Scholar]
- Dini-Andreote F., Jia X., Salles J.F. Molecular methods to study microbial succession in soil. Microb. Ecol. 2019 doi: 10.21775/9781912530021.03. [DOI] [Google Scholar]
- Dini-Andreote F., Stegen J.C., Van Elsas J.D., Salles J.F. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. U S A. 2015;11:1326–1332. doi: 10.1073/pnas.1414261112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditterich F., Poll C., Pronk G.J., Heister K., Chandran A., Rennert T., Kögel-Knabner I., Kandeler E. Succession of soil microbial communities and enzyme activities in artificial soils. Pedobiologia. 2016;59:93–104. [Google Scholar]
- Dumbrell A.J., Nelson M., Helgason T., Dytham C., Fitter A.H. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 2010;4:337–345. doi: 10.1038/ismej.2009.122. [DOI] [PubMed] [Google Scholar]
- Eisenhauer N., Schulz W., Scheu S., Jousset A. Niche dimensionality links biodiversity and invasibility of microbial communities. Funct. Ecol. 2013;27:282–288. [Google Scholar]
- Emmerson M., Morales M.B., Oñate J.J., Batáry P., Berendse F., Liira J., Aavik T., Guerrero I., Bommarco R., Eggers S., et al. How agricultural intensification affects biodiversity and ecosystem services. Adv. Ecol. Res. 2016;55:43–97. [Google Scholar]
- Evans S., Martiny J.B.H., Allison S.D. Effects of dispersal and selection on stochastic assembly in microbial communities. ISME J. 2017;11:176–185. doi: 10.1038/ismej.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine S., Mariotti A., Abbadie L. The priming effect of organic matter: a question of microbial competition? Soil Biol. Biochem. 2003;35:837–843. [Google Scholar]
- Fu L., Penton C.R., Ruan Y., Shen Z., Xue C., Li R., Shen Q. Inducing the rhizosphere microbiome by biofertilizer application to suppress banana Fusarium wilt disease. Soil Biol. Biochem. 2017;104:39–48. [Google Scholar]
- Girvan M.S., Campbell C.D., Killham K., Prosser J.I., Glover L.A. Bacterial diversity promotes community stability and functional resilience after perturbation. Environ. Microbiol. 2005;7:301–313. doi: 10.1111/j.1462-2920.2005.00695.x. [DOI] [PubMed] [Google Scholar]
- Glasl B., Smith C.E., Bourne D.G., Webster N.S. Exploring the diversity-stability paradigm using sponge microbial communities. Sci. Rep. 2018;8:1–9. doi: 10.1038/s41598-018-26641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl D.M., Vangay P., Garbe J., MacLean A., Hauge A., Becker A., Gould T.J., Clayton J.B., Johnson T.J., Hunter R., et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 2016;34:942–949. doi: 10.1038/nbt.3601. [DOI] [PubMed] [Google Scholar]
- Handa I.T., Aerts R., Berendse F., Berg M.P., Bruder A., Butenschoen O., Chauvet E., Gessner M.O., Jabiol J., Makkonen M., et al. Consequences of biodiversity loss for litter decomposition across biomes. Nature. 2014;509:218–221. doi: 10.1038/nature13247. [DOI] [PubMed] [Google Scholar]
- Ho A., Di Lonardo D.P., Bodelier P.L.E. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 2017;93:1–14. doi: 10.1093/femsec/fix006. [DOI] [PubMed] [Google Scholar]
- Johnsen K., Jacobsen C.S., Torsvik V., Sørensen J. Pesticide effects on bacterial diversity in agricultural soils - a review. Biol. Fertil. Soils. 2001;33:443–453. [Google Scholar]
