Abstract
Calcium homeostasis modulator 1 (CALHM1) is a voltage- and Ca2+-gated ATP channel that plays an important role in neuronal signaling. However, as the previously reported CALHM structures are all in the ATP-conducting state, the gating mechanism of ATP permeation is still elusive. Here, we report cryo-EM reconstructions of two Danio rerio CALHM1 heptamers with ordered or flexible long C-terminal helices at resolutions of 3.2 Å and 2.9 Å, respectively, and one D. rerio CALHM1 octamer with flexible long C-terminal helices at a resolution of 3.5 Å. Structural analysis shows that the heptameric CALHM1s are in an ATP-nonconducting state with a central pore diameter of approximately 6.6 Å. Compared with those inside the octameric CALHM1, the N-helix inside the heptameric CALHM1 is in the “down” position to avoid steric clashing with the adjacent TM1 helix. Molecular dynamics simulations show that as the N-helix moves from the “down” position to the “up” position, the pore size of ATP molecule permeation increases significantly. Our results provide important information for elucidating the mechanism of ATP molecule permeation in the CALHM1 channel.
Keywords: calcium homeostasis modulator, CALHM1, ATP permeation, gating mechanism, assembly, cryo-EM
Abbreviations: CALHM, calcium homeostasis modulator; CHS, cholesteryl hemisuccinate; DDM, dodecyl-β-D-maltoside; LCH, long C-terminal helix; MD, molecular dynamics; POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; RMSD, root mean square deviation; TM, transmembrane helix
Adenosine triphosphate (ATP) release channels play an important role in various cellular signaling events (1). Consequently, their malfunctions are associated with many pathophysiological processes, including neurological disorders, inflammation, and cancer progression (2). To date, extensive research has identified five family proteins as human ATP release channels, although some of them need to be further verified. These five family proteins are connexins, pannexins, calcium homeostasis modulators (CALHMs), volume-regulated anion channels, and maxi-anion channels (2). Convergent evolution analysis showed that they all have four transmembrane helices (TMs) in the monomer that form an oligomer with a central pore (3). These oligomers are usually called hemichannels which release ATP to mediate the subsequent purinergic signal transduction of neighboring cells (4). Hemichannels formed by some isoforms of connexin and CALHM (such as connexin 26, connexin 43, connexin 50, CALHM2, CALHM4, etc.) can dock with each other and further form gap junction channels that allow direct exchange of small metabolites between cells (5, 6, 7).
CALHM1 is a large-pore nonselective ion channel gated by voltage and extracellular Ca2+ concentration (8). In addition, CALHM1 is a voltage-gated ATP release channel that mediates purinergic neurotransmission of sweet, bitter, and umami tastes from type II taste bud cells to the taste nerve (9, 10). Compared with the slow activation kinetics of CALHM1 homo-oligomerized channels, CALHM1/CALHM3 can form hetero-oligomerized ion channels with rapid voltage-gated activation kinetics that are closer to physiological states in vivo (2). Structural studies of CALHM family proteins have recently made great progress (11). Different CALHM isoforms form diverse oligomeric assemblies, ranging from 8 monomers to 13 monomers (12, 13, 14). The CALHM1 structures of vertebrate species have been reported to be an octamer and form a hemichannel (12, 13, 14), while the Caenorhabditis elegans CLHM1 can assemble as a nonamer, decamer, or undecamer and form two conformations of hemichannels and gap junctions (12, 13). Recent research on the CALHM1–CALHM2 chimera structure suggested that interactions between the long C-terminal helices (LCHs) and the TM4-LCH linker may determine the oligomeric state of the CALHM channels (14). Ren et al. reported a Danio rerio CALHM1 octamer with ordered long C-terminal helices (octamer+LCH) and proposed that the extracellular loop 1 region within the dimer interface may contribute to oligomeric assembly (13).
In this research, we reported cryo-EM reconstructions of two D. rerio CALHM1 heptamers with ordered long C-terminal helices (heptamer+LCH) or flexible long C-terminal helices (heptamer-noLCH) and one D. rerio CALHM1 octamer with flexible long C-terminal helices (octamer-noLCH). The comparison between different CALHM1 oligomers from the same species suggests that the CALHM1 channel may have multiple conformational states.
Results
Structure determination of drCALHM1 channels
The structures of the drCALHM1 channels were determined by cryo-EM. drCALHM1 channels were purified in the presence or absence of Ca2+ and concentrated for cryo-EM sample preparation. All datasets were collected using a Titan Krios electron microscope operated at 300-kV accelerating voltage with a K2 Summit direct electron-counting detector. Details on the data collection and data processes are shown in Figs. S1–S4. The heptamer+LCH structure was reconstructed from the dataset in the presence of Ca2+. The heptamer-noLCH and octamer-noLCH structures were reconstructed from datasets in the absence of Ca2+. The final 3D reconstructions of heptamer+LCH, heptamer-noLCH, and octamer-noLCH were determined at overall resolutions of 3.2 Å, 2.9 Å, and 3.5 Å, respectively (Figs. S1 and S2). The cryo-EM maps are sufficient to resolve most amino acid side chains (Figs. S3 and S4). Due to the quality of density, the N-helix of heptamer+LCH, heptamer-noLCH, or octamer-noLCH structures only builds an alanine model (Figs. S3 and S4).
Overall structure of the heptamer+LCH channel
The overall structure of heptameric drCALHM1 is shaped like a barrel and can be divided into three parts: the extracellular loop, transmembrane domain, and cytoplasmic domain (Fig. 1). From the top view, its extracellular region forms a ring with a diameter of approximately 98 Å, and seven small helices in the middle of the ring can be clearly observed (Fig. 1, A and D). The side view shows that the height of the drCALHM1 heptamer is approximately 92 Å. There are multiple lipid molecules that exist inside a protomer and within the interface of two protomers (Fig. 1, B and E). From the cytosolic view, it can be observed that the cytoplasmic domain mainly consists of one long helix. Seven LCHs intercross each other (Fig. 1, C and F), resulting in a ring diameter of approximately 100 Å. The overall drCALHM1 heptamer is quite similar to that of a previously reported octamer (13) except for the different oligomerization states.
