Abstract
Physically effective neutral detergent fiber (peNDF) is a concept that accounts for the particle length of NDF in a feed, sustaining the normal chewing behavior and rumen fermentation of ruminants. This study aimed to elucidate the effects of dietary peNDF on growth performance and bacterial communities in the rumen of goats through a high-throughput sequencing technique. A total of 30 male Lezhi black goats were randomly assigned to five groups, corresponding to five diets with identical compositions and nutrient levels but with varying forage lengths (the peNDF1.18 contents of the diets were 33.0, 29.9, 28.1, 26.5, and 24.8%, respectively). The whole trial lasted for 44 days. As results show, feed intake and average daily gain were highest when peNDF1.18 content was 26.5%, in which the papilla length of the dorsal sac in rumen was the highest. Chao1 and ACE indexes were similar among the treatments, while Shannon and Simpson indexes of the peNDF1.18 = 28.1% group were the highest (p < 0.05). As the level of dietary peNDF1.18 decreased, the dominant phylum transitioned from Bacteroidetes to Firmicutes. The top three dominant genera of rumen bacteria were Prevotella 1, Ruminococcaceae NK4A214 group, and Christensenellaceae R-7 group. They all showed a quadratic correlation with dietary peNDF1.18 level (p < 0.05). The relative abundance of Ruminococcaceae UCG-011 was positively correlated, while that of Prevotella 1 was negatively correlated, with amino acid metabolism and energy metabolism (p < 0.01). In conclusion, dietary peNDF level influenced goat growth performance, rumen development, and rumen bacterial community structures, and a peNDF1.18 level between 26.5 and 28.1% was considered optimal for goat diet.
Keywords: goat, particle size, peNDF, rumen bacteria, growth performance
Introduction
Physically effective neutral detergent fiber (peNDF) is a concept proposed by Mertens (1997), which reflects the particle size (PS) of feed and the impact of feed on chewing activities and the ability to stimulate rumen stratification (Zhao et al., 2010). Ruminants need a suitable level and length of fiber to maintain the normal rumen pH. Effects of peNDF on the growth performance and nutrient digestibility of goats have been assessed in a previous study (Jang et al., 2017). Park et al. (2014) found that feeding growing steers with a low-peNDF diet could increase feed intake, thereby increasing daily gain. Likewise, Cao et al. (2021) found that nutrient digestibility and milk production dramatically improved with increased peNDF content. Through in situ assessment, Yang and Beauchemin (2007) found that the ruminal degradation rates of dry matter (DM), organic matter (OM), NDF, acid detergent fiber (ADF), starch, and nitrogen in the high-peNDF group were higher than those in the low-peNDF group. It was documented that reducing the PS of diet expanded the contact area between microorganisms and feed (Zhao et al., 2010) and therefore increased nutrient digestibility and reduced chewing time in dairy goats (Li et al., 2014). Beauchemin and Yang (2005) found that the long fiber particles (> 1 cm) promoted chewing and saliva secretion of dairy cows, which were essential for maintaining the stability of the rumen environment. Zebeli et al. (2012) proposed that the suitable level of peNDF1.18 in the diet of dairy cattle should be around 31.2%. However, Zhao et al. (2010) found that 22.4% peNDF1.18 was more conducive to the nutrient digestion of goats than was 43.1%. The difference in the optimal dietary peNDF level between cattle and goats suggested the inconsistent requirements for dietary peNDF level.
Ruminal microorganisms help their host in degrading fiber more successfully than do monogastric animals, which is of great value (Li et al., 2018b). Gastrointestinal microbes are affected by many factors, including diet, host genetics, living environments, and age (Jin et al., 2016). Studies have proved that changes in diet affect the gastrointestinal microbes in cattle (Klieve et al., 2007) and dairy cows (Valizadeh et al., 2010). However, few reports concerning the impact of peNDF on the rumen bacterial community in goats have ever been reported. Previous studies revealed that the counts of total bacteria (Valizadeh et al., 2010) and Ruminococcus albus (Zebeli et al., 2008) were not affected by dietary PS. Huo et al. (2014) discovered that a high-grain diet influenced the liquid- and solid-associated rumen bacteria of goats, but a high-forage diet increased the relative abundance of Prevotella.
Until now, there has been no research concerning the effects of the PS of fiber in diets on solid-associated bacteria in the rumen of goats. We assumed that changes in dietary peNDF levels could affect growth performance, rumen development, and rumen microbial structure in goats with the same dietary compositions and nutrient contents. The main objective of this study was to evaluate the effects of dietary peNDF level on the diversity and composition of rumen bacteria, which could explain the differences in growth performance in goats, based on which a reasonable range of dietary peNDF was proposed.
Materials and Methods
Ethics Statement
The experiment was approved by the Animal Policy and Welfare Committee of Animal Nutrition Institute, Sichuan Agriculture University, and followed the current laws of animal protection (Ethics Approval Code SCAUAC201408-3).
Animals and Diets
The study was carried out at the Experimental Base of Animal Nutrition Institute (30.3°N, 103.0°E), Sichuan Agricultural University. Thirty male 4-month-old Lezhi black goats (purchased from the farm of Tianlong Agriculture and Animal Husbandry Technology Co. Ltd.) with similar body weights (BWs; 21.4 ± 0.24 kg) were randomly divided into five groups corresponding to five dietary treatments. The diets (Supplementary Table 1) were formulated according to the nutrient requirements of a 20-kg goat with a 150-g daily gain based on National Research Council (NRC) (2007) and were identical in composition and nutrient content but varied in length of roughage. The forages (alfalfa hay, peanut vine, and Leymus chinensis) were cut into 1, 5, 1, 4, and 7 cm, respectively, by a forage cutter-FS60, purchased from Nongfengli Machinery Equipment Co., Ltd (Jining, China). Forage PS distribution was determined using the Penn State Particle Separator, as reported by Kononoff et al. (2003). Pef8.0 and pef1.18 were calculated as the proportions of the DM of forage particles retained on 19-mm and 8-mm sieves and those on 19-mm, 8-mm, and 1.18-mm sieves of total DM content, respectively. The measured peNDF1.18 contents of the diets were 24.8% (1 mm, L), 26.5% (5 mm, ML), 28.1% (1 cm, M), 29.9% (4 cm, MH), and 33.0% (7 cm, H), respectively (Table 1).
TABLE 1.
PS distribution and peNDF content of five groups.
| Items | Content of peNDFa |
||||
| H (33.0%) | MH (29.9%) | M (28.1%) | ML (26.5%) | L (24.8%) | |
| % DM retained on sieves | |||||
| 19 mm | 27.3 | 11.2 | 7.0 | 0.0 | 0.0 |
| 8 mm | 9.7 | 11.8 | 9.3 | 6.3 | 3.1 |
| 1.18 mm | 39.9 | 46.8 | 49.4 | 55.5 | 54.6 |
| Pan | 23.1 | 30.2 | 34.4 | 38.3 | 42.3 |
| Physically effective factorb | |||||
| pef8.0 | 37.0 | 23.0 | 16.3 | 6.3 | 3.1 |
| pef1.18 | 76.9 | 69.8 | 65.6 | 61.7 | 57.7 |
| peNDF content (%, DM)c | |||||
| peNDF8.0 | 15.9 | 9.8 | 7.0 | 2.7 | 1.3 |
| peNDF1.18 | 33.0 | 29.9 | 28.1 | 26.5 | 24.8 |
aThe different peNDF1.18 (PS > 1.18 mm) contents of 33.0, 29.9, 28.1, 26.5, and 24.8% were obtained by crushing the forage into 7 cm (H), 4 cm (MH), 1 cm (M), 5 mm (ML), or 1 mm (L).
bPhysical effectiveness factor determined as the proportion of the DM of particles retained on two sieves (19 mm and 8 mm) or on three sieves (19 mm, 8 mm, and 1.18 mm), respectively.
cpeNDF content, physically effective factor * NDF; peNDF8.0 and peNDF1.18, physically effective NDF determined as NDF content of ration sample multiplied by pef8.0 and pef1.18, respectively.
During the whole period, each goat was reared in a single metabolic cage in the same barn. The concentrate and the forage were evenly mixed and then fed to goats at 08:00 and 20:00 at equal amounts, allowing about 10% orts. Goats were free to access fodder and water. The leftovers were weighed at 8:00 every morning to calculate dry matter intake (DMI). The experiment lasted for 44 days, including a 14-day adaptation period and a 30-day experimental period. The BW of each goat was recorded on the 1st and 30th days of the formal trial using an electronic scale. Average daily gain (ADG) was calculated as ADG = (Final BW – Initial BW)/30 (kg/day).
Sample Collection
At the end of the formal trial and before morning feeding, 15 goats were selected for slaughter after electric shocks. For each sampling, 300 g of whole rumen contents was obtained. A portion (∼50 g) of the whole rumen sample was homogenized on ice for three 1-min cycles at 1-min intervals using a Polytron grinding mill (Thermo Fischer Scientific, France). Approximately 0.5 g was transferred into 2-ml Eppendorf tubes and stored at −80°C until molecular biology analyses (Silberberg et al., 2013). After the experiment, the rest of the goats continued to be cultivated normally. The dorsal sac and ventral sac of rumen were collected from the same site and fixed in 4% paraformaldehyde for determination of tissue morphology.
