Abstract
Whether and how innate antiviral response is regulated by humoral metabolism remains enigmatic. We show that viral infection induces progesterone via the hypothalamic-pituitary-adrenal axis in mice. Progesterone induces downstream antiviral genes and promotes innate antiviral response in cells and mice, whereas knockout of the progesterone receptor PGR has opposite effects. Mechanistically, stimulation of PGR by progesterone activates the tyrosine kinase SRC, which phosphorylates the transcriptional factor IRF3 at Y107, leading to its activation and induction of antiviral genes. SARS-CoV-2-infected patients have increased progesterone levels, and which are co-related with decreased severity of COVID-19. Our findings reveal how progesterone modulates host innate antiviral response, and point to progesterone as a potential immunomodulatory reagent for infectious and inflammatory diseases.
Subject terms: Innate immunity, Inflammation
Introduction
Innate immunity is the first line of host defense against viral infection. Upon infection, viral components particularly their genomes or replication intermediates are sensed by distinct pattern-recognition receptors (PRRs) including Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), and/or intracellular DNA sensors.1 Sensing of viral nucleic acids by PRRs triggers signaling cascades to induce transcription of type I interferons (IFNs), pro-inflammatory cytokines, and other downstream antiviral effector genes. The downstream cytokines and effectors inhibit viral replication or induce apoptosis of infected cells, leading to innate antiviral response.2–8
Different types of viruses are sensed by distinct PPRs. Upon infection of RNA virus, the invaded cytosolic viral RNA is sensed by the RNA helicase RIG-I or MDA5.9,10 Binding of RIG-1 or MDA5 to viral RNA triggers its recruitment to VISA (also called MAVS), a mitochondrion-associated adapter protein.11,12 VISA then recruits the adapter protein TRAF3, the TBK1 kinase, and the transcription factor IRF3. In this process, TBK1 phosphorylates IRF3 at multiple sites including S386 and S396, which causes IRF3 dimerization and translocation into the nucleus, leading to the ultimate induction of downstream cytokines and other antiviral effector genes.13
Upon infection of DNA virus, the invaded cytosolic viral DNA is sensed by the cyclic GMP-AMP (cGAMP) synthase (cGAS).14,15 cGAS can also recognize mis-located cellular genomic DNA or mitochondrial DNA released into the cytosol upon mitostress triggered by viral infection or other stress conditions.16,17 Upon sensing of DNA, cGAS utilizes GTP and ATP as substrates to synthesize cGAMP, which binds to the ER-located adapter protein MITA (also called STING).18–20 This triggers the translocation of MITA from the ER via Golgi apparatus to perinuclear punctate structures. In this process, MITA recruits TBK1 and IRF3, leading to IRF3 activation and induction of downstream antiviral genes.20–22
As a central transcriptional factor for induction of innate antiviral response, IRF3 needs to be fully activated in the early phase of viral infection, and then timely terminated to avoid excessive and harmful innate immune response.23,24 So far, various post-translational modifications, such as ubiquitination, sumoylation, and acetylation, have been reported to restrain IRF3 activity.25–27 In contrast, how IRF3 is optimally activated upon viral infection is not fully understood.
Viral infection not only triggers innate antiviral response, but also alters metabolic homeostasis in hosts.28–30 For example, viral infection changes the expression levels of rate-limiting enzymes involved in oxidative phosphorylation, sterol biosynthesis and redox homeostasis, leading to subsequent changes of various metabolic reactions in infected cells.28–30 Viral infection also causes changes of serum levels of neuroendocrine and other hormones, thereby affecting various physiological processes of the host.31–33 In recent years, several studies have revealed the mechanisms on how innate antiviral response is regulated by cellular metabolism such as cholesterol depletion,34 bile acid synthesis,35 lactate accumulation,36 and lipid oxidation.37 So far, how humoral metabolism regulates innate antiviral immunity is largely unknown.
Progesterone is an important humoral steroid hormone in mammals.38 In addition to its role in maintaining pregnancy of females, progesterone is also produced and functions in males. It has been reported that progesterone regulates neural development and T-cell differentiation regardless of genders.39–42 Progesterone is biosynthesized in specialized tissues, including adrenal gland and reproductive organs, and progesterone production is tightly regulated by the neuroendocrine hypothalamic-pituitary-adrenal (HPA) axis.43,44 Though progesterone is produced in specialized cells and tissues, it targets various types of cells in different tissues and organs to exert its diverse biological roles. Progesterone signals through PGR, a nuclear hormone receptor that directly induces transcription of downstream effector genes.45 In addition to its function as a nuclear hormone receptor, PGR can activate the proto-oncogene tyrosine kinase SRC rapidly in the cytoplasm of breast cancer and mammary epithelial cells upon progesterone stimulation.46–48
In this study, we identified progesterone as the mostly elevated humoral steroid hormone in virus-infected mice. Our results indicate that progesterone promotes innate antiviral response in vitro and in vivo. Mechanistically, the progesterone-PGR axis activates SRC, which then mediates phosphorylation of IRF3 at Y107. This phosphorylation of IRF3 primes for its association with TBK1 and self-association, leading to its activation and induction of downstream antiviral genes. Additionally, our studies also indicate that SARS-CoV-2-infected patients have increased serum progesterone levels, which are oppositely co-related with severity of the disease. Our findings reveal mechanisms on how progesterone metabolism modulates host innate antiviral response, and point to progesterone as a potential immunomodulatory reagent for infectious and inflammatory diseases.
Results
Viral infection increases serum progesterone level via the HPA axis in mice
How innate antiviral immune response is regulated by humoral metabolites is largely unknown. Therefore, we performed a series of targeted metabolomic analysis of the sera from both control and virus-infected mice by liquid chromatography mass spectrometry. In these experiments, we used male mice due to their steadier humoral metabolic homeostasis which is affected by the menstrual cycle in female mice.49 These experiments led to our identification of progesterone (P4) as the mostly elevated steroid hormone in the sera of mice infected by Sendai virus (SeV), which is a RNA virus (Fig. 1a and Fig. S1a). We confirmed the increase of P4 in the sera of mice infected with SeV as well as the RNA virus encephalomyocarditis virus (EMCV) and the DNA virus herpes simplex virus-1 (HSV-1) (Fig. 1b), suggesting that humoral progesterone levels in mice are increased following infection of different types of viruses.
Fig. 1.
Viral infection increases progesterone level in cells and mice. a Targeted metabolomic analysis of steroid hormones in sera of SeV-infected mice. C57BL/6 male mice (n = 6 in each group) were left un-infected or infected intravenously with SeV for 3 h before the sera were collected and target metabolomic analysis was performed by liquid chromatography mass spectrometry (LC-MS). The data were then displayed by heatmap (the left panel), and the individual serum progesterone (P4) intensity in each sample was shown in the right histogram. MRM, multi reaction monitoring. b Measurement of neuroendocrine hormones in mice infected with different types of viruses. C57BL/6 male mice (n = 4 in each group) were left uninfected or infected intravenously with SeV for 3 h, EMCV for 2 h or HSV-1 for 6 h before sera were collected for measurements of progesterone (P4), GnRH and LH as indicated by ELISA assays. c Measurement of the mRNA levels of Cyp11a1 and Hsd3b2 genes. C57BL/6 male mice (n = 7 in each group) were left uninfected or infected intravenously with SeV for 3 h or HSV-1 for 6 h before their adrenal glands were collected for qPCR analysis of mRNA levels of Cyp11a1 and Hsd3b2 genes. d Measurement of the mRNA levels of CYP11A1 and HSD3B2 in human progesterone-producing cells. JEG-3 cells were left uninfected or infected with the indicated viruses for the indicated times before qPCR analysis of the mRNA levels of CYP11A1 and HSD3B2 genes. e. Effects of SeV infection on progesterone secretion in JEG-3 cells. JEG-3 cells were cultured in basic RPMI medium and then left uninfected or infected with SeV for the indicated times. The culture media were then collected for progesterone measurement by ELISA assays. *P < 0.05; **P < 0.01; ***P < 0.001; ns not significant
In mammals, secretion of P4 is regulated by the neuroendocrine HPA axis.43,44 The hypothalamus-secreted gonadotropin-releasing hormone (GnRH) stimulates the pituitary to produce luteinizing hormone (LH), which then targets the adrenal and reproductive organs to promote transcription of CYP11A1, the rate-limiting metabolic enzyme for P4 biosynthesis. We examined the levels of GnRH and LH in sera of both control and virus-infected mice, and found that viral infection increased the levels of both GnRH and LH (Fig. 1b). Additionally, the mRNA level of Cyp11a1 but not Hsd3b2 (a non-rate-limiting metabolic enzyme for P4 biosynthesis) gene was increased in the adrenal gland of virus-infected in comparison to control mice (Fig. 1c and S1b). In the same experiments, the mRNA levels of enzyme genes involved in biosynthesis of other sex steroid hormones was also examined50,51 (Fig. S1b). The results showed that transcription of Cyp17a1 (the rate-limiting metabolic enzyme for testosterone and estradiol biosynthesis) gene was induced upon viral infection (Fig. S1c), but the mRNA level of Cyp19a1 (another rate-limiting metabolic enzyme for estradiol biosynthesis) gene was hardly affected by virus (Fig. S1c). Interestingly, the mRNA level of Hsd17b1 gene (a non-rate-limiting metabolic enzyme for both testosterone and estradiol biosynthesis) was markedly down-regulated following viral infection (Fig. S1c), which might account for the unchanged serum levels of testosterone and estradiol in SeV-infected mice (Figs. 1a and S1a). Furthermore, we found that viral infection had no marked effects on transcription of CYP11A1 and HSD3B2 genes (Fig. 1d) as well as P4 production (Fig. 1e) in cultured chorionic epithelial cell line JEG-3, which naturally produces P4.52 In these experiments, viral infection induced transcription of IFNB1 gene in these cells (Fig. 1d), suggesting that the cells are successfully infected. Taken together, these results suggest that viral infection does not directly induces P4 production but increases its humoral levels via the HPA axis in mice.