Figure 1.
The overall architecture of heptamer+LCH. The surface (A) and cartoon (D) representations of the cryo-EM density map of heptameric drCALHM1 viewed from the extracellular side of the membrane. The seven subunits are represented by different colors. The surface (B) and cartoon (E) representations of the cryo-EM density map of heptamer+LCH viewed parallel to the membrane. The lipid-like density is gray. The surface (C) and cartoon (F) representations of the cryo-EM density map of heptamer+LCH viewed from the cytosolic side of the membrane. LCH, long C-terminal helix.
ATP-nonconducting state of the heptamer+LCH
As shown in Figure 1, the heptamer has smaller pores than the octamer, and we further calculated the exact pore size with the N-helix having all alanines in the current model (Fig. 2, A and B). The narrowest diameter of the heptamer+LCH channel is approximately 14 Å (Fig. 2C). If the putative side chain was built for the N-helix, the pore diameter was reduced to 6.6 Å, which blocked ATP permeation through the pore but allowed ion flux. To further validate this result, we took advantage of molecular dynamics (MD) simulation methodology. Coarse-grained simulations of heptamer+LCH in the presence of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipids were carried out. We added hydrated Na+, hydrated K+, and Cl− ions on either side of the POPC bilayer to the Martini, version 2.0, forcefield (15). Ca2+ ions were added on the extracellular side. During replicates of 10-μs coarse-grained MD simulations, we observed that multiple Na+ cations, K+ cations, and Cl− anions crossed the membrane through the heptamer+LCH channel. In contrast, Ca2+ ions were still on the extracellular side of the POPC bilayer after simulations and were unable to pass through the heptameric channel (Fig. 2D). These simulation results indicated that the heptamer+LCH channel favors the permeation of monovalent ions (Na+, K+, and Cl−) but repulses Ca2+ ions. Moreover, its pore size is too narrow for bulky ATP molecules.
Figure 2.
The center pore is in an ATP-nonconductive state.A, the stereo image for the fit of the N-helix (a polyalanine model) in the map. B, yellow dots represent the permeation path of the heptamer, where the N-helix is built as a polyalanine model. C, the pore radius along the central axis of drCALHM1 in different oligomerization states is marked as follows: Orange represents heptamer, where the N helix has a putative side chain; blue represents heptamer, where the N helix is a polyalanine model; gray represents octamer, where the N helix is a polyalanine model. D, the position of the ion in the heptamer channel in the coarse-grained simulations. Two opposing subunits of the heptamer are shown in ribbon. Na+, K+, Cl−, and Ca2+ ions are shown in the sphere model and colored purple, red, green, and orange, respectively. The color changes from transparent to opaque with the simulation time.
We also calculated the electrostatic surface potential distribution of the heptamer+LCH channel and found that the positive potential at the center of the pores of the heptamer is stronger than that of the octamer (Fig. S5), which may be due to the change in the oligomerization state to make the heptameric channel more compact. The strong positive potential distribution inside pores may indicate the voltage-dependent characteristics of ion flux through the CALHM1 channel, as shown by electrophysiological studies (16).
Structural comparison of a heptamer and octamer
To address the underlying mechanism of the formation of two different oligomers from the same CALHM1 protomer, we first compared the heptamer and octamer by superimposing one protomer of each assembly (Fig. 3A). The entire heptamer forms a smaller circle than the octamer. The protomer is composed of four TMs (TM1, TM2 plus TM2b, TM3 plus TM3b, and TM4), one extracellular helix, and one LCH. The protomers from the heptamer and octamer are extremely similar except for the position of the N-helix. Compared with the N-helix of the octamer, the N-helix of the heptamer moves toward the cytosolic direction by approximately 6 Å (Fig. 3B) (defined as the “down” position in the heptamer and the “up” position in the octamer hereafter). In the heptamer, the N-helix is aligned parallel to and interacts with TM1 (Fig. 3C). The side chain of Leu28 has hydrophobic interactions with Ala10. Moreover, the N-helix from one protomer has multiple interactions with TM1 from the neighboring protomer. One hydrogen bond was formed between the side chain of Gln32 and the main chain carboxyl oxygen atom of Ala9. Additionally, the side chain of Ile25 has hydrophobic interactions with Ala16. We anticipated more interactions between the N-helix and TM1 if the side chain of the N-helix could be built.
Figure 3.
Comparison of the structures of heptamer and octamer.A, cartoon representation of the drCALHM1 heptamer (rainbow) and octamer (gray), viewed from the top of the membrane after superimposing the protomer of each oligomer. B, the superposition of the transmembrane domain of the drCALHM1 heptamer (orange) and octamer (gray), viewed from the side of the membrane. The N-helix of the heptamer is marked in red for better contrast. C, the interaction between the N-helix and TM1. The orange TM1 and the red N-helix are in the same protomer, and the blue TM1 is in another adjacent protomer. The dashed lines represent hydrogen bonds. D, the comparison of the dimers of different oligomerization states by superimposing a protomer. The protomer of the heptamer is superimposed in orange, the other is blue, and the N-helix is marked red for contrast. The octamer is gray. E, comparison with trimers of the drCALHM1 heptamer and octamer (gray) by superimposition of a protomer, viewed from the top of the membrane. The three protomers of the heptamer are represented by orange, blue, and magenta. The red arrows indicate the steric clashes between the N-helix of the octamer and adjacent TM1 subunits.
