Abstract
In this study, we systematically examined in vitro frequencies and spectra of the spontaneous mutations in Helicobacter pylori that confer resistance to clarithromycin (Clar), metronidazole (Mtzr), amoxicillin (Amxr), ciprofloxacin (Cipr), and rifampin (Rifr). The mutation rate of Rifr or Cipr determined in a fluctuation assay is 1 × 10−8 to 2 × 10−8 per cell per division. In contrast, the mutation rates of Clar, Mtzr, and Amxr are much lower (<10−9). However, Mtzr mutants could be readily selected in vitro by using the serial passage method, suggesting that the mutagenic effect and selective effect of a sublethal dose of metronidazole contribute to the rapid development of Mtzr. Analysis of spontaneous Rifr, Clar, and Cipr mutants confirmed previous results indicating that mutations within the rpoB gene, the 23S rRNA gene, and the gyrA gene, respectively, are responsible; also, several new mutant alleles were identified. Mtzr mutants resulted most frequently, but not always, from mutations in the rdxA gene. DNA fragments containing each mutant allele could readily transform susceptible H. pylori strains to resistance, confirming that each mutant allele is responsible for the resistance phenotype.
Antibiotic resistance is an increasing problem for the treatment of infectious diseases. Bacteria have evolved diverse mechanisms (pathways) of resistance to antimicrobial agents, including control of uptake and efflux of drugs, modification and detoxification of drugs, alteration and protection of the target sites, and acquisition of heterologous resistance genes from external sources. In Helicobacter pylori, the etiological agent of a wide range of gastric diseases, genetic determinants for resistance to several antibiotics, including clarithromycin, metronidazole, ciprofloxacin, and rifampin, have been determined. Remarkably, the known mechanisms of antibiotic resistance in H. pylori are all due to mutations in chromosomal genes. Clarithromycin resistance is associated with mutations in the 23S rRNA gene (22, 25), which inhibit the binding of clarithromycin to the ribosome. Ciprofloxacin resistance is due to mutations in the gyrA gene, which encodes the A subunit of DNA gyrase (16), and rifampin resistance results from mutations in the rpoB gene, encoding the β subunit of RNA polymerase (7). For metronidazole resistance, although several different mechanisms may exist, the predominant determinant has been shown to be the mutational inactivation of the rdxA gene that encodes an oxygen-insensitive NADPH nitroreductase (4, 9, 10, 13, 21).
The importance of de novo mutation in developing antibiotic resistance prompted us to ask how mutations occur in H. pylori (28). The first step in elucidating the mechanisms of mutagenesis is to define the background of frequency and specificity of spontaneous mutations. From the pioneering works of Luria and Delbruck (14) and recent developments in determining mutation rates of bacterial populations, it is known that determination of mutation rates is not simple (15, 17), and determination of mutation spectra is particularly tedious. In this study we systematically examined the in vitro frequencies and spectra of spontaneous mutations in H. pylori that confer resistance to clarithromycin, metronidazole, amoxicillin, ciprofloxacin, and rifampin.
MATERIALS AND METHODS
H. pylori strains, growth medium, and antibiotics.
H. pylori reference strains 26695, NCTC11639, and UA802 (26), as well as some isolates from University of Alberta Hospital, were used; all are susceptible to the antibiotics tested in this study. H. pylori strains were grown on BHI-YE broth (3.7% brain heart infusion with 0.3% yeast extract and 5% animal serum) or agar plates at 37°C under microaerobic conditions (5% CO2, 5% H2, and 90% N2). Antibiotics used in this study include clarithromycin (Bayer), metronidazole (Sigma), ciprofloxacin (Bayer), rifampin (Sigma), and amoxicillin (Sigma).
MIC test.
H. pylori cells were grown for 2 days and suspended in sterile BHI-YE liquid medium, and the turbidity of the suspensions was adjusted to that of a 2.0 McFarland standard. The suspended cells were inoculated (8 μl/spot) onto BHI-YE agar plates containing different concentrations of antibiotics obtained by serial twofold dilution. The plates were incubated as described above, and the growth was examined after 3 days.
Determination of mutant frequency and mutation rate.
An H. pylori strain that is susceptible to an antibiotic was grown in BHI-YE broth to late log phase (about 3 days), with the viable cell number being around 109/ml. This culture was diluted 10−4 in BHI-YE broth (∼105 cells/ml) and divided into 0.5-ml aliquots. The number of aliquots was 12 to 30 (see Table 2). These aliquots were allowed to grow for 3 days to obtain parallel, independent cultures. The number of resistant mutants that emerged in each culture was determined by plating the entire culture on BHI-YE agar plates containing a selective antibiotic. The total number of cells was determined by plating an appropriate (10−5, 10−6, and 10−7) dilution of three cultures on nonselective medium. Colonies on both selective and nonselective plates were counted after incubation for 4 days. The frequency of resistant mutants was expressed as the mean number of resistant cells divided by the total number of viable cells per culture. For calculation of the mutation rate, the most likely number of mutations per culture (m) was first calculated from the distribution of numbers of resistant mutants in the independent cultures by using an appropriate estimator (17). A number of different estimators (equations developed by mathematicians) are available, each of which is valid for a particular range of the m value. Then the mutation rate (μ) per cell division was calculated as μ = m/Nt, where Nt is the total cell number per culture (17).
