Abstract
The retinal pigment epithelium (RPE) resides at the back of the eye and performs functions essential for maintaining the health and integrity of adjacent retinal and vascular tissues. At present, the limited reparative capacity of mammalian RPE, which is restricted to small injuries, has hindered progress to understanding in vivo RPE regenerative processes. Here, a detailed methodology is provided to facilitate the study of in vivo RPE repair utilizing the zebrafish, a vertebrate model capable of robust tissue regeneration. This protocol describes a transgenic nitroreductase/metronidazole (NTR/MTZ)-mediated injury paradigm (rpe65a:nfsB-eGFP), which results in ablation of the central two-thirds of the RPE after 24 h treatment with MTZ, with subsequent tissue recovery. Focus is placed on RPE ablations in larval zebrafish and methods for testing the effects of pharmacological compounds on RPE regeneration are also outlined. Generation and validation of RpEGEN, a MATLAB script created to automate quantification of RPE regeneration based on pigmentation, is also discussed. Beyond active RPE repair mechanisms, this protocol can be expanded to studies of RPE degeneration and injury responses as well as the effects of RPE damage on adjacent retinal and vascular tissues, among other cellular and molecular processes. This zebrafish system holds significant promise in identifying genes, networks, and processes that drive RPE regeneration and RPE disease-related mechanisms, with the long-term goal of applying this knowledge to mammalian systems and, ultimately, toward therapeutic development.
Introduction
The methodology described herein details a protocol to genetically ablate the retinal pigment epithelium (RPE) utilizing larval zebrafish. The RPE extends over the back of the eye and resides between the stratified layers of the neural retina and the layer of vasculature constituting the choroid. Trophic support, absorption of phototoxic light, and maintenance of visual cycle proteins are only some of the critical functions the RPE performs that are essential for sustaining the health and integrity of these adjacent tissues1. Damage to mammalian RPE is reparable when lesions are small2; however, damage suffered by larger injuries or progressive degenerative disease is irreversible. In humans, RPE degenerative diseases (e.g., age-related macular degeneration (AMD) and Stargardt disease) lead to permanent vision loss and, with few treatment options available, decreased patient quality of life. The limited ability for mammalian RPE to self-repair has created a knowledge gap in the field of RPE regenerative processes. Given the robust regenerative capacity of the zebrafish across many different tissue types, this protocol was developed to establish an in vivo vertebrate system to facilitate studies on intrinsically regenerating RPE and uncover mechanisms that drive that response. Using the ablation paradigm outlined here, the canonical Wnt signaling pathway3, the mTOR pathway4, and immune-related responses5 have been identified as critical mediators of RPE regeneration, likely with overlapping functions.
In this genetic ablation paradigm, Tg(rpe65a:nfsB-eGFP)3 zebrafish express the bacterial-derived nitroreductase (NTR/nfsB) gene6 fused to eGFP under control of the RPE enhancer element, rpe65a7. Ablation is achieved by adding the prodrug, metronidazole (MTZ), to system water housing zebrafish. Intracellular activation of MTZ by nitroreductase results in DNA crosslinking and apoptosis in NTR/nfsB-expressing cells8, 9. This technology has been widely used in zebrafish to ablate cells of the retina10, 11, 12, 13 and other tissues8. Together, these elements enable targeted expression (rpe65a) of an inducible cell ablation methodology (NTR/MTZ)8, 9 and a fluorescent marker (eGFP) for visualization.
Other interesting in vivo models also exist that can be used to study the regenerative potential of the RPE14. These are broad and include RPE-to-retina transdifferentiation post-retinectomy in amphibians, in which RPE cells lost to retinal regrowth are replaced15, 16; RPE restoration post-injury in the “super healing” MRL/MpJ mouse17; and exogenous stimulation of RPE proliferation in a rat model of spontaneous RPE and retinal degeneration18, among others. In vitro models, such as adult human RPE stem cells (RPESCs)19 have also been developed. These models are all valuable tools working to uncover the cellular processes related to RPE regeneration (e.g., proliferation, differentiation, etc.); however, the zebrafish is unique in its capacity for intrinsic RPE repair post-ablation.
While the methodology here is written to focus on understanding the mechanisms driving RPE regeneration, the Tg(rpe65a:nfsB-eGFP) line and this genetic ablation protocol could be utilized to study other cellular processes such as RPE apoptosis, RPE degeneration, and the effect of RPE injury on adjacent retinal and vascular tissues. The ablation protocol can also be modified to include pharmacological manipulation, which is a convenient preliminary strategy to screen signaling pathways of interest. For example, blocking the canonical Wnt pathway using Inhibitor of Wnt Response-1 (IWR-1)20, has been shown to impair RPE regeneration3. This was repeated here to guide users through a pharmacological manipulation experiment and serve as proof-of-concept to validate a MATLAB script (RpEGEN) created to quantify RPE regeneration based on recovery of pigmentation. Like the transgenic line and ablation protocol, the RpEGEN scripts are adaptable and could be used to quantify other markers/cellular processes within the RPE.
Protocol
All methodologies outlined herein are compliant with the Institutional Animal Care and Use Committee (IACUC) of the University of Pittsburgh.
1. Preparation prior to zebrafish embryo collection
Set embryo incubator to 28.5°C.
-
Prepare a 25x stock solution of the melanogenesis inhibitor, N-phenylthiourea (PTU)21, 22. This stock solution is scaled from a common recipe22 and 1x is equal to 0.003% weight per volume (% w/v) (e.g., 0.003 g of PTU powder into 100 mL of liquid solvent).
-
To make a large 25x PTU stock solution, add 0.75 g of PTU powder to 1 L of purified deionized water (hereafter referred to as dH2O) and mix thoroughly at room temperature (~25 °C) using a stir bar and stir plate. Store at 4°C for up to 3 months protected from light.
NOTE: It is difficult to get PTU to go into aqueous solution and extended stirring overnight may be necessary.
CAUTION: PTU is hazardous, and care should be taken to prevent ingestion, inhalation, and/or contact with the skin or eyes. PTU powder and all PTU liquid derivatives described herein may need to be disposed of as chemical waste depending on state and institutional regulations. Confirmation of proper PTU waste disposal methods, if any, is recommended prior to use.
-
To make a 1.5x PTU working solution (hereafter referred to as 1.5x PTU), add 60 mL of 25x PTU stock solution to 940 mL of zebrafish housing facility water (hereafter referred to as system water). Optimal water quality parameters for zebrafish have been described23 and aquatics facilities should have standard water monitoring procedures in place. Store 1.5x PTU at 28.5°C for 1-2 weeks protected from light.
NOTE: This protocol is routinely performed using the PTU concentrations, solvents, and storage parameters described in step 1.2. As a precaution, embryos/larvae should be observed every 1-2 days while in PTU to validate efficacy and confirm sustained depigmentation. Dissolution and/or storage conditions should be optimized if decreases in PTU solubility/efficacy are suspected.
-
Prepare a stock solution of 0.05% w/v methylene blue, a fungal growth inhibitor, by adding 0.05 g of methylene blue powder to 100 mL of dH2O. Mix thoroughly using a stir bar and stir plate. Store at 4°C.
-
Prepare pipettes for embryo/larva manipulation (e.g., moving embryos/larvae between Petri dishes, separating larvae during fluorescence screening, collecting euthanized larvae into microcentrifuge tubes for fixation, etc.) by cutting back the tapered end of a glass Pasteur pipette using a diamond tip scribing pen. Etch around the circumference of the pipette with the diamond pen and gently pull or snap the end off to make a clean break.
NOTE: The mouth of the pipette should be smooth and wide enough to easily take up an embryo still inside the chorion without shearing. Use a bulb draw transfer pipette as an alternative. Prepared pipettes can also be used to remove liquid during water changes (rather than pouring) to minimize embryo/larva loss.
-
Prepare a 4% paraformaldehyde (PFA) solution in 1x phosphate buffered saline (PBS) (e.g., add 10 mL of 16% PFA to 4 mL of 10x PBS and 26 mL of dH2O). Store at 4°C for up to 4 weeks protected from light.
CAUTION: Paraformaldehyde is a hazardous chemical and should be handled in a chemical fume hood and disposed of properly. Care should be taken, and personal protective equipment (PPE) should be worn, to prevent ingestion, inhalation, and/or contact with the skin or eyes.
2. Zebrafish embryo collection and maintenance prior to genetic ablation (0-5 days post-fertilization)
Maintain adult zebrafish as described previously3, 4, 5. The afternoon/evening prior to embryo collection, separate adult zebrafish into breeding tanks for spawning.
-
The following morning (0 days post-fertilization (dpf)), collect embryos into 10 cm diameter Petri dishes in system water and remove all nonviable or unfertilized eggs, which will appear opaque and/or show irregular cytoplasm and failed cleavage24.
NOTE: Normal cleavage and developmental staging events will be apparent in healthy embryos as described25. Petri dishes should be kept three-fourths full (~30 mL for a 10 cm diameter dish) throughout the protocol.
Add two drops of 0.05% w/v methylene blue to each Petri dish, mix gently, and store embryos at 28.5°C for the remainder of the protocol.
-
Around 6 h post-fertilization (hpf)4, 5, 26, replace system water in embryo Petri dishes with 1.5x PTU (working solution made in step 1.2.2) and replenish methylene blue.
NOTE: PTU must be added to embryos prior to the onset of pigmentation (i.e., before 24 hpf)25 as already pigmented tissues will not depigment upon addition of PTU21. It should be noted, however, that reduced eye size, ocular and craniofacial deficits, and disruption of some signaling pathways (e.g., thyroid signaling) have been reported in PTU-treated zebrafish27, 28, 29. The developmental toxicity of PTU appears to be dependent on concentration and timing of PTU addition27, 29. As mentioned above for validating PTU efficacy (step 1.2), signs of PTU toxicity should also be carefully monitored and, if suspected, the working concentration and/or time of PTU addition should be optimized.
-
On 2-3 dpf, dechorionate embryos using freshly made pronase solution.
-
Dissolve pronase in 1.5x PTU at a concentration of 2 mg/mL by vortexing.
CAUTION: Pronase is packaged as a very fine powder and is an irritant. Take measures to avoid inhalation and/or contact with skin, eyes, etc.
Separate hatched embryos from unhatched embryos and pronase-treat only unhatched embryos.
Replace 1.5x PTU with 2 mg/mL pronase solution made in step 2.4.1 and leave on unhatched embryos for 4-5 min with gentle agitation (e.g., on a tabletop rotator/shaker or by manual swirling).
Pour off the pronase solution and immediately rinse with fresh 1.5x PTU. Gently triturate 1.5x PTU rinse over embryos with a bulb-draw transfer pipette.
-
Repeat a second 1.5x PTU rinse to discard all chorion debris, and then replenish 1.5x PTU for maintenance.
NOTE: Embryos can also be dechorionated manually using fine-tipped forceps. In this case, chorion debris should be removed and 1.5x PTU replenished after manual dechorionation.
-
-
Monitor embryo/larval health and replenish 1.5x PTU every 1-2 days. Embryos/larvae are kept in 1.5x PTU until ablation at 5 dpf.
NOTE: The importance of steps 2.3 and 2.4 are addressed further in the Discussion section.
3. Screening zebrafish larvae for rpe65a:nfsB-eGFP and genetic ablation of the retinal pigment epithelium (5-6 days post-fertilization)
-
Make a fresh 10 mM metronidazole (MTZ) solution on 5 dpf (day of ablation). This process takes 2 h to complete.
