Abstract
As a partner of tetraspanins, EWI2 suppresses glioblastoma, melanoma, and prostate cancer; but its role in lung cancer has not been investigated. Bioinformatics analysis reveals that EWI2 gene expression is up regulated in lung adenocarcinoma and higher expression of EWI2 mRNA may predict poorer overall survival. However, experimental analysis shows that EWI2 protein is actually downregulated constantly in the tissues of lung adenocarcinoma and lung squamous cell carcinoma. Forced expression of EWI2 in human lung adenocarcinoma cells reduces total cellular and cell surface levels of various integrins and growth factor receptors, which initiates the outside-in motogenic and mitogenic signaling. These reductions result in the decreases in 1) cell-matrix adhesion, cell movement, and cell transformation in vitro and 2) tumor growth, burden, and metastasis in vivo, and result from the increases in lysosomal trafficking and proteolytic degradation of theses membrane receptors. EWI2 elevates lysosome formation by promoting nuclear retention of TFEB, the master transcription factor driving lysosomogenesis. In conclusion, EWI2 as a lung cancer suppressor attenuates lung cancer cells in a comprehensive fashion by inhibiting both tumor growth and tumor metastasis; EWI2 as an endolysosome regulator promotes lysosome activity to enhance lysosomal degradation of growth factor receptors and integrins and then reduce their levels and functions; and EWI2 can become a promising therapeutic candidate given its accessibility at the cell surface, dual inhibition on growth factor receptors and integrins, and broad-spectrum anti-cancer activity. More importantly, our observations also provide a novel therapeutic strategy to bypass the resistance to EGFR inhibitors.
Keywords: EGFR, integrin, lysosome, TFEB, tumor metastasis
Introduction
EWI2 (IgSF8 or CD316) is an immunoglobulin superfamily (IgSF) protein, physically associates with tetraspanins such as CD9, CD81, and CD82, and affects molecular interactions in and cellular functions of tetraspanin-enriched membrane domains. [1–4] For example, high stoichiometric interaction between EWI2 and CD81 influences CD81 function as a co-receptor of hepatitis C virus and subsequently cellular sensitivity to hepatitis C virus infection. [5] EWI2 interaction with CD9, another primary partner of EWI2, reduces homoclustering of CD9 and subsequently availability of CD9 to other membrane proteins such as laminin-binding integrins and TGFβ receptor. [6] Similar to some of its associated tetraspanins acting as tumor suppressors, [7] EWI2 inhibits melanoma growth and metastasis, [8] glioblastoma proliferation and invasion, [9] and prostate cancer cell migration, proliferation, and metastasis. [4, 10–11] Roles of EWI2 in other major cancers, especially those with high morbidity and mortality like lung cancer, are unknown. Moreover, beyond the understanding that EWI2 regulates tumor cell behaviors through tetraspanin-enriched membrane domains, [12] how EWI2 inhibits cancer still remains elusive at the molecular, cellular, and organism levels.
Cancer bioinformatics analyses can link gene expression, at the transcriptional and translation levels, to cancer initiation and progression and predict cancer patient prognosis. EWI2 roles in cancer have not been examined with bioinformatics systematically. Experimental analyses can establish causal or correlative links of a given gene to cancers and their related events. Integration of bioinformatics and experiment approaches, e.g., examining EWI2 in lung cancer, will draw more cogent and complete pictures for the relationships between molecules and cancers.
In the present study, we compared the mRNA and protein expression levels of EWI2 between lung cancer and normal lung tissues and assessed the prognostic value of their expressions for lung cancer, through integrated bioinformatics analyses and/or through clinical specimen examination. By discovering EWI2 diminution in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) and reconstituting EWI2 expression in human lung cancer cells, we revealed that EWI2, as a motogenicity and mitogenicity suppressor of lung cancer and an enhancer of lysosome, inhibited growth factor receptors and/or integrins in lung cancer cells by reducing their presence at the plasma membrane and enhancing their degradation through intracellular proteolytic machinery.
Materials and Methods
Oncomine and The Cancer Genome Atlas (TCGA) analyses
We used Oncomine (https://www.oncomine.org/) to analyze the differences in EWI2 mRNA expression between lung cancer tissues and normal lung tissues. For each analysis, the thresholds were set as follows: p-value: 0.05; fold change: 1.5; gene rank: 10%; analysis type: cancer vs. normal analysis; data type: mRNA. We used UALCAN (http://ualcan.path.uab.edu/analysis.html), cBioPortal (http://www.cbioportal.org) and MEXPRESS (https://mexpress.be/) to perform TCGA data (https://www.cancer.gov/) analyses. For the analyses on cBioPortal, our sets were as follows: selected studies: TCGA provisional; select genomic profiles: mutation and copy-number alterations; select patient/case set: cases with both mutations and copy-number alterations data. p-values < 0.05 were considered statistically significant.
Prognosis analyses
We used Kaplan-Meier (KM)-Plotter (https://kmplot.com/analysis/) to evaluate the prognostic values of EWI2 in lung cancer. For each analysis, patients were split into high and low expression groups by the median values of mRNA expression. Survival analyses were carried out to achieve KM plots. We also acquired prognostic information from Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/) based on TCGA. p-values < 0.05 were considered statistically significant.
Cancer Cell Line Encyclopedia (CCLE) analyses
We performed analyses on EWI2 mRNA expression level among different types of human lung cancer cell lines by the CCLE online tool (https://portals.broadinstitute.org/ccle). p-values < 0.05 were considered statistically significant.
Clinical Proteomic Tumor Analysis Consortium (CPTAC) analysis
The protein level of EWI2 in human lung adenocarcinoma was acquired from UALCAN (http://ualcan.path.uab.edu/analysis-prot). p-value < 0.05 was considered statistically significant.
Patients and tissue collection
We collected human lung cancer tissues and adjacent normal lung tissues from Department of Thoracic Surgery, Tongji Hospital of Huazhong University of Science and Technology. The sample collection was approved by the institutional ethical review board of Tongji Hospital. Informed consent was obtained from subjects before their inclusion in the study. Samples, including tumor tissues and tumor-free lung tissues that were at least 5cm away from the tumors, were collected from 13 patients, including 8 cases of LUAD (Stage III: 3 cases; Stage II: 3 cases; and Stage I: 2 cases) and 5 cases of LUSC (Stage III: 2 cases; Stage II: 2 cases; and Stage I: 1 case).
Antibodies and reagents
Supplemental Tables 1–3 provide information about all antibodies and reagents used in this study.
Cell culture and transfectant establishment
HCC827 cells kindly provided by Dr. Rajagopal Ramesh of the University of Oklahoma Health Science Center were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. A549 cells and H358 cells were friendly offered by Dr. Jie Wu of the University of Oklahoma Health Science Center and cultured in F12K and RPMI 1640, respectively, supplemented with 10% FBS and 1% penicillin-streptomycin. NIH3T3-EWI2-GFP cells were established in our earlier study [13] and cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. PC3 cells were originally purchased from ATCC and also cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. Cells were detached with 2 mM EDTA in PBS. All experiments were performed with mycoplasma-free cells.
One day prior to the transfection, 1×106 cells were seeded into each well of a 6-well plate. Transfection was performed according to the instructions of the Lipofectamine 3000 Reagent Kit. Briefly, 7.5 μl Lipofectamine 3000 Reagent was diluted into 125 μl Opti-MEM medium, and 2.5 μg plasmids pcDNA3.1-EWI2 and 5 μl P3000 Reagent were mixed in 125 μl Opti-MEM medium. After incubating the two kinds of dilution together for 15 minutes, the mixture was added directly into the culture medium. Transfected cells were selected by G418 for 14 days. The selected cells were stained with primary antibodies against EWI2 and secondary antibodies sequentially and sorted by Aria Fusion (BD Biosciences) to collect EWI2-overexpressed cells.
Cells were treated with lysosome inhibitor bafilomycin A1 (Baf A1) (0.5 μM) for 8 hours prior to other experiments.
Flow cytometry
Detached cells were washed by ice-cold PBS twice and incubated with 15% goat serum on ice for 1 hour. The cells were neither fixed nor permeabilized. Then, the cells were stained with primary Abs against the extracellular domain of the target protein for 1 hour and secondary Abs for 1 hour sequentially on ice, followed by washes with ice-cold PBS and centrifugation at 4°C. When lysosome was investigated, detached cells were incubated with 1μM Lysotracker at 37°C for 30 minutes. Expression levels were detected by using the Stratedigm S1200Ex or Stratedigm S1400Exi flow cytometer platform (Stratedigm Inc.) and analyzed by FlowJo software (BD Science).
Cell-matrix adhesion assay
The 96-well plate was coated with 10 μg/ml of different matrix (Table S3, fibronectin [FN] and vitronectin [VN] in 0.01M NaHCO3, collagen-I [Col I] and laminin [LN] in PBS) at 4°C overnight, then was blocked with 0.1% thermally denatured BSA at 37°C for one hour. Then, 1×105 cells in 100 μl FBS-free culture medium were added to each well. Non-adherent cells were removed by gently washing each well with pre-warmed FBS-free culture medium with a multi-channel pipette after 30 minutes. Adherent cells were fixed with 4% paraformaldehyde (PFA) for 10 minutes and stained with 0.1% crystal violet for 10 min. Then, 100 μl of 2% SDS was added to each well to dissolve the crystal violet. Optical density (OD) was measured with an EnVision® Multilabel Reader (Perkin Elmer).
Cell movement assays
For chemo-haptotaxis migration assay, the lower side of the 8-μm Transwell insert (Corning) filter membrane was coated with 50 μl FN (10 μg/ml in 0.01M NaHCO3) or LN-511 (10 μg/ml in PBS) overnight at 4°C. After washing with PBS and dehydration, the inserts were put in complete medium. 1×105 cells in 100 μl FBS-free culture medium were added on top of the filter membrane. The culture medium and remaining cells that were on the top side of the filter membrane were carefully removed with cotton-tipped applicators after 6-hour incubation at 37°C. The migrated cells were fixed with 4% PFA for 10 minutes and washed with PBS. Then, 0.1% crystal violet was used to stain cells on the lower side of the filter membrane for 10 minutes and was then removed with ddH2O. The number of cells on the lower side of the filter membrane was counted by taking the average of cell counts in 7 different randomly chosen views under an inverted light microscope (Olympus). For chemotaxis migration, the inserts were not coated with FN or LN-511. For haptotaxis migration, the inserts were coated with 50 μl FN (10 μg/ml in 0.01M NaHCO3), and cells in the upper chamber were in complete medium.
For cell invasion assay, 50 μl Matrigel (3 mg/ml) or 30 μl collagen-I gel (2 mg/ml) was added to the top side of the filter membrane to form a thin gel layer by incubating at 37°C for 30 minutes before adding cells. Cells were counted either 8 or 24 hours after the starts of experiments for the Matrigel or collagen-I gel invasion, respectively.
For collective movement or wound healing assay, 1.5×105 cells were added in each well of a 96-well plate to form a confluent monolayer. The gaps or wounds were made in Incucyte system by its woundmaker tool (Essen BioScience). [14] Then, 100 μl FBS-free culture medium was added to each well after the detached cells were removed gently. Relative cell density in the wound (cell density in the wound area / cell density outside of the wound area) was measured in the Incucyte Live Cell Imaging System (Essen BioScience). Collective movement was also examined with the conventional wound healing assay. Briefly, 1.5×106 cells were seeded in each well of a 6-well plate in complete media to form a confluent monolayer. Wounds were generated with pipet tips, and then the detached cells were gently removed by FBS-free medium. Images of the wound at 3 different locations in each well were taken at various time points by an inverted light microscope (Olympus). Areas of the wounds were analyzed and assessed by ImageJ software. Ratios of remaining wound areas/original wound areas were calculated.
