Summary
The protein kinase complex target of rapamycin complex 1 (TORC1) is a critical mediator of nutrient sensing that has been widely studied in cultured cells and yeast, yet our understanding of the regulatory activities of TORC1 in the context of a whole, multi-cellular organism is still very limited. Using Caenorhabditis elegans, we analyzed the DAF-15/Raptor-dependent phosphoproteome by quantitative mass spectrometry and characterized direct kinase targets by in vitro kinase assays. Here, we show new targets of TORC1 that indicate previously unknown regulation of transcription and autophagy. Our results further show that DAF-15/Raptor is differentially expressed during postembryonic development, suggesting a dynamic role for TORC1 signaling throughout the life span. This study provides a comprehensive view of the TORC1 phosphoproteome, reveals more than 100 DAF-15/Raptor-dependent phosphosites that reflect the complex function of TORC1 in a whole, multi-cellular organism, and serves as a rich resource to the field.
Subject areas: Cell biology, Functional aspects of cell biology, Developmental biology, Omics, Proteomics
Graphical abstract
Highlights
-
•
Detailed, unbiased analysis of the Caenorhabditis elegans TORC1 phosphoproteome
-
•
Characterization of DAF-15 expression and localization
-
•
TORC1 direct targets with roles in transcription machinery and autophagy
-
•
Unique technical approach and datasets provide rich resource to the field
Cell biology; Functional aspects of cell biology; Developmental biology; Omics; Proteomics
Introduction
Protein phosphorylation is an important post-translational modification that impacts protein activity, localization, and stability. Among the vast array of proteins within a cell, protein kinases specifically recognize and phosphorylate target phosphosites and thereby regulate numerous cellular processes (Ubersax and Ferrell, 2007). Identifying the substrates of a kinase is fundamental to understanding the cellular function, regulatory range, and specificity of each kinase; this important characterization should not be assumed or inferred.
The target of rapamycin complex 1 (TORC1) is a conserved protein kinase that has been extensively studied as a master regulator of cellular activities in response to changes in nutrient and metabolite signals, yet our understanding of TORC1 function in a whole, multi-cellular organism under physiological conditions is still limited and highly desired (Wang and Proud, 2011; Sabatini, 2017; González and Hall, 2017; Blackwell et al., 2019; Goberdhan et al., 2016). The TORC1 complex is comprised of two core components that are well conserved in C. elegans, the kinase LET-363/Tor and the scaffold protein DAF-15/Raptor (Long et al., 2002; Hara et al., 2002). Because LET-363/Tor can complex with DAF-15/Raptor (forming TORC1) or RICT-1/Rictor (forming TORC2), knockdown of let-363 would disable both complexes. Instead, perturbation of DAF-15 expression alone would cause TORC1-specific changes.
Genetic studies in C. elegans have indicated the roles of TORC1 in regulating postembryonic development, germline proliferation, lifespan, stress responses, and other physiological events (Blackwell et al., 2019). In a series of studies initiated from the analysis of lipid functions, we found that TORC1 acts downstream of the glucosylceramide biosynthesis pathway to critically regulate postembryonic development in response to changes in the availability of nutrients in the intestine (Zhu et al., 2013, 2015, 2021) (Kniazeva et al., 2015). This lipid-mediated TORC1 signaling pathway has recently been shown to sense the availability of all amino acids (Zhu et al., 2021). However, the factors acting downstream of TORC1, including its direct targets, are still unclear for much of the regulatory functions in C. elegans. Factors such as 4E-BP and S6K have been shown in yeast and cultured mammalian cells to be TORC1-specific phosphorylation targets that mediate the regulation of TORC1 on translation (Saxton and Sabatini, 2017). However, efforts in C. elegans have not produced direct evidence that 4E-BP and S6K are direct targets of TORC1 (Blackwell et al., 2019); TORC1 may act on different targets for these functions. Even in mammals, there are likely other targets of TORC1 that play significant roles during animal development that remain to be identified. Recognizing this significant knowledge gap, we took an unbiased, proteome-wide approach to identify the TORC1 kinase targets in C. elegans. Through mass spectrometry-based phosphoproteomics and targeted in vitro kinase assays, we have identified new direct targets of TORC1 in C. elegans.
Results
DAF-15 localization and expression
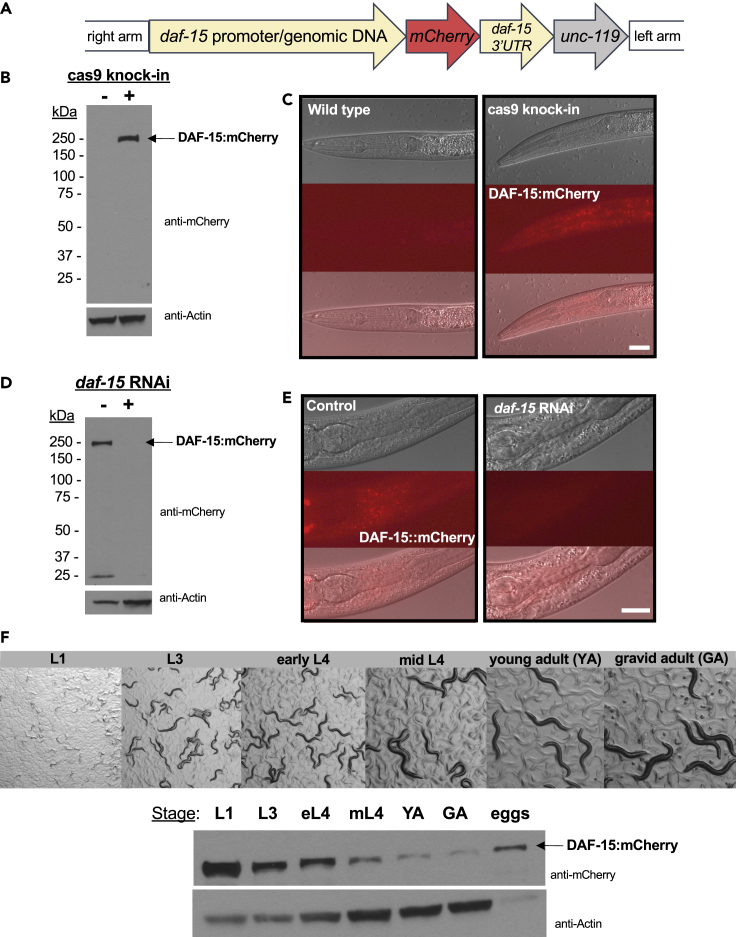
Given the specific role of DAF-15/Raptor in TORC1, but not TORC2, disrupting DAF-15 function is expected to block the activity of TORC1. In order to investigate DAF-15/TORC1 activities in C. elegans, we constructed a single copy insertion of the full length, genomic sequence of the daf-15 gene (with the endogenous promoter and 3′ UTR) fused to mCherry at the C terminus (termed DAF-15::mCherry hereafter), using the CRISPR/Cas9 genome engineering method (Dickinson et al., 2013) (Figure 1A). Western analysis of the DAF-15::mCherry strain revealed a single band of the expected size migrating at ∼250kD (Figure 1B). The weak signal of this single copy reporter was broadly detectable throughout the body by fluorescence microscopy, and most clearly distinguished in the head (Figure 1C). Our efforts to visualize DAF-15::mCherry expression in the intestine were unfortunately obscured by high background autofluorescence, similar to the observation in a recent report (Duong et al., 2020). daf-15(RNAi) treatment was sufficient to cause L3 larval arrest in these animals, and to strongly reduce the DAF-15::mCherry signal based on western analysis (Figure 1D) and fluorescence microscopy (Figure 1E). Importantly, we also found that this translational reporter was functional and sufficient to rescue the function in daf-15 loss-of-function mutant (m81) animals. Taken together, these data suggest that our CRISPR/Cas9 DAF-15::mCherry strain expresses a functional, tagged protein that may closely reflect the level and localization of endogenous DAF-15.
Figure 1.
DAF-15:mCherry expression during C. elegans development
(A) A cartoon diagram of the DAF-15::mCherry CRISPR/Cas9 construct used in this study.
(B and C) Western analysis and representative microscopy images of mixed population samples of control (N2 WT) and cas9 knock-in (DAF-15::mCherry) worms. Probing with anti-mCherry revealed a single band (B) corresponding to DAF-15::mCherry at ∼250kD, and a visible red signal in the head region (C). Scale bar is 100um.
(D and E) Western analysis and representative microscopy images of DAF-15::mCherry worms fed either HT115 or daf-15(RNAi) (arrested at L3 stage). The single band (D) detected in the control sample was strongly reduced by daf-15(RNAi); the visible red signal detected in the head region of the control sample was strongly reduced by daf-15(RNAi). Scale bar is 100um.
(F) Microscopy images and western analysis of synchronized, staged populations of DAF-15::mCherry worms revealed the DAF-15:mCherry level decreased with age.
Analysis of TORC1 binding proteins
To identify the proteins bound to DAF-15/Raptor, we used the DAF-15::mCherry strain to perform immunoprecipitations (IPs) from whole worm lysates using an anti-mCherry antibody. The precipitates were analyzed by LC-MS/MS and found to include 15 proteins that were independently identified in two or more biological replicates (Table 1). The resulting peptide counts were modest, perhaps due to the low level of endogenous expression of DAF-15::mCherry (Figure 1). The most abundant of these interactors was LET-363/Tor, indicating that our DAF-15::mCherry reporter may form a functional TORC1 complex. MLST-8, the C. elegans ortholog of mammalian Lst8 (Jones et al., 2009), was also identified. While Lst8 was previously identified in complex with mTORC1 and mTORC2 in mammalian cells (Kim et al., 2003), the C. elegans MLST-8 had only been indicated to play a role in both TORC1 and TORC2-related pathways by genetic analysis (Jones et al., 2009; Nukazuka et al., 2011; Ruf et al., 2013). Our finding of the physical association of MLST-8 with DAF-15 and LET-363 further confirms that MLST-8 acts within the TORC1 complex. We did not identify RHEB-1/Rheb in our IP, consistent with other reports that Rheb is not tightly associated with mTORC1 (Sancak et al., 2007). Several other proteins were identified in two or more replicates with lesser peptide counts, which may warrant further investigation of their relationship with TORC1.
