Abstract
Background
Zika virus (ZIKV) is an emerging flavivirus of global concern. ZIKV infection of the central nervous system has been linked to a variety of clinical syndromes, including microcephaly in fetuses and rare but serious neurologic disease in adults. However, the potential for ZIKV to influence brain physiology and host behavior following apparently mild or subclinical infection is less well understood. Furthermore, though deficits in cognitive function are well-documented after recovery from neuroinvasive viral infection, the potential impact of ZIKV on other host behavioral domains has not been thoroughly explored.
Methods
We used transcriptomic profiling, including unbiased gene ontology enrichment analysis, to assess the impact of ZIKV infection on gene expression in primary cortical neuron cultures. These studies were extended with molecular biological analysis of gene expression and inflammatory cytokine signaling. In vitro observations were further confirmed using established in vivo models of ZIKV infection in immunocompetent hosts.
Results
Transcriptomic profiling of primary neuron cultures following ZIKV infection revealed altered expression of key genes associated with major psychiatric disorders, such as bipolar disorder and schizophrenia. Gene ontology enrichment analysis also revealed significant changes in gene expression associated with fundamental neurobiological processes, including neuronal development, neurotransmission, and others. These alterations to neurologic gene expression were also observed in the brain in vivo using several immunocompetent mouse models of ZIKV infection. Mechanistic studies identified TNF-α signaling via TNFR1 as a major regulatory mechanism controlling ZIKV-induced changes to neurologic gene expression.
Conclusions
Our studies reveal that cell-intrinsic innate immune responses to ZIKV infection profoundly shape neuronal transcriptional profiles, highlighting the need to further explore associations between ZIKV infection and disordered host behavioral states.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12974-022-02460-8.
Introduction
Zika virus (ZIKV) is a mosquito-borne pathogen of global concern [1]. Like many other members of the genus Flaviviridae, ZIKV is both neuroinvasive and neurotropic [2]. Infection of the central nervous system has been linked to diverse clinical syndromes, including severe congenital neurodevelopmental abnormalities in infants following vertical infection in utero [3–5]. Severe neurologic disease is less frequent in adults, though cases of encephalitis, myelitis, and, more commonly, peripheral neuropathy have been reported [2, 6]. While research on ZIKV pathogenesis to date has heavily focused on severe neurologic disease, it remains unclear whether ZIKV accesses the central nervous system during mild and/or subclinical infection, though data from animal models suggest this is probable [7–9]. Even in the case of established neuroinvasive infection, the long-term neurologic consequences that follow viral clearance and recovery remain poorly understood. However, recent evidence suggests that a range of potential neurologic sequelae may occur in the postinfectious brain, including changes to host behavior [10–13].
In particular, recovery from neuroinvasive infection by flaviviruses, including ZIKV, has been associated with neurocognitive deficits [14–17]. These effects have been attributed in part to the activities of immune cells, including both T cells and microglia, which act in concert to aberrantly prune neuronal synapses following flavivirus recovery [18–20]. Flavivirus infection has also been shown to alter neurodevelopmental programs [21–24], including adult neurogenesis [25–27], a feature of flavivirus infection that contributes to altered learning and memory following recovery in rodent models [28]. Cognitive decline, including persistent memory loss, is also a common occurrence in human patients recovering from flavivirus encephalitis [29–31].
Despite these insights into the cognitive consequences of flavivirus infection, the potential impact of these viruses on other behavioral domains remains relatively unexplored. The multifaceted impact of neurotropic flaviviruses on a diverse array of neurologic functions suggests that such infections may also promote or exacerbate neuropsychiatric conditions, including mood and psychotic disorders. Indeed, depression is another common behavioral symptom reported in patients recovering from flavivirus infection [32–34]. Case reports have also documented the appearance of psychotic symptoms, including hallucinations, in adult patients infected with ZIKV [35, 36]. However, the cellular and molecular mechanisms that underlie these effects remain unknown. In particular, the potential for ZIKV infection to impact host behavior due to cell intrinsic effects on neuronal gene expression has been relatively unexplored.
In this study, we examined how ZIKV infection in neurons impacted the expression of key neurologic genes that promote homeostatic neural function, as well as genes associated with disordered behavioral states. Transcriptomic profiling of primary cortical neurons following ZIKV infection revealed altered expression of many genes associated with psychiatric disorders, including autism, depression, and schizophrenia. Moreover, unbiased gene ontology enrichment analysis revealed that ZIKV infection disproportionately impacted expression of genes associated with neurotransmission and neurodevelopment. These patterns of altered gene expression were also observed in vivo, using an established model of central nervous system (CNS) ZIKV infection in immunocompetent mice. Observed changes in gene expression were due, at least in part, to innate cytokine signaling via tumor necrosis factor receptor-1 (TNFR1). Our data describe a mechanism linking the cell intrinsic innate immune response to ZIKV with dysregulation of a diverse array of neurologic gene pathways, opening new avenues of inquiry into the effect of flavivirus infection on host behavior.
Materials and methods
Viruses
ZIKV strain MR766 was originally provided by Dr. Andrew Oberst (University of Washington, Seattle, WA, USA). ZIKV-DAKAR-MA was first generated [7] and generously provided by Dr. Michael Diamond (Washington University, St. Louis, MO, USA). Viral stocks were generated by infecting Vero cells (MOI 0.01) and harvesting supernatants at 72hpi. Viral titers of stocks were determined via plaque assay on Vero cells (ATCC, #CCL-81). Cells were maintained in DMEM (Corning #10-013-CV) supplemented with 10% Heat Inactivated FBS (Gemini Biosciences #100-106), 1% Penicillin–Streptomycin-Glutamine (Gemini Biosciences #400-110), 1% Amphotericin B (Gemini Biosciences #400–104), 1% Non-Essential Amino Acids (Cytiva #SH30238.01), and 1% HEPES (Cytiva SH30237.01). Plaque assay basal media was 10X EMEM (Lonza # 12-684F) adjusted to 1X and supplemented with 2% Heat Inactivated FBS (Gemini Biosciences #100-106), 1% Penicillin–Streptomycin-Glutamine (Gemini Biosciences #400-110), 1% Amphotericin B (Gemini Biosciences #400-104), 1% Non-Essential Amino Acids (Cytiva #SH30238.01), and 1% HEPES (Cytiva SH30237.01), 0.75% Sodium Bicarbonate (VWR #BDH9280) and 0.5% Methyl Cellulose (VWR #K390). Plaque assays were developed 4dpi by removal of overlay media and staining/fixation using 10% neutral buffered formalin (VWR #89370) and 0.25% crystal violet (VWR #0528).
Cell culture experiments
Primary cerebral cortical neurons were generated using E15 C57BL/6 J embryos as described [37]. Cells were maintained on cell culture treated multiwell dishes supplemented by coating with 20 μg/mL Poly-L-Lysine (Sigma-Aldrich, #9155). Neurobasal Plus + B-27 supplement was used for all experiments (Thermo-Fisher Scientific, #A3582901). All primary mouse cells were generated using pooled tissues derived from both male and female animals. For ZIKV infection experiments, primary neuron cultures were infected at an MOI of 0.1.
Quantitative real-time PCR
Total RNA from cultured cells was isolated with Qiagen RNeasy mini extraction kit (Qiagen, #74106) following the manufacturer’s protocol. RNA concentration was measured with a Quick Drop device (Molecular Devices). cDNA was subsequently synthesized with qScript cDNA Synthesis Kit (Quantabio, #95048). qPCR was performed with SYBR Green Master Mix (Applied Biosystems, #A25742) using a QuantStudio5 instrument (Applied Biosystems). Cycle threshold (CT) values for analyzed genes were normalized to CT values of the housekeeping gene 18S (CTTarget − CT18S = ΔCT). Data were further normalized to baseline control values (ΔCTexperimental − ΔCTcontrol = ΔΔCT). Primers were designed using Primer3 (https://bioinfo.ut.ee/primer3/) against murine genomic sequences. A list of primer sequences used in the study appears in Additional file 1: Table S1.
Murine models of ZIKV infection
C57BL/6 J mice were bred in-house for all experiments. hSTAT2 knockin mice were originally obtained from Jackson Laboratories (strain 031630) and subsequently bred in-house. All animals were housed under pathogen-free conditions in the animal facilities in Nelson Biological Laboratories at Rutgers University. Both male and female mice were inoculated intracranially (10 µl) with 104 PFU of ZIKV-MR766, intracranially with 101 PFU ZIKV-DAK-MA, or subcutaneously (50 µl) in a rear footpad with 103 PFU ZIKV-DAK-MA, as described previously [7, 8].
Tissue preparation
All tissues harvested from mice were extracted following extensive cardiac perfusion with 30 mL of sterile PBS. Extracted tissues were weighed and homogenized using 1.0 mm diameter zirconia/silica beads (Biospec Products, #11079110z) in sterile PBS for ELISA (VWR #L0119) or TRI Reagent (Zymo, #R2050-1) for gene expression analysis. Homogenization was performed in an Omni Beadrupter Elite for 2 sequential cycles of 20 s at a speed of 4 m/s. Total RNA was extracted using Zymo Direct-zol RNA Miniprep kit, as per manufacturer instructions (Zymo, #R2051).
Flow cytometric analysis
Leukocytes were isolated from whole brains after digestion in 0.05% collagenase A (Sigma-Aldrich, #C0130) and 10 mg/ml DNase I (Sigma-Aldrich, #D4527), then purified via centrifugation in 37% isotonic Percoll (VWR, # 89428-524) as described [38]. Isolated leukocytes from brains were stained as described [39] with fluorescently conjugated antibodies to CD3e (Biolegend, clone 17A2), CD44 (Biolegend, clone IM7), CD19 (Biolegend, clone 6D5), CD107a (Biolegend, clone 1D4B), CD8a (Biolegend, clone 53-6.7), CD4 (Biolegend, clone RM4-5), CD45.2 (Biolegend, clone 104), MHC-II (Biolegend, clone M5/114.15.2), NK1.1 (Biolegend, clone PK136), CD11c (Biolegend, clone N418), F4/80 (Biolegend, clone BM8), CD11b (Biolegend, clone M1/70), Ly6G (Biolegend, clone 1A8), Ly6C (Biolegend, clone HK1.4), CD80 (Biolegend, clone 16-10A1), and Zombie NIR (Biolegend, 423105). Data collection and analysis were performed using an Cytek Northern Lights Cytometer and FlowJo software (Treestar).
ELISA
A TNF-α sandwich ELISA kit (EBioscience, #MTA00B) was used for detection of cytokine levels in cell culture supernatants and brain tissue homogenates. Colorimetric reading of ELISA plates was performed with a microplate reader and Gen5 software (BioTek Instruments, Inc.).
