Abstract
Three different resistance factors from the avilamycin biosynthetic gene cluster of Streptomyces viridochromogenes Tü57, which confer avilamycin resistance when expressed in Streptomyces lividans TK66, were isolated. Analysis of the deduced amino acid sequences showed that AviABC1 is similar to a large family of ATP-binding transporter proteins and that AviABC2 resembles hydrophobic transmembrane proteins known to act jointly with the ATP-binding proteins. The deduced amino acid sequence of aviRb showed similarity to those of other rRNA methyltransferases, and AviRa did not resemble any protein in the databases. Independent expression in S. lividans TK66 of aviABC1 plus aviABC2, aviRa, or aviRb conferred different levels of resistance to avilamycin: 5, 10, or 250 μg/ml, respectively. When either aviRa plus aviRb or aviRa plus aviRb plus aviABC1 plus aviABC2 was coexpressed in S. lividans TK66, avilamycin resistance levels reached more than 250 μg/ml. Avilamycin A inhibited poly(U)-directed polyphenylalanine synthesis in an in vitro system using ribosomes of S. lividans TK66(pUWL201) (GWO), S. lividans TK66(pUWL201-Ra) (GWRa), or S. lividans TK66(pUWL201-Rb) (GWRb), whereas ribosomes of S. lividans TK66 containing pUWL201-Ra+Rb (GWRaRb) were highly resistant. aviRa and aviRb were expressed in Escherichia coli, and both enzymes were purified as fusion proteins to near homogeneity. Both enzymes showed rRNA methyltransferase activity using a mixture of 16S and 23S rRNAs from E. coli as the substrate. Coincubation experiments revealed that the enzymes methylate different positions of rRNA.
Avilamycins (Fig. 1), produced by Streptomyces viridochromogenes Tü57, belong to the orthosomycin class of antibiotics (7). Avilamycins as well as other orthosomycins inhibit the growth of multidrug-resistant gram-positive bacteria (7, 30). One member of this class, the compound everninomicin (also called SCH27899 or ziracin), may provide an alternative to vancomycin and other antibiotics for the treatment of many infectious diseases (8, 12, 13, 17, 19, 28). Avilamycin A has been proposed to be a translation inhibitor binding to the 30S ribosomal subunit, and it was suggested that the drug acts by preventing the attachment of tRNA to the ribosomes (29). In contrast, everninomicin and avilamycin A were shown to bind exclusively to the 50S ribosomal subunit (20). Recently a clinical isolate of Streptococcus pneumoniae that showed decreased susceptibility to everninomicin was detected, and it was shown that the resistance was due to a point mutation that causes an Ile52-Ser substitution in ribosomal protein L16 (2). It was speculated that everninomicin interacts with rRNA and that this interaction is potentiated by an additional interaction with protein L16. Since S. viridochromogenes Tü57 is also resistant to the inhibitory action of its own antibiotic, an understanding of its mechanism of resistance may give further information on the mode of action of orthosomycin antibiotics. We have reported the cloning of a 65-kb region of DNA from S. viridochromogenes Tü57 containing genes (aviD, aviE, and aviM) encoding proteins involved in the biosynthesis of avilamycin A (14). Here we present the cloning and characterization of DNA fragments from the avilamycin biosynthetic gene cluster which, after transformation into Streptomyces lividans, conferred avilamycin resistance. Sequencing and expression of the corresponding genes revealed that they code for an ATP-binding cassette (ABC) transporter system and two rRNA methyltransferases.
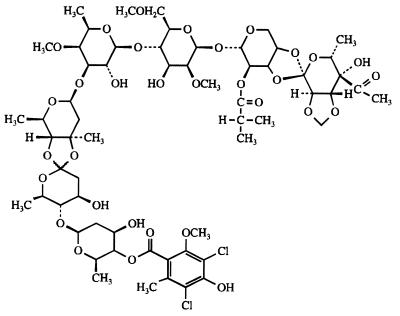
FIG. 1.
Structure of avilamycin A.
MATERIALS AND METHODS
Bacterial strains, plasmids, and materials.
S. viridochromogenes Tü57 and S. lividans TK66 were obtained from the culture collection of H. Zähner and W. Wohlleben, University of Tübingen (Tübingen, Germany). Escherichia coli XLIBlue and pBluescript SK(−) were from Stratagene (Heidelberg, Germany), and E. coli BL21(DE3)pLysS and pRSETb were from Invitrogen (Leek, The Netherlands). Plasmids pMunI and pMunII were generated in our lab, pUWL201 (25) was obtained from U. Wehmeier and W. Piepersberg (Wuppertal, Germany), pWHM3 (27) was obtained from H. Decker (Aventis, Frankfurt, Germany), and pLitmus 28 was from New England Biolabs (Beverly, Mass.). Medium components were purchased from Hartge Ingredients (Hamburg, Germany), and restriction enzymes and Sephadex G-25 columns were from Amersham-Pharmacia (Freiburg, Germany). Avilamycin A was a gift from Eli Lilly, carbenicillin was from Roth (Karlsruhe, Germany), and thiostrepton was from Sigma (Deisenhofen, Germany). l-[2,3,4,5,6-3H]phenylalanine (specific activity, 137 Ci/mmol) and S-adenosyl-l-[methyl-3H]methionine ([methyl-3H]AdoMet) (specific activity, 15 Ci/mmol) were from Amersham Life Science (Buckinghamshire, United Kingdom). Poly(U), tRNA (phenylalanine specific), pyruvate kinase, ATP, GTP, S-adenosyl-l-methionine (AdoMet), and sodium salt phosphoenolpyruvate were from Sigma (Deisenhofen, Germany), and all other chemicals were from Roth.
