Abstract
Sexual reproduction is ubiquitous throughout the eukaryotic kingdom, but the capacity of pathogenic fungi to undergo sexual reproduction has been a matter of intense debate. Pathogenic fungi maintained a complement of conserved meiotic genes but the populations appeared to be clonally derived. This debate was resolved first with the discovery of an extant sexual cycle and then unisexual reproduction. Unisexual reproduction is a distinct form of homothallism that dispenses with the requirement for an opposite mating-type. Pathogenic and non-pathogenic fungi previously thought to be asexual are able to undergo robust unisexual reproduction. We review here recent advances in our understanding of the genetic and molecular basis of unisexual reproduction throughout fungi and the impact of unisex on the ecology and genomic evolution of fungal species.
Keywords: Unisexual reproduction, Cryptococcus neoformans, Candida albicans, Neurospora, pathogenicity, mating type, homothallic, heterothallic, fungal meiosis
One of the defining characteristics of eukaryotic life is the ability to reproduce sexually. Sexual reproduction was likely an attribute of the most recent common ancestor of all extant eukaryotic lineages (Baldauf, 2003; Baldauf, Roger, Wenk-Siefert, & Doolittle, 2000; Cavalier-Smith, 2004; Dacks & Roger, 1999; Derelle et al., 2006; Fritz-Laylin et al., 2010; Patterson, 1999; Ramesh, Malik, & Logsdon Jr, 2005; Simpson & Roger, 2004). Less than 1% of angiosperm plants and just 0.1% of characterized animals are thought to lack the ability to undergo meiosis and indeed, since the first sexual fungus Syzygites megalocarpus was described, eukaryotic microorganisms have been discovered to be not just sexual or asexual but homothallic, that is self-fertile, as well as heterothallic, self-incompatible (Asker & Jerling, 1992; Blakeslee, 1904; Vrijenhoek, 1998; White, 1978; Whitton, Sears, Baack, & Otto, 2008). In fungi, heterothallic species are self-sterile and require two partners with compatible mating types to initiate the sexual cycle, while homothallic species are self-fertile and can initiate sexual reproduction between cells of the same mating type and, in some species, even from a single cell or between cells descended from a single cell.
Fungal organisms possess diverse sexual strategies for both homothallic reproduction and maintaining heterothallic incompatibility. Fungi establish cell identify using either a bipolar mating type system, that is one locus that produces two mating types, or tetrapolar mating type system, that is two loci that are able to produce many mating types. The basidiomycetous fungus Ustilago maydis has a tetrapolar mating system with the two mating type loci, a and b, located on different chromosomes. The a mating locus encodes pheromones and pheromone receptors involved in mate recognition and cell fusion during mating (Bölker, Urban, & Kahmann, 1992). The b locus encodes bE and bW, homeodomain proteins, that heterodimerize to regulate the expression of genes required for filamentous growth and completion of meiosis (Kämper, Reichmann, Romeis, Bölker, & Kahmann, 1995; Kronstad & Leong, 1990; Schulz et al., 1990). There are two alleles of the a locus and at least 25 alleles of the b locus; mating requires cells to carry different alleles at both the a and b loci. As a result, meiotic segregants can mate with only 25% of the products of the cross. In contrast, the budding yeast Saccharomyces cerevisiae possesses a mating type system with features common to bipolar fungi; cells that inherit the MATα idiomorph are α-cells, and those that inherit MATa are a-cells. MATα and MATa are idiomorphs as they encode non-allelic genes, α1 and α2 in MATα and a1 in MATa. The two idiomorphes of the MAT locus encode critical DNA binding proteins that regulate expression of cell identity genes throughout the genome. Mating between α- and a-cells rsults in cellular and nuclear fusion to generate an α/a diploid that can undergo meiosis in which any given segregants can mate with 50% of the progeny of the cross.
Mating type systems enable self-recognition and incompatibility in heterothallic fungi, however homothallic species overcome self-incompatibility through three general mechanisms. S. cerevisiae is an example of the first homothallic mechanism, as only one allele is expressed from the active MAT locus, while two additional silent MAT cassettes are present and enable mating-type switching through gene conversion of the MAT locus. Thus, mating type switching enables even a clonal population to mate but only after conversion to opposite mating type haploid cells. More recent work has revealed a second mechanism of homothallism, the presence of both mating types in a single genome, at either linked or unlinked loci. Sordaria macrospora, a filamentous ascomycete, contains tandem copies of the MATA and MATa loci allowing expression of both mating type loci in its genome and self-fertility (Debuchy & Turgeon, 2006; Pöggeler, Risch, Kück, & Osiewacz, 1997). The third type of homothallism, unisex, in which the requirement for an opposite mating type is dispensed with entirely, has most recently been discovered. While the molecular mechanisms and phylogenetic extent of unisex are not yet fully elucidated, new research has revealed that unisexual fungi are an important and medically relevant subset of homothallic fungi.
Homothallism in Ascomycetes and Basidiomycetes
Of the two major fungal divisions, the relative frequency of homothallism is higher in the Ascomycetes than in the Basidiomycetes (Whitehouse, 1949). This higher frequency of homothallism in Ascomycetes could be a reflection of the differences in the self-incompatibility systems in the two groups of fungi. In Ascomycetes, self-sterility is governed by a bipolar (unifactoral) system, while for the majority of the Basidiomycetous species self-incompatibility is controlled by a tetrapolar (bifactoral) system. Thus, if self-fertility evolved from ancestral self-sterility, theoretically this transition would require less change in the incompatibility system in Ascomycetes than in Basidiomycetes. However, it should be noted that homothallism has also been identified in Basidiomycetous species that are close relatives of self-sterile species with tetrapolar mating systems. In addition, the debate about whether the ancestral state of the mating system in Ascomycetes and Basidiomycetes is homothallism or heterothallism is yet to be settled, with incongruent and inconclusive results generated by studies of different groups of species (Galagan et al., 2005; Gioti, Mushegian, Strandberg, Stajich, & Johannesson, 2012; Inderbitzin, Harkness, Turgeon, & Berbee, 2005; Nygren et al., 2011; Rydholm, Dyer, & Lutzoni, 2007).
Remarkably, there are fungal species that have the ability to transition between heterothallism and homothallism. For example, S. cerevisiae Ho+ strains can undergo mating type switching and are homothallic, while naturally occurring ho− strains are heterothallic. Among the fungal species that show plasticity in their reproductive modes are two important human pathogenic fungi, Candida albicans and Cryptococcus neoformans. Both species have bipolar mating systems and can undergo heterothallic reproduction between cells with opposite mating types. However, if appropriate conditions are present, unisexual reproduction can also be initiated and completed involving cells of the same mating type in the absence of cells of the opposite mating type.
S. cerevisiae is the classic example of homothallism using mating type switching. It had been long debated whether S. cerevisiae is homothallic or heterothallic, as both modes of reproduction were observed in this species (Hicks & Herskowitz, 1977; Hicks, Strathern, & Herskowitz, 1977; X. Lin & Heitman, 2007; Lindegren & Lindegren, 1943a, 1943b, 1944; Winge, 1935; Winge & Laustsen, 1937; Winge & Roberts, 1949). It turned out that both arguments are correct, and it depends on which strain is under examination. S. cerevisiae has a unifactorial self-incompatibility system, and the mating type is governed by the allele at the active mating type (MAT) locus. In addition to the active MAT locus, there are also two silent MAT cassettes, HML and HMR, in the genome that contain two compatible MAT alleles. HML and HMR are normally kept silent by a modified chromatin structure that requires the action of the Sir protein complex. However, during mitosis, a special endonuclease encoded by the gene HO initiates a gene conversion process that replaces the existing allele at the active MAT locus with the opposite allele from one of the two silent MAT cassettes. Colonies established by an Ho+ strain will contain cells with compatible MAT alleles at the active MAT locus. Therefore Ho+ strains are homothallic. On the other hand, there are naturally occurring ho− S. cerevisiae strains that are unable to undergo mating type switching and are heterothallic (McCusker, 2006).
A similar mating type switching system has been identified in another model yeast, the fission yeast Schizosaccharomyces pombe. The two mating type switching systems in S. cerevisiae and S. pombe share similarities in that: 1) there are three mating type cassettes present on the same chromosome, with one of them being active and the other two silenced, and 2) efficient unidirectional switching through gene conversion is initiated by a DNA lesion during mitosis. However, significant differences also exist between the two systems. First, the two species differ in the structure, sequence, as well as the nature of the genes encoded within MAT. In addition, the DNA lesion that initiates switching differs between the two systems. While a DSB is provoked by the Ho endonuclease in S. cerevisiae, the switch-initiating lesion in S. pombe appears to require a certain unusual type of replication-induced break that involves a nick and possibly retention of ribonucleotides from the primer of an Okazaki fragment (Arcangioli & de Lahondes, 2000; Dalgaard & Klar, 1999; Kaykov & Arcangioli, 2004; Vengrova & Dalgaard, 2004, 2006). Furthermore, the underlying mechanisms for the silencing of the two silent MAT cassettes also appear to be different between S. cerevisiae and S. pombe. It is thus apparent that the similar mating type switching systems have evolved independently in the two lineages.
Remarkably, it has recently been shown that another budding yeast, Kluyveromyces lactis, also undergoes mating type switching through yet another mechanism that is distinct from S. cerevisiae and S. pombe (Barsoum, Martinez, & Åström, 2010). K. lactis has lost the Ho endonuclease gene and was thought to switch stochastically by mitotic gene conversion. However, recently it was discovered that a special protein, α3, is essential for switching in K. lactis. α3 is homologous to transposases and the amino acids conserved among transposases are required for successful mating type switching in K. lactis, suggesting α3 originated through domestication of a transposase (Barsoum et al., 2010). The fact that several similar yet distinct homothallic mating systems involving mating type switching have evolved independently in different fungal lineages suggests there are selection pressures that favor selfing, at least under certain circumstances.
Homothallism in fungi could also be achieved through the co-existence of compatible MAT alleles within one individual. The compatible MAT alleles could be located in different nuclei (i.e. bi-nuclei spore, so-called pseudo-homothallism), within the same nucleus but separated from each other (e.g. A. nidulans), or within the same nucleus and fused together (e.g. Cochliobolus sp.). Several basidiomycetous species produce heterokaryotic spores, such as Agrocybe semiorbicularis, Conocybe tenera for. bispora, Coprinus ephemerus, and Aleurodiscus canadensis (Raper, 1966). These species produce 2-spored basidia. Following meiosis, two nuclei migrate into each of the two spores. Germination of each individual spore can establish dikaryotic mycelia, and in most cases, form clamp connections (Raper, 1966). In these pseudo-homothallic species, occasionally there are spores that produce stable mycelia that lack clamp connections. Further mating analyses using these “non-homothallic” individuals suggest the existence of an underlying bipolar self-incompatibility system in each of these species (Raper, 1966). Thus, it is likely that homothallism by the production of heterokaryotic spores is derived from an ancestral bipolar mating system. Similar examples of pseudo-homothallism based on the production of heterokaryotic spores with underlying bipolar incompatibility also occur in several Ascomycetous species, such as Neurospora tetrasperma and Podospora anserina (Ames, 1934; Dodge, 1927; Dodge, Singleton, & Rolnick, 1950; Raper, 1966).
There are also a few Basidiomycetous species that are homothallic without clamp connections, such as Calocera cornea, Coprinus ephemeroides, Octojuga pleurotelloides, Octojuga pseudopinsitus, and Filobasidiella depauperata (Raper, 1966; Rodriguez-Carres, Findley, Sun, Dietrich, & Heitman, 2010). F. depauperata appears to be an obligate sexual species that is closely related to the human pathogenic Cryptococcus species complex. F. depauperata spores are uninucleate, and each germinated spore establishes a monokaryotic mycelia that lacks clamp connections but produces basidia decorated with four long spore chains (Kwon-Chung et al., 1995; Rodriguez-Carres et al., 2010). It is likely that the obligate selfing in F. depauperata evolved from an ancestral bipolar self-incompatibility system. However, it is not yet clear how this transition was achieved, although analyses of genes located within the MAT locus of C. neoformans suggest they have been involved in extensive chromosomal rearrangements, such as translocations and inversions (Fraser et al., 2004; Rodriguez-Carres et al., 2010).
Unisexual Reproduction in Pathogenic Fungi
The numerous and distinct fungal life cycles illustrate the plasticity of reproductive strategies fungi employ in nature to preserve the benefits of sexual reproduction. The transition between the traditional heterothallic sexual cycle requiring mating to take place between different mating type cells and homothallic cycles that allow more promiscuous mating are common and occur throughout the fungal kingdom. Individual isolates of the same species are capable of choosing between a homothallic or a heterothallic lifestyle that will either ameliorate the cost of mating or will fit the environmental needs of the species and allow them to expand and survive in hostile niches. The frequent transitions illustrate the balance between outcrossing and inbreeding, or between vegetative growth and increased frequency of sexual reproduction, are likely a response to specific environmental cues that favor one or the other strategy.
Pathogens such as C. neoformans and C. albicans highlight a conundrum for mycologists; both have extant sexual cycles yet they generate largely clonal populations in nature. C. neoformans has an opposite α-a sexual cycle that induces a dimorphic transition from yeast growth to hyphae (Kwon-Chung, 1975, 1976a, 1976b). However, the predominance of the α mating type in clinical and environmental isolates led to the hypothesis that this species might have been largely asexual. Recent studies revealed that α mating type isolates of C. neoformans evolved a self-fertile strategy and are able to complete a unisexual cycle in the absence of an opposite mating partner (X. Lin, Hull, & Heitman, 2005). Population studies show that unisexual reproduction occurs in several of the lineages of the Cryptococcus species and it is likely an important strategy for reproduction in nature (Bui, Lin, Malik, Heitman, & Carter, 2008; Fraser et al., 2005; X. Lin et al., 2007; X. Lin et al., 2009; Ni et al., 2013).
Similarly, C. albicans was considered to be a strictly asexual species for over a century. Yet the discovery of the mating-type like (MTL) loci, MTLα and MTLa, the presence of mating- and meiosis-specific genes in the genome, the isolation of α and a mating competent strains, and finally the discovery that the white-opaque transition dramatically enhances mating established the presence of a parasexual cycle (Bennett & Johnson, 2003; Hull & Johnson, 1999; Hull, Raisner, & Johnson, 2000; Lockhart et al., 2002; Magee & Magee, 2000; Miller & Johnson, 2002). C. albicans also undergoes unisexual reproduction in the absence of the Bar1 protease (Alby, Schaefer, & Bennett, 2009). However, despite retaining many meiotic genes, meiosis has yet to be observed in C. albicans (Sherwood & Bennett, 2009). Other pathogenic Candida species are missing key meiotic regulators, and yet undergo extant meiotic sexual cycles (Reedy, Floyd, & Heitman, 2009; Sherwood & Bennett, 2009). Cryptococcus and Candida species serve as excellent examples of reproductive plasticity that allows cryptic sexual cycles to enable outcrossing and inbreeding, generating genetic diversity or preserving genetic configurations that confer a fitness advantage in a specific environmental niche.
C. neoformans is able to undergo unisexual reproduction and generate genetic diversity, even in clonal populations of pathogenic fungi (Ni et al., 2013). From a medical perspective, unisexual reproduction poses a challenge for confronting and controlling emerging new strains that have been found to be either hypervirulent or resistant to current antifungal treatments (Fraser et al., 2005; Ni et al., 2013). Similar studies on pathogenic parasites reveal that unisexual reproduction is widespread in pathogenic microbes, and can generate “superbugs” that are resistant to current treatments and responsible for local outbreaks (Heitman, 2006, 2010; Wendte et al., 2010).
Cryptococcus neoformans
The basidiomycetous fungi Cryptococcus neoformans and its sibling species are the most common fungal agents of meningoencephalitis, which is fatal if untreated. The pathogenic species of Cryptococcus comprise four serotypes based on capsular antigens: C. neoformans (serotype D), Cryptococcus neoformans var. grubii (serotype A), and the sister species Cryptococcus gattii (serotypes B and C) (Hull & Heitman, 2002). C. neoformans serotypes A and D are prevalent worldwide but most commonly infect immunocompromised individuals while C. gattii is usually restricted to tropical and subtropical regions and is able to infect immunocompetent individuals (Ellis & Pfeiffer, 1990; Speed & Dunt, 1995). These pathogenic Cryptococcus species are typically found as haploid yeasts in nature and inside the host. They comprise two mating types, α and a, and display a well-defined sexual cycle. In response to nutrient limitation, cells of opposite mating type secrete pheromones that are sensed by the pheromone receptors and initiate the formation of conjugation tubes expanding towards the pheromone source. Pheromone production triggers a cell-cell fusion event; however, the nuclei remain separate, producing a dikaryon that undergoes a dimorphic transition to filamentous hyphae. The hyphae grow and produce yeast cells, called blastospores, via budding, which harbor the same DNA content as the hyphal compartment. At the apex of the hyphae specialized structures (basidia) are formed where nuclear fusion occurs, followed by meiosis. Multiple rounds of mitosis and budding produce four chains of basidiospores [reviewed in (Heitman, 2006, 2010; Heitman, Sun, & James, 2013; Idnurm et al., 2005)].
Sexual reproduction and virulence are linked as the cycle can generate and disperse Cryptococcus spores in the environment. Infection is thought to be acquired by inhalation of spores, which are readily aerosolized and of an ideal size to lodge in the alveoli of the lung to cause pulmonary infection. Both spores and yeast are capable of causing fatal infections in mice, and particles small enough to be spores are found in air samples from Cryptococcus outbreak locations on Vancouver Island. Taken together, these observations support the hypothesis that spores are infectious propagules of Cryptococcus (Giles, Dagenais, Botts, Keller, & Hull, 2009; Kidd, Bach, et al., 2007; Kidd, Chow, et al., 2007; Springer, Saini, Byrnes, Heitman, & Frothingham, 2013; Velagapudi, Hsueh, Geunes-Boyer, Wright, & Heitman, 2009).