Neutralizing-antibody studies
Neutralizing-antibody studies were performed after 30 min of pretreatment with purified anti-mouse TNFR1 (Invitrogen, # 16-1202-85) and anti-mouse IFNAR1 (Leinco, # I-400) antibodies. IgG isotype antibodies (eBioscience, # eBio299Arm; Leinco, # I-443) were used as controls.
Curation of psychiatric disorder-associated gene list
Genes associated with autism spectrum disorder (ASD) for our bioinformatics study were identified using the Sfari Gene database (https://gene.sfari.org) [40]. The Sfari database includes a ranked list of genes with known associations to ASD. We included genes within the top 3 levels of evidential strength of association (syndromic, category 1, and category 2). All genes within this curation have at least two reported de novo likely-gene-disrupting mutations. We were unable to identify similar database resources for other psychiatric disorders. We thus assembled gene lists for additional disorders by consultation of recent and/or highly cited literature in these areas, including metanalyses and systematic reviews. More information about our gene list can be found in Additional file 1: Table S2.
Microarray analysis
Microarray studies were performed on primary cortical neurons derived from C57BL/6 J mouse embryos following 24 h exposure to PBS or ZIKV-MR766 (MOI 0.1), as described [8]. Microarray analysis was conducted using Agilent Whole Mouse Genome arrays (Agilent Technologies) according to manufacturer’s instructions. Expression values and additional sample metadata are publicly available through the NCBI Gene Expression Omnibus Database (accession: GSE122121).
Bioinformatics and statistical analysis
Secondary analysis of microarray data was performed in GEO2R and the GO Enrichment Analysis tool (geneontology.org). Biological pathways were defined using the PANTHER (Protein Analysis Through Evolutionary Relationships) classification system. Corrected p values (false discovery rate) were determined using the Benjamini & Hochberg procedure. For molecular biology assays, two-way analysis of variance (ANOVA) with Sidak’s correction for multiple comparisons was performed using GraphPad Prism Software v8 (GraphPad Software, San Diego, CA). P < 0.05 was considered statistically significant. Data points in all experiments represent biological replicates unless otherwise noted.
Results
ZIKV infection in neurons dysregulates expression of genes associated with abnormal or pathologic behavioral states
We first assessed whether ZIKV infection in neurons resulted in altered expression of genes associated with abnormal and/or disordered behavioral states. To do so, we first generated a list of 676 genes that have been linked in previous studies to psychiatric disorders, including autism, attention deficit hyperactivity disorder (ADHD), bipolar disorder, major depressive disorder, and schizophrenia [40–87] (Additional file 1: Table S2). This list of genes includes a combination of known risk genes as well as genes associated with behavioral abnormalities in each of the above human disorders and related animal models. While we stress that the list is not designed to be comprehensive or definitive, it serves as a starting point for probing the behavioral consequences of neuronal ZIKV infection. We assessed the impact of ZIKV infection on the expression of these genes using a data set previously published by our group and others in which primary cortical neurons derived from C57BL/6 J mice were infected with 0.01 MOI ZIKV-MR766 [8]. Gene expression in this study was profiled via microarray analysis at 24 h following infection.
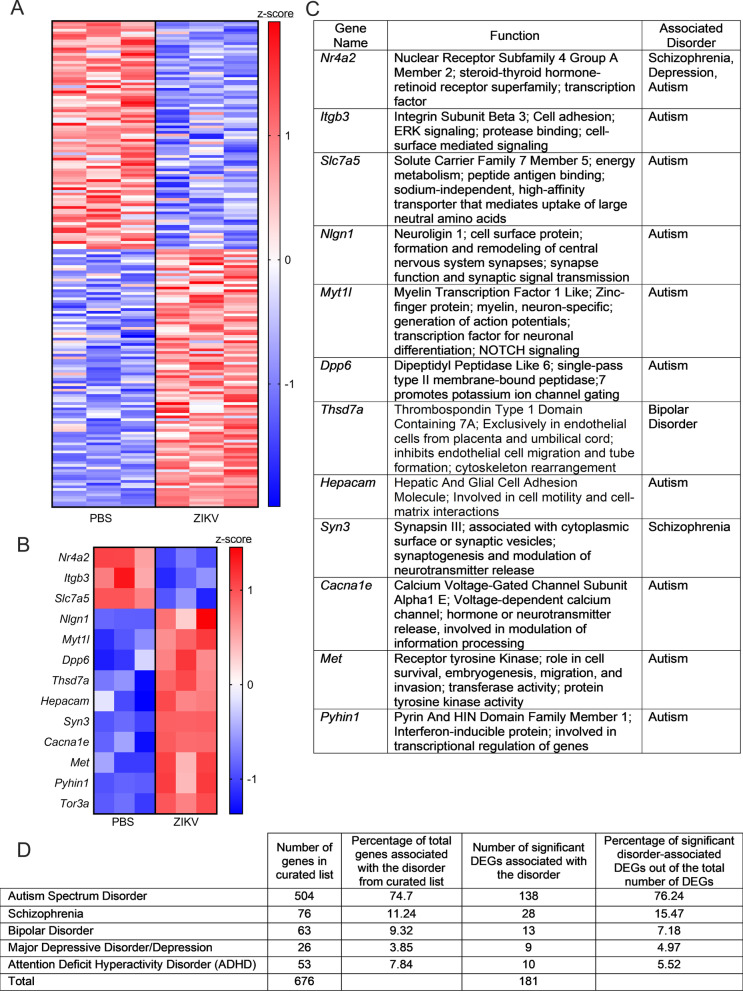
Cross-comparison of the differentially expressed genes (DEGs) from the microarray analysis with our curated gene list revealed that 181 out of 676 (26.8%) genes exhibited significant differential expression following ZIKV infection (Fig. 1A). Of these significant DEGs, 85 (46.96%) were downregulated by ZIKV infection, while 96 (53.04%) were upregulated by ZIKV infection. The 13 genes with the lowest p value in this analysis included Nr4a2, Itgb3, and Slc7a5, which were downregulated by ZIKV infection, along with Nlgn1, Myt1l, Dpp6, Thsd7a, Hepcam, Syn3, Cacna1e, Met, Pyhin1, and Tor3a, which were upregulated by ZIKV infection (Fig. 1B). The functions of these genes are summarized in Fig. 1C, and generally include synaptic function, ion channel physiology, and neurodevelopmental processes. None of the specific psychiatric disorder gene lists were significantly overrepresented among the list of significant DEGs (Fig. 1D). Together, these data suggest that ZIKV infection in neurons dysregulates expression of a broad set of genes associated with abnormal or pathologic behavioral states.
Fig. 1.
ZIKV infection in neurons dysregulates expression of a broad set of genes associated with abnormal or pathologic behavioral states. A Heatmap depicting relative expression values of 181 candidate genes associated with psychiatric disorders. Values are derived from microarray analysis of primary cortical neurons 24 h following ZIKV-MR766 infection (MOI 0.1) or PBS control treatment. B, C Expression values (B) of the top 13 significant differentially expressed genes (DEGs) in our microarray analysis (identified by lowest p values). Table in C lists known functions and associated disorders for these genes. D Descriptive statistics for the curated psychiatric disorder-associated gene list and the significant DEGs observed for each disorder. Data in A and B represent normalized and z-transformed expression values. Data in A include all genes with a False Discovery Rate (FDR) < 0.1
ZIKV infection results in age-dependent changes in gene pathways associated with fundamental neurobiological processes
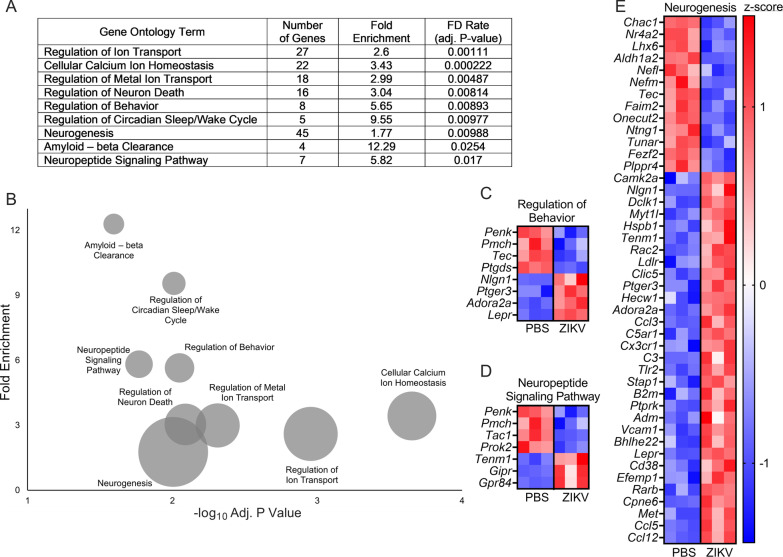
To better understand the consequences of ZIKV infection on neurologic gene expression, we next took an unbiased approach by performing gene ontology (GO) enrichment analysis on the DEGs derived from our microarray data set. Similar to our results using the curated psychiatric disorder-associated gene list, this unbiased analysis revealed significant enrichment of several GO terms related to neurotransmission, neuronal stress responses, and neurodevelopment, a subset of which are highlighted in Fig. 2A, B. Notably, regulatory pathways influencing ion transport and ion homeostasis were particularly enriched in our data set. We next questioned whether there were clear patterns in the direction of differential expression among the significantly enriched GO terms. Heatmaps depicting the expression of genes associated with several representative GO terms are shown in Fig. 2C–E, each of which revealed a mixed set of both up- and down-regulated genes. These findings suggest that the impact of ZIKV infection on the transcriptional state of neurons likely involves complex alterations to a variety of fundamental neurobiological processes.
Fig. 2.