We then superimposed the dimers from the heptamer and the octamer. These two dimers are similar except for two N-helices (Fig. 3D). Similar to the protomer comparison result, two N-helices in the dimer adapted from the heptamer obviously move down. After superimposing one protomer together, the neighboring protomer from the heptamer occupies a closer position to the pore than that from the octamer (Fig. 3E), resulting in the N-helix from the octamer having a steric clash with TM1 from the heptamer (as shown by the red arrow). Therefore, the N-helix is mandatory in the “down” position in the heptamer.
Movement of the N-helix increases the pore diameter
In order to check whether drCALHM1 maintains a functional channel in human cells, we transfected DNA encoding GFP-tagged full-length drCALHM1 into HEK293T cells and conducted electrophysiological studies. A large voltage-dependent outward current was observed using the whole-cell configuration (Fig. S6), indicating that the expression of drCALHM1 in HEK293T cells forms a channel similar to the previously reported Homo sapiens CALHM1 channel (17), although the oligomeric states of drCALHM1 are unknown.
To test the possibility that the movement of the N-helix can be involved in the pore size of CALHM1, we carried out MD simulations. With the help of unbiased supervised MD and biased targeted MD simulations for heptamer+LCH, we accelerated the movement of the N-helix in one protomer of the heptamer from the “down” position to the “up” position on a time scale accessible to MD simulations. During the supervised MD simulations, we monitored the root mean square deviation (RMSD) of the N-helix of protomer 1 (P1) in the current structure compared with that of P1 in the octamer to represent the movement of the N-helix. To monitor the pore size, we calculated the minimum distance of any atom between two N-helices from P1 and P4 protomers (denoted as P1−P4 hereafter), from P2 and P5 protomers (P2−P5), from P3 and P6 protomers (P3−P6), and from P4 and P7 protomers (P4−P7) (Fig. S7A). There was obvious deformation of the 7-fold symmetry structure in the N-helix region caused by the movement of the N-helix in P1 during the supervised MD simulations. Then, we calculated the potential of mean force for the RMSD of the N-helix versus. the P4−P7 distance (Fig. 4A). The potential of mean force map shows the transition process of the heptamer and clearly depicts three different conformational states (state 1, state 2, and state 3) of the heptamer starting from the initial state (state 0) (Fig. 4B). In initial state 0, the RMSD of the N-helix is approximately 6.66 Å, and the P4–P7 distance is approximately 9.57 Å. In states 1, 2, and 3 transformed from state 0, the RMSD of the N-helix is 5.63, 4.80, and 4.70 Å, and the P4−P7 distances are approximately 9.79, 11.89, and 13.26 Å, respectively. The conformational state of the N-helix moves closer to that of P1 in the octamer. Meanwhile, the pore size gradually increased from 9.79 Å to 13.26 Å. Furthermore, the targeted MD also observed that the pore size increased due to the movement of the N-helix (Fig. S7B). Conventional MD simulations were performed as controls, and the position of the N-helix and the pore size of the heptamer remained similar to those in the initial state throughout the simulation time (Fig. S7C). Additionally, we performed conventional MD simulations to show the time-dependent RMSD values for the Cα atoms of the heptamer+LCH or octamer+LCH structures relative to conformations obtained from the cryo-EM structures (Fig. S7, D and E). The RMSDs exhibited small variations during the atomic simulations, implying that the two structures were conformationally stable.
Figure 4.
The updown motion of the N-helix increases the pore diameter.A, two-dimensional PMF profile of the RMSD of the P1 N-helix versus the pore radius in the heptameric channel. The initial structure and three low-energy conformational states during supervised MD simulations are labeled state 0, state 1, state 2, and state 3. B, representative structures of P4 and P7 in state 0, state 1, state 2 and state 3 of the heptameric channel. Two opposing protomers of the heptamer are shown in cartoon. PMF, potential of mean force.
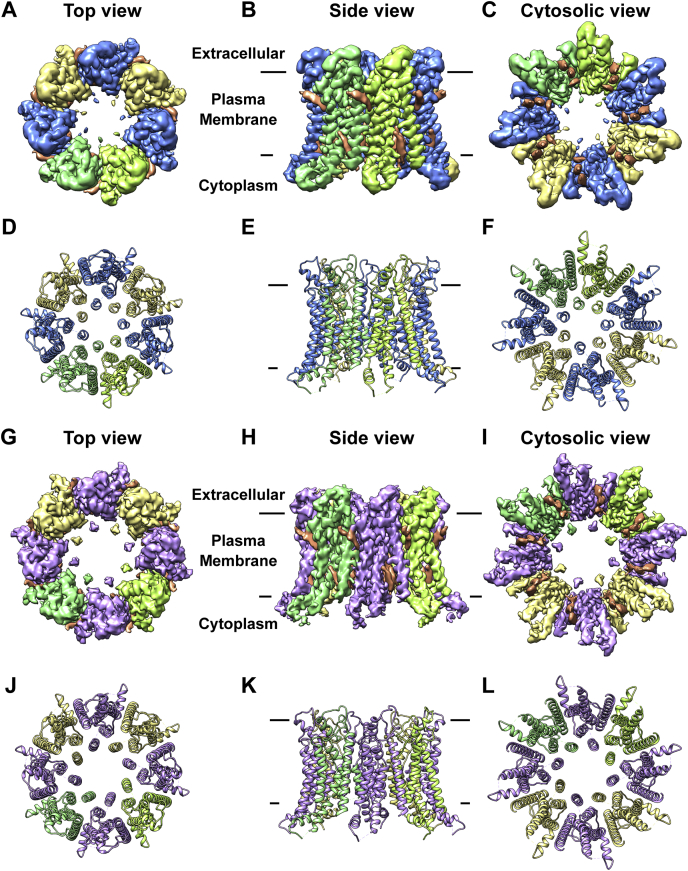
Structures of heptamer-noLCH and octamer-noLCH
Since the heptamer+LCH structure was determined using the dataset in the presence of Ca2+, we tried to understand whether Ca2+ can induce different oligomerization states of drCALHM1. Reprocessing the dataset in the absence of Ca2+, in addition to the previously reported octamer+LCH structure (13), we were able to identify two new conformation states of the drCALHM1 channel, in which the LCH is disordered (Fig. S2). We named them heptamer-noLCH (Fig. 5, A–F) and octamer-noLCH (Fig. 5, G–L). These results suggest that Ca2+ is unlikely to be the cause of the different oligomerization states of the drCALHM1 channel.