Cryptoccoccus exhibits an extant sexual cycle under laboratory conditions with readily observed production of hyphae and spores. However, natural populations exhibit such a marked bias towards mating type α that it is not clear how relevant these laboratory conditions are for environmental populations. A small number of environmental and clinical isolates are diploid hybrids of serotype A and D (aADα, αADa isolates) that harbor both mating types and serve as indirect evidence of mating in nature (Lengeler, Cox, & Heitman, 2001; Litvintseva, Lin, Templeton, Heitman, & Mitchell, 2007). However, the population of environmental and clinical isolates sampled thus far is almost exclusively of mating type α, raising doubts about the frequency of α-a heterosexual reproduction that occurs in nature (Lengeler et al., 2001; Litvintseva et al., 2007). The first isolate of mating type a was isolated from a clinical sample belonging to the highly pathogenic serotype A group (Lengeler, Wang, Cox, Perfect, & Heitman, 2000). Further genotyping of ~3,000 strains showed that only 3 were MATa isolates that also exhibited low competence for mating (Keller, Viviani, Esposto, Cogliati, & Wickes, 2003; Viviani, Nikolova, Esposto, Prinz, & Cogliati, 2003). Frequencies of mating type alleles do vary considerably between geographically isolated populations. For instance, a sub-Saharan African population of C. neoformans is comprised of ~25% mating type a individuals (Litvintseva et al., 2003). MATa isolates from this population exhibit evidence of recombination, and are robustly fertile under laboratory conditions (Litvintseva et al., 2003). The presence of these MATa isolates is geographically restricted and the opportunities for α-a heterosexual reproduction beyond this locale seems likely to be severely limited in nature.
As the vast majority of Cryptococcus isolates are mating type α, it was thought that the organism was asexual and reproduced mitotically. The absence of a sexual cycle was hypothesized to have evolved concomitantly with the emergence of the highly pathogenic isolates. The same notion was advanced and popularized for many other pathogenic eukaryotic microorganisms, including C. albicans, and eukaryotic parasites. This theory was fundamentally challenged by evidence of sexual recombination in natural populations that are exclusively of mating type α (Brandt, Hutwagner, Kuykendall, & Pinner, 1995; Xu, Vilgalys, & Mitchell, 2000). Extensive genotyping and sequencing revealed evidence for both clonal expansion and recombination in serotype A and D isolates from sub-Saharan Africa and the United States (Litvintseva, Kestenbaum, Vilgalys, & Mitchell, 2005; Litvintseva et al., 2003).
Mating type α isolates were observed to form hyphae, basidia, and spores upon culture on mating media; however due to the absence of an opposite mating partner this process was considered to be strictly mitotic and asexual, and was termed monokaryotic or haploid fruiting (Wickes, Mayorga, Edman, & Edman, 1996). Monokaryotic fruiting shares many characteristic features with sexual reproduction, including the production of infectious spores, and was eventually discovered to be a sexual cycle involving cells of one mating type (X. Lin et al., 2005). In a process similar to heterosexual reproduction, nutrient limitation induces a dimorphic transition from yeast to hyphae in cells of one mating type. Unisexual hyphae grow to form basidia at the tips, where meiosis and multiple rounds of mitosis produce abundant spores. In response to nutrient limitation, haploid cells can generate hyphae that grow to produce the basidium where a late endoreplication event occurs and produces a transient diploid nucleus that undergoes meiosis and sporulation. Diploidization can also occur early during α-α mating, either through endoreplication or cell-cell fusion between genetically different or clonal cells. In this case the diploid cells initiate the formation of diploid hyphae that lead to basidia where meiosis and budding occurs to produce haploid spores (X. Lin et al., 2005). Unisexual reproduction generates a mixture of haploid and diploid hyphae, reflecting either a late or an early diploidization event, indicating considerable plasticity in unisexual reproduction (Feretzaki & Heitman, 2013).
Unisexual reproduction is a sexual cycle that involves ploidy changes (1N→2N→1N) and hyphal development. Unisexual rates of recombination are similar to those observed in heterosexual reproduction, suggesting that both sexual programs are meiotic cycles (X. Lin et al., 2005). Deletion of the highly conserved meiotic factors Spo11 or Dmc1 severely impairs sporulation during unisexual and heterosexual reproduction (Feretzaki & Heitman, 2013; X. Lin et al., 2005). Spo11 induces double strand breaks on the homologous chromosomes that initiate recombination, while Dmc1 is responsible for repairing DNA breaks by facilitating the invasion of DNA strands leading to the formation of Holliday junctions. Both genes are dispensable for hyphal and basidia development but critical for meiosis and sporulation, providing further evidence that unisexual reproduction is an extant sexual meiotic cycle.
Unisexual reproduction was initially observed only in α mating type cells, and thus it was thought the cycle was restricted to the α mating type. Further analysis revealed that some MATa isolates also possess the ability to undergo unisexual reproduction (Hull & Heitman, 2002; X. Lin, Huang, Mitchell, & Heitman, 2006; Tscharke, Lazera, Chang, Wickes, & Kwon-Chung, 2003). Subsequent studies revealed that hyphal development is a quantitative trait and a scan of ~25% of the genome identified five major QTLs that orchestrate hyphal initiation and elongation during unisexual reproduction. The MAT locus is the most prominent of the five QTLs, and the MATα allele is linked to increased unisexual reproduction (X. Lin et al., 2006). This is perhaps not surprising as numerous regulatory molecules of unisexual and heterosexual reproduction are encoded by the mating type locus (Feretzaki & Heitman, 2013; X. Lin et al., 2005; X. Lin, Jackson, Feretzaki, Xue, & Heitman, 2010). Pheromone production and sensing genes activate the pheromone-sensing pathway, highly conserved in fungi, which controls unisexual reproduction through the G-protein activated MAP kinase cascade (Figure 1) (Hsueh, Lin, Kwon-Chung, & Heitman, 2011). The pheromone receptors are coupled to G-protein signaling subunits that transfer the signal to the downstream kinases. Upon pheromone stimulation the G-proteins activate the signal transduction protein Ste20α/a that then triggers the three-tiered phosphorylation cascade of the major kinases Ste11α/a (MAPKKK), Ste7 (MAPKK), and Cpk1 (MAPK) (Davidson, Nichols, Cox, Perfect, & Heitman, 2003; Hsueh et al., 2011).
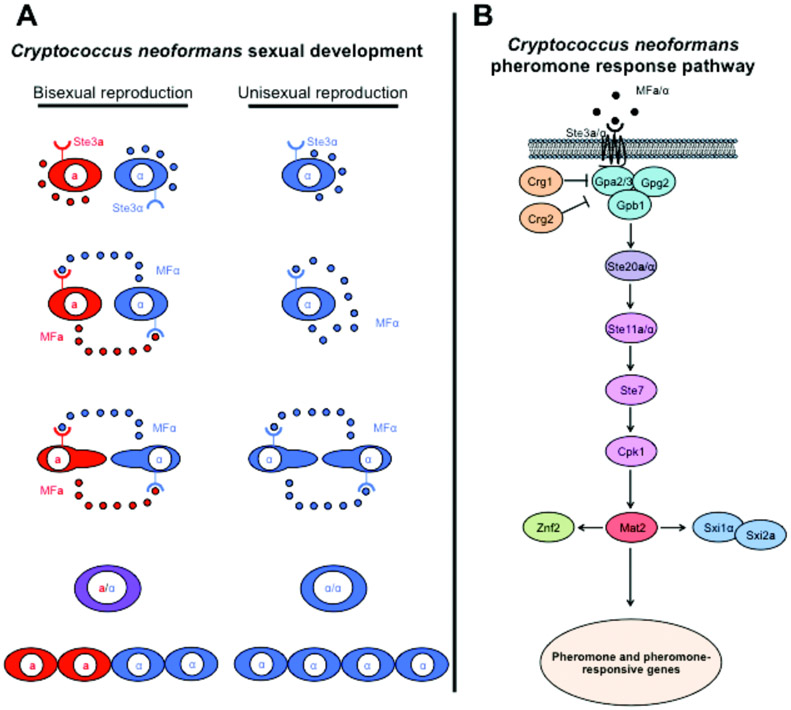
Figure 1. Sexual development of Cryptococcus neoformans.
(A) During bisexual reproduction cells of opposite-mating type secrete pheromones that are sensed by the pheromone receptor Ste3α/a. Pheromone sensing leads to cell-cell fusion producing a dikaryotic and then diploid intermediate that undergoes meiosis to generate recombinant progeny. During unisexual reproduction nutrient-limiting conditions trigger pheromone production that enhances cell-cell fusion or endoreplication and the diploid intermediate undergoes meiosis, similar to bisexual reproduction. (B) The pheromone-signaling pathway governs sexual development in C. neoformans. Binding of pheromone to the pheromone receptor stimulates the pathway through a G-protein coupled receptor complex that activates the Ste20α/a kinase and the three tiered MAPK phosphorelay system. The transcription factor target of the pathway is Mat2, which regulates the expression of sexual- and filamentation-specific genes. Although the signaling-cascade is highly conserved among fungi, the downstream transcriptional network exhibits extensive rewiring in different species.
Few genetic differences between unisexual and heterosexual reproduction have been identified thus far. The first pathway specific elements to be discovered were the mating-type-specific SXI1α and SXI2a genes, which encode homeodomain transcription factors that control mating-type identify and sexual development. Sxi1α and Sxi2a are required for heterosexual reproduction but are dispensable for unisexual reproduction (Hull, Boily, & Heitman, 2005; Hull, Davidson, & Heitman, 2002). These differences suggest divergent unisexual and heterosexual pathways with distinct downstream effectors of cell identity or sexual development.
The pheromone-signaling pathway is a critical regulator of mating in fungi and plays an essential role in recognizing and responding to opposite mating type cells (reviewed in (Jones & Bennett, 2011)). The pathway is structurally and functionally conserved among fungi closely (Ustilago maydis) and distantly (S. cerevisiae and C. albicans) related to Cryptococcus and thus the majority of these components were identified in Cryptococcus through homology-based approaches. However, the downstream pathway of the pheromone-signaling cascade exhibits significant rewiring since its divergence from the most recent common ancestor shared with S. cerevisiae. The major transcription factor target of the pheromone signaling cascade is Ste12 in S. cerevisiae; however, in Cryptococcus Ste12 is dispensable for hyphal development during both unisexual and heterosexual reproduction (Fields & Herskowitz, 1985; Hartwell, 1980; Yue et al., 1999).
The major transcription factor target of pheromone sensing in Cryptococcus is the High Mobility Group (HMG) protein Mat2 (X. Lin et al., 2010). Mat2, along with other components of the pathway, is required for cell-cell fusion indicating an early role during hyphal development. Mat2 induces pheromone production and subsequently triggers pheromone evoked responses by binding directly to a cis-regulatory sequence in the promoter region of the pheromone genes known as the pheromone-response element (PRE) (Kruzel, Giles, & Hull, 2012). The transcriptional circuit of the pheromone cascade became more complex with the identification of Znf2, a novel zinc finger transcription factor. Znf2 is required for hyphal development, however it is dispensable for cell-cell fusion. Surprisingly, deletion of the gene increases the efficiency of cell fusion events during heterosexual reproduction and stimulates pheromone expression during unisexual reproduction (X. Lin et al., 2010). All of these components are essential for hyphal development during both unisexual and heterosexual reproduction. The only known exceptions are Sxi1α and Sxi2a, which are required for heterosexual but not unisexual reproduction. It is surprising that no other components of the pheromone-signaling or response pathway have been identified as uniquely essential for either the unisexual or heterosexual pathways, especially given these pathways’ role in restricting heterothallic systems to the heterosexual cycle. It may be that there are additional components that act downstream of the mating pathway to specifically regulate unisexual but not heterosexual reproduction. Further experimentation will allow the identification of these hypothesized factors and further elucidate the pathways.
The components of the mating pathway are highly conserved in C. neoformans and the sibling species C. gattii. Unisexual reproduction was first directly observed for laboratory strains of the C. neoformans serotype D lineage (X. Lin et al., 2005). More recent evidence suggests that unisexual reproduction also occurs in C. neoformans serotype A. Genotypic analysis of diploid serotype AD hybrid strains revealed a homozygous α/α mating type locus (αADα) produced via mating between mating type α isolates of different serotypes (X. Lin et al., 2007). Moreover, independent population genetic studies revealed evidence of recombination in serotype A isolates from trees in India and infected animals in Australia that were derived exclusively from mating type α populations (Bui et al., 2008; Hiremath et al., 2008). Under laboratory conditions, αAAα diploids generate abundant hyphae but few spores when cultured solo on mating media, indicating that unisexual reproduction occurs in this population. Robust hyphal growth but a paucity of spore production could suggest either a limited unisexual cycle or a defect of these isolates, which have been growing for extended periods in the intermediate diploid state. This diploid intermediate or unisexual product is relatively common in the population; screening ~500 environmental and clinical isolates found that ~8% were diploid, the majority of which were αAAα diploids (X. Lin et al., 2009)
C. gattii, the sibling species of C. neoformans, thought to be restricted to tropical and subtropical regions, is emerging as a pathogen of significant global public health importance (Byrnes, Bartlett, Perfect, & Heitman, 2011; Datta et al., 2009). Of particular interest is the link between sexual cycles with both the increasing geographic range of outbreaks and virulence (Byrnes et al., 2010; Fraser et al., 2005; Voelz et al., 2013). Although C. gattii undergoes heterosexual reproduction under laboratory conditions, like C. neoformans the natural population is predominantly MATα, providing further evidence of a correlation between unisexual reproduction and pathogenesis (Byrnes et al., 2010; Fraser et al., 2005; Fraser, Subaran, Nichols, & Heitman, 2003). Geographically isolated populations from Eucalyptus trees in Australia exhibit similar evidence of recombination in both α-a mixed populations and exclusively α mating type populations (Saul, Krockenberger, & Carter, 2008). Moreover, the strains associated with the ongoing outbreak of meningoencephalitis on Vancouver Island and in the Pacific Northwest are exclusively mating type α haploids that are fertile under laboratory conditions. Genotypic analysis suggests that the genotype associated with the outbreak could be the progeny of unisexual reproduction between two α mating type parents (Fraser et al., 2005; Fraser et al., 2003). In addition, the identification of an α/α diploid intermediate and the isolation of particles small enough to be spores present in air samples from Vancouver Island further supports that C. gattii undergoes unisexual reproduction in nature (Fraser et al., 2005; Kidd, Chow, et al., 2007). These studies show that unisexual reproduction occurs in nature in several independent lineages, and may facilitate the expansion of the geographic range of pathogens and contribute to the production of infectious spores.
Candida albicans
C. albicans is the most common human fungal pathogen, normally associated with asymptomatic commensal colonization of the gastrointestinal tract and oral and vaginal mucosa of most of the world’s population. C. albicans is not commonly a burden in immunocompetent hosts but candidiasis of the oral cavity and candidemia leading to colonization of internal organs and central nervous system are prevalent and cause serious infections in immunocompromised individuals (Klein et al., 1984). Diagnosis of candidemia is frequently too late for antifungal treatments to be effective, leading to mortality rates between 30% and 50% (Kibbler et al., 2003; Pfaller, Jones, Messer, Edmond, & Wenzel, 1998). Until recently C. albicans was thought to be an obligate diploid organism with an asexual lifecycle. However, the identification of the MTL locus and the isolation of mating competent partners led to the discovery of a parasexual cycle (Hull & Johnson, 1999; Hull et al., 2000; Xie et al., 2013) Two diploid cells of the opposite mating type fuse to produce a transient tetraploid that undergoes stochastic chromosome loss to return to a diploid or aneuploid state (Bennett & Johnson, 2003; Legrand et al., 2004; Magee & Magee, 2000).
Sequencing of C. albicans clinical isolates revealed that the MTL locus contains highly conserved genes involved in mating, including the idiomorphic transcriptional regulators of cell identity a1, a2, α1, and α2 (Hull & Johnson, 1999). The C. albicans MTL locus is larger than closely the related ascomycetous fungus S. cerevisiae MAT locus, harboring additional genes that encode phosphatidylinositol kinases (PIK), oxysterol binding proteins (OBP), and poly A polymerases (PAP). Almost 90% of environmental and clinical diploid isolates are heterozygous at the MTL, harboring both mating types (α/a) and do not mate (Miller & Johnson, 2002; Tavanti et al., 2005). Genetic manipulation to induce gene deletion or homozygosis of the chromosome containing the MTL locus generates mating competent strains (MTLα/α, MTLa/a, MTLΔ/α, MTLΔ/a) and these strains mate successfully in infected animals and the laboratory (Hull & Johnson, 1999; Magee & Magee, 2000).
A further breakthrough in understanding mating was the discovery of a link to the phenotypic white-opaque cell-type switch. The switch from a white to an opaque cell enhances mating efficiency one million-fold (Miller & Johnson, 2002). The white-opaque switch was initially described two decades ago, where virulent white cells spontaneously switched to avirulent opaque cells (Slutsky et al., 1987). The phenotypic switch from white to opaque is driven by the transcription factor Wor1 (white-opaque regulator 1) through a positive feedback loop, during which Wor1 binds its own promoter to increase its transcription 40-fold (Huang et al., 2006; Zordan, Galgoczy, & Johnson, 2006). WOR1 transcription is repressed by the MTL a1–α2 heterodimer in heterozygous diploid cells, which causes MTLα/a clinical isolates to repress white-opaque switching and mating (Figure 2) (Miller & Johnson, 2002). Other transcriptional regulators promote white-opaque switching, including Czf1 and Wor2, which act in concert with Wor1 to down-regulate the expression of Efg1, a promoter of the white phenotype (Lachke, Srikantha, & Soll, 2003; Vinces, Haas, & Kumamoto, 2006; Zordan, Miller, Galgoczy, Tuch, & Johnson, 2007). In addition to genetic control, numerous environmental cues affect white-opaque switching. Opaque cells, but not white cells, colonize skin where mating occurs, perhaps because the opaque phenotype is unstable at higher physiological temperatures (Kvaal et al., 1999; Lachke et al., 2003; Slutsky et al., 1987). In addition, despite the instability of opaque cells at high temperatures, elevated CO2 and high concentration of N-acetylglucosamine promote white-opaque switching, conditions that mimic the environment of the GI tract where mating is likely to occur (Dumitru et al., 2007; Huang, Srikantha, Sahni, Yi, & Soll, 2009; Huang et al., 2010; Hull et al., 2000).
