Abstract
Purpose:
Increased incidence of ductal carcinoma in situ (DCIS) associated with mammographic screening for breast cancer has emphasized the challenges of managing this condition. The aim of this study was to identify informative clinical indicators of DCIS biology by molecular profiling.
Experimental Design:
Areas of in situ carcinoma, atypical ductal hyperplasia, and benign epithelium were microdissected from 46 invasive breast cancers. Oligonucleotide probes showing differential expression between DCIS associated with grade 1 and 3 invasive cancer were identified by microarray-based gene expression profiling. Expression at these probes was used to define a "molecular grade" subcategorization of all samples. The genomic basis of molecular grade was examined by array-based comparative genomic hybridization. Clinical course was examined in a cohort of 134 patients with DCIS treated by surgery alone.
Results:
DCIS samples were designated as low or high molecular grade based on expression at 173 probes. The low molecular grade subgroup included low (n = 10) and intermediate (n =11) nuclear grade DCIS as well as all samples of atypical ductal hyperplasia (n = 4) and benign epithelium (n = 7). The high molecular grade subgroup included DCIS of intermediate (n = 7) and high (n = 19) nuclear grade. The character and degree of genomic aberration were distinct between molecular grade subgroups. A classification tree model including nuclear grade and Ki67 score accurately predicted molecular grade for 95.7% of samples. In an independent cohort, this showed a pattern of rapid disease recurrence for high molecular grade DCIS.
Conclusions:
Molecular profiling indicates a binary grading scheme for DCIS. This practical approach has potential to improve clinical evaluation of DCIS.
In the current era of breast cancer diagnosis, ductal carcinoma in situ (DCIS) has become an important entity. This is because of a dramatic increase in incidence associated with mammographic breast cancer screening and has two principal implications: DCIS is a more common clinical problem (1), and the ability to appropriately diagnose and manage DCIS has become a factor determining the effectiveness of screening programs (2).
The breast cancer–associated mortality of DCIS is very low compared with invasive breast cancer (3), and therefore the relative benefit of any clinical intervention requires close consideration. For example, there is concern that a proportion of screen detected DCIS may not have given rise to clinically detected cancer in the lifetime of the patient (4). Moreover, following a DCIS diagnosis, the risk of disease progression needs to be aligned to a range of treatment options, which currently includes mastectomy or breast conserving surgery, sentinel lymph node biopsy, radiotherapy, and adjuvant endocrine therapy (5). In this circumstance, it is widely acknowledged that improved predictors of DCIS clinical course are needed.
In contrast to invasive breast cancer for which histopathologic grade, individual biomarkers, and, more recently, gene expression signatures are proven indicators of prognosis (6–8), it has been remarkably difficult to identify clinically informative features of DCIS biology. In this regard, a major impediment has been the absence of a definitive histopathologic grading system. A number of DCIS grading schemes have been proposed, variously using architectural patterns, nuclear morphology, mitoses, and necrosis to distinguish low-, intermediate–, and high-grade cases (9). However, there is a wide range of appearances of DCIS, and combinations of high and low grade features are not uncommon (10, 11). Consequently, whereas classification of lesions with uniformly high or low grade features poses no difficulty, "intermediate grade" DCIS can be particularly heterogeneous.
Despite a pressing need for robust clinical indicators of DCIS biology, studies aiming to identify such markers are difficult. In particular, there is an absence of accessible clinical end points to which candidate markers can be compared. Local disease recurrence might seem the best indicator of clinically aggressive DCIS; however, this may be entirely determined by the extent to which the lesion is removed rather than its inherent biology (12). An alternative approach is to use a surrogate end point that is intermediate in the relationship between the variable of interest (DCIS biology) and the true clinical end point (breast cancer – specific mortality; ref. 13). The histopathologic grade of invasive cancer coexisting with DCIS fulfills these criteria on the basis that (a) the biology of DCIS and concomitant invasive breast cancer are closely related as evidenced by strongly concordant expression of biological indicators (14); (b) the biology of invasive cancer is reflected by histopathologic grade (15); and (c) grade is correlated with survival (16).
The aim of this study was to establish a clinically applicable and informative classification of DCIS. To do this, gene expression profiles were generated from the in situ component of invasive breast cancers and a DCIS molecular classification was determined by reference to concomitant invasive cancer grade. The genomic basis of this "molecular grade" was examined by array-based comparative genomic hybridization (CGH). The clinical significance of molecular grade was then further evaluated in a cohort of 134 patients with DCIS uniformly treated by local excision alone.
Materials and Methods
Patient samples for microdissection.
The discovery phase study cohort consisted of 46 cases of invasive breast cancer identified from a collection of frozen tumor samples taken from therapeutic excisions done at Westmead Hospital Australia between 1989 and 1998. Cancers with lobular carcinoma in situ only were not included; however, one case judged initially as containing DCIS that was reassigned lobular carcinoma in situ following detailed review was retained. All patient information and materials were de-identified and the study was conducted with institutional Human Research Ethics Committee approval.
Histopathology review.
Histopathologic features of each case were documented by review of archival diagnostic tissue sections using a dual-observer protocol (R.L.B./E.L.S.). A standardized reporting format was used including invasive cancer grade according to the criteria of Elston and Ellis (16) and individual features of DCIS following reference to criteria listed in the 1997 Consensus Conference Committee report (17). In 14 of 46 (30.4%) tumors, two distinct morphologic subtypes of DCIS were identified and separately described.
Determination of tumor estrogen receptor, progesterone receptor, human epidermal growth factor receptor-2, and p53 expression.
Results from clinical enzyme immunoassay or immunoperoxidase staining assessment of tumor estrogen receptor and progesterone receptor content were used. Tumor human epidermal growth factor receptor-2 (HER2) and p53 expression were determined following immunohistochemical staining (see Supplementary Methods).
DCIS Ki67 scoring.
Immunohistochemical staining for Ki67 was done on both frozen and paraffin-embedded tissue sections with a rabbit polyclonal antibody (Novocastra) at 1:1,000 dilution. The Ki67 score (percentage of positive cells) was determined by a single observer (L.R.W.) by manual counting of positive and negative in situ carcinoma cell nuclei using the manual tag function in the ImagePro Plus 4.0 software (Image Processing Solutions). In tumors with two morphologic subtypes of DCIS present, a separate Ki67 score was determined for each subtype. There was a high level of concordance between Ki67 scores in frozen and paraffin sections (r = 0.70), but to maintain consistency with areas sampled for molecular analysis as far as possible, scores from frozen sections were used except in 3 cases where DCIS had been cut out of the frozen tissue block.
Laser capture microdissection.
DCIS foci were isolated from 10-μm serial frozen tissue sections by laser capture microdissection (PALM Microlaser Technologies AG). In addition, lobular carcinoma in situ and coexisting areas of atypical ductal hyperplasia, proliferative disease without atypia, and benign epithelium were sampled from a proportion of cases. For in situ lesions, care was taken to capture pure intraduct cell populations. Benign epithelium samples were lobular tissue collected with intralobular stroma.
Microarray analysis.
Oligonucleotide microarrays used for both gene expression and CGH experiments were the Array-Ready Oligo Set for the Human Genome Version 3.0 (Qiagen, Inc.) printed onto glass slides. This consisted of 34,580 60-mer probes representing 24,650 genes and 37,123 gene transcripts.