Table 1.
List of proteins identified in the DAF-15:mCherry IP by LC/MS/MS, ranked by peptide count, and listed with closest mammalian homolog by sequence similarity
| Protein | Peptide count range | Mammalian Homolog |
|---|---|---|
| DAF-15 | 22–75 | Raptor |
| LET-363 | 4–19 | Tor |
| MLST-8 | 3–10 | mLST8, mTor-associated protein |
| USP-5 | 2–3 | Ubiquitin carboxyl-terminal hydrolase 5 (USP5) |
| PGES-2 | 2–3 | Prostaglandin E synthase 2 (PTGES2) |
| MCM-7 | 1–3 | DNA replication licensing factor (MCM7) |
| B0513.5 | 2 | Proline dehydrogenase 1 (PRODH) |
| F38A1.8 | 2 | Signal recognition particle receptor subunit alpha (SRPRA) |
| HMG-11 | 2 | Uncharacterized |
| CCT-6 | 2 | T-complex protein 1 subunit zeta-2 isoform 3 (CCT6B) |
| F37C4.5 | 2 | None |
| CCT-1 | 2 | T-complex protein 1 subunit alpha (TCP1) |
| CHIN-1 | 2 | Beta-chimaerin (CHN2) |
| RHO-1 | 2 | Transforming protein RhoA |
| DHS-6/DHS-18 | 2 | Hydroxsteroid dehydrogenase-like protein 2 (HSDL2) |
DAF-15 expression at different developmental stages
Based on studies using Raptor knockout mutations, TORC1 is clearly required for early development in various animals (Guertin et al., 2006; Hara et al., 2002; Jia et al., 2004), but the stage-specific activities of TORC1 for developmental control are not clear. In our pursuit to identify potential targets of TORC1 during postembryonic development, it was important for us to analyze DAF-15 protein expression throughout development in the context of a whole organism. Western analysis of DAF-15::mCherry in synchronized, stage-matched populations revealed that DAF-15::mCherry protein levels varied throughout development, with expression highest in eggs and young larvae and lower at advanced stages, relative to actin (Figure 1F). Independent analyses by others agree with these results (Duong et al., 2020). These results confirm DAF-15 expression at all stages, and this protein expression pattern over development may indicate stage-specific regulation of TORC1 targets.
The daf-15-dependent phosphoproteome
While many phosphoproteomic studies employ potent and specific inhibitors of the kinase of interest, the C. elegans field lacks such an inhibitor for TORC1. In yeast and cultured cells, rapamycin inhibits TORC1 and causes strong cell cycle arrest (Blackwell et al., 2019). In C. elegans, the impact of rapamycin is milder and perhaps limited (Robida-Stubbs et al., 2012; Blackwell et al., 2019; Long et al., 2002). As an alternative, we took an unbiased genetic approach to determine the daf-15-dependent phosphorylation changes as a proxy for TORC1 activity, reasoning that loss of DAF-15/Raptor would result in decreased activity of TORC1, but not that of TORC2.
To effectively disrupt TORC1 activity to reveal direct targets, we first determined a minimal RNAi treatment that was sufficient to reduce DAF-15 protein before causing strong developmental phenotypes that may indirectly cause confounding pleiotropic effects. Prolonged exposure to daf-15(RNAi) from hatching produces the well-known L3 larval arrest phenotype in the second generation, but we found that this treatment also produced defective embryos (Figure S1). By testing several treatments, we found that feeding daf-15(RNAi) from the L4 stage was sufficient to strongly reduce the DAF-15::mCherry protein level in L1 larvae of the next generation (Figure 2A). This timepoint also had tight developmental synchrony that would eliminate some variability based on developmental stage, making this condition ideal for our analysis.
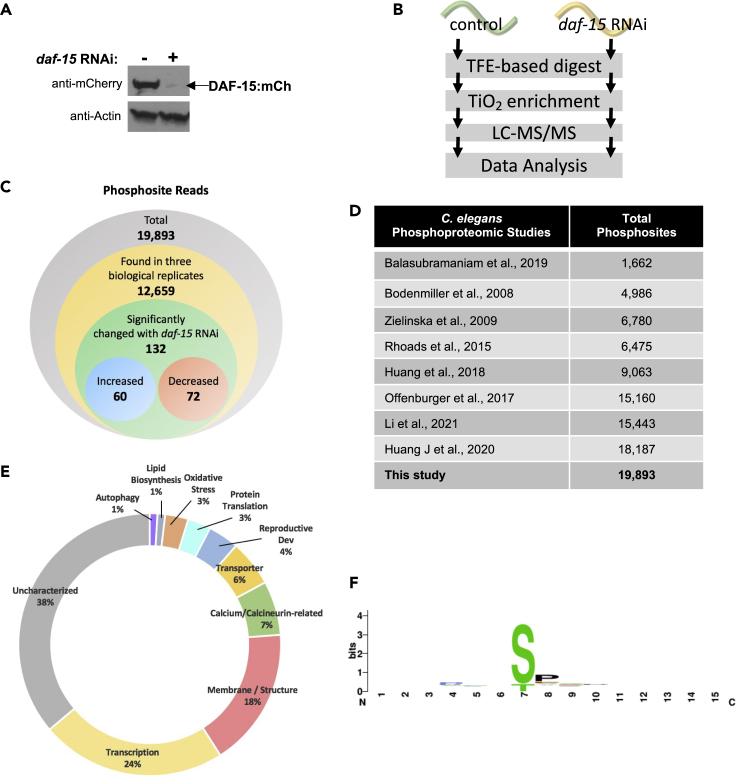
Figure 2.
The DAF-15-dependent phosphoproteome
(A) Western analysis of DAF-15::mCherry worms with or without daf-15(RNAi) treatment, from P0/L4 stage until F1/L1 stage showed a drastic reduction of the DAF-15::mCherry band.
(B) Work-flow diagram for label-free phosphoproteomic analysis.
(C) Illustration of the tiers of phosphoproteomic data described in the text.
(D) A comparison table of total phosphosites reported from published C. elegans phosphoproteomic studies.
(E) Functional categorization of the proteins containing the 72 phosphosites that were decreased with daf-15(RNAi) treatment.
(F) Calculated TORC1 kinase motif based on the 72 high-confidence phosphosites that showed decreased phosphorylation with daf-15(RNAi). See also Figure S1.
RNAi-treated (daf-15 or control) DAF-15::mCherry L1 larvae were pelleted and processed through a phosphoproteomic workflow (Figure 2B). Label-free phosphopeptides were isolated from L1 larvae by titanium (TiO2) enrichment, followed by peptide detection by single-run LC-MS/MS, based on a previously described method (Humphrey et al., 2015). By this method, 19,893 phosphosites were detected, with 12,659 phosphosites found in at least three biological replicates (for each condition) (Figure 2C and Table S1). This approach yielded many more phosphosites than previous phosphoproteomic studies in C. elegans (Figure 2D) (Balasubramaniam et al., 2019; Bodenmiller et al., 2008; Zielinska et al., 2009; Rhoads et al., 2015; Huang et al., 2018, 2020; Offenburger et al., 2017; Li et al., 2021).
The data were analyzed by the following criteria: non-zero reads in at least 3 out of 10 biological replicates, reproducible fold changes across replicates, and a significant change in phosphorylation with daf-15(RNAi) treatment as determined by either of three significance calculations. This resulted in a list of 132 peptides that were significantly changed by daf-15(RNAi) (Table S2). We identified 72 phosphosites, represented in 55 proteins, that showed significant decreases in phosphorylation with daf-15(RNAi) treatment (Figure 2C, Tables 2 and S3). A DAF-15 phosphopeptide was also found to be significantly reduced, serving as an internal control for RNAi efficiency. By the same criteria, we found 60 phosphosites that were significantly increased with daf-15(RNAi) treatment (Figure 2C and Table S4). As the substrate binding element of the TORC1 kinase, we expect that loss of daf-15/raptor reduces the phosphorylation levels of TORC1 substrates. We therefore focused subsequent analyses on the 72 phosphosites that showed significant decreases in phosphorylation in the daf-15(RNAi)-treated samples.
Table 2.
