Abstract
To assess the role of interferon regulatory factor (IRF) 8 in B-cell development and lymphomagenesis, we studied its expression in reactive lymphoid tissues, its relationship to other B-cell transcription factors, and its expression in a series of 232 B-cell tumors and 30 cell lines representing a variety of B-cell developmental stages. We found that although IRF8 was detectable in most reactive B-cells, its expression levels differed with developmental stage. Germinal center B cells contained the highest levels of IRF8, with lower levels seen in mantle and marginal zone B cells and none in plasma cells. IRF8 was coexpressed with PAX-5, Pu.1, and B-cell lymphoma (BCL)-6, and similar to BCL-6, was absent from the small population of IRF4-positive germinal center B cells thought to be committed to postgerminal center developmental programs. Similarly, IRF8 was most strongly expressed in lymphomas of germinal center origin with lower levels present in mantle cell lymphomas, chronic lymphocytic leukemia, and marginal zone lymphomas, and no expression observed in plasmacytic/plasmablastic neoplasms. The reciprocal expression pattern with IRF4 in reactive tissues was generally maintained in lymphomas with some exceptions. These results suggest an important role for IRF8 during germinal center B-cell development and in related lymphomas, and provide a new diagnostic marker helpful in distinguishing B-cell non-Hodgkin lymphoma subtypes.
Keywords: IRF8/ICSBP-1, IRF4/MUM1, lymphoma, germinal centers
The interferon regulatory factors (IRFs) are a family of related transcription factors involved in the regulation of the target genes in response to interferons (IFNs). The human family contains at least 10 members with an identical helix-loop-helix DNA-binding motif.26,34 Although IRFs are widely expressed, IRF8/interferon consensus sequence-binding protein-1 (ICSBP-1) and IRF4/multiple myeloma oncogene-1 (MUM-1) also designated as lymphocyte-specific interferon regulatory factor/interferon consensus sequence binding protein for activated T-cells/Pip are almost exclusively expressed in the cells of hematopoietic lineage.16,22 IRF8 and IRF4 are structurally homologous, exhibit a low DNA-binding affinity, and require the interaction and cooperation of an E26 transformation-specific transcription factor family member, such as Pu.1, to regulate target promoters.
IRF4 is primarily expressed in B cells and activated T lymphocytes.22 Unlike other members of the IRF family, IRF4 is not IFN responsive, but instead, seems to be modulated by stimuli known to induce lymphocyte activation and differentiation, such as conconavalin A, staphylococcal enterotoxin A, lipopolysaccharide, interleukin (IL)-4, engagement of the CD40 receptor, and engagement of the antigen receptor.10,18,20 In B cells, IRF4 is expressed in a small subset of germinal center cells committed to plasmacytic or memory cell differentiation, and in plasma cells and lymphomas of putative postgerminal center derivation.8 IRF4 is an important regulator of B-cell development and IRF4-deficient mice lack germinal centers, plasma cells, and exhibit profound hypogammaglobulinemia.19
In contrast to IRF4, IRF8 has been shown to play major roles in myeloid and monocytic lineage development and function, by stimulating macrophage differentiation and inhibiting granulocyte differentiation.33 Mice deficient in IRF8 develop a chronic myeloid leukemia (CML)-like syndrome,13 and decreased expression of IRF8 has also been also found in human CML, suggesting that IRF8 may function as a tumor suppressor in this disease.29 More recent studies have shown that IRF8 also has a major role in the development and function of dendritic cells.1,27,28 Few efforts have focused on the role of IRF8 in lymphoid cells. Early studies of IRF8 indicated that in mice it was constitutively expressed in mature B cells and could be induced in T cells by activation with mitogens.7,20 In addition, mouse B-cell lymphoma (BCL) but not myeloma cell lines were also shown to express IRF8.7 Data from Lu and colleagues17 suggested a cooperative role of IRF8 and IRF4 in the coordination of the transition from pre-B to mature B cell during early B-cell development in mice. Intriguingly, the IRF8-deficient mice discussed above showed an excess of terminally differentiated B cells and plasma cells, suggesting that IRF8 was also important in modulating later stages of B-cell development. Consistent with a role in late B-cell development, Lee and colleagues15 recently demonstrated that IRF8 is expressed in germinal center B cells and capable of up-regulating BCL-6, a critical germinal center transcription factor.
To characterize the role of IRF8 in human B-cell development and lymphomagenesis, we studied its expression in normal reactive B-cells, its relationship to other well-known B-cell transcription factors, such as PAX-5, BCL-6, and Pu.1, and its expression in a series of 232 primary B-cell tumors and 30 cell lines representing a broad range of B-cell developmental stages. We found that IRF8 was highly expressed in most germinal center B cells, with lower levels of expression seen in mantle and marginal zone B cells, and an absence of expression in plasma cells. Similar to its expression pattern in reactive lymphoid tissue, IRF8 was most highly expressed in lymphomas of follicle center origin, at lower levels in mantle cell lymphomas (MCL), marginal zone lymphomas (MZL), and chronic lymphocytic leukemia (CLL), and was absent in postgerminal center plasmacytic lymphomas. Although IRF8 was coexpressed with PAX-5, Pu.1, and BCL-6 in germinal centers, a reciprocal expression pattern was observed with IRF4 in reactive B cells and in the majority of B-cell neoplasms.
MATERIALS AND METHODS
Tissue Specimens
Lymphoma/leukemia specimens from 232 patients were obtained from the files of the Hematopathology Section, Laboratory of Pathology of the National Cancer Institute (NCI) (Bethesda, MD). Tumors were diagnosed according to the World Health Organization classification as precursor B-lymphoblastic leukemia/lymphoma (LB) (5 cases), Burkitt lymphoma (BL) (8 cases), follicular lymphoma (FL) (46 cases), CLL (30 cases), lymphoplasmacytic lymphoma (LPL) (8 cases), MCL (28 cases), MZL (21 cases), diffuse large B-cell lymphoma (DLBCL) (58 cases), plasmablastic lymphoma (PBL) (14 cases including 1 primary effusion lymphoma), and multiple myeloma (MM) (14 cases). The DLBCLs were further divided into germinal center-like B-cell lymphomas (GCB) and non-GCB–like lymphomas using CD10, BCL-6, and IRF4 immunohistochemistry, according to the criteria of Hans et al.11 By these criteria, there were 39 GCB, 13 non-GCB cases, and 6 were unclassifiable because of insufficient (immunostain) classifiers. In addition, reactive lymphoid tissue from lymph node, spleen, and tonsil were also studied. At least 3 parts of each reactive tissue were studied. All tissues used were fixed in 10% buffered formalin and processed routinely for paraffin embedding. Of the 232 tissues analyzed, 62 samples were part of a tissue microarray prepared by the NCI Tissue Array Research Program facility.
Cell Lines and Culture Conditions
The following 32 human tumor cell lines (30 of B-cell origin) were studied for IRF8 and IRF4 expression: RPMI-9837 (B-cell acute lymphoblastic leukemia); SUDHL 4 to 7 and 10 (DLBCL, GCB-like), OCI-Ly3, and OCI-Ly10 (DLBCL, ABC-like); ST-486, Ramos, Raji, BJAB, and JD38 (BL); REC-1, Granta-519, Z-138, and NCEB (MCL); ESKOL (hairy cell leukemia); U937 (myelomonocytic leukemia); KMM-1, KMS-11, KMS-12-BM, KMS-12-PE, KMS-18 and KMS-20, JIM-3, and OPM-2 (MM); BCLB-1, BC-1, BC-2, BC-3 (primary effusion lymphoma), and MCF-7 (breast carcinoma). All cell lines were grown in RPMI 1640 supplemented with 10% fetal bovine serum, 2-mM glutamine and 50-mg/mL penicillin–streptomycin (Gibco BRL, Gaithersburg, MD) at 37°C in a humidified atmosphere containing 5% CO2.