TABLE 2.
Frequency of spontaneous mutations
| Selective antibiotic (concn [μg/ml])a | H. pylori strain | No. of independent cultures | No. of cells per culture (108) | Resistant bacteria
|
Frequency of mutants | Mutation rate per cell divisionb | ||
|---|---|---|---|---|---|---|---|---|
| Zero fractionc | Mean | Median | ||||||
| RIF (20) | UA802 | 30 | 5.0 | 2/30 | 33d | 27 | 6.6 × 10−8 | 1.6 × 10−8 |
| RIF (20) | 26695 | 15 | 4.8 | 2/15 | 18.1 | 15 | 3.8 × 10−8 | 1.1 × 10−8 |
| RIF (20) | 11639 | 15 | 4.5 | 1/15 | 20.8 | 20 | 4.6 × 10−8 | 1.4 × 10−8 |
| CIP (1) | UA802 | 15 | 4.0 | 2/15 | 15.2 | 12 | 3.8 × 10−8 | 1.1 × 10−8 |
| CLA (0.5) | UA802 | 12 | 5.0 | 8/12 | 1.5 | 0 | 3 × 10−9 | 8 × 10−10 |
| MTZ (8) | 11639 | 15 | 4.5 | 11/15 | 2.3 | 0 | 5.1 × 10−9 | 6.9 × 10−10 |
| AMX (1) | 11639 | 12 | 4.3 | 12/12 | 0 | 0 | ?e | ? |
RIF, rifampin; CIP, ciprofloxacin; CLA, clarithromycin; MTZ, metronidazole; AMX, amoxicillin.
For calculation of mutation rate, see reference 28. The most likely number of mutations per culture was calculated by the p0 method for Clar and Mtzr and by the Lea-Coulson method of median for Cipr and Rifr.
Proportion of cultures without mutants. For example, 2 of 30 cultures of UA802 used for selection of Rifr gave rise to no resistant mutants.
A jackpot (∼1,000) was removed for calculation of the mean number of resistant mutants.
?, undetectable.
Selection and identification of spontaneous mutations.
Independent cultures were grown and plated as described above. To ensure that all mutations represent independent events (but are not the descendents of the same mutation), only one mutant colony was picked from each culture. From the selected independent mutants, a DNA fragment containing the respective gene of interest was PCR amplified using the primers listed in Table 1. By DNA sequencing, the mutations in the gene that are responsible for resistance were identified by comparison with the nucleotide sequence of the wild-type susceptible strain. When a distinct mutation in the gene was identified, it was designated a specific allele of that gene (i.e., a specific variant of the gene that conferred resistance).
TABLE 1.
Oligonucleotide primers
| Primer | Sequence (5′→3′) | Application |
|---|---|---|
| 23SP1 | ACGGCGGCCGTAACTATA | PCR of a 307-bp fragment of 23S rRNA gene; sequencing of the PCR fragment from both directions |
| 23SP2 | ACAGGCCAGTTAGCTA | |
| gyrAP1 | ATGCATGAATTAGGCCTTACT | PCR of a 360-bp fragment of gyrA gene; sequencing of the PCR fragment from both directions |
| gyrAP2 | TTCTTCACTCGCCTTAGTCAT | |
| rpoBP1 | TTTGATTCGCTCATGCCCCAT | PCR of a 330-bp fragment of rpoB gene; sequencing of the PCR fragment from both directions |
| rpoBP2 | CACAACCTTTTTATAAGGGGC | |
| rdxAP1 | TTTGAGCATGGGGCAGATT | PCR of an 850-bp fragment covering the entire rdxA gene with primer rdxAP1 and rdxAP12; sequencing of the PCR fragment using all 5 primers |
| rdxAP2 | GAAGAAATCGCTGAAATCG | |
| rdxAP3 | GTTAATGGTGGTATGCTCT | |
| rdxAP4 | CAAATTTGCATGGGCGTGA | |
| rdxAP12 | AGAGCGATTAAAACCATTCT |
Selection of resistant mutants by the serial passage method.
A serial passage technique as described by Haas et al. (5) was used to select Mtzr mutants. A 3-day-grown H. pylori strain was transferred onto agar plates containing one-half the MIC of metronidazole. After 3 days of incubation, the surviving cells were transferred onto the medium containing twice the prior selective concentration of the antibiotic. These plates were then incubated for 3 days. The process was repeated serially until no growth occurred or a predefined antimicrobial concentration was reached.
