Abstract
Background:
The risk factors determining short- and long-term morbidity following acute respiratory infection (ARI) due to respiratory syncytial virus (RSV) in infancy remain poorly understood.
Objectives:
To examine the associations of the upper respiratory tract (URT) microbiome during RSV ARI in infancy with the acute local immune response and short- and long-term clinical outcomes.
Methods:
We characterized the URT microbiome by 16S ribosomal RNA sequencing and assessed the acute local immune response by measuring 53 immune mediators with high-throughput immunoassays in 357 RSV-infected infants. Our short- and long-term clinical outcomes included several markers of disease severity and the number of wheezing episodes in the 4th year of life, respectively.
Results:
We found several specific URT bacterial-immune mediator associations. In addition, the Shannon α-diversity index of the URT microbiome was associated with a higher respiratory severity score (RSS) (β [95% CI]=0.50 [0.13–0.86]), greater odds of a lower ARI (OR [95% CI]=1.63 [1.10–2.43]), and higher number of wheezing episodes in the 4th year of life (β [95% CI]=0.89 [0.37–1.40]). The Jaccard β-diversity index of the URT microbiome differed by level of care required (p=0.04). Furthermore, we found an interaction between the Shannon α-diversity index of the URT microbiome and the first principal component of the acute local immune response on the RSS (p=0.048).
Conclusions:
The URT microbiome during RSV ARI in infancy is associated with the acute local immune response, the disease severity, and the number of wheezing episodes in the 4th year of life. Our results also suggest complex URT bacterial-immune interactions that can impact the severity of the RSV ARI.
Keywords: Airway, bronchiolitis, chemokines, cytokines, growth factors, immune response, infancy, mediation, microbiome, nasopharynx, respiratory syncytial virus, severity, wheezing
Capsule Summary:
The upper respiratory tract microbiome during infant respiratory syncytial virus infection is associated with the acute local immune response, the disease severity, and the number of wheezing episodes in the 4th year of life.
Introduction:
Respiratory syncytial virus (RSV) is one of the most common causes of early-life acute respiratory infections (ARIs) worldwide.1 The majority of infants with RSV ARIs have relatively mild disease; however, RSV continues to be a leading cause of hospitalizations in infancy globally and the risk factors that predispose some RSV-infected infants to develop more severe disease are still unknown.1, 2 To date, there is limited and conflicting evidence on the effect of the URT microbiome on the acute local immune response to RSV and the severity of RSV ARIs in the pediatric population.3–6 Furthermore, although we have previously shown that certain URT microbiome signatures in RSV-infected infants are associated with the risk of recurrent wheeze by age 2 years,7 whether the URT microbiome during RSV ARI in infancy is associated with childhood wheezing illnesses beyond age 2 years is unknown.
Based on our prior studies,6–9 we hypothesized that specific URT microbial signatures in RSV-infected infants are associated with the acute local immune response and short- and long-term clinical outcomes (including several markers of disease severity and the number of wheezing episodes in the 4th year of life). Using a combination of next-generation sequencing, high-throughput immunoassays, novel bioinformatics, and a causal mediation framework,10 we tested this hypothesis in 357 RSV-infected infants enrolled the Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Infection in Infancy study (INSPIRE).
Methods:
Full details are available in the E-Methods section of the Online Supplement.
Overview of the Study Population and Design
INSPIRE is a large (n=1,949), population-based, birth cohort of previously healthy, term, non-low-birth-weight infants born between June and December of 2012 and 2013 in the middle Tennessee region. The Institutional Review Board of Vanderbilt University Medical Center approved this study. The detailed methods for INSPIRE have been previously reported.10
To capture symptomatic RSV ARIs throughout infancy (i.e., between 0–12 months of age) in children enrolled in INSPIRE —including the first, which is usually the most severe11—, we conducted intensive passive and active surveillance during each infant’s first RSV season (November to March in our region2, 12). If an infant met pre-specified criteria for an ARI, we then conducted an in-person respiratory illness visit that included a parental questionnaire, a physical exam, collection of a nasal wash and other biospecimens, and —for infants who required a health care encounter— a structured medical chart review.
For the current study and based on our hypothesis of interest, we only included infants enrolled in INSPIRE who had ≥1 RSV ARI during their first RSV season (Figure 1). The detection of RSV, human rhinovirus, and human enterovirus was performed by quantitative RT-PCR, as previously described.13 For infants with multiple RSV ARIs, only data from the first RSV ARI available was included in statistical analyses.
Figure 1.
