Abstract
Between 1994 and 1998, 97 imipenem-resistant Acinetobacter isolates were identified at the Prince of Wales Hospital, Hong Kong, China. A blaIMP PCR product was obtained from 23 of 35 viable cultures; 12 isolates belonged to genomic DNA group 3, 8 belonged to group 2 (Acinetobacter baumannii), 2 belonged to group 13TU, and 1 belonged to group 1. The blaIMP homologues were sequenced from two isolates from genomic DNA group 2 and one isolate each from groups 3 and 13TU. The four sequences included an identical 738-bp open reading frame, predicted to encode a polypeptide of 246 amino acids, with 95.6% homology to IMP-1 and 89.3% homology to IMP-2. The new enzyme, designated IMP-4, was partially purified. It had a pI of 8.0 and was strongly active against imipenem and meropenem, with Vmax values 53 and 8% of that for penicillin G, respectively. Strong activity was also seen against oxyimino-aminothiazolyl cephalosporins but not against aztreonam. Hydrolytic activity was inhibited by EDTA but not by clavulanate or tazobactam. Carbapenem MICs for most blaIMP-positive isolates were 4 to 32 μg/ml, but one isolate with the intact gene was susceptible, with imipenem and meropenem MICs of 0.25 and 0.5 μg/ml, respectively. The latter isolate did not produce the band with a pI of 8.0, and gene expression was inferred to have been lost. None of the isolates studied in detail contained extrachromosomal DNA, and carbapenem resistance was not transmissible to Escherichia coli. Nevertheless, the presence of blaIMP-4 in different genomic DNA groups implies horizontal transfer, and sequences resembling a GTTRRRY integrase-dependent recombination motif were identified in the flanking regions of blaIMP-4.
Acinetobacter spp. are important opportunistic pathogens, with Acinetobacter baumannii being the predominant species in clinical settings. Infection can involve virtually any body site in compromised patients, but acinetobacters are particularly associated with invasion of burn wounds and with nosocomial pneumonias in ventilated patients (5). The therapy of Acinetobacter infections is complicated by multidrug resistance: aminoglycosides, extended-spectrum cephalosporins, and fluoroquinolones were active against many Acinetobacter isolates during the early 1980s, but many clinical isolates are now resistant. Carbapenems have retained better activity than other antimicrobial agents, and resistance to carbapenems is still rare (15). Nevertheless, reports of carbapenem resistance among Acinetobacter spp. are accumulating steadily, with three types of mechanisms being encountered. Most carbapenem-resistant acinetobacters have OXA-type β-lactamases with weak activity against carbapenems; such enzymes have been found in A. baumannii isolates from Argentina, Belgium, France, Kuwait, Scotland, Spain, and Singapore (1, 2, 3, 6, 11, 13, 24). Several of these enzymes have been sequenced and are found to form a subgroup among class D β-lactamases, presently comprising the OXA-23, -24, -25, -26, and -27 types (3, 6, 11). A smaller (or less reported) group of acinetobacters owe their carbapenem resistance to β-lactamase-independent mechanisms (8, 12, 31). Finally, resistance mediated by metallo-β-lactamases has been reported in acinetobacters from Cuba, Italy, and Japan (9, 23, 30). The enzyme from the Italian isolates, designated IMP-2, has 84.9% amino acid homology with IMP-1 (26), a metallo-β-lactamase that is scattered in Pseudomonas aeruginosa, Serratia marcescens, and acinetobacters in Japan and that has been recorded from single isolates of Klebsiella pneumoniae in Japan and Singapore (17, 21, 28, 29). We report here a further IMP-type β-lactamase that confers carbapenem resistance in acinetobacters collected at the Prince of Wales Hospital, Hong Kong, China, between 1994 and 1998.
MATERIALS AND METHODS
Strain selection and identification.
Acinetobacters were obtained from clinical specimens at the Prince of Wales Hospital, Shatin, Hong Kong, and all those (n = 97) reported by the clinical diagnostic laboratory to be resistant to imipenem between September 1994 and October 1998 were selected. This categorization was based on the British Society for Antimicrobial Chemotherapy disk method (33) from 1994 to 1997 and on the NCCLS disk method (20) from January 1998 onward. Viable cultures were revived from nutrient agar slants, which had been stored at room temperature for up to 5 years. Transformation (16) was used to confirm the genus identification, and genomic DNA groups were determined by amplified 16S rRNA gene restriction analysis (ARDRA) (10).