Immunohistochemistry and Confocal Microscopy
Immunohistochemistry was performed on formalin-fixed paraffin-embedded tissue sections, as previously described.25 Briefly, paraffin sections on silane-coated slides were dewaxed and subjected to antigen retrieval for 8 minutes in a buffer solution at pH 6.1 (Target Retrieval Solution, Dako, Carpinteria, CA) in a microwavable pressure cooker. Primary antibodies were incubated for 2 hours at room temperature, using an automated immunostainer (Dakoautostainer, Dako) and a peroxidase-based detection system (Envision Plus System, Dako) and 3,3’-diaminobenzidine (DAB) as a chromogen. The slides were counterstained in Gill hematoxylin and mounted in Pertex (Histolab GmbH, Göteborg, Germany). A rabbit polyclonal antibody (1:200) raised against human recombinant ICSBP-1 [ICSBP (C-19); sc-6058] (Santa Cruz Biotechnology, CA) was used to detect IRF8 and a mouse monoclonal (1:200) antihuman MUM-1p (Dako, Carpinteria, CA; 1:200 dilution) was used for detection of IRF4, as previously described.8 Cases were scored as positive for IRF8 and IRF4 when more than 20% of the tumor cells were positive at any intensity level. A minimum of 500 tumor cells per case were counted in representative areas of the tumors, except in rare cases from the tissue microarray in which there were an insufficient number of tumor cells. Additional antibodies used in this study were CD20 (clone L26) (Dako, Carpinteria, CA; 1:1000 dilution), CD3 (clone F7.2.38) (Dako; 1:20 dilution), CD10 (clone 56C6) (Dako; 1:40 dilution), CD68 (clone KP-1) (Dako; 1:50 dilution), BCL-6 (clone PG-B6p) (Dako; 1:400 dilution), PAX-5 (clone 24) (Becton-Dickinson Biosciences Pharmingen, San Jose, CA; 1:100 dilution), and Pu.1 (clone G148–74) (Becton-Dickinson Biosciences Pharmingen; 1:50 dilution).
Double-staining immunohistochemistry for coexpression studies was performed in representative reactive and tumor samples. Sequential primary incubations and sequential detections using a peroxidase-based detection kit (Envision Plus System, Dako) were used in all cases with DAB as the first chromogen and VIP (Vector Laboratories, Burlingame, CA) as the second chromogen, followed by a methyl green (Vector Laboratories) counterstain. An additional peroxidase block was added after the first detection.
Immunofluorescence
Immunofluorescence was performed on formalin-fixed paraffin-embedded tissue sections. After antigen retrieval, the slides were blocked with 10% normal goat serum and incubated overnight at 4°C with anti–ICSBP-1. After washing, the slides were incubated at room temperature for 1 hour with a Cy3-labeled goat antirabbit (Jackson ImmunoResearch Laboratories Inc, PA). The slides were rinsed and reblocked with 10% normal goat serum and a secondary incubation with anti–MUM-1p, PAX-5, or Pu.1 was performed for 2 hours at room temperature. A Cy2-labeled goat antimouse was applied for 1 hour in the dark and mounted with an aqueous mounting media (Fluorescent Mounting Medium, Dako). A Bio-Rad MRC 1024 Krypton/Argon laser confocal microscope (Nikon OPTiHOT-2, Japan) and LaserSharp 2000, 5.1 build 775 image software analysis was used to evaluate antigen localization (Bio-Rad Cell Science Division, Hemel Hempstead, UK).
For quantitative analysis of relative IRF8 content of follicular B-cell subpopulations, paraffin-embedded formalin fixed tissue sections were stained as above, except that a fluorescein (fluorescein isothiocyanate)-conjugated goat antirabbit or rhodamine (tetramethylrhodamine isothiocyanate)-conjugated goat antirabbit secondary (Jackson ImmunoResearch Laboratories Inc) was substituted for the Cy3-labeled antirabbit secondary. Images were acquired using a Zeiss LSM 510 NLO confocal system mounted on an Axiovert 200 M microscope with a Zeiss Plan-Apochromat 63 × /1.4 NA oil DIC objective (Carl Zeiss Inc, Thornwood, NY). All acquired images used the helium neon 543-nm laser and a 2-photon laser tuned to 760 nm for detection of IRF8 and 4’,6-diamidino-2-phenylindole, respectively. Confocal images were acquired using the optimal focal plane for the brightest signal-to-noise ratio without including saturated pixels. Acquired images of mantle and germinal center B-cells were further analyzed using Image-Pro Plus software version 5.1.2 (Media Cybernetics Inc Silver Spring, MD). Each image, using identical settings for all, was processed using a fluorescent density picker to outline each cell. All outlined cells were then counted and sorted by fluorescent densities (25 to 254 arbitrary units) and finally binned into their respective mean density classes.
Western Blot
Exponentially growing cells (about 106 cells/mL) were lysed by sonication (Branson Sonicator, Cell Disrupter 185, Branson Ultrasonics, Danbury, CT) in ice-cold phosphate-buffered saline buffer containing protease inhibitors (Complete Mini, Roche, Manheim, Germany). The protein content was determined using a BCA Protein Assay kit (Pierce, Rockford, IL) according to the manufacturer’s instructions. In all, 10 μg of protein was boiled in Laemmli buffer and separated by electrophoresis on 4% to 20% (wt/vol) SDS-polyacrylamide gel. After transfer to a 0.45 μ pore-size nitrocellulose membrane (Bio-Rad Laboratories), they were blocked overnight at 4°C in Tween—tris-buffered saline (50 mmol/L Tris-buffered saline, pH 7.6, with 0.05% Tween-20) containing 5% nonfat dry milk. The membranes were then incubated with anti-ICSBP and MUM-1 antibodies for 2 hours at room temperature and binding was detected using the appropriate donkey secondary antibodies conjugated to horseradish peroxidase (ECL, Amersham Biosciences, Little Chaflont Buckinghamshire, UK) and an enhanced chemiluminescence SuperSignal West Pico detection kit (Pierce Biotechnology Inc, Rockville, IL). Equal protein loading was confirmed with an α-tubulin antibody (Oncogene Science Inc, Cambridge, MA).
RESULTS
IRF8/ICSBP-1 Expression in Reactive Lymphoid Tissues
The pattern and distribution of IRF8 expression in reactive lymph nodes and tonsils was assessed by immunohistochemistry using a polyclonal antibody from Santa Cruz Biotechnology on paraffin-embedded tissue sections. IRF8 was most strongly expressed in the nuclei of scattered mononuclear cells throughout the paracortical T-cell zones and diffusely within the follicular B-cell areas (Fig. 1A). Dual-label immunohistochemistry using antibodies to CD68, CD20, and CD3 to identify cells of histiocyte/macrophage, B cell, and T-cell lineages, respectively, revealed that the majority of the intensely stained IRF8-positive cells within the parafollicular and paracortical T-cell zones coexpressed CD68 and had few to extensive dendritic processes, indicating origin from histiocyte/macrophage lineages (Fig. 1B; upper inset). This is consistent with the known role of IRF8 in macrophage development. Interestingly, the mature macrophages ("tingible body macrophages") of the germinal centers, which strongly expressed CD68, showed only weak expression of IRF8 (Fig. 1B; lower inset), and CD68-negative antigen presenting follicular dendritic cells expressed no discernable IRF8 (not shown).
FIGURE 1.
