Abstract
Large-scale SARS-CoV-2 molecular testing coupled with whole genome sequencing in the diagnostic laboratories is instrumental for real-time genomic surveillance. The extensive genomic, laboratory, and clinical data provide a valuable resource for understanding cases of reinfection versus prolonged RNA shedding and protracted infections. In this study, data from a total of 22,292 clinical specimens, positive by SARS-CoV-2 molecular diagnosis at Johns Hopkins clinical virology laboratory between March 11th 2020 to September 23rd 2021, were used to identify patients with two or more positive results. A total of 3,650 samples collected from 1,529 patients who had between 2 and 20 positive results were identified in a time frame that extended up to 403 days from the first positive. Cycle threshold values (Ct) were available for 1,622 samples, the median of which was over 30 by 11 days after the first positive. Extended recovery of infectious virus on cell culture was notable for up to 70 days after the first positive in immunocompromised patients. Whole genome sequencing data generated as a part of our SARS-CoV-2 genomic surveillance was available for 1,027 samples from patients that had multiple positive tests. Positive samples collected more than 10 days after initial positive with high quality sequences (coverage >90% and mean depth >100), were more likely to be from unvaccinated, or immunosuppressed patients. Reinfections with viral variants of concern were found in 3 patients more than 130 days from prior infections with a different viral clade. In 75 patients that had 2 or more high quality sequences, the acquisition of more substitutions or deletions was associated with lack of vaccination and longer time between the recovered viruses. Our study highlights the value of integrating genomic, laboratory, and clinical data for understanding the biology of SARS-CoV-2 as well as for setting a precedent for future epidemics and pandemics.
Keywords: SARS-CoV-2, variants, prolonged infection, prolonged shedding, reinfection
Introduction
The molecular detection of SARS-CoV-2 has been the gold standard for COVID-19 diagnosis since the beginning of the pandemic. The infrequency of diagnostic tests in March 2020 that limited testing to symptomatic patients under investigation was quickly replaced with large scale screening for both symptomatic and asymptomatic individuals (Anonymous and Centers for Disease Control and Prevention). Although a single positive result is sufficient for making a COVID-19 diagnosis, repeated testing of patients with positive test results has been a notable practice since the beginning of the pandemic. Hospitalized positive patients were re-tested to make infection control related decisions (Gniazdowski et al., 2020). In addition, re-testing is common in immunocompromised patients with symptoms, patients scheduled for certain procedures, as well as patients who develop new symptoms after resolution of COVID-19 (Dong et al., 2021).
Repeat testing revealed that detection of SARS-CoV-2 RNA can be prolonged for up to several months after the start of symptoms (Fontana et al., 2021). The significance of prolonged RNA shedding is dependent on the patient population and the clinical context. In patients with severe disease, the median duration of RNA shedding was longer than mild/asymptomatic cases (Liu et al., 2020; Xu et al., 2020; Zheng et al., 2020). Most cases of prolonged RNA shedding were associated with non-infectious viral recovery. Recovery of infectious virus from patients with extended RNA shedding was mainly reported from immunocompromised patients (Avanzato et al., 2020; Aydillo et al., 2020; Choi et al., 2020; Baang et al., 2021; Tarhini et al., 2021; Truong et al., 2021). A few cases of confirmed reinfections were reported; however, those cases are difficult to differentiate from prolonged infections or RNA shedding without whole genome sequencing. The Centers for Disease Control and Prevention (CDC) has defined the criteria of probable reinfections to either (a) a repeat positive molecular test 90 days after the initial infection regardless of symptoms; or (b) a repeat positive molecular test 45 days after the initial infection in the presence of symptoms consistent with COVID-19 (CDC. Centers for Disease Control and Prevention).
The Johns Hopkins clinical virology Laboratory started molecular testing for SARS-CoV-2 on March 11th 2020, and SARS-CoV-2 whole genome sequencing for understanding the genomic diversity started with the diagnosis of the first positives (Uhteg et al., 2020; Thielen et al., 2021). As of October 9th, 2021, the laboratory has tested a total of 555,983 specimens, identified a total of 28,904 positives, and sequenced a total of 8,027 genomes. In this study, we identified patients who had more than one positive result in our laboratory. The time between the first and subsequent tests, cycle threshold (Ct) values, clinical data, and genomic data were examined in addition to cell culture of selected samples to differentiate cases of prolonged RNA shedding, persistent infection, and reinfections. The impact of vaccination and immune suppression on prolonged shedding was evaluated.
Materials and Methods
Ethical Considerations and Data Availability
Ethical approval for this study was obtained from the Johns Hopkins Institutional Review Board (IRB00221396) with a waiver of consent. Whole viral genomes were made publicly available at GISAID.
Data and Sample Selection
Molecular diagnosis for SARS-CoV-2 at Johns Hopkins diagnostic laboratory is performed by different assays that include the NeuMoDx (Qiagen) (Mostafa et al., 2020a; Mostafa et al., 2020b), cobas (Roche) (Mostafa et al., 2020a), Aptima (Hologic), the Xpert Xpress SARS-CoV-2/Flu/RSV (Cepheid) (Mostafa et al., 2020c), the ePlex respiratory pathogen panel 2 (Roche) (Jarrett et al., 2021), the Accula (Hogan et al., 2020), and the RealStar SARS-CoV-2 assays (altona diagnostics) (Uhteg et al., 2020). Testing was performed in accordance with the manufacturer instructions and the Johns Hopkins laboratory’s validated protocols. Patients with more than one positive result were identified via the laboratory information system (SOFT). Only Ct values collected from the NeuMoDx assay were used in analysis as the majority of testing is performed using this system (target used, NSP2 gene). Samples with whole genome sequencing data were identified through our surveillance database. Whole genome sequencing and genomic data analysis were performed as we described previously (Morris et al., 2021; Thielen et al., 2021).
Post Consensus Analysis of Genomes
High quality genomes were defined as genomes with >90% coverage and a mean depth of >100. For analysis of acquired substitutions and deletions, only high-quality genomes were used and genomic areas with poor coverage were excluded. Manual reviews were performed on the acquired substitutions and deletions using the integrated genomic viewer.
Cell Culture
Aliquots of swab specimens were cultured on Vero-TMPRSS2 cells as previously described for VeroE6 cells (Gniazdowski et al., 2020). Cultures with cytopathic effect were confirmed for the presence of SARS-CoV-2 by reverse transcriptase PCR.
Clinical Data Analysis
Clinical data for the cohort were retrieved via bulk extraction from a data warehouse that contains all encounter-related information from hospital and outpatient visits to any Johns Hopkins Medical Institutions Facilities in addition to manual reviews of electronic medical charts. Codes associated with immunosuppression are listed in Table S1 .
Statistical Analysis
Chi squared or Welch’s t-tests were performed to show associations depending on type and number of results evaluated. Linear regression was performed with Scipy and visualized with Seaborn (Waskom, 2021).