Figure 5.
Overall structure of heptamer-noLCH and octamer-noLCH. Density maps (A–C) and cartoons (D–F) of heptamer-noLCH in different directions. Blue and yellow are interlaced between adjacent subunits. Density maps (G–I) and cartoons (J–L) of different sides of octamer-noLCH. The adjacent subunits are interlaced with purple and yellow. To clearly show the changes in subunit assembly during the transition of the oligomeric state, the key subunits are marked with different degrees of green. The lipid-like densities are in orange.
The overall structures of heptamer-noLCH versus heptamer+LCH and octamer-noLCH versus octamer+LCH are highly similar except for LCHs (Fig. S8, A and B). Moreover, the pore diameters remained similar in these structures with or without LCHs (Fig. S8, C and D), indicating that the LCH did not regulate the pore size. We also compared the buried surface area of the protomer in different conformational states. With the LCH, the buried surface areas are 948.6 Å2 and 959.6 Å2 for the monomer in the heptamer+LCH structure and octamer+LCH structure, respectively. However, without the LCH, the buried surface areas are 627.6 Å2 and 603.3 Å2 for the protomer in the heptamer-noLCH structure and octamer-noLCH structure, respectively. These results suggest that the LCH may serve as a scaffold to stabilize oligomerized CALHM1 channels.
Discussion
In this study, we reported a heptameric drCALHM1 channel with an ATP nonconducting conformation. Compared with the octameric drCALHM1 channel (13), the heptameric drCALHM1 channel undergoes two major changes. One is a much smaller channel ring, leading to a smaller pore and thus possibly blocking ATP permeation. Since the narrowest pore diameter is approximately 6.6 Å, it is likely that the ions can pass through the pores of the heptamer. Indeed, MD results showed that Na+, K+, and Cl− can pass through the pore. However, Ca2+ was precluded from the pore, which may be caused by the positively charged environment inside the pore. Nevertheless, such narrow pores will unlikely allow ATP molecule permeation. The other difference is that the N-helix moves toward the cytosolic direction by approximately 6 Å to avoid the steric clash with neighboring TM1 helices, suggesting that the change in the oligomer state may correlate with the movement of the N-helix. Supervised MD simulations and targeted MD simulations indeed showed that the movement of the N-helix to the “up” position led to the deformation of the 7-fold symmetric structure and significantly increased pore size.
We also determined two conformational states of heptameric and octameric drCALHM1 channels with flexible LCHs. Although the current structures of heptamer-noLCH and octamer-noLCH were reconstructed from the Ca2+-free dataset, these two conformational states can be observed from the dataset in the presence of Ca2+. Moreover, it has been noted that the final models of heptamer-noLCH and octamer-noLCH do not include LCHs due to the intrinsic flexibility in these specific conformational states instead of protein degradation. The purified protein sample did not show obvious degraded bands from the SDS-PAGE gel. Therefore, it is reasonable to hypothesize that for the same type of oligomer, the drCALHM1 channel may exhibit equilibrium between two conformational states (ordered versus flexible LCHs).
Together with previously published results, we proposed a molecular mechanism of ATP permeation through the CALHM1 channel (Fig. S9). In the resting state, the CALHM1 channel may form a heptamer and equilibrium between ordered LCHs and flexible LCHs. Upon sensing stimulus, the N-helix inside the pore moves up toward the extracellular direction, driving conformational changes and oligomeric rearrangement to facilitate ATP molecule permeation.
Compared with the published ATP permeation mechanism of another ATP release channel, pannexin 1 (18), our proposed mechanism of ATP permeation has a common point: both use the C-terminal tail to regulate ATP permeation. The major difference is that the C-terminal tail has to be cleaved during ATP permeation in pannexin 1, while the C-terminal part in CALHM1 may switch to the disordered region for further assembly into higher-order oligomers to permeate ATP molecules.
To date, several CALHM subtype structures have been reported to form various oligomers (12, 13, 14, 19, 20). Consequently, the “protein gate” or “lipid gate” model has been proposed to explain ATP permeation (14, 19, 20). Our results are unfavorable for these two models. We did not observe any TM1 movement when compared with the closed and open states of drCALHM1 channels. Additionally, MD simulations did not support the argument that lipids enter the pore. During the 10-μs coarse-grained MD simulations, the lipids assembled into clearly defined upper and lower leaflets around both proteins but could not be accommodated within both channel pores of proteins (Fig. S10). Although the molecular mechanism of ATP permeation in CALHM channels requires further investigation, the feature of dynamic assembly in CALHM channels is certainly quite unique. The correlation between dynamic assembly and biological function remains to be ascertained in the future.
