Summary
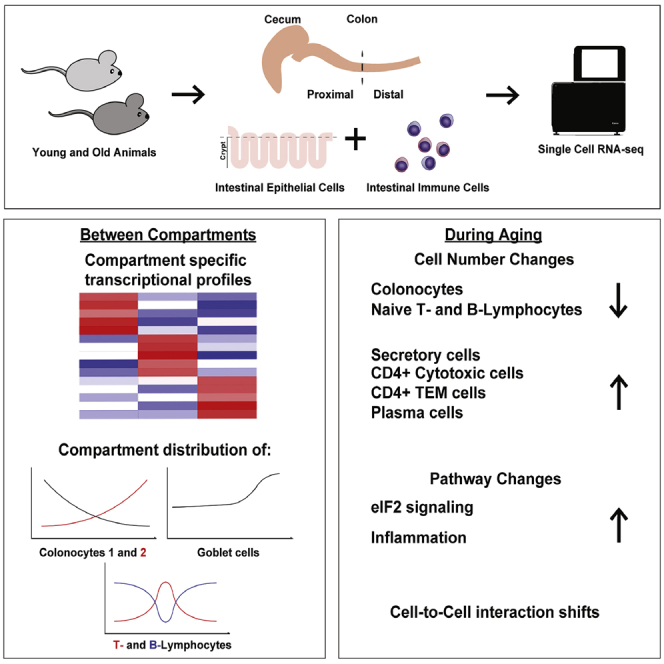
We performed massive single-cell sequencing in the aging mouse colonic epithelium and immune cells. We identified novel compartment-specific markers as well as dramatic aging-associated changes in cell composition and signaling pathways, including a shift from absorptive to secretory epithelial cells, depletion of naive lymphocytes, and induction of eIF2 signaling. Colon cancer is one of the leading causes of death within the western world, incidence of which increases with age. The colonic epithelium is a rapidly renewing tissue, tasked with water and nutrient absorption, as well as hosting intestinal microbes. The colonic submucosa is populated with immune cells interacting with and regulating the epithelial cells. However, it is unknown whether compartment-specific changes occur during aging and what impact this would cause. We show that both epithelial and immune cells differ significantly between colonic compartments and experience significant age-related changes in mice. We found a shift in the absorptive-secretory cell balance, possibly linked to age-associated intestinal disturbances, such as malabsorption. We demonstrate marked changes in aging immune cells: population shifts and interactions with epithelial cells, linking cytokines (Ifn-γ, Il1B) with the aging of colonic epithelium. Our results provide new insights into the normal and age-associated states of the colon.
Subject areas: Immunology, Cell biology, Omics, Transcriptomics
Graphical abstract
Highlights
-
•
Mouse colon shows compartment-specific transcriptional and population differences
-
•
Old animal colon switches to a pro-inflammatory state
-
•
Changes in epithelium linked to changes in tissue-resident immune cells
Immunology ; Cell biology; Omics; Transcriptomics
Introduction
Colon cancer is one of the leading causes of death within the western world and is linked to the aging of the colon. The disease presents differently between men and women, developing in different parts of the colon and often with a different morphology (Schmuck et al., 2020). In addition, it is known that colon cancer tumors differ in responsiveness to treatment, susceptibility to mutations, and expected patient survival rates according to the tumor’s anatomic location (Cannon and Buechler, 2019; Liao et al., 2018; Phipps et al., 2013). It is thus imperative to determine the innate and aging-associated differences in different parts of the colon to better treat or prevent the development of colon cancer.
The colon (a.k.a. large intestine) is the terminal segment of the gastrointestinal tract and consists of an epithelial layer with an underlying connective tissue known as the lamina propria (which houses various immune cells) and the smooth muscle layer. It is tasked with the absorption of water and minerals as well as beneficial bacterial products. The colon holds the bulk of all microorganisms that reside within the gastrointestinal tract and is an important modulator of the immune system and its response to the microbiome. During embryonic development, different parts of the hindgut contribute to the formation of the large intestine (Garriock et al., 2020). Different compartments of the large intestine are more specialized to perform different functions, as seen by differential expression of nutrient-absorption, immunity, and hormonal genes in specific cell types (Wang et al., 2019). It has also been documented that different compartments react differently to inflammation (Lyons et al., 2018).
The colonic epithelium is a rapidly renewing tissue, organized into crypts. The bottom of the colonic crypt is populated by intestinal stem cells (ISC) (Barker et al., 2007) in between deep secretory cells (DSCs) that provide them with necessary niche factors (Sasaki et al., 2016). The differentiation gradient increases from the crypt base toward the colonic lumen, populating the crypt with transit-amplifying (TA) cells that then differentiate into absorptive colonocytes or secretory cells (goblet, tuft, or enteroendocrine (EE) cells). It is known that epithelial homeostasis is linked with ulcerative colitis (Parikh et al., 2019): barrier function maintenance being reliant on goblet cell-produced protease inhibitor WAP Four-Disulfide Core Domain Protein two, colonocyte pH regulation, and uroguanylin production. However, it is completely unknown whether epithelial homeostasis becomes altered during aging and whether it contributes to aging-related disorders.
Colonic immune cells reside in the lamina propria and the gut associated lymphoid tissue (GALT). Lamina propria is populated with plasma cells, neutrophils, dendritic cells, macrophages, and T cells (Gui et al., 2018; Lee et al., 1988), whereas GALT is mainly made up of T lymphocytes, macrophages, dendritic Cells, innate lymphoid cells, and B cells (Chassaing et al., 2014). Colonic immune cells interact with epithelial cells directly or through excreted cytokines (Biton et al., 2018; Geuking and Burkhard, 2020; Gronke et al., 2019; Miyata et al., 2018; Thelemann et al., 2014); therefore, changes in the immune compartment could play a major role in the aging of the colonic epithelium. Indeed, it has been shown that during ulcerative colitis, T cells form interactions (not present in normal tissue) with epithelial cells (Smillie et al., 2019) and in general that inflammation and immune infiltration promote colonic tumorigenesis (Alteber et al., 2018; Girondel et al., 2020; Zhang et al., 2020).
It is known that the number of CD4 T cells drop during aging in secondary lymphoid organs, except the GALT, where CD4 (as well as CD8) T cells accumulate with increased age (Martinet et al., 2014). However, not much is known about whether compartment-specific changes occur during aging and how said changes could impact the epithelium.
Utilizing single-cell RNA sequencing (scRNAseq), we show that the mouse colonic epithelium has distinct transcriptional and cell population differences between compartments (cecum, proximal (ProxCol) and distal colon (DistCol)) during aging and characterized the changes to the GALT immune cells.
Results
Epithelial cell differences between compartments
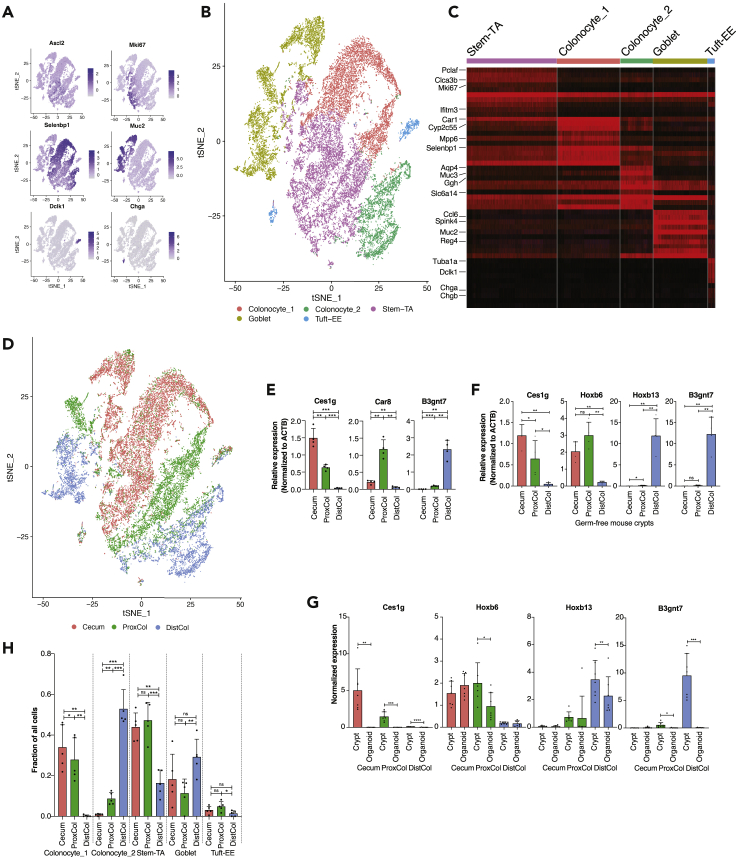
By using scRNASeq, we profiled a total of 23,762 epithelial cells from the whole large intestinal (including cecum, ProxCol and DistCol) crypts of five young and six old female C57BL/6J mice (Figure S1A). We employed a cell hash-tagging approach (Stoeckius et al., 2018) that allowed us to downstream demultiplex cells according to the mouse and large intestine’s compartment of origin. The t-SNE plot separated cells comprehensively according to expression of known colon cell markers, such as Ascl2 for stem, Mki67 for TA, and Muc2 for Goblet cells (Figure 1A). K-means clustering, an unsupervised algorithm that groups cells according to similarly expressed genes, separated cells into 5 clusters, characterized by known cell markers (Figures 1B and 1C). Remarkably, k-means clustering robustly separated two colonocyte subpopulations while maintaining stem and TA cells as well as tuft and EE cells together, suggesting pronounced transcriptional differences between colonocytes 1 and 2. Interestingly, the stem-TA, colonocyte, and goblet cell clusters were visually separated in the t-Stochastic Neighbor Embedding (tSNE) plot according to colon compartment, suggesting a robust difference in the transcriptome between the three large intestine compartments in these cell types (Figure 1D). We identified and validated genes that were specific for each compartment, such as Ces1g for cecum, Car8 for ProxCol, and B3gnt7 for DistCol (Figures 1E, S1B, and S1C). We were also able to find genes that had overlap for cecum and ProxCol (e.g., Cyp2c55, Car1, and Hoxb6), whereas others had significantly higher expression in DistCol (e.g., Slc6a14, Muc3), suggesting cecum and ProxCol are more closely related either from a developmental or functional point of view. However, several other genes (e.g., Agr2 and Stmn1) showed a stronger overlap between ProxCol and DistCol than with cecum, showing that colonic compartments have both distinct differences and commonalities.
Figure 1.
Compartment differences of young colonic epithelium
(A–C) tSNE plot of epithelial cells (young and old together) overlaid with cell type markers (n = 5 young, six old biologically independent animals) (B) tSNE plot of epithelial cells overlaid with clusters from 5k-means clustering. (n = 5 young, six old biologically independent animals) (C) Top 10 marker gene heatmap for 5k-means clusters.
(D) tSNE plot of epithelial cells overlaid with origin compartment (n = 5 young, six old biologically independent animals).
(E) qPCR performed on crypts isolated from different compartments of young female mice. (n = 4 biologically independent animals; 1-tailed paired t-test; Data are represented as mean +SD).
(F) qPCR performed on crypts isolated from different compartments of young female germ-free mice. (n = 4 biologically independent animals; 1-tailed paired t-test; Data are represented as mean +SD).
(G and H) qPCR performed on crypt and organoid pairs from different compartments of young female mice. (n = 7 biologically independent animals; 1-tailed paired t-test; Data are represented as mean +SD) (H) Bar chart of cell fractions of different cell types in different compartments from scRNAseq. (n = 5 biologically independent animals; 1-tailed paired t-test; Data are represented as mean +SD). ns, p>=0.05; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001.
We then wanted to see whether these transcriptional differences between compartments were intrinsic or caused by extrinsic factors, such as the gut microbiome. Quantitative PCR (qPCR) on germ-free mice showed that the expression of Ces1g, Hoxb6, Hoxb13, and B3gnt7 followed the same compartment expression pattern as wild type mice housed under specific pathogen free conditions (Figure 1F), suggesting that gut microbiome is not responsible for these differences. We performed organoid culture to separate the epithelium from all other cell-extrinsic factors like interactions with non-epithelial cells (e.g., mesenchymal and immune cells) or extracellular matrix. Ces1g and B3gnt7 were completely lost at day 9 of organoid culture, whereas Hoxb6 and Hoxb13 remained expressed at a similar level compared to crypts (Figure 1G). This effect was also observed in organoids from male mouse crypts (Figure S1D). It is thus evident that compartmentalization of the large intestine is dependent on both cell-intrinsic (developmental) factors as well as cell-extrinsic factors but independent of the gut microbiome (at least with regard to our validated markers).
Differentially expressed genes (DEG) between young and old cells of each compartment were run through the Ingenuity Pathway Analysis (IPA) software to perform a functional and ontological classification. We were able to see some overlap in top enriched canonical pathways. EIF2 signaling was strongly enriched (and predicted to be hyperactivated) in cecum and ProxCol, whereas mitochondrial dysfunction/oxidative phosphorylation was mainly enriched (and predicted hyperactivated) in ProxCol and DistCol (Figure S1E). Compartments had common predicted upstream regulators, such as Stk11 (activating) and Rictor (inhibiting) while also having more compartment-specific ones, such as Mlxipl, Myc (activating) and Kdm5a, Hnf1a (inhibiting) for cecum and ProxCol; Nrf1 (activating) for ProxCol and DistCol (Figure S1F). This suggests that the large intestinal epithelium has both global and compartment-specific alteration of gene pathways during aging. Interestingly, ileum-adjacent large intestinal tissue seems to have an enhanced alteration of EIF2 signaling translation machinery (unbalanced proteostasis has been found in aged cells (Steffen and Dillin, 2016), whereas the terminal (rectum-adjacent) tissue suffers more from mitochondria-related dysfunctions (also a common aging phenotype) (Christian and Shadel, 2014).
The eIF2 pathway is linked to the development of colorectal cancer (Schmidt et al., 2020); therefore, we performed qPCR on Atf3 (transcribed upon phosphorylation of eIF2-alpha) for confirmation. Atf3 had a significantly reduced expression in old ProxCol and DistCol (Figure S1G), as predicted by IPA software. Calorie restriction (CR) is a well-known intervention, known to ameliorate the aging phenotype (Liang et al., 2018; Speakman and Mitchell, 2011). Therefore, we performed the experiment on old mice kept in the CR regime. Interestingly, Atf3 was not rescued in old-CR mice, suggesting EIF2 signaling alterations are not ameliorated by CR. Conversely, Atf3 was upregulated in aged germ-free mice, suggesting that the microbiome plays a role in these aging differences.