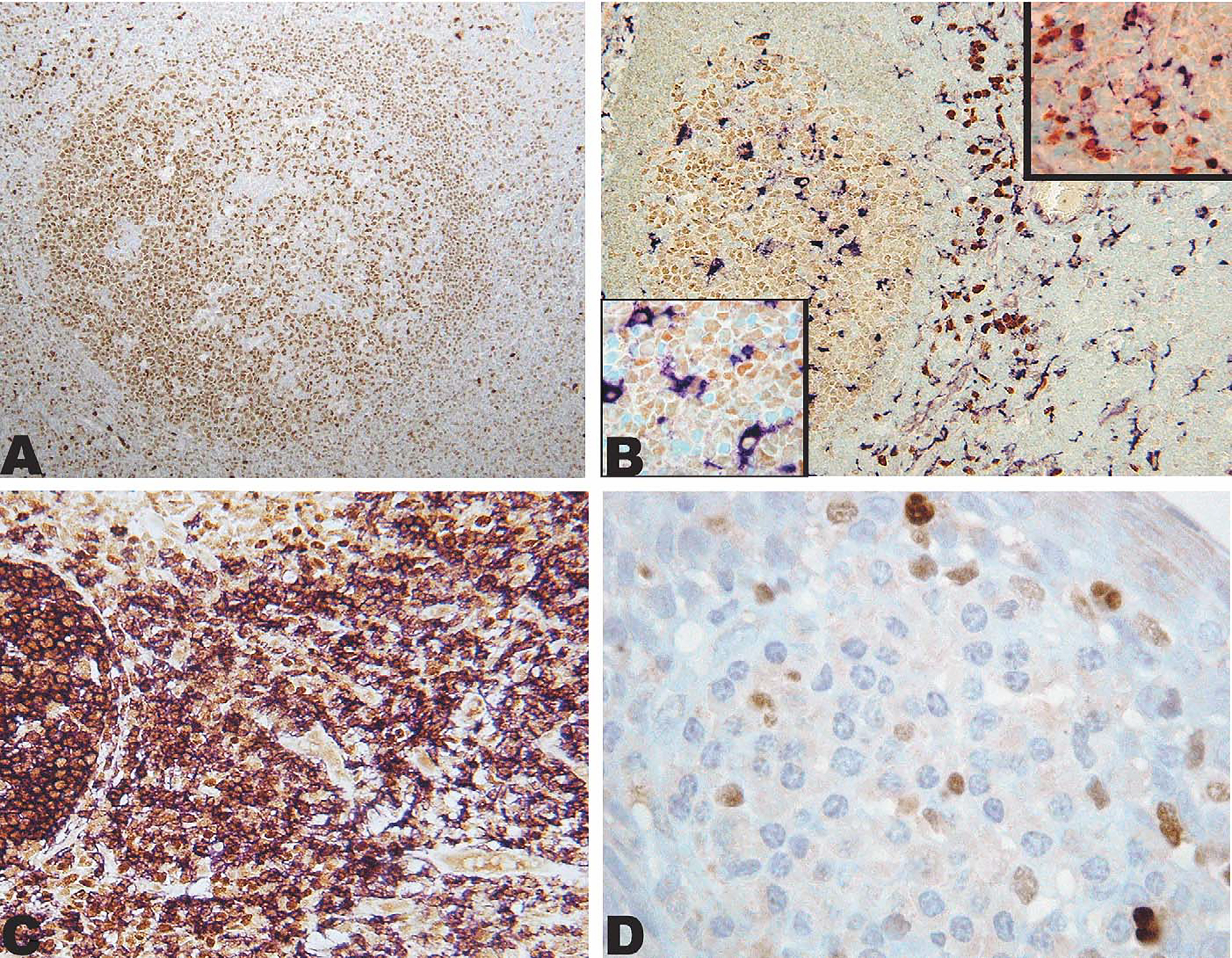
IRF8 expression in reactive lymphoid tissue. A, IRF8 is expressed in a reactive polarized follicle with stronger staining of the germinal center in comparison with the mantle. Strong staining can be seen in scattered interfollicular mononuclear cells. B, Double immunostain showing IRF8 (brown; DAB) expression in CD68 (purple; VIP) positive cells (macrophage/dendritic cells) in the extrafollicular area (upper inset) and weakly in CD68-positive cells (macrophages) in the germinal center (lower inset). C, Double immunostain showing nearly all B cells CD20-positive (purple; VIP) expressing IRF8 (brown; DAB). D, Cluster of plasma cells in the submucosa of a reactive tonsil, rare mononuclear cells are positive with IRF8, whereas plasma cells are negative.
Of particular interest to the current study, IRF8 was also expressed variably in the majority of CD20-positive B cells (Fig. 1C). The strongest expression of IRF8 in B-cell follicles was in the germinal centers, with weaker expression in the mantle and marginal zones (best seen in Fig. 1A). Plasma cells, identified by their distinctive morphology and submucosal location in the tonsil (Fig. 1D), and T cells, identified by dual-label immunostaining with CD3, were uniformly negative for IRF8 (not shown).
To quantify the relative expression of IRF8, we employed an image analysis program on captured IRF8-stained tonsillar tissue images, as described in the Materials and Methods section. Thresholds for fluorescent image capture were set to enable comparison of the follicle B-cell subsets (mantle and germinal center B cells). Mean nuclear IRF8 intensities were calculated from 264 germinal center cells and 200 mantle cells, which were taken from 9 different follicles of 4 reactive tonsils. The relative mean intensity for the mantle cells was 39 arbitrary fluorescent units, and for the germinal center cells, 69 U. Assuming a linear relationship between fluorescent mean intensity and IRF8 content, these data indicate a 1.75-fold increase in the expression of IRF8 in germinal center cells as compared with mantle cells. (Table 1; Fig. 2). The relative increase in IRF8 was reproducible between different staining runs and tonsils, and was not influenced by the type of secondary antibody labeling (rhodamine or fluorescein) (data not shown). IRF8 content in macrophages was not included in the analyses owing to oversaturation under conditions used to capture B-cell fluorescent staining. However, separate experiments using lower detector gain indicated that macrophages have approximately 2 to 3 times as much IRF8 content as the strongest expressing follicle center cells (data not shown).
TABLE 1.
IRF8 Expression in Follicular B-cell Compartments as Assessed by Quantitative Fluorescence Confocal Microscopy
| Follicle | Mean F.U. GC | Mean F.U. MC | Ratio GC/MC |
|---|---|---|---|
| F1-T1 | 52.3 | 30.9 | 1.69 |
| F2-T1 | 74.3 | 36.9 | 2.01 |
| F3-T1 | 78.0 | 50.9 | 1.53 |
| F4-T1 | 72.4 | 42.6 | 1.70 |
| F5-T1 | 60.9 | 30.2 | 2.01 |
| F6-T1 | 69.3 | 41.6 | 1.67 |
| F7-T2 | 60.0 | 37.8 | 1.59 |
| F8-T3 | 92.9 | 52.5 | 1.77 |
| Mean | 70.01 | 40.42 | 1.75 |
F.U. indicates fluorescence units; GC, germinal center B-cells; MC, mantle zone B-cells.
FIGURE 2.
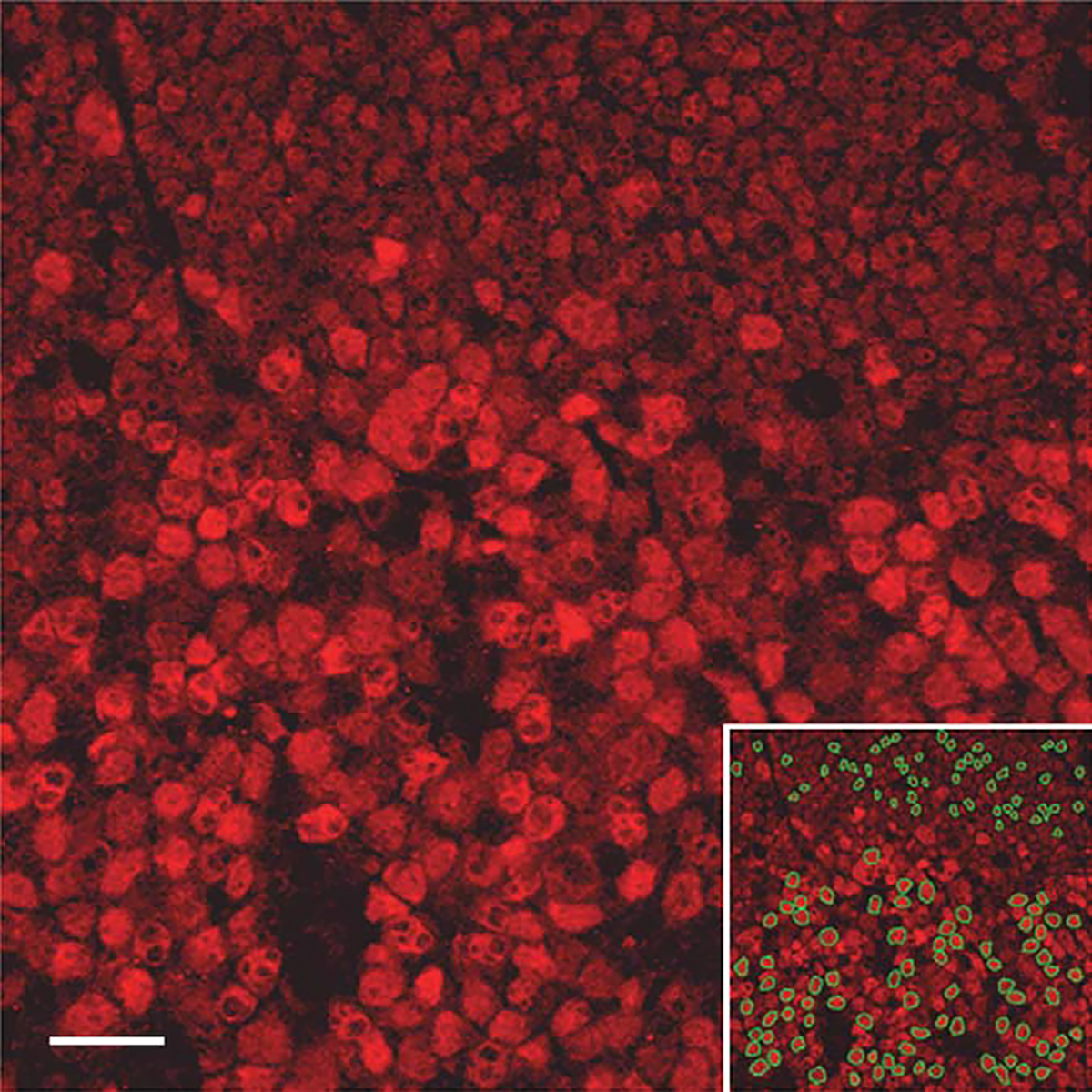
IRF8 expression in follicular B cells by quantitative confocal microscopy: Reactive follicle stained with IRF8 by immunofluorescence (Rhodamine) showing germinal center cells (bottom two-thirds of image) and adjacent mantle cells (top third of image). Lower right inset identifies cells (outlined in green) selected for fluorescence quantitation in each compartment.