The progesterone-PGR axis modulates innate antiviral response in vivo
Since viral infection causes increase of humoral progesterone, we investigated whether progesterone regulates innate antiviral response in vivo. We found that administration of exogenous P4 by tail vein injection potentiated both SeV- and EMCV-induced serum production of IFN-β, a central cytokine in innate antiviral immune response (Figs. 2a and S2a). In the same experiments, serum TNFα and IL-6 in virus-infected mice were not markedly changed by P4 treatment (Fig. S2b–d), suggesting that P4 has a selective role in promoting IFN-β production. Exogenous P4 administration dramatically suppressed SeV and EMCV replication in mice (Fig. 2b). Injection of mice with an antagonistic antibody for type I IFN receptor IFNAR-1 potentiated viral replication and rescued P4-induced inhibition of viral replication (Fig. 2b). These results suggest that P4 inhibits viral replication in mice in a type I IFN-dependent manner. In addition, exogenous P4 protected mice from EMCV-induced death (Fig. 2c). These results suggest that progesterone promotes innate antiviral response in mice.
Fig. 2.
The progesterone-PGR axis is required for efficient innate antiviral response in vivo. a Effects of progesterone on virus-induced IFN-β production in the sera of mice. Mice (male, n = 6 in each group) were injected intravenously with the indicated doses of P4 for half an hour, and then infected with SeV or EMCV (1 × 108 pfu) for 6 h before measurement of serum IFN-β by ELISA assays. b Effects of progesterone on viral replication in mice. Mice (8-week-old male, n = 3) were intraperitoneally (i.p.) treated with a neutralizing antibody (nAb) against IFNAR-1 (500 μg/mouse) or PBS, and then injected intravenously with DMSO or P4 (30 μg/kg) for half an hour, followed by intraperitoneal injection of SeV or EMCV (1 × 108 pfu) and repeated antibody treatments as described in the Materials and Methods section. Viral genomic copy numbers in the spleens of SeV-infected mice or the brains of EMCV-infected mice were quantified by qPCR. c Effects of progesterone on virus-induced death of mice. Mice (8-week-old male, n = 6) were injected intravenously with DMSO or P4 (30 μg/kg) for half an hour, and then infected intraperitoneally with EMCV (1 × 107 pfu). The survival rates were monitored for twelve days. d Effects of Pgr-deficiency on virus-induced production of serum cytokines. Pgr+/+ and Pgr−/− male mice were infected intravenously with SeV (n = 6 mice in each group) or EMCV (1 × 108 pfu, n = 4 mice in each group) for 6 h before serum cytokines were measured by ELISA. e Effects of Pgr-deficiency on EMCV-induced death of mice. Pgr+/+ and Pgr−/− mice (n = 8 per strain, 8-week-old male) were infected intraperitoneally with EMCV (1 × 107 pfu per mice), and the survival rates of mice were monitored for ten days. f Progesterone levels in pregnant mice. Sera from control and two-week pregnant female mice (n = 4 in each group) were collected for progesterone measurement by ELISA assays. g Levels of serum IFN-β and CXCL10 in virus-infected pregnant mice. The control and two-week pregnant female mice (n = 4 in each group) were infected intravenously with SeV for the indicated times before serum IFN-β and CXCL10 were measured by ELISA assays. h The mRNA levels of antiviral genes in different tissues of virus-infected pregnant mice. The control and two-week pregnant female mice (n = 4 in each group) were infected intravenously with SeV for 6 h, and then the indicated tissues were collected for qPCR analysis of mRNA levels of the indicated genes. *P < 0.05; **P < 0.01; ***P < 0.001; ns not significant
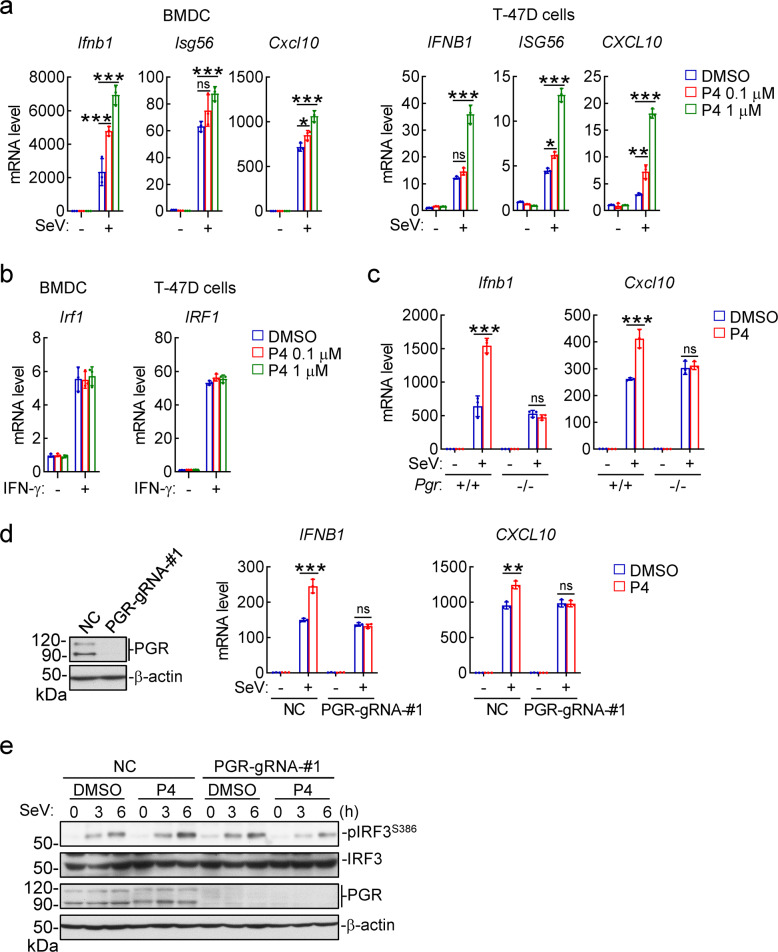
Progesterone exerts most of its biological functions via the progesterone receptor (PGR) in mammals.53 To investigate whether endogenous progesterone-PGR axis plays a role in innate antiviral response in vivo, we generated Pgr-knockout mice (Fig. S3a) and confirmed by both genotyping (Fig. S3b) and qPCR analysis of Pgr mRNA in mouse primary cells including bone marrow-derived dendric cells (BMDCs) and bone marrow-derived macrophages (BMDMs) (Fig. S3c). These experiments also indicated that Pgr mRNA was detected in BMDCs but not BMDMs (Fig. S3c). Furthermore, we observed decrease of serum P4 level in Pgr-knockout mice regardless of the genders (Fig. S3d), suggesting that PGR is involved in transcriptional regulation of genes related to P4 biosynthesis.54,55 Next, we found that Pgr-deficiency markedly inhibited serum levels of IFN-β induced by either SeV or EMCV in male mice (Fig. 2d), though the basal serum levels of IFN-β were comparable between wide-type and Pgr-knockout male and female mice (Fig. S3e). Pgr-deficiency also caused an earlier on-set of death and a lower survival rate of mice infected with EMCV (Fig. 2e). These results suggest that the progesterone-PGR axis plays an important role in efficient host defense against viral infection in vivo.