Body Weight and Average Daily Gain
The BW of each goat was recorded on the 1st and 30th days of the formal trial using an electronic scale. ADG was calculated as ADG = (Final BW - Initial BW)/30 (kg/day).
Determination of Morphology of Rumen Epithelium
The rumen epithelial tissue was sent to Wuhan Service Biotechnology Co., Ltd. (Wuhan, China) for H&E staining and sectioning. Image-Pro Plus 6.0 was used to verify the qualified sample, with millimeters as the standard unit. The thickness of the five muscle layers, the width of the nipple, and the height of the nipple in the rumen epithelial section were measured at a 20-fold scale. Photographs were recorded at the same time.
DNA Extraction and PCR Amplification and Sequencing
The DNA extraction kit (Tiangen Biochemical Technology, Peking, China) was used to extract the total DNA from the rumen chyme samples of goats, as described previously (Wang et al., 2019). The purity and concentration of the extracted DNA were detected by agarose gel electrophoresis. With the extracted DNA (1 ng/μl) as template, the V4–V5 region of the 16S rRNA gene was amplified by PCR with bacterial universal primers 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 926R (5′-CCGYCAATTYMTTTRAGTTT-3′) (Parada et al., 2016). The PCR system (25 μl) consisted of 1 ng/μl DNA template (10 μl), 1 × PCR buffer (2.5 μl), 1.5 mM MgCl2 (1.5 μl), 0.4 μM dNTPs (2.5 μl), 1 μM upstream and downstream primers (1.5 μl each), 0.5 U of KOD-Plus-Neo enzyme (TOYOBO) (0.5 μl), and water (5 μl, added up to 25 μl). PCR procedure was as follows: predenaturation at 94°C for 1 min for 30 cycles (denaturation at 94°C for 20 s, annealing at 54°C for 30 s, and extension at 72°C for 30 s) and extension at 72°C for 5 min. PCR products were electrophoresed using a 2% agarose gel, recovered (using a gel recovery kit, Qiagen), and purified. 16S rRNA high-throughput sequencing (Rhonin Biotechnology Ltd., Chengdu, China) was performed after the tests were qualified by the Hiseq 2500 PE 250 sequencing platform.
Sequencing Data Analyses
The sample data were distinguished according to the Barcode sequence, and the chimera was filtered by the Uchime algorithm to obtain clean data (effective data) (Knight, 2011). Uparse (V7.0.1001)1 was used to cluster all samples with 97% identity for operational taxonomic units (OTUs) (Edgar, 2013). The sequence with the highest frequency in OTUs was selected as the representative sequence of OTUs. Using UCLUST (DiTullio et al., 2000) to process the representative sequences of OTUs and comparing them with the SILVA1322 database (Quast et al., 2012), the taxonomy annotation of microbial classification levels was carried out, and PyNAST was used to perform multiple alignments of representative sequences. Vegan and Picante packages in the R software (version 2.15.3) were used to calculate the values of observed species, as well as the Shannon, Simpson, Chao1, ACE, Goods coverage, and PD indexes, and to draw the rarefaction curve (Oksanen et al., 2010; Webb, 2010). Combining the count of the same OTUs and the relative abundance of OTUs, the Bray–Curtis distance was calculated by Vegan software. Principal coordinates analysis (PCoA) was drawn by ape software. Tax4Fun was used to predict functional features based on 16S rRNA gene sequencing data (Aßhauer et al., 2015), and secondary metabolic pathways were clustered.
Statistical Analyses
The data of this study were analyzed using one-way ANOVA of SPSS 25.0 (IBM, Armonk, NY, United States), in which the relative abundances of microbial phyla and genera were compared using Kruskal–Wallis test. The regression relationship between the relative abundance of bacteria and the level of dietary peNDF was analyzed, and Spearman correlation analysis between the relative abundance and the function of rumen bacteria was also executed. Tukey’s multiple test was used to compare differences among the treatment groups. Statistical significance was defined as p < 0.05, and trends were discussed at 0.05 < p < 0.10. Results are expressed as mean and standard error of the mean.
Results
Effects of Dietary Physically Effective Neutral Detergent Fiber Level on Feed Intake and Growth Performance
The effects of differing peNDF1.18 contents in diets on the BW, DMI, and ADG of goats are presented in Table 2. DMI and ADG increased first and then decreased with decreasing peNDF1.18 content in the diets (p < 0.05), where the DMI of the 26.5% peNDF1.18 treatment was significantly more than those of the 33.0, 29.9, and 24.8% peNDF1.18 treatments (p < 0.05); the DMI of the 28.1% peNDF1.18 treatment was not significantly different from those of other treatments. The ADG of the 26.5% peNDF1.18 treatment was greater than that of 33.0%,and the ADG of the 24.8% treatment was significantly lower than those of other treatments.
TABLE 2.
Effects of different contents of peNDF1.18 in diets on the BW, ADG, and DMI of goats.
| Items | Groups1 |
SEM | P-value | ||||
| H (33.0%) | MH (29.9%) | M (28.1%) | ML (26.5%) | L (24.8%) | |||
| Initial BW (kg) | 21.4 | 21.3 | 21.5 | 21.4 | 21.5 | 0.249 | 1.000 |
| Final BW (kg) | 24.6 | 24.9 | 25.2 | 26.0 | 24.1 | 0.332 | 0.480 |
| DMI (g/day) | 678b | 704b | 737ab | 831a | 702b | 17.55 | 0.032 |
| ADG (g/day) | 70.6b | 75.1ab | 83.0ab | 98.5a | 58.7c | 3.720 | 0.043 |
a–cMeans within a row with different superscripts differ significantly (p < 0.05).
1Groups: different peNDF1.18 (PS > 1.18 mm) contents of 33.0, 29.9, 28.1, 26.5, and 24.8% were obtained by chopping or crusher crushing the forage into the following lengths: H (7 cm), MH (4 cm), M (1 cm), ML (5-mm sieve), and L (1-mm sieve).
Effects of Dietary Physically Effective Neutral Detergent Fiber Level on the Morphology of Goat Rumen
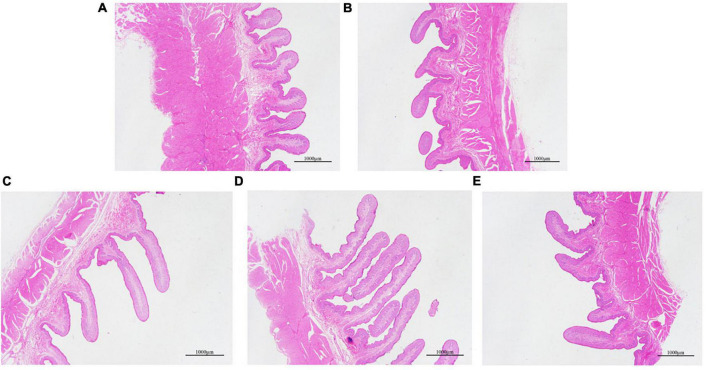
As shown in the results in Table 3, the level of dietary peNDF has a significant quadratic effect on the length of the rumen dorsal papilla and the width of the abdominal papilla, in which the values of 26.5 and 28.1% peNDF1.18 treatments are the highest, respectively. The photographs of each are shown in Figure 1, where we choose five representative images of the dorsal sac of the rumen from each group.
TABLE 3.
Effects of dietary peNDF level on the morphology of goat rumen.
| Items | Groups1 |
SEM |
P-value |
||||||
| H (33.0%) | MH (29.9%) | M (28.1%) | ML (26.5%) | L (24.8%) | ANOVA linear quadratic |
||||
| Dorsal sac | |||||||||
| Papilla length (mm) | 1.015c | 1.427bc | 1.642b | 2.281a | 1.652b | 0.205 | < 0.001 | < 0.001 | 0.003 |
| Papilla width (mm) | 0.560ab | 0.487b | 0.595a | 0.533ab | 0.588a | 0.034 | 0.046 | 0.194 | 0.349 |
| Muscle thickness (mm) | 1.470a | 1.327b | 1.130c | 1.446ab | 1.210c | 0.125 | 0.014 | 0.159 | 0.330 |
| Ventral sac | |||||||||
| Papilla length (mm) | 1.405 | 1.657 | 1.594 | 1.554 | 1.850 | 0.271 | 0.374 | 0.198 | 0.875 |
| Papilla width (mm) | 0.558b | 0.583ab | 0.664a | 0.584ab | 0.449c | 0.031 | < 0.001 | 0.003 | < 0.001 |
| Muscle thickness (mm) | 1.237 | 1.155 | 1.423 | 1.199 | 1.493 | 0.131 | 0.069 | 0.061 | 0.452 |
a–cMeans within a row with different superscripts differ significantly (p < 0.05).
1Groups: different peNDF1.18 (PS > 1.18 mm) contents of 33.0, 29.9, 28.1, 26.5, and 24.8% were obtained by chopping or crusher crushing the forage into the following lengths: H (7 cm), MH (4 cm), M (1 cm), ML (5-mm sieve), and L (1-mm sieve).