Cell proliferation assays
For anchorage-dependent cell proliferation, HCC827 cells at different densities (2500, 5000, and 10000 cells per well) in 100-μl culture media were seeded in 96-well plates. Cell proliferation was monitored by analyzing the occupied area of cell images over time in the Incucyte Live Cell Imaging System (Essen BioScience) [15] and presented as the folds of increased confluence. Cell proliferation was also examined in a conventional way. [16] Briefly, cells were plated in the wells of 96-well plates at different densities, cultured in complete medium, and fixed by 4% PFA and stained with 0.1% crystal violet at different time points. Then, 100 μl of 2% SDS was added the cells per well to dissolve crystal violet. OD values of crystal violet were measured by EnVision® Multilabel Reader (Perkin Elmer).
Anchorage-independent cell proliferation was measured with soft agar assay. 0.75 ml 2×DMEM culture medium mixed with 0.75 ml 1% noble agar in deionized water was plated into each well of a 6-well plate. After the agar solidified, each well received another mixture containing 0.75 ml 2×DMEM culture medium, 0.75 ml 0.6% noble agar in deionized water, and 5,000 cells. After 2 or 4 weeks, colonies were stained with 0.005% crystal violet. The number and size of colonies was determined microscopically using an inverted microscope (Olympus).
Cell survival and death assays
Cell survival was examined with ApoTox-Glo™ triplex assay. A 96-well assay plate was set with 1×105 cells in each well. According to the kit, 20 μl viability/cytotoxicity reagent containing GF-AFC substrate and bis-AAF-R110 substrate was added into all wells and mixed by orbital shaking. After incubation at 37°C for 30 minutes, fluorescence was measured in an EnVision® Multilabel Reader (Perkin Elmer) at the following two wavelength sets: 400Ex/505Em for viability, and 485Ex/520Em for cytotoxicity. Then, 100 μl of Caspase-Glo 3/7 reagent was added to each well and mixed by orbital shaking. Luminescence was detected in an EnVision® Multilabel Reader (Perkin Elmer) after incubation for 30 minutes at room temperature.
Apoptosis was examined in TUNEL assay. Sections of tumor tissues were de-paraffinized and rehydrated sequentially. After antigen retrieval, the sections were stained for DNA strand breaks with TUNEL technology and observed and photographed with an SP8 Confocal White Light Laser system (Leica Microsystems).
Anoikis was examined as described elsewhere. [17] Cells were detached by using 2mM EDTA then seeded in FBS free medium onto low adherence dishes for 24 hours. Cells were labelled with PI and percentages of PI+ cells were evaluated by flow cytometry.
Extracellular vesicle (EV) preparation and analyses
15 ml harvested medium from cultured 1.5×107 cells for 2 days was centrifuged at 2000g for 10 minutes at 4°C.The cleared supernatant was centrifuged at 10,000g for 30 minutes to receive the 10k EV fraction. Subsequently, the 10,000g cleared supernatant was ultracentrifuged (Type 45 Ti rotor, Beckman XPN) with 100,000g for 3 hours, yielding the 100k EV fractions.
Immunofluorescence
Cells were fixed by 4% PFA in PBS for 10 minutes, permeabilized with 0.1% TritonX-100 in PBS for 5 minutes and blocked by 15% goat serum in PBS for 1 hour at room temperature. Sequentially, cells were incubated with primary antibodies diluted in the blocking solution at 4°C overnight. The cells were washed three times by PBS and treated with secondary antibodies for 1 hour and DAPI for 10 minutes at room temperature. Phalloidin staining was processed together with secondary antibodies. Cells were incubated with 1 μM Lysotracker at 37°C for 30 minutes before fixation when lysosomes were investigated. Cover slips were coated with FN (50 μg/ml) before seeding cells when focal adhesion was analyzed. Stained cells were observed with an SP8 Confocal White Light Laser system (Leica Microsystems).
Immunohistochemistry (IHC)
Immunochemistry assessment was performed with a DAB substrate kit. Briefly, slides with 4-μm-thick sections or tissue microarray (TMA, LC992a, US Biomx Inc.) were deparaffinized and rehydrated by soaking in xylene, 100% ethanol, 95% ethanol, 75% ethanol, and ddH2O sequentially. Antigen retrieval was carried out by boiling slides in 10 mM sodium citrate buffer (pH 6.0) for 20 minutes and cooling the slides to room temperature. After incubation with 3% hydrogen peroxide for 10 minutes, samples were blocked with 1% BSA for 1 hour. Then, slides were incubated with primary antibodies overnight at 4°C and secondary antibodies for 1 hour at room temperature. Samples were treated with DAB kit reagents to obtain an acceptable staining intensity. Sections were counterstained with hematoxylin to provide a contrast to the brown color of the DAB chromogen and better visualization of tissue morphology. Dehydration was performed by soaking in 75% ethanol, 95% ethanol, 100% ethanol, and xylene sequentially. For immunohistochemistry, 3 tissue sections were cut from each tumor and 5 pairs of tumors were processed. For MMP9 and vimentin staining, intensity of each image was analyzed by ImageJ software. [18] For Ki67, percentage of positive cells was calculated. [19] Averaged result from 3 randomly selected images/microscopic fields of each section was considered as one individual readout. Images were taken under the Nikon microscope (Nikon). Tissue microarray (TMA) was scanned by Zeiss Axio Scan.Z1 (Zeiss) and analyzed by ZEN 3.2 (Zeiss).
Western blot
Proteins were extracted by RIPA buffer (0.05M Tris-HCl [pH 8.0], 0.1% SDS, 2 mM EDTA, 1% TritonX-100, 0.15M NaCl) supplemented with 1 mM PMSF, 1 mM NaF, 1 mM Na4P2O7, 1 mM Na3VO4, 1 μg/ml aprotinin, and 1 μg/ml leupeptin from the collected tissues or cells. NE-PER™ nuclear and cytoplasmic extraction reagents were used to isolate cytoplasm protein and nuclei protein separately. Proteins were analyzed by SDS-PAGE gels, followed by electrotransfer onto the nitrocellulose membranes. After blocking the membranes in 5% skim milk in PBST for 1 hour, the membranes were incubated with primary antibodies overnight at 4°C. Then, the membranes were washed thrice with PBST and incubated with secondary antibodies for 1 hour. Beta-actin and GAPDH were used as housekeeping genes. The bands were developed with ECL immunoblotting detection reagents.
Quantitative reverse transcription PCR (qRT-PCR)
RNA was extracted for cells by TRIzol™ Reagent, and reverse-transcribed into cDNA using an iScript™ cDNA Synthesis Kit. qRT-PCR was achieved with an iQ™ SYBR® Green Supermix kit in a Bio-Rad CFX96 Real-Time System (Bio-Rad) to examine fluorescence intensity. The program was set as follows: 95°C for 3 minutes, 40 cycles of 95°C for 10 seconds, 55°C for 30 seconds, and 72°C for 30 seconds. The primers were shown in Supplemental Table 4. The relative level of mRNA for each gene was calculated by using the ΔΔCT method with β-actin as the housekeeping gene.
Tumor initiation and growth in vivo
HCC827 transfectant cells (5×106 in 100 μl PBS) were injected subcutaneously into 8-week-old male NU/J nude mice (Foxn1nu). Mice body weights and tumor sizes were measured every four days. The volume was calculated as length×width2×0.52. Tumor tissues were collected five weeks after injection. The final tumor weight was measured.
Experimental metastasis
HCC827 transfectant cells (5×106 in 100 μl PBS) were injected via the tail vein into 8-week-old NU/J nude mice (Foxn1nu). Lung tissues were collected at four weeks after the injection. Sections were produced from five different locations of each lung, followed by histochemistry analysis and metastatic lesion examination.
Statistical analyses
Data were processed using GraphPad Prism 7 (GraphPad Software Inc.). The t-test or ANOVA was applied for comparison of measurement data. Fisher’s test was used for enumeration data. The significance was set at p<0.05.
Results
Bioinformatics analysis predicts that higher expression of EWI2 gene correlate with poorer prognosis of LUAD patients.
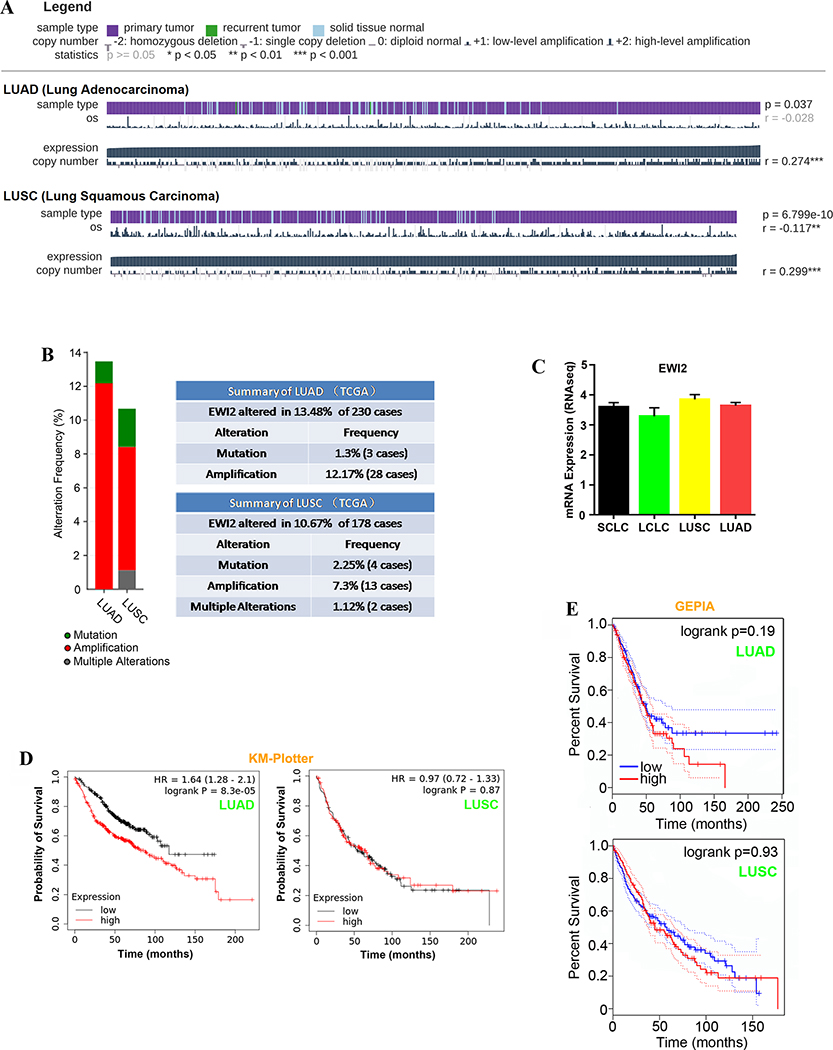
Non-small cell lung cancer (NSCLC) as the most frequently seen lung cancer in clinical settings consists mainly of LUAD and LUSC. EWI2 mRNA levels increased in LUAD, based on all studies listed in Oncomine; while its mRNA levels in LUSC were not different between normal and tumor tissues, as shown by Hou’s dataset (Table 1). However, based on TCGA analyses, EWI2 mRNA levels increased in both LUAD and LUSC groups comparing with normal lung tissues (Figure 1A). By using cBioPortal, we confirmed amplification as the major change for EWI2 gene in LUAD and LUSC (Figure 1B). Besides lung cancer, EWI2 gene is also amplified in several other adenocarcinomas such as the ones from breast, esophagus, and stomach (data not shown). Further, no significant difference in EWI2 mRNA level was found among the four major types of lung cancer cell lines, based on CCLE database (Figures 1C).
Table 1.
Comparison of EWI2 Gene Expressions between Lung Cancer and Normal Lung Tissues
| Database | Cancer Type | Datasets | Normal Cases | Cancer Cases | Fold Change | p-value |
|---|---|---|---|---|---|---|
| Oncomine | LUAD | Wei | 25 | 25 | 1.148 | 0.025 |
| Hou | 65 | 45 | 1.137 | 0.019 | ||
| Okayama | 20 | 226 | 1.366 | 5.01×10−6 | ||
| Selamat | 58 | 58 | 1.249 | 7.29×10−15 | ||
| LUSC | Hou | 65 | 27 | 1.026 | 0.353 | |
| UALCAN | LUAD | TCGA | 59 | 515 | upregulated | 1.63×10−12 |
| LUSC | TCGA | 52 | 503 | upregulated | 1.21×10−20 |
Figure 1. EWI2 gene expression in human lung cancer.