Add MTZ powder to system water without PTU and mix thoroughly by vigorous shaking (e.g., 250 rotations per min) for 1 h at 37 °C.
-
Cool 10 mM MTZ solution for an additional 1 h at room temperature on a tabletop rotator/shaker and ensure complete dissolution prior to adding to Petri dishes with larvae.
NOTE: Fluorescence screening and separation of eGFP+ larvae (step 3.2) can be performed during the 37 °C and room temperature incubations.
CAUTION: MTZ is hazardous, and care should be taken to prevent ingestion, inhalation, and/or contact with the skin or eyes. MTZ powder and all liquid derivatives described herein may need to be disposed of as chemical waste depending on state and institutional regulations. Confirmation of proper MTZ waste disposal methods, if any, is recommended prior to use.
-
Screen zebrafish larvae for the rpe65a:nfsB-eGFP transgene.
-
Anesthetize larvae with 0.168 g/L of tricaine (MS-222) and separate transgenic (eGFP+) larvae (Figure 1) from non-transgenic (eGFP−) larvae using a fluorescence stereo microscope with a 488 nm excitation laser/filter.
NOTE: Tricaine should be added to 1.5x PTU and/or pharmacological compound solutions as larvae should remain immersed in the treatment while being screened for eGFP. Incubate larvae in tricaine only for the duration of screening (e.g., ≤10 min for a single 10 cm Petri dish containing 50 larvae).
Wake screened larvae up immediately by pipetting directly into a Petri dish with fresh 1.5x PTU without tricaine.
Upon completion of screening, further separate the eGFP+ larvae into two groups of Petri dishes: one group to receive MTZ treatment (ablated/MTZ+) and one group to be the unablated (MTZ−) control.
-
-
Ablate the retinal pigment epithelium.
Remove 1.5x PTU from the unablated (MTZ−) control dishes and add fresh system water without PTU.
Remove 1.5x PTU from the ablated (MTZ+) treatment dishes and add the freshly made 10 mM MTZ solution (step 3.1.).
-
Remove the 10 mM MTZ solution after exactly 24 h (designated 1 day post-injury (dpi)) and add fresh system water without PTU. Change out the fresh system water without PTU on the MTZ− dish(es). Larvae will not be exposed to PTU again for the remainder of the protocol.
NOTE: It may be difficult to pipette or pour off all 1.5x PTU (steps 3.3.1 and 3.3.2) or 10 mM MTZ (step 3.3.3) between solution exchanges without larval loss as animals are actively swimming around. In this event, wash(es) of system water without PTU can be added to ensure successful solution exchange.
Figure 1: rpe65a:nfsB-eGFP transgene expression on 5 days post-fertilization.
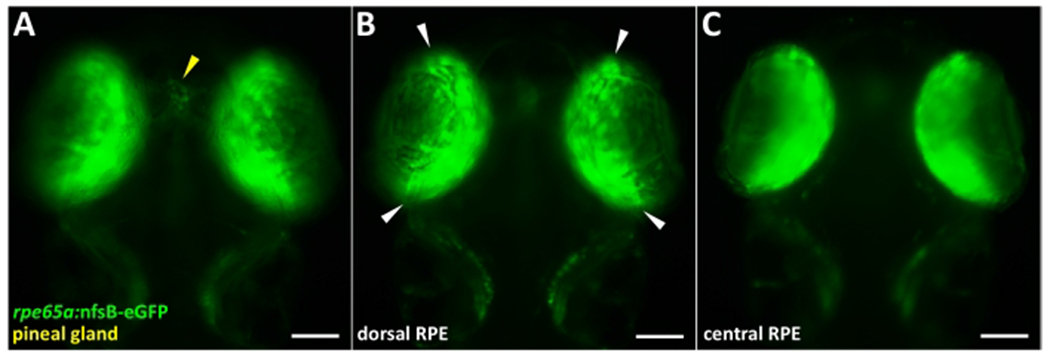
(A-C) Wholemount images of a PTU-treated 5 dpf larva showing dim transgene expression in the pineal gland (A; yellow arrowhead) and bright transgene expression in the RPE (B,C) at the time of screening for genetic ablation. (B) Transgene expression is visibly localized to the central two-thirds of the RPE, and white arrowheads highlight the boundary between peripheral (immature) and central (mature) RPE. Green = eGFP. Anterior is up. Scale bars = 100 μm. Please click here to view a larger version of this figure.
4. Larval maintenance post-genetic ablation (6+ days post-fertilization)
Check larvae and replenish system water without PTU daily until euthanasia (step 5.6) or return to zebrafish housing facility.
Monitor the success and extent of ablation in vivo on 2 dpi (7 dpf) using transmitted light illumination on a stereo microscope (Figure 2).
Figure 2: Verification of successful genetic ablation of the RPE in vivo.
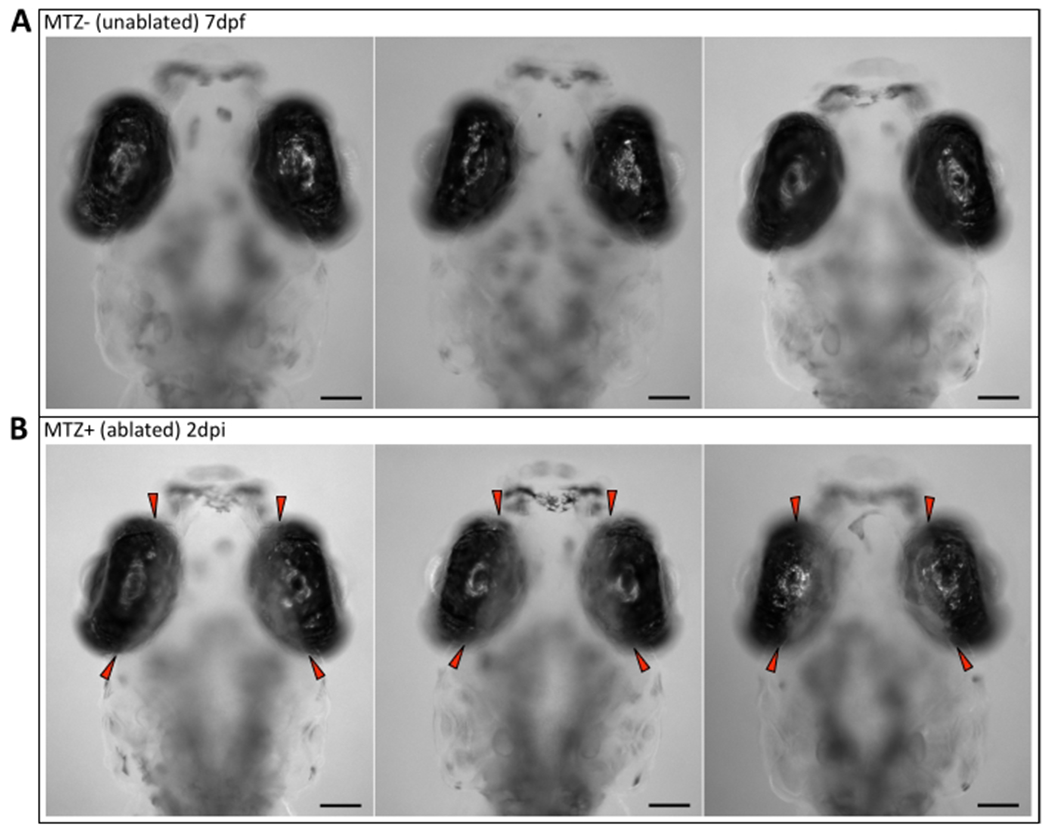
Wholemount images of (A) three unablated (MTZ−) 7 dpf larvae showing RPE pigmentation throughout the eye and (B) three ablated (MTZ+) 2 dpi larvae showing an ablation zone where there is an absence of pigment in the central two-thirds of the RPE (red arrowheads). Anterior is up. Scale bars = 100 μm. Please click here to view a larger version of this figure.
5. Incorporating pharmacological treatment into zebrafish retinal pigment epithelium ablation protocol
NOTE: As performed previously3, treatment with 15 μM IWR-1 or volume-matched dimethyl sulfoxide (DMSO) vehicle control starting at 4 dpf is outlined here as an example experiment to test RpEGEN. Concentrations and timelines may vary with different pharmacological compounds and recommendations for dose-response validation, treatment duration, screening, and other aspects of experimental design for pharmacological manipulation studies are addressed in the Discussion section. Follow steps 6 and 7 if image analysis is required.
Collect and maintain embryos as described in step 2. Dechorionate embryos on 2 dpf.
-
Screen eGFP+ larvae on 4 dpf as described in step 3.2. Rather than Petri dishes, place eGFP+ larvae into 6-well plates at a density of n ≤ 10 larvae per 6-well for pharmacological treatment. Designate separate 6-well plates for larvae that will be unablated (MTZ−) and larvae that will be ablated (MTZ+).
NOTE: The eGFP signal is visible on 4 dpf but appears dimmer than signal intensity on 5 dpf.
-
Pretreat 4 dpf eGFP+ larvae with 15 μM IWR-1 or volume-matched DMSO vehicle control for exactly 24 h prior to genetic ablation with 10 mM MTZ solution.
NOTE: Often, very little compound is needed for pharmacological treatment experiments. To avoid weighing out small quantities of IWR-1 powder, this pharmacological compound is purchased already in DMSO solution, at a concentration of 25 mM, and aliquoted into smaller volumes upon arrival to avoid repeated freeze-thaw cycles.
Determine the volume of pharmacological and vehicle control treatments needed and aliquot 1.5x PTU into conical tubes accordingly. A volume of 5 mL/well of a 6-well plate is recommended.
-
Add IWR-1 stock to 1.5x PTU for a final concentration of 15 μM IWR-1 (e.g., 3 μL of 25 mM IWR-1 per 5 mL of 1.5x PTU). Add a matched volume of DMSO stock to 1.5x PTU (e.g., 3 μL ≥ 99.7% DMSO per 5 mL of 1.5x PTU). Here, this will end up yielding a final concentration of 0.06% volume/volume (% v/v) DMSO. Mix well by vortexing and visually confirm dissolution of compounds.
CAUTION: DMSO, IWR-1, and other pharmacological compounds and solvents may need to be disposed of as chemical waste depending on state and institutional regulations. Confirmation of hazard level and proper waste disposal methods for these compounds, if any, is recommended prior to use.
Remove 1.5x PTU from the eGFP+ larvae in 6-well plates and add 5 mL/well of freshly made 0.06% v/v DMSO or 15 μM IWR-1 treatments from step 5.3.2.
-
On 5 dpf, ablate the RPE. In addition to the 24 h pretreatment (step 5.3), larvae will remain immersed in pharmacological and vehicle control treatments during 24 h of genetic ablation with 10 mM MTZ and during post-ablation recovery, until fixation (e.g., from 4-9 dpf).
Make 10 mM MTZ solution 2 h prior to performing genetic ablation (step 3.1).
Determine the volume of pharmacological and vehicle control treatments needed for both unablated (MTZ−) and ablated (MTZ+) 6-well plates and aliquot appropriate volumes of either fresh system water without PTU (MTZ−) or 10 mM MTZ solution (MTZ+) into conical tubes. This will yield four treatment conditions: 1) 0.06% v/v DMSO, MTZ−; 2) 15 μM IWR-1, MTZ−; 3) 0.06% v/v DMSO, MTZ+; and 4) 15 μM IWR-1, MTZ+.
Add IWR-1 and DMSO stock solutions to respective conical tubes as performed in step 5.3.2. Mix well by vortexing and visually confirm dissolution of compounds.