We were able to identify cell population differences along the compartment axis (Figure 1H). Colonocyte_1 was significantly more enriched in cecum (Cell fraction: 0.34 ± 0.11) than in both ProxCol (0.28 ± 0.10) and DistCol (0.002 ± 0.004), with ProxCol having significantly more than DistCol. Conversely, colonocyte_2 was significantly more enriched in DistCol (0.53 ± 0.10) than in both cecum (0.01 ± 0.005) and ProxCol (0.09 ± 0.03), with ProxCol having significantly more than cecum. Both cecum and ProxCol had significantly more Stem-TA cells than DistCol (0.44 ± 0.07; 0.47 ± 0.09 and 0.16 ± 0.06, respectively). DistCol has significantly more goblet cells than ProxCol but significantly less Tuft-EE cells (0.01 ± 0.01 and 0.05 ± 0.02, respectively).
Changes in aging epithelial proliferative cells
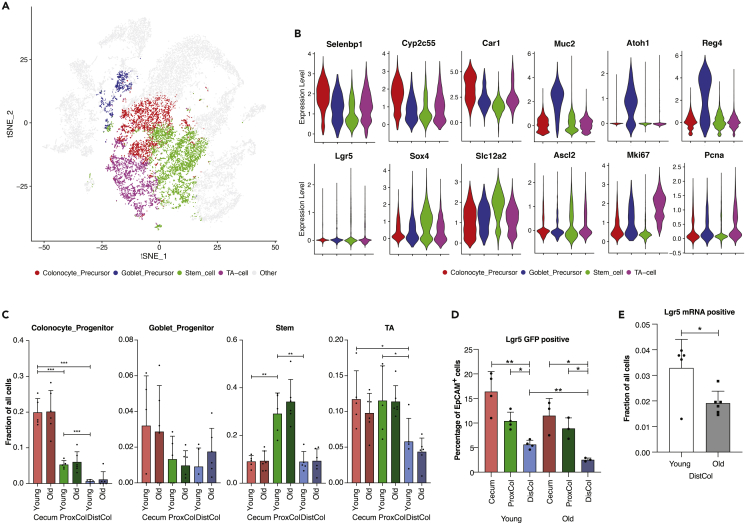
We performed k-means re-clustering of the Stem-TA cluster in order to clarify the cell types. 4 k-means were arbitrarily chosen to best re-cluster the cells according to known marker gene expression (Figure 2A). The colonocyte precursors had a higher expression of Selenbp1, Cyp2c55, and Car1 genes; goblet precursors had a higher expression of Muc2, Atoh1, and Reg4; stem cells had a higher expression of Lgr5, Sox4, Scl12a2, and Ascl2; TA cells had a higher expression of Mki67 and Pcna genes (Figure 2B). Within re-clustered cell types, we found compartment-dependent differences (Figure 2C). Colonocyte precursors were significantly more enriched in the cecum (0.20 ± 0.04) than both ProxCol (0.05 ± 0.01) and DistCol (0.006 ± 0.004). Goblet precursors did not differ significantly between compartments but still had a downward trend from cecum (0.03 ± 0.01) toward the DistCol (0.009 ± 0.01). Stem cells were significantly enriched in the ProxCol (0.29 ± 0.04) compared with both cecum (0.09 ± 0.01) and DistCol (0.09 ± 0.02). There was a similar number of TA cells in cecum (0.12 ± 0.04) and ProxCol (0.12 ± 0.02), which was significantly more than in DistCol (0.06 ± 0.01). We were not able to observe any statistically significant changes in either cell type or compartment between young and old animals.
Figure 2.
Changes in the proliferative compartment of the colon during aging
(A and B) tSNE plot of epithelial cells overlaid with the 4k-means re-cluster of the initial Stem-TA cluster. (n = 5 young, 6 old biologically independent animals) (B) Violin plots of cell-type-associated gene expression in 4k-means re-cluster (n = 5 young, 6 old biologically independent animals).
(C and D) Bar chart of cell fractions of re-clustered cell types in different compartments in young and old mice. (n = 5 young, 6 old biologically independent animals, 1-tailed paired t-test used between young compartments, Data are represented as mean +SD) (D) FACS sorted Lgr5-GFP-positive cells in different compartments from young and old female Lgr5-eGFPki/+ mice. (n = 4 young, 3 old biologically independent animals, 1-tailed paired t-test used between young compartments, 1-tailed unpaired t-test used between young and old animals; Data are represented as mean +SD).
(E) Comparison of cell fraction of Lgr5 mRNA expressing cells from Single-Cell RNAseq. (n = 5 young, 6 old biologically independent animals, 1-tailed unpaired t-test; Data are represented as mean +SD). ns, p>=0.05; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001.
To investigate the apparent enrichment of progenitors and proliferative cells in the cecum and ProxCol, we analyzed cells from Lgr5-EGFP-IRES-creERT2ki/+ mice (Barker et al., 2007) using flow cytometry. We confirmed that both cecum and ProxCol had significantly more Lgr5-eGFP+ cells than DistCol (16.40% ± 4.10%; 10.43% ± 1.79 and 5.63% ± 0.87%, respectively) in young mice, with an identical pattern maintained in old mice as well (Figures 2D and S2A). Interestingly, we found a statistically significant decrease in Lgr5-EGFP+ cells in old DistCol (Figure 2D), which we also saw in scRNAseq (Figure 2E).
IPA of DEGs of the Stem-TA cluster during aging showed a statistically significant enrichment (and predicted activation) of the EIF2 Signaling pathway as well as predicted upstream regulators: Mlxipl, Myc, Sim1, and Arnt2 (activating) and Rictor (inhibiting) (Figure S2B).
Changes in aging colonocytes
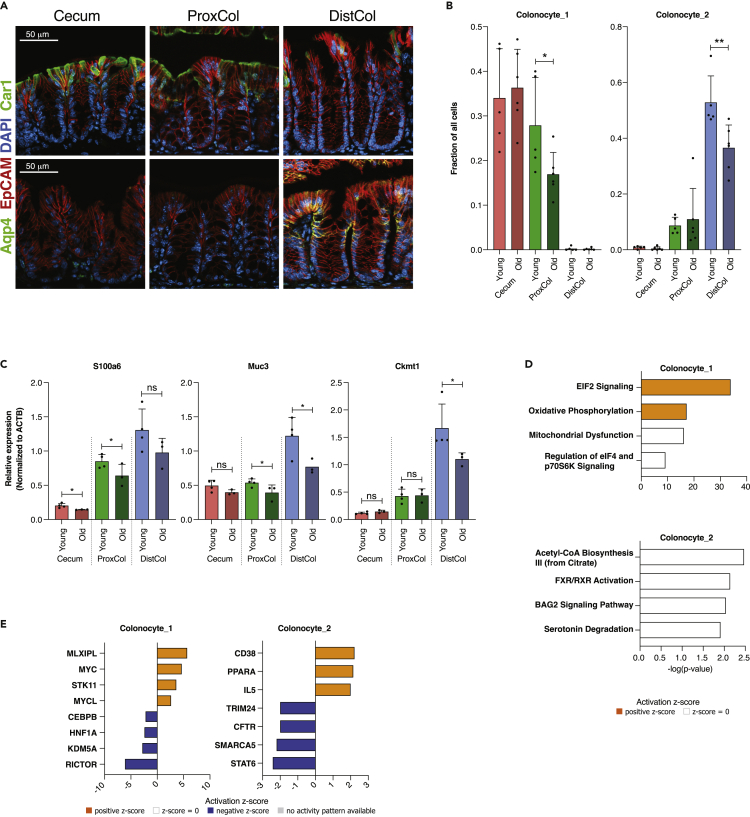
We confirmed the existence of Colonocyte_1 and Colonocyte_2 subpopulations and their differential distribution along the colonic tract by immunofluorescent staining for the two most expressed population-specific markers (Car1 and Aqp4). Colonocyte_1 marker, Car1, was present in Cecum, to a lesser extent in ProxCol, and absent in DistCol; conversely, Colonocyte_2 marker, Aqp4, was present in DistCol but absent in Cecum and ProxCol (Figure 3A) in accordance with the single-cell clustering. Old mice had significantly less Colonocyte_1 cells in ProxCol (Cell fraction: 0.28 ± 0.11 and 0.17 ± 0.05, respectively) and less Colonocyte_2 cells in DistCol (0.53 ± 0.10 and 0.37 ± 0.08, respectively) (Figure 3B), indicating a reduction of epithelial cells with absorptive function in aging colon. RT-qPCR of colonocyte marker expression between young and old mice confirmed the reduced colonocyte numbers in aged ProxCol and DistCol (Figure 3C).
Figure 3.
Changes in colonocytes during aging
(A) Immunofluorescent staining of different compartments for Car1 (upper) and Aqp4 (lower). EpCAM staining used to stain epithelial cells, DAPI used to stain nuclei. Scale bar 50 μm.
(B) Bar charts of colonocyte_1 and _2 fractions in different compartments in young and old. (n = 5 young, six old biologically independent animals, 1-tailed unpaired t-test used between young and old animals; Data are represented as mean +SD).
(C) qPCR of colonocyte-specific genes between young and old female mice. Young samples are taken from Figure S1 to be used as controls. (n = 4 young, 3 old biologically independent animals; 1-tailed unpaired t-test used between young and old animals; Data are represented as mean +SD).
(D) Enriched Canonical Pathways from IPA for colonocytes (n = 5 young, 6 old biologically independent animals).
(E) Predicted Upstream Regulators from IPA for colonocytes (n = 5 young, 6 old biologically independent animals). ns, p>=0.05; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001.
Though colonocytes from cecum and ProxCol were grouped together in the Colonocyte_1 k-means cluster (Figure 1B), likely because of similarities in transcriptome, according to the tSNE plot, colonocytes could be further separated into compartment-specific subtypes. For this, we utilized an alternative approach: a graph-based clustering that clusters cells according to a proximity graph and is capable of generating 3 clusters with specific markers for each subpopulation (Figures S2C–S2E), which were already validated by qPCR (Figures 1E and S1C). Thus, it highlights that even though some similarities exist between compartments, they are still transcriptionally distinct. The age-associated changes in colonocyte numbers in the ProxCol and DistCol remained under this new separation (Figures 3B and S2F).
IPA analysis showed that aging Colonocyte_1 cells were enriched (and predicted activated) for EIF2 signaling and Oxidative phosphorylation pathways (Figure 3D). Colonocyte_2 cells were enriched for Acetyl-CoA Biosynthesis III, FXR/RXR Activation, BAG2 Signaling, and Serotonin degradation pathways (Figure 3D). Most of the upstream regulators for Colonocyte_1 were shared with upstream regulators for cecum and ProxCol, whereas Colonocyte_2 had different upstream regulators than the whole DistCol: Cd38, Ppara, and Il5 (activating) and Trim24, Cftr, Smarca5, and Stat6 (inhibiting) (Figure 3E). Because Acetyl-CoA synthesis and FXR/RXR activation pathways are associated with lipid/cholesterol synthesis/metabolism, it is tempting to link Colonocyte_2 aging with lipid/cholesterol metabolism dysfunction. However, it is noteworthy that both FXR/RXR activation and Bag2 activation pathways have been shown to have roles in carcinogenesis (Vaquero et al., 2013; Yue et al., 2015) and even intestinal barrier function (Inagaki et al., 2006); therefore, further inquiry would be required to reach any conclusions. Even so, the results suggest that the DistCol ages differently from cecum or ProxCol.
Changes in aging epithelial secretory cells
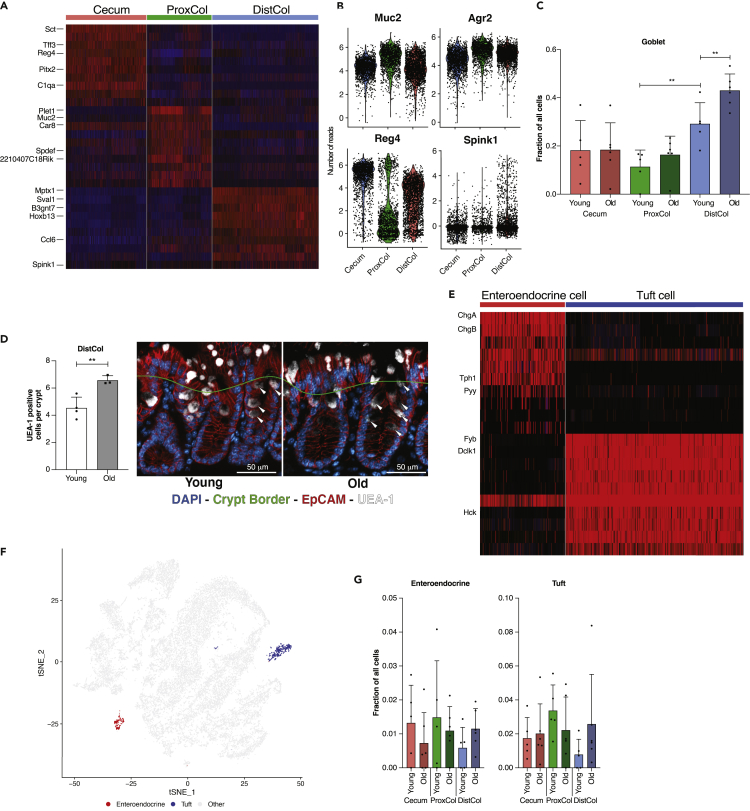
Gene markers for goblet cells of different compartments are shown in the heatmap (Figure 4A), notably C1qa, Car8, and Sval1, as validated previously by PCR (Figure S1C). Generally, goblet cells had a comparable level of Muc2 and Agr2 expression between compartments (Figure 4B). ProxCol had a lower number of Reg4-positive goblet cells than Cecum and DistCol, as reported previously (Sasaki et al., 2016). DistCol had a higher number of Spink1 positive goblet cells. As Spink1 is expressed at the bottom of the colonic crypt and has been shown to act as a growth factor, especially for colon cancer (Ida et al., 2015; Tiwari et al., 2015), it could be a marker for a DistCol specific DSC cell (Figure 4B). We found that the fraction of DistCol goblet cells increased significantly during aging (0.29 ± 0.09 in young and 0.43 ± 0.07 in old) (Figure 4C), probably at the expense of the absorptive lineage cells. To validate the aging increase, we performed IF staining and cell counting for Ulex Europaeus Agglutinin 1 (UEA-1) (staining goblet granules) positive cells. We were able to find a statistically significant increase of UEA-1-positive cells per crypt (white arrows) in the old DistCol (Figure 4D).