IRF8/ICSBP-1 is Coexpressed With BCL-6, PAX-5, and Pu.1, but not With IRF4 in Germinal Center B cells
Within the germinal centers, the highly proliferative centroblasts in the dark zone showed the strongest staining for IRF8. The light zone showed a lower density of cellular staining and there seemed to be a small subpopulation of B cells that were negative for IRF8 (Fig. 3, panel A). Sequential immunostaining for IRF8 and IRF4 suggested that the population of IRF8-negative light zone B cells were, instead, positive for IRF4 (Fig. 3, panel B). Dual-staining immunofluorescence confirmed this impression and also showed that germinal center B cells express either IRF8, or in a small minority of cells, IRF4, but not both (Fig. 3, panel C). The population of IRF8-negative, IRF4-positive B cells seems to correspond to terminal differentiation by committed B cells, as described by Falini et al.8 The mutually exclusive expression pattern for IRF8 and IRF4 was observed in the B cells of all reactive lymphoid tissues examined (tonsil, lymph node, and spleen). On the contrary, IRF8 was coexpressed with other transcription factors that play an important role in B-cell differentiation and germinal center formation such as PAX-5, BCL-6, and Pu.1 (Fig. 4). The expression of BCL-6, a transcriptional repressor required for germinal center formation, has been shown to be limited to the germinal center and scattered parafollicular large cells.5 Dual-label immunofluorescence revealed coexpression between IRF8 and BCL-6 in the germinal center but not in the mantle cells that were weakly positive for IRF8 and negative for BCL-6 (Fig. 4, top panel). PAX-5, known to be expressed in B cells from pre-B to mature B cells, but not in terminally differentiated B cells, was coexpressed with IRF8 in the follicle center B cells (Fig. 4, middle panel). As expected, there was also colocalization between IRF8 and Pu.1 with the most intense staining in the macrophages surrounding the germinal centers in keeping with the biology and functional interaction between these 2 factors (Fig. 4, lower panel).
FIGURE 3.
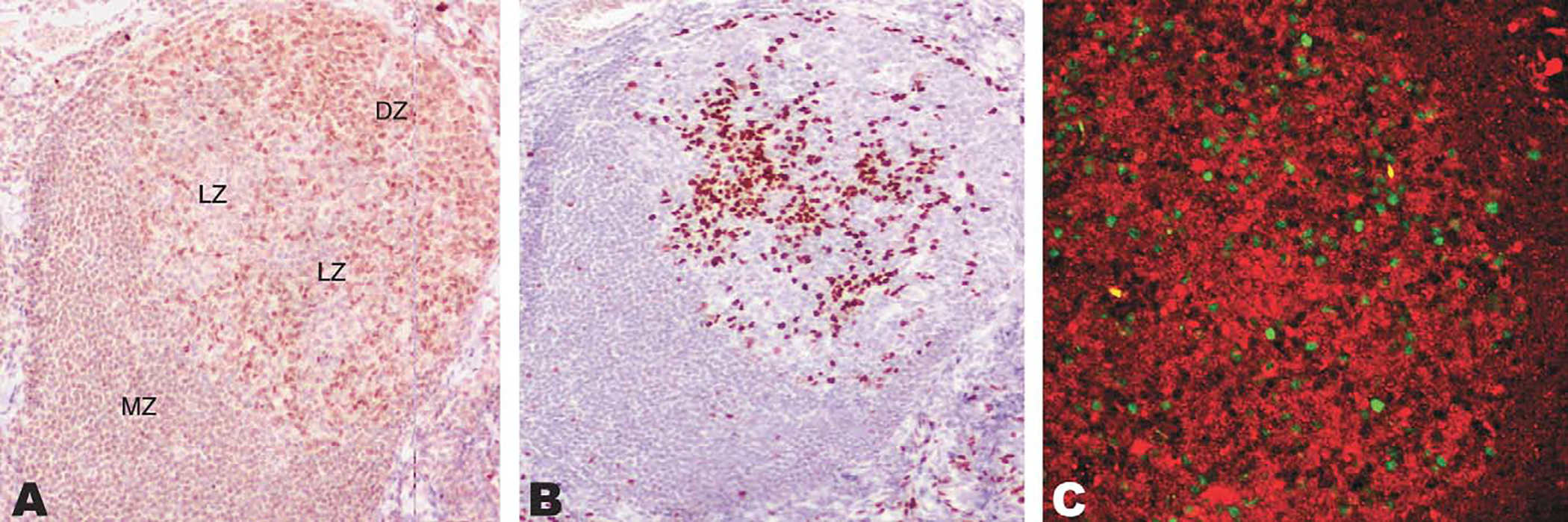
Exclusion between IRF8 and IRF4 in germinal centers shown by sequential immunohistochemical stains and immunoflurorescence. A, The dark zone (DZ) of the germinal center is densely packed by IRF8-positive cells, whereas less numerous positive cells are noted in the light zone (LZ). The mantle zone of the follicle is labeled MZ for reference. B, IRF4-positive cells are present in the light zone of the germinal center. C, Confocal immunofluorescence for IRF8 (red) and IRF4 (green) confirms a mutually exclusive pattern in reactive follicles.
FIGURE 4.