Details of RNA and DNA extraction, amplification, labeling, and array hybridization and analysis methods are included in Supplementary Methods. The complete microarray raw data are available through the Gene Expression Omnibus data repository, accession no. GSE 7882.
Gene expression grade index.
Expression at 173 oligonucleotide probes that showed differential expression with a false discovery rate <0.05 between DCIS associated with grade 1 and grade 3 invasive cancer were used to calculate a gene expression grade index (GGI) according to the formula of Sotiriou et al. (18). Note that the GGI is standardized such that the grade 1 associated DCIS cases had a mean GGI of −1, and grade 3 associated DCIS a mean score of 1. Each sample was left out of the standardization process to determine its own GGI.
Pathway analysis.
The probes showing differential expression between DCIS associated with grade 1 and grade 3 invasive cancer (false discovery rate <0.05) were imported into Ingenuity Pathway Analysis software (Ingenuity Systems). Probes without a valid Entrez Gene ID were excluded as were probes that did not have information available in Ingenuity, resulting in 115 unique genes. Core analysis was done, resulting in a number of high-scoring networks. The top two networks are represented in Supplementary Fig. S3A and B.
CGH analysis.
Data were segmented (19) to provide a list of discrete regions across the genome each with an associated copy number estimate (see Supplementary Methods). The randomForest package (20) in the R statistical programming language was applied to the segmented data to calculate the random forest classification and importance measures. “High level amplification” was defined as a discrete amplification exceeding a threshold corresponding to the 95% of the distribution of segment means (for all tumors, all segments) from the segmentation algorithm.
DCIS cohort.
The relationship of molecular grade to the clinical course of pure DCIS was examined by analysis of existing data from a cohort of 156 patients with DCIS treated by surgery alone with clear surgical excision margins. This cohort and its evaluation have previously been described in detail (21). Data were available to predict molecular grade in 134 of 156 cases with high molecular grade assigned on the basis of either nuclear grade 3 or an estimated percentage of cells positive for Ki67 ≥17. In 42 of 134 patients, ipsilateral recurrence of invasive or in situ breast cancer was diagnosed between 11 and 97 mo following initial surgery (median, 28.5 mo). In this report, follow-up for cases without recurrence ranged from 2 to 170 mo (median, 76.0 mo).
Statistical analysis of pathologic feature and subgroup covariates.
Most statistical analyses were done using SPSS for Windows version 12.0 (SPSS Science). A Mann-Whitney or Student’s t test was used to test for association between categorical and continuous variables. Spearman rank correlation (r) was used to quantify the degree of association between ordered categorical or continuous variables. Odds ratios were calculated using exact logistic regression analysis (LogXact 4, Cytel Software Corporation). A regression tree model (22) was used to study the relationship between gene expression groupings and DCIS histopathologic and biological predictor variables, and a 10-fold cross-validation was used to obtain an estimated error rate. The regression tree model was fitted using S-Plus version 6.2 (Insightful Corporation). Kaplan-Meier survival curves were used to illustrate the recurrence distributions by molecular grade group, and log-rank tests were used to test for significant differences in recurrence between the groups. Due to nonproportionate hazards of disease recurrence for molecular grade in the first 48 mo following DCIS surgery compared with subsequently, a time-dependent Cox model was fitted to recurrence data.
Results
Invasive breast cancers for microdissection.
This discovery phase cohort included 46 cases of invasive breast cancer: 45 with concomitant DCIS and 1 with lobular carcinoma in situ only. Tumor characteristics are summarized in Table 1 and listed in detail in Supplementary Table S1.
Table 1.
Discovery phase cohort tumor characteristics
| n (%) | |
|---|---|
|
| |
| Invasive tumor size (mm), n = 46 | |
| <5 | 5 (10.9) |
| 5–10 | 5 (10.9) |
| 10–20 | 16 (34.8) |
| 20–50 | 17 (37.0) |
| >50 | 3 (6.5) |
| Lymph node status (no. positive nodes), n = 45* | |
| 0 | 13 (28.9) |
| 1–3 | 23 (51.1) |
| 4–9 | 6 (13.3) |
| >10 | 3 (6.7) |
| Histologic subtype, n = 46 | |
| Ductal NOS | 41 (89.1) |
| Lobular, pleomorphic | 1 (2.2) |
| Cribriform | 1 (2.2) |
| Mixed † | 2 (4.3) |
| Too small to type | 1 (2.2) |
| Grade, n = 43 ‡ | |
| 1 | 14 (32.6) |
| 2 | 19 (44.2) |
| 3 | 10 (23.3) |
| Lymphovascular invasion, n = 46 | |
| Absent | 10 (21.7) |
| Present | 36 (78.3) |
| % Overall tumor comprised by in situ carcinoma, n = 46 | |
| <25 | 17 (37.0) |
| 25–50 | 15 (32.6) |
| 50–75 | 4 (8.7) |
| >75 | 10 (21.7) |
| Uniform/mixed DCIS morphology, n = 45§ | |
| Uniform | 32 (69.6) |
| Mixed | 14 (30.4) |
Abbreviation: NOS, not otherwise specified.
For 1 case, lymph node status not available.
Mixed subtypes included a mixture of NOS/tubular/cribriform and NOS/mucinous.
Three cases were too small to be graded.
Excludes one sample from a tumor with lobular carcinoma in situ only.
Gene expression profiling of microdissected DCIS.
RNA was extracted for gene expression profiling from microdissected areas of in situ carcinoma and, in a proportion of cases, adjacent atypical ductal hyperplasia, proliferative disease without atypia, and benign epithelium (Supplementary Fig. S1). Of the 14 tumors with two distinct morphologic subtypes of DCIS, both of these were present and separately captured from frozen sections in two cases. In the remaining 12 cases, one subtype only was available for molecular analysis. Histopathologic features of samples analyzed are summarized in Table 2A.
Table 2.