High humoral progesterone is a biomarker for pregnancy of females.56 We examined the potential difference in innate antiviral immunity between pregnant and non-pregnant mice after viral infection. As expected, high serum progesterone levels were identified in pregnant (Yes) but not non-pregnant (No) mice (Fig. 2f). In addition, the levels of serum IFN-β and CXCL10 induced by SeV infection in pregnant mice were markedly higher than in non-pregnant mice (Fig. 2g). The mRNA levels of downstream antiviral genes Isg56 and Cxcl10 in the livers and adrenal glands of pregnant mice were markedly higher than that of non-pregnant mice after SeV infection (Fig. 2h). These results suggest that pregnant mice have heightened innate antiviral response.
The progesterone-PGR axis modulates innate antiviral response via SRC activation
We next investigated how progesterone promotes innate antiviral response. We found that P4 treatment dose-dependently potentiated SeV-induced transcription of antiviral IFNB1, ISG56 and CXCL10 genes in mouse primary bone marrow-derived dendric cells (BMDCs) and human breast cancer T-47D cells (Fig. 3a), both expresses PGR (Fig. S3c).57 In similar experiments, P4 had no marked effects on IFN-γ-induced transcription of IRF1 gene in these cells (Fig. 3b). Knockout of PGR impaired the ability of P4 to potentiate SeV-induced transcription of IFNB1 and CXCL10 genes in both BMDCs (Fig. 3c) and T-47D cells (Fig. 3d). In addition, P4 treatment potentiated SeV-induced phosphorylation of IRF3 at Ser386 (p-IRF3S386, a hallmark for its activation) in control but not PGR-knockout T-47D cells (Fig. 3e). These results suggest that the progesterone-PGR axis promotes innate antiviral signaling in both human and mouse cells.
Fig. 3.
The progesterone-PGR axis promotes innate antiviral signaling. a Effects of progesterone on transcription of antiviral genes. BMDCs or T-47D cells were treated with the indicated doses of P4 for 1 h and then infected with SeV for 6 h before qPCR analysis of mRNA levels of the indicated antiviral genes. b Effects of progesterone on IFN-γ-induced transcription of IRF1 gene. BMDCs or T-47D cells were treated with the indicated doses of P4 for 1 h and then stimulated with IFN-γ (100 ng/ml) for 6 h before qPCR analysis of IRF1 mRNA level. c Effects of progesterone on transcription of antiviral genes in Pgr+/+ and Pgr−/− BMDCs. Pgr+/+ and Pgr−/− BMDCs were treated with P4 (1 μM) for 1 h and then infected with SeV for 6 h before qPCR analysis of mRNA levels of the indicated genes. d Effects of progesterone on transcription of antiviral genes in PGR-knockout cells. The control and PGR-knockout T-47D cells were treated with P4 (1 μM) for 1 h and then infected with SeV for 6 h before qPCR analysis of mRNA levels of the indicated genes. e Effects of progesterone treatment on virus-induced activation of IRF3 in control and PGR-knockout T-47D cells. The control and PGR-knockout T-47D cells were treated with P4 (1 μM) for 1 h, and then left uninfected or infected with SeV for the indicated times before immunoblotting analysis with the indicated antibodies. Data shown in a-d are mean ± SD (n = 3) from one representative experiment, which was repeated for at least two times with similar results. *P < 0.05; **P < 0.01; ***P < 0.001
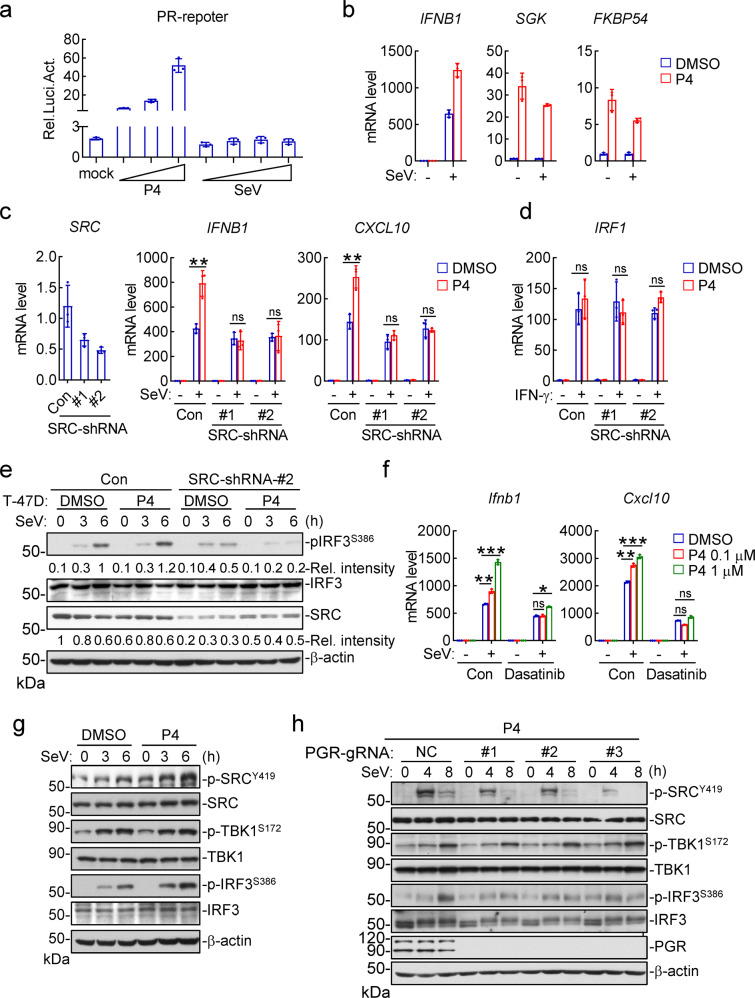
It has been well established that PGR acts either as a nuclear steroid receptor for transcriptional induction or as a cytoplasmic activator of the tyrosine kinase SRC.45,46 In reporter assays, P4 activated transcription driven by the PGR-responsive element (PR) in a dose-dependent manner (Fig. 4a). In similar experiments, SeV infection had no marked effects on PR activity (Fig. 4a). Consistently, P4 treatment potentiated SeV-induced transcription of IFNB1 gene, whereas SeV infection failed to potentiate P4-induced transcription of nuclear PGR-targeted genes including SGK and FKBP5454,58,59 (Fig. 4b). These results suggest that viral infection has no marked effects on the transcriptional activity of PGR.
Fig. 4.