Culture conditions.
S. lividans TK66 was maintained on HA medium containing 1% malt extract, 0.4% yeast extract, 1.6% agar, 0.4% glucose, and 1 mM CaCl2. Protoplast formation, transformation, and regeneration of protoplasts of S. lividans TK66 were done according to standard procedures (16). Plasmids were passed through E. coli ET12567 (dam dcm hsdS Cm+) (11) to generate unmethylated DNA before their use to transform S. lividans TK66. General methods for the cultivation of E. coli strains were as previously described (24). Thiostrepton (25 μg/ml), carbenicillin (50 μg/ml), and avilamycin (5 μg/ml) were used for selective growth of recombinant strains.
DNA manipulation.
Isolation of E. coli plasmid DNA, digestion of DNA with restriction endonucleases, and Southern hybridization were carried out according to the directions of the manufacturer (Amersham, Braunschweig, Germany). Restriction mapping, other routine molecular methods, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Coomassie blue staining) were performed as described previously (24).
DNA sequencing and computer analysis.
DNA was sequenced by the dideoxynucleotide chain termination method with Thermo Sequenase. Universal and reverse primers were used. Sequencing reactions were performed on an automated sequencer (Vistra 725) from Molecular Dynamics (Krefeld, Germany) and on an ABI sequencer from 4-Base Lab GmbH (Reutlingen, Germany). Computer-aided sequence analysis was done with the DNASIS software package (version 2.1, 1995; Hitachi Software Engineering), and database searches were performed with the BLAST 2.0 program on the server of the National Center for Biotechnology Information, Bethesda, Md.
Construction of plasmids for heterologous gene expression in S. lividans TK66.
For cloning experiments, pMunII and pMunI, which had been derived from pBluescript Sk− by removing the BamHI, SmaI, PstI, EcoRI, and EcoRV sites between the SpeI and HindIII sites of the polylinker and introducing EcoRI, BglII, NdeI, NsiI, NcoI, and MunI (pMunII) and BamHI, NsiI, EcoRI, EcoRV, and MunI (pMunI) sites instead, were generated (26). Restriction sites 5′ to the ribosome-binding site and 3′ to the termination codon (aviRa, BamHI and EcoRI; aviRb, BglII and EcoRI; aviABC1 plus aviABC2, MunI and EcoRI) were introduced using PCR. Templates for the PCR were F4-B3 (fragment a in Fig. 2) for aviRa, F4-Bg10 (fragment b in Fig. 2) for aviRb, and P2-E8 (fragment c in Fig. 2) for aviABC1 plus aviABC2. The synthetic oligonucleotides used for the amplification of aviRa were primer A3 (5′ CTATGATGGATCCAGGGCTGCGACGGAGGC 3′) and primer A4 (5′ GTGAAAACACTGGAATTCACACTGGGGACA 3′), those for the amplification of aviRb were primer B3 (5′ CAGTCATTTTGTAGATCTCAGGAGCGGAAG 3′) and primer B4 (5′ GGCACGCGGAATTCGTGGGGCGGATCCAGG 3′), and those for the amplification of aviABC1 plus aviABC2 were primer ABC1 (5′ CGCTGAGCCAATTGATGAACGCCGCTGACC 3′) and primer ABC2 (5′ GGTCCGGAATTCCCGGACGGCACGCGGTTC 3′) (restriction sites are underlined). PCR fragments containing aviRa were restricted with BamHI and EcoRI and ligated into the BglII and EcoRI sites of pMunII to create pMunII-Ra. PCR fragments containing aviRb were ligated into the BglII and EcoRI sites of pLitmus28 to generate pLitmus28-Rb. A BglII - XbaI fragment containing aviRb was ligated into the corresponding site of pMunII to generate pMunII-Rb. PCR fragments containing aviABC1 plus aviABC2 were cloned into the EcoRI site of pMunI to generate pMunI-ABC. For expression the resistance genes were cloned into the HindIII and XbaI sites of pUWL201 to create the corresponding plasmids pUWL201-Ra, pUWL201-Rb, and pUWL201-ABC. For the coexpression of aviRa and aviRb, pMunII-Ra was restricted with MunI and XbaI, and the fragment containing aviRa was ligated into pMunII-Rb restricted with EcoRI and XbaI, generating pMunII-Rb+Ra. After HindIII and XbaI restriction, both genes were cloned into pUWL201 to create pUWL201-Rb+Ra. The construction of pUWL201-Rb+Ra+ABC was performed in a similar way. pMunI-ABC was restricted with MunI and XbaI, and the fragment containing aviABC1 plus aviABC2 was ligated into pMunII-Rb+Ra restricted with EcoRI and XbaI to create pMunII-Rb+Ra+ABC. From this construct, the fragment containing all four genes was ligated into the HindIII and XbaI sites of pUWL201, generating pUWL201+Rb+Ra+ABC (Table 1).