- Jurburg S.D., Nunes I., Stegen J.C., Le Roux X., Priemé A., Sørensen S.J., Salles J.F. Autogenic succession and deterministic recovery following disturbance in soil bacterial communities. Sci. Rep. 2017;7:1–11. doi: 10.1038/srep45691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpouzas D.G., Karatasas A., Spiridaki E., Rousidou C., Bekris F., Omirou M., Ehaliotis C., Papadopoulou K.K. Impact of a beneficial and of a pathogenic Fusarium strain on the fingerprinting-based structure of microbial communities in tomato (Lycopersicon esculentum Milll.) rhizosphere. Eur. J. Soil Biol. 2011;47:400–408. [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijn D., Kohler F., Báldi A., Batáry P., Concepción E.D., Clough Y., Díaz M., Gabriel D., Holzschuh A., Knop E., et al. On the relationship between farmland biodiversity and land-use intensity in Europe. Proc. R. Soc. B Biol. Sci. 2012;276:903–909. doi: 10.1098/rspb.2008.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner A., Herschkovitz Y., Baudoin E., Nazaret S., Moenne-Loccoz Y., Okon Y., Jurkevitch E. Effect of Azospirillum brasilense inoculation on rhizobacterial communities analyzed by denaturing gradient gel electrophoresis and automated ribosomal intergenic spacer analysis. Soil Biol. Biochem. 2006;38:1212–1218. [Google Scholar]
- Lin Q., De Vrieze J., Li C., Li J., Li J., Yao M., Hedenec P., Li H., Li T., Rui J., et al. Temperature regulates deterministic processes and the succession of microbial interactions in anaerobic digestion process. Water Res. 2017;123:134–143. doi: 10.1016/j.watres.2017.06.051. [DOI] [PubMed] [Google Scholar]
- Ma M., Jiang X., Wang Q., Guan D., Li L., Ongena M., Li J. Isolation and identification of PGPR strain and its effect on soybean growth and soil bacterial community composition. Int. J. Agric. Biol. 2018;20:1289–1297. [Google Scholar]
- Mallon C.A., Le Roux X., Van Doorn G.S., Dini-Andreote F., Poly F., Salles J.F. The impact of failure: unsuccessful bacterial invasions steer the soil microbial community away from the invader’s niche. ISME J. 2018;12:728–741. doi: 10.1038/s41396-017-0003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon C.A., Poly F., Le Roux X., Marring I., Van Elsas J.D., Salles J.F. Resource pulses can alleviate the biodiversity-invasion relationship in soil microbial communities. Ecology. 2015;4:915–926. doi: 10.1890/14-1001.1. [DOI] [PubMed] [Google Scholar]
- Mallon C.A., Van Elsas J.D., Salles J.F. Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol. 2015;23:719–729. doi: 10.1016/j.tim.2015.07.013. [DOI] [PubMed] [Google Scholar]
- Mar Vázquez M., César S., Azcón R., Barea J.M. Interactions between arbuscular mycorrhizal fungi and other microbial inoculants (Azospirillum, Pseudomonas, Trichoderma) and their effects on microbial population and enzyme activities in the rhizosphere of maize plants. Appl. Soil Ecol. 2000;15:261–272. [Google Scholar]
- Martinez Arbizu P. R. Package Version 0.4; 2020. pairwiseAdonis: pairwise multilevel comparison using adonis.https://github.com/pmartinezarbizu/pairwiseAdonis [Google Scholar]
- Mawarda P.C., Le Roux X., van Elsas J.D., Salles J.F. Deliberate introduction of invisible invaders: a critical appraisal of the impact of microbial inoculants on soil microbial communities. Soil Biol. Biochem. 2020;148:1–13. [Google Scholar]
- McCann K.S. The diversity-stability. Nature. 2000;405:228–233. doi: 10.1038/35012234. [DOI] [PubMed] [Google Scholar]
- McDonald D., Price M.N., Goodrich J., Nawrocki E.P., Desantis T.Z., Probst A., Andersen G.L., Knight R., Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012 doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P.J., Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddas J.J., Roller G.B., Long J.W., Saah D.S., Moritz M.A., Stark D.T., Schmidt D.A., Buchholz T., Freed T.J., Alvey E.C., et al. California Natural Resources Agency; 2018. Fuel Treatment for Forest Resilience and Climate Mitigations: a Critical Review for Coniferous Forests of the Sierra Nevada, Southern Cascade, Klamath, and Transverse Ranges. CCCA4-CNRA-2018-017. [Google Scholar]
- Moorhead D.L., Sinsabaugh R.L. A theoretical model of litter decay and microbial interaction. Ecol. Monogr. 2006;76:151–174. [Google Scholar]