(A) MEXPRESS was used to analyze the relationship between EWI2 mRNA levels and LUAD and LUSC based on TCGA data after excluding samples without expression data (LUAD: n=574 cases, LUSC: n=550 cases). (B) Genetic alterations of EWI2 in some types of human cancers analyzed through cBioPortal (TCGA provisional dataset). (C) EWI2 mRNA expression levels in cell lines among four major types of lung cancer were compared based on CCLE (small cell lung cancer [SCLC]: n=51 cases, large cell lung cancer [LCLC]: n=16, LUSC: n=39, and LUAD: n=84). (D) Association between EWI2 expression and lung cancer survival was evaluated by using microarray datasets from lung cancer patients and the software package KMPlotter. Patients were split by EWI2 median expression into high and low and overall survival was calculated (LUAD: n=719 cases, LUSC: n=524 cases). (E) Kaplan-Meier survival curves for EWI2 in TCGA data were plotted by using GEPIA (LUAD: n=239 cases, LUSC: n=241 cases). Patients were divided by median EWI2 expression levels.
We then examined whether EWI2 associated with the prognosis of patients with lung cancers via KM-Plotter. Increased expression of EWI2 projected poor overall survival for LUAD patients, but not for LUSC patients (Figures 1D). We also examined the prognostic values of EWI2 for LUAD and LUSC with the TGCA database via GEPIA. There was a trend of higher expression of EWI2 with shorter survival, but it did not reach statistical significance (Figure 1E).
Hence, it appears that EWI2 gene expression is upregulated in LUAD and LUSC and predicts poorer prognosis of LUAD patients.
Level of EWI2 protein is reduced in tumor tissue of human lung cancer.
We used CPTAC-based proteomics analytic tool to confirm those findings based on mRNA level above. Surprisingly, EWI2 protein level was reduced in LUAD tumor tissues (Figure 2A), which was just opposite to the finding from genomics analyses.
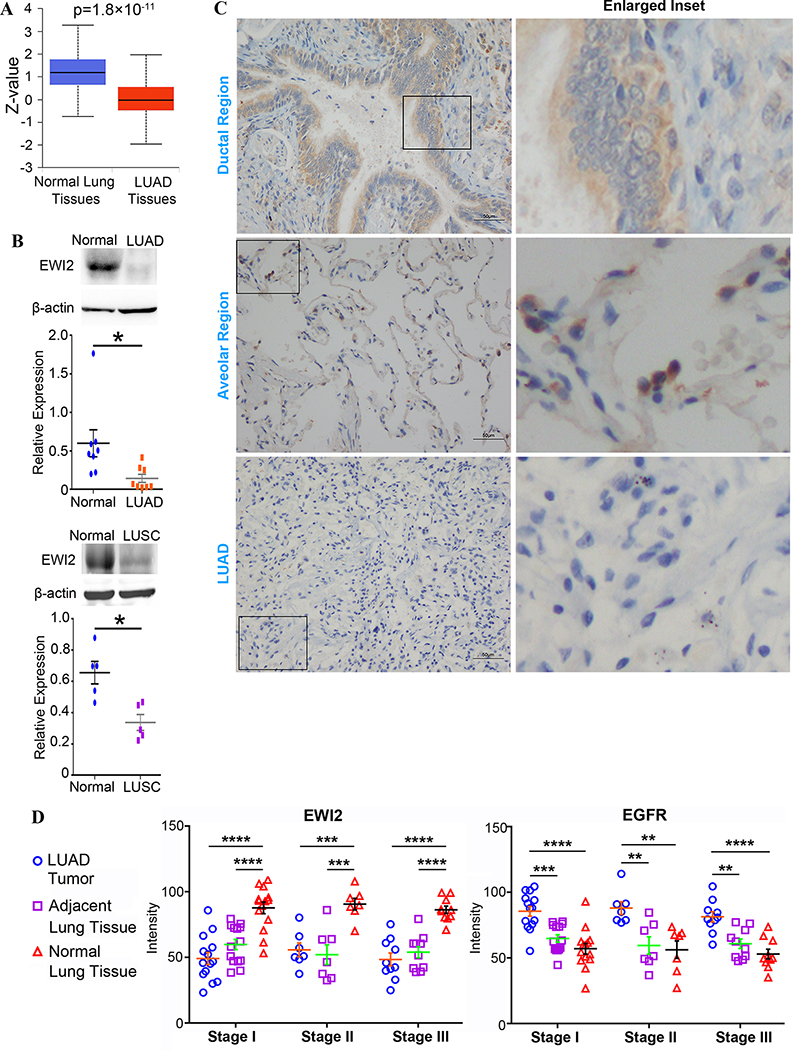
Figure 2. EWI2 protein levels in human lung cancer.
(A) EWI2 protein levels in LUAD tumor tissues and normal lung tissues were assessed with CPTAC database (n=111 cases). (B) Western blot analyses on EWI2 protein in tumor tissues of patients with LUAD (n=8 cases) and LUSC (n=5 cases), and normal lung tissues. Beta-actin served as a loading control. The normalized band densities were quantified with ImageJ software, analyzed by t-test, and presented as mean±SD. * p<0.05. (C) Distribution of EWI2 in normal lung tissues and lung adenocarcinoma was examined with immunohistochemistry. Scale bar: 50 μm. (D) Tissue microarrays of different stages of human lung adenocarcinoma, adjacent lung tissues and normal lung tissues were stained with EWI2 and EGFR. The intensities were quantified by ZEN 3.2, analyzed by t-test, and presented as mean ± SD (Stage I: n=14 cases; Stage II: n=7 cases; and Stage III: n=10 cases) ** p<0.01; *** p<0.001; **** p<0.0001.
We collected LUAD tumor tissues and corresponding normal lung tissues from LUAD patients and examined EWI2 protein with Western blot. Indeed, EWI2 protein level was drastically diminished in the tumor tissues of stages I, II, and III LUAD patients, compared to control lung tissues (Figure 2B and S1A). The same change was also observed in stages I, II, and III LUSC patients (Figure 2B and S1A). Thus, to this end, we found largely reduced or almost abolished EWI2 protein expression in LUAD and LUSC tumors from both bioinformatics and experimental analyses.
Immunohistochemistry showed that EWI2 highly distributed on bronchial epithelial cells in normal lung tissues (Figure 2C). EWI2 was also found at lower density within cells at the respiratory portion of lungs. However, EWI2 was expressed only in a tiny fraction of LUAD tumor cells. Using TMAs of stages I, II, and III human lung adenocarcinoma, we confirmed EWI2 expression in tumor and tumor-adjacent tissues were significantly lower than normal lung tissues throughout all stages (Figure 2D and S1B), while EGFR expression displayed an opposite trend.
Given the high relevance of EWI2 with LUAD, we investigated how EWI2 is linked to LUAD. HCC827 is a human LUAD cell line and doesn’t express endogenous EWI2, as expected (Figure 3A). We expressed EWI2 in HCC827 line and established the stable transfectants (Figure 3A).
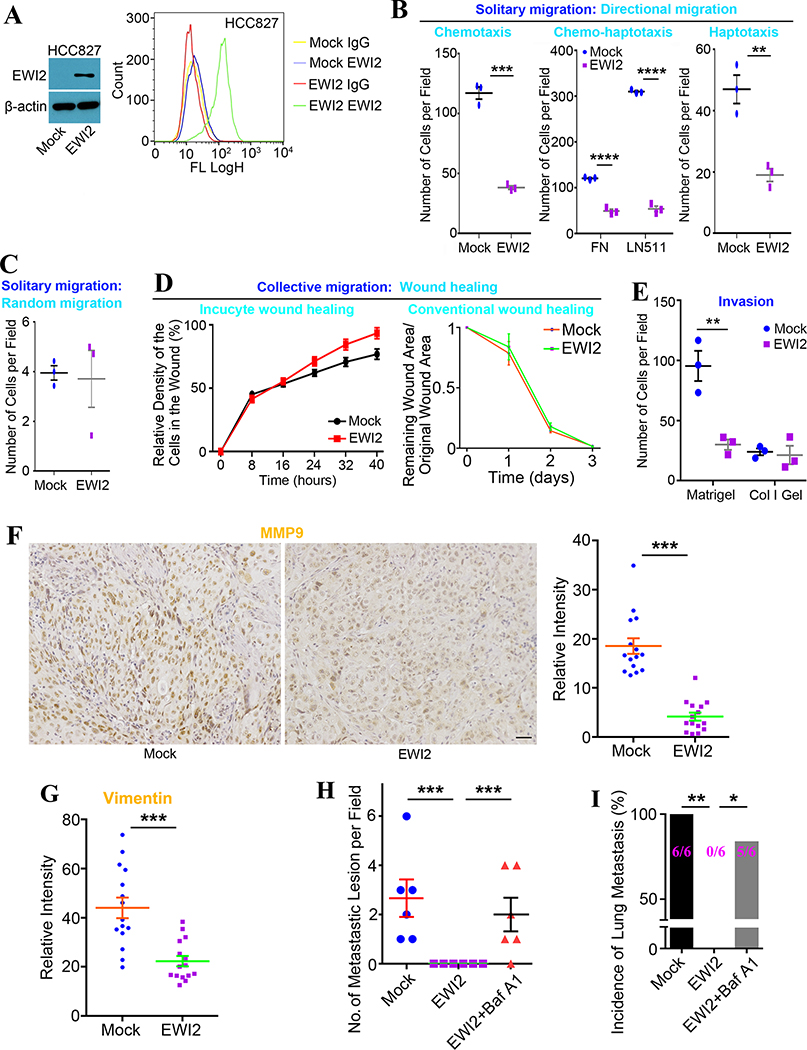
Figure 3. EWI2 suppresses tumor cell migration and invasion.
(A) Establishment of EWI2 stable transfectants in HCC827 cell line. Western blot and flow cytometry analyses on total cellular and cell surface expressions of EWI2, respectively. (B) Solitary migration assay was performed in Transwell system. 1×105 tumor cells in 100 μl medium were added on the upper chamber. In chemotaxis migration assay, the lower chamber was filled with complete medium, and cells with FBS free medium were on the upper side. One more step for chemo-haptotaxis migration was coating the membrane with FN or LN-511 at 4°C overnight before adding cells. In haptotaxis migration, the membrane was coated with FN, and tumor cells were incubated in complete medium on the upper side. Number of migrated cells was compared by t-test and presented as mean±SD (n=3 individual experiments). ** p<0.01; *** p<0.001; and **** p<0.0001. (C) Cells in FBS free medium on the upper side migrated to the lower chamber which was also filled with FBS free medium through non-coated membrane when random migration was investigated. Cell numbers were determined at 6 hours after seeding cells. Number of migrated cells was compared by t-test and presented as mean±SD (n=3 individual experiments). (D) Wound healing assay Left panel: Incucyte Live Cell Imaging System was applied in wound healing assay. 1.5×105 cells were added in each well to form a confluent monolayer in a 96-well plate. The gap was made by the woundmaker tool. Relative cell density within the wound (cell density within the wound area/cell density outside of the wound area) was measured over time, compared by t-test at each time point, and shown as mean±SD (n=3 individual experiments). Right panel: Wound was made by the pipet tip. The ratio of wound area at different time points and original wound area was measured and presented as mean±SD (n=3 individual experiments). (E) Invasion assay was also performed in Transwell system. The membrane was coated with FN. Matrigel or collagen-I gel was added before seeding 1×105 tumor cells in 100 μl FBS free medium. The lower chamber was filled with complete medium. Cell numbers on the bottom side of the membrane were counted at 8 hours and 22 hours, respectively, after seeding cells, compared by t-test, and presented as mean±SD (n=3 individual experiments). **: p<0.01. (F) MMP9 and (G) vimentin were stained on tumor tissues formed on nude mice and analyzed by ImageJ. 3 different sections were cut from each tumor tissues. 5 pairs of tumor tissues were processed. Intensities of images were analyzed by ImageJ. Averaged data from 3 random images of each section was considered as an individual readout. Data was compared by t-test and shown as mean±SEM. ***: p<0.001. Scale bar: 50 μm. (H) Three groups of NU/J mice were injected with 5×105 HCC827-Mock cells, HCC827-EWI2 cells, and Baf A1-treated HCC827-EWI2 cells via tail veins, respectively. Each group contained 3 male and 3 female mice. Lung tissues were collected 4 weeks after injection. Five random sections were cut for each lung tissue. 10 random fields were observed in each section. Pulmonary metastatic lesions were counted in each field. The average number in a field for each mouse was compared by t-test and shown as mean±SEM. ***: p<0.001. (I) Number of mice with pulmonary metastasis was counted in each group. The incidence was calculated and compared by Fisher’s test. *: p<0.05 and **: p<0.01.