Figure 4.
Changes in secretory cells of the colon during aging
(A) Top 10 marker gene heatmap for goblet cells in different compartments (n = 5 young, 6 old biologically independent animals).
(B) Expression levels of different goblet cell genes in different compartments from Single-Cell RNAseq (n = 5 young, 6 old biologically independent animals).
(C) Fractions of goblet cells in different compartments (n = 5 young, 6 old biologically independent animals; 1-tailed paired t-test used between young compartments, 1-tailed unpaired t-test used between young and old animals; Data are represented as mean +SD).
(D) Quantification of goblet cells in DistCol utilizing UEA-1 (left) (n = 4 young, 3 old biologically independent animals; 1-tailed unpaired t-test; Data are represented as mean +SD) with representative pictures (right) Crypt border designated manually to have at least one cell separation from the lumen. Scale bar 50 μm.
(E) Top 10 marker gene heatmap for 2k-means re-cluster of initial Tuft-EE cluster (n = 5 young, six old biologically independent animals).
(F) 2k-means re-clustered Tuft-EE cells overlaid on initial tSNE (n = 5 young, six old biologically independent animals).
(G) Fractions of initial and re-clustered clusters in different compartments between young and old (n = 5 young, 6 old biologically independent animals). ns, p>=0.05; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001.
We re-clustered the Tuft-EE population using 2 k-means and were able to generate separate clusters for EE (expressing e.g., Chga, Chgb, and Pyy) and Tuft cells (expressing e.g., Fyb, Dclk1, and Hck) (Figures 4E and 4F). No significant changes were observed in the number of either cell type between compartments or in aging (Figure 4G), probably because of the high inter-sample variance. However, it was possible to observe a trend: both EE and tuft cell average numbers were reduced in aged ProxCol but increased in DistCol (Figure 4G).
By plotting tuft cell gene expression according to compartment, we found a different transcriptional profile of DistCol tuft cells, compared with cecum and ProxCol (Figure S2I).
EIF2 signaling pathway was again the most commonly enriched (and predicted activated) canonical pathway in goblet and Tuft-EE cells from all compartments (Figures S2G and S2J). The predicted upstream regulators were also common with other compartments/cell types: Myc and Mlxipl (activating) and Rictor (inhibiting) (Figures S2H and S2K). These data, together with the functional analysis in colonocytes, strongly suggest that alteration of the mRNA translation process occurs in all the epithelial cells of the large intestine in old mice, promoting colon stem cells as the cell of origin for this general aging phenotype.
Changes in aging immune cells
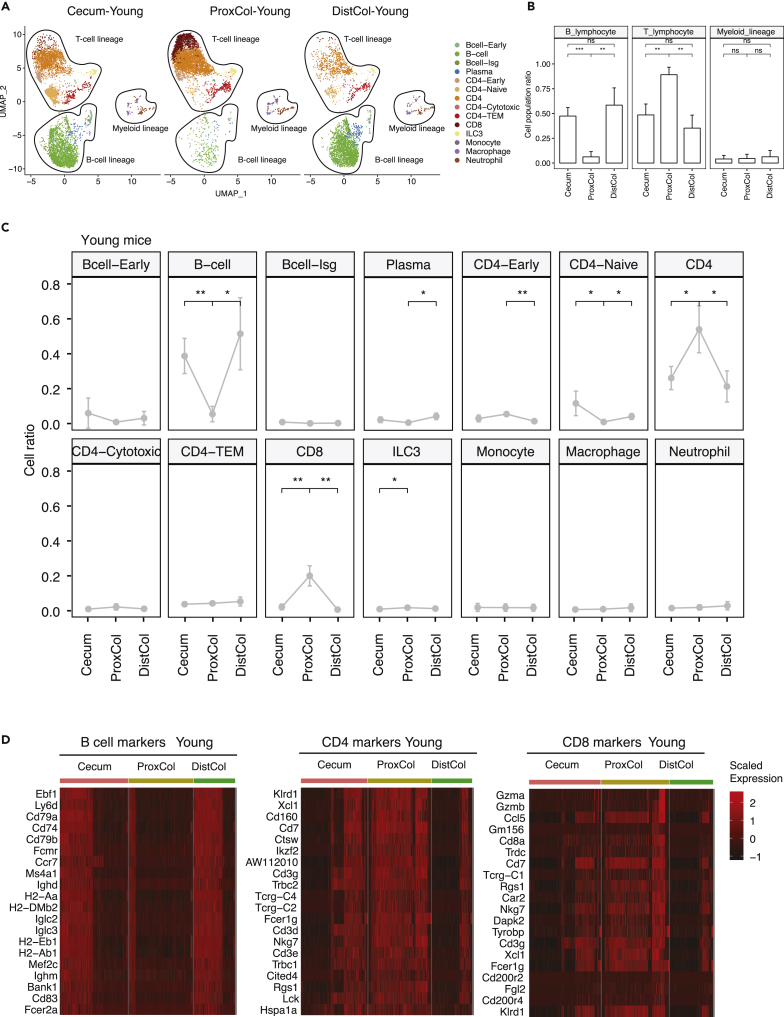
Intestinal epithelial crypt cells strongly interact with the immune cells located in the lamina propria region just below the crypt zone (Peterson and Artis, 2014). We isolated immune cells from this region by FACS and profiled a total of 25,788 CD45+ cells from four young and four old female C57BL/6J mice by using scRNAseq. The Uniform Manifold Approximation and Projection (UMAP) separated cells into the three major immune cell lineages: T cell, B cell, and myeloid cells (Figures 5A and 5B), reproducing the known lymphocyte-enrichment in the colon. Each separate cluster produced unique transcriptome profiles with some known markers expressed in one specific cluster or shared in some clusters (Figure S3A). By comparing all the specific markers with those found in literature (Blecher-Gonen et al., 2019; Elyahu et al., 2019; Zhao et al., 2020), we assigned the clusters to their cell types (Figure 5A). B cells and CD4+-cells were identified in a general way, which could not be further identified effectively by curated marker sets. Some cell types expressed specific marker genes, such as Pou2af1 for Bcell-Early; Ifit3, Stat1, and Isg15 for Bcell-Isg; Igfbp4, Lef1, Sell, and Tcf7 for CD4-Naïve; Ctla2a and Gzmk for CD4-Cytotoxic; Gzma, Gzmb, and Jaml for CD8; Rorc for ILC3; Adgre1, Lgmn, and Hmox1 for Macrophage; Msrb1 for Neutrophil (Figure S3B). In contrast, some cells in a stage of early development expressed multiple cell-type marker genes, such as Bcell-Early expressed both the plasma-cell marker, Herpud1, and CD4 marker, Cd3g; CD4-Early also expressed both CD4 markers, Cd3d and Cd3e, as well as Bcell marker, Bank1 (Figure S3B). We also found that Ccr2 was expressed in Monocytes and Neutrophils, whereas Ccrl2 was expressed in both Macrophages and Neutrophils. This indicates the role of Ccrl2 in the process of differentiation from monocytes to macrophages (Galligan et al., 2004). Interestingly, the ProxCol was predominantly populated with T cells, whereas the cecum and DistCol had a more balanced T cell to B cell ratio (Figure 5B). ProxCol showed significantly less B cells but more CD4 and CD8 T lymphocytes than either cecum or DistCol (Figure 5C). Other cell types did not have substantial differences between compartments. Consistently, marker genes from B cells were less expressed in ProxCol cells, with more cells expressing CD4 and CD8 markers (Figure 5D). Apart from some genes involved in T cell development (Ikzf2, Cd3d, Cd3e, Cd3g, Cd7, and Lck) and binding proteins (Tcrg family, Fcer1g, and Cd160), highly expressed genes in ProxCol also contained those with specific immune functions, such as Xcl1 (inflammatory and immunological responses), Ctsw (microbial infection), Trbc family (adaptive immunity), Nkg7 (infection), Cd8a (recognition of antigens), and Ccl5 (inflammation). Altogether, these results suggest a different enrichment of immune cells and immune functions in different compartments of the large intestine in young female mice.
Figure 5.
Immune cell population enrichment in different compartments of young mouse colon
(A) Scatterplots of UMAP of immune single cells in different compartments of the large intestine from young mice (n = 4 biologically independent animals).
(B) Boxplots of cell population regarding the immune cell lineages in different compartments in the same dataset as in (A). (n = 4 biologically independent animals; Welch’s two-tailed t-test; Data are represented as mean +SD).
(C) Line plots indicating the population changes of different cell types in different compartments in the same dataset as in (A). (n = 4 biologically independent animals; Welch’s two-tailed t-test; Data are represented as mean +SD).
(D) Hierarchical clustering and heatmap of scaled expression of marker genes (B cell, CD4, CD8) identified in the scRNAseq (n = 4 biologically independent animals). ns, p≥0.05; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001.
The scRNA-seq indicates ProxCol in young mice requires less B cell-dependent adaptive immune functions. Instead, it relies on T cells and expression of functional genes like Xcl1, Ctsw, Nkg7, Cd8a, and Ccl5 to defend against infections, tumor cells, or other invaders. We also observed that this immune difference in ProxCol occurs mainly in young mice, as the enrichment difference between marker genes of B cells and T cells was reduced in aged ProxCol (Figures S3C and S3D). This implies that some factors, either extrinsic or intrinsic, could have an impact on ProxCol-specific immune characteristics during aging.
In addition to the change in B cell and T cell populations of ProxCol in aging, we further investigated how immune cell populations changed during aging in general. Cell population change of different immune cell types in aging was displayed by compartment (Figure 6A).
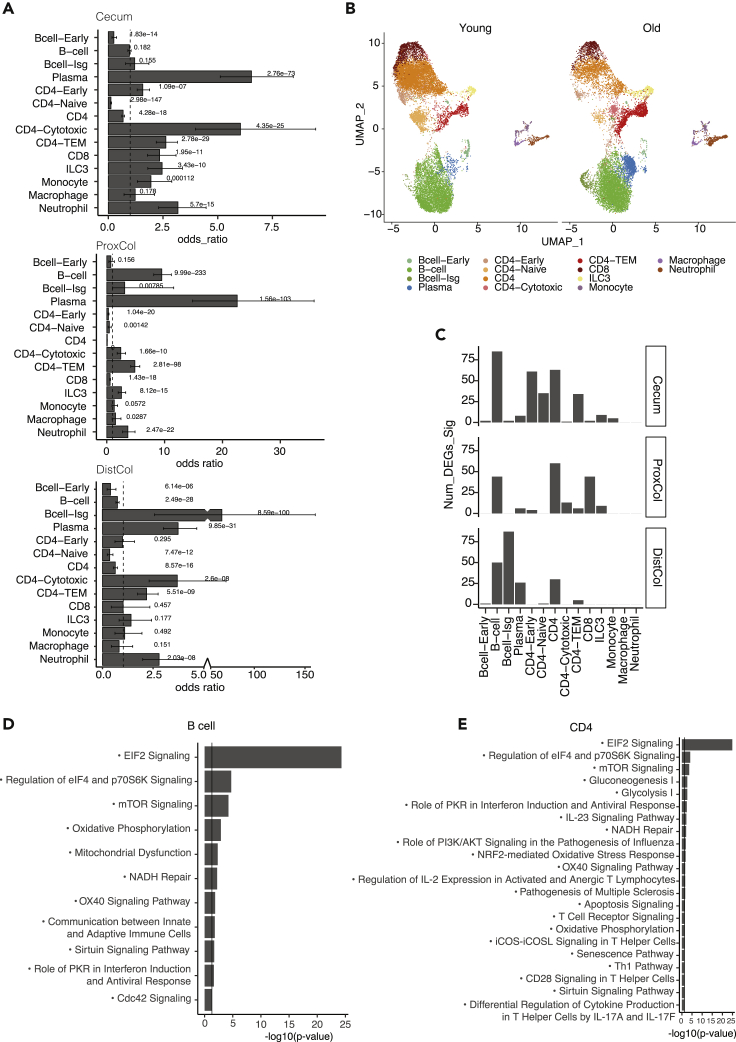
Figure 6.
Different aging characteristics of immune cells from different compartments of large intestine
(A) Boxplots of cell population change of immune sub cell-types in aging in cecum (top panel), proximal colon (middle panel), and distal colon (bottom panel). (n = 4 young, four old biologically independent animals; p value was calculated by hypergeometric distribution, confidence intervals for the odds ratio were calculated by Fisher’s test; Data are represented as OR ± CI).
(B) UMAP scatterplot of identified immune cell types in young and old mice from scRNAseq analysis (n = 4 young, four old biologically independent animals).
(C) Bar charts showing the number of DEGs of the 14 cell types in aging mice’s large intestine: cecum (top panel), proximal colon (middle panel), and distal colon (bottom panel) (n = 4 young, four old biologically independent animals).
(D and E) Bar charts showing the -log10 enrichment p value (x axis) of the GO pathways enriched in B cell aging (D) and CD4 aging (E) of the large intestine. (n = 4 young, four old biologically independent animals; p-value was calculated by the right-tailed Fisher’s Exact Test).
No significant changes were observed at the cell lineage level during aging (Figures 6A and S4A), probably because of intra-lineage compensations. However, when comparing aging population changes at cell-type level in a compartment-specific manner, many differences were observed (Figures 6A, 6B, and S4B). The aged cecum had significantly more CD4-Early, CD4-Cytotoxic, CD4-TEM, CD8, and ILC3 from T lymphocytes as well as more monocytes, neutrophils, and plasma cells. Conversely, aged cecum had significantly less B-cell-Early, CD4, and CD4-Naïve cells (Figure 6A). Aged ProxCol had significantly more B-cells, B-cell-Isg, Plasma cells, CD4-Cytotoxic, CD4-TEM, and ILC3 as well as more macrophages and neutrophils but less CD4, CD4-Early, CD4-Naïve, and CD8 T lymphocytes (Figure 6A). Aged DistCol had a significant increase in CD4-Cytotoxic, CD4-TEM, B cell-Isg, plasma cells, and neutrophils but less B cell, B cell-Early, CD4, CD4-TEM cells (Figure 6A). Altogether, these results indicate that immune cells in the lamina propria of old mice switch from a more naïve to a more activated state as previously observed for other compartments (Blecher-Gonen et al., 2019; Elyahu et al., 2019) likely as the result of a pro-inflammatory environment.