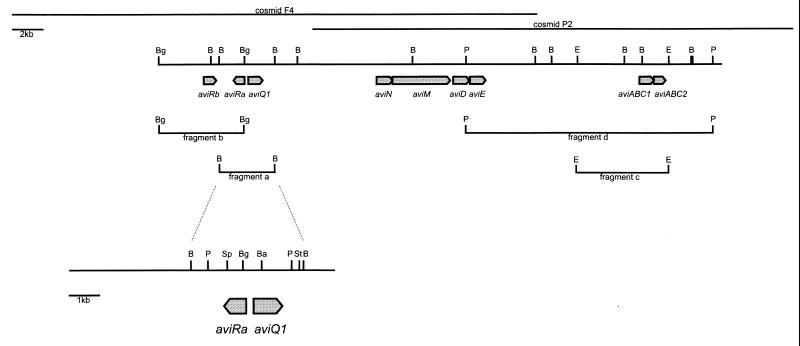
FIG. 2.
Genetic organization of the avilamycin biosynthetic gene cluster. Genes are indicated as arrows. Fragments a to c were used in constructing plasmids for gene expression experiments. B, BamHI; Ba, BalI; Bg, BglII; E, EcoRI; P, PstI; Sp, SphI; St, StuI. The restriction map is complete for BamHI; for the other sites, only those relevant for subcloning and gene deletion experiments are shown.
TABLE 1.
Most important strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| S. viridochromogenes Tü57 | Wild-type strain, avilamycin producer | 7, 14 |
| S. lividans | ||
| TK66 | Strain devoid of plasmid | 16 |
| TK66 (pUWL201) (GWO) | TK66 harboring plasmid pUWL201 | This study |
| TK66 (pUWL201-Ra) (GWRa) | TK66 harboring plasmid pUWL201-Ra | This study |
| TK66 (pUWL201-Rb) (GWRb) | TK66 harboring plasmid pUWL201-Rb | This study |
| TK66 (pUWL201-ABC) (GWABC) | TK66 harboring plasmid pUWL201-ABC | This study |
| TK66 (pUWL201-RbRa) (GWRbRa) | TK66 harboring plasmid pUWL201-RbRa | This study |
| TK66 (pUWL201-RbRaABC) (GWRbRaABC) | TK66 harboring plasmid pUWL201-RbRaABC | This study |
| E. coli | ||
| XLIBlue | Host strain for cloning experiments | Stratagene |
| BL21 (DE3)pLysS | Host strain for expression experiments | Invitrogen |
| Plasmids | ||
| pWHM3 | E. coli-Streptomyces shuttle vector, used for expression experiments in Streptomyces | 27 |
| pSG1 | 16-kb PstI fragment cloned into the vector pWHM3 | This study |
| pUWL201 | E. coli-Streptomyces shuttle vector, used for expression experiments in Streptomyces | 26 |
| pUWL201-Ra | Plasmid containing the cloned aviRa gene | This study |
| pUWL201-Rb | Plasmid containing the cloned aviRb gene | This study |
| pUWL201-ABC | Plasmid containing the cloned aviABC1/2 genes | This study |
| pUWL201-RbRa | Plasmid containing aviRa and aviRb | This study |
| pUWL201-RbRaABC | Plasmid containing aviRa, aviRb, and aviABC1/2 | This study |
| pRSETb | Vector used for expression experiments in E. coli | Invitrogen |
| pRSETb-Ra | Plasmid containing the cloned aviRa gene | This study |
| pRSETb-Rb | Plasmid containing the cloned aviRb gene | This study |
Avilamycin susceptibility testing.
Spores of S. lividans TK66 containing the single resistance genes or combinations were streaked on minimal medium (5) containing 25 μg of thiostrepton per ml and different concentrations of avilamycin A. Growth was determined after 4 days of incubation at 28°C.
Expression of aviRa and aviRb in E. coli and purification of the gene products.
An NdeI restriction site spanning the ATG start codon, a BamHI (or BglII for aviRb) site 5′ to the start codon, and an EcoRI site 3′ to the termination codon were introduced into aviRa or aviRb using PCR. The templates for PCR were F4-B3 (aviRa) and F4-Bg10 (aviRb). The primer sequences were primer A1 (5′ ACTGGGGGATCCACATATGAGTGCGTACCG 3′), primer A2 (5′ GTGAAAACACTGGAATTCACACTGGGGACA 3′), primer B1 (5′ GATAGAGATCTTCATATGGCAAGATCACGT 3′), and primer B2 (5′ GGCACGCGGAATTCGTGGGGCGGATCCAGG 3′) (the BamHI [BglII for aviRb], NdeI, and EcoRI restriction sites are underlined). The conditions for PCR were similar to those described previously (3). Reactions were performed using the Genetic Thermal Cycler system GeneAmp 2400 (Perkin-Elmer, Norwalk, Conn.). The PCR products were restricted with BamHI (or BglII) and EcoRI and were ligated into the expression vector pRSETb, generating pRSETb-Ra and pRSETb-Rb. E. coli BL21 (DE3)pLysS cells harboring plasmids pRSETb-Ra, pRSETb-Rb, and pRSETb (control) were grown at 37°C in NZCYM medium to an A600 of 0.6. Expression was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to give a final concentration of 1 mM. Growth was allowed to continue for 4 h. Cells were harvested by centrifugation and were washed and resuspended in RS buffer (10 mM Tris-HCl [pH 7.6], 10 mM MgSO4, 50 mM NH4Cl, 10 mM dithiothreitol). Cells were lysed by freezing to −20°C followed by thawing and treatment with lysozyme (2 mg/ml). Cell debris were removed by centrifugation (30 min, 15,000 × g), and the supernatant was applied to Ni-nitrilotriacetic acid agarose columns. Washing and elution steps were performed as described by the manufacturer. For removing the imidazole, extracts were applied to gel filtration columns (Sephadex G-25). The purity of both proteins was analyzed by electrophoresis on sodium dodecyl sulfate-polyacrylamide gels. The protein concentration was estimated as described by Bradford using bovine serum albumin as a standard (4).