Antibiotics and susceptibility tests.
MICs were determined by the British Society for Antimicrobial Chemotherapy's agar dilution method (33). The sources of the antibiotics were as follows: ampicillin, benzylpenicillin, cephaloridine, cephalothin, nalidixic acid, oxacillin, and rifampin, Sigma Chemical Co. (St. Louis, Mo.); aztreonam, Bristol-Myers Squibb (Syracuse, N.Y.); cefotaxime, Aventis (Wembley, United Kingdom); ceftazidime and cefuroxime, GlaxoWellcome (Stevenage, United Kingdom); clavulanic acid, SmithKline Beecham (Brentford, United Kingdom); imipenem, Merck Sharp & Dohme (Hoddesdon, United Kingdom); meropenem, AstraZeneca (Macclesfield, United Kingdom); nitrocefin, BBL (Cockeysville, Md.); piperacillin and tazobactam, Wyeth (Taplow, United Kingdom); and sulbactam, Pfizer (Sandwich, United Kingdom).
Detection of blaIMP-related sequences.
PCR was used to detect blaIMP-related sequences. Each reaction mixture (50 μl) contained 2 μl of genomic DNA, prepared by boiling a single colony in 200 μl of sterile distilled water; 25 pmol of each of the two primers (5′-ATG AGC AAG TTA TCT GTA TTC T [IMP11] and 5′-AGT GTG TCC CGG GCC ACC [IMP12]); 0.5 U of Taq DNA polymerase (Amersham-Pharmacia Biotech, Uppsala, Sweden); and the reaction buffer, supplied by the manufacturer. The mixtures were heated to 94°C for 2 min and were then subjected to 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s, followed by a final extension for 3 min at 72°C.
Cloning and sequencing of the blaIMP open reading frame.
Primers 5′-ATC CAA GCA GCA AGC GCG TTA (IMP13) and 5′-AGG CGT GCT GCT GCA ACG ACT TGT (IMP14) were used to amplify an 879-bp fragment containing the entire blaIMP gene. The PCR conditions were those used for detection of blaIMP (see above). The fragment was then subcloned into the expression vector pT-Adv with an AdvanTage PCR cloning kit (CLONTECH Laboratories, Palo Alto, Calif.). Recombinant plasmids were purified from selected clones with alkaline lysis minipreps (27). The DNA sequences and orientations of the inserts in these plasmids were determined on an ALFexpress DNA sequencer (Amersham Pharmacia Biotech) by using a cycle sequencing kit (SequiTherm Excel II; Epicentre Technologies, Madison, Wis.). The Cy-5-labeled M13 universal and reverse sequencing primers were used.
Plasmid detection, transfer, and curing.
Plasmids were extracted for electrophoresis by alkaline lysis or by boiling of the minipreps (27). Conjugative plasmid transfer was attempted by plate mating with Escherichia coli K-12 J53-1 pro Nalr or J53-2 pro Rifr as the recipients, with counterselection on Diagnostic Sensitivity Test Agar (Oxoid, Basingstoke, United Kingdom) containing imipenem (1 μg/ml) plus nalidixic acid or rifampin (250 μg/ml), as appropriate. Curing was attempted by growing cultures in the presence of ethidium bromide at 0.25 or 0.5 time the MIC, recovering the cells on nutrient agar plates, and replica plating onto Iso-Sensitest agar (Oxoid) with and without imipenem (2 μg/ml).
Isoelectric focusing.
Crude cell extracts were prepared by sonicating the overnight growth from nutrient agar in 0.1 M phosphate buffer (pH 7.0) and were clarified by centrifugation at 12,000 × g. Electrofocusing was performed at 15 W of constant current on gels with a pH range of 3.5 to 10, prepared by the method of Livermore and Williams (19), or on Phastgels at pH 3.5 to 9.0 run by using on automated electrophoresis system (Phastsystem; Amersham Pharmacia Biotech, Milton Keynes, United Kingdom). β-Lactamase bands were located with 0.5 mM nitrocefin. In some experiments duplicate gels were run, one of which was overlaid with 3 mM EDTA for 10 min before being developed with nitrocefin.