EWI2 expression inhibits tumor cell movement.
EWI2 inhibits cell movement. [4, 20] We first examined the effects of EWI2 forced expression on various modes of cell movement. Using Transwell migration assay, we found that EWI2 inhibited i) chemotaxis of HCC827 cells toward FBS, ii) chemo-haptotaxis toward FBS and immobilized FN or LN511, and iii) haptotaxis toward immobilized FN, which represent three types of directional solitary cell migration (Figure 3B and S2A). EWI2 didn’t affect random migration of HCC827 cells through non-coated Transwell inserts toward no-gradient, a type of non-directional solitary cell migration (Figure 3C and S2B), suggesting that basic machinery for cell movement such as cytoskeleton was not obviously affected by EWI2 expression. EWI2 didn’t change the ability of HCC827 cells to heal wounds, a type of collective cell migration (Figures 3D and S2C). EWI2 forced expression also reduced HCC827 cell invasion through Matrigel, but not through collagen-I gel (Figures 3E and S2D).
We stained primary tumors formed in nude mice with MMP9, an indicator of tumor invasiveness, [21] and found lower expression of MMP9 in the tumor tissues formed by HCC827-EWI2 cells (Figure 3F). Vimentin, a marker for invasive tumors that underwent epithelial-to-mesenchymal transition, [22] was also markedly down-regulated in HCC827-EWI2 tumors (Figures 3G and S3A). These observations strongly suggest that EWI2 attenuates tumor invasiveness. We also performed experimental metastasis assay by injecting HCC827 transfectant cells through tail veins of nude mice and found that the formation of pulmonary metastatic lesions became lost in HCC827-EWI2 group. Pulmonary metastasis was observed in all mice injected with HCC827-Mock cells (6/6), while none of mice injected with HCC827-EWI2 cells (0/6) formed lung metastasis (Figures 3H–I and S3B).
EWI2 expression reduces the presence of integrins at cell surface and inhibits cell-matrix adhesion.
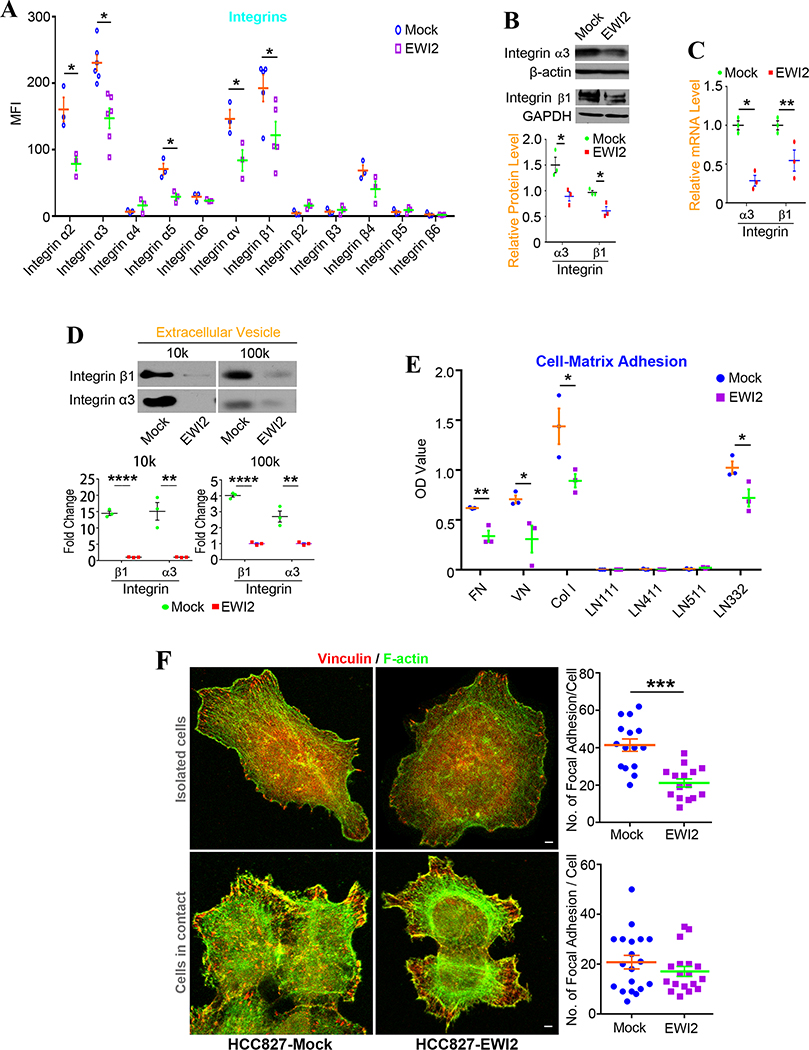
As EWI2 interacts with integrins [20] and integrin-mediated cell-matrix adhesion determines cell movement [23–24], we examined the effect of EWI2 expression on integrins and observed significantly fewer integrins α2, α3, α5, and αV and integrin β1 at the surface of HCC827-EWI2 transfectant cells (Figure 4A). We chose integrin α3 and integrin β1 as examples in HCC827 cells for further analyses and found both of their protein (Figure 4B) and mRNA (Figure 4C) level were downregulated by EWI2. Integrins α3 and β1 released from EVs were also decreased upon EWI2 expression (Figure 4D).
Figure 4. Effects of EWI2 on integrins and tumor cell-matrix adhesion.
(A) Integrins expressed on HCC827 cell surfaces were analyzed by flow cytometry. t-test was used for statistical analyses on MFI (mean±SD, n=5 individual experiments for integrins α3 and β1 subunits, n=3 individual experiments for other integrins). *: p<0.05. (B) Western blot analyses of integrin α3 and β1 in HCC827 cells. GAPDH and β-actin acted as house keeping controls. The band densities were measure by ImageJ and compared by t-test. Data was presented as mean±SD (n=3 individual experiments). *: p<0.05. (C) q-RT PCR was performed to analyze integrin α3 and β1 mRNA levels in HCC827 cells. Beta-actin was used as a house keeping gene. The relative level of mRNA for each gene was calculated by using the ΔΔCT method, compared with t-test, and shown as mean±SD (n=3 individual experiments). *: p<0.05 and **: p<0.01. (D) Integrin α3 and β1 expression in EVs were investigated with Western blot analyses. The densities of bands were measured by ImageJ, normalized by the ones of HCC827-EWI2 group, compared by t-test and presented as mean±SD (n=3 individual experiments). **: p<0.01 and ****: p<0.0001. (E) HCC827 cell adhesion on different types of extracelluar matrix was measured. The plates were coated with 10 μg/ml of different matrix (FN and VN in 0.01M NaHCO3, Col-I and LN in PBS) at 4°C overnight. 105 cells in 100 μl FBS free medium were added in each well and incubated at 37°C for 30 minutes. OD values for each type of matrix was subtracted OD value of non-coated wells. t-test was applied for statistical analyses. Data were presented as mean±SD (n=3 individual experiments). *: p<0.05 and **: p<0.01. (F) Immunofluorescence staining of vinculin in HCC827 cells. The cover slips were coated with FN (50 μg/ml in 0.01M NaHCO3) before seeding cells. After the cells were fixed, permeabilized and stained with vinculin mAb, F-actin with phalloidin. Images were captured with confocal fluorescence microscopy. Number of focal adhesions in each cell was counted, compared by t-test between the groups, and presented as mean±SEM (n=15~19 cells per group from 3 individual experiments). ***: p<0.001. Scale bar for upper panel is 2.5 μm and for lower panel is 3 μm.
Consistently, cell adhesions onto collagen-I, laminin-332, fibronectin, and vitronectin, the matrix ligands for integrins α2, α3, α5, and αV, respectively, were reduced upon EWI2 expression in HCC827 cells (Figure 4E). Focal adhesions, revealed by immunofluorescence staining of vinculin, became decreased in number in isolated HCC827-EWI2 cells compared with individualized HCC827-Mock cells (Figures 4F and S4), but remained equivalent in the clustered cells or cells in contact with each other between two groups (Figures 4F and S4), which supports the observations that EWI2 inhibits solitary migration but not collective migration of HCC827 cells.
EWI2 inhibits tumor cell colony formation in vitro and tumor growth in vivo.
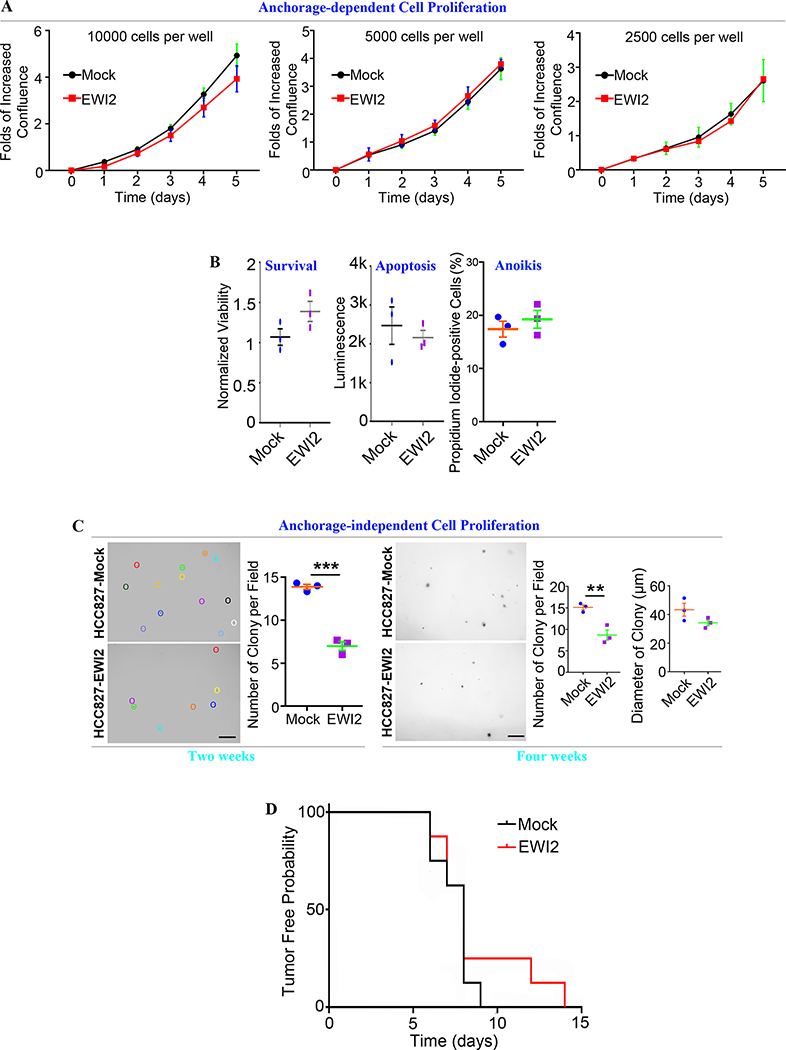
To determine the effect of EWI2 on LUAD, we examined cell proliferation and survival of HCC827 transfectants. No difference in anchorage-dependent cell proliferation was observed between 2 groups (Figures 5A and S5A). Necrosis reflected by cell survival, apoptosis, and anoikis were not affected by EWI2 in HCC827 cells (Figure 5B and S5B). We also performed a soft agar assay to assess EWI2 role in tumor cell transformation and carcinogenesis. Fewer clones were formed in HCC827-EWI2 group, compared to HCC827-Mock group. But EWI2 didn’t affect the clone size (Figures 5C and S5C). In addition to these in vitro studies, we injected HCC827 cells subcutaneously into NU/J nude mice to observe tumor initiation and growth. Palpable and visible masses could be detected earlier in HCC827-Mock group, but the difference didn’t reach statistical significance (Figure 5D). Tumors formed by HCC827-Mock cells grew faster and more aggressively than tumors of HCC827-EWI2 cells, evaluated by tumor volume (Figure 5E) and tumor burden (Figures 5F and S5D). EWI2 expression elevated apoptosis, reflected by TUNEL staining, within tumor masses (Figure 5G) but didn’t alter proliferation, shown by Ki67 staining (Figures 5H and S5E). Hence, the reduced growth of EWI2-expressing tumors results likely from increased apoptosis of HCC827-EWI2 group in vivo.