Remove 0.06% v/v DMSO and 15 μM IWR-1 treatments in 1.5x PTU (step 5.3.2) from designated unablated (MTZ−) and ablated (MTZ+) 6-well plates and replenish with the appropriate treatments made in step 5.4.3.
Remove 0.06% v/v DMSO and 15 μM IWR-1 treatments in 10 mM MTZ solution after exactly 24 h and replenish with treatments in fresh system water without PTU. Replenish 0.06% v/v DMSO and 15 μM IWR-1 treatments in fresh system water without PTU on the unablated (MTZ−) 6-well plate(s).
Follow larval maintenance post-ablation as outlined in step 4 and replenish 0.06% v/v DMSO or 15 μM IWR-1 treatments daily in system water without PTU.
Euthanize larvae on 9 dpf (4 dpi for age-matched MTZ-treated siblings) by immersing animals in 0.3 g/L tricaine solution (lethal overdose) coupled with rapid chilling (e.g., place Petri dishes on ice) for at least 20 min30. Check to make sure that larvae are unresponsive to touch and fix in 4% PFA (step 1.5) for 3 h at room temperature or at 4 °C overnight.
Process post-fixation larval tissue for z-stack image acquisition on a confocal microscope as described previously5, 31 and here in the Representative Results section. Analysis in steps 6 and 7 will require, at minimum, acquisition of nuclear marker (e.g., DAPI) and brightfield z-stack images.
6. Confocal microscope z-stack image preprocessing in FIJI (ImageJ)
-
Import and format confocal microscope z-stack images using FIJI32.
Open microscope images using the Bio-Formats Import Options set to View stack with: Hyperstack and Color mode: Grayscale.
Generate a maximum intensity projection of the imported microscope image by choosing Image | Stacks | Z Project. In the ZProjection window, set Start Slice and Stop Slice to include all slices for that image. For example, set Start Slice: 1 and Stop Slice: 18 for a z-stack that has a total of 18 slices. Choose Projection Type: Max Intensity and click on OK.
Convert the maximum intensity projection file to an 8-bit image (if not already) by choosing Image | Type | 8-bit.
-
Reorient the image so that the dorsal side is up and distal (i.e., the lens) is left by choosing Image | Transform | Flip Horizontally (for the last command, choose the option that best suits the directionality needed for that image). For a multichannel image, click on Yes in the Process Stack? window to reorient all channels.
NOTE: This step is critical, but not needed if the image is already in the dorsal up, distal left orientation. See Figure 3A–D and Figure 4 for images with eyes in the correct directionality for processing.
Save the 8-bit maximum intensity projection as a tagged image file format (TIF) file by choosing File | Save As | Tiff….
-
Generate RPE region of interest (ROI) using FIJI.
Open an 8-bit TIF image generated as described in step 6.1. Start the ROI Manager by choosing Analyze | Tools | ROI Manager.
Use Image | Zoom and toggle between the DAPI and brightfield channels to identify the point(s) at which the apical side of the RPE is adjacent to the tip of the outer limiting membrane (OLM) (Figure 3B’, B”, D’, D”; blue arrowheads). Use this anatomical landmark as the ROI starting point.
-
Create the RPE ROI with the Polygon Selections tool (in the FIJI toolbar) using both the DAPI and brightfield image channels and zoom function (Image | Zoom) to identify apical and basal RPE boundaries. Bring the dorsal and ventral ends of the ROI (i.e., where the ROI transitions from the apical to basal side of the RPE) to a sharp point rather than blunting or rounding off (Figure 3B’, B”, D’, D”; magenta lines).
NOTE: Creating a pointed ROI end is a critical step for optimizing endpoint detection using RpEGEN and is addressed in the Discussion section.
Add the ROI by clicking on Add in the ROI Manager. Adjust the ROI as needed and click on Update in the ROI Manager.
Save the ROI file by choosing More» | Save… within the ROI Manager. Use identical names for matched ROI and TIF image files (e.g., [file name].tif and [file name].roi).
Combine 8-bit TIF and ROI files for each condition into a single folder. For example, the folder, DMSO_9dpf, will contain all the matched TIF and ROI files for the 9 dpf unablated (MTZ−) 0.06% v/v DMSO-treated larval group.
Figure 3: RPE regions of interest (ROIs) for automated quantification using RpEGEN.
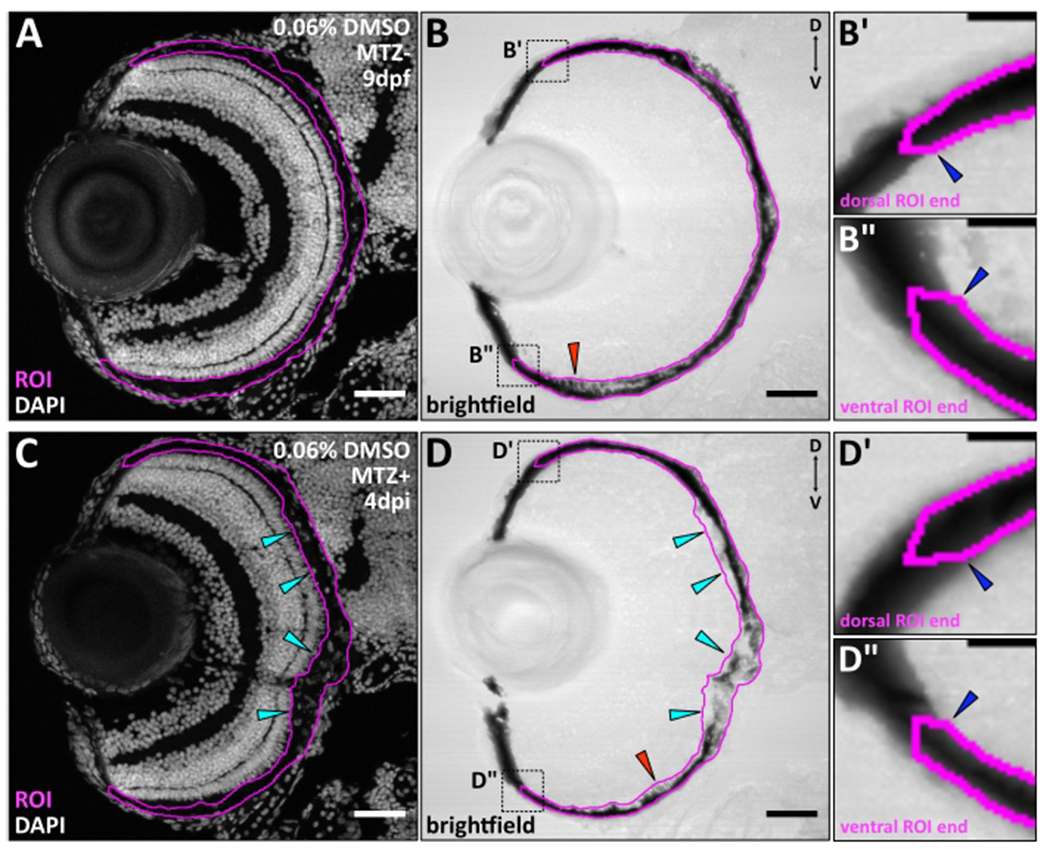
Transverse cryosections of (A,B) an unablated (MTZ−) 9 dpf larva and (C,D) an ablated (MTZ+) 4 dpi larva with the RPE ROIs highlighted in magenta. (B,D) Red arrowheads highlight regions where visibly pigmented apical microvilli were included in the ROI. (C,D) Cyan arrowheads point to example regions of injury site-localized DAPI+ debris, which was used to include RPE cell debris in the ROI. (B’,B”,D’,D”) Digital zooms of both the dorsal and ventral ROI regions (black dotted boxes in B and D) show suggested ROI starting points (blue arrowheads) and the pointed ROI ends that are critical for endpoint detection in MATLAB. Brightfield images are also shown in Figure 9E,G. White/gray = nuclei. Dorsal is up and distal is left. (A-D) Scale bars = 40 μm. (B’,B”,D’,D”) Scale bars = 10 μm. Please click here to view a larger version of this figure.
Figure 4: RpEGEN processing workflow.
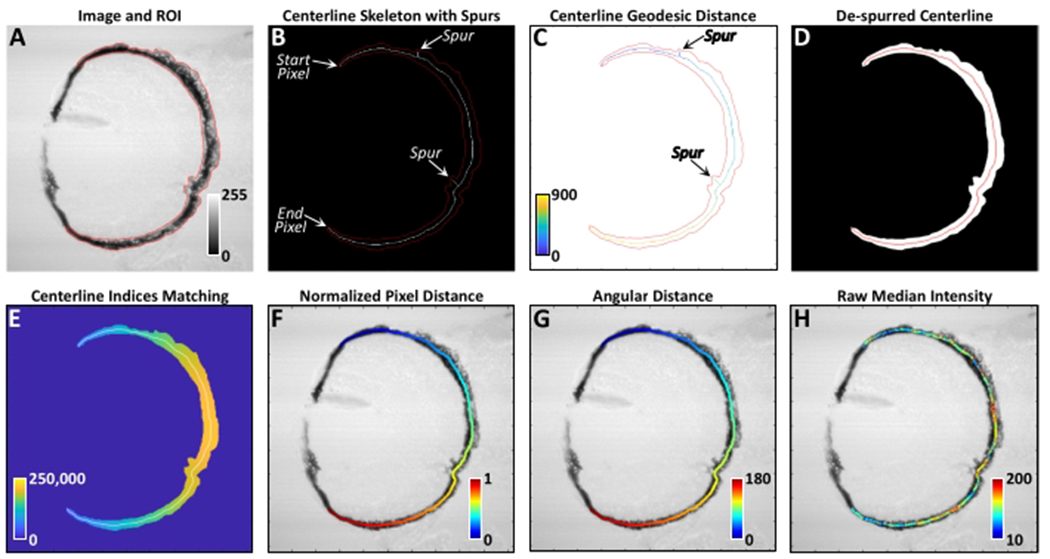
(A) Example of a properly oriented 8-bit brightfield image and FIJI-generated ROI (red) imported into MATLAB. Dorsal is up and distal is left. Color bar represents 8-bit grayscale intensity values where 0 = black and 255 = white. (B) Initial binary skeletonization (white) of the ROI mask (red) showing the presence of erroneous spurs and the delineation of the start and end points for geodesic pixel distance delineation as shown in (C). Color bar in (C) represents Euclidean pixel distance. (D) Spurs are removed using a simplified least-cost pathway from the end pixel back to the start pixel, resulting in a continuous centerline devoid of spurs (red). (E) Nearest linear centerline indices based on a distance transform between all pixels in the ROI mask and the centerline pixels (white). These index values permit each image pixel within the ROI mask to be assigned to the closest centerline pixel for analysis. Color bar represents the linear centerline pixel index values. (F) An example of the normalized pixel distance along the centerline, where 0 (blue, distal-most dorsal pixel) is the start pixel and 1 (red, distal-most ventral pixel) is the end pixel. (G) Similar to (F) but using the angular distance, where 0 degrees (blue) is the start pixel and 180 degrees (red) is the end pixel. (H) Example of the median pixel intensity values calculated for each centerline pixel. Color bar represents the median grayscale intensity value of all ROI pixels contributing to each given centerline pixel. This figure shows imagery from a test dataset and not larvae from the 0.06% v/v DMSO or 15 μM IWR-1 treatment groups. Please click here to view a larger version of this figure.
7. Quantification and visualization of RPE regeneration using RpEGEN scripts
-
Install and prepare RpEGEN scripts.