Results
Repeat Positives From March 11th 2020 to September 23rd 2021
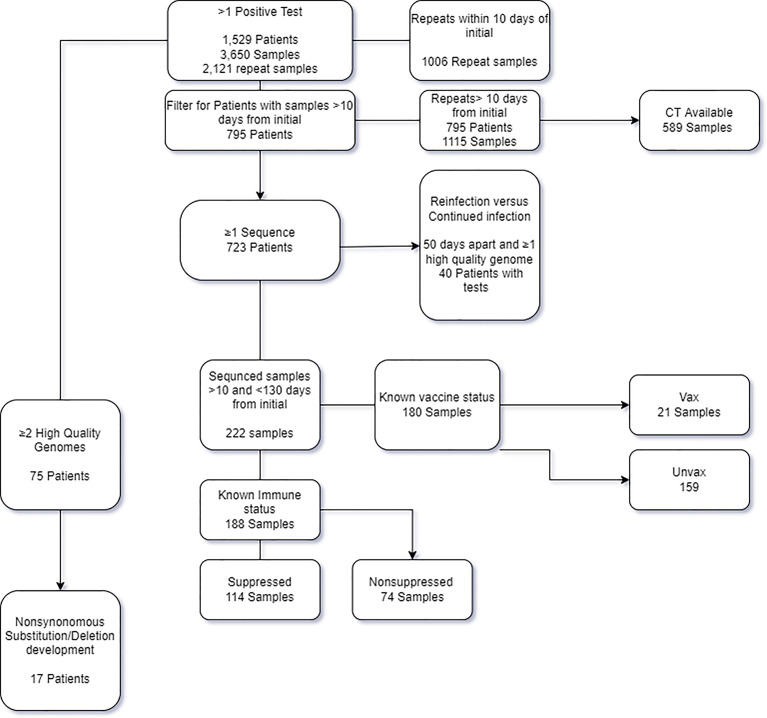
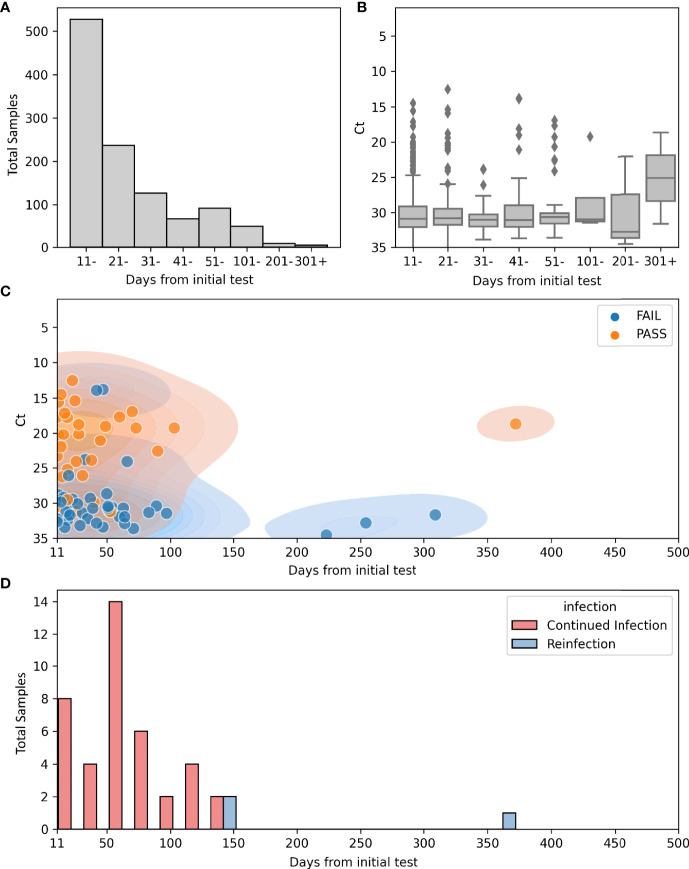
In the time frame between March 11th 2020 and September 23rd 2021, a total of 542,948 samples were tested at the Johns Hopkins clinical virology laboratory, of which 28,521 tested positive with a total positivity rate of 5.3%. A total of 1,529 patients had more than one SARS-CoV-2 positive result for during this timeframe, for a total of 3,650 samples (7% of the total positives, Figure 1 ). Repeat positive samples were collected between 0 and 403 days from the original positive sample ( Figure 2A ). a total of 943 of the 2,057 repeat samples were tested within the first 10 days of the first positive sample. The majority of the rest were testing that was primarily repeated between 11-20 days (N = 528) after the first positive and only 63 were tested 100 days or more after the first positive ( Figure 2A ).
Figure 1.
Flow chart of the patients and samples included in the study and analysis.
Figure 2.
Repeat positives from March 11th 2020 to September 23rd 2021. (A) Count plot of total numbers of repeat positives >10 days from the initial positive and the relation to the time after the first positive (N = 1115): 528 (11-20), 237 (21-30), 127 (31-40), 67 (41-50), 92 (51-100), 49 (101- 200), 9 (201-300), 6 (301-400). (B) Boxplot for Ct values in repeat positive samples and the relation to the time after the first positive (N = 589): 306 (11-20), 140 (21 -30), 60 (31- 40), 34 (41-50), 40 (51-100), 4 (101-200), 3 (201- 300), 2 (301-400). (C) Scatterplot of sequenced repeat samples >10 days from initial positive overlying kernel density estimate (KDE plot) of the same information. Hue indicates quality of sequences. N=222 Total, 155 failed quality metrics, 67 High quality genomes. (D) Histogram of days from initial positive sample in continued infection (Red, N = 40) or reinfection (Blue, N = 3).
As SARS-CoV-2 continued RNA positivity during the first 10 days of symptoms is expected (Sethuraman et al., 2020), and to exclude repeat testing that was performed for some patients in the same day, we limited the majority of our analysis to repeat positive samples that occurred more than 10 days from the original positive. Ct values were available for 589 of the 1,115 repeat samples > 10 days from the initial positive result (52%; Figures 2B, C ). Although Ct values were lower prior to 11 days from the initial sample (data not shown), the majority of samples collected after 11 days from the first positive showed CT values greater than 30 ( Figures 2B, C ). The majority of samples were collected prior to 100 days, and only two Ct values were available for samples collected 300 days after the first positive ( Figure 2B ).
Of the 1,151 repeat samples more than 10 days from the initial positive, whole genome sequencing was attempted on 222 samples collected from 173 of the 795 patients from this cohort ( Figure 2C ). Of the sequenced samples in this cohort, 69.8% did not meet the quality cut-off scores (155 of 222) ( Table 1 ). The mean Ct values of samples with high quality sequences was 22.1 in contrast to a mean Ct of 30.4 for those with low quality (data detailed in Table S2 and summarized in Table 1 ).
Table 1.
Samples with whole genome sequencing data in our cohort of repeats collected 11 days or more after the first positive.
| Days after the first positive | Genomes with high quality | Genomes with low quality | p value | ||||
|---|---|---|---|---|---|---|---|
| Number | Average Ct | Stdev | Number | Average Ct | Stdev | ||
| 11- 20 | 28 | 21.1 | 5.7 | 43 | 31.2 | 2.3 | 0.00006 |
| 21-50 | 25 | 21.1 | 5.7 | 46 | 30.4 | 6.5 | 0.02 |
| 51+ | 14 | 19.3 | 4.9 | 66 | 31.4 | 2.3 | 0.0009 |
| total | 67 | 20.24 | 5.4 | 155 | 31.24 | 4.2 | 1.00E-09 |
High quality genomes were defined as sequences with coverage >90% and a mean depth of >100. Ct, cycle threshold; stdev, standard deviation.