Purification and characterization of IMP-4 β-lactamase.
Isolate 74510 was used as a source of β-lactamase for kinetic studies. Logarithmic-phase cells were harvested from 10 liters of nutrient broth culture, washed, and resuspended in 10 mM sodium phosphate buffer (pH 6.8) and were then disrupted by three passes through a French pressure cell (SLM Aminco, Urbana, Ill.) at 12,000 lb/in2. After ultracentrifugation at 100,000 × g to remove the debris, the supernatant was loaded onto a column (40 by 2.6 cm) of carboxymethyl Sephadex C-50 equilibrated with 10 mM sodium phosphate buffer (pH 6.8). This was washed with the equilibration buffer and was then eluted with a linear gradient of 0 to 0.5 M NaCl, also prepared in 10 mM sodium phosphate buffer (pH 6.8). Eluent fractions were screened for activity against 0.1 mM imipenem by spectrophotometry at 297 nm and were subjected to isoelectric focusing. Imipenem-hydrolyzing fractions containing a single β-lactamase band were retained at −20°C. Enzyme activity was assayed by spectrophotometry at 37°C in 0.1 M sodium phosphate buffer (pH 6.8) containing 0.1 mM ZnCl2. The assay wavelengths were those listed by Livermore and Williams (19), and the kinetic parameters were calculated from Hanes plots (s/v versus s) of initial velocity data (v) obtained with 10 or more different substrate (s) concentrations. Inhibition assays with clavulanate, tazobactam, and disodium EDTA were performed under conditions in which either (i) the enzyme was added to a mixture of inhibitor and 1 mM benzylpenicillin or (ii) the inhibitor and enzyme were incubated for 10 min before addition of benzylpenicillin to a concentration of 1 mM.
Nucleotide sequence accession number.
The nucleotide sequence containing the blaIMP-4 open reading frame has been assigned the EMBO/GenBank accession number AF244145.
RESULTS
Confirmation of resistance and detection of blaIMP.
Between September 1994 and October 1998, the clinical diagnostic laboratory reported 97 Acinetobacter isolates resistant to imipenem on the basis of the results of disk diffusion tests. Of these isolates, 35 remained viable and 23 gave 474-bp PCR products with the primers IMP11 and IMP12. By ARDRA, 12 of the 23 blaIMP-positive isolates were found to belong to genomic DNA group 3, eight were found to belong to group 2 (A. baumannii), two were found to belong to group 13TU, and one was found to belong to group 1 (Acinetobacter calcoaceticus). The MICs for the blaIMP-positive isolates are summarized in Table 1: one isolate (isolate 116665, Table 1) was fully susceptible to both imipenem and meropenem; the others were resistant or had reduced susceptibility compared with the susceptibilities of typical acinetobacters. Of the 12 PCR-negative isolates, 5 isolates were nevertheless confirmed to be imipenem resistant by NCCLS disk diffusion tests (20), whereas resistance was not confirmed for the remaining seven isolates.
TABLE 1.
MICs for blaIMP-4-positive isolates of Acinetobacter spp.
| Isolate type and lab no. | Sample | DNA group | Date isolated (mo/yr) | β-Lactamase pI(s) | MIC (μg/ml)a
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IMP | MEM | ATM | PIP | TZP | CTX | SUL | |||||
| Isolates from which blaIMP-4 was sequenced | |||||||||||
| 116665 | Blood | 2 | 10/94 | 5.4 | 0.5 | 0.25 | 16 | >128 | 32 | >128 | 16 |
| 127091 | Blood | 2 | 9/96 | 7.6, 8.0b | 16 | 16 | 64 | >128 | 64 | >128 | 4 |
| 74510 | Wound | 13TU | 5/95 | 5.7, 7.6, 8.0b | >32 | >32 | 32 | >128 | 32 | >128 | 16 |
| 104680 | Blood | 3 | 8/98 | 5.7, 7.6, 8.0b | 8 | 16 | 32 | >128 | 64 | >128 | 1 |
| Collection of 23 blaIMP-4 positive isolates | |||||||||||
| MIC range | 0.25–16 | 0.25–16 | 2–64 | 64–256 | 4–64 | 32–256 | 1–64 | ||||
| MIC50c | 4 | 8 | 32 | 256 | 32 | 128 | 2 | ||||
| MIC90d | 16 | 16 | 64 | 256 | 64 | 256 | 16 | ||||
ATM, aztreonam; CTX, cefotaxime; IMP, imipenem; MEM, meropenem; PIP, piperacillin; SUL, sulbactam; TZP, piperacillin-tazobactam (tazobactam at 4 μg/ml).