ZIKV infection impacts transcriptional pathways associated with fundamental neurobiological processes. A, B Selected overrepresented GO terms obtained from GO enrichment analysis of DEGs resulting ZIKV-MR766 infection in primary cortical neurons. Tabular results (A) are graphically represented in a bubble plot (B) to demonstrate associations between fold enrichment, FDR, and number of associated genes for each GO term. C–E Heatmaps depicting expression values for genes associated with regulation of behavior (C), neuropeptide signaling pathway (D), and neurogenesis (E). Data in (C), (D) and (E) represent normalized and z-transformed expression values
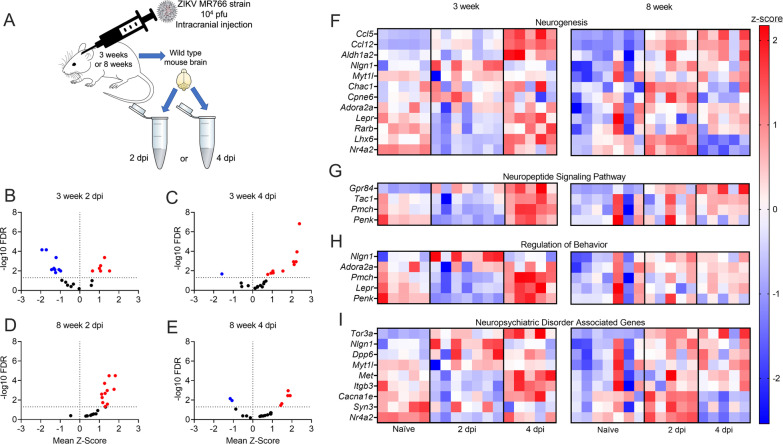
While our microarray data revealed profound alterations to neurologic gene expression in primary neuronal culture following ZIKV infection, we next wanted to assess whether similar changes to gene expression occur in vivo. To do so, we inoculated male and female wildtype (C57BL/6 J) mice intracranially with 104 PFU ZIKV-MR766. We performed these studies in separate cohorts of adolescent (3 weeks) and adult (8 weeks) animals to account for potential differences in the expression of neurologic genes across development (Fig. 3A). On days 2 and 4 following infection, we harvested brains and used qRT-PCR to assess the expression of a panel of genes derived from the top DEGs identified in our microarray analysis. This candidate gene panel included genes from both our curated psychiatric disorder-associated gene list, as well as DEGs from the highly enriched GO terms identified in Fig. 2.
Fig. 3.
Gene expression associated with disease-relevant neurologic pathways is significantly dysregulated following ZIKV infection of the brain in vivo. A Schematic of experimental design of in vivo murine model. C57/BL/6 J WT mice were infected with 104 PFU ZIKV-MR766 via intracranial inoculation at 3 and 8 weeks of age. Whole brain homogenates were collected at 2 or 4 day post-infection (dpi). B-E Expression profiles of 23 candidate genes assessed by qRT-PCR analysis are described and separated into 3-week mice at 2dpi (B), 3-week mice at 4dpi (C), 8-week mice at 2dpi (D), and 8-week mice at 4dpi (E). Significant differences (p < 0.05) are noted in red (upregulated DEGs) or blue (downregulated DEGs). F–I Heatmaps showing individual expression values per mouse for candidate genes from microarray and GO enrichment analyses in 3-week-old adolescent and 8-week-old adult mouse brains following ZIKV-MR766 infection. Relative gene expression values are reported for naïve, 2 dpi, or 4 dpi groups. Data in (F), (G), (H) and (I) represent normalized and z-transformed values of qRT-PCR expression data. n = 5–7 mice/group
These experiments revealed that a majority of genes in our panel did exhibit differential expression at the whole-brain level following intracranial ZIKV infection in vivo. However, the magnitude and direction of differential expression exhibited complex patterns that differed across time post-infection and between 3-week-old and 8-week-old animals. In particular, 3-week-old animals exhibited a mix of significantly downregulated and upregulated expression of selected neurologic genes 2 day post-infection (dpi), with a marked shift to primarily upregulated expression at 4dpi (Fig. 3B, C). In contrast, 8-week-old animals exhibited an essentially inverse pattern, with nearly uniform upregulation of significant DEGs at 2dpi, but a mixture of down- and up-regulated DEGs at 4dpi (Fig. 3D, E). These patterns of differential expression were evident across GO terms, including neurogenesis (Fig. 3F), neuropeptide signaling pathway (Fig. 3G), and regulation of behavior (Fig. 3H), as well as genes taken from our curated list of psychiatric disorder-associated genes (Fig. 3I). Together, these data confirm that gene expression associated with disease-relevant neurologic pathways is significantly dysregulated following ZIKV infection of the brain in vivo, but the mechanisms that control these transcriptional responses are under complex regulation by factors that vary with host age.
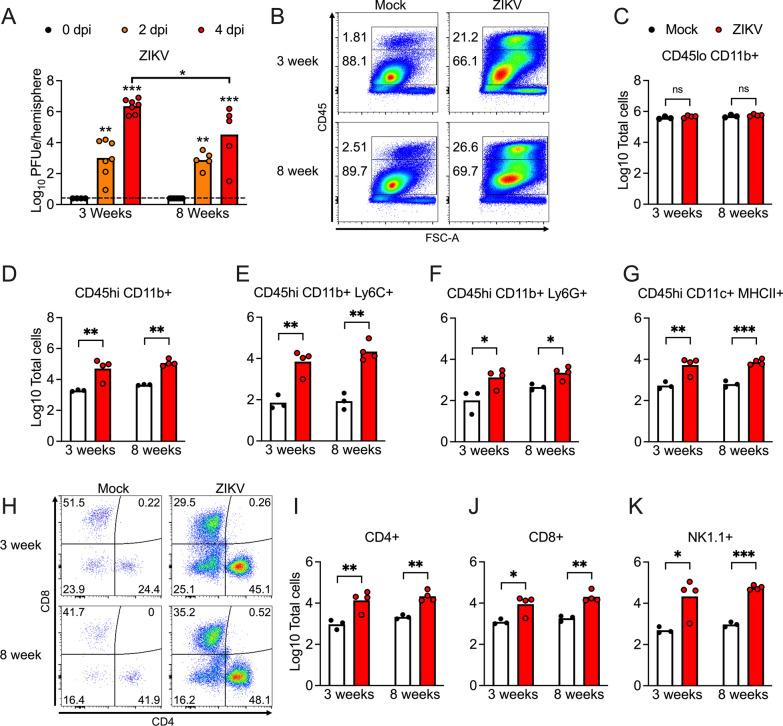
To assess whether differences in virologic burden or inflammation might correspond to age-dependent changes in neurologic gene expression, we profiled viral burden and inflammatory infiltrates following intracranial ZIKV-MR766 infection. Mice of both ages exhibited similar levels of viral RNA in the brain following infection, with 8-week-old mice showing a modest decrease in brain viral burden at 4dpi compared to 3-week-old animals (Fig. 4A). We next performed flow cytometric analysis of infiltrating leukocytes in the brain at 4dpi. While numbers of CD45lo CD11b+ microglia were unchanged between groups, mice of both ages exhibited similarly robust recruitment of CD45hi CD11b+ peripheral immune cells (Fig. 4B–D), including myeloid subsets expressing Ly6C (Fig. 4E) and Ly6G (Fig. 4F). Numbers of CD11c+ MHC-II+ antigen presenting cells were also increased similarly in both age cohorts following infection (Fig. 4G). Mice of both ages also exhibited similar recruitment of lymphocytes to the infected CNS (Fig. 4H), including CD4 T cells (Fig. 4I), CD8 T cells (Fig. 4J), and NK cells (Fig. 4K). Together, these data suggest that differences in neurologic gene expression between adolescent and adult mice following ZIKV infection were not driven by overt differences in CNS viral burden or infiltrating immune cells, suggesting that other age-dependent factors underlie this phenotype.
Fig. 4.
3-week-old and 8-week-old mice exhibit similar neuroinflammatory responses to intracranial ZIKV infection. A qRT-PCR analysis of ZIKV RNA in the brains of mice at indicated day post-infection (dpi) following intracranial infection with ZIKV-MR766 (104 PFU). Values are reported in plaque forming unit-equivalents (PFUe) per hemisphere of brain tissue. n = 5–7 mice/group. B–K Flow cytometric analysis of infiltrating immune cells in brains of mice at 4dpi following intracranial ZIKV-MR766 infection. B Representative flow cytometry plots showing percentages of infiltrating (CD45hi) vs. resident (CD45lo) immune cells. C–G) Quantification of absolute numbers of myeloid cell populations expressing indicated markers. H) Representative flow cytometry plots showing CD45hi CD3+ cells stratified by expression of T cell markers CD4 and CD8. I–K Quantification of absolute numbers of CD45hi CD3+ cell populations expressing indicated markers. n = 3–4 mice/group. * p < 0.05, ** p < 0.01, *** p < 0.001. Bars represent group means
Expression of candidate neurologic genes is significantly dysregulated in in vivo models of mild/asymptomatic ZIKV infection
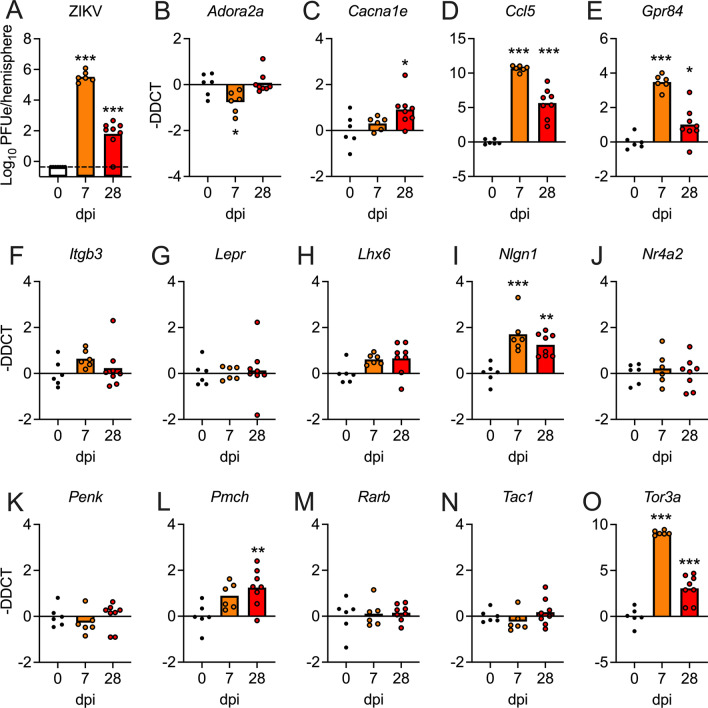
While our transcriptional findings to this point were promising, a limitation of the intracranial ZIKV-MR766 infection model is a high degree of virulence, including uniform mortality around 1 week following infection (data not shown). To better model the mild disease course experienced by most human hosts, we thus repeated key analyses using two separate models of infection. In the first, we performed intracranial inoculation with a low dose (10 PFU) of a mouse adapted African lineage strain (DAK-41525) [7], here referred to as ZIKV-DAK-MA, in 8-week-old C57BL/6 J mice. All mice survive this challenge with no observation of neurologic signs of disease. Infected mice exhibited substantial levels of viral RNA in the brain at 7dpi, with greatly diminished but still detectable levels of viral RNA at 28dpi (Fig. 5A). We next analyzed a panel of our candidate neurologic genes in whole brain homogenates using qRT-PCR (Fig. 5B–O). While the extent and magnitude of gene expression changes were not as robust as the virulent ZIKV-MR766 infection model, we nevertheless saw significant changes in the expression of 7 of 14 genes analyzed, including Adora2 (Fig. 5B), Cacna1e (Fig. 5C), Ccl5 (Fig. 5D), Gpr84 (Fig. 5E), Nlgn1 (Fig. 5I), Pmch (Fig. 5L), and Tor3a (Fig. 5O). Remarkably, differential expression of 6 of these genes persisted out to day 28 following infection. These data suggest that differential expression of neurologic genes following intracranial ZIKV infection was not dependent on severe clinical disease or uncontrolled viral replication within the CNS.