Prolonged Shedding Versus Reinfections
To differentiate cases of prolonged shedding from reinfections based on viral genomics analyses, genomes from the same patients were compared. For this analysis, we focused primarily on the 40 patients in our cohort who had at least two positive SARS-CoV-2 PCR tests >50 days apart and with at least one high quality genome ( Table S3 ). Patients from this group tested positive between 2 and 16 times and the time between the first and last positive tests was between 51 and 372 days. In total, there were 162 positive samples of which 109 were sequenced and all available genomes were analyzed regardless of time from initial positive. Despite low quality sequences of genomes from many of the samples collected at later time points, 14 of these patients showed evidence of prolonged shedding of RNA, consistent with the initial infecting virus, and only 3 showed genomic evidence of reinfection ( Table 2 and Table S3 ). Two were initially infected with one clade, [20G or 20I (Alpha)], followed by a later infection with the Delta variant. The third patient was considered a reinfection because the second infection was with the Delta variant which was not circulating when the patient first tested positive 372 days earlier ( Table 2 ). The earliest positive test from a reinfection occurred 133 days following the initial infection ( Figure 2D ), and Continued RNA detection was noted as far as 139 days following the initial positive.
Table 2.
Prolonged shedding, persistent infections, versus reinfections.
| PID | Vaccinated | Immunosuppressed | Days from the first positive | HPID | Cell culture | Ct | % Coverage | Depth | Lineage | Clade |
|---|---|---|---|---|---|---|---|---|---|---|
| Reinfections | ||||||||||
| 1 | Yes | No | 0 | |||||||
| 372 | HP08786 | 18.7 | 98.68 | 400.00 | B.1.617.2 | 21A (Delta) | ||||
| 13 | Yes | Yes | 0 | HP05124 | 99.60 | 400.00 | B.1.2 | 20G | ||
| 11 | HP04658 | 17.73 | 98.60 | 385.09 | B.1.2 | 20G | ||||
| 144 | HP08875 | 86.10 | 172.40 | AY.7.1 | 21A (Delta) | |||||
| 28 | Yes | No | 0 | HP05001 | 98.87 | 333.78 | B.1.1.7 | 20I (Alpha, V1) | ||
| 133 | HP09101 | 95.54 | 381.78 | B.1.617.2 | 21A (Delta) | |||||
| Persistent infection | ||||||||||
| 10 | No | Yes | 0 | 24.19 | ||||||
| 27 | 23.65 | |||||||||
| 33 | HP13078 | Positive | 23.83 | 0.00 | 3.00 | None | Attempted | |||
| 38 | HP13068 | Negative | 23.93 | 97.25 | 349.00 | A.3 | 19B | |||
| 53 | Positive | 20.69 | ||||||||
| 81 | HP13137 | Negative | 95.50 | 378.49 | A.3 | 19B | ||||
| 12 | No | Yes | 0 | |||||||
| 6 | HP13142 | Positive | 16.99 | 99.60 | 400.00 | B.1 | 20C | |||
| 42 | HP13143 | Positive | 13.93 | 2.69 | 9.15 | None | Attempted | |||
| 46 | ||||||||||
| 60 | HP13074 | Positive | 17.71 | 98.73 | 400.00 | B.1 | 20C | |||
| 73 | HP13075 | Negative | 19.28 | 98.75 | 400.00 | B.1 | 20C | |||
| 84 | 22.47 | |||||||||
| 16 | No | Yes | 0 | HP13076 | Positive | 16.5 | 99.01 | 400.00 | B.1.520 | 20C |
| 39 | HP13144 | Positive | 99.59 | 400.00 | B.1.520 | 20C | ||||
| 65 | ||||||||||
| 70 | HP13077 | Positive | 16.95 | 99.57 | 400.00 | B.1 | 20C | |||
| Prolonged shedding | ||||||||||
| 15 | No | Yes | 0 | HP12091 | Positive | 18.62 | 0.00 | 400.00 | B.1.1.434 | 20B |
| 20 | ||||||||||
| 26 | HP12092 | Positive | 24.1 | 0.00 | 378.28 | B.1.1.434 | 20B | |||
| 33 | HP12093 | Negative | 235.88 | B.1.1.434 | 20B | |||||
| 38 | HP12094 | Negative | 400.00 | B.1.1.434 | 20B | |||||
| 103 | HP02090 | 19.27 | 99.57 | 400.00 | B.1.1.434 | 20B | ||||
| 117 | HP02480 | 98.59 | 142.00 | B.1.1.434 | 20B | |||||
| 126 | HP02621 | 64.60 | 63.50 | None | 20B | |||||
| 132 | HP03033 | 32.65 | 23.28 | None | 20B | |||||
| 139 | HP03236 | 9.04 | 5.85 | None | Attempted | |||||
| 19 | Yes | Yes | 0 | HP02155 | 13.69 | 99.60 | 400.00 | B.1.409 | 20A | |
| 19 | ||||||||||
| 57 | HP01654 | 99.59 | 400.00 | B.1.409 | 20A | |||||
| 61 | HP01599 | 99.59 | 400.00 | B.1.409 | 20A | |||||
| 77 | HP02479 | 13.62 | 9.00 | None | Attempted | |||||
| 20 | No | Yes | 0 | 19.15 | ||||||
| 1 | HP03788 | 14.16 | 99.60 | 400.00 | B.1.526 | 21F (Iota) | ||||
| 7 | 15.31 | |||||||||
| 14 | HP04418 | 14.52 | 98.60 | 400.00 | B.1.526 | 21F (Iota) | ||||
| 19 | HP04748 | 17.78 | 99.60 | 400.00 | B.1.526 | 21F (Iota) | ||||
| 21 | HP04804 | 98.60 | 400.00 | B.1.526 | 21F (Iota) | |||||
| 25 | HP04922 | 15.41 | 99.60 | 400.00 | B.1.526 | 21F (Iota) | ||||
| 28 | HP05155 | 18.78 | 97.75 | 400.00 | B.1.526 | 21F (Iota) | ||||
| 31 | HP05251 | 26.11 | 97.89 | 354.79 | B.1.526 | 21F (Iota) | ||||
| 35 | ||||||||||
| 39 | ||||||||||
| 64 | HP06212 | 32.91 | 56.72 | 172.78 | None | 21F (Iota) | ||||
| 3 | No | 0 | HP13038 | Positive | 20.22 | 99.60 | 400.00 | B.1 | 20A | |
| 50 | 25.13 | |||||||||
| 50 | HP10152 | 87.25 | 165.00 | B.1 | 20A | |||||
| 54 | HP13039 | Negative | 30.61 | 89.76 | 102.34 | B.1 | 20A | |||
| 4 | No | Yes | 0 | HP13053 | Positive | 21.41 | 99.60 | 400.00 | B.1.494 | 20C |
| 36 | 29.45 | |||||||||
| 37 | ||||||||||
| 64 | HP13054 | Negative | 76.25 | 40.85 | B.1.446 | 20C | ||||
| 124 | 31.53 | |||||||||
| 11 | No | No | 0 | HP13040 | Negative | 25.43 | 99.60 | 400.00 | B.1.369 | 20C |
| 38 | 31.51 | |||||||||
| 53 | HP13041 | Negative | 31.21 | 90.19 | 227.39 | B.1.369 | 20C | |||
| 27 | No | No | 0 | HP13044 | Positive | 14.47 | 99.60 | 400.00 | B.1.369 | 20C |
| 25 | 33.14 | |||||||||
| 51 | HP13045 | Negative | 30.73 | 67.59 | 227.37 | None | 20C | |||
| 29 | Yes | Yes | 0 | HP02004 | 99.60 | 400.00 | B.1.2 | 20G | ||
| 3 | HP02005 | 15.53 | 99.60 | 400.00 | B.1.2 | 20G | ||||
| 12 | HP02006 | 99.60 | 387.00 | B.1.2 | 20G | |||||
| 17 | HP02007 | 98.60 | 400.00 | B.1.2 | 20G | |||||
| 23 | ||||||||||
| 24 | 20.48 | |||||||||
| 30 | 21.19 | |||||||||
| 37 | HP02008 | 29.34 | 37.73 | 26.00 | None | 20C | ||||
| 44 | HP02009 | 81.92 | 117.00 | B.1.2 | 20G | |||||
| 51 | HP02010 | 30.48 | 4.47 | 4.00 | None | Attempted | ||||
| 34 | Yes | No | 0 | HP13046 | Negative | 29.6 | 99.60 | 400.00 | B.1 | 20C |
| 92 | HP13047 | Negative | 46.98 | 52.01 | None | 20C | ||||
| 35 | Yes | No | 0 | HP13036 | Negative | 32.86 | 99.59 | 212.41 | B.1.110 | 20A |
| 16 | 29.14 | |||||||||
| 57 | HP13037 | Negative | 87.83 | 64.10 | B.1 | 20A | ||||
| 37 | Yes | Yes | 0 | HP02713 | Positive | 99.60 | 400.00 | B.1.2 | 20G | |
| 23 | HP03492 | 99.60 | 368.98 | B.1.2 | 20G | |||||
| 38 | ||||||||||
| 39 | ||||||||||
| 45 | HP04440 | 21.11 | 98.60 | 387.16 | B.1.2 | 20G | ||||
| 50 | HP04626 | 28.69 | 31.43 | 18.08 | None | 20G | ||||
| 57 | HP04882 | 29.1 | 94.15 | 314.68 | B.1.2 | 20G | ||||
PID, patient identifier; HPID, identification of samples with whole genome sequencing; Ct, cycle threshold.