High-confidence DAF-15-dependent changes in phosphorylation identified by combining daf-15(RNAi) with quantitative phosphoproteomics
| Protein ID | Gene Name | Phosphosite | Localization Probability | Log fold change |
|---|---|---|---|---|
| Q18409 | rsp-6 | S99 | 1.00 | −4.545 |
| P53806 | rcn-1 | S202 | 1.00 | −3.221 |
| O62183 | aly-2 | S173 | 0.99 | −3.105 |
| P30640 | R08D7.1 | S193 | 1.00 | −2.792 |
| Q18003 | C15B12.2 | T18 | 1.00 | −2.538 |
| G5ED33 | eps-8 | T267 | 0.73 | −2.389 |
| Q9U2D6 | cnp-2 | S678 | 0.97 | −2.388 |
| Q6A2D2 | hpo-34 | S1228 | 1.00 | −2.367 |
| Q9BL69 | smg-6 | S69 | 1.00 | −2.180 |
| G5ED33 | eps-8 | S594 | 1.00 | −2.103 |
| G5ED33 | eps-8 | S596 | 1.00 | −2.103 |
| Q20626 | F49E2.5 | S268 | 0.88 | −1.919 |
| Q93238 | nra-3 | T373 | 0.94 | −1.862 |
| Q17490 | unc-44 | S3306 | 1.00 | −1.774 |
| G5EGN2 | fat-6 | S216 | 1.00 | −1.715 |
| Q9NEN6 | rps-6 | S203 | 1.00 | −1.714 |
| G5EGD6 | ipla-3 | S44 | 1.00 | −1.659 |
| O61967 | let-413 | S664 | 1.00 | −1.568 |
| Q17490 | unc-44 | S2406 | 0.90 | −1.541 |
| G5EE72 | mrp-5 | S1357 | 1.00 | −1.450 |
| Q56VY4 | Y87G2A.19 | S16 | 1.00 | −1.438 |
| Q68T18 | daf-15 | S1703 | 1.00 | −1.413 |
| Q17490 | unc-44 | S2411 | 0.91 | −1.406 |
| G5EFG4 | abcf-2 | S41 | 1.00 | −1.394 |
| Q20733 | saeg-1 | T91 | 1.00 | −1.357 |
| Q9BKR9 | Y28E9BR.16 | S560 | 0.89 | −1.326 |
| Q19994 | F34D10.4 | S634 | 1.00 | −1.238 |
| P34428 | pqn-38 | S270 | 1.00 | −1.205 |
| H2KZA3 | pqn-22 | S927 | 0.97 | −1.176 |
| Q17490 | unc-44 | S3372 | 1.00 | −1.168 |
| P34706 | sdc-3 | T914 | 1.00 | −1.167 |
| P34706 | sdc-3 | S926 | 1.00 | −1.167 |
| G5EGK6 | din-1 | S1472 | 0.98 | −1.135 |
| H2KZ09 | srsx-34 | S248 | 0.98 | −1.129 |
| P91136 | C37H5.5 | S166 | 1.00 | −1.088 |
| P55326 | F13E6.1 | S25 | 1.00 | −1.043 |
| P10567 | unc-15 | S44 | 1.00 | −0.994 |
| P91156 | acin-1 | S156 | 0.91 | −0.944 |
| Q95XQ8 | mcm-4 | S4 | 1.00 | −0.913 |
| H2L003 | lmd-3 | T226 | 1.00 | −0.908 |
| H2L003 | lmd-3 | S230 | 1.00 | −0.908 |
| P10567 | unc-15 | S32 | 0.99 | −0.897 |
| Q7Z1P8 | R11G1.6 | S510 | 0.98 | −0.890 |
| Q6A2D2 | hpo-34 | S1346 | 1.00 | −0.884 |
| Q22836 | T27F2.1 | S227 | 1.00 | −0.858 |
| Q22836 | T27F2.1 | S235 | 1.00 | −0.858 |
| H2KYP2 | ncx-2 | S251 | 1.00 | −0.829 |
| Q7JLB1 | ret-1 | S3 | 1.00 | −0.815 |
| Q11114 | C03B1.7 | S113 | 1.00 | −0.795 |
| E1NZ12 | Y40B1A.3 | S318 | 1.00 | −0.791 |
| E1NZ12 | Y40B1A.3 | S324 | 1.00 | −0.791 |
| P91240 | F08D12.1 | S604 | 1.00 | −0.789 |
| P90900 | ifa-4 | S53 | 1.00 | −0.784 |
| Q20626 | F49E2.5 | S940 | 0.94 | −0.759 |
| O44895 | ZK484.3 | S168 | 0.68 | −0.757 |
| Q23206 | W06E11.1 | S7 | 1.00 | −0.734 |
| P34675 | ZK688.5 | S1251 | 1.00 | −0.688 |
| Q1XFY2 | tag-343 | S294 | 0.90 | −0.686 |
| Q9BKQ7 | Y67D2.7 | S69 | 1.00 | −0.680 |
| Q9BKQ7 | Y67D2.7 | S71 | 1.00 | −0.680 |
| P35449 | nhx-9 | S576 | 0.88 | −0.658 |
| Q9XWU8 | epg-3 | S43 | 1.00 | −0.652 |
| Q10020 | T28D9.1 | S109 | 1.00 | −0.651 |
| Q966L6 | let-504 | S373 | 1.00 | −0.637 |
| Q966L6 | let-504 | S375 | 1.00 | −0.634 |
| Q09936 | C53C9.2 | S328 | 0.96 | −0.596 |
| Q09936 | C53C9.2 | S327 | 1.00 | −0.596 |
| Q20626 | F49E2.5 | T1134 | 0.98 | −0.590 |
| O76407 | T10B5.3 | S17 | 1.00 | −0.585 |
| Q9GRZ3 | dpy-21 | T902 | 1.00 | −0.549 |
| Q45FX5 | vav-1 | S195 | 1.00 | −0.493 |
| Q20626 | F49E2.5 | S961 | 1.00 | −0.434 |
The 72 phosphosites that showed significant reductions in phosphorylation with daf-15(RNAi) included two proteins with mammalian homologs that have been previously shown to be regulated by mTORC1, ACIN-1/Acinus L, and RPS-6/rpS6, perhaps validating our approach. Acinus L was shown to be a direct target of TORC1, and rpS6 was shown to be phosphorylated by S6K, downstream of TORC1 (Schwarz et al., 2015; Biever et al., 2015), and neither of these have previously been demonstrated as TORC1 targets in C. elegans.
These 72 phosphosites fall into several generalized functional categories (Figure 2E). Transcription (24%: RSP-6, ALY-2, SMG-6, SAEG-1, SDC-3, DIN-1, ACIN-1, MCM-4, TAG-343, LET-504, and W06E11.1) and membrane/structural functions (18%: LET-413, EPS-8, UNC-44, IPLA-3, UNC-15, RET-1, and IFA-4) contained the most peptides/proteins with characterized functions. While there were a few peptides belonging to well-known TORC1-related functions like autophagy (1%: EPG-3) and protein translation (3%: RPS-6 and F08D12.1), they were in the minority, suggesting that this approach may reveal previously unknown TORC1-regulated functions. Several proteins contained multiple phosphosites that were significantly reduced with daf-15(RNAi) (four phosphosites: UNC-44, three phosphosites: EPS-8, two phosphosites: UNC-15, LMD-3, SDC-3, LET-504, C53C9.2, T27F2.1, Y40B1A.3, and Y67D2.7).
As expected, DAF-15 was found in both the IP/MS analysis (bait) (Table 1) and the significantly changed phosphoproteomic list (RNAi target) (Table S2). However, no other proteins were found in both analyses. This lack of overlap is consistent with the transient nature of kinase associations with their targets.
The C. elegans TORC1 kinase motif was determined by analyzing the 72 high-confidence phosphosites that had decreased phosphorylation with daf-15i, using WebLogo (weblogo.Berkley.edu/logo.cgi). The analysis revealed that the majority of phosphosites contained an S/T-P motif, with higher incidence of serine (S) phosphorylation than threonine (T) (Figure 2F). This motif is common to many protein kinases and does not reveal a unique phosphorylation motif for TORC1 in C. elegans, consistent with previous analyses of mTOR in mammals (Avruch et al., 2006). Other pathway-based analyses (GO terms, tissue enrichment) could not be performed due to the small size of this dataset.
Previous functional studies from our lab and others have indicated an important role for intestine-specific TORC1 activity in regulating postembryonic development in C. elegans (Zhu et al., 2013, 2015; Kniazeva et al., 2015; Qi et al., 2017; Duong et al., 2020). In order to better understand TORC1 activity in the intestine, the data were further filtered for phosphorylation changes in intestine-expressed proteins, and for conservation of the protein in mammals (to exclude nematode-specific proteins), resulting in 18 peptides represented in 14 proteins (Table 3). These 18 peptides were investigated further as potential direct phosphorylation targets of TORC1.
Table 3.
Candidate TORC1 targets that are expressed in the intestine and conserved in mammals
| Protein | Sequence window |
|---|---|
| ABCF-2 | MSSQHPSVNSLCCLDEELD |
| ALY-2 | IRGTPNRRQSSGKPINRKV |
| CNP-2 | AYQQSHRPSSPKRRPSGRT |
| DIN-1 | TSTSSTKRSSISDHENLIS |
| EPG-3 | AGRGMKPSPSQNTLNRMER |
| FAT-6 | CDEIIGRQVSNHGCDIQRG |
| IFA-4 | GGSNLTSGMSPFGGHAASA |
| LET-413 | TVELVVLRRSPSPVSRTSV |
| LMD-3 | DIFDPDHLRTPEDSPKKQI |
| LMD-3 | PDHLRTPEDSPKKQIKQNG |
| NHX-9 | RSNLSAMFRSTEQLPSETP |
| RSP-6 | GRGGGGRDRSPYRGDRGRS |
| SMG-6 | TMSGNSRRRSNDTDSVTSR |
| UNC-44 | RESLLRQERSIDSPHASEQ |
| UNC-44 | HPACDSERLSEPAQSPEPV |
| UNC-44 | SERLSEPAQSPEPVETHAE |
| UNC-44 | EGEKGSPADSEKSLPHVVE |
| VAV-1 | DRIVTNRKPSMNENDLQNT |
The DAF-15-dependent phosphosite identified in this study is marked in bold.