Fig. 5.
Expression of candidate neurologic genes is significantly dysregulated following low dose intracranial inoculation with ZIKV-DAK-MA. A qRT-PCR analysis of ZIKV RNA in the brains of 8-week-old C57BL/6 J mice at indicated day post-infection (dpi) following intracranial inoculation with ZIKV-DAK-MA (101 PFU). Values are reported in plaque forming unit-equivalents (PFUe) per hemisphere of brain tissue. n = 6–8 mice/group. B–O Expression profiles of 14 candidate genes assessed by qRT-PCR analysis on indicated days following intracranial ZIKV-DAK-MA infection. n = 6–8 mice/group. * p < 0.05, ** p < 0.01, *** p < 0.001. Bars represent group means
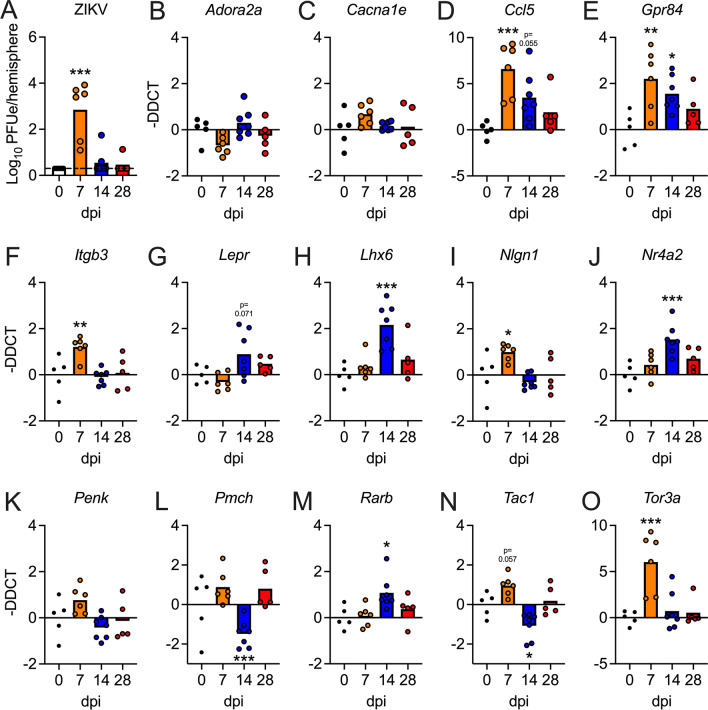
We next assessed whether our target neurologic genes would be impacted using a model of subcutaneous ZIKV infection, which most faithfully models the route of infection experienced by human hosts following the bite of an infected mosquito. As WT mice strongly resist ZIKV infection in the periphery due to an inability of ZIKV to antagonize murine STAT2 signaling, we used a mouse line harboring a homozygous replacement of the murine Stat2 gene with human STAT2 (hSTAT2 knockin) [7]. These mice are normal and immunocompetent but are susceptible to ZIKV-mediated antagonism of STAT2-mediated antiviral signaling, more faithfully modeling the human innate immune response to ZIKV-infection. 5-week-old hSTAT2 mice were infected with 103 PFU ZIKV-DAK-MA in a rear footpad. All mice exhibited detectable viral RNA in the brain at 7dpi, confirming that neuroinvasion occurs in this model of peripheral ZIKV inoculation (Fig. 6A). All mice survived this challenge with no clear exhibition of clinical signs of neurologic disease; moreover, most mice completely cleared infection in the brain by 14dpi (Fig. 6A). Remarkably, qRT-PCR analysis of our neurologic gene panel (Fig. 6B–O) revealed significant differential expression of 10 of 14 genes analyzed, including Ccl5 (Fig. 6D), Gpr84 (Fig. 6E), Itgb3 (Fig. 6F), Lhx6 (Fig. 6H), Nlgn1 (Fig. 6I), Nr4a2 (Fig. 6J), Pmch (Fig. 6L), Rarb (Fig. 6M), Tac1 (Fig. 6N), and Tor3a (Fig. 6O). Many of these genes exhibited differential expression at 14dpi, a timepoint when viral RNA was undetectable in the majority of animals, though all genes had returned to baseline expression by 28dpi. These data suggest that genes associated with neurologic function and neuropsychiatric disorders are impacted in the brain in a physiologic model of subcutaneous ZIKV infection, and that changes to neurologic gene expression persist for some time following the resolution of active CNS infection.
Fig. 6.
Expression of candidate neurologic genes is significantly dysregulated following subcutaneous inoculation with ZIKV-DAK-MA in hSTAT2 knockin mice. A qRT-PCR analysis of ZIKV RNA in the brains of 5-week-old hSTAT2 knockin mice at indicated day post-infection (dpi) following subcutaneous inoculation with ZIKV-DAK-MA (103 PFU). Values are reported in plaque forming unit-equivalents (PFUe) per hemisphere of brain tissue. n = 5–7 mice/group. B-O Expression profiles of 14 candidate genes assessed by qRT-PCR analysis on indicated days following intracranial ZIKV-DAK-MA infection. n = 5–7 mice/group. * p < 0.05, ** p < 0.01, *** p < 0.001. Bars represent group means
Neuronal TNF-α signaling downstream of ZIKV infection dysregulates expression of genes relevant to neuronal function
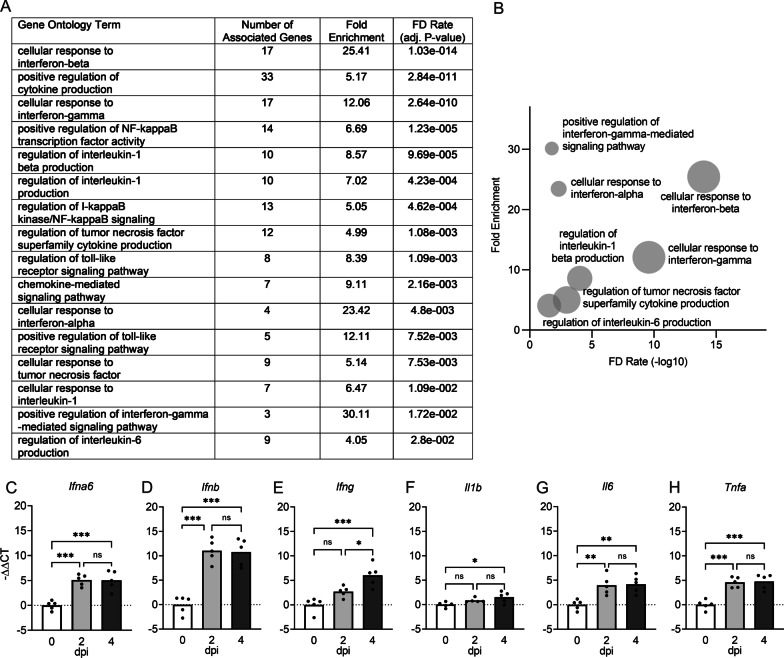
We next questioned whether innate immune activation following ZIKV infection in neurons may be linked to observed changes in neurologic gene expression. To answer this, we returned to our gene ontology enrichment analysis to identify innate immune signaling pathways that were most significantly impacted by neuronal ZIKV infection. Well-known antiviral cytokine responses were among the most enriched pathways in this analysis, particularly those related to innate cytokines, including type I interferon (IFN), interleukin (IL)-1 and IL-6, and tumor necrosis factor (TNF)-α (Fig. 7A, B). Pathways related to signaling by the inflammatory transcription factor nuclear factor kappa B (NF-κB) were also particularly enriched within the list of significant DEGs. We next confirmed that each of these major cytokine responses was induced in the brains of 8-week-old mice following intracranial infection with ZIKV. Expression analysis via qRT-PCR showed that ZIKV infection induced significant upregulation of each of the cytokines analyzed, including Ifna6, Ifnb, Ifng, Il1b, Il6, and Tnfa (Fig. 7C–H). Together, these data confirm that wildtype neurons mount a robust innate immune cytokine response to ZIKV infection, including several cytokines previously established to influence brain function and behavior.
Fig. 7.
Neurons mount a robust innate immune cytokine response to ZIKV infection. A, B Results of GO enrichment analysis of microarray data derived from primary cortical neurons following ZIKV-MR766 infection (MOI 0.1) compared to PBS-treated controls after 24 h. Selected GO terms focus on cytokine activation or inflammatory transcription factor responses. Tabular results (A) are graphically represented in a bubble plot (B) to demonstrate associations between fold enrichment, FDR, and number of associated genes for each GO term. C–H qRT-PCR analysis was performed measuring cytokine genes Ifna6 (C), Ifnb (D), Ifng (E), Il1b (F), Il6 (G), and Tnfa (H) at indicated timepoints in 8-week-old adult mouse brains following intracranial ZIKV-MR766 infection. n = 5. ns not significant, * p < 0.05, ** p < 0.01, *** p < 0.001. Bars represent group means
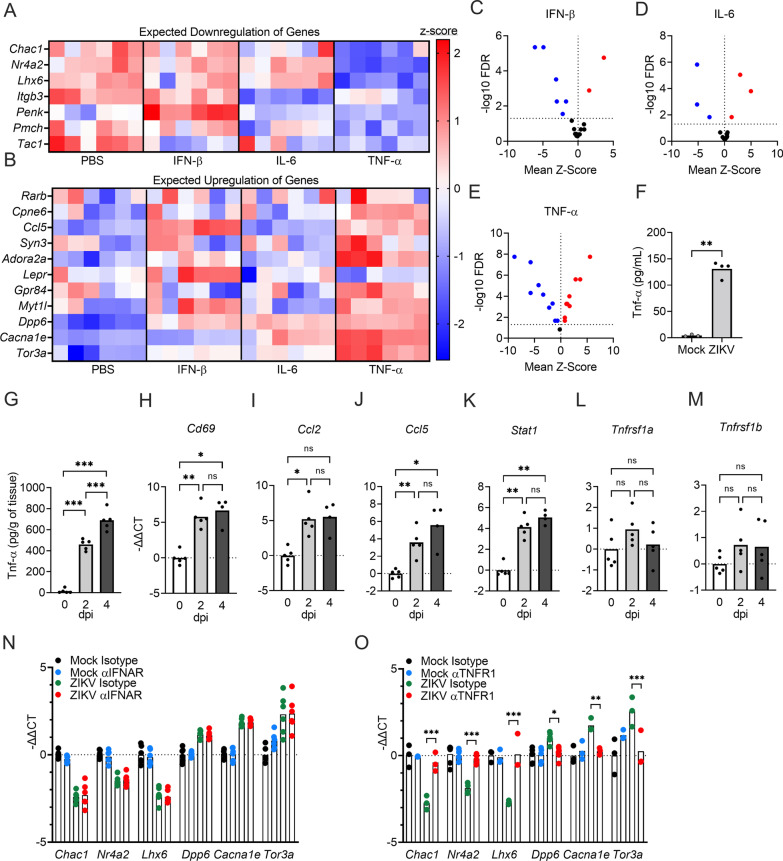
To examine whether neuronal cytokines may be implicated in ZIKV-induced changes in neurologic gene expression, we generated primary cultures of cortical neurons and examined expression of targets in our candidate gene panel following treatment with exogenous cytokines, including IFN-β, IL-6, and TNF-α (each at 10 pg/ml). We then compared the direction of differential expression for each gene to that induced by ZIKV to see which, if any, cytokine most closely phenocopied the pattern of gene expression induced by ZIKV infection. While IFN-β and IL-6 did significantly alter the expression of some target genes, significant DEGs following these treatments did not closely follow the pattern of downregulation (Fig. 8A) or upregulation (Fig. 8B) induced by ZIKV infection. In contrast, TNF-α treatment induced a strikingly similar pattern of differential expression to that induced by ZIKV infection. Moreover, while IFN- β and IL-6 only induced significant changes in expression of a handful of neurologic genes in our analysis (Fig. 8C, D), TNF-α significantly altered 17 out of 18 genes in our panel (Fig. 8E), and of these, all but 2 matched the pattern of up- or downregulated expression observed in ZIKV-infected neuronal cultures. These data identified TNF-α signaling as a promising candidate mechanism for the altered neurologic gene expression observed in the setting of neuronal ZIKV infection.