Histopathologic features and molecular grade of microdissected samples
| Characteristic | (A) |
(B) |
OR (95% CI) | P | |
|---|---|---|---|---|---|
| Total |
Low MG group |
High MG group |
|||
| n (%) | n (%) | n (%) | |||
|
| |||||
| Sample type (n = 61) | |||||
| Benign epithelium | 7 (11.5) | 7 (20.6) | 0 (0) | ||
| PDWA | 1 (1.6) | 1 (2.9) | 0 (0) | ||
| ADH | 4 (6.6) | 4 (11.8) | 0 (0) | ||
| LCIS* | 2 (3.3) | 1 (2.9) | 1 (3.7) | ||
| DCIS | 47 (77.0) | 21 (61.8) | 26 (96.3) | ||
| DCIS histopathologic features | |||||
| DCIS nuclear grade (n = 47 †) | |||||
| Low | 10 (21.3) | 10 (47.6) | 0 (0) | 1 | – |
| Intermediate | 18 (38.3) | 11 (52.4) | 7 (26.9) | 7.6 (0.97 to >100) | 0.05 |
| High | 19 (40.4) | 0 (0) | 19 (73.1) | 226 (22.1 to >100) | <0.0001 |
| Necrosis (n = 47 †) | |||||
| Absent/punctate | 11 (23.4) | 8 (38.1) | 3 (11.5) | 1 | – |
| Comedo | 36 (76.6) | 13 (61.9) | 23 (88.5) | 4.6 (0.89–31.4) | 0.07 |
| Architectural pattern ‡ (n = 47 †) | |||||
| Polarized | 15 (31.9) | 12 (57.1) | 3 (11.5) | 1 | – |
| Mixed | 24 (51.1) | 9 (42.9) | 15 (57.7) | 6.3 (1.2–44.6) | 0.02 |
| Nonpolarized | 8 (17.0) | 0 (0) | 8 (30.8) | 30.7 (3.7 to >100) | <0.0001 |
| Cell polarization (n = 47 †) | |||||
| Present | 39 (83.0) | 21 (100) | 18 (69.2) | 1 | – |
| Absent | 8 (17.0) | 0 (0) | 8 (30.8) | 11.9 (1.7 to >100) | 0.001 |
| Calcification (n = 47 †) | |||||
| Absent | 14 (29.8) | 8 (38.1) | 6 (23.1) | 1 | – |
| Present§ | 33 (70.2) | 13 (61.9) | 20 (76.9) | 2.0 (0.48–8.92) | 0.42 |
| Whole tumor features | |||||
| Estrogen receptor (n = 45) | |||||
| Positive | 38 (84.4) | 20 (100) | 18 (72) | 1 | – |
| Negative | 7 (15.6) | 0 (0) | 7 (28) | 9.8 (1.3 to >100) | 0.02 |
| Progesterone receptor (n = 45) | |||||
| Positive | 35 (77.8) | 19 (95.0) | 16 (64) | 1 | – |
| Negative | 10 (22.2) | 1 (5.0) | 9 (36) | 10.2 (1.2 to >100) | 0.03 |
| HER2 (n = 44∥) | |||||
| Negative (0, +, ++) | 35 (79.5) | 19 (100) | 16 (64) | 1 | – |
| Positive (+++) | 9 (20.5) | 0 (0) | 9 (36) | 13.5 (1.9 to >100) | 0.006 |
| p53 (n = 43¶) | |||||
| Negative | 35 (81.4) | 19 (100) | 16 (66.7) | 1 | – |
| Positive | 8 (18.6) | 0 (0) | 8 (33.3) | 12.0 (1.7 to >100) | 0.01 |
NOTE: (A) Features of samples microdissected from invasive breast cancers assessed by gene expression microarray analysis. (B) Features of DCIS samples in low MG and high MG groups. Odds ratios were calculated by exact logistic regression analysis.
Abbreviations: OR, odds ratio; 95% CI, 95% confidence interval; PDWA, proliferative disease without atypia; ADH, atypical ductal hyperplasia; LCIS, lobular carcinoma in situ.
Includes one sample from a tumor with LCIS only and one sample of LCIS adjacent to DCIS that was separately sampled.
Represents all DCIS lesions that were laser microdissected, including two morphologic subtypes separately sampled from two cases.
Polarized architectural patterns include cribriform, papillary, and micropapillary; nonpolarized architectural patterns include solid and comedo.
Includes coarse and fine/punctate calcification.
For 1 case, no HER2 result was obtained.
For 2 cases, no p53 result was obtained.
In support of the reliability of the gene expression data, there was a significant correlation between protein levels and relative expression at oligonucleotide probes corresponding to estrogen receptor (P < 0.0001), progesterone receptor (P < 0.0001), HER2 (P < 0.0001), and Ki67 (P < 0.0001, r = 0.83; Supplementary Fig. S2).
Grade-associated gene expression profile of DCIS.
To determine a gene expression profile that could distinguish DCIS cases according to malignant potential, supervised analysis of gene expression in DCIS associated with grade 1 invasive cancer (n = 14) versus DCIS associated with grade 3 invasive cancer (n = 9) was done. In this analysis, the predominant DCIS type only was included for cases with multiple types separately sampled (n = 2) and the single tumor with lobular carcinoma in situ only was excluded.
There was a marked difference between the two groups at the gene expression level, with significant differential expression detected at 173 oligonucleotide probes, corresponding to 146 individual genes and 13 expressed sequence tags (false discovery rate <0.05; Supplementary Table S2). Moreover, differential expression at many oligonucleotides was highly significant (false discovery rate for the top 50 probes, 0.0002–0.0078).
The molecular grade of DCIS.
Clustering of all samples according to expression at the top 100 grade-associated probes showed two principal clusters (Fig. 1A). In addition, the GGI was calculated for each sample as a standardized representation of combined expression at all 173 grade-associated probes (Fig. 1B). A GGI cutoff of 0 delineated cluster membership of all samples, with the exception of a single proliferative disease without atypia case, and was therefore set to define low molecular grade (low MG) and high molecular grade (high MG) subgroups. Histopathologic features of molecular grade subgroups are listed in Table 2B.
Fig. 1.
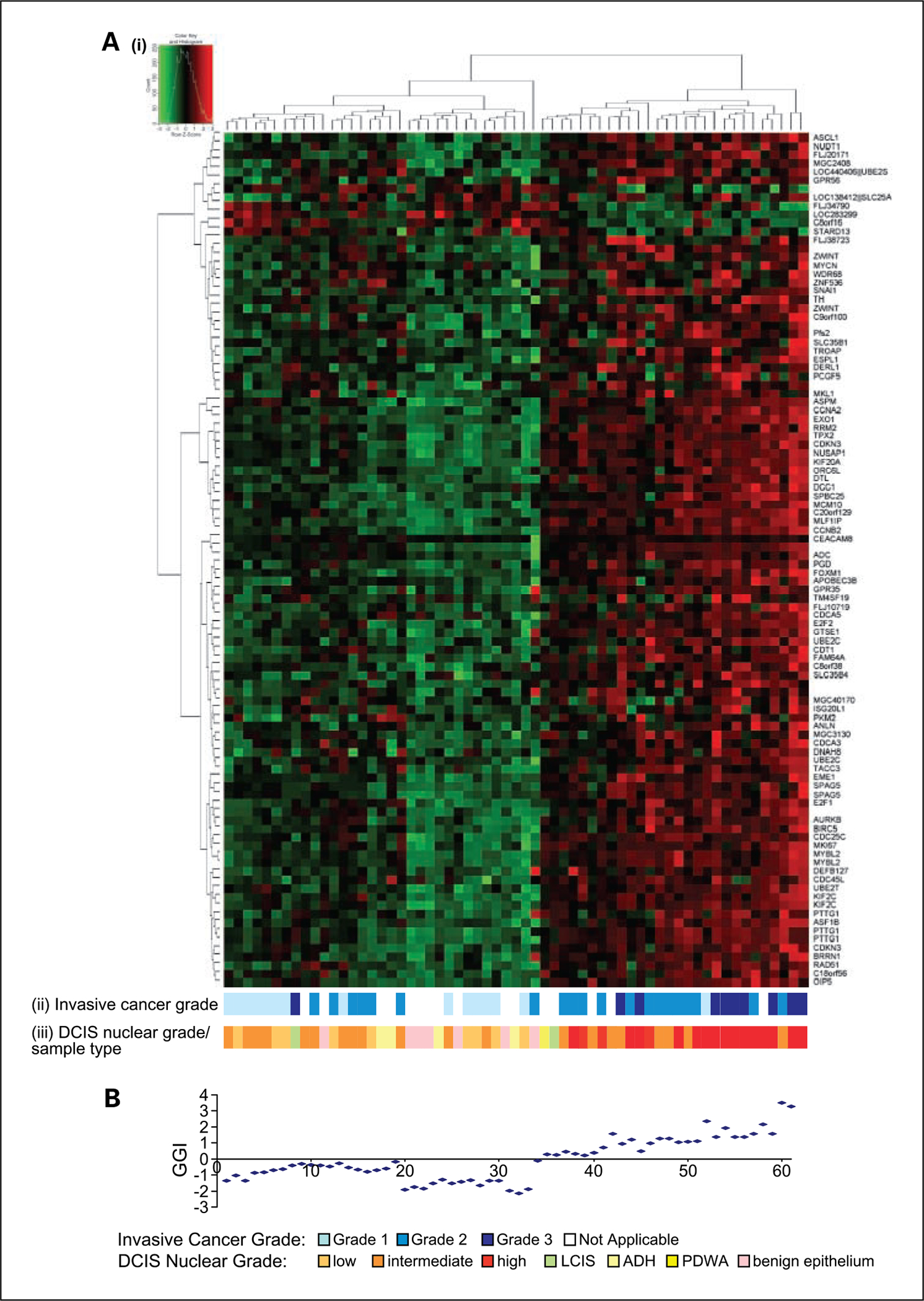
A, i, hierarchical clustering of 61 samples according to expression of the top 100 differentially expressed oligonucleotide probes (selected by supervised analysis of DCIS lesions associated with grade 1 and 3 invasive cancer). Columns represent samples; rows represent individual probes. Heatmap depicts high (red) and low (green) relative levels of gene expression. ii, grade of associated invasive breast cancer. iii, DCIS nuclear grade or sample type. B, GGI calculated for each sample in the corresponding heatmap column.