In vitro kinase assays reveal several direct kinase targets of TORC1
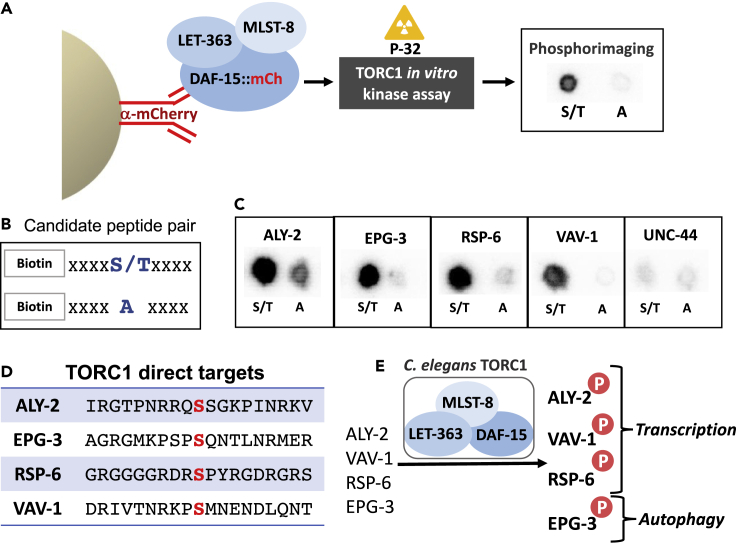
In order to determine the direct kinase targets of TORC1, each of the 18 candidates were tested by an in vitro kinase assay with the C. elegan’ TORC1 complex, using a method adapted from a previous study (Hsu et al., 2011). The TORC1 complex was isolated by our IP method used for the in vitro kinase assays. The limited number of proteins identified by our IP method (Table 1) indicates that the resulting complex is an enriched preparation of TORC1. Importantly, no other kinases were identified in the IP preparations. Briefly, the C. elegans TORC1 complex was isolated by immunoprecipitation from whole worm lysates and incubated in the presence of peptide containing the phosphosite of interest; kinase activity for a given peptide was quantified by measuring incorporation of 32P from [γ-32P]ATP into candidate peptides by autoradiography (Figure 3A). For each candidate phosphosite, two peptides were analyzed. The test peptide contained the endogenous phosphosite of interest (S/T residue) and the control peptide contained an alanine (A) residue (that cannot be phosphorylated) in place of the S/T phosphosite (Figure 3B). Using small peptides (19AAs) and phospho-specific amino acid substitutions for this analysis reduced the phosphorylation “noise” that may result from a whole protein. Each pair was analyzed side-by-side to determine phosphorylation of the candidate peptide relative to its own “phospho-dead” control (Table S5). If a candidate phosphosite showed 2-fold greater P-32 intensity than the corresponding control (confirmed by at least three independent biological replicates), then it was deemed a direct in vitro target of C. elegans’ TORC1. Among the eighteen candidates tested, four peptides were identified as direct TORC1 targets by this strategy (Figures 3C and 3D) (Table S6).
Figure 3.
In vitro kinase assays reveal direct, intestine-expressed kinase targets of TORC1
(A) A schematic diagram of the immunoprecipitation of the TORC1 complex followed by radiolabeled in vitro kinase assays.
(B) An illustration of the synthesized peptide pair tested for each candidate peptide. “S/T” corresponds to the serine or threonine daf-15-dependent phosphosite. The phosphosite residue was replaced with alanine (A) to serve as a phospho-dead control.
(C) Representative autoradiographs of candidate peptide pairs that showed over 2-fold greater P-32 intensity for the “S/T” peptide versus each “A” phospho-dead control. The UNC-44 peptide pair showed no significant difference in P-32 intensity and serves as an example of a candidate peptide revealed by our MS analysis that is not likely a direct TORC1 target by the in vitro kinase assay.
(D) List of TORC1 direct targets with corresponding phosphosites and flanking peptide sequence identified in this study. TORC1-specific phosphosites (S) are marked in red.
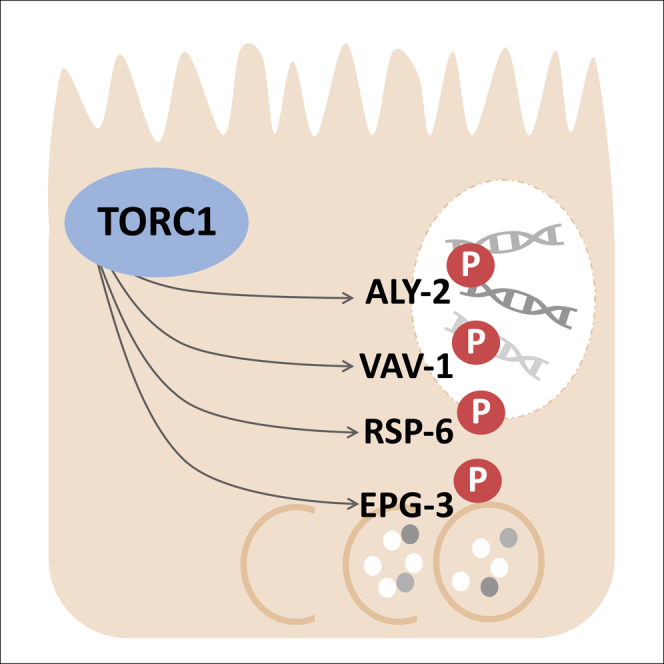
(E) A cartoon model depicting the proteins found bound to DAF-15 to form the C. elegans TORC1 complex, and the phosphorylation of ALY-2, RSP-6, VAV-1, and EPG-3 by TORC1 in C. elegans. Phosphorylation is indicated by the letter P in a red circle. Known protein functions are noted.
UNC-44 is presented here as an example of a negative result, with no difference between intensity of peptides pairs. This result may indicate that UNC-44 is not a direct TORC1 target at this phosphosite and that UNC-44 has phosphorylated residues that are indirectly affected by loss of DAF-15, or that the UNC-44 short peptides lack necessary 3D structure for binding and phosphorylation by TORC1. Candidate phosphosites that failed by the in vitro kinase assay should be interpreted carefully and analyzed further.
The positive results by the in vitro kinase assay are very encouraging. The four targets of TORC1 identified by this study (Figures 3D and 3E) are unique and have not been previously connected to regulation by TORC1. Due to conservation of these proteins in mammals, further investigation is warranted.
RSP-6 is an RNA-binding protein that has a predicted role in pre-mRNA splicing and plays roles in transcription termination (WormBase). RSP-6::GFP expression begins at the 20-30-cell stage of embryogenesis and localizes to the nucleus (WormBase). rsp-6(lf) suppresses the SynMuv phenotype of lin-15(n765) (Cui et al., 2008). RSP-6 is the ortholog of SRp20/SRSF3 in humans, also known as an RNA export adaptor (Kim et al., 2018; Walsh et al., 2010). SRp20 has many known functions in transcription, translation, and embryogenesis, and phosphorylation of the protein is expected to affect its function in the splicesome and enhance transport into the nucleus (Corbo et al., 2013; Ghosh and Adams, 2011), but this regulation has not previously been connected to TORC1. The RSP-6 S99 phosphosite identified in this study was previously found in a proteome-wide search for phosphorylated proteins in C. elegans (Zielinska et al., 2009) but was not previously connected to TORC1.
ALY-2 is an RNA export adaptor, required for export of TRA-1/tra-2 mRNA complexes, and thought to promote recruitment of mRNA export factors to mRNAs in C. elegans (Kuersten et al., 2004; Castellano-Pozo et al., 2012). ALY-2 is the ortholog of AlyRef/THOC4 in humans (Kim et al., 2018). The role of AlyRef in mRNA export activity has been shown to be regulated by inositol polyphosphate multi-kinase (IPMK) (Wickramasinghe et al., 2013). It would be intriguing to further explore the regulation of ALY-2 by TORC1. It is worth highlighting that RSP-6 and ALY-2 are both RNA export adaptors in C. elegans. Based on the respective orthologs, it is expected that RSP-6/SRp20 and ALY-2/AlyRef both may bind directly to mRNA and increase the binding affinity of the Tap protein for the mRNA, to aid in nuclear export. A phosphorylation-dependent role for SRp20 in the export of mRNA from the nucleus has already been indicated, although not previously connected to TORC1 regulation (Walsh et al., 2010).
VAV-1 is a Rho/Rac-family guanine exchange factor with a variety of functions, known to regulate intracellular calcium for rhythmic behavior (pharyngeal pumping, defecation, and ovulation) with prominent roles in development (WormBase) (Norman et al., 2005). VAV-1 is the ortholog of Vav1 in humans (Kim et al., 2018), a proto-oncogene. Vav1 is phosphorylated by Syk- and Src-related kinases at Tyr residues to turn on GEF function (Baylis, 2005; Katzav, 2004). Vav1 has been shown to play roles in cytoskeletal reorganization, transcription, development, and immune response (Katzav, 2004). The VAV-1 S227 phosphosite identified in this study was previously found in a proteome-wide search for phosphorylated proteins in C. elegans (Bodenmiller et al., 2008), but was not previously connected to TORC1.
The fourth phosphorylation target of TORC1 identified by this study, EPG-3, is the ortholog of vacuole membrane protein 1 (VMP1) in humans (Kim et al., 2018), an ER transmembrane protein that mediates contact between the ER and isolation membranes, endosomes, lipid droplets, and mitochondria (Tian et al., 2010; Wang et al., 2020). VMP1 has also been shown to play a role in autophagy, as overexpression of VMP1 induces autophagy, and depletion blocks autophagosome formation, in mammalian cells (Wang et al., 2020). This role in autophagosome formation is conserved and has been attributed to loss of EPG-3 in C. elegans (Zhao et al., 2017). It has also been shown that human VMP1 was sufficient to rescue the defects in epg-3(−) C. elegans, demonstrating strong functional conservation between these two proteins (Tian et al., 2010). Interestingly, VMP1 has not been previously identified as a TORC1 phosphorylation target, nor is VMP1 known to be regulated by phosphorylation. Further analysis of EPG-3 in C. elegans and VMP1 in mammalian models may reveal the TORC1-regulated functions of these proteins. In C. elegans, functions downstream of EPG-3 (phase separation and P granule assembly) have been shown to be regulated by LET-363/Tor phosphorylation (perhaps TORC1) (Zhang et al., 2018). The EPG-3 S43 phosphosite identified in this study was previously found in a proteome-wide search for phosphorylated proteins in C. elegans (Bodenmiller et al., 2008), but was not previously connected to TORC1. It is worth noting that the EPG-3 S43 short peptide also included an SP motif (often recognized and phosphorylated without specificity by kinases under in vitro conditions) just two residues away (Figure 3D), yet the in vitro kinase assay still showed a strong change in intensity when S43 alone was modified (Figure 3C). This further highlights the specificity of TORC1 for this target phosphosite.