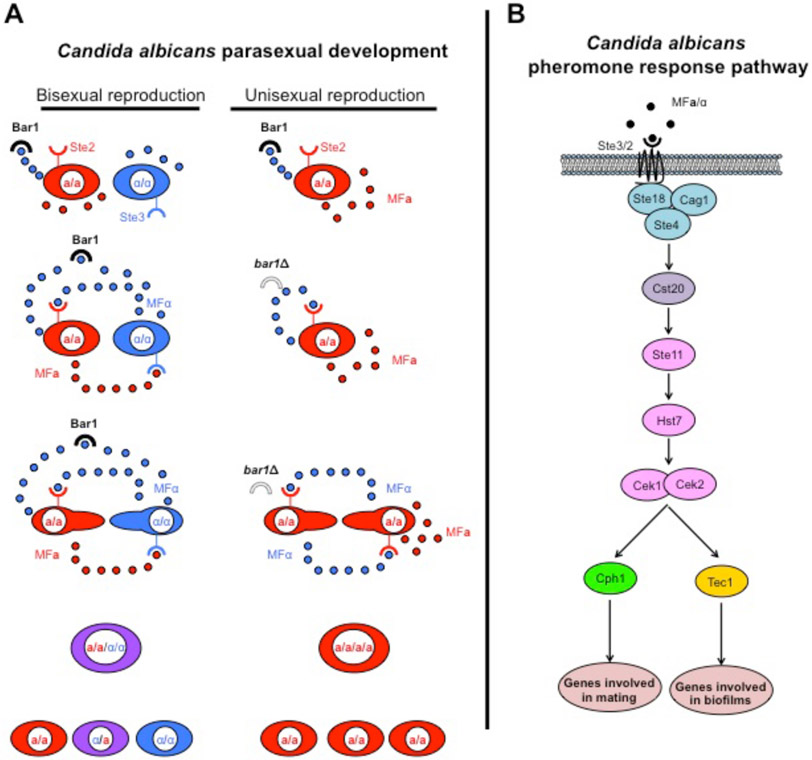
Figure 2. Parasexual development of Candida albicans.
(A) Homozygous opaque MTLα/α cells secrete α pheromone that is sensed by the Ste2 pheromone receptor on the surface of MTLa/a cells. During bisexual reproduction pheromone sensing induces polarized growth towards the pheromone source and initiates cell-cell fusion. The intermediate tetraploid MTLα/α/a/a cell undergoes stochastic chromosome loss that generates MTL homozygous and MTL heterozygous diploid progeny with significant rates of aneuploidy. Surprisingly, homozygous opaque MTLa/a cells can secrete both a and α pheromones. The Bar1 protease degrades α pheromone to prevent autocrine activation of the mating pathway. In the absence of the Bar1 protease MTLa/a cells secrete α pheromone that accumulates and binds to the Ste2 pheromone receptor on the same or neighboring cell. The autocrine pheromone signaling pathway drives unisexual reproduction that leads to cell-cell fusion and results in a tetraploid intermediate. The MTLa/a/a/a cell undergoes stochastic chromosome loss to return to the diploid state, generating significant aneuploidy in the process. (B) The pheromone response pathway is highly conserved in the Saccharomycotina subphylum and all of the components of C. albicans are orthologs of the S. cerevisiae pheromone pathway. The pathway consists of the G-protein coupled receptor complex and the Cst20 kinase followed by the MAPK signaling cascade. In opaque cells the transcription factor target of the pathway is Cph1 that governs the expression of mating-specific genes, while in white cells the Tec1 transcription factor regulates the abundance of biofilm-specific factors.
Sexual development of C. albicans is driven by pheromones that activate the highly conserved pheromone signaling cascade. Just as in S. cerevisiae, pheromones bind to the receptors (Ste2 or Ste3), which activate the coupled trimeric G protein complex and the downstream Cst20 kinase (Magee, Legrand, Alarco, Raymond, & Magee, 2002). Subsequently, the signal triggers the sequential phosphorylation of the MAP kinase cascade components Ste11 (MAPKKK), Hst7 (MAPKK), and Cek1/Cek2 (MAPKs) (Chen, Chen, Lane, & Liu, 2002; Magee et al., 2002).
The same MAPK pathway operates in both white and opaque cells, but the response to pheromones is quite different. In opaque cells, pheromone binding leads to the formation of conjugation tubes and cell-cell fusion, while white cells respond to pheromone by increasing adhesion and forming biofilms (Daniels, Srikantha, Lockhart, Pujol, & Soll, 2006; Lockhart, Daniels, Zhao, Wessels, & Soll, 2003). The differential response to pheromones led to the hypothesis that, although opaque and white cells share the same pathway, the main transcription factor target is different. It was initially proposed that MAPK signaling activates the Cph1 transcription factor, the homolog of S. cerevisiae Ste12, which mediates the expression of mating specific genes in opaque cells, while in white cells pheromone signaling was proposed to activate Tec1, which orchestrates the expression of genes involved in adhesion and biofilm formation (Sahni et al., 2010; Yi et al., 2008). Although Tec1 is required for pheromone-induced biofilm formation, evidence has been presented that Cph1 is the sole transcriptional target of the pheromone signaling cascade in both white and opaque cells. The downstream targets of Cph1 diverge in white and opaque cells, mediating the expression of genes required for biofilm formation and mating (including pheromone secretion and sensing), respectively (C. H. Lin et al., 2013). All of these components are essential for pheromone production in C. albicans and deletion of the genes severely impairs mating; however, their function is unknown during unisexual reproduction. Marked strains of α/α or a/a opaque cells co-cultured with opposite mating type cells produce marked unisexual mating products as well as heterosexual mating products, indicating that unisexual reproduction can occur in both α/α and a/a backgrounds in the presence of high pheromone concentrations (Alby et al., 2009). Based on these and other findings we can infer that the same pheromone signaling cascade likely operates in both α/α and a/a opaque cells during heterosexual and unisexual reproduction.
The specialized mating competent opaque cells respond to pheromones to produce conjugation tubes that lead to cell-cell fusion and nuclear fusion to generate a uninucleate tetraploid cell (Bennett, Miller, Chua, Maxon, & Johnson, 2005). In contrast, white cells, which are unable to mate, respond to pheromone sensing by activating a specialized transcriptional pathway regulating biofilm formation (Sahni et al., 2009). If white cells are mating incompetent, it raises the question of the purpose of the white cell response to pheromones. The pheromones secreted have limited diffusion range, particularly the prenylated 14 amino acid MFa secreted by a cells. Opaque cells are rare in natural Candida populations; thus the majority of the population, comprised of white cells, forms biofilms in response to pheromones produced by minority opaque cells, facilitating a pheromone gradient that enables distant mating competent cells to locate opposite mating partners (Daniels et al., 2006). The initial steps of mating between opaque cells closely mirrors S. cerevisiae sexual reproduction and putative regulators of cell fusion may have conserved functions in C. albicans (Bennett, Uhl, Miller, & Johnson, 2003; Lockhart et al., 2003).
Unlike S. cerevisiae, which undergoes meiosis and sporulation to produce asci with four spores, C. albicans can mate to produce an a/a/α/α tetraploid but neither meiosis nor asci with spores have been observed. Although tetraploid C. albicans cells are stable, certain conditions induce a parasexual cycle. During parasex ploidy is reduced through stochastic chromosome loss that returns tetraploid cells to the diploid or near diploid state, generating considerable aneuploidy in the process (Bennett & Johnson, 2003). Although meiosis is absent, the parasexual cycle generates genetic recombination and gene conversion in the progeny (Forche et al., 2008). Moreover, SPO11, the meiosis specific gene whose product is responsible for initiating recombination by inducing DNA double strand breaks, is dispensable for parasexual ploidy reduction; however, it is required for the observed recombination during concerted chromosome loss. Given that SPO11 is a highly conserved meiotic “toolkit” gene conserved across eukaryotes it suggests that a meiosis-like process may be operating during the parasexual cycle, although it has also been suggested that Spo11 may have been reconfigured to play a novel mitotic recombination role, as it is expressed in mitotically dividing cells (Forche et al., 2008).
C. albicans has well-established heterosexual mating between α/α and a/a cells followed by parasexual reduction; however, MLST analysis indicated that the populations of C. albicans are predominantly clonal, hinting that outbreeding is rare in nature (Odds et al., 2007). This raised the question of how prevalent white-opaque switching and heterosexual mating is in natural populations. However, in addition to opposite-sex mating, C. albicans utilizes an alternate mating pathway between cells of the same mating type (Alby et al., 2009). Unlike S. cerevisiae, where the cells express only one of the two pheromones, opaque a/a cells are able to express and secrete both α- and a-mating pheromones (Alby et al., 2009; Bennett & Johnson, 2006). Typically, the aspartyl protease Bar1 degrades the α pheromone produced by a/a cells and prevents auto-activation of the mating pathway in the absence of the α/α mating partner (Figure 2) (Schaefer, Cote, Whiteway, & Bennett, 2007). However, in the absence of this protease, a/a cells produce and respond to α pheromone and initiate mating through autocrine pheromone signaling. The α pheromone secreted by a/a cells binds the Ste2 pheromone receptor, stimulating the mating pathway and leading to pheromone responses. Cell and nuclear fusion occurs producing an a/a/a/a tetraploid that undergoes stochastic chromosome loss to return to the a/a diploid or aneuploid state, while undergoing some limited recombination (Alby et al., 2009). Cell fusion of a/a cells can also be induced by a minority of α/α cells that serve as donors of α pheromone in a ménage a trois mating. Inactivation of Bar1 might also occur in certain environmental niches such as the acidic environment of the vaginal mucosa, where lower pH could inhibit Bar1 and induce unisexual reproduction in vivo. A striking morphological difference between the heterosexual and unisexual cycle is that the α/α/a/a tetraploid undergoes an opaque-white switch due to the expression of the a1/α2 heterodimer (that inhibits opaque-specific transcriptional regulators) generating white sterile cells, while the a/a/a/a tetraploid remains opaque. The production of mating pheromones is crucial to the induction of unisexual reproduction, as in Cryptococcus species, however the specific mechanisms of pheromone production and sensing are significantly different, emphasizing the diversity and independent origin of unisexual reproduction in the two species.
C. albicans, long thought to be an obligate diploid, was recently discovered to be capable of producing haploid cells (Hickman et al., 2013). The obligate diploid hypothesis was proposed and historically suggested to result from balanced recessive lethal mutations that may be present throughout the genome. Haploid C. albicans cells have been observed only under selective laboratory conditions and in some chromosomes contained alleles from only one parental homolog, consistent with a limited number of haploid deleterious or lethal mutations. These haploids are proposed to arise through a concerted chromosome loss mechanism, similar to that of the tetraploid parasexual cycle (Bennett & Johnson, 2003; Hickman et al., 2013). As with diploids homozygous at the MTL locus, the haploid cells, which have a single copy of either the MTLa or MTLα idiomorphs, efficiently switch from white to the opaque state and undergo heterosexual but not unisexual mating (Hickman et al., 2013). C. albicans haploids are inherently unstable and undergo autodiploidization during propagation. The mechanism of autodiploidization is as yet unknown, although it is not thought to involve homothallic mating, as neither haploid white or opaque cells of the same mating type were able to mate on mating media. Haploids exhibit a reduced growth rate and attenuated virulence, although this fitness defect was rescued by mating but not autodiploidization, leading to the hypothesis that a burden of recessive mutations may be the cause of slow growth, and not the ploidy state of the cells.
Although recent studies revealed genetic evidence of recombination in clonal populations of C. albicans, the frequency of unisexual reproduction in nature remains unclear. Unisexual reproduction has been observed only in fertile opaque homozygous a/a or α/α cells, while the majority of clinical and environmental isolates are sterile white heterozygous α/a cells unable to switch to the opaque state in vitro. However, it is possible that certain conditions may stimulate white-opaque switching of α/a cells in vivo. Lower levels of hemoglobin in an in vitro model alter the expression of a1, a2, α1, and α2, which cause white α/a cells to behave phenotypically as a/a cells, inducing white-opaque switching and heterosexual reproduction (Pendrak, Yan, & Roberts, 2004). These white α/a cells are mating competent, leading to the hypothesis that unisexual reproduction can also occur under conditions in the mammalian host. When an α/a cell switches to a/a, Bar1 may function to allow the new a cell to escape the cell-cycle arrest invoked by the pheromone produced by the α progenitor, however unisex is also repressed by the activity of Bar1 (Chan & Otte, 1982). Unisexual and heterosexual reproduction are both maintained in C. albicans and Bar1 may regulate the balance between inbreeding and outbreeding with each strategy generating genetic diversity in response to environmental cues in respective niches.
It is now appreciated that pheromones govern unisexual reproduction and general intercellular communication. Pheromones are implicated in both heterothallic and homothallic reproduction in a variety of fungi (Paoletti et al., 2007; Spellig, Bolker, Lottspeich, Frank, & Kahmann, 1994). Surprisingly, C. albicans pheromone/receptor binding, which regulates the pheromone cascade, exhibits considerable plasticity. Mutations in much of the 13 amino acid sequence of the α mating pheromone did not affect signaling or unisexual reproduction of a/a cells. Moreover, C. albicans can respond to a variety of pheromone analogs, including pheromones from the related species C. dubliniensis, C. parapsilosis, and C. tropicalis, which are able to bind to the receptor and induce unisexual reproduction in C. albicans WT opaque cells and biofilm formation in white cells (Alby & Bennett, 2011; Chen et al., 2002; Magee et al., 2002). This suggests that alternative pheromones may activate the pathway and promote homothallism in niches inhabited by diverse flora. C. albicans, C. dubliniensis, C. parapsilosis, and C. tropicalis co-inhabit the oral cavity of healthy individuals, providing just such a diverse niche in nature (Daniels et al., 2006; Ghannoum et al., 2010; Lockhart et al., 2003; Martins et al., 2010; Melton et al., 2010). It is hypothesized that similar signals from the host or other microbiota may stimulate C. albicans’ promiscuous pheromone receptor and signaling cascade to drive biofilm formation and unisexual reproduction in WT cells, as interspecies pheromone-signaling may ameliorate the repression of the Bar1 protease if these cross-species pheromones are resistant to Bar1 cleavage.
Neurospora
The Neurospora genus provides an interesting case study for the evolution of mating systems and unisex. The genus contains species with multiple mating systems including heterothallism, homothallism, and pseudohomothallism (Wik, Karlsson, & Johannesson, 2008). By examining the synteny of the mat loci of homothallic Neurospora species and comparing them to the mat loci of closely related heterothallic species, it was discovered that the mat locus is conserved among heterothallic species belonging to distinct phylogenetic clades (Figure 3) (Gioti et al., 2012). However, the mat loci in the homothallic species have undergone distinct chromosomal rearrangements, suggesting the ancestral state in Neurospora is heterothallism and homothallism has evolved independently in different lineages (Gioti et al., 2012). Of four transitions from heterothallism to homothallism examined in Neurospora species, three involve acquisition of compatible mat alleles into the same haploid genome. In two species, N. pannonica and N terricola, unequal crossover events or chromosomal translocations are hypothesized to have led to the linkage of the mat A and mat a loci on a single chromosome. In the N. sublineolata genome, conversely, the mat loci are unlinked but both present in a haploid. Interestingly, the N. sublineolata matA locus contains a novel retrotransposon, suggesting the transition from heterothallism to homothallism might have been facilitated by these selfish genetic elements (Gioti et al., 2012).
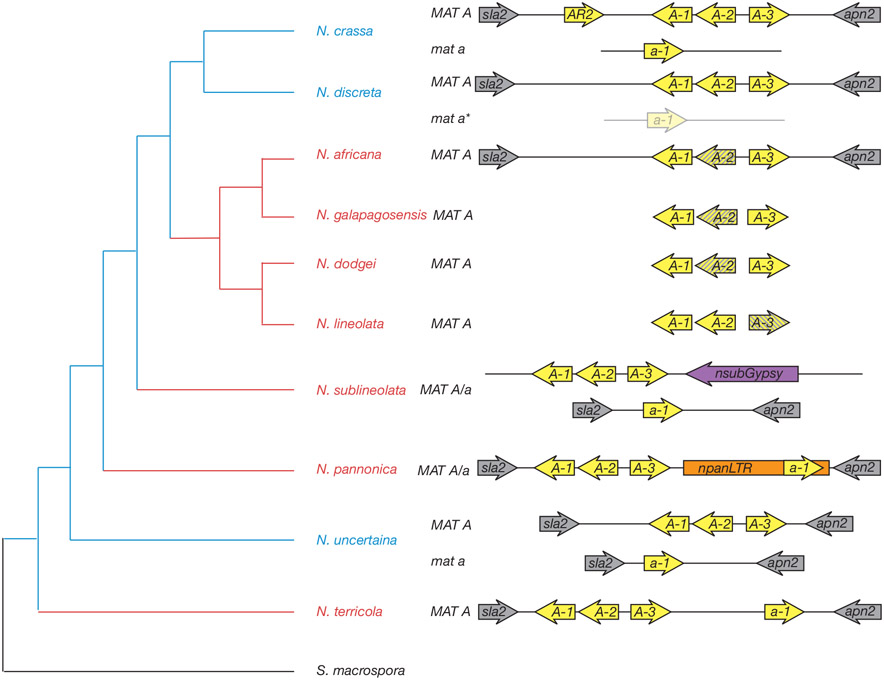
Figure 3. Neurospora genus mat locus organization.
The organization of the mat locus of selected Neurospora species rooted with the homothallic outgroup species Sordaria macrospora (adapted from (Gioti et al., 2012)). Blue branches of the phylogram represent heterothallic species and red branches depict homothallic species. Grey arrows represent genes flanking the mat locus and their transcriptional orientation. Yellow arrows indicate mat genes, with A-1, A-2, A-3, and a-1 corresponding to mat A-1, mat A-2, mat A-3, and mat a-1 respectively. Intact transposable elements are represented by purple and orange elements. The N. discreta mat a idiomorph has not yet been sequenced and is inferred phylogenetically. Genes truncated by stop codons are striped arrows (Wik et al., 2008). Sequencing of the coding regions and cDNA of mat A-1, A-2 and A-3 revealed independent mutations that may result in pseudogenization. Mat A-2, an HPG domain protein, suffered nonsense mutations at codon 39 in N. dodgei and codon 288 in N. africana and N. galapagosensis; however, mat A-2 is still transcribed in N. dodgei and N. galapagosensis. The HMG transcription factor mat A-3 is highly divergent in the homothallic species related to N. africana; N. lineolata has a stop-codon at position 215, amino acids 100-177 are deleted in N. africana and N. galapagosensis, and N. dodgei has a mutation of the canonical start codon.