We then wondered whether aging also leads to alterations of the transcriptome for the different immune cell types. By comparing the number of DEGs in aging mice, we found that CD4 cells and B cells change in all compartments, whereas other cell types changed only in specific compartments: CD4-Early, CD4-Naïve and CD-TEM in Cecum, CD8 in ProxCol, B cell-Isg, and plasma cells in DistCol (Figure 6C).
Pathway Enrichment of B cells and CD4 cells in aging can be categorized into three different groups of processes: translational regulation (EIF2, regulation of eIF4 and p70S6K signaling, and mTOR), oxidative stress (oxidative phosphorylation, NADH repair, mitochondrial dysfunction in B cell, and NRF2-mediated oxidative stress response in CD4), and immune development process (e.g., OX40 signaling, role of PKR in interform induction and antiviral response). The top enrichment was for general aging pathways: EIF2 and mTOR signaling, which were common with epithelial cells (Figures 6D and 6E). Metabolic oxidative stress, like oxidative phosphorylation and mitochondrial dysfunction, were also both enriched in immune as well as epithelial cells (Figure 6D). There were also upstream regulators that were in common with epithelial cells, such as Mlxipl, Rictor, and Myc (Figure S4D).
CD4-Early and CD4-Naïve cells in the cecum had the aging pathway profile of translational regulation (Figures S4E and S4F). In cecum CD4 cell aging, additional pathways of antigen presentation and HIF1α were also noticeable. ProxCol CD8 cells had by far the highest number of enriched pathways during aging, with many signals associated with interleukin-related cellular immune responses and kinase signaling (Figure S4G). Some kinase pathways play a role in regulating CD8 T cell proliferation and survival, such as ERK/MAPK, SAPK/JNK (Conze et al., 2002), and p38 MAPK (Henson et al., 2015), whereas some contribute to CD8 T cell differentiation and development, such as Tec kinase (Readinger et al., 2009). IL-17 signaling (Boari et al., 2018) and CD40 signaling (Bhadra et al., 2013) could modulate CD8+ T cell survival or exhaustion in chronic infection, whereas IL-7 (Schluns et al., 2000) could mediate the homeostasis of naïve and memory CD8 T cells.
Changes in cell-to-cell interactions in aging
Cell-to-cell interactions are important for the maintenance of homeostasis, dysregulation of which leads to pathological phenotypes (e.g., ulcerative colitis (Smillie et al., 2019)); therefore, we wanted to investigate what happens to these interactions during aging. To determine the possible interactions between cells, we applied the CellPhoneDB software (Efremova et al., 2020) on the scRNAseq data from both epithelial and immune cells to map the produced ligands with their respective receptors. Sums of significant interactions were determined between epithelial cells, T lymphocytes, B cell lymphocytes, and Myeloid lineages versus epithelial cells (Figure 7A).
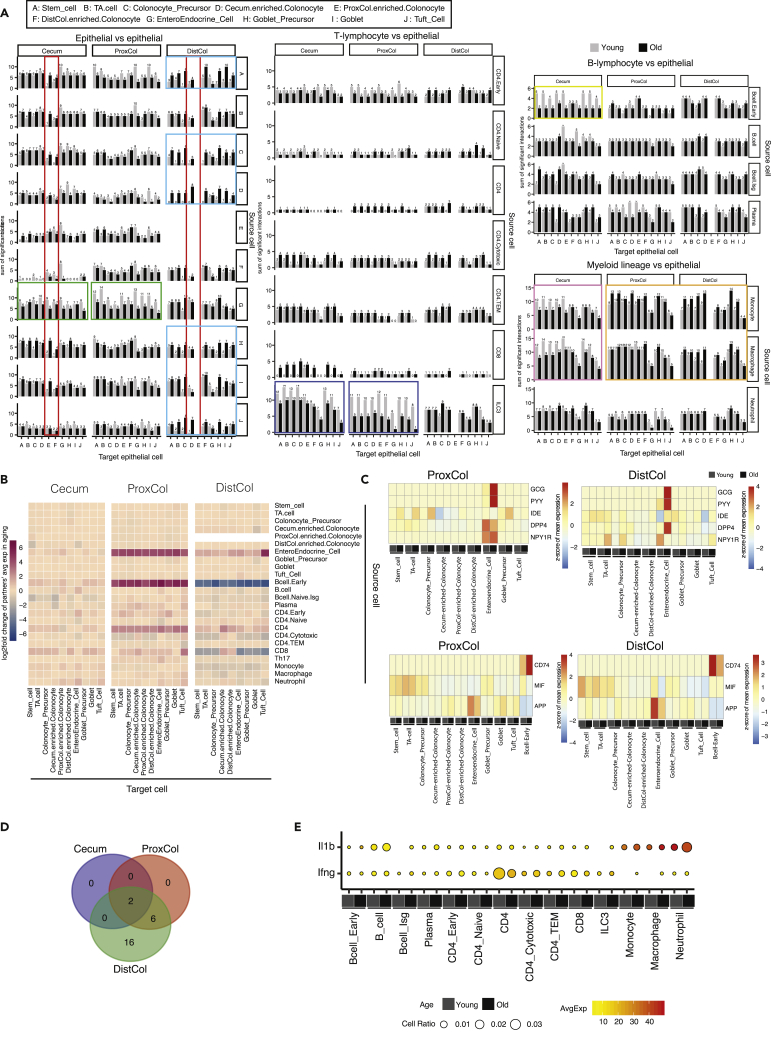
Figure 7.
Changes in cell-type interaction during aging, associated with interaction between immune and epithelial cells
(A–D) Bar charts indicating the number of ligand-receptor pairs found in the interaction between epithelial cells/immune cells and epithelial cells. Interacting ligand-receptor pairs were detected by the software of CellphoneDB which used a null-distribution based permutation test on the corresponding expression of simple/complex ligands/receptors curated by CellphoneDB. Row - source cell types; column - target cell types. (n = 4 young, four old biologically independent animals) (B) Heatmap of log2 fold change of ligand-receptor’s average expression in aging for the interaction between epithelial/immune cells and epithelial cells. Row - source cell types; column - target cell types. (n = 4 young, four old biologically independent animals) (C) Heatmap of the scaled expression of the pronounced interacting partners (ligand and receptor) between EE cells (top panels), B cell early (bottom panels) and epithelial cells in proximal colon (left panels) and distal colon (right panels) of young and old mice. (n = 4 young, four old biologically independent animals) (D) Venn diagram of the upstream cytokines predicted to be regulating epithelial cells’ aging in cecum, proximal colon and distal colon (n = 4 young, four old biologically independent animals).
(E) Bubble plot of expression of Il1b and Ifng in immune cells, which are shared in cecum and colon as upstream cytokines for epithelial aging. (n = 4 young, four old biologically independent animals).
The analysis seems to be functional; for example, epithelial cells from one compartment do not show (or have very few) potential interactions with epithelial cells of other compartments, reflecting the actual physical separation (see cecum epithelial cells vs DistCol-enriched colonocytes or DistCol epithelial cells vs cecum- or ProxCol-enriched colonocytes - Figure 7A, red squares).
Young EE cells produced more ligands in the ProxCol (and to a lesser extent in cecum) that interacted with other epithelial cells than old epithelial cells (Figure 7A). Conversely, several old epithelial cell types in DistCol acquired new interactions, such as ligands from Stem, Colonocyte precursor, Cecum-enriched-colonocyte, Goblet precursor, and Goblet and Tuft cell interacting with other epithelial cells (Figure 7A). Considering interactions between T lymphocytes ligands and epithelial cells, it was most noticeable concerning ProxCol (and to a lesser extent in cecum) ILC3 cells: aged ILC3 cells had a reduced number of interactions with epithelial cells (Figure 7A). A noticeable loss of interactions was also evident between aged B cell-early and the cecum epithelium (Figure 7A). Aged macrophages and monocytes showed a similar pattern: loss of interactions with cecum epithelium but gain of interactions with ProxCol and DistCol epithelium (Figure 7A). Altogether, these results illustrated a loss of cell-to-cell interactions in cecum and ProxCol and an increase in the DistCol during aging (Figure S5A).
Because the gain or loss of interactions is not the only way in which interactions could change during aging, we also investigated the log2fold change in aging (Figure 7B) on the mean expression of ligand-receptor partners (Figure S5B) to detect hypoactivation or hyperactivation of stable ligand-receptor pairs. With this method, we saw a clear increase in ligand-receptor partner expression between aged EE or CD4 cells and other epithelial cells in ProxCol and DistCol (Figures 7B and S5B). Interestingly, B cell-early and CD8 cells had the opposite change between ProxCol and DistCol: interaction intensity increased in aged ProxCol, whereas decreased in DistCol (Figures 7B and S5B). Decreased interaction partner number and increased partner expression in aging of ProxCol implied that interaction between different cell types (especially between EE/immune cells and epithelial cells) in ProxCol become more dedicated during aging. Increased interaction number and decreased partner expression in aging of DistCol suggested that interaction between immune cells and epithelial cells in DistCol become more universal during aging.
Because a large increase in one ligand could mask a decrease in other ligand interactions, we separated and focused on important ligand-receptor pairs. It was evident that EE cell secreted Glucagon (Gcg) and Peptide YY (Pyy), and their interaction with Insulin Degrading Enzyme (Ide), DipeptidylPeptidase 4 (Dpp4), and Neuropeptide Y Receptor Y1 (Npy1r) were the main factors responsible for the hyper-activation of EE cell interactions with other epithelial cells in ProxCol and DistCol (Figures 7C and S5C). Two of the peptides translated from Gcg are secreted from gut endocrine cells and promote nutrient absorption. Peptides translated from Pyy have a vasoconstrictory effect and inhibit jejunal and colonic motility. These important interactions between EE and epithelial cells demonstrate that the function of enteroendocrine cells on insulin regulation by stimulating glycogenolysis/gluconeogenesis (Gcg-Ide/Dpp4) and on the colonic motility by neurotransmission (Pyy-Npy1r) is enhanced in aging.
Interactions between CD74 and Macrophage Migration Inhibitory Factor (Mif)/Amyloid Beta Precursor Protein (App) were mainly responsible for the different B cell-Early pattern between ProxCol and DistCol (Figures 7C and S5D). Interaction by CD74-Mif/App between B-early cells and epithelial cells revealed that antibacterial activities involved by MHCII antigen-presentation were strengthened in aging.
We took the list of IPA predicted aging-related upstream regulators of epithelial cells and isolated only cytokines/growth factors. The DistCol had the biggest number of cytokines predicted to be responsible for changes in aging (Figures 7D and 7E). ProxCol and DistCol had six common upstream cytokines/growth factors (Il22, Il5, Il4, Fam3b, Csf1, and Wnt1), while all three compartments had two common upstream cytokines (IFN- γ and IL1b) (Figures 7E and S5D), of which IL1b was detected to be increased in aging from our immune scRNAseq data (Fig, S5E). By checking the expression of the upstream cytokines in different immune cell types, we were able to find that CD4 cells (and to a lesser degree CD8 in ProxCol) are the likely source of IFN-γ, whereas neutrophils are the likely source of IL1b (Figures 7F and S5D) in the aged lamina propria. This supports the notion that the aging immune system is at least in part responsible for the changes in aged epithelial cells.
Discussion
Our work proposes new colonic compartment marker genes, which could help with further inquiries into disorders of the large intestine, such as Ces1g for Cecum, Car8 for ProxCol, and B3gnt7 for DistCol. Along with our new marker genes, we captured genes that had been described previously to demonstrate some compartment delineation, such as Hoxb6 and Hoxb13 (Schaum et al., 2018). We believe this strengthens the validity of the novel marker genes.
As the microbiota is known to have important interactions with gut immune cells and is proposed as a driver of multiple aging-associated changes, it was surprising to see no impact of microbiome on the compartmentalization of the large intestine. This could be an important message, emphasizing that no one factor is solely responsible for gastrointestinal health or lack thereof. However, it would be erroneous to dismiss the microbiota overall, as the massively enlarged cecum of germ-free mice and disrupted cecum stem cell niche of antibiotic-treated mice demonstrate their crucial role in the normal function of the colon (Savage and Dubos, 1968; Zaborin et al., 2016). Interestingly, it has been reported that microbial composition differs between different parts of the colon (James et al., 2020) and changes after senolytic treatment, which along with our findings support that either the epithelium or immune cells are causing this change. Unfortunately, because James et al. separated immune cells even during processing, losing information about original cell ratios, we were unable to compare the differences between compartments we observed with their data.
Our result of increasing number of goblet cells during aging clashes with previously reported data (Sovran et al., 2019), which showed a decrease in viable colonic goblet cells in aged mice. We postulate that several factors could be responsible for such a discrepancy. Because our single-cell dataset had very few numbers of Krt20 expressing goblet cells (data not shown), we were generally not profiling the most mature (crypt top) goblet cells (Schaum et al., 2018), which were probably counted by Sovran et al. In addition, the use of different staining and imaging protocols (immunofluorescent staining of UEA-1 versus Alcian blue staining) could account for some changes, which would explain why including crypt-top UEA-1-positive cells in our calculations resulted in a decrease that did not reach statistical significance (data not shown). Lastly, we think that the different results could actually supplement each other, as a decrease in the number of fully mature, functional goblet cells could either be caused by defects in aged goblet cell maturation (thus accumulating immature goblet cells) or explain the increased need for production of new goblet cells. However, additional experiments would be needed to substantiate these claims.
It is interesting to note just how distinct the ProxCol is, even if some features are shared with the cecum. Our data suggests that the ProxCol is in a very different immunological state, compared to both Cecum and DistCol. As Reg4 and Reg3g are both antimicrobial genes, the lack of Reg4 cells in ProxCol could be linked to the expression of Reg3g in the ProxCol-enriched-colonocytes as an adaptive or causative mechanism. Perhaps the amount of Reg3g that could be produced by colonocytes is higher than the possible Deep Secretory Cell Reg4 amounts.
We found that ProxCol has few B lymphocytes. It has been demonstrated that B lymphocytes are produced in the cecal patch, subsequently migrating toward the colon (Masahata et al., 2014); however, it is unclear why so few remain in the ProxCol. In addition, the ProxCol’s very low B lymphocyte number would mean a decreased production of secreted IgA, perhaps linked to a unique niche for specific microbiota. If so, the increasing B cell numbers in aged ProxCol could influence the microbe composition and contribute to age-related intestinal issues.