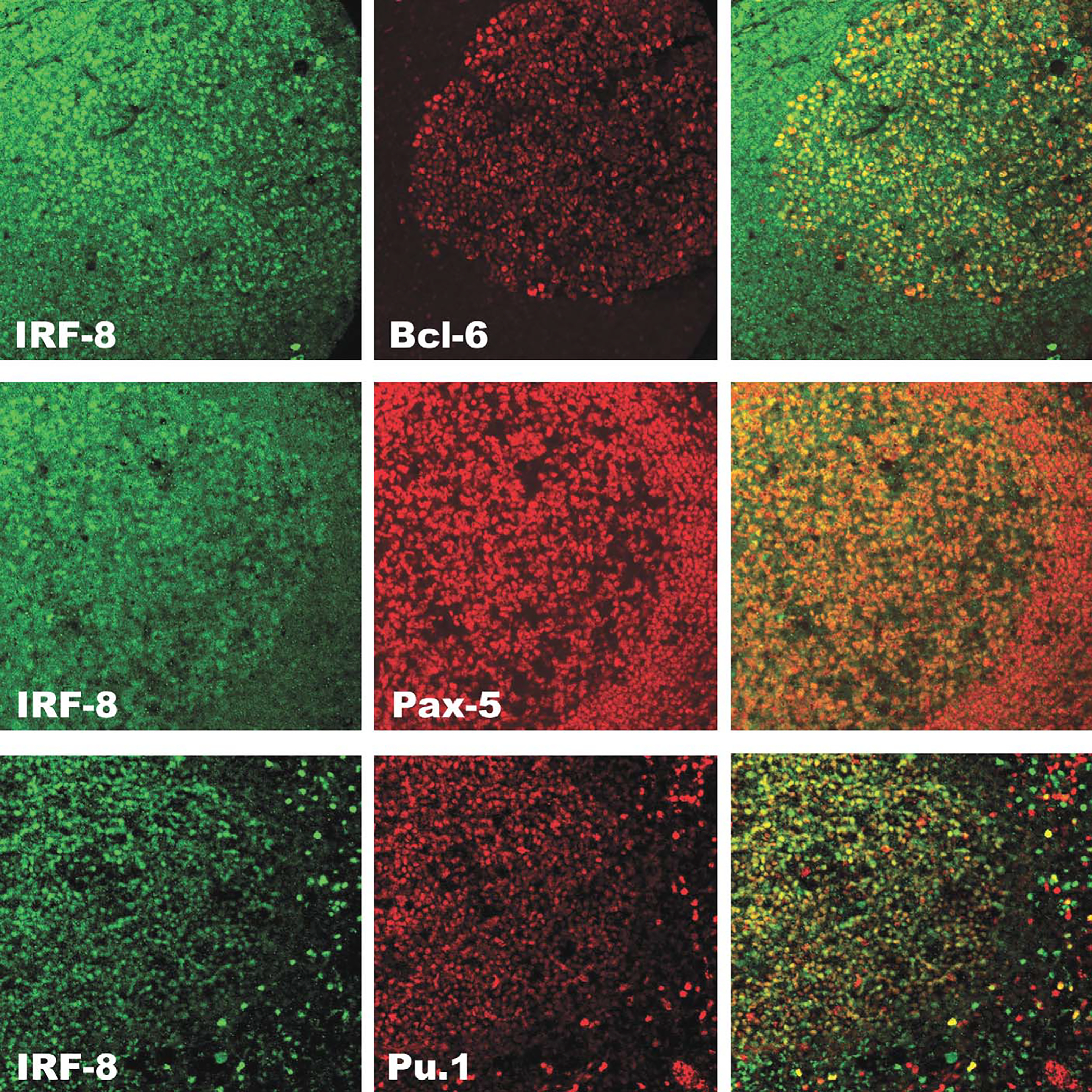
Dual-label confocal immunofluorescence of IRF8 with BCL-6, PAX-5, and Pu.1 in a reactive B-cell follicle. Upper panel. IRF8 (green) is coexpressed with BCL-6 (red) in the germinal center, but not in the mantle as shown in the overlay (yellow-green/yellow-orange). Middle panel. IRF8 (green) is coexpressed with PAX-5 (red) in germinal center and mantle as shown in the overlay (yellow). Lower panel. IRF8 (green) is coexpressed with Pu.1 (red) in the germinal center, mantle and in scattered is interfollicular macrophages (overlay, yellow).
IRF8/ICSBP-1 Expression in Hematolymphoid Cell Lines
We next analyzed the expression pattern of IRF8 in 30 human B-cell non-Hodgkin lymphoma (B-NHL) cell lines by a combination of Western blot analysis (representative cell lines are shown in Fig. 5) and immunohistochemistry (not shown). The histiocytic lymphoma cell line U-937 and the human carcinoma cell line MFC-7 served as positive and negative controls, respectively. The expression of IRF8 was confined to the hematologic cell lines. The 53-kDa IRF8-specific band was present in protein extracts from 1 of 1 B-lymphoblastic leukemia cell line, 5 of 5 DLBCL cell lines GCB-type, 2 of 2 DLBCL ABC-type, 5 of 5 BL cell lines, 3 of 4 MCL cell lines, and 1 of 1 hairy cell leukemia line. IRF8 was not identified in the 8 MM and 4 primary effusion lymphomas cell lines studied. These data suggest that IRF8 expression in lymphoma cell lines generally corresponds with the expression of IRF8 seen in normal cellular counterparts.
FIGURE 5.
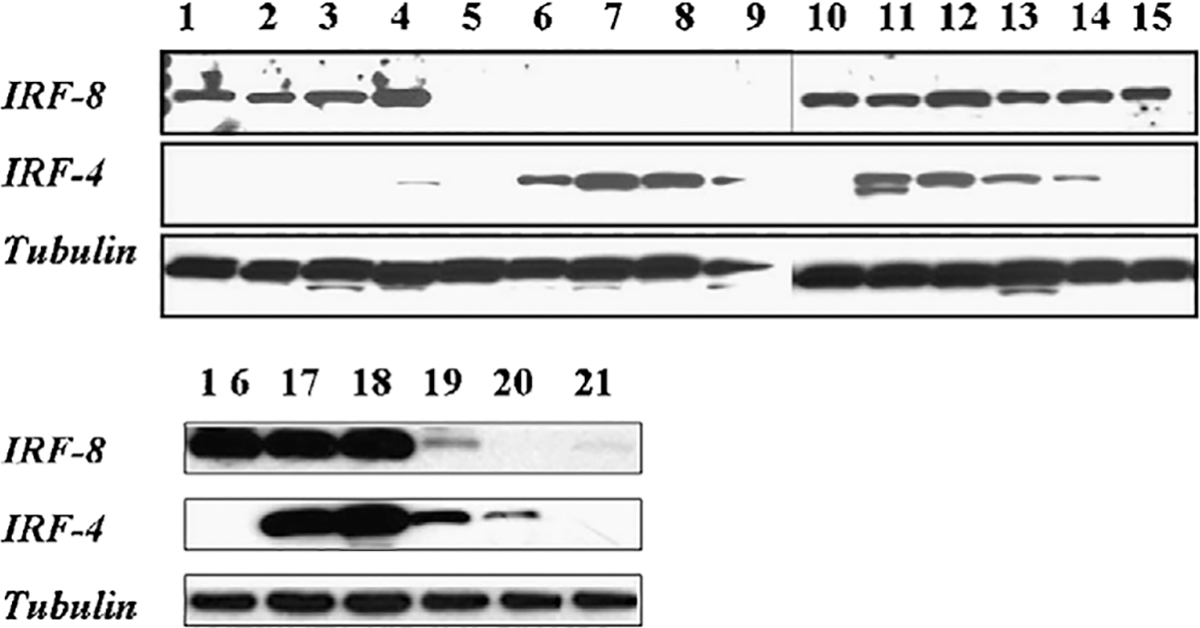
Western blot for IRF8 and IRF4 in cell lines. Western blot analysis of representative cell lines for IRF8 (panel A), IRF4 (panel B), and tubulin as a loading control (panel C). IRF8 is present in DLBCL GCB-like cell lines SUDHL-4, SUDHL-5, SUDHL-6, SUDHL-7 (lanes 1, 2, 3, 16) and negative KMS-12, KMS-20 myeloma (lanes 6, 7), and BC-2, BC-3 primary effusion lymphoma cell lines (lanes 8, 9). In contrast OCI-Ly3 and OCY-Ly10 DLBCL non-GCB–like (lanes 17, 18), Granta and Z-138 MCL (lanes 19, 20), and the JD38 BL cell lines (lane 4) show expression of both IRF8 and IRF4.
Because IRF4 seemed to be expressed in a mutually exclusive manner with IRF8 in reactive tissues, we also examined its expression in the cell lines. The IRF4 specific 42-kDa band was identified in lysates from all 12 MM and primary effusion lymphomas cell lines, as expected. The germinal center-like B-cell (GCB)-like DLBCL cell lines of GCB-type showed no (SUDHL 4, 5, 6, and 10) or low (SUDHL 7) expression of IRF4, whereas the DLBCL cell lines of ABC-type, the BL cell lines, and 3 of 4 MCL cell lines expressed IRF4 and IRF8. Double immunolabeling for IRF4 and IRF8 on formalin-fixed paraffin-embedded cell blocks from these latter cell lines demonstrated that a subpopulation of cells variably expressed both transcription factors (not shown). Thus, although the mutually exclusive pattern of IRF expression that was seen in the reactive tissues tended to be maintained in the terminally differentiated cell lines (ie, myeloma and primary effusion lymphoma), it was not maintained in MCLs, BLs, and DLBCLs, particularly of the ABC-type.