In vitro methylation of rRNA with AviRa and AviRb.
Methyltransferase activity was investigated as described previously (3) using a mixture of 16S rRNA and 23S rRNA from E. coli as the substrate. In a final volume of 100 μl, each incubation mixture contained 3.1 μM rRNA, 125 μM AdoMet, 12.75 μCi of [methyl-3H]AdoMet, 1 μg of protein in 10 μl of RSG buffer, 25 mM HEPES-KOH (pH 7.5), 5 mM MgCl2, 25 mM NH4Cl, 5 mM dithiothreitol, and 1 U of RNase inhibitor. After 10 min at 37°C, the incubation mixture was extracted with phenol-chloroform, 2.5 μg of 5S rRNA and 10 μl of 3 M sodium acetate (pH 5.6) were added to the aqueous phase, and RNA was precipitated with 5 volumes of ethanol. The pellet was washed with 70% ethanol to remove most of the unincorporated labeled AdoMet and was redissolved in 1 ml of 10 mM Tris–1 mM EDTA (pH 7.6). The RNA was purified over NAP-5 columns, and radioactivity was determined by liquid scintillation counting. One picomole of methylated substrate was represented by 180 cpm.
Isolation of ribosomes.
The isolation of ribosomes was performed as described previously (22) with some modifications. Cultures of GWO, GWRa, GWRb, and GWRaRb were grown in TSB medium for about 60 h at 30°C. Mycelia were harvested by centrifugation and washed twice by resuspension in, and sedimentation from, water and subsequently from R buffer (25 mM HEPES-KOH [pH 7.5], 10 mM MgCl2, 50 mM NH4Cl, 3 mM dithiothreitol). The mycelium was then resuspended in the same buffer and passed through a chilled French pressure cell twice. The resultant extract was centrifuged at 30,000 × g for 30 min at 4°C, and the supernatant (designated S30) was retained. To prepare salt-washed ribosomes, S30 was layered over an equal volume of high-salt buffer (25 mM HEPES-KOH [pH 7.5], 30 mM MgCl2, 1 M NH4Cl, 3 mM dithiothreitol, 40% [wt/vol] sucrose) and centrifuged at 100,000 × g overnight at 4°C. The ribosomes were resuspended in R buffer, subdivided, and stored at −70°C. The postribosomal supernatant (designated S100) was dialyzed against R buffer at 4°C.
Poly(U)-dependent protein synthesis.
The sensitivity of ribosomes to avilamycin was determined using an in vitro translation system dependent on poly(U), as previously described (22), with the following minor modifications. As a source of soluble factors, S100 from the avilamycin-susceptible strain GWO was used. Before starting the assay, ribosomes (50 pmol) were preincubated (30°C, 5 min) with different concentrations of avilamycin A.
Nucleotide sequence accession numbers.
The nucleotide sequences reported here have been deposited in the GenBank database under accession numbers AF317788, AF317789, AF317790, and AF317791.
RESULTS
Cloning and sequencing of aviABC1 and aviABC2.