Fig. 8.
Induction of TNF-α following neuronal ZIKV infection is a major regulatory mechanism that alters expression of genes relevant to neuronal function. A, B Expression values of candidate genes previously shown to be either downregulated (A) or upregulated (B) by ZIKV infection were measured via qRT-PCR in neuronal cultures treated for 24 h with 10 pg/ml exogenous cytokines: IFN-β, IL-6, or TNF-α. n = 6. C–E Volcano plots depicting mean z-score and -log10FDR of representative DEGs in neurons treated with IFN-β (C), IL-6 (D), or TNF-α (E). Significant differences (p < 0.05) are noted in red (upregulated DEGs) or blue (downregulated DEGs). n = 6. F, G) Concentrations of TNF-α in supernatants of in vitro neuronal cell cultures infected with ZIKV for 24 h (n = 4) (F) and brains harvested following in vivo intracranial infection (n = 5) (G). Cytokine concentrations were quantified via ELISA assay. H–M) qRT-PCR analysis was performed for known TNF-α transcriptional target genes, Cd69 (H), Ccl2 (I), Ccl5 (J), Stat1 (K), and TNF-α receptor genes, Tnfrsf1a (L) and Tnfrsf1b (M), in 8-week-old adult mouse brains following ZIKV-MR766 infection at 2 or 4 dpi. n = 5. N, O qRT-PCR analysis of representative DEGs associated with neurological functions (Chac1, Nr4a2, Lhx6, Dpp6, Cacna1e, and Tor3a) following pretreatment with neutralizing antibodies against IFN α/β receptor (IFNAR) (N) or TNFR1 (O) and subsequent 24 h infection with ZIKV-MR766. n = 3–6 biological replicates per group. Data in (A) and (B) represent normalized and z-transformed values of qRT-PCR expression data. ns not significant, * p < 0.05, ** p < 0.01, *** p < 0.001. Bars represent group means
While we previously confirmed that Tnfa was induced at the transcriptional level in the brain in vivo following ZIKV infection, we next wanted to confirm that a robust TNF-α-dependent signature could indeed be observed following infection. We thus performed enzyme-linked immunosorbent assay (ELISA) to confirm that TNF-α was upregulated at the protein level in both supernatants of primary neuronal cultures at 24 h following infection (Fig. 8F) and whole brain homogenates derived from 8-week-old animals on days 2 and 4 following intracranial ZIKV inoculation (Fig. 8G). We also confirmed upregulation of known TNF-α transcriptional targets, including Cd69, Ccl2, Ccl5, and Stat1, in the brains of infected 8-week-old mice (Fig. 8H–K). In contrast, transcript expression of the TNF-α receptors TNFR1 (Tnfrsf1a) and TNFR2 (Tnfrsf1b) were not altered in the brain following infection (Fig. 8L, M), suggesting that enhanced TNF-α signaling in this setting is mediated primarily through induction of cytokine expression. Together, these data confirm that TNF-α signaling is active in both cultured neurons and the brain in vivo following ZIKV infection.
To more carefully assess whether TNF-α signaling was required for ZIKV-mediated alterations to neurologic gene expression, we cultured primary cortical neurons and pretreated with neutralizing antibodies against cytokine receptors for 2 h prior to infection. After 24 h, we then performed qRT-PCR analysis of major DEGs from our microarray analysis to assess the impact of cytokine signaling on ZIKV-induced gene expression. These experiments revealed that blockade of type I IFN signaling via neutralization of the IFN α/β receptor (IFNAR) had no impact on ZIKV-induced changes in expression of the neurologic genes we analyzed (Fig. 8N), findings which mirrored our previous result showing that exogenous IFN-β treatment did not phenocopy ZIKV-induced patterns of expression in our target gene list (Fig. 8A, B). In contrast, blockade of TNFR1 rescued ZIKV-induced changes in each of the 6 genes we analyzed, including Chac1, Nr4a2, Lhx6, Dpp6, Cacna1e, and Tor3a (Fig. 8O). Taken together, these data suggest that the induction of TNF-α following neuronal ZIKV infection is a major regulatory mechanism that alters expression of genes relevant to neuronal function.
Discussion
Emerging flaviviruses represent a significant and growing challenge to global public health. While most famously associated with rare but severe clinical manifestations, including encephalitis, congenital abnormalities, etc., the consequences of apparently mild and/or asymptomatic infection by neuroinvasive flaviviruses remain poorly understood [2, 88]. The observation of behavioral sequelae following recovery from severe flavivirus infections raises the possibility that subclinical neuroinvasive infection may also impact brain function in ways that promote or exacerbate psychiatric disorders. This idea is supported by some case reports [15, 35, 89–91], though, to our knowledge, this hypothesis has not been rigorously tested in the clinical literature. The prevalence of psychiatric sequelae following ZIKV infection, in particular, may be hard to discern due to the relatively low neurovirulence of ZIKV compared to other flaviviruses, resulting in symptoms that may not be severe enough to warrant clinical attention and the documentation of infection status. Our study highlights the need for increased attention to behavioral symptoms in patients who are seropositive for ZIKV and other neuroinvasive flaviviruses, as well as further mechanistic investigation into the cellular and molecular impacts of flavivirus infection on brain physiology and function.
In our study, we show that neurons mount a robust innate cytokine response to ZIKV infection, including a number of cytokines with previously established effects on behavior. A large body of evidence has established that neuroinflammation and inflammatory cytokine signaling is associated with psychiatric disorders, including major depressive disorder [92–95] and schizophrenia [96–98]. TNF-α is a major pleiotropic cytokine induced strongly in the CNS by ZIKV and other flaviviruses [99–102]. Notably, recent work has described complex neuromodulatory effects of TNF-α signaling, including direct effects on glutamatergic neurotransmission [103–105], neuronal differentiation [106–108], and other fundamental neurologic processes [109–111]. In our study, ZIKV-mediated changes to neurologic gene expression greatly overlapped those induced by TNF-α, and gene expression changes induced by ZIKV could be rescued in part by blockade of TNFR1 signaling. These data identify TNF-α as a candidate for further mechanistic investigation of the potential impacts of flavivirus infection on neuronal function.
To date, the most well-described behavioral outcomes of neuroinvasive flavivirus infection in animal models are changes to learning and memory [18, 19, 27, 112–114]. While it is clear that a variety of pathogenic processes related both to viral infection and neuroinflammation can impact cognition, comparatively less attention has been devoted to how flavivirus infection impacts other behavioral domains, including mood, affect, and emotional regulation. This discrepancy is likely due, in part, to technical limitations, including difficulty modeling these behavioral domains in rodents and containment issues related to using ABSL2 and ABSL3 pathogens within behavioral laboratories. Nevertheless, our data identify a need for more robust assessment of behavioral changes in models of flavivirus infection, particularly measures of anxiety, fear/avoidance, and other paradigms with relevance to human psychiatric disorders.
Finally, our data add to a growing body of evidence suggesting that the impact of flavivirus infection varies across the lifespan. In our study, intracranial ZIKV infection resulted in very different impacts on neurologic gene expression in adolescent compared to adult animals, suggesting that developmental factors may significantly influence the neurologic outcomes of flavivirus infection. Recent work has shown that neuroimmune responses to other flaviviruses are impacted by aging [115, 116], and thus differential engagement of cytokine signaling, adaptive immune priming, and blood–brain barrier function may all be relevant variables in determining how flavivirus infection might impact behavior differentially across life stages. Moreover, while the potential for ZIKV to induce severe congenital abnormalities following vertical transmission in utero has now been well established, it remains less clear what the impact of ZIKV infection is on apparently developmentally normal fetuses, including those who are exposed to ZIKV late in gestation, when rates of microcephaly and severe birth defects are exceedingly rare [114, 117, 118]. Further work will be needed to assess whether ZIKV infection in this context may result in changes to neurodevelopment and brain function that impact behavior postnatally and beyond.
Conclusions
Our study describes the impact of ZIKV infection on both immunologic and neurologic transcriptional responses in neurons, adding new insight into how neuronal physiology and function may be altered in the setting of neuroinvasive flavivirus infection. The apparent dysregulation of numerous genes with known associations with human psychiatric disorders, in particular, suggests that more robust analyses of the impact of flavivirus infection on host behavior are warranted.
Supplementary Information
Additional file 1: Table S1. Primer sequences for qRT-PCR. Table S2. Curated psychiatric disorder-associated gene list.
Acknowledgements
Not applicable.
