Abstract
Long non-coding RNAs (lncRNAs) act as important biological regulators in human cancers. The purpose of this study was to identify promising biomarkers for improved diagnosis and prognosis of papillary thyroid cancer (PTC). We analyzed the lncRNA expression profile of PTC patients and identified five upregulated and three downregulated lncRNAs as diagnostic biomarkers for PTC in our cohorts, which were confirmed using The Cancer Genome Atlas (TCGA) data. Several lncRNAs have been linked with lymph node (LN) metastasis in patients with PTC. A nomogram combining two lncRNAs, lnc-MPEG1-1:1 and lnc-ABCA12-5:2, with age, T stage, histological type, and predicted LN metastasis was developed. The area under the curve of the developed nomogram was 0.77 (0.73–0.81) in the TCGA training cohort and 0.88 (0.79–0.96) in our validation cohort. In particular, in vivo and in vitro experiments showed that overexpression of lnc-MPEG1-1:1 in PTC cell lines promoted the proliferation and migration of PTC. lnc-MPEG1-1:1 is overexpressed in the cytoplasm of PTC cells and functionally promotes cellular proliferation and migration and functions as a competitive endogenous RNA (ceRNA) by competitively occupying the shared binding sequences of miR-766-5p. lnc-MPEG1-1:1 knockdown suppressed epithelial-mesenchymal transition by miR-766-5p in PTC cells. Collectively, these results revealed a lnc-MPEG1-1:1/miR-766-5p pathway for thyroid cancer progression and suggest that a nomogram effectively predicted the LN metastasis in PTC.
Keywords: MT, non-coding RNAs, long non-coding RNA, papillary thyroid carcinoma, biomarker, lymph node metastasis, nomogram
Graphical abstract
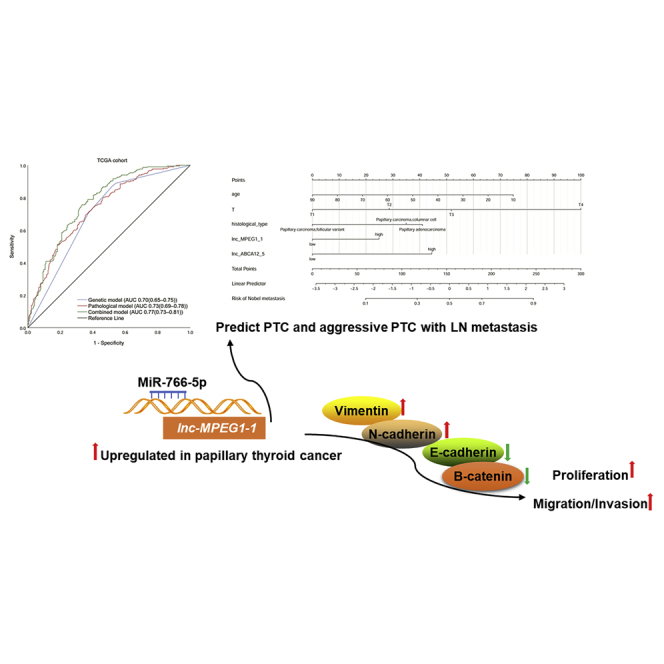
A nomogram combining lnc-MPEG1-1:1 and lnc-ABCA12-5:2 with age, T stage, and histological type predicted well the metastasis of the lymph node metastasis of PTC. Moreover, lnc-MPEG1-1:1 was highlighted to promote the proliferation and migration of PTC by occupying miR-766-5p.
Introduction
Thyroid cancer is the most common endocrine malignancy, and more than 80% of thyroid cancers are papillary thyroid carcinomas (PTCs).1 Although PTC grows slowly and has a good prognosis, it is prone to cervical lymph node (LN) metastasis in 30%–80% of patients at their first diagnosis.2 LN metastasis may increase the risk of local recurrence and distant metastasis and reduce the quality of life and long-term prognosis of patients.3 An increasing number of studies have focused on the pathogenesis of PTC growth and LN metastasis.
Long non-coding RNAs (lncRNAs) are known to be altered in a variety of tumors, allowing their potential use as a feasible biomarker for various cancers.4 In thyroid cancer, several lncRNAs, such as MALAT1, H19, BANCR, and PTCSC3, have been identified as contributing factors to cancer development.5, 6, 7, 8 However, research on lncRNAs in thyroid cancer is still in progress. Genomic profiling techniques and bioinformatics allow the screening of novel candidate biomarkers that may be involved in the pathogenesis of PTC.9 There is room for improvement in the current measures for thyroid cancer management, especially for more aggressive subgroups, and highlight the need to identify specific molecular diagnostic/prognostic biomarkers for personalized medicine.
Current diagnostic biomarkers in thyroid cancer include point mutations in the BRAF, NRAS, KRAS, and HRAS genes and rearrangements in PAX8/PPARG and RET. However, they did not increase the specificity of such diagnostic methods.10 Moreover, the performance of these kits may also differ in different populations due to the different prevalence of genetic alterations of interest. In the Australian urban population, 68% of patients with PTC have the BRAF V600E mutation.11 The mutation rate of the BRAF V600E in PTC patients in Greece was 17%.12 The mutation rate in Asian countries is higher than that in Western countries.13 The mutation rate of the BRAF V600E in Chinese patients with PTC was 72.4%, which is higher than the rate (59.7%) in the United States.14 These findings suggest that more molecular tools are needed, especially for the timely diagnosis of advanced thyroid cancers. The identification of novel molecular biomarkers to facilitate early diagnosis and predict LN metastasis would be invaluable in improving the survival rate and quality of life of patients with thyroid cancer. A series of lncRNAs have been identified as abnormally regulated and expressed in thyroid cancers.
In this study, we used lncRNA expression microarray to screen differentially expressed biomarkers between PTC and adjacent noncancerous samples, validate the candidate lncRNA in enlarged samples, and assess their diagnostic and prognostic value in PTC. Finally, we identified the molecular function of a lncRNA, lnc-MPEG1-1:1 (Ensembl: ENSG00000214797), which is highly expressed in PTC and promotes cellular growth and metastasis.
Results
Overview of the differential expressed lncRNAs in PTC
To identify candidate lncRNAs that potentially participate in PTC progression, we screened differentially expressed lncRNAs between PTC cancerous and adjacent normal thyroid tissues using lncRNA microarray. A total of 715 differentially expressed lncRNAs were determined with a fold change value ≥2.0 and a p < 0.05, 298 of which were upregulated and 417 were downregulated (Table S1). Of the genes that were located in the intergenic region, five upregulated and five downregulated lincRNAs (Table 1) in the discovery stage were subsequently validated in The Cancer Genome Atlas (TCGA) cohorts (Table 1 and Figure S1). To obtain the hub genes, we further detected these candidate lncRNAs by real-time PCR in 73 pairs of PTC primary cancerous tissues and adjacent normal thyroid tissues in our cohorts from the Sun Yat-sen University Cancer Center (SYSUCC). We confirmed that five lncRNAs were upregulated and three lncRNAs were significantly decreased in PTC samples from patients compared with non-tumor tissues (Table 1). Two downregulated lncRNAs, lnc-ARHGAP10-2:2 and lnc-PDLIM3-5:1, did not show statistical significance.
Table 1.
Discovery and validation of differential expressed lncRNAs between PTC tumorous and normal tissues
| lncRNA ID | Regulation | Discovery dataseta |
TCGA dataset |
Validation dataseta |
|||
|---|---|---|---|---|---|---|---|
| Fold change | p | Fold change | p | Fold change | p | ||
| lnc-LRRK2-1:1 | up | 39.48 | 1.70 × 10−4 | 17.73 | 1.53 × 10−28 | 70.99 | 1.14 × 10−9 |
| FAM230B | up | 20.43 | 1.95 × 10−4 | 5.08 | 4.89 × 10−11 | 62.43 | 6.80 × 10−32 |
| lnc-RXRG-1:10 | up | 19 | 0.001 | 36.07 | 1.13 × 10−37 | 183.12 | 1.44 × 10−20 |
| lnc-MPEG1-1:1 | up | 30.33 | 0.003 | 28.08 | 1.44 × 10−23 | 56.54 | 2.79 × 10−14 |
| lnc-ABCA12-5:2 | up | 40.44 | 9.36 × 10−4 | 23.06 | 3.58 × 10−29 | 6.67 | 1.29 × 10−5 |
| lnc-CENPJ-1:4 | down | −14.67 | 0.019 | −4.42 | 1.79 × 10−30 | −4.76 | 0.0012 |
| lnc-AADACL2-1:3 | down | −7.95 | 0.023 | −16.48 | 2.33 × 10−72 | −8.33 | 3.59 × 10−8 |
| lnc-TCF4-5:18 | down | −8.72 | 0.002 | −12.21 | 3.75 × 10−54 | −5.55 | 0.0003 |
| lnc-ARHGAP10-2:2 | down | −2.38 | 0.01 | −5.87 | 2.31 × 10−11 | −0.01 | 0.963 |
| lnc-PDLIM3-5:1 | down | −4.96 | 0.018 | −3.96 | 2.08 × 10−17 | −2.38 | 0.182 |
The study samples were enrolled from SYSUCC.