Download the latest RpEGEN scripts from the GitHub repository (https://github.com/burchfisher/RpEGEN) by clicking on Code | Download ZIP.
Unzip the folder and place it in the desired workspace location (e.g., Desktop).
Open MATLAB.
Navigate to the RpEGEN folder in the Current Folder pane (usually on the left side).
Right click on the RpEGEN folder and choose Add to Path | Selected Folders and Subfolders. This adds the folder to the MATLAB path so it can automatically find and run any scripts in the folder.
Double click on the RpEGEN folder in the Current Folder pane to show all the subfolders and M files.
Double click on the RpEGEN.m file to open in the Editor pane.
Under the USER-DEFINED VARIABLES section of the RpEGEN.m file, enter the directory locations for the folders containing the ROI files (.roi), image files (.tif), and where output files should be saved. Enter the group name for the .mat file to be exported (e.g., DMSO_9dpf, DMSO_4dpi, etc.) and the location of the brightfield channel in the TIF image stack (e.g., 3, if brightfield is the third channel in an image stack). If the image file only contains the brightfield image, then this should be equal to 1.
-
Run RpEGEN.m script and validate the results.
NOTE: RpEGEN requires the Image Processing Toolbox, the Curve Fitting Toolbox, and the Statistics and Machine Learning Toolbox to be activated on the user MATLAB license in order to run. In addition, the freely available ReadlmageJROI toolbox33 is required to import FIJI ROIs into MATLAB; however, it is provided in the RpEGEN folder along with other function M files that do not require any activation.
-
Run the script by clicking on the Run button in the Editor menu at the top of MATLAB.
NOTE: Once initiated, the Command window will provide verbose output indicating the progression of the script. After saving the MAT file containing the extracted data, a three-panel figure will appear and be saved as a PDF to the output directory for each image run. These are quality control (QC) figures to make sure that everything has run properly and include: 1) the brightfield image overlaid by the ROI (Figure 4A); 2) the ROI with centerline and associated angular distance (degrees) (Figure 4G); and 3) the ROI with the centerline median intensity values (0-255, 8-bit color scale) (Figure 4H). Wait until the QC PDFs have been saved to the output folder and the last figure has disappeared before proceeding to the next step.
-
Open the individual PDFs exported to the output folder in any PDF viewer and verify that all ROIs match the brightfield images, that centerline values are reasonable approximations of the center of the ROIs, and that median intensity values are appropriately populated with data (i.e., not all the same value across the entire centerline).
NOTE: A detailed description and structure of each variable saved in the MAT file by RpEGEN.m can be found in Table 1.
-
-
Run the RpEGEN_PermPlot.m script.
NOTE: The RpEGEN_PermPlot.m script uses the output of RpEGEN.m to run statistical comparisons using a permutation simulation of the medians of two groups and also provides the code for reproducing the plots in this paper using the freely available GRAMM toolbox34, which has also been included in the RpEGEN folder.
Double click on the RpEGEN_PermPlot.m file to open in a new Editor tab.
Under the SECTION 1 - USER-DEFINED VARIABLES in the RpEGEN_PermPlot.m file, enter the directory location for the output folder containing the MAT files from running RpEGEN.m and enter each MAT file name to be loaded (e.g., DMSO_4dpi.mat, IWR1_4dpi.mat).
Run this section of the script by clicking on the Run Section button in the Editor menu at the top of MATLAB.
-
In Section 2, enter the names of the two groups for statistical comparison in the data_A and data_B variables (these are the groups from which medians will be derived using the permutation simulation). In the bin_sz variable, enter the number of degrees over which to integrate the median intensity values for the datasets (default is 1-degree bins).
NOTE: The reps variable indicates the number of permutations to use to build a probability distribution and can be set to any number (default value is 20,000). In general, a higher number of repetitions will be more statistically robust but will increase processing time.
Run this section of the script by clicking on the Run Section button in the Editor menu at the top of MATLAB. This section may take some time to complete depending on the number of repetitions specified but does provide a continuous status update in the command window.
-
Run HEATMAP FIGURE and GROUP RESULTS AND P-VALUES sections independently using the Run Section button. Edit data variables in any sections commented with “ENTER DATA HERE”. PDFs of these figures are automatically saved for each and can be easily modified in any vector software post processing.
NOTE: The plots generated in the RpEGEN_PermPlot.m file are ad hoc and will likely require modifications based on each user’s specific data and visualization needs. However, the figures do provide a solid foundation that can be easily individualized using both MATLAB and GRAMM websites.
Table 1:
Description of variables exported from RpEGEN.m script to the MAT file.
| Variable Name | Variable Type | Variable Size | Variable Description |
|---|---|---|---|
| ROIname | Cell | {N x 1} | Names of the ROI files processed |
| ROIxy | Cell | {N x 1} | ROI vertices for each ROI file processed |
| IMGname | Cell | {N x 1} | Names of the TIF image files processed |
| IMG | Cell | {N x 1} | 8-bit brightfield image data for each TIF processed |
| ROIBW | Cell | {N x 1} | Logical (0 or 1) ROI mask with the same dimensions as IMG images |
| CLine | Cell w/ Tables | {N x 1} (n x 16) | Centerline data for individual images including x, y, distance, angle, index, # of points, and statistics |
| Cldx | Cell | {N x 1} | Centerline indice data relating ROI mask points to the closest centerline point for calculating CLine |
| CLineBW | Cell | {N x 1} | Logical (0 or 1) matrix with the same dimensions as IMG images indicating the location of the centerline (=1) |
| RAW_data | Table | N x 3 | Table combining all the raw data for each pixel in the ROIs across all the different IMG images |
| BIN_1_deg_all | Table | N x 9 | Table containing 1-degree binned data and statistics using the data from RAW_data |
| BIN_5_deg_all | Table | N x 9 | Table containing 5-degree binned data and statistics using the data from RAW_data |
| BIN_10_deg_all | Table | N x 9 | Table containing 10-degree binned data and statistics using the data from RAW_data |
Definitions are as follows: ROI(s) = region(s) of interest; TIF = tagged image file format.
Representative Results
Inhibiting the canonical Wnt signaling pathway is known to significantly impair zebrafish RPE regeneration using the genetic ablation paradigm (rpe65a:nfsB-eGFP) and pharmacological manipulation methodology (IWR-1) described in the protocol3. This experiment was repeated here to validate an automated method for quantifying zebrafish RPE regeneration based on pigmentation. The results summarized below encompassed all steps of the protocol, from the day of fertilization (0 dpf) to quantification of RPE regeneration using RpEGEN.
Implementing the RPE ablation protocol (with pharmacological manipulation) in larval zebrafish
Embryos collected from three separate parent groups (N = 3) began treatment with 1.5x PTU around 6 hpf and absence of pigmentation in the RPE and surface melanocytes was visually confirmed on 1 dpf. Embryos were enzymatically dechorionated on 2 dpf using 2 mg/mL of pronase (step 2.4). On 4 dpf, larvae were anesthetized using tricaine (MS-222) and screened for eGFP using a fluorescence stereo microscope (steps 3.2 and 5.2). eGFP+ larvae were moved into 6-well plates (n = 10 larvae/well) and were treated with either 0.06% v/v DMSO (vehicle control) or 15 μM IWR-1 (step 5.3). The larval density reported here was initially based on the approximation of 1 larva/cm2 growth area and was validated through careful health monitoring (e.g., swim bladder development) over time. Intensity of eGFP appeared dim on 4 dpf, so larvae were re-screened on 5 dpf to confirm bright eGFP expression (Figure 1). RPE65 (rpe65a in zebrafish) is a marker of mature RPE7 and, in teleost fish, retinal and RPE cells are continually generated from the ciliary marginal zone (CMZ), a stem cell niche at the distal tip of the retina, adjacent to the lens35. Thus, in the rpe65a:nfsB-eGFP transgenic line, the more immature RPE exist at the periphery and appear eGFP− (approximately one-third of the total RPE tissue) while the mature, central two-thirds of the RPE expresses eGFP (Figure 1B; white arrowheads). The transgene was also visible in the pineal gland (Figure 1A; yellow arrowhead), which was expected as rpe65a shows pineal expression in zebrafish36. Comparatively dim or eGFP− larvae were pulled from 6-well plates and euthanized on 5 dpf. At exactly 24 h post-treatment with DMSO or IWR-1, 5 dpf larvae were treated with 10 mM MTZ to ablate the RPE (steps 3.1, 3.3, and 5.4). MTZ was washed out after exactly 24 h (i.e., on 6 dpf/1 dpi) to allow RPE regeneration to take place.
Larvae were observed daily on a stereo microscope using transmitted light illumination from 6 dpf/1 dpi to the time of euthanasia on 9 dpf/4 dpi. Success of genetic ablation was confirmed in vivo on 2 dpi by the absence of pigment in the central two-thirds of the eye in MTZ+ larvae (Figure 2B; red arrowheads) compared to 7 dpf MTZ− control siblings (Figure 2A) (step 4.2). As anticipated, the area devoid of pigment on 2 dpi appeared analogous to the region of rpe65a:nfsB-eGFP transgene expression observed on 5 dpf (Figure 1B). Assessment was performed on 2 dpi and not earlier as RPE undergo repigmentation after removal of PTU and depending on the time of observation in vivo, a marked difference between the ablated central RPE and spared (unablated) peripheral RPE may be difficult to discern if the latter has not fully repigmented. Despite the possibility for macroscopic ambiguity, RPE ablation at 1 dpi was previously shown to be readily apparent in sectioned tissue, revealing a loss of central RPE and outer nuclear layer (ONL; i.e., photoreceptor) tissue integrity along with evidence of cell death (pyknotic nuclei) (Figure 5)3. Counter to tissue loss, robust proliferation was determined to be a key driver during tissue regeneration in the rpe65a:nfsB-eGFP zebrafish model3. Both the early apoptotic response, where RPE ablation leads to degeneration of adjacent tissues (e.g., photoreceptors and Bruch’s membrane; Figure 6A–C), and the peripheral-to-central proliferative response that follows, which yields heterogeneous regeneration “zones” moving inward into the injury site (Figure 6D–F), have been extensively characterized and discussed previously3.
Figure 5: Evidence of RPE ablation and photoreceptor degeneration.