Most antibiotic gene clusters in Streptomyces contain an ABC transporter system that confers self-resistance to the antibiotic by secreting the antibiotic through the cell membrane. Based on this fact, we tried to identify a possible ABC transporter in the avilamycin gene cluster. Two oligonucleotide primers were designed from well-conserved amino acid sequences present in the so-called Walker A and B motifs that are present in the ATP-binding domains of antibiotic-producing actinomycetes (21). These primers were used to amplify DNA fragments using either chromosomal or cosmid DNA containing part of the avilamycin biosynthetic gene cluster as a template. PCR fragments were subcloned and sequenced. Two types of fragments with sizes of 450 and 402 bp were obtained using chromosomal DNA as a template. The deduced amino acid sequence of the 450-bp fragment showed homology to those of different amino acid transporters (1) while the deduced amino acid sequence of the 402-bp fragment was very similar to those of different proteins involved in antibiotic efflux (21). PCR amplification using cosmid P2 (Fig. 2) containing part of the avilamycin biosynthetic gene cluster as the template gave one type of PCR product which was identical to the 402-bp fragment. In order to clone the entire transporter gene, the 402-bp fragment was used as a probe in Southern hybridization experiments. A 16-kb PstI fragment (fragment d in Fig. 2) hybridized to the probe. This fragment was ligated into pWHM3 to create pSG1. This construct and pWHM3 were used to transform S. lividans TK66. Spores of S. lividans cultures containing plasmid pSG1 or pWHM3 were streaked on plates containing thiostrepton (25 μg/ml) plus avilamycin A (5 μg/ml). After incubation at 28°C for 72 h, transformants containing pSG1 were growing on the plates. No growth was observed on plates inoculated with spores of S. lividans TK66 containing pWHM3. Approximately 2 kb of DNA of a 5-kb EcoRI fragment (P2-E8) containing the ABC transporter gene was sequenced. Two open reading frames (ORFs) were identified by their predicted codon usage. The deduced amino acid sequence of ORF1 (AviABC1) showed high similarity to those of different ATP-binding proteins. The highest similarities were found to DrrA from Streptomyces peucetius (15) (51% identical amino acids) and to OleC from Streptomyces antibioticus (23) (49% identical amino acids). The deduced amino acid sequence of ORF2 (AviABC2) was similar to those of Ole-ORF5, a transmembrane protein from S. antibioticus (23) (28% identical amino acids), and DrrB, a transmembrane protein from S. peucetius (15) (27% identical amino acids). These proteins are thought to interact with the ATP-binding proteins, forming part of the ABC transporter system (21).
Cloning and sequencing of aviRa.
S. viridochromogenes Tü57 is highly resistant to avilamycin, but aviABC1 and aviABC2 only conferred low levels of resistance (see above). Consequently, it was assumed that other resistance determinants should be present in the avilamycin biosynthetic gene cluster. In order to isolate these resistance genes, BamHI and EcoRI fragments from cosmid F4 and cosmid P2 were ligated into pWHM3 and transformed into S. lividans TK66. Clones were subsequently selected on plates containing avilamycin A (5 μg/ml) plus thiostrepton (25 μg/ml). This selection scheme yielded several colonies. Restriction analysis of plasmids isolated from these colonies indicated three types of inserts: type A, with EcoRI inserts of 5 kb; type B, with BamHI inserts of 3.7 kb; and type C, with EcoRI inserts of 5 kb. Type B and type C plasmids contained overlapping DNA. Restriction mapping and Southern analysis revealed that the AviABC transporter genes were located on type A plasmids, and therefore these clones were discarded for further analysis. A clone from the second region containing a 3.7-kb BamHI fragment (fragment a in Fig. 2) was used for further investigations. Analysis of the nucleotide sequence revealed two ORFs, reading in opposite orientations. One ORF, aviQ1, consisted of nucleotides encoding a protein with 306 amino acids. The deduced amino acid sequence of aviQ1 showed similarity to those of UDP-glucose 4-epimerases. The highest similarities were found with a UDP-glucose 4-epimerase from Methanococcus jannaschii (6) (42% identical amino acids), ExoB from Azospirillum brasiliense (9) (38% identical amino acids), and ExoB from Rhizobium leguminosarum (25) (38% identical amino acids). The second ORF, aviRa, comprises 753 nucleotides encoding a protein with 251 amino acids. Searching available protein databases with the deduced amino acid sequence failed to reveal significant similarities. The size of the resistance-conferring fragment could be reduced to a 2,809-bp PstI fragment by subcloning experiments. This fragment was used to introduce deletions into either aviRa or aviQ1. A fragment containing a 1,255-bp BalI-StuI deletion still conferred avilamycin resistance. A fragment containing a frameshift mutation in aviRa which was obtained by SphI restriction and treatment with T4 polymerase and T4 DNA ligase failed to grow on avilamycin plates. This clearly limited the resistance determinant to aviRa (Fig. 2).
Cloning and sequencing of aviRb.
Another avilamycin resistance gene, aviRb, was found by sequencing approximately 3 kb DNA downstream from aviRa (fragment b in Fig. 2). The deduced amino acid sequence of aviRb was similar to those of several rRNA methyltransferases belonging to the SpoU family (18). aviRb was cloned into pUWL201 to create pUWL201-Rb. GWRb was shown to grow on avilamycin-containing plates (see below).
Avilamycin susceptibility testing.
Susceptibility to avilamycin was determined for S. lividans TK66 containing the avilamycin resistance genes alone or in combination. For coexpression of several genes in our mutants, special cloning vectors (pMunI and pMunII) were created. These vectors allow the construction of gene sets in a convenient way, taking advantage of the compatibility of MunI and EcoRI. The different genes were expressed individually (aviRa or aviRb) or in different combinations (aviABC1 plus aviABC2, aviRa plus aviRb, or aviRa plus aviRb plus aviABC1 plus aviABC2) under the control of the ermE∗ promoter. As shown in Table 2, GWO was susceptible to avilamycin even at a very low concentration (5 μg/ml). The MIC of avilamycin against GWABC was 10 μg/ml, that against GWRa was 20 μg/ml, and that for GWRb was >250 μg/ml. S. lividans TK66 containing either aviRa plus aviRb or all four resistance genes was even more resistant to avilamycin than GWRb.
TABLE 2.