FIGURE 1.
Representative image of the dorsal sac of rumen from five groups. (A) 33.0%, 7 cm, H group. (B) 29.9%, 4 cm, MH group. (C) 28.1%, 1 cm, M group. (D) 26.5%, 5 mm, ML group. (E) 24.8%, 1 mm, L group.
Data Acquired From Sequencing and Operational Taxonomic Unit Diversity
The number of raw sequences in the rumen was 535,650 and the effective sequences were 509,933 based on the high-throughput sequencing analysis of 16S rRNA genes. The average effective ratio reached 90.36%. Clustering was based on the 97% sequence similarity from the effective sequences, where a total of 9,659 OTUs were obtained. The average OTUs in the 33.0, 29.9, 28.1, 26.5, and 24.8% peNDF1.18 treatments were 1,491, 2,011, 2,134, 1,991, and 2,032, respectively. A total of 689 OTUs were shared across the five treatments, and the number of sequences in shared OTUs accounted for 88.19% of the total number of sequences (Figure 2).
FIGURE 2.
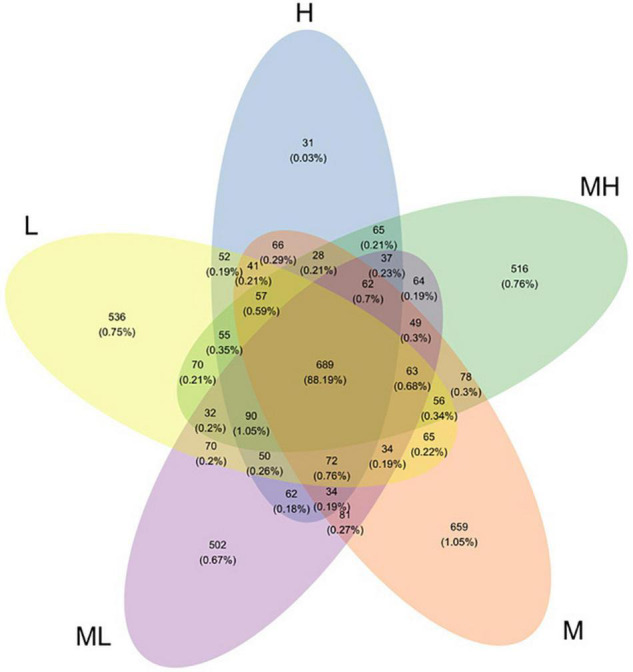
Venn diagram at the 97% similarity level. H group: 33.0% peNDF1.18 treatment. MH group: 29.9% peNDF1.18 treatment. M group: 28.1% peNDF1.18 treatment. ML group: 26.5% peNDF1.18 treatment. L group: 24.8% peNDF1.18 treatment.
The rarefaction curve (Supplementary Figure 1) was inclined to reach a plateau, and the value of coverage (Table 4) reached 0.96, both of which indicated that the sampling had sufficient sequence coverage to detect the majority of rumen bacteria.
TABLE 4.
Effects of dietary peNDF1.18 level on alpha diversity indexes of rumen bacteria in goats.
| Items | Groups1 |
SEM | P-value | ||||
| H (33.0%) | MH (29.9%) | M (28.1%) | ML (26.5%) | L (24.8%) | |||
| Chao1 | 1,179.535 | 1,473.747 | 1,485.104 | 1,544.153 | 1,535.871 | 189.289 | 0.344 |
| ACE | 1,203.733 | 1,524.217 | 1,538.947 | 1,593.639 | 1,584.490 | 204.956 | 0.350 |
| Simpson | 0.985c | 0.990ab | 0.993a | 0.989ab | 0.987bc | 0.001 | 0.001 |
| Shannon | 5.391b | 5.711ab | 5.939a | 5.672ab | 5.702ab | 0.144 | 0.043 |
| PD | 54.047 | 67.621 | 71.232 | 67.648 | 69.662 | 10.585 | 0.529 |
| Coverage | 0.972 | 0.963 | 0.965 | 0.960 | 0.961 | 0.006 | 0.392 |
a–cMeans within a row with different superscripts differ significantly (p < 0.05).
1Groups: different peNDF1.18 (PS > 1.18 mm) contents of 33.0, 29.9, 28.1, 26.5, and 24.8% were obtained by chopping or crusher crushing the forage into the following lengths: H (7 cm), MH (4 cm), M (1 cm), ML (5-mm sieve), and L (1-mm sieve).
Alpha Diversity Analysis of Rumen Bacteria
Alpha diversity indexes include Shannon, Simpson, Chao1, ACE, and PD indexes. Chao1 and ACE indexes were similar among the treatments (Table 4), which meant that the total number of rumen bacteria did not differ. Shannon and Simpson indexes of the 28.1% peNDF1.18 treatment were the highest (p < 0.05), indicating the highest richness and most even distribution of bacteria in the 28.1% peNDF1.18 treatment. Dietary peNDF1.18 levels had significant impacts on the diversity of goat rumen bacterial flora.
Beta Diversity Analysis of Rumen Bacteria
A PCoA plot with the Bray–Curtis distance matrix was drawn to visualize the differences among the five groups (Figure 3). The percentage of variation was represented by PCo1 (25.9%) and PCo2 (23.1%); the closer the distance in the figure, the more similar the bacterial community composition of the sample. There was separation among groups, with the different structures of rumen bacteria shown. Permutational multivariate analysis of variance (PerMANOVA) was used to test the significance of differences between peNDF levels (Table 5), with significant differences (p < 0.001) between the five groups.
FIGURE 3.
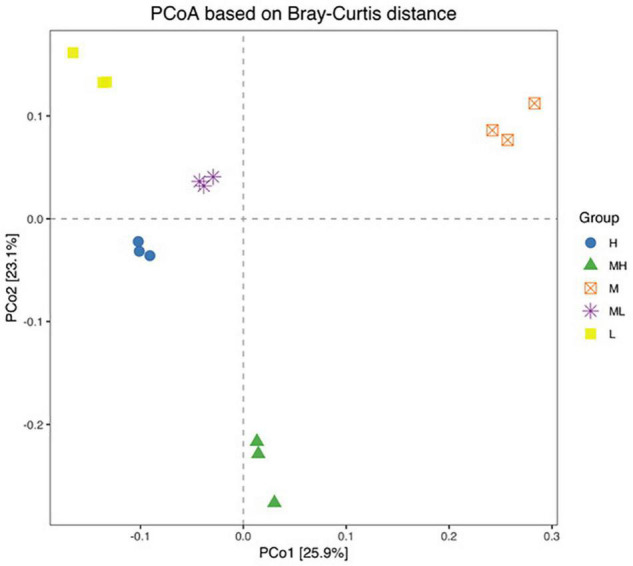
Bray–Curtis distance matrix PCoA of rumen bacterial samples. H group: 33.0% peNDF1.18 treatment. MH group: 29.9% peNDF1.18 treatment. M group: 28.1% peNDF1.18 treatment. ML group: 26.5% peNDF1.18 treatment. L group: 24.8% peNDF1.18 treatment.
TABLE 5.
PerMANOVA of rumen bacterial samples based on Bray–Curtis distances.
| Items | Df | SumsOfSqs | MeanSqs | F.model | R 2 | Pr (> F) |
| Group | 4 | 0.901 | 0.225 | 8.440 | 0.771 | < 0.001 |
| Residuals | 10 | 0.267 | 0.027 | NA | 0.228 | NA |
| Total | 14 | 1.168 | NA | NA | 1 | NA |
Core Rumen Bacteria at Phylum and Genus Levels
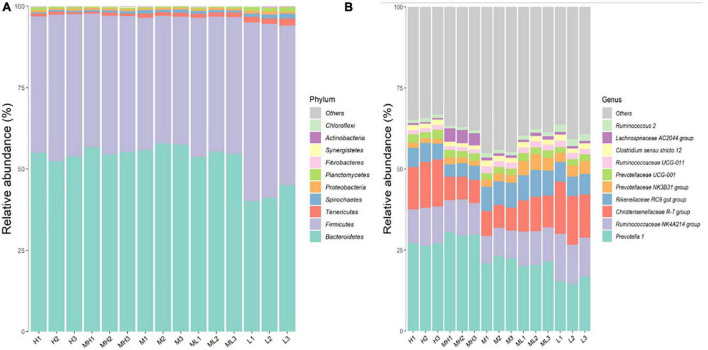
In this experiment, 22 phyla and 242 genera were taxonomically classified. The dominant phyla of goat rumen bacteria were Bacteroidetes (42.07–57.01%) and Firmicutes (39.77–52.51%) (Figure 4). The dominant phylum transitioned from Bacteroidetes to Firmicutes with the decrease of dietary peNDF1.18 level. In other words, the relative abundance of Bacteroidetes decreased with the decrease of dietary peNDF1.18 level, while the trend of changes in Firmicutes was the opposite. The relative abundances of Tenericutes, Spirochetes, and Planctomycetes were the highest in the 24.8% peNDF1.18 treatment (p < 0.05).
FIGURE 4.