Abbreviations
- ZIKV
Zika virus
- TNF-α
Tumor necrosis factor-alpha
- TNFR1
Tumor necrosis factor-alpha receptor 1
- CNS
Central nervous system
- DMEM
Dulbecco’s modified eagle medium
- FBS
Fetal bovine serum
- HEPES
4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid
- DPI
Day post-infection
- MOI
Multiplicity of infection
- PFU
Plaque forming unit
- PBS
Phosphate buffered saline
- ELISA
Enzyme-linked immunosorbent assay
- IFNAR1
Interferon-alpha receptor 1
- ASD
Autism spectrum disorder
- GO
Gene ontology
- PANTHER
Protein analysis through evolutionary relationships
- ANOVA
Analysis of variance
- ADHD
Attention deficit hyperactivity disorder
- DEG
Differentially expressed gene
- IFN
Interferon
- IL
Interleukin
- NF-κB
Nuclear factor kappa B
- ABSL
Animal biosafety level
- FDR
False discovery rate
Author contributions
Conceptualization: PLK and BPD; Investigation and analysis: PLK, TWC, ML, NPC, IE, BDB, CA, and BPD.; Writing- original draft: PLK and BPD; Writing- review and editing: PLK, TWC, NPC, CA, and BPD; Supervision: CA and BPD; Funding acquisition: BPD. All authors read and approved the final manuscript.
Funding
This work was supported by NIH Grant R21 MH125034 and startup funds from Rutgers University (to BPD). PLK was supported in part by a Division of Life Sciences Summer Undergraduate Research Fellowship from Rutgers University. IE was supported in part by NIH Grant R25 GM055145 and an NIH Research Supplement to Promote Diversity (R01 NS120895-S2).
Availability of data and materials
All data are available upon reasonable request to the corresponding author. Microarray data used in this study are deposited in NCBI’s Gene Expression Omnibus and can be accessed under accession number GSE122121.
Declarations
Ethics approval and consent to participate
All animal experiments were performed with approval of the Rutgers University Institutional Animal Care and Use Committee (IACUC).
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pierson TC, Diamond MS. The continued threat of emerging flaviviruses. Nat Microbiol. 2020;5(6):796–812. doi: 10.1038/s41564-020-0714-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munoz LS, Parra B, Pardo CA. Neuroviruses emerging in the Americas S. neurological implications of zika virus infection in adults. J Infect Dis. 2017;216(suppl_10):S897–S905. doi: 10.1093/infdis/jix511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freitas DA, Souza-Santos R, Carvalho LMA, Barros WB, Neves LM, Brasil P, et al. Congenital zika syndrome: a systematic review. PLoS ONE. 2020;15(12):e0242367. doi: 10.1371/journal.pone.0242367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marques VM, Santos CS, Santiago IG, Marques SM, Nunes Brasil MDG, Lima TT, et al. Neurological complications of congenital zika virus infection. Pediatr Neurol. 2019;91:3–10. doi: 10.1016/j.pediatrneurol.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Coyne CB, Lazear HM. Zika virus—reigniting the TORCH. Nat Rev Microbiol. 2016;14(11):707–715. doi: 10.1038/nrmicro.2016.125. [DOI] [PubMed] [Google Scholar]
- 6.Schwartzmann PV, Ramalho LN, Neder L, Vilar FC, Ayub-Ferreira SM, Romeiro MF, et al. Zika virus meningoencephalitis in an immunocompromised patient. Mayo Clin Proc. 2017;92(3):460–466. doi: 10.1016/j.mayocp.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 7.Gorman MJ, Caine EA, Zaitsev K, Begley MC, Weger-Lucarelli J, Uccellini MB, et al. An immunocompetent mouse model of zika virus infection. Cell Host Microbe. 2018;23(5):672–85 e6. doi: 10.1016/j.chom.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniels BP, Kofman SB, Smith JR, Norris GT, Snyder AG, Kolb JP, et al. The nucleotide sensor ZBP1 and kinase RIPK3 induce the enzyme IRG1 to promote an antiviral metabolic state in neurons. Immunity. 2019;50(1):64–764. doi: 10.1016/j.immuni.2018.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berry N, Ferguson D, Ham C, Hall J, Jenkins A, Giles E, et al. High susceptibility, viral dynamics and persistence of South American Zika virus in New World monkey species. Sci Rep. 2019;9(1):14495. doi: 10.1038/s41598-019-50918-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raper J, Kovacs-Balint Z, Mavigner M, Gumber S, Burke MW, Habib J, et al. Long-term alterations in brain and behavior after postnatal Zika virus infection in infant macaques. Nat Commun. 2020;11(1):2534. doi: 10.1038/s41467-020-16320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mavigner M, Raper J, Kovacs-Balint Z, Gumber S, O'Neal JT, Bhaumik SK, et al. Postnatal Zika virus infection is associated with persistent abnormalities in brain structure, function, and behavior in infant macaques. Sci Transl Med. 2018;10(435):eaao6975. doi: 10.1126/scitranslmed.aao6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nem de Oliveira Souza I, Frost PS, Franca JV, Nascimento-Viana JB, Neris RLS, Freitas L, et al. Acute and chronic neurological consequences of early-life Zika virus infection in mice. Sci Transl Med. 2018;10(444):eaar2749. doi: 10.1126/scitranslmed.aar2749. [DOI] [PubMed] [Google Scholar]
- 13.Adams Waldorf KM, Olson EM, Nelson BR, Little ME, Rajagopal L. The aftermath of zika: need for long-term monitoring of exposed children. Trends Microbiol. 2018;26(9):729–732. doi: 10.1016/j.tim.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel H, Sander B, Nelder MP. Long-term sequelae of West Nile virus-related illness: a systematic review. Lancet Infect Dis. 2015;15(8):951–959. doi: 10.1016/S1473-3099(15)00134-6. [DOI] [PubMed] [Google Scholar]
- 15.Zucker J, Neu N, Chiriboga CA, Hinton VJ, Leonardo M, Sheikh A, et al. Zika virus-associated cognitive impairment in adolescent, 2016. Emerg Infect Dis. 2017;23(6):1047–1048. doi: 10.3201/eid2306.162029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ripamonti E, Gaffuri M, Molteni F. Cognitive, neuropsychiatric, and motor profile in post tick-borne flaviviral encephalomyelitis. Neurol Sci. 2020;41(12):3759–3760. doi: 10.1007/s10072-020-04531-1. [DOI] [PubMed] [Google Scholar]
- 17.Belaunzaran-Zamudio PF, Ortega-Villa AM, Mimenza-Alvarado AJ, Guerra-De-Blas PDC, Aguilar-Navarro SG, Sepulveda-Delgado J, et al. Comparison of the impact of zika and dengue virus infection, and other acute illnesses of unidentified origin on cognitive functions in a prospective cohort in Chiapas Mexico. Front Neurol. 2021;12:631801. doi: 10.3389/fneur.2021.631801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasek MJ, Garber C, Dorsey D, Durrant DM, Bollman B, Soung A, et al. A complement-microglial axis drives synapse loss during virus-induced memory impairment. Nature. 2016;534(7608):538–543. doi: 10.1038/nature18283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garber C, Soung A, Vollmer LL, Kanmogne M, Last A, Brown J, et al. T cells promote microglia-mediated synaptic elimination and cognitive dysfunction during recovery from neuropathogenic flaviviruses. Nat Neurosci. 2019;22(8):1276–1288. doi: 10.1038/s41593-019-0427-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figueiredo CP, Barros-Aragao FGQ, Neris RLS, Frost PS, Soares C, Souza INO, et al. Zika virus replicates in adult human brain tissue and impairs synapses and memory in mice. Nat Commun. 2019;10(1):3890. doi: 10.1038/s41467-019-11866-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayless NL, Greenberg RS, Swigut T, Wysocka J, Blish CA. Zika virus infection induces cranial neural crest cells to produce cytokines at levels detrimental for neurogenesis. Cell Host Microbe. 2016;20(4):423–428. doi: 10.1016/j.chom.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosa-Fernandes L, Cugola FR, Russo FB, Kawahara R, de Melo Freire CC, Leite PEC, et al. Zika virus impairs neurogenesis and synaptogenesis pathways in human neural stem cells and neurons. Front Cell Neurosci. 2019;13:64. doi: 10.3389/fncel.2019.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brault JB, Khou C, Basset J, Coquand L, Fraisier V, Frenkiel MP, et al. comparative analysis between flaviviruses reveals specific neural stem cell tropism for zika virus in the mouse developing neocortex. EBioMedicine. 2016;10:71–76. doi: 10.1016/j.ebiom.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon KJ, Song G, Qian X, Pan J, Xu D, Rho HS, et al. Zika-virus-encoded NS2A disrupts mammalian cortical neurogenesis by degrading adherens junction proteins. Cell Stem Cell. 2017;21(3):349–58 e6. doi: 10.1016/j.stem.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Saucedo-Cuevas L, Regla-Nava JA, Chai G, Sheets N, Tang W, et al. Zika virus infects neural progenitors in the adult mouse brain and alters proliferation. Cell Stem Cell. 2016;19(5):593–598. doi: 10.1016/j.stem.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016;18(5):587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soung AL, Dave VA, Garber C, Tycksen ED, Vollmer LL, Klein RS. IL-1 reprogramming of adult neural stem cells limits neurocognitive recovery after viral encephalitis by maintaining a proinflammatory state. Brain Behav Immun. 2021 doi: 10.1016/j.bbi.2021.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garber C, Vasek MJ, Vollmer LL, Sun T, Jiang X, Klein RS. Astrocytes decrease adult neurogenesis during virus-induced memory dysfunction via IL-1. Nat Immunol. 2018;19(2):151–161. doi: 10.1038/s41590-017-0021-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray KO, Garcia MN, Rahbar MH, Martinez D, Khuwaja SA, Arafat RR, et al. Survival analysis, long-term outcomes, and percentage of recovery up to 8 years post-infection among the Houston West Nile virus cohort. PLoS ONE. 2014;9(7):e102953. doi: 10.1371/journal.pone.0102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehta R, Soares CN, Medialdea-Carrera R, Ellul M, da Silva MTT, Rosala-Hallas A, et al. The spectrum of neurological disease associated with Zika and chikungunya viruses in adults in Rio de Janeiro, Brazil: a case series. PLoS Negl Trop Dis. 2018;12(2):e0006212. doi: 10.1371/journal.pntd.0006212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carson PJ, Konewko P, Wold KS, Mariani P, Goli S, Bergloff P, et al. Long-term clinical and neuropsychological outcomes of West Nile virus infection. Clin Infect Dis. 2006;43(6):723–730. doi: 10.1086/506939. [DOI] [PubMed] [Google Scholar]