Growth of S. lividans TK66 harboring different avilamycin resistance genes on avilamycin containing plates
| Gene(s) | Growth (%) on plates with the following avilamycin concn (μg/ml):
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 5 | 10 | 20 | 50 | 100 | 150 | 250 | |
| Control | 100 ± 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| aviABC1 plus aviABC2 | 100 ± 10 | 17 ± 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| aviRa | 100 ± 15 | 21 ± 0.5 | 10 ± 1 | 0 | 0 | 0 | 0 | 0 |
| aviRb | 100 ± 15 | 78 ± 4 | 68 ± 5 | 47 ± 7 | 44 ± 5 | 39 ± 4 | 32 ± 3 | 6 ± 1.5 |
| aviRa plus aviRb | 100 ± 15 | 100 ± 15 | 100 ± 12 | 100 ± 10 | 100 ± 15 | 100 ± 13 | 100 ± 9 | 51 ± 11 |
| aviRa plus aviRb plus aviABC1/2 | 100 ± 1 | 100 ± 16 | 100 ± 16 | 100 ± 12 | 100 ± 10 | 100 ± 13 | 100 ± 8 | 68 ± 13 |
Expression of AviRa and AviRb in E. coli and purification of both enzymes by affinity chromatography.
The endogenous promoters of aviRa and aviRb were removed, and the genes were placed in the T7 RNA polymerase-dependent expression vector pRSETb to create pRSETb-Ra and pRSETb-Rb. These plasmids were used to transform E. coli BL21(DE3)pLysS. After induction with IPTG, the production of soluble fusion proteins was observed. Recombinant proteins were purified using Ni-nitrilotriacetic acid columns. In both cases a nearly homogenous protein could be obtained.
Identification of AviRa and AviRb as rRNA methyltransferases.
rRNA methyltransferase activity on a mixture of 16S and 23S rRNAs from E. coli was detected using E. coli BL21(DE3)pLysS extracts containing either AviRa or AviRb. In a first incubation step mixtures of 21 pmol of 16S rRNA and 21 pmol of 23S rRNA were incubated for 60 min using 2.0 μg of AviRa or 2.0 μg of AviRb. In a second step, either AviRa or AviRb was added to the reaction mixtures and incubations were carried out for a second time. No rRNA methylation was detected using E. coli BL21(DE3)pLysS plus pRSETb (control). rRNA previously methylated by AviRa was accepted as a substrate by AviRb and vice versa, indicating that the enzymes methylate different positions of the rRNA (values for methyl groups incorporated into rRNA [picomoles per incubation] were as follows: AviRb plus AviRb, 16; AviRb plus AviRa, 33; AviRa plus AviRa, 17; AviRa plus AviRb, 34).
Sensitivity of ribosomes to avilamycin.
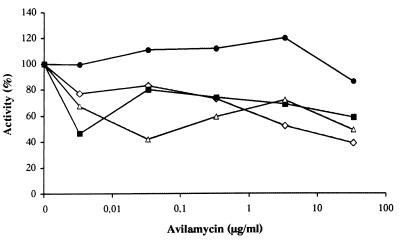
To investigate if methylation of rRNA by AviRa and AviRb confers resistance to avilamycin, 70S ribosomes of S. lividans TK66 harboring either pUWL201, pUWL201-Ra, pUWL201-Rb, or pUWL201-Rb+Ra were isolated. The sensitivity of the isolated ribosomes to avilamycin A was determined using an in vitro translation system. As shown in Fig. 3, avilamycin inhibited the poly(U)-directed polyphenylalanine synthesis using ribosomes of GWO to the extent of approximately 62%. Ribosomes of GWRaRb were highly resistant. Inhibition of the poly(U)-directed polyphenylalanine synthesis by avilamycin A was also observed when ribosomes isolated from GWRa and GWRb were used.
FIG. 3.
Effect of different concentrations of avilamycin on polyphenylalanine synthesis directed by poly(U). Activity is expressed as the percentage of activity obtained without antibiotic. Assay mixtures contained S100∗ from GWO and 70S ribosomes isolated from different strains. ◊, GWO; ▵, GWRb; ■, GWRa; ●, GWRbRa.