IRF8/ICSBP-1 and IRF4 Expression in B-NHL
To assess the distribution of IRF8 in B-NHLs, 232 lymphomas representing a variety of pregerminal center, germinal center, and postgerminal center-derived B-NHLs were studied (Table 2, Fig. 6). The majority of pregerminal center and germinal center derived B-NHLs, including 5 of 5 LB, 27 of 28 MCL, 46 of 46 FL, 8 of 8 BL, and 52 of 58 DLBCLs were positive for IRF8 (39 of 39 GCB-like; 7 of 13 non-GCB–like, 6 of 6 unclassified), whereas plasmacytic tumors of postgerminal center derivation, including 7 of 8 LPL, 14 of 14 PBLs (includes 1 PEL), and 14 of 14 MMs, were negative for IRF8. MZLs and CLL, both believed to be derived from postgerminal center memory cells, were positive for IRF8 in 20 of 21 cases and 33 of 35 cases, respectively.9,14 The intensity of IRF8 staining was consistent with the putative cellular origin with germinal center-derived lymphomas (FLs and BLs) staining with greatest intensity, and the pregerminal center lymphomas (LBs and MCLs) showing weaker staining for IRF8. MZL stained with similar intensity to the MCL, as did CLL except in the proliferation centers and in scattered prolymphocytes and paraimmunoblasts, which stained strongly for IRF8.
TABLE 2.
Frequency of IRF8 and IRF4 Expression in B-cell Malignancies
| Lymphoma Type | n | IRF8 |
IRF4 |
Coexpression |
|||
|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | ||
| Lymphoblastic | 5 | 5 | 100 | 0 | 0 | ||
| Burkitt | 8 | 8 | 100 | 0 | 0 | ||
| Follicular | 46 | 46 | 100 | 0 | 0 | ||
| Small lymphocytic | |||||||
| Small cells | 30 | 29 | 97 | 1 | 3 | 0 | |
| Proliferation centers | (19/30) | 19 | 100 | 19 | 100 | 19 | 100 |
| Mantle cell | 28 | 27 | 96 | 3 | 11 | 3 | 11 |
| Diffuse large B-cell | |||||||
| GCB | 39 | 39 | 100 | 5 | 13 | 5 | 13 |
| Non-GCB | 13 | 7 | 54 | 6 | 46 | 6 | 46 |
| Other | 6 | 6 | 100 | 0 | 0 | ||
| Marginal zone | 21 | 20 | 95 | 6 | 29 | 6 | 29 |
| Lymphoplasmacytic | 8 | 1 | 13 | 3 | 43 | 0 | |
| Plasmablastic | 14 | 0 | 9 | 100 | 0 | ||
| Multiple myeloma | 14 | 0 | 14 | 100 | 0 | ||
| Total | 232 | 188 | — | 47 | — | 20* | — |
Excluding proliferation centers of small lymphocytic lymphoma.
FIGURE 6.
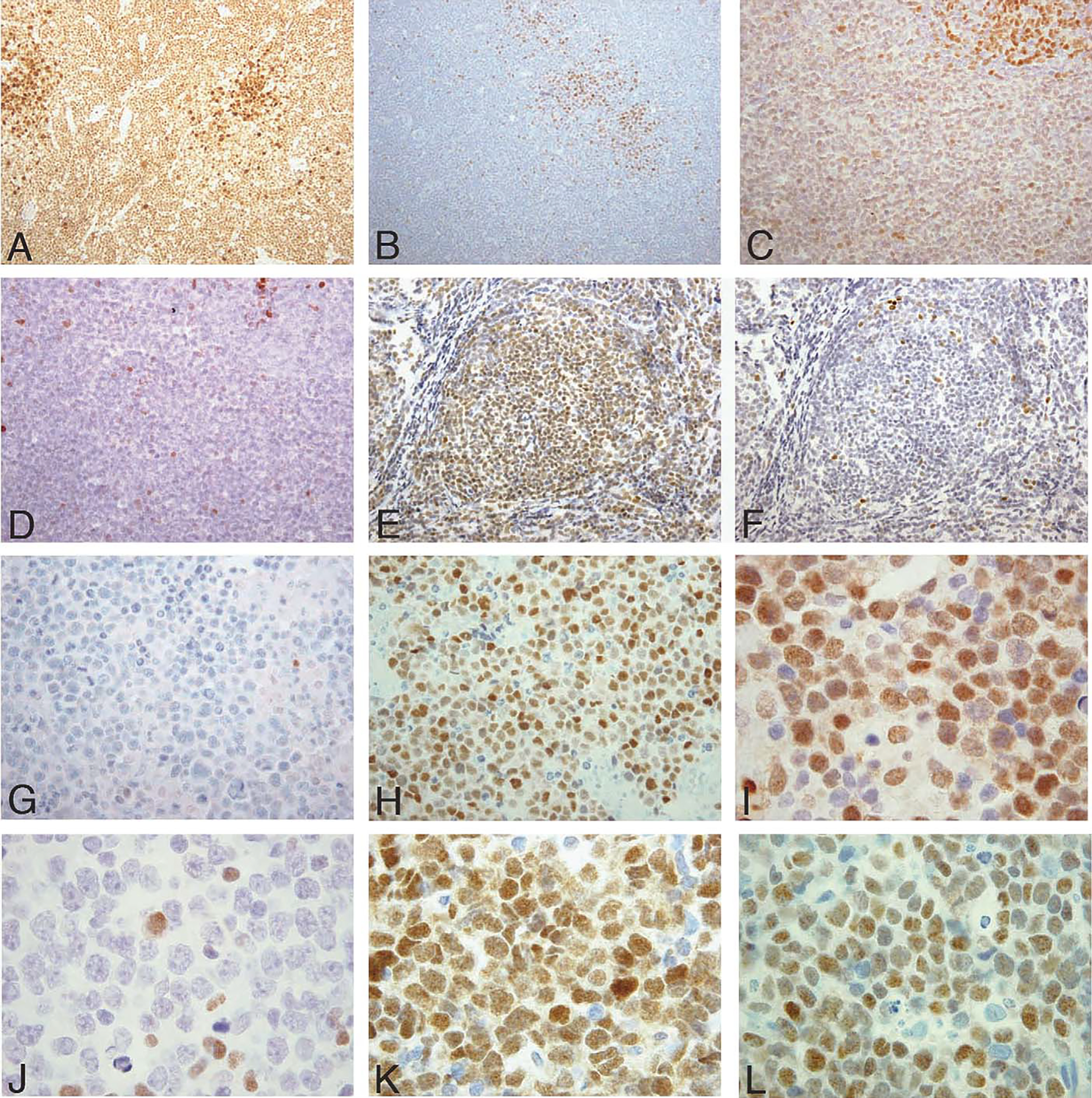
IRF8 and IRF4 expression by immunohistochemistry in B-cell non-Hodgkin lymphomas. A and B, CLL ( × 20 HPF). A, Neoplastic cells express IRF8 with strongest intensity in proliferation centers. B, IRF4 is negative in most cells with the exception of proliferation centers. C and D, MCL ( × 20 HPF). C, IRF8 is weakly expressed in the neoplastic cells. Note the stronger IRF8 expression in a residual reactive germinal center (top right). D, Scattered IRF4-positive cells with the majority of neoplastic cells negative. E and F, FL ( × 20 HPF). E, Neoplastic cells show strong expression for IRF8, and (F) are negative for IRF4. G and H, MM ( × 40 HPF). G, Myeloma cells are negative for IRF8 and (H) positive for IRF4. I and J, DLBCL of GCB type ( × 60 HPF) showing expression of IRF8 (I), but not IRF4 (J). K and L, DLBCL of GCB type ( × 60 HPF) with coexpression of IRF8 (K) and IRF4 (L). [This latter DLBCL was classified as GCB type on the basis on a positive CD10 stain (not shown), according to the criteria of Hans et al11].
In contrast, IRF4 was primarily, but not exclusively, detected in lymphomas of postgerminal center plasmacytic derivation. IRF4 was detected in all 14 myelomas, in 9 of 9 PBLs (includes 1 PEL), and in 3 of 7 LPLs examined. Pregerminal and germinal center-derived lymphomas expressed either no IRF4 (B-cell LB, 0 of 4; FL, 0 of 35), or expressed IRF4 in a low-to-moderate percentage of cases (MCL, 3 of 28; BL, 1 of 6; DLBCL, 19 of 58). Within the DLBCL, IRF4 was expressed in 6 of 39 GCB and in 13 of 19 non-GCB, consistent with previously published data.11 Six of 21 MZL cases, lymphomas thought to be of postgerminal center memory B-cell derivation, expressed IRF4. In CLL, only 1 of 30 CLL cases expressed IRF4 in the small tumor B-cells; however, all cases expressed IRF4 in the proliferation centers when present (19 of the 30 cases).