- 32.Murray KO, Resnick M, Miller V. Depression after infection with West Nile virus. Emerg Infect Dis. 2007;13(3):479–481. doi: 10.3201/eid1303.060602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nolan MS, Hause AM, Murray KO. Findings of long-term depression up to 8 years post infection from West Nile virus. J Clin Psychol. 2012;68(7):801–808. doi: 10.1002/jclp.21871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.John CC, Carabin H, Montano SM, Bangirana P, Zunt JR, Peterson PK. Global research priorities for infections that affect the nervous system. Nature. 2015;527(7578):S178–S186. doi: 10.1038/nature16033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Correa-Oliveira GE, do Amaral JL, da Fonseca BAL, Del-Ben CM. Zika virus infection followed by a first episode of psychosis: another flavivirus leading to pure psychiatric symptomatology. Braz J Psychiatry. 2017;39(4):381–2. [DOI] [PMC free article] [PubMed]
- 36.Carteaux G, Maquart M, Bedet A, Contou D, Brugieres P, Fourati S, et al. Zika virus associated with meningoencephalitis. N Engl J Med. 2016;374(16):1595–1596. doi: 10.1056/NEJMc1602964. [DOI] [PubMed] [Google Scholar]
- 37.Daniels BP, Snyder AG, Olsen TM, Orozco S, Oguin TH, 3rd, Tait SWG, et al. RIPK3 restricts viral pathogenesis via cell death-independent neuroinflammation. Cell. 2017;169(2):301–13 e11. doi: 10.1016/j.cell.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szretter KJ, Daniels BP, Cho H, Gainey MD, Yokoyama WM, Gale M, Jr, et al. 2'-O methylation of the viral mRNA cap by West Nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog. 2012;8(5):e1002698. doi: 10.1371/journal.ppat.1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durrant DM, Daniels BP, Klein RS. IL-1R1 signaling regulates CXCL12-mediated T cell localization and fate within the central nervous system during West Nile Virus encephalitis. J Immunol. 2014;193(8):4095–4106. doi: 10.4049/jimmunol.1401192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180(3):568–84 e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol Autism. 2013;4(1):36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arcos-Burgos M, Vélez JI, Solomon BD, Muenke M. A common genetic network underlies substance use disorders and disruptive or externalizing disorders. Hum Genet. 2012;131(6):917–929. doi: 10.1007/s00439-012-1164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arias B, Fabbri C, Serretti A, Drago A, Mitjans M, Gastó C, et al. DISC1-TSNAX and DAOA genes in major depression and citalopram efficacy. J Affect Disord. 2014;168:91–97. doi: 10.1016/j.jad.2014.06.048. [DOI] [PubMed] [Google Scholar]
- 44.Balu DT, Li Y, Puhl MD, Benneyworth MA, Basu AC, Takagi S, et al. Multiple risk pathways for schizophrenia converge in serine racemase knockout mice, a mouse model of NMDA receptor hypofunction. Proc Natl Acad Sci USA. 2013;110(26):E2400–E2409. doi: 10.1073/pnas.1304308110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barkus C, Sanderson DJ, Rawlins JN, Walton ME, Harrison PJ, Bannerman DM. What causes aberrant salience in schizophrenia? A role for impaired short-term habituation and the GRIA1 (GluA1) AMPA receptor subunit. Mol Psychiatry. 2014;19(10):1060–1070. doi: 10.1038/mp.2014.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13(2):197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beaulieu JM, Gainetdinov RR. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol Rev. 2011;63(1):182–217. doi: 10.1124/pr.110.002642. [DOI] [PubMed] [Google Scholar]
- 48.Bhat S, Dao DT, Terrillion CE, Arad M, Smith RJ, Soldatov NM, et al. CACNA1C (Cav1.2) in the pathophysiology of psychiatric disease. Prog Neurobiol. 2012;99(1):1–14. doi: 10.1016/j.pneurobio.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Charney AW, Ruderfer DM, Stahl EA, Moran JL, Chambert K, Belliveau RA, et al. Evidence for genetic heterogeneity between clinical subtypes of bipolar disorder. Transl Psychiatry. 2017;7(1):e993-e. doi: 10.1038/tp.2016.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaste P, Leboyer M. Autism risk factors: genes, environment, and gene-environment interactions. Dialogues Clin Neurosci. 2012;14(3):281–292. doi: 10.31887/DCNS.2012.14.3/pchaste. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen DT, Jiang X, Akula N, Shugart YY, Wendland JR, Steele CJ, et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol Psychiatry. 2013;18(2):195–205. doi: 10.1038/mp.2011.157. [DOI] [PubMed] [Google Scholar]
- 52.Chen Q, Che R, Wang X, O'Neill FA, Walsh D, Tang W, et al. Association and expression study of synapsin III and schizophrenia. Neurosci Lett. 2009;465(3):248–251. doi: 10.1016/j.neulet.2009.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christensen JH, Børglum AD. Modeling the cooperativity of schizophrenia risk genes. Nat Genet. 2019;51(10):1434–1436. doi: 10.1038/s41588-019-0508-6. [DOI] [PubMed] [Google Scholar]
- 54.Cichon S, Mühleisen TW, Degenhardt FA, Mattheisen M, Miró X, Strohmaier J, et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. 2011;88(3):372–381. doi: 10.1016/j.ajhg.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51(1):63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Escamilla MA, Zavala JM. Genetics of bipolar disorder. Dialogues Clin Neurosci. 2008;10(2):141–152. doi: 10.31887/DCNS.2008.10.2/maescamilla. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gordovez FJA, McMahon FJ. The genetics of bipolar disorder. Mol Psychiatry. 2020;25:544+. doi: 10.1038/s41380-019-0634-7. [DOI] [PubMed] [Google Scholar]
- 58.Hawi Z, Segurado R, Conroy J, Sheehan K, Lowe N, Kirley A, et al. Preferential transmission of paternal alleles at risk genes in attention-deficit/hyperactivity disorder. Am J Hum Genet. 2005;77(6):958–965. doi: 10.1086/498174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayman V, Fernandez TV. Genetic insights into ADHD biology. Front Psychiatry. 2018 doi: 10.3389/fpsyt.2018.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hou L, Heilbronner U, Degenhardt F, Adli M, Akiyama K, Akula N, et al. Genetic variants associated with response to lithium treatment in bipolar disorder: a genome-wide association study. Lancet. 2016;387(10023):1085–1093. doi: 10.1016/S0140-6736(16)00143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang J, Perlis RH, Lee PH, Rush AJ, Fava M, Sachs GS, et al. Cross-disorder genomewide analysis of schizophrenia, bipolar disorder, and depression. Am J Psychiatry. 2010;167(10):1254–1263. doi: 10.1176/appi.ajp.2010.09091335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kandaswamy R, McQuillin A, Sharp SI, Fiorentino A, Anjorin A, Blizard RA, et al. Genetic association, mutation screening, and functional analysis of a kozak sequence variant in the metabotropic glutamate receptor 3 gene in bipolar disorder. JAMA Psychiat. 2013;70(6):591–598. doi: 10.1001/jamapsychiatry.2013.38. [DOI] [PubMed] [Google Scholar]
- 63.Kerner B. Genetics of bipolar disorder. Appl Clin Genet. 2014;7:33–42. doi: 10.2147/TACG.S39297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kerner B, Lambert CG, Muthén BO. Genome-wide association study in bipolar patients stratified by co-morbidity. PLoS ONE. 2011;6(12):e28477. doi: 10.1371/journal.pone.0028477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.LeDoux MS. The genetics of dystonias. Adv Genet. 2012;79:35–85. doi: 10.1016/B978-0-12-394395-8.00002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu C, Kanazawa T, Tian Y, Mohamed Saini S, Mancuso S, Mostaid MS, et al. The schizophrenia genetics knowledgebase: a comprehensive update of findings from candidate gene studies. Transl Psychiatry. 2019;9(1):205. doi: 10.1038/s41398-019-0532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lohoff FW. Overview of the genetics of major depressive disorder. Curr Psychiatry Rep. 2010;12(6):539–546. doi: 10.1007/s11920-010-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McMahon F, Detera-Wadleigh S. Genetics of bipolar disorder. 2020. pp. 735–43.