Our IPA results pointed to possible issues with oxidative phosphorylation in the colonic epithelium (possibly at a higher level in DistCol). This falls in line with published data, showing a decrease in oxidative phosphorylation capacity of colonic epithelium (Greaves et al., 2010; Özsoy et al., 2020). Interestingly, Özsoy et al. reported that different oxidative phosphorylation complexes change during aging according to colonic compartment, which would support our findings and highlight the different aging possibilities of different colonic compartments.
It has been reported that B cells respond to antigens by promptly increasing their metabolic activity, including both oxidative phosphorylation and glycolysis. When absent from signals of T helper cells, B cells gradually lose mitochondrial function and glycolytic capacity, which leads to apoptosis (Akkaya et al., 2018). This indicates that oxidative phosphorylation and mitochondrial dysfunction could be aging characteristics in B cells, which is supported by our data. Cdc42, which is required for antiviral humoral immunity, is a crucial regulator for B cell differentiation and is required for B cells to present antigen to T cells (Burbage et al., 2015); thus, enrichment of Cdc42 signaling suggests an increased necessity of B cell activation during aging. Interestingly, elevated Cdc42 activity has recently been shown to result in impaired and aged ISC function in the small intestine (Nalapareddy et al., 2021). This could hint at a link between chronically increased antiviral response and aging-related impairments.
Pathway enrichment of various interleukin cytokines, including IL-23 signaling, IL-2 expression, and IL-17 regulation, in aged CD4 suggests increased inflammatory immunity during aging (Glosson-Byers et al., 2014). Apart from regular immune aging pathways, gluconeogenesis I and glycolysis I are enriched in CD4 aging. It has been demonstrated that CD4 T cells have higher basal glycolysis level (Jones et al., 2017). This alteration of glucose homeostasis in aged CD4 could result from T cell senescence with systemic inflammation (Yi et al., 2019). OX40 pathways with OX40 (CD134) and its binding partner, OX40L (CD252), as members of the TNFR/TNF superfamily, play an important role in modulatory functions (regulating cytokine production and modulating cytokine receptor signaling) in CD4 cells as well as in other lymphoid and nonlymphoid cells. OX40 signaling is also involved in the autoactivation of CD4+ CD28− T cells (Jiang et al., 2017; Linton et al., 2003), thus could be at least in part responsible for the changes in aging immune cells we observed.
Our results showed an increase of IFN-γ, produced by CD4 and CD8 cells, during aging. Interestingly, we did not observe expression of MHC class II molecules on colonic epithelial cells, which is induced by IFN-γ. Conflicting findings can be found in literature on the effects of IFN-γ on the colon, as some demonstrate protective anti-inflammatory effects (Thelemann et al., 2014), whereas others show IFN-γ to exacerbate inflammation (Nava et al., 2010). Thelemann et al. proposed that small amounts of IFN-γ could maintain intestinal homeostasis during infection, which could mean the increased production in aging is an adaptive response to restore homeostasis but is not high enough to mimic the exacerbation of inflammation in Dextran sulfate sodium (DSS)-treated mice, shown by Nava et al.
This study provides a resource for further insight into the aging changes of the colonic epithelium and immune system. We provide compartment-specific epithelial marker genes, transcriptional changes, and cell population changes in both epithelial and immune cells as well as their shifts during aging. Finally, we propose interaction changes between immune and epithelial cells in aging.
Limitations of the study
In this study, we performed single-cell RNAseq on young and old female mouse colonic epithelium and immune cells, which allows pairwise comparison of the two cell populations during aging. One limitation of the study is the lack of a comparable single-cell RNAseq dataset of young and old male animals, which could demonstrate some sex-dependent differences during colonic aging. In addition, though our dataset includes important players of the colon, there is possibility for expansion with the inclusion of, for example, microbiome data or single-cell RNAseq of connective tissue cells. Finally, more validation experiments would help provide more solid proof of the detected changes.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Aquaporin 4 (Aqp4) polyclonal antibody | Thermo Fisher Scientific | Cat# PA5-77716; RRID: AB_2735469 |
| CD45 anti-mouse antibody, FITC coupled | Biolegend | Cat# 103108; RRID: AB_312973 |
| Carbonic Anhydrase I (CAR1) polyclonal antibody | Thermo Fisher Scientific | Cat# PA5-97527; RRID: AB_2812143 |
| EpCAM antibody, PE-Cy7 coupled | Thermo Fisher Scientific | Cat# 25-5791-80; RRID: AB_1724047 |
| Secondary donkey anti-rabbit antibody, AlexaFluor 488 coupled | Thermo Fisher Scientific | Cat# A21206; RRID: AB_2535792 |
| Secondary goat anti-rat antibody, AlexaFluor 568 | Thermo Fisher Scientific | Cat# A11077; RRID: AB_2534121 |
| TotalSeq anti-mouse antibody, hashtag-1 coupled | Biolegend | Cat# 155801; RRID: AB_2750032 |
| TotalSeq anti-mouse antibody, hashtag-2 coupled | Biolegend | Cat# 155803; RRID: AB_2750033 |
| TotalSeq anti-mouse antibody, hashtag-3 coupled | Biolegend | Cat# 155805; RRID: AB_2750034 |
| TotalSeq anti-mouse antibody, hashtag-4 coupled | Biolegend | Cat# 155807; RRID: AB_2750035 |
| TotalSeq anti-mouse antibody, hashtag-5 coupled | Biolegend | Cat# 155809; RRID: AB_2750036 |
| TotalSeq anti-mouse antibody, hashtag-6 coupled | Biolegend | Cat# 155811; RRID: AB_2750037 |
| TotalSeq anti-mouse antibody, hashtag-7 coupled | Biolegend | Cat# 155813; RRID: AB_2750039 |
| TotalSeq anti-mouse antibody, hashtag-8 coupled | Biolegend | Cat# 155815; RRID: AB_2750040 |
| TotalSeq anti-mouse antibody, hashtag-9 coupled | Biolegend | Cat# 155817; RRID: AB_2750042 |
| TotalSeq anti-mouse antibody, hashtag-10 coupled | Biolegend | Cat# 155819; RRID: AB_2750043 |
| TotalSeq anti-mouse antibody, hashtag-11 coupled | Biolegend | Cat# 155821; RRID: AB_2750136 |
| TotalSeq anti-mouse antibody, hashtag-12 coupled | Biolegend | Cat# 155823; RRID: AB_2750137 |
| TruStain FcX anti-mouse antibody | Biolegend | Cat# 101319; RRID: AB_1574973 |
| Chemicals, peptides, and recombinant proteins | ||
| Na2HPO4 x2H2O | Sigma-Aldrich | Cat# 4984.3 |
| KH2PO4 | Roth | Cat# 3904.1 |
| NaCl | Roth | Cat# 9265.2 |
| KCl | Roth | Cat# 6781.1 |
| Sucrose | Sigma-Aldrich | Cat# 84097-250G |
| D-sorbitol | Roth | Cat# 6213.1 |
| PBS | VWR | Cat# E404-200TABS |
| EDTA | Roth | Cat# X986.2 |
| DTT | VWR | Cat# 0281-25G |
| MgCl2 | Sigma-Aldrich | Cat# 63069-100ML |
| Fetal Bovine Serum | Sigma-Aldrich | Cat# F9665-500ML |
| Collagenase-D | Merc | Cat# 11088866001 |
| RPMI medium | Thermo Fisher Scientific | Cat# 21875091 |
| Percoll | Sigma-Aldrich | Cat# P1644-1L |
| BSA | Life technologies | Cat# 15260037 |
| Qiazol Lysis Reagent | Qiagen | Cat# 79306 |
| OCT Q PATH® Mounting Media for Cryotomy | VWR | Cat# 361603E |
| TrypLE Express Enzyme | Thermo Fisher Scientific | Cat# 12604021 |
| DNase I grade II, f. bov. pancreas | Sigma-Aldrich | Cat# 10104159001 |
| Ulex Europaeus Agglutinin I (UEA-1), Dylight® 649 coupled | Vector Labs | Cat# DL-1068-1 |
| Y27632 | Abcam | Cat# ab120129 |
| DAPI | Sigma-Aldrich | Cat# D9542 |
| Paraformaldehyde | Roth | Cat# 0335.2 |
| Sodium citrate | Roth | Cat# 3580.1 |
| Proteinase-K | Thermo Fisher Scientific | Cat# AM2548 |
| Formamide | Roth | Cat# P040.1 |
| Dextran sulfate | Sigma-Aldrich | Cat# D8906-5G |
| Vanadyl-ribonucleoside complex | NEB | Cat# S1402S |
| DEPEX | Serva | Cat# 18243.01 |
| Matrigel | Corning | Cat# 356231 |
| Recombinant human Rspondin-1 | Lab-made | N/A |
| Recombinant murine Noggin | PeproTech | Cat# 250-38 |
| Recombinant murine EGF | PeproTech | Cat# 315-09 |
| CHIR99021 | Tocris | Cat# 4423/10 |
| Recombinant murine Wnt-3a | PeproTech | Cat# 315-20 |
| N2 supplement | Thermo Fisher Scientific | Cat# 15410294 |
| B27 supplement | Thermo Fisher Scientific | Cat# 11500446 |
| Penicillin-Streptomycin | Thermo Fisher Scientific | Cat# 15140122 |
| Advanced DMEM/F12 | Thermo Fisher Scientific | Cat# 11550446 |
| Chloroform | Sigma-Aldrich | Cat# 32211-1L-M |
| Critical commercial assays | ||
| TruSeq™ RNA Library Preparation Kit v2 | illumina | Cat# RS-122-2001 |
| Qubit® dsDNA HS Assay Kit | Thermo Fisher Scientific | Cat# Q32854 |
| High Sensitivity RNA Analysis Kit (15 nt) | Agilent | Cat# DNF-472-0500 |
| Agencourt AMPure XP Kit | Beckman Coulter | Cat# A63881 |
| iScript™ cDNA Synthesis Kit | Bio-Rad Laboratories | Cat# 1708891 |
| SYBR GreenER™ qPCR SuperMix for iCycler® Instrument | Thermo Fisher Scientific | Cat# 11761500 |
| Chromium™ Single Cell 3' Library & Gel Bead Kit v2 | 10X Genomics | Cat# 120237 |
| Chromium™ Single Cell 3' Library & Gel Bead Kit v3.1 | 10X Genomics | Cat# 1000121 |
| Chromium™ Single Cell A Chip Kit | 10X Genomics | Cat# 1000009 |
| Chromium™ Next GEM Chip G Single Cell kit | 10X Genomics | Cat# 1000120 |
| Chromium™ i7 Multiplex kit | 10X Genomics | Cat# 120262 |
| Dynabeads™ MyOne™ Silane | Thermo Fisher Scientific | Cat# 37002D |
| SYTOX™ Blue Dead Cell Stain | Thermo Fisher Scientific | Cat# S34857 |
| Zombie Aqua Fixable Viability Kit | Biolegend | Cat# 423101 |
| NextSeq® 500/550 High Output Kit v2.5 | Illumina | Cat# 20024906 |
| Deposited data | ||
| Single cell RNA sequencing data: Epithelial cells | this paper | GEO: GSE168448 |
| Single cell RNA sequencing data: Immune cells | this paper | GEO: GSE169351 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL6/J | Fritz Lipmann Institute | N/A |
| Mouse: Lgr5-ki-e-GFP-creER | Fritz Lipmann Institute | N/A |
| Oligonucleotides | ||
| See Materials and methods table for Primers for Sequencing Library preparation and Real-time PCR | this paper | |
| Software and algorithms | ||
| ImageJ | (Schneider et al., 2012) | RRID: SCR_003070 |
| GraphPad Prism 7 | GraphPad Software | RRID: SCR_002798 |
| ZEN Digital Imaging for Light Microscopy | Carl Zeiss Microscopy | RRID: SCR_013672 |
| Primer-BLAST | NCBI | RRID: SCR_006472 |
| FastQC | Babraham Bioinformatics, UK | RRID: SCR_014583 |
| R Project for Statistical Computing | RRID: SCR_001905 | |
| CellPhoneDB | Vento-Tormo et al. (2018) | RRID: SCR_017054; https://www.cellphonedb.org/ |
| Ingenuity Pathway Analysis | RRID: SCR_008653 | |
| Seurat (version 3.1.1) | Stuart et al. (2019) | RRID: SCR_016341; https://satijalab.org/seurat/get_started.html |
| Cell Ranger (version 3.1.0) | 10xGenomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/using/count |
| Custom Scripts | in house developed | https://github.com/Dsirvinskas/Aging_Colon_Atlas |
| Other | ||
| Falcon 70 μm filters | Corning | Cat# 352,350 |
| Falcon 40 μm filters | Corning | Cat# 352,340 |
| Incubator shaker | VWR | |
| Nanodrop 8000 | Thermo Fisher Scientific | |
| Qubit 3.0 | Thermo Fisher Scientific | |
| 5200 Fragment Analyzer | Agilent | |
| Bioanalyzer 2100 | Agilent | |
| illumina NextSeq 500 | illumina | |
| Corbett RotorGene 6000 | Qiagen | |
| CFX96 Touch Real-Time PCR | Bio-Rad | |
| Primers | Eurofins Genomics | |
| FACS LSRII | BD Biosciences | |
| Axiovert 200 inverted microscope, with ApoTome slider module | Zeiss | |
| 10X Chromium Controller | 10X Genomics | |
| TT Cryomolds | Science Services | Cat# SA62534-15 |
| Mm10-3.0.0 mouse genome assembly | NCBI | RefSeq assembly accession: GCF_000001635.20 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Francesco Neri (francesco.neri@leibniz-fli.de)
Materials availability
This study did not generate new unique reagents. All unique/stable reagents generated in this study are available from the Lead contact without restriction.
Experimental model and subject details
Mice
All mouse work was in accordance to protocols approved by the state government of Thuringia (licenses number: TG/J-0002858/A; TG/J-0003616/A; TG/J-0003681/A; FLI-17-109; FLI-18-005).
Mice were housed and maintained in a Specific Opportunist Pathogen Free (SOPF) animal facility in Fritz Lipmann Institute with 12 h of light/dark cycle and fed with a standard mouse chow. Mice were assigned to age groups: Young 8 to 18 weeks and Old 89 to 117 weeks of age (Germ-free old mice were 56-57 weeks of age). Male and female mice were grouped separately. According to experimental needs, mouse lines used: wild-type C57BL/6 (Jackson Laboratory or Charles River Laboratories), germ-free C57BL/6J and Lgr5-EGFP-IRES-CreER (Lgr5-GFP).