Correlation of the hub lncRNAs with TNM stage of PTC
To explore the clinical significance of the 10 candidate lncRNAs in PTC, we analyzed the correlation between their expression levels and TNM staging of PTC patients in the TCGA cohorts (Table 2). These lncRNAs were positively correlated with LN metastasis, except for lnc-ARHGAP10-2:2, which is rarely expressed in PTC cancerous tissues. lnc-RXRG-1:10, lnc-MPEG1-1:1, and lnc-ABCA12-5:2 were significantly increased and lnc-CENPJ-1:4, lnc-AADACL2-1:3, lnc-TCF4-5:18, and lnc-PDLIM3-5:1 were decreased in patients with advanced T stage and advanced TNM staging. We also explored whether these lncRNAs were differently expressed between N1a and N1b, and therefore we divided the N1 into N1a and N1b subgroups. It was found that lnc-LRRK2-1:1 was upregulated in N1b while downregulated in N1a (p value for N1b versus N1a: 3.13 × 10−4). However, the other nine lncRNA did not show statistical difference between N1a and N1b (Table 2).
Table 2.
Correlation of hub lncRNAs with TNM staging in TCGA dataset
| Target lncRNA | T Stage |
N stage |
N stage |
M Stage |
TNM staging |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1–2 (305) | T3–4 (187) | N0 (226) | N1 (219) | N0 (226)a | N1a (88)b | N1b(73)c | M0 (278) | M1 (8) | I–II (329) | III–IV (163) | ||
| lnc-LRRK2-1:1 | median | 89.69 | 89.4 | 87.23 | 91.94 | 87.23 | 84.69 | 91.02 | 88.69 | 87.57 | 87.56 | 90.32 |
| p | 0.54 | 0.04 | 0.95 | 1.66 × 10−3 | 3.13 × 10−4 | 0.59 | 0.09 | |||||
| FAM230B | median | 0.38 | 0.35 | 0.29 | 0.5 | 0.29 | 0.53 | 0.34 | 0.43 | 0.94 | 0.36 | 0.38 |
| p | 0.44 | 1.51 × 10−4 | 1.97 × 10−3 | 5.31 × 10−2 | 0.22 | 0.18 | 0.46 | |||||
| lnc-RXRG-1:10 | median | 26.55 | 37.24 | 20.82 | 45.92 | 20.82 | 48.99 | 31.29 | 37.57 | 44.12 | 29.62 | 43.48 |
| p | 0.01 | 2.61 × 10−9 | 7.64 × 10−7 | 3.59 × 10−5 | 0.64 | 0.64 | 0.01 | |||||
| lnc-MPEG1-1:1 | median | 18.37 | 32.99 | 12.1 | 36.8 | 12.1 | 34,.54 | 19.49 | 30.61 | 26.01 | 19.67 | 36.88 |
| p | 2.04 × 10−6 | 4.95 × 10−14 | 3.85 × 10−9 | 1.06 × 10−6 | 0.88 | 0.97 | 4.31 × 10−6 | |||||
| lnc-ABCA12-5:2 | median | 186.87 | 217.49 | 174.53 | 222.17 | 174.53 | 218.97 | 196.82 | 211.05 | 165.81 | 192.87 | 226.05 |
| p | 1.19 × 10−9 | 6.88 × 10−15 | 4.97 × 10−9 | 2.70 × 10−9 | 0.22 | 0.77 | 4.70 × 10−9 | |||||
| lnc-CENPJ-1:4 | median | 27.72 | 24.53 | 27.65 | 24.21 | 27.65 | 24.58 | 25.79 | 25.74 | 23.97 | 27.99 | 24 |
| p | 0.01 | 6.43 × 10−3 | 0.08 | 5.15 × 10−3 | 0.26 | 0.75 | 2.53 × 10−4 | |||||
| lnc-AADACL2-1:3 | median | 2.19 | 1.57 | 2.31 | 1.44 | 2.31 | 1.51 | 2.10 | 2.06 | 1.59 | 2.08 | 1.38 |
| p | 2.78 × 10−4 | 1.40 × 10−3 | 7.97 × 10−3 | 0.16 | 0.37 | 0.29 | 1.70 × 10−4 | |||||
| lnc-TCF4-5:18 | median | 2.65 | 1.43 | 2.86 | 1.27 | 2.86 | 1.47 | 2.20 | 1.8 | 0.79 | 2.4 | 1.35 |
| p | 6.63 × 10−4 | 1.02 × 10−3 | 9.00 × 10−3 | 3.62 × 10−2 | 0.82 | 0.39 | 4.61 × 10−3 | |||||
| lnc-ARHGAP10-2:2 | median | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| p | 0.88 | 0.17 | 0.83 | 0.06 | 0.16 | 0.68 | 0.78 | |||||
| lnc-PDLIM3-5:1 | median | 62.46 | 54.6 | 65.85 | 51.27 | 65.85 | 51.57 | 60.90 | 57.53 | 41.31 | 60.59 | 51.91 |
| p | 9.76 × 10−6 | 3.78 × 10−13 | 1.6 × 10−7 | 1.59 × 10−6 | 0.93 | 0.08 | 5.98 × 10−8 | |||||
p value between N1a versus N0.
p value between N1b versus N0.
p value between N1b versus N1a.
Diagnostic and predictive performance of a single lncRNA for PTC
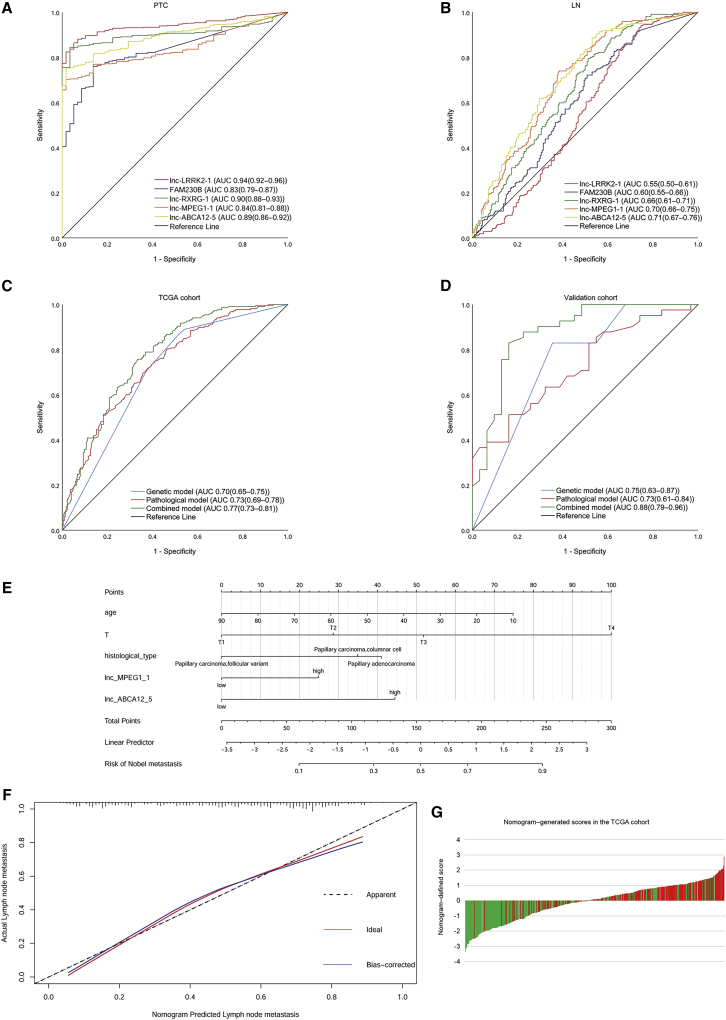
We further investigated the diagnostic and predictive role of the hub lncRNAs and mainly focused on upregulated lncRNAs, given that these lncRNAs may serve as improved diagnostic or predictive biomarkers. In the receiver operating characteristic (ROC) curve analysis of a single lncRNA (Figures 1A and 1B ; Table 3), almost all of the area under the curve (AUC) for PTC diagnosis were larger than 0.80 in the TCGA cohort and in our cohort, except lnc-ABCA12-5:2, indicating that these lncRNAs may serve as improved diagnostic biomarkers for PTC. In addition, three lncRNAs showed potential in predicting LN metastasis in PTC, particularly for lnc-MPEG1-1:1, the AUC of which exceeded 0.70 in both the TCGA cohort and our validation cohort.
Figure 1.
Diagnostic and predictive performance of lncRNA for papillary thyroid cancer (PTC)
(A) Receiver operating characteristic curve (ROC) curves of a single lncRNA for the diagnosis of PTC in the Sun Yat-sen University Cancer Center (SYSUCC) cohort. (B) ROC curves of a single lncRNA for predicting the lymph node (LN) metastasis of PTC in the SYSUCC cohort. (C) ROC curves for the models in The Cancer Genome Atlas (TCGA) cohort. (D) ROC curves for the models in the SYSUCC cohort. (E) Nomogram for predicting LN metastasis. (F) Calibration curve for the nomogram. (G) Nomogram-generated scores for each patient in the TCGA cohort.
Table 3.
Performance of a single lncRNA for PTC diagnosis and LN metastasis
| Event | Lnc-LRRK2-1:1 | FAM230B | Lnc-MPEG1-1:1 | Lnc-ABCA12-5:2 | Lnc-RXRG-1:10 |
|---|---|---|---|---|---|
| Predicting PTC | |||||
| TCGA cohort | |||||
| AUC (95% CI) | 0.94 (0.92–0.96) | 0.83 (0.79–0.87) | 0.84 (0.81–0.88) | 0.89 (0.86–0.92) | 0.90 (0.88–0.93) |
| p | 5.05 × 10−28 | 1.07 × 10−16 | 2.11 × 10−17 | 1.57 × 10−22 | 7.25 × 10−24 |
| SYSUCC cohort | |||||
| AUC (95% CI) | 0.80 (0.72–0.87) | 0.89 (0.84–0.95) | 0.84 (0.77–0.90) | 0.76 (0.68–0.84) | 0.90 (0.84–0.95) |
| p | 3.32 × 10−10 | 60.2 × 10−17 | 7.65 × 10−13 | 2.22 × 10−8 | 3.64 × 10−17 |
| Predicting LN metastasis | |||||
| TCGA cohort | |||||
| AUC (95% CI) | 0.55 (0.50–0.60) | 0.60 (0.55–0.65) | 0.70 (0.66–0.75) | 0.71 (0.67–0.76) | 0.66 (0.61–0.71) |
| p | 0.07 | 2.00 × 10−4 | 9.56 × 10−14 | 7.60 × 10−15 | 6.01 × 10−9 |
| SYSUCC cohort | |||||
| AUC (95% CI) | 0.56 (0.42–0.70) | 0.68 (0.56–0.80) | 0.70 (0.57–0.84) | 0.65 (0.52–0.78) | 0.60 (0.46–0.73) |
| p | 0.38 | 7.64 × 10−3 | 2.52 × 10−3 | 0.02 | 0.15 |
Nomogram development for predicting LN metastasis in PTC patients
We analyzed the patients’ general variables (age and sex), pathological parameters, and gene expression levels between the LN+ and LN− groups in the TCGA data. Univariate logistic analysis showed an association between age, tumor size, T stage, histological type, and three upregulated lncRNAs (lnc-RXRG-1:10, lnc-MPEG1-1:1, and lnc-ABCA12-5:2) with LN metastasis. The data from TCGA database included columnar and follicular variants but did not involve the tall cell variant, which precludes us from the analysis of the tall cell variant of PTC. Multivariate logistic analysis constructed a model combining age, T stage, and histological type with lnc-MPEG1-1:1 and lnc-ABCA12-5:2 for predicting LN metastasis (Table S2). The AUC for this model (95% confidence interval [CI]) in the TCGA cohort was 0.77 (0.73–0.81) and the AUC (95% CI) in the SYSUCC cohort was 0.88 (0.79–0.96). These results indicated that the combined model outperformed the gene expression alone and the clinical pathological parameters alone (Figures 1C and 1D). Moreover, similar predicting efficacies were observed between N1a and N1b (Figure S2). A nomogram was derived from this model based on the resulting coefficients from the multivariate logistic analysis. The calibration curve revealed excellent agreement between the estimates derived from the nomogram and the actual probabilities. Within the five variables that construct the nomogram, each model covariate was assigned a score by drawing a vertical line straight down to the axis-labeled points. By summing the total score and locating it on the total point scale, the individual probabilities of LN metastasis can be determined. The nomogram-generated scores for each patient in the TCGA data are depicted in Figures 1E–1G. A high score indicated a high risk of LN metastasis. We assessed the external validity of the nomogram constructed using the SYSUCC cohort. ROC curve analysis was performed using the TCGA dataset and the validation dataset (Table 4). To assess the accuracy of the nomogram, the patients were classified into four groups according to the quartile of the predicted score as low risk, intermediate risk, high risk, and very high risk (Table 5).
Table 4.
Performance of prediction models for LN metastasis
| Dataset | AUC(95%CI) | Cutoff value | Sen | Spe | PPV | NPV | p Value |
|---|---|---|---|---|---|---|---|
| For TCGA cohort | |||||||
| Genetic model | 0.70 (0.65–0.75) | 0.41 | 0.87 | 0.48 | 0.62 | 0.79 | 3.23 × 10−13 |
| Pathological model | 0.73 (0.69–0.78) | 0.47 | 0.75 | 0.59 | 0.64 | 0.71 | 1.99 × 10−17 |
| Combined model | 0.77 (0.73–0.81) | 0.49 | 0.76 | 0.67 | 0.69 | 0.74 | 9.81 × 10−23 |
| For validation cohort | |||||||
| Genetic model | 0.75 (0.63–0.87) | 0.58 | 0.83 | 0.65 | 0.76 | 0.74 | 3.33 × 10−4 |
| Pathological model | 0.73 (0.61–0.84) | 0.53 | 0.51 | 0.84 | 0.81 | 0.57 | 1.10 × 10−3 |
| Combined model | 0.88 (0.79–0.96) | 0.49 | 0.83 | 0.84 | 0.87 | 0.79 | 5.45 × 10−8 |
LN, lymph node.
Table 5.
Number of events in the nomogram-defined risk groups
| Dataset | Risk group 1 (n = 111) | Risk group 2 (n = 111) | Risk group 3 (n = 111) | Risk group 4 (n = 110) |
|---|---|---|---|---|
| TCGA cohort (n = 443) | 15 (13.5%) | 52 (46.8%) | 66 (59.5%) | 85 (77.3%) |
| Risk group 1 (n = 18) | Risk group 2 (n = 18) | Risk group 3 (n = 18) | Risk group 4 (n = 18) | |
|---|---|---|---|---|
| Validation cohort (n = 73) | 2 (11.1%) | 8 (44.4%) | 15 (83.3%) | 16 (88.9%) |
Effects of lnc-MPEG1-1:1 overexpression on the proliferation and viability of PTC cells in vitro and in vivo
Intriguingly, lnc-MPEG1-1:1 (referred to as MPEG1 in subsequent functional analysis) exhibited an AUC of 0.70 in the TCGA cohorts and in our cohorts, which is also implicated in a nomogram to better predict the LN metastasis of PTC. It is suggested that MPEG1 plays an important role in the development and metastasis of PTC. MPEG1 is located on chromosome 11 in humans and is composed of two exons with a full length of 2,805 nt. There was no protein-coding gene between its upstream and downstream 20 kb (http://asia.ensembl.org/) (Figure S3). Compared with normal thyroid epithelial cells (Hrori-3), the expression of MPEG1 in PTC cell lines (TPC1 and BCPAP) was significantly higher (p < 0.001; Figure S4A). We then overexpressed MPEG1-1 in TPC1 and silenced MPEG1-1 in BCPAP cells through transfection. As shown in Figures S4B and S4C, MPEG1-1 expression was notably increased in the MPEG1-1 overexpression group and markedly decreased in the MPEG1-1 silencing groups (especially for the sh-MPEG1-1#2 group) compared with that in the control group (all p < 0.001).
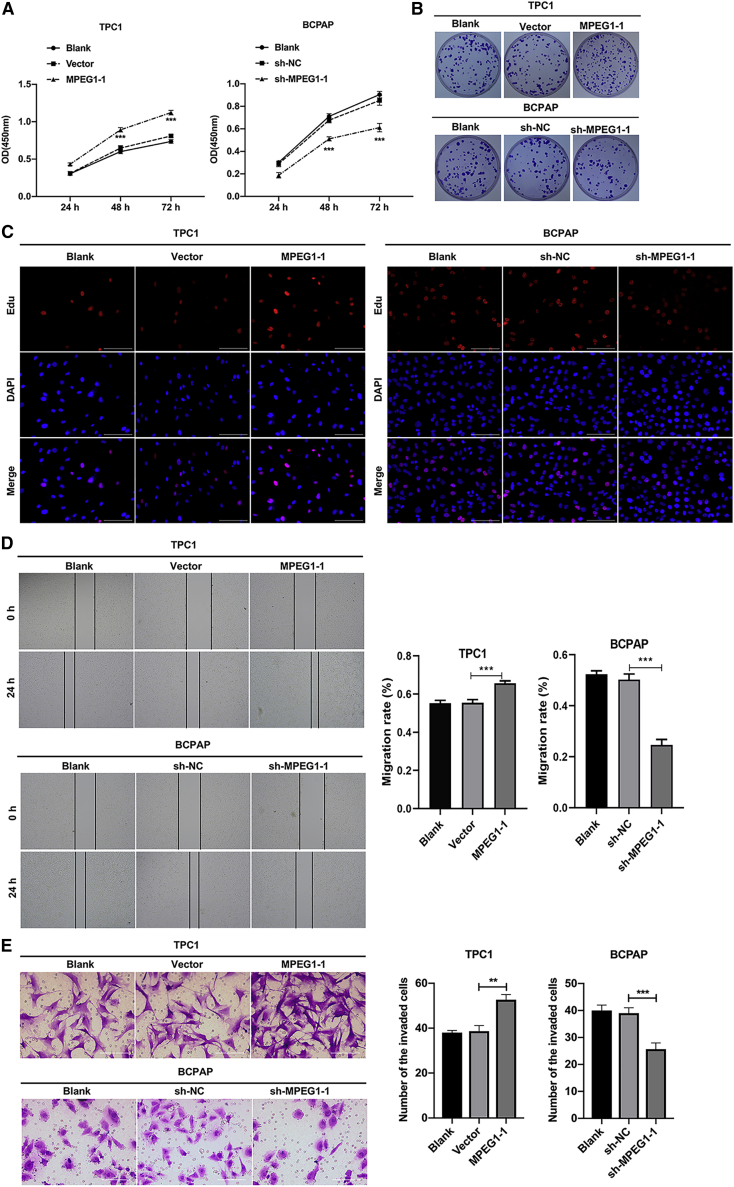
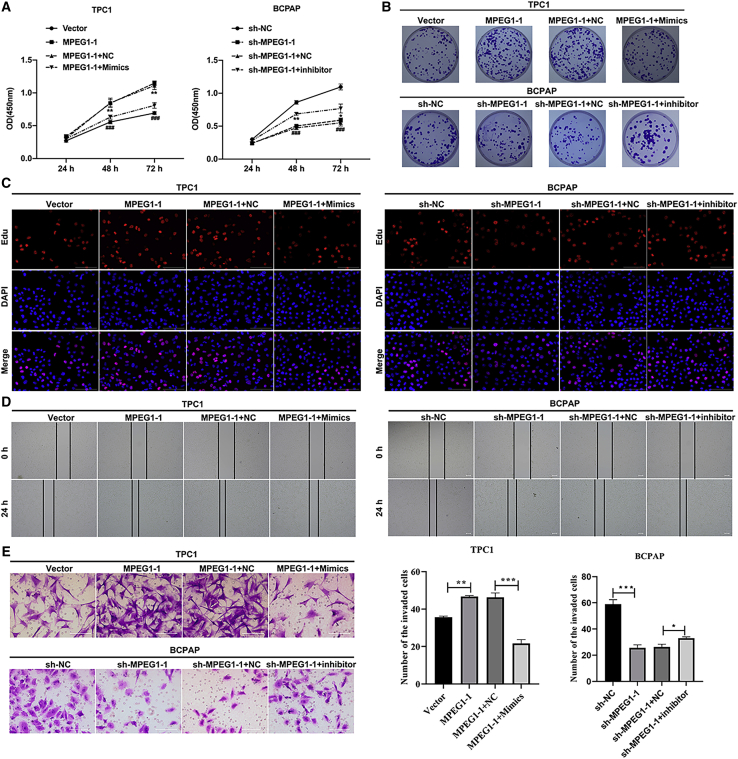
Subsequently, we studied the influence of MPEG1-1 overexpression or knockdown on the malignant biological properties of PTC cells. Cell counting kit-8 (CCK-8) data showed that the viability of TPC1 cells was significantly reduced in the MPEG1-1 overexpression group compared to that in the vector group, and the viability of BCPAP cells was prominently diminished in the MPEG1-1 silencing group versus that in the short hairpin negative control (sh-NC) group (p < 0.001; Figure 2A). Meanwhile, colony formation results indicated that the overexpression of MPEG1-1 enhanced the colony-forming capacity of TPC1 cells, and silencing of MPEG1-1 restrains the colony-forming capacity of BCPAP cells (Figures 2B and S5A). Similarly, EdU staining also revealed the proliferation induction of TPC1 cells mediated by the overexpression of MPEG1-1 and proliferation suppression of BCPAP cells mediated by knockdown of MPEG1-1 (Figure 2C). In addition, the wound healing data indicated that the migration ability of TPC1 cells was dramatically strengthened in the MPEG1-1 overexpression group relative to that in the vector group, and the migration ability of BCPAP cells was notably reduced in the MPEG1-1 knockdown group compared to that in the sh-NC group (Figure 2D). Moreover, Transwell assays showed that MPEG1-1 overexpression remarkably enhanced the invasiveness of TPC1 cells, and MPEG1-1 knockdown resulted in a noteworthy reduction in the invasiveness of BCPAP cells (Figure 2E). In short, the current results confirmed that an increase in MPEG1-1 could accelerate the malignant progression of PTC cells.
Figure 2.
MPEG1-1 observably enhanced PTC cell proliferation, migration, and invasion
(A) CCK-8 assay was adopted to analyze the effects of MPEG1-1 overexpression or knockdown on the proliferation of TPC1 or BCPAP cells at the appointed time. (B) Clony formation assay was used to evaluate the colony-forming capacity of MPEG1-1-overexpressed TPC1 cells and MPEG1-1-silenced BCPAP cells. (C) After MPEG1-1 overexpression or knockdown, cell proliferation was tested using EdU staining in TPC1 and BCPAP cells. (D) Wound healing displayed the change in cell migration in MPEG1-1-upregulated TPC1 cells and MPEG1-1-downregulated BCPAP cells. (E) Cell invasion was evaluated via Transwell assay in TPC1 or BCPAP cells, which were transfected with MPEG1-1-overexpressed plasmid or shRNAs. ∗p < 0.05, ∗∗∗p < 0.001.
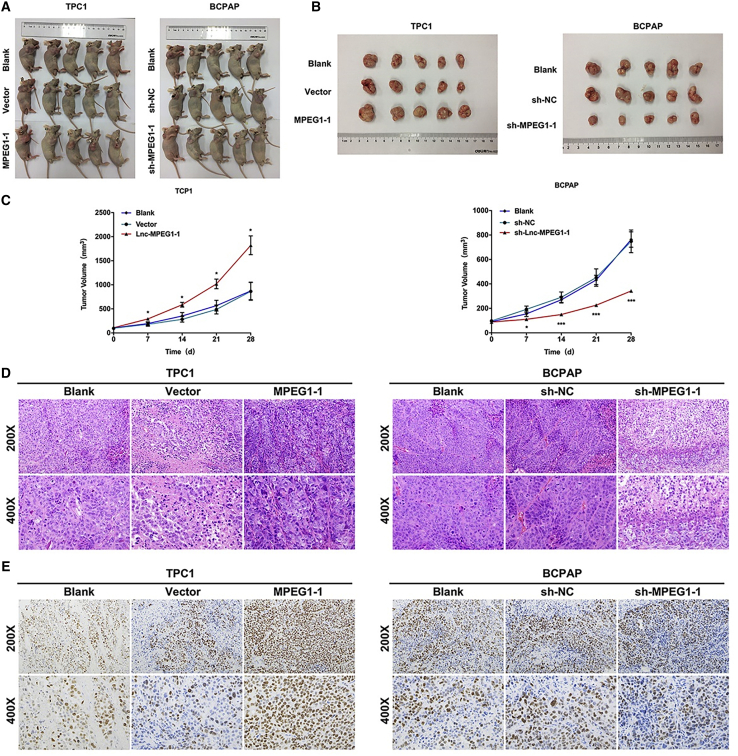
Based on the above results in PTC cells, we further speculated that MPEG1-1 could also significantly facilitate the tumor growth of PTC in vivo. To verify this hypothesis, nude mice were subcutaneously injected with MPEG1-1-overexpressed TPC1 and MPEG1-1-silenced BCPAP cells for 28 days. Moreover, tumor-forming nude mice and the subcutaneous tumors were presented, and we discovered that the size of the subcutaneous tumors was larger in the MPEG1-1 overexpression group than that in the vector group, and the size was smaller in the MPEG1-1 knockdown group than that in the sh-NC group (Figures 3A and 3B). Likewise, the growth curve of the tumor showed that the overexpression of MPEG1-1 markedly increased the tumor volume, and silencing of MPEG1-1 significantly degraded the tumor volume (Figure 3C). In addition, H&E staining results indicated that in the blank and control groups, there were abundant necrotic cells in the tumor along with nuclear fragmentation and pyknosis. While the overexpression of MPEG1-1 could dramatically aggravate this pathological change, the silencing of MPEG1-1 could memorably alleviate this pathological change in the tumors of mice (Figure 3D). More important, immunohistochemistry (IHC) results also showed that Ki67 expression was prominently elevated in the MPEG1-1 overexpression group compared to that in the vector group, and Ki67 expression was significantly lowered in the MPEG1-1 knockdown group relative to that in the sh-NC group (Figure 3E). Consequently, our data suggest that the upregulation of MPEG1-1 is relevant to the promotion of tumor growth and exacerbation of the pathological structure of PTC in vivo.
Figure 3.
MPEG1-1 prominently accelerated tumor growth in the mice xenograft model with PTC cells
After MPEG1-1 overexpression or knockdown, 2 × 106 TPC1 and BCPAP cells were subcutaneously injected into nude mice for 28 days. (A) Tumor formation was demonstrated in each group of nude mice. (B) Four weeks later, the subcutaneous tumors were excised and presented. (C) The length and width of the tumor in each group were measured and its volume was calculated at 7, 14, 21, and 28 days. (D) Pathological construction of tumors was observed by H&E staining in each group. (E) Expression level of Ki67 was examined by immunohistochemistry assay in each group. All of the experiments were performed in triplicate, and data are presented as the means ± SDs. ∗p < 0.05. ∗∗∗p < 0.001.
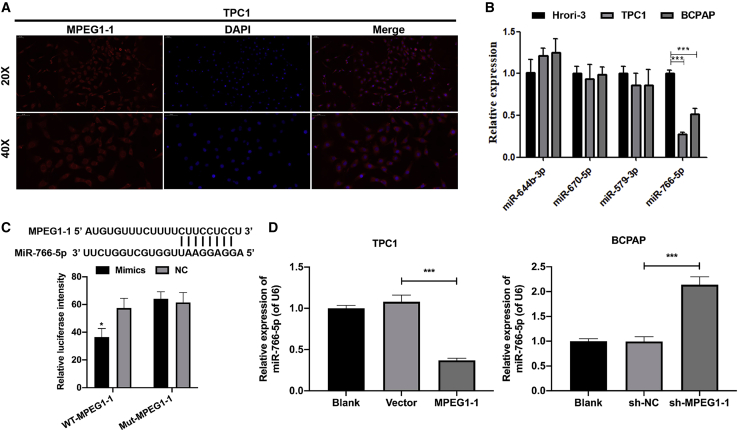
MPEG1 function as a miR-766-5p sponge in PTC cells
We predicted the subcellular localization of MPEG1-1 using IncLocator (http://www.csbio.sjtu.edu.cn/bioinf71ncLocator/) and discovered that MPEG1-1 is located in the cytoplasm. The FISH results also showed that MPEG1-1 was mainly located in the cytoplasm (Figure 4A). Thus, we further explored the underlying microRNAs (miRNAs) that may be targeted by MPEG1-1. Through bioinformatics software predictions, we found that MPEG1-1 has potential binding sites with miR-644-3p, miR-670-5p, miR-579-3p, and miR-766-5p. We confirmed that miR-766-5p was downregulated in PTC cells (TPC1 and BCPAP) relative to Nthy-ori 3-1 cells by real-time PCR (Figure 4B). Thus, we further verified the regulatory effect of MPEG1-1 on miR-766-5p in PTC cells. The luciferase reporter results showed that the luciferase activity of WT-MPEG1-1 was significantly reduced in the miR-766-5p overexpression group compared to the control group, while the overexpression of miR-766-5p had no effect on the luciferase activity of MUT-MPEG1-1, indicating the targeted regulation of MPEG1-1 on miR-766-5p (p < 0.05; Figure 4C). Simultaneously, real-time PCR data showed that the overexpression of MPEG1-1 downregulated miR-766-5p in TPC1 cells and thatsilencing of MPEG1-1 significantly upregulated miR-766-5p in BCPAP cells (p < 0.001; Figure 4D). Overall, we demonstrated that the overexpression of MPEG1-1 prominently downregulated miR-766-5p by targeting regulation in PTC cells.
Figure 4.
MPEG1-1 notably inhibited the synthesis of miR-766-5p in PTC cells
(A) Fluorescence in situ hybridization (FISH) assay was applied to detect the co-localization of lncRNA. (B) Expression levels of miR-644-3p, miR-670-5p, miR-579-3p, and miR-766-5p in Nthy-ori 3-1, TPC1, and BCPAP cells, respectively, were analyzed by real-time PCR. (C) TPC1 cells were co-transfected with WT-MPEG1-1 or MUT-MPEG1-1 together with miR-766-5p mimics, and the luciferase intensity was determined by luciferase reporter assay. (D) real-time PCR assay was adopted to monitor the expression level of miR-766-5p in MPEG1-1-upregulated TPC1 cells and MPEG1-1-downregulated BCPAP cells. ∗p < 0.05, ∗∗∗p < 0.001.
In accordance with the result that miR-766-5p could be targeted by MPEG1-1 in PTC, we further investigated the effects of MPEG1-1 and miR-766-5p on the biological processes of PTC cells using rescue experiments. MPEG1-1-overexpressed plasmid and miR-766-5p mimics were co-transfected into TPC1 cells, and MPEG1-1 and miR-766-5p inhibitors were co-transfected into BCPAP cells. After successful co-transfection, we verified the influence of MPEG1-1 and miR-766-5p on the functions of TPC1 and BCPAP cells. CCK-8 data first signified that the overexpression of MPEG1-1 signally aggrandized the viability of TPC1 cells, while this change could be notably reversed by miR-766-5p; the knockdown of MPEG1-1 restrained the viability of BCPAP cells, while this change could also be partly reversed by an miR-766-5p inhibitor (p < 0.001; Figure 5A). Similarly, colony formation results denoted that the overexpression of miR-766-5p could prominently reduce the number of clones, which has been promoted by MPEG1-1 overexpression in TPC1 cells, and the inhibition of miR-766-5p could markedly increase the number of clones, which has been lowered by MPEG1-1 knockdown BCPAP cells (Figures 5A and S5B). Likewise, EdU staining results also showed that the overexpression of miR-766-5p dramatically reversed the promotion of proliferation mediated by the MPEG1-1 overexpression in TPC1 cells and that the inhibition of miR-766-5p markedly reversed the inhibition of proliferation mediated by MPEG1-1 knockdown in BCPAP cells (Figure 5C). Next, wound healing data indicated that the induction of cell migration mediated by MPEG1-1 overexpression in TPC1 cells could be attenuated by miR-766-5p overexpression, and the inhibition of cell migration mediated by MPEG1-1 knockdown in BCPAP cells could be weakened by the miR-766-5p inhibitor (Figure 5D). In addition, Transwell data also showed that cell invasion accelerated by MPEG1-1 overexpression in TPC1 cells could be receded significantly by miR-766-5p overexpression and that cell invasion suppressed by MPEG1-1 knockdown in BCPAP cells could be notably subsided by the miR-766-5p inhibitor (Figure 5E). We demonstrated that miR-766-5p is required for MPEG1-1 to repress the proliferation, migration, and invasion of PTC cells.
Figure 5.
Cancer progression mediated by MPEG1-1 was partially reversed by miR-766-5p in PTC cells
TPC1 cells were co-transfected with MPEG1-1-overexpressed plasmid and miR-766-5p mimics, and BCPAP cells were co-transfected with MPEG1-1 shRNAs and miR-766-5p inhibitor. (A) Change of cell proliferation was assessed by CCK-8, ∗p < 0.05, ∗∗p < 0.01 versus vector group; ###p < 0.001 vs. MPEG1-1+NC group. (B) Colony formation assay was performed to evaluate the change in cell viability. (C) Change of cell viability was also analyzed via EdU staining. (D) Wound healing was performed to confirm the change in migration ability. (E) Transwell assay showed the change in cell invasion ability. ∗p < 0.05, ∗∗∗p < 0.001.
Inhibition of miR-766-5p reversed the changes of EMT mediated by MPEG1-1 knockdown in BCPAP cells to a certain extent
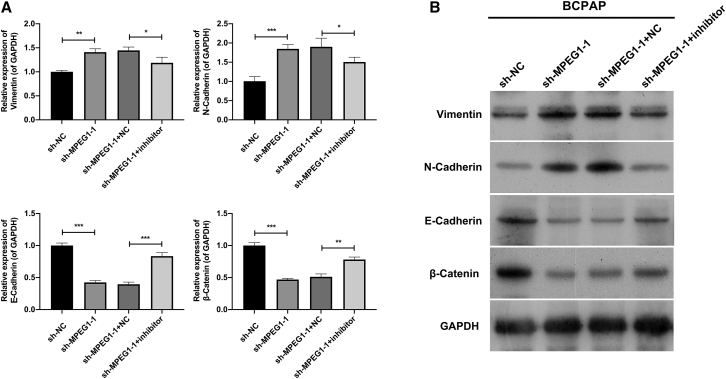
Subsequently, we explored the potential mechanism of MPEG1-1 and miR-766-5p in PTC progression, especially metastasis. Through real-time PCR screening, we found that epithelial-mesenchymal transition (EMT)-related biomarkers were altered. Vimentin and N-cadherin were markedly downregulated, and E-cadherin and β-catenin were observably upregulated in the MPEG1-1 knockdown group relative to that in the sh-NC group, while the changes in the expression of these genes mediated by MPEG1-1 knockdown could be partially reversed by miR-766-5p inhibitor in BCPAP cells (Figure 6). Thus, our findings revealed that MPEG1-1 knockdown suppressed EMT progression by miR-766-5p in PTC cells.
Figure 6.
Inhibition of miR-766-5p reversed the changes of EMT mediated by MPEG1-1 knockdown in BCPAP cells to a certain extent
(A and B) MPEG1-1 shRNAs and miR-766-5p inhibitor were co-transfected into BCPAP cells. real-time PCR assay and western blot were applied to determine the changes in vimentin, E-cadherin, N-cadherin, and β-catenin expressions.
Discussion
In recent years, the incidence of PTC has shown a significant upward and younger trend.15 This disease is often asymptomatic but aggressive in its early stages. In some patients, surgery does not result in the destruction of the neoplasm and inhibition of the natural course of the tumor.16 These cases may exhibit recurrence and LN metastasis.17 Therefore, it is important to identify the factors that may indicate a more aggressive course of PTC. Such information should affect treatment decisions and the effectiveness of treatment to ultimately achieve PTC remission.
The biological role of lncRNAs has attracted increasing attention in cancer progression research.18 Recent research indicates that lncRNAs play an important role in the biology of thyroid cancers. For instance, studies have shown a significant overexpression of lncRNA GAS5, MALAT1, and PTCSC3 in thyroid cancer and revealed an association with LN metastasis.5,8,19 Wang et al. also confirmed that PTCSC3 has a tumor-suppressive function in thyroid cancer cells.8 However, research on lncRNAs in thyroid cancer is still in progress. In this study, we screened associated lncRNAs for PTC in only three pairs of classical variant PTC, with a risk of data loss of certain lncRNAs and fluctuation in screening results. To avoid false-positive results due to the limited sequencing samples, we subsequently verified 10 hub lncRNAs by real-time PCR methods. The upregulated lncRNAs showed good diagnostic performance for the diagnosis of PTC. Among these, two lncRNAs, lnc-MPEG1-1:1 and lnc-ABCA12-5:2, are closely related to LN metastasis. Therefore, a nomogram was developed combining these lncRNAs and pathological factors to predict LN metastasis. The nomogram identified in the present study demonstrated an AUC of 0.77 in the TCGA cohort and 0.88 in the validation cohort, showing certain potential in identifying patients presenting with LN metastasis. The training and validation cohorts may ensure the reliability of the model developed in this study. Effective treatment to ultimately achieve PTC remission is of utmost importance for patients with LN metastasis.
lnc-MPEG1-1:1 is a novel diagnostic and predictive biomarker for PTC. This finding motivated our further investigation of the putative role of lnc-MPEG1-1:1 as an oncogene PTC. There is currently no report of lnc-MPEG1-1:1-related functions and mechanisms in cancer on the basis of literature retrieval. Therefore, in vitro experiments were conducted targeting lnc-MPEG1-1:1. In this study, lnc-MPEG1-1:1 was confirmed to be highly expressed in PTC cells. Meanwhile, its overexpression could dramatically accelerate the proliferation and promote the metastasis of PTC cells, while knockdown experiments also demonstrated an opposite change in PTC cells. In vivo experiments further confirmed that it had a significant promoting effect on tumor growth in a mouse xenograft model. Overall, we demonstrated for the first time that, as an oncogene, the dysfunction of lnc-MPEG1-1:1 is likely to be a risk factor for PTC progression.
In 2011, the Pandolfi research group of the Harvard Medical School proposed the competitive endogenous RNA (ceRNA) hypothesis.20 Various RNA molecules, such as lncRNA, mRNA, and pseudo-gene transcripts, can act as ceRNAs to shield the inhibition or degradation of another mRNA by a miRNA. Using cell cytoplasmic/nuclear fractionation and RNA fluorescence in situ hybridization (FISH) assays, we found that lnc-MPEG1-1:1 was preferentially localized in the cytoplasm, indicating its potential for functioning as a miRNA sponge. Using bioinformatics, we further found that lnc-MPEG1-1:1 is likely to be a sponge of miR-766-5p. In vitro, we revealed for the first time that lnc-MPEG1-1:1 can serve as an miR-766-5p sponge and that miR-766-5p could also be prominently downregulated by lnc-MPEG1-1:1 overexpression in PTC cells. In addition, we demonstrated that the blocking effect of lnc-MPEG1-1:1 on PTC progression is achieved through the targeted regulation of miR-766-5p. Based on reports in the literature, we also discovered that miR-766-5p, a tumor suppressor gene, is crucial in a variety of cancers.21, 22, 23, 24 Together, our results revealed that lnc-MPEG1-1:1 could serve as a ceRNA by sponging miR-766-5p in thyroid cancer.
In summary, we determined that the uncharacterized lnc-MPEG1-1:1 was significantly increased in PTC tissues and participated in a monogram combining several other risk factors that predict LN metastasis in PTC patients. Functional studies revealed that lnc-MPEG1-1:1, as an miR-766-5p sponge, could promote the proliferation and mobility of PTC cells in vitro and in vivo, indicating a tumor-promoter role in PTC. Although several dysregulated lncRNAs have been identified, more studies are needed to elucidate their functions.
Materials and methods
Study design and study samples
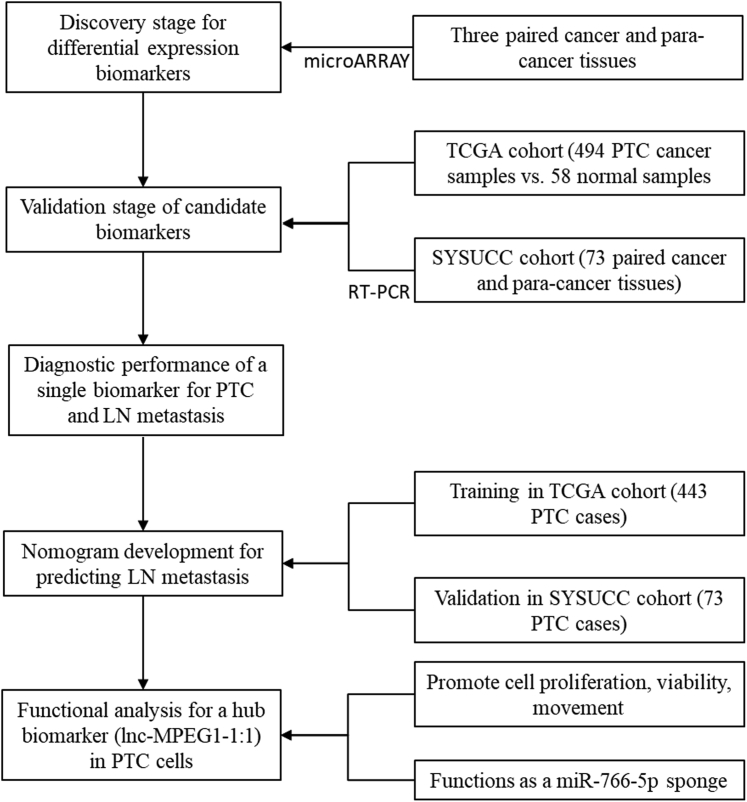
A flowchart of this study is shown in Figure 7. As depicted, we used lncRNA expression microarray to screen differentially expressed biomarkers between paired PTC and adjacent noncancerous tissues that were recruited from the SYSUCC. The expression of candidate lncRNAs was validated in the TCGA and SYSUCC cohorts. The diagnostic value for PTC and LN metastasis was explored, and accordingly, a predictive nomogram for LN metastasis was developed and validated. Finally, we identified the molecular function of a hub lncRNA, lnc-MPEG1-1, which is significantly overexpressed in PTC.
Figure 7.
Study design flowchart
In the SYSUCC cohort, a total of 76 PTC patients were recruited, and postoperative cancer and adjacent noncancerous tissues were collected during 2014 and 2017. Three paired tissues were detected by microarray in the discovery stage, and 73 paired tissues were detected by real-time PCR in the validation stage. In the TCGA cohorts, RNA sequencing (RNA-seq) data of 552 samples, including 494 PTC cancer samples and 58 para-cancer samples, and their clinicopathological information were downloaded from the University of California, Santa Cruz (UCSC) Cancer Genomics Browser (https://genome-cancer.ucsc.edu).
Microarray experiment and data analysis
An Agilent Human lncRNA (4∗ 180 K, Design: 076500) was used in this experiment. Microarray analysis was performed using BIO MIAO Biological. Total RNA was quantified using a NanoDrop ND-2000 (Thermo Scientific), and RNA integrity was assessed using Agilent Bioanalyzer 2100 (Agilent Technologies). Sample labeling, microarray hybridization, and washing were performed according to the manufacturer’s standard protocols. Briefly, total RNA was transcribed to double-stranded cDNA, then synthesized into cRNA, and labeled with cyanine-3-CTP. The labeled cRNAs were hybridized to the microarrays. After washing, the arrays were scanned using an Agilent Scanner G2505C (Agilent Technologies).
Feature Extraction software (version 10.7.1.1, Agilent Technologies) was used to analyze array images to obtain raw data. GeneSpring (version 13.1, Agilent Technologies) was used to complete the basic analysis of the raw data. The raw data were normalized with the quantile algorithm. Probes with at least one of the two conditions with flags in “P” were chosen for further data analysis. Differentially expressed genes or lncRNAs were then identified through fold change as well as the p value calculated using the t test. The threshold set for up- and downregulated genes was a fold change ≥2.0 and a p ≤ 0.05. Subsequently, hierarchical clustering was performed to display the distinguishable gene expression patterns among the samples (Figure S6).
Quantitative real-time PCR assay
Total RNA was extracted from the tissues or cultured cells using the TRIzol Reagent (cat. no. 15596026, Thermo Fisher). Total RNA (2 μg) was reverse transcribed into cDNA using a PrimeScript RT reagent kit (cat. no. RR047A; TaKaRa Biotech). The relative expression levels were determined by qPCR using a SYBR Premix Ex Taq II kit (cat. no. RR820A; TaKaRa Biotech) on an Applied Biosystems 7500 qPCR system according to the manufacturer’s instructions. The conditions used for qPCR analysis were as follows: 95°C for 30 s, followed by 45 cycles of 95°C for 5 s and 60°C for 30 s. The relative expression of lncRNAs and mRNAs was normalized to that of the internal control, glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The relative expression levels of lncRNAs and mRNAs were calculated using the 2−ΔΔCt method. The primer sequences are presented in Table S3.
Western blot
Total proteins were also extracted from the transfected PTC cells after cell lysis with radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime). After quantification, 40μg protein was subjected to electrophoresis with sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes. After blocking, the membranes were exposed to specific primary antibodies (vimentin: Cell Signaling, 5741S, diluted as 1:1,000; E-cadherin: Cell Signaling, 14472S, diluted as 1:1,000; N-cadherin: Abcam, ab29952, diluted as 1:1,000; β-catenin: Cell Signaling, 25362S, diluted as 1:1,000; GAPDH: Abcam, ab8245, diluted as 1:10,000) overnight at 4°C and corresponding secondary antibodies for 2 h at room temperature. The results were visualized with the chemiluminescence reagent (Millipore).
Datasets and model construction
In the TCGA cohort, we focused on the data of 10 differentially expressed lncRNAs that were screened in our study sample at the discovery stage using microarray. Their correlation with TNM staging was analyzed. Five significantly upregulated lncRNAs were analyzed for PTC diagnosis and LN metastasis.
To develop a predictive model for LN metastasis in patients with PTC, a multivariable logistic regression model was applied to select the variables for predicting LN metastasis in the training dataset (TCGA cohort). The lncRNA expression was converted into categorical variables (high- and low-expression subgroups) based on the optimal cutoff values. We then derived a nomogram for the constructed pathological- and lncRNA-based classifier for LN metastasis; the predictive accuracy and discrimination ability of the nomogram risk scores were determined using a calibration plot and AUC, respectively. Furthermore, independent validation was conducted using our dataset (SYSUCC cohort).
Cell culture
Nthy-ori 3-1 and 2 PTC cell lines (TPC1 and BCPAP) were purchased from the American Type Culture Collection (ATCC). Nthy-ori 3-1 cells were grown in DMEM:F12 medium (Life Technologies) at 37°C and 5% CO2; TPC1 and BCPAP cells were cultured in DMEM medium (Gibco) at 37°C and 5% CO2. These two media were supplemented with 10% fetal bovine serum (FBS; Sigma).
Cell transfection
NC, miR-766-5p mimics, and inhibitors were purchased from GenePharma (Suzhou). Vector (control), lnc-MPEG1-1:1 (referred to as MPEG-1 in subsequent functional analysis), NC small hairpin RNAs (shRNAs) (sh-NC), and MPEG1-1 shRNAs (sh-MPEG1-1) were also purchased from HanBio Biotechnology. The sequences of sh-MPEG1-1#1 were 5′-AAGUGUUUGCUAAUUGUUC-3′ and 5′-GAACAAUUAGCAAACACUU-3′; the sequences of sh-MPEG1-1#2 were 5′-UGUGAAAUGGGUAAAAAGG-3′ and 5′-CCUUUUUACCCAUUUCACA-3′; and the sequences of sh-MPEG1-1#3 were 5′-UAAUUAUACAUAAACACUG-3′ and 5′-CAGUGUUUAUGUAUAAUUA-3′. For the MPEG1-1-overexpressed plasmid, MPEG1-1 was amplified and inserted into the pcDNA3.0 vector, and the primer sequence of MPEG1-1 was 5′-GGGGTACCACTGGACATCTGCAGAGAGGACAAG-3′ (forward) and 5′-CCCTCGAGGGCTTATAAACAGAAATTTATTCTCACAATTC-3′ (reverse). Overexpressed MPEG1-1 plasmid was constructed by Lipofectamine 2000 according to the manufacturer’s instruction. The sequence with the highest efficiency for gene silencing was selected as the shRNA (sh-MPEG1-1) and ligated with the lentivirus LV003 vector. A negative control sequence was used as the Scramble (shNC) sequence. The above vectors were used for transfecting HEK293T cells together with packaging vectors for preparation of recombinant virus vectors, which were then transfected into TPC1 or BCPAP cells. The stable clones for MPEG1-1 shRNA-expressing cells and overexpressed MPEG1-1 cells were generated by using the lentiviral vectors and selection in puromycin.
CCK-8 assay
TPC1 and BCPAP in each group were added evenly into 96-well plates at a density of 1×103 cells/well. The cells in each well were treated with CCK-8 reagent (Dojindo) after 24, 48, and 72 h of culture. A microplate analyzer was used to determine the absorbance at 450 nm.
Clony formation assay
The transfected TPC1 and BCPAP cells (1×103 cells) were routinely cultured in a 6-well plate for 14 days. After fixation, the clones were dyed using 0.1% crystal violet.
EdU staining
The transfected TPC1 and BCPAP cells (1.0×105 cells/well) in 6-well plates were processed with EdU reagent (10 μM, Life Technologies) for 2 h at 37°C. After fixing with 4% formaldehyde, EdU+ cells were monitored and photographed using a fluorescence microscope (Olympus).
Wound healing assay
The transfected TPC1 and BCPAP cells (1 × 106 cells/well) in 24-well plates were incubated for 18 h at 37°C. A 100-μL pipette tip was used to make scratches. After washing, the cells were grown for 24 h in a serum-free medium, and the images were captured under a light microscope.
Transwell assay
The chamber inserts were first pretreated with 3 mg/mL Matrigel (BD Biosciences) for 30 min at 37°C. The transfected TPC1 and BCPAP cells (5 × 105 cells/well) were inoculated evenly into the upper chamber, and the corresponding medium with 20% FBS was added to the lower chamber. After incubation for 48 h, the invaded cells were fixed, stained, and observed under a microscope.
FISH assay
Cell slides including the transfected TPC1 cells (5 × 103 cells/well) were placed at the bottom of a 24-well plate and cultured for 24 h. After washing, the cells were fixed with 4% paraformaldehyde and treated with 0.5% Triton X-100. The cells were sealed with prehybridization solution at 37°C and hybridized with the MPEG1-1 probe overnight at 37°C. After washing, the cells were nucleated with DAPI. The location of MPEG1-1 was observed using a fluorescence microscope. The probe sequence of MPEG1-1 was 5′-CAAGCGGCACATACTGGACAAGTGTTAGGTTTC-3′.
Tumor xenograft model
A total of 30 BALB/c nude mice (specific pathogen free [SPF], 21–28 days, SYXK [Yue] 2016-0112) were purchased from Jingyuan Biotechnology. All of the mice were raised in a sterile environment. The right hind legs of the mice were subcutaneously injected with stably transfected TPC1 or BCPAP cells (2 × 106 cells/mL) in each group. The length and width of the tumors were tested, and the tumor volume was calculated based on the formula length × width 2/2 at 7, 14, 21, and 28 days. After 28 days, xenograft tumors were collected and used in subsequent experiments. This animal experiment has passed ethical review and been strictly conducted based on the Institutional Animal Protection and Use Committee of Sun Yat-sen University.
H&E staining
Tumor tissues in nude mice were first subjected to a series of treatments, including fixation (4% paraformaldehyde), dehydration (gradient ethanol), and embedding in paraffin. Then, the tissues were continuously cut into 4-μm slices. Afterward, the slices were dewaxed with xylene I and xylene II; dehydrated with 95%, 90%, 80%, and 70% ethanol; and addressed with distilled water. Finally, the slices were processed with Harris hematoxylin, 1% hydrochloric acid alcohol, 0.6% ammonia, and eosin. After dehydration (gradient ethanol) and immersion (xylene), the pathologic structure was observed with a microscope.
Luciferase reporter assay
In line with the predicted binding sites between MPEG1-1 and the miR-766-5p promoter region, the wild type (WT) and mutant (Mut) MPEG1-1 plasmids were constructed with psiCHECK-2. The primer sequences of WT-MPEG1-1 were 5′-CCCTCGAGATCCTCTGAATTTCGCTTGCTCG-3′ (forward) and 5′-ATTTGCGGCCGCGATACACCCTTGGCATCTCCTGG-3′ (reverse); the primer sequences of Mut-MPEG1-1 were 5′-CCGGTTTTCAGTGCTCATCTCTGGGAATCTGCTGCCAATG-3′ (forward) and 5′-CATTGGCAGCAGATTCCCAGAGATGAGCACTGAAAACCGG-3′ (reverse). TPC1 cells were co-transfected with miR-766-5p mimics and the corresponding plasmids using Lipofectamine 3000 (Invitrogen). A dual luciferase assay kit (Promega) was used to determine luciferase activity.
Statistical analysis
All of the statistical tests were conducted using R statistical software version 4.0.3 (R Foundation for Statistical Computing). An independent t test was performed to compare the continuous variables between the two groups. Categorical data were analyzed using the chi-square test. Multivariable logistic regression was used to screen for significant risk factors and to construct a pathological-and-lncRNA-based classifier for LN metastasis. Differential expression visualization was implemented by the “edgR” package. The pheatmap was made with “pheatmap” packages. The ROC curves were plotted with the “pROC” package. The nomogram and calibration plots were generated with the “rms” package. Statistical significance was set at p < 0.05.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no. 81802950).
Author contributions
Conception & design, C.-Y.H., H.C., X.S., and Z.-L.Y. Development of methodology, C.H., X.S., D.-L.Z., and Z.-L.Y. Data acquisition of data, D.-L.Z., Z.-L.Y., Q.L., X.Z., T.T., and B.-H.X. Data analysis & interpretation, C.H., X.S., and X.-H.Y. Administrative, technical, or material support, Z.-L.Y., X.S., and C.H. Writing, review, and/or revision of the manuscript, all of the authors.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.03.023.
Contributor Information
Zu-Lu Ye, Email: yezl@sysucc.org.cn.
Cai-Yun He, Email: hecy@sysucc.org.cn.
Supplemental information
References
- 1.Cabanillas M.E., McFadden D.G., Durante C. Thyroid cancer. Lancet (London, England) 2016;388:2783–2795. doi: 10.1016/S0140-6736(16)30172-6. [DOI] [PubMed] [Google Scholar]
- 2.Roh J.L., Kim J.M., Park C.I. Central lymph node metastasis of unilateral papillary thyroid carcinoma: patterns and factors predictive of nodal metastasis, morbidity, and recurrence. Ann. Surg. Oncol. 2011;18:2245–2250. doi: 10.1245/s10434-011-1600-z. [DOI] [PubMed] [Google Scholar]
- 3.Nie X., Tan Z., Ge M., Jiang L., Wang J., Zheng C. Risk factors analyses for lateral lymph node metastases in papillary thyroid carcinomas: a retrospective study of 356 patients. Arch. Endocrinol. Metab. 2016;60:492–499. doi: 10.1590/2359-3997000000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao A., Rajkumar T., Mani S. Perspectives of long non-coding RNAs in cancer. Mol. Biol. Rep. 2017;44:203–218. doi: 10.1007/s11033-017-4103-6. [DOI] [PubMed] [Google Scholar]
- 5.Huang J.K., Ma L., Song W.H., Lu B.Y., Huang Y.B., Dong H.M., Ma X.K., Zhu Z.Z., Zhou R. MALAT1 promotes the proliferation and invasion of thyroid cancer cells via regulating the expression of IQGAP1. Biomed. Pharmacother. Biomedecine pharmacotherapie. 2016;83:1–7. doi: 10.1016/j.biopha.2016.05.039. [DOI] [PubMed] [Google Scholar]
- 6.Rolla M., Jawiarczyk-PrzybyŁowska A., KolaČkov K., Bolanowski M. H19 in endocrine system tumours. Anticancer Res. 2021;41:557–565. doi: 10.21873/anticanres.14808. [DOI] [PubMed] [Google Scholar]
- 7.Yu X., Zheng H., Chan M.T., Wu W.K.K. BANCR: a cancer-related long non-coding RNA. Am. J. Cancer Res. 2017;7:1779–1787. [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Lu X., Geng Z., Yang G., Shi Y. LncRNA PTCSC3/miR-574-5p governs cell proliferation and migration of papillary thyroid carcinoma via wnt/β-catenin signaling. J. Cell. Biochem. 2017;118:4745–4752. doi: 10.1002/jcb.26142. [DOI] [PubMed] [Google Scholar]
- 9.Haugen B.R., Alexander E.K., Bible K.C., Doherty G.M., Mandel S.J., Nikiforov Y.E., Pacini F., Randolph G.W., Sawka A.M., Schlumberger M., et al. 2015 American thyroid association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: the American Thyroid Association guidelines task force on thyroid nodules and differentiated thyroid cancer. Thyroid. 2016;26:1–133. doi: 10.1089/thy.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eszlinger M., Hegedüs L., Paschke R. Ruling in or ruling out thyroid malignancy by molecular diagnostics of thyroid nodules. Best Pract. Res. Clin. Endocrinol. Metab. 2014;28:545–557. doi: 10.1016/j.beem.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Mond M., Alexiadis M., Fuller P.J., Gilfillan C. Mutation profile of differentiated thyroid tumours in an Australian urban population. Intern. Med. J. 2014;44:727–734. doi: 10.1111/imj.12476. [DOI] [PubMed] [Google Scholar]
- 12.Argyropoulou M., Veskoukis A.S., Karanatsiou P.M., Manolakelli A., Kostoglou-Athanassiou I., Vilaras G., Karameris A., Liadaki K. Low prevalence of TERT promoter, BRAF and RAS mutations in papillary thyroid cancer in the Greek population. Pathol. Oncol. Res. 2020;26:347–354. doi: 10.1007/s12253-018-0497-2. [DOI] [PubMed] [Google Scholar]
- 13.Bychkov A. Prevalence of BRAF(V600E) mutation in Asian patients with thyroid cancer. Malaysian J. Pathol. 2017;39:95–96. [PubMed] [Google Scholar]
- 14.Liang J., Cai W., Feng D., Teng H., Mao F., Jiang Y., Hu S., Li X., Zhang Y., Liu B., et al. Genetic landscape of papillary thyroid carcinoma in the Chinese population. J. Pathol. 2018;244:215–226. doi: 10.1002/path.5005. [DOI] [PubMed] [Google Scholar]
- 15.Seib C.D., Sosa J.A. Evolving understanding of the epidemiology of thyroid cancer. Endocrinol. Metab. Clin. North America. 2019;48:23–35. doi: 10.1016/j.ecl.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Lewiński A., Adamczewski Z. Papillary thyroid carcinoma: a cancer with an extremely diverse genetic background and prognosis. Polish Arch. Intern. Med. 2017;127:388–389. doi: 10.20452/pamw.4058. [DOI] [PubMed] [Google Scholar]
- 17.Yu J., Deng Y., Liu T., Zhou J., Jia X., Xiao T., Zhou S., Li J., Guo Y., Wang Y., et al. Lymph node metastasis prediction of papillary thyroid carcinoma based on transfer learning radiomics. Nat. Commun. 2020;11:4807. doi: 10.1038/s41467-020-18497-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhan A., Soleimani M., Mandal S.S. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo L.J., Zhang S., Gao B., Jiang Y., Zhang X.H., Tian W.G., Hao S., Zhao J.J., Zhang G., Hu C.Y., et al. Low expression of long non-coding RNA GAS5 is associated with poor prognosis of patients with thyroid cancer. Exp. Mol. Pathol. 2017;102:500–504. doi: 10.1016/j.yexmp.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai Y., Zhang G., Cheng R., Yang R., Chu H. CASC15 contributes to proliferation and invasion through regulating miR-766-5p/KLK12 axis in lung cancer. Cell Cycle (Georgetown, Tex) 2019;18:2323–2331. doi: 10.1080/15384101.2019.1646562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai Y., Zhang K., Cao L., Sun H., Wang H. Inhibition of microrna-766-5p attenuates the development of cervical cancer through regulating SCAI. Tech. Cancer Res. Treat. 2020;19 doi: 10.1177/1533033820980081. 1533033820980081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia B., Xia L., Cao F. The role of miR-766-5p in cell migration and invasion in colorectal cancer. Exp. Ther. Med. 2018;15:2569–2574. doi: 10.3892/etm.2018.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao Y., Shen G., Su Z., Du J., Xu F., Yu Y. RAD21 inhibited transcription of tumor suppressor MIR4697HG and led to glioma tumorigenesis. Biomed. Pharmacother. Biomedecine pharmacotherapie. 2020;123:109759. doi: 10.1016/j.biopha.2019.109759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.