The progesterone-PGR axis promotes innate antiviral response via activation of SRC. a Effects of progesterone or viral infection on PR activation. The PGR-overexpressed HEK293 cells were transfected with PR reporter for 24 h, and then left untreated or treated with increased doses of progesterone (0.01, 0.1, 1 μM) or infected with different doses of SeV for 10 h before luciferase assays. b Effects of progesterone and viral infection on transcription of nuclear PGR-targeted genes. T-47D cells were treated with DMSO or P4 (1 μM) for 1 h and then left uninfected or infected with SeV for 6 h before qPCR analysis of mRNA levels of the indicated genes. c Effects of progesterone on virus-induced transcription of antiviral genes in SRC-knockdown cells. The control and SRC-knockdown T-47D cells were treated with P4 (1 μM) for 1 h and then left uninfected or infected with SeV for 6 h before qPCR analysis of mRNA levels of the indicated genes. The knockdown efficiency of SRC was shown by qPCR analysis of mRNA level in the left panels. d Effects of progesterone on IFN-γ-induced transcription of IRF1 genes in SRC-knockdown cells. The control and SRC-knockdown T-47D cells were treated with P4 (1 μM) for 1 h and then left untreated or treated with IFN-γ (100 ng/ml) for 6 h before qPCR analysis of IRF1 mRNA level. e Effects of progesterone on virus-induced phosphorylation events in SRC-knockdown T-47D cells. The control and SRC-knockdown T-47D cells were treated with P4 (1 μM) for 1 h, and then left uninfected or infected with SeV for the indicated times before immunoblotting analysis with the indicated antibodies. The relative intensities of p-IRF3S386 and SRC (normalized to β-actin) were analyzed by ImageJ. f Effects of SRC inhibitor on progesterone-potentiated transcription of antiviral genes. BMDCs were pretreated with DMSO or Dasatinib (SRC inhibitor, 10 μM) and together with the indicated doses of P4 for 1 h. The cells were then left uninfected or infected with SeV for 6 h before qPCR analysis of the mRNA levels of Ifnb1 and Cxcl10 genes. g Effects of progesterone on virus-induced activation of SRC, TBK1 and IRF3. T-47D cells were treated with P4 (1 μM) for 1 h, and then left un-infected or infected with SeV for the indicated times before immunoblotting analysis with the indicated antibodies. h Effects of PGR-deficiency on SeV-induced activation of SRC, TBK1, and IRF3. The control and PGR-KO T-47D cells were cultured in P4-containing medium, and then left uninfected or infected with SeV for the indicated times before immunoblotting analysis with the indicated antibodies. Data shown in a-d, f are mean ± SD (n = 3) from one representative experiment, which was repeated for at least two times with similar results. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant
We next examined whether PGR regulates innate antiviral response via its downstream tyrosine kinase SRC.46 We found that P4 potentiated SeV-induced transcription of IFNB1 and CXCL10 genes in control but not SRC-knockdown T-47D cells (Fig. 4c). In similar experiments, P4 had no marked effects on IFN-γ-induced transcription of IRF1 in these cells (Fig. 4d). Consistently, P4 potentiated SeV-induced phosphorylation of IRF3S386 in control but not SRC-knockdown T-47D cells (Fig. 4e). Inhibition of SRC kinase activity with a selective inhibitor Dasatinib60 impaired the potentiation effects of P4 on SeV-induced transcription of the antiviral genes Ifnb1 and Cxcl10 in BMDCs (Fig. 4f). In addition, we found that SeV infection induced phosphorylation of SRC at Y419 (p-SRCY419, a hallmark for its activation) to a limited degree, which was potentiated by P4 treatment in T-47D cells (Fig. 4g). In these experiments, SeV-induced phosphorylation of IRF3S386 was also potentiated by P4 (Fig. 4g). Furthermore, knockout of PGR inhibited phosphorylation of SRCY419 and IRF3S386 induced by SeV infection and P4 stimulation in T-47D cells (Fig. 4h). Coimmunoprecipitation experiments indicated that endogenous PGR was weakly associated with SRC in P4- but not DMSO-treated T-47D cells, which was further enhanced after SeV infection (Fig. S4). Taken together, these results suggest that P4 induces PGR-dependent activation of SRC, which promotes virus-triggered activation of IRF3 as well as induction of downstream antiviral genes.
SRC mediates tyrosine phosphorylation of IRF3 to promote its activation
In the above experiments, we noticed that SeV-induced phosphorylation of IRF3S386 but not phosphorylation of TBK1S172 (p-TBK1S172, a hallmark for TBK1 activation) was increased by P4 treatment (Fig. 4g) or decreased by knockout of PGR (Fig. 4h). Since TBK1 acts at upstream of IRF3 in innate antiviral signaling, these results suggest that the progesterone-PGR-SRC axis regulates innate antiviral signaling at IRF3 level. Endogenous coimmunoprecipitation experiments indicated that viral infection induced association of IRF3 with SRC, and P4 treatment further enhanced their association (Fig. 5a). In addition, overexpression of SRC catalyzed tyrosine phosphorylation of IRF3 (Fig. 5b). We next analyzed potential phosphorylation sites of IRF3 in both SRC-overexpressing and control cells by mass spectrometry. The results indicated that Y107 and Y292 of human IRF3 were two candidate residues targeted by SRC (Fig. S5a), and sequence alignments indicated that Y107 but not Y292 is conserved from murine to human (Fig. S5b). Mutation of IRF3 Y107 to phenylalanine (F) impaired SRC-mediated tyrosine phosphorylation of IRF3 in mammalian overexpression system (Fig. 5c). We made an antibody specific for Y107-phosphorylated IRF3 (Fig. S5c). With this antibody, we detected a signal of p-IRF3Y107 when wild-type human IRF3 but not IRF3Y107F was co-transfected with SRC (Fig. 5d). In these experiments, SRC also caused phosphorylation of S386 in wild-type IRF3 but not IRF3Y107F (Fig. 5d), indicating that SRC-mediated tyrosine phosphorylation of IRF3Y107 is required for its activation. Furthermore, P4 increased phosphorylation of endogenous IRF3Y107 and SeV-induced phosphorylation of IRF3S386, which was not seen in SRC-knockdown T-47D cells (Fig. 5e). Functionally, overexpression of wild-type IRF3 potentiated SeV-induced activation of the IFN-β promoter, whereas overexpression of IRF3Y107F inhibited it (Fig. 5f). P4 treatment also potentiated SeV-induced activation of the IFN-β promoter, whereas overexpression of IRF3Y107F inhibited it (Fig. 5g). Collectively, these results suggest that the P4-PGR-SRC axis mediates tyrosine phosphorylation of IRF3Y107 to promote its activation upon viral infection.
Fig. 5.
SRC mediates phosphorylation of IRF3 at Y107 and its activation. a Effects of progesterone on endogenous association of IRF3 with SRC. T-47D cells were treated with P4 (1 μM) for 1 h, and then left uninfected or infected with SeV for the indicated times before coimmunoprecipitation and immunoblotting analysis with the indicated antibodies. b Effects of SRC on tyrosine phosphorylation of IRF3. HEK293 cells were transfected with the indicated plasmids for 24 h before coimmunoprecipitation and immunoblotting analysis with the indicated antibodies. c SRC does not mediate IRF3Y107F phosphorylation. HEK293 cells were transfected with the indicated plasmids for 24 h before coimmunoprecipitation and immunoblotting analysis with the indicated antibodies. d SRC mediates phosphorylation of IRF3Y107. HEK293 cells were transfected with the indicated plasmids for 24 h. The cell lysates were denatured by 1% SDS, which were then diluted with regular lysis buffer till the final concentration of SDS reached to 0.1%. The lysates were then immunoprecipitated with a Flag antibody, and then the immunoprecipitation were analyzed by immunoblots with the indicated antibodies. e Effects of progesterone on virus-induced phosphorylation of IRF3Y107 in SRC-knockdown cells. The control and SRC-knockdown T-47D cells were treated with P4 (1 μM) for 1 h, and then left uninfected or infected with SeV for the indicated times before immunoblotting analysis with the indicated antibodies. f Effects of IRF3 and IRF3Y107F on SeV-induced activation of the IFN-β promoter. HEK293 cells were transfected with the indicated plasmids for 24 h, and then infected with SeV for 12 h before luciferase assays. Data shown are mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. g Effects of IRF3Y107 mutant on progesterone-potentiated activation of the IFN-β promoter. The PGR-overexpressed HEK293 cells were transfected with the indicated plasmids for 24 h, and then left treated or untreated with P4 (1 μM) for 1 h followed by SeV infection for 12 h before luciferase assays. Data shown are mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. h. Effects of IRF3 Y107 mutation on its association with VISA and TBK1 or self-association. HEK293 were transfected with the indicated plasmids for 24 h before coimmunoprecipitation and immunoblotting analysis with the indicated antibodies. i Effects of IRF3 Y107 mutation on its dimerization. HEK293 were transfected with the indicated plasmids for 24 h, and then left treated or untreated with P4 (1 μM) for 1 h followed by SeV infection for 6 h. The cells were lysed for native PAGE and immunoblotting analysis with the indicated antibodies
We further investigated how phosphorylation of IRF3Y107 regulates its activity. Structural studies have shown that IRF3 is consisted of the N-terminal DNA-binding domain (DBD, aa1-104), a middle flexible linker (MFL, aa105-198), an IRF3-associated domain (IAD, aa199-375) and the C-terminal repressor domain (RD, aa376-427).61 It has also been demonstrated that IRF3 is activated via three steps: unlock of its intramolecular autoinhibition to associate with other signal molecules (eg, VISA and TBK1), its dimerization and translocation into nucleus for transcriptional induction of antiviral genes.62 Since Y107 is located at the adjacent site of DBD and MFL of IRF3, we hypothesized that phosphorylation of IRF3Y107 may cause its conformational changes which affect its activation. Coimmunoprecipitation experiments showed that IRF3Y107F lost its ability to associate with VISA and TBK1 (Fig. 5h). IRF3Y107F also had dramatically reduced ability to self-associate in comparison to wild-type IRF3 (Fig. 5h). Consistently, IRF3Y107F had reduced dimerization after SeV infection either with or without P4 treatment (Fig. 5i). These results suggest that phosphorylation of IRF3Y107 unlocks its intramolecular autoinhibition and promotes its activation. Additionally, we found that SRC interacted with the C-terminus (aa141-427) of IRF3 by domain mapping experiments (Fig. S6a). Based on our study and the previous reports on the VISA-TBK1-IRF3 complex,63,64 we propose a model on how SRC-mediated tyrosine phosphorylation of IRF3 at Y107 unlocks its intermolecular autoinhibition and primes its activation (Fig. S6b).