Rumen bacterial compositions (A) at the phylum level (only the top 10 abundant phyla were presented) and (B) at the genus level (only the top 10 abundant genera were presented). H group: 33.0% peNDF1.18 treatment. MH group: 29.9% peNDF1.18 treatment. M group: 28.1% peNDF1.18 treatment. ML group: 26.5% peNDF1.18 treatment. L group: 24.8% peNDF1.18 treatment.
The top three dominant genera of goat rumen bacteria were Prevotella 1 (average, 23.00%), Ruminococcaceae NK4A214 group (average, 10.70%), and Christensenellaceae R-7 group (average, 10.61%). The relative abundance of Prevotella 1 was the highest in the 29.9% peNDF1.18 treatment and the lowest in the 24.8% peNDF1.18 treatment (p < 0.001). The relative abundances of Ruminococcaceae NK4A214 group and Christensenellaceae R-7 group decreased when peNDF level increased from 24.8 to 28.1%, and then increased with dietary peNDF level. An uncultured genus, f_F082| g_uncultured, subordinated to Bacteroidetes, was also one of the predominant genera.
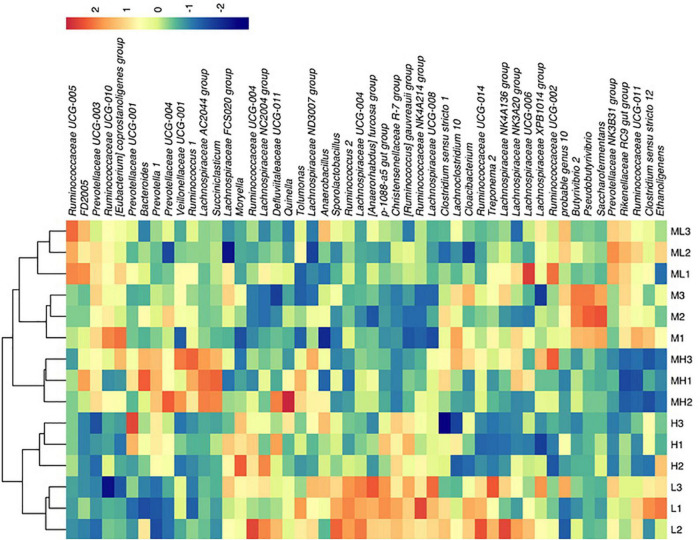
According to the species annotation and relative abundance information of all samples at the genus level, the top 50 abundant genera were selected for clustering and plotted into a heat map (Figure 5). The 28.1 and 26.5 peNDF1.18 treatments were grouped into one cluster first and then grouped into one cluster with the 29.9% peNDF1.18 treatment, which meant that the composition of these three groups was different from the 24.8 to 33.0% peNDF1.18 treatments.
FIGURE 5.
Heat map of species abundance at the genus level (top 50 bacteria). H group: 33.0% peNDF1.18 treatment. MH group: 29.9% peNDF1.18 treatment. M group: 28.1% peNDF1.18 treatment. ML group: 26.5% peNDF1.18 treatment. L group: 24.8% peNDF1.18 treatment. The abscissa is sorted according to the order of the samples, and the ordinate is arranged according to the total average abundance from top to bottom. The relative abundance of the genus is not converted, and the legend shows the percentage. The redder color represents higher abundance, and the bluer color represents lower abundance.
Comparisons of Rumen Bacterial Composition Among the Five Groups
Bacteria with a phylum level greater than 1% and a genus level more than 0.1% were selected for differential analysis (Zened et al., 2013). We found that the bacterial flora of the rumen solid contents was significantly affected by dietary peNDF1.18 levels (Table 6). At the phylum level, the relative abundances of Bacteroidetes, Firmicutes, Tenericutes, Spirochetes, and Planctomycetes were affected by dietary peNDF1.18 levels; besides, the relative abundance of Proteobacteria (average, 0.654%) was also different (p = 0.04) and that in the 24.8% peNDF1.18 treatment was the highest. At the genus level, in addition to the three dominant genera, there were also another 20 genera that significantly differed among groups, and most of them belong to Firmicutes. The relative abundance of many uncultured bacteria genera (belonging to Bacteroidetes) also differed greatly among treatments.
TABLE 6.
Effects of dietary peNDF1.18 level on the relative abundance of rumen bacteria in goats.
| Items | Groups1 |
SEM | P-value | ||||
| H (33.0%) | MH (29.9%) | M (28.1%) | ML (26.5%) | L (24.8%) | |||
| Phylum (%) | |||||||
| Bacteroidetes | 53.713a | 55.471a | 57.010a | 54.551a | 42.073b | 1.224 | < 0.001 |
| Firmicutes | 43.572b | 41.801b | 39.772b | 42.100b | 52.510a | 1.321 | 0.019 |
| Tenericutes | 0.779b | 0.789b | 1.173b | 1.275b | 1.821a | 0.154 | < 0.001 |
| Spirochetes | 0.470b | 0.621b | 0.887ab | 0.763b | 1.259a | 0.136 | 0.002 |
| Planctomycetes | 0.680b | 0.355b | 0.414b | 0.421b | 1.157a | 0.117 | 0.033 |
| Genus (%) | |||||||
| Prevotella 1 | 26.776b | 29.889a | 22.167c | 20.669c | 15.518d | 0.696 | < 0.001 |
| Rikenellaceae RC9 gut group | 5.571b | 4.105c | 7.484a | 7.905a | 6.117b | 0.276 | < 0.001 |
| Prevotellaceae NK3B31 group | 1.482c | 1.795c | 2.347c | 4.510a | 3.468b | 0.291 | < 0.001 |
| Prevotellaceae UCG-001 | 2.449a | 2.225a | 1.992b | 2.146ab | 1.910b | 0.206 | 0.037 |
| Prevotellaceae UCG-003 | 0.881b | 1.479a | 1.505a | 1.426a | 0.937b | 0.084 | < 0.001 |
| Ruminococcus 1 | 0.651b | 1.308a | 0.920b | 0.917b | 0.631b | 0.106 | 0.001 |
| Ruminococcus 2 | 1.091bc | 0.812c | 1.131bc | 1.236b | 2.209a | 0.108 | 0.003 |
| Ruminococcaceae UCG-002 | 0.230b | 0.427a | 0.345ab | 0.437a | 0.342ab | 0.086 | 0.022 |
| Ruminococcaceae UCG-004 | 0.289ab | 0.197ab | 0.141b | 0.194ab | 0.306a | 0.046 | 0.022 |
| Ruminococcaceae UCG-005 | 0.513b | 0.457b | 0.529b | 1.440a | 0.306b | 0.070 | < 0.001 |
| Ruminococcaceae UCG-010 | 0.569ab | 0.638ab | 0.818a | 0.746ab | 0.450b | 0.099 | 0.029 |
| Ruminococcaceae UCG-011 | 1.335b | 1.167b | 1.897a | 1.831a | 1.828a | 0.113 | < 0.001 |
| Ruminococcaceae UCG-014 | 0.559c | 1.052b | 0.894bc | 0.703bc | 1.538a | 0.116 | < 0.001 |
| Ruminococcaceae NK4A214 group | 11.212ab | 10.255bc | 8.576c | 10.476bc | 12.957a | 0.674 | 0.001 |
| Lachnospiraceae AC2044 group | 0.519c | 3.990a | 0.861bc | 0.914b | 0.786bc | 0.108 | < 0.001 |
| Lachnospiraceae UCG-004 | 0.424bc | 0.486b | 0.293c | 0.299c | 1.206a | 0.055 | < 0.001 |
| Lachnospiraceae UCG-008 | 0.615b | 0.631b | 0.319c | 0.440bc | 0.927a | 0.124 | < 0.001 |
| Christensenellaceae R-7 group | 13.871a | 7.129c | 7.240c | 10.002b | 14.791a | 0.608 | < 0.001 |
| Butyrivibrio 2 | 0.789b | 1.009b | 1.969a | 1.049b | 0.976b | 0.144 | 0.049 |
| Moryella | 0.595a | 0.273b | 0.309b | 0.273b | 0.437ab | 0.072 | 0.005 |
| Pseudobutyrivibrio | 0.171b | 0.247b | 1.788a | 0.342b | 0.174b | 0.103 | < 0.001 |
| Sporolactobacillus | 1.022ab | 0.874b | 0.878b | 0.904b | 1.512a | 0.156 | 0.010 |
| Treponema 2 | 0.595a | 0.273b | 0.309b | 0.273b | 0.437ab | 0.072 | 0.005 |
| p-1088-a5 gut group | 0.289ab | 0.197ab | 0.141c | 0.194ab | 0.306a | 0.046 | 0.036 |
| Uncultured (%) | |||||||
| f_F082| g_uncultured | 10.111a | 6.853c | 10.397a | 8.507b | 7.774bc | 0.284 | < 0.001 |
| f_Muribaculaceae| g_uncultured | 3.389b | 5.085a | 6.045a | 5.877a | 3.100b | 0.292 | 0.015 |
| f_Bacteroidales RF16 group| g_uncultured | 0.483bc | 0.772a | 0.667ab | 0.546b | 0.306c | 0.057 | < 0.001 |
| f_Bacteroidales BS11 gut group| g_uncultured | 0.243b | 0.782a | 0.713a | 0.332b | 0.358b | 0.065 | < 0.001 |
| f_Paludibacteraceae| g_uncultured | 0.365b | 0.427ab | 0.546a | 0.460ab | 0.398ab | 0.046 | 0.034 |
| f_p-251-o5| g_uncultured | 0.463ab | 0.197c | 0.634a | 0.289bc | 0.424b | 0.055 | < 0.001 |
| f_Lachnospiraceae| g_uncultured | 0.930b | 1.134ab | 1.292a | 0.832b | 1.134ab | 0.093 | 0.004 |
| o_Mollicutes RF39| f_uncultured | 0.772b | 0.782b | 1.150b | 1.256b | 1.814a | 0.153 | < 0.001 |
a–cMeans within a row with different superscripts differ significantly (p < 0.05).