Expression of both IRF8 and IRF4 in a significant fraction of cells was observed in 11 of 58 DLBCLs, 6 of 21 MZLs, the latter of which are believed to originate from postgerminal center, memory cells, and in 3 of 28 MCLs (3 of 28), and the proliferation centers of CLL (19 of 19). In DLBCL coexpressing IRF4 and IRF8, IRF4 was always more intense than IRF8. Five cases were GCB-like DLBCLs and 6 were non-GCB–like DLBCLs. Thus, coexpression was present in 5 of the 39 GCB lymphomas and 6 of the 13 non-GCB lymphomas. Sequential immunohistochemical studies indicated that in all but 1 of these, individual tumor cells expressed both IRF8 and IRF4. Dual-immunostaining using confocal immunofluorescence was also performed and confirmed coexpression at the individual cell level, with the exception of 1 case in which there were 2 separate neoplastic subpopulations expressing either IRF4 or IRF8. Coexpression at the individual cell level seemed to be present in other IRF4 and IRF8 coexpressing lymphomas as well, although this evaluation in the MZLs was difficult because of the intermingling of reactive lymphoid cells in these tumors.
DISCUSSION
IRF8 is a hematopoietic cell restricted regulator of IFN-inducible gene transcription that is constitutively expressed in murine B cells, but its role in human B-cell development and in lymphomagenesis is largely unknown.7 Here we show that the IRF8 expression in human B cells is not constitutive, rather is modulated during late B-cell development being expressed at moderate levels in the marginal and mantle zones, strongly expressed in the germinal center B cells, and absent in mature plasma cells. Quantitative fluorescent analysis of IRF8 content in the reactive lymphoid tissue was consistent, with a dynamic increase in IRF8 during the germinal center reaction.
In the secondary lymphoid follicles, IRF8 was coexpressed with BCL-6 in the germinal centers, and like the latter transcription factor, it was excluded from a small population of IRF4-positive centroblasts located in the light zones. These BCL-6-negative, IRF4-positive B cells are believed to be committed to postgerminal center differentiation programs.8 Pu.1, an ETS family transcription factor capable of dimerizing with both IRF8 and IRF4 to form active complexes, was also up-regulated in the germinal center, but expressed in both the IRF8 and IRF4 germinal center B-cell subpopulations. Although, IRF4 has been demonstrated to be involved in postgerminal center plasma cell differentiation, the coexpression of IRF8 with BCL-6 suggests that IRF8 is more closely related to the germinal center transcriptional program.
BCL-6 is a transcriptional repressor and is considered to be a master regulator of the germinal center reaction.6,31,32 It is up-regulated in germinal center B cells, and its expression is extinguished when these cells enter the postgerminal center differentiation program. The extinction of BCL-6 expression releases its repression of Blimp-1, a critical transcription factor required for transitioning from the germinal center program into the plasma cell differentiation program.36 The signals responsible for the initial up-regulation of BCL-6 in the germinal center are unclear; however, recently Lee et al15 have shown that IRF8 is capable of activating the transcription of BCL-6 in mouse and human germinal center B-cells, suggesting that IRF8 functions upstream of BCL-6 in the germinal center program. Previously, Pu.1 binding sequences had been shown to be present in the BCL-6 promoter,35 which would be consistent with activation of BCL-6 through IRF8/Pu.1 complexes. Moreover, recent evidences have shown a role of IRF8 together with BCL-6 in the down-regulation of p53 dependent genomic DNA repair responses that allow the receptor editing and class switch during the germinal center reaction.15,24 These data, along with our own, suggest that IRF8 plays a major role in the germinal center reaction.
Unlike BCL-6, however, IRF8 is not essential for germinal center development, as IRF8-deficient mice form germinal centers and develop humoral immune responses.13 In contrast, these mice present hypergammaglobulinemia and have abnormal numbers of plasmablasts. This suggests that IRF8 may act as a modulator of the germinal center reaction by maintaining adequate transcription levels of BCL-6. The premature reduction in BCL-6 might lead to the derepression of Blimp-1 and entry into the postgerminal center plasma cell program before the development of a mature humoral immune response. The signals responsible for down-regulation of BCL-6 involve CD40-CD40L interactions, signaling through the B-cell receptor, and the production of cytokines such as IL-4, IL-6, and IL-10.3,12,21 Some of these same signaling pathways have also been shown to initiate the transcription of IRF4.22 It will be of interest to determine how these signals might influence the expression of IRF8.
The pattern of IRF8 expression in normal tissues, its association with BCL-6, and its exclusion with IRF4, both implicated in lymphomagenesis, suggested that IRF8 may also participate in B-cell lymphomagenesis, and be a useful diagnostic target in differentiating subtypes of B-NHLs. In fact, B-cell tumors and cell lines representing various stages of B-cell development mirrored the differential expression pattern seen in the lymph node, with germinal center-derived lymphomas showing the strongest IRF8 expression, lymphomas derived from pregerminal center B cells showing weaker IRF8 expression, and lymphomas derived from postgerminal center plasmacytic B cells showing no expression of IRF8. The mutual exclusion of IRF8 and IRF4 expression was generally maintained in the primary B-NHLs, with some exceptions. Weak-to-moderate expression of IRF8 in lymphomas of mantle and marginal zone derivation is consistent with their cell of origin, as the B cells of the normal mantle and marginal zones showed weaker intensities of IRF8 expression. Similarly, the lack of IRF8 expression in plasmacytic neoplasms is in accord with IRF8 absence in non-neoplastic plasma cells. The presence of moderate-to-strong IRF8 staining in FLs and BLs is again consistent with their origin from germinal center B cells, which showed the strongest staining in the reactive lymphoid tissues.
DLBCL has been shown to be a heterogeneous subgroup of lymphomas.2,11 In fact, although 90% of these cases expressed IRF8 at variable intensity, 33% stained strongly for IRF4 as well, with 19% of cases staining for both transcription factors. Coexpression of both IRF4 and IRF8 occurred in 5 GCB-like lymphomas, and in 6 non-GCB–like lymphomas. The coexpression observed of both IRFs in DLBCL may reflect unstable transitional states that are normally not apparent in normal follicle center cells as they traverse the follicular B-cell program and enter into the plasmacytic differentiation program.
The nodal MZLs, which are believed to be related to memory B cells, are another B lymphoma subgroup that showed coexpression of both IRF8 and IRF4, occurring in about 29% of cases. This is somewhat lower than the 52% IRF4 positivity reported by Camacho et al.4 It has recently been suggested that IRF4 expression in MZL is related to the degree of plasmacytic differentiation.23 The high rate of coexpression of both IRF8 and IRF4 might suggest that these lymphomas, thought to be derived from memory B cells, exist in a transcriptionally unstable state, poised to reexpress genes (such as IRF4) involved in effector B-cell programs when restimulated.
A similar argument could be made for the coexpression of IRF4 and IRF8 in CLL. Although the cell of origin of CLL is debated, there is accumulating evidence that at least a portion of these represent neoplastic memory B cells. Although the small cells of CLL exclusively expressed IRF8, the paraimmunoblasts and proliferation centers showed moderately strong staining with IRF4 as well. Immunostains of sequential slides revealed that most of these cells expressed both, IRF8 and IRF4, making proliferation centers of CLL another exception to the mutual exclusion model.
A small percentage of MCLs (11%; 3 of 28) showed focal IRF4 staining. The percentage of positive cells was always relatively low (20% to 25%), and the predominant IRF was IRF8. This is consistent with the results from the cell lines as well in which IRF4 coexpression was seen. It is difficult to surmise why these tumors occasionally express IRF4, as normal mantle cells are believed to transition into the follicle B-cell program upon stimulation, a transition that does not involve the induction of IRF4. It is possible that IRF4 expression may reflect an earlier pregerminal center/germinal center transition state, as IRF4 has also been shown to be expressed during early B-cell development.17
It is unclear what specific role IRF8 plays in lymphomagenesis. Mice deficient in IRF8 develop a CML-like syndrome, suggesting that IRF8 may function as a tumor suppressor gene.13 Moreover, IRF8 mRNA expression seems to be suppressed in human CML, and the degree of this suppression functions as an independent prognostic factor in this disease.29,30 However, there are no previous reports linking overexpression of IRF8 with tumors. Overexpression of BCL-6 is believed to contribute to lymphomagenesis by maintaining the proliferative germinal center phenotype.32 Whether IRF8 plays a similar or cooperative role with BCL-6 in helping to maintain the proliferative germinal center phenotype in tumors is yet to be determined.