- Nassal D., Spohn M., Eltlbany N., Jacquiod S., Smalla K., Marhan S., Kandeler E. Effects of phosphorus-mobilizing bacteria on tomato growth and soil microbial activity. Plant Soil. 2018;427:17–37. [Google Scholar]
- Nemergut D.R., Knelman J.E., Ferrenberg S., Bilinski T., Melbourne B., Jiang L., Violle C., Darcy J.L., Prest T., Schmidt S.K., Townsend A.R. Decreases in average bacterial community rRNA operon copy number during succession. ISME J. 2016;10:1147–1156. doi: 10.1038/ismej.2015.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut D.R., Schmidt S.K., Fukami T., O’Neill S.P., Bilinski T.M., Stanish L.F., Knelman J.E., Darcy J.L., Lynch R.C., Wickey P., Ferrenberg S. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 2013;77:342–356. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen, J., Blanchet, F.G., Friendly, M., Kindt, R., Legendre, P., Mcglinn, D., Minchin, P.R., O’hara, R.B., Simpson, G.L., Solymos, P. et al., 2020. Package “Vegan” Title Community Ecology Package Version 2.5-7. http://CRAN.Rproject.org/package=vegan
- Owen S.M., Sieg C.H., Johnson N.C., Gehring C.A. Exotic cheatgrass and loss of soil biota decrease the performance of a native grass. Biol. Invasions. 2013;15:2503–2517. [Google Scholar]
- Pearson D.E., Ortega Y.K., Eren Ö., Hierro J.L. Community assembly theory as a framework for biological invasions. Trends Ecol. Evol. 2018;33:313–325. doi: 10.1016/j.tree.2018.03.002. [DOI] [PubMed] [Google Scholar]
- Pennekamp F., Pontarp M., Tabi A., Altermatt F., Alther R., Choffat Y., Fronhofer E.A., Ganesanandamoorthy P., Garnier A., Griffiths J.I., et al. Biodiversity increases and decreases ecosystem stability. Nature. 2018;563:109–112. doi: 10.1038/s41586-018-0627-8. [DOI] [PubMed] [Google Scholar]
- Philippot L., Spor A., Hénault C., Bru D., Bizouard F., Jones C.M., Sarr A., Maron P.A. Loss in microbial diversity affects nitrogen cycling in soil. ISME J. 2013;7:1609–1619. doi: 10.1038/ismej.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashar P., Shah S. In: Sustainable Agriculture Reviews. Lichtfouse E., Navarrete M., Debaeke P., Veronique S., editors. Springer; 2016. Impact of fertilizers and pesticides on soil microflora in agriculture; pp. 331–361. [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:1–10. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riah-Anglet W., Trinsoutrot-Gattin I., Martin-Laurent F., Laroche-Ajzenberg E., Norini M.P., Latour X., Laval K. Soil microbial community structure and function relationships: a heat stress experiment. Appl. Soil Ecol. 2015;86:121–130. [Google Scholar]
- Shade A., McManus P.S., Handelsman J. Unexpected diversity during community succession in the apple flower microbiome. mBio. 2013;4:1–12. doi: 10.1128/mBio.00602-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A., Peter H., Allison S.D., Baho D.L., Berga M., Bürgmann H., Huber D.H., Langenheder S., Lennon J.T., Martiny J.B.H., et al. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 2012 doi: 10.3389/fmicb.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegen J.C., Lin X., Fredrickson J.K., Chen X., Kennedy D.W., Murray C.J., Rockhold M.L., Konopka A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013;7:2069–2079. doi: 10.1038/ismej.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D., Reich P.B., Knops J.M.H. Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature. 2006;441:629–632. doi: 10.1038/nature04742. [DOI] [PubMed] [Google Scholar]
- Tsiafouli M.A., Thébault E., Sgardelis S.P., de Ruiter P.C., van der Putten W.H., Birkhofer K., Hemerik L., de Vries F.T., Bardgett R.D., Brady M.V., et al. Intensive agriculture reduces soil biodiversity across Europe. Glob. Change Biol. 2015;21:973–985. doi: 10.1111/gcb.12752. [DOI] [PubMed] [Google Scholar]