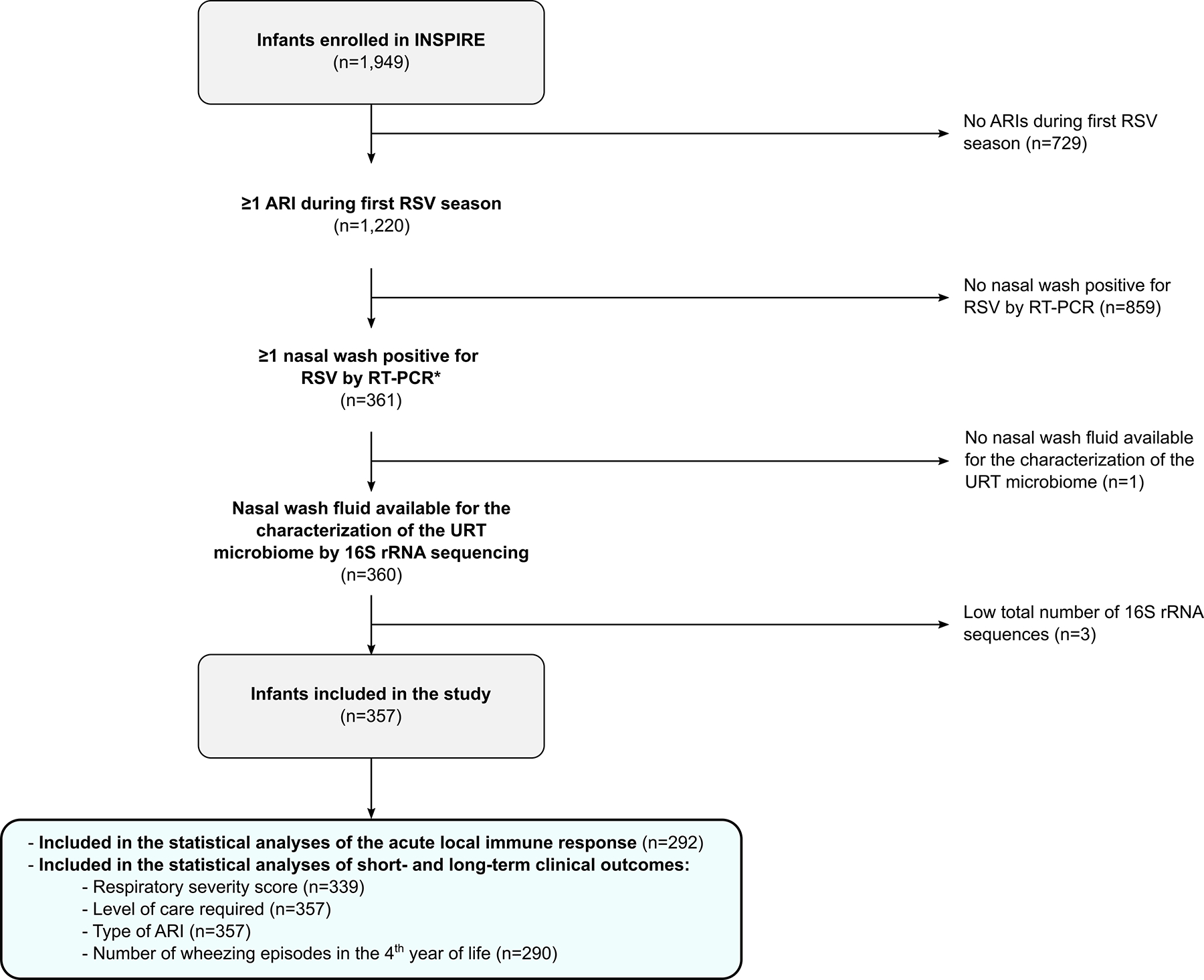
Flow chart diagram of infants with RSV ARI included in the study. The total sample size at each step of the flow chart diagram (n) is shown. *For infants with multiple RSV ARIs, only data from the first RSV ARI available were included in statistical analyses. Definition of abbreviations: ARI = Acute respiratory infection, INSPIRE = Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure Study, rRNA = Ribosomal ribonucleic acid, RSV = Respiratory syncytial virus, URT = Upper respiratory tract.
Characterization of the Upper Respiratory Tract Microbiome
The methods used to characterize the URT microbiome in infants enrolled in INSPIRE have been previously described in detail.6–8, 14 In brief, following bacterial DNA extraction from the nasal wash obtained during the RSV ARI, we amplified the V4 region of the 16S ribosomal ribonucleic acid (rRNA) gene using universal primers. The libraries were then sequenced on an Illumina MiSeq platform with 2×300 base pair reads. Negative and positive controls (with known taxonomic composition) were amplified and sequenced concurrently for quality control. Next, we processed the 16S rRNA sequences using the R package dada2 by following its standard operating procedure.15 To this end, we grouped sequences into amplicon sequence variants and assigned taxonomy using the SILVA reference database.16 Low-quality sequences, chimeras, and non-bacterial sequences were discarded as part of the dada2 pipeline. We subsequently processed the remaining sequences using the R package decontam to remove any suspected contaminants that were found in the negative controls.17 We also discarded samples with <200 sequences (n=3). Last, we calculated the relative abundances of individual taxa using simple proportions.
Characterization of the Acute Local Immune Response
To characterize the acute local immune response, we measured the levels of 53 immune mediators (cytokines, chemokines, and growth factors [Table E1]) in the nasal wash obtained during the RSV ARI using 2 multiplex magnetic bead-based assays, as previously described.6, 9 The assays were all conducted in duplicate using a Luminex MAGPIX platform with the xMAP technology as per manufacturers’ instructions. The Luminex xMAP data was processed using the method described by Won et al,18 which uses median fluorescence intensities (MFIs) of individual beads instead of the usual standard curve-based data-processing method to increase the sensitivity and accuracy of high-throughput immunoassays.6, 9, 18 To improve data distribution, we applied generalized log and inverse normal transformations prior to statistical analyses.18
Definition of Clinical Outcomes
Our short- (infant) and long-term (4-year) clinical outcomes included 1) markers of RSV ARI severity, such as the respiratory severity score (RSS),19, 20 type of ARI (upper vs. lower), and level of care required (none, office sick visit, urgent care or emergency department, or hospitalization), and 2) parental report of the number of wheezing episodes in the 4th year of life. The RSS ranges from 0 to 12, with higher scores indicating more severe disease.19, 20
Statistical Analyses
The framework for our statistical analyses is shown in Figure 2. Statistical analyses were mostly performed in R version 3.6.2.21 First, we rarefied the processed dataset to the lowest library size of all samples (n=221) and calculated pre-specified, common richness (Chao1 estimator), α-diversity (Shannon and inverse Simpson indices), and β-diversity (Bray-Curtis [abundance-based] and Jaccard [presence/absence-based] indices) metrics of the URT microbiome at the amplicon sequence variant level.22 In brief, the α-diversity takes into account the richness and evenness of a community, whereas the β-diversity reflects its overall structure.