Progesterone levels in SARS-CoV-2-infected patients
Recently, it has been reported that pregnant women have increased rates of progressing into severe COVID-19 symptoms after SARS-CoV-2 infection.65 We collected and analyzed sex hormone-testing reports of SARS-CoV-2-infected male patients (n = 104) and the healthy males (n = 107). As shown in Fig. 6a, progesterone levels (1.93 ± 0.99 ng/mL) in SARS-CoV-2-infected patients were ~3-fold higher to the healthy individuals (0.76 ± 0.34 ng/mL), whereas the other tested sex hormones including testosterone (T) and estradiol (E2) were comparable between the two groups (Fig. 6a). SARS-CoV-2 infection increased serum P4 levels in all age groups (Fig. 6b). In addition, patients infected by SARS-CoV-2 had increased serum LH levels (Fig. 6c), which was similar to our observation in virus-infected mice (Fig. 1c). Furthermore, statistical results showed that patients with severe symptoms (n = 13) displayed significant lower levels of serum P4 than those with moderate symptoms (n = 89) (Fig. 6d), suggesting a negative correlation between serum P4 level and the severity of COVID-19 symptoms. These results are consistent with an important role of progesterone in host innate antiviral response.
Fig. 6.
SARS-CoV-2 infection increases serum progesterone levels in COVID-19 patients. a–c Analysis of sex-related hormones in healthy individuals and SARS-CoV-2-infected patients. The measurements of testosterone (T), estradiol (E2) and progesterone (P4) are shown in (a). The age-classified data of P4 are shown in (b). The measurements of LH and FSH levels are shown in (c). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. d Correlation analysis between progesterone levels and severity of symptoms of COVID-19. e Effects of progesterone on replication of SARS-CoV-2 in Calu-3 cells. Calu-3 cells were treated with P4 (1 μM) for 1 h and then infected with SARS-CoV-2 (MOI = 0.01) for 48 h. Total RNAs in both cell lysate and the supernatant was collected to detect the envelope (e) and nucleocapsid (N) RNA of SARS-CoV-2 via TaqMan-qRT-PCR. f Effects of progesterone on replication of SARS-CoV-2 in mice. The K18-hACE2 transgenic mice (n = 3 in each group, 8-week-old male) were injected intraperitoneally (i.p.) with DMSO or P4 (30 μg/kg) for half an hour, and then intranasally infected with 250 PFU of SARS-CoV-2 in a total volume of 50 μl DMEM. Mice were then continued to be administered with DMSO or P4 daily until five days post-challenge. Viral genomic copy numbers in the lungs of infected mice were quantified by qPCR. g A work model for modulation of innate antiviral response by humoral progesterone
To confirm the potential role of P4 in modulating SARS-CoV-2 infection, we examined the effects of P4 treatment on SARS-CoV-2 replication by detecting the copy numbers of the viral genome with two specific probes for either envelope (E) or nucleocapsid (N) RNA of SARS-CoV-2.66 The results showed that P4 treatment markedly down-regulated the copy numbers of SARS-CoV-2 in cultured Calu-3 lung epithelial cells (Fig. 6e). Administration of P4 also down-regulated SARS-CoV-2 replication in the lung of K18-hACE2 transgenic mice (Fig. 6f). These results suggest that P4 has an inhibitory role on SARS-CoV-2 replication.
Discussion
Several studies have revealed that viral infection causes changes of intracellular metabolic pathways, which in turn regulate innate antiviral response.34–37 However, how humoral metabolism regulates host innate antiviral response remains enigmatic. In this study, we found that viral infection induces humoral progesterone, which plays a critical role in promoting efficient innate antiviral response.
To investigate whether and how humoral hormones regulate innate antiviral response, we performed targeted metabolomics analysis of the sera of infected mice. These efforts identified progesterone as the mostly increased among tested humoral hormones after viral infection in mice. Viral infection also caused increases of GnRH and LH, which acts upstream to regulate the level of progesterone in vivo.67,68 Analysis of the sex-related hormone-testing reports of SARS-CoV-2-infected patients and healthy individuals indicated that SARS-CoV-2 infection also increased serum progesterone and LH levels. These results suggest that increase of humoral progesterone level via the HPA axis following viral infection is a conserved biological function from mouse to human. Future studies are needed to determine how viral infection activates the HPA axis. It is possible that the HPA axis is directly activated by viral infection of certain peripheral or central neurons in hypothalamus, or indirectly activated by virus-induced effects.
By studying with both mouse and human cells, as well as gene-knockout mice, we demonstrate that humoral progesterone facilitates innate antiviral response, and progesterone inhibits viral replication in a type I IFN-dependent manner and protects mice from virus-induced death. Several lines of evidences suggest that progesterone promotes innate antiviral response via PGR-mediated activation of SRC, which subsequently phosphorylates and activates IRF3 as illustrated in Fig. 6g. Firstly, either PGR- or SRC-deficiency impaired progesterone-triggered potentiation of IRF3 activation and antiviral gene expression following viral infection. Second, SRC interacted with and mediated phosphorylation of IRF3Y107. Third, phosphorylation of IRF3Y107 by SRC was important for IRF3 association with TBK1 and its self-association, as well as induction of IFN-β upon viral infection. Sequence analysis suggests that IRF3 Y107 is conserved from mouse to human, and located at the junction of the N-terminal DNA-binding domain and the middle flexible linker of IRF3. It is possible that phosphorylation of IRF3Y107 causes its conformational changes which unlock its autoinhibition and promote its phosphorylation at S386/S396 and activation by TBK1. Our current study suggests for the first time that phosphorylation of IRF3Y107 is essential for priming its activation. We noticed that IRF3Y107 is basally phosphorylated in un-infected cells and which is increased following viral infection and progesterone stimulation. It is possible that other stimuli that can activate SRC are also able to modulate innate antiviral response.