- Valentín L., Rajala T., Peltoniemi M., Heinonsalo J., Pennanen T., Mäkipää R. Loss of diversity in wood-inhabiting fungal communities affects decomposition activity in Norway spruce wood. Front. Microbiol. 2014;5:1–11. doi: 10.3389/fmicb.2014.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellend M. Conceptual synthesis in community ecology. Q. Rev. Biol. 2010;85:183–206. doi: 10.1086/652373. [DOI] [PubMed] [Google Scholar]
- Wagg C., Bender S.F., Widmer F., Van Der Heijden M.G.A. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. U S A. 2014;111:5266–5270. doi: 10.1073/pnas.1320054111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li X., Li X., Wang H., Su Z., Wang X., Zhang H. Dynamic changes in microbial communities during the bioremediation of herbicide (chlorimuron-ethyl and atrazine) contaminated soils by combined degrading bacteria. PLoS One. 2018;13:1–14. doi: 10.1371/journal.pone.0194753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Yang T., Friman V.P., Xu Y., Shen Q., Jousset A. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 2015;6:1–9. doi: 10.1038/ncomms9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz S., Degrange V., Prosser J.I., Poly F., Commeaux C., Guillaumaud N., Le Roux X. Decline of soil microbial diversity does not influence the resistance and resilience of key soil microbial functional groups following a model disturbance. Environ. Microbiol. 2007;9:2211–2219. doi: 10.1111/j.1462-2920.2007.01335.x. [DOI] [PubMed] [Google Scholar]
- Wu B., Wang X., Yang L., Yang H., Zeng H., Qiu Y., Wang C., Yu J., Li J., Xu D., et al. Effects of Bacillus amyloliquefaciens ZM9 on bacterial wilt and rhizosphere microbial communities of tobacco. Appl. Soil Ecol. 2016;103:1–12. [Google Scholar]
- Xing J., Jia X., Wang H., Ma B., Salles J.F., Xu J. The legacy of bacterial invasions on soil native communities. Environ. Microbiol. 2020;23:1–13. doi: 10.1111/1462-2920.15086. [DOI] [PubMed] [Google Scholar]
- Xiong W., Guo S., Jousset A., Zhao Q., Wu H., Li R., Kowalchuk G.A., Shen Q. Bio-fertilizer application induces soil suppressiveness against Fusarium wilt disease by reshaping the soil microbiome. Soil Biol. Biochem. 2017;114:238–247. [Google Scholar]
- Yeates G.W., Wardle D.A., Watson R.N. Responses of soil nematode populations, community structure, diversity and temporal variability to agricultural intensification over a seven-year period. Soil Biol. Biochem. 1999;31:1721–1733. [Google Scholar]
- Yi Y., de Jong A., Spoelder J., Elzenga J.T.M., van Elsas J.D., Kuipers O.P. Draft genome sequence of Bacillus mycoides M2E15, a strain isolated from the endosphere of potato. Genome Announc. 2016;4:1–16. doi: 10.1128/genomeA.00031-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D., Wang N., Xia F., Li Q., Wang W. Impact of biocontrol agents Pseudomonas fluorescens 2P24 and CPF10 on the bacterial community in the cucumber rhizosphere. Eur. J. Soil Biol. 2013;59:36–42. [Google Scholar]
- Zhang Q.G., Zhang D.Y. Resource availability and biodiversity effects on the productivity, temporal variability and resistance of experimental algal communities. Oikos. 2006;114:385–396. [Google Scholar]
- Zhou J., Ning D. Stochastic community assembly: does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 2017;81:1–17. doi: 10.1128/MMBR.00002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw reads of DNA-based sequences used in this study are available in the Sequence Read Archive of the National Center for Biotechnology Information under the accession number PRJNA721368.
-
•
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.