DNA manipulation.
Chromosomal DNA from H. pylori strains was isolated as previously described (3). DNA sequencing was carried out using the thermocycling sequencing system with Thermo-Sequenase purchased from Amersham Life Sciences, Cleveland, Ohio. Other DNA manipulations including PCR and gel electrophoresis were performed by standard methods (18).
Natural transformation.
Antibiotic-susceptible H. pylori strains were transformed using chromosomal DNA or a specific PCR fragment from the resistant strains by the method described previously (3). Briefly, recipient cells were heavily inoculated on cold BHI-YE agar plates and grown for 5 h, followed by addition of DNA (300 ng for chromosomal DNA or 30 ng for the PCR fragment) onto the bacterial lawn. After incubation for 20 h under microaerobic conditions the cells were streaked onto BHI-YE plates containing the selective antibiotic for selecting the transformants (the concentrations of antibiotics are the same as those used for selection of spontaneous mutations). A small aliquot of cells (after serial dilution) was plated on drug-free plates to determine the total cell number. After 4 days of incubation, transformants (single colonies) were obtained, and the transformation frequency was calculated as the fraction of the transformants compared with the total number of viable cells. In our experience, the amount of DNA used for transformation is at saturation.
RESULTS AND DISCUSSION
Frequencies of spontaneous mutations.
It is known that there is huge variation in determining the frequency of spontaneous mutants (average fraction of mutant bacteria in a few replicate cultures). Fluctuation assays have been developed to determine the mutation rate (probability of mutation event per cell per generation). Following the methods described by Rosche and Foster (17), we determined both the mutant frequency and mutation rate of H. pylori strains that become resistant to different antibiotics (Table 2). Based on previous reports on the susceptibility of the majority of H. pylori clinical isolates, we choose the selective concentrations of antibiotics as follows: 20 μg/ml for rifampin, 1 μg/ml for ciprofloxacin, 0.5 μg/ml for clarithromycin, 8 μg/ml for metronidazole, and 1 μg/ml for amoxicillin.
Firstly, we examined the rifampin resistance mutations in three different H. pylori strains. Each independent culture (∼5 × 108 cells) usually produces a few to 100 Rifr colonies, although a few cultures may produce no resistant colonies or too many colonies (jackpot). A jackpot is most probably due to a mutation that occurred at very early stage rather than to many mutation events. Out of the 30 cultures of UA802, one culture gave rise to a jackpot (∼1,000 Rifr colonies), which was removed for calculation of the mutant frequency. Jackpot does not affect the calculation of the mutation rate, as the median number is used. A mutant frequency of ∼5 × 10−8 and a mutation rate of ∼1.5 × 10−8 were observed for the three H. pylori strains tested (Table 2). This is a level comparable to that found to occur in Escherichia coli (11, 17). Determination of the frequency and rate of ciprofloxacin resistance (Cipr) mutations in UA802 indicated that it is at levels similar to those of Rifr.
We proceeded to examine the occurrence of mutations that confer resistance to clarithromycin, metronidazole, and amoxicillin, the antibiotics that are frequently used in triple therapy for H. pylori infection. No Clar mutants were observed in 8 out of 12 independent cultures (strain UA802), whereas a few mutants were obtained in the other four cultures. For metronidazole resistance, we had expected a relatively higher frequency. However, no mutants were observed in 11 out of 15 independent cultures (strain 11639), while a few mutants were obtained in the other 4 cultures. The calculated mutation rates for Clar and for Mtzr were <10−9 per cell division, which is about 20-fold lower than that of Rifr. For amoxicillin resistance, an extremely low frequency was observed: no single resistant colony was obtained from 12 cultures of H. pylori 11639. As H. pylori strains exhibit high genetic diversity, the mutant frequencies of Clar, Mtzr, and Amxr were examined using several other strains, and similar results were observed (data not shown).
The low frequency of Clar is not surprising, because mutations at only two particular bases (adenines at positions 2142 and 2143) in the 23S rRNA gene can confer resistance (26). In addition, A-to-C and A-to-T mutants, although conferring resistance, have defects in ribosomal function and cannot compete with the wild type and A-to-G mutant for growth (27). Moreover, the majority of H. pylori Clar isolates contain mutations in both copies of the 23S rRNA gene. Most possibly, this resulted from a spontaneous mutation in one gene followed by gene conversion of the other copy by homologous recombination. Thus, the observed mutation frequency may be the product of the spontaneous mutation rate and the frequency of homologous recombination.