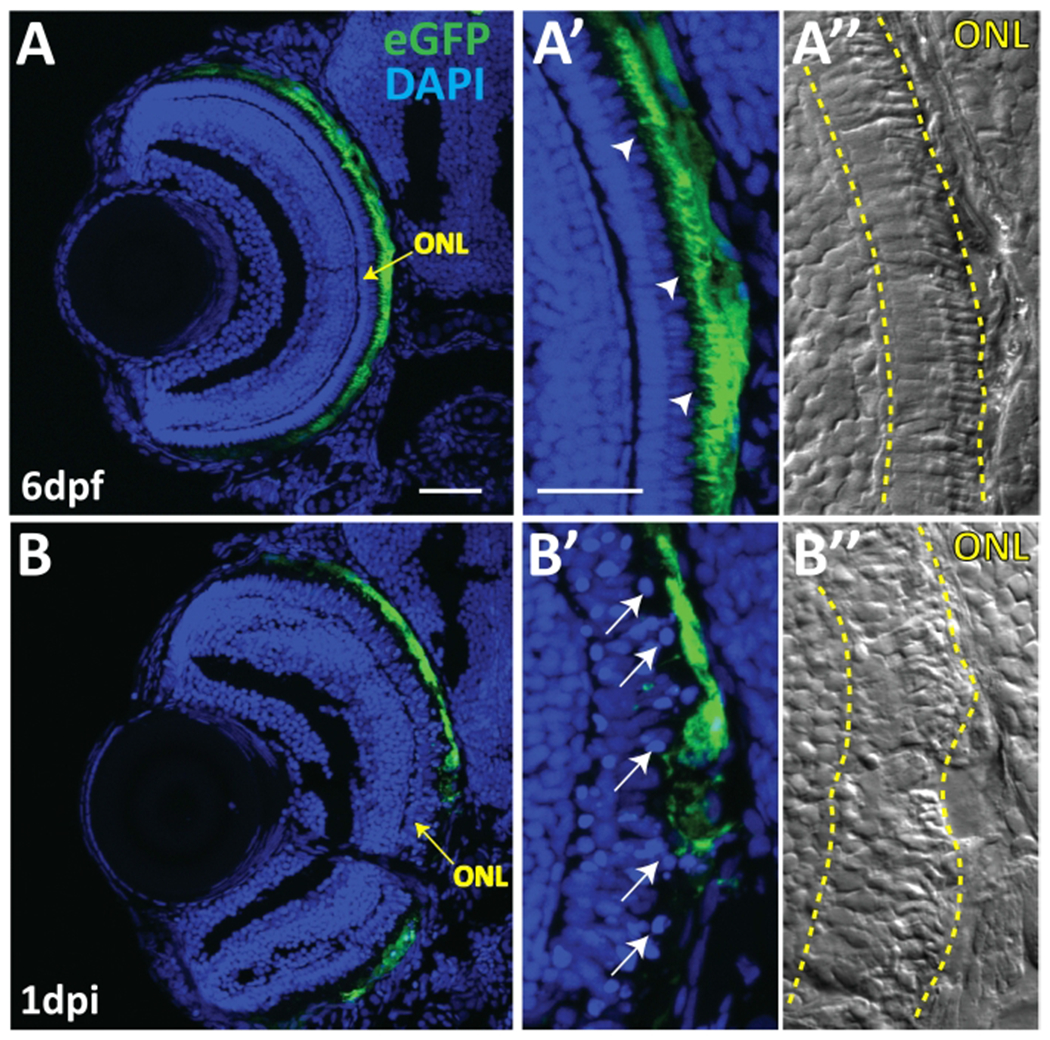
(A) Transverse cryosections of an unablated 6 dpf larva. (A,A’) After exposure to PTU, transgene expression is restricted to mature RPE cells, with the brightest expression confined to the central two-thirds of the RPE. Arrowheads indicate apical microvilli. (A”) Differential interference contrast (DIC) images reveal normal outer nuclear layer (ONL; i.e., photoreceptor) architecture. (B,B’) Transverse cryosections of a 1 dpi larva reveal significant disruption of eGFP+ cell morphology and disorganization in ONL lamination. Arrows indicate delaminated and pyknotic nuclei. (B”) DIC images further reveal the marked disruption of ONL architecture. Green = eGFP, blue = nuclei, yellow = ONL. Dorsal is up and distal is left. Scale bar in (A) represents 40 μm and can be applied to (B). Scale bar in (A’) represents 40 μm and can be applied to (A”,B’,B”). This figure and figure legend text have been modified from Hanovice et al. 2019 (Figure 1)3. Please click here to view a larger version of this figure.
Figure 6: Model of RPE regeneration in larval zebrafish.
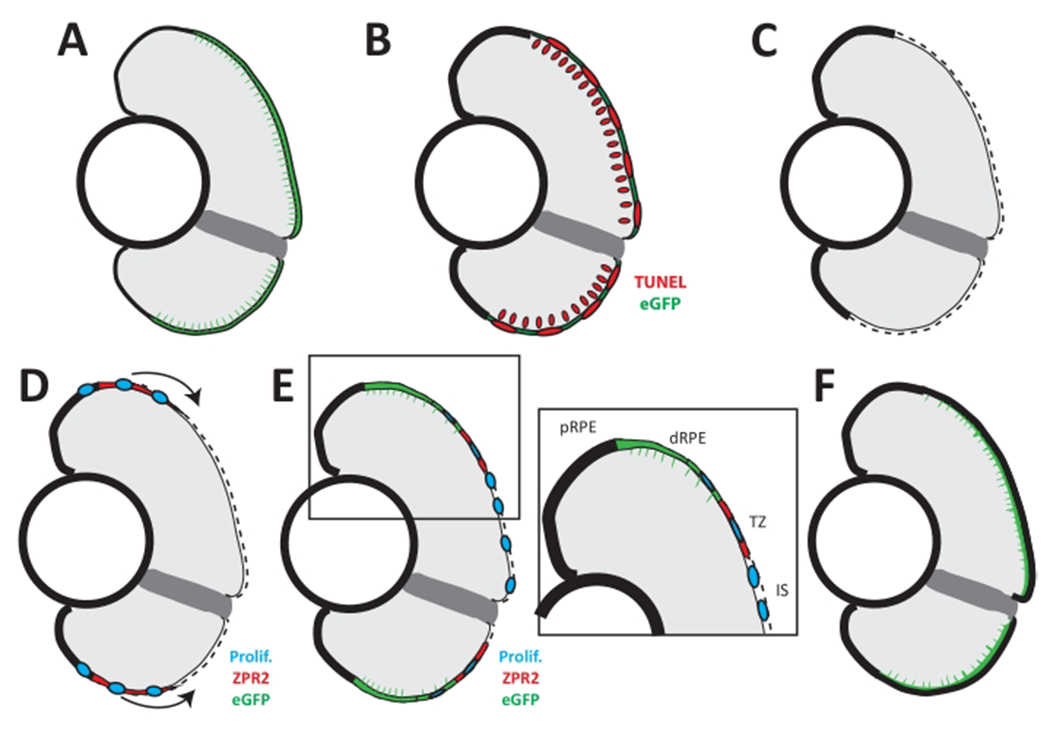
(A) nfsB-eGFP is expressed in mature RPE in the central two-thirds of the eye. (B) Application of MTZ leads to apoptosis (TUNEL, red) of RPE and photoreceptors. (C) RPE ablation leads to degeneration of photoreceptors and Bruch’s membrane (dotted line). (D) Unablated RPE in the periphery begin to proliferate and extend into the injury site (blue). (E) As regenerated eGFP+ RPE appear in the periphery, the RPE can be divided into four zones: peripheral RPE (pRPE), differentiated RPE (dRPE), transition zone (TZ), and injury site (IS). (E, inset) Regenerated differentiated RPE (green) appears in the periphery proximal to the unablated peripheral RPE, and contains proliferative cells adjacent to the transition zone. The transition zone consists of still-differentiating RPE cells (ZPR2, red) and proliferative cells (blue). The injury site comprises unpigmented proliferative cells that do not express any RPE differentiation markers. (F) Regeneration of a functional RPE layer and Bruch’s membrane is complete by 14 dpi. This figure and figure legend text have been modified from Hanovice et al. 2019 (Figure 14)3. Please click here to view a larger version of this figure.
Tissue preparation, confocal image acquisition, and image preprocessing for automated quantification based on RPE pigmentation using RpEGEN
On 9 dpf/4 dpi, DMSO- and IWR-1-treated larvae were euthanized by tricaine overdose and fixed in 4% PFA for 3 h at room temperature (step 5.6). Four larvae from each independent parent group were randomly chosen for subsequent tissue processing (N = 3; n = 12 larvae per treatment). Cryoprotection, cryosectioning (at 12 μm thickness), nuclear counterstaining with DAPI, and coverslip mounting were performed as referenced in step 5.75, 31. A central section, with visible optic nerve, from each larva was imaged on a confocal laser scanning microscope using a 40x oil immersion objective (numerical aperture = 1.30) to acquire 512 x 512-pixel z-stack images with a 1 μm z-step interval. Each image contained data from three channels: channel 1 = 405 nm excitation for DAPI, channel 2 = 488 nm excitation for eGFP, channel 3 = transmitted light for brightfield. As pixel intensity was quantified in brightfield images, transmitted light lamp voltage settings were kept constant and all data collected for statistical comparisons were imaged on the same day.
Confocal microscope z-stack images were preprocessed for automated quantification as described in step 6, using FIJI. Images were excluded from preprocessing and quantification if the tissue was compromised in a way that would make ROI generation difficult (e.g., from tearing (Figure 7A; magenta arrowheads) or landmark obstruction (Figure 7B; magenta ovals)) or skew ROI intensity measurements (e.g., from folding (Figure 7C; magenta ovals)). Despite omitting a few larvae with these exclusion criteria, all datasets herein represent N = 3 with the following number of biological replicates (larvae): n = 11, DMSO MTZ−; n = 10, IWR-1 MTZ−; n = 11, DMSO MTZ+; n = 12, IWR-1 MTZ+. Using the DAPI channel, RPE ROIs were initiated by first identifying the point at which the dorsal apical side of the RPE (retinafacing) was directly adjacent the dorsal tip of the OLM (Figure 3B’,D’; blue arrowheads) (step 6.2.2). As distal RPE extension can vary between sections, the OLM was used as an anatomical landmark to standardize RPE ROI endpoints and normalize angular distance measurements in MATLAB (where 0° = dorsal ROI endpoint and 180° = ventral ROI endpoint). When performing RPE ablations with pharmacological manipulation or in a mutant background, an anatomical landmark should be identified and validated prior to preprocessing and quantification. Here, the OLM was apparent in all larvae quantified and was not compromised by tearing (as in Figure 7B) or treatment with DMSO or IWR-1. After identifying the dorsal starting point, ROIs were generated following the apical side of the RPE (dorsal-to-ventral) until the point adjacent the tip of the ventral OLM was reached (Figure 3B”,D”; blue arrowheads). Then, a ventral ROI endpoint was made between the apical and basal (choroid-facing) sides of the RPE as discussed in step 6.2.3 (Figure 3B”,D”; magenta line). The ROI was continued, following the basal side of the RPE (ventral-to-dorsal) until reaching the basal side adjacent to the starting point at the dorsal OLM. The ROI was then enclosed by creating a pointed dorsal end (Figure 3B’,D’; magenta line) as done ventrally. Genetic ablation has been shown to result in a loss of central eGFP expression that is recovered as regeneration proceeds (summarized in Figure 6)3; thus, depending on the extent of regeneration and the intensity of eGFP at the time point of interest, the eGFP channel may only be useful for ROI generation in the MTZ− group. Therefore, while confocal acquisitions also contained eGFP channel images, ROI generation was completed using the DAPI and brightfield channels as mentioned in step 6.2.3. RPE ROI generation was straightforward in DMSO- and IWR-1-treated MTZ− larval groups and encompassed regions with visibly pigmented apical microvilli (Figure 3B; red arrowhead) along with the prominently pigmented cell bodies. The same parameters were applied to MTZ+ larval groups (Figure 3D; red arrowhead); however, due to residual damaged tissue within the injury site, apical microvilli and pigmentation boundaries were difficult to delineate in the brightfield channel alone in some cases. In these areas, the DAPI channel was also used to identify cellular debris within the RPE injury site, which was included in the ROI (Figure 3C,D; examples of injury site-localized cellular debris are indicated with cyan arrowheads). Immunostaining with markers of immature/mature RPE (e.g., ZPR2; Figure 6D,E)37 and/or photoreceptors (e.g., ZPR1)37, 38 could also be employed to facilitate outlining RPE ROIs in ablated larvae.
Figure 7: Compromised tissue sections excluded from image analysis.