Neurospora also contains a homothallic species with a single mating-type locus in the population. N. africana retains only the mat A genes: a mat a idiomorph has not been discovered in the population and is not required for mating (Glass & Smith, 1994). The mat A idiomorph is conserved in sequence and arrangement between homothallic N. africana and heterothallic N. crassa. It is not yet known what genetic mechanism enabled self-fertility but N. africana lacks any requirement for a contribution from an opposite mating-type cell, similar to other unisexual organisms. Intriguingly, N. africana is not unique. Three closely related species that share with N. africana a monophyletic origin, N. galapagosensis, N. dodgei, and N. lineolata, are also homothallic and have only one mating type idiomorph (Gioti, Stajich, & Johannesson, 2013; Glass, Metzenberg, & Raju, 1990; Nygren et al., 2011; Wik et al., 2008). This raises the possibility that the ancestor to these species was unisexual and this sexual cycle has persisted over significant evolutionary timescales.
Cryptic Unisexual Species
It is difficult to ignore the pattern of historical thought about the lifecycles of fungi thought to be asexual. It was often observed that each populations were asexual and clonal, often with just a single idiomorph at the mating type locus but retaining well-conserved meiotic genes. Further investigation of population genetics revealed evidence of recombination before discovery of a cryptic sexual cycle. We hypothesize that this pattern may be relevant to additional fungal species, including pathogenic species, which are thought to be clonal and lack a recognized sexual cycle. T. rubrum has long been thought to be clonal, however signatures of recombination were detected (Gräser, Kühnisch, & Presber, 1999). Only one mating type idiomorph has been reported, but both mating and meiosis genes are well conserved, suggesting that there might be an extant unisexual cycle (Kano et al., 2013; Li, Metin, White, & Heitman, 2010; Martinez et al., 2012). Alternaria species are important agricultural pathogens that were first reported as clonal, however evidence now suggests there may be a cryptic sexual or unisexual cycle (Peever et al., 1999; Simmons, 1999). Two highly conserved mating type idiomorphs are found in Alternaria alternata, in addition to ample evidence of recombination both between opposite mating type populations and in populations with a single mating type (Berbee, Payne, Zhang, Roberts, & Turgeon, 2003; Stewart, Kawabe, Abdo, Arie, & Peever, 2011; Stewart et al., 2013). Following a similar trajectory, evidence of recombination in populations of Batrachochytrium dendrobatidis, a global zoonotic pathogen thought to be asexual, was discovered (Farrer et al., 2013; Farrer et al., 2011; James et al., 2009; J. A. T. Morgan et al., 2007). While the mating type locus has not yet been identified, meiotic genes are present and conserved in the B. dendrobatidis genome (Halary et al., 2011). Ashbya gossypii is a preduplication filamentous ascomycete closely related to S. cerevisiae (Dietrich et al., 2004; Simmons, 1986, 1999). The A. gossypii genome contains three identical unlinked copies of MATa, a truncated MATα, and a single complete MATα loci (Dietrich, Voegeli, Kuo, & Philippsen, 2013; Wendland & Walther, 2011). The standard lab strain contains only MATa loci, but pheromone sensing is not required for A. gossypii to sporulate, as strains deleted for the α-pheromone receptor STE2 or the a-pheromone receptor STE3 still are able to sporulate (Wendland & Walther, 2011). Strains deleted for the mating transcription factor STE12 exhibited increased sporulation, further complicating the question of what, if any sexual or unisexual cycle A. gossypii engages in (Wendland, Dünkler, & Walther, 2011). A detailed investigation may yield evidence of heretofore-unrecognized heterothallic or homothallic sexual cycles in these species.
Evolutionary Origin of Unisex
Transitions between heterothallism to homothallism, and an increased potential for inbreeding, are common throughout the fungal kingdom (Debuchy & Turgeon, 2006; Gioti et al., 2012; Inderbitzin et al., 2005; Yun, Berbee, Yoder, & Turgeon, 1999). Homothallism may be hypothesized to confer long-term evolutionary costs, primarily because the selfing population will inevitably experience reduced effective recombination rates and population size, and consequently have reduced efficacy of purifying selection and become more prone to genetic drift (Charlesworth & Wright, 2001; Hill & Robertson, 1966; Otto & Lenormand, 2002; Pollak, 1987), which could lead to genomic maladaptation, including the spread of deleterious selfish genetic elements (e.g. transposable elements) as well as accelerated rates of protein evolution or decay. However, given the frequency with which species have become homothallic, we must consider that homothallism could be a neutral mutational event (Lynch, 2007). When a mutation occurs that reduces self-incompatibility, there may be no immediate negative or positive selection on that mutation. Mitotically dividing fungal populations show a spectrum of high frequency mutations, including both adaptive and neutral changes (Lang et al., 2013). Mutations conferring self-compatibility may occur in species and a homothallic sexual cycle could then be fixed stochastically by drift or carried to fixation by linked adaptive mutations.
Against this neutral hypothesis, it must be considered that changes from a heterothallic to homothallic sexual cycle impact a species’ life cycle and genome in remarkable ways. As predicted by population genetic theory, homothallic Neurospora species show relaxation of purifying selection in protein-coding genes, reduced efficiency in silencing transposable elements, as well as reduced codon usage bias in highly expressed genes (Gioti et al., 2013). Additionally, a study of genome sequences from 16 yeast species in the family Saccharomycetaceae found that the MAT locus appears to be a deletion hotspot, in that the distance between MAT and HML is gradually eroding over evolutionary time as genes near MAT are repeatedly deleted, truncated, and transposed (Gordon et al., 2011). The authors proposed that this evolutionary erosion of the yeast sex chromosomes is caused by accidents occurring during mating type switching, coupled with the selection pressure to keep MAT and HML on the same chromosome. There is ample evidence supporting the hypothesis that selfing can have long-term negative effects. On the other hand, self-fertility assures sexual reproduction, which could provide short-term and even long-term benefits to the species such as generation of spores that can better cope with harsh environments and generation of novel genotypes through meiosis (Table 1). The reproductive strategy being selected for in a particular species depends on interactions among many factors, including both environmental (e.g. nutrient availability) and biological (e.g. population structure). Given all of the long-term disadvantages potentially associated with selfing, the fact that many species maintain the ability to undergo homothallic reproduction and selfing suggests the conditions that favor homothallism may be widespread.
Table 1.
Putative Impacts of Unisexual Reproduction
| Access to Recombination |
|---|
| Avoid Muller's Ratchet |
| Recombination within the mating type locus |
| Increased mating |
| Universal mating partner |
| Unisexual mating improves fitness for heterosexual mating |
| Meiotic rejuvenation (Ünal, Kinde, & Amon, 2011) |
| Production of hyphae and spores to explore new environments |
| Increased Genetic Diversity |
| Access to the diploid state as a capacitor for evolution |
| Generation of epimutations through Sex Induced Silencing (SIS) |
| Meiotic de novo generation of phenotypic and genotypic diversity |
| Biparential mitochondrial inheritance and mitochondrial recombination |
| Other Impacts |
| Toggle ploidy to enhance survival in environments where the haploid or diploid is more fit |
| Suppression of transposons |
Unisex Increases the Opportunities for Meiosis
That most eukaryotes reproduce sexually during their life cycle is puzzling when considering the two-fold cost of sex (Maynard Smith, 1978; Williams, 1975). The first cost is genome dilution; each allele will be passed on to each daughter during clonal division but has just a 50% chance to be passed on during sexual reproduction. The second cost is that each daughter requires contributions from two parents while a single mother can produce a daughter mitotically. This is typically thought of as a cost of males but other mating systems have similar costs (Lloyd, 1988). Homothallic reproductive strategies such as unisex may have initially evolved, in part, to avoid the two-fold cost of modern sex.
The ubiquity of sex must be the result of significant advantages to counterbalance the costs of sex (Bell, 1982; Williams, 1975). Hermann Muller recognized that asexually dividing populations faced a fundamental challenge; their genomes would gradually accumulate deleterious mutations in an irreversible manner, termed Muller’s Ratchet (Figure 4) (Felsenstein, 1974; Haldane, 1937; Muller, 1932, 1964). Every individual in a population would eventually suffer a harmful mutation, and selection will not be sufficient to maintain fitness without the ability to recombine and purge deleterious mutations. Sexual reproduction allows organisms to use recombination to purge the genome of these deleterious mutations to avoid Muller’s Ratchet. In the few species with characterized unisexual pathways, both sexual and unisexual cycles utilize the same post cell-fusion meiotic and recombinational pathways (Bui et al., 2008; Feretzaki & Heitman, 2013; X. Lin et al., 2005). Therefore, unisexual reproduction, like heterosexual sexual reproduction, can be hypothesized to allow populations to avoid Muller’s Ratchet but this remains to be documented experimentally. Using unisex to access recombination is especially important for pathogenic species, where a host might be colonized by a small number of cells of a single mating type.
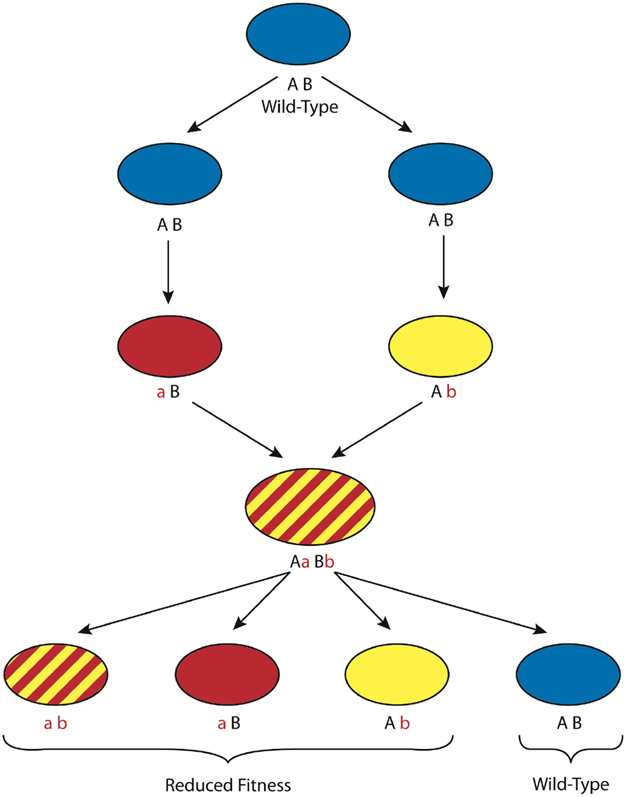
Figure 4. Muller’s Ratchet.
In asexually growing organisms every cell or lineage will eventually suffer a deleterious mutation. There is no mechanism for selection to remove all of the deleterious mutations (“a”, “b”) from the population, and therefore fitness will irreversibly decline in mitotically dividing populations. Heterosexual and unisexual cycles give access to recombination that may restore wild type (“AB”) fitness to some lineages in the population.
Sex increases genetic and phenotypic diversity of populations and species through recombination (Dobzhansky, Levene, Spassky, & Spassky, 1959; Levene, 1959; Spassky, Spassky, Levene, & Dobzhansky, 1958; Spiess, 1959). Recombination is not the only mechanism for unisex to promote fungal genetic diversity; however, diversity is also generated through aneuploidy in fungi, which can be deleterious in many eukaryotes (Pavelka et al., 2010; Selmecki, Forche, & Berman, 2010; Torres et al., 2010; Torres et al., 2007). Aneuploidy is a facilitator of fungal responses to changing or stressful environmental conditions (Dunham et al., 2002; Rancati et al., 2008; Yona et al., 2012). In addition, in fungal pathogens such as C. neoformans and C. albicans, aneuploid chromosomes commonly confer antifungal drug resistance (Selmecki, Dulmage, Cowen, Anderson, & Berman, 2009; Selmecki, Forche, & Berman, 2006; Sionov, Lee, Chang, & Kwon-Chung, 2010). Unisexual reproduction is able to generate de novo genetic diversity and aneuploidy (Ni et al., 2013). In C. neoformans, changes in pathogenic phenotypes were observed in ~7% of the progeny of unisexual reproduction and two-thirds of these variant progeny were aneuploid for at least one chromosome and aneuploidy was the cause of their variant phenotype. Understanding how unisex generates genetic and phenotypic diversity, and how this enables pathogens to adapt to new environments and medical interventions, is of particular importance in dealing with emerging and established fungal pathogens.
Transitioning from a heterothallic to homothallic sexual cycle changes the gene flow and selection dynamics of a population, in particular presenting new barriers to the invasion of selfish genetic elements. Transposons have been found in all organisms with sequenced genomes but there is still some debate as to the way they invade populations. Transposons, like all selfish genetic elements, are fundamentally sexually transmitted parasites of a genome. Invasion of these selfish elements relies on being transmitted to offspring at a frequency higher than expected under Mendelian segregation. Population genetic models predict that a change from outcrossing to selfing will reduce the effectiveness of this selfish strategy in diploid organisms, as increased homozygosity of these deleterious transposons will increase the strength of selection in the population until the elements are purged from the genome (Boutin, Le Rouzic, & Capy, 2012; Wright, Ness, Foxe, & Barrett, 2008; Wright & Schoen, 1999). Comparisons between selfing and outcrossing species of plants, as well as hermaphroditic and outcrossing nematodes, found that selfing lineages had significantly fewer transposable elements in their genomes (Dolgin, Charlesworth, & Cutter, 2008; M. T. Morgan, 2001). A change to a unisexual cycle may leverage increased selection to reduce the number of transposons in the genome.
The switch to homothallism is likely to lead to increased opportunities for meiosis and the significant meiotic transcriptional changes and marshaling of genomic defenses. Some fungal species recognize repeated sequences and utilize a repeat induced point-mutation (RIP) or methylation induced premeiotically (MIP) mechanism to disable transposons that are highly active during the sexual cycle (Graia et al., 2001; Hamann, Feller, & Osiewacz, 2000; Hua-Van, Hericourt, Capy, Daboussi, & Langin, 1998; Ikeda et al., 2002; Nakayashiki, Nishimoto, Ikeda, Tosa, & Mayama, 1999; Neuveglise, Sarfati, Latge, & Paris, 1996; E. U. Selker & Stevens, 1985). RIP detects linked and unlinked sequences that are repeated two or more times in the genome and causes G-C to A-T transition mutations, efficiently degrading duplicated genetic elements, although RIP is less efficient in selfing populations (Cambareri, Jensen, Schabtach, & Selker, 1989; E. U. Selker, Cambareri, Jensen, & Haack, 1987). MIP targets similar duplicated sequence for methylation and silencing prior to meiosis (Barry, Faugeron, & Rossignol, 1993; Faugeron, Rhounim, & Rossignol, 1990; Freedman & Pukkila, 1993; Goyon & Faugeron, 1989).
In addition to silencing repeated sequences, unpaired DNA is targeted for modification and repression during meiosis. A new insertion of a transposon in a haploid will have no match on the homologous chromosome during mating. This unpaired DNA activates the RNAi response to silence all homologs of the unpaired DNA (Aramayo & Metzenberg, 1996; Janbon et al., 2010; Shiu & Metzenberg, 2002; Shiu, Raju, Zickler, & Metzenberg, 2001; Son, Min, Lee, Raju, & Lee, 2011). RNAi silencing is significantly upregulated during heterosexual cycles suppressing new transposon insertions in meiosis. In addition to suppressing transposable elements, RNAi can increase phenotypic diversity during environmental challenges. Cryptococcus uses RNAi to epigenetically silence repeated genes and generate phenotypic diversity. This silencing can be activated during mitotic or meiotic growth, however the silencing is 250 times more efficiently during heterosexual and unisexual reproduction cycles (Wang, Darwiche, & Heitman, 2013; Wang et al., 2010; Wang et al., 2012).
Outcrossing enhances the advantages of recombination, through increasing the genetic diversity of populations while decreasing the risk of a diploid carrying a homozygous deleterious mutation that recombination cannot restore. Plants and animals often have molecular and developmental systems to impede diploid selfing, such as dioecy. Fungi such as Cryptococcus determine sexual compatibility by their haploid genotypes and typically cannot inhibit diploid selfing. Though predominately diploid, Candida species can grow and are competent to mate as haploids or diploids. Restriction of mating to opaque, homozygous MTL locus cells combined with a paucity of haploids in the natural population is likely to effectively promote outcrossing during heterosex because MTL α/a cells do not undergo meiosis to produce mating compatible haploids. In addition to genetic or molecular mechanisms, outcrossing can be promoted through developmental strategies, such as wide dispersal of spores (Murphy & Zeyl, 2010; Wright et al., 2008). Fungal heterosexual reproduction promotes genetic diversity and outcrossing through incompatibility with cells of the same mating type and the depression of haploid selfing. One cost of self-incompatibility is a relative dearth of potential mating partners in the populations. In bipolar species with equal frequencies of MAT alleles the chance of any two cells being compatible is 50% but for those species with skewed MAT distributions the chances that any two cells are mating compatible is much lower. These tradeoffs make heterothallism, or obligate outcrossing, a high risk and high reward strategy. Some portion of the population may not be able to find a compatible mating partner but because there is no chance of selfing, the progeny of successful matings will have higher genetic diversity.
Unisexual reproduction has arisen in fungal species that are both predominantly diploid, such as Candida species, or haploid, such as Cryptococcus species. The costs posed by incompatibility with same mating type partners is reduced when all cells are available for mating through unisex for predominantly haploid species (Iwasa & Sasaki, 1987). If an opposite-mating type partner is not available, a same mating type partner or even a daughter can be a unisexual partner. Haploids expend considerable resources signaling, responding, and interacting with potential mating partners in the often stressful conditions that prompt mating (Xu, 2005). Unisexual haploids may be able to expend less energy finding suitable mates, and any haploid encountered will be compatible. Unisexual reproduction may have contradictory effects on the genetic diversity of species by increasing the number of successful matings with unrelated same mating type partners while also making daughters available for mating, increasing inbreeding depression. The benefits of increased outcrossing are even more pronounced in a species like C. neoformans, a species with uneven mating type ratios (Halliday et al., 1999; Kwon-Chung & Bennett, 1978; Litvintseva et al., 2005; Yan, Li, & Xu, 2002). The changes in outcrossing rate will be more modest in species that grow predominately as diploid, undergoing meiosis and returning quickly to the diploid state, as even in diploids that are strictly heterothallic spores produced by a diploid are able to mate with sisters. A unisexual cycle does increase the proportion of other siblings from a single meiotic event that a spore is compatible from 50% to 100%. When diploids carry lethal or deleterious recessive mutations, as C. albicans does, this promiscuity may be even more critical to enable unfit haploids to return to the favored diploid state. After sporulation, these deleterious mutations can render a portion of the opposite mating type haploids inviable or unsuitable for mating. Unisex allows a greater number of haploids to return to the favored diploid condition in such circumstances.