Figure 5. EWI2 inhibits anchorage-independent tumor cell proliferation in vitro and tumor growth in vivo.
(A) Different densities of cells were seeded in each well of the 96-well plate. Incucyte Live Cell Imaging System was used to observe the confluence of each well at different time points. The ratio of increased confluence to confluence at the very beginning was measured and compared by t-test at each time point. Data was presented as mean±SD (n=3 individual experiments). (B) Cell survival was examined by ApoTox-Glo™ triplex assay, in which GF-AFC substrate, bis-AAF-R110 substrate, and Caspase-Glo 3/7 reagent were used to report viability, necrosis, and apoptosis, respectively. Results were shown as mean±SD (n=3 individual experiments). Anoikis was reflected by percentage of PI+ cells and measured by flow cytometry analysis on detached cells. Results were shown as mean±SD (n=3 individual experiments). (C) Soft agar assay was applied to analyze anchorage-independent proliferation. Number and size of colonies were measured in 7 randomly selected images in each group, compared between groups by t-test, and presented as mean±SD (n=3 individual experiments). Colonies were stained and photographed 2 weeks (left panel) or 4 weeks (right panel) after the onset of the experiments. **: p<0.01 and ***: p<0.001. Scale bar: 300 μm. (D) Eight pairs of NU/J mice were injected with 5×106 HCC827-Mock or -EWI2 transfectant cells. Injection areas were monitored on a daily basis to detect palpable mass. Tumor-free probability was calculated and represented as KM curve, to report tumor initiation. (E) Mice body weight and primary tumor size were recorded every 4 days. Tumor volume was calculated as length×width2×0.52, presented as mean±SD and compared by t-test at each time point or by ANOVA for whole data. *: p<0.05; **: p<0.01. (F) Mice body weight and tumor weight was measured 5 weeks post injection. Tumor burden was shown as the ratio of tumor weight to body weight and was compared by t-test. Data was presented as mean±SD. * p<0.05. (G) Primary tumor tissue sections were stained by TUNEL assay. Three randomly selected images from each tissue section were examined, three tissue sections were cut discontinuously from each primary tumor, and five individual tumors were processed for each group. Fluorescence intensity was measured with ImageJ software and shown as mean±SEM. *: p<0.05. Scale bar: 50 μm. (H) Ki67 staining of primary tumors formed in nude mice was analyzed by ImageJ software and quantified as the percentage of Ki67+ cells (mean±SEM). Three randomly selected fields/images were analyzed from each tissue section, three tissue sections were cut discontinuously from each tumor, and five individual tumors were examined for each group.
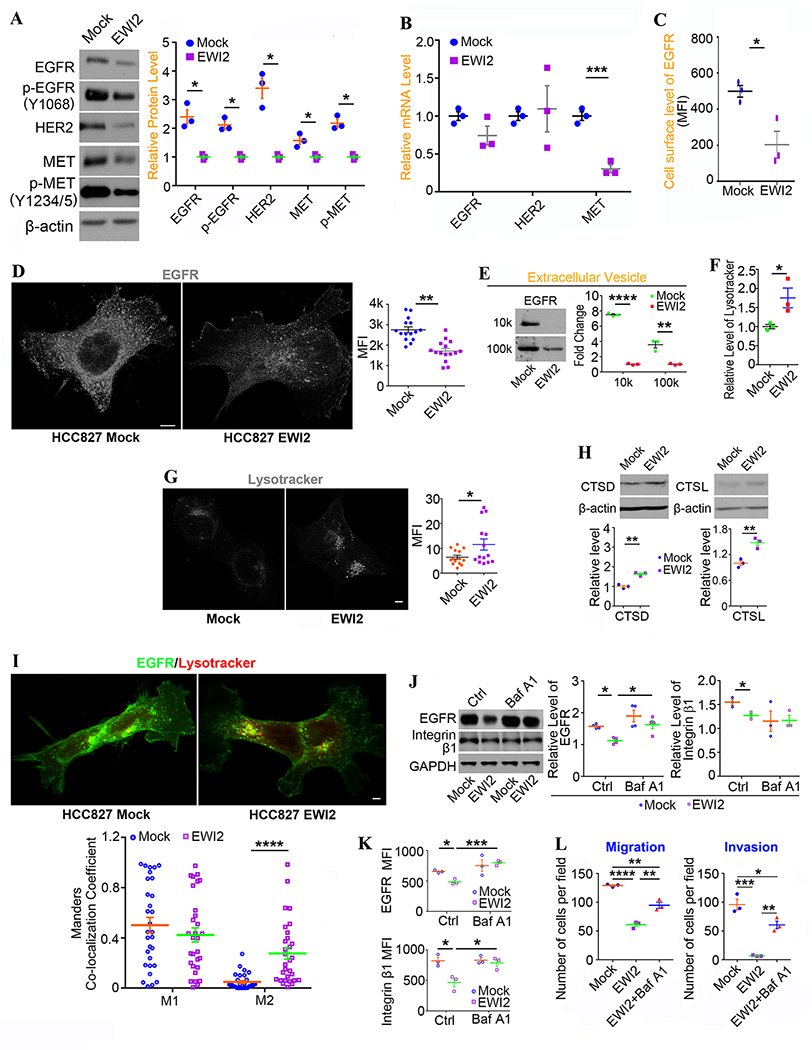
EWI2 promotes lysosomal trafficking and proteolysis of EGFR.
As EGFR is crucial for lung cancer progression, we examined EGFR and its phosphorylated form (pTyr1068-EGFR) and found they both were reduced in HCC827-EWI2 cells (Figure 6A). Such reductions were also observed in A549 and H358 human lung adenocarcinoma cells when EWI2 is over-expressed (Figure S6A), and reciprocally increases in total and phosphorylated EGFR were found when EWI2 is silenced or ablated in other cancer cells (our unpublished data), underlining EGFR regulation as a common effect and mechanism of EWI2. The reduction of total EGFR level was not caused by less transcription (Figure 6B). The cell surface level of EGFR was also reduced upon EWI2 forced expression (Figure 6C), so was the fluorescence intensity of EGFR immuno-staining in HCC827-EWI2 cells (Figure 6D). EGFR in EVs was also decreased in EWI2-overexpressed group (Figure 6E), indicating that decrease in cellular EGFR was not caused by increase in its discharge through EVs.
Figure 6. EWI2 reduces growth factor receptors by accelerating their degradation.
(A) EGFR, HER2, and MET proteins and their phosphorylation in HCC827 cells were detected by Western blot, with β-actin as the loading control. Band densities were quantified with ImageJ software, normalized by the ones of HCC827-EWI2 group, and presented as mean±SD (n=3 individual experiments). *: p<0.05. (B) qRT-PCR analysis on EGFR, HER2 and MET mRNA levels. Beta-actin was used as a housekeeping gene. Relative level of mRNA for each gene was calculated with the ΔΔCT method, normalized by the one of HCC827-Mock group, and shown as mean±SD (n=3 individual experiments). ***: p<0.001. (C) Levels of EGFR at the cell surface were measured with flow cytometry and reported as MFI (mean±SD, n=3 individual experiments). *: p<0.05 (D) Immunofluorescence of EGFR in HCC827 cells were imaged with confocal microscopy and quantified with ImageJ software as MFI per cell (mean±SEM, n=15 randomly selected images from 3 individual experiments). **: p<0.01. Scale bar: 7 μm. (E) Released EGFR in extracellular vesicles was investigated with Western blot. Band densities were normalized by the ones from HCC827-EWI2 group and presented as fold change (mean±SD, n=3 individual experiments). **: p<0.01 and ****: p<0.0001. (F) Lysosomes labeled by lysotracker were detected by flow cytometry. The ratio of EWI2 group MFI to mock group MFI was compared by t-test (mean±SD, n=3 individual experiments). *: p<0.05. (G) Lysotracker staining in HCC827 cells was imaged with fluorescence microscopy and quantified by ImageJ software as MFI per cell (mean±SEM, n=15 randomly selected images from 3 individual experiments). *: p<0.05. Scale bar: 6.5 μm. (H) CTSD and CTSL were investigated by Western blot, with β-actin as the loading control. The band densities were quantified with ImageJ. Fold changes were compared between groups by t-test and presented as mean±SD. **: p<0.01. (I) Colocalization of EGFR and Lysotracker was examined with ImageJ software. Manders coefficients (M1: EGFR-colocalized Lysotracker/total Lysotracker, and M2: Lysotracker-colocalized EGFR/total EGFR) were assessed for individual cells and compared between groups by t-test (mean±SEM, n=30 randomly selected cells per group from 3 individual experiments). ****: p<0.0001. Scale bar: 10 μm. (J) EGFR and integrin β1 from the cells treated with DMSO or Baf A1 were measured by Western blot, with GAPDH as the loading control. Band densities were quantified with ImageJ software and presented as mean±SD (n=3~4 individual experiments). *: p<0.05. (K) Levels of EGFR and integrin β1 at the cell surface after Baf A1 or DMSO treatment were measured by flow cytometry and presented as MFI (mean±SD, n=3 individual experiments). *: p<0.05 and ***: p<0.001. (L) EWI2-expressing HCC827 cells were treated by Baf A1 for 8 hours prior to the assays. For migration assay Transwell inserts were coated with 50 μl of FN (10 μg/ml) per insert at 4°C overnight, and for invasion assay 50 μl of Matrigel (3 mg/ml) per insert was added to the upper side of the filter membrane and incubated at 37°C for 30 minutes for gel formation. Then, each upper chamber was loaded with 1×105 cells in 100 μl of serum-free media, and the lower chamber was filled with complete media. Numbers of migrated or invaded cells were compared by t-test between groups and presented as mean±SD (n=3 individual experiments). *: p<0.05; ** p<0.01; ***: p<0.001; and ****: p<0.0001.
Since less EGFR in HCC827-EWI2 cells was not caused by changes in its synthesis and release, we postulate that EWI2 promotes EGFR degradation. By staining HCC827 cells with Lysotracker, we observed increased lysosomes upon EWI2 expression, based on both flow cytometry (Figure 6F) and immunofluorescence (Figure 6G) analyses. Cathepsin-D (CTSD) and cathepsin-L (CTSL), markers of lysosome, [25–26] was also increased in EWI2-expressed HCC827 cells (Figure 6H), supporting the upregulation of lysosome upon EWI2 expression. As we expected, EGFR co-localization with lysotracker was also increased in HCC827-EWI2 cells (Figure 6I), indicating that EWI2 promotes EGFR trafficking to lysosomes.