Low and high DCIS nuclear grade corresponded to molecular grade although intermediate nuclear grade DCIS was divided between low MG and high MG subgroups. Moreover, despite a statistical correlation between high MG and recognized high grade features of breast cancer, such as estrogen receptor and progesterone receptor negativity (P = 0.02, P = 0.03) and HER2 and p53 positivity (P = 0.006, P = 0.01), there was considerable variation within the high MG subgroup with respect to these features (Table 2B).
Molecular grade and proliferation.
Ranking of overrepresented functional gene categories among the 173 grade-associated probes indicated a dominant influence of cell cycle and proliferation. For example, the top three overrepresented gene ontology categories were mitotic cell cycle (EASE score P = 1.36 × 10−18), cell cycle (P = 3.71 × 10−17), and mitosis (P = 4.91 × 10−15; a full list is given in Supplementary Table S3). In keeping with this observation, there was a strong correlation between GGI and DCIS Ki67 scores (P < 0.0001, r = 0.79; Fig. 2A), and Ki67 scores were significantly different between DCIS in low MG and high MG subgroups (P < 0.0001; Fig. 2B). When the grade-associated genes were mapped to functional pathways, two major networks emerged with nodes, which are critical components of the cell cycle machinery (Supplementary Fig. S3A and B).
Fig. 2.
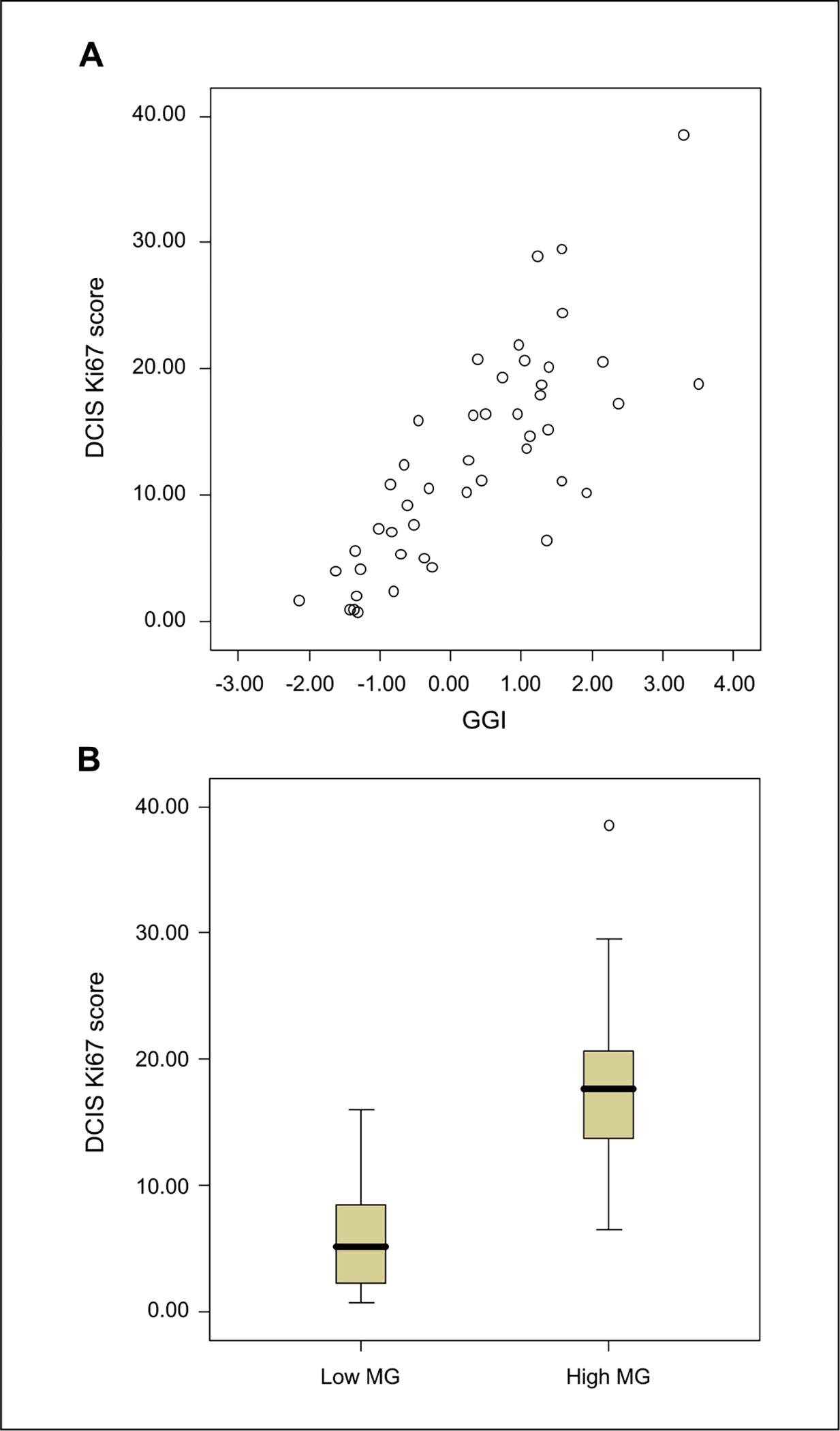
A, correlation between GGI and DCIS Ki67 score (P < 0.0001, r = 0.79, n = 46). B, Ki67 scores for DCIS samples in low MG and high MG groups (P < 0.0001, n = 46).
DNA copy number profiles of low MG and high MG DCIS.
A genomic profile correlate of the grade-associated gene expression profile was sought by comparing array-CGH profiles of in situ carcinoma associated with grade 1 (DCIS, n = 14) and grade 3 (DCIS, n = 10; lobular carcinoma in situ, n = 1) invasive breast cancer (Fig. 3A). This showed some striking differences with distinct regions of the genome differentially altered between the two groups. A random forest algorithm used to determine the importance of each CGH probe to the distinction between grade 1 and grade 3 associated lesions showed particular influence of regions on chromosomes 8, 11, and 17 with smaller contributions from other chromosomes (Fig. 3A). In addition, the number of high-level DNA amplifications was both positively correlated with GGI (P < 0.0001, r = 0.62; Fig. 3B) and significantly different between low MG and high MG DCIS subgroups (P = 0.003; Fig. 3C). These data indicated that molecular grade was related to both the character and degree of genomic aberration.