Several fungi with identified unisexual cycles grow mitotically primarily as haploids. While the full impact of ploidy on evolution and response to selection is not yet clear, a diploid cell clearly has several evolutionary advantages over the haploid (Gerstein & Otto, 2009; Haldane, 1937). First, diploids simply have twice the genetic material for mutation to act upon. Each gene is present twice and each copy is replicated during cell division providing additional substrate on which mutation can act (Orr & Otto, 1994). This doubling of mutational targets is especially important if the population facing selection is small, such as during an incipient infection (Zeyl, Vanderford, & Carter, 2003). Secondly, haploids face a challenge when confronting selective pressure that diploids avoid. Any genetic changes a haploid suffers are immediately expressed and potentially impact the phenotype. It may not be possible for haploids to acquire certain phenotypes or genetic changes directly. A new phenotype may require several genetic changes, one or more of which are detrimental but act epistatically or combinatorially to be advantageous. Any one of the changes may be detrimental, and subject to strong negative selection in haploids, but once several mutations occur, the changes are advantageous. It is difficult for haploids to cross these selective valleys to reach a global peak. On the other hand, diploids are able to complement initially deleterious heterozygous mutations with a dominant wild-type allele and carry those mutations for generations until the epistatic or compensatory mutation occurs. Thus, diploids can serve as a capacitor for evolution, by increasing the quantity of DNA substrate for mutation to act on and buffering mutations from selection. As diploids complete a sexual cycle and return to the haploid state mutations and new phenotypes are exposed to selection. The unisexual cycle allows haploid cells to reach the diploid state more frequently and without the need to find an opposite mating type cell, expanding the evolutionary potential of a haploid.
While sex is nearly universal in the eukaryotic lineage, it is not clear how meiotic ability is maintained in facultative sexual organisms such as fungi. Conditions suitable for mating, low nutrient conditions, and the presence of a compatible mating partners may not arise for many generations, resulting in relaxed selection on mating pathway genes. While a population may be able to divide mitotically indefinitely, continual mitotic growth results in a rapid decrease in fecundity (Lang, Murray, & Botstein, 2009; Xu, 2002; Xu, Ali, et al., 2000). Unisexual reproduction is an intermittent substitute when conditions are unfavorable for heterosexual mating or compatible mating type partners are not available. Unisexual reproduction utilizes the same, or very similar, genetic pathways as heterosexual reproduction (Feretzaki & Heitman, 2013). Frequent unisexual mating continually selects for functional or increasingly functional alleles of mating pathway genes, acting as practice for heterosexual mating. Thus, unisexual mating may help increase heterosexual fecundity through maintenance of shared pathways.
Unisex Alters the Life Cycle of Fungi
The transition from haploid to diploid or diploid to haploid brings about a large number of transcriptional, phenotypic, and morphological changes. These changes can be environmentally dependent, or specific to the lifecycle of a species. In general, under limiting growth conditions diploid cells are larger, increasing the ratio of volume to surface area. The change in surface area changes the number of transporters and receptors the cell can utilize while a change in volume can affect the protein content and number of organelles. Changes in the ratio of these components interact with specific environmental conditions. In addition, there are species-specific morphological changes that are dependent on changes in ploidy state as well, such as the transition from yeast to hyphae during the sexual cycle in Cryptococcus and Ustilago maydis (Snetselaar, Bölker, & Kahmann, 1996). The unisexual cycle can give access to these advantageous morphological and transcriptional changes.
Many unisexual species undergo significant morphological changes during the sexual cycle. C. neoformans grows mitotically as a haploid yeast and only produces hyphae, basidia, diploid cells, and haploid spores during the unisexual or heterosexual cycles. These cell types have significant morphological changes that can be advantageous. C. neoformans uses the hyphae produced by the unisexual cycle to explore the environment and forage for nutrients (Phadke, Feretzaki, & Heitman, 2013). The exploration of the environment by hyphae may also have other salutary effects such as bringing mating ready cells into contact with relatively distant mating partners. In this way, unisex mating can increase the number and frequency of outcrossing by enabling foraging for mates, akin to courtship in S. cerevisiae (Jackson & Hartwell, 1990a, 1990b).
Perhaps the most important morphological state that a sexual cycle gives access to is the spore. Sporulation is a response to adverse environmental conditions and is essential for dispersal to new niches. Species from the Cryptococcus genus are able to produce spores only via a unisexual or heterosexual cycle. A unisexual cycle may be essential for lineage survival in the case of a relative absence of opposite mating type partners. Other species, such as Neurospora, are able to generate mitotic spores called conidia that are able to effectively disperse and fill many roles of a sexual spore. However, even in these species there can be significant differences between the survival rates between mitotically generated conidia and meiotically generated spores (Trapero-Casas & Kaiser, 2007).
In addition to morphological changes, unisex changes the ploidy from haploid to diploid and allows the cell to return to the haploid state. Significant effort has been made to model under what environmental conditions a diploid or haploid state would be favored. Factors that have been considered include nutrient conditions, mutational load, and life cycle but experimental work had thus far been unable to support general conditions that favor a particular ploidy state (Adams & Hansche, 1974; Anderson, Sirjusingh, & Ricker, 2004; Crow & Kimura, 1965; Lewis, 1985; Mable, 2001; Mable & Otto, 2001; Orr & Otto, 1994; Weiss, Kukora, & Adams, 1975). Recently an extensive study of the interactions between ploidy and environment in the Saccharomyces group found that an impact of ploidy on growth is common. Under some environmental conditions ploidy had no impact on growth but for those conditions where the growth rates of different ploidy states differ, nearly equal numbers of conditions favored diploids and haploids (Zörgö et al., 2013). Advantages in an environmental condition of a particular ploidy were consistent over significant evolutionary time but there were no consistent growth advantages across environmental conditions for a ploidy state. Even in DNA damaging conditions, where an additional copy of each gene was hypothesized to be advantageous, there was no consistent growth advantage for diploids. Phleomycin, an inducer of DNA lesions, strongly favored diploids while hydroxyurea, which impedes DNA repair, favored haploids. Nutrient limiting conditions did not clearly favor either the haploid or diploid state, with some limiting nutrients favoring haploids and others favoring diploids. Thus, each ploidy state confers advantages depending on the environment. A unisexual organism can easily access both the haploid and diploid state by mating quickly in response to conditions that favor diploids and sporulating when conditions are favorable for haploid growth. A unisexual cycle that simply toggles a cell from haploid to diploid and then back to haploid could provide benefits to the organism in a fluctuating environment with no need to admix genetic diversity, or recombine.
Cellular and organelle genomic interactions are common and fraught with implications for sexual reproduction. Multicellular organisms uniparentally inherit mitochondrial and chloroplast genomes to limit organelle heteroplasmy. While uniparental inheritance is common in fungal species, it is by no means the rule. The model yeasts S. cerevisiae and S. pombe inherit mitochondrial genomes from either parent but Cryptococcus species uniparentially inherit their genome from the mating type a parent (Callen, 1974; Seitz-Mayr, Wolf, & Kaudewitz, 1978; Strausberg & Perlman, 1978; Xu, Ali, et al., 2000). Mitochondrial heteroplasmy causes reduced metabolic activity and accentuated stress responses in multicellular eukaryotes and is a common cause of disease (Sharpley et al., 2012) but it is not yet clear what affect mitochondrial heteroplasmy has on fungi. C. neoformans uniparentally inherits the mitochondria from the mating type a parent but mating type α is the prevalent unisexual mating type (Kwon-Chung & Bennett, 1978; Yan & Xu, 2003). Unisex is likely to disrupt the uniparental inheritance of mitochondria. In C. neoformans, mating type α will retain its mitochondria instead of receiving a mating type a derived mitochondria. There is no known mechanism to differentiate between two mating type α mitochondria during unisex, potentially introducing mitochondrial heteroplasmy and enabling mitochondrial genomic recombination (Yan, Hull, Sun, Heitman, & Xu, 2007). In addition, it would allow mitochondria that had evolved an epistatic, commensal, or selfish interaction with a mating type α genome to be transmitted through the unisexual cycle.
Medical Impacts of Unisex
Despite the ubiquity of sexual reproduction in complex eukaryotes and early observance in fungal lineages, asexuality was thought to be common among pathogenic eukaryotes. The widespread clonal reproduction among pathogenic and parasitic microorganisms engaged in an evolutionary arms race with hosts was even more surprising given the “Red Queen” arms race that pathogens must engage in with their hosts (Van Valen, 1974a, 1974b). A host-pathogen arms race places special emphasis on an organism’s ability to rapidly evolve and take advantage of genetic diversity (Delsing, Bleeker-Rovers, Kullberg, & Netea, 2012; Maor & Shirasu, 2005; Maynard Smith, 1978). Indeed, genetic conflict between hosts and pathogens is one of the leading evolutionary hypotheses for the ubiquity of sex in eukaryotic hosts (Ladle, Johnstone, & Judson, 1993; Otto & Michalakis, 1998; West, Lively, & Read, 1999). Selection on pathogens leads to intense specialization for common host genotypes, leading to the reduction of those hosts’ fitness relative to rare host genotypes (Haldane, 1949; Williams, 1975). Sex and recombination in hosts helps to increase genetic diversity and generate new and rare genotypes (Hamilton, 1980; Hamilton, Axelrod, & Tanese, 1990). Eukaryotic pathogens, such as C. albicans and C. neoformans, can employ sexual reproduction to maintain diversity and produce genotypes that are competent to infect new hosts (Byrnes et al., 2009; Carriconde et al., 2011; Kidd et al., 2004).
It was first noted that although there was little evidence of recombination, the genomes of these pathogenic eukaryotes retained a full complement of meiotic genes (Bougnoux et al., 2008; Loftus et al., 2005; Ramesh et al., 2005; Sturm, Vargas, Westenberger, Zingales, & Campbell, 2003; Tzung et al., 2001). Only in the last decade has it been considered possible that eukaryotic microorganisms are both clonal and sexually reproducing (Heitman, 2006, 2010). The discovery of both canonical sexual and a new novel unisexual cycle in multiple pathogenic fungi, and also parasites, revealed how eukaryotic microbial pathogens are equipped to engage in the arms race with their hosts (Alby et al., 2009; X. Lin et al., 2005). Thus, we now appreciate that rather than being asexual, clonal, and strictly mitotic as proposed as recently as a decade ago, eukaryotic microbial pathogens are unisexual, or cryptically sexual, resolving this decades long paradox.
Unisex plays a particularly important role in C. neoformans virulence as the infectious propagule is thought to be the spores produced by sexual reproduction (Giles et al., 2009; Sukroongreung, Kitiniyom, Nilakul, & Tantimavanich, 1998; Velagapudi et al., 2009). The C. neoformans population is >95% α dramatically limiting the opportunities for heterosexual spore generation leaving unisexual spores as the likely major infectious propagule (Halliday et al., 1999; Kwon-Chung & Bennett, 1978; Litvintseva et al., 2005). In addition, C. neoformans deploys unisex to generate de novo genetic variation through mutation and aneuploidy, after infection bottleneck (Ni et al., 2013; Sionov et al., 2010). These factors highlight how unisex may be integral to the virulence of this medically important fungal pathogen.
Homothallism is widespread in the fungal kingdom and the full extent of alternative homothallic mechanisms such as unisex are just beginning to be investigated. It is particularly intriguing that extant unisexual cycles were discovered in phylogenetically divergent human pathogens only relatively recently. Unisex may be a particularly advantageous means of homothallism for animal pathogens. Further research into the molecular mechanisms underlying unisex, how outcrossing versus inbreeding is regulated, the diversity and extent of unisexual species, and the environmental impacts of unisex will increase our understanding of fungal diversity in addition to its impact and evolutionary origin.
Acknowledgements
We thank Dr. Eve Chow, Blake Billmyre and Shelly Clancey for critical reading and thoughtful discussions. K.C.R. is supported by the NIH Molecular Mycology and Pathogenesis Training Program (AI52080). This work was supported by NIH/NIAID R37 grant AI39115-16 and R01 grant AI50113-10 to J.H.
Bibliography
- Adams J, & Hansche PE (1974). Population studies in microorganisms I. Evolution of diploidy in Saccharomyces cerevisiae. Genetics, 76(2), 327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alby K, & Bennett RJ (2011). Interspecies pheromone signaling promotes biofilm formation and same-sex mating in Candida albicans. Proceedings of the National Academy of Sciences, USA, 108(6), 2510–2515. doi: 10.1073/pnas.1017234108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alby K, Schaefer D, & Bennett RJ (2009). Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature, 460(7257), 890–893. doi: 10.1038/nature08252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames LM (1934). Hermaphroditism involving self-sterility and cross-fertility in the Ascomycete Pleurage anserina. Mycologia, 26(5), 392–414. doi: 10.2307/3754255 [DOI] [Google Scholar]
- Anderson JB, Sirjusingh C, & Ricker N (2004). Haploidy, diploidy and evolution of antifungal drug resistance in Saccharomyces cerevisiae. Genetics, 168(4), 1915–1923. doi: 10.1534/genetics.104.033266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramayo R, & Metzenberg RL (1996). Meiotic transvection in fungi. Cell, 86(1), 103–113. doi: 10.1016/S0092-8674(00)80081-1 [DOI] [PubMed] [Google Scholar]
- Arcangioli B, & de Lahondes R (2000). Fission yeast switches mating type by a replication-recombination coupled process. The EMBO Journal, 19(6), 1389–1396. doi: 10.1093/emboj/19.6.1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asker SE, & Jerling L (1992). Apomixis in Plants. Boca Raton, FL: CRC Press. [Google Scholar]
- Baldauf SL (2003). The deep roots of eukaryotes. Science, 300(5626), 1703–1706. doi: 10.1126/science.1085544 [DOI] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, & Doolittle WF (2000). A kingdom-level phylogeny of eukaryotes based on combined protein data. Science, 290(5493), 972–977. doi: 10.1126/science.290.5493.972 [DOI] [PubMed] [Google Scholar]
- Barry C, Faugeron G, & Rossignol JL (1993). Methylation induced premeiotically in Ascobolus: coextension with DNA repeat lengths and effect on transcript elongation. Proceedings of the National Academy of Sciences, USA, 90(10), 4557–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum E, Martinez P, & Åström SU (2010). α3, a transposable element that promotes host sexual reproduction. Genes & Development, 24(1), 33–44. doi: 10.1101/gad.557310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G (1982). The Masterpiece of Nature. The Evolution and Genetics of Sexuality. Los Angeles, CA: University of California Press. [Google Scholar]
- Bennett RJ, & Johnson AD (2003). Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. The EMBO Journal, 22(10), 2505–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, & Johnson AD (2006). The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of C. albicans. Molecular Microbiology, 62(1), 100–119. doi: 10.1111/j.1365-2958.2006.05367.x [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Miller MG, Chua PR, Maxon ME, & Johnson AD (2005). Nuclear fusion occurs during mating in Candida albicans and is dependent on the KAR3 gene. Molecular Microbiology, 55(4), 1046–1059. doi: 10.1111/j.1365-2958.2005.04466.x [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Uhl MA, Miller MG, & Johnson AD (2003). Identification and characterization of a Candida albicans mating pheromone. Molecular and Cellular Biology, 23(22), 8189–8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbee ML, Payne BP, Zhang G, Roberts RG, & Turgeon BG (2003). Shared ITS DNA substitutions in isolates of opposite mating type reveal a recombining history for three presumed asexual species in the filamentous ascomycete genus Alternaria. Mycological Research, 107(Pt 2), 169–182. [DOI] [PubMed] [Google Scholar]
- Blakeslee AF (1904). Sexual reproduction in the Mucorineae. Proceedings of the American Academy of Arts and Sciences, 40(4), 205–319. doi: 10.2307/20021962 [DOI] [Google Scholar]
- Bölker M, Urban M, & Kahmann R (1992). The a mating type locus of U. maydis specifies cell signaling components. Cell, 68(3), 441–450. [DOI] [PubMed] [Google Scholar]