Enzymes with pIs of 8.0 ceased to be detectable if the gel was overlaid with 3 mM EDTA for 10 min before addition of nitrocefin.
MIC50, MIC at which 50% of isolates are inhibited.
MIC90, MIC at which 90% of isolates are inhibited.
Sequencing of blaIMP homologues.
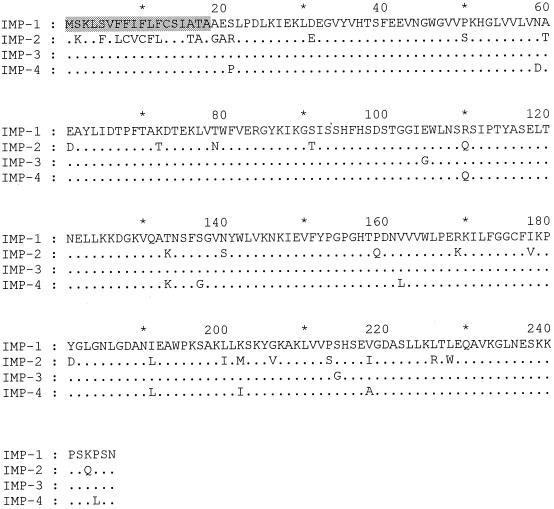
An 879-bp fragment was cloned from four representative PCR-positive isolates (Table 1). These were selected (i) as two representatives from genomic DNA groups 2 and one each from DNA groups 3 and 13TU and (ii) as being from 4 of the 5 years in which PCR-positive isolates were collected. Three of the organisms were resistant to imipenem and meropenem, whereas one (isolate 116665) was the highly susceptible organism mentioned earlier. The cloned fragments from each of the four isolates were identical and contained the entire blaIMP-related open reading frame and its flanking DNA. This open reading frame comprised 738 bases with 95.5% nucleotide identity to the blaIMP-1 nucleotide sequence, as represented by the published sequences for P. aeruginosa (18) and S. marcescens (21). A total of 31 base differences from blaIMP-1 were identified. These translated into 10 substitutions in the deduced amino acid sequence, designated IMP-4 (Fig. 1). The amino acid replacements reflected 11 of the base changes, whereas 20 further base changes were silent. A total of 96 nucleotide differences (13.0% divergence) and 37 amino acid differences (15.0% divergence) were observed compared with the nucleotide sequence of blaIMP-2 and the amino acid sequence of its protein product, IMP-2 (Fig. 1). However, 10 of the differences from the IMP-2 protein lay in the putative signal peptide, which comprises the first 18 amino acids in the IMP-1 β-lactamase (21).
FIG. 1.
Comparison of the amino acid sequences of IMP-1, IMP-2, IMP-3, and IMP-4 β-lactamases. The 18 residues that comprise the IMP-1 signal peptide (21) are shaded.
A GTTRRRY motif, also present upstream of blaIMP-1 in S. marcescens isolates (4), was similarly seen upstream of the four blaIMP-4 sequences and may be involved in integrase-dependent recombinations (see Discussion).
Plasmid transfer, detection, and curing.
Both the alkaline lysis and the boiling miniprep methods failed to detect plasmids in any of the four strains used as sources of DNA for sequencing. Resistance was not conjugatively transferred to E. coli K-12 derivatives, and attempts to cure resistance with ethidium bromide were unsuccessful.
β-Lactamase characterization.
Electrofocusing was performed with extracts of the four isolates from which blaIMP-4 was sequenced. All except strain 116665 yielded bands with pIs of 8.0 that ceased to be detectable if the gels were overlaid with 3 mM EDTA for 10 min before nitrocefin was added as the reporter substrate and which therefore were deduced to be zinc dependent (Table 1). Isolates 104680 and 74510 additionally had β-lactamases with pIs of ca. 5.7 and 7.6; isolate 127091 had a β-lactamase with a pI of ca. 7.6, and isolate 116665 had a β-lactamase with a pI of 5.4. These enzymes with pI values of 5.4, 5.7, and 7.6 were not inhibited by EDTA.