1Groups: different peNDF1.18 (PS > 1.18 mm) contents of 33.0, 29.9, 28.1, 26.5, and 24.8% were obtained by chopping or crusher crushing the forage into the following lengths: H (7 cm), MH (4 cm), M (1 cm), ML (5–0 mm sieve), and L (1-mm sieve).
Regression Analysis Between peNDF1.18 Level and Relative Abundance of Rumen Bacteria
The relationships between dietary peNDF1.18 levels (x) and the relative abundance (y) of rumen bacteria at the phylum or genus level are summarized in Table 7. We found that there was a significant quadratic correlation (p < 0.001) between most of the bacterial flora and dietary peNDF1.18 levels, and that the level of dietary peNDF1.18 had the greatest impact on the relative abundance of Tenericutes and Christensenellaceae R-7 group, with high fitness (R2 was 0.804 and 0.958, respectively).
TABLE 7.
Regression relationship between dietary peNDF1.18 level (x) and relative abundance of rumen bacteria (y) (n = 15, R2 > 0.6).
| Items | Regression equation | R 2 | P-value |
| Phylum | |||
| Bacteroidetes | y = –55.129x2 + 32.985x–4.327 | 0.795 | < 0.001 |
| Firmicutes | y = 47.844x2–28.484x + 4.657 | 0.745 | < 0.001 |
| Tenericutes | y = 2.009x2–1.287x + 0.215 | 0.804 | < 0.001 |
| Spirochetes | y = 0.848x2–0.576x + 0.104 | 0.622 | 0.001 |
| Planctomycetes | y = 3.534x2–2.087x + 0.313 | 0.729 | < 0.001 |
| Genus | |||
| Prevotella 1 | y = –29.128x2 + 18.351x–2.590 | 0.851 | < 0.001 |
| Ruminococcaceae NK4A214 group | y = 16.860x2–9.899x + 1.555 | 0.604 | 0.002 |
| Christensenellaceae R-7 group | y = 44.347x2–25.763x + 3.818 | 0.958 | < 0.001 |
| Ruminococcus 2 | y = 4.394x2–2.665x + 0.414 | 0.885 | < 0.001 |
| Treponema 2 | y = 0.486x2–0.357x + 0.071 | 0.638 | 0.001 |
| Moryella | y = 1.452x2–0.820x + 0.119 | 0.660 | 0.001 |
| p-1088-a5 gut group | y = 3.644x2–2.140x + 0.318 | 0.765 | < 0.001 |
| Prevotellaceae UCG-003 | y = –3.735x2 + 2.147x–0.292 | 0.884 | < 0.001 |
| Uncultured | |||
| o_Mollicutes RF39| f_uncultured | y = 2.068x2–1.319x–0.220 | 0.807 | < 0.001 |
| f_Muribaculaceae| g_uncultured | y = –15.459x2 + 8.895x–1.226 | 0.763 | < 0.001 |
| f_Bacteroidales RF16 group| g_uncultured | y = –2.003x2 + 1.182x–0.166 | 0.846 | < 0.001 |
Prediction of Rumen Microbial Flora Function
According to the prediction of microbial function by the Tax4Fun program and the SILVA database, the gene functions of each group in annotation level 2 were selected to perform differential analysis. We found that dietary peNDF1.18 levels had significant effects on the prediction of rumen bacterial function (Table 8). Table 8 shows that the main gene functions of rumen bacteria are associated with carbohydrate metabolism, amino acid metabolism, and membrane transport.
TABLE 8.
Effects of dietary peNDF1.18 level on the prediction of rumen bacterial function (%).
| Functions | Groups1 |
SEM | P-value | ||||
| H (33.0%) | MH (29.9%) | M (28.1%) | ML (26.5%) | L (24.8%) | |||
| Metabolism | |||||||
| Nucleotide metabolism | 7.190a | 7.103b | 7.064b | 7.112ab | 7.095b | 0.025 | 0.006 |
| Amino acid metabolism | 10.654d | 10.693cd | 10.962b | 10.836bc | 11.196a | 0.044 | < 0.001 |
| Metabolism of other amino acids | 2.146ab | 2.154a | 2.138b | 2.151ab | 2.110c | 0.004 | < 0.001 |
| Carbohydrate metabolism | 15.868ab | 15.778b | 15.811b | 16.026a | 15.511c | 0.062 | < 0.001 |
| Glycan biosynthesis and metabolism | 4.415a | 4.228bc | 4.196c | 4.397ab | 3.979d | 0.056 | < 0.001 |
| Lipid metabolism | 2.810b | 2.815b | 2.822ab | 2.853a | 2.752c | 0.011 | < 0.001 |
| Energy metabolism | 6.636c | 6.648bc | 6.681ab | 6.681abc | 6.706a | 0.014 | 0.003 |
| Genetic information processing | |||||||
| Translation | 6.651a | 6.526b | 6.425bc | 6.497b | 6.320c | 0.033 | < 0.001 |
| Replication and repair | 6.310a | 6.202b | 6.078c | 6.170bc | 5.951d | 0.032 | < 0.001 |
| Folding, sorting, and degradation | 2.947a | 2.902ab | 2.879b | 2.914ab | 2.828c | 0.015 | < 0.001 |
| Environmental information processing | |||||||
| Membrane transport | 8.197c | 8.644b | 8.455bc | 8.056c | 9.085a | 0.125 | < 0.001 |
| Signal transduction | 6.024c | 6.165bc | 6.346ab | 6.170bc | 6.479a | 0.067 | < 0.001 |
| Cellular processes | |||||||
| Cell motility | 1.724bc | 1.881a | 1.828ab | 1.669c | 1.962a | 0.046 | 0.001 |
| Cell growth and death | 1.808a | 1.799ab | 1.791b | 1.790b | 1.796b | 0.003 | 0.002 |
| Organismal systems | |||||||
| Endocrine system | 0.422a | 0.415ab | 0.404b | 0.420a | 0.378c | 0.004 | < 0.001 |
| Immune system | 0.143ab | 0.133c | 0.135bc | 0.144a | 0.123d | 0.003 | < 0.001 |
| Digestive system | 0.534a | 0.484c | 0.487bc | 0.530ab | 0.427d | 0.013 | < 0.001 |
| Human diseases | |||||||
| Infectious diseases | 1.645a | 1.641a | 1.616b | 1.617b | 1.564c | 0.004 | 0.043 |
a–dMeans within a row with different superscripts differ significantly (p < 0.05).
1Groups: different peNDF1.18 (PS > 1.18 mm) contents of 33.0, 29.9, 28.1, 26.5, and 24.8% were obtained by chopping or crusher crushing the forage into the following lengths: H (7 cm), MH (4 cm), M (1 cm), ML (5-mm sieve), and L (1-mm sieve).
Discussion
Growth Performance and Rumen Development
Compared with pigs and poultry, goats, as ruminants, can better use fiber in feed as energy source for their growth and development. Therefore, evaluation of dietary fiber, especially NDF (peNDF), is crucial for the nutritional value of goats or other ruminants. In this study, hay was ground into different lengths to obtain five peNDF1.18 levels in diets, which have an impact on goat DMI. When peNDF1.18 > 26.5%, as peNDF declined, goat DMI increased, which is consistent with the research of Park et al. (2014). Wang et al. (2017) observed that body growth improved with increasing peNDF8.0. Although heifers’ DMI did not change significantly among different peNDF8.0 contents in the study of Wang et al. (2017) and in another study concluded by Beauchemin and Yang (2005), in the present work and in this study, the DMI and ADG of the 26.5% peNDF1.18 treatment were significantly greater than those of 33.0 and 24.8% peNDF1.18 treatments (Jang et al., 2017). The different results of the study may be attributed to the differences in the animals and in the diets: goats are smaller than cows, so their sensitivity to peNDF may be higher than that of cattle. In a research conducted by Yang et al. (2017), they found that the differences in the ADG of goats among the treatment groups are generally higher than those in this study, which is also due to the smaller body size of goats in the study of Jang et al. (2017). Particles with a length of 10 mm were retained in the reticulo-rumen 19–28 h longer than 1-mm-long particles of the same density. Multiple regression analysis indicated that particle density and PS accounted for 59 and 28% of the total variation of mean retention time in the reticulo-rumen, respectively (Ehle et al., 1984; Kaske and Engelhardt, 1990).