DISCUSSION
We have previously reported the isolation of a biosynthetic gene cluster from S. viridochromogenes Tü57 containing genes involved in the biosynthesis of avilamycin (14). We now report the cloning, sequencing and characterization of DNA fragments isolated from this cluster which confer avilamycin resistance when expressed in S. lividans. One of these fragments, identified by PCR amplification, confers low level of resistance to avilamycin (MIC, 10 μg/ml) in S. lividans. It contains two genes (aviABC1 and aviABC2) that resemble those encoding ABC transporter systems from antibiotic-producing actinomycetes. ABC transporters are characterized by a highly conserved domain comprising the ATP-binding site and a hydrophobic membrane domain. Three types of ABC transporter systems have been identified in antibiotic gene clusters (21). The AviABC1-AviABC2 transporter would belong to type I, where two independent genes code for the ATP-binding and membrane proteins, respectively. ABC transporter genes of this type have been isolated from several antibiotic producers such as S. peucetius (15), the producer of doxorubicin and daunorubicin, S. antibioticus, the producer of oleandomycin (23), and Streptomyces argillaceus, the producer of mithramycin (10). Since this ABC transporter confers a low level of resistance to avilamycin and since S. viridochromogenes Tü57, the avilamycin producer, is resistant to its own antibiotic at much higher concentrations, we thought that the strain should possess more than one resistance determinant. By subcloning DNA fragments from cosmid F4 (which contains avilamycin biosynthetic genes) into S. lividans, a second resistance gene (aviRa) that confers a low level of resistance (MIC, 20 μg/ml) was identified. Additionally, by sequencing flanking regions of cosmids F4 and P2, a third resistance gene (aviRb) was isolated. The AviRb protein resembles rRNA methyltransferases and confers a high level of resistance to avilamycin (250 μg/ml). S. lividans harboring either aviRa and aviRb or all four avilamycin resistance genes on one expression plasmid was tolerant to even higher avilamycin concentrations than S. lividans harboring aviRb, indicating an additive effect of the resistance proteins. Although AviRa did not resemble any protein in the databases, we were able to demonstrate that, like AviRb, it is an AdoMet-dependent rRNA methyltransferase. Both enzymes accept a mixture of 23S and 16S rRNAs as a substrate, and both enzymes act on rRNA previously methylated by either enzyme. Therefore, we assume that the two enzymes methylate different positions of the rRNA. We could confirm that methylation of the rRNA by AviRa and AviRb together is responsible for avilamycin resistance by demonstrating that polyphenylalanine synthesis directed by poly(U) was not inhibited by avilamycin A when 70S ribosomes isolated from GWRaRb were used. Ribosomes isolated from S. lividans TK66 containing either aviRa, aviRb, or no resistance gene were sensitive to avilamycin A. This was in contrast to the results of the in vivo studies. Recently it was shown that everninomicin as well as avilamycin binds exclusively to the 50S subunits, and this was contradictory to results of a previous study in which avilamycin was shown to act on 30S subunits. Protein L16 has also been implicated in the mechanism of action for everninomicin. Based on the known functions of protein L16, it was concluded that everninomicin inhibits protein synthesis either by altering the conformation of the A site and preventing the correct positioning of the bound tRNA or by competing with the tRNA molecule for a position within the A site (2). We now demonstrate that ribosomes from GWRaRb are completely resistant to avilamycin. We suggest that avilamycin binds to rRNA and that at least two sites of the rRNA are involved in this interaction. Further studies will be performed to elucidate which nucleotides of the rRNA are methylated by AviRa and AviRb.
ACKNOWLEDGMENTS
This work was supported by a grant from Baden Württemberg (Strukturfond), a grant from the Deutschen Forschungsgemeinschaft (Be 1389/4-1), and a grant from Eli Lilly and Company Limited, United Kingdom (to A.B.).
We especially thank F. Burke for supporting this study.
REFERENCES
- 1.Adams M D, Wagner L M, Graddis T J, Landick R, Antonucci T K, Gibson A L, Oxender D L. Nucleotide sequence and genetic characterization reveal six essential genes for the LIV-I and LS transport systems of Escherichia coli. J Biol Chem. 1990;265:11436–11443. [PubMed] [Google Scholar]
- 2.Adrian P V, Zhao W, Black T A, Shaw K J, Hare R S, Klugman K P. Mutations in ribosomal protein L16 conferring reduced susceptibility to everninomicin ( SCH27899): implications for mechanism of action. Antimicrob Agents Chemother. 2000;44:732–738. doi: 10.1128/aac.44.3.732-738.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bechthold A, Floss H G. Overexpression of the thiostrepton-resistance gene from Streptomyces azureus in Escherichia coli and characterization of recognition sites of the 23S rRNA A1067 2′-methyltransferase in the guanosine triphosphatase center of 23S ribosomal RNA. Eur J Biochem. 1994;224:431–437. doi: 10.1111/j.1432-1033.1994.00431.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–252. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Brawner M E, Auerbach J I, Fornwald J A, Rosenberg M, Taylor D P. Characterization of Streptomyces promoter sequences using the Escherichia coli galactokinase gene. Gene. 1985;40:191–201. doi: 10.1016/0378-1119(85)90042-3. [DOI] [PubMed] [Google Scholar]