Figure 2.
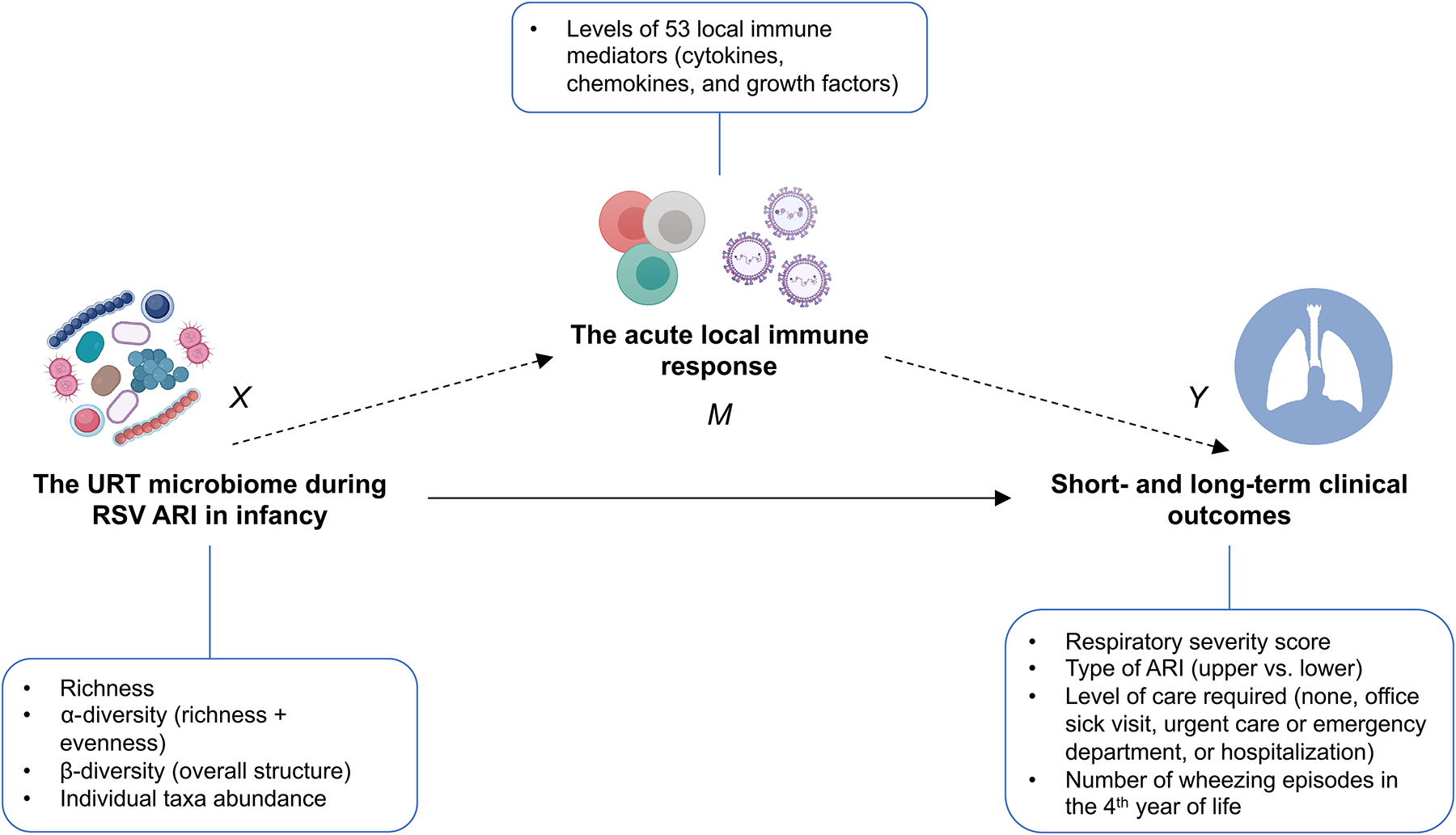
Graphical representation of the study design. First, we examined the total effect of the URT microbiome during RSV ARI in infancy on the acute local immune response (path X→M) and on short- and long-term clinical outcomes (all paths X→Y). Based on our results and those of our prior studies showing an effect of the acute local immune on short- and long-term clinical outcomes (path M→Y), we the used a causal mediation framework to assess whether the effect of the URT microbiome during RSV ARI on short- and long-term clinical outcomes (all paths X→Y) could be explained —at least in part— by the acute local immune response (path X→M→Y [indirect effect]). Our causal mediation models also took into account the interaction between the exposure and the mediator (X and M, respectively). Definition of abbreviations: ARI = Acute respiratory infection, RSV = Respiratory syncytial virus, URT = Upper respiratory tract.