The level of dietary fiber is extremely important for the growth and development of the gastrointestinal tract of young ruminants, while the length and width of the rumen papilla are especially vital indicators of the degree of rumen development (Lesmeister et al., 2004). The experimental results of Beiranvand et al. (2014) showed that 2.6-mm alfalfa hay could increase the thickness of the rumen muscle of calves and reduce the width of the nipple. Xu et al. (2010) found that feeding whole corn reduced the height and width of the rumen papilla of goats compared to when feeding goat with fine, coarse, and whole corn. The results of this study show that the 28.1 and 26.5% peNDF1.18 treatments are more beneficial to the development of rumen papilla. In other words, moderate peNDF level is conducive to the development of the gastrointestinal tract of goats.
Rumen Bacterial Community
In this study, we attempted to study the impact of dietary peNDF1.18 level on the rumen bacterial flora of goat and discovered that rumen bacteria could be affected by just changing the PS of forage. Results in Shannon and Simpson indexes indicated that the level of dietary peNDF1.18 was a strong determinant of rumen microbial community composition in goats. Previous studies reported that the high diversity of microorganisms was beneficial for the production performance of animals (Arrazuria et al., 2016; Yuan et al., 2019). Similarly, high microbial diversity is linked to good meat and wool production in sheep (Zeng et al., 2017), which is the common ruminant. In this experiment, the microbial diversity indexes between peNDF1.18 treatments implied that the peNDF1.18 levels between 26.5 and 29.9% were perhaps more beneficial to goat health. Zebeli et al. (2008) and Valizadeh et al. (2010) also found that dietary peNDF level did not affect the number of rumen bacteria by absolute quantification of rumen bacteria. Moreover, the level of dietary peNDF1.18 was a strong determinant of rumen microbial community structure, according to the results of PCoA profiling and PerMANOVA. Previous studies on cow and dairy goat also showed that the PS of the feedstuff influenced the relative abundance of rumen (Tajima et al., 2001; Li et al., 2018a).
Previous studies reported that the dominant phyla in the rumen were usually Bacteroidetes, Firmicutes, and Proteobacteria (Shabat et al., 2016). The fiber in diets has recently been proved to be a factor affecting microbiota in the gastrointestinal tract. Liu et al. (2017) discovered that the relative abundance of Proteobacteria in goat rumen would increase with the decrease of feed fiber content. Sika deer fed oak-leave-based diets had a higher relative abundance of Bacteroidetes than deer fed corn-stalk-based diets (Li et al., 2018b). Crowley et al. (2017) found that the relative abundance of Bacteroidetes increased when rabbits were fed a diet with a higher fiber content. However, there were only limited studies on the effects of roughage length on rumen microbiota. One study conducted by Li et al. (2018a) found that both roughage and concentration length influence the rumen microbiota. Specifically, this study found that as the level of dietary peNDF1.18 decreased, the dominant phylum transitioned from Bacteroidetes to Firmicutes. The shorter the forage, the higher was the relative abundance of Proteobacteria, which was consistent with the results in rabbits (Yuan et al., 2019). Existing studies have shown that increased relative abundance of Proteobacteria is a common feature of health impairment (Perea et al., 2017), suggesting that the peNDF1.18 level of goat diet should not be less than 24.8%.
Shen et al. (2017) reported that the relative abundance of Tenericutes in goat rumen increased with the level of concentrate. Yuan et al. (2019) found that the relative abundance of Tenericutes in the caecum of rabbits was negatively correlated to the PS of alfalfa (Yuan et al., 2019), which was similar to our study. Spirochetes, a kind of fiber-degrading phylum, was negatively affected by the dietary addition of starch or oil (Zened et al., 2013). Our results proved that the relative abundance of Spirochetes was highest in the 24.8% peNDF1.18 treatments. More studies are needed to explore the reasons for the changes in the relative abundance of phyla with dietary peNDF and to explore the functions of these predominant phyla.
As for the dominant genera of rumen bacteria, a large number of reports point out that Prevotella is the dominant genus in the rumen (Ramírez-Restrepo et al., 2016) and that its main function is protein degradation (Myer et al., 2015). Moreover, Satoshi et al. (2010) found that Prevotella was also involved in fiber degradation. Bekele et al. (2010) found that Prevotella accounted for 56 or 60% of goat rumen bacteria and that its relative abundance was even higher when goats were fed hay diet than when fed concentrate diet. Huo et al. (2014) found that Prevotella accounted for less than 20% of goat rumen bacteria, which was close to our results. Huo et al. (2014) also discovered that a high-grain diet was not good for the growth of Prevotella. Our study, for the first time, disclosed that the growth of Prevotella was sensitively affected by forage length; fine-crushed forage (1 mm) was not conducive to the growth of Prevotella 1 and Prevotellaceae UCG-001; and the optimal level of peNDF1.18 for the growth of Prevotellaceae NK3B31 group and Prevotellaceae UCG-003 was 26.5% as judged by the relative abundance. Thus, we could conclude that dietary peNDF affects the growth of Prevotella at different patterns according to the different Prevotellaceae isolates. Spearman correlation analysis showed that the relative abundance of Prevotellaceae UCG-001 was negatively related, but that of Prevotellaceae NK3B31 group was positively related, to amino acid metabolism. This implied that rumen bacteria at the genus level may be too rough to study the function of the bacteria, and isolates under the genus would be accurate enough to investigate the functions. A previous study has shown that Prevotella is related to protein degradation (Myer et al., 2015); however, we found that the relative abundance of Prevotella 1 (the dominant Prevotella in goat rumen) was negatively related to amino acid metabolism, and that the apparent digestibility of crude protein and the relative abundance of Prevotella 1 were similarly the lowest in the 24.8% peNDF1.18 treatments. This result shows that the function of a single strain cannot represent the role of bacteria at the genus level. Moreover, there is still a gap between the predicted functions and the actual situation, and more technical methods are needed to study the specific functions of bacteria.
Ruminococcus is a major cellulolytic genus (Bryant, 1959). In our study, the relative abundance of Ruminococcaceae isolates did not change orderly with dietary peNDF1.18 levels, but the relative abundances of Ruminococcaceae NK4A214 group and Ruminococcus 2 were quadratically related to the level of dietary peNDF1.18 (Table 6). Pitta et al. (2014) found that increasing the proportion of fiber in buffalo diet could increase the relative abundance of Ruminococcus. Zebeli et al. (2008) found that the count of Ruminococcus was not affected by dietary PS via real-time quantitative PCR. However, our experiment showed that the count of Ruminococcus was affected by peNDF by high-throughput sequencing, and different Ruminococcus isolates (Ruminococcus 1 and 2) responded differently to peNDF levels (Table 5), reminding us to be species-specific when studying dietary effects on bacteria. Christensenella was also a kind of dominant genus in goat rumen, and its relative abundance varied with the different treatments, but Spearman correlation analysis showed that it was not significantly related to the top 10 functions of bacteria (p > 0.05). In other words, their functions might be minor for host metabolism. Zhang et al. (2017) also concluded that Christensenella possessed little function in the rumen of Holstein heifer, but they attributed this to the low relative abundances.
Butyrivibrio, Clostridium, and Fibrobacter are the main fiber-degrading bacteria in the rumen (Klieve et al., 2007; Zhang et al., 2017), and many members of Lachnospiraceae also have cellulolytic activities (Nyonyo et al., 2014). Zhang et al. (2017) found that the relative abundance of Fibrobacter (0.36–2.35%) in the rumen of Holstein heifer elevated with the increase in the concentrate level of their diet. Our result showed that the relative abundance of Fibrobacter in goat rumen was less than 0.1% and reached the peak when peNDF1.18 was 24.8% (24.8% peNDF1.18 treatment). This difference might be caused by the differences in animal species and diet (Metzler-Zebeli et al., 2013; Jin et al., 2016), but it could also be caused by differences in sequencing technology and primers. In this study, the relative abundance of Clostridium in the rumen of goat ranged from 1.27 to 1.97%, very similar to that in the rumen of sheep (Nyonyo et al., 2014), and was not affected by the level of peNDF1.18. We found that the function of Clostridium sensu stricto 12 was most likely in the metabolism of cofactors and vitamins instead of carbohydrates (Figure 5). Li et al. (2019) found that Clostridium could be involved in fiber fermentation. The different functions between the two reports were possibly due to the specific strain of Clostridium, but the deeper reason remains to be studied. The relative abundance of Butyrivibrio 2 reached the summit at 28.1% peNDF1.18 treatment (p = 0.049), which signified that dietary peNDF1.18 deviating from 28.1% might reduce fiber digestion. Previous studies have shown that Butyrivibrio 2 was negatively related to immune factors (IL-6 and IL-1β) (Li et al., 2019) and was also a considerable butyrate producer (Nyonyo et al., 2014) and a regulator of hemicellulose-degrading enzyme secretion (Dunne et al., 2012). Thus, it is necessary to study the relationship between relative abundance and host health, as well as production performance.