- Bougnoux M-E, Pujol C, Diogo D, Bouchier C, Soll DR, & d’Enfert C (2008). Mating is rare within as well as between clades of the human pathogen Candida albicans. Fungal Genetics and Biology, 45(3), 221–231. doi: 10.1016/j.fgb.2007.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin TS, Le Rouzic A, & Capy P (2012). How does selfing affect the dynamics of selfish transposable elements? Mobile DNA, 3(1), 1–9. doi: 10.1186/1759-8753-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt ME, Hutwagner LC, Kuykendall RJ, & Pinner RW (1995). Comparison of multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for molecular subtyping of Cryptococcus neoformans. Journal of Clinical Microbiology, 33(7), 1890–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui T, Lin X, Malik R, Heitman J, & Carter DA (2008). Isolates of Cryptococcus neoformans from infected animals reveal genetic exchange in unisexual, alpha mating type populations. Eukaryotic Cell, 7(10), 1771–1780. doi: 10.1128/EC.00097-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes EJ 3rd, Bartlett KH, Perfect JR, & Heitman J (2011). Cryptococcus gattii: an emerging fungal pathogen infecting humans and animals. Microbes and Infection, 13(11), 895–907. doi: 10.1016/j.micinf.2011.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes EJ 3rd, Bildfell RJ, Frank SA, Mitchell TG, Marr KA, & Heitman J (2009). Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the Pacific Northwest in the United States. Journal of Infectious Diseases, 199(7), 1081–1086. doi: 10.1086/597306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes EJ 3rd, Li W, Lewit Y, Ma H, Voelz K, Ren P, … Heitman J (2010). Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the Northwest United States. PLoS Pathogens, 6(4), e1000850. doi: 10.1371/journal.ppat.1000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen DF (1974). Recombination and segregation of mitochondrial genes in Saccharomyces cerevisiae. Molecular and General Genetics, 134(1), 49–63. doi: 10.1007/BF00332812 [DOI] [PubMed] [Google Scholar]
- Cambareri EB, Jensen BC, Schabtach E, & Selker EU (1989). Repeat-induced G-C to A-T mutations in Neurospora. Science, 244(4912), 1571–1575. [DOI] [PubMed] [Google Scholar]
- Carriconde F, Gilgado F, Arthur I, Ellis D, Malik R, van de Wiele N, … Meyer W (2011). Clonality and α-a recombination in the Australian Cryptococcus gattii VGII population - an emerging outbreak in Australia. PLoS ONE, 6(2), e16936. doi: 10.1371/journal.pone.0016936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T (2004). Only six kingdoms of life. Proceedings of the Royal Society of London. Series B: Biological Sciences, 271(1545), 1251–1262. doi: 10.1098/rspb.2004.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RK, & Otte CA (1982). Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and α factor pheromones. Molecular and Cellular Biology, 2(1), 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, & Wright SI (2001). Breeding systems and genome evolution. Current Opinion in Genetics & Development, 11(6), 685–690. doi: 10.1016/S0959-437X(00)00254-9 [DOI] [PubMed] [Google Scholar]
- Chen J, Chen J, Lane S, & Liu H (2002). A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Molecular Microbiology, 46(5), 1335–1344. [DOI] [PubMed] [Google Scholar]
- Crow JF, & Kimura M (1965). Evolution in sexual and asexual populations. The American Naturalist, 99(909), 439–450. doi: 10.2307/2459132 [DOI] [Google Scholar]
- Dacks J, & Roger AJ (1999). The first sexual lineage and the relevance of facultative sex. Journal of Molecular Evolution, 48(6), 779–783. doi: 10.1007/PL00013156 [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, & Klar AJS (1999). Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature, 400(6740), 181–184. [DOI] [PubMed] [Google Scholar]
- Daniels KJ, Srikantha T, Lockhart SR, Pujol C, & Soll DR (2006). Opaque cells signal white cells to form biofilms in Candida albicans. The EMBO Journal, 25(10), 2240–2252. doi: 10.1038/sj.emboj.7601099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Bartlett KH, Baer R, Byrnes EJ 3rd, Galanis E, Heitman J, …. Marr KA (2009). Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerging Infectious Diseases, 15(8), 1185–1191. doi: Doi 10.3201/Eid1508.081384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Nichols CB, Cox GM, Perfect JR, & Heitman J (2003). A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Molecular Microbiology, 49(2), 469–485. [DOI] [PubMed] [Google Scholar]
- Debuchy R, & Turgeon BG (2006). Mating-type structure, evolution, and function in euascomycetes. In Kües U & Fischer R (Eds.), Growth, Differentiation and Sexuality (Vol. 1, pp. 293–323): Springer Berlin Heidelberg. [Google Scholar]
- Delsing CE, Bleeker-Rovers CP, Kullberg B-J, & Netea MG (2012). Treatment of candidiasis: insights from host genetics. Expert Review of Anti-infective Therapy, 10(8), 947–956. doi: 10.1586/eri.12.79 [DOI] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouzé P, Worden AZ, Robbens S, … Moreau H (2006). Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proceedings of the National Academy of Sciences, USA, 103(31), 11647–11652. doi: 10.1073/pnas.0604795103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, Steiner S, … Philippsen P (2004). The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science, 304(5668), 304–307. doi: 10.1126/science.1095781 [DOI] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Kuo S, & Philippsen P (2013). Genomes of Ashbya fungi isolated from insects reveal four mating-type loci, numerous translocations, lack of transposons, and distinct gene duplications. G3: Genes, Genomes, Genetics, 3(8), 1225–1239. doi: 10.1534/g3.112.002881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T, Levene H, Spassky B, & Spassky N (1959). Release of genetic variability through recombination. III. Drosophila prosaltans. Genetics, 44(1), 75–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge BO (1927). Nuclear phenomena associated with heterothallism and homothallism in the Ascomycete Neurospora. Journal of Agricultural Research, 35, 289–305. [Google Scholar]
- Dodge BO, Singleton JR, & Rolnick A (1950). Studies on lethal E gene in Neurospora tetrasperma, including chromosome counts also in races of N. sitophila. Proceedings of the American Philosophical Society, 94(1), 38–52. doi: 10.2307/3143250 [DOI] [Google Scholar]
- Dolgin ES, Charlesworth B, & Cutter AD (2008). Population frequencies of transposable elements in selfing and outcrossing Caenorhabditis nematodes. Genetics Research, 90(04), 317–329. doi: doi: 10.1017/S0016672308009440 [DOI] [PubMed] [Google Scholar]
- Dumitru R, Navarathna DH, Semighini CP, Elowsky CG, Dumitru RV, Dignard D, … Nickerson KW (2007). In vivo and in vitro anaerobic mating in Candida albicans. Eukaryotic Cell, 6(3), 465–472. doi: 10.1128/EC.00316-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, & Botstein D (2002). Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences, USA, 99(25), 16144–16149. doi: 10.1073/pnas.242624799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis DH, & Pfeiffer TJ (1990). Natural habitat of Cryptococcus neoformans var. gattii. Journal of Clinical Microbiology, 28(7), 1642–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer RA, Henk DA, Garner TWJ, Balloux F, Woodhams DC, & Fisher MC (2013). Chromosomal copy number variation, selection and uneven rates of recombination reveal cryptic genome diversity linked to pathogenicity. PLoS Genetics, 9(8), e1003703. doi: 10.1371/journal.pgen.1003703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer RA, Weinert LA, Bielby J, Garner TWJ, Balloux F, Clare F, … Fisher MC (2011). Multiple emergences of genetically diverse amphibian-infecting chytrids include a globalized hypervirulent recombinant lineage. Proceedings of the National Academy of Sciences, USA, 108(46), 18732–18736. doi: 10.1073/pnas.1111915108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faugeron G, Rhounim L, & Rossignol JL (1990). How does the cell count the number of ectopic copies of a gene in the premeiotic inactivation process acting in Ascobolus immersus? Genetics, 124(3), 585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (1974). The evolutionary advantage of recombination. Genetics, 78(2), 737–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, & Heitman J (2013). Genetic circuits that govern bisexual and unisexual reproduction in Cryptococcus neoformans. PLoS Genetics, 9(8), e1003688. doi: 10.1371/journal.pgen.1003688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, & Herskowitz I (1985). The yeast STE12 product is required for expression of two sets of cell-type-specific genes. Cell, 42(3), 923–930. doi: 10.1016/0092-8674(85)90288-0 [DOI] [PubMed] [Google Scholar]
- Forche A, Alby K, Schaefer D, Johnson AD, Berman J, & Bennett RJ (2008). The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biology, 6(5), e110. doi: 10.1371/journal.pbio.0060110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Diezmann S, Subaran RL, Allen A, Lengeler KB, Dietrich FS, & Heitman J (2004). Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biology, 2(12), e384. doi: 10.1371/journal.pbio.0020384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, … Heitman J (2005). Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature, 437(7063), 1360–1364. doi: 10.1038/nature04220 [DOI] [PubMed] [Google Scholar]
- Fraser JA, Subaran RL, Nichols CB, & Heitman J (2003). Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans variety gattii: implications for an outbreak on Vancouver Island. Eukaryotic Cell, 2, 1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman T, & Pukkila PJ (1993). De novo methylation of repeated sequences in Coprinus cinereus. Genetics, 135(2), 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, … Dawson SC (2010). The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell, 140(5), 631–642. doi: 10.1016/j.cell.2010.01.032 [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo CA, Ma L-J, Wortman JR, Batzoglou S, … Birren BW (2005). Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature, 438(7071), 1105–1115. [DOI] [PubMed] [Google Scholar]
- Gerstein AC, & Otto SP (2009). Ploidy and the causes of genomic evolution. Journal of Heredity, 100(5), 571–581. doi: 10.1093/jhered/esp057 [DOI] [PubMed] [Google Scholar]
- Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, & Gillevet PM (2010). Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathogens, 6(1), e1000713. doi: 10.1371/journal.ppat.1000713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles SS, Dagenais TRT, Botts MR, Keller NP, & Hull CM (2009). Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infection and Immunity, 77(8), 3491–3500. doi: 10.1128/IAI.00334-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioti A, Mushegian AA, Strandberg R, Stajich JE, & Johannesson H (2012). Unidirectional evolutionary transitions in fungal mating systems and the role of transposable elements. Molecular Biology and Evolution, 29(10), 3215–3226. doi: 10.1093/molbev/mss132 [DOI] [PubMed] [Google Scholar]
- Gioti A, Stajich JE, & Johannesson H (2013). Neurospora and the dead-end hypothesis: Genomic consequences of selfing in the model genus. Evolution, 67(12), 3600–3616. doi: 10.1111/evo.12206 [DOI] [PubMed] [Google Scholar]
- Glass NL, Metzenberg RL, & Raju NB (1990). Homothallic Sordariaceae from nature: The absence of strains containing only the a mating type sequence. Experimental Mycology, 14(3), 274–289. doi: 10.1016/0147-5975(90)90025-O [DOI] [Google Scholar]
- Glass NL, & Smith ML (1994). Structure and function of a mating-type gene from the homothallic species Neurospora africana. Molecular and Cellular Biology, 244(4), 401–409. [DOI] [PubMed] [Google Scholar]
- Gordon JL, Armisén D, Proux-Wéra E, ÓhÉigeartaigh SS, Byrne KP, & Wolfe KH (2011). Evolutionary erosion of yeast sex chromosomes by mating-type switching accidents. Proceedings of the National Academy of Sciences, USA, 108(50), 20024–20029. doi: 10.1073/pnas.1112808108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C, & Faugeron G (1989). Targeted transformation of Ascobolus immersus and de novo methylation of the resulting duplicated DNA sequences. Molecular and Cellular Biology, 9(7), 2818–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graia F, Lespinet O, Rimbault B, Dequard-Chablat M, Coppin E, & Picard M (2001). Genome quality control: RIP (repeat-induced point mutation) comes to Podospora. Molecular Microbiology, 40(3), 586–595. [DOI] [PubMed] [Google Scholar]
- Gräser Y, Kühnisch J, & Presber W (1999). Molecular markers reveal exclusively clonal reproduction in Trichophyton rubrum. Journal of Clinical Microbiology, 37(11), 3713–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halary S, Malik S-B, Lildhar L, Slamovits CH, Hijri M, & Corradi N (2011). Conserved meiotic machinery in Glomus spp., a putatively ancient asexual fungal lineage. Genome Biology and Evolution, 3, 950–958. doi: 10.1093/gbe/evr089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS (1937). The effect of variation of fitness. The American Naturalist, 71(735), 337–349. doi: 10.2307/2457289 [DOI] [Google Scholar]
- Haldane JBS (1949). Disease and evolution. Supplement to La Ricerca Scientifica(19), 68–76. [Google Scholar]
- Halliday CL, Bui T, Krockenberger M, Malik R, Ellis DH, & Carter DA (1999). Presence of α and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia. Journal of Clinical Microbiology, 37(9), 2920–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann A, Feller F, & Osiewacz HD (2000). The degenerate DNA transposon Pat and repeat-induced point mutation (RIP) in Podospora anserina. Molecular and Cellular Biology, 263(6), 1061–1069. [DOI] [PubMed] [Google Scholar]
- Hamilton WD (1980). Sex versus non-sex versus parasite. Oikos, 35(2), 282–290. doi: 10.2307/3544435 [DOI] [Google Scholar]
- Hamilton WD, Axelrod R, & Tanese R (1990). Sexual reproduction as an adaptation to resist parasites (a review). Proceedings of the National Academy of Sciences, USA, 87(9), 3566–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH (1980). Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. The Journal of Cell Biology, 85(3), 811–822. doi: 10.1083/jcb.85.3.811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J (2006). Sexual reproduction and the evolution of microbial pathogens. Current Biology, 16(17), R711–R725. doi: 10.1016/j.cub.2006.07.064 [DOI] [PubMed] [Google Scholar]
- Heitman J (2010). Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host & Microbe, 8(1), 86–99. doi: 10.1016/j.chom.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Sun S, & James TY (2013). Evolution of fungal sexual reproduction. Mycologia, 105(1), 1–27. doi: 10.3852/12-253 [DOI] [PubMed] [Google Scholar]
- Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, Harrison BD, … Berman J (2013). The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature, 494(7435), 55–59. doi: 10.1038/nature11865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JB, & Herskowitz I (1977). Interconversion of yeast mating types II. Restoration of mating ability to sterile mutants in homothallic and heterothallic strains. Genetics, 85(3), 373–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JB, Strathern JN, & Herskowitz I (1977). Interconversion of yeast mating types III. Action of the homothallism (HO) gene in cells homozygous for the mating type locus. Genetics, 85(3), 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, & Robertson A (1966). The effect of linkage on limits to artificial selection. Genetical Research, 8(3), 269–294. [PubMed] [Google Scholar]
- Hiremath SS, Chowdhary A, Kowshik T, Randhawa HS, Sun S, & Xu J (2008). Long-distance dispersal and recombination in environmental populations of Cryptococcus neoformans var. grubii from India. Microbiology, 154(Pt 5), 1513–1524. doi: 10.1099/mic.0.2007/015594-0 [DOI] [PubMed] [Google Scholar]
- Hsueh Y-P, Lin X, Kwon-Chung KJ, & Heitman J (2011). Sexual reproduction of Cryptococcus. In Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR & Casadevall A (Eds.), Cryptococcus: From Human Pathogen to Model Yeast (pp. 81–96). Washington, DC: ASM Press. [Google Scholar]
- Hua-Van A, Hericourt F, Capy P, Daboussi MJ, & Langin T (1998). Three highly divergent subfamilies of the impala transposable element coexist in the genome of the fungus Fusarium oxysporum. Molecular and Cellular Biology, 259(4), 354–362. [DOI] [PubMed] [Google Scholar]
- Huang G, Srikantha T, Sahni N, Yi S, & Soll DR (2009). CO(2) regulates white-to-opaque switching in Candida albicans. Current Biology, 19(4), 330–334. doi: 10.1016/j.cub.2009.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Wang H, Chou S, Nie X, Chen J, & Liu H (2006). Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proceedings of the National Academy of Sciences, USA, 103(34), 12813–12818. doi: 10.1073/pnas.0605270103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, & Soll DR (2010). N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathogens, 6(3), e1000806. doi: 10.1371/journal.ppat.1000806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Boily M-J, & Heitman J (2005). Sex-specific homeodomain proteins Sxi1α and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryotic Cell, 4(3), 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Davidson RC, & Heitman J (2002). Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1α. Genes & Development, 16(23), 3046–3060. doi: 10.1101/gad.1041402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, & Heitman J (2002). Genetics of Cryptococcus neoformans. Annual Review of Genetics, 36, 557–615. doi: 10.1146/annurev.genet.36.052402.152652 [DOI] [PubMed] [Google Scholar]
- Hull CM, & Johnson AD (1999). Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science, 285(5431), 1271–1275. [DOI] [PubMed] [Google Scholar]
- Hull CM, Raisner RM, & Johnson AD (2000). Evidence for mating of the "asexual" yeast Candida albicans in a mammalian host. Science, 289(5477), 307–310. [DOI] [PubMed] [Google Scholar]
- Idnurm A, Bahn Y-S, Nielsen K, Lin X, Fraser JA, & Heitman J (2005). Deciphering the model pathogenic fungus Cryptococcus neoformans. Nature Reviews Microbiology, 3(10), 753–764. doi: 10.1038/nrmicro1245 [DOI] [PubMed] [Google Scholar]
- Ikeda K, Nakayashiki H, Kataoka T, Tamba H, Hashimoto Y, Tosa Y, & Mayama S (2002). Repeat-induced point mutation (RIP) in Magnaporthe grisea: implications for its sexual cycle in the natural field context. Molecular Microbiology, 45(5), 1355–1364. [DOI] [PubMed] [Google Scholar]
- Inderbitzin P, Harkness J, Turgeon BG, & Berbee ML (2005). Lateral transfer of mating system in Stemphylium. Proceedings of the National Academy of Sciences, USA, 102(32), 11390–11395. doi: 10.1073/pnas.0501918102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa Y, & Sasaki A (1987). Evolution of the number of sexes. Evolution, 41(1), 49–65. doi: 10.2307/2408972 [DOI] [PubMed] [Google Scholar]
- Jackson CL, & Hartwell LH (1990a). Courtship in S. cerevisiae: Both cell types choose mating partners by responding to the strongest pheromone signal. Cell, 63(5), 1039–1051. doi: 10.1016/0092-8674(90)90507-B [DOI] [PubMed] [Google Scholar]