Method details
Crypt isolation, cell dissociation and preparation for sorting
Crypt isolation: the colon (with cecum) was isolated by cutting the intestine along the illeo-cecal junction. The cecum was collected, the remaining colon split in half into the ProxCol and DistCol. The tissue was rinsed with ice-cold PBS and cleaned of connective tissue, cut longitudinally and chopped into small pieces. Two protocols for crypt separation from tissue were used, with no apparent differences in yield or cell viability: (Ahmad et al., 2014) and (Schaum et al., 2018).
Ahmad et al.: Tissue was rinsed once in ice-cold Rinse Buffer (5.6 mM Na2HPO4 x2H2O (Sigma-Aldrich, Cat. 4984.3); 8.0 mM KH2PO4 (Roth, Cat. 3904.1); 96.2 mM NaCl (Roth, Cat. 9265.2); 1.6 mM KCl (Roth, Cat. 6781.1); 43.4 mM Sucrose (Sigma-Aldrich, Cat. 84097-250G); 54.9 mM D-sorbitol (Roth, Cat. 6213.1) in PBS (VWR, Cat. E404-200TABS)), then incubated for 30 min in room-temperature (RT) Isolation Buffer (2 mM EDTA (Roth, Cat. X986.2); 0.5 mM DTT (VWR, Cat. 0281-25G) in Rinse Buffer). Tissue then transferred to a falcon tube with RT Rinse Buffer and shaken for 20s (repeated once). Tissue then transferred to PBS for immune-cell isolation. Resulting 2 supernatant aliquots were combined and centrifuged for 10 min at 450 g, 4°C. Supernatant decanted, crypt pellet subsequently used for single-cell isolation.
Schaum et al. “Tabula Muris”: Tissue was placed into ice-cold 5 mM EDTA (in PBS) and shaken briefly. The tissue was then transferred into a pre-warmed 5 mM EDTA (in PBS) and incubated for 8 min at 37°C. Tissue then transferred into ice-cold PBS and shaken vigorously for 30 s. The tissue was discarded, crypt suspension centrifuged for 10 min at 450 g, 4°C. Supernatant decanted, crypt pellet subsequently used for single-cell isolation.
Crypt pellet was re-suspended in Single-Cell-Isolation-Buffer (1 mg/ml DNase I (Sigma-Aldrich, Cat. 10104159001); 5 mM MgCl2 (Sigma-Aldrich, Cat. 63069-100 ML); 80 μM Y27 (Abcam, Cat. ab120129) in TrypLE (Thermo Fisher Scientific, Cat. 12604021)) and incubated for 10 min at 37°C. Suspension was briefly vortexed and incubated for another 10 min at 37°C. Suspension filtered from clumps with Falcon 70 μm filters (Corning, Cat. 352350), digestion was quenched by dilution at 1:10 with ice-cold PBS. Cell suspension centrifuged for 5 min at 1000 g, 4°C. Supernatant carefully decanted. Cells re-suspended in ice-cold FACS-Staining-Medium (FSM: 2% Fetal Bovine Serum (Sigma-Aldrich, Cat. F9665-500 ML); 3 mM EDTA; 80 μM Y27 in PBS).
Immune cells were isolated from lamina propria: tissue (cleared of crypts) was chopped finely with a scalpel and then incubated in 3 ml of 1 mg/ml collagenase-D (Merck, Cat. 11088866001) and 1 mg/ml DNaseI in RPMI medium (ThermoFisherScientific, Cat. 21875091) in 6 well plates in an incubator shaker (80 rpm) (VWR, Cat. 444–0732) for 20 min at 37°C. Tissue was pipetted up and down several times with a cut p1000 tip. The supernatant was passed through a 100 μm strainer into 5% FBS in RPMI. Leftover tissue incubated again twice in 3 ml collagenase-D/DNaseI RPMI solution in an incubator shaker (80 rpm) for 30 min at 37°C, collecting filtered supernatant. After the third incubation, the remaining tissue was smashed with a syringe plunger on a Falcon 40 μm cell strainer (Corning, Cat. 352340) and washed with 5% FBS/RPMI to collect the maximum number of cells. The supernatant was centrifuged at 300 g, 4°C for 5 min. The pellet was re-suspended in 40% percoll (Sigma-Aldrich, Cat. P1644-1L) in RPMI. The cell suspension was carefully pipetted over 80% percoll in a falcon tube in order to create a gradient. The falcon tubes were centrifuged at 1600 g, RT for 20 min (centrifuge break disabled). The immune cells formed a white ring on the border of the two percoll concentrations: they were collected carefully and washed with FSM (2% FBS in PBS). The suspension was centrifuged at 450 g, 4°C for 5 min. Resulting pellet was resuspended in FSM and transferred for staining.
Cells were blocked with TruStain FcX anti-mouse antibody according to manufacturer specifications. Staining and hash-tagging performed at the same time with PE-Cy7 coupled EpCAM and different TotalSeq anti-mouse Hashtag antibodies for each compartment.
Cell sorting
FACS sorting performed using BD FACSAria Fusion. Cells sorted for single-cell sequencing: viable (SYTOX™ Blue Dead Cell Stain (Thermo Fisher Scientific, Cat. S34857) Epithelial Cell Adhesion Molecule (EpCAM) positive cells. Cells were sorted into one tube at up to 10 000 cells per compartment. Cell suspension subsequently concentrated by centrifugation and re-suspension in ice-cold Re-Suspension-Buffer (0.04% BSA (Lifetechnologies, Cat. 15260037) in PBS) up to 1000 cell/μl. Cells for UEA-1+ quantification were fixed with 0.1% PFA. UEA-1+ cells analyzed on Zombie Aqua (Biolegend, Cat. 423101) negative (viable), EpCAM positive cells. Lgr5-eGFP cells analyzed on viable cells.
Droplet-based scRNAseq
Single-cell suspension processed with 10X Single-cell 3’ Gene Expression kit (ver. 2 and ver. 3.1) (10X Genomics, Cat. 120237 and Cat. 1000121), using the Chromium Single Cell A chip (10X Genomics, Cat. 1000009) (ver.2) and Chromium Next GEM Chip G Single Cell kit (10X Genomics, Cat. 1000120) (ver. 3.1) on the 10X Chromium Controller (10X Genomics, Cat. 1000202) according to manufacturer specifications. Library construction performed with 10X Single-cell 3’ Library Construction Kit, utilizing the Chromium i7 Multiplex kit (10X Genomics, Cat. 120262). Hashtag library recovery performed according to Biolegend protocol: https://www.biolegend.com/en-us/protocols/totalseq-a-antibodies-and-cell-hashing-with-10x-single-cell-3-reagent-kit-v3-3-1-protocol. Library quantification performed with Qubit dsDNA HS assay (Thermo Fisher Scientific, Cat. Q32854), fragment profiles checked using the High Sensitivity NGS Fragment Analysis kit (Advanced Analytical Technologies GmbH, Cat. DNF-474-0500) on the 5200 Fragment Analyzer (Advanced Analytical Technologies GmbH, Cat. FSv2-CE2). Libraries sequenced with NextSeq® 500/550 High Output Kit v2.5 (75 cycles) (Illumina, Cat. 20024906) on the NextSeq 500.
Immunofluorescence
Tissue for staining was fixed in 4% Paraformaldehyde (PFA) (Roth, Cat. 0335.2) overnight at 4°C, dehydrated in 20% Sucrose at least overnight at 4°C. Tissue was embedded in OCT (VWR, Cat. 361603E) in TT Cryomolds (Science Services, Cat. SA62534-15) and frozen. Staining performed utilizing smRNA FISH protocol by Shalev Itzkovitz http://shalevlab.weizmann.ac.il/wp-content/uploads/2016/02/Tissue-smFISH1.pdf: sections allowed to warm to RT, incubated with 4% PFA for 10 min, washed with RT PBS and placed into 4°C 70% Ethanol for at least 2 h. Sections washed in 2X SSC (0.3 M NaCl, 0.03 M Sodium citrate (Roth, Cat. 3580.1)) for 5 min, then incubated at RT with Proteinase-K (Thermo Fisher Scientific, Cat. AM2548) in 2X SSC for 10 min. Sections washed twice for 5 min with 2X SSC. Sections then incubated with Wash Buffer (15% Formamide (Roth, Cat. P040.1) in 2X SSC). Primary antibodies were diluted in Hybridization Buffer (10% Dextran sulfate (Sigma-Aldrich, Cat. D8906-5G), 15% Formamide, 0.02% BSA, 2 mM Vanadyl-ribonucleoside complex (NEB, Cat. S1402 S) in 2X SSC) at 1:100 ratio. Sections incubated with primary antibodies at 37°C over-night. Sections then washed for 30 min with Wash Buffer at 37°C. Secondary antibodies and DAPI were diluted in pre-warmed Wash Buffer at 1:500 and 1:1000 ratios respectively. Sections incubated with secondary antibodies for 30 min at 37°C. Sections then washed once for 30 min in Wash Buffer with 1:1000 DAPI (Sigma-Aldrich, Cat. D9542-1MG) at 37°C. Sections were then mounted with DEPEX (Serva, Cat. 18243.01) and imaged the same day.
Image analysis
Immunofluorescent slides imaged on the Zeiss Axio Imager 2 microscope at 400X magnification, using Apotome optical sectioning. Deconvolution performed with weak Fourier correction. Cell number/crypt counting done manually, utilizing ImageJ FIJI software (ver. 2.1.0/1.53 c) for region of interest tracking.
Antibodies
EpCAM-PE-Cy7 (Santa Cruz Biotechnology, Cat. 25-5791-80)
FITC anti-mouse CD45 Antibody (Biolegend, Cat. 103108)
Carbonic Anhydrase I (Car1) polyclonal antibody (Thermo Fisher Scientific, Cat. PA5-97527)
Aquaporin 4 (Aqp4) polyclonal antibody (Thermo Fisher Scientific, Cat. PA5-77716)
Ulex Europaeus Agglutinin I (UEA-1) Dylight® 649 (Vector Labs, Cat. DL-1068-1)
Secondary Goat anti Rat AlexaFluor 568 (Thermo Fisher Scientific, Cat. A11077)
Secondary Donkey anti Rabbit AlexaFluor 488 (Thermo Fisher Scientific, Cat. A21206)
TruStain FcX anti-mouse antibody (Biolegend, Cat. 101319)
TotalSeq anti-mouse hashtag antibodies
Hashtag 1 (Biolegend, Cat. 155801)
Hashtag 2 (Biolegend, Cat. 155803)
Hashtag 3 (Biolegend, Cat. 155805)
Hashtag 4 (Biolegend, Cat. 155807)
Hashtag 5 (Biolegend, Cat. 155809)
Hashtag 6 (Biolegend, Cat. 155811)
Hashtag 7 (Biolegend, Cat. 155813)
Hashtag 8 (Biolegend, Cat. 155815)
Hashtag 9 (Biolegend, Cat. 155817)
Hashtag 10 (Biolegend, Cat. 155819)
Hashtag 11 (Biolegend, Cat. 155821)
Hashtag 12 (Biolegend, Cat. 155823)
Colon organoid culture
Colon crypts were resuspended in ice-cold Matrigel (Corning, Cat. 356231) and allowed to solidify at 37°C, then were supplied with culture medium (750 ng/ml recombinant human Rspondin-1 (lab-made), 100 ng/ml recombinant murine Noggin (PeproTech, Cat. 250-38), 50 ng/ml recombinant murine EGF (PeproTech, Cat. 315-09), 10 μM CHIR99021 (Tocris, Cat. 4423/10), 100 ng/ml recombinant murine Wnt-3a (PeproTech, Cat. 315-20), N2 (Thermo Fisher Scientific, Cat. 15410294), B27 (Thermo Fisher Scientific, Cat. 11500446), 1X Penicillin-Streptomycin (Thermo Fisher Scientific, Cat. 15140122) in Advanced DMEM/F12 (ThermoFisherScientific, Cat. 11550446)). Organoids were cultured for 7 days, passaged via re-suspension in ice-cold PBS, then cultured for an additional 3 days before being collected in QIAzol (Qiagen, Cat. 79306) for RNA isolation.
qPCR
All RNA samples were isolated utilizing QIAzol (Qiagen, Cat. 79306), Chloroform (Sigma-Aldrich, Cat. 32211-1L-M). Reverse-transcription reaction performed with iScript cDNA Synthesis Kit (Bio-Rad Laboratories GmbH, Cat. 1708891). Quantitative PCR performed using SYBR GreenER™ qPCR SuperMix (Thermo Fisher Scientific, Cat. 11761500) on Rotor-Gene Q 6000 (Qiagen, Cat. 9001560)or CFX96 Touch Real-Time PCR (Bio-Rad, Cat. 1855195). Primers designed on the NCBI primer-BLAST online software: https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi.