Although beta-lactams have been extensively used for treating other infectious diseases, emergence of resistance to amoxicillin in H. pylori has been reported rarely (1, 6, 23). Using the serial passage procedure in vitro, no mutations that confer resistance to >0.25 μg of amoxicillin/ml have been identified (5). Here we show that the frequency of Amxr mutants is below the level that can be determined in the present assay system (≪10−9). Currently, the mechanism of amoxicillin resistance in H. pylori is not very clear, although some reports suggested that mutation or modification of penicillin binding proteins may be responsible (2, 12). The extremely low frequency of Amxr (observed both in vivo and in vitro) suggests that cooperative mutations in more than one target may be required for Amxr in H. pylori.
The low frequency of Mtzr determined in vitro is in sharp contrast to the high incidence of Mtzr in H. pylori clinical isolates as well as in the H. pylori mouse model experiment (8). As suggested by Martinez and Baquero (15), mutation frequencies are probably much higher in the course of an infective process than those determined in vitro, because bacteria growing in vivo are frequently under environmental stress and challenge. In addition, the actual concentration of metronidazole in vivo to which the bacteria are exposed may not reflect the dose used in vitro. This may account for part of the discrepancy between the observations in vivo and in vitro, but this cannot explain the low frequency of Mtzr compared to that of Rifr, as both were determined in vitro under the same conditions.
Given that the mutational inactivation of the rdxA gene is the major determinant for Mtzr (4), theoretically any mutation in the rdxA gene that leads to the defect of RdxA enzyme renders H. pylori Mtzr. Thus, a spontaneous mutation frequency would be expected to be similar to or even higher than that of Rifr. This prompted us to consider the unique mechanism of metronidazole resistance. Wild-type H. pylori cells have a functional RdxA that reduces metronidazole (nontoxic) to hydroxylamine (toxic), which is responsible for killing the bacterium, whereas the rdxA mutant cannot reduce metronidazole (4). We hypothesized that a few rdxA mutations had occurred but the mutant cells had been killed due to the following possibilities. First, the hydroxylamine was produced from the wild-type cells and penetrated the mutant cells. In other words, when only a very small fraction of rdxA mutants exist in a huge population of the wild-type cells, these mutant cells cannot survive exposure to the drug, even though they themselves do not reduce metronidazole to hydroxylamine. Second, within a short time after the rdxA gene mutation occurs (in the same or the next generation), the functional RdxA enzymes may remain in the cell and produce hydroxylamine. In other words, the new rdxA mutants may still have an Mtzs phenotype (phenotypic lag).
In a reconstruction experiment, we tested the first possibility using an Mtzr 11639 mutant selected in the previous experiment. The culture of these cells (∼109 cells/ml) was diluted 10−7 and 10−8 so that there would be only a few (1–10) cells in a 0.1 ml aliquot. These cells, either alone or together with ∼108 wild-type 11639 (Mtzs) cells, were plated on medium containing 8 μg of metronidazole/ml. After 3 days of incubation, a similar number of colonies were observed for both conditions (i.e., with or without Mtzs cells). As a control, plating ∼108 wild-type cells alone did not give rise to any Mtzr colonies. This result indicated that the presence of a huge population of the wild-type cells does not affect the survival of a few Mtzr mutants in the metronidazole-containing medium. Currently, we suspect that the second possibility, in which the rdxA mutation may not result immediately in Mtzr (phenotypic lag), is most likely responsible for the low frequency observed in vitro.
It was reported that Mtzr H. pylori can be readily selected out in vitro by serial passages on increasing sublethal doses of metronidazole (5, 24). We performed similar experiments to select Mtzr mutants by the serial passage method. The MIC of metronidazole for both strains 26695 and 11639 is 2 μg/ml. Consistent with previous results (5, 24), we were successful in obtaining Mtzr mutants from the both strains by the serial passage method. The mutants can be obtained by one of the following passage procedures: 1 → 2 → 4 → 8 μg/ml, 1 → 2 → 2 → 8 μg/ml, 2 → 4 → 8 μg/ml, 2 → 2 → 8 μg/ml, or (the simplest) 2 → 8 μg/ml.
Selection of resistant mutants by the serial passage procedure may be more similar to the situation in vivo. At a particular site of infection, there could exist a low concentration of metronidazole (metronidazole is usually used to treat parasitic and anaerobic bacterial infections) and thus a correspondingly low level of hydroxylamine, which may increase the mutation rate (hydroxylamine is a DNA-damaging agent) but may not kill the bacteria. We observed that pretreatment of H. pylori cells with 2 μg of metronidazole/ml increased the frequency of Rifr mutants about 10-fold (data not shown), which is in agreement with the recent finding that metronidazole activation is mutagenic (20). Jenks et al. (8) determined the proportion of the Mtzr mutants that emerged in the mice originally infected with a single Mtzs strain, and the results indicated that repeated exposure to sublethal doses of metronidazole in vivo encouraged the development of resistance. Similarly, we observed the drastic increase in the frequency of Mtzr mutants (resistant to 8 μg of metronidazole/ml) in the cell population that was preincubated with 2 μg of metronidazole/ml (data not shown). Therefore, the mutagenic and selective effects of the low doses of metronidazole account for the high frequency of the Mtzr mutants in vivo as well as in the serial passages in vitro.