IRF4 is expressed in tumors of plasmacytic/plasmablastic differentiation and is part of the molecular signature that characterizes the activated subtype of DLBCLs.2 Lack of IRF4 expression has been used in combination with other phenotypic markers to distinguish the germinal center DLBCL subtype from the activated B-cell subtype of DLBCL.11 Our study suggests that strong IRF8 expression may complement the use of IRF4 in distinguishing subtypes of B-cell neoplasms.
In conclusion, we have shown that IRF8 is expressed variably in nearly all mature B cells, but is most highly expressed during the germinal center reaction, before its expression is extinguished in B-cells committed to postgerminal center programs, particularly the plasma cell program. Within the germinal centers, IRF8 shares a similar expression pattern with BCL-6, and is excluded in the IRF4-positive light zone B cells. Similar to the expression pattern in normal B cells, our data in lymphomas suggests that strong expression of IRF8 is exclusive to lymphomas of follicle center origin, whereas tumors of pregerminal center origin and postgerminal center origin show weaker or no IRF8 expression, respectively. These data suggest an important role for IRF8 during the germinal center reaction and indicate that IRF8 may be a useful marker to assist in distinguishing IRF8-negative postgerminal center B-cell neoplasms from other subtypes of IRF8-positive B-NHLs.
AKNOWLEDGMENTS
The authors thank Alan Epstein for providing SUDHL 1, 4, 5, 7, and 10 cell lines; Michael Kuehl for Jim3 and OPM2; Ian Magrath for BL41; Takemi Otsuki for the KMS cell lines; Louis Staudt for OCI-Ly7 and OCI-Ly10; and Edward F. Srour for the ESKOL cell line. Cell lines not mentioned above were obtained from the American Type Culture Collection (ATCC, Rockville, MD). The authors also thank Dr Herbert Morse for sharing unpublished results with us and for many helpful discussions. Special thanks to Mr Stephen M. Wincovitch Sr and Ms Susan Garfield of the Confocal Microscopy Core Facility, Laboratory of Experimental Carcinogenesis, NCI.
This work was funded through the NIH/NCI intramural program.
REFERENCES
- 1.Aliberti J, Schulz O, Pennington DJ, et al. Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood. 2003;101:305–310. [DOI] [PubMed] [Google Scholar]
- 2.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. [DOI] [PubMed] [Google Scholar]
- 3.Allman D, Jain A, Dent A, et al. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–5268. [PubMed] [Google Scholar]
- 4.Camacho FI, Algara P, Mollejo M, et al. Nodal marginal zone lymphoma: a heterogeneous tumor: a comprehensive analysis of a series of 27 cases. Am J Surg Pathol. 2003;27:762–771. [DOI] [PubMed] [Google Scholar]
- 5.Cattoretti G, Shaknovich R, Smith PM, et al. Stages of germinal center transit are defined by B cell transcription factor coexpression and relative abundance. J Immunol. 2006;177:6930–6939. [DOI] [PubMed] [Google Scholar]
- 6.Chang CC, Ye BH, Chaganti RS, et al. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci USA. 1996;93:6947–6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Driggers PH, Ennist DL, Gleason SL, et al. An interferon gamma-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc Natl Acad Sci USA. 1990;87:3743–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falini B, Fizzotti M, Pucciarini A, et al. A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood. 2000;95:2084–2092. [PubMed] [Google Scholar]
- 9.Ghia P, Caligaris-Cappio F. The origin of B-cell chronic lymphocytic leukemia. Semin Oncol. 2006;33:150–156. [DOI] [PubMed] [Google Scholar]
- 10.Gupta S, Jiang M, Anthony A, et al. Lineage-specific modulation of interleukin-4 signaling by interferon regulatory factor 4. J Exp Med. 1999;190:1837–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103: 275–282. [DOI] [PubMed] [Google Scholar]
- 12.Harris MB, Chang CC, Berton MT, et al. Transcriptional repression of Stat6-dependent interleukin-4–induced genes by BCL-6: specific regulation of epsilon transcription and immunoglobulin E switching. Mol Cell Biol. 1999;19:7264–7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holtschke T, Lohler J, Kanno Y, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. [DOI] [PubMed] [Google Scholar]
- 14.Klein U, Dalla-Favera R. New insights into the phenotype and cell derivation of B cell chronic lymphocytic leukemia. Curr Top Microbiol Immunol. 2005;294:31–49. [DOI] [PubMed] [Google Scholar]
- 15.Lee CH, Melchers M, Wang H, et al. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J Exp Med. 2006;203: 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levi BZ, Hashmueli S, Gleit-Kielmanowicz M, et al. ICSBP/IRF8 transactivation: a tale of protein-protein interaction. J Interferon Cytokine Res. 2002;22:153–160. [DOI] [PubMed] [Google Scholar]
- 17.Lu R, Medina KL, Lancki DW, et al. IRF4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 2003;17: 1703–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuyama T, Grossman A, Mittrucker HW, et al. Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE). Nucleic Acids Res. 1995;23:2127–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mittrucker HW, Matsuyama T, Grossman A, et al. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. [PubMed] [Google Scholar]
- 20.Nelson N, Kanno Y, Hong C, et al. Expression of IFN regulatory factor family proteins in lymphocytes. Induction of Stat-1 and IFN consensus sequence binding protein expression by T cell activation. J Immunol. 1996;156:3711–3720. [PubMed] [Google Scholar]
- 21.Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 1998;12:1953–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pernis AB. The role of IRF4 in B and T cell activation and differentiation. J Interferon Cytokine Res. 2002;22:111–120. [DOI] [PubMed] [Google Scholar]
- 23.Petit B, Chaury MP, Le Clorennec C, et al. Indolent lymphoplasmacytic and marginal zone B-cell lymphomas: absence of both IRF4 and Ki67 expression identifies a better prognosis subgroup. Haematologica. 2005;90:200–206. [PubMed] [Google Scholar]
- 24.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. [DOI] [PubMed] [Google Scholar]
- 25.Quintanilla-Martinez L, Thieblemont C, Fend F, et al. Mantle cell lymphomas lack expression of p27Kip1, a cyclin-dependent kinase inhibitor. Am J Pathol. 1998;153:175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato M, Taniguchi T, Tanaka N. The interferon system and interferon regulatory factor transcription factors — studies from gene knockout mice. Cytokine Growth Factor Rev. 2001;12:133–142. [DOI] [PubMed] [Google Scholar]
- 27.Schiavoni G, Mattei F, Sestili P, et al. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J Exp Med. 2002;196:1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiavoni G, Mattei F, Borghi P, et al. ICSBP is critically involved in the normal development and trafficking of Langerhans cells and dermal dendritic cells. Blood. 2004;103:2221–2228. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt M, Nagel S, Proba J, et al. Lack of interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemias. Blood. 1998;91:22–29. [PubMed] [Google Scholar]
- 30.Schmidt M, Hochhaus A, Nitsche A, et al. Expression of nuclear transcription factor interferon consensus sequence binding protein in chronic myeloid leukemia correlates with pretreatment risk features and cytogenetic response to interferon-alpha. Blood. 2001; 97:3648–3650. [DOI] [PubMed] [Google Scholar]
- 31.Seyfert VL, Allman D, He Y, et al. Transcriptional repression by the proto-oncogene BCL-6. Oncogene. 1996;12:2331–2342. [PubMed] [Google Scholar]
- 32.Staudt LM, Dent AL, Shaffer AL, et al. Regulation of lymphocyte cell fate decisions and lymphomagenesis by BCL-6. Int Rev Immunol. 1999;18:381–403. [DOI] [PubMed] [Google Scholar]
- 33.Tamura T, Ozato K. ICSBP/IRF8: its regulatory roles in the development of myeloid cells. J Interferon Cytokine Res. 2002;22: 145–152. [DOI] [PubMed] [Google Scholar]
- 34.Taniguchi T, Ogasawara K, Takaoka A, et al. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. [DOI] [PubMed] [Google Scholar]
- 35.Torlakovic E, Malecka A, Myklebust JH, et al. PU.1 protein expression has a positive linear association with protein expression of germinal centre B cell genes including BCL-6, CD10, CD20 and CD22: identification of PU.1 putative binding sites in the BCL-6 promotor. J Pathol. 2005;206:312–319. [DOI] [PubMed] [Google Scholar]
- 36.Tunyaplin C, Shaffer AL, Angelin-Duclos CD, et al. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–1165. [DOI] [PubMed] [Google Scholar]