Experimental procedures
Expression of the drCALHM1 channel
Full-length D. rerio CALHM1 (UniProtKB number: E7F2J4, synthesized by Genewiz Inc) was ligated into the EcoRI-NotI restriction sites of the pEG-BacMam vector, with a tobacco etch virus protease cleavage site and enhanced green fluorescent protein at its N-terminus. The BacMam viruses were generated and amplified following the standard Bac-to-Mam Baculovirus expression system (Invitrogen) in Spodoptera frugiperda (Sf9) cells. The bacmid produced by DH10Bac cells was transfected into Sf9 cells using X-tremeGENE HP DNA Transfection Reagent (Roche) and then cultured at 27 °C. HEK293S GnTi- cells were cultured in Freestyle 293 expression medium (Thermo Fisher Scientific) at 37 °C with 5% CO2. When the cell density reached 2 × 106 cells per mL, the cells are transfected with the second-generation virus (P2). After transfection, the cells were incubated at 37 °C for 24 h, and then sodium butyrate was added to a final concentration of 10 mM to facilitate protein expression. The cells were cultured at 30 °C for another 60 h before harvest.
Purification of the drCALHM1 channel
The cells were collected by centrifugation at 800g, resuspended in lysis buffer consisting of 20 mM Tris-HCl pH 7.5, 200 mM NaCl, 1 mM PMSF, and then lysed through sonication for 15 min. The membrane pellets were collected by ultracentrifugation at 180,000g for 1 h and then homogenized in the buffer consisting of 20 mM Tris-HCl pH 7.5, 200 mM NaCl, and 1 × protease inhibitor cocktail (Roche), and solubilized in 1% (w/v) n-dodecyl-β-D-maltoside (DDM, Anatrace), 0.2% (w/v) cholesteryl hemisuccinate (CHS, Sigma–Aldrich) at 4 °C for 3 h. Insoluble membrane debris was removed by centrifugation at 110,000g for 40 min. The supernatant was applied to agarose beads conjugated with anti-GFP nanobody and rotated at 4 °C for 2.5 h. The beads were rinsed three times in 20 mM Tris-HCl pH 7.5, 500 mM NaCl, 0.025% (w/v) DDM, and 0.005% (w/v) CHS and then rinsed three times in 10 mM ATP (Macklin), 10 mM MgCl2, 20 mM Tris-HCl pH 7.5, 200 mM NaCl, 0.025% (w/v) DDM, and 0.005% (w/v) CHS. After that, the beads were washed with W buffer, which contained 20 mM Tris-HCl pH 7.5, 200 mM NaCl, and 0.0063% (w/v) glycol-diosgenin (Anatrace). drCALHM1 protein was eluted with W buffer containing tobacco etch virus protease overnight at 4 °C and concentrated for further purification by size-exclusion chromatography on a Superose 6 Increase 10/300 Gl column (GE Healthcare) in 20 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 0.0063% glycol-diosgenin with 2 mM Ca2+. The peak fractions were pooled, concentrated to 6 mg/ml by a 100-kDa cutoff Centricon (Merck Millipore), and flash-frozen for further cryo-EM grid preparation.
Cryo-EM sample preparation and imaging
An aliquot of 2.5 μl of purified drCALHM1 sample was applied onto a glow discharged holey carbon film grid (200 mesh, R2/1, Quantifoil). The grid was blotted and flash-frozen in liquid ethane with FEI Vitrobot Mark IV. The grid was loaded onto an FEI Titan Krios electron microscope operated at 300-kV accelerating voltage. Image stacks were recorded on a K2 Summit direct electron counting detector (Gatan) set in counting mode using SerialEM (http://bio3d.colorado.edu/SerialEM/). The defocus range is −1.5 to −2.5 μm, and the pixel size is 1.014 Å/pixel. A total of 40 frames were acquired in 8 s for each image stack, giving a total electron dose of 51.2 e−/Å2. Finally, 2292 image stacks were acquired in an imaging session of 24 h.
Image processing
The recorded image stacks were processed by MotionCor2 (21) for a 5 × 5 patch drift correction with dose weighting. The non–dose-weighted images were used for CTF estimation by CTFFIND 4 (22). Poor-quality images (no protein particles or ice-breaking streaks) are removed before the particles are picked up. A total of 801,380 particles were semiautomatically picked from dose-weighted images by Gautomatch (https://www.mrc-lmb.cam.ac.uk/kzhang/) and extracted by RELION-3 (23) in a box size of 260 pixels. A round of 2D classification was performed in RELION-3 to remove contaminations, ice, and bad particles, yielding 482,632 good-quality particles. The selected particles were then used to generate the initial model with an ab initio method with C1 symmetry in CryoSPARC-2 (https://cryosparc.com/). The initial model was low-pass filtered to 30 Å resolution and applied to the first round of 3D classification in RELION-3. The particles were divided into 3 subsets, and only 1 of the 3 subsets showed a 7-fold symmetry structure. Particles in this subset were selected for 3D autorefine with C7 symmetry. Two more rounds of 3D classification by local angular search with C7 symmetry yielded 23,073 particles in a well-defined class that showed the LCH domain in the map. Another round of 3D autorefining was performed with C7 symmetry and generated reconstruction at 3.4 Å resolution. Per-particle motion correction was carried out using Bayesian polishing in RELION-3. The shiny, polished particles were then transferred to CryoSPARC-2 for 3D classification using heterogenous refinement with C7 symmetry. A total of 12,671 particles were selected and then refined to 3.2 Å resolution in CryoSPARC-2 using nonuniform refinement. The stated resolutions were evaluated using the “gold_standard” FSC = 0.143 criterion. The local resolution was calculated by ResMap (24) or using two cryo-EM maps independently refined from halves of the data. For the structures of heptamer-noLCH and octamer-noLCH, details of the data processing are listed in Fig. S2. Data collection and reconstruction statistics are presented in Table S1.
Model building and refinement
Initially, the monomer structure of the drCALHM1 octamer was a rigid body fitted into the map obtained at 3.2 Å resolution. The fitted model was rebuilt manually using COOT optimizing the fit and subjected to global refinement and minimization in real space using the module “real_space_refinement” in PHENIX (25). The quality of the model was assessed with MolProbity (25). The final model exhibited good geometry, as indicated by the Ramachandran plot. The pore radius was calculated using HOLE (http://www.holeprogram.org/). The refinement statistics of the three drCALHM1 structures are shown in Table S1.