Spectra of spontaneous mutations. (i) Rifampin resistance.
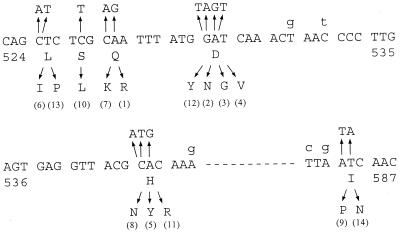
In many bacteria, rifampin resistance is due to mutations in the rpoB gene encoding the β subunit of RNA polymerase, and the Rifr mutations are mainly located in a short region (200 to 300 bp) of the gene. We proceeded to analyze a large number of Rifr mutations for a detailed analysis similar to that done with E. coli (11, 19). We isolated 60 independent Rifr mutants of strain UA802. The MIC of rifampin for these mutants ranges from 32 to 256 μg/ml (Table 3), whereas the MIC for the wild-type UA802 is 0.1 μg/ml. Of the 60 independent mutants analyzed, 57 were identified to have single base substitutions at 14 different alleles of the rpoB gene (designated rpoB1 to rpoB14) (Table 3). The nature of the other three mutants has not yet been determined because we sequenced only the 330-bp fragment of the rpoB gene (Table 1). For these three mutants, no mutation was found in this region, and the mutations are probably in another region of the rpoB gene. The sequence change of each of the Rifr mutations within the rpoB gene is shown in Fig. 1 and listed in Table 3. Many of these spontaneous Rifr mutations are identical to those identified previously by Heep et al. (7). In addition, several new Rifr alleles have been identified, including those encoding D530G, L525I, I586P, S526L, H540R, and D530Y. All mutations identified were located in a short region of the rpoB gene corresponding to cluster I and cluster II of the Rifr-determining region in E. coli (11, 19). The codon corresponding to Asp530 was shown to be the most frequently mutated site. Substitutions at amino acid residues 525, 527, 540, and 586 were also identified multiple times. Four spontaneous Rifr mutants were also isolated from strain 26695, which contained the rpoB2, rpoB3, or rpoB4 alleles (Table 3). This confirms that Asp530 is the most important rifampin-binding site.
TABLE 3.
Spectra of spontaneous mutations
| Mutant strain | No. of independent mutations analyzed
|
Mutant allelesa
|
MIC (μg/ml) | |||
|---|---|---|---|---|---|---|
| Total | Distribution | Designation | Nucleotide change | Amino acid change | ||
| UA802 Rifr | 60 | 6 | rpoB1 | A to G | Q527R | 256 |
| 8 | rpoB2 | G to A | D530N | 128 | ||
| 7 | rpoB3 | A to G | D530G | 256 | ||
| 8 | rpoB4 | A to T | D530V | 128 | ||
| 5 | rpoB5 | C to T | H540Y | 256 | ||
| 1 | rpoB6 | C to A | L525I | 256 | ||
| 3 | rpoB7 | C to A | Q527K | 128 | ||
| 3 | rpoB8 | C to A | H540N | 64 | ||
| 2 | rpoB9 | A to T | I586P | 64 | ||
| 2 | rpoB10 | C to T | S526L | 32 | ||
| 3 | rpoB11 | A to G | H540R | 128 | ||
| 2 | rpoB12 | G to T | D530Y | 32 | ||
| 4 | rpoB13 | C to T | L525P | 128 | ||
| 3 | rpoB14 | T to A | I586N | 128 | ||
| 3 | ? | Unknown | Unknown | 64 | ||
| 26695 Rifr | 4 | 1 | rpoB2 | G to A | D530N | 128 |
| 2 | rpoB3 | A to G | D530G | 256 | ||
| 1 | rpoB4 | A to T | D530V | 128 | ||
| UA802 Clar | 4 | 2 | 23S1 | A2142G | NA | 16 |
| 2 | 23S2 | A2143G | NA | 4 | ||
| UA802 Cipr | 12 | 1 | gyrA1 | A to C | D91A | 4 |
| 4 | gyrA2 | G to T | D91Y | 8 | ||
| 5 | gyrA3 | G to A | D91N | 8 | ||
| 2 | gyrA4 | A to G | D91G | 8 | ||
| 11639 Mtzr | 4 | 1 | rdxA1 | C to T | R16C | 32 |
| 2 | rdxA2 | +A | Shift at 64, stop at 73 | 64 | ||
| 1 | rdxA3 | C to T | P51 L | 64 | ||
| 11639 Mtzr (serial passage) | 18 | 5 | rdxA2 | +A | Shift at 64, stop at 73 | 64 |
| 2 | rdxA4 | −T | Shift at 72, stop at 76 | 64 | ||
| 1 | rdxA5 | +AG | Shift at 41, stop at 55 | 128 | ||
| 1 | rdxA6 | −G | Shift at 133, stop at 137 | 64 | ||
| 1 | rdxA7 | G to T | E175 stop | 32 | ||
| 1 | rdxA8 | −A | Shift at 35, stop at 55 | 64 | ||
| 1 | rdxA9 | G to A | C87Y | 64 | ||
| 1 | rdxA10 | +A | Shift at 9, stop at 23 | 128 | ||
| 1 | rdxA11 | C to T | A67V | 16 | ||
| 1 | rdxA12 | +14 nt | Shift at 70, stop at 80 | 128 | ||
| 1 | rdxA13 | T to C | Y47H | 64 | ||
| 2 | ? | Unknown | Unknown | 64 | ||
When a distinct mutation was identified, it was given an allele name (designation), which represents a specific change at the indicated nucleotide (nt) in a gene and a corresponding amino acid change in the gene product.