According to the correlation analysis between the relative abundance of bacteria, changing trends in Bacteroidetes and Firmicutes, and that of the genera under the two phyla, were inverse. In addition, the regression relationship between peNDF1.18 level and the relative abundance of Ruminococcaceae NK4A214 group and Prevotella 1 was inverse. Functional prediction explored that Prevotella 1 and Ruminococcaceae UCG-011 were in opposing camps, suggesting that the two bacteria may alter host nutrition in the opposite direction, but further studies are needed to explore the optimal relative abundance of the two genera and their functions in nutrient metabolism in goat rumen. Moreover, three uncultured bacteria were quadratically related to dietary peNDF1.18 level, especially f_Muribaculaceae| g_uncultured, but this genus showed no correlation with the predicted function, so the specific function remains confirmatory. Overall, although a quadratic correlation exists, follow-up research experiments should be carried out on the premise of animal health and welfare. One of the shortcomings of this study was the small number of repetitions, which is worth noting in future experiments.
Existing studies have shown that gastrointestinal microbes have a strong correlation with the metabolism of the host (Nieuwdorp et al., 2014; Shi et al., 2017). In this study, we detected the main functions of rumen bacteria, and most genes were closely related to carbohydrate metabolism. The lowest function percentage in the 24.8% peNDF1.18 treatment suggested that low dietary peNDF may not be beneficial to carbohydrate metabolism activities of rumen microorganisms. In terms of the metabolism of carbohydrates, amino acids, and energy, the range of dietary peNDF1.18 between 26.5 and 28.1% seemed more beneficial to the metabolic activity of the host. We also found that the relative abundances of Prevotella 1, Prevotellaceae UCG-001, and Ruminococcaceae UCG-011 were significantly related to the majority of the dominant functions, so it is important to study their functions in the future.
Conclusion
This study proved that the differences in growth performance and rumen development were associated with changes in the rumen bacterial community. Based on the results of growth performance, rumen development, and rumen bacterial community, a peNDF level of between 26.5 and 28.1% in the diet was optimal for goats.
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih. gov/, PRJNA693130.
Ethics Statement
The animal study was reviewed and approved by the Animal Policy and Welfare Committee of Animal Nutrition Institute, Sichuan Agriculture University.
Author Contributions
BCX and MW performed the experiments, analyzed the statistical data, prepared the figures and tables, reviewed earlier versions of the manuscripts, and approved the final manuscript. AH and XL carried out the experiments and gave tremendous help in data analysis. BX and QH designed and supervised the experiments. BX and SY were also in charge of preparing the manuscript. ZW, LW, and QP gave guidance on statistical analysis and on the plan of this study. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the staff at our laboratory for their assistance.
Footnotes
Funding
This work was supported by the Key Technologies Research and Development Projects for Modernized Livestock and Poultry of China (2017YFD0502005).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.820509/full#supplementary-material
References
- Arrazuria R., Elguezabal N., Juste R. A., Derakhshani H., Khafipour E. (2016). Mycobacterium avium Subspecies paratuberculosis infection modifies gut microbiota under different dietary conditions in a rabbit model. Front. Microbiol. 7:446. 10.3389/fmicb.2016.00446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aßhauer K. P., Wemheuer B., Daniel R., Meinicke P. (2015). Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics (Oxford, England) 31 2882–2884. 10.1093/bioinformatics/btv287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin K. A., Yang W. Z. (2005). Effects of physically effective fiber on intake, chewing activity, and ruminal acidosis for dairy cows fed diets based on corn silage. J. Dairy Sci. 88 2117–2129. 10.3168/jds.S0022-0302(05)72888-5 [DOI] [PubMed] [Google Scholar]
- Beiranvand H., Ghorbani G. R., Khorvash M., Nabipour A., Dehghan-Banadaky M., Homayouni A., et al. (2014). Interactions of alfalfa hay and sodium propionate on dairy calf performance and rumen development. J. Dairy Sci. 97 2270–2280. 10.3168/jds.2012-6332 [DOI] [PubMed] [Google Scholar]
- Bekele A. Z., Koike S., Kobayashi Y. (2010). Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis. FEMS Microbiol. Lett. 305 49–57. 10.1111/j.1574-6968.2010.01911.x [DOI] [PubMed] [Google Scholar]
- Bryant M. P. (1959). Bacterial species of the rumen. Bacteriol. Rev. 23 125–153. 10.1128/br.23.3.125-153.1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang D., Wang L., Wei X., Li X., Cai C., et al. (2021). Physically effective neutral detergent fiber improves chewing activity, rumen fermentation, plasma metabolites, and milk production in lactating dairy cows fed a high-concentrate diet. J. Dairy Sci. 104 5631–5642. 10.3168/jds.2020-19012 [DOI] [PubMed] [Google Scholar]
- Crowley E. J., King J. M., Wilkinson T., Worgan H. J., Huson K. M., Rose M. T., et al. (2017). Comparison of the microbial population in rabbits and guinea pigs by next generation sequencing. PLoS One 12:e0165779. 10.1371/journal.pone.0165779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTullio G. R., Grebmeier J. M., Arrigo K. R., Lizotte M. P., Robinson D. H., Leventer A., et al. (2000). Rapid and early export of Phaeocystis antarctica blooms in the Ross Sea, Antarctica. Nature 404 595–598. 10.1038/35007061 [DOI] [PubMed] [Google Scholar]
- Dunne J. C., Li D., Kelly W. J., Leahy S. C., Bond J. J., Attwood G. T., et al. (2012). Extracellular polysaccharide-degrading proteome of Butyrivibrio proteoclasticus. J. Proteome Res. 11 131–142. 10.1021/pr200864j [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10:996. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Ehle F. R., Bas F., Barno B., Martin R., Leone F. (1984). Particulate rumen turnover rate measurement as influenced by density of passage marker. J. Dairy Sci. 67 2910–2913. 10.3168/jds.S0022-0302(84)81654-9 [DOI] [Google Scholar]
- Huo W., Zhu W., Mao S. (2014). Impact of subacute ruminal acidosis on the diversity of liquid and solid-associated bacteria in the rumen of goats. World J. Microbiol. 30 669–680. 10.1007/s11274-013-1489-8 [DOI] [PubMed] [Google Scholar]
- Jang S. Y., Kim E. K., Park J. H., Oh M. R., Tang Y. J., Ding Y. L., et al. (2017). Effects of physically effective neutral detergent fiber content on dry matter intake, digestibility, and chewing activity in Korean native goats (Capra hircus coreanae) fed with total mixed ration. Asian-Australasian J. Animal Sci. 30 1405–1409. 10.5713/ajas.16.0868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Fan H., Ye H., Zhao J., Zhou Z. (2016). Characterization of the microbial communities along the gastrointestinal tract of sheep by 454 pyrosequencing analysis. Asian-Australas J. Anim. Sci. 30 100–110. 10.5713/ajas.16.0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaske M., Engelhardt W. V. (1990). The effect of size and density on mean retention time of particles in the gastrointestinal tract of sheep. Br. J. Nutr. 63 457–465. 10.1079/BJN19900133 [DOI] [PubMed] [Google Scholar]
- Klieve A. V., O’Leary M. N., McMillen L., Ouwerkerk D. (2007). Ruminococcus bromii, identification and isolation as a dominant community member in the rumen of cattle fed a barley diet. J. Appl. Microbiol. 103 2065–2073. 10.1111/j.1365-2672.2007.03492.x [DOI] [PubMed] [Google Scholar]
- Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononoff P. J., Heinrichs A. J., Buckmaster D. R. (2003). Modification of the Penn State forage and total mixed ration particle separator and the effects of moisture content on its measurements. J. Dairy Sci. 86 1858–1863. 10.3168/jds.S0022-0302(03)73773-4 [DOI] [PubMed] [Google Scholar]
- Lesmeister K. E., Tozer P. R., Heinrichs A. J. (2004). Development and analysis of a rumen tissue sampling procedure. J. Dairy Sci. 87 1336–1344. 10.3168/jds.S0022-0302(04)73283-X [DOI] [PubMed] [Google Scholar]
- Li F., Li Z., Li S., Ferguson J. D., Yang T. (2014). Effect of dietary physically effective fiber on ruminal fermentation and the fatty acid profile of milk in dairy goats. J. Dairy Sci. 97 2281–2290. 10.3168/jds.2013-6895 [DOI] [PubMed] [Google Scholar]