In our study, we found that except for the RNA viruses SeV and EMCV, the DNA virus HSV-1 also induced progesterone. In addition, progesterone also potentiated HSV-1-induced transcription of downstream antiviral genes. This is consistent with the notion that IRF3 is also a critical mediator of innate immune response to DNA virus, and suggests that progesterone modulates innate immune response to both RNA and DNA viruses. Our findings reveal a conserved and common regulatory mechanism of innate antiviral response by humoral progesterone. Systemic lupus erythematosus (SLE) is characterized by multi-system immune-mediated injury in the setting of autoimmunity to nuclear antigens with strong female bias.69,70 Since enhanced type I IFN signal is associated with SLE, it would be interesting to explore whether progesterone-mediated potentiation of type I IFN production is related to the higher risk of SLE in females.71
Sex differences in COVID-19 vulnerability and clinical manifestations have been observed by various studies. It has been well established that SARS-CoV-2-infected males have significantly higher rates of progressing into severe diseases, and male-to-female fatality ratios obtained from different countries are between 1.7-1.8.72,73 Notably, the male fatality predominance is mostly pronounced in the middle age group and linearly declined in the higher age groups,72 suggesting that the potential female’s protective factors for COVID-19 decline during aging. Our discovery that progesterone is induced following viral infection and which promotes innate antiviral response provides an explanation on the observations that SARS-CoV-2-infected females have reduced severity and fatality in comparison to males. Our study indicates that the progesterone levels are negatively co-related with the severity of COVID-19. It is possible that heightened innate immune response in SARS-CoV-2-infected patients with higher levels of progesterone contributes to the alleviated symptoms of COVID-19. It has been reported that pregnant women, who have a very high level of progesterone, have an increased requirement of intensive care unit admission after infection of SARS-CoV-2.65 Previously, it has been demonstrated that high level of progesterone has anti-inflammatory effects by inhibiting NF-κB pathways and T-cell-mediated adaptive immune responses.42,74 Since viral diseases are caused by both viral factors and improperly regulated inflammatory/immune responses, and progesterone has pleiotropic roles in modulating physiological functions and immunological responses, the eventual beneficial or harmful effects of progesterone on viral-infected patients may be dictated by its levels, host status and timing of viral infection. Nevertheless, our study suggests that progesterone may serve as a potential immunomodulatory agent for infectious diseases such as COVID-19 by both promoting innate antiviral response and restraining excessive inflammatory response.
Materials and methods
Reagents, antibodies, viruses, and cells
Progesterone (HY-N0437) and Dasatinib (HY-10181) (MedChemExpress); IFN-γ (300-02, Peptech); Anti-mouse IFNAR-1-in vivo (clone MAR1-5A3; Selleck A2121); ELISA kits for progesterone (582601, Cayman Chemical), GnRH (MM-0506M1) and LH (MM-44039M1) (MEIMAIN), murine IFN-β(42400, PBL), murine CXCL10 (EMC121, Neobioscience), murine TNFα (430904, BioLegend), murine IL-6 (431304, BioLegend); Mouse monoclonal antibodies against HA (TA180128, OriGene), Flag (F3165) and β-actin (A2228) (Sigma), IRF3 (sc-33641, Santa Cruz Biotechnology); Rabbit polyclonal antibodies against PGR (8757), SRC (2109), p-SRCY419 (2101) and p-Tyrosine (9411S) (Cell signaling technology), TBK1 (ab40676), p-TBK1S172 (ab109272) and p-IRF3S386 (ab76493) (Abcam) were purchased from the indicated companies. SeV, EMCV and HSV-1 were previously described.75,76 HEK293 cells were obtained from ATCC. The T-47D cells were provided by Dr. Jing Zhang (Wuhan University). The Calu-3 cells were provided by Dr. Ke Lan (Wuhan University).
Constructs
Flag- and HA-tagged PGR, SRC, IRF3 or its mutants; HA-tagged VISA, TBK1, CRISPR-Cas9 gRNA plasmids for PGR, pSuper-Retro-shRNA plasmids for SRC were constructed by standard molecular biology techniques. The target sequences of genes for gRNA and shRNA construction were listed in Table S1.
Mice
Pgr-knockout mice on the C57BL/6 background were generated utilizing the CRISPR/Cas9 method by GemPharmatech. In Pgr−/− mice, the entire fragment containing the first exon of the Pgr gene is removed. The strategy for construction of the targeting vector is illustrated in Supplemental Fig. 1a.
Genotyping by PCR was performed using the following pairs of primers.
Primer pair 1: 5′-ATGACACTCAATGTCCAAACTCCC-3′, 5′-TTCCTCAATGCTGGGTCACAAC-3′;
Primer pair 2: 5′-TAGAGCGCCAACGCTTGCTAGA-3′, 5′-TGGTCAGCTCCTGTCCTTACCCTC-3′.
Amplification of the disrupted allele with Primer pair 1 results in a 442-bp fragment, whereas amplification of the WT allele with Primer pair 2 results in a 510-bp fragment.
Mice were maintained in the special pathogen-free facility of Medical Research Institute at Wuhan University. Six- to eight-week-old male mice were randomly allocated for each experimental group and littermates were used as controls. All animal experiments were performed in accordance with the Wuhan University Medical Research Institute Animal Care and Use Committee guidelines.
K18-hACE2 transgenic mice, which express human ACE2 driven by the human epithelial cell cytokeratin-18 (K18) promoter used as an infection model for both SARS-CoV and SARS-CoV-2,77–79 were purchased from Gempharmatech and housed in ABSL-3 pathogen-free facilities. All animal experiments were approved by the Animal Care and Use Committee of Wuhan University.
Transfection and reporter assays
HEK293 cells were transfected by standard calcium phosphate precipitation method. In reporter assays, pRL-TK (Renilla luciferase) reporter plasmid (0.01 μg) was added to each transfection for normalization of transfection efficiency. Luciferase assays were performed using a Dual-Specific Luciferase Assay Kit (Promega). Firefly luciferase activities were normalized on the basis of Renilla luciferase activities.
qPCR
Total RNA was isolated for qPCR analysis to measure mRNA levels of the indicated genes. Data shown are the relative abundance of the indicated mRNA normalized to that of GAPDH. qPCR primers were listed in Table S1.
Coimmunoprecipitation and immunoblotting analysis
Cells were lysed in NP-40 lysis buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40,10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride). For each immunoprecipitation, a 0.4 ml aliquot of the lysate was incubated with the indicated antibody or control IgG (0.5 μg, or 0.5 μl for antiserum) and 25 μl of a 1:1 slurry of Protein G Sepharose (GE Healthcare) for 2 h. Sepharose beads were washed three times with 1 ml of lysis buffer containing 0.5 M NaCl. Proteins were separated by 8% SDS–PAGE, followed by immunoblotting analysis with the indicated antibodies.
Phosphorylation site identification by mass spectrometry
HEK293 cells were transfected with Flag-IRF3 and HA-SRC or Vector. Flag-IRF3 was immunoprecipitated and desalted. The mass spectrometry analysis was performed as previously described by SpecAlly (Wuhan) Life Science and Technology Company.80
Preparation of anti-phospho-IRF3Y107
Preparation of anti-Phospho-IRF3Y107 was conducted by ABclonal Technology Company. Briefly, a peptide with DPHKI(Y-P)EF-C sequence was synthesized and conjugated to KLH for immunization of the experimental Japanese white-eared rabbits. Rabbit anisera were passed through affinity columns conjugated with the synthesized control peptide (DPHKIYEF-C) for two times. The sera were then purified by affinity columns conjugated with the modified peptide DPHKI(Y-P)EF-C. The final eluted anti-phospho-IRF3Y107 was validated by dot blotting analysis with both the synthesized un-modified and modified peptides.
In vivo IFNAR blockade
For SeV replication experiments, WT mice were intraperitoneally (i.p.) administrated with anti-mouse IFNAR-1 (500 μg/mouse) or PBS, and then injected intravenously with DMSO or P4 (30 μg/kg). Half an hour later, mice were intraperitoneally injected with SeV. Fifteen hours after the first injection of anti-mouse IFNAR-1, the mice were administrated with anti-mouse IFNAR-1 or PBS again (500 μg/mouse). Four hours later, the mice were killed by cervical dislocation for analysis.
For EMCV replication experiments, WT mice were i.p. administrated with anti-mouse IFNAR-1 (500 μg/mouse) or PBS, and then injected intravenously with DMSO or P4 (30 μg/kg). Half an hour later, mice were i.p. injected with EMCV (1 × 108 pfu). The mice were administrated with anti-mouse PBS or anti-mouse IFNAR-1 for two more times, one (500 μg/mouse) or two days (250 μg/mouse) after EMCV infection. The mice were also administrated intravenously with DMSO or P4 (30 μg/kg) for one more time at 2 days after EMCV infection. The mice were killed by cervical dislocation one day later.