Fig. 3.
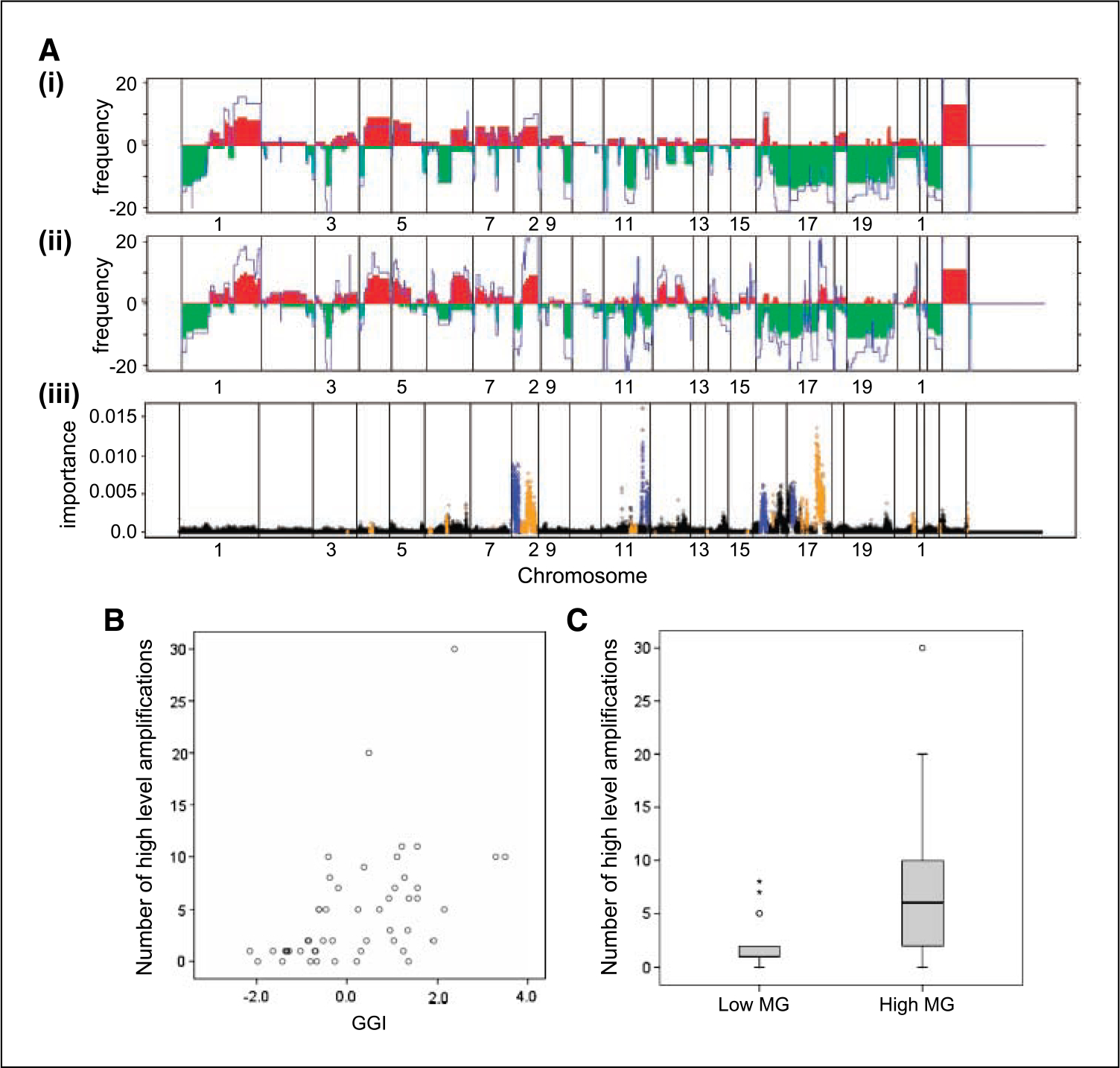
A, frequency of DNA copy number gains (red) and losses (green) across the genome (plotted from chromosome 1pter to 22qter, X and Y) in in situ carcinoma associated with grade 1 (i) and grade 3 (ii) invasive cancer. Average log 2 ratio copy number compared with normal male reference DNA is shown in blue. iii, representation of random forest algorithm applied to determine the importance measure for each probe in distinguishing lesions associated with grade 1 or grade 3 invasive cancer. Points colored in blue highlight regions that have a much higher copy number in grade 1 cases than in grade 3; points in orange have a higher copy number in grade 3 than grade 1. Higher copy number is defined as a difference of 0.25 mean log 2 ratio or larger. B, correlation between the number of regions of high-level DNA amplification and GGI (P < 0.0001, r = 0.62, n = 50). C, number of regions of high-level DNA amplification in low MG and high MG subgroups of DCIS (P = 0.003, n = 46).
A combination of histopathologic and biomarker features can predict DCIS molecular grade.
Histopathologic and biomarker features of DCIS were examined to determine whether routinely accessible information could be used to predict molecular grade. DCIS nuclear grade, necrosis, cell polarization, estrogen receptor, progesterone receptor, HER2, Ki67, and p53 were considered for inclusion in a classification tree model (22). The tree based on DCIS nuclear grade and Ki67 score (Fig. 4A) was an accurate predictor, with 44 of 46 (95.7%) cases correctly reassigned to their original MG subgroup; the 10-fold cross-validated error rate for this predictor was 6.52%. These data indicated that a combination of DCIS nuclear grade and Ki67 score could be used to assign a DCIS sample as either low MG or high MG.
Fig. 4.