| Primer | Target |
|---|---|
| GTGACTGGAGTTCAGACGTGTGC∗T∗C | HTO cDNA additional primer |
| CAAGCAGAAGACGGCATACGAGATCGAG TAATGTGACTGGAGTTCAGACGTGT∗G∗C |
Illumina TruSeq D701_s i7 index 1 |
| CAAGCAGAAGACGGCATACGAGATTCTC CGGAGTGACTGGAGTTCAGACGTGT∗G∗C |
Illumina TruSeq D702_s i7 index 1 |
| CAAGCAGAAGACGGCATACGAGATAATG AGCGGTGACTGGAGTTCAGACGTGT∗G∗C |
Illumina TruSeq D703_s i7 index 1 |
| CAAGCAGAAGACGGCATACGAGATGGAA TCTCGTGACTGGAGTTCAGACGTGT∗G∗C |
Illumina TruSeq D704_s i7 index 1 |
| TCTTTGCAGCTCCTTCGTTG | Mouse ActB |
| ACGATGGAGGGGAATACAGC | |
| GGCAGGCCTACCCAATTCTT | Mouse Ces1g |
| TCTGTCATTGTGGCAGGGTC | |
| CTGGTGGTTGTCAAGTCGGT | Mouse B3gnt7 |
| CTGCTGATAGTGGGTTCGCT | |
| TGACACACTGTCAGCTTTGAG | Mouse St3gal6 |
| CTTCACGTGTGCGTGATGC | |
| GGCTGGGTTAAAGACTCAAGC | Mouse Oit1 |
| AAGAATGGGTTGCGGTCAGG | |
| CAGACAAGATGCTTCCCCGT | Mouse Reg3g |
| GCAACTTCACCTTGCACCTG | |
| TTCTCAGAGCCTGGAGCAAC | Mouse Fabp2 |
| GATGACGAATGAGCCTGGCA | |
| GTCAAGACCAGCACTACCCC | Mouse Hoxb13 |
| TGTCATCTCTTGGGGTCCCT | |
| GGCTATACTACTAATAGGCGCGG | Mouse Hoxb6 |
| TAGGAACTCATCGGGAGGGG | |
| GGTGTAAGTGCAGTTAGTCATTT | Mouse Car1 |
| TAGTTCCATCCACGGTGTGC | |
| CCCAAGGGCACAGAGTTAGT | Mouse Cyp2c55 |
| AAGGTGGTACACGGCCAAAA | |
| TGTCACGGAGTTTGCTGTGA | Mouse 1810065E05Rik |
| GTACCCATGGGCTGACAGAG | |
| GGCAGGCCTACCCAATTCTT | Mouse Ces1g |
| TCTGTCATTGTGGCAGGGTC | |
| TGGCTCACTTTGGCAGGTAG | Mouse Slc6a14 |
| TCGGGCACTTCAATCTGTCC | |
| AGTGTATGCTGTCCAGTGGC | Mouse Ggh |
| TGGTGACTGTTCTTCCGAGC | |
| CCAGGAGTATGTCGCCTTCC | Mouse S100a6 |
| GATTTGACCGAGAGAGGCCC | |
| GCTGGCTTTCATCCTCCACT | Mouse Muc3 |
| CCTCCATCCCACACACTTCC | |
| CTCACCATTCCCTGCCCAAC | Mouse Ckmt1 |
| ATAGACTGCTGGGGTCAGGT | |
| CTCCCGATTCGTGGTGAACA | Mouse Nxpe2 |
| GGTGCATGGGCTCTTTACCT | |
| CTCCTCCGTCAAACTCCGTC | Mouse Acsm3 |
| ATGTTGGCCAAGTCCATTGC | |
| GAAGGGCGTGAAAGGCAATC | Mouse C1qa |
| CAAGCGTCATTGGGTTCTGC | |
| TGCAGCGAAGGAGTTACCTG | Mouse Car8 |
| ACATGTGTCCTCAGCCTTCG | |
| CTCCCAGCCCCAAGCTGTTA | Mouse Sval1 |
| ATCCTGACCAGAGGCAATCTC | |
| AACGATTCAGTCAGTGCCGT | Mouse Plet1 |
| TGACTTTGAGGCTGTGCGAT | |
| CTCCCCTAAAGTGCACAGGG | Mouse 2210407C18Rik |
| TAACTGGGAGTGATGCAGCG | |
| CAAATGCAAGTGCAAGGGCT | Mouse Mt3 |
| TGGCACACTTCTCACATCCG | |
| CCTCAGAAGTCAGTGCGACC | Mouse Atf3 |
| CATCCGATGGCAGAGGTGTT | |
| CCTATAAAGGCTTGCGGCCA | Mouse Atf4 |
| ACTGCTGCTGGATTTCGTGA | |
| GGAGCCAAAAAGGACCCAAAG | Mouse Agr2 |
| CTGTTGCTTGTCTTGGATCTGT | |
| GGAGGGGTGGACTTTTCGTT | Mouse Adh1 |
| TACGACGACGCTTACACCAC | |
| AGATCTCGGAATGGACCCCA | Mouse Mt1 |
| AGGAGCAGCAGCTCTTCTTG | |
| AGCAGGATTCCTATGGGGGA | Mouse Mptx1 |
| ATAGAGCAATTAGCTTAAGGCCAC | |
| GCTGGCTTTCATCCTCCACT | Mouse Muc3 |
| CCTCCATCCCACACACTTCC | |
| AGACACTCAGACGGAATGTTCA | Mouse Sct |
| CTGGTCCTCTAAGGGCTTGGA |
Quantification and statistical analysis
Computational analysis
The Cell Ranger Software Suite (Version 3.1.0) was used to perform sample de-multiplexing, barcode processing, and single-cell 3’ Unique Molecular Identifier (UMI) counting with mm10–3.0.0 as the reference genome. Cells with less than 10 total reads mapped to hashtags were discarded. Only cells that had the majority of hashtag reads on one hashtag were used for analysis. Analysis performed with the “Seurat” R package (ver. 3.1.1). Epithelial and immune cells had to be analyzed separately.
Epithelial
Cells were filtered by percentage of mitochondrial genes (less than 15%) and feature number (more than 1000/cell). In order to remove batch variation, samples were integrated using the reciprocal Principal Component Analysis (rPCA) method implemented in the “Seurat” package, using mouse Young-1 as reference. Dimension reduction was applied by PCA. The t-distributed Stochastic Neighbor Embedding (t-SNE) was utilized for visualizing the cells. K-means clustering utilized all genes and performed with different numbers of k-means (4:10). 5 k-means was picked manually due to some known markers appearing as top-10 markers of the clusters. Further re-clustering performed to refine cell types. Stem-TA and Tuft-EE clusters re-clustered with k-means, Colonocyte_1 and 2 re-clustered with Graph-based clustering (FindNeighbors(Colonocyte, dims = 1:10); FindClusters (Colonocyte, resolution = 0.035)) in order to separate colonocytes by compartment.
Immune
Effective reads - UMI per cell were scaled to the same level (median UMI counts per cell: 1330) in each sample by down sampling raw reads. Cells were filtered by percentage of mitochondrial genes (less than 8%), gene number (200–1500 genes), UMI (less than 5000). A subset of features (7,067 genes) that exhibit high cell-to-cell variation in the dataset are selected with the method of “mvp” in Seurat (with mean cutoff between 0.0125 and 2; dispersion cut off more than 0.9). We used these variable genes for dimension reduction. Dimension reduction was applied by PCA. Then UMAP was performed on the top 50 principal components for visualizing the cells. Graph-based clustering was executed on the dimension-reduced data.
For hierarchical clustering, pair-wise correlation between each cluster was calculated, and centered expression of each gene was used for visualization by heatmap. Classication of immune cell subsets was inferred from the annotation of cluster-specic genes. Different cell types were designated manually by referring to known markers. The identication markers for each subset are summarized in Table S1.
Statistical analysis
Data represented as ‘mean ± SD’. Results were considered signicant at p < 0.05. Differential expression analysis performed based on the Wilcoxon rank sum test, as implemented in the “Seurat” package. Cell fraction and gene expression comparisons performed with either paired or unpaired t test, according to circumstance using GraphPad Prism (ver. 8.4.3 (471)) or R.
Shifts in cell proportions of immune cells calculated by odds ratio. The probability of observing these values was calculated using the R function ‘phyper’ from the ‘stats’ package, using the command: p = phyper(q, k, m, n), reported as a hypergeometric p value. Confidence intervals for the odds-ratio were computed using the R function ‘fisher.test’.
Pathway analysis
Statistically significant DEGs were uploaded to QIAGEN Ingenuity Pathway Analysis (IPA) software for Canonical Pathway and Upstream Regulator analysis (Qiagen 2020, ver. 01-18-06).
Interaction analysis by using receptor-ligand pairs
Normalized gene expression from both immune and epithelial cells was taken to perform cell-cell interactions. CellPhoneDB (Vento-Tormo et al., 2018) was utilized to filter all integrins (HUGO ‘‘Integrin’’ gene group), which were involved in many non-specific cell-cell interactions.
We further required input genes from cell subsets to be expressed in at least 10% of all cells within the subset. To identify highly specific and unbiased interactions, we used only those variable genes defined above (2000 variable genes in epithelial cells and 7067 variable genes in immune cells). Young sample (20 weeks) in epithelial group was not included in the analysis to increase the interaction specificity in aging. Statistical model of null-distribution based permutation test on the corresponding expression of simple/complex ligand-receptor partner was applied (1000 permutations). For network visualization two kinds of cell-cell interaction interpretations were used: number of significant interacting partners in each cell-cell interaction and the average expression of single partners (ligand-receptor) in each cell-cell interaction. All receptor-ligand interactions were included in Table S2.
Cytokine expression analysis
We simulated pseudo bulk RNAseq from scRNAseq, normalized by condition. Cytokines, predicted as epithelial upstream regulators, were checked in immune cells during aging.
Additional resources
Online supplemental material
Supplementary material contains 5 Supplementary Figures and 2 Supplementary Tables.
Figure S1 provides additional visualization of compartment differences of mouse colon epithelial cells as well as IPA analysis results. Figure S2 shows additional information and IPA analysis results for the major colonic epithelium cell types. Figure S3 provides extensive, immune cell type specific gene expression, used for cell type determination and compartment specificity determination. Figure S4 provides immune cell population changes in different compartments during aging as well as IPA analysis results. Figure S5 shows more extensive information about cell-to-cell interactions during aging. Table S1 provides a full list of marker genes used for the designation of immune cell types. Table S2 provides a table of all cell-to-cell interaction counts.
Acknowledgments
We would like to thank our colleagues from the Fritz Lipmann Institute Core Facilities and Services: Flow Cytometry, DNA Sequencing, Imaging, and Animal Facilities for their assistance throughout the course of this study. We also thank all the members of AG Neri laboratory for fruitful discussions and Dr. George Garside for manuscript proofreading. This work was supported by funding from Alexander von Humboldt foundation (1164767-ITA-SKP) and the Leibniz Institute on Aging (FLI).
Author contributions
F.N. and D.S. conceived and designed the study. D.S. and O.O. carried out the experiments with the support of A.K., L.A., S.K. and F.S.. D.S., M.R. and J.L. analyzed the data and prepared the figures. F.N., D.S., and J.L.wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104202.
Supplemental information
Data and code availability
Sequencing data has been deposited to the GEO repository with accession numbers GSE168448 (Epithelial cells) and GSE169351 (Immune cells). Custom codes used in this manuscript can be found here: https://github.com/Dsirvinskas/Aging_Colon_Atlas
References
- Ahmad A.A., Wang Y., Gracz A.D., Sims C.E., Magness S.T., Allbritton N.L. Optimization of 3-D organotypic primary colonic cultures for organ-on-chip applications. J. Biol. Eng. 2014;8:9. doi: 10.1186/1754-1611-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkaya M., Traba J., Roesler A.S., Miozzo P., Akkaya B., Theall B.P., Sohn H., Pena M., Smelkinson M., Kabat J., et al. Second signals rescue B cells from activation-induced mitochondrial dysfunction and death. Nat. Immunol. 2018;19:871–884. doi: 10.1038/s41590-018-0156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alteber Z., Sharbi-Yunger A., Pevsner-Fischer M., Blat L., Roitman D., Tzehoval L., Elinav E., Eisenbach L. The anti-inflammatory IFITM genes ameliorate colitis and partially protect from tumorigenesis by changing immunity and microbiota. Immunol. Cell Biol. 2018;96:284–297. doi: 10.1111/imcb.12000. [DOI] [PubMed] [Google Scholar]
- Barker N., Es J.H., van, Kuipers J., Kujala P., Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Bhadra R., Cobb D.A., Khan I.A. CD40 signaling to the rescue: a CD8 exhaustion perspective in chronic infectious diseases. Crit. Rev. Immunol. 2013;33:361–378. doi: 10.1615/critrevimmunol.2013007444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton M., Haber A.L., Rogel N., Burgin G., Beyaz S., Schnell A., Ashenberg O., Su C.-W., Smillie C., Shekhar K., et al. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell. 2018;175:1307–1320.e22. doi: 10.1016/j.cell.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blecher-Gonen R., Bost P., Hilligan K.L., David E., Salame T.M., Roussel E., Connor L.M., Mayer J.U., Halpern K.B., Tóth B., et al. Single-cell analysis of diverse pathogen responses defines a molecular roadmap for generating antigen-specific immunity. Cell Syst. 2019;8:109–121.e6. doi: 10.1016/j.cels.2019.01.001. [DOI] [PubMed] [Google Scholar]
- Boari J.T., Furlan C.L.A., Vernengo F.F., Rodriguez C., Ramello M.C., Vesely M.C.A., Serrán M.G., Nuñez N.G., Richer W., Piaggio E., et al. IL-17RA-Signaling modulates CD8+ T cell survival and exhaustion during trypanosoma cruzi infection. Front Immunol. 2018;9:2347. doi: 10.3389/fimmu.2018.02347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbage M., Keppler S.J., Gasparrini F., Martínez-Martín N., Gaya M., Feest C., Domart M.-C., Brakebusch C., Collinson L., Bruckbauer A., et al. Cdc42 is a key regulator of B cell differentiation and is required for antiviral humoral immunityCdc42 in B cell differentiation and function. J. Exp. Med. 2015;212:53–72. doi: 10.1084/jem.20141143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon E., Buechler S. Colon cancer tumor location defined by gene expression may disagree with anatomic tumor location. Clin. Colorectal Canc. 2019;18:149–158. doi: 10.1016/j.clcc.2019.02.002. [DOI] [PubMed] [Google Scholar]
- Chassaing B., Kumar M., Baker M.T., Singh V., Vijay-Kumar M. Mammalian gut immunity. Biomed. J. 2014;37:246–258. doi: 10.4103/2319-4170.130922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian B.E., Shadel G.S. Aging: it’s SIRTainly possible to restore mitochondrial dysfunction. Curr. Biol. Cb. 2014;24:R206–R208. doi: 10.1016/j.cub.2014.01.027. [DOI] [PubMed] [Google Scholar]
- Conze D., Krahl T., Kennedy N., Weiss L., Lumsden J., Hess P., Flavell R.A., Gros G.L., Davis R.J., Rincón M. c-Jun NH2-terminal kinase (JNK)1 and JNK2 have distinct roles in CD8+ T cell activation. J. Exp. Med. 2002;195:811–823. doi: 10.1084/jem.20011508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- Elyahu Y., Hekselman I., Eizenberg-Magar I., Berner O., Strominger I., Schiller M., Mittal K., Nemirovsky A., Eremenko E., Vital A., et al. Aging promotes reorganization of the CD4 T cell landscape toward extreme regulatory and effector phenotypes. Sci. Adv. 2019;5:eaaw8330. doi: 10.1126/sciadv.aaw8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galligan C.L., Matsuyama W., Matsukawa A., Mizuta H., Hodge D.R., Howard O.M.Z., Yoshimura T. Up-regulated expression and activation of the orphan chemokine receptor, CCRL2, in rheumatoid arthritis. Arthritis Rheum. 2004;50:1806–1814. doi: 10.1002/art.20275. [DOI] [PubMed] [Google Scholar]