Patients
This study was reviewed and approved by the Medical Ethical Committee of Zhongnan Hospital of Wuhan University (Approval# 2020033-1 and 2020068). The clinical sex-related hormone-testing reports were collected from Wuhan Leishenshan Hospital or Zhongnan Hospital. A convenient sampling strategy was employed, in which all testing reports from consecutive patients meeting inclusion criteria were collected for analysis at the day of their initial visits to the hospitals. We collected sex-related hormone-testing reports from 104 reproductive-aged (median age 40 years, range 31–49) male COVID-19 patients, who were admitted to Wuhan Leishenshan Hospital or Zhongnan Hospital from March 5 to March 31, 2020. The control group were from males who were admitted for evaluation of fertilization ability. A total of 107 age-matched men (median age 40, range 32–49) were randomly selected and the data of their sex hormones were obtained. The diagnosis of COVID-19 and the degree of severity (mild, moderate, severe, critical) were determined according to the New Coronavirus Pneumonia Prevention and Control Program (7th edition) published by the National Health Commission of China.
Statistics
For mouse experiments, no specific blinding method was used, but mice in each group were selected randomly. The sample size (n) of each experimental group is described in the corresponding figure legend. GraphPad Prism software was used for all statistical analyses. Quantitative data in histograms are shown as means ± s.d. Data were analyzed using a one-way ANOVA, Student’s unpaired t-test or Log-rank (Mantel-Cox) test. The number of asterisks represents the degree of significance with respect to P values. Statistical significance was set at P < 0.05.
Supplementary information
Acknowledgements
This work was supported by grants from the State Key R&D Program of China (2017YFA0505800 and 2021YFA1302400), the National Natural Science Foundation of China (31922021, 32170713, 31830042, and 31771555), and the CAMS Innovation Fund for Medical Sciences (2019-I2M-5-071).
Author contributions
M.M.H., S.S., and H.B.S. conceived and designed the study; S.S., D.H., L.B.C., X.N.Z., and Y.G. performed the experiments except for the SARS-CoV-2 infection experiments; J.P.L. and M.Z. collected the clinical testing reports and analyzed the data; L.B, J.D., X.L. and K.L. performed the SARS-CoV-2 infection experiments and analyzed the data. M.M.H., S.S., and H.B.S. analyzed all the other data. M.M.H., S.S., and H.B.S. wrote the manuscript. All authors have read and approved the article.
Data availability
All the data shown in this paper are available from the corresponding authors upon reasonable request.
Competing interests
The authors declare no competing interests. H.-B.S. is one of the Associate Editors of Signal Transduction and Targeted Therapy, but he has not been involved in the process of the manuscript handling.
Footnotes
These authors contributed equally: Shan Su, Duo Hua, Jin-Peng Li
Contributor Information
Ming-Ming Hu, Email: mmhu@whu.edu.cn.
Hong-Bing Shu, Email: shuh@whu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41392-022-00981-5.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Hu MM, Shu HB. Cytoplasmic mechanisms of recognition and defense of microbial nucleic acids. Annu Rev. Cell Dev. Biol. 2018;34:357–379. doi: 10.1146/annurev-cellbio-100617-062903. [DOI] [PubMed] [Google Scholar]
- 3.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 4.Hu MM, Shu HB. Innate immune response to cytoplasmic DNA: mechanisms and diseases. Annu. Rev. Immunol. 2020;38:79–98. doi: 10.1146/annurev-immunol-070119-115052. [DOI] [PubMed] [Google Scholar]
- 5.Zevini A, Olagnier D, Hiscott J. Crosstalk between cytoplasmic RIG-I and STING sensing pathways. Trends Immunol. 2017;38:194–205. doi: 10.1016/j.it.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motwani M, Pesiridis S, Fitzgerald KA. DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 2019;20:657–674. doi: 10.1038/s41576-019-0151-1. [DOI] [PubMed] [Google Scholar]
- 7.Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020;20:537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopfner KP, Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 2020;21:501–521. doi: 10.1038/s41580-020-0244-x. [DOI] [PubMed] [Google Scholar]
- 9.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 10.Kang DC, et al. Expression analysis and genomic characterization of human melanoma differentiation associated gene-5, mda-5: a novel type I interferon-responsive apoptosis-inducing gene. Oncogene. 2004;23:1789–1800. doi: 10.1038/sj.onc.1207300. [DOI] [PubMed] [Google Scholar]
- 11.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. Viral RNA detection by RIG-I-like receptors. Curr. Opin. Immunol. 2015;32:48–53. doi: 10.1016/j.coi.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Li XD, et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He WR, et al. VRK2 is involved in the innate antiviral response by promoting mitostress-induced mtDNA release. Cell Mol. Immunol. 2021;18:1186–1196. doi: 10.1038/s41423-021-00673-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ablasser, A & Chen, Z. J. cGAS in action: expanding roles in immunity and inflammation. Science363, eaat8657 (2019). [DOI] [PubMed]
- 18.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobbs N, et al. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe. 2015;18:157–168. doi: 10.1016/j.chom.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo WW, et al. iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nat. Immunol. 2016;17:1057–1066. doi: 10.1038/ni.3510. [DOI] [PubMed] [Google Scholar]
- 23.Chiang C, Gack MU. Post-translational control of intracellular pathogen sensing pathways. Trends Immunol. 2017;38:39–52. doi: 10.1016/j.it.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jefferies CA. Regulating IRFs in IFN driven disease. Front. Immunol. 2019;10:325. doi: 10.3389/fimmu.2019.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang P, Zhao W, Zhao K, Zhang L, Gao C. TRIM26 negatively regulates interferon-beta production and antiviral response through polyubiquitination and degradation of nuclear IRF3. PLoS Pathog. 2015;11:e1004726. doi: 10.1371/journal.ppat.1004726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubota T, et al. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J. Biol. Chem. 2008;283:25660–25670. doi: 10.1074/jbc.M804479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huai W, et al. KAT8 selectively inhibits antiviral immunity by acetylating IRF3. J. Exp. Med. 2019;216:772–785. doi: 10.1084/jem.20181773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varanasi SK, Rouse BT. How host metabolism impacts on virus pathogenesis. Curr. Opin. Virol. 2018;28:37–42. doi: 10.1016/j.coviro.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Thaker SK, Ch’ng J, Christofk HR. Viral hijacking of cellular metabolism. BMC Biol. 2019;17:59. doi: 10.1186/s12915-019-0678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buck MD, Sowell RT, Kaech SM, Pearce EL. Metabolic instruction of immunity. Cell. 2017;169:570–586. doi: 10.1016/j.cell.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayer KA, Stockl J, Zlabinger GJ, Gualdoni GA. Hijacking the supplies: metabolism as a novel facet of virus-host interaction. Front. Immunol. 2019;10:1533. doi: 10.3389/fimmu.2019.01533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu K, et al. SARS-CoV-2 effects in the genitourinary system and prospects of sex hormone therapy. Asian J. Urol. 2021;8:303–314. doi: 10.1016/j.ajur.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rousseau GG. Fifty years ago: the quest for steroid hormone receptors. Mol. Cell Endocrinol. 2013;375:10–13. doi: 10.1016/j.mce.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 34.York AG, et al. Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell. 2015;163:1716–1729. doi: 10.1016/j.cell.2015.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu MM, et al. Virus-induced accumulation of intracellular bile acids activates the TGR5-beta-arrestin-SRC axis to enable innate antiviral immunity. Cell Res. 2019;29:193–205. doi: 10.1038/s41422-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang W, et al. Lactate is a natural suppressor of RLR signaling by targeting MAVS. Cell. 2019;178:176–189. doi: 10.1016/j.cell.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia M, et al. Redox homeostasis maintained by GPX4 facilitates STING activation. Nat. Immunol. 2020;21:727–735. doi: 10.1038/s41590-020-0699-0. [DOI] [PubMed] [Google Scholar]
- 38.Lange CA, Gioeli D, Hammes SR, Marker PC. Integration of rapid signaling events with steroid hormone receptor action in breast and prostate cancer. Annu. Rev. Physiol. 2007;69:171–199. doi: 10.1146/annurev.physiol.69.031905.160319. [DOI] [PubMed] [Google Scholar]
- 39.Bashour NM, Wray S. Progesterone directly and rapidly inhibits GnRH neuronal activity via progesterone receptor membrane component 1. Endocrinology. 2012;153:4457–4469. doi: 10.1210/en.2012-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hierweger AM, et al. Progesterone modulates the T-cell response via glucocorticoid receptor-dependent pathways. Am. J. Reprod. Immunol. 2019;81:e13084. doi: 10.1111/aji.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schumacher M, et al. Revisiting the roles of progesterone and allopregnanolone in the nervous system: resurgence of the progesterone receptors. Prog. Neurobiol. 2014;113:6–39. doi: 10.1016/j.pneurobio.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 42.Lee JH, Ulrich B, Cho J, Park J, Kim CH. Progesterone promotes differentiation of human cord blood fetal T cells into T regulatory cells but suppresses their differentiation into Th17 cells. J. Immunol. 2011;187:1778–1787. doi: 10.4049/jimmunol.1003919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheng JA, et al. The hypothalamic-pituitary-adrenal axis: development, programming actions of hormones, and maternal-fetal interactions. Front. Behav. Neurosci. 2020;14:601939. doi: 10.3389/fnbeh.2020.601939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Handa RJ, Weiser MJ. Gonadal steroid hormones and the hypothalamo-pituitary-adrenal axis. Front. Neuroendocrinol. 2014;35:197–220. doi: 10.1016/j.yfrne.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peluso JJ. Multiplicity of progesterone’s actions and receptors in the mammalian ovary. Biol. Reprod. 2006;75:2–8. doi: 10.1095/biolreprod.105.049924. [DOI] [PubMed] [Google Scholar]
- 46.Boonyaratanakornkit V, et al. Progesterone receptor contains a proline-rich motif that directly interacts with SH3 domains and activates c-Src family tyrosine kinases. Mol. Cell. 2001;8:269–280. doi: 10.1016/S1097-2765(01)00304-5. [DOI] [PubMed] [Google Scholar]
- 47.Skildum A, Faivre E, Lange CA. Progesterone receptors induce cell cycle progression via activation of mitogen-activated protein kinases. Mol. Endocrinol. 2005;19:327–339. doi: 10.1210/me.2004-0306. [DOI] [PubMed] [Google Scholar]
- 48.Castoria G, et al. Non-transcriptional action of oestradiol and progestin triggers DNA synthesis. EMBO J. 1999;18:2500–2510. doi: 10.1093/emboj/18.9.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia NM, Walker RS, Zoellner LA. Estrogen, progesterone, and the menstrual cycle: A systematic review of fear learning, intrusive memories, and PTSD. Clin. Psychol. Rev. 2018;66:80–96. doi: 10.1016/j.cpr.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 50.Kelemen LE, Sellers TA, Vachon CM. Can genes for mammographic density inform cancer aetiology? Nat. Rev. Cancer. 2008;8:812–823. doi: 10.1038/nrc2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sathish V, Martin YN, Prakash YS. Sex steroid signaling: implications for lung diseases. Pharm. Ther. 2015;150:94–108. doi: 10.1016/j.pharmthera.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drwal E, Rak A, Gregoraszczuk E. Co-culture of JEG-3, BeWo and syncBeWo cell lines with adrenal H295R cell line: an alternative model for examining endocrine and metabolic properties of the fetoplacental unit. Cytotechnology. 2018;70:285–297. doi: 10.1007/s10616-017-0142-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jakovac H. Sex differences in COVID-19 course and outcome: progesterone should not be neglected. J. Appl Physiol. 2020;129:1007–1008. doi: 10.1152/japplphysiol.00740.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richer JK, et al. Differential gene regulation by the two progesterone receptor isoforms in human breast cancer cells. J. Biol. Chem. 2002;277:5209–5218. doi: 10.1074/jbc.M110090200. [DOI] [PubMed] [Google Scholar]
- 55.Jacobsen BM, Horwitz KB. Progesterone receptors, their isoforms and progesterone regulated transcription. Mol. Cell Endocrinol. 2012;357:18–29. doi: 10.1016/j.mce.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franco HL, Jeong JW, Tsai SY, Lydon JP, DeMayo FJ. In vivo analysis of progesterone receptor action in the uterus during embryo implantation. Semin Cell Dev. Biol. 2008;19:178–186. doi: 10.1016/j.semcdb.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 57.Szmyd M, et al. The effects of black cohosh on the regulation of estrogen receptor (ERalpha) and progesterone receptor (PR) in breast cancer cells. Breast Cancer (Dove Med Press) 2018;10:1–11. doi: 10.2147/BCTT.S144865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jeong JW, et al. Identification of murine uterine genes regulated in a ligand-dependent manner by the progesterone receptor. Endocrinology. 2005;146:3490–3505. doi: 10.1210/en.2005-0016. [DOI] [PubMed] [Google Scholar]
- 59.Daniel AR, Knutson TP, Lange CA. Signaling inputs to progesterone receptor gene regulation and promoter selectivity. Mol. Cell Endocrinol. 2009;308:47–52. doi: 10.1016/j.mce.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee D, Gautschi O. Clinical development of SRC tyrosine kinase inhibitors in lung cancer. Clin. Lung Cancer. 2006;7:381–384. doi: 10.3816/CLC.2006.n.020. [DOI] [PubMed] [Google Scholar]
- 61.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 62.Takahasi K, et al. X-ray crystal structure of IRF-3 and its functional implications. Nat. Struct. Biol. 2003;10:922–927. doi: 10.1038/nsb1001. [DOI] [PubMed] [Google Scholar]
- 63.Liu S, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 64.Zhao B, et al. Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins. Proc. Natl Acad. Sci. USA. 2016;113:E3403–E3412. doi: 10.1073/pnas.1603269113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khan, D. S. A. et al. The differences in clinical presentation, management, and prognosis of laboratory-confirmed COVID-19 between pregnant and non-pregnant women: a systematic review and meta-analysis. Int. J. Environ. Res. Public Health18, 5613 (2021). [DOI] [PMC free article] [PubMed]
- 66.Bai L, et al. Coinfection with influenza A virus enhances SARS-CoV-2 infectivity. Cell Res. 2021;31:395–403. doi: 10.1038/s41422-021-00473-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zirkin BR, Papadopoulos V. Leydig cells: formation, function, and regulation. Biol. Reprod. 2018;99:101–111. doi: 10.1093/biolre/ioy059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaprara A, Huhtaniemi IT. The hypothalamus-pituitary-gonad axis: tales of mice and men. Metabolism. 2018;86:3–17. doi: 10.1016/j.metabol.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 69.Vincent FB, Morand EF, Schneider P, Mackay F. The BAFF/APRIL system in SLE pathogenesis. Nat. Rev. Rheumatol. 2014;10:365–373. doi: 10.1038/nrrheum.2014.33. [DOI] [PubMed] [Google Scholar]
- 70.Souyris, M. et al. TLR7 escapes X chromosome inactivation in immune cells. Sci. Immunol.3, eaap8855 (2018). [DOI] [PubMed]
- 71.Crow MK, et al. Type I interferons in autoimmune disease. Annu. Rev. Pathol. 2019;14:369–393. doi: 10.1146/annurev-pathol-020117-043952. [DOI] [PubMed] [Google Scholar]
- 72.Gebhard C, Regitz-Zagrosek V, Neuhauser HK, Morgan R, Klein SL. Impact of sex and gender on COVID-19 outcomes in Europe. Biol. Sex. Differ. 2020;11:29. doi: 10.1186/s13293-020-00304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kragholm K, et al. Association between male sex and outcomes of coronavirus disease 2019 (COVID-19)-a Danish Nationwide, Register-based Study. Clin. Infect. Dis. 2021;73:e4025–e4030. doi: 10.1093/cid/ciaa924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cui L, et al. Progesterone inhibits inflammatory response in E. coli- or LPS-stimulated bovine endometrial epithelial cells by NF-kappaB and MAPK pathways. Dev. Comp. Immunol. 2020;105:103568. doi: 10.1016/j.dci.2019.103568. [DOI] [PubMed] [Google Scholar]
- 75.Hu MM, Liao CY, Yang Q, Xie XQ, Shu HB. Innate immunity to RNA virus is regulated by temporal and reversible sumoylation of RIG-I and MDA5. J. Exp. Med. 2017;214:973–989. doi: 10.1084/jem.20161015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu MM, et al. Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity. 2016;45:555–569. doi: 10.1016/j.immuni.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 77.McCray PB, Jr., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng J, et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589:603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Winkler ES, et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shang, J. et al. Quantitative proteomics identified TTC4 as a TBK1 interactor and a positive regulator of SeV-induced innate immunity. Proteomics18, 1700403 (2018). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data shown in this paper are available from the corresponding authors upon reasonable request.