Discussion
This study set out to discover the direct targets of TORC1 in a whole multi-cellular organism. Our findings should make a significant contribution toward understanding the specific mechanisms by which TORC1 regulates cellular functions in animals under physiological conditions and serve as a rich resource to the field (Goberdhan et al., 2016; Wang and Proud, 2011; Chao and Avruch, 2019; Blackwell et al., 2019). Previous TORC1 phosphoproteomic studies in cultured cells were conducted under starved, hyperactivated, and inhibited conditions, with very limited analysis of steady-state conditions (Hsu et al., 2011; Yu et al., 2011). Here, we have probed the DAF-15-dependent phosphoproteome in the complex context of a whole, multi-cellular organism using only genetic manipulation and have made fresh discoveries. Follow-up analyses of these findings in mammals may reveal unique aspects of TORC1 function.
mTORC1 is well known for regulating metabolism, translation, and autophagy (Condon and Sabatini, 2019), and our intriguing identification of TORC1 regulatory targets in transcriptional machinery may reveal new functions for this popular kinase. Recent studies have revealed a role for mTOR activity within the nucleus (nmTOR) to regulate transcription (Giguère, 2018). The mechanism for this activity is not known and therefore raises the question of whether this regulation is carried out through TORC1-mediated phosphorylation, and if so, what are the substrates (Giguère, 2018)? The three proteins (RSP-6, ALY-2, and VAV-1) identified in this study with known functions in transcription, may shed light on an answer. RSP-6/SRp20 is known to regulate many genes (Corbo et al., 2013), and it would be interesting to determine if these downstream functions are also impacted by TORC1 regulation of RSP-6/SRp20.
Perhaps it is equally interesting to note the direct targets that were not identified in this study, namely S6K and 4E-BP. The s6 kinase (S6K) has been shown to be a direct phosphorylation target of TORC1 in yeast and cultured cells, and this phosphorylation activity and regulation have been expected to be conserved in C. elegans (Blackwell et al., 2019). Yet, there is an absence of direct biochemical evidence for this claim. The S6K homolog in C. elegans is RSKS-1. Previous global phosphoproteomic studies in C. elegans did not identify phosphorylation of RSKS-1 at the predicted TORC1-regulated site (T389 mammals = T404 C. elegans) (Bodenmiller et al., 2008; Zielinska et al., 2009), suggesting that this regulatory phosphosite is not conserved. Furthermore, no significant changes in the phosphorylation of RSKS-1 peptides were found in our unbiased study (Table S2). Instead, our data suggest that RSKS-1 phosphorylation is not significantly altered by daf-15(RNAi) and may not be a direct target of TORC1 in C. elegans. Although S6K has been regarded as one of the major downstream targets of TORC1, our finding is consistent with other studies that have shown RSKS-1/S6K is dispensable for viability in both C. elegans and mammals (Shima et al., 1998; Pende et al., 2004; Long et al., 2002; Pan et al., 2007) and therefore may not be essential for TORC1-mediated development. Similarly, eukaryotic initiation factor 4E-BP has been regarded as a direct downstream target of mTORC1 phosphorylation, based on mammalian cell culture (Ma and Blenis, 2009), but the C. elegans functional homolog IFET-1 (Li et al., 2009) was not found to be significantly changed in our phosphoproteomic analysis (Table S2).
Instead of S6K or 4E-BP, one of the validated TORC1 targets from this study (Figure 3E) may prove to be a useful tool for measuring TORC1 activity, both in C. elegans and other organisms. Perhaps, the best next step to evaluate TORC1 signaling in C. elegans would be to design tagged, endogenous proteins for these TORC1 targets (ALY-2, EPG-3, RSP-6, and VAV-1), then edit the phosphosites (phospho-dead, constitutively phosphorylated, etc) and monitor the impact on protein function in vivo (expression, localization, and binding). Controlled manipulation of TORC1 activity by genetic means would serve to validate the direct target proteins as in vivo reporters for TORC1 activity. Such tools would allow rapid examination of numerous inputs (nutrient conditions, genetic interactions, aging, etc) to TORC1 activity in C. elegans. Our results highlight the importance of direct confirmation of a kinase for a target protein, helping avoid the pitfalls of assumption.
It is worth noting that this study also revealed a large number of phosphorylation events that increased with daf-15(RNAi) (Figure 2C and Table S4). Although not pursued by this TORC1-focused study, it is an interesting observation that may be related to TORC2 activity. It has been reported that loss of daf-15 resulted in an increase in the association of LET-363/Tor and RICT-1/Rictor (TORC2 formation), tipping the balance toward formation of the TORC2 complex (Nukazuka et al., 2011). Further analysis of this subset of phosphosites for TORC2 regulation would be worthwhile.
Previous work by our lab showed that a putative intestine-specific raga-1(gf) TORC1 hyperactivating transgene was sufficient to cause partial suppression of larval arrest caused by deficiency of monomethyl branched-chain fatty acids (mmBCFAs) (Zhu et al., 2013). This suggested that TORC1 activity in the intestine, but not in other tissues tested, was critical and sufficient for postembryonic growth, and may indicate that the intestine is the primary tissue of action. Following this finding, the present study filtered genome-wide phosphoproteomic results to focus on proteins known to be expressed in the C. elegans intestine in order to understand these regulatory properties of TORC1. Given the intestine-specific functional rescue seen with the raga-1 transgene, it would be logical to expect that loss of TORC1 activity specifically in the intestine may result in developmental defects. Surprisingly, we found that intestine-specific RNAi of daf-15 did not cause Lva (Figure S2), consistent with a recent study that used an auxin-induced degron system to create conditional depletion of daf-15 (Duong et al., 2020). Indeed, qualitative tissue-specific RNAi tests showed there was no single tissue (intestine, hypodermis, and neuron) where loss of daf-15 caused larval arrest. It is therefore likely that TORC1 signaling has broad impacts across tissues. A recent study in C. elegans proposed that TORC1 may have both unique and overlapping roles in the intestine and neurons as an AA/FA sensor (Zhu et al., 2021), further adding to the complexity of the mechanisms.
Our data also indicate that there are stage-specific regulations and roles for DAF-15/TORC1. We observed that protein expression decreased sharply over development (Figure 1F). The high expression seen at early developmental stages may indicate critical, stage-specific roles. Moreover, we also found that daf-15(RNAi) from hatching strongly reduces DAF-15 in L4/adults and impacts the next generation as dead embryos or L3-arrested animals (Figure S1). This embryonic lethality suggests that there are critical roles for DAF-15 in L4/Adult to support embryonic viability (possible maternal effect). Further investigation into these stage-specific roles for TORC1 is needed. Such observations can only be appreciated in the complex environment of a multi-cellular organism.
One initial goal of this characterization of TORC1 was to reveal the mechanism by which hyperactivation of TORC1 [by nprl-3(−)] suppresses developmental arrest caused by mmBCFA deficiency [elo-5(−)](Zhu et al., 2013). How hyperactivation of TORC1 recovers development of an FA-deficient animal without recovering the FA/lipid (Zhu et al., 2013) remains a fascinating and challenging question. Additional phosphoproteomic and biochemical analyses of the nprl-3(−) mutant may reveal the critical downstream activities that mediate these phenotypes. Although the kinase targets revealed by this phosphoproteomic study have implicated new roles for TORC1 function beyond previous connections to protein translation and autophagy, the regulatory connection to the lipid-mediated nutrient-sensing activity of TORC1 is not yet apparent. It will be very interesting to investigate how these TORC1 targets (ALY-2, EPG-3, RSP-6, and VAV-1) may act to ultimately execute the regulatory function by the lipid-TORC1 pathway.
Limitations of the study
This study provides a snapshot of TORC1 activity in C. elegans. Because our dataset focuses on phosphorylation changes at the L1 larval stage and relies on the effectiveness of RNAi to knockdown daf-15 (perhaps with limited effectiveness in the nervous system), other protein targets of TORC1 may have been missed.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-mCherry | Chromotek | Cat#5F8; RRID: AB_2336064 |
| Mouse anti-rat HRP | Abcam | Cat#ab106783; RRID:AB_10866498 |
| Rabbit anti-actin | Sigma | Cat#A2066; RRID:AB_476693 |
| Goat anti-rabbit HRP | Jackson Labs | Cat#111035003; RRID:AB_2313567 |
| RFP-Trap agarose | Chromotek | Cat#rta-20; RRID:AB_2631362 |
| Bacterial and virus strains | ||
| E. coli OP50 | Caenorhabditis Genetics Center (CGC) | |
| E. coli RNAi feeding strain HT115(DE3) | Dharmacon | N/A |
| E. coli daf-15 RNAi in HT115(DE3) background | (Zhu et al., 2013) | N/A |
| E. coli nhx-2 RNAi in HT115(DE3) background | Dharmacon | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| CHAPS | Sigma | Cat#3023 |
| Agarose beads | Chromotek | Cat#bab-20 |
| Custom peptides with N-terminal Biotin (>95% purity) | GenScript | N/A |
| [Gamma-P32]ATP | Perkin Elmer | BLU002A250UC |
| Critical commercial assays | ||
| ECL Prime | GE Healthcare | Cat#RPN2232 |
| Pierce BCA protein assay kit | Life Tech | Cat#23227 |
| Deposited data | ||
| C. elegans phosphoproteomics – DAF-15 knockdown | This study | PXD032260 |
| Experimental models: Organisms/strains | ||
| C. elegans strain: EG6699: ttTi5605 II; unc-119(ed3) III; oxEx1578 [eft-3p::GFP + Cbr-unc-119(+)] | Caenorhabditis Genetics Center (CGC) | N/A |
| C. elegans strain: MH5015: DAF-15::mCherry | This study | N/A |
| C. elegans strain: OD95: unc-119(ed3) III; ltIs37 [(pAA64) pie-1p::mCherry::his-58 + unc-119(+)] IV. ltIs38 [pie-1p::GFP::PH(PLC1delta1) + unc-119(+)] | Caenorhabditis Genetics Center (CGC) | N/A |
| C. elegans strain: DR732: daf-15(m81) unc-22(s7)/nT1 IV; +/nT1 V | Caenorhabditis Genetics Center (CGC) | N/A |
| C. elegans strain: WM27: rde-1(ne219) V | Caenorhabditis Genetics Center (CGC) | N/A |
| C. elegans strain: VP303: rde-1(ne219) V; kbIs7 [nhx-2p::rde-1 + rol-6(su1006)] | Caenorhabditis Genetics Center (CGC) | N/A |
| C. elegans strain: NL3321: sid-1(pk3321) V | Caenorhabditis Genetics Center (CGC) | N/A |
| Software and algorithms | ||
| AxioVision 4.8.2 | Carl Zeiss | |
| MaxQuant/Andromeda Version 1.5.2.8 | (Cox et al., 2011) | |
| R Bioconductor VSN package | (Välikangas et al., 2018) | https://bioconductor.org/packages/release/bioc/html/vsn.html |
| R package Limma Empirical Bayes | (Ritchie et al., 2015) | http://www.bioconductor.org |
| ImageQuantTL | GE Healthcare | |
| Other | ||
| SAM2 biotin capture membrane | Promega | V2861 |
Resource availability
Lead contact
Further information and requests for reagents should be directed to the lead contact, Min Han (mhan@colorado.edu).