On the basis of these inhibition experiments it was deduced that the band with a pI of 8.0 corresponded to IMP-4, and this enzyme was purified from isolate 74510. The final preparation was free of the β-lactamases with pIs of 5.7 and 7.6 produced by the isolate and contained only two major protein species when examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. One of these, which accounted for 80% of a total protein, had a molecular mass of ca. 30 kDa and was deduced to be IMP-4; the other was considerably smaller. This preparation of IMP-4 enzyme had a very broad spectrum of activity (Table 2), encompassing penicillins, cephalosporins, and carbapenems. Vmax values for imipenem and meropenem were 53 and 8% of those for benzylpenicillin, respectively, and hydrolysis of oxyimino-aminothiazolyl cephalosporins also was rapid, whereas aztreonam was stable. The 50% inhibitory concentrations of EDTA, clavulanate, and tazobactam were <0.1, 1, and 1.5 mM, respectively, when the inhibitor and enzyme were incubated together for 10 min before addition of 1 mM benzylpenicillin as the reporter substrate and <0.1, 0.85, and 2.7 mM, respectively, when no preincubation was allowed before addition of substrate.
TABLE 2.
Kinetic properties of IMP-4 β-lactamase from isolate 74510
| Substrate | Relative Vmax (%)a | Km (μM) |
|---|---|---|
| Benzylpenicillin | 100 | 365 |
| Ampicillin | 52 | 575 |
| Piperacillin | 8 | 144 |
| Carbenicillin | 62 | 1,000 |
| Oxacillin | 307 | 420 |
| Cephaloridine | 81 | 32 |
| Cephalothin | 111 | 71 |
| Cefuroxime | 46 | 52 |
| Cefotaxime | 9 | 18 |
| Ceftazidime | 7 | 28 |
| Imipenem | 53 | 42 |
| Meropenem | 8 | 12 |
| Aztreonam | 0.001 | Not measured |
Based on a predicted molecular mass of 27,087 and a purity of 80%; based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, a kcat of 27.5 s−1 was estimated for IMP-4 against benzylpenicillin.
Behavior of cloned blaIMP-4 in E. coli.
One E. coli TOP10F′ transformant, which resulted from the TA cloning of the 879-bp blaIMP-4-coding fragment from isolate 104680, was tested for imipenem susceptibility. This organism had the insert correctly oriented with respect to the orientation of the lac promoter in the vector pT-Adv. The imipenem MIC rose from 0.38 μg/ml in the absence of isopropyl-β-d-thiogalactopyranoside (IPTG) to 1.5 μg/ml in its presence, whereas IPTG had no effect on the imipenem MIC (also 0.38 μg/ml) for another transformant, which had the insert in the reverse orientation with respect to the orientation of the lac promoter. An analogous effect was seen for meropenem in disk diffusion tests, but this was not investigated in detail.
DISCUSSION
Carbapenem resistance in Acinetobacter spp. is an emerging concern and is disturbing because many nosocomial acinetobacters are already resistant to most other antibiotics (1, 5). The present isolates were from among carbapenem-resistant Acinetobacter spp. at the Prince of Wales Hospital, Hong Kong, that had been a long-standing (5-year) problem. Of 35 recoverable isolates reported to be imipenem resistant by the diagnostic laboratory, 23 had blaIMP homologues, 5 were confirmed to be imipenem resistant but lacked blaIMP and 7 lacked or had lost their resistance. Sequencing of the blaIMP homologues from four representative PCR-positive isolates revealed that they had blaIMP-4, a new variant in the growing blaIMP family. The imipenem-resistant isolates that failed to give PCR products remain to be investigated further.