FIG. 1.
Sequence alterations corresponding to Rifr mutations. Changes in the nucleotide sequence and the corresponding amino acid residue for the representative Rifr allele are indicated by the arrows, respectively, above and below the sequence of the wild-type UA802 rpoB gene (numbers in parentheses are alleles corresponding to those listed in Table 3). The sequence covers the region of amino acid residues 524 to 587. The lowercase letters above the sequence indicate the nucleotides in the 26695 rpoB gene that diverge from the rpoB gene of UA802. Note that the majority of divergence between these two wild-type strains is due to the base substitutions in the third positions of the codons (silent mutations). In contrast, the mutations that confer Rifr are all in the first or second positions of codons and lead to an amino acid change.
(ii) Clarithromycin resistance.
Because of the low frequency of Clar mutations, only four independent Clar mutants were obtained and analyzed. The sequencing results showed that two of them are due to the mutation A2142G and the other two are due to A2143G (Table 3). This result is in agreement with the observations that these two types of mutation are predominantly associated with Clar in clinical isolates (22, 25).
(iii) Ciprofloxacin resistance.
We analyzed 12 independent ciprofloxacin-resistant (Cipr) mutants of UA802, and four different alleles (base substitutions) of gyrA were found (Table 3). The GyrA protein contains a quinolone resistance-determining region (QRDR) (about 40 amino acids long) at the amino terminus. Mutations in this region of GyrA in many bacteria gave a high level of resistance to quinolones. There was a single report of ciprofloxacin-resistant H. pylori clinical isolates (16), in which several types of base substitutions leading to the amino acid changes in the QRDR of the GyrA were identified. The four Cipr alleles identified in our study all affect the codon Asp91 (changed to different amino acids) in the QRDR of the GyrA, indicating that this residue is the most important target site for ciprofloxacin binding. Two mutations, D91Y and D91N, are frequently found, and these were also identified by Moore et al. (16) in clinical isolates. The D91A mutation was not found in that study.
(iv) Metronidazole resistance.
We obtained only four independent Mtzr mutants from strain 11639 by direct selection on 8 μg of metronidazole/ml. All of them were found to have a mutation in the rdxA gene (Table 3). One of them has a C-to-T mutation that leads to the amino acid change of Arg16 to Cys. A single base substitution mutation (C to T) was also found in another mutant that results in a change of Pro51 to Leu. The other two mutants were due to insertion of an adenine in a run of seven adenines, causing the shift of the reading frame (at position 64) that encounters a stop codon at position 73 (truncation of the protein).
Using the serial passage method, we were able to select many more Mtzr mutants. We analyzed the spectrum of the mutations in 18 independent mutants that were obtained from 18 independent passage experiments. An 850-bp DNA fragment covering the entire rdxA gene coding region as well as ∼100 bp each of its upstream and downstream regions was sequenced (Fig. 2). In 2 of 18 mutants, no mutations were found; the sequence is identical to that of the wild type. The other 16 mutants contain mutations in the rdxA gene, which are shown in Fig. 2 and listed in Table 3. In total, 10 new rdxA alleles (rdxA4 to rdxA13) have been identified from this collection of Mtzr mutants. Mutations that occurred most frequently are frameshifts in simple nucleotide repeat sequences. The addition of an A in a run of seven A's (rdxA2) and the deletion of a T in a run of four T's (rdxA4) were observed five and two times, respectively. The deletion of an A in a run of three A's (rdxA8) and the addition of an A in a run of six A's (rdxA10) occurred once each. The other three insertion/deletion mutations that cause frameshifts are the addition of an A and a G (rdxA5), the deletion of a G (rdxA6), and the addition of 14 bp (rdxA12). The remaining four mutations are base substitutions, including three transitions (rdxA9, rdxA11, and rdxA13) and one transversion (rdxA7). Except for alleles encoding the frameshifts at simple nucleotide repeats, many rdxA alleles identified here are different from those identified in previous studies (4, 9, 10, 13, 21), indicating that these mutations occurred randomly in the rdxA gene and led to its inactivation.