The treatment with lysosome inhibitor Baf A1 eliminated the differences of EGFR in total protein level (Figure 6J) and cell surface level (Figure 6K) between two groups, supporting the notion that EWI2 facilitates lysosomal degradation of EGFR. Likewise, the differences of integrin β1 also became diminished after Baf A1 treatment. We measured movement and metastasis of the Baf A1-treated cells and found Baf A1 partially or largely rescued or canceled the inhibitory effects of EWI2 on chemo-haptotaxis migration, Matrigel invasion (Figures 6L and S4B), and experimental metastasis (5/6 mice formed lung metastasis) (Figures 3H–I and S3B), underlining a key role of lysosome in EWI2 function.
Besides EGFR, EGF receptor HER2 and HGF receptor c-MET were also downregulated in HCC827-EWI2 cells (Figure 6A). Like EGFR, HER2 wasn’t altered at the mRNA level upon EWI2 expression, but c-MET mRNA was reduced (Figure 6B). Notably, EWI2 expression lowered c-MET protein levels in HCC827, H358, and A549 cells, indicating that c-Met is another common target of EWI2. Since i) EWI2 didn’t change HER2 level in A549 (Figure S6A) and other cancer cells (our unpublished data) and ii) EGFR is closely relevant to lung cancer, we focused on EGFR in this study.
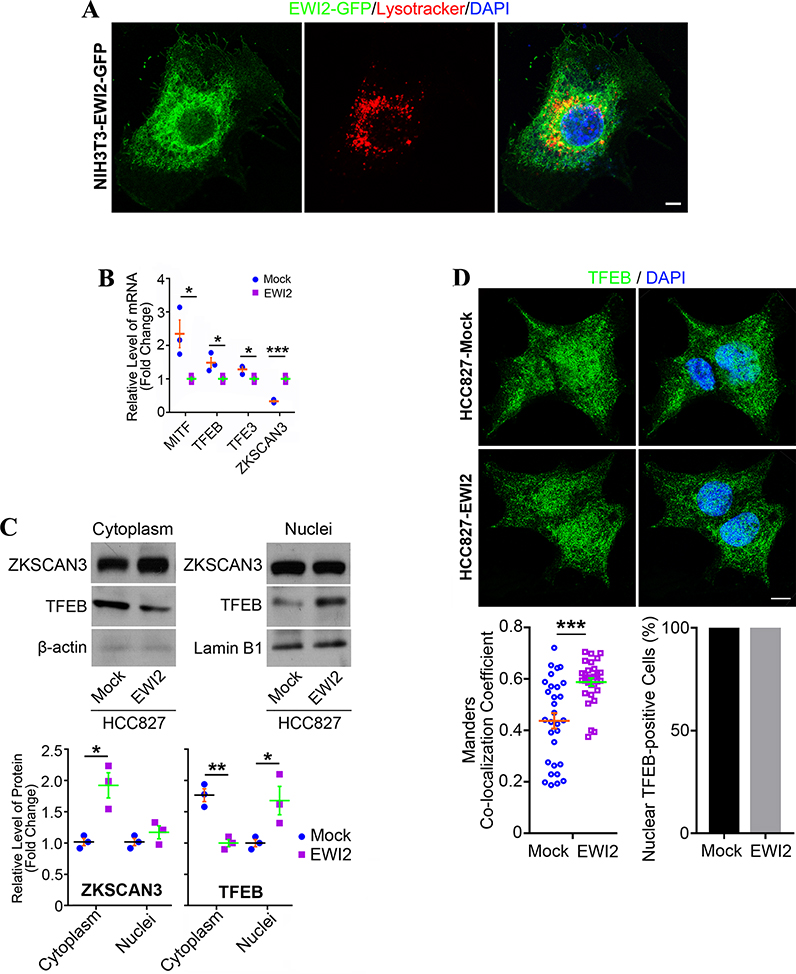
EWI2 promotes the nuclear retention of TFEB to upregulate lysosome formation.
Given the importance of lysosome in EWI2 function, we questioned if EWI2 traffics to lysosomes. As EWI2 Abs didn’t work well in immunofluorescence, we used EWI2-GFP fusion, which was generated in our earlier study, for the analysis. Indeed, in NIH3T3-EWI2 stable transfectant cells, EWI2-GFP fusions were partially colocalized with Lysotracker (Figure 7A), suggesting that EWI2 proteins per se traffic into lysosomes.
Figure 7. EWI2 enhances lysosome capacity by promoting TFEB nuclear translocation.
(A) NIH3T3-EWI2-GFP cells were stained with Lysotracker and imaged with confocal microscopy. Scale bar: 10 μm. (B) Analysis on MITF, TFEB, TFE3, and ZKSCAN3 gene expression with qRT-PCR. Beta-actin serves as a housekeeping gene. Relative levels of the mRNA for each gene were calculated by using the ΔΔCT method, compared between groups with t-test, and presented as mean±SD (n=3 individual experiments). *: p<0.05; ***: p<0.001. (C) ZKSCAN3 and TFEB in cytoplasm and nuclei were examined with Western blot. Beta-actin and lamin-B1 were used as loading controls for cytoplasmic and nuclear proteins, respectively. The band densities were quantified with ImageJ software, analyzed by t-test, and presented as mean±SD (n=3 individual experiments). *: p<0.05; **: p<0.01. (D) Colocalization of TFEB with DAPI was analyzed with ImageJ software, presented as Manders coefficient, and compared between groups by t-test (mean±SEM, n=30 randomly selected cells per group from 3 individual experiments). Percentages of the cells positive in nuclear TFEB were compared between groups by Fisher’s test. Scale bar: 20 μm.
More lysosomes upon EWI2 expression also drove us to examine the transcription factors that determine and regulate lysosome formation. Based on qRT-PCR analysis, gene expressions of MITF [27], TFE3 [28] and TEFB [29], the key transcription factors that promote lysosome genesis, were decreased in HCC827-EWI2 cells, compared to the Mock cells. Meanwhile, mRNA level of ZKSCAN3, a transcription repressor of lysosome genesis, [30] was increased in HCC827-EWI2 cells (Figure 7B). These alterations at mRNA level imply a cellular feedback mechanism in HCC827-EWI2 transfectant to reduce its already elevated formation of lysosome compensatively. As the master activator of lysosome formation, TFEB was reduced in cytoplasm but became accumulated in nuclei in HCC827-EWI2 cells, compared to HCC827-Mock cells (Figure 7C), fitting well with the lysosome upregulation upon EWI2 expression. Immunofluorescence analysis of the TFEB-DAPI co-localization levels also supported more accumulation of TFEB per nucleus in EWI2-expressed cells, despite no difference in nuclear presence of TFEB between two groups (Figure 7D). ZKSCAN3 functions within nucleus as a transcription repressor, but ZKSCAN3 proteins in nuclei were not altered by EWI2 expression although they became increased in cytoplasm (Figures 7C and S6C).
Since PC3 human prostate cancer cells express EWI2, we established EWI2-ablated PC3 cells by CRISPR-Cas9 technique (Figure S7A). As shown by immunofluorescence (Figure S7B), increased EGFR but decreased lysotracker staining upon EWI2 ablation mirrored well with the observations from EWI2 overexpression. EGFR colocalization with lysotracker was reduced in PC3-EWI2 knockout cells, suggesting less lysosmal targeting of EGFR. Levels of lysotracker (Figure S7C) and lysosome markers CTSD and CTSL (Figure S7D), examined respectively by flow cytometry and Western blot, were also reduced upon EWI2 knockout. Then we investigated transcription factors that regulate lysosome biogenesis. Lysosome promoters MITF and TFE3 were elevated at mRNA level upon EWI2 knockout, probably due to compensation, and lysosome suppressor ZKSCAN3 mRNA was upregulated (Figure S7E). Importantly, less nuclear TFEB proteins in PC3-EWI2 knockout than PC3-Mock cells but equivalent nuclear ZKSCAN3 proteins between two groups (Figure S7F) are consistent with the observation of fewer lysosomes upon EWI2 removal.
Discussion
IgSF members regulate tumor formation and progression. [31] Mounting evidence points to IgSF protein EWI2 as a suppressor of various solid malignant tumors. The mechanisms by which EWI2 inhibits tumor cells in vitro and tumors in vivo remain to be further and fully unveiled. Our study expands the anti-tumor spectrum of EWI2 to LUAD and also reveals the connection of EWI2 tumor-inhibitory activity to endosomal/lysosomal trafficking and turnover of growth factor receptors and integrins.
Polarized expressions of EWI2 mRNA and protein in LUAD patients
Bioinformatics analyses on the relationship of EWI2 with lung cancer suggest relatively robust and consistent links of EWI2 to LUAD. The higher their gene expressions are, the poorer LUAD patient prognoses appear to be, based on KM-plotter. Although we could not draw the same conclusion from GEPIA, inconsistent readouts from different bioinformatics analytic platforms can be attributed to the differences in dataset composition, as KM-plotter is based on gene microarray datasets while GEPIA is based on TCGA RNAseq datasets, and split point of these platforms. [32] Because the majority of LUAD cases in KM-plotter are the stage-I LUAD patients, the prognostic value of EWI2 may be limited to the stage-I LUAD patients and remains to be evaluated for the stages II-IV LUAD patients.
Surprisingly, EWI2 protein level seems inconsistent with its mRNA level in LUAD. Both CPTAC database analyses and our experimental examination indicate diminished or lost expression of full-length EWI2 proteins in the tumor tissues from patients with stages I, II, and III LUAD and LUSC, compared to their non-tumorous lung tissues.
Shedding of EWI2 membrane-distal IgG domain by furin-mediated cleavage forms EWI2win isoform, [5, 33] while multiple protein species in EWI2 immunoprecipitations from some cells suggest that other proteolytic forms of EWI2 also exist. [4] Antigen epitope(s) of the EWI2 pAb for Western blot analyses on human tissues located in the ectodomain of EWI2 but was not well-defined (https://www.rndsystems.com/products/human-mouse-rat-igsf8-cd316-antibody_af3117) (Table S1). Hence, the observation of “undetectable” EWI2 proteins in LUAD and LUSC tumor tissues may actually reflect extensive shedding of the EWI2 ectodomain in the tumor cells. While higher expression of EWI2 gene in lung cancer could correlate with more truncated EWI2 proteins remained in the tumor cells, implying functionally contrast roles of intact and truncated EWI2 proteins in tumor cells, given that intact and truncated EWI2 proteins function oppositely in supporting CD81 as a co-receptor for hepatitis C virus. [5, 33]
Our study also underpins the importance of taking a comprehensive bioinformatics approach, instead of using a single approach like genomics analyses or a single bioinformatics analytic platform/tool like KM-plotter, to obtain a complete, high-fidelity picture of cancer-related molecules.
EWI2 inhibits tumor growth and metastasis by reducing the levels and/or signaling of growth factor receptors and integrins.
Forced expression of EWI2 in HCC827 lung adenocarcinoma cells reduces cell adhesion onto not only basement membrane matrix laminin but also the interstitial tissue matrices fibronectin, vitronectin, and collagen-I, by down-regulating both cell surface and total cellular levels of their integrin receptors. Fewer integrin proteins appear to be caused by lower mRNA levels based on integrin α3 and integrin β1 analyses.
EWI2 selectively inhibits various types of growth factor-stimulated and/or integrin-mediated cell movement, consistent with the reductions of these two classes of receptors. No difference in collective migration suggests that EWI2 effect on cell-cell adhesion limits or cancels its effect on cell motility during collective migration, which is also reflected by its differential effects on focal adhesions in individualized versus contacted cells. While no difference in random migration suggests that EWI2 doesn’t significantly affect basic machinery for cell motility. Since invasion through tumor microenvironment requires dynamic interactions of tumor cells with various matrices in tumor microenvironment, global downregulation of major integrins upon EWI2 expression predict and also can explain the attenuated invasiveness of HCC827-EWI2 cells. Indeed, the inhibitory effect of EWI2 on invasiveness is confirmed by invasion through Matrigel in vitro and MMP9 staining in tumor tissues in vivo.