For the evaluation of the relations between richness or α-diversity metrics with all outcomes we used linear or logistic regression, as appropriate. To examine the association of β-diversity metrics with all outcomes, we used the PERMANOVA test, as implemented in the R package vegan.23 For taxa abundance testing, we conducted statistical analyses at the genus level using a non-rarified dataset. To identify whether the abundances of particular genera were associated with a local immune mediator MFI, we performed variable selection using a regression method based on the sparse linear log-contrast model.24 For each immune mediator, we built a separate model including its MFI as the dependent variable and the log-transformed relative abundances of the top 30 most abundant genera (which together represented 94.78% of the sequences) as the independent variables. To obtain robust results, we generated 100 bootstrapped samples and used a 10-fold cross-validation procedure to select the genera related to an individual local immune mediator MFI. The genera reported to be associated with a particular immune mediator MFI are those selected over 70 times out of the 100 bootstrap replicates. To identify whether the abundances of particular genera were associated with short- and long-term clinical outcomes, we used the R package miLineage.25
Based on our results and those of our prior studies,6, 7, 26 we then assessed whether the acute local immune response underlies the association between the URT microbiome and short- and long-term clinical outcomes following RSV ARI in infancy. For this, we built causal mediation models that included the Shannon index of the URT microbiome as the exposure, the first 2 components from principal component (PC) analysis of the acute local immune response data as joint mediators, and each short- and long-term clinical outcome as the outcome.27 To examine the interplay between the URT microbiome and the acute local immune response, the mediation analyses also took into account the interaction between the exposure and the mediator (Figure 2).27
For all statistical analyses, we included infant- (age at the time of the RSV ARI, sex, race and ethnicity, ever breastfeeding, exposure to antibiotics in utero or prior to enrollment, exposure to tobacco smoke in utero or prior to enrollment, and maternal asthma) and RSV-related (viral load, number of days from symptom onset to nasal wash collection, and detection of other viruses) covariates in the models. Statistical significance was defined as p<0.05 after controlling for multiple comparisons using the Benjamini-Hochberg procedure, as appropriate (see full details in the E-Methods section of the Online Supplement).28
Results:
Baseline Characteristics of the Study Population
Three hundred fifty-seven (98.89%) of the 361 infants with ≥1 RSV ARI enrolled in INSPIRE had 16S rRNA data that passed our quality control metrics and were thus included in statistical analyses (Figure 1). The baseline characteristics of these infants are shown in Table 1. The median (interquartile range [IQR]) age at the time of the RSV ARI was 4.67 (2.73–6.02) months. The majority of infants had RSV-only ARIs and were males, White non-Hispanic, and born by vaginal delivery. The median (IQR) RSS was 3 (2–4) and 168 (47.06%) infants had a lower ARI. The number (%) of infants requiring no health care encounter, an office sick visit, an urgent care or emergency department visit, or hospitalization was 78 (21.85%), 198 (55.46%), 28 (7.84%), and 53 (14.85%), respectively. Of the 53 RSV-infected infants requiring hospitalization, 2 (5.00%) required observation (i.e., hospital admission for <24 hours), 25 (48.08%) required supplemental oxygen, and 4 (7.55%) required admission to the pediatric intensive care unit. The median (IQR) length of stay in those hospitalized was 2 (1–4) days. The RSS was positively associated with the level of care required (p<0.001) (Figure E1), as expected.
Table 1.
| Age at enrollment (days) | 59 (17–83) |
| Age at the time of the RSV ARI (months) | 4.67 (2.73–6.02) |
| Female sex | 161 (45.10%) |
| Race or ethnicity | |
| Black non-Hispanic | 57 (15.97%) |
| White non-Hispanic | 239 (66.95%) |
| Hispanic | 32 (8.96%) |
| Other | 29 (8.12%) |
| Gestational age (weeks) | 39.00 (38.30–40.00) |
| Birth weight (grams) | 3,433.15 (3,120.90–3,745.40) |
| Birth by cesarean section | 119 (33.33%) |
| Ever breastfeeding | 270 (75.63%) |
| Daycare attendance | 148 (44.31%) |
| Presence of another child age ≤6 years living in the same home | 209 (58.54%) |
| Exposure to tobacco smoke in utero or prior to enrollment | 83 (23.25%) |
| Exposure to antibiotics in utero or prior to enrollment | 193 (54.06%) |
| Maternal asthma | 69 (19.33%) |
| Type of insurance | |
| Federal or state | 181 (50.70%) |
| Private | 173 (48.46%) |
| Other | 3 (0.84%) |
| RSV cycle threshold value by RT-PCR | 24.37 (20.63–27.87) |
| Number of days from symptom onset to nasal wash collection | 3 (2–4) |
| HRV or HEV co-infection by RT-PCR | |
| RSV only | 302 (84.59%) |
| RSV + HRV | 50 (14.01%) |
| RSV + HEV | 1 (0.28%) |
| RSV + HRV + HEV | 4 (1.12%) |
The data are presented as median (interquartile range) for continuous variables or number (%) for categorical variables.
The estimates were calculated for infants with complete data.
Definition of abbreviations: ARI = Acute respiratory infection, HEV = Human enterovirus, HRV = Human rhinovirus, RSV = Respiratory syncytial virus.
Two hundred ninety (81.23%) infants had 4-year data available and 62 of these (21.38%) had ≥1 wheezing episode in the 4th year of life. In comparison to those without 4-year data, infants with 4-year data were more likely to have ever breastfed, to have attended daycare, and to have private insurance, and less likely to have been exposed to tobacco smoke in utero or prior to enrollment. Infants with 4-year data also had higher viral load (i.e., lower cycle threshold values by RT-PCR) at the time of the RSV ARI (Table E2).
The Association of the Upper Respiratory Tract Microbiome with the Acute Local Immune Response
The median (IQR) high-quality sequence count per sample was 10,513 (6,579–14,692) among all RSV-infected infants. The richness of the URT microbiome was not associated with the levels of any of the 53 local immune mediators (p>0.05 for all comparisons) (Table E3). In contrast, its α-diversity (both the Shannon and inverse Simpson indices) was negatively associated with, and its β-diversity (particularly the Jaccard index) differed according to, the levels of multiple immune mediators (Table E3 and Table E4). In taxa abundance testing, we found several specific taxa-immune mediator associations (Figure 3 and Table E5). The abundances of Klebsiella and Moraxella were positively associated with the levels of type 2 (IL-13 and IL-4, respectively), type 3 (IL-17A), and pro-inflammatory (TNF-α, IL-1β, IL-6, IL-8, and MCP-1) immune mediators. We found similar —albeit less consistent— direct associations for other respiratory pathogens, such as Haemophilus (positively associated with the levels of IL-17A, TNF-α, IL-1β, IL-6, and MCP-1) and Streptococcus (positively associated with the levels of TNF-α and IL-8). In contrast, we found the abundance of other genera to be inversely associated with the levels of multiple immune mediators, such as Methylobacterium, which was negatively associated with the levels of type 1 (e.g., IFN-γ, IL-2, and IL-12), type 2 (e.g., IL-4, IL-5, and IL-13), type 3 (e.g., IL-17A), and pro-inflammatory (e.g., TNF-α, IL-1β, IL-6, IL-8, MCP-1, and MIP-1α) immune mediators.