- Jackson CL, & Hartwell LH (1990b). Courtship in Saccharomyces cerevisiae: an early cell-cell interaction during mating. Molecular and Cellular Biology, 10(5), 2202–2213. doi: 10.1128/mcb.10.5.2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TY, Litvintseva AP, Vilgalys R, Morgan JAT, Taylor JW, Fisher MC, … Longcore JE (2009). Rapid global expansion of the fungal disease chytridiomycosis into declining and healthy amphibian populations. PLoS Pathogens, 5(5), e1000458. doi: 10.1371/journal.ppat.1000458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon G, Maeng S, Yang D-H, Ko Y-J, Jung K-W, Moyrand F, … Bahn Y-S (2010). Characterizing the role of RNA silencing components in Cryptococcus neoformans. Fungal Genetics and Biology, 47(12), 1070–1080. doi: 10.1016/j.fgb.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SK JR, & Bennett RJ (2011). Fungal mating pheromones: Choreographing the dating game. Fungal Genetics and Biology, 48(7), 668–676. doi: 10.1016/j.fgb.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämper J, Reichmann M, Romeis T, Bölker M, & Kahmann R (1995). Multiallelic recognition: Nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell, 81(1), 73–83. doi: 10.1016/0092-8674(95)90372-0 [DOI] [PubMed] [Google Scholar]
- Kano R, Isizuka M, Hiruma M, Mochizuki T, Kamata H, & Hasegawa A (2013). Mating type gene (MAT1-1) in Japanese isolates of Trichophyton rubrum. Mycopathologia, 175(1-2), 171–173. doi: 10.1007/s11046-012-9603-2 [DOI] [PubMed] [Google Scholar]
- Kaykov A, & Arcangioli B (2004). A programmed strand-specific and modified nick in S. pombe constitutes a novel type of chromosomal imprint. Current Biology, 14(21), 1924–1928. doi: 10.1016/j.cub.2004.10.026 [DOI] [PubMed] [Google Scholar]
- Keller SM, Viviani MA, Esposto MC, Cogliati M, & Wickes BL (2003). Molecular and genetic characterization of a serotype A MATa Cryptococcus neoformans isolate. Microbiology, 149(Pt 1), 131–142. [DOI] [PubMed] [Google Scholar]
- Kibbler CC, Seaton S, Barnes RA, Gransden WR, Holliman RE, Johnson EM, … Wilson JA (2003). Management and outcome of bloodstream infections due to Candida species in England and Wales. Journal of Hospital Infection, 54(1), 18–24. doi: 10.1016/S0195-6701(03)00085-9 [DOI] [PubMed] [Google Scholar]
- Kidd SE, Bach PJ, Hingston AO, Mak S, Chow Y, MacDougall L, … Bartlett KH (2007). Cryptococcus gattii dispersal mechanisms, British Columbia, Canada. Emerging Infectious Diseases, 13(1), 51–57. doi: 10.3201/eid1301.060823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd SE, Chow Y, Mak S, Bach PJ, Chen H, Hingston AO, … Bartlett KH (2007). Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Applied and Environmental Microbiology, 73(5), 1433–1443. doi: 10.1128/AEM.01330-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, … Meyer W (2004). A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proceedings of the National Academy of Sciences, USA, 101(49), 17258–17263. doi: 10.1073/pnas.0402981101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RS, Harris CA, Small CB, Moll B, Lesser M, & Friedland GH (1984). Oral candidiasis in high-risk patients as the initial manifestation of the acquired immunodeficiency syndrome. New England Journal of Medicine, 311(6), 354–358. doi: doi: 10.1056/NEJM198408093110602 [DOI] [PubMed] [Google Scholar]
- Kronstad JW, & Leong SA (1990). The b mating-type locus of Ustilago maydis contains variable and constant regions. Genes & Development, 4(8), 1384–1395. doi: 10.1101/gad.4.8.1384 [DOI] [PubMed] [Google Scholar]
- Kruzel EK, Giles SS, & Hull CM (2012). Analysis of Cryptococcus neoformans sexual development reveals rewiring of the pheromone-response network by a change in transcription factor identity. Genetics, 191(2), 435–449. doi: 10.1534/genetics.112.138958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvaal C, Lachke SA, Srikantha T, Daniels KJ, McCoy J, & Soll DR (1999). Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infection and Immunity, 67(12), 6652–6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ (1975). A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia, 67, 1197–1200. [PubMed] [Google Scholar]
- Kwon-Chung KJ (1976a). Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia, 68(4), 821–833. [PubMed] [Google Scholar]
- Kwon-Chung KJ (1976b). A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia, 68(4), 943–946. [PubMed] [Google Scholar]
- Kwon-Chung KJ, & Bennett JE (1978). Distribution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. American Journal of Epidemiology, 108(4), 337–340. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Chang YC, Bauer R, Swann EC, Taylor JW, & Goel R (1995). The characteristics that differentiate Filobasidiella depauperata and Filobasidiella neoformans. Studies of Mycology, 38, 67–79. [Google Scholar]
- Lachke SA, Srikantha T, & Soll DR (2003). The regulation of EFG1 in white-opaque switching in Candida albicans involves overlapping promoters. Molecular Microbiology, 48(2), 523–536. [DOI] [PubMed] [Google Scholar]
- Ladle RJ, Johnstone RA, & Judson OP (1993). Coevolutionary dynamics of sex in a metapopulation: escaping the Red Queen. Proceedings: Biological Sciences, 253(1337), 155–160. doi: 10.2307/49803 [DOI] [Google Scholar]
- Lang GI, Murray AW, & Botstein D (2009). The cost of gene expression underlies a fitness trade-off in yeast. Proceedings of the National Academy of Sciences, USA, 106(14), 5755–5760. doi: 10.1073/pnas.0901620106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Botstein D, & Desai MM (2013). Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature, 500(7464), 571–574. doi: 10.1038/nature12344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand M, Lephart P, Forche A, Mueller F-MC, Walsh T, Magee PT, & Magee BB (2004). Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Molecular Microbiology, 52(5), 1451–1462. doi: 10.1111/j.1365-2958.2004.04068.x [DOI] [PubMed] [Google Scholar]
- Lengeler KB, Cox GM, & Heitman J (2001). Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infection and Immunity, 69(1), 115–122. doi: 10.1128/IAI.69.1.115-122.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Wang P, Cox GM, Perfect JR, & Heitman J (2000). Identification of the MATa mating-type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct. Proceedings of the National Academy of Sciences, USA, 97(26), 14555–14460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene H (1959). Release of genetic variability through recombination. IV. Statistic theory. Genetics, 44(1), 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis WM Jr. (1985). Nutrient scarcity as an evolutionary cause of haploidy. The American Naturalist, 125(5), 692–701. doi: 10.2307/2461479 [DOI] [Google Scholar]
- Li W, Metin B, White TC, & Heitman J (2010). organization and evolutionary trajectory of the mating type (MAT) locus in dermatophyte and dimorphic fungal pathogens. Eukaryotic Cell, 9(1), 46–58. doi: 10.1128/ec.00259-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Kabrawala S, Fox EP, Nobile CJ, Johnson AD, & Bennett RJ (2013). Genetic control of conventional and pheromone-stimulated biofilm formation in Candida albicans. PLoS Pathogens, 9(4), e1003305. doi: 10.1371/journal.ppat.1003305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, & Heitman J (2007). Mechanisms of homothallism in fungi and transitions between heterothallism and homothallism. In Heitman J, Kronstad J, Taylor J & Casselton L (Eds.), Sex in Fungi (pp. 35–58). Washington, DC: ASM Press. [Google Scholar]
- Lin X, Huang JC, Mitchell TG, & Heitman J (2006). Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATα allele enhances filamentation. PLoS Genetics, 2(11), e187. doi: 10.1371/journal.pgen.0020187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Hull CM, & Heitman J (2005). Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature, 434(7036), 1017–1021. doi: 10.1038/nature03448 [DOI] [PubMed] [Google Scholar]
- Lin X, Jackson JC, Feretzaki M, Xue C, & Heitman J (2010). Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genetics, 6(5), e1000953. doi: 10.1371/journal.pgen.1000953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, & Heitman J (2007). αADα hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genetics, 3(10), 1975–1990. doi: 10.1371/journal.pgen.0030186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Patel S, Litvintseva AP, Floyd A, Mitchell TG, & Heitman J (2009). Diploids in the Cryptococcus neoformans serotype A population homozygous for the α mating type originate via unisexual mating. PLoS Pathogens, 5(1), e1000283. doi: 10.1371/journal.ppat.1000283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindegren CC, & Lindegren G (1943a). Legitimate and illegitimate mating in Saccharomyces cerevisiae. Genetics, 28(81). [Google Scholar]
- Lindegren CC, & Lindegren G (1943b). Segregation, mutation, and copulation in Saccharomyces cerevisiae. Annals of the Missouri Botanical Garden, 30(4), 453–468. doi: 10.2307/2394308 [DOI] [Google Scholar]
- Lindegren CC, & Lindegren G (1944). Instability of the mating type alleles in Saccharomyces. Annals of the Missouri Botanical Garden, 31(2), 203–217. doi: 10.2307/2394338 [DOI] [Google Scholar]
- Litvintseva AP, Kestenbaum L, Vilgalys R, & Mitchell TG (2005). Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. Journal of Clinical Microbiology, 43(2), 556–564. doi: 10.1128/jcm.43.2.556-564.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Lin X, Templeton I, Heitman J, & Mitchell TG (2007). Many globally isolated AD hybrid strains of Cryptococcus neoformans originated in Africa. PLoS Pathogens, 3(8), e114. doi: 10.1371/journal.ppat.0030114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Marra RE, Nielsen K, Heitman J, Vilgalys R, & Mitchell TG (2003). Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryotic Cell, 2(6), 1162–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd DG (1988). Benefits and costs of biparental and uniparental reproduction in plants. In Michod RE & Levin BR (Eds.), The Evolution of Sex. (pp. 233–252). Sunderland, Massachusetts: Sinauer Associates. [Google Scholar]
- Lockhart SR, Daniels KJ, Zhao R, Wessels D, & Soll DR (2003). Cell biology of mating in Candida albicans. Eukaryotic Cell, 2(1), 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart SR, Pujol C, Daniels KJ, Miller MG, Johnson AD, Pfaller MA, & Soll DR (2002). In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics, 162(2), 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, … Hyman RW (2005). The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science, 307(5713), 1321–1324. doi: 10.1126/science.1103773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M (2007). The frailty of adaptive hypotheses for the origins of organismal complexity. Proceedings of the National Academy of Sciences, USA, 104(suppl 1), 8597–8604. doi: 10.1073/pnas.0702207104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable BK (2001). Ploidy evolution in the yeast Saccharomyces cerevisiae: A test of the nutrient limitation hypothesis. Journal of Evolutionary Biology, 14(1), 157–170. doi: 10.1046/j.1420-9101.2001.00245.x [DOI] [PubMed] [Google Scholar]
- Mable BK, & Otto SP (2001). Masking and purging mutations following EMS treatment in haploid, diploid and tetraploid yeast (Saccharomyces cerevisiae). Genetical Research, 77(1), 9–26. [DOI] [PubMed] [Google Scholar]
- Magee BB, Legrand M, Alarco AM, Raymond M, & Magee PT (2002). Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Molecular Microbiology, 46(5), 1345–1351. [DOI] [PubMed] [Google Scholar]
- Magee BB, & Magee PT (2000). Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science, 289(5477), 310–313. [DOI] [PubMed] [Google Scholar]
- Maor R, & Shirasu K (2005). The arms race continues: battle strategies between plants and fungal pathogens. Current Opinion in Microbiology, 8(4), 399–404. doi: 10.1016/j.mib.2005.06.008 [DOI] [PubMed] [Google Scholar]
- Martinez DA, Oliver BG, Gräser Y, Goldberg JM, Li W, Martinez-Rossi NM, … White TC (2012). Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. mBio, 3(5), e00259–00212. doi: 10.1128/mBio.00259-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M, Henriques M, Ribeiro AP, Fernandes R, Goncalves V, Seabra A, … Oliveira R (2010). Oral Candida carriage of patients attending a dental clinic in Braga, Portugal. Revista Iberoamericana De Micologia, 27(3), 119–124. doi: 10.1016/j.riam.2010.03.007 [DOI] [PubMed] [Google Scholar]
- Maynard Smith J (1978). The Evolution of Sex. Cambridge: Cambridge University Press. [Google Scholar]
- McCusker JH (2006). Saccharomyces cerevisiae: an emerging and model pathogenic fungus. In Heitman J, Filler SG, Edwards JE & Mitchell AP (Eds.), Molecular Principles of Fungal Pathogenesis. (pp. 245–259). Washington, DC: ASM Press. [Google Scholar]
- Melton JJ, Redding SW, Kirkpatrick WR, Reasner CA, Ocampo GL, Venkatesh A, & Mealey BL (2010). Recovery of Candida dubliniensis and other Candida species from the oral cavity of subjects with periodontitis who had well-controlled and poorly controlled type 2 diabetes: a pilot study. Special Care in Dentistry, 30(6), 230–234. doi: 10.1111/j.1754-4505.2010.00159.x [DOI] [PubMed] [Google Scholar]
- Miller MG, & Johnson AD (2002). White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell, 110(3), 293–302. [DOI] [PubMed] [Google Scholar]
- Morgan JAT, Vredenburg VT, Rachowicz LJ, Knapp RA, Stice MJ, Tunstall T, … Taylor JW (2007). Population genetics of the frog-killing fungus Batrachochytrium dendrobatidis. Proceedings of the National Academy of Sciences, USA, 104(34), 13845–13850. doi: 10.1073/pnas.0701838104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MT (2001). Transposable element number in mixed mating populations. Genetics Research, 77(03), 261–275. doi: 10.1017/S0016672301005067 [DOI] [PubMed] [Google Scholar]
- Muller HJ (1932). Some genetic aspects of sex. The American Naturalist, 66(703), 118–138. doi: 10.2307/2456922 [DOI] [Google Scholar]
- Muller HJ (1964). The relation of recombination to mutational advance. Mutation Research, 1(1), 2–9. doi: 10.1016/0027-5107(64)90047-8 [DOI] [PubMed] [Google Scholar]
- Murphy HA, & Zeyl CW (2010). Yeast sex: Surprisingly high rates of outcrossing between asci. PLoS ONE, 5(5), e10461. doi: 10.1371/journal.pone.0010461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki H, Nishimoto N, Ikeda K, Tosa Y, & Mayama S (1999). Degenerate MAGGY elements in a subgroup of Pyricularia grisea: a possible example of successful capture of a genetic invader by a fungal genome. Molecular and General Genetics, 261(6), 958–966. [DOI] [PubMed] [Google Scholar]
- Neuveglise C, Sarfati J, Latge JP, & Paris S (1996). Afut1, a retrotransposon-like element from Aspergillus fumigatus. Nucleic Acids Research, 24(8), 1428–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Feretzaki M, Li W, Floyd-Averette A, Mieczkowski P, Dietrich FS, & Heitman J (2013). Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biology, 11(9), e1001653. doi: 10.1371/journal.pbio.1001653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygren K, Strandberg R, Wallberg A, Nabholz B, Gustafsson T, García D, … Johannesson H (2011). A comprehensive phylogeny of Neurospora reveals a link between reproductive mode and molecular evolution in fungi. Molecular Phylogenetics and Evolution, 59(3), 649–663. doi: 10.1016/j.ympev.2011.03.023 [DOI] [PubMed] [Google Scholar]
- Odds FC, Bougnoux M-E, Shaw DJ, Bain JM, Davidson AD, Diogo D, … d'Enfert C (2007). Molecular phylogenetics of Candida albicans. Eukaryotic Cell, 6(6), 1041–1052. doi: 10.1128/EC.00041-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, & Otto SP (1994). Does diploidy increase the rate of adaptation? Genetics, 136(4), 1475–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, & Lenormand T (2002). Resolving the paradox of sex and recombination. Nature Reviews Genetics, 3(4), 252–261. doi: 10.1038/nrg761 [DOI] [PubMed] [Google Scholar]
- Otto SP, & Michalakis Y (1998). The evolution of recombination in changing environments. Trends in Ecology & Evolution, 13(4), 145–151. doi: 10.1016/S0169-5347(97)01260-3 [DOI] [PubMed] [Google Scholar]
- Paoletti M, Seymour FA, Alcocer MJC, Kaur N, Calvo AM, Archer DB, & Dyer PS (2007). Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Current Biology, 17(16), 1384–1389. doi: 10.1016/j.cub.2007.07.012 [DOI] [PubMed] [Google Scholar]
- Patterson DJ (1999). The diversity of eukaryotes. The American Naturalist, 154(S4), S96–S124. doi: 10.1086/303287 [DOI] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, … Li R (2010). Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature, 468(7321), 321–325. doi: 10.1038/nature09529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peever TL, Canihos Y, Olsen L, Ibanez A, Liu YC, & Timmer LW (1999). Population genetic structure and host specificity of Alternaria spp. causing brown spot of Minneola tangelo and rough lemon in Florida. Phytopathology, 89(10), 851–860. doi: 10.1094/PHYTO.1999.89.10.851 [DOI] [PubMed] [Google Scholar]
- Pendrak ML, Yan SS, & Roberts DD (2004). Hemoglobin regulates expression of an activator of mating-type locus alpha genes in Candida albicans. Eukaryotic Cell, 3(3), 764–775. doi: 10.1128/EC.3.3.764-775.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Jones RN, Messer SA, Edmond MB, & Wenzel RP (1998). National surveillance of nosocomial blood stream infection due to Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE program. Diagnostic Microbiology and Infectious Disease, 31(1), 327–332. doi: 10.1016/s0732-8893(97)00240-x [DOI] [PubMed] [Google Scholar]
- Phadke SS, Feretzaki M, & Heitman J (2013). Unisexual reproduction enhances fungal competitiveness by promoting habitat exploration via hyphal growth and sporulation. Eukaryotic Cell, 12(8), 1155–1159. doi: 10.1128/ec.00147-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöggeler S, Risch S, Kück U, & Osiewacz HD (1997). Mating-type genes from the homothallic fungus Sordaria macrospora are functionally expressed in a heterothallic ascomycete. Genetics, 147(2), 567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak E (1987). On the theory of partially inbreeding finite populations. I. Partial selfing. Genetics, 117(2), 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh MA, Malik S-B, & Logsdon JM Jr (2005). A phylogenomic inventory of meiotic genes: Evidence for sex in Giardia and an early eukaryotic origin of meiosis. Current Biology, 15(2), 185–191. doi: 10.1016/j.cub.2005.01.003 [DOI] [PubMed] [Google Scholar]
- Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, … Li R (2008). Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell, 135(5), 879–893. doi: 10.1016/j.cell.2008.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper JR (1966). Genetics of Sexuality in Higher Fungi. New York: Ronald Press Co. [Google Scholar]