FIG. 2.
Sequence alterations of Mtzr mutations. The DNA sequence of strain 11639 covers the entire rdxA coding region and extends slightly upstream (top line) and downstream (bottom line). The rdxA gene encodes 210 amino acids, and the positions of codons are on the left. The lowercase letters above the sequence indicate the nucleotides in the 26695 rdxA gene that diverge from the rdxA gene of 11639. Nucleotide changes in the Mtzr mutants are indicated by the arrows below the wild-type sequence. The corresponding allele numbers and amino acid changes are given in parentheses (fs, frameshift). Note that in the allele rdxA12, 14 nucleotides (TTGCAGCACACAGC) are inserted, which is the duplicate of the overlined sequence.
While the MIC of metronidazole for the wild-type strain 11639 is 2 μg/ml, the MICs for the isogenic Mtzr mutants varied, ranging from 16 to 128 μg/ml (Table 3). Recent studies (10, 13) demonstrated that rdxA mutation alone results in an MIC up to 32 μg/ml, whereas additional mutations (frequently in the frxA gene) give rise to a higher level of resistance. We have not yet determined what, if any, additional mutations exist in the Mtzr mutants we selected in vitro.
Transformation with mutant DNA.
The chromosomal DNAs were isolated from all mutant (resistant) strains listed in Table 3, except for Clar mutants, which have been studied previously (26). All the DNAs have the ability to transform the susceptible strain to generate a resistant isolate (data not shown). To test if the transformation ability is attributable to the specific mutant allele identified, DNA fragments containing each mutant allele (i.e., PCR fragments of 360 bp for gyrA, of 330 bp for rpoB, and of 850 bp for rdxA) (Table 1) were used for transformation. Again, each mutant allele (i.e., gyrA1 to gyrA4, rdxA1 to rdxA13, and rpoB1 to rpoB14) (Table 3) was able to readily transform the susceptible parental strain to generate resistance to the corresponding antibiotic. The transformation frequencies are in the range of 10−6 to 10−4 transformants per viable cell, which is at least several hundred-fold higher than the frequency of spontaneous mutation. As controls, the recipient cells (total cell number was ∼107 in a typical transformation experiment) that received an aliquot of water did not yield any resistant colonies. Therefore, each resistance phenotype is indeed attributable to the corresponding mutant allele identified. Transformation and recombination of the mutant allele from the resistant cells to susceptible cells (of the same or other strains) at a rate that is at least several hundred-fold higher than spontaneous mutation frequency could contribute to the rapid spread of the mutant allele in the bacterial population. Therefore, the emergence of antibiotic resistance observed in vivo is probably due to the combined effects of spontaneous mutation and recombination.
ACKNOWLEDGMENTS
This work was supported in part by funding from the Canadian Bacterial Diseases Network (Centers of Excellence Program) to D.E.T., who is a Medical Scientist with the Alberta Heritage Foundation for Medical Research (AHFMR).
We thank Yong Leng for calculating the mutation rate.
REFERENCES
- 1.Dore M P, Osato M S, Realdi G, Mura I, Graham D Y, Sepulveda A R. Amoxycillin tolerance in Helicobacter pylori. J Antimicrob Chemother. 1999;43:47–54. doi: 10.1093/jac/43.1.47. [DOI] [PubMed] [Google Scholar]
- 2.Dore M P, Graham D Y, Sepulveda A R. Different penicillin-binding protein profiles in amoxicillin-resistant Helicobacter pylori. Helicobacter. 1999;4:154–161. doi: 10.1046/j.1523-5378.1999.99310.x. [DOI] [PubMed] [Google Scholar]
- 3.Ge Z, Taylor D E. H. pylori DNA transformation by natural competence and electroporation. In: Clayton C L, Mobley H L T, editors. Methods in molecular medicine, Helicobacter pylori protocols. Totowa, N.J: Humana Press Inc.; 1997. pp. 145–152. [DOI] [PubMed] [Google Scholar]
- 4.Goodwin A, Kersulyte D, Sisson G, van Zanten S J O V, Berg D E, Hoffman P S. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 5.Haas C E, Nix D E, Schentag J J. In vitro selection of resistant Helicobacter pylori. Antimicrob Agents Chemother. 1990;34:1637–1641. doi: 10.1128/aac.34.9.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han S-R, Bhakdi S, Maeurer M J, Schneider T, Gehring S. Stable and unstable amoxicillin resistance in Helicobacter pylori: should antibiotic resistance testing be performed prior to eradication therapy? J Clin Microbiol. 1999;37:2740–2741. doi: 10.1128/jcm.37.8.2740-2741.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heep M, Beck D, Bayerdorffer E, Lehn N. Rifampin and rifabutin resistance mechanism in Helicobacter pylori. Antimicrob Agents Chemother. 1999;43:1497–1499. doi: 10.1128/aac.43.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenks P J, Labigne A, Ferrero R L. Exposure to metronidazole in vivo readily induces resistance in Helicobacter pylori and reduces the efficacy of eradication therapy in mice. Antimicrob Agents Chemother. 1999;43:777–781. doi: 10.1128/aac.43.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenks P J, Ferrero R L, Labigne A. The role of the rdxA gene in the evolution of metronidazole resistance in Helicobacter pylori. J Antimicrob Chemother. 1999;43:753–758. doi: 10.1093/jac/43.6.753. [DOI] [PubMed] [Google Scholar]