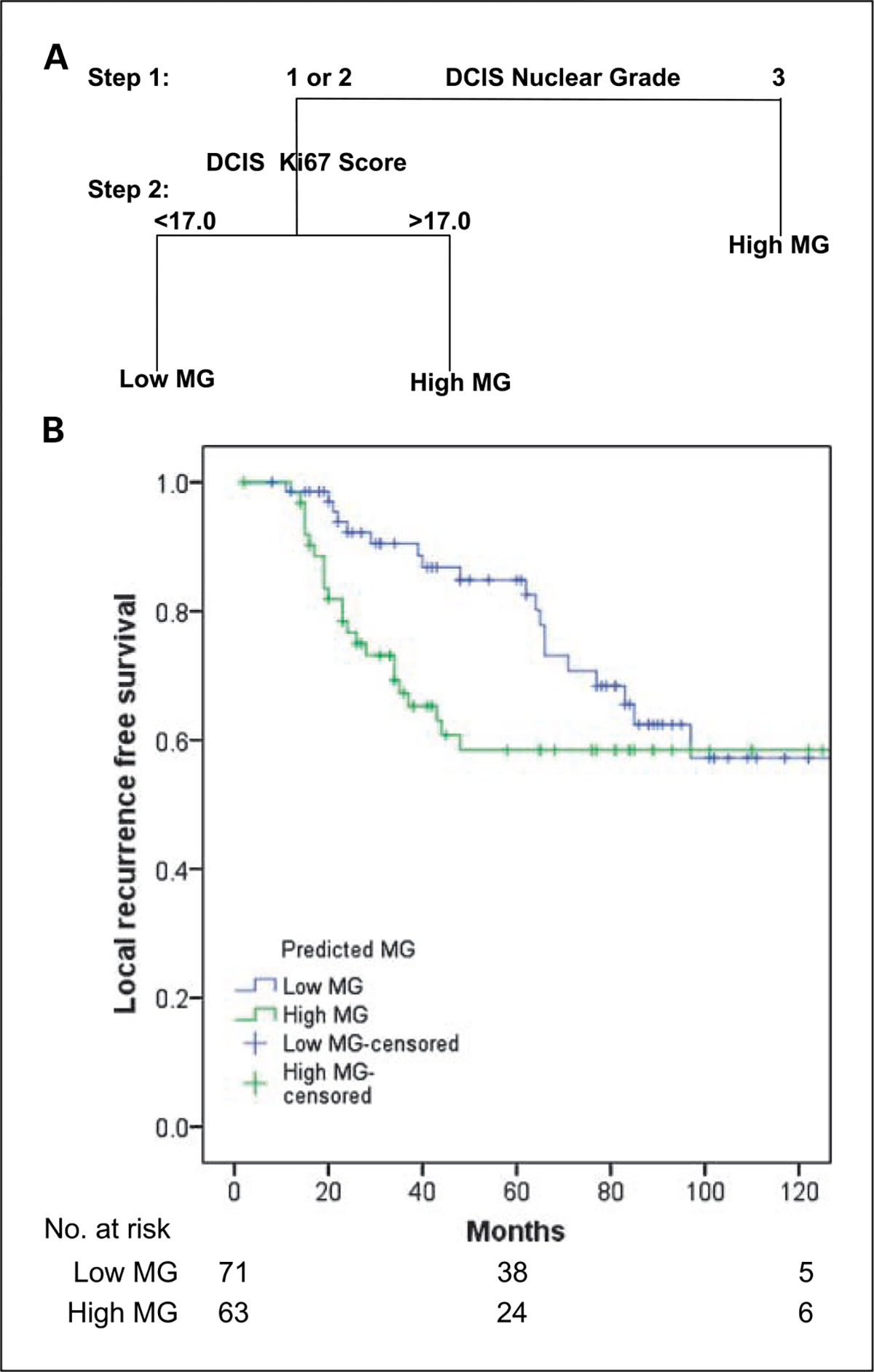
A, two-step classification tree model illustrating prediction of DCIS molecular grade by histopathologic and biomarker features. B, local-recurrence -free survival for patients with DCIS of low MG and high MG predicted by nuclear grade and Ki67 score (n = 134).
Molecular grade and ipsilateral disease recurrence in DCIS treated by surgery alone.
To examine the relationship between DCIS molecular grade and clinical course, ipsilateral recurrence of in situ or invasive breast cancer was examined in a cohort of 134 DCIS cases treated by surgery alone. Molecular grade was assigned to these cases on the basis of existing nuclear grade and Ki67 score data.
Overall, recurrence was observed in 19 of 71 (26.8%) low MG cases and 23 of 63 (36.5%) high MG. There was no significant difference between the distribution of recurrence in the two subgroups (P = 0.1; Fig. 4B). However, a striking difference in the shape of the two local-recurrence–free survival curves indicated nonproportionate hazards over time with behavior in the first 48 months different from the subsequent period. Consistent with this, a time-dependent Cox model showed that the hazard for disease recurrence was 3.5 times higher for high MG versus low MG in the first 48 months (95% confidence interval, 1.6–7.9; P = 0.002). In contrast, after 48 months, recurrence risk was reversed, with the hazard ratio for high MG versus low MG being 0.14 (95% confidence interval, 0.02–1.0; P = 0.056). These results were virtually unchanged when adjusted for lesion size (data not shown).
Discussion
In this study, gene expression profiling was used to determine genes differentially expressed between DCIS associated with grade 1 and grade 3 invasive breast cancer. A binary low MG/high MG classification based on expression at grade-associated oligonucleotide probes classified benign epithelium and atypical ductal hyperplasia as low grade and importantly divided DCIS into low MG and high MG subgroups. Different regions of DNA aberration and rates of high-level amplification associated with molecular grade indicated that the gene-expression – based classification was reflective of essential differences in malignant phenotype.
The use of invasive cancer grade as a surrogate end point in the study of DCIS biology has some acknowledged limitations. In particular, use of specimens in which DCIS and invasive cancer are both present precludes examination of DCIS that would never have progressed to invasion. However, it is reasonable to expect that such cases would be classified as low MG as was atypical ductal hyperplasia in the current report. Further evidence in support of the applicability of the grade-associated gene expression profile determined in this study to pure DCIS comes from three cases with extensive DCIS associated with only a minor invasive cancer component. One of these with intermediate DCIS nuclear grade clustered with the low-grade cases and the other two, one intermediate and the other high nuclear grade, clustered with the high-grade group. Classification of these cases by gene expression profiling was both appropriate relative to other cases in the cohort and potentially informative given the distinction of two intermediate nuclear grade cases.
Discrimination of DCIS into low MG and high MG clearly shows the feasibility of an informative biological classification of DCIS, and omission of the intermediate grade category is a major improvement on other proposed DCIS grading schemes (23, 24). Moreover, the difficulty of arriving at such a classifier by histopathologic assessment is apparent from the diversity of individual pathologic features in each MG subgroup. For example, the presence of comedo type necrosis, which has been an influential indicator of high grade in many proposed histopathologic DCIS grading schemes (9), was present in 61.9% of cases in the low MG subgroup. Area-to-area morphologic heterogeneity is a further characteristic of DCIS that has frustrated attempts to devise a robust histopathologic classification scheme (25, 26). This was evident in the current study with two distinct forms of DCIS identified in 14 of 46 (30.4%) tumors. Delineation of individual subtypes and their separate sampling by laser capture microdissection greatly increased the precision of molecular analysis, which proved to be a major advantage in formulation of a DCIS classifier. This is especially apparent by comparison to the report of Hannemann et al. (27) in which DCIS gene expression profiles derived from whole tissue sections gave only limited resolution of intermediate grade cases.
Relatively few gene expression profiling studies of DCIS have been published to date and most have focused on identification of progression-associated genes by comparison of in situ and invasive disease (28–30). In a study by Ma et al. (30), the existence of a grade-associated gene expression profile was clearly shown in microdissected samples of atypical ductal hyperplasia, DCIS, and invasive cancer. The list of grade-associated genes identified by these investigators showed little overlap with our own and did not clearly resolve intermediate grade cases. However, these differences may be attributable to the fact that Ma et al. restricted the analysis to genes showing distinct expression in abnormal compared with normal breast to emphasize aspects of disease progression. In contrast, our list of 173 grade-associated probes shows striking concordance with 128 probe sets showing differential expression between grade 1 and grade 3 invasive cancer reported by Sotiriou et al. (18). In this instance, there were 50 common elements, including 18 of the 35 (51.4%) most significant differentially expressed from our study. Analogous to our findings in DCIS, Sotiriou et al. reported that a grade-associated gene expression profile provided a clinically meaningful resolution of intermediate grade invasive cancer; a finding that has been supported by a subsequent report (31). CGH data provided additional evidence of biological differences between low MG and high MG DCIS, which differed with respect to both the specific loci altered and the frequency of high-level amplifications.