Materials availability
Unique and stable reagents generated in this study are available from the Lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
C. elegans strains and maintenance
C. elegans strains were grown and maintained on nematode growth media (NGM) plates seeded with E. coli OP50 at 20°C (Brenner, 1974). The following strains were obtained from the Caenorhabditis Genetics Center (CGC): N2 Bristol (wild type), EG6699: ttTi5605; unc-119(ed3); oxEx1578 [eft-3p::GFP + Cbr-unc-119(+)], OD95: ltIs37 [(pAA64) pie-1p::mCherry::his-58 + unc-119(+)] IV. ltIs38 [pie-1p::GFP::PH (PLC1delta1) + unc-119(+)], DR732: daf-15(m81) unc-22(s7)/nT1, WM27: rde-1(ne219), VP303: rde-1(ne219) nhx-2p:rde-1(+) rol-6(su1006), and NL3321: sid-1(pk3321). MH5015: DAF-15::mCherry was constructed for this study.
Method details
Construction of the cas9 DAF-15::mCherry strain
The full daf-15 genomic sequence with the endogenous promoter, mCherry coding sequence, and 3′UTR, were integrated into ttTi5605 (EG6699), and confirmed by loss of array and presence of mCherry protein. To evaluate the function of the DAF-15::mCherry strain (MH5015), the strain was crossed into DR732 [daf-15(m81) unc-22(s7)/nT1]. Functional rescue was scored by observing growth past dauer in Unc animals carrying DAF-15:mCherry.
RNAi
The RNAi construct targeting daf-15 was previously described (Zhu et al., 2013, 2015). The RNAi construct targeting nhx-2 came from the C. elegans ORF RNAi library (Dharmacon). Data in Figures 1D and 1E resulted from RNAi feeding from the time of hatching. Worms were collected and analyzed after reaching L3 arrest in the second generation.
Western analysis
Protein samples were generated as whole worm lysates for the indicated stage and treatment. Worm pellets (pellet volumes ∼20uL or more) were boiled for 10 min in SDS loading buffer. The sample was cleared by centrifugation (5,000xg for 1min) and then loaded and separated on 7.5% SDS-PAGE gels (Bio-Rad).
DAF-15::mCherry and Actin were analyzed using standard wet western transfer conditions. mCherry was probed with rat anti-mCherry (Chromotek 5F8) 1:1,000, and mouse anti-rat HRP (Abcam ab106783) 1:10,000. Actin was probed with rabbit anti-Actin (Sigma A2066) 1:2,000, and goat anti-rabbit HRP (Jackson Labs 111-035-003) 1:20,000. Bands were detected with ECL Prime (GE Healthcare) and X-ray film. The strongest detection of mCherry was obtained using excess ECL and fresh blocking buffer.
Microscopy
Fluorescent microscopy was performed using a Zeiss Axioplan2 equipped with DIC (Nomarski) optics, AxioVision 4.8.2 software, and a Zeiss Axiocam MRm. Plate images were captured on a Leica M165 FC with Leica IC80 HD camera.
Immunoprecipitation
Mixed populations of well-fed DAF-15::mCherry or N2 worms were collected in M9, washed in M9 (3 × 10mL), pelleted (∼100uL worm pellet), and frozen at −70°C until analyzed. The immunoprecipitation was based on the manufacturer’s protocol for RFP-Trap beads (Chromotek, rta-20). 0.3% CHAPS (Sigma, C3023) was used in the lysis buffer instead of NP40. Total protein concentration of sonicated worm lysates was determined by BCA (Pierce), normalized for input, pre-cleared with agarose beads (Chromotek, bab-20), and subsequently incubated with RFP-Trap beads (Chromotek, rta-20) to pull down the mCherry-tagged protein. Beads were extensively washed (1× TE buffer, 150mM NaCl), then further analyzed for DAF-15::mCherry binding partners by on-bead digest and MS/ID, or used directly for TORC1 in vitro kinase assays. Preliminary evaluation of a daf-15p::mCherry strain showed unregulated, high expression of mCherry that could not be used for a meaningful comparison.
Mass spec analysis for DAF-15::mCherry interacting proteins
Beads were submitted to the University of Colorado Mass Spec Facility for analysis. Samples from immunoprecipitation were resuspended in 0.1M ammonium bicarbonate (ABC), reduced using 5mM TCEP at 60°C for 30 min, and alkylated using 15mM chloroacetamide at room temperature for 20min in darkness. Samples were then treated with 0.25 μg of trypsin at 37°C for 4 h. The resulting tryptic peptides were desalted using StageTips C18 (Thermo Scientific) and dried using vacuum centrifugation. The peptides were reconstituted in 15μL of Buffer A (0.1% formic acid in water), of which 5μL was subjected to LC-MS/MS analysis.
The tryptic peptides were resolved using a Waters nanoACQUITY UPLC system in a single pump trap mode. The peptides were loaded onto a nanoACQUITY 2G-V/MTrap 5μm Symmetry C18 column (180 μm × 20 mm) with 99.5% Buffer A and 0.5% Buffer B (0.1% formic acid in acetonitrile) at 15 μL/min for 3min. The trapped peptides were eluted and resolved on a BEH C18 column (130 Å, 1.7 μm × 75 μm × 250mm) using gradients of 3-5% B (0-3 min) and 8-28% B (3-185 min) at 0.3 μL/min. MS/MS was performed on an LTQ Orbitrap Velos mass spectrometer, scanning precursor ions between 300 and 1800 m/z (1 × 106 ions, 60,000 resolution) and selecting the 20 most intense ions for MS/MS with 180s dynamic exclusion, 10 ppm exclusion width, repeat count = 1, and 30s repeat duration. Ions with unassigned charge state and MH+1 were excluded from the MS/MS. Maximal ion injection times were 500ms for FT (one microscan) and 250ms for LTQ, and the AGC was 1 × 104. The normalized collision energy was 35% with activation Q 0.25 for 10ms.
MaxQuant/Andromeda (version 1.5.2.8) was used to process raw files from LTQ-Orbitrap, and search the peak lists against database consisting of Uniprot C. elegans proteome (total 27,217 entries, downloaded 12/03/2014). The search allowed trypsin specificity with a maximum of two missed-cleavages and set carbamidomethyl modification on cysteine as a fixed modification and protein N-terminal acetylation and oxidation on methionine as variable modifications. MaxQuant used 4.5 ppm main search tolerance for precursor ions, 0.5 Da MS/MS match tolerance, searching top 8 peaks per 100 Da. False discovery rates for both protein and peptide were 0.01 with minimum seven amino acid peptide length. A label-free quantification was enabled with minimum 2 LFQ ratio counts and a fast LFQ option. Table 1 reports the list of proteins identified in the DAF-15:mCherry IP by LC/MS/MS, ranked by peptide count, and listed with closest mammalian homolog by sequence similarity, as determined by OrthoList 2 (Kim et al., 2018).
RNAi treatment for phosphoproteomics
This method was used to generate data in Figures 2 and 3 and Tables S1, S2, S3, and S4. Briefly, worms were fed RNAi from P0 L4 stage until F1 L1 3h fed. Stage synchronized MH5015 worms were fed HT115 empty vector or daf-15(RNAi) E. coli from the mid-L4 until gravid adult stage. The P0 gravid adults were then bleached to release the F1 eggs; the eggs were incubated and hatched in M9 overnight for a synchronized F1 population. The synchronized L1s were added to plates with the corresponding RNAi fed to the parent (HT115 or daf-15i), recovered on food for 3hrs, then collected in M9 and washed (3 × 10mL). Residual M9 was removed and the worm pellets were frozen (−70C) for subsequent analysis by western blotting, immunoprecipitation or phosphoproteomics.
Ten 100uL worm pellets (10 biological replicates) of each treatment were used for phosphoproteomic mass spec analysis. RNAi efficiency was confirmed by observing daf-15i L3 arrest in the second generation on test plates.