- 6.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;23:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 7.Buzzetti F, Eisenberg F, Grant H N, Keller-Schierlein W, Voser W, Zähner H. Avilamycin. Experimentia. 1968;24:320–324. doi: 10.1007/BF02140794. [DOI] [PubMed] [Google Scholar]
- 8.Champney W S, Tober C L. Everninomicin ( SCH27899) inhibits both translation and 50S ribosomal subunit formation in Staphylococcus aureus cells. Antimicrob Agents Chemother. 2000;44:1413–1417. doi: 10.1128/aac.44.6.1413-1417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Troch P, Keijers V, Vanderleyden J. Sequence analysis of the Azospirillum brasilense exoB gene, encoding UDP-glucose 4′-epimerase. Gene. 1994;144:143–144. doi: 10.1016/0378-1119(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez E, Lombo F, Mendez C, Salas J A. An ABC transporter is essential for resistance to the antitumor agent mithramycin in the producer Streptomyces argillaceus. Mol Gen Genet. 1996;26:692–698. doi: 10.1007/BF02174118. [DOI] [PubMed] [Google Scholar]
- 11.Flett F, Mersinias V, Smith C P. High efficiency intergenic conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett. 1997;155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 12.Foster D R, Rybak M J. Pharmacologic and bacteriologic properties of SCH-27899 (Ziracin), an investigational antibiotic from the everninomicin family. Pharmacotherapy. 1999;19:1111–1117. doi: 10.1592/phco.19.15.1111.30576. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs P C, Barry A L, Brown S D. In vitro activities of SCH27899 alone and in combination with 17 other antimicrobial agents. Antimicrob Agents Chemother. 1999;43:2996–2997. doi: 10.1128/aac.43.12.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaisser S, Trefzer A, Stockert S, Kirschning A, Bechthold A. Cloning of an avilamycin biosynthetic gene cluster from Streptomyces viridochromogenes Tu57. J Bacteriol. 1997;179:6271–6278. doi: 10.1128/jb.179.20.6271-6278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guilfoile P G, Hutchinson C R. A bacterial analog of the mdr gene of mammalian tumor cells is present in Streptomyces peucetius, the producer of daunorubicin and doxorubicin. Proc Natl Acad Sci USA. 1991;88:8553–8557. doi: 10.1073/pnas.88.19.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 17.Jones R N, Marshall S A, Erwin M E. Antimicrobial activity and spectrum of SCH27899 (ziracin) tested against gram-positive species including recommendations for routine susceptibility testing methods and quality control. Diagn Microbiol Infect Dis. 1999;34:103–110. doi: 10.1016/s0732-8893(98)00093-5. [DOI] [PubMed] [Google Scholar]
- 18.Koonin, E. V., and K. E. Rudd. SpoU protein of Escherichia coli belongs to a new family of putative rRNA methylases. Nucleic Acids Res. 21:5519. [DOI] [PMC free article] [PubMed]
- 19.Lin C, Gupta S, Loebenberg D, Cayen M N. Pharmacokinetics of an everninomicin (SCH 27899) in mice, rats, rabbits, and cynomologus monkeys following intravenous administration. Antimicrob Agents Chemother. 2000;44:916–919. doi: 10.1128/aac.44.4.916-919.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNicholas P M, Najarian D J, Mann P A, Hesk D, Hare R S, Shaw K J, Black T A. Everninomicin binds exclusively to the 50S ribosomal subunit and inhibits translation in cell-free systems derived from both gram-positive and gram-negative bacteria. Antimicrob Agents Chemother. 2000;44:1121–1126. doi: 10.1128/aac.44.5.1121-1126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendez C, Salas J A. ABC transporters in antibiotic-producing actinomycetes. FEMS Microbiol Lett. 1998;158:1–8. doi: 10.1111/j.1574-6968.1998.tb12792.x. [DOI] [PubMed] [Google Scholar]
- 22.Quiros L M, Fidalgo S, Mendez F J, Hardisson C, Salas J A. Novel mechanisms of resistance to lincosamides in Staphylococcus and Arthrobacter spp. Antimicrob Agents Chemother. 1988;32:420–425. doi: 10.1128/aac.32.4.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez A M, Olano C, Vilches C, Mendez C, Salas J A. Streptomyces antibioticus contains at least three oleandomycin-resistance determinants, one of which shows similarity with proteins of the ABC-transporter superfamily. Mol Microbiol. 1993;8:571–582. doi: 10.1111/j.1365-2958.1993.tb01601.x. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Sanchez-Andujar B, Coronado C, Philip-Hollingsworth S, Dazzo F B, Palomares A J. Structure and role in symbiosis of the exoB gene of Rhizobium leguminosarum bv trifolii. Mol Gen Genet. 1997;255:131–140. doi: 10.1007/s004380050481. [DOI] [PubMed] [Google Scholar]
- 26.Trefzer A, Hoffmeister D, Kunzel E, Stockert S, Weitnauer G, Westrich L, Rix U, Fuchser J, Bindseil K U, Rohr J, Bechthold A. Function of glycosyltransferase genes involved in urdamycin A biosynthesis. Chem Biol. 2000;7:133–142. doi: 10.1016/s1074-5521(00)00079-x. [DOI] [PubMed] [Google Scholar]
- 27.Vara J, Lewandowska-Skarbek M, Wang Y G, Donadio S, Hutchinson C R. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus) J Bacteriol. 1989;171:5872–5881. doi: 10.1128/jb.171.11.5872-5881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang E, Simard M, Bergeron Y, Beauchamp D, Bergeron M G. In vivo activity and pharmacokinetics of ziracin ( SCH27899), a new long-acting everninomicin antibiotic, in a murine model of penicillin-susceptible or penicillin-resistant pneumococcal pneumonia. Antimicrob Agents Chemother. 2000;44:1010–1018. doi: 10.1128/aac.44.4.1010-1018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolf H. Avilamycin, an inhibitor of the 30S ribosomal subunit function. FEBS Lett. 1973;36:181–186. doi: 10.1016/0014-5793(73)80364-3. [DOI] [PubMed] [Google Scholar]
- 30.Wright E D. The orthosomycins, a new family of antibiotics. Tetrahedron. 1979;35:1207–1236. [Google Scholar]