Overexpression of genes involved in cellular proliferation in high-grade cases was a dominant feature of our grade-associated gene list and, furthermore, is consistent with findings from studies that have compared high- and low-grade cancers from a variety of tissues (32). This feature enabled a simple approach to prediction of MG subgrouping because a classification tree model showed that 44 of 46 (95.7%) DCIS samples in the discovery phase cohort could be accurately assigned to the low MG or high MG subgroup on the basis of Ki67 score and/or nuclear grade. In practice, molecular profiling approaches to determination of molecular grade have the clear advantage of objective measurement and applicability to small biopsy samples. However, prediction of molecular grade by a combination of routinely accessible markers offers a practical clinical reporting alternative that may be especially useful for assessment of therapeutic excision specimens.
A limitation of the discovery phase of this study was that all of the DCIS samples were associated with invasive breast cancer. To verify that the molecular grade classification was applicable to pure DCIS, it was examined in a historic cohort of 134 patients, allocated to low MG and high MG subgroups on the basis of previously reported nuclear grade and Ki67 scores. Particular advantages of this cohort were uniform treatment by local excision alone and a long follow-up period (21). Overall, 71 of 134 (53%) cases were designated low MG, and the remaining 63 (47%) high MG. A striking finding was dramatic differences in the pattern of disease recurrence over time between the two subgroups. All recurrences for high MG DCIS occurred within 48 months of initial surgery. In contrast, time to disease recurrence in low MG DCIS was prolonged to the extent that, after 48 months, the hazard for recurrence was somewhat higher for low MG compared with high MG cases. Overall, these data suggest that the molecular grade classification is indicative of essential differences in the natural history and clinical course of DCIS with particular implications for interpretation of recurrence risk over time.
This result indicating different rates of progression for high MG and low MG DCIS is consistent with previous studies. For example, in 1996 Solin et al. reported a shorter median time to recurrence following treatment by local excision and radiotherapy for DCIS that was nuclear grade 3 with comedo necrosis compared with all other cases (33). In addition, the slow progression of low MG DCIS is consistent with data from long-term follow-up of untreated low-grade DCIS that has shown persistence and progression of disease over a period of decades (34). In consideration of these data, the clear advantage of the molecular grade classification is apparent because it allocated all cases of DCIS to a subgroup with distinctive outcome, including not only straightforward high- and low-grade cases but also those with intermediate pathology.
A robust indicator of DCIS at high risk of recurrence following local excision would be of considerable benefit in clinical practice. In this study, disease recurrence was recorded for 42 of 134 (31.3%) patients with DCIS, and this proportion was similar in both the low MG and high MG subgroups by the end of the follow-up period. This finding is consistent with earlier studies showing that the incidence of recurrence for DCIS with high grade features becomes equivalent to other types with long-term follow-up (33, 35). It is also consistent with data suggesting that ipsilateral disease recurrence following breast conserving surgery for DCIS is related to the completeness of local eradication irrespective of the pathology of the lesion. For example, in both the National Surgical Adjuvant Breast Project B-17 and European Organization for Research and Treatment of Cancer 10853 randomized controlled trials of radiotherapy following local excision for treatment of DCIS, the rate of ipsilateral recurrence was reduced in all pathologic subcategories of DCIS by addition of radiotherapy (36–38). Furthermore, the importance of surgical margin clearance as a predictor of recurrence was shown in both of these studies. On the basis that disease recurrence following local treatment of DCIS may be a largely physical rather than a biological consequence, a robust biological predictor of recurrence may not be feasible. Moreover, studies such as the recent report by Gauthier et al., comparing DCIS cases with and without recurrence to identify such markers, need to be interpreted with a view to the particular confounding effects of treatment and unmeasurable residual disease on this end point (39).
A means to predict the clinical course of DCIS has important implications for the interpretation and management of this increasingly important condition. In our study of gene expression profiles, we determined a signature that clearly distinguishes between DCIS subgroups of differing malignant potential. The ability of histopathologic nuclear grade and Ki67 score to essentially recapitulate this molecular classification suggests a novel DCIS grading scheme for routine use that has the advantage of practical application and the support of an objective and fully integrated analysis of biology.
Supplementary Material
Translational Relevance.
In this study, we have used both gene expression and genomic profiling of microdissected ductal carcinoma in situ (DCIS) samples to devise a novel system of "molecular grading" that clearly distinguished high- and low-grade lesions. We also present an accurate predictor of molecular grade based on routinely accessible pathologic features of DCIS that could facilitate application in a clinical setting. In an independent cohort of 134 cases of DCIS treated by surgical excision alone, this molecular grade predictor revealed distinct patterns of disease progression for high and low molecular grade DCIS with relatively rapid recurrence of high molecular grade lesions.
Acknowledgments
We thank Michael Bilous, Paul Harnett, and Stan Lipkowitz for helpful comments on different aspects of this work.
Sections of this article appear in a patent specification detailing the invention entitled "Molecular Grading Methods." The patent specification has been submitted to the U.S. Patent and Trademark Office as both a Provisional Patent Application and as a Patent Cooperation Treaty Application (U.S. Patent Application Serial No. 60/ 936,526). Co-owners of the invention are the NIH, Sydney West Area Health Service, and the University of Sydney. The specification has not been published and is currently not in the public domain.
As a co-owner of the copyright of sections of the article that appear in the above named patent specification, Sydney West Area Health Service hereby grants the American Association for Cancer Research Inc. an exclusive license to reproduce this material in the journal Clinical Cancer Research for the purpose of publishing the article “Molecular grading of ductal carcinoma in situ of the breast.”