Phosphoproteomics
The phosphoproteomic platform used in this study was based on the EasyPhos method for single-run LC-MS/MS analysis that has been successfully applied to samples with diverse cells and tissues (Humphrey et al., 2015). However, our analysis did not use stable isotope labeling (label-free). Briefly, L1 larvae pellets (100uL worm pellet volume x 10 per condition) were lysed and subjected to protein digestion by Lys-C and trypsin in TFE buffer. Phosphopeptides were isolated by titanium (TiO2) enrichment, followed by peptide detection by single-run LC-MS/MS. Phospho-enriched tryptic peptide samples were suspended in 0.1% (v/v) trifluoracetic acid with 3% (v/v) acetonitrile and 1 ug of tryptic peptides were directly injected onto a C18 1.7 μm, 130 Å, 75 μm × 250 mm M-class column (Waters), using a Waters M-class UPLC. Peptides were eluted at 300 nL/min using a gradient from 3% to 20% acetonitrile over 100 min into an Orbitrap Fusion mass spectrometer (Thermo Scientific). Precursor mass spectra (MS1) were acquired at a resolution of 120,000 from 380-1,500 m/z with an AGC target of 2.0 × 105 and a maximum injection time of 50ms. Dynamic exclusion was set for 20 s with a mass tolerance of +/− 10 ppm. Precursor peptide ion isolation width for MS2 fragment scans was 1.6 Da using the quadrupole, and the most intense ions were sequenced using Top Speed with a 3-s cycle time. All MS2 sequencing was performed using higher energy collision dissociation (HCD) at 35% collision energy and scanning in the linear ion trap. An AGC target of 1.0 × 104 and 35-s maximum injection time was used. Raw files were searched against the Uniprot C. elegans database using Maxquant with cysteine carbamidomethylation as a fixed modification. Methionine oxidation, protein N-terminal acetylation and phosphoserine, threonine, and tyrosine were searched as variable modifications. All peptides and proteins were thresholded at a 1% false discovery rate (FDR). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository with the dataset identifier PXD032260.
WebLogo
The 72 high-confidence phosphosites were analyzed using the WebLogo tool (weblogo.Berkley.edu/logo.cgi). Three peptides were dropped from analysis because the phosphosite was located too close to the termini for analysis to be possible, resulting in 69 phosphosites analyzed. The MS sequence window was trimmed to 15 amino acids (7 amino acids flanking each phosphosite).
Peptide synthesis
Custom peptides analyzed in this study (Table S5) were synthesized by GenScript. Each peptide included 19AAs centered on the S/T phosphosite of interest, with an N-terminal Biotin modification (>95% purity).
In vitro kinase assay
The in vitro kinase assay method was adapted from previous studies (Hsu et al., 2011). The C. elegans TORC1 complex was precipitated from whole worm lysates (see IP method above) and used for in vitro kinase assays. TORC1 kinase activity for a given substrate was quantified by measuring incorporation of 32P radioactivity from [γ-32P]ATP (Perkin Elmer, BLU002A250UC) into candidate peptides. For each candidate phosphosite (Table 3) a pair of custom 19AA peptides (conjugated to biotin) were synthesized (GenScript) (Table S5). “S/T” indicates the Serine or Threonine residue identified in the phosphoproteomic analysis. “A” indicates Alanine, a non-phosphorylatable amino acid used as the “phospho-dead” control peptide. In addition, peptides that showed no fold change (NFC) by phosphoproteomic analysis were chosen randomly from the dataset and used as negative controls. A 250mM stock in DMSO was prepared for each peptide. Briefly, each kinase reaction included the TORC1 complex on beads (IP), 25mM HEPES (pH 7.4), 50mM KCl, 10mM MgCl2, 90uM cold ATP, 3uCi P32-ATP, and peptide (0.5mM in 50mM HEPES pH 7.4). The reaction was terminated by addition of 8M guanidine HCl. Equal volumes of each reaction were pipetted onto a SAM2 biotin capture membrane (Promega, V2861) with 2-3 technical replicates and washed thoroughly (2M NaCl, 1% H3PO4). The dry membrane was exposed to a phosphor screen and imaged with a Typhoon Imager (GE Amersham). P32 intensity was quantified with ImageQuant (GE Healthcare). 2-3 technical replicates for each reaction were averaged and fold change was determined between the “S/T” intensity and “A” intensity for each peptide pair. Any S/T peptide that demonstrated a fold change greater than 2 (S/T intensity/A intensity), and was confirmed by 3 or more biological replicates, was considered a direct TORC1 target.
Some candidate peptides (FAT-6, LET-413, ABCF-2, DIN-1, VAV-1) had poor solubility under the standard assay conditions and had to be extensively diluted. Thus, the kinase assay was performed under very dilute conditions and failure to observe phosphorylation change may have been a consequence of this dilute condition. Although these peptides were not confirmed as TORC1 targets by this in vitro kinase assay (with the exception of VAV-1), they should be investigated further.
Quantification and statistical analysis
Ten replicates of label-free intensity measurements were compared between treatment (daf-15 mutant) and wild type control. After log2 intensity conversion, normalization was performed using the R Bioconductor VSN (Variance stabilization and calibration for microarray data) package (Välikangas et al., 2018). Empirical Bayes from the R Limma package (Ritchie et al., 2015) was implemented to calculate t-statistics, p-values and Benjamini-Hochberg adjusted FDR. For the ttest, the python scipy stats.ttest_ind independent t-test package was used for p-value calculation and Benjamini-Hochberg was subsequently applied for multiple hypothesis false discovery rate calculation. The final candidate list included all phosphosites that had at least 3 non-zero reads in both datasets (daf-15i and control) and a p < 0.05 by either empirical bayes, t-test or exact test for significance.
P32 intensity was quantified with ImageQuant (GE Healthcare).
Acknowledgments
We are sincerely grateful to H Zhu, B Weaver, Y Weaver, L Edgar, J Avruch, PM Triller, and PBJ Sewell for helpful discussions and technical expertise. We thank the CGC (funded by NIH P40OD010440) for strains, the University of Colorado Boulder Central Analytical Mass Spectrometry Facility and W.M. Keck Foundation Proteomics Resource core facility for services, and the MCDB BioCore (SCR_019302) for equipment. This work was funded by NIH grants R35GM139631 (MH) and R01AR074503 (MH), a DARPA cooperative agreement 13-34-RTA-FP-007 (WMO), and partly by the Howard Hughes Medical Institute at the early stage of the study (MH and AKS).
Author contributions
AKS conceived the project, designed and performed the experiments, analyzed the data, and wrote the paper.
ZCP, CCE, and JJ performed MS sample prep and analysis.
MH and WMO secured the funding, provided the resources and supervision.
Declaration of interests
The authors declare no competing interests. Some authors have changed professional affiliations since making contributions to this work: ZCP – Strategic Analysis, Inc; JJ – Bioloomics; WMO - Pfizer.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104186.
Supplemental information
Data and code availability
-
•
Phosphoproteomic data have been deposited at ProteomeXchange and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table.
-
•
All other data reported in this paper will be shared by the lead contact upon request. This paper does not report original code.
References
- Avruch J., Hara K., Lin Y., Liu M., Long X., Ortiz-Vega S., Yonezawa K. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene. 2006;25:6361–6372. doi: 10.1038/sj.onc.1209882. [DOI] [PubMed] [Google Scholar]
- Balasubramaniam B., Vinitha T., Deepika S., Jebamercy G., Venkatakrishna L.M., Balamurugan K. Analysis of Caenorhabditis elegans phosphoproteome reveals the involvement of a molecular chaperone, HSP-90 protein during Salmonella enterica Serovar Typhi infection. Int. J. Biol. Macromol. 2019;137:620–646. doi: 10.1016/j.ijbiomac.2019.06.085. [DOI] [PubMed] [Google Scholar]
- Baylis H.A. VAV's got rhythm. Cell. 2005;123:5–7. doi: 10.1016/j.cell.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Biever A., Valjent E., Puighermanal E. Ribosomal protein S6 phosphorylation in the nervous system: from regulation to function. Front. Mol. Neurosci. 2015;8:75. doi: 10.3389/fnmol.2015.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T.K., Sewell A.K., Wu Z., Han M. TOR signaling in Canorhabditis elegans development, metabolism, and aging. Genetics. 2019;213:329–360. doi: 10.1534/genetics.119.302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B., Campbell D., Gerrits B., Lam H., Jovanovic M., Picotti P., Schlapbach R., Aebersold R. PhosphoPep--a database of protein phosphorylation sites in model organisms. Nat. Biotechnol. 2008;26:1339–1340. doi: 10.1038/nbt1208-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Pozo M., García-Muse T., Aguilera A. The Caenorhabditis elegans THO complex is required for the mitotic cell cycle and development. PLoS One. 2012;7:e52447. doi: 10.1371/journal.pone.0052447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L.H., Avruch J. Cryo-EM insight into the structure of MTOR complex 1 and its interactions with Rheb and substrates. F1000Res. 2019;8:14. doi: 10.12688/f1000research.16109.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon K.J., Sabatini D.M. Nutrient regulation of mTORC1 at a glance. J. Cell Sci. 2019;132:jcs222570. doi: 10.1242/jcs.222570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo C., Orrù S., Salvatore F. SRp20: an overview of its role in human diseases. Biochem. Biophys. Res. Commun. 2013;436:1–5. doi: 10.1016/j.bbrc.2013.05.027. [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Cui M., Allen M.A., Larsen A., Macmorris M., Han M., Blumenthal T. Genes involved in pre-mRNA 3'-end formation and transcription termination revealed by a lin-15 operon Muv suppressor screen. Proc. Natl. Acad. Sci. U S A. 2008;105:16665–16670. doi: 10.1073/pnas.0807104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D.J., Ward J.D., Reiner D.J., Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong T., Rasmussen N.R., Ballato E., Mote F.S., Reiner D.J. The Rheb-TORC1 signaling axis functions as a developmental checkpoint. Development. 2020;147:dev181727. doi: 10.1242/dev.181727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh G., Adams J.A. Phosphorylation mechanism and structure of serine-arginine protein kinases. FEBS J. 2011;278:587–597. doi: 10.1111/j.1742-4658.2010.07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguère V. Canonical signaling and nuclear activity of mTOR-a teamwork effort to regulate metabolism and cell growth. FEBS J. 2018;285:1572–1588. doi: 10.1111/febs.14384. [DOI] [PubMed] [Google Scholar]