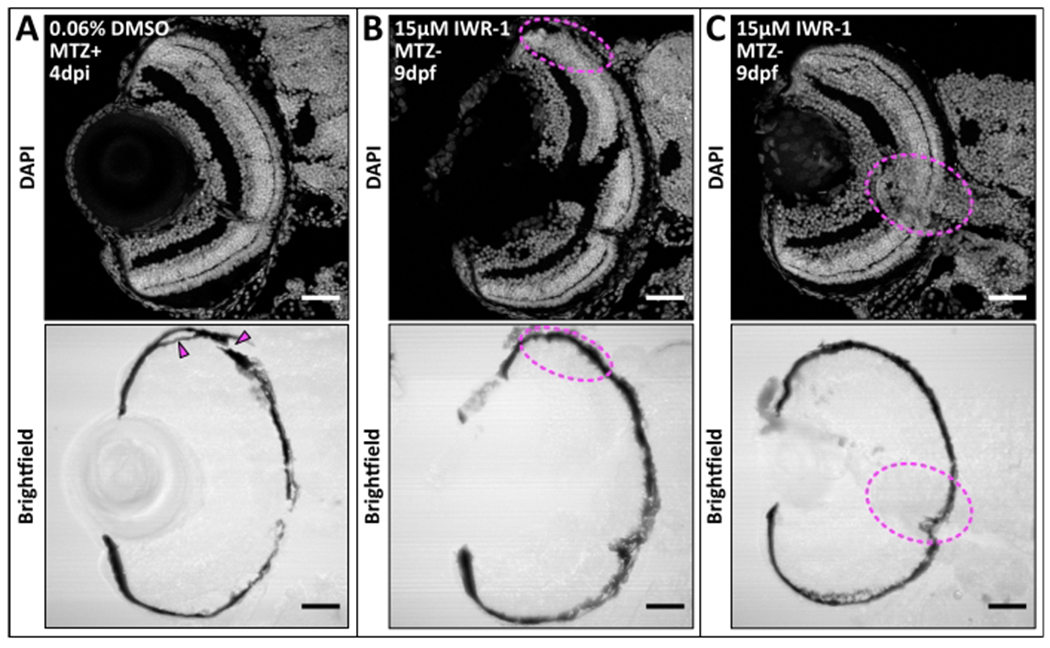
(A-C) Transverse cryosections of larvae from the 0.06% v/v DMSO and 15 μM IWR-1 datasets that met the exclusion criteria. One larva shows dorsal RPE tissue tearing (A; magenta arrowheads) and two other larvae show tissue folding that either obstructs an anatomical landmark (dorsal OLM) in the DAPI channel (B; magenta dotted ovals) or may skew RPE intensity measurements from the brightfield channel (C; magenta dotted ovals). White/gray = nuclei. Dorsal is up and distal is left. Scale bars = 40 μm. Please click here to view a larger version of this figure.
Previously, ablated larvae treated with IWR-1 from 4 dpf to 4 dpi showed significant impairment of RPE regeneration compared to DMSO-treated sibling controls (Figure 8C)3. This was reported as the percent of central pigment recovery, which required substantial prior experience with the model and manual measurements of angular distance to designate regeneration boundaries (Figure 8B; black arrowheads). RpEGEN was created to automate quantification of RPE regeneration and reduce intrinsic biases. Not only that, RpEGEN also enabled generation of highly robust datasets; for example, 10 data points were previously generated from manual quantification of n = 10 DMSO-treated MTZ+ larvae (Figure 8C; % pigment recovery per section measurements)3, whereas 174,801 data points were generated from n = 11 DMSO-treated MTZ+ larvae/ROIs here using RpEGEN (Figure 9C; pixel intensity measurements).
Figure 8: Pharmacological inhibition using IWR-1 impairs RPE regeneration.
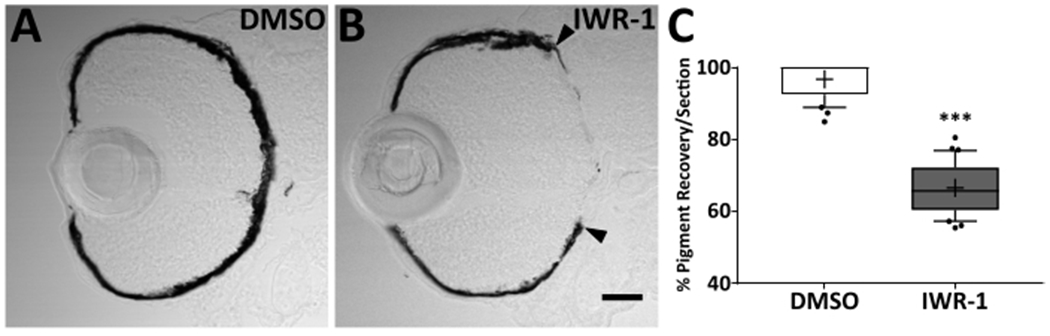
Transverse cryosections of ablated (MTZ+) 4 dpi larvae from (A) 0.06% v/v DMSO and (B) 15 μM IWR-1 treatment groups. Brightfield images (A,B) and quantification of percent RPE recovery/section (C) shows a significant delay in recovery of a pigmented monolayer in IWR-1 treated larvae (Student’s unpaired t-test, *** p < 0.0001). (B) Black arrowheads indicate the central-most edge of the regenerating RPE. Dorsal is up and distal is left. Scale bar in (B) represents 40 μm and can be applied to (A). This figure and figure legend text have been modified from Hanovice et al. 2019 (Figure 13)3. Please click here to view a larger version of this figure.
Figure 9: Raw data output from automated quantification of RPE regeneration in RpEGEN.
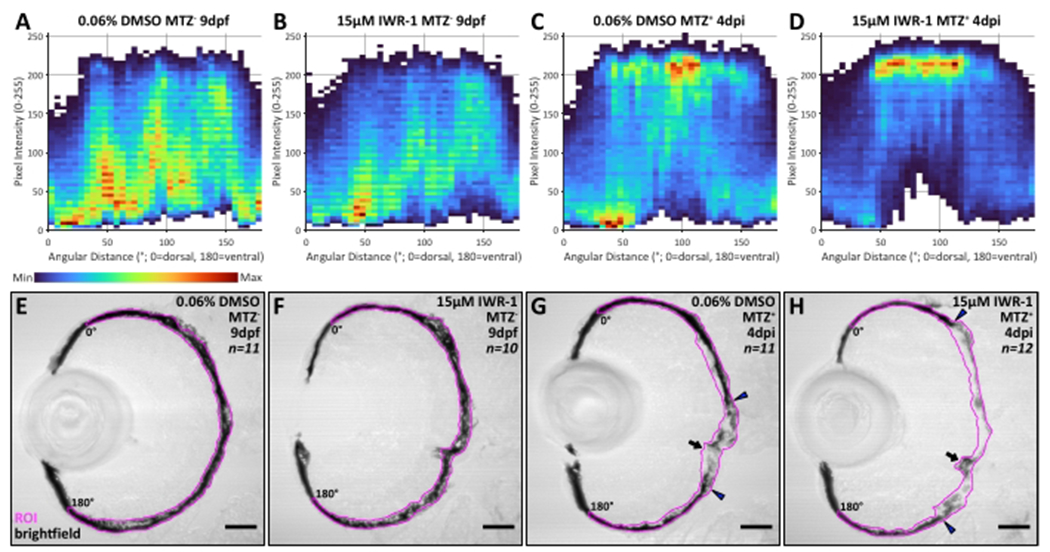
(A-D) Heatmaps showing brightfield pixel intensity distributions compiled from the entire RPE ROI region, from dorsal (x-axes; angular distance = 0°) to ventral (x-axes; angular distance = 180°), for all larvae in each dataset: (A) n = 11, DMSO MTZ−; (B) n = 10, IWR-1 MTZ−; (C) n = 11, DMSO MTZ+; (D) n = 12, IWR-1 MTZ+ (from three independent parent groups, N = 3). For example, (A) displays data from 177,460 pixels across 11 ROIs. On the y-axes, pixel intensity is shown based on an 8-bit color scale where 0 = black and 255 = white. Raw data is displayed in 5-degree (x-axis) by 5-8-bit intensity value (y-axis) bins where red = maximum bin count and dark blue = minimum bin count. (E-H) Transverse cryosections of representative (E,F) unablated (MTZ−) 9 dpf larvae and (G,H) ablated (MTZ+) 4 dpi larva from 0.06% v/v DMSO and 15 μM IWR-1 treatment groups. RPE ROIs are highlighted in magenta and angular distance extremes are indicated (0° = dorsal, 180° = ventral). Broadly, these data show distribution of darker pixels (intensity values between 0-150) in (A,B,E,F) unablated ROIs when compared to (C,D,G,H) ablated ROIs, regardless of treatment. Data from ablated ROIs shows (C,G) centralized (100° ± 15°) distribution of lighter pixels (intensity values between 150-250) in the DMSO treatment group that (D,H) expands dorsally (to ~50°) and slightly ventrally (by ~5°) in IWR-1-treated larvae. (G,H) These central ablation zones absent pigment are highlighted by blue arrowheads. Black arrows indicate location of the optic nerve. Images from DMSO-treated larvae are also shown in Figure 3. Scale bars = 40 μm. Please click here to view a larger version of this figure.
RpEGEN was developed to incrementally assess regional changes in pigmentation over the entirety of the RPE by deriving a skeletonized centerline (1-pixel width) from the original ROI generated using FIJI (Figure 4A,B). To incorporate all pixels within the ROI for analysis (i.e., not just the centerline pixels), a Euclidean distance transform was performed on each pixel from the centerline to create a map of the closest centerline index for each pixel in the ROI mask. This map of indices allowed each pixel outside the centerline to be attributed to a single centerline pixel, creating a comprehensive dataset across the entire RPE for each larva (Figure 4E). The dorsal-to-ventral length of the RPE was represented by angular distance (Figure 4G; 0-180°) as opposed to normalized pixel distance (Figure 4F; 0-1 arbitrary units), as this was more intuitive based on previous measurements of RPE regeneration3, 4, 5 and the fact that angular distance does not vary with morphological differences. Median intensities derived from 5-degree bins were the metric chosen to highlight large-scale trends and minimize high-frequency variability observed in the 1-degree binned data (Table 1, Figure 10A). Permutation simulation was chosen to test the null hypothesis to see whether the medians of the two treatment groups (here, DMSO MTZ+ and IWR-1 MTZ+ (both 4 dpi), Figure 10B) are similar39. This resampling technique lacks strict assumptions about the distribution of the data and can be used on a range of test statistics (e.g., mean, median, etc.) to generate robust p-value estimates. A number of these parameters (e.g., bin size of raw and median data, permutation simulation repetitions, etc.) can be adapted and modified to fit user needs. For the latter, 20,000 repetitions were chosen to generate a statistically robust comparison, however, care should be taken to balance computational efficiency with statistical robustness as too few repetitions may produce erroneous p-value estimates. It is recommended that multiple repetition values be run (10,000, 20,000, etc.) to assure that the p-value distribution and values are relatively stable before commencing interpretations. Increasing permutation repetitions will increase statistical power (but will also take longer to run), which may be beneficial for some datasets.
Figure 10: Group results and statistical comparison derived from automated quantification of RPE regeneration in RpEGEN.