Electrophysiology
HEK293T cells were transiently transfected with GFP-tagged drCALHM1 for 16 to 24 h at 37 °C with 5% CO2. Transfected cells were then digested by trypsin and plated onto 35-mm dishes for cultivation for at least 3 h before electrophysiology. The glass pipettes were pulled to a suitable shape using a P-97 glass microelectrode puller (Sutter Instrument, Novato) and polished with an MF-830 (Narishige, Tokyo, Japan). For the whole-cell configuration of recording drCALHM1-3C-GFP currents, all internal pipette solutions were 140 mM CsF, 6 mM MgCl2, 1 mM CaCl2, 11 mM EGTA, 2 mM TEA+, and 10 mM Hepes (pH adjusted to 7.3 with NaOH and methanesulfonic acid, ∼310 mOsm). The resistance of the pipette was 3 to 5 MΩ after being filled with the internal recording solution. For the whole-cell configuration of recording currents, the external bath solution was 145 mM NaCl, 5.4 mM KCl 10 mM TEA+, 10 mM glucose, 1.5 mM CaCl2, 1 mM MgCl2, and 10 Hepes (pH adjusted to 7.4 with NaOH and methanesulfonic acid, ∼330 mOsm). All experiments were conducted at room temperature with the stimulation voltage: 25-ms voltage step to −100 mV from a potential of 0 mV, followed by a ramp voltage increasing from −100 to +60 mV in 5 s. Whole-cell currents were amplified with an Axopatch 700B and digitized with a Digidata 1550A system (Molecular Devices). All currents were sampled at 10 kHz and low-pass filtered at 2 kHz through pCLAMP software (Molecular Devices). Origin 9.0 software (OriginLab Corp.) was also used for data analysis.
MD simulations
The molecular structure of the heptamer was prepared for the MD simulations. The missing residues (21–24, 93–94, and 205–207) were built by homology modeling using Modeller 9.24 (26). The residues are protonated at neutral pH. The heptamer was inserted into 160 Å × 160 Å POPC bilayers with their pore axis aligned parallel to the z-axis through visualized operations in VMD (27). The system was then solvated in TIP3P water boxes (28) and neutralized by 0.15 M NaCl. The final system of the heptamer consisted of 343,897 atoms. Simulations were performed with Amber 2018 (http://ambermd.org/AmberMD.php) by using AMBER force field FF14SB (29) for proteins and LIPID14 (30) for POPC. A 12-Å cutoff was set for the nonbonded interactions. The SHAKE algorithm (31) integration was used to constrain the covalent bonds involving hydrogen atoms, and the particle mesh Ewald algorithm (32) was applied to treat long-range electrostatic interactions. The time step was set to 2 fs.
The initial energy minimization, thermalization, and a series of equilibrations were performed for the systems. First, each system was minimized for 10,000 steps. Second, the thermalization of each system heating from 0 K to 310 K was carried out in 500 ps using the Langevin thermostat (33). The proteins and lipid head groups were fixed with a constraint of 50 kcal mol−1 Å−2. Third, a series of equilibrations were performed for each system. The POPC bilayer was equilibrated for 30 ns with the proteins constrained (50 kcal mol−1 Å−2). After that, the missing residues were optimized for 30 ns, and the other residues of proteins were constrained (50 kcal mol−1 Å−2). Finally, all atoms in the heptamer system were released and equilibrated for 20 ns with no constraints. The frames were saved every 500 steps for analysis.
Supervised MD simulation incorporates a tabu-like supervision algorithm on the reaction coordinate into a conventional MD simulation (34, 35), accelerating the process of the “updown” motion of the N-helix. Protomer 1 in the heptamer system was replaced with the monomer of the octamer (Target0) to obtain the reference structure. Then, we calculated the RMSD of the N-helix of protomer 1 in the current structure compared with Target0. The in-house Python script was employed to monitor the RMSD and process the MD simulation with the Amber engine (http://ambermd.org/AmberMD.php). The supervised MD was carried out in three main steps: (i) the 600-ps conventional MD simulations were performed and arrived at the checkpoint; (ii) 8 snapshots in 75-ps intervals were extracted from the trajectory, and the RMSD of each frame was calculated and collected; and (iii) if the slope of 8 values of RMSD was negative, the next 600-ps simulation was performed to reach the next checkpoint. Otherwise, the supervised MD simulation restarts from the checkpoint using the velocities randomly assigned. We performed an 80-ns supervised MD simulation, and then the supervised MD procedure was stopped. Only the productive steps were saved for analysis.
Targeted MD simulations (36, 37) can also accelerate the transition process by using a constraint. We carried out the targeted MD in the isothermal-isobaric (NPT) ensemble. Here, the coordinates of the N-helix in the “up” and “down” positions of protomer 1 were used as the initial and target positions, respectively. The targeted MD simulations were performed for 1 ns with force constants of 0.02 kcal mol−1 Å−2. The frames were saved every 500 steps for analysis. As controls, a 100-ns conventional MD simulation was performed.