Of the 14 patients with genomic evidence of prolonged shedding, Cell culture was attempted for 28 samples from this cohort to assess prolonged active infection versus prolonged RNA shedding. Three immunocompromised patients showed prolonged recovery of infectious virus on cell culture ( Table 2 , patients 10, 12, and 16) for 53, 60, and 70 days after the first positive, indicating persistent infection. Notably, three additional immunocompromised patients had prolonged shedding (patients 15, 19, and 20, Table 2 ) with a notable low Ct values or recovery of complete genomes for up to 117, 61, and 31 days after the first positive. The rest of the 14 patients from this cohort had extended shedding at higher Ct values, and 6 patients had negative cell culture results suggesting protracted RNA shedding rather than persistent infection ( Table 2 ).
Impact of Vaccination on Prolonged Viral Shedding
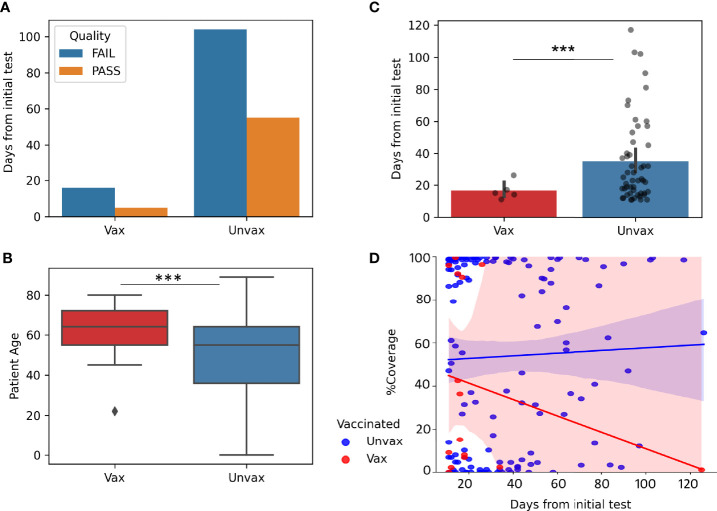
To study the impact of vaccination on prolonged viral shedding, we compared the genomic data of samples from vaccinated compared to unvaccinated patients. We limited this analysis to sequenced samples that were positive 11 to 130 days from the initial positive test (in order to remove possible repeat infections, as repeat infections started to be seen at 130 days). Patients who had received the full vaccination series at least 2 weeks prior to the first positive test were classified as vaccinated in this cohort. Only 21 samples from vaccinated patients with repeated positive tests met these criteria ( Figure 3A ) compared to 159 repeat positive samples from unvaccinated patients. Only 5 of 21(23%) genomes within this timeframe met our quality scores in vaccinated patients compared to 55 of 159(35%) in unvaccinated patients, despite older median age in the vaccinated patients (64 compared to 55, p=0.00065) ( Figure 3B ), and lower median days from the initial test for all samples where sequencing was attempted (17 days vaccinated, 32 days unvaccinated, p=0.0003, Figure 3C ).In general, genomes of repeat positives from vaccinated patients after 20 days from the first positives were very few which caused a notable trend of a decrease in coverage with time when compared to the unvaccinated group ( Figure 3D ).
Figure 3.
Impact of Vaccination on prolonged viral shedding. Genomes from repeat positive samples within the first 130 days after the initial positives (N = 21 Vax “vaccinated” and 159 Unvax “unvaccinated”). (A) Quality of genomes in samples from vaccinated and unvaccinated patients. (B) Boxplot of Patient Age in samples from vaccinated (Median 64 years) and unvaccinated (median 55 years). (C) Barplots with overlayed strip plots of the recovery of high-quality genomes compared in relation to days after the first positive. (D) Implots of genome coverage compared to days from the initial positive. ***p < 0.0001.
Impact of Immunosuppression on Prolonged Viral Shedding
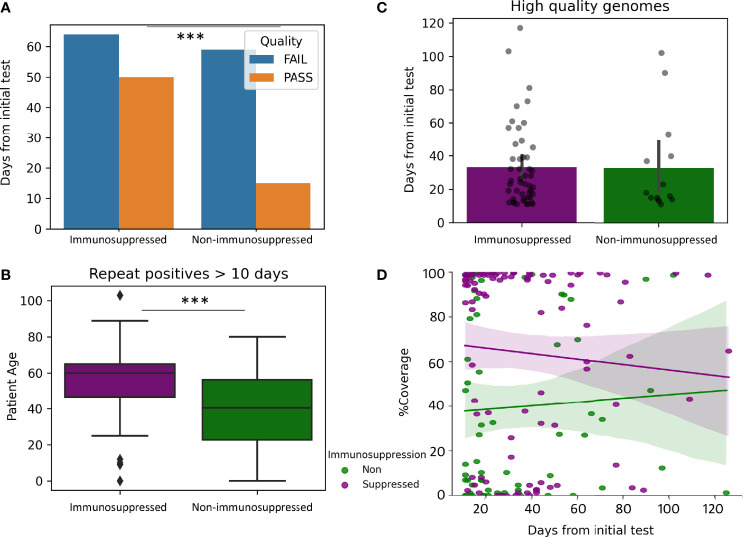
To evaluate the impact of immunosuppression the coverage of the SARS-CoV-2 genome in 74 samples from patients without immunosuppression were compared to 114 samples from patients with immunosuppression (greater than 10 days and less than 130 days from the initial positive). High quality genomes were more represented at a higher percentage in Immunosuppressed compared to non-immunosuppressed patients (43% compared to 20%, p-value 0.0009, Figure 4A ). Immunosuppressed patients were older than non-immunosuppressed patients (median years 60 compared to 40.5, p-value >0.0001, Figure 4B ). However, high quality genomes were recovered for similar mean days between immunosuppressed and non-immunosuppressed patients (33.1, compared to 31.5 days, Figure 4C ) with similar coverage over-time ( Figure 4D ). Additional analysis was performed to see how vaccination impacted Immunosuppressed patients. Of the sampled from immunosuppressed patients, 13 were from vaccinated and 94 were from unvaccinated patients. The percentages of samples that passed quality control from vaccinated and unvaccinated immunosuppressed were similar, but high-quality genomes were obtained for shorter periods of time in samples from vaccinated compared to unvaccinated patients (16.6 days compared to 35.7 days, p=0.009) (data not shown).