- Goberdhan D.C., Wilson C., Harris A.L. Amino acid sensing by mTORC1: intracellular transporters mark the spot. Cell Metab. 2016;23:580–589. doi: 10.1016/j.cmet.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A., Hall M.N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017;36:397–408. doi: 10.15252/embj.201696010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin D.A., Stevens D.M., Thoreen C.C., Burds A.A., Kalaany N.Y., Moffat J., Brown M., Fitzgerald K.J., Sabatini D.M. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Hsu P.P., Kang S.A., Rameseder J., Zhang Y., Ottina K.A., Lim D., Peterson T.R., Choi Y., Gray N.S., Yaffe M.B., et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Wu Z., Wang J., Zhang X. Quantitative phosphoproteomics reveals GTBP-1 regulating C. elegans lifespan at different environmental temperatures. Biochem. Biophys. Res. Commun. 2018;503:1962–1967. doi: 10.1016/j.bbrc.2018.07.142. [DOI] [PubMed] [Google Scholar]
- Huang J., Wu Z., Zhang X. Short-term mild temperature-stress-induced alterations in the. Int. J. Mol. Sci. 2020;21:6409. doi: 10.3390/ijms21176409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey S.J., Azimifar S.B., Mann M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 2015;33:990–995. doi: 10.1038/nbt.3327. [DOI] [PubMed] [Google Scholar]
- Jia K., Chen D., Riddle D.L. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Jones K.T., Greer E.R., Pearce D., Ashrafi K. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 2009;7:e60. doi: 10.1371/journal.pbio.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzav S. Vav1: an oncogene that regulates specific transcriptional activation of T cells. Blood. 2004;103:2443–2451. doi: 10.1182/blood-2003-08-2834. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Sarbassov D.D., Ali S.M., Latek R.R., Guntur K.V., Erdjument-Bromage H., Tempst P., Sabatini D.M. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- Kim W., Underwood R.S., Greenwald I., Shaye D.D. OrthoList 2: a new comparative genomic analysis of human and Caenorhabditis elegans genes. Genetics. 2018;210:445–461. doi: 10.1534/genetics.118.301307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeva M., Zhu H., Sewell A.K., Han M. A lipid-TORC1 pathway promotes neuronal development and foraging behavior under both fed and fasted conditions in C. elegans. Dev. Cell. 2015;33:260–271. doi: 10.1016/j.devcel.2015.02.015. [DOI] [PubMed] [Google Scholar]
- Kuersten S., Segal S.P., Verheyden J., Lamartina S.M., Goodwin E.B. NXF-2, REF-1, and REF-2 affect the choice of nuclear export pathway for tra-2 mRNA in C. elegans. Mol. Cell. 2004;14:599–610. doi: 10.1016/j.molcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Li W., Debella L.R., Guven-Ozkan T., Lin R., Rose L.S. An eIF4E-binding protein regulates katanin protein levels in C. elegans embryos. J. Cell Biol. 2009;187:33–42. doi: 10.1083/jcb.200903003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.J., Wang C.W., Tao L., Yan Y.H., Zhang M.J., Liu Z.X., Li Y.X., Zhao H.Q., Li X.M., He X.D., et al. Insulin signaling regulates longevity through protein phosphorylation in Caenorhabditis elegans. Nat. Commun. 2021;12:4568. doi: 10.1038/s41467-021-24816-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X., Spycher C., Han Z.S., Rose A.M., Müller F., Avruch J. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr. Biol. 2002;12:1448–1461. doi: 10.1016/s0960-9822(02)01091-6. [DOI] [PubMed] [Google Scholar]
- Ma X.M., Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Norman K.R., Fazzio R.T., Mellem J.E., Espelt M.V., Strange K., Beckerle M.C., Maricq A.V. The Rho/Rac-family guanine nucleotide exchange factor VAV-1 regulates rhythmic behaviors in C. elegans. Cell. 2005;123:119–132. doi: 10.1016/j.cell.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Nukazuka A., Tamaki S., Matsumoto K., Oda Y., Fujisawa H., Takagi S. A shift of the TOR adaptor from Rictor towards Raptor by semaphorin in C. elegans. Nat. Commun. 2011;2:484. doi: 10.1038/ncomms1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offenburger S.L., Bensaddek D., Murillo A.B., Lamond A.I., Gartner A. Comparative genetic, proteomic and phosphoproteomic analysis of C. elegans embryos with a focus on ham-1/STOX and pig-1/MELK in dopaminergic neuron development. Sci. Rep. 2017;7:4314. doi: 10.1038/s41598-017-04375-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan K.Z., Palter J.E., Rogers A.N., Olsen A., Chen D., Lithgow G.J., Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende M., Um S.H., Mieulet V., Sticker M., Goss V.L., Mestan J., Mueller M., Fumagalli S., Kozma S.C., Thomas G. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5'-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., Kundu D.J., Prakash A., Frericks-Zipper A., Eisenacher M., et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi B., Kniazeva M., Han M. A vitamin-B2-sensing mechanism that regulates gut protease activity to impact animal's food behavior and growth. Elife. 2017;6:e26243. doi: 10.7554/eLife.26243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads T.W., Prasad A., Kwiecien N.W., Merrill A.E., Zawack K., Westphall M.S., Schroeder F.C., Kimble J., Coon J.J. NeuCode labeling in nematodes: proteomic and phosphoproteomic impact of Ascaroside treatment in Caenorhabditis elegans. Mol. Cell Proteomics. 2015;14:2922–2935. doi: 10.1074/mcp.M115.049684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robida-Stubbs S., Glover-Cutter K., Lamming D.W., Mizunuma M., Narasimhan S.D., Neumann-Haefelin E., Sabatini D.M., Blackwell T.K. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012;15:713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf V., Holzem C., Peyman T., Walz G., Blackwell T.K., Neumann-Haefelin E. TORC2 signaling antagonizes SKN-1 to induce C. elegans mesendodermal embryonic development. Dev. Biol. 2013;384:214–227. doi: 10.1016/j.ydbio.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini D.M. Twenty-five years of mTOR: uncovering the link from nutrients to growth. Proc. Natl. Acad. Sci. U S A. 2017;114:11818–11825. doi: 10.1073/pnas.1716173114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Thoreen C.C., Peterson T.R., Lindquist R.A., Kang S.A., Spooner E., Carr S.A., Sabatini D.M. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- Schwarz J.J., Wiese H., Tölle R.C., Zarei M., Dengjel J., Warscheid B., Thedieck K. Functional proteomics identifies Acinus L as a direct insulin- and amino acid-dependent mammalian target of rapamycin complex 1 (mTORC1) substrate. Mol. Cell Proteomics. 2015;14:2042–2055. doi: 10.1074/mcp.M114.045807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima H., Pende M., Chen Y., Fumagalli S., Thomas G., Kozma S.C. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Li Z., Hu W., Ren H., Tian E., Zhao Y., Lu Q., Huang X., Yang P., Li X., et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–1055. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- Ubersax J.A., Ferrell J.E. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- Välikangas T., Suomi T., Elo L.L. A systematic evaluation of normalization methods in quantitative label-free proteomics. Brief. Bioinf. 2018;19:1–11. doi: 10.1093/bib/bbw095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M.J., Hautbergue G.M., Wilson S.A. Structure and function of mRNA export adaptors. Biochem. Soc. Trans. 2010;38:232–236. doi: 10.1042/BST0380232. [DOI] [PubMed] [Google Scholar]
- Wang P., Kou D., Le W. Roles of VMP1 in autophagy and ER-membrane contact: potential implications in neurodegenerative disorders. Front. Mol. Neurosci. 2020;13:42. doi: 10.3389/fnmol.2020.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Proud C.G. mTORC1 signaling: what we still don't know. J. Mol. Cell Biol. 2011;3:206–220. doi: 10.1093/jmcb/mjq038. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe V.O., Savill J.M., Chavali S., Jonsdottir A.B., Rajendra E., Grüner T., Laskey R.A., Babu M.M., Venkitaraman A.R. Human inositol polyphosphate multikinase regulates transcript-selective nuclear mRNA export to preserve genome integrity. Mol. Cell. 2013;51:737–750. doi: 10.1016/j.molcel.2013.08.031. [DOI] [PubMed] [Google Scholar]
- Yu Y., Yoon S.O., Poulogiannis G., Yang Q., Ma X.M., Villén J., Kubica N., Hoffman G.R., Cantley L.C., Gygi S.P., Blenis J. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Wang Z., Du Z., Zhang H. mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell. 2018;174:1492–1506.e22. doi: 10.1016/j.cell.2018.08.006. [DOI] [PubMed] [Google Scholar]
- Zhao Y.G., Chen Y., Miao G., Zhao H., Qu W., Li D., Wang Z., Liu N., Li L., Chen S., et al. The ER-localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control ER-isolation membrane contacts for autophagosome formation. Mol. Cell. 2017;67:974–989.e6. doi: 10.1016/j.molcel.2017.08.005. [DOI] [PubMed] [Google Scholar]
- Zhu H., Sewell A.K., Han M. Intestinal apical polarity mediates regulation of TORC1 by glucosylceramide in C. elegans. Genes Dev. 2015;29:1218–1223. doi: 10.1101/gad.263483.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Shen H., Sewell A.K., Kniazeva M., Han M. A novel sphingolipid-TORC1 pathway critically promotes postembryonic development in Caenorhabditis elegans. Elife. 2013;2:e00429. doi: 10.7554/eLife.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Teng F., Li N., Zhang L., Zhang S., Xu F., Shao J., Sun H., Zhu H. Monomethyl branched-chain fatty acid mediates amino acid sensing upstream of mTORC1. Dev. Cell. 2021;56:2692–2702.e5. doi: 10.1016/j.devcel.2021.09.010. [DOI] [PubMed] [Google Scholar]
- Zielinska D.F., Gnad F., Jedrusik-Bode M., Wiśniewski J.R., Mann M. Caenorhabditis elegans has a phosphoproteome atypical for metazoans that is enriched in developmental and sex determination proteins. J. Proteome Res. 2009;8:4039–4049. doi: 10.1021/pr900384k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Phosphoproteomic data have been deposited at ProteomeXchange and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table.
-
•
All other data reported in this paper will be shared by the lead contact upon request. This paper does not report original code.