- 10.Jeong J Y, Mukhopadhyay A K, Dailidiene D, Wang Y, Velapatino B, Gilman R H, Parkinson A J, Nair G B, Wong B C, Lam S K, Mistry R, Segal I, Yuan Y, Gao H, Alarcon T, Brea M L, Ito Y, Kersulyte D, Lee H K, Gong Y, Goodwin A, Hoffman P S, Berg D E. Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori. J Bacteriol. 2000;182:5082–5090. doi: 10.1128/jb.182.18.5082-5090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin D J, Gross C A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 12.Kusters J G, Schuijffel D F, Gerrits M M, van Zwet A A, Vandenbroucke-Grauls C M J E. A single amino acid change in PBP-1A causes amoxicillin resistance in Helicobacter pylori. Gut. 1999;45(Suppl. III):A5. [Google Scholar]
- 13.Kwon D H, El-Zaatari F A, Kato M, Osato M S, Reddy R, Yamaoka Y, Graham D Y. Analysis of rdxA and involvement of additional genes encoding NAD(P)H flavin oxidoreductase (FrxA) and ferredoxin-like protein (FdxB) in metronidazole resistance of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44:2133–2142. doi: 10.1128/aac.44.8.2133-2142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luria S E, Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez J L, Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother. 2000;44:1771–1777. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore R A, Beckthold B, Wong S, Kureishi A, Bryan L E. Nucleotide sequence of the gyrA gene and characterization of ciprofloxacin-resistant mutants of Helicobacter pylori. Antimicrob Agents Chemother. 1995;39:107–111. doi: 10.1128/aac.39.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosche W A, Foster P L. Determining mutation rates in bacterial populations. Methods. 2000;20:4–17. doi: 10.1006/meth.1999.0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Severinov K, Soushko M, Goldfarb A, Nikiforov V. Rifampicin region revisited. New rifampicin-resistant and streptolydigin-resistant mutants in the β subunit of Escherichia coli RNA polymerase. J Biol Chem. 1993;268:14820–14825. [PubMed] [Google Scholar]
- 20.Sisson G, Jeong J Y, Goodwin A, Bryden L, Rossler N, Lim-Morrison S, Raudonikiene A, Berg D E, Hoffman P S. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori RdxA+ (nitroreductase) gene. J Bacteriol. 2000;182:5091–5096. doi: 10.1128/jb.182.18.5091-5096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tankovic J, Lamarque D, Delchier J C, Soussy C J, Labigne A, Jenks P J. Frequent association between alteration of the rdxA gene and metronidazole resistance in French and North African isolates of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44:608–613. doi: 10.1128/aac.44.3.608-613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor D E, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of the two copies of 23S rRNA genes from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997;41:2621–2628. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Zwet A A, Vandenbroucke-Grauls C M J E, Thijs J C, van der Wouden E J, Gerrits M M, Kusters J G. Stable amoxicillin resistance in Helicobacter pylori. Lancet. 1998;352:1595. doi: 10.1016/s0140-6736(98)00064-6. [DOI] [PubMed] [Google Scholar]
- 24.Van Zwet A A, Thijs J C, Vries W S, Schiphuis J, Snijder J A M. In vitro studies on stability and development of metronidazole resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1994;38:360–362. doi: 10.1128/aac.38.2.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Versalovic J, Shortridge D, Kibler K, Griffy M V, Bryer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G, Taylor D E. Site-specific mutations in the 23S rRNA gene of Helicobacter pylori confer two types of resistance to macrolide-lincosamide-streptogramin B antibiotics. Antimicrob Agents Chemother. 1998;42:1952–1958. doi: 10.1128/aac.42.8.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G, Rahman M S, Humayun M Z, Taylor D E. Multiplex sequence analysis demonstrates the competitive growth advantage of the A-to-G mutants of clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother. 1999;43:683–685. doi: 10.1128/aac.43.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang G, Humayun M Z, Taylor D E. Mutation as an origin of genetic variability in Helicobacter pylori. Trends Microbiol. 1999;7:488–493. doi: 10.1016/s0966-842x(99)01632-7. [DOI] [PubMed] [Google Scholar]