Figure 4.
Impact of Immunosuppression on prolonged viral shedding. Genomes from repeat positive samples within the first 130 days after the initial positives (N = 114 immunosuppressed and 74 non-immunosuppressed). (A) Quality of genomes in samples from immunosuppressed and non-immunosuppressed patients. (B) Boxplot of patient age in samples from immunosuppressed (Median 60 years) and non-immunosuppressed (median 40.5 years). (C) Barplots with overlayed strip plots of the recovery of high-quality genomes compared in relation to days after the first positive. (D) Implots of genome coverage compared to days from the initial positive in immunosuppressed and non-immunosuppressed patients. ***p < 0.00001.
Prolonged Viral Shedding and Genomic Changes
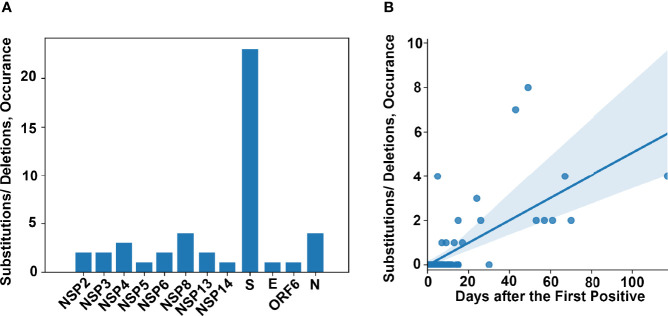
In order to study the development of new substitutions or deletions within the genome of SARS-CoV-2 over-time, within the same patient, we identified all patients for whom we had at least 2 high quality genomes from different samples and whose genomic data was consistent with prolonged shedding or persistent infection but not reinfection. This analysis was not limited to a specific timeframe between positive samples. We identified 75 patients that met these criteria. The timeframe between the initial and subsequent high-quality genomes was between 0 and 117 days (detailed in Table S4 ). A total of 48 amino acid (AA) substitutions or deletions developed in 17 of these patients ( Table 3 ). Of the 17 patients that acquired substitutions or deletions, 16 were unvaccinated and 14 were immunosuppressed. The substitutions and deletions which developed and the number of instances in our cohort can be seen in Table S5 , along with information on how many times these specific substitutions or deletions have been seen among all the genomes sequenced in our laboratory and lineages associated with these changes in multiple instances. Of the 48 substitutions or deletions noted, three spike protein deletions (L141del, G142del, and V143del) and one spike protein substitution (E484K) occurred in two instances. Each of these substitutions or deletions that occurred more than once in this dataset was a part of disparate lineages, especially S:E484K which was seen in at least 19 lineages by our group ( Table S5 ). The most common protein to develop substitutions or deletions was the Spike protein ( Figure 5A ) A correlation between AA changes and days between recoverable genomes was noted ( Figure 5B , p < 0.00001). Recoverable genomes over 24 days from the original positive showed a minimum of two non-synonymous substitutions or deletions ( Table 3 ).
Table 3.
Prolonged viral shedding and genomic changes.
| PID | Positives | Sequenced | HQ Genomes | Days between first and last positives | Days between first and last HQ Genomes | First HQ genome lineage | last HQ genome lineage | Number Acquired subs/dels | Acquired AA substitutions/deletions | Vaccinated | Immunosuppressed | Monoclonal antibody | Convalescent Plasma |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PIDVWVGDCM | 2 | 2 | 2 | 5 | 5 | B.1.2 | B.1.2 | 4 | S:G142del,S:V143del,S:Y144del,S:L141del | No | Yes | Yes | |
| PIDWUGBIZH | 5 | 4 | 3 | 14 | 7 | B.1.637 | B.1.637 | 1 | S:H146Q | No | Yes | Yes | |
| PIDCXIYIPP | 6 | 5 | 4 | 20 | 9 | B.1.243 | B.1.243 | 1 | NSP8:F6L | No | Yes | Yes | |
| PIDLUEEQYV | 5 | 4 | 2 | 13 | 13 | B.1.1.7 | B.1.1.7 | 1 | NSP14:D126Y | No | Yes | ||
| PIDEAUSZWF | 2 | 2 | 2 | 15 | 15 | B.1.2 | B.1.2 | 2 | NSP2:E490K,S:E484K | No | |||
| PIDRSHNQNS | 12 | 8 | 5 | 51 | 17 | B.1.2 | B.1.2 | 1 | E:V70F | No | Yes | Yes | |
| PIDGBIBYYQ | 7 | 4 | 4 | 24 | 24 | Q.4 | Q.4 | 3 | NSP4:H313Y,N:F307L,S:R102G | No | Yes | Yes | |
| PIDBXQZZLV | 5 | 4 | 4 | 26 | 26 | B.1.526 | B.1.526 | 2 | S:G446V,S:E406Q | Yes | Yes | Yes | |
| PIDLCYVIUV | 2 | 2 | 2 | 37 | 37 | B.1.564 | B.1 | 2 | S:L270F,NSP2:T85I | No | |||
| PIDFTUQFAV | 7 | 3 | 2 | 81 | 43 | A.3 | A.3 | 7 | S:R190S,S:L244del,NSP4:F317S,S:T1006I,NSP3:F1646I,NSP4:G232V,S:A243del | No | Yes | ||
| PIDSDHALQC | 5 | 4 | 4 | 49 | 49 | B.1.637 | B.1.637 | 8 | S:T95I,S:G142del,NSP13:T115K,S:V143del,S:L141del,S:E484Q,NSP8:T123I,S:W152R | No | Yes | ||
| PIDHCPHTBW | 3 | 2 | 2 | 53 | 53 | B.1.369 | B.1.369 | 2 | NSP5:Q192stop,N:T271I | No | |||
| PIDXTTUNJN | 9 | 5 | 4 | 57 | 57 | B.1.2 | B.1.2 | 2 | S:E484K,NSP6:L37F | No | Yes | Yes | |
| PIDNXQFFZG | 6 | 4 | 3 | 77 | 61 | B.1.409 | B.1.409 | 2 | NSP8:I156L,NS6:E13K | No | Yes | Yes | Yes |
| PIDHNGFDXK | 7 | 4 | 3 | 84 | 67 | B.1 | B.1 | 4 | NSP8:T148I,S:P1079S,NSP3:T820I,N:P326L | No | Yes | ||
| PIDLYROPCD | 4 | 3 | 3 | 70 | 70 | B.1.520 | B.1 | 2 | NSP6:M86I,N:T205I | No | Yes | ||
| PIDLVBEHYS | 11 | 9 | 6 | 139 | 117 | B.1.1.434 | B.1.1.434 | 4 | NSP2:K534R,S:N334K,NSP13:D56G,S:S939F | No | Yes | Yes |
PID, patient identifier; HPID, identification of samples with whole genome sequencing; HQ, High quality genomes were defined as sequences with coverage >90% and a mean depth of >100. Subs/dels, substitutions/deletions; AA, amino acid.