- Garriock R.J., Chalamalasetty R.B., Zhu J., Kennedy M.W., Kumar A., Mackem S., Yamaguchi T.P. A dorsal-ventral gradient of Wnt3a/β-catenin signals controls mouse hindgut extension and colon formation. Dev. Camb Engl. 2020;147:dev185108. doi: 10.1242/dev.185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking M.B., Burkhard R. Microbial modulation of intestinal T helper cell responses and implications for disease and therapy. Mucosal Immunol. 2020;13:855–866. doi: 10.1038/s41385-020-00335-w. [DOI] [PubMed] [Google Scholar]
- Girondel C., Lévesque K., Langlois M.-J., Pasquin S., Saba-El-Leil M.K., Rivard N., Friesel R., Servant M.J., Gauchat J.-F., Lesage S., et al. Loss of interleukin-17 receptor D promotes chronic inflammation-associated tumorigenesis. Oncogene. 2020;40:452–464. doi: 10.1038/s41388-020-01540-4. [DOI] [PubMed] [Google Scholar]
- Glosson-Byers N.L., Sehra S., Kaplan M.H. STAT4 is required for IL-23 responsiveness in Th17 memory cells and NKT cells. Jak-Stat. 2014;3:e955393. doi: 10.4161/21623988.2014.955393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves L.C., Barron M.J., Plusa S., Kirkwood T.B., Mathers J.C., Taylor R.W., Turnbull D.M. Defects in multiple complexes of the respiratory chain are present in ageing human colonic crypts. Exp. Gerontol. 2010;45:573–579. doi: 10.1016/j.exger.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronke K., Hernández P., Zimmermann J., Klose C., Kofoed-Branzk M., Guendel F., Witkowski M., Tizian C., Amann L., Schumacher F., et al. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature. 2019;566:249–253. doi: 10.1038/s41586-019-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui X., Li J., Ueno A., Iacucci M., Qian J., Ghosh S. Histopathologic features of inflammatory bowel disease are associated with different CD4+ T cell subsets in colonic mucosal lamina propria. J. Crohns Colitis. 2018;12:1448–1458. doi: 10.1093/ecco-jcc/jjy116. [DOI] [PubMed] [Google Scholar]
- Henson S.M., Macaulay R., Riddell N.E., Nunn C.J., Akbar A.N. Blockade of PD-1 or p38 MAP kinase signaling enhances senescent human CD8+ T-cell proliferation by distinct pathways. Eur. J. Immunol. 2015;45:1441–1451. doi: 10.1002/eji.201445312. [DOI] [PubMed] [Google Scholar]
- Ida S., Ozaki N., Araki K., Hirashima K., Zaitsu Y., Taki K., Sakamoto Y., Miyamoto Y., Oki E., Morita M., et al. SPINK1 status in colorectal cancer, impact on proliferation, and role in colitis-associated cancer. Mol. Cancer Res. 2015;13:1130–1138. doi: 10.1158/1541-7786.MCR-14-0581. [DOI] [PubMed] [Google Scholar]
- Inagaki T., Moschetta A., Lee Y.-K., Peng L., Zhao G., Downes M., Yu R.T., Shelton J.M., Richardson J.A., Repa J.J., et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl. Acad. Sci. U S A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James K.R., Gomes T., Elmentaite R., Kumar N., Gulliver E.L., King H.W., Stares M.D., Bareham B.R., Ferdinand J.R., Petrova V.N., et al. Distinct microbial and immune niches of the human colon. Nat. Immunol. 2020;21:343–353. doi: 10.1038/s41590-020-0602-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Liu C., Liu M., Shen Y., Hu X., Wang Q., Wu J., Wu M., Fang Q., Zhang X. OX40 signaling is involved in the autoactivation of CD4+CD28− T cells and contributes to the pathogenesis of autoimmune arthritis. Arthritis Res. Ther. 2017;19:67. doi: 10.1186/s13075-017-1261-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N., Cronin J.G., Dolton G., Panetti S., Schauenburg A.J., Galloway S.A.E., Sewell A.K., Cole D.K., Thornton C.A., Francis N.J. Metabolic adaptation of human CD4+ and CD8+ T-cells to T-cell receptor-mediated stimulation. Front Immunol. 2017;8:1516. doi: 10.3389/fimmu.2017.01516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Schiller L.R., Fordtran J.S. Quantification of colonic lamina propria cells by means of a morphometric point-counting method. Gastroenterology. 1988;94:409–418. doi: 10.1016/0016-5085(88)90430-1. [DOI] [PubMed] [Google Scholar]
- Liang Y., Liu C., Lu M., Dong Q., Wang Z., Wang Z., Xiong W., Zhang N., Zhou J., Liu Q., et al. Calorie restriction is the most reasonable anti-ageing intervention: a meta-analysis of survival curves. Sci. Rep. 2018;8:5779. doi: 10.1038/s41598-018-24146-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C., Chiang J., Tsai W., You J., Hsieh P., Hung H., Chen H., Tang R., Chen J., Yeh C. Primary tumor location in stage III colon cancer has prognostic impact on subsequent liver metastasis. J. Surg. Oncol. 2018;118:1301–1310. doi: 10.1002/jso.25270. [DOI] [PubMed] [Google Scholar]
- Linton P.-J., Bautista B., Biederman E., Bradley E.S., Harbertson J., Kondrack R.M., Padrick R.C., Bradley L.M. Costimulation via OX40L expressed by B cells is sufficient to determine the extent of primary CD4 cell expansion and Th2 cytokine secretion in vivo. J. Exp. Med. 2003;197:875–883. doi: 10.1084/jem.20021290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons J., Ghazi P.C., Starchenko A., Tovaglieri A., Baldwin K.R., Poulin E.J., Gierut J.J., Genetti C., Yajnik V., Breault D.T., et al. The colonic epithelium plays an active role in promoting colitis by shaping the tissue cytokine profile. PLoS Biol. 2018;16:e2002417. doi: 10.1371/journal.pbio.2002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinet K.Z., Bloquet S., Bourgeois C. Ageing combines CD4 T cell lymphopenia in secondary lymphoid organs and T cell accumulation in gut associated lymphoid tissue. Immun. Ageing. 2014;11:8. doi: 10.1186/1742-4933-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masahata K., Umemoto E., Kayama H., Kotani M., Nakamura S., Kurakawa T., Kikuta J., Gotoh K., Motooka D., Sato S., et al. Generation of colonic IgA-secreting cells in the caecal patch. Nat. Commun. 2014;5:3704. doi: 10.1038/ncomms4704. [DOI] [PubMed] [Google Scholar]
- Miyata N., Morris L.L., Chen Q., Thorne C., Singla A., Zhu W., Winter M., Melton S.D., Li H., Sifuentes-Dominguez L., et al. Microbial sensing by intestinal myeloid cells controls carcinogenesis and epithelial differentiation. Cell Rep. 2018;24:2342–2355. doi: 10.1016/j.celrep.2018.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalapareddy K., Hassan A., Sampson L.L., Zheng Y., Geiger H. Suppression of elevated Cdc42 activity promotes the regenerative potential of aged intestinal stem cells. Iscience. 2021;24:102362. doi: 10.1016/j.isci.2021.102362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava P., Koch S., Laukoetter M.G., Lee W.Y., Kolegraff K., Capaldo C.T., Beeman N., Addis C., Gerner-Smidt K., Neumaier I., et al. Interferon-γ regulates intestinal epithelial homeostasis through converging β-catenin signaling pathways. Immunity. 2010;32:392–402. doi: 10.1016/j.immuni.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özsoy M., Zimmermann F.A., Feichtinger R.G., Mayr J.A., Kofler B., Neureiter D., Klieser E., Schütz S., Weghuber D., Schneider A.M. Changes in the expression of oxidative phosphorylation complexes in the aging intestinal mucosa. Exp. Gerontol. 2020;135:110924. doi: 10.1016/j.exger.2020.110924. [DOI] [PubMed] [Google Scholar]
- Parikh K., Antanaviciute A., Fawkner-Corbett D., Jagielowicz M., Aulicino A., Lagerholm C., Davis S., Kinchen J., Chen H.H., Alham N.K., et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 2019;567:49–55. doi: 10.1038/s41586-019-0992-y. [DOI] [PubMed] [Google Scholar]
- Peterson L.W., Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- Phipps A.I., Lindor N.M., Jenkins M.A., Baron J.A., Win A.K., Gallinger S., Gryfe R., Newcomb P.A. Colon and rectal cancer survival by tumor location and microsatellite instability: the Colon Cancer Family Registry. Dis. Colon Rectum. 2013;56:937–944. doi: 10.1097/DCR.0b013e31828f9a57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Readinger J.A., Mueller K.L., Venegas A.M., Horai R., Schwartzberg P.L. Tec kinases regulate T-lymphocyte development and function: new insights into the roles of Itk and Rlk/Txk. Immunol. Rev. 2009;228:93–114. doi: 10.1111/j.1600-065X.2008.00757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki N., Sachs N., Wiebrands K., Ellenbroek S.I.J., Fumagalli A., Lyubimova A., Begthel H., van den Born M., van Es J.H., Karthaus W.R., et al. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc. Natl. Acad Sci. 2016;113:E5399–E5407. doi: 10.1073/pnas.1607327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage D.C., Dubos R. Alterations IN the mouse cecum and its flora produced by antibacterial drugs. J. Exp. Med. 1968;128:97–110. doi: 10.1084/jem.128.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaum N., Karkanias J., Neff N.F., May A.P., Quake S.R., Wyss-Coray T., Darmanis S., Batson J., Botvinnik O., Chen M.B., et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluns K.S., Kieper W.C., Jameson S.C., Lefrançois L. Interleukin-7 mediates the homeostasis of naïve and memory CD8 T cells in vivo. Nat. Immunol. 2000;1:426–432. doi: 10.1038/80868. [DOI] [PubMed] [Google Scholar]
- Schmidt S., Denk S., Wiegering A. Targeting protein synthesis in colorectal cancer. Cancers. 2020;12:1298. doi: 10.3390/cancers12051298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuck R., Gerken M., Teegen E.-M., Krebs I., Klinkhammer-Schalke M., Aigner F., Pratschke J., Rau B., Benz S. Gender comparison of clinical, histopathological, therapeutic and outcome factors in 185,967 colon cancer patients. Langenbecks Arch. Surg. 2020;405:71–80. doi: 10.1007/s00423-019-01850-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C., Rasband W., Eliceiri K. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie C.S., Biton M., Ordovas-Montanes J., Sullivan K.M., Burgin G., Graham D.B., Herbst R.H., Rogel N., Slyper M., Waldman J., et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 2019;178:714–730.e22. doi: 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sovran B., Hugenholtz F., Elderman M., Beek A.A.V., Graversen K., Huijskes M., Boekschoten M.V., Savelkoul H.F.J., Vos P.D., Dekker J., et al. Age-associated impairment of the mucus barrier function is associated with profound changes in microbiota and immunity. Sci. Rep. 2019;9:1437. doi: 10.1038/s41598-018-35228-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman J.R., Mitchell S.E. Caloric restriction. Mol. Aspects Med. 2011;32:159–221. doi: 10.1016/j.mam.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Steffen K.K., Dillin A. A ribosomal perspective on proteostasis and aging. Cell Metab. 2016;23:1004–1012. doi: 10.1016/j.cmet.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Stoeckius M., Zheng S., Houck-Loomis B., Hao S., Yeung B.Z., Mauck W.M., Smibert P., Satija R. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018;19:224. doi: 10.1186/s13059-018-1603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., III, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;7:1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelemann C., Eren R.O., Coutaz M., Brasseit J., Bouzourene H., Rosa M., Duval A., Lavanchy C., Mack V., Mueller C., et al. Interferon-γ induces expression of MHC class II on intestinal epithelial cells and protects mice from colitis. PLoS One. 2014;9:e86844. doi: 10.1371/journal.pone.0086844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari R., Pandey S.K., Goel S., Bhatia V., Shukla S., Jing X., Dhanasekaran S.M., Ateeq B. SPINK1 promotes colorectal cancer progression by downregulating Metallothioneins expression. Oncogenesis. 2015;4:e162. doi: 10.1038/oncsis.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero J., Briz O., Herraez E., Muntané J., Marin J.J.G. Activation of the nuclear receptor FXR enhances hepatocyte chemoprotection and liver tumor chemoresistance against genotoxic compounds. Biochim. Biophys. Acta. 2013;1833:2212–2219. doi: 10.1016/j.bbamcr.2013.05.006. [DOI] [PubMed] [Google Scholar]
- Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.-E., Stephenson E., Polański K., Goncalves A., et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Song W., Wang J., Wang T., Xiong X., Qi Z., Fu W., Yang X., Chen Y.-G. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J. Exp. Med. 2019;217:e20191130. doi: 10.1084/jem.20191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H.-S., Kim S.Y., Kim J.T., Lee Y.-S., Moon J.S., Kim M., Kang Y.E., Joung K.H., Lee J.H., Kim H.J., et al. T-cell senescence contributes to abnormal glucose homeostasis in humans and mice. Cell Death Dis. 2019;10:249. doi: 10.1038/s41419-019-1494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X., Zhao Y., Liu J., Zhang C., Yu H., Wang J., Zheng T., Liu L., Li J., Feng Z., et al. BAG2 promotes tumorigenesis through enhancing mutant p53 protein levels and function. Elife. 2015;4:e08401. doi: 10.7554/eLife.08401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborin A., Krezalek M., Hyoju S., DeFazio J.R., Setia N., Belogortseva N., Bindokas V.P., Guo Q., Zaborina O., Alverdy J.C. The critical role of microbiota within cecal crypts on the regenerative capacity of the intestinal epithelium following surgical stress. Am. J. Physiol. Gastrointest Liver Physiol. 2016;312:G112–G122. doi: 10.1152/ajpgi.00294.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhao H., Shi X., Jia X., Yang Y. Identification and validation of an immune-related gene signature predictive of overall survival in colon cancer. Aging. 2020;12:26095–26120. doi: 10.18632/aging.202317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhang S., Liu Y., He X., Qu M., Xu G., Wang H., Huang M., Pan J., Liu Z., et al. Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human. Cell Discov. 2020;6:22. doi: 10.1038/s41421-020-0157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data has been deposited to the GEO repository with accession numbers GSE168448 (Epithelial cells) and GSE169351 (Immune cells). Custom codes used in this manuscript can be found here: https://github.com/Dsirvinskas/Aging_Colon_Atlas