Figure 3:
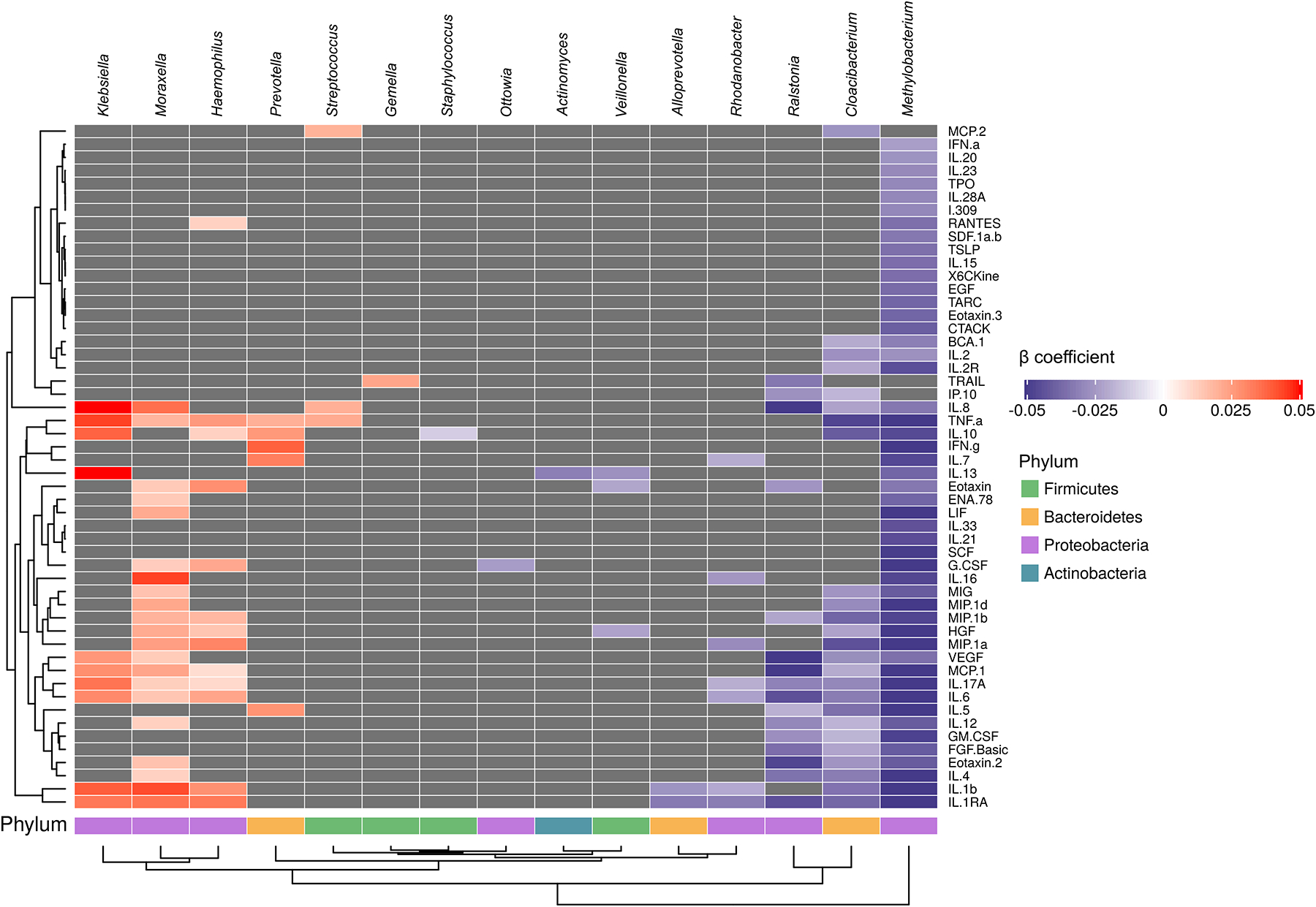
The association of upper respiratory tract microbiome genera during an acute respiratory infection due to respiratory syncytial virus in infancy with the acute local immune response. To identify relevant associations, we used a variable selection method. For each immune mediator, we built a separate model including its median fluorescence intensity as the dependent variable and the log-transformed relative abundances of the top 30 most abundant genera as the independent variables. Every model also included infant- and respiratory syncytial virus-related (see main text for more details). Only genera that were selected for ≥1 immune mediator and immune mediators for which ≥1 genera were selected are shown. The color of each cell represents the regression coefficient from the models (blue = negative association, red = positive association, dark grey = no association). The heatmap columns (genera) and rows (immune mediators) were hierarchically clustered. The corresponding phyla are also shown.