After discounting the 18 N-terminal residues, which are believed to constitute a signal peptide (21), the deduced sequence for the mature blaIMP-4 product had 95.6% amino acid homology to the amino acid sequence of the IMP-1 enzyme and 89.3% homology to that of the IMP-2 enzyme. The six residues that are believed to hold zinc ions in the active center of metallo-β-lactamases (His 95, His 97, Asp 99, His 157, Cys 176, and His 215 [22]) were conserved in the IMP-4 enzyme as well as in the IMP-1, -2, and -3 enzymes. The signal peptides of IMP-1 and -4 were also deduced to be identical, although blaIMP-4 had a silent C→T change at codon 13. IMP-2 has a substantially different signal peptide, with 10 amino acid substitutions compared with the sequences of both IMP-1 and -4 (26). Although all these data indicate that IMP-4 is closer to IMP-1 than to IMP-2, the new enzyme did share some of the amino acid changes that distinguish IMP-2 from IMP-1, specifically, Arg(110)→Gln, Thr(133)→Lys, and Ile(191)→Leu. IMP-3, which was recently described from Shigella flexneri in Japan, is a minor variant of IMP-1 with two amino acid substitutions (14).
The four isolates from which the sequence of blaIMP-4 was confirmed were collected in 1994, 1995, 1996, and 1998, indicating long-term stability. They included representatives of genomic DNA groups 2, 3, and 13TU (Table 1), indicating horizontal spread; nevertheless, none of the isolates contained detectable plasmids, and none could transfer carbapenem resistance by conjugation. It is therefore inferred that blaIMP-4 had become chromosomally integrated. Significantly, a GTTRRRY motif that is known to be involved in integrase-dependent recombination was found upstream of the blaIMP-4 reading frame, and a related sequence was identified downstream (data not shown). The same sequence was previously found in blaIMP-1-carrying elements from S. marcescens in Japan (4, 18). Transfer of blaIMP-encoding integrons among Acinetobacter strains, with subsequent chromosomal integration, is therefore a plausible explanation for the spread of these genes. blaIMP-1 likewise is often nontransmissible (28) and is inferred to become chromosomally inserted. Takahashi et al. (30) have recently reported the transfer of a blaIMP determinant, possibly on a plasmid, to an Acinetobacter isolate which had lost the gene on storage.
Multiple sets of kinetic data have been published for the IMP-1 enzyme, with relative Vmax (kcat) rates for ampicillin, cephaloridine, and imipenem variously reported as 100, 6, and 5, respectively (18); 100, 30, and 7, respectively (21); and 100, 46, and 11, respectively (32). This scatter may reflect assay conditions rather than fundamental differences among the IMP enzymes. Nevertheless, it is evident that IMP-4, like IMP-1, -2, and -3, hydrolyzes imipenem more rapidly than it hydrolyzes meropenem, has strong activity against oxyimino-aminothiazolyl cephalosporins and carboxy- and amino-penicillins, but spares monobactams (14, 18, 21, 32).
Expression of resistance correlated imperfectly with carriage of blaIMP-4. Of 23 isolates positive for blaIMP by PCR, MICs were at least 4 μg of meropenem per ml for 22 isolates and at least 4 μg of imipenem per ml for 20 isolates. One isolate, however, was inhibited by these carbapenems at 0.25 or 0.5 μg/ml and thus was no less susceptible than Acinetobacter isolates without carbapenem-hydrolyzing β-lactamases (25). This isolate (isolate 116665, Table 1) was among the four from which blaIMP-4 was sequenced, and since no activity of the β-lactamase with a pI of 8.0 was detectable on isoelectric focusing, it is deduced that the organism had little or no expression of its blaIMP-4 gene. Carbapenem susceptibility is also observed in some blaIMP-1-positive P. aeruginosa isolates from Japan (28, 29), but it is not clear whether this behavior is because resistance demands secondary changes to permeability (via the loss of OprD in the case of P. aeruginosa) or because blaIMP-1 is not always expressed. Cloned blaIMP-4 gave only a very low level of imipenem resistance in E. coli, even when it was linked to a lac promoter and induced with IPTG. Similar results were obtained with cloned bla-IMP-1 (21), and it seems that IMP enzymes confer carbapenem resistance only in members of the family Enterobacteriaceae with concomitant permeability lesions.
Although the origins of the growing family of IMP β-lactamases remain uncertain, it is evident that multiple Acinetobacter lineages with blaIMP-4 have been prevalent at the Prince of Wales Hospital for a protracted period. This, taken together with the growing worldwide catalogue of reports of carbapenem-resistant acinetobacters, presents a disturbing situation. Clinicians treating infections caused by these organisms are forced to use ampicillin-sulbactam or cefoperazone-sulbactam so as to exploit the inherent activity that sulbactam has against many Acinetobacter strains (7, 31) or to use polymyxins, which are almost universally active against Acinetobacter spp. in vitro but which have questionable clinical efficacy.