Grant support:
Project grant support from the Cure Cancer Australia Foundation and National Health and Medical Research Council project grant 306700 (R.L. Balleine, L.R. Webster, and C.L. Clarke). L.R. Webster was recipient of a National Breast Cancer Foundation Scholarship (supported by the Esteée Lauder group of companies) and travel grants from The University of Sydney (Grants-in-Aid) and The Australian Federation of University Women-South Australia Inc. Trust Fund (Jean Gilmore Bursary). She was a Cancer Institute New South Wales Research Scholar. R.L. Balleine is a Cancer Institute New South Wales Fellow, and C.L. Clarke a Principal Research Fellow of the National Health and Medical Research Council.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
References
- 1.Li CI, Daling JR, Malone KE. Age-specific incidence rates of in situ breast carcinomas by histologic type, 1980 to 2001. Cancer Epidemiol Biomarkers Prev 2005;14:1008–11. [DOI] [PubMed] [Google Scholar]
- 2.Ernster VL, Ballard-Barbash R, Barlow WE, et al. Detection of ductal carcinoma in situ in women undergoing screening mammography. J Natl Cancer Inst 2002;94:1546–54. [DOI] [PubMed] [Google Scholar]
- 3.Ernster VL, Barclay J, Kerlikowske K, Wilkie H, Ballard-Barbash R. Mortality among women with ductal carcinoma in situ of the breast in the population-based surveillance, epidemiology and end results program. Arch Intern Med 2000;160:953–8. [DOI] [PubMed] [Google Scholar]
- 4.Duffy SW, Agbaje O, Tabar L, et al. Overdiagnosis and overtreatment of breast cancer: estimates of overdiagnosis from two trials of mammographic screening for breast cancer. Breast Cancer Res 2005;7:258–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burstein HJ, Polyak K, Wong JS, Lester SC, Kaelin CM. Ductal carcinoma in situ of the breast. N Engl J Med 2004;350:1430–41. [DOI] [PubMed] [Google Scholar]
- 6.Elledge RM, McGuire WL, Osborne CK. Prognostic factors in breast cancer. Semin Oncol 1992;19:244–53. [PubMed] [Google Scholar]
- 7.Elston CW, Ellis IO, Pinder SE. Pathological prognostic factors in breast cancer. Crit Rev Oncol Hematol 1999;31:209–23. [DOI] [PubMed] [Google Scholar]
- 8.van deVijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 2002;347:1999–2009. [DOI] [PubMed] [Google Scholar]
- 9.Shoker BS, Sloane JP. DCIS grading schemes and clinical implications. Histopathology 1999;35:393–400. [DOI] [PubMed] [Google Scholar]
- 10.Patchefsky AS, Schwartz GF, Finkelstein SD, et al. Heterogeneity of intraductal carcinoma of the breast. Cancer 1989;63:731–41. [DOI] [PubMed] [Google Scholar]
- 11.Bobrow LG, Happerfield LC, Gregory WM, Millis RR. Ductal carcinoma in situ: assessment of necrosis and nuclear morphology and their association with biological markers. J Pathol 1995; 176:333–41. [DOI] [PubMed] [Google Scholar]
- 12.Silverstein MJ, Lagios MD, Groshen S, et al. The influence of margin width on local control of ductal carcinoma in situ of the breast. N Engl J Med 1999;340: 1455–61. [DOI] [PubMed] [Google Scholar]
- 13.Schatzkin A, Gail M. The promise and peril of surrogate end points in cancer research. Nat Rev Cancer 2002;2:19–27. [DOI] [PubMed] [Google Scholar]
- 14.Leong AS, Sormunen RT, Vinyuvat S, Hamdani RW, Suthipintawong C. Biologic markers in ductal carcinoma in situ and concurrent infiltrating carcinoma. A comparison of eight contemporary grading systems. Am J Clin Pathol 2001;115:709–18. [DOI] [PubMed] [Google Scholar]
- 15.Webster LR, Bilous AM, Willis L, et al. Histopathologic indicators of breast cancer biology: insights from population mammographic screening. Br J Cancer 2005;92:1366–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 1991;19:403–10. [DOI] [PubMed] [Google Scholar]
- 17.Consensus Conference Committee. Consensus Conference on the classification of ductal carcinoma in situ. The Consensus Conference Committee. Cancer 1997;80:1798–802. [DOI] [PubMed] [Google Scholar]
- 18.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 2006;98:262–72. [DOI] [PubMed] [Google Scholar]
- 19.Lipson D, Aumann Y, Ben-Dor A, Linial N, Yakhini Z. Efficient calculation of interval scores for DNA copy number data analysis. J Comput Biol 2006;13:215–28. [DOI] [PubMed] [Google Scholar]
- 20.Liaw A, Wiener M. Classification and regression by randomForest. R News 2002;2:18–22. [Google Scholar]
- 21.Cornfield DB, Palazzo JP, Schwartz GF, et al. The prognostic significance of multiple morphologic features and biologic markers in ductal carcinoma in situ of the breast: a study of a large cohort of patients treated with surgery alone. Cancer 2004; 100:2317–27. [DOI] [PubMed] [Google Scholar]
- 22.Brieman L, Friedman JH, Olshen RA, Stone CJ. Classification and regression trees. Belmont (CA): Wadsworth; 1984. [Google Scholar]
- 23.Holland R, Peterse JL, Millis RR, et al. Ductal carcinoma in situ: a proposal for a new classification. Semin Diagn Pathol 1994;11:167–80. [PubMed] [Google Scholar]
- 24.Silverstein MJ, Poller DN, Waisman JR, et al. Prognostic classification of breast ductal carcinoma-in-situ. Lancet 1995;345:1154–7. [DOI] [PubMed] [Google Scholar]
- 25.Quinn CM, Ostrowski JL. Cytological and architectural heterogeneity in ductal carcinoma in situ of the breast. J Clin Pathol 1997;50:596–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison M, Coyne JD, Gorey T, Dervan PA. Comparison of cytomorphological and architectural heterogeneity in mammographically-detected ductal carcinoma in situ. Histopathology 1996;28:445–50. [DOI] [PubMed] [Google Scholar]
- 27.Hannemann J, Velds A, Halfwerk JB, Kreike B, Peterse JL, van de Vijver MJ. Classification of ductal carcinoma in situ by gene expression profiling. Breast Cancer Res 2006;8:R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishidate T, Katagiri T, Lin ML, et al. Genome-wide gene-expression profiles of breast-cancer cells purified with laser microbeam microdissection: identification of genes associated with progression and metastasis. Int J Oncol 2004;25:797–819. [PubMed] [Google Scholar]
- 29.Schuetz CS, Bonin M, Clare SE, et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res 2006;66:5278–86. [DOI] [PubMed] [Google Scholar]
- 30.Ma XJ, Salunga R, Tuggle JT, et al. Gene expression profiles of huma nbreast cancer progression. Pro cNatl Acad Sci U S A 2003;100:5974–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivshina AV, George J, Senko O, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res 2006;66: 10292–301. [DOI] [PubMed] [Google Scholar]
- 32.Rhodes DR, Yu J, Shanker K, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptiona lprofiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A 2004;101:9309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solin LJ, Kurtz J, Fourquet A, et al. Fifteen-year results of breast-conserving surgery and definitive breast irradiation for the treatment o fductal carcinoma in situ of the breast. J Clin Oncol 1996;14:754–63. [DOI] [PubMed] [Google Scholar]
- 34.Sanders ME, Schuyler PA, Dupont WD, Page DL. The natural history of low-grade ductal carcinoma in situ of the breast in women treated by biopsy only revealed over 30 years of long-term follow-up. Cancer 2005;103:2481–4. [DOI] [PubMed] [Google Scholar]
- 35.Silverstein MJ, Cohlan BF, Gierson ED, et al. Duct carcinoma in situ: 227 cases without microinvasion. Eur J Cancer 1992;28:630–4. [DOI] [PubMed] [Google Scholar]
- 36.Fisher ER, Dignam J,Tan-Chiu E, et al. Pathologic findings from the National Surgical Adjuvant Breast Project (NSABP) eight-year update of Protocol B-17: intraductal carcinoma. Cancer 1999;86:429–38. [DOI] [PubMed] [Google Scholar]
- 37.Fisher B, Dignam J,Wolmark N, et al. Lumpectomy and radiation therapy for the treatment of intraductal breast cancer: findings from National Surgical Adjuvant Breast and Bowel Project B-17. J Clin Oncol 1998;16:441–52. [DOI] [PubMed] [Google Scholar]
- 38.Bijker N, Meijnen P, Peterse JL, et al. Breast-conserving treatment with or without radiotherapy in ductal carcinoma-in-situ: ten-year results of European Organisation for Research and Treatment of Cancer randomized phase III Trial 10853-A study by the EORTC Breast Cancer Cooperative Group and EORTC Radiotherapy Group. J Clin Oncol 2006;24: 3381–7. [DOI] [PubMed] [Google Scholar]
- 39.Gauthier ML, Berman HK, Miller C, et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell 2007;12:479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