The Association of the Upper Respiratory Tract Microbiome During Respiratory Syncytial Virus Infection in Infancy with Short- and Long-term Clinical Outcomes
In linear or logistic regression analyses, the richness of the URT microbiome was not associated with either markers of RSV ARI severity or the number of wheezing episodes in the 4th year of life (p>0.05 for all comparisons) (Figure E2). In contrast, its α-diversity was positively associated with an increased RSS (linear regression coefficient [95% CI, p-value] for Shannon and inverse Simpson indices = 0.50 [0.13 to 0.86, p=0.02] and 0.15 [0.04 to 0.25, p=0.01], respectively), greater odds of a lower ARI (OR [95% CI, p-value] for Shannon and inverse Simpson indices = 1.63 [1.10 to 2.43, p=0.02] and 1.18 [1.05 to 1.34, p=0.01]), and higher number of wheezing episodes in the 4th year of life (linear regression coefficient [95% CI, p-value] for Shannon and inverse Simpson indices = 0.89 [0.37 to 1.40, p=0.003] and = 0.31 [0.15 to 0.47, p<0.001], respectively) (Figure 4 and Figure E3). To explore the possibility of non-linear associations, we also included the square term of each α-diversity metric in every model, but none of these were statistically significant (p>0.05 for all terms). We also obtained similar results in sensitivity analyses using linear regression and applying an inverse-normal transformation to the RSS, utilizing ordinal regression for the level of care required, or using Poisson regression for the number of wheezing episodes (data not shown).
Figure 4:
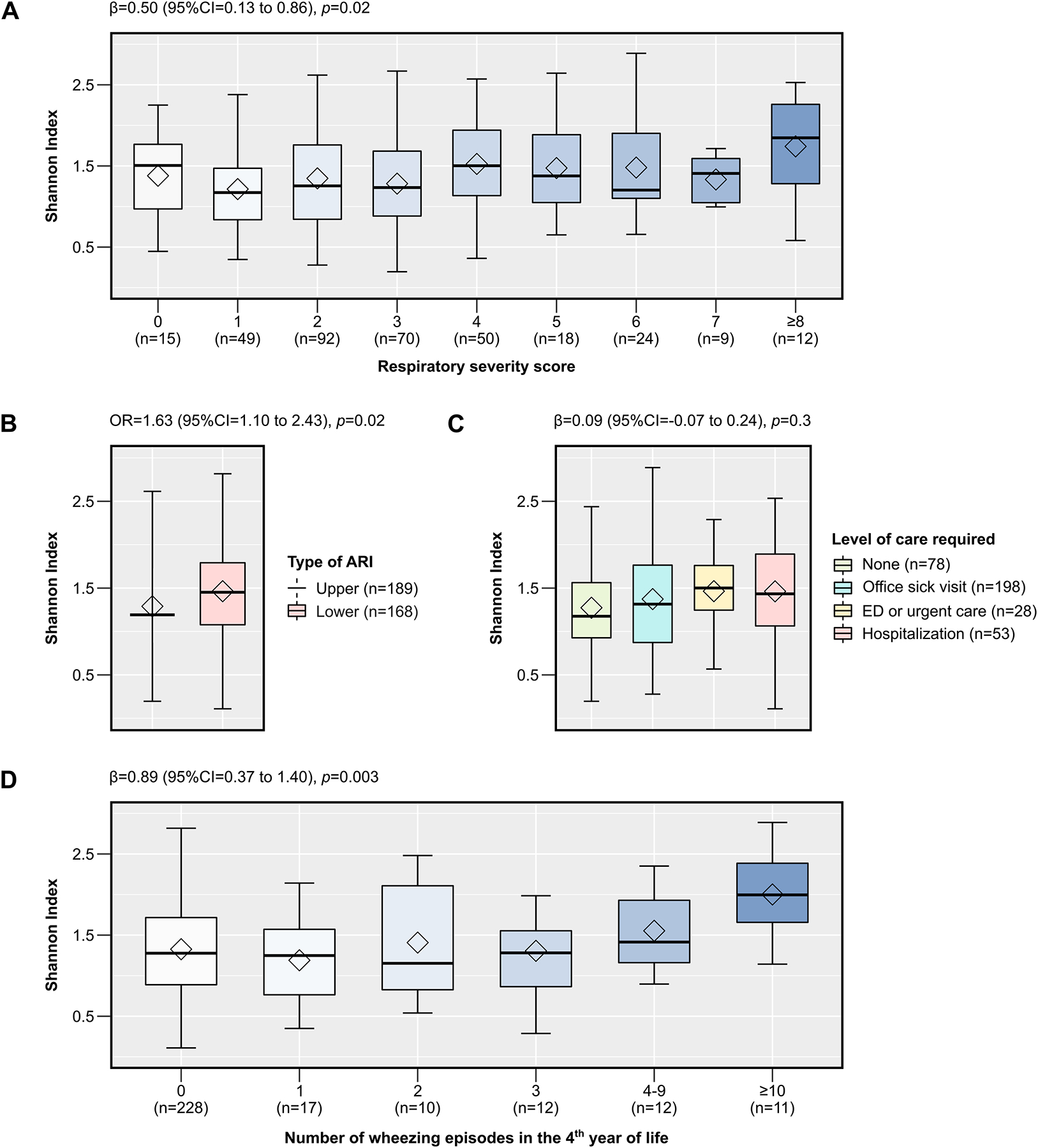
The association of the α-diversity of the upper respiratory tract microbiome during RSV ARI in infancy with short- and long-term clinical outcomes, including the respiratory severity score (A), type of ARI (B), level of care required (C), and number of wheezing episodes in the 4th year of life (D). The box-and-whisker plots show the mean (diamond), median (middle bar), 1st quartile (lower bar), 3rd quartile (upper bar), minimum observation above the lowest fence (lower whisker), and maximum observation below the upper fence (upper whisker) of the Shannon index for each outcome. The total sample size for each outcome (n) and the estimates from corresponding linear or logistic regression models (regression coefficient [β] or OR, 95% CI, and p-value) are also shown, as appropriate. Every model also included infant- and RSV-related covariates (see main text for more details). The Benjamini-Hochberg procedure was used to control for multiple comparisons by adjusting the p-values for the number of clinical outcomes assessed (i.e., controlling for a total of 4 tests). Definition of abbreviations: ARI = Acute respiratory infection, RSV = Respiratory syncytial virus.