- 70.Mei L, Nave KA. Neuregulin-ERBB signaling in the nervous system and neuropsychiatric diseases. Neuron. 2014;83(1):27–49. doi: 10.1016/j.neuron.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mei L, Xiong W-C. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9(6):437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mill J, Xu X, Ronald A, Curran S, Price T, Knight J, et al. Quantitative trait locus analysis of candidate gene alleles associated with attention deficit hyperactivity disorder (ADHD) in five genes: DRD4, DAT1, DRD5, SNAP-25, and 5HT1B. Am J Med Genet B Neuropsychiatr Genet. 2005;133B(1):68–73. doi: 10.1002/ajmg.b.30107. [DOI] [PubMed] [Google Scholar]
- 73.Mühleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J, et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun. 2014;5(1):3339. doi: 10.1038/ncomms4339. [DOI] [PubMed] [Google Scholar]
- 74.Neves FS, Silveira G, Romano-Silva MA, Malloy-Diniz L, Ferreira AA, De Marco L, et al. Is the 5-HTTLPR polymorphism associated with bipolar disorder or with suicidal behavior of bipolar disorder patients? Am J Med Genet B Neuropsychiatr Genet. 2008;147b(1):114–116. doi: 10.1002/ajmg.b.30563. [DOI] [PubMed] [Google Scholar]
- 75.Ni X, Trakalo JM, Mundo E, Macciardi FM, Parikh S, Lee L, et al. Linkage disequilibrium between dopamine D1 receptor gene (DRD1) and bipolar disorder. Biol Psychiatry. 2002;52(12):1144–1150. doi: 10.1016/s0006-3223(02)01433-6. [DOI] [PubMed] [Google Scholar]
- 76.Pagnamenta AT, Khan H, Walker S, Gerrelli D, Wing K, Bonaglia MC, et al. Rare familial 16q21 microdeletions under a linkage peak implicate cadherin 8 (CDH8) in susceptibility to autism and learning disability. J Med Genet. 2011;48(1):48. doi: 10.1136/jmg.2010.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Palladino VS, McNeill R, Reif A, Kittel-Schneider S. Genetic risk factors and gene–environment interactions in adult and childhood attention-deficit/hyperactivity disorder. Psychiatr Genet. 2019 doi: 10.1097/YPG.0000000000000220. [DOI] [PubMed] [Google Scholar]
- 78.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 79.Poquet H, Faivre L, Chehadeh S, Morton J, McMullan D, Hamilton S. Further evidence for Dlgap2 as strong autism spectrum disorders/intellectual disability candidate gene. Autism-Open Access. 2016 doi: 10.4172/2165-7890.1000197. [DOI] [Google Scholar]
- 80.Schmidt-Kastner R, Guloksuz S, Kietzmann T, van Os J, Rutten BPF. Analysis of GWAS-derived schizophrenia genes for links to ischemia-hypoxia response of the brain. Front Psychiatry. 2020 doi: 10.3389/fpsyt.2020.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shadrina M, Bondarenko EA, Slominsky PA. Genetics factors in major depression disease. Front Psychiatry. 2018;9:334. doi: 10.3389/fpsyt.2018.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51(5):793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thomson PA, Malavasi ELV, Grünewald E, Soares DC, Borkowska M, Millar JK. DISC1 genetics, biology and psychiatric illness. Front Biol. 2013;8(1):1–31. doi: 10.1007/s11515-012-1254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Verrall L, Burnet PWJ, Betts JF, Harrison PJ. The neurobiology of D-amino acid oxidase and its involvement in schizophrenia. Mol Psychiatry. 2010;15(2):122–137. doi: 10.1038/mp.2009.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weber H, Kittel-Schneider S, Heupel J, Weißflog L, Kent L, Freudenberg F, et al. On the role of NOS1 ex1f-VNTR in ADHD—allelic, subgroup, and meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2015;168(6):445–458. doi: 10.1002/ajmg.b.32326. [DOI] [PubMed] [Google Scholar]
- 86.Yoshikawa A, Kushima I, Miyashita M, Toriumi K, Suzuki K, Horiuchi Y, et al. Dysregulation of post-transcriptional modification by copy number variable microRNAs in schizophrenia with enhanced glycation stress. Transl Psychiatry. 2021;11(1):331. doi: 10.1038/s41398-021-01460-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zamanian Azodi M, Rezaei-Tavirani M. Identification of the key genes of autism spectrum disorder through protein-protein interaction network. Galen Med J; 2019; 8(2019): 2019. [DOI] [PMC free article] [PubMed]
- 88.Burger-Calderon R, Bustos Carrillo F, Gresh L, Ojeda S, Sanchez N, Plazaola M, et al. Age-dependent manifestations and case definitions of paediatric Zika: a prospective cohort study. Lancet Infect Dis. 2020;20(3):371–380. doi: 10.1016/S1473-3099(19)30547-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simoes ESAC, Moreira JM, Romanelli RM, Teixeira AL. Zika virus challenges for neuropsychiatry. Neuropsychiatr Dis Treat. 2016;12:1747–1760. doi: 10.2147/NDT.S113037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tucci V, Moukaddam N, Meadows J, Shah S, Galwankar SC, Kapur GB. The forgotten plague: psychiatric manifestations of ebola, zika, and emerging infectious diseases. J Glob Infect Dis. 2017;9(4):151–156. doi: 10.4103/jgid.jgid_66_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Joob B, Wiwanitkit V. Zika virus infection and psychosis. Braz J Psychiatry. 2018;40(1):113. doi: 10.1590/1516-4446-2017-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Euteneuer F, Dannehl K, Del Rey A, Engler H, Schedlowski M, Rief W. Peripheral immune alterations in major depression: the role of subtypes and pathogenetic characteristics. Front Psychiatry. 2017;8:250. doi: 10.3389/fpsyt.2017.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu JJ, Wei YB, Strawbridge R, Bao Y, Chang S, Shi L, et al. Peripheral cytokine levels and response to antidepressant treatment in depression: a systematic review and meta-analysis. Mol Psychiatry. 2020;25(2):339–350. doi: 10.1038/s41380-019-0474-5. [DOI] [PubMed] [Google Scholar]
- 94.Pedrotti Moreira F, Wiener CD, Jansen K, Portela LV, Lara DR, Souza LDM, et al. Childhood trauma and increased peripheral cytokines in young adults with major depressive: population-based study. J Neuroimmunol. 2018;319:112–116. doi: 10.1016/j.jneuroim.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 95.Dowlati Y, Herrmann N, Swardfager W, Liu H, Sham L, Reim EK, et al. A meta-analysis of cytokines in major depression. Biol Psychiatry. 2010;67(5):446–457. doi: 10.1016/j.biopsych.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 96.Fineberg AM, Ellman LM. Inflammatory cytokines and neurological and neurocognitive alterations in the course of schizophrenia. Biol Psychiatry. 2013;73(10):951–966. doi: 10.1016/j.biopsych.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rodrigues-Amorim D, Rivera-Baltanas T, Spuch C, Caruncho HJ, Gonzalez-Fernandez A, Olivares JM, et al. Cytokines dysregulation in schizophrenia: a systematic review of psychoneuroimmune relationship. Schizophr Res. 2018;197:19–33. doi: 10.1016/j.schres.2017.11.023. [DOI] [PubMed] [Google Scholar]
- 98.Reale M, Costantini E, Greig NH. Cytokine imbalance in schizophrenia. From research to clinic: potential implications for treatment. Front Psychiatry. 2021;12:536257. doi: 10.3389/fpsyt.2021.536257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leis AA, Grill MF, Goodman BP, Sadiq SB, Sinclair DJ, Vig PJS, et al. Tumor necrosis factor-alpha signaling may contribute to chronic west nile virus post-infectious proinflammatory state. Front Med (Lausanne) 2020;7:164. doi: 10.3389/fmed.2020.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Daniels BP, Jujjavarapu H, Durrant DM, Williams JL, Green RR, White JP, et al. Regional astrocyte IFN signaling restricts pathogenesis during neurotropic viral infection. J Clin Invest. 2017;127(3):843–856. doi: 10.1172/JCI88720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Olmo IG, Carvalho TG, Costa VV, Alves-Silva J, Ferrari CZ, Izidoro-Toledo TC, et al. Zika virus promotes neuronal cell death in a non-cell autonomous manner by triggering the release of neurotoxic factors. Front Immunol. 2017;8:1016. doi: 10.3389/fimmu.2017.01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen Z, Wang X, Ashraf U, Zheng B, Ye J, Zhou D, et al. Activation of neuronal N-methyl-D-aspartate receptor plays a pivotal role in Japanese encephalitis virus-induced neuronal cell damage. J Neuroinflamm. 2018;15(1):238. doi: 10.1186/s12974-018-1280-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen W, Sheng J, Guo J, Gao F, Zhao X, Dai J, et al. Tumor necrosis factor-alpha enhances voltage-gated Na(+) currents in primary culture of mouse cortical neurons. J Neuroinflamm. 2015;12:126. doi: 10.1186/s12974-015-0349-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Clark IA, Vissel B. Excess cerebral TNF causing glutamate excitotoxicity rationalizes treatment of neurodegenerative diseases and neurogenic pain by anti-TNF agents. J Neuroinflamm. 2016;13(1):236. doi: 10.1186/s12974-016-0708-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pickering M, Cumiskey D, O'Connor JJ. Actions of TNF-alpha on glutamatergic synaptic transmission in the central nervous system. Exp Physiol. 2005;90(5):663–670. doi: 10.1113/expphysiol.2005.030734. [DOI] [PubMed] [Google Scholar]
- 106.Bernardino L, Agasse F, Silva B, Ferreira R, Grade S, Malva JO. Tumor necrosis factor-alpha modulates survival, proliferation, and neuronal differentiation in neonatal subventricular zone cell cultures. Stem Cells. 2008;26(9):2361–2371. doi: 10.1634/stemcells.2007-0914. [DOI] [PubMed] [Google Scholar]
- 107.Keohane A, Ryan S, Maloney E, Sullivan AM, Nolan YM. Tumour necrosis factor-alpha impairs neuronal differentiation but not proliferation of hippocampal neural precursor cells: role of Hes1. Mol Cell Neurosci. 2010;43(1):127–135. doi: 10.1016/j.mcn.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 108.Borsini A, Zunszain PA, Thuret S, Pariante CM. The role of inflammatory cytokines as key modulators of neurogenesis. Trends Neurosci. 2015;38(3):145–157. doi: 10.1016/j.tins.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 109.Heir R, Stellwagen D. TNF-mediated homeostatic synaptic plasticity: from in vitro to in vivo models. Front Cell Neurosci. 2020;14:565841. doi: 10.3389/fncel.2020.565841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Maggio N, Vlachos A. Tumor necrosis factor (TNF) modulates synaptic plasticity in a concentration-dependent manner through intracellular calcium stores. J Mol Med (Berl) 2018;96(10):1039–1047. doi: 10.1007/s00109-018-1674-1. [DOI] [PubMed] [Google Scholar]
- 111.Pozniak PD, Darbinyan A, Khalili K. TNF-alpha/TNFR2 regulatory axis stimulates EphB2-mediated neuroregeneration via activation of NF-kappaB. J Cell Physiol. 2016;231(6):1237–1248. doi: 10.1002/jcp.25219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cornelius ADA, Hosseini S, Schreier S, Fritzsch D, Weichert L, Michaelsen-Preusse K, et al. Langat virus infection affects hippocampal neuron morphology and function in mice without disease signs. J Neuroinflamm. 2020;17(1):278. doi: 10.1186/s12974-020-01951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu YH, Cui XY, Yang W, Fan DY, Liu D, Wang PG, et al. Zika virus infection in hypothalamus causes hormone deficiencies and leads to irreversible growth delay and memory impairment in mice. Cell Rep. 2018;25(6):1537–47 e4. doi: 10.1016/j.celrep.2018.10.025. [DOI] [PubMed] [Google Scholar]
- 114.Stanelle-Bertram S, Walendy-Gnirss K, Speiseder T, Thiele S, Asante IA, Dreier C, et al. Male offspring born to mildly ZIKV-infected mice are at risk of developing neurocognitive disorders in adulthood. Nat Microbiol. 2018;3(10):1161–1174. doi: 10.1038/s41564-018-0236-1. [DOI] [PubMed] [Google Scholar]
- 115.Funk KE, Arutyunov AD, Desai P, White JP, Soung AL, Rosen SF, et al. Decreased antiviral immune response within the central nervous system of aged mice is associated with increased lethality of West Nile virus encephalitis. Aging Cell. 2021;20(8):e13412. doi: 10.1111/acel.13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Montgomery RR. Age-related alterations in immune responses to West Nile virus infection. Clin Exp Immunol. 2017;187(1):26–34. doi: 10.1111/cei.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pecanha PM, Gomes Junior SC, Pone SM, Pone M, Vasconcelos Z, Zin A, et al. Neurodevelopment of children exposed intra-uterus by Zika virus: a case series. PLoS ONE. 2020;15(2):e0229434. doi: 10.1371/journal.pone.0229434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vouga M, Pomar L, Panchaud A, Musso D, Baud D. A critical analysis of the neurodevelopmental and neurosensory outcomes after 2 years for children with in utero Zika virus exposure. Nat Med. 2019;25(11):1641–1642. doi: 10.1038/s41591-019-0630-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primer sequences for qRT-PCR. Table S2. Curated psychiatric disorder-associated gene list.
Data Availability Statement
All data are available upon reasonable request to the corresponding author. Microarray data used in this study are deposited in NCBI’s Gene Expression Omnibus and can be accessed under accession number GSE122121.