Proliferation acceleration and apoptosis resistance are major characteristics of cancer cells. [34] Despite no impact on survival and anchorage-dependent proliferation of HCC827 cells in vitro, EWI2 expression i) reduces anchorage-independent tumor clone formation, which reflects cell transformation [35] and ii) induces apoptosis in xenograft tumor tissue. Slower tumor growth and less tumor burden in vivo upon EWI2 expression is caused possibly by the increased apoptosis, which could be the result of diminished signaling from growth factor receptors and integrins in the tumor cells as EWI2 diminishes these membrane proteins. Discrepancy in apoptosis between in vitro and in vivo observations suggests that EWI2-expressing tumor cells are more susceptible to the death-promoting signal(s) from tumor microenvironment in mice and need the non-tumor cell autonomous signal(s) to commit death. Although in vivo impact of the diminished signaling from growth factor receptors and integrins on tumor cell proliferation cannot be excluded, equivalent numbers of Ki67-positive cells between the two groups strongly suggest that tumor cell proliferation is not a major determinant for the contrast tumor growth seen in this study. Moreover, EWI2 could affect the tumors by altering other membrane proteins beyond growth factor receptors and integrins, through tetraspanin-enriched membrane domains. [36–37] The mechanisms remain to be investigated in future study.
EWI2 promotes endosomal/lysosomal trafficking of EGFR and integrin.
EGFR activation triggered by ligands induces EGFR endocytosis, which in turn regulates downstream signaling. The fates of EGFR after endocytosis include recycling, degradation, as well as atypical trafficking to nucleus or mitochondria. [38] Decreased EGFR upon EWI2 expression is not caused by less gene expression or more EV-mediated release but results from increased targeting to lysosomes in EWI2-expressing HCC827 cells. Endosomal/lysosomal trafficking of membrane receptors such as EGFR can cause their proteolytic degradation by lysosomal enzymes such as CTSD and CTSL or by proteasome after their ubiquitination. [39–40] Indeed, lysosomal inhibitor Bafilomycin blocks EWI2-induced decreases in the protein levels of EGFR and integrin β1, supporting that EWI2 promotes proteolytic degradation of EGFR and integrin β1 in lysosomes. Lysosome inhibition also partially or largely attenuates EWI2-induced inhibitions of tumor cell movement and metastasis, respectively.
A weakness of our study is that we didn’t analyze the roles of tetraspanins CD81 and CD9, which typically coordinate with or regulate EWI2 functionally, in endolysosomal trafficking of growth factor receptors and integrins. Since tetraspanins undergo endolysosomal trafficking and interact with these membrane proteins, [7, 36–37] tetraspanin contributions to the EWI2 impact on lysosome are imaginable and need to be determined.
EWI2 promotes the formation of lysosomes by upregulating nuclear accumulation of TFEB.
We demonstrated herein that EWI2 overexpression and knockout results in the expansion and diminution of lysosome compartment and/or activity, respectively. Because of its existence in lysosomes, EWI2 likely regulates lysosome directly. However, the observation of nuclear retention of master transcription factor TFEB, which drives lysosomogenesis, upon EWI2 expression argues against the notion of EWI2 as a direct regulator of lysosome. How to reconcile such a discrepancy and explain EWI2 impact on TFEB remain to be investigated in future. Nevertheless, EWI2 appears to promote lysosome formation instead of simply pumping up lysosomal activity, given the facts that it elevates i) cellular capacity to absorb low-pH sensor Lysotracker, ii) cellular levels of lysosomal markers CTSD and CTSL, and iii) nuclear buildup of pro-lysosomal TFEB and/or cytoplasmic retention of anti-lysosomal ZKSCAN3.
In summary, EWI2 inhibits trans-plasma membrane signaling of lung cancer cells to suppress tumor cell movement, clone formation in vitro, and tumor growth in vivo by down-regulating growth factor receptor and integrin through their endosomal/lysosomal trafficking and/or turnover. Because of the utmost importance of EGFR for lung cancer, EWI2 inhibition of EGFR is apparently clinically connected, in addition to its multiple layers of anti-cancer activities. Because of frequent resistance of lung cancers to the EGFR inhibitor therapy, our observation provides a plausible therapeutic strategy to direct EGFR for degradation and then bypass the resistance to EGFR inhibitors.
Supplementary Material
Supplementary Figure 1 EWI2 in human lung cancer and normal lung tissues (A) Western blot analyses on EWI2 protein in tumor tissues of patients (except the one represented in Figure 2) with LUAD (n=7) and LUSC (n=4), and normal lung tissues. Beta-actin served as a loading control. (B) EWI2 and EGFR were stained on tissue microarrays. Scale bar: 300 μm.
Supplementary Figure 2 Role of EWI2 in cell movement (A) Solitary directional migration assay was performed on HCC827 cells in Transwell system. Scale bar: 50 μm. (B) Random migration assay was performed on HCC827 cells in Transwell system. Scale bar: 50 μm. (C) Wound healing dynamics was photographed at different time points. Scale Bar: 200 μm. (D) Invasion assay in Transwell for HCC827 cells. Scale bar: 50 μm.
Supplementary Figure 3 Histological and immunohistochemical analyses (A) Vimentin was stained with the sections of the primary tumor tissues formed in nude mice. Scale bar: 50 μm. (B) H&E staining of lung tissue sections of experimental metastasis models. Scale bar: 700 μm. Black arrows indicate metastatic lesions.
Supplementary Figure 4 Effect of EWI2 overexpression on focal adhesions in HCC827 cells Vinculin and F-actin staining of HCC827 cells, which were spread on fibronectin (50 μg/ml)-coated substratum in complete medium overnight, were imaged with confocal fluorescence microscopy. Scale bar: 4 μm.
Supplementary Figure 5 EWI2 overexpression and tumor cell proliferation (A) Different densities of cells were seeded in each well of the 96-well plate. Cells were fixed by 4% PFA and stained with 0.1% crystal violet. 100μl 2% SDS was added to dissolve crystal violet. OD values were measured by EnVision® Multilabel Reader. Increased folds of OD values were calculated at different time points and presented as mean±SD (n=3 individual experiments). (B) Anoikis was reflected by percentage of PI+ cells and measured by flow cytometry. (C) Higher magnification of colonies staining 4 weeks after seeding in soft agar assay. Number and size of colonies were quantified in 7 randomly selected images from each group and presented as mean±SD (n=3 individual experiments). *: p<0.05. Scale bar: 20 μm. (D) Subcutaneous tumor formed on nude mice. (E) Ki67 staining of primary tumor tissues formed in nude mice. Left images were from 2 different HCC827-Mock tumors, and right images were from 2 different HCC827-EWI2 tumors. Scale bar: 50 μm.
Supplementary Figure 6 Other molecular and cellular effects of EWI2 forced expression (A) EGFR, HER2, and MET proteins and their phosphorylation in H358 and A549 cells were detected by Western blot. GAPDH serves as a loading control. Band densities were quantified with ImageJ software, normalized by the ones of EWI2 expression groups, compared between groups by t-test, and presented as mean±SD (n=3 individual experiments). *: p<0.05; **: p<0.01; and ***: p<0.001. (B) Light microscopic images of the cells that moved onto bottom side of Transwell inserts. Upper panel: chemo-haptotaxis migration assay was performed with HCC827 cells in Transwell system after Baf A1 treatment. Scale bar: 50 μm. Lower panel: Matrigel invasion assay was performed with HCC827 cells in Transwell system after Baf A1 treatment. Scale bar: 150 μm. (C) Immunofluorescence staining of ZKSCAN3 in HCC827 cells, with DAPI as the marker for nucleus. Images were captured with confocal microscopy. Scale bar: 20 μm.
Supplementary Figure 7 EWI2 knockout downregulates lysosome activity and formation (A) Western blot analysis on EWI2 in PC3-Mock and PC3-EWI2 KO cells, with β-actin as the loading control. Band densities were quantified with ImageJ software and presented as mean±SD (n=3 individual experiments). ** p<0.01. (B) PC3 cells were stained with EGFR Ab and Lysotracker and photographed with confocal microscopy. EGFR-Lysotracker colocalization was analyzed by ImageJ software and presented as Pearson coefficient (mean±SEM, n=24 randomly selected cells from 3 individual experiments). **** p<0.0001. (C) The cells labeled with Lysotracker and examined with flow cytometry. The ratio of EWI2-KO group MFI to mock group MFI was compared by t-test (mean±SD, n=3 individual experiments). * p<0.05. (D) Western blot analysis on CTSD and CTSL, with β-actin as the loading control. Band densities were quantified with ImageJ software and presented as mean±SD (n=3 individual experiments). * p<0.05; ** p<0.01. (F) Cytoplasmic and nuclear ZKSCAN3 and TFEB proteins were examined with Western blot. Beta-actin and lamin B1 serve as loading controls for cytoplasmic and nuclear proteins, respectively. Band densities were quantified with ImageJ software and presented as mean±SD (n=3 individual experiments). * p<0.05; ** p<0.01.
Highlights.
EWI2 acts as a lung cancer suppressor to inhibit the tumor growth and metastasis.
EWI2 reduces the activities of membrane receptors by enhancing their degradation.
EWI2 promotes lysosomes formation by upregulating nuclear accumulation of TFEB.
Our finding provides an alternative therapeutic strategy against EGFR.
Acknowledgments
This work was supported by National Institutes of Health grants HL137819 and GM135547, OCAST grant HR20–055, and research grants from OCASCR, a program of TSET, to XAZ. XAZ is an Oklahoma TSET Cancer Research Scholar. We thank Ms. Kathy Kyler for English editing. We acknowledge the functional genomics core and tissue pathology core of Stephenson Cancer Center at the University of Oklahoma Health Sciences Center.
Abbreviation:
- ANOVA
analysis of variance
- BSA
bovine serum albumin
- CCLE
Cancer Cell Line Encyclopedia
- CPTAC
Clinical Proteomic Tumor Analysis Consortium
- CTSD
cathepsin D
- CTSL
cathepsin L
- EGF
epidermal growth factor
- EV
extracellular vesicle
- FBS
fetal bovine serum
- HGF
hepatocyte growth factor
- HRP
horseradish peroxidase
- IgSF
immunoglobulin superfamily
- LCLC
large cell lung carcinoma
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- MFI
mean fluorescence intensity
- MITF
microphthalmia-associated transcription factor
- NSCLC
non-small-cell lung cancer
- OS
overall survival
- PBS
phosphate buffered solution
- PBST
phosphate buffered solution with 0.1% Tween-20
- PFA
paraformaldehyde
- PI
propidium iodide
- PRAD
prostate adenocarcinoma
- SCLC
small-cell lung cancer
- SDS
sodium dodecyl sulphate
- TCGA
The Cancer Genome Atlas
- TFE3
transcription factor binding to IGHM enhancer 3
- TFEB
transcription factor EB
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- ZKSCAN3
zinc finger protein with KRAB and SCAN domains 3
Footnotes
Conflict of Interest
The authors declare no competing financial interests.