In regard to the β-diversity of the URT microbiome, the Bray-Curtis index of the URT microbiome was not associated with any of the short- or long-term clinical outcomes, whereas the Jaccard index only differed by level of care required (p=0.04) (Figure 5 and Table E6). In taxa abundance testing, no particular genera were found to be associated with any of the short- or long-term clinical outcomes (p>0.05 for all genera).
Figure 5:
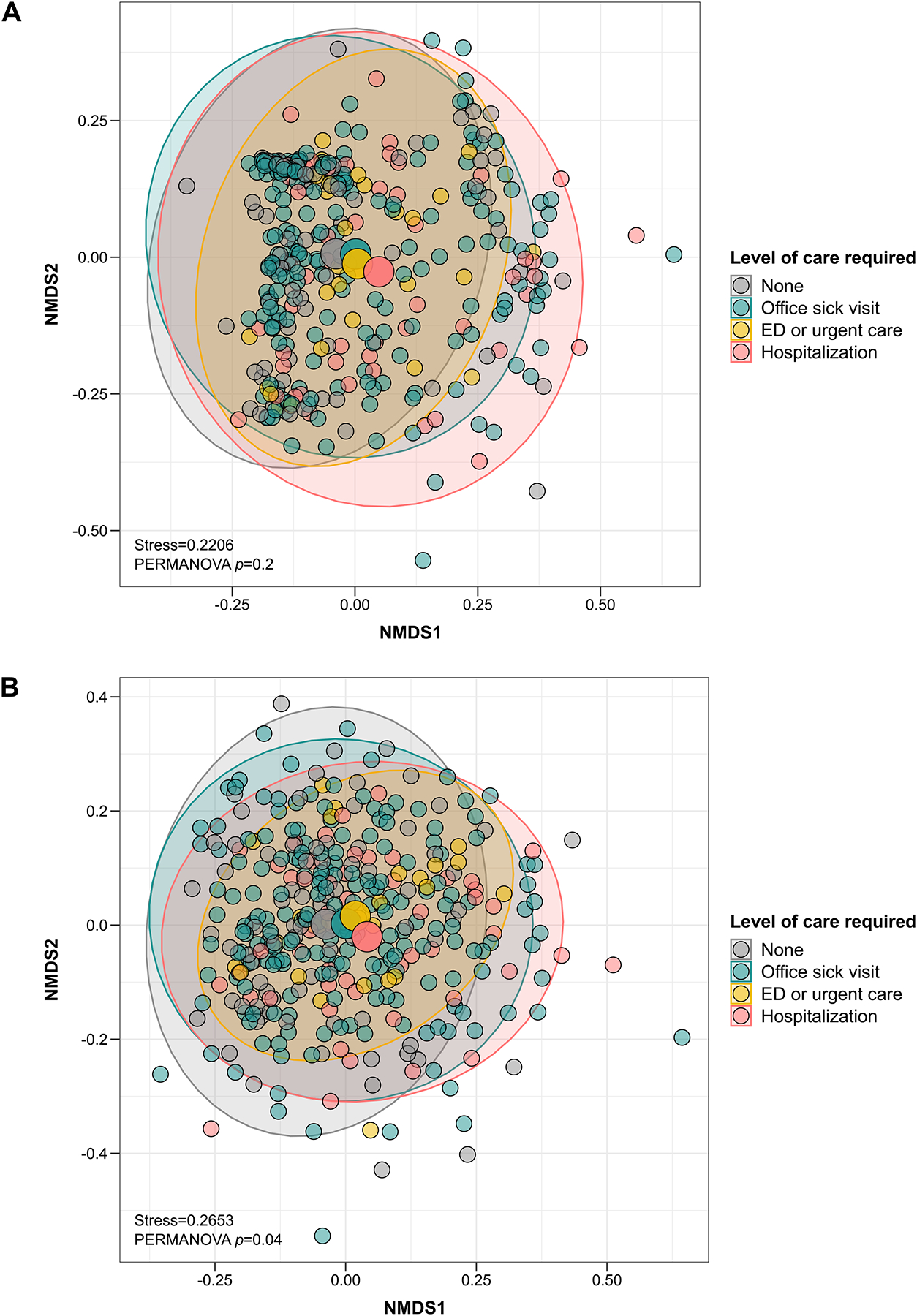
The association of the β-diversity of the URT microbiome during an acute respiratory infection due to respiratory syncytial virus in infancy with level of care required. The scatter plots show each infant’s microbial community composition (small circles) by level of care required, as well as each group’s centroid (large circles) and 95% CI ellipses. The scatter plots were generated using NMDS ordination based on Bray-Curtis (A) or Jaccard (B) indices. For ease of visualization, only 2 dimensions were used; however, the stress values of ≥0.2 suggest that a higher number of dimensions may be needed to accurately ordinate and represent the multidimensional data. The p-values for the comparison between groups from PERMANOVA tests are also shown. Every model also included infant- and respiratory syncytial virus-related covariates (see main text for more details). For each β-diversity index, the Benjamini-Hochberg procedure was used to control for multiple comparisons by adjusting the p-values for the number of clinical outcomes assessed (i.e., controlling for a total of 4 tests). Definition of abbreviations: NMDS = Non-metric multidimensional scaling.