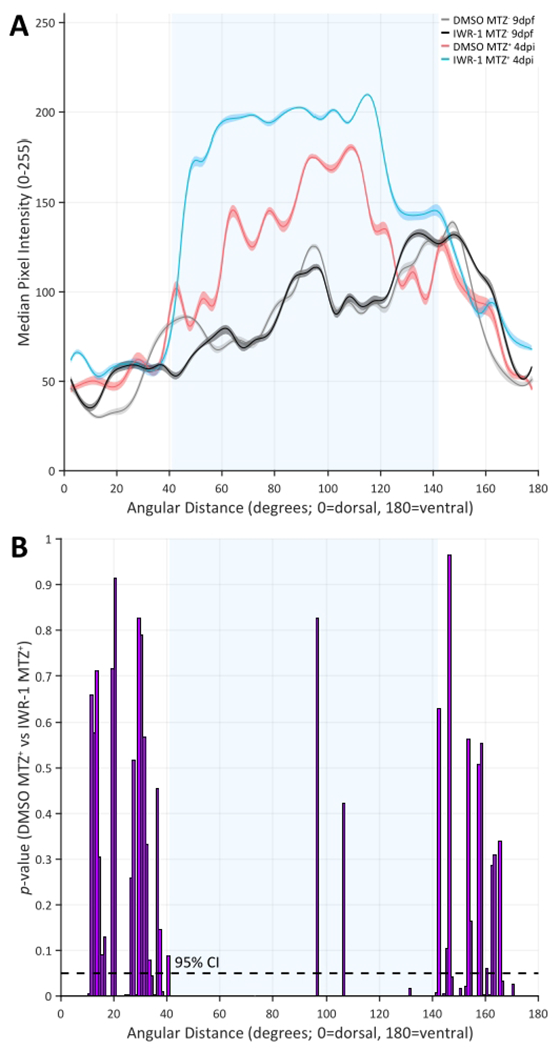
(A) 5-degree binned median values and 95% confidence envelopes derived from the raw data for each group. The plot shows similarity between the MTZ− groups (black and gray lines) across the dorsal (0°) to ventral (180°) length of the RPE with notable differences between and among the MTZ+ groups (blue and red lines) in the central RPE (light blue shaded area between ~40-140°). Specifically, median pixel intensity appears lighter (0 = black and 255 = white) in the IWR-1 MTZ+ 4 dpi when compared to any other treatment group, which corresponds to decreased central RPE pigmentation. (B) A statistical comparison of the median values derived from 1-degree bins across the dorsal (0°) to ventral (180°) length of the RPE for DMSO MTZ+ 4 dpi and IWR-1 MTZ+ 4 dpi reinforces the observation in (A). p-Values were calculated using a permutation simulation with 20,000 repetitions and a two-sided test for each 1-degree bin. Under the null hypothesis that the two groups have similar median values for any corresponding 1-degree bin, a p-value ≤ 0.05 indicates a statistically significant difference between the group medians (dashed black line = 95% confidence interval (CI)). The presence of p-values ≤ 0.05 across the central RPE (light blue shaded area between ~40-140°) indicates significantly less pigmentation in ablated (MTZ+) IWR-1-treated larvae when compared to ablated (MTZ+) DMSO-treated sibling controls. Please click here to view a larger version of this figure.
Confocal image datasets were quantified using RpEGEN as described in step 7. The annotated RpEGEN and support scripts are available at GitHub (https://github.com/burchfisher/RpEGEN) along with the 8-bit TIF and corresponding ROI files for interested users to test. Heatmaps displaying raw data from MTZ− larval ROIs showed an overall distribution of darker pixel intensities (where 0 = black and 255 = white, 8-bit color scale) spanning the dorsal (0°) to ventral (180°) length of the RPE, regardless of treatment with 0.06% v/v DMSO or 15 μM IWR-1 (Figure 9A,B). Plotting median pixel intensities facilitated visualization among all groups and showed similarities between the MTZ− groups across the RPE (Figure 10A; black and gray lines). These data supported the presence of an intact pigmented RPE monolayer (Figure 9E,F) and provided baseline median pixel intensity values (i.e., below 150) for unablated RPE (Figure 10A; black and gray lines). Comparatively, raw (Figure 9C,D) and median (Figure 10A; blue and red lines) data from MTZ+ larval ROIs showed an overall distribution of lighter pixel intensities in the central RPE, again, regardless of treatment condition. Ablated DMSO-treated larvae showed centralized distribution of light pixels at approximately 100° (± 15°) (Figure 9C; orange-red bins). This corresponded to a visible absence of pigmentation in the central RPE injury site in/around the optic nerve (Figure 9G; blue arrowheads). Ablated IWR-1-treated larvae also showed centralized distribution of light pixels that were expanded both dorsally (to around 50°) and ventrally (by approximately 5°) when compared to MTZ+ DMSO-treated sibling controls (Figure 9D; orange-red bins). The presence of an expanded injury site was clearly visible in sectioned tissue (Figure 9H; blue arrowheads). Except for two 1-degree bins, the observed difference between the MTZ+ groups was statistically significant in the central RPE (Figure 10B; light blue shaded area between ~40-140°; p-value ≤ 0.05), indicating significantly less central RPE pigmentation in MTZ+ IWR-1-treated larvae when compared to MTZ+ DMSO-treated sibling controls. Interpretation of these raw (Figure 9C,D) and median (Figure 10A) data with statistical comparison (Figure 10B) using RpEGEN was validated not only by observing sectioned tissue (Figure 9G,H), but also supported previous findings from manual quantification (Figure 8)3.
Discussion
This protocol describes methodology to genetically ablate the RPE and study mechanisms of degeneration and regeneration in larval-aged zebrafish. This protocol has also been successfully performed in adult zebrafish3 but with less extensive characterization, which is why larvae are the focus here. Critical aspects of this part of the protocol (steps 1-4) include: 1) adding 1.5x PTU to embryos prior to the onset of melanogenesis, 2) dechorionating PTU-treated embryos on 2-3 dpt, 3) careful screening for eGFP, and 4) timing of water changes during genetic ablation with MTZ. PTU inhibits melanin synthesis21, 22 and has been utilized in this RPE ablation paradigm to facilitate screening for rpe65a:nfsB-eGFP transgene expression and to enable characterization of RPE degenerative and regenerative processes based on the absence and subsequent return, respectively, of pigmented tissue3. As PTU will inhibit further pigmentation, but not depigment tissues once melanogenesis has begun21, PTU must be added before the onset of pigmentation, which begins around 24 hpf in zebrafish25. Here, and in prior studies4, 5, PTU has been added at approximately 6 hpf26 to ensure inhibition before melanin synthesis begins. While PTU enables visualization of key protocol steps, PTU treatment can affect some aspects of development (as discussed in step 2.3) and impair embryo hatching. The latter process, if delayed too long, can lead to morphology and motility deficits and affect viability40; thus, dechorionating (hatching) embryos either enzymatically with pronase or manually using forceps on 2-3 dpf is important for maintaining and standardizing larval health prior to genetic ablation. Another important consideration to avoid issues with older larval health and viability (unrelated to PTU-treatment), is that larvae should begin standardized feeding regimens if remaining out of an aquatic facility system past 9 dpf/4 dpi for experiments41.
As explained, rpe65a drives transgene expression in mature RPE in the central two-thirds of the posterior eye. Expression of eGFP in PTU-treated 5 dpf larvae is normally easily detected and visibly intense (bright), as shown in Figure 1. Larvae that dimly express eGFP at 5 dpf compared to siblings with bright expression may also be observed. Variation in expression intensity ratios (i.e., bright vs. dim) between generations may also exist, with some generations having more dimly expressing larvae than others. In any case, larvae with dim eGFP expression usually represent a minority of the animals in each clutch. While it is unknown if dimly expressing larvae show less severe RPE injury phenotypes, ablations have been performed in the bright eGFP+ cohort from each clutch and comparatively dim siblings have been euthanized at screening and excluded from analysis. Similarly, extensive characterization of the zebrafish RPE ablation and regeneration phenotypes has been performed in larvae heterozygous for the rpe65a:nfsB-eGFP transgene, by outcrossing rpe65a:nfsB-eGFP heterozygous adults to wildtype adults3. It is unlikely, however, not known, whether larvae homozygous for rpe65a:nfsB-eGFP show more severe ablation phenotypes. Other efforts to standardize the ablation protocol include addition of equal, measured volumes (e.g., 30 mL per 10 cm Petri dish) to MTZ− and MTZ+ dishes during the 24 h genetic ablation period. Likewise, larvae in pharmacological treatment dishes have received equal, measured volumes (e.g., 5 mL per 6-well) and timely replenishment of fresh compounds (e.g., every 24 h). When handling dishes with different MTZ and/or pharmacological compound treatments, careful measures should be taken to prevent spilling and inadvertent cross-contamination during transport and water changes.
The zebrafish RPE ablation protocol can be modified to include pharmacological manipulation (step 5). This has been done previously to identify immune response5, mTOR4, and Wnt3 signaling pathways as critical regulators of RPE regeneration; the latter has been shown to be repeatable here and was used to validate RpEGEN. In addition to illuminating critical RPE regeneration pathways, pharmacological manipulation has been widely employed for zebrafish retinal regeneration studies10, 42, 43, 44, 45, 46, 47. Prior to incorporation into the RPE ablation protocol, pharmacological agents should be rigorously evaluated for toxicity, efficacy, and treatment duration. This can be done by: performing dose response experiments to assess toxicity48; assessing expression of known target genes/proteins4, 5; and evaluating known functional consequences of pharmacological manipulation, for example, depletion of leukocytes with PLX3397 treatment48, 49, 50, 51. Here, DMSO (vehicle control) and IWR-1 were added 24 h prior to genetic ablation and larvae were screened immediately prior to pharmacological pretreatment on 4 dpf. While eGFP+ larvae are distinguishable from eGFP− larvae on 4 dpf, intensity of eGFP can appear quite dim, which is why larvae were rescreened for eGFP expression on 5 dpf (Representative Results). Screening for eGFP prior to 4 dpf may be difficult as expression may be so low as to be undetectable to the eye. Thus, pretreatment with pharmacological agents prior to 4 dpf (i.e., on 2 dpf)4 can be added to unscreened larvae that will be separated on 5 dpf when eGFP is clearly visible. In any scenario where larvae need to be manipulated (e.g., during screening, embedding for imaging, etc.), pharmacological treatment conditions should be maintained and, regardless of screening age, RPE should be ablated with MTZ on 5 dpf.
In addition to the genetic ablation protocol, step-by-step instructions for confocal microscope image preprocessing and automated quantification of RPE regeneration using RpEGEN are outlined (steps 6-7). RpEGEN was created to standardize RPE regeneration quantification across users and increase the robustness of the output dataset, while minimizing intrinsic biases that may come with the tedious manual quantification done previously. Critical aspects of this part of the protocol are performed during ROI generation (step 6). First, the image/eye must be in the dorsal up, distal left orientation (e.g., Figure 3A–D, Figure 4) as RpEGEN has been optimized for this directionality. It is also critical that the dorsal and ventral terminal ends of the RPE ROIs taper to a pointed tip as shown in Figure 3B’,B”,D’,D” (magenta lines). If these ends are instead squared or rounded off, the RpEGEN script may have difficulty determining the centerline skeleton start and end pixels and may generate spurs at terminal ROI ends rather than a centralized line (Figure 4B). Spurs at the terminal ROI ends could lead to shortening of the centerline during de-spurring (Figure 4D) and/or issues with angular distance measurements in the peripheral RPE (Figure 4G). Identical naming systems for the TIF and ROI files corresponding to each individual larva is also a significant step and imperative for pairing the 8-bit TIF file with the correct ROI in RpEGEN. Mismatched TIF and ROI file names will result in failure to generate the RpEGEN processing workflow outlined in the Representative Results section (Figure 4) and, ultimately, prevent RPE regeneration quantification using this script.