ACKNOWLEDGMENTS
We are grateful to G. Rossolini for prepublication information on the IMP-2 β-lactamase.
The work was supported by the Research Grants Council, Hong Kong (grant ID 4290/99M).
REFERENCES
- 1.Afzal-Shah M, Livermore D M. Worldwide emergence of carbapenem-resistant Acinetobacter spp. J Antimicrob Chemother. 1998;41:576–577. doi: 10.1093/jac/41.5.576. [DOI] [PubMed] [Google Scholar]
- 2.Afzal-Shah M, Villar H E, Livermore D M. Biochemical characteristics of a carbapenemase from an Acinetobacter baumannii isolate collected in Buenos Aires, Argentina. J Antimicrob Chemother. 1999;43:127–131. doi: 10.1093/jac/43.1.127. [DOI] [PubMed] [Google Scholar]
- 3.Afzal-Shah M, Woodford N, Livermore D M. Antimicrob, Agents Chemother. 583–588. 2001. Characterization of OXA-25, OXA-26 and OXA-27, molecular class D β-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arakawa Y, Murakami M, Susuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N, Ohta M. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergogne-Berezin E, Towner K J. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bou G, Oliver A, Martínez-Beltrán J. OXA-24, a novel class D β-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob Agents Chemother. 2000;44:1556–1561. doi: 10.1128/aac.44.6.1556-1561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown S, Bantar C, Young H K, Amyes S G B. Limitation of Acinetobacter treatment by plasmid mediated carbapenemase ARI-2. Lancet. 1998;351:186–187. doi: 10.1016/S0140-6736(05)78210-6. [DOI] [PubMed] [Google Scholar]
- 8.Clark R B. Imipenem resistance among Acinetobacter baumannii: association with reduced expression of a 33–36 KDa outer membrane protein. J Antimicrob Chemother. 1996;38:245–251. doi: 10.1093/jac/38.2.245. [DOI] [PubMed] [Google Scholar]
- 9.Cornaglia G, Riccio M L, Mazzariol A, Lauretti L, Fontana R, Rossolini G M. Appearance of IMP-1 metallo-β-lactamase in Europe. Lancet. 1999;353:899–900. doi: 10.1016/s0140-6736(98)05954-6. [DOI] [PubMed] [Google Scholar]
- 10.Dijkshoorn L, Van Harsselaar B, Tjernberg I, Bouvet P J, Vaneechoutte M. Evaluation of amplified ribosomal DNA restriction analysis for identification of Acinetobacter genomic species. Syst Appl Microbiol. 1998;21:33–39. doi: 10.1016/S0723-2020(98)80006-4. [DOI] [PubMed] [Google Scholar]
- 11.Donald H M, Scaife W, Amyes S G B, Young H K. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob Agents Chemother. 2000;44:196–199. doi: 10.1128/aac.44.1.196-199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gehrlein M, Leyling H, Cullmann W, Wendt S, Opferkuch W. Imipenem resistance in Acinetobacter baumannii is due to altered penicillin binding protein. Chemotherapy (Basel) 1991;37:405–412. doi: 10.1159/000238887. [DOI] [PubMed] [Google Scholar]
- 13.Hornstein M, Sautjeau-Rostoker C, Peduzzi J, Vessieres A, Hong L T H, Barthelemy M, Scavizzi M, Labia R. Oxacillin-hydrolyzing β-lactamase involved in resistance to imipenem in Acinetobacter baumannii. FEMS Microbiol Lett. 1997;153:333–339. doi: 10.1111/j.1574-6968.1997.tb12593.x. [DOI] [PubMed] [Google Scholar]
- 14.Iyobe S, Kusadokoro H, Ozaki J, Matsumara N, Minami S, Haruta S, Sawai T, O'Hara K. Amino acid substitutions in a variant of IMP-1 metallo-β-lactamase. Antimicrob Agents Chemother. 2000;44:2023–2027. doi: 10.1128/aac.44.8.2023-2027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones M E, Thornsberry C, Livermore D M, Sahm D F. Prevalence of Acinetobacter spp. isolates with reduced sensitivity to imipenem, as determined by a USA-wide electronic surveillance network. J Antimicrob Chemother. 1999;43:429–431. doi: 10.1093/jac/43.3.429. [DOI] [PubMed] [Google Scholar]