Ethics Statement
All procedures involving animals were performed according to protocols approved by the Institutional Animal Care and Use Committee (IACUC).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Stipp CS, Kolesnikova TV, Hemler ME. EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J Biol Chem. 2001; 276(44):40545–54 [DOI] [PubMed] [Google Scholar]
- 2.Charrin S, Le Naour F, Oualid M, et al. The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J Biol Chem. 2001; 276(17):14329–37 [DOI] [PubMed] [Google Scholar]
- 3.Sala-Valdés M, Ursa A, Charrin S, Rubinstein E, et al. EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J Biol Chem. 2006; 281(28):19665–75 [DOI] [PubMed] [Google Scholar]
- 4.Zhang XA, Lane WS, Charrin S, et al. EWI2/PGRL associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. Cancer Res. 2003; 63(10):2665–74 [PubMed] [Google Scholar]
- 5.Montpellier Claire, Tews Birke Andrea, Julien Poitrimole, et al. Interacting regions of CD81 and two of its partners, EWI-2 and EWI-2wint, and their effect on hepatitis C virus infection. J Biol Chem. 2011; 286(16):13954–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Xiuwei H, Kovalenko Oleg V, Kolesnikova Tatiana V, et al. Contrasting effects of EWI proteins, integrins, and protein palmitoylation on cell surface CD9 organization. J Biol Chem. 2006; 281(18):12976–85 [DOI] [PubMed] [Google Scholar]
- 7.Richardson Mekel M, Jennings Lisa K, Zhang Xin A. Tetraspanins and tumor progression. Clin Exp Metastasis. 2011; 28(3):261–70 [DOI] [PubMed] [Google Scholar]
- 8.Wang HX, Sharma C, Knoblich K, et al. EWI-2 negatively regulates TGF-β signaling leading to altered melanoma growth and metastasis. Cell Res. 2015; 25(3):370–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolesnikova TV, Kazarov AR, Lemieux ME, et al. Glioblastoma inhibition by cell surface immunoglobulin protein EWI-2, in vitro and in vivo. Neoplasia. 2009; 11(1):77–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levina E, Ji H, Chen M, et al. Identification of novel genes that regulate androgen receptor signaling and growth of androgen-deprived prostate cancer cells. Oncotarget. 2015; 6(15):13088–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Chenying, Zhang Qing, Wang Ani, et al. EWI-2 controls nucleocytoplasmic shuttling of EGFR signaling molecules and miRNA sorting in exosomes to inhibit prostate cancer cell metastasis. Mol Oncol. 2021; 15(5):1543–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Hong-Xing, Hemler Martin E. Novel impact of EWI-2, CD9, and CD81 on TGF-β signaling in melanoma. Mol Cell Oncol. 2015; 2(1): e1030536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Bo, Zhang Yanhui H, Richardson Mekel M, et al. Differential functions of phospholipid binding and palmitoylation of tumour suppressor EWI2/PGRL. Biochem J. 2011; 437(3):399–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Somchai Parinyachat, Phongkitkarun Kriengkrai, Kueanjinda Patipark, et al. Novel Analytical Platform for Robust Identification of Cell Migration Inhibitors. Sci Rep. 2020; 10(1):931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang Mihwa, Jun Dong Wha, Kang Eun Hye, et al. EI24, as a Component of Autophagy, Is Involved in Pancreatic Cell Proliferation. Front Oncol. 2019; 9:652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vandersickel Veerle, Slabbert Jacobus, Thierens Hubert, et al. Comparison of the colony formation and crystal violet cell proliferation assays to determine cellular radiosensitivity in a repair-deficient MCF10A cell line. Radiation Measurements. 2011; 46(1): 72–75 [Google Scholar]
- 17.Joussaume Aurélie, Karayan-Tapon Lucie, Omar Benzakour, et al. A Comparative Study of Anoikis Resistance Assays for Tumor Cells. Biomedical Journal of Scientific & Technical Research. 2020; 29(2): pp 22255–22262 [Google Scholar]
- 18.Crowe Alexandra R, Wei Yue. Semi-quantitative Determination of Protein Expression using Immunohistochemistry Staining and Analysis: An Integrated Protocol. Bio Protoc. 2019; 9(24): e3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prall Friedrich, Maja Hühns. Quantitative evaluation of TP53 immunohistochemistry to predict gene mutations: lessons learnt from a series of colorectal carcinomas. Hum Pathol. 2019; 84:246–253 [DOI] [PubMed] [Google Scholar]
- 20.Stipp Christopher S, Kolesnikova Tatiana V, Hemler Martin E. EWI-2 regulates alpha3beta1 integrin-dependent cell functions on laminin-5. J Cell Biol. 2003; 163(5):1167–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farina Antonietta Rosella, Mackay Andrew Reay. Gelatinase B/MMP-9 in Tumour Pathogenesis and Progression. Cancers. 2014; 6(1): 240–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richardson Alessandra M, Havel Lauren S, Koyen Allyson E, et al. Vimentin Is Required for Lung Adenocarcinoma Metastasis via Heterotypic Tumor Cell-Cancer-Associated Fibroblast Interactions during Collective Invasion. Clin Cancer Res. 2018; 24(2):420–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar CC. Signaling by integrin receptors. Oncogene. 1998; 17(11 Reviews): 1365–73 [DOI] [PubMed] [Google Scholar]
- 24.Friedl P, Bröcker EB, Zänker KS. Integrins, cell matrix interactions and cell migration strategies: fundamental differences in leukocytes and tumor cells. Cell Adhes Commun. 1998; 6(2–3):225–36 [DOI] [PubMed] [Google Scholar]
- 25.Benes Petr, Vetvicka Vaclav, Fusek Martin. Cathepsin D--many functions of one aspartic protease. Crit Rev Oncol Hematol. 2008; 68(1):12–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Qizhu, Cai Jun, Shen Difei, et al. Lysosomal cysteine peptidase cathepsin L protects against cardiac hypertrophy through blocking AKT/GSK3beta signaling. J Mol Med (Berl). 2009; 87(3):249–60 [DOI] [PubMed] [Google Scholar]
- 27.Ploper Diego, Taelman Vincent F, Lidia Robert, et al. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci USA. 2015; 112(5): E420–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Hongsheng, Zhu Chumeng, Ruan Yanyun, et al. TFE3 Regulates the Function of the Autophagy-Lysosome Pathway to Drive the Invasion and Metastasis of Papillary Thyroid Carcinoma. Anal Cell Pathol (Amst). 2021; 2021:3081491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Settembre Carmine, Malta Chiara Di, Polito Vinicia Assunta, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011; 332(6036):1429–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chauhan Santosh, Goodwin Jinesh G, Swati Chauhan, et al. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013; 50(1):16–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong Chee Wai, Dye Danielle E., Coombe Deirdre R. The Role of Immunoglobulin Superfamily Cell Adhesion Molecules in Cancer Metastasis. Int J Cell Biol. 2012; 2012: 340296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng Hong, Zhang Guosen, Zhang Lu, et al. Comprehensive Review of Web Servers and Bioinformatics Tools for Cancer Prognosis Analysis. Front Oncol. 2020; 10:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potel Julie, Rassam Patrice, Montpellier Claire, et al. EWI-2wint promotes CD81 clustering that abrogates Hepatitis C Virus entry. Cell Microbiol. 2013; 15(7):1234–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanahan Douglas, Weinberg Robert A. Hallmarks of cancer: the next generation. Cell. 2011; 144(5):646–74 [DOI] [PubMed] [Google Scholar]
- 35.Taddei ML, Giannoni E, Fiaschi T, et al. Anoikis: an emerging hallmark in health and diseases. J Pathol. 2012; 226(2):380–9 [DOI] [PubMed] [Google Scholar]
- 36.Zöller Margot. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer. 2009; 9(1):40–55 [DOI] [PubMed] [Google Scholar]
- 37.Charrin Stéphanie, Jouannet Stéphanie, Boucheix Claude, et al. Tetraspanins at a glance. J Cell Sci. 2014; 127(Pt 17): 3641–8 [DOI] [PubMed] [Google Scholar]
- 38.Tomas Alejandra, Futter Clare E, Eden Emily R. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 2014; 24(1):26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kreitman Matthew, Noronha Ashish, Yarden Yosef. Irreversible modifications of receptor tyrosine kinases. FEBS Lett. 2018; 592(13):2199–2212 [DOI] [PubMed] [Google Scholar]
- 40.Weissman Allan M, Nitzan Shabek, Ciechanover Aaron. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat Rev Mol Cell Biol. 2011; 12(9):605–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 EWI2 in human lung cancer and normal lung tissues (A) Western blot analyses on EWI2 protein in tumor tissues of patients (except the one represented in Figure 2) with LUAD (n=7) and LUSC (n=4), and normal lung tissues. Beta-actin served as a loading control. (B) EWI2 and EGFR were stained on tissue microarrays. Scale bar: 300 μm.
Supplementary Figure 2 Role of EWI2 in cell movement (A) Solitary directional migration assay was performed on HCC827 cells in Transwell system. Scale bar: 50 μm. (B) Random migration assay was performed on HCC827 cells in Transwell system. Scale bar: 50 μm. (C) Wound healing dynamics was photographed at different time points. Scale Bar: 200 μm. (D) Invasion assay in Transwell for HCC827 cells. Scale bar: 50 μm.
Supplementary Figure 3 Histological and immunohistochemical analyses (A) Vimentin was stained with the sections of the primary tumor tissues formed in nude mice. Scale bar: 50 μm. (B) H&E staining of lung tissue sections of experimental metastasis models. Scale bar: 700 μm. Black arrows indicate metastatic lesions.
Supplementary Figure 4 Effect of EWI2 overexpression on focal adhesions in HCC827 cells Vinculin and F-actin staining of HCC827 cells, which were spread on fibronectin (50 μg/ml)-coated substratum in complete medium overnight, were imaged with confocal fluorescence microscopy. Scale bar: 4 μm.
Supplementary Figure 5 EWI2 overexpression and tumor cell proliferation (A) Different densities of cells were seeded in each well of the 96-well plate. Cells were fixed by 4% PFA and stained with 0.1% crystal violet. 100μl 2% SDS was added to dissolve crystal violet. OD values were measured by EnVision® Multilabel Reader. Increased folds of OD values were calculated at different time points and presented as mean±SD (n=3 individual experiments). (B) Anoikis was reflected by percentage of PI+ cells and measured by flow cytometry. (C) Higher magnification of colonies staining 4 weeks after seeding in soft agar assay. Number and size of colonies were quantified in 7 randomly selected images from each group and presented as mean±SD (n=3 individual experiments). *: p<0.05. Scale bar: 20 μm. (D) Subcutaneous tumor formed on nude mice. (E) Ki67 staining of primary tumor tissues formed in nude mice. Left images were from 2 different HCC827-Mock tumors, and right images were from 2 different HCC827-EWI2 tumors. Scale bar: 50 μm.
Supplementary Figure 6 Other molecular and cellular effects of EWI2 forced expression (A) EGFR, HER2, and MET proteins and their phosphorylation in H358 and A549 cells were detected by Western blot. GAPDH serves as a loading control. Band densities were quantified with ImageJ software, normalized by the ones of EWI2 expression groups, compared between groups by t-test, and presented as mean±SD (n=3 individual experiments). *: p<0.05; **: p<0.01; and ***: p<0.001. (B) Light microscopic images of the cells that moved onto bottom side of Transwell inserts. Upper panel: chemo-haptotaxis migration assay was performed with HCC827 cells in Transwell system after Baf A1 treatment. Scale bar: 50 μm. Lower panel: Matrigel invasion assay was performed with HCC827 cells in Transwell system after Baf A1 treatment. Scale bar: 150 μm. (C) Immunofluorescence staining of ZKSCAN3 in HCC827 cells, with DAPI as the marker for nucleus. Images were captured with confocal microscopy. Scale bar: 20 μm.
Supplementary Figure 7 EWI2 knockout downregulates lysosome activity and formation (A) Western blot analysis on EWI2 in PC3-Mock and PC3-EWI2 KO cells, with β-actin as the loading control. Band densities were quantified with ImageJ software and presented as mean±SD (n=3 individual experiments). ** p<0.01. (B) PC3 cells were stained with EGFR Ab and Lysotracker and photographed with confocal microscopy. EGFR-Lysotracker colocalization was analyzed by ImageJ software and presented as Pearson coefficient (mean±SEM, n=24 randomly selected cells from 3 individual experiments). **** p<0.0001. (C) The cells labeled with Lysotracker and examined with flow cytometry. The ratio of EWI2-KO group MFI to mock group MFI was compared by t-test (mean±SD, n=3 individual experiments). * p<0.05. (D) Western blot analysis on CTSD and CTSL, with β-actin as the loading control. Band densities were quantified with ImageJ software and presented as mean±SD (n=3 individual experiments). * p<0.05; ** p<0.01. (F) Cytoplasmic and nuclear ZKSCAN3 and TFEB proteins were examined with Western blot. Beta-actin and lamin B1 serve as loading controls for cytoplasmic and nuclear proteins, respectively. Band densities were quantified with ImageJ software and presented as mean±SD (n=3 individual experiments). * p<0.05; ** p<0.01.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.