Collectively, this protocol provides instructions to successfully ablate the RPE in larval zebrafish, to manipulate signaling pathways that may be of interest pre-, during, or post-RPE injury, and to quantify RPE regeneration in a standardized way with limited inherent biases. In the context of available in vivo and in vitro models in which to study the regenerative potential of the RPE, the zebrafish is unique in its capacity for intrinsic RPE regeneration14. However, this is an acute model of RPE injury, not chronic as are the RPE degenerative diseases targeted for therapeutic development (e.g., AMD). While this is a limitation of the model, it remains an excellent platform in which to study the mechanisms of RPE regeneration, which are largely unknown, and may be adaptable in the future to study chronic injury and disease. The tools and methodology described herein are versatile and could also be applied to studies of cellular processes involved in the degenerative response and to study the fate of RPE-adjacent tissues post-ablation. Likewise, the RpEGEN script could be modified to fit user data output needs; for example, to perform spatial analyses of markers other than pigment (e.g., in situ hybridization probes, protein expression, etc.).
Supplementary Material
Acknowledgments
Work described herein was supported by the National Institutes of Health (RO1-EY29410 to J.M.G, and NIH CORE Grant P30-EY08098 to the Department of Ophthalmology); the UPMC Immune Transplant & Therapy Center (to L.L.L. and J.M.G.); and the E. Ronald Salvitti Chair in Ophthalmology Research (to J.M.G.). Additional support was received from the Wiegand Fellowship in Ophthalmology (to L.L.L), the Eye & Ear Foundation of Pittsburgh, and an unrestricted grant from Research to Prevent Blindness, New York, NY. Authors also wish to thank Amanda Platt for technical assistance and Dr. Hugh Hammer and the aquatics staff for excellent animal care support.
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/63658.
Disclosures
L.L.L. is the co-inventor on US Patent #9,458,428, which describes an expedited method to derive retinal pigment epithelium from human pluripotent stem cells; this is unrelated to the content herein. J.M.G. and G.B.F. have nothing to disclose.
References
- 1.Strauss O The retinal pigment epithelium in visual function. Physiological Reviews. 85 (3), 845–881 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Grierson I et al. Development, repair and regeneration of the retinal pigment epithelium. Eye. 8 (2), 255–262 (1994). [DOI] [PubMed] [Google Scholar]
- 3.Hanovice NJ et al. Regeneration of the zebrafish retinal pigment epithelium after widespread genetic ablation. PLoS Genetics. 15 (1), e1007939 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu F, Leach LL, Gross JM mTOR activity is essential for retinal pigment epithelium regeneration in zebrafish. bioRxiv. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leach LL, Hanovice NJ, George SM, Gabriel AE, Gross JM The immune response is a critical regulator of zebrafish retinal pigment epithelium regeneration. Proceedings of the National Academy of Sciences of the United States of America. 118 (21), e2017198118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zenno S, Koike H, Tanokura M, Saigo K Gene cloning, purification, and characterization of nfsb, a minor oxygen-insensitive nitroreductase from escherichia coli, similar in biochemical properties to frase I, the major flavin reductase in vibrio fischeri. The Journal of Biochemistry. 120 (4), 736–744 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Hamel CP et al. Molecular cloning and expression of rpe65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. Journal of Biological Chemistry. 268 (21), 15751–15757 (1993). [PubMed] [Google Scholar]
- 8.Curado S et al. Conditional targeted cell ablation in zebrafish: A new tool for regeneration studies. Developmental Dynamics. 236 (4), 1025–1035 (2007). [DOI] [PubMed] [Google Scholar]
- 9.White DT, Mumm JS The nitroreductase system of inducible targeted ablation facilitates cell-specific regenerative studies in zebrafish. Methods. 62 (3), 232–240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White DT et al. Immunomodulation-accelerated neuronal regeneration following selective rod photoreceptor cell ablation in the zebrafish retina. Proceedings of the National Academy of Sciences of the United States of America. 114 (18), E3719–E3728 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshimatsu T et al. Presynaptic partner selection during retinal circuit reassembly varies with timing of neuronal regeneration in vivo. Nature Communications. 7, 10590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montgomery JE, Parsons MJ, Hyde DR A novel model of retinal ablation demonstrates that the extent of rod cell death regulates the origin of the regenerated zebrafish rod photoreceptors. The Journal of Comparative Neurology. 518 (6), 800–814 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagerman GF et al. Rapid recovery of visual function associated with blue cone ablation in zebrafish. PLoS One. 11 (11), e0166932 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.George SM, Lu F, Rao M, Leach LL, Gross JM The retinal pigment epithelium: Development, injury responses, and regenerative potential in mammalian and non-mammalian systems. Progress in Retinal and Eye Research. 85, 100969 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiba C et al. Visual cycle protein rpe65 persists in new retinal cells during retinal regeneration of adult newt. The Journal of Comparative Neurology. 495 (4), 391–407 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Yoshii C, Ueda Y, Okamoto M, Araki M Neural retinal regeneration in the anuran amphibian xenopus laevis post-metamorphosis: Transdifferentiation of retinal pigmented epithelium regenerates the neural retina. Developmental Biology. 303 (1), 45–56 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Xia H, Krebs MP, Kaushal S, Scott EW Enhanced retinal pigment epithelium regeneration after injury in mrl/mpj mice. Experimental Eye Research. 93 (6), 862–872 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGill TJ et al. Subretinal transplantation of human central nervous system stem cells stimulates controlled proliferation of endogenous retinal pigment epithelium. Translational Vision Science and Technology. 8 (3), 43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salero E et al. Adult human rpe can be activated into a multipotent stem cell that produces mesenchymal derivatives. Cell Stem Cell. 10 (1), 88–95 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Chen B et al. Small molecule-mediated disruption of wnt-dependent signaling in tissue regeneration and cancer. Nature Chemical Biology. 5 (2), 100–107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whittaker JR An analysis of melanogenesis in differentiating pigment cells of ascidian embryos. Developmental Biology. 14 (1), 1–39 (1966). [DOI] [PubMed] [Google Scholar]
- 22.Westerfield M Zebrafish Book, 5th Edition; A Guide for the Laboratory Use of Zebrafish(Danio rerio). University of Oregon Press, Eugene, Oregon, USA: (2007). [Google Scholar]
- 23.Hammer HS Water quality for zebrafish culture in The Zebrafish in Biomedical Research., 321–335. Elsevier, Amsterdam, Netherlands: (2020). [Google Scholar]
- 24.Avdesh A et al. Regular care and maintenance of a zebrafish (danio rerio) laboratory: An introduction. Journal of Visualized Experiments: JoVE. (69), e4196 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF Stages of embryonic development of the zebrafish. Developmental Dynamics. 203 (3), 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Camp E, Lardelli M Tyrosinase gene expression in zebrafish embryos. Development Genes and Evolution. 211 (3), 150–153 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Baumann L, Ros A, Rehberger K, Neuhauss SC, Segner H Thyroid disruption in zebrafish (danio rerio) larvae: Different molecular response patterns lead to impaired eye development and visual functions. Aquatic Toxicology. 172, 44–55 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Li Z et al. Phenylthiourea specifically reduces zebrafish eye size. PloS One. 7 (6), e40132 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bohnsack BL, Gallina D, Kahana A Phenothiourea sensitizes zebrafish cranial neural crest and extraocular muscle development to changes in retinoic acid and igf signaling. PLoS One. 6 (8), e22991 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leary S et al. Avma Guidelines for the Euthanasia of Animals: 2020 Edition. American veterinary medical association, Schaumburg, Illinois, USA: (2020). [Google Scholar]
- 31.Uribe RA, Gross JM Immunohistochemistry on cryosections from embryonic and adult zebrafish eyes. Cold Spring Harbor Protocols. 2007, 4779 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Schindelin J et al. Fiji: An open-source platform for biological-image analysis. Nature Methods. 9 (7), 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muir D. GitHub - ReadlmageJROI at <https://github.com/DylanMuir/ReadlmageJROI>. (2021).
- 34.Morel P Gramm: Grammar of graphics plotting in matlab. Journal of Open Source Software. 3 (23), 568 (2018). [Google Scholar]
- 35.Reinhardt R et al. Sox2, tlx, gli3, and her9 converge on rx2 to define retinal stem cells in vivo. The EMBO Journal. 34 (11), 1572–1588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schonthaler HB et al. Evidence for rpe65-independent vision in the cone-dominated zebrafish retina. European Journal of Neuroscience. 26 (7), 1940–1949 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yazulla S, Studholme KM Neurochemical anatomy of the zebrafish retina as determined by immunocytochemistry. Journal of Neurocytology. 30 (7), 551–592 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Larison KD, Bremiller R Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development. 109 (3), 567–576 (1990). [DOI] [PubMed] [Google Scholar]
- 39.Dwass M Modified randomization tests for nonparametric hypotheses. The Annals of Mathematical Statistics. 28 (1), 181–187 (1957). [Google Scholar]
- 40.Karlsson J, von Hofsten J, Olsson PE Generating transparent zebrafish: A refined method to improve detection of gene expression during embryonic development. Marine Biotechnology (NY). 3 (6), 522–527 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Hernandez RE, Galitan L, Cameron J, Goodwin N, Ramakrishnan L Delay of initial feeding of zebrafish larvae until 8 days postfertilization has no impact on survival or growth through the juvenile stage. Zebrafish. 15 (5), 515–518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyers JR et al. B-catenin/wnt signaling controls progenitor fate in the developing and regenerating zebrafish retina. Neural Development. 7, 30 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tappeiner C et al. Inhibition of the tgfβ pathway enhances retinal regeneration in adult zebrafish. PLoS One. 11 (11), e0167073 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bailey TJ, Fossum SL, Fimbel SM, Montgomery JE, Hyde DR The inhibitor of phagocytosis, o- phospho-l-serine, suppresses müller glia proliferation and cone cell regeneration in the light-damaged zebrafish retina. Experimental Eye Research. 91 (5), 601–612 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramachandran R, Zhao XF, Goldman D Ascl1a/dkk/beta-catenin signaling pathway is necessary and glycogen synthase kinase-3beta inhibition is sufficient for zebrafish retina regeneration. Proceedings of the National Academy of Sciences of the United States of America. 108 (38), 15858–15863 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemmens K et al. Matrix metalloproteinases as promising regulators of axonal regrowth in the injured adult zebrafish retinotectal system. The Journal of Comparative Neurology. 524 (7), 1472–1493 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Elsaeidi F, Bemben MA, Zhao XF, Goldman D Jak/stat signaling stimulates zebrafish optic nerve regeneration and overcomes the inhibitory actions of socs3 and sfpq. The Journal of Neuroscience. 34 (7), 2632–2644 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Dyck A et al. Müller glia-myeloid cell crosstalk accelerates optic nerve regeneration in the adult zebrafish. Glia. 69 (6), 1444–1463 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Conedera FM, Pousa AMQ, Mercader N, Tschopp M, Enzmann V Retinal microglia signaling affects müller cell behavior in the zebrafish following laser injury induction. Glia. 67 (6), 1150–1166 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Chen S, Lathrop KL, Kuwajima T, Gross JM Retinal ganglion cell survival after severe optic nerve injury is modulated by crosstalk between jak/stat signaling and innate immune responses in the zebrafish retina. Development. 149 (8), dev199694 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Preux Charles AS, Bise T, Baier F, Marro J, Jaźwińska A Distinct effects of inflammation on preconditioning and regeneration of the adult zebrafish heart. Open Biology. 6 (7), 160102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