- Li F., Shah A. M., Wang Z., Peng Q., Hu R., Zou H., et al. (2019). Effects of land transport stress on variations in ruminal microbe diversity and immune functions in different breeds of cattle. Animals 9:599. 10.3390/ani9090599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Liu N., Cao Y., Jin C., Li F., Cai C., et al. (2018a). Effects of fumaric acid supplementation on methane production and rumen fermentation in goats fed diets varying in forage and concentrate particle size. J. Animal Sci. Biotechnol. 9:21. 10.1186/s40104-018-0235-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wang X., Zhang T., Si H., Nan W., Xu C., et al. (2018b). The development of microbiota and metabolome in small intestine of sika deer (Cervus nippon) from birth to weaning. Front. Microbiol. 9:4. 10.3389/fmicb.2018.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Xu Q., Wang L., Wang J., Zhou M. (2017). The impact of diet on the composition and relative abundance of rumen microbes in goat. Asian-Australas J. Anim. Sci. 30 531–537. 10.5713/ajas.16.0353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens D. R. (1997). Creating a system for meeting the fiber requirements of dairy cows: symposium: meeting the fiber requirements of dairy cows. J. Dairy Sci. 80 1463–1481. 10.3168/jds.s0022-0302(97)76075-2 [DOI] [PubMed] [Google Scholar]
- Metzler-Zebeli B. U., Schmitz-Esser S., Klevenhusen F., Podstatzky-Lichtenstein L., Wagner M., Zebeli Q. (2013). Grain-rich diets differently alter ruminal and colonic abundance of microbial populations and lipopolysaccharide in goats. Anaerobe 20 65–73. 10.1016/j.anaerobe.2013.02.005 [DOI] [PubMed] [Google Scholar]
- Myer P. R., Wells J. E., Smith T., Kuehn L. A., Freetly H. C. (2015). Cecum microbial communities from steers differing in feed efficiency. J. Anim. Sci. 93:5327. 10.2527/jas.2015-9415 [DOI] [PubMed] [Google Scholar]
- Nieuwdorp M., Gilijamse P. W., Pai N., Kaplan L. M. (2014). Role of the microbiome in energy regulation and metabolism. Gastroenterology 146 1525–1533. 10.1053/j.gastro.2014.02.008 [DOI] [PubMed] [Google Scholar]
- Nyonyo T., Shinkai T., Mitsumori M. (2014). Improved culturability of cellulolytic rumen bacteria and phylogenetic diversity of culturable cellulolytic and xylanolytic bacteria newly isolated from the bovine rumen. FEMS Microbiol. Ecol. 88 528–537. 10.1111/1574-6941.12318 [DOI] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F. G., Kindt R., Legendre P., O’Hara R. G., Simpson G. L., et al. (2010). vegan: community ecology package 1.18-2. Time Int. 1997 15–17. [Google Scholar]
- Parada A. E., Needham D. M., Fuhrman J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18 1403–1414. 10.1111/1462-2920.13023 [DOI] [PubMed] [Google Scholar]
- Park J. H., Kim K. H., Park P. J., Jeon B. T., Oh M. R., Jang S. Y., et al. (2014). Effects of physically effective neutral detergent fibre content on dry-matter intake, digestibility and chewing activity in beef cattle fed total mixed ration. Animal Production Ence 55:166. 10.1071/an14241 [DOI] [Google Scholar]
- Perea K., Perz K., Olivo S. K., Williams A., Yeoman C. J. (2017). Feed efficiency phenotypes in lambs involve changes in ruminal, colonic, and small-intestine-located microbiota. J. Anim. Sci. 95:2585. 10.2527/jas.2016.1222 [DOI] [PubMed] [Google Scholar]
- Pitta D. W., Kumar S., Vei, Cc Harelli B., Parmar N., Reddy B., et al. (2014). Bacterial diversity associated with feeding dry forage at different dietary concentrations in the rumen contents of Mehshana buffalo (Bubalus bubalis) using 16S pyrotags. Anaerobe 25 31–41. 10.1016/j.anaerobe.2013.11.008 [DOI] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Glckner F. O. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Restrepo C. A., Cui T., O’Neill C. J., Lopez-Villalobos N., Padmanabha J., Wang J. K., et al. (2016). Methane production, fermentation characteristics, and microbial profiles in the rumen of tropical cattle fed tea seed saponin supplementation. Animal Feed Sci. Technol. 216 58–67. 10.1016/j.anifeedsci.2016.03.005 [DOI] [Google Scholar]
- Satoshi K., Sayo Y., Yasuo K., Keiichi T. (2010). Phylogenetic analysis of fiber-associated rumen bacterial community and PCR detection of uncultured bacteria. FEMS Microbiol. Lett. 229 23–30. 10.1016/S0378-1097(03)00760-2 [DOI] [PubMed] [Google Scholar]
- Shabat S. K. B., Sasson G., Doron-Faigenboim A., Durman T., Yaacoby S., Berg Miller M. E., et al. (2016). Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10 2958–2972. 10.1038/ismej.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Lu Z., Xu Z., Chen Z., Shen Z. (2017). Associations among dietary non-fiber carbohydrate, ruminal microbiota and epithelium G-protein-coupled receptor, and histone deacetylase regulations in goats. Microbiome 5:123. 10.1186/s40168-017-0341-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi N., Li N., Duan X., Niu H. (2017). Interaction between the gut microbiome and mucosal immune system. Military Med. Res. 4:14. 10.1186/s40779-017-0122-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg M., Chaucheyras-Durand F., Commun L., Mialon M. M., Monteils V., Mosoni P., et al. (2013). Repeated acidosis challenges and live yeast supplementation shape rumen microbiota and fermentations and modulate inflammatory status in sheep. Animal Int. J. Animal Biosci. 7 1910–1920. 10.1017/S1751731113001705 [DOI] [PubMed] [Google Scholar]
- Tajima K., Aminov R. I., Nagamine T., Matsui H., Nakamura M., Benno Y. (2001). Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl. Environ. Microbiol. 67 2766–2774. 10.1128/AEM.67.6.2766-2774.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valizadeh R., Behgar M., Mirzaee M., Naserian A. A., Ghovvati S. (2010). The effect of physically effective fiber and soy hull on the ruminal cellulolytic bacteria population and milk production of dairy cows. Asian Australasian J. Animal Sci. 23 1325–1332. 10.5713/ajas.2010.90651 [DOI] [Google Scholar]
- Wang H. R., Chen Q., Chen L. M., Ge R. F., Wang M. Z., Yu L. H., et al. (2017). Effects of dietary physically effective neutral detergent fiber content on the feeding behavior, digestibility, and growth of 8- to 10-month-old Holstein replacement heifers. J. Dairy Sci. 100 1161–1169. 10.3168/jds.2016-10924 [DOI] [PubMed] [Google Scholar]
- Wang L., Liu K., Wang Z., Bai X., Peng Q., Jin L. (2019). Bacterial community diversity associated with different utilization efficiencies of nitrogen in the gastrointestinal tract of goats. Front. Microbiol. 10:239. 10.3389/fmicb.2019.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C. O. (2010). picante: R tools for integrating phylogenies and ecology. Bioinformatics 26 1463–1464. 10.1093/bioinformatics/btq166 [DOI] [PubMed] [Google Scholar]
- Xu M., Dong Y., Du S., Hao Y. S., Yao J. H. (2010). Effect of corn particle size on mucosal morphology and digesta pH of the gastrointestinal tract in growing goats. Livestock Sci. 123 34–37. 10.1016/j.livsci.2008.10.003 [DOI] [Google Scholar]
- Yang L., Gao L., Bai Y., Zhang K., Meng M., Song Q. (2017). Effects of pelleted diets with different physically effective neutral detergent fiber levels on nutrient apparent digestibility of goats. Chinese J. Animal Nutrition 29 2996–3003. [Google Scholar]
- Yang W. Z., Beauchemin K. A. (2007). Altering physically effective fiber intake through forage proportion and particle length: chewing and ruminal pH. J. Dairy Sci. 90 2826–2838. 10.3168/jds.2007-0032 [DOI] [PubMed] [Google Scholar]
- Yuan M., Liu S., Wang Z., Wang L., Peng Q. (2019). Effects of particle size of ground alfalfa hay on caecal bacteria and archaea populations of rabbits. PeerJ 7:e7910. 10.7717/peerj.7910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebeli Q., Aschenbach J. R., Tafaj M., Boguhn J., Ametaj B. N., Drochner W. (2012). Invited review: role of physically effective fiber and estimation of dietary fiber adequacy in high-producing dairy cattle. J. Dairy Sci. 95 1041–1056. 10.3168/jds.2011-4421 [DOI] [PubMed] [Google Scholar]
- Zebeli Q., Tafaj M., Junck B., Oelschlaeger V., Ametaj B. N., Drochner W. (2008). Evaluation of the response of ruminal fermentation and activities of nonstarch polysaccharide-degrading enzymes to particle length of corn silage in dairy cows. J. Dairy Sci. 91 2388–2398. 10.3168/jds.2007-0810 [DOI] [PubMed] [Google Scholar]
- Zened A., Combes S., Cauquil L., Mariette J., Klopp C., Bouchez O., et al. (2013). Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol. Ecol. 83 504–514. 10.1111/1574-6941.12011 [DOI] [PubMed] [Google Scholar]
- Zeng Y., Zeng D., Ni X., Zhu H., Jing B. (2017). Microbial community compositions in the gastrointestinal tract of Chinese Mongolian sheep using Illumina MiSeq sequencing revealed high microbial diversity. AMB Exp. 7:75. 10.1186/s13568-017-0378-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Shi H., Wang Y., Li S., Cao Z., Ji S., et al. (2017). Effect of dietary forage to concentrate ratios on dynamic profile changes and interactions of ruminal microbiota and metabolites in holstein heifers. Front. Microbiol. 8:2206. 10.3389/fmicb.2017.02206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. H., Zhang T., Xu M., Yao J. H. (2010). Effects of physically effective fiber on chewing activity, ruminal fermentation, and digestibility in goats. J. Anim. Sci. 89 501–509. 10.2527/jas.2010-3013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih. gov/, PRJNA693130.