The coarse-grained MARTINI models (15, 38) for the heptamer and the octamer were built, respectively, with the ElNeDyn elastic network (39) applied to proteins. The CG protein coordinates of the heptamer and the octamer were separately positioned in the center of the simulation box of size 16 × 16 × 18 nm3 and 17 × 17 × 18 nm3, respectively, with their pore axis aligned parallel to the z-axis and embedded in a POPC bilayer using the insane script (40). Each system was solvated at 0.15 M NaCl concentration on either side and 0.02 M CaCl2 concentration on the extracellular side. Coarse-grained simulations were carried out using Gromacs, version 2020.2. The Martini, version 2.2, force field was used for protein (15, 38), and the Martini, version 2.0, force field was used for POPC and ions (41). The simulations were run in the NPT ensemble. The time step was set to 20 fs. By using the velocity-rescaling thermostat (42) with coupling constants of τT = 1.0 ps, the temperature was maintained at 310 K. The pressure was controlled at 1 bar by a semi-isotropic Parrinello-Rahman barostat (43) with a coupling constant of τP = 12.0 ps. The type of constraint applied to bonds was the LINCS algorithm (44). A Verlet cutoff scheme was used, and the particle mesh Ewald method (32) was applied to calculate long-range electrostatic interactions. Periodic boundary conditions were used in the x- and y-axes. Ten microliters of equilibrium simulation trajectory data was collected for each system. The all-atom and coarse-grained MD simulations in this work were repeated 3 times.
Data availability
Atomic coordinates have been deposited in the Protein Data Bank under accession numbers 7DSE, 7DSD, and 7DSC for heptamer+LCH, heptamer-noLCH, and octamer-noLCH, respectively. The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-30832, EMD-30831, and EMD-30830 for heptamer+LCH, heptamer-noLCH, and octamer-noLCH, respectively.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are grateful to Ms K. Tang in the Center of Cryo-Electron Microscopy (CCEM), Zhejiang University for their technical assistance on Cryo-EM data acquisition and Dr A. Li and Dr J. Lu for the support of electron microscopy at Nankai University.
Author contributions
Y. R., Y. W., T. W., and X. L. investigation; Y. R. verification; Y. R. and Y. L. writing-original draft; Y. L. software; Y. L. formal analysis; Y. L., Y. W., and Y. S. validation; Y. L., S. C., and X. Z. data curation; S. C. resources; Y. S. conceptualization; Y. S. review and editing; Y. S. supervision; X. Y. methodology; X. Y. funding acquisition; X. Y. writing-review and editing.
Funding and additional information
This work was supported by National Key Research and Development Program of China (grant 2017YFA0504801 to Y. S.; 2017YFA0504803 and 2018YFA0507700 to X. Z.), National Natural Science Foundation of China (grants 91954119 and 31870736 to X. Y.; grants 32071231 and 31870834 to Y. S.), the Fundamental Research Funds for the Central Universities (2018XZZX001–13 to X. Z.; 035–63201110 to X. Y.; and 035–63201109 to Y. S.), and the Talent Training project in Nankai University (035-BB042112).
Edited by Wolfgang Peti
Contributor Information
Yuequan Shen, Email: yshen@nankai.edu.cn.
Xue Yang, Email: yangxue@nankai.edu.cn.
Supporting information
References
- 1.Abbracchio M.P., Burnstock G., Verkhratsky A., Zimmermann H. Purinergic signalling in the nervous system: An overview. Trends Neurosci. 2009;32:19–29. doi: 10.1016/j.tins.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Ma Z., Taruno A., Ohmoto M., Jyotaki M., Lim J.C., Miyazaki H., Niisato N., Marunaka Y., Lee R.J., Hoff H., Payne R., Demuro A., Parker I., Mitchell C.H., Henao-Mejia J., et al. CALHM3 is essential for rapid ion channel-mediated purinergic neurotransmission of GPCR-mediated tastes. Neuron. 2018;98:547–561. doi: 10.1016/j.neuron.2018.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siebert A.P., Ma Z., Grevet J.D., Demuro A., Parker I., Foskett J.K. Structural and functional similarities of calcium homeostasis modulator 1 (CALHM1) ion channel with connexins, pannexins, and innexins. J. Biol. Chem. 2013;288:6140–6153. doi: 10.1074/jbc.M112.409789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Contreras J.E., Saez J.C., Bukauskas F.F., Bennett M.V. Gating and regulation of connexin 43 (Cx43) hemichannels. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11388–11393. doi: 10.1073/pnas.1434298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldberg G.S., Lampe P.D., Nicholson B.J. Selective transfer of endogenous metabolites through gap junctions composed of different connexins. Nat. Cell Biol. 1999;1:457–459. doi: 10.1038/15693. [DOI] [PubMed] [Google Scholar]
- 6.Kumar N.M., Gilula N.B. The gap junction communication channel. Cell. 1996;84:381–388. doi: 10.1016/s0092-8674(00)81282-9. [DOI] [PubMed] [Google Scholar]
- 7.Taruno A. ATP release channels. Int. J. Mol. Sci. 2018;19:808. doi: 10.3390/ijms19030808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Z., Tanis J.E., Taruno A., Foskett J.K. Calcium homeostasis modulator (CALHM) ion channels. Pflugers Arch. 2016;468:395–403. doi: 10.1007/s00424-015-1757-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taruno A., Vingtdeux V., Ohmoto M., Ma Z., Dvoryanchikov G., Li A., Adrien L., Zhao H., Leung S., Abernethy M., Koppel J., Davies P., Civan M.M., Chaudhari N., Matsumoto I., et al. CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes. Nature. 2013;495:223–226. doi: 10.1038/nature11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romanov R.A., Lasher R.S., High B., Savidge L.E., Lawson A., Rogachevskaja O.A., Zhao H., Rogachevsky V.V., Bystrova M.F., Churbanov G.D., Adameyko I., Harkany T., Yang R., Kidd G.J., Marambaud P., et al. Chemical synapses without synaptic vesicles: Purinergic neurotransmission through a CALHM1 channel-mitochondrial signaling complex. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aao1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foskett J.K. Structures of CALHM channels revealed. Nat. Struct. Mol. Biol. 2020;27:227–228. doi: 10.1038/s41594-020-0391-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demura K., Kusakizako T., Shihoya W., Hiraizumi M., Nomura K., Shimada H., Yamashita K., Nishizawa T., Taruno A., Nureki O. Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba8105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren Y., Wen T., Xi Z., Li S., Lu J., Zhang X., Yang X., Shen Y. Cryo-EM structure of the calcium homeostasis modulator 1 channel. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syrjanen J.L., Michalski K., Chou T.H., Grant T., Rao S., Simorowski N., Tucker S.J., Grigorieff N., Furukawa H. Structure and assembly of calcium homeostasis modulator proteins. Nat. Struct. Mol. Biol. 2020;27:150–159. doi: 10.1038/s41594-019-0369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monticelli L., Kandasamy S.K., Periole X., Larson R.G., Tieleman D.P., Marrink S.-J. The MARTINI coarse-grained force field: Extension to proteins. J. Chem. Theor. Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 16.Tanis J.E., Ma Z., Foskett J.K. The NH2 terminus regulates voltage-dependent gating of CALHM ion channels. Am. J. Physiol. Cell Physiol. 2017;313:C173–C186. doi: 10.1152/ajpcell.00318.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Z., Siebert A.P., Cheung K.H., Lee R.J., Johnson B., Cohen A.S., Vingtdeux V., Marambaud P., Foskett J.K. Calcium homeostasis modulator 1 (CALHM1) is the pore-forming subunit of an ion channel that mediates extracellular Ca2+ regulation of neuronal excitability. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E1963–E1971. doi: 10.1073/pnas.1204023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruan Z., Orozco I.J., Du J., Lu W. Structures of human pannexin 1 reveal ion pathways and mechanism of gating. Nature. 2020;584:646–651. doi: 10.1038/s41586-020-2357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi W., Clemente N., Sun W., Du J., Lu W. The structures and gating mechanism of human calcium homeostasis modulator 2. Nature. 2019;576:163–167. doi: 10.1038/s41586-019-1781-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drozdzyk K., Sawicka M., Bahamonde-Santos M.I., Jonas Z., Deneka D., Albrecht C., Dutzler R. Cryo-EM structures and functional properties of CALHM channels of the human placenta. Elife. 2020;9 doi: 10.7554/eLife.55853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohou A., Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez-Leiro R., Scheres S.H.W. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol. 2017;73:496–502. doi: 10.1107/S2059798316019276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swint-Kruse L., Brown C.S. Resmap: Automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics. 2005;21:3327–3328. doi: 10.1093/bioinformatics/bti511. [DOI] [PubMed] [Google Scholar]
- 25.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 2016;86:2.9.1–2.9.37. doi: 10.1002/cpps.20. [DOI] [PubMed] [Google Scholar]
- 27.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. Model. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 28.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 29.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickson C.J., Madej B.D., Skjevik A.A., Betz R.M., Teigen K., Gould I.R., Walker R.C. Lipid14: The amber lipid force field. J. Chem. Theory Comput. 2014;10:865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of N-alkanes. J. Chem. Phys. 1977;23:327–341. [Google Scholar]
- 32.Darden T., York D., Pedersen L. Particle mesh Ewald: An Nlog (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 33.Pastor R.W., Brooks B.R., Szabo A. An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol. Phys. 1988;65:1409–1419. [Google Scholar]
- 34.Salmaso V., Sturlese M., Cuzzolin A., Moro S. Exploring protein-peptide recognition pathways using a supervised molecular dynamics approach. Structure. 2017;25:655–662. doi: 10.1016/j.str.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 35.Deganutti G., Moro S., Reynolds C. A supervised molecular dynamics approach to unbiased ligand-protein unbinding. J. Chem. Inf. Model. 2020;60:1804–1817. doi: 10.1021/acs.jcim.9b01094. [DOI] [PubMed] [Google Scholar]
- 36.Schlitter J., Engels M., Krüger P. Targeted molecular-dynamics – a new approach for searching pathways of conformational transitions. J. Mol. Graph. 1994;12:84–89. doi: 10.1016/0263-7855(94)80072-3. [DOI] [PubMed] [Google Scholar]
- 37.Xiao X., Zeng X., Yuan Y., Gao N., Guo Y., Pu X., Li M. Understanding the conformation transition in the activation pathway of β2 adrenergic receptor via a targeted molecular dynamics simulation. Phys. Chem. Chem. Phys. 2015;17:2512–2522. doi: 10.1039/c4cp04528a. [DOI] [PubMed] [Google Scholar]
- 38.Marrink S.J., Risselada H.J., Yefimov S., Tieleman D.P., de Vries A.H. The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 39.Periole X., Cavalli M., Marrink S.J., Ceruso M.A. Combining an elastic network with a coarse-grained molecular force field: Structure, dynamics, and intermolecular recognition. J. Chem. Theory Comput. 2009;5:2531–2543. doi: 10.1021/ct9002114. [DOI] [PubMed] [Google Scholar]
- 40.Wassenaar T.A., Ingolfsson H.I., Boeckmann R.A., Tieleman D.P., Marrink S.J. Computational lipidomics with insane: A versatile tool for generating custom membranes for molecular simulations. J. Chem. Theor. Comput. 2015;11:2144–2155. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- 41.Marrink S.J., de Vries A.H., Mark A.E. Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B. 2004;108:750–760. [Google Scholar]
- 42.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126 doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 43.Parrinello M.R.A., Rahman A.J. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1982;52:7182–7190. [Google Scholar]
- 44.Hess B., Bekker H., Berendsen H., Fraaije J. Lincs: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates have been deposited in the Protein Data Bank under accession numbers 7DSE, 7DSD, and 7DSC for heptamer+LCH, heptamer-noLCH, and octamer-noLCH, respectively. The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-30832, EMD-30831, and EMD-30830 for heptamer+LCH, heptamer-noLCH, and octamer-noLCH, respectively.