Interplay between the Upper Respiratory Tract Microbiome and Acute Local Immune Response in Impacting Clinical Outcomes
Because the Shannon index was the microbial ecology metric most strongly and consistently associated with all outcomes, we included it as the exposure in our mediation models. Together, the first 2 PCs of the acute local immune response data accounted for ~70% of the data variance (PC1=58.80% and PC2=16.80%), whereas other PCs individually accounted for <5% of the data variance (Figure E4). Based on this, we only included PC1 and PC2 as mediators in the mediation analyses. The immune mediators were all negatively correlated with PC1, while their correlations with PC2 differed according to the specific immune mediator (Figure E5). The top 5 immune mediators contributing to PC1 were IL-4, IL-2R, IL-17A, IL-12, and IL-2, whereas the top 5 immune mediators contributing to PC2 were IL-23, IL-21, TSLP, IL-28A, and SCF (Figure E6).
The association of the Shannon index of the URT microbiome with the RSS during RSV ARI in infancy was partly mediated by the acute local immune response, although this mediation effect was only borderline significant (p=0.09). The first 2 PCs jointly explained ~20% of the indirect effect (PC1=2.74% and PC2 =16.68%) (Table E7). However, there was a significant interaction between the Shannon index and the first PC of the acute local immune response (p=0.048). In exploratory analyses, the direction of the association of PC1 with the RSS was positive at high Shannon indices, whereas at low Shannon indices it was negative (Figure 6). We performed similar mediation analyses using the other short- or long-term clinical outcomes. The mediation effects of the first 2 PCs explained a larger proportion of the total effect of the URT microbiome on the other short-term clinical outcomes (i.e., type of ARI and level of care required); however, these mediation effects were not significant, probably because the total effect size was already small (Table E7).
Figure 6.
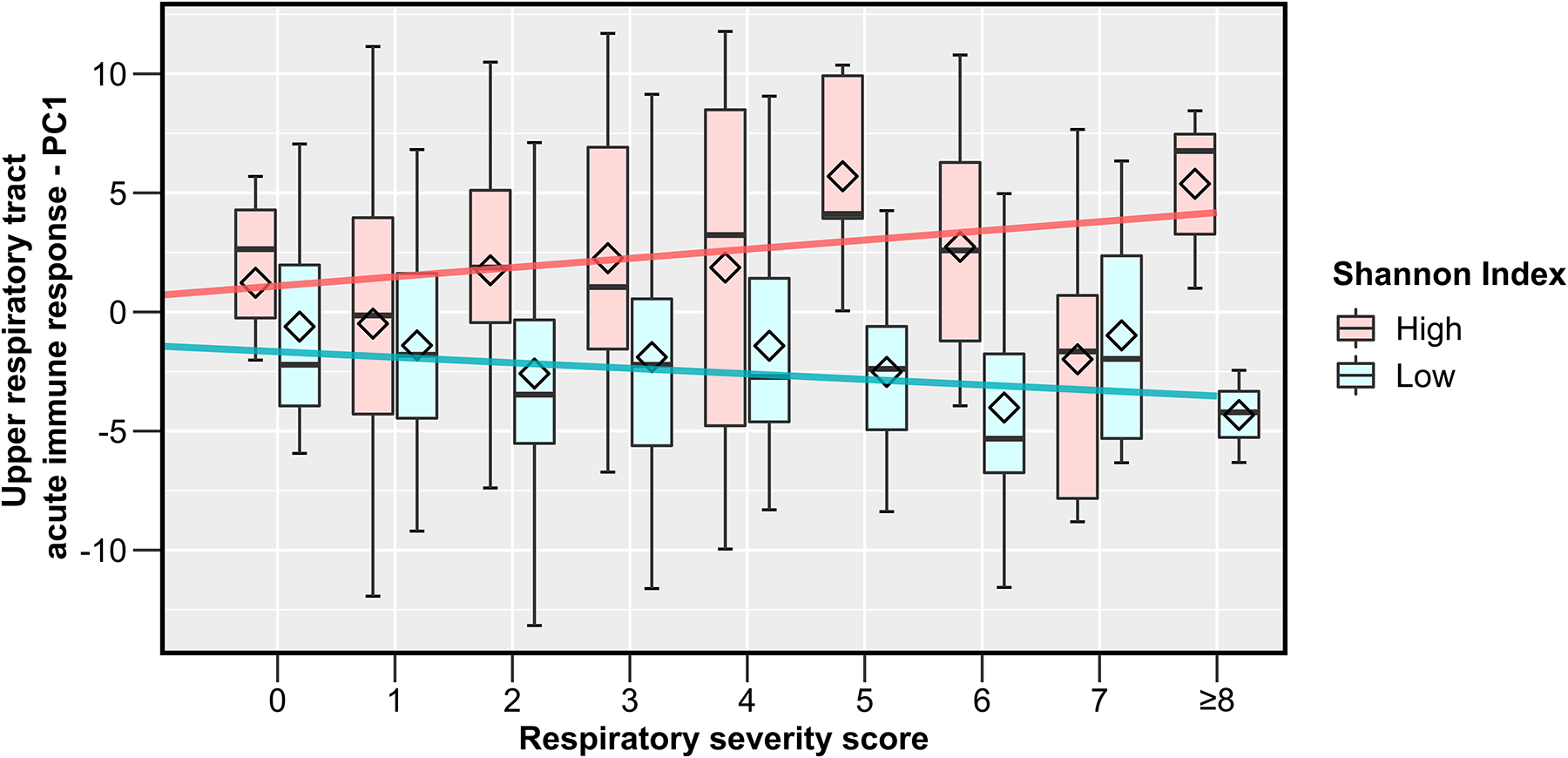
The modification of the effect of the acute local immune response on the respiratory severity score by the α-diversity of the upper respiratory tract microbiome during an acute respiratory infection due to respiratory syncytial virus in infancy. The box-and-whisker plots show the