Solvik et al. reveal that the autophagy machinery facilitates the secretion of autophagy cargo receptors and other cellular components via extracellular vesicles and particles in response to lysosome inhibition or impaired autophagosome maturation.
Abstract
The endolysosome system plays central roles in both autophagic degradation and secretory pathways, including the release of extracellular vesicles and particles (EVPs). Although previous work reveals important interconnections between autophagy and EVP-mediated secretion, our understanding of these secretory events during endolysosome inhibition remains incomplete. Here, we delineate a secretory autophagy pathway upregulated in response to endolysosomal inhibition, which mediates EVP-associated release of autophagic cargo receptors, including p62/SQSTM1. This secretion is highly regulated and dependent on multiple ATGs required for autophagosome formation, as well as the small GTPase Rab27a. Furthermore, disrupting autophagosome maturation, either via genetic inhibition of autophagosome-to-autolysosome fusion or expression of SARS-CoV-2 ORF3a, is sufficient to induce EVP secretion of autophagy cargo receptors. Finally, ATG-dependent EVP secretion buffers against the intracellular accumulation of autophagy cargo receptors when classical autophagic degradation is impaired. Thus, we propose secretory autophagy via EVPs functions as an alternate route to clear sequestered material and maintain proteostasis during endolysosomal dysfunction or impaired autophagosome maturation.
Introduction
Although autophagy is classically viewed as a lysosomal degradation pathway, increasing evidence implicates autophagy-related genes (ATGs) in the secretion of inflammatory molecules (Dupont et al., 2011; Lock et al., 2014), tissue repair factors (DeSelm et al., 2011), bactericidal enzymes (Bel et al., 2017), and extracellular vesicles (EVs; Guo et al., 2017; Leidal et al., 2020; Murrow et al., 2015). Recently, we described an autophagy-related secretory pathway called LC3-dependent EV loading and secretion (LDELS), which captures proteins at late endosomes and facilitates their secretion outside the cell (Leidal et al., 2020). These expanding roles for ATGs in secretion, collectively termed secretory autophagy, poignantly suggest that cellular catabolism and secretion are functionally coordinated within cells. Nevertheless, the mechanisms orchestrating secretory autophagy versus classical degradative autophagy, as well as how this interplay contributes to proteostasis and cellular quality control remain poorly understood.
Notably, recent studies demonstrate that endolysosome impairment profoundly impacts the dynamics of both autophagy-dependent degradation and EV secretion. Lysosomal inhibition via Bafilomycin A1 and chloroquine (CQ) treatment impairs autophagy and stimulates EV secretion (Cashikar and Hanson, 2019; Mauthe et al., 2018; Ortega et al., 2019). Moreover, chemical inhibitors targeting PIKfyve and VPS34, two PI3K kinases important for endosomal trafficking and autophagy, disrupt autophagy-dependent protein turnover and promote secretion of various autophagy components in EV fractions (Hessvik et al., 2016; Miranda et al., 2018). These observations suggest that the status of the endolysosomal system influences secretory autophagy via EV release, which bears fundamental significance for understanding various pathological conditions and therapies that impair endolysosome function. Importantly, many pathogens specifically impede endolysosome function to prevent clearance by the autophagy pathway. For example, lysosomal disruption during uropathogenic Escherichia coli infection triggers autophagy-dependent expulsion of bacteria in EVs, whereas many RNA viruses exploit autophagic membranes that accrue in response to impaired lysosomal degradation for viral replication and exocytosis (Miao et al., 2015; Teo et al., 2021). Endolysosome deregulation, also common in age-related diseases including neurodegeneration and autophagy-dependent secretion in diseased neurons, may contribute to the spread of aggregation-prone proteins (Ejlerskov et al., 2013; Minakaki et al., 2018; Nilsson et al., 2013). Finally, the lysosomal inhibitor hydroxychloroquine has been extensively employed in multiple clinical oncology trials to target the heightened dependence of cancer cells on the autophagy–lysosome pathway for growth and survival (Amaravadi et al., 2019). Nevertheless, whether and how endolysosome impairment in these contexts contributes to secretory autophagy, and more specifically to EV-associated secretion, remains largely unknown.
Here, we demonstrate that lysosomal inhibition robustly induces ATG-dependent secretion of autophagy components and autophagy cargo receptors in EV fractions both in vitro and in vivo. Remarkably, autophagy cargo receptors secreted in EV-associated fractions during lysosome inhibition are largely unprotected from proteolytic cleavage, suggesting that, in contrast to the LDELS pathway, these proteins are not selectively loaded into EVs. Rather, they are released outside the cell in tight association with EVs but as part of a broader fraction of nanoparticles, collectively termed extracellular vesicles and particles (EVPs; Zhang et al., 2018). This extracellular secretion is highly regulated and critically dependent on multiple ATGs required for the progressive steps in autophagosome formation. Furthermore, disruption of autophagosome maturation, due to genetic loss of the autophagosome-to-autolysosome fusion machinery or blockade via the SARS-CoV-2 viral protein ORF3a, promotes EVP-associated secretion of autophagy cargo receptors. Finally, the suppression of this secretory pathway causes intracellular accumulation of individual cargo receptors, suggesting that when autophagy-dependent degradation is impaired due to endolysosome inhibition, the autophagy machinery diverts autophagic cargo receptors for secretion outside the cell via EVPs.
Results
Inhibition of lysosome acidification promotes the EVP-associated release of core autophagy machinery and degradative cargo
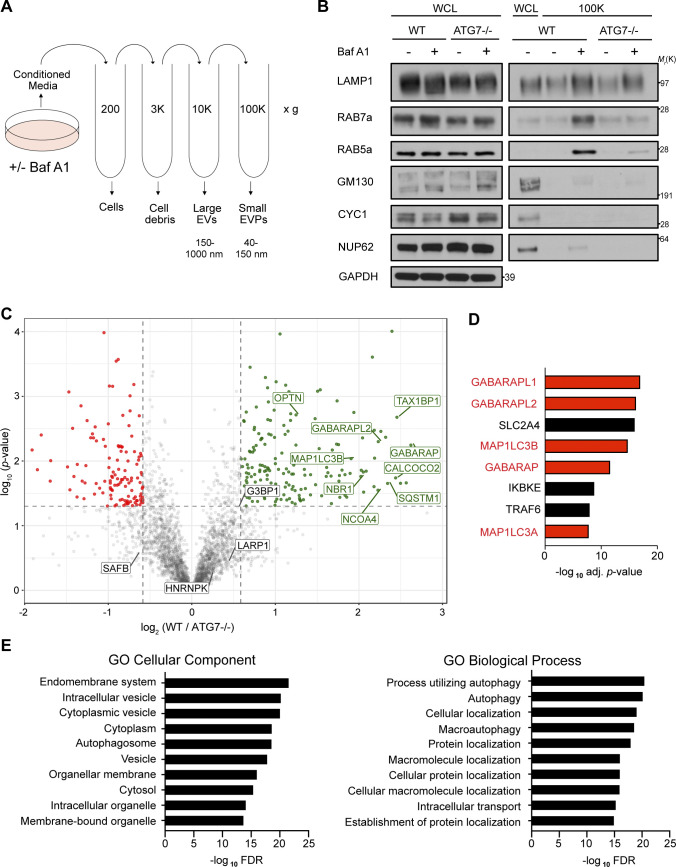
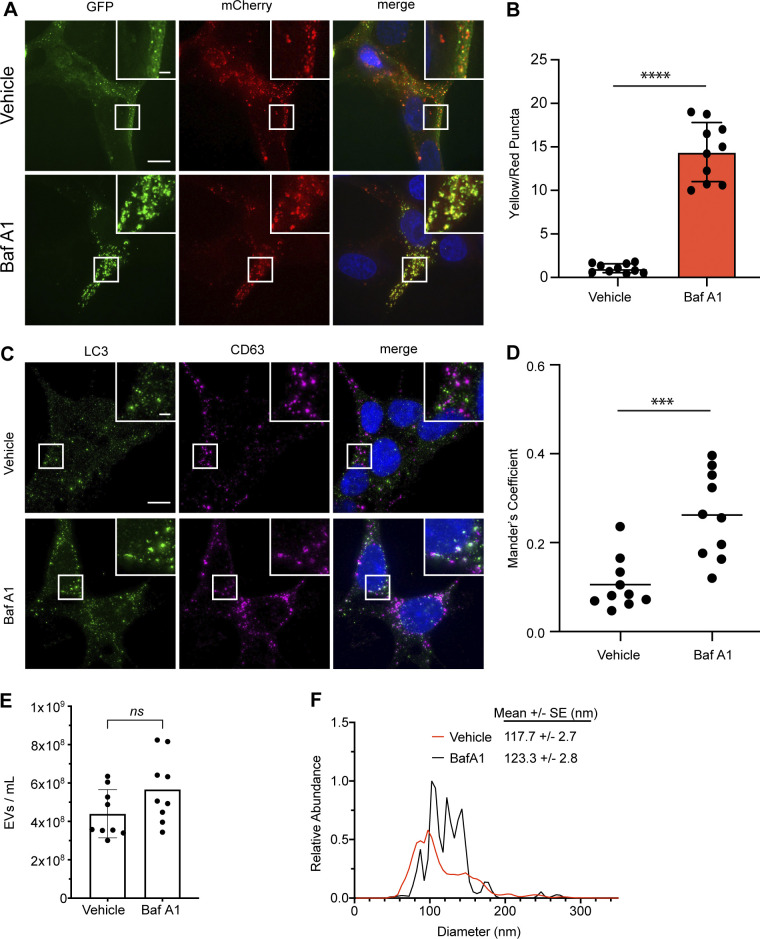
To ascertain how lysosome impairment affects ATG-dependent EV secretion, we treated WT and ATG7-deficient HEK293T cells with the vacuolar-type ATPase (v-ATPase) inhibitor Bafilomycin A1 (BafA1), which disrupts lysosome acidification, and evaluated its impact on protein cargo released in small EV fractions (Fig. 1 A). Treatment of serum-starved WT cells with BafA1 resulted in robust increases in the secretion of endosomal proteins in EV fractions, including the Rab GTPases Rab5a and Rab7a, markers of early and late endosomes, respectively (Fig. 1 B). In contrast, endosomal protein secretion was largely unaltered in BafA1-treated, ATG7-deficient cells, which are defective for the lipid conjugation of MAP1LC3B (LC3) family proteins that is essential for both classical autophagy and emerging secretory autophagy pathways, including LDELS (Nieto-Torres et al., 2021). These results broached that the autophagy machinery facilitates the diversion of endocytic intermediates for extracellular secretion during lysosome inhibition. Importantly, Rab5a and Rab7a secretion during BafA1 treatment correlated with impaired autophagosome maturation, supported by BafA1-induced accumulation of yellow puncta (mCherry+; GFP+) delineating immature autophagosomes in cells expressing the mCherry-GFP-LC3B autophagic flux reporter (Fig. S1, A and B; Pankiv et al., 2007). We also observed that BafA1 significantly increased the co-location of LC3 and the late endosome marker CD63 (Fig. S1, C and D), suggesting that impaired lysosome acidification results in increased numbers of amphisomes compared with untreated controls. Although nanoparticle tracking analysis of conditioned media revealed that BafA1 did not significantly impact the overall levels or mean particle size of secreted EVPs, we noted increases in specific nanoparticle populations measuring 110, 125, and 140 nm in diameter (Fig. S1, E and F).
Figure 1.
BafA1-induced disruption of lysosome acidification promotes ATG-dependent secretion of autophagy cargo receptors via EVPs. (A) Schematic for the differential centrifugation protocol used to isolate small EVP-enriched fractions from cells treated with BafA1 or vehicle. (B) Cell lysate (WCL; left) and 100,000 g EVP fractions (100K; right) from WT and ATG7−/− serum starved HEK293T cells treated with 20 nM BafA1 or vehicle were collected, normalized and blotted to detect the indicated proteins (n = 3). (C) A volcano plot of the proteins identified in 100K EVP-enriched fractions from BafA1 treated WT and ATG7−/− HEK293T cells quantified by TMT MS. TMT-labeled proteins are plotted according to their −log10 P values as determined by two-tailed t test and log2 fold enrichment (WT/ATG7−/−; n = 3). Gray dots: proteins not enriched in EVPs from BafA1 treated WT or ATG7−/− cells identified with P > 0.05 and/or log2 fold change between −0.5 and 0.5. Green dots: proteins significantly enriched in EVPs from BafA1 treated WT cells relative to treated ATG7−/− cells. Red dots: proteins significantly enriched in EVPs from BafA1 treated ATG7−/− cells relative to treated WT cells. (D) A ranked list of the proteins with the greatest connectivity to the 182 proteins enriched in EVPs from BafA1 treated WT cells relative to treated ATG7−/− cells. Statistical significance was calculated in Enrichr by a one-way Fisher’s exact test. LC3 family members are highlighted in red. PPI, protein–protein interaction. (E) GO enrichment analysis of the 182 proteins enriched in EVPs from BafA1 treated WT cells relative to treated ATG7−/− cells with the top terms for cellular component (left) and biological processes (right) plotted according to −log10 FDR. Source data are available for this figure: SourceData F1.
Figure S1.
BafA1 treatment inhibits autophagic flux and modulates EV secretion. (A) Representative images of WT HEK293T cells stably expressing the mCherry-EGFP-LC3 reporter were treated with 20 nM BafA1 or vehicle in serum-free media (BafA1) for 16 h. Scale bar, 10 μm; inset scale bar, 2 μm. (B) Quantification of the ratio of double-positive (mCherry+/GFP+) to mCherry-only (mCherry+/GFP−) LC3 puncta per cell. Statistical significance was calculated by unpaired two-tailed t test (mean ± SEM; vehicle, n = 10; BafA1, n = 10; ****, P < 0.001). (C) Representative images of WT HEK293T cells treated with 20 nM BafA1 or vehicle in serum-free media (BafA1) for 16 h and immunostained for endogenous LC3 and CD63. Scale bar, 10 μm; inset scale bar, 2 μm. (D) A scatter plot of Mander’s coefficients for the co-occurrence of LC3 with CD63 in the immunostained cells in C. Statistical significance was calculated by unpaired two-tailed t-test (mean ± SEM; Vehicle, n = 10; BafA1, n = 10; ***, P < 0.005). (E) Nanoparticle tracking analysis of conditioned media from equal numbers of WT cells treated with vehicle in serum-free media or 20 nM BafA1 (mean ± SEM; n = 3). Statistical significance calculated by unpaired two-tailed t test. (F) EV size distribution from indicated cell treatments in E (mean ± SEM; n = 3).
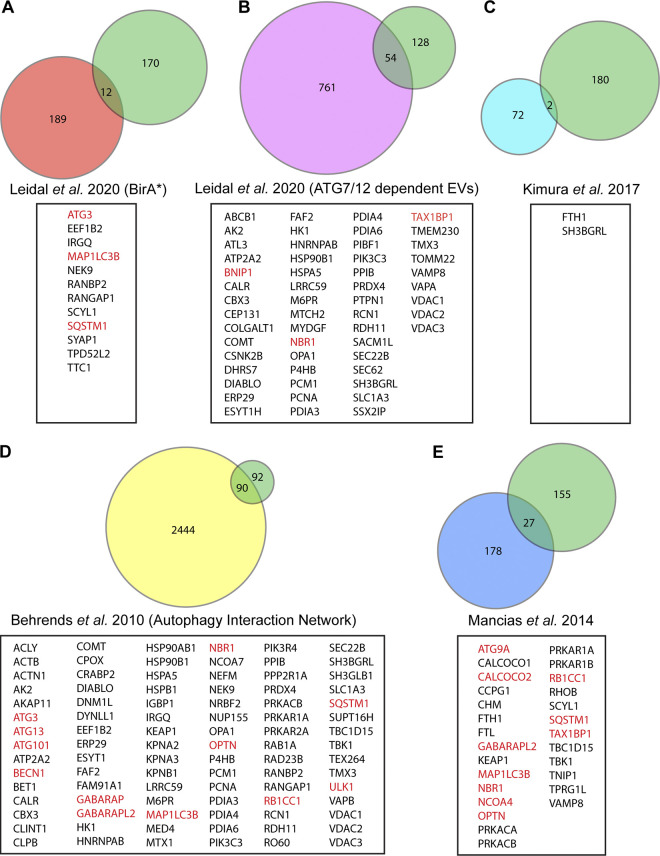
To further define the ATG-dependent secreted proteome during lysosome inhibition in an unbiased manner, we performed tandem mass tag (TMT)–based quantitative proteomics comparing small EV fractions purified via differential centrifugation (100,000 g; 100K) from the conditioned media of WT and ATG7-deficient cells treated with BafA1. Importantly, while these 100K fractions are enriched for small EVs, they contain a broader array of nanoparticles and proteins, collectively termed extracellular vesicles and particles (EVPs; Zhang et al., 2018). Overall, we identified 182 proteins enriched in the EVP fractions from BafA1-treated WT versus ATG7-deficient cells (Fig. 1 C and Table S1). Many were core autophagy proteins including LC3/ATG8 family members and components of complexes regulating discrete steps of autophagosome formation. Importantly, we also identified numerous autophagy cargo receptors, proteins critical for selectively capturing cargo for autophagic degradation as well as serving as scaffolds for cellular signaling (Johansen and Lamark, 2020). These included p62/SQSTM1, NBR1, OPTN, and NDP52 (Fig. 1 C). Notably, five LC3/ATG8 family members ranked amongst the top proteins connected to the 182 proteins enriched in the BafA1-induced ATG7-dependent EV proteome (Fig. 1 D). Moreover, gene ontology (GO) analyses highlighted a profound enrichment in proteins that localize to the endomembrane system and proteins with roles in autophagy (Fig. 1 E). Interestingly, the ATG7-dependent EVP proteome from BafA1 treated cells was distinct from the recently defined LDELS proteome; there was no evidence for functional enrichment of RBPs, including SAFB, HNRNPK, LARP1, and G3BP1, in control versus ATG7 deficient cells treated with BafA1 (Fig. 1 C). Remarkably, proteomic analyses of EV fractions from WT BafA1 treated cells identified specific regulators of endosome-associated Rab GTPases, including TBC1D15 and TBC1D17, suggesting this secretory mechanism may involve specific endosomal subpopulations or endocytic machinery (Fig. S1, E and F). Overall, the BafA1-induced ATG7-dependent EVP proteome shared 12 proteins in common with the BirA*-LC3B–labeled secretome, 54 proteins with the ATG7 and ATG12-dependent EV proteome previously described for LDELS, and two proteins with the ATG5-dependent secretome from murine macrophages (Fig. 2, A–C; Kimura et al., 2017; Leidal et al., 2020). Remarkably, the ATG7-dependent EVP proteome secreted by BafA1-treated cells shared numerous common proteins with proteomic datasets derived from studies of degradative autophagy (Fig. 2, D and E; Behrends et al., 2010; Mancias et al., 2014). Based on these results, we hypothesized that lysosome impairment diverts proteins residing in autophagosomes, but originally destined for degradation, for secretion outside the cell via EVPs.
Figure 2.
The ATG-dependent EVP secretome from BafA1 treated cells overlaps with previous proteomic analyses of the autophagy pathway. Venn diagrams showing: (A) The overlap of ATG-dependent EVP secretion candidates from BafA1 treated cells with the class I and class II BirA*-LC3B labeled secretome in Leidal et al. (2020). (B) The overlap of ATG-dependent EVP secretion candidates from BafA1 treated cells with the ATG7 and ATG12-dependent EV secretome in Leidal et al. (2020). (C) The overlap of ATG-dependent EVP secretion candidates from BafA1 treated cells with the ATG5-dependent bone-marrow–derived macrophage secretome in Kimura et al. (2017). (D) The overlap of ATG-dependent EVP secretion candidates from BafA1 treated cells with the autophagy interaction network defined in Behrends et al. (2010). (E) The overlap of ATG-dependent EVP secretion candidates from BafA1 treated cells with the autophagosome enriched proteome in Mancias et al. (2014). Core autophagy machinery and autophagy cargo receptors are highlighted in red.
Lysosome inhibition promotes EVP secretion of autophagy cargo receptors in vitro and in vivo
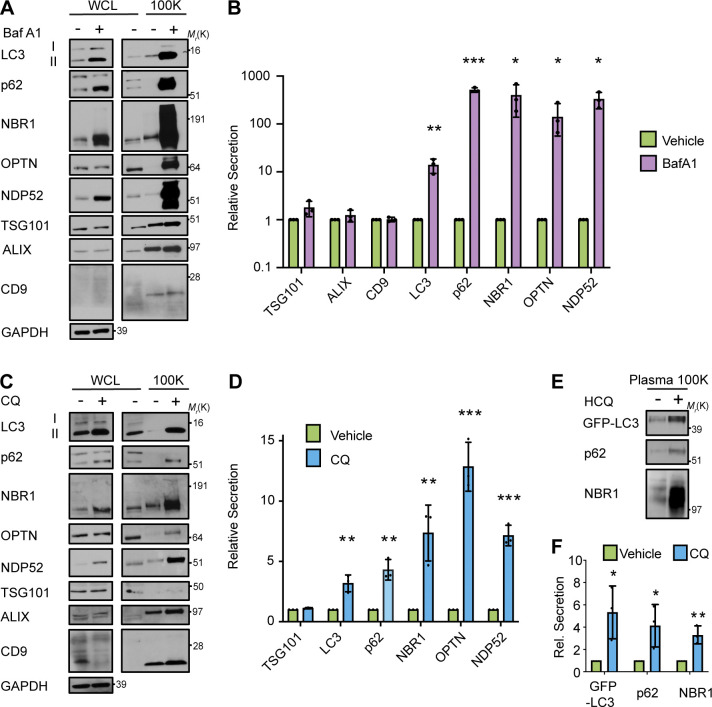
We next assessed whether specific ATG7-dependent secretion candidates identified via TMT-mass spectrometry (MS) were released in association with EVPs in response to lysosome inhibition. Small EVP fractions from serum-starved WT cells treated in the absence and presence of BafA1 were isolated via differential centrifugation and probed for autophagy pathway components and markers of EVs (Fig. S2, A and B). Importantly, the 100K fraction from BafA1-treated cells was highly enriched in lipidated LC3 (LC3-II) and multiple autophagy cargo receptors, including p62/SQSTM1 (p62), NBR1, OPTN, and NDP52. In contrast, BafA1 treatment had a negligible impact on the secretion of TSG101, ALIX, and CD9, classical markers of EVs (Fig. 3, A and B). To ascertain whether this was a general response to lysosome inhibition and impaired autophagosome maturation, we treated cells with CQ, a lysosomotropic anti-malarial agent, used to therapeutically target the autophagy–lysosome pathway (Amaravadi et al., 2019). CQ, similar to BafA1, increased extracellular secretion of LC3-II and autophagy cargo receptors in EV fractions isolated from conditioned media without significantly altering the secretion of EV marker proteins (Fig. 3, C and D). Importantly, enhanced secretion of LC3-II and autophagy cargo receptors during lysosome inhibition did not correlate with increased cell death and was observed in a diverse array of BafA1 treated cells, including murine and human breast cancer and neuronal cell lines (Fig. S2 and Fig. S3, A and B). Finally, upon treatment of GFP-LC3 transgenic mice with CQ and purification of blood plasma EVP fractions from these animals, we observed increased in vivo extracellular secretion of both GFP-LC3 and the cargo receptors p62 and NBR1 in CQ-treated mice compared with untreated controls (Fig. 3, E and F). Overall, these findings confirm that pharmacological inhibition of the lysosome results in increased extracellular secretion of LC3 and autophagy cargo receptors both in vitro and in vivo.
Figure S2.
Lysosomal inhibition has a negligible impact on cell death. (A) Quantification of cell death in WT cells prior to treatment (0 h) or treated with vehicle in serum-free media, or 20 nM BafA1 in serum-free media for 8 h using trypan blue staining (mean ± SEM; n = 3). (B) Quantification of cell death in WT cells prior to treatment (0 h) or treated with vehicle in serum-free media, or 25 µM CQ in serum-free media (BafA1) for 8 h using trypan blue staining (mean ± SEM; n = 3).
Figure 3.
Lysosomal inhibition broadly promotes secretion of autophagy cargo receptors via EVPs. (A) Cell lysate (WCL; left) and 100,000 g EVP fractions (100K; right) from serum-starved HEK293T cells treated with vehicle or 20 nM BafA1 for 16 h were collected and blotted for the indicated proteins (n = 3). (B) Quantification of the proteins in EVP fractions from BafA1 treated cells relative to controls in A. (mean ± SEM; n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.005). Statistical significance calculated by unpaired, two-tailed t test. (C) WCL (left) and 100K fractions (right) from serum starved HEK293T cells treated with vehicle or 25 µM CQ for 16 h were collected and blotted for the indicated proteins (n = 3). (D) Quantification of the proteins in EVP fractions from CQ treated cells relative to controls in C (mean ± SEM; n = 3; **, P < 0.01; ***, P < 0.005). Statistical significance calculated unpaired, two-tailed t test. (E) Plasma EVPs from mice treated with vehicle or 60 mg/kg hydroxychloroquine (HCQ) for three consecutive days were collected and blotted for the levels of GFP-LC3, p62, and NBR1. (F) Quantification of the indicated proteins in plasma EV fractions from mice treated with vehicle or CQ from E (mean ± SEM; n = 3; *, P < 0.05; **, P < 0.01). Statistics were calculated by unpaired, two-tailed t-test. Source data are available for this figure: SourceData F3.
Figure S3.
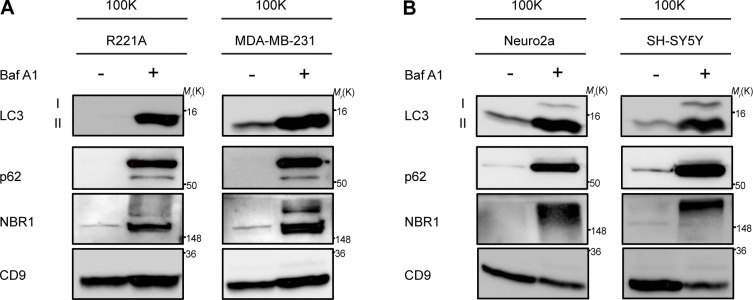
LC3 and autophagy cargo receptors are secreted in EVPs from diverse cell-types in response to lysosome inhibition. (A) 100,000 g EVP fractions (100K) from WT mouse R221A and human MDA-MB-231 breast cancer cell lines treated with vehicle or 20 nM BafA1 in serum-free media for 18 h were collected and blotted for indicated proteins (n = 3). (B) 100,000 g EVP fractions (100K) from WT mouse Neuro2a and human SH-SY5Y neuronal cell lines treated with vehicle or 20 nM BafA1 in serum free media for 18 h were collected and blotted for indicated proteins (n = 3). Source data are available for this figure: SourceData FS3.
Autophagy cargo receptors are not extracellularly secreted within protease-protected vesicular intermediates
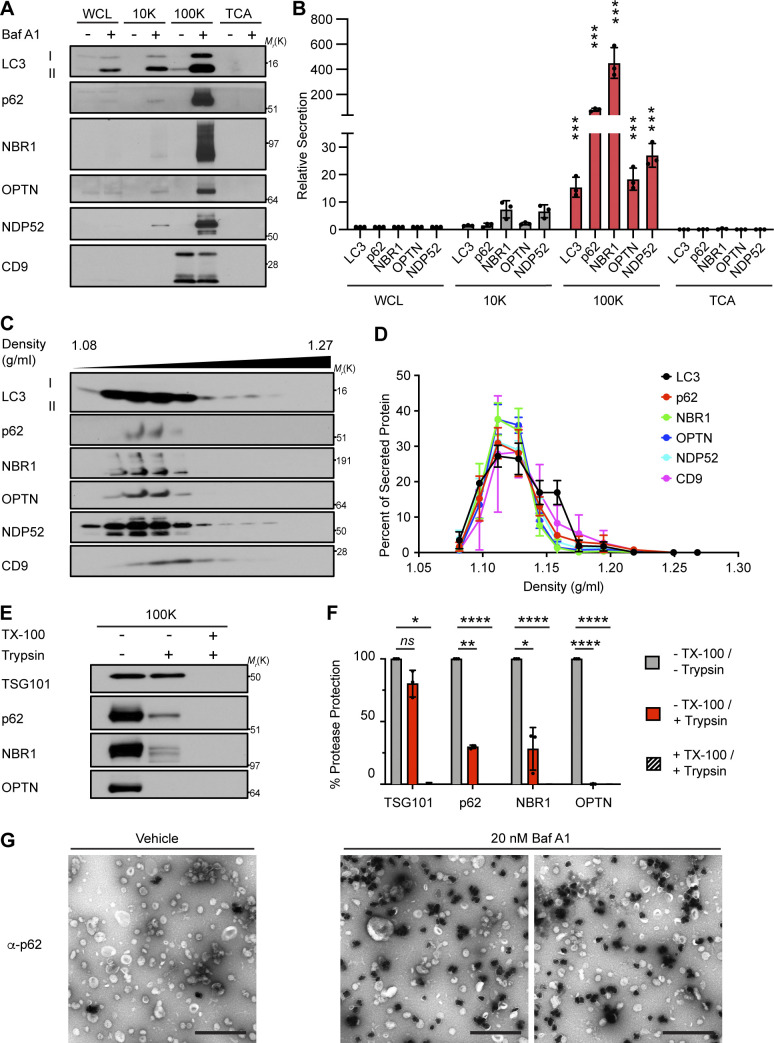
Our group recently revealed a novel role for the autophagy conjugation machinery in specifying RNA-binding proteins for secretion within small EVs (Leidal et al., 2020). To more precisely determine whether LC3 and autophagy cargo receptors are specifically released via small EVs during lysosome impairment, we performed serial differential centrifugation of conditioned media from serum-starved, BafA1-treated cells to isolate fractions enriched in large EVs (10,000 g; 10K), small EVs and particles (EVPs, 100K), and soluble proteins precipitated from the remaining sample. Notably, the 100K fraction from BafA1 treated cells was highly enriched in LC3, p62, NBR1, OPTN, and NDP52, whereas the other fractions contained significantly less LC3 and near undetectable levels of autophagy cargo receptors (Fig. 4, A and B). Upon further purifying EVs from BafA1 treated cells via linear sucrose density gradient ultracentrifugation, we found that LC3-II and autophagy cargo receptors in these isolates co-fractionated with the EV-associated tetraspanin CD9 (Fig. 4, C and D).
Figure 4.
Autophagy cargo receptors are secreted as EVP-associated proteins in response to lysosome inhibition. (A) Cell lysate (WCL) and fractionated conditioned media (CM) were collected from serum-starved HEK293Ts treated with vehicle or 20 nM BafA1 for 16 h. CM subjected to serial differential ultracentrifugation to recover large EVs (10,000 g; 10K), small EVPs (100,000 g; 100K), and precipitated free soluble protein (TCA). Equal protein from WCL and fractionated CM were probed for the indicated targets (n = 3). (B) Quantification of LC3 and autophagy cargo receptors in the indicated fractions of CM from serum-starved BafA1 treated cells relative to WCL (mean ± SEM; n = 3; ***, P < 0.005). Statistical significance between CM fractions was calculated by nonparametric one-way ANOVA with Dunnett’s post hoc test. (C) Small EVPs from CM separated via linear sucrose density gradient ultracentrifugation, fractionated, and blotted to detect LC3, autophagy cargo receptors, and CD9 (n = 3). (D) Percent of total secreted LC3, cargo receptors, and CD9 detected in gradient fractions (mean ± SEM; n = 4). (E) Representative blots of indicated proteins from untreated EVPs or EVPs incubated with 100 μg/ml trypsin and/or 1% Triton X-100 (TX-100) for 30 min at 4°C (n = 3). (F) Percent protease protection for indicated proteins in EVPs incubated with 100 μg/ml trypsin and/or 1% TX-100 (mean ± SEM; n = 3; *, P < 0.05; **, P < 0.01; ****, P < 0.001). Statistics calculated by nonparametric one-way ANOVA with Dunnett’s post hoc test. (G) Representative TEM of small EVPs collected from vehicle or 20 nM BafA1 treated cells immunostained with anti-p62 primary antibody and detected using ultrasmall gold-conjugated secondary antibody with silver enhancement (scale bar, 500 nm). Source data are available for this figure: SourceData F4.
We next performed protease protection of 100K fractions to assess whether autophagy cargo receptors released during lysosome impairment were localized within small EVs. However, in contrast to the EV luminal marker TSG101, the autophagy cargo receptors p62, NBR1, and OPTN within EV fractions were almost completely digested by trypsin in the absence of detergent (Fig. 4, E and F). To further corroborate this result, we performed EM of EVP fractions with anti-p62 silver-enhanced immunogold labeling, which revealed that p62 was located either on the exterior surface of EVs or in secreted nanoparticles, rather than inside the lumen of individual EVs (Fig. 4 G). Together, these results indicate that autophagy cargo receptors released during lysosome inhibition are predominantly associated with the exterior surface of small EVs or secreted as nanoparticles (EVPs), rather than being directly packaged into the lumen of EVs.
Autophagy cargo receptor secretion functionally requires multiple steps in classical autophagosome formation
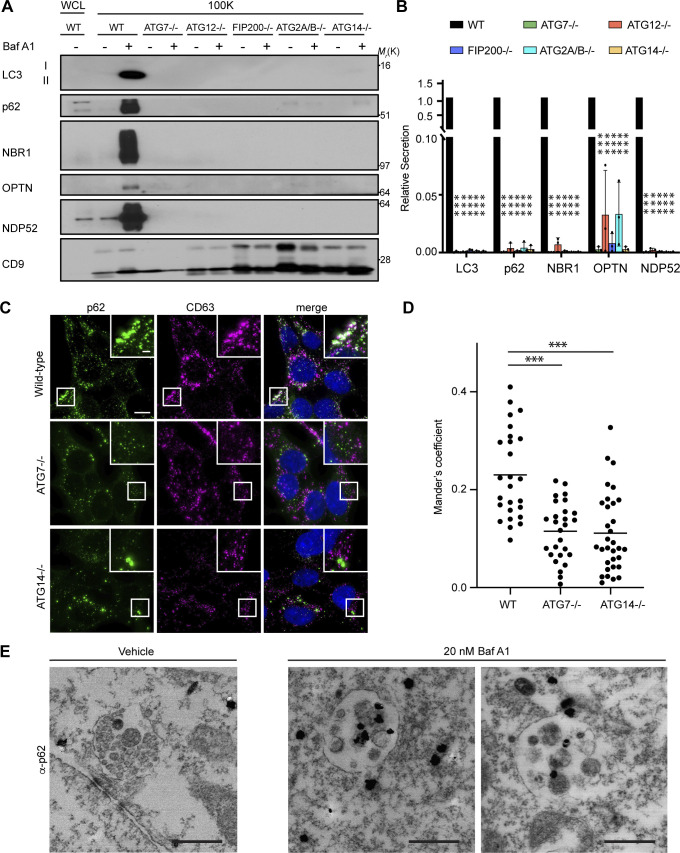
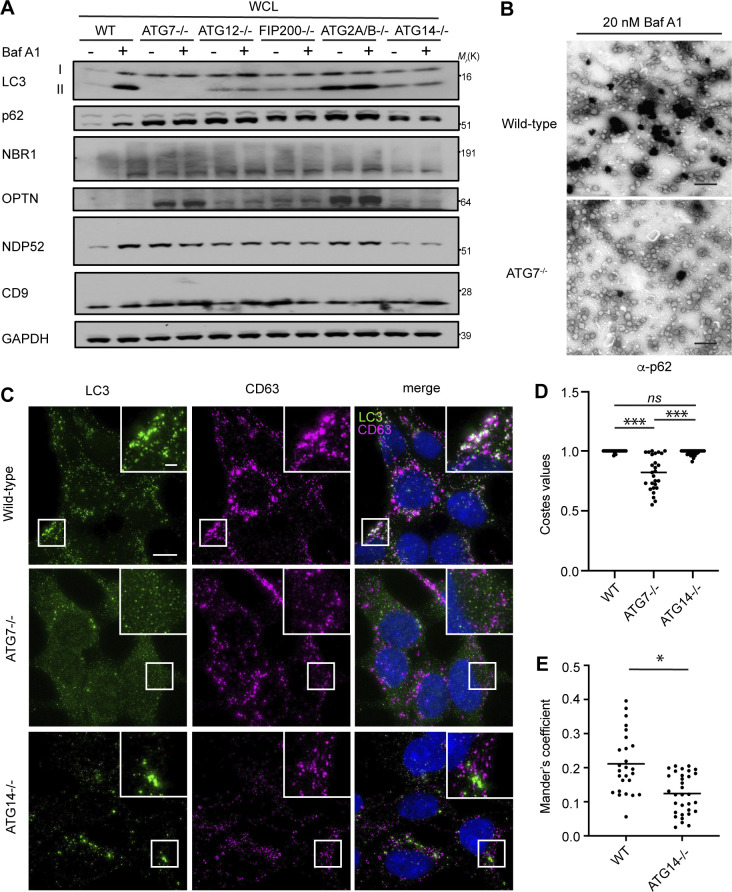
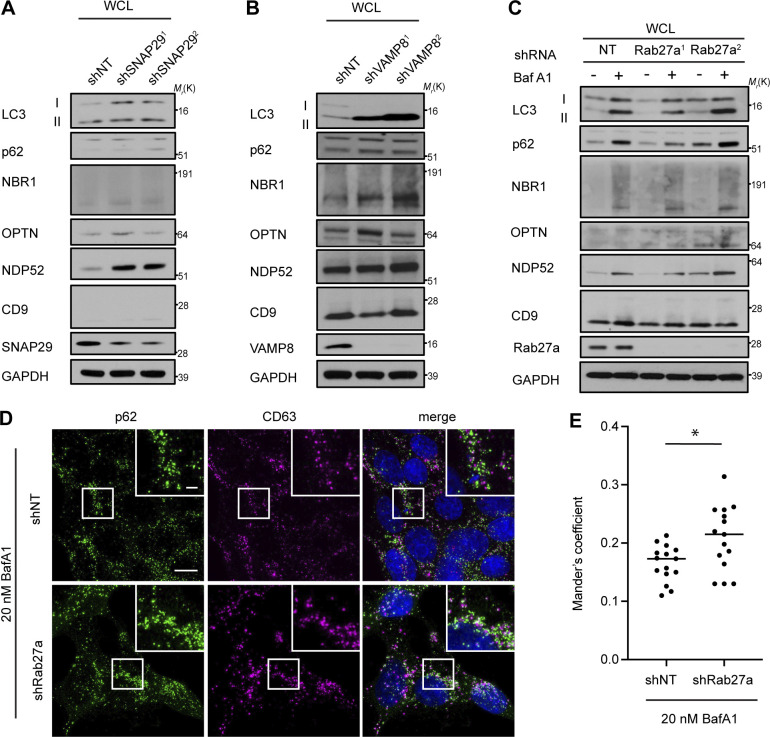
The secretion of autophagy cargo receptors during lysosomal inhibition may be mediated by vesicular intermediates derived from classical double-membrane autophagosomes or proceed through emerging pathways distinct from classical autophagy. To distinguish between these possibilities, we evaluated LC3 and autophagy cargo receptor secretion using a panel of ATG deficient cells that individually disrupt discrete steps required for autophagosome formation. Importantly, these analyses revealed that components required for phagophore formation (FIP200/RB1CC1, ATG14), LC3 conjugation (ATG7, ATG12), and sealing of the double-membrane autophagosome (ATG2A/B) were all critical for autophagy cargo receptor secretion in response to BafA1 (Fig. 5, A and B; and Fig. S4 A). Moreover, EM confirmed dramatically reduced anti-p62 immunogold labeling in small EV-enriched fractions from ATG7 deficient cells treated with BafA1 relative to controls (Fig. S4 B). Thus, in contrast to LDELS, the progressive steps of autophagosome formation are critical for the delivery of LC3 and autophagy cargo receptors outside the cell during lysosome impairment. In further support, BafA1-treated cells deficient for ATG14, which are defective for both autophagosome formation and cargo receptor secretion, showed significantly reduced co-localization of LC3 and p62 with the late endosome and EV marker CD63 compared with controls (Fig. 5, C and D; and Fig. S4, C–E). Moreover, immuno-gold EM for p62 showed robust staining of this cargo receptor within late endosomes of BafA1-treated WT cells (Fig. 5 E). These results demonstrate that all of the key steps required for double-membrane autophagosome formation are necessary for the capture and secretion of autophagy cargo receptors during lysosome impairment, and this material may transit through late endosomes along its itinerary to the extracellular space.
Figure 5.
Autophagy cargo receptor secretion during lysosome inhibition requires autophagosome formation. (A) Cell lysate (WCL; left) and 100,000 g EV fractions (100K; right) from serum-starved WT and ATG-deficient HEK293T cells treated with vehicle or 20 nM BafA1 for 16 h were blotted to detect LC3, autophagy cargo receptors, and CD9 (n = 4). (B) Quantification of the indicated proteins in EVP fractions from BafA1 treated cells relative to treated WT controls. Statistics were calculated by nonparametric one-way ANOVA coupled with Dunnett’s post hoc test (mean ± SEM; n = 4; ***, P < 0.005). (C) Representative fluorescence micrographs from serum-starved WT, ATG7−/−, and ATG14−/− cells treated with 20 nM BafA1 and immunostained for endogenous p62 (green), CD63 (magenta), and LC3. Immunofluorescence micrographs of endogenous LC3 and CD63 from these exact cell samples and corresponding co-occurrence data can be found in Fig. S4, C–E. Scale bar, 10 μm; inset scale bar, 2 μm. (D) Scatter plot of Mander’s coefficients for the co-occurrence of p62 and CD63 in C. Statistics were calculated by nonparametric one-way ANOVA with Dunnett’s post hoc test (mean ± SEM; WT n = 26; ATG7−/− n = 26, ATG14−/−, n = 32; ***, P < 0.005). (E) Representative images from TEM of late endosomes from vehicle or 20 nM BafA1 treated cells that were immunostained with anti-p62 primary antibody and detected using ultrasmall gold-conjugated secondary antibody with silver enhancement (scale bar, 500 nm). Source data are available for this figure: SourceData F5.
Figure S4.
Autophagosome formation is required for autophagy cargo receptor secretion in response to lysosome inhibition. (A) WCL from serum-starved HEK293Ts of the indicated genotypes treated with or without 20 nM BafA1 and corresponding to secretion experiments in Fig. 5, A and B, were collected and immunoblotted for LC3, autophagy cargo receptors, CD9, and GAPDH. (B) Representative images from transmission EM of small EVPs collected from 20 nM BafA1 treated WT and ATG7−/− cells that were immunostained with primary antibody against p62/SQSTM1 and detected with ultrasmall gold-conjugated secondary antibody with silver enhancement (scale bar, 500 nm). (C) Representative fluorescence micrographs from serum-starved WT, ATG7−/−, and ATG14−/− cells treated with 20 nM BafA1 and immunostained for endogenous LC3 (green), CD63 (magenta), and p62. Immunofluorescence micrographs of endogenous p62 and CD63 from these exact cell samples and corresponding co-occurrence data can be found in Fig. 5, C and D. Scale bar, 10 μm; inset scale bar, 2 μm. (D) A scatter plot of the P values obtained from Costes significance tests to assess whether the overlap of LC3 and CD63 staining observed in C exceeds thresholds of random co-occurrence. Statistical significance was calculated by nonparametric two-way ANOVA with Tukey’s post hoc test (mean ± SEM; WT, n = 26; ATG7−/−, n = 26; ATG14−/−, n = 32; ***, P < 0.005). (E) A scatter plot of Mander’s coefficients for the co-occurrence of LC3 with CD63 in BafA1 treated WT and ATG14−/− cells in C. Statistical significance was calculated by an unpaired two-tailed t test (mean ± SEM; WT, n = 26; ATG14−/−, n = 32; *, P < 0.05). Source data are available for this figure: SourceData FS4.
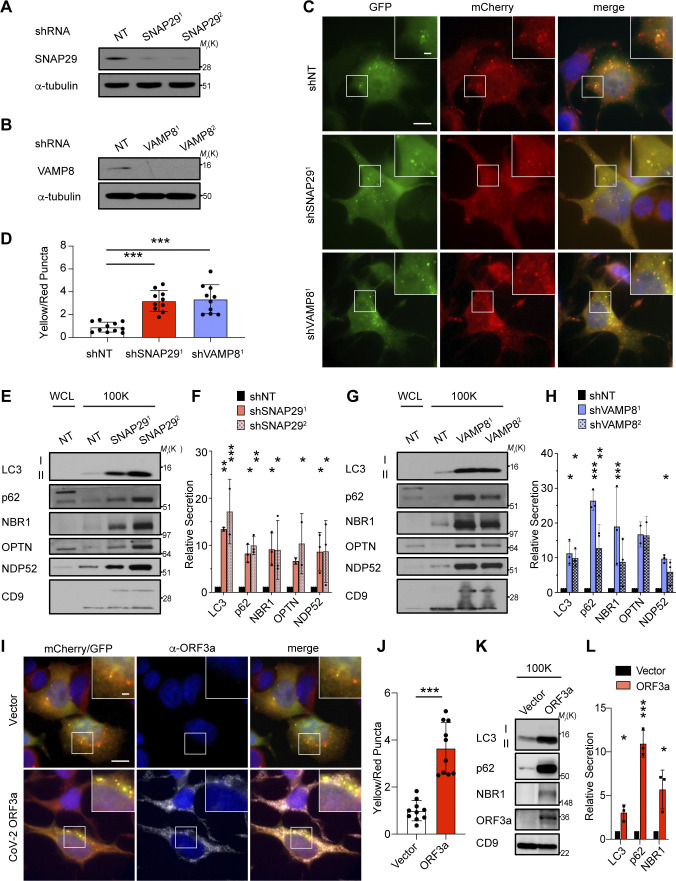
Evidence supports that lysosomal inhibitors perturb autophagosome maturation into autolysosomes, resulting in the impaired turnover of autophagic material, which may subsequently be diverted for secretion outside the cell (Mauthe et al., 2018). Since our results with BafA1 treatment demonstrated that autophagosomes are required for autophagy cargo receptor release, we interrogated whether the genetic disruption of autophagosome maturation was able to specifically promote this secretory phenotype. We evaluated autophagy cargo receptor secretion in cells deficient for SNAP29 or VAMP8, two soluble N-ethylmaleimide-sensitive factor attachment receptors required for autophagosome–lysosome fusion (Itakura et al., 2012; Fig 6, A and B). Consistent with prior studies, genetic depletion of SNAP29 or VAMP8 led to impaired autophagosome maturation, evidenced by the accumulation of yellow autophagic puncta (mCherry+, GFP+) within cells that co-expressed the mCherry-GFP-LC3 flux reporter along with the intracellular accumulation of autophagy cargo receptors (Fig. 6, C and D; and Fig. S5, A and B). In addition to these impairments, we observed both increased LC3-II and autophagy cargo receptors secreted in the EVPs isolated from the conditioned media of SNAP29 and VAMP8 deficient cells compared with controls (Fig. 6, E–H). Thus, similar to pharmacological lysosomal inhibition, the genetic inhibition of autophagosome–lysosome fusion is sufficient to drive the secretion of autophagy cargo receptors via EVPs.
Figure 6.
Genetic inhibition of autophagosome–lysosome fusion promotes the secretion autophagy cargo receptors via EVPs. (A) Lysate from cells stably expressing control shRNA or SNAP29 shRNA (SNAP291; SNAP292) was blotted for SNAP29 and α-tubulin. (B) Lysate from cells expressing control shRNA or VAMP8 shRNA (VAMP81; VAMP82) blotted for VAMP8 and α-tubulin. (C) Representative images of cells stably expressing the mCherry-EGFP-LC3 reporter and control shRNA (shNT), SNAP29 shRNA (shSNAP291), or VAMP8 shRNA (shVAMP81). Scale bar, 10 μm; inset scale bar, 2 μm. (D) Quantification of the ratio of double-positive (mCherry+;EGFP+) yellow puncta to mCherry only (mCherry+;EGFP−) red puncta in cells stably co-expressing the mCherry-EGFP-LC3 reporter and the indicated shRNAs from C. (mean ± SEM; shNT, n = 10; shSNAP291, n = 10, shVAMP81, n = 10; ***, P < 0.005). Statistics were calculated by nonparametric one-way ANOVA with Dunnett’s post hoc test. (E) Cell lysate (WCL; left) and 100,000 g EVP fractions (100K; right) from serum-starved cells expressing control shRNA (NT) or SNAP29 shRNA (shSNAP291; shSNAP292) was blotted for the indicated proteins. (F) Quantification of the proteins in EVPs from cells expressing SNAP29 shRNA relative to control shRNA (shNT) in E. Statistics were calculated by nonparametric one-way ANOVA with Dunnett’s post hoc test (mean ± SEM; n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.005). (G) WCL (left) and 100K fractions (right) from serum starved cells expressing control shRNA (NT) or VAMP8 shRNA (shVAMP81; shVAMP82) was blotted for the indicated proteins. (H) Quantification of the proteins in EVPs from cells expressing VAMP8 shRNA relative to control shRNA (shNT) in G. Statistics were calculated by nonparametric one-way ANOVA with Dunnett’s post hoc test (mean ± SEM; n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.005). (I) Representative images of cells that stably co-express the mCherry-EGFP-LC3 reporter and SARS-CoV-2 ORF3a (2xStrepTag) or vector controls stained with anti-StrepTag antibody. Scale bar, 10 μm; inset scale bar, 2 μm. (J) Quantification of the ratio of double-positive (mCherry+;EGFP+) yellow puncta to mCherry only (mCherry+;EGFP−) red puncta in cells stably co-expressing the mCherry-EGFP-LC3 reporter and ORF3a or vector in I. Statistical significance was calculated by unpaired two-tailed t-test (mean ± SEM; vector, n = 10; ORF3a, n = 10; ***, P < 0.005). (K) 100K fractions from serum starved cells expressing ORF3a (2xStrepTag) or vector were collected and blotted for the indicated proteins. (L) Quantification of LC3, p62, and NBR1 in EVPs from cells stably expressing ORF3a or vector in K. Statistics were calculated by unpaired two-tailed t test (mean ± SEM; n = 3; *, P < 0.05; ***, P < 0.005). Source data are available for this figure: SourceData F6.
Figure S5.
Rab27a is required for autophagy cargo receptor secretion in response to lysosome inhibition. (A) WCL from serum-starved HEK293 Ts that stably express shRNAs targeting SNAP29 (shSNAP291; shSNAP292) or control shRNA (shNT) and corresponding to secretion experiments in Fig. 6, E and F, were immunoblotted for the indicated proteins. (B) WCL from serum-starved HEK293Ts that stably express shRNAs targeting VAMP8 (shVAMP81; shVAMP82) or control shRNA (shNT) and corresponding to secretion experiments in Fig. 6, E and F were immunoblotted for the indicated proteins. (C) WCL from serum-starved HEK293Ts that stably express shRNAs targeting Rab27a (Rab27a1; Rab27a2) or control shRNA (NT) and corresponding to secretion experiments in Fig. 6, E and F, were immunoblotted for the indicated proteins. (D) Representative fluorescence micrographs from cells expressing control shRNA (shNT) or shRNA targeting Rab27a (shRab27a) treated with 20 nM BafA1 and immunostained for endogenous p62 (green) and CD63 (magenta). Scale bar, 10 μm; inset scale bar, 2 μm. (E) Scatter plot of Mander’s coefficients for the co-occurrence of p62 with CD63 in BafA1 treated cells expressing control shRNA (shNT) or Rab27a shRNA (shRab27a) in C. Statistical significance was calculated by an unpaired two-tailed student’s t test (mean ± SEM; shNT, n = 10; shRab27a, n = 10; *, P < 0.05). Source data are available for this figure: SourceData FS5.
SARS-CoV-2 ORF3a inhibits autophagosome–lysosome fusion and promotes cargo receptor secretion via EVPs
Multiple small RNA viruses, including SARS-CoV-2, impair autophagosome–lysosome fusion to evade clearance as well as to generate intracellular membranes important for viral replication and packaging (Miao et al., 2021; Zhang et al., 2021). Therefore, we tested a recently identified viral inhibitor of autophagosome maturation, SARS-CoV-2 ORF3a, for its impact on the secretion of autophagy cargo receptors. Consistent with recent reports, cells stably expressing ORF3a had marked accumulation of early autophagosomes relative to controls in mCherry-GFP-LC3 reporter assays, characteristic of impaired autophagosome maturation (Fig. 6, I and J). Moreover, EVPs from the conditioned media secreted by ORF3a expressing cells were enriched in LC3-II, p62, and NBR1; notably, these fractions also contained ORF3a itself (Fig. 6, K and L). These results further corroborate that impaired autophagosome-to-autolysosome maturation promotes the diversion of autophagic vesicular intermediates containing LC3 and autophagy cargo receptors for EVP-mediated secretion.
Rab27a promotes autophagy cargo receptor secretion via EVPs
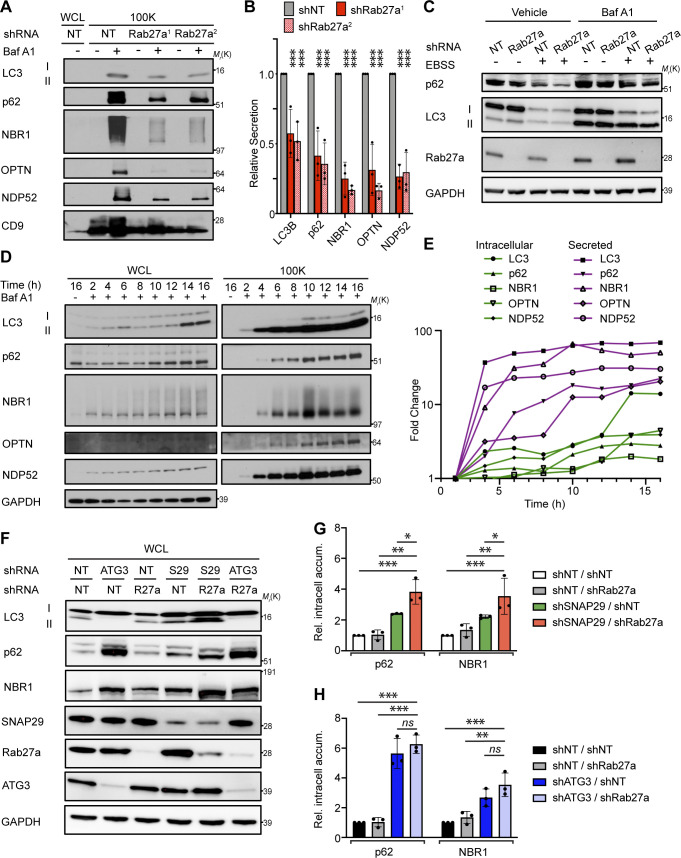
A subpopulation of EVPs is formed at late endosomes termed multivesicular endosomes (MVEs). Autophagosomes are also known to deliver material to MVEs, suggesting that LC3 and autophagy cargo receptors may be diverted through MVEs when autophagosome maturation is impaired to facilitate extracellular release. Rab27a, a small GTPase that promotes the fusion of endolysosomal intermediates with the plasma membrane, has been implicated in multiple regulated lysosomal and granule exocytosis pathways, including the extracellular release of EVPs originating from MVEs (Desnos et al., 2003; Ostrowski et al., 2010; Stinchcombe et al., 2001). Thus, we tested whether Rab27a was required for autophagy cargo receptor secretion; indeed, stable Rab27a knockdown suppressed the secretion of LC3, p62, NBR1, OPTN, and NDP52 via EVPs in BafA1-treated cells (Fig. 7, A and B; and Fig. S5 C). Importantly, assays of autophagic flux confirmed that lysosomal turnover of LC3-II and p62 was unaltered in Rab27a deficient cells, indicating that Rab27a is specifically required for the secretion of cargo receptors in response to BafA1, rather than modulating the autophagic degradation of these components (Fig. 7 C). Furthermore, the inhibition of Rab27a in BafA1 treated cells promoted increased co-localization between p62 and CD63, suggesting the intracellular accumulation of cargo receptors within late endosomes when Rab27a-dependent exocytosis was blocked (Fig. S5, D and E). Altogether, these results demonstrate that Rab27a is functionally required for the secretion of LC3 and cargo receptors in EVP populations. Moreover, these results broach that during lysosome impairment, early autophagic intermediates are diverted to MVEs, which require Rab27a-dependent exocytosis for extracellular release.
Figure 7.
Rab27a is required for EVP secretion of autophagy cargo receptors in response to lysosome inhibition and impaired autophagosome maturation. (A) WCL (left) and 100,000 g EVP fractions (100K; right) from serum-starved cells expressing control shRNA (NT) or Rab27a shRNA (shRab27a1; shRab27a2) were collected and blotted for indicated proteins. (B) Quantification of indicated proteins in EVPs from cells expressing SNAP29 shRNA relative to control shRNA (shNT) in A. Statistics were calculated by nonparametric one-way ANOVA with Dunnett’s post hoc test (mean ± SEM; n = 3; ***, P < 0.005). (C) Cells expressing control shRNA (NT) or Rab27a shRNA were EBSS starved for 12 h, lysed, and blotted for the indicated proteins. BafA1 = 50 nM, BafA1 added 1 h before lysis (n = 3). (D) WCL (left) and 100K fractions (right) from serum starved cells treated with 20 nM BafA1 were collected at the indicated times after treatment and blotted for LC3, autophagy cargo receptors, and GAPDH. (E) Quantification of the fold change in intracellular levels (green lines) and EVP-mediated secretion (secreted; shapes with magenta lines) for the indicated proteins relative to levels at 2 h after treatment with BafA1 in D. (F) WCL from cells stably expressing the indicated combinations of control shRNA (NT) and shRNAs targeting ATG3, Rab27a (R27a), and SNAP29 (S29) collected 8 h after starvation and blotted for indicated proteins. (G) Quantification of p62 and NBR1 in WCL from starved cells co-expressing control shRNA and Rab27a shRNA (shNT/shRab27a), SNAP29 shRNA and control shRNA (shSNAP29/shNT), or SNAP29 shRNA and Rab27a shRNA (shSNAP29/shRab27a) relative to shRNA controls (shNT/shNT) in F. Statistics were calculated by nonparametric one-way ANOVA coupled with Dunnett’s post hoc test (mean ± SEM; n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.005). (H) Quantification of p62 and NBR1 in WCL from starved cells co-expressing control shRNA and Rab27a shRNA (shNT/shRab27a), ATG3 shRNA and control shRNA (shATG3/shNT) or ATG3 shRNA and Rab27a shRNA (shATG3/shRab27a) relative to shRNA controls (shNT/shNT) in F. Statistics were calculated by nonparametric one-way ANOVA coupled with Dunnett’s post hoc test (mean ± SEM; n = 3; **, P < 0.01; ***, P < 0.005). Source data are available for this figure: SourceData F7.
Secretory autophagy reduces intracellular accumulation of autophagy cargo receptors when degradative autophagy is impaired
In the setting of autophagy or lysosomal deficiency, the aberrant intracellular accumulation of autophagy cargo receptors, including p62 and NBR1, is associated with numerous detrimental effects ranging from perturbations in intracellular signaling, changes in cellular differentiation, and the formation of intracellular inclusions (Bjorkoy et al., 2005; Mancias and Kimmelman, 2016; Marsh et al., 2020). Since cells may control the intracellular levels of these proteins through multiple routes, including extracellular secretion, we postulated that the diversion of autophagy cargo receptors for secretion via EVPs serves as a mechanism to control intracellular accumulation of these important effector proteins when autophagosome maturation is impaired. To test this hypothesis, we first assayed the kinetics of autophagy cargo receptor secretion and accumulation in cells following acute treatment with BafA1 over 16 h. Notably, the secretion of autophagy cargo receptors in EVP fractions was rapidly induced within 4 h of BafA1 treatment and continued to 8–10 h, at which time it reached a steady-state (Fig. 7, D and E; magenta lines). In contrast, intracellular accumulation of LC3 and cargo receptors was modest from 2 to 10 h after treatment; thereafter, more robust intracellular accumulation of these targets was observed between 10 and 16 h of lysosome inhibition (Fig. 7, D and E; green lines). These observations highlighted that BafA1-induced extracellular secretion of autophagy cargo receptors precedes their robust intracellular accumulation and suggests that secretory autophagy via EVPs buffers against the intracellular accumulation of these targets when lysosomal degradation is impaired.
To determine whether secretory autophagy directly impacts the intracellular accumulation of autophagy cargo receptors in response to impaired autophagy-dependent degradation, we scrutinized the effects of the combined genetic inhibition of degradative and secretory autophagy on intracellular levels of p62 and NBR1. Indeed, our results above demonstrated that genetic depletion of SNAP29 impaired autophagosome maturation, resulting in increased autophagy cargo receptor secretion. In contrast, Rab27a was functionally required for cargo receptor secretion in response to lysosomal inhibition but had minimal effects on degradative autophagy. Accordingly, if secretory autophagy is important for mitigating the effects of impaired autophagy-dependent cargo receptor degradation, cells doubly deficient for SNAP29 and Rab27a would be predicted to intracellularly accumulate autophagy cargo receptors at increased levels. To test this prediction, we compared intracellular cargo receptor levels in cells deficient for SNAP29 versus those doubly deficient for SNAP29 and Rab27a. Cells genetically depleted of both SNAP29 and Rab27a demonstrated increased intracellular accumulation of p62 and NBR1 relative to cells depleted of SNAP29 alone or WT controls (Fig. 7, F and G). In contrast, Rab27a loss did not influence cargo receptor proteostasis when early autophagosome formation was impaired. The combined knockdown of ATG3 and Rab27a did not significantly promote autophagy cargo receptor accumulation compared with ATG3 depletion alone (Fig. 7, F and H). Thus, the secretion of autophagy cargo receptors via EVPs serves as a homeostatic mechanism for balancing the intracellular levels of these autophagy targets in response to impaired autophagosome maturation and lysosomal degradation.
Discussion
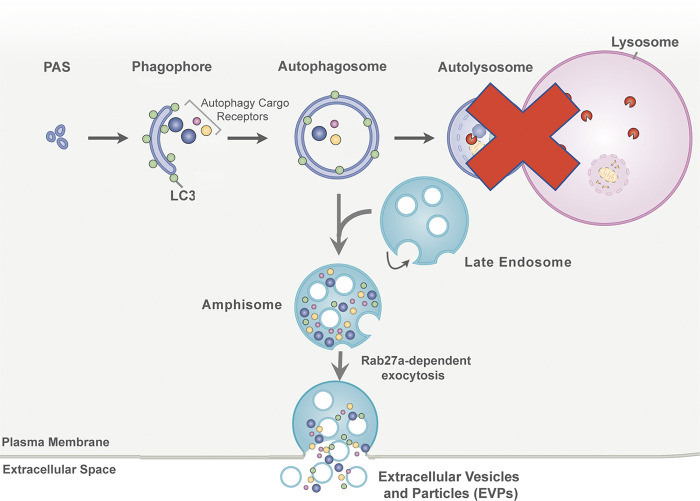
Here, we delineate a secretory autophagy process that targets autophagic cargo receptors for release via EVP-associated secretion pathways when autophagy-dependent degradation is impaired, a process that we term secretory autophagy during lysosome inhibition (SALI; Fig. 8). Previous work has revealed important interconnections between autophagy and EV secretion. For example, our group demonstrated that LC3-conjugation components regulate the loading and release of small EVs (Leidal et al., 2020). In addition, infectious pathogens and pharmacological agents that disrupt endolysosome function and autophagic turnover also facilitate the secretion of autophagy components and autophagy cargo receptors in association with EVs (Hessvik et al., 2016; Miao et al., 2015; Miranda et al., 2018). These connections have led both us and others to conjecture that autophagy and EVP secretion pathways function cooperatively to maintain cellular homeostasis (Leidal and Debnath, 2021; Xu et al., 2018). In this study, we illuminate that impaired autophagosome maturation promotes the rerouting of autophagic material through late endosomal intermediates, which results in its secretion in EVP-associated fractions via Rab27a-dependent exocytosis.
Figure 8.
SALI. In response to lysosome inhibition or impaired autophagosome maturation, autophagic intermediates containing autophagy cargo receptor proteins are diverted to late endosomes. Within these autophagosome–late endosome hybrid organelles, termed amphisomes, autophagy cargo receptors intermix with intraluminal vesicles that are formed through intraluminal budding mechanisms. The contents of amphisomes are subsequently released as EVPs via Rab27a-dependent exocytosis at the plasma membrane.
Importantly, SALI is distinct from the recently defined LDELS pathway, which specifies the loading of RNA-binding proteins into intraluminal vesicles and their subsequent secretion within EVs (Leidal et al., 2020). LDELS represents one of a growing number of autophagy-related processes, including LC3-associated phagocytosis and LC3-associated endocytosis, in which the autophagy conjugation machinery mediates the conjugation of LC3 to single membranes organelles of the endolysosomal system via pathways distinct from classical autophagy (Nieto-Torres et al., 2021). In contrast, autophagy cargo receptor secretion in response to impaired lysosome function or autophagosome maturation is more closely akin to classical autophagy in that it requires multiple ATGs that mediate the progressive steps in autophagosome formation. Overall, our results support that defects in autophagosome-to-autolysosome maturation promote the accumulation of immature autophagosomes, which fuse with late endosomes and MVEs, forming amphisomes. As a result, autophagic material such as autophagy cargo receptors can mix with intraluminal vesicles and particles, and this material is subsequently released with EVPs upon amphisome fusion with the plasma membrane via a Rab27a-dependent process (Fig. 8). In support, we observe strong immuno-gold labeling of p62 within the MVEs of BafA1 treated cells, and the co-localization of LC3 and p62 with late endosomal markers during lysosome inhibition is reduced in cells deficient for ATG7 or ATG14 compared with WT controls.
We propose that SALI utilizes EVP-based secretion pathways as an alternate route to clear sequestered material when degradation is impaired. As a result, blocking Rab27a-mediated exocytosis not only reduces the extracellular secretion of autophagy cargo receptors but also exacerbates the accumulation of these proteins when autophagosome-to-autolysosome maturation is simultaneously impaired. Defects in autophagic flux and lysosomal function are observed both during aging and various age-related disorders, and secretory autophagy may serve important roles in maintaining proteostasis in these settings (Leidal et al., 2018). An exciting question for future study is determining whether lysosomal dysfunction in neurodegenerative disorders, such as Parkinson’s disease, Alzheimer’s disease, and frontotemporal dementia, elicits increased secretion of autophagy cargo receptors, and if so, scrutinizing how secretory autophagy impacts proteostasis in such patients (Leidal et al., 2018). Nevertheless, despite these potential cell-autonomous benefits of secretory autophagy in mitigating intracellular protein accumulation during lysosomal inhibition, the extracellular secretion of autophagy cargo receptors may harbor deleterious effects in vivo. Notably, recent work demonstrates that extracellular p62 promotes sepsis-induced death in mice due to disruptions in immune metabolism and increased systemic coagulation (Zhou et al., 2020). Hence, further understanding whether and how the EVP secretion of p62 and other cargo receptors modulates inflammatory and immune function in various human pathologies due to defective lysosome function remains an important question for future study. Interestingly, we demonstrate that the SARS-CoV-2 ORF3a viral protein not only disrupts autophagosome-to-autophagosome maturation but potently promotes the extracellular secretion of p62 via EVPs (Miao et al., 2021; Zhang et al., 2021). Accordingly, further scrutinizing whether and how extracellular p62 secreted from SARS-CoV-2-infected cells contributes to COVID19-associated immune pathologies and coagulopathies remains an important future direction (Fajgenbaum, 2020).
Finally, we show that treatment with the antimalarial CQ in vivo results in the increased EVP secretion of autophagy cargo receptors in murine plasma. Over the past several years, hydroxychloroquine has been actively pursued to therapeutically inhibit the autophagy–lysosome pathway in human oncology trials (Amaravadi et al., 2019). However, a critical barrier for assessing the efficacy of such strategies has been non-invasively monitoring changes in autophagic flux in humans, particularly in response to the therapeutic modulation of autophagy and lysosomal pathways (Mizushima and Murphy, 2020). Given the continued interest in targeting autophagy and lysosomal pathways for cancer treatment combined with the growing excitement regarding EVPs proteins as non-invasive liquid biopsy tools, measuring the autophagy-dependent EVP secretion of cargo receptors in human plasma potentially represents a powerful biomarker for monitoring the efficacy of next-generation lysosomal inhibitors in cancer treatment, and more broadly, for monitoring lysosomal dysfunction in diverse therapeutic settings.
Materials and methods
Cell culture
HEK-293T (CRL-3216; American Type Culture Collection [ATCC]), MDA-MB-231 (CRM-HTB-26; ATCC), Neuro2a (CCL-131; ATCC), and R221A cells (Gift from Barbara Fingleton, Vanderbilt University, Nashville, TN) were cultured in DMEM, high glucose, pyruvate (11995040; Gibco) supplemented with 10% FBS (F-0500-D; Atlas Biologicals), 25 mM Hepes (H1030; Teknova), 100 U/ml penicillin, and 100 μl/ml streptomycin (14140163; Gibco). SH-SY5Y cells (CRL-2266; ATCC) were cultured in DMEM/F12 (11330032; Gibco) supplemented with 10% FBS (F-0500-D; Atlas Biologicals), 100 U/ml penicillin, and 100 μl/ml streptomycin (14140163; Gibco). All cell lines were tested for mycoplasma contamination (MP0035; Sigma-Aldrich).
CRISPR/Cas9 gene deletion
HEK-293T knockout cell lines were generated by transient transfection of pSpCas9(BB)-2A-Puro (48139; Addgene) encoding U6 driven expression of sgRNAs (Scramble Guide: 5′-GCACTACCAGAGCTAACTCA-3′; ATG7 Guide: 5′-ACACACTCGAGTCTTTCAAG-3′; ATG12 Guide: 5′-CCGTCTTCCGCTGCAGTTTC-3′; ATG14 Guide: 5′-CTACTTCGACGGCCGCGACC-3′; FIP200 Guide: 5′-AGAGTGTGTACCTACAGTGC-3′; ATG2A Guide: 5′-CGCTGCCCTTGTACAGATCG-3′; ATG2B Guide: 5′-ATGGACTCCGAAAACGGCCA-3′). Cells were selected 48–72 h after transfection with 1 g/ml puromycin for 48 h. Polyclonal populations were collected for surveyor analysis (706020; IDT) and were sorted into single-cell populations by limiting dilution at 1.5 cells/well per 96-well plate. For DNA analysis, genomic DNA samples were prepared using QuickExtract (Epicentre). The PCR products were column purified and analyzed with surveyor mutation detection kit (IDT). For genotyping of single-sorted cells, PCR amplified products encompassing the edited region (ATG7 Fwd: 5′-TGGGGGACAGTAGAACAGCA-3′, ATG7 Rev: 5′-CCTGGATGTCCTCTCCCTGA-3′; ATG12 Fwd: 5′-AGCCGGGAACACCAAGTTT-3′, ATG12 Rev: 5′-GTGGCAGCCAAGTATCAGGC-3′; ATG14 Fwd: 5′-AAAATCCCACGTGACTGGCT-3′, ATG14 Rev: 5′-AATGGCAGCAACGGGAAAAC-3′; FIP200 Fwd: 5′-ATTCTCTGGCTTGACAGGACAG-3′, FIP200 Rev: 5′-AAATACTGAGCGTGCACATTGC-3′; ATG2A Fwd: 5′-GAGCCGGACGGGGATCGC-3′, ATG2A Rev: 5′-CTGCAAGGTGAGCTGGAGGC-3′; ATG2B Fwd: 5′-ATAGGGATGGAGGGGCCGC-3′, Rev: 5′-TCATTGAGACACCATTTGTC-3′) were cloned into pCR4-TOPO TA vector using the TOPO-TA cloning kit (450030; Thermo Fisher Scientific) and sequence verified.
Plasmid constructs
The following vectors are available or were obtained from Addgene: pBABE-puro mCherry-EGFP-LC3 (#22418), pBABE-hygro (#1765), psPAX2 (#12260), pMD2.G (Plasmid #12259), pLKO.1-blast (#26655), and pLVX-EF1alpha-SARS-CoV-2-orf3a-2xStrep-IRES-Puro (#141383). pBABE-hygro mCherry-EGFP-LC3 was generated by subcloning mCherry-EGFP-LC3 from pBABE-puro mCherry-EGFP-LC3 into shared AgeI and SalI restriction sites in pBABE-hygro. pLKO.1 lentiviral vectors for shRNAs targeting SNAP29 (#1 TRCN0000083650; #2 TRCN0000231850), VAMP8 (#1 TRCN0000219911; #2 TRCN0000181123), Rab27a (#1 TRCN0000005295; #2 TRCN0000005296), and non-targeting control (SHC002) are available from Sigma-Aldrich. pLKO.1-blast shRab27a was generated by subcloning the blasticidin resistance gene from pLKO.1-blast into shared BamHI and KpnI restriction sites in pLKO.1 shRab27a (#TRCN0000005295).
Retroviral and lentiviral packaging, infection, and selection
Retroviral pBABE expression vectors were packaged and target cells were transduced according to established protocols. Briefly, Phoenix amphotropic packaging cells (gift from C. McCormick, Dalhousie University, Halifax, Canada) were seeded and transfected with retroviral vectors using polyethylenimine. Virus-containing conditioned media was collected 2 d after transfection and clarified using a 0.45-μM filter. Prior to infection, virus-containing medium was diluted 1:4 in DMEM growth medium, and the mix was supplemented with Polybrene at 8 μg/ml final concentration. Subsequently, the viral transduction mix (5 ml total volume/10 cm culture dish) was incubated with HEK293T cells for 24 h. Cells were selected 24 h after transduction with 1 μg ml−1 puromycin for 2 d or 100 μg ml−1 hygromycin for 5 d. To package lentivirus, HEK293T cells were seeded and co-transfected with the packaging vectors psPAX2 and pMD2.G, and individual pLKO.1 transfer vectors or pLVX-EF1alpha-SARS-CoV-2-orf3a-2xStrep-IRES-Puro. Virus collection and infection were carried out as above. Stably transduced cell pools were selected with 1 μg/ml puromycin for 2 d or 10 μg/ml hygromycin for 4 d.
Antibodies
The following antibodies were used for immunoblotting: rabbit anti-ATG14 (PD026; MBL; 1:1,000), rabbit anti-LAMP1 (9091; Cell Signaling Technology; 1:1,000), rabbit anti-Rab7a (9367; Cell Signaling Technology; 1:1,000), rabbit anti-Rab5a (3547; Cell Signaling Technology; 1:1,000), mouse anti-GM130 (610823; BD Biosciences; 1:1,000), rabbit anti-CYC1 (NBP1-86872; Novus Biologicals; 1:1,000), mouse anti-NUP62 (610497; BD Biosciences; 1:1,000), mouse anti-GAPDH (MAB374; Millipore; 1:10,000), rabbit anti-LC3 (ABC232; Millipore; 1:1,000), guinea pig anti-p62/SQSTM1 (GP62-C; Progen; 1:1,000), rabbit anti-p62/SQSTM1 (5114; Cell Signaling Technology; 1:1,000), mouse anti-NBR1 (sc-130380; Santa Cruz Biotechnology; 1:500), rabbit anti-OPTN (ab23666; Abcam; 1:1,000), mouse anti-NDP52/CALCOCO2 (H00010241-B01P; Abnova; 1:1,000), mouse anti-TSG101 (612696; BD Biosciences; 1:1,000), mouse anti-ALIX (2171; Cell Signaling Technology; 1:1,000), mouse anti-CD9 (CBL162; Sigma-Aldrich; 1:1,000), mouse anti-GFP (sc-9996; Santa Cruz Biotechnology), rabbit anti-Rab27a (69295; Cell Signaling Technology; 1:1,000), rabbit anti-SNAP29 (ab138500; Abcam; 1:1,000), rabbit anti-VAMP8 (ab76021; Abcam; 1:1,000), rabbit anti-StrepTag II (ab76949; Abcam; 1:500), ATG3 (3415S; Cell Signaling Technology; 1:1,000), Peroxidase-AffiniPure Donkey Anti-Rabbit IgG (H + L; 711-035-152; Jackson; 1:5,000), Peroxidase-AffiniPure Donkey Anti-Guinea Pig IgG (H + L; 706-035-148; Jackson; 1:5,000), Peroxidase-AffiniPure Donkey Anti-Goat IgG (H + L; 705-035-147; Jackson; 1:5,000), and Peroxidase-AffiniPure Donkey Anti-Mouse IgG (H + L; 715-035-150; Jackson; 1:5,000).
For immunofluorescence, we employed: rabbit anti-LC3B (#3868T; Cell Signaling Technology; 1:200), mouse anti-CD63 (#ab8219; Abcam; 1:200), guinea pig anti-p62/SQSTM1 (#GP62-C; Progen/Cedarlane; 1:1,000), rabbit anti-StrepTag II (ab76949; Abcam; 1:200), AlexaFluor goat anti-rabbit 488 ( #A-11034; Thermo Fisher Scientific; 1:500), goat anti-mouse 568 (#A-11031; Thermo Fisher Scientific; 1:500), and goat-anti guinea pig (A-21450; Thermo Fisher Scientific, 1:1,000).
Immunoblotting
For whole cell lysate (WCL), cells were lysed in radioimmunoprecipitation assay buffer (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) supplemented with 10 mM NaF, 10 mM b-glycerophosphate, 1 mM Na3VO4, 10 nM calcyulin A, 0.5 mM phenylmethyl sulphonyl fluoride, 0.1 mM E-64-D, 10 μg/ml pepstatin A, and protease inhibitor cocktail. Lysates were cleared by centrifugation at 15,000 g for 10 min at 4°C. For EV lysate, pelleted EVs were resuspended in urea buffer (50 mM Tris-HCl, pH 8.0, 8 M urea, 2% SDS, 10 mM NaF, 5 mM EDTA). WCL and EV lysates were quantified by bicinchoninic acid assay (23225; Thermo Fisher Scientific), mixed with sample buffer, resolved by SDS-PAGE, and transferred to polyvinylidene fluoride membrane. Membranes were then blocked in 5% milk in PBS with 0.1% Tween 20 (PBST), incubated in primary antibody overnight at 4°C in blocking buffer, washed with PBST, incubated in HRP-conjugated secondary antibodies for 1 h at RT in blocking buffer, washed with PBST, and visualized by enhanced chemiluminescence (32106; Thermo Fisher Scientific) on film. Immunoblots were quantified by densitometry using Fiji (ImageJ v.2.0.0-rc-69/1.52p).
Fluorescence and immunofluorescence microscopy
To perform mCherry-GFP-LC3 fluorescence microscopy, cells were stably transduced with retroviral vectors that encoded mCherry-GFP-LC3 and puromycin or hygromycin resistance genes for 24 h and then selected in 1 μg/ml puromycin or 100 μg/ml hygromycin B (#10687010; Thermo Fisher Scientific) for 2 or 5 d, respectively. Subsequently, cells were seeded onto coverslips coated with 10 μg/ml fibronectin, and 24 h later treated with vehicle or 20 nM BafA1 in serum-free DMEM for 16 h. Cells were fixed with 4% PFA in PBS for 15 min at RT, washed with PBS, and mounted onto slides with Prolong Gold Antifade with DAPI (P36935; Thermo Fisher Scientific). Cells were visualized at 25°C using a DeltaVision microscope (Applied Precision Ltd.) fitted with a 100×, 1.4-NA objective in the presence of immersion oil and captured on a CoolSnap HQ camera (Photometrics). Images were acquired using softWoRx software (Applied Precision Ltd.) and prepared in Fiji and Adobe Photoshop. To quantify mCherry-EGFP-LC3 puncta, cells were outlined manually, and each channel was independently autothresholded using the same settings across all images from each channel. mCherry and GFP puncta were then quantified using the Analyze Particles plugin in ImageJ. Double-positive puncta were identified and counted using the Colocalization and Analyze Particles plugins in ImageJ.
For immunofluorescence of endogenous p62 and CD63 or LC3 and CD63, cells were seeded on coverslips coated with 10 µg/ml fibronectin and 24 h later treated with vehicle or 20 nM BafA1 in serum-free DMEM for 16 h. Subsequently, cells were fixed with 4% PFA in PBS for 15 min at RT, permeabilized with ice-cold methanol, and incubated at −20°C for 5 min before quenching with PBS/glycine. Samples were then blocked in blocking buffer (PBS + 0.1% Tween + 10% goat serum) for 1 h at RT, incubated with guinea pig anti-p62 (#GP62-C; Progen/Cedarlane; 1:200) and mouse anti-CD63 (ab8219; Abcam; 1:200) or rabbit anti-LC3B (PM036; MBL; 1:200), and mouse anti-CD63 (ab8219; Abcam; 1:200) antibodies for primary staining. Cells were visualized at 25°C using a DeltaVision microscope (Applied Precision) fitted with a 100×, 1.4-NA objective in the presence of immersion oil and captured on a CoolSnap HQ camera (Photometrics). Images were acquired using softWoRx software (Applied Precision) and prepared in Fiji and Adobe Photoshop. Costes significance tests for co-occurrence and the Mander’s overlap coefficient for p62 and CD63 or LC3 and CD63 were performed by drawing a region of interest around individual cells and then employing the Coloc2 analysis function within Fiji (PSF: 20, Costes randomizations: 10).
Cell viability assay
To quantify cell viability, cells were seeded in 12-well plates, and after 24 h were treated with vehicle, 20 nM BafA1 or 25 μM CQ in serum-free DMEM for 8 h. Cells were collected and stained with 0.4% trypan blue (T10282; Thermo Fisher Scientific). Live and dead cells were then enumerated using the Countess II Automated Cell Counter (AMQAX1000; Thermo Fisher Scientific).
EV preparation and characterization
EVs were isolated according to standard differential centrifugation protocol as described previously (Théry et al., 2006). Briefly, HEK-239T cells were seeded in 15-cm tissue culture dishes and, when confluent, incubated with serum-free DMEM ± treatment (20 nM BafA1, 25 μM CQ) for 16 or 24 h. Conditioned media was then collected and serially centrifuged: 200 g for 10 min at 4°C to pellet cells, 2,000 g for 10 min at 4°C to pellet cellular debris and apoptotic bodies, 10,000 g for 30 min at 4°C in an ultracentrifuge to pellet large EVs, and 110,000 g for 70 min or 3 h at 4°C in an ultracentrifuge to pellet small EVs. The 110,000 g pellet was then gently resuspended in 1 ml PBS by pipetting, diluted in 12 ml PBS, and ultracentrifuged at 110,000 g for 70 min at 4°C. The final EV pellets were resuspended in urea lysis buffer for immunoblotting analysis, 10% sucrose for sucrose density gradient analysis, or PBS for protease protection analysis. For analysis of secreted soluble protein, the supernatant from the first 110,000 g spin was collected and precipitated by the addition of 15% TCA, then incubated for 1 h on ice before centrifugation at 180,000 g for 2 h at 4°C. The 180,000 g pellet was then resuspended in 10 ml ice-cold 100% acetone, centrifuged at 180,000 g for 1 h at 4°C, and finally resuspended in urea lysis buffer. For comparison of EVs between experimental conditions in vitro, EV quantification or EV protein quantification were corrected as indicated in figure legends based on total cell number or WCL protein quantification to correct for cell seeding differences between conditions.
Sucrose density gradient separation was used to analyze co-fractionation between LC3, ATG cargo receptors, and canonical EV markers. 10 and 60% sucrose solutions in PBS were used to prepare continuous 10–60% sucrose gradients on a gradient station (Biocomp Instruments 153). EV pellets were generated by differential centrifugation as described previously, then resuspended in 100 μl 10% sucrose solution, gently layered on top of the 10–60% gradient, and ultracentrifuged at 210,000 g for 18 h at 4 C. After ultracentrifugation, 1-ml fractions of the gradient were carefully top-unloaded, weighed, and diluted in 12 ml PBS. The diluted sucrose gradient fractions were ultracentrifuged at 110,000 g for 70 min at 4°C, and pellets were resuspended in urea lysis buffer for immunoblotting analysis.
Protease protection analysis was used to determine membrane protection of secreted cargo receptors. EV pellets were generated by differential centrifugation as described previously, resuspended in 60 μl PBS, and divided equally into three fractions for resuspension in PBS, 100 μg/ml trypsin in PBS, or 1% NP-40 and 100 μg/ml trypsin in PBS. After incubation and occasional mixing for 30 min at 4°C, protease reactions were stopped with the addition of 10 μl protease inhibitor cocktail and 2× sample buffer before immunoblotting analysis.
Nanoparticle tracking analysis (NTA) technology was used to determine EV concentration and size. Conditioned media clarified of cell debris and large EVs by 0.22 μm filter was normalized by cell number, then loaded on a nanoparticle analyzer (Malvern Panalytical NanoSight NS300), and analyzed using NTA software (NTA 3.3 Dev Build 3.3.104). Camera level was set at 15 for all recordings. Camera focus was adjusted to make the particles appear as individual dots with surrounding refractory rings. Three 30-s videos were recorded for each sample with a delay of 5 s between each recording.
MS of extracellular vesicle and particle proteins
WT and ATG7−/− HEK293T cells were treated with 20 nM BafA1 or vehicle in serum-free media for 16 h. After 16 h, conditioned media was collected and EVs were isolated using the differential centrifugation protocol outlined previously. Pelleted EVs were lysed in 800 μl radioimmunoprecipitation assay buffer (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) supplemented with 2% SDS, then sonicated with a probe sonicator at amplitude 8 for 10 pulses of 10 s each. Samples were then diluted in 4 ml ice-cold 100% acetone and incubated at −20°C overnight. Samples were spun in an ultracentrifuge at 200,000 g for 18 h at 4°C, the acetone was decanted, and the samples air-dried for 1 h at 25°C before being stored at −80°C until further processing.
Three biological replicates of precipitated EVs from WT and ATG7−/− BafA1-treated cells were resuspended in 30 μl 6 M guanidinium-chloride, 100 mM Tris, pH 8.0, 10 mM tris(2-carboxyethyl)phosphine, 40 mM 2-chloroacetamide. EV proteins were denatured for 1 h at 37°C and then quantified with 660 nm Protein Assay Reagent (22660; Thermo Fisher Scientific). Samples were then diluted sixfold in 150 μl 100 mM Tris, pH 8.0. About 125–150 μg of protein for each sample was trypsinized with 15 μg trypsin (90057; Thermo Fisher Scientific) in an orbital shaker at 250 rpm, 37°C for 20 h. Trypsin digestion was then stopped by adding 10% trifluoracetic acid (TFA) to a final concentration of 0.5% TFA. Samples were desalted with SOLA solid-phase extraction cartridges (60109; Thermo Fisher Scientific). Briefly, columns were washed with 500 μl 100% acetonitrile (ACN), then equilibrated with 500 μl 0.1% TFA twice before adding sample. Samples were washed with 500 μl 0.1% TFA three times, then 500 μl 0.1% formic acid (FA), 2% ACN, and eluted with 450 μl 0.1% FA, 50% ACN. Samples were dried by speed-vac, then resuspended in 10 μl 50 mM Hepes, pH 8.5, and quantified by Pierce Quantitative Colorimetric Peptide Assay (23275; Thermo Fisher Scientific).
For tandem mass tag (TMT) labeling, 800 μg of TMT10-126, 127N, 128C, 129N, 130C, and 131 (90110; Thermo Fisher Scientific) was reconstituted with 41 μl 100% anhydrous ACN. 15 μg of peptides from each replicate were individually combined with 7.69 μl (150 μg) of the TMT isobaric tags. Samples were incubated at 25°C for 1 h before quenching the reaction with 8 μl 5% hydroxylamine for 15 min. After labeling, the six individually labeled samples were pooled, dried by speed-vac, and then resuspended in 300 μl 0.1% TFA. The pooled samples were then fractionated into eight using Pierce High pH Reversed-Phase Peptide Fractionation Kit (84868; Thermo Fisher Scientific) and then dried by speed-vac. The fractions were then resuspended in 0.1% FA, 2% ACN before liquid chromatography (LC)–MS/MS analysis.
For LC-MS/MS analysis, 1 μg TMT-labeled peptide per fraction was analyzed on a 15 cm C18 analytical column, in-line with a Q-Exactive Plus mass spectrometer. The peptides were separated on a multi-slope, 100 min gradient (6.4–27.2% ACN with 0.1% FA for 80 min at 0.2 μl/min, then 27.2–40% ACN with 0.09% FA for 15 min at 0.3 μl/min, then 40–56% ACN with 0.09% FA for 5 min at 0.3 μl/min, and then washed for 3 min). Data dependent acquisition with MS1 resolution of 70,000, top15 method, and HCD normalized collision energy of 32 was used, with MS2 resolution of 35,000 and an isolation window of 0.7 m/z. Dynamic exclusion was activated for 30 s after initial parent ion selection.
Eight injections of the different fractions of TMT-labeled EV peptides were analyzed together via MaxQuant (v1.6.0.16). Search parameters for peptide search tolerance was 4.5 ppm, centroid match tolerance was 8 ppm, and two missed tryptic cleavages were permitted. Constant modification of carbamidomethylation of cysteines and variable modifications of N-terminal acetylation, methionine oxidation, and Ser/Thr/Tyr phosphorylation were allowed. Peptide spectrum match false discovery rate (FDR) and protein FDR were set at 1%. “Match between runs” was enabled to increase peptide identification. Type was set to “Reporter ion MS2,” and the six labels used in sample preparation were selected. The resulting quantifications were then median-normalized for each channel, and statistical analysis (two-sample t test) was applied in R with a statistical significance threshold of P < 0.05.
Bioinformatic analyses
For MS, the top eight proteins with the highest connectivity to candidates statistically enriched in EVs from WT cells treated with BafA1 versus EVs from ATG7−/− cells treated with BafA1 (P-value < 0.05; log2 WT/ATG7−/− > 0.585; n = 3) were identified using the protein–protein interaction hub protein tool in the Enrichr gene set enrichment analysis web server (Chen et al., 2013; Kuleshov et al., 2016) and plotted according to their −log10 adjusted P-values. GO analysis (database released 2019-12-09) of the secretome was performed with PANTHER Overrepresentation Test (released 2019-07-11; Mi et al., 2019) and the top ten terms for biological process and cellular component were plotted according to their −log10 FDR. The overlaps between the defined secretome and the LDELS secretome (Leidal et al., 2020), ATG7- and ATG12-dependent EV secretome (Leidal et al., 2020), ATG5-dependent macrophage secretome (Kimura et al., 2017), autophagy interaction network (Behrends et al., 2010), and the autophagosome proteome from chloroquine treated cells (Mancias et al., 2014) were performed using Biovenn (Hulsen et al., 2008).
In vivo extracellular vesicle isolation from mice
All experimental procedures and treatments were conducted in compliance with University of California, San Francisco Institutional Animal Care and Use Committee guidelines under an approved animal protocol (#AN170608). C57BL/6JJc1 homozygous transgenic mice expressing GFP-LC3B (Mizushima et al., 2004; Kuma and Mizushima, 2008; Riken Bioresource Center No. 00806) received either 60 mg/kg CQ or vehicle via intraperitoneal injection for three consecutive days. After 6 h of final CQ injection, whole blood was collected in heparin-coated capillary tubes (CB 300 Lithium Heparin; Sarstedt) and centrifuged at 2,000 g for 5 min at RT. The plasma phase was collected and 1 ml was pooled (from three to four mice per condition), diluted in 10 ml PBS, clarified by 0.22 μm filter, and ultracentrifuged at 110,000 g for 3 h at 4°C. The pellet was resuspended in 10 ml PBS, ultracentrifuged at 110,000 g for 70 min at 4°C, and resuspended in urea buffer for immunoblotting analysis. For comparison of EVPs between experimental groups in vivo, samples were normalized by collection from equivalent volumes of plasma from experimental groups.
Transmission EM
For immunogold labeling of intracellular p62/SQSTM1, HEK293T cells treated with vehicle or BafA1 were fixed in 4% PFA for 20 min and permeabilized and blocked in blocking buffer (0.1% saponin, 4% normal goat serum, PBS buffer) for 1 h. Subsequently, cells were incubated with antibody against p62/SQSTM1 (1:100; #GP GP62-C; Progen) in blocking buffer with 1% normal goat serum overnight. The next day, cells were incubated with 0.2% BSAc (Electron Microscopy Sciences) and 0.2% fish gelatin (Electron Microscopy Sciences) in PBS for 10 min and then goat-anti-guinea pig ultrasmall gold nanoparticle-conjugated IgG (1:100; #25321; Electron Microscopy Sciences) for 2 h. After washing with PBS, the cells were fixed with 1% glutaraldehyde for 10 min. The signal for immunogold particles was enhanced by a silver enhancement kit (#25521; Electron Microscopy Sciences) at 25°C for 1 h. Cells were postfixed with 3% glutaraldehyde and 1% PFA in 0.1 M cacodylate buffer for 1 h and then 2% osmium tetroxide for 30 min. Finally, the cells were stained in 5% uranyl acetate at 40°C for 1 h, dehydrated, and embedded in resin. A Leica UC6 Ultramicrotome (Leica) was used to cut thin sections (100 nm) from the surface of the block until reaching a depth of 4–5 μm to ensure full cell coverage. Formvar-coated 50-mesh copper grids were used to collect serial sections.
For immunogold labeling for secreted p62/SQSTM1, EVPs from BafA1 or vehicle-treated cells were fixed to nickel formvar carbon-coated grids (#FCF200-NI; Electron Microscopy Sciences) with PBS containing 2% PFA and 0.1% glutaraldehyde. Grids were then blocked in 2% normal goat serum for 30 min and incubated with antibody against p62/SQSTM1 (1:100; #GP GP62-C; Progen) in blocking buffer overnight. Unbound antibody was washed out and grids were incubated with goat-anti-guinea pig ultrasmall gold nanoparticle-conjugated IgG (1:100; #25321; Electron Microscopy Sciences) for 1 h. After washing with PBS, the EVPs and bound antibody were postfixed with 1% glutaraldehyde for 10 min. The signal for immunogold particles was enhanced by a silver enhancement kit (#25521; Electron Microscopy Sciences) at 4°C for 1 h and subjected to negative staining with 5% aqueous uranyl acetate for 5 min. Grids were then left to dry overnight.
Imaging was performed using a Tecnai 12 120-kV transmission electron microscope (FEI), and data were recorded using UltraScan 1000 with Digital Micrograph 3 software (Gatan Inc.).
Statistical analyses
Statistical analyses were performed using Prism GraphPad 5 software. Groups were compared using unpaired two-tail Student’s t test or nonparametric ANOVA followed by Dunnett’s or Tukey’s post-hoc test for multiple comparisons where indicated. The sample size was chosen on the basis of the size of the effect and variance for the different experimental approaches. Details regarding the statistical analysis of the proteomic data and the bioinformatics analysis of the BafA1 induced ATG7-dependent EVP proteome are provided in the corresponding figure legends and the relevant methods sections above. P values < 0.05 are considered to be significant.
Data availability
The MS proteomics data associated with this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the identifier PXD031527. Furthermore, the data and/or reagents that support the findings of this study are available from the corresponding authors, A.M. Leidal and J. Debnath, upon reasonable request. Source data for Figs. 1, 3, 4, 5, 6, 7, S3, S4, and S5 are provided online.
Online supplemental material
Fig. S1 shows that BafA1 inhibits autophagosome maturation and promotes enhanced co-localization of LC3 with late endosomes. Fig. S2 shows that BafA1 treatment negligibly affects cell viability. Fig. S3 shows BafA1 promotes autophagy cargo receptor secretion in diverse cell types. Fig. S4 shows autophagy cargo receptor secretion requires ATGs necessary for autophagosome formation. Fig. S5 shows p62 accumulates within late endosomes of Rab27a deficient cells. Table S1 contains TMT MS data and a list of proteins secreted via ATG-dependent mechanisms upon lysosome inhibition.
Supplementary Material
contains TMT MS data and a list of proteins secreted via ATG-dependent mechanisms upon lysosome inhibition.
contains original blots for Fig. 1.
contains original blots for Fig. 3.
contains original blots for Fig. 4.
contains original blots for Fig. 5.
contains original blots for Fig. 6.
contains original blots for Fig. 7.
contains original blots for Fig. S3.
contains original blots for Fig. S4.
contains original blots for Fig. S5.
Acknowledgments
We thank Ivy Hsieh and Yu-Hsin Huang for assistance with immuno-EM sample preparation and data collection.
Grant support includes the National Institutes of Health (CA201849, CA126792, CA213775 to J. Debnath, AG057462 to J. Debnath and E.J. Huang, CA226851 to A.P. Wiita, and F31CA217015 to T. Marsh), Samuel Waxman Cancer Research Foundation (to J. Debnath), Mark Foundation for Cancer Research (Endeavor Award to J. Debnath), University of California, San Francisco QB3 Calico Longevity Fellowship (to J. Debnath and A.M. Leidal), and Dale Frey Breakthrough Award from the Damon Runyon Cancer Research Foundation (DFS 14–15 to A.P. Wiita). Fellowship support includes a Banting Postdoctoral Fellowship (201409BPF-335868) and Cancer Research Society Award (22805) to A.M. Leidal, and a National Science Foundation Graduate Student Fellowship (1650113) to T.A. Solvik.
J. Debnath is a member of the Scientific Advisory Board of Vescor Therapeutics, LLC. No other disclosures were reported.
Author contributions: J. Debnath, A.M. Leidal, E.J. Huang, and T.A. Solvik conceived the study and designed the experiments. T.A. Solvik, T.A. Nguyen, and Y.-H.T. Lin collected samples, performed quantitative MS, and analyzed the resulting LC-MS/MS data with input from A.M. Leidal, A.P. Wiita, E.J. Huang, and J. Debnath. T.A. Solvik, T.A. Nguyen, and A.M. Leidal performed biochemical and cell biological experiments. T.A. Solvik, A.M. Leidal, and T. Marsh performed mouse experiments. A.M. Leidal, T.A. Solvik, and J. Debnath wrote the paper, with input from all other authors. J. Debnath, E.J. Huang, and A.M. Leidal supervised the study.
References
- Amaravadi, R.K., Kimmelman A.C., and Debnath J.. 2019. Targeting autophagy in cancer: recent advances and future directions. Cancer Discov. 9:1167–1181. 10.1158/2159-8290.CD2159-19-0292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends, C., Sowa M.E., Gygi S.P., and Harper J.W.. 2010. Network organization of the human autophagy system. Nature. 466:68–76. 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bel, S., Pendse M., Wang Y., Li Y., Ruhn K.A., Hassell B., Leal T., Winter S.E., Xavier R.J., and Hooper L.V.. 2017. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science. 357:1047–1052. 10.1126/science.aal4677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy, G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., and Johansen T.. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614. 10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashikar, A.G., and Hanson P.I.. 2019. A cell-based assay for CD63-containing extracellular vesicles. PLoS One. 14:e0220007. 10.1371/journal.pone.0220007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., and Ma’ayan A.. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 14:128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSelm, C.J., Miller B.C., Zou W., Beatty W.L., van Meel E., Takahata Y., Klumperman J., Tooze S.A., Teitelbaum S.L., and Virgin H.W.. 2011. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev. Cell. 21:966–974. 10.1016/j.devcel.2011.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnos, C., Schonn J.S., Huet S., Tran V.S., El-Amraoui A., Raposo G., Fanget I., Chapuis C., Menasche G., de Saint Basile G., et al. 2003. Rab27A and its effector MyRIP link secretory granules to F-actin and control their motion towards release sites. J. Cell Biol. 163:559–570. 10.1083/jcb.200302157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont, N., Jiang S., Pilli M., Ornatowski W., Bhattacharya D., and Deretic V.. 2011. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 30:4701–4711. 10.1038/emboj.2011.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejlerskov, P., Rasmussen I., Nielsen T.T., Bergstrom A.-L., Tohyama Y., Jensen P.H., and Vilhardt F.. 2013. Tubulin polymerization-promoting protein (TPPP/p25α) promotes unconventional secretion of α-synuclein through exophagy by impairing autophagosome-lysosome fusion. J. Biol. Chem. 288:17313–17335. 10.1074/jbc.m112.401174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajgenbaum, D.C., and June C.H.. 2020. Cytokine storm. N. Engl. J. Med. 383:2255–2273. 10.1056/NEJMra2026131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Chitiprolu M., Roncevic L., Javalet C., Hemming F.J., Trung M.T., Meng L., Latreille E., Tanese de Souza C., McCulloch D., et al. 2017. Atg5 disassociates the V1V0-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Dev. Cell. 43:716–730.e7. 10.1016/j.devcel.2017.11.018 [DOI] [PubMed] [Google Scholar]
- Hessvik, N.P., Øverbye A., Brech A., Torgersen M.L., Jakobsen I.S., Sandvig K., and Llorente A.. 2016. PIKfyve inhibition increases exosome release and induces secretory autophagy. Cell Mol. Life Sci. 73:4717–4737. 10.1007/s00018-016s0002309-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen, T., de Vlieg J., and Alkema W.. 2008. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 9:488. 10.1186/1471-2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura, E., Kishi-Itakura C., and Mizushima N.. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269. 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Johansen, T., and Lamark T.. 2020. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. 432:80–103. 10.1016/j.jmb.2019.07.016 [DOI] [PubMed] [Google Scholar]
- Kimura, T., Jia J., Kumar S., Choi S.W., Gu Y., Mudd M., Dupont N., Jiang S., Peters R., Farzam F., et al. 2017. Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J. 36:42–60. 10.15252/embj.201695081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov, M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44:W90-7. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma, A., and Mizushima N.. 2008. Chromosomal mapping of the GFP-LC3 transgene in GFP-LC3 mice. Autophagy. 4:61–62. 10.4161/auto.4846 [DOI] [PubMed] [Google Scholar]
- Leidal, A.M., and Debnath J.. 2021. Emerging roles for the autophagy machinery in extracellular vesicle biogenesis and secretion. FASEB Bioadv. 3:377–386. 10.1096/fba.2020-00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidal, A.M., Huang H.H., Marsh T., Solvik T., Zhang D., Ye J., Kai F., Goldsmith J., Liu J.Y., Huang Y.H., et al. 2020. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 22:187–199. 10.1038/s41556-019-0450-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidal, A.M., Levine B., and Debnath J.. 2018. Autophagy and the cell biology of age-related disease. Nat. Cell Biol. 20:1338–1348. 10.1038/s41556-018-0235-8 [DOI] [PubMed] [Google Scholar]
- Lock, R., Kenific C.M., Leidal A.M., Salas E., and Debnath J.. 2014. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov. 4:466–479. 10.1158/2159-8290.CD2159-13-0841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias, J.D., and Kimmelman A.C.. 2016. Mechanisms of selective autophagy in normal physiology and cancer. J. Mol. Biol. 428:1659–1680. 10.1016/j.jmb.2016.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias, J.D., Wang X., Gygi S.P., Harper J.W., and Kimmelman A.C.. 2014. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 509:105–109. 10.1038/nature13148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, T., Kenific C.M., Suresh D., Gonzalez H., Shamir E.R., Mei W., Tankka A., Leidal A.M., Kalavacherla S., Woo K., et al. 2020. Autophagic degradation of NBR1 restricts metastatic outgrowth during mammary tumor progression. Dev. Cell. 52:591–604.e6. 10.1016/j.devcel.2020.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauthe, M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K.J., Coppes R.P., Engedal N., Mari M., and Reggiori F.. 2018. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 14:1435–1455. 10.1080/15548627.2018.1474314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H., Muruganujan A., Ebert D., Huang X, and Thomas P.D.. 2019. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47:D419–D426. 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, G., Zhao H., Li Y., Ji M., Chen Y., Shi Y., Bi Y., Wang P., and Zhang H.. 2021. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev. Cell. 56:427–442.e5. 10.1016/j.devcel.2020.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, Y., Li G., Zhang X., Xu H., and Abraham S.N.. 2015. A TRP channel senses lysosome neutralization by pathogens to trigger their expulsion. Cell. 161:1306–1319. 10.1016/j.cell.2015.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakaki, G., Menges S., Kittel A., Emmanouilidou E., Schaeffner I., Barkovits K., Bergmann A., Rockenstein E., Adame A., Marxreiter F., et al. 2018. Autophagy inhibition promotes SNCA/alpha-synuclein release and transfer via extracellular vesicles with a hybrid autophagosome-exosome-like phenotype. Autophagy. 14:98–119. 10.1080/15548627.2017.1395992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda, A.M., Lasiecka Z.M., Xu Y., Neufeld J., Shahriar S., Simoes S., Chan R.B., Oliveira T.G., Small S.A., and Di Paolo G.. 2018. Neuronal lysosomal dysfunction releases exosomes harboring APP C-terminal fragments and unique lipid signatures. Nat. Commun. 9:291. 10.1038/s41467-017-02533-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima, N., and Murphy L.O.. 2020. Autophagy assays for biological discovery and therapeutic development. Trends Biochem. Sci. 45:1080–1093. 10.1016/j.tibs.2020.07.006 [DOI] [PubMed] [Google Scholar]
- Mizushima, N., Yamamoto A., Matsui M., Yoshimori T., and Ohsumi Y.. 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 15:1101–1111. 10.1091/mbc.e03-09-0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrow, L., Malhotra R., and Debnath J.. 2015. ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat. Cell Biol. 17:300–310. 10.1038/ncb3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres, J.L., Leidal A.M., Debnath J., and Hansen M.. 2021. Beyond autophagy: the expanding roles of ATG8 proteins. Trends Biochem. Sci. 46:673–686. 10.1016/j.tibs.2021.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, P., Loganathan K., Sekiguchi M., Matsuba Y., Hui K., Tsubuki S., Tanaka M., Iwata N., Saito T., and Saido T.C.. 2013. Aβ secretion and plaque formation depend on autophagy. Cell Rep. 5:61–69. 10.1016/j.celrep.2013.08.042 [DOI] [PubMed] [Google Scholar]
- Ortega, F.G., Roefs M.T., de Miguel Perez D., Kooijmans S.A., de Jong O.G., Sluijter J.P., Schiffelers R.M., and Vader P.. 2019. Interfering with endolysosomal trafficking enhances release of bioactive exosomes. Nanomedicine. 20:102014. 10.1016/j.nano.2019.102014 [DOI] [PubMed] [Google Scholar]
- Ostrowski, M., Carmo N.B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C.F., Schauer K., Hume A.N., Freitas R.P., et al. 2010. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12:19–30; sup pp 1-13. 10.1038/ncb2000 [DOI] [PubMed] [Google Scholar]
- Pankiv, S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Overvatn A., Bjorkoy G., and Johansen T.. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145. 10.1074/jbc.m702824200 [DOI] [PubMed] [Google Scholar]
- Stinchcombe, J.C., Barral D.C., Mules E.H., Booth S., Hume A.N., Machesky L.M., Seabra M.C., and Griffiths G.M.. 2001. Rab27a is required for regulated secretion in cytotoxic T lymphocytes. J. Cell Biol. 152:825–834. 10.1083/jcb.152.4.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo, Q.W., van Leur S.W., and Sanyal S.. 2021. Escaping the Lion’s Den: redirecting autophagy for unconventional release and spread of viruses. FEBS J. 288:3913–3927. 10.1111/febs.15590 [DOI] [PubMed] [Google Scholar]
- Théry, C., Amigorena S., Raposo G., and Clayton A.. 2006. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3:Unit 3.22. 10.1002/0471143030.cb0322s30 [DOI] [PubMed] [Google Scholar]
- Xu, J., Camfield R., and Gorski S.M.. 2018. The interplay between exosomes and autophagy - partners in crime. J. Cell Sci. 131:jcs215210. 10.1242/jcs.215210 [DOI] [PubMed] [Google Scholar]
- Zhang, H., Freitas D., Kim H.S., Fabijanic K., Li Z., Chen H., Mark M.T., Molina H., Martin A.B., Bojmar L., et al. 2018. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 20:332–343. 10.1038/s41556-018-0040-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Sun H., Pei R., Mao B., Zhao Z., Li H., Lin Y., and Lu K.. 2021. The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes. Cell Discov. 7:31. 10.1038/s41421-021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B., Liu J., Zeng L., Zhu S., Wang H., Billiar T.R., Kroemer G., Klionsky D.J., Zeh H.J., Jiang J., et al. 2020. Extracellular SQSTM1 mediates bacterial septic death in mice through insulin receptor signalling. Nat. Microbiol. 5:1576–1587. 10.1038/s41564-020s4100795-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains TMT MS data and a list of proteins secreted via ATG-dependent mechanisms upon lysosome inhibition.
contains original blots for Fig. 1.
contains original blots for Fig. 3.
contains original blots for Fig. 4.
contains original blots for Fig. 5.
contains original blots for Fig. 6.
contains original blots for Fig. 7.
contains original blots for Fig. S3.
contains original blots for Fig. S4.
contains original blots for Fig. S5.
Data Availability Statement
The MS proteomics data associated with this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the identifier PXD031527. Furthermore, the data and/or reagents that support the findings of this study are available from the corresponding authors, A.M. Leidal and J. Debnath, upon reasonable request. Source data for Figs. 1, 3, 4, 5, 6, 7, S3, S4, and S5 are provided online.
Online supplemental material
Fig. S1 shows that BafA1 inhibits autophagosome maturation and promotes enhanced co-localization of LC3 with late endosomes. Fig. S2 shows that BafA1 treatment negligibly affects cell viability. Fig. S3 shows BafA1 promotes autophagy cargo receptor secretion in diverse cell types. Fig. S4 shows autophagy cargo receptor secretion requires ATGs necessary for autophagosome formation. Fig. S5 shows p62 accumulates within late endosomes of Rab27a deficient cells. Table S1 contains TMT MS data and a list of proteins secreted via ATG-dependent mechanisms upon lysosome inhibition.