Figure 5.
Genomic changes in prolonged viral shedding. (A) Frequency of amino acid changes (substitutions and deletions) per protein in a cohort of 75 patients with two or more complete genomes from samples collected 0 - 117 days after the first positive. (B) Amino acid changes as a factor of time between collected positive samples.
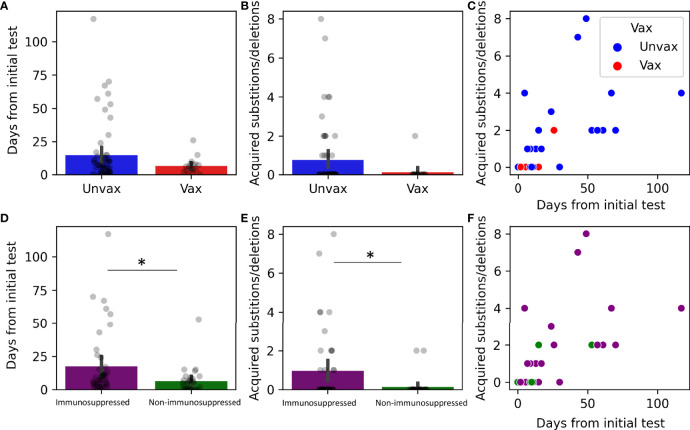
We next examined the impact of vaccination and immune suppression on the accumulation of nonsynonymous mutations. Of the 75 patients that had multiple recoverable genomes, 17 were vaccinated ( Figures 6A – C ). Only one vaccinated patient developed nonsynonymous mutations ( Table 3 ). Although vaccinated patients that had multiple recoverable genomes showed a lower mean time of recoverable genomes, and a lower mean number of developed mutations, this did not reach significance ( Figures 6A – C ). Immunocompromised status was associated with longer periods between the recoverable genomes ( Figure 6D , p=0.022) and a higher number of substitutions or deletions ( Figures 6E, F , p=0.022).
Figure 6.
Genomic changes in prolonged viral shedding by vaccination and immune status. Substitutions and deletions in 75 patients by status: vaccinated (N =17, A–C) and immunosuppressed (N=44, D–F). (A, D) Barplots with overlayed strip plots of mean time of prolonged shedding of complete genomes in each group, (B, E) Barplots with overlayed strip plots of amino acid change frequency in each group, (C, F) Scatterplots of the correlation of amino acid change and the days after the first positive in each group. *p < 0.05. Unvax, unvaccinated; Vax, vaccinated.
Discussion
In this study, we combined large scale laboratory diagnostic data with whole genome sequencing data for surveillance to analyze prolonged SARS-CoV-2 shedding and reinfections. Clinical data analysis was performed to study the impact of vaccination and immunocompromised status on genomic changes over-time. Cell culture was performed in certain cases with extended viral shedding combined with low Ct values to characterize cases of prolonged active infection. Our data showed that the majority of patients with prolonged positive SARS-CoV-2 molecular tests have their subsequent positives collected within the first 50 days of the initial positive with Ct values that are likely higher than 30. Prolonged shedding after 50 days was largely associated with low quality genomes reflecting low viral loads except in 17 patients, 14 of whom had subsequent genomes matching the initial, indicating prolonged shedding and 3 had a second genome of a different clade, indicating reinfection. Vaccination reduced the likelihood of the recovery of good quality genomes but immunocompromised status contributed to the increased duration of viral RNA shedding and the accumulation of genomic changes. The spike gene was the region of the genome most prone to nonsynonymous changes in our cohort, but overall, the development of nonsynonymous mutations and AA changes was infrequent.
Prolonged SARS-CoV-2 shedding was shown to extend to multiple months after the onset of symptoms (Fontana et al., 2021). Prolonged shedding though might not correlate with prolonged infectiousness or recovery of viable virus. Previously we showed that shedding of infectious virus can extend to more than 3 weeks after the initial positive (Gniazdowski et al., 2020). In this study, we show the recovery of infectious virus for up to 70 days after the first positive in a subset of immunocompromised patients. This data is consistent with previous reports that showed prolonged SARS-CoV-2 replication and infectiousness in immunocompromised patients (Aydillo et al., 2020; Baang et al., 2021; Tarhini et al., 2021). In general, the recovery of infectious virus correlated with lower Ct values consistent with higher viral loads (Basile et al., 2020; Bullard et al., 2020; Gniazdowski et al., 2020). The value of differentiating prolonged shedding from prolonged infection include controlling the transmission of infection in patients with actively replicating virus as well as optimizing antiviral intervention strategies. Our data is consistent with previous reports that show that prolonged active infection is remarkably less frequent than prolonged shedding of viral RNA and are primarily associated with immunocompromised patient populations.
Reinfection with SARS-CoV-2 was previously reported and a time frame of 45 days was proposed to suggest possible reinfections when a positive test is associated with symptoms consistent with COVID-19. Genomic sequencing is the only method that can conclusively characterize reinfection cases when the initial genomes are compared to subsequent positives. When initial isolates are not available, it might be possible to characterize reinfections if the lineage identified is an emerging variant that was not circulating when the patient was previously diagnosed. Interestingly, in the time frame when the Delta variant became predominant (Morris et al., 2021), we only identified three patients who had a previous positive in our institution. Even though this data might be largely impacted by the availability of other testing sites outside of the Johns Hopkins system, within the population of patients tested in our laboratory, we identified breakthrough cases after vaccination in rates that were much higher than reinfections (Luo et al., 2021).
The evolution of SARS-CoV-2 variants associated with increased transmissibility or escape from vaccine induced or natural immune responses has been globally concerning. Specific variants showed significant reduction in neutralization by monoclonal antibodies and convalescent plasma (Hoffmann et al., 2021; Planas et al., 2021). Those variants attracted the attention to specific changes within SARS-CoV-2 genome that could be of therapeutic concern. Certain spike changes were shown to impact the action of monoclonal antibodies and convalescent sera including L452R, E484K, K417N, and K417T. In our cohort, the spike changes that we observed more than once included the E484K and the deletions 141-143. The E484K in particular was previously reported to reduce the neutralization efficacy and was identified as an escape change the could develop after treatment with certain monoclonal antibodies or convalescent plasma (Baum et al., 2020; Weisblum et al., 2020; Greaney et al., 2021). The deletions 141-144 are within the NTD and were reported to reduce binding to the monoclonal antibody 4A8 (recovered from the convalescent plasma of patients with COVID-19) as do deletions 243-244 (which occurred once in our cohort) (McCarthy et al., 2021). Other changes we detected in our cohort are within epitopes or associated with escape from convalescent sera including W152R, F157S, and G446V (Liu et al., 2021; Suryadevara et al., 2021). Although 9 of the 17 patients in our cohort whom infecting virus developed nonsynonymous mutations received monoclonal or convalescent sera treatments ( Table 3 ), associations between those treatments and the rate of developing mutations were not performed due to the incompleteness of this data in some of the clinical charts. In our cohort, the accumulation of genomic changes associated with the prolonged shedding time that was most remarkable in immunosuppressed individuals.