- Reedy JL, Floyd A, & Heitman J (2009). Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Current Biology, 19(11), 891–899. doi: 10.1016/j.cub.2009.04.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Carres M, Findley K, Sun S, Dietrich FS, & Heitman J (2010). Morphological and genomic characterization of Filobasidiella depauperata: a homothallic sibling species of the pathogenic Cryptococcus species complex. PLoS ONE, 5(3), e9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydholm C, Dyer P, & Lutzoni F (2007). DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryotic Cell, 6(5), 868–874. doi: 10.1128/EC.00319-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni N, Yi S, Daniels KJ, Huang G, Srikantha T, & Soll DR (2010). Tec1 mediates the pheromone response of the white phenotype of Candida albicans: insights into the evolution of new signal transduction pathways. PLoS Biology, 8(5), e1000363. doi: 10.1371/journal.pbio.1000363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni N, Yi S, Daniels KJ, Srikantha T, Pujol C, & Soll DR (2009). Genes selectively up-regulated by pheromone in white cells are involved in biofilm formation in Candida albicans. PLoS Pathogens, 5(10), e1000601. doi: 10.1371/journal.ppat.1000601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul N, Krockenberger M, & Carter DA (2008). Evidence of recombination in mixed-mating-type and α-only populations of Cryptococcus gattii sourced from single eucalyptus tree hollows. Eukaryotic Cell, 7(4), 727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer D, Cote P, Whiteway M, & Bennett RJ (2007). Barrier activity in Candida albicans mediates pheromone degradation and promotes mating. Eukaryotic Cell, 6(6), 907–918. doi: 10.1128/EC.00090-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz B, Banuett F, Dahl M, Schlesinger R, Schäfer W, Martin T, … Kahmann R (1990). The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell, 60(2), 295–306. doi: 10.1016/0092-8674(90)90744-Y [DOI] [PubMed] [Google Scholar]
- Seitz-Mayr G, Wolf K, & Kaudewitz F (1978). Extrachromosomal inheritance in Schizosaccharomyces pombe. Molecular and General Genetics, 164(3), 309–320. doi: 10.1007/BF00333162 [DOI] [PubMed] [Google Scholar]
- Selker EU, Cambareri EB, Jensen BC, & Haack KR (1987). Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell, 51(5), 741–752. doi: 10.1016/0092-8674(87)90097-3 [DOI] [PubMed] [Google Scholar]
- Selker EU, & Stevens JN (1985). DNA methylation at asymmetric sites is associated with numerous transition mutations. Proceedings of the National Academy of Sciences, USA, 82(23), 8114–8118. doi: 10.1073/Pnas.82.23.8114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki AM, Dulmage K, Cowen LE, Anderson JB, & Berman J (2009). Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genetics, 5(10), e1000705. doi: 10.1371/journal.pgen.1000705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki AM, Forche A, & Berman J (2006). Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science, 313(5785), 367–370. doi: 10.1126/science.1128242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki AM, Forche A, & Berman J (2010). Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryotic Cell, 9(7), 991–1008. doi: 10.1128/ec.00060-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpley MS, Marciniak C, Eckel-Mahan K, McManus M, Crimi M, Waymire K, … Wallace DC (2012). Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell, 151(2), 333–343. doi: 10.1016/j.cell.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RK, & Bennett RJ (2009). Fungal meiosis and parasexual reproduction--lessons from pathogenic yeast. Current Opinion in Microbiology, 12(6), 599–607. doi: 10.1016/j.mib.2009.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu PKT, & Metzenberg RL (2002). Meiotic silencing by unpaired DNA: properties, regulation and suppression. Genetics, 161(4), 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu PKT, Raju NB, Zickler D, & Metzenberg RL (2001). Meiotic silencing by unpaired DNA. Cell, 107(7), 905–916. doi: 10.1016/S0092-8674(01)00609-2 [DOI] [PubMed] [Google Scholar]
- Simmons EG (1986). Alternaria themes and variations (22-26). Mycotaxon, 25(1), 287–308. [Google Scholar]
- Simmons EG (1999). Alternaria themes and variations (226-235) - Classification of citrus pathogens. Mycotaxon, 70, 263–323. [Google Scholar]
- Simpson AGB, & Roger AJ (2004). The real ‘kingdoms’ of eukaryotes. Current Biology, 14(17), R693–R696. doi: 10.1016/j.cub.2004.08.038 [DOI] [PubMed] [Google Scholar]
- Sionov E, Lee H, Chang YC, & Kwon-Chung KJ (2010). Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathogens, 6(4), e1000848. doi: 10.1371/journal.ppat.1000848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller MA, & Soll DR (1987). "White-opaque transition": a second high-frequency switching system in Candida albicans. Journal of Bacteriology, 169(1), 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snetselaar KM, Bölker M, & Kahmann R (1996). Ustilago maydis mating hyphae orient their growth toward pheromone sources. Fungal Genetics and Biology, 20(4), 299–312. doi: 10.1006/Fgbi.1996.0044 [DOI] [PubMed] [Google Scholar]
- Son H, Min K, Lee J, Raju NB, & Lee YW (2011). Meiotic silencing in the homothallic fungus Gibberella zeae. Fungal Biology, 115(12), 1290–1302. doi: 10.1016/j.funbio.2011.09.006 [DOI] [PubMed] [Google Scholar]
- Spassky B, Spassky N, Levene H, & Dobzhansky T (1958). Release of genetic variability through recombination. I. Drosophila pseudoobscura. Genetics, 43(5), 844–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed B, & Dunt D (1995). Clinical and host differences between infections with the two varieties of Cryptococcus neoformans. Clinical Infectious Diseases, 21(1), 28–34. doi: 10.1093/clinids/21.1.28 [DOI] [PubMed] [Google Scholar]
- Spellig T, Bolker M, Lottspeich F, Frank RW, & Kahmann R (1994). Pheromones trigger filamentous growth in Ustilago maydis. The EMBO Journal, 13(7), 1620–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess EB (1959). Release of genetic variability through recombination. II. Drosophila persimilis. Genetics, 44(1), 43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer DJ, Saini D, Byrnes EJ 3rd, Heitman J, & Frothingham R (2013). Development of an aerosol model of Cryptococcus reveals humidity as an important factor affecting the viability of Cryptococcus during aerosolization. PLoS ONE, 8(7), e69804. doi: 10.1371/journal.pone.0069804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JE, Kawabe M, Abdo Z, Arie T, & Peever TL (2011). Contrasting codon usage patterns and purifying selection at the mating locus in putatively asexual Alternaria fungal species. PLoS ONE, 6(5), e20083. doi: 10.1371/journal.pone.0020083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JE, Thomas KA, Lawrence CB, Dang H, Pryor BM, Timmer LMP, & Peever TL (2013). Signatures of recombination in clonal lineages of the citrus brown spot pathogen, Alternaria alternata sensu lato. Phytopathology, 103(7), 741–749. doi: 10.1094/phyto-08-12-0211-r [DOI] [PubMed] [Google Scholar]
- Strausberg RL, & Perlman PS (1978). The effect of zygotic bud position on the transmission of mitochondrial genes in Saccharomyces cerevisiae. Molecular and General Genetics, 163(2), 131–144. doi: 10.1007/BF00267404 [DOI] [PubMed] [Google Scholar]
- Sturm NR, Vargas NS, Westenberger SJ, Zingales B, & Campbell DA (2003). Evidence for multiple hybrid groups in Trypanosoma cruzi. International Journal for Parasitology, 33(3), 269–279. doi: 10.1016/S0020-7519(02)00264-3 [DOI] [PubMed] [Google Scholar]
- Sukroongreung S, Kitiniyom K, Nilakul C, & Tantimavanich S (1998). Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans. Medical Mycology, 36(6), 419–424. [PubMed] [Google Scholar]
- Tavanti A, Davidson AD, Fordyce MJ, Gow NAR, Maiden MCJ, & Odds FC (2005). Population structure and properties of Candida albicans, as determined by multilocus sequence typing. Journal of Clinical Microbiology, 43(11), 5601–5613. doi: 10.1128/jcm.43.11.5601-5613.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, … Amon A (2010). Identification of aneuploidy-tolerating mutations. Cell, 143(1), 71–83. doi: 10.1016/j.cell.2010.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, & Amon A (2007). Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science, 317(5840), 916–924. doi: 10.1126/science.1142210 [DOI] [PubMed] [Google Scholar]
- Trapero-Casas A, & Kaiser WJ (2007). Differences between ascospores and conidia of Didymella rabiei in spore germination and infection of chickpea. Phytopathology, 97(12), 1600–1607. doi: 10.1094/PHYTO-97-12-1600 [DOI] [PubMed] [Google Scholar]
- Tscharke RL, Lazera M, Chang YC, Wickes BL, & Kwon-Chung KJ (2003). Haploid fruiting in Cryptococcus neoformans is not mating type α-specific. Fungal Genetics and Biology, 39(3), 230–237. doi: Doi 10.1016/S1087-1845(03)00046-X [DOI] [PubMed] [Google Scholar]
- Tzung K-W, Williams RM, Scherer S, Federspiel N, Jones T, Hansen N, … Agabian N (2001). Genomic evidence for a complete sexual cycle in Candida albicans. Proceedings of the National Academy of Sciences, USA, 98(6), 3249–3253. doi: 10.1073/pnas.061628798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ünal E, Kinde B, & Amon A (2011). Gametogenesis eliminates age-induced cellular damage and resets life span in yeast. Science, 332(6037), 1554–1557. doi: 10.1126/science.1204349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L (1974a). Molecular evolution as predicted by natural selection. Journal of Molecular Evolution, 3(2), 89–101. [DOI] [PubMed] [Google Scholar]
- Van Valen L (1974b). Two modes of evolution. Nature, 252(5481), 298–300. [DOI] [PubMed] [Google Scholar]
- Velagapudi R, Hsueh Y-P, Geunes-Boyer SG, Wright JR, & Heitman J (2009). Spores as infectious propagules of Cryptococcus neoformans. Infection and Immunity, 77(10), 4345–4355. doi: 10.1128/iai.00542-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengrova S, & Dalgaard JZ (2004). RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes & Development, 18(7), 794–804. doi: 10.1101/gad.289404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengrova S, & Dalgaard JZ (2006). The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO reports, 7(1), 59–65. doi: 10.1038/sj.embor.7400576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinces MD, Haas C, & Kumamoto CA (2006). Expression of the Candida albicans morphogenesis regulator gene CZF1 and its regulation by Efg1p and Czf1p. Eukaryotic Cell, 5(5), 825–835. doi: 10.1128/EC.5.5.825-835.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viviani MA, Nikolova R, Esposto MC, Prinz G, & Cogliati M (2003). First European case of serotype A MATa Cryptococcus neoformans infection. Emerging Infectious Diseases, 9(9), 1179–1180. doi: 10.3201/eid0909.020770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelz K, Ma H, Phadke S, Byrnes EJ 3rd, Zhu P, Mueller O, … May RC (2013). Transmission of hypervirulence traits via sexual reproduction within and between lineages of the human fungal pathogen Cryptococcus gattii. PLoS Genetics, 9(9), e1003771. doi: 10.1371/journal.pgen.1003771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijenhoek RC (1998). Animal clones and diversity. Bioscience, 48(8), 617–628. doi: 10.2307/1313421 [DOI] [Google Scholar]
- Wang X, Darwiche S, & Heitman J (2013). Sex-induced silencing operates during opposite-sex and unisexual reproduction in Cryptococcus neoformans. Genetics, 193(4), 1163–1174. doi: 10.1534/genetics.113.149443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hsueh Y-P, Li W, Floyd A, Skalsky R, & Heitman J (2010). Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes & Development, 24(22), 2566–2582. doi: 10.1101/gad.1970910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang P, Sun S, Darwiche S, Idnurm A, & Heitman J (2012). Transgene induced co-suppression during vegetative growth in Cryptococcus neoformans. PLoS Genetics, 8(8), e1002885. doi: 10.1371/journal.pgen.1002885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RL, Kukora JR, & Adams J (1975). The relationship between enzyme activity, cell geometry, and fitness in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences, USA, 72(3), 794–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland J, Dünkler A, & Walther A (2011). Characterization of α-factor pheromone and pheromone receptor genes of Ashbya gossypii. FEMS Yeast Research, 11(5), 418–429. doi: 10.1111/j.1567-1364.2011.00732.x [DOI] [PubMed] [Google Scholar]
- Wendland J, & Walther A (2011). Genome evolution in the Eremothecium clade of the Saccharomyces complex revealed by comparative genomics. G3: Genes, Genomes, Genetics, 1(7), 539–548. doi: 10.1534/g3.111.001032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendte JM, Miller MA, Lambourn DM, Magargal SL, Jessup DA, & Grigg ME (2010). Self-mating in the definitive host potentiates clonal outbreaks of the apicomplexan parasites Sarcocystis neurona and Toxoplasma gondii. PLoS Genetics, 6(12), e1001261. doi: 10.1371/journal.pgen.1001261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Lively CM, & Read AF (1999). A pluralist approach to sex and recombination. Journal of Evolutionary Biology, 12(6), 1003–1012. doi: 10.1046/j.1420-9101.1999.00119.x [DOI] [Google Scholar]
- White MJD (1978). Modes of Speciation. San Francisco, CA: WH Freeman. [Google Scholar]
- Whitehouse HLK (1949). Heterothallism and sex in the fungi. Biological Reviews of the Cambridge Philosophical Society, 24(4), 411–447. doi: 10.1111/J.1469-185x.1949.Tb00582.X [DOI] [PubMed] [Google Scholar]
- Whitton J, Sears Christopher J., Baack Eric J., & Otto Sarah P. (2008). The dynamic nature of apomixis in the angiosperms. International Journal of Plant Sciences, 169(1), 169–182. doi: 10.1086/523369 [DOI] [Google Scholar]
- Wickes BL, Mayorga ME, Edman U, & Edman JC (1996). Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the α-mating type. Proceedings of the National Academy of Sciences, USA, 93(14), 7327–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wik L, Karlsson M, & Johannesson H (2008). The evolutionary trajectory of the mating-type (mat) genes in Neurospora relates to reproductive behavior of taxa. BMC Evolutionary Biology, 8, 109. doi: 10.1186/1471-2148-8-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GC (1975). Sex and Evolution. Princeton, NJ: Princeton University Press. [Google Scholar]
- Winge Ö (1935). On haplophase and diplophase in some Saccharomycetes. Comptes-Rendus des travaux du Laboratoire Carlsberg. Série Physiologique, 21, 34. [Google Scholar]
- Winge Ö, & Laustsen O (1937). On two types of spore germination, and on genetic segregations in Saccharomyces: demonstrated through single-spore cultures. Comptes-Rendus des travaux du Laboratoire Carlsberg. Série Physiologique, 22, 99–117. [Google Scholar]
- Winge Ö, & Roberts C (1949). A gene for diploidization in yeasts. Comptes-Rendus des travaux du Laboratoire Carlsberg. Série Physiologique, 24, 5. [PubMed] [Google Scholar]
- Wright SI, Ness Rob W., Foxe John P., & Barrett Spencer C. H. (2008). Genomic consequences of outcrossing and selfing in plants. International Journal of Plant Sciences, 169(1), 105–118. doi: 10.1086/523366 [DOI] [Google Scholar]
- Wright SI, & Schoen DJ (1999). Transposon dynamics and the breeding system. Genetica, 107(1-3), 139–148. doi: 10.1023/A:1003953126700 [DOI] [PubMed] [Google Scholar]
- Xie J, Tao L, Nobile CJ, Tong Y, Guan G, Sun Y, … Huang G (2013). White-opaque switching in natural MTLa/α Isolates of Candida albicans: Evolutionary implications for roles in host adaptation, pathogenesis, and sex. PLoS Biology, 11(3), e1001525. doi: 10.1371/journal.pbio.1001525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J (2002). Estimating the spontaneous mutation rate of loss of sex in the human pathogenic fungus Cryptococcus neoformans. Genetics, 162(3), 1157–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J (2005). Cost of interacting with sexual partners in a facultative sexual microbe. Genetics, 171(4), 1597–1604. doi: 10.1534/genetics.105.045302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Ali RY, Gregory DA, Amick D, Lambert SE, Yoell HJ, … Mitchell TG (2000). Uniparental mitochondrial transmission in sexual crosses in Cryptococcus neoformans. Current Microbiology, 40(4), 269–273. doi: 10.1007/s002849910053 [DOI] [PubMed] [Google Scholar]
- Xu J, Vilgalys R, & Mitchell TG (2000). Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Molecular Ecology, 9(10), 1471–1481. doi: 10.1046/j.1365-294x.2000.01021.x [DOI] [PubMed] [Google Scholar]
- Yan Z, Hull C, Sun S, Heitman J, & Xu J (2007). The mating type-specific homeodomain genes SXI1α and SXI2a coordinately control uniparental mitochondrial inheritance in Cryptococcus neoformans. Current Genetics, 51(3), 187–195. doi: 10.1007/s00294-006-0115-9 [DOI] [PubMed] [Google Scholar]
- Yan Z, Li X, & Xu J (2002). Geographic distribution of mating type alleles of Cryptococcus neoformans in four areas of the United States. Journal of Clinical Microbiology, 40(3), 965–972. doi: 10.1128/jcm.40.3.965-972.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, & Xu J (2003). Mitochondria are inherited from the MATa parent in crosses of the basidiomycete fungus Cryptococcus neoformans. Genetics, 163(4), 1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Sahni N, Daniels KJ, Pujol C, Srikantha T, & Soll DR (2008). The same receptor, G protein, and mitogen-activated protein kinase pathway activate different downstream regulators in the alternative white and opaque pheromone responses of Candida albicans. Molecular Biology of the Cell, 19(3), 957–970. doi: 10.1091/mbc.E07-07-0688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona AH, Manor YS, Herbst RH, Romano GH, Mitchell A, Kupiec M, … Dahan O (2012). Chromosomal duplication is a transient evolutionary solution to stress. Proceedings of the National Academy of Sciences, USA, 109(51), 21010–21015. doi: 10.1073/pnas.1211150109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue C, Cavallo LM, Alspaugh JA, Wang P, Cox GM, Perfect JR, & Heitman J (1999). The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics, 153(4), 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S-H, Berbee ML, Yoder OC, & Turgeon BG (1999). Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proceedings of the National Academy of Sciences, USA, 96(10), 5592–5597. doi: 10.1073/pnas.96.10.5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyl C, Vanderford T, & Carter M (2003). An evolutionary advantage of haploidy in large yeast populations. Science, 299(5606), 555–558. doi: 10.1126/science.1078417 [DOI] [PubMed] [Google Scholar]
- Zordan RE, Galgoczy DJ, & Johnson AD (2006). Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proceedings of the National Academy of Sciences, USA, 103(34), 12807–12812. doi: 10.1073/pnas.0605138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, & Johnson AD (2007). Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biology, 5(10), e256. doi: 10.1371/journal.pbio.0050256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zörgö E, Chwialkowska K, Gjuvsland AB, Garré E, Sunnerhagen P, Liti G, … Warringer J (2013). Ancient evolutionary trade-offs between yeast ploidy states. PLoS Genetics, 9(3), e1003388. doi: 10.1371/journal.pgen.1003388 [DOI] [PMC free article] [PubMed] [Google Scholar]