- 16.Juni E. Interspecies transformation of Acinetobacter: genetic evidence for a ubiquitous genus. J Bacteriol. 1972;112:917–931. doi: 10.1128/jb.112.2.917-931.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koh T H, Babini G S, Woodford N, Sng L H, Hall L M C, Livermore D. Carbapenem-hydrolysing IMP-1 β-lactamase in Klebsiella pneumoniae from Singapore. Lancet. 1999;353:2162. doi: 10.1016/s0140-6736(05)75604-x. [DOI] [PubMed] [Google Scholar]
- 18.Laraki N, Galleni M, Thamm I, Riccio M L, Amicosante G, Frère J M, Rossolini G M. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob Agents Chemother. 1999;43:891–901. doi: 10.1128/aac.43.4.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livermore D M, Williams J D. β-Lactams: mode of action and mechanisms of bacterial resistance. In: Lorian V, editor. Antibiotics in laboratory medicine. 4th ed. Baltimore, Md: The Williams & Wilkins Co.; 1996. pp. 502–578. [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. 6th ed. Approved standard M2–A6. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 21.Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Yoshimura F, Kato N. Molecular characterization of an enterobacterial metallo-β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother. 1994;38:71–78. doi: 10.1128/aac.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul-Soto R, Bauer R, Frère J-M, Galleni M, Meyer-Klaucke W, Nolting H, Rossolini G M, de Seny D, Hernandez-Valladares M, Zeppezauer M, Adolph H W. Mono- and binuclear Zn2+ β-lactamase. J Biol Chem. 1999;274:13242–13249. doi: 10.1074/jbc.274.19.13242. [DOI] [PubMed] [Google Scholar]
- 23.Perez A N, Bonet I G, Robledo E H, Abascal R D, Plous C V. Metallo-β-lactamases in Acinetobacter calcoaceticus? Med Sci Res. 1996;24:315–317. [Google Scholar]
- 24.Poirel L, Karin A, Mercat A, Le Thomas I, Vahaboglu H, Richard C, Nordmann P. Extended-spectrum β-lactamase-producing strain of Acinetobacter baumannii isolated from a patient in France. J Antimicrob Chemother. 1999;43:157–165. [PubMed] [Google Scholar]
- 25.Rasmussen B A, Bush K. Carbapenem-hydrolyzing β-lactamases. Antimicrob Agents Chemother. 1997;41:223–232. doi: 10.1128/aac.41.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riccio M L, Franceschini N, Boschi L, Carravelli B, Cornaglia G, Fontana R, Amicosante G, Rossolini G M. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob Agents Chemother. 2000;44:1229–1235. doi: 10.1128/aac.44.5.1229-1235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Senda K, Arakawa Y, Nakashima K, Ito H, Ichiyama S, Shimokata K, Kato N, Ohta M. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams including carbapenems. Antimicrob Agents Chemother. 1996;40:349–353. doi: 10.1128/aac.40.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, Shimokata K, Kato N, Ohta M. PCR detection of metallo-β-lactamase gene (blaIMP) in gram-negative rods resistant to broad spectrum β-lactams. J Clin Microbiol. 1996;34:2909–2913. doi: 10.1128/jcm.34.12.2909-2913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi A, Yomoda S, Kobayashi I, Okubo T, Tsunoda M, Iyobe S. Detection of carbapenemase-producing Acinetobacter baumannii in a hospital. J Clin Microbiol. 2000;38:526–529. doi: 10.1128/jcm.38.2.526-529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Urban C, Go E, Meyer K S, Mariano N, Rahal J J. Interactions of sulbactam, clavulanic acid and tazobactam with penicillin-binding proteins of imipenem-resistant and -susceptible Acinetobacter baumannii. FEMS Microbiol Lett. 1995;125:193–197. [Google Scholar]
- 32.Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:147–151. doi: 10.1128/aac.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Working Party of the British Society for Antimicrobial Chemotherapy. A guide to sensitivity testing. J Antimicrob Chemother. 1991;27(Suppl. D):1–50. [PubMed] [Google Scholar]