A main finding in our study was the correlation between vaccination and a lower likelihood of prolonged shedding or recovery of good quality genomes after the first 20 days of the initial positive result. Even though, vaccine breakthrough infections correlated with a bias to genomes carrying the S: E484K (Feder et al., 2021; Mostafa et al., 2021), a limited infectious virus shedding was notable when samples from patients were longitudinally analyzed (Ke et al., 2021). SARS-CoV-2 genomic diversity was also shown to decline after widespread vaccination (Niesen et al., 2021). The selected cohort for our analysis was restricted to individuals who had an initial positive when fully vaccinated. This data indicate that vaccination is interrupting the extended prolonged shedding observed since the start of the pandemic with unvaccinated individuals. Our data also show that vaccinated individuals are less likely to accumulate significant genomic changes over time.
In summary, the COVID-19 pandemic challenged the diagnostic laboratories to not only ramp up testing, but to also assist with the nationwide genomic surveillance. The diagnostic laboratories are capable of providing real-time epidemiological and clinical data that are essential for a better understanding of the biology of SARS-CoV-2. Diagnostic laboratories with limited resources can also assist by coordinating sharing real-time positive samples with public health laboratories for surveillance. The workflow of diagnosis, surveillance, and basic research we established at Johns Hopkins laboratory for characterizing SARS-CoV-2 provides a template for other evolving pathogens of concern and emphasizes the power of generating real-time data amid a quickly evolving pandemic.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Ethics Statement
The studies involving human participants were reviewed and approved by the Johns Hopkins Institutional Review Board (IRB00221396) with a waiver of consent. Written informed consent from the participants’ legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author Contributions
CM data collection and analysis, data interpretation, figures, writing. CL, JS and ML cell culture. DG, VG, NG, AA, JN, MS, and WW. data collection. EK. Clinical data collection and analysis. AP. cell culture and scientific revision. HM study design, data collection and analysis, data interpretation, writing, and fund acquisition. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Centers for Disease Control (contract 75D30121C11061), Johns Hopkins University President’s Fund Research Response, the Johns Hopkins Department of Pathology, the Maryland Department of Health, and the Johns Hopkins Center of Excellence in Influenza Research and Surveillance (HHSN272201400007C). HM is supported by the HIV Prevention Trials Network (HPTN) sponsored by the National Institute of Allergy and Infectious Diseases (NIAID), National Institute on Drug Abuse, National Institute of Mental Health, and Office of AIDS Research of the NIH, DHHS (UM1 AI068613), the NIH RADx-Tech program (3U54HL143541-02S2), and National Institute of Health RADx-UP initiative (Grant R01 DA045556-04S1). The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Institute of Biomedical Imaging and Bioengineering; the National Heart, Lung, and Blood Institute; the National Institutes of Health, or the U.S. Department of Health and Human Services.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This study was only possible with the unique efforts of the Johns Hopkins Clinical Microbiology Laboratory faculty and staff.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.809407/full#supplementary-material
References
- Anonymous. Centers for Disease Control and Prevention Overview of Testing for SARS-CoV-2 (COVID-19). Available at: https://wwwcdcgov/coronavirus/2019-ncov/hcp/testing-overviewhtml. (Updates as of August 2, 2021).
- Avanzato V. A., Matson M. J., Seifert S. N., Pryce R., Williamson B. N., Anzick S. L., et al. (2020). Case Study: Prolonged Infectious SARS-CoV-2 Shedding From an Asymptomatic Immunocompromised Individual With Cancer. Cell 183, 1901–1912.e9. doi: 10.1016/j.cell.2020.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydillo T., Gonzalez-Reiche A. S., Aslam S., van de Guchte A., Khan Z., Obla A., et al. (2020). Shedding of Viable SARS-CoV-2 After Immunosuppressive Therapy for Cancer. N. Engl. J. Med. 383, 2586–2588. doi: 10.1056/NEJMc2031670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baang J. H., Smith C., Mirabelli C., Valesano A. L., Manthei D. M., Bachman M. A., et al. (2021). Prolonged Severe Acute Respiratory Syndrome Coronavirus 2 Replication in an Immunocompromised Patient. J. Infect. Dis. 223, 23–27. doi: 10.1093/infdis/jiaa666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basile K., McPhie K., Carter I., Alderson S., Rahman H., Donovan L., et al. (2020). Cell-Based Culture of SARS-CoV-2 Informs Infectivity and Safe De-Isolation Assessments During COVID-19. Clin. Infect. Dis. 73 (9), e2952–e2959. doi: 10.1093/cid/ciaa1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B. O., Wloga E., Copin R., Pascal K. E., Russo V., et al. (2020). Antibody Cocktail to SARS-CoV-2 Spike Protein Prevents Rapid Mutational Escape Seen With Individual Antibodies. Science 369, 1014–1018. doi: 10.1126/science.abd0831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard J., Dust K., Funk D., Strong J. E., Alexander D., Garnett L., et al. (2020). Predicting Infectious Severe Acute Respiratory Syndrome Coronavirus 2 From Diagnostic Samples. Clin. Infect. Dis. 71, 2663–2666. doi: 10.1093/cid/ciaa638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Centers for Disease Control and Prevention Investigative Criteria for Suspected Cases of SARS-CoV-2 Reinfection (ICR). Available at: https://www.cdc.gov/coronavirus/2019-ncov/php/invest-criteria.html (Accessed October 29, 2020).
- Choi B., Choudhary M. C., Regan J., Sparks J. A., Padera R. F., Qiu X., et al. (2020). Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N. Engl. J. Med. 383, 2291–2293. doi: 10.1056/NEJMc2031364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Zhou Y., Shu X. O., Bernstam E. V., Stern R., Aronoff D. M., et al. (2021). Comprehensive Characterization of COVID-19 Patients With Repeatedly Positive SARS-CoV-2 Tests Using a Large U.S. Electronic Health Record Database. Microbiol. Spectr. 9, e0032721. doi: 10.1128/Spectrum.00327-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder K. A., Patel A., Vepachedu V. R., Dominguez C., Keller E. N., Klein L., et al. (2021). Association of E484K Spike Protein Mutation With SARS-CoV-2 Infection in Vaccinated Persons—Maryland, January - May 2021. Clin. Infect. Dis. ciab762. doi: 10.1093/cid/ciab762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L. M., Villamagna A. H., Sikka M. K., McGregor J. C. (2021). Understanding Viral Shedding of Severe Acute Respiratory Coronavirus Virus 2 (SARS-CoV-2): Review of Current Literature. Infect. Control Hosp. Epidemiol. 42, 659–668. doi: 10.1017/ice.2020.1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gniazdowski V., Morris C. P., Wohl S., Mehoke T., Ramakrishnan S., Thielen P., et al. (2020). Repeat COVID-19 Molecular Testing: Correlation of SARS-CoV-2 Culture With Molecular Assays and Cycle Thresholds. Clin. Infect. Dis. 73 (4), e860–e869. doi: 10.1093/cid/ciaa1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A. J., Loes A. N., Crawford K. H. D., Starr T. N., Malone K. D., Chu H. Y., et al. (2021). Comprehensive Mapping of Mutations in the SARS-CoV-2 Receptor-Binding Domain That Affect Recognition by Polyclonal Human Plasma Antibodies. Cell Host Microbe 29, 463–476.e6. doi: 10.1016/j.chom.2021.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Hofmann-Winkler H., Kruger N., Kempf A., Nehlmeier I., Graichen L., et al. (2021). SARS-CoV-2 Variant B.1.617 Is Resistant to Bamlanivimab and Evades Antibodies Induced by Infection and Vaccination. Cell Rep. 36, 109415. doi: 10.1016/j.celrep.2021.109415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C. A., Garamani N., Lee A. S., Tung J. K., Sahoo M. K., Huang C., et al. (2020). Comparison of the Accula SARS-CoV-2 Test With a Laboratory-Developed Assay for Detection of SARS-CoV-2 RNA in Clinical Nasopharyngeal Specimens. J. Clin. Microbiol. 58 (8), e01072–20. doi: 10.1128/JCM.01072-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett J., Uhteg K., Forman M. S., Hanlon A., Vargas C., Carroll K. C., et al. (2021). Clinical Performance of the GenMark Dx Eplex Respiratory Pathogen Panels for Upper and Lower Respiratory Tract Infections. J. Clin. Virol. 135, 104737. doi: 10.1016/j.jcv.2021.104737 [DOI] [PubMed] [Google Scholar]
- Ke R., Martinez P. P., Smith R. L., Gibson L. L., Achenbach C. J., McFall S., et al. (2021). Longitudinal Analysis of SARS-CoV-2 Vaccine Breakthrough Infections Reveal Limited Infectious Virus Shedding and Restricted Tissue Distribution. medRxiv 2021.08.30.21262701. doi: 10.1101/2021.08.30.21262701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L. A., Bloyet L. M., Rothlauf P. W., Chen R. E., Stumpf S., et al. (2021). Identification of SARS-CoV-2 Spike Mutations That Attenuate Monoclonal and Serum Antibody Neutralization. Cell Host Microbe 29, 477–488.e4. doi: 10.1016/j.chom.2021.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yan L. M., Wan L., Xiang T. X., Le A., Liu J. M., et al. (2020). Viral Dynamics in Mild and Severe Cases of COVID-19. Lancet Infect. Dis. 20, 656–657. doi: 10.1016/S1473-3099(20)30232-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C. H., Morris C. P., Sachithanandham J., Amadi A., Gaston D., Li M., et al. (2021). Infection With the SARS-CoV-2 Delta Variant Is Associated With Higher Infectious Virus Loads Compared to the Alpha Variant in Both Unvaccinated and Vaccinated Individuals. medRxiv ciab986. doi: 10.1101/2021.08.15.21262077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K. R., Rennick L. J., Nambulli S., Robinson-McCarthy L. R., Bain W. G., Haidar G., et al. (2021). Recurrent Deletions in the SARS-CoV-2 Spike Glycoprotein Drive Antibody Escape. Science 371, 1139–1142. doi: 10.1126/science.abf6950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C. P., Luo C. H., Amadi A., Schwartz M., Gallagher N., Ray S. C., et al. (2021). An Update on SARS-CoV-2 Diversity in the United States National Capital Region: Evolution of Novel and Variants of Concern. Clin. Infect. Dis. ciab636. doi: 10.1093/cid/ciab636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa H. H., Carroll K. C., Hicken R., Berry G. J., Manji R., Smith E., et al. (2020. c). Multi-Center Evaluation of the Cepheid Xpert(R) Xpress SARS-CoV-2/Flu/RSV Test. J. Clin. Microbiol. 59 (3). doi: 10.1128/JCM.02955-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa H. H., Hardick J., Morehead E., Miller J. A., Gaydos C. A., Manabe Y. C. (2020. a). Comparison of the Analytical Sensitivity of Seven Commonly Used Commercial SARS-CoV-2 Automated Molecular Assays. J. Clin. Virol. 130, 104578. doi: 10.1016/j.jcv.2020.104578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa H. H., Lamson D. M., Uhteg K., Geahr M., Gluck L., de Cardenas J. N. B., et al. (2020. b). Multicenter Evaluation of the NeuMoDx SARS-CoV-2 Test. J. Clin. Virol. 130, 104583. doi: 10.1016/j.jcv.2020.104583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa H. H., Luo C. H., Morris C. P., Li M., Swanson N. J., Amadi A., et al. (2021). SARS-CoV-2 Infections in mRNA Vaccinated Individuals Are Biased for Viruses Encoding Spike E484K and Associated With Reduced Infectious Virus Loads That Correlate With Respiratory Antiviral IgG Levels. medRxiv 135, 104737. doi: 10.1016/j.jcv.2021.104737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesen M. J. M., Anand P., Silvert E., Suratekar R., Pawlowski C., Ghosh P., et al. (2021). COVID-19 Vaccines Dampen Genomic Diversity of SARS-CoV-2: Unvaccinated Patients Exhibit More Antigenic Mutational Variance. medRxiv doi: 10.1101/2021.07.01.21259833 [DOI] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M. M., et al. (2021). Reduced Sensitivity of SARS-CoV-2 Variant Delta to Antibody Neutralization. Nature 596, 276–280. doi: 10.1038/s41586-021-03777-9 [DOI] [PubMed] [Google Scholar]
- Sethuraman N., Jeremiah S. S., Ryo A. (2020). Interpreting Diagnostic Tests for SARS-CoV-2. JAMA 323, 2249–2251. doi: 10.1001/jama.2020.8259 [DOI] [PubMed] [Google Scholar]
- Suryadevara N., Shrihari S., Gilchuk P., VanBlargan L. A., Binshtein E., Zost S. J., et al. (2021). Neutralizing and Protective Human Monoclonal Antibodies Recognizing the N-Terminal Domain of the SARS-CoV-2 Spike Protein. Cell 184, 2316–2331.e15. doi: 10.1016/j.cell.2021.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarhini H., Recoing A., Bridier-Nahmias A., Rahi M., Lambert C., Martres P., et al. (2021). Long-Term Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infectiousness Among Three Immunocompromised Patients: From Prolonged Viral Shedding to SARS-CoV-2 Superinfection. J. Infect. Dis. 223, 1522–1527. doi: 10.1093/infdis/jiab075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielen P. M., Wohl S., Mehoke T., Ramakrishnan S., Kirsche M., Falade-Nwulia O., et al. (2021). Genomic Diversity of SARS-CoV-2 During Early Introduction Into the Baltimore-Washington Metropolitan Area. JCI Insight 6 (6), e144350. doi: 10.1172/jci.insight.144350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong T. T., Ryutov A., Pandey U., Yee R., Goldberg L., Bhojwani D., et al. (2021). Increased Viral Variants in Children and Young Adults With Impaired Humoral Immunity and Persistent SARS-CoV-2 Infection: A Consecutive Case Series. EBioMedicine 67, 103355. doi: 10.1016/j.ebiom.2021.103355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhteg K., Jarrett J., Richards M., Howard C., Morehead E., Geahr M., et al. (2020). Comparing the Analytical Performance of Three SARS-CoV-2 Molecular Diagnostic Assays. J. Clin. Virol. 127, 104384. doi: 10.1016/j.jcv.2020.104384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskom M. L. (2021). Seaborn: Statistical Data Visualization. J. Open Source Softw. 6 (60), 3021. doi: 10.21105/joss.03021 [DOI] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J. C., et al. (2020). Escape From Neutralizing Antibodies by SARS-CoV-2 Spike Protein Variants. Elife 9, 2020.07.21.214759. doi: 10.7554/eLife.61312.sa2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Chen Y., Yuan J., Yi P., Ding C., Wu W., et al. (2020). Factors Associated With Prolonged Viral RNA Shedding in Patients With Coronavirus Disease 2019 (COVID-19). Clin. Infect. Dis. 71, 799–806. doi: 10.1093/cid/ciaa351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., et al. (2020). Viral Load Dynamics and Disease Severity in Patients Infected With SARS-CoV-2 in Zhejiang Province, China, January-March 2020: Retrospective Cohort Study. BMJ 369, m1443. doi: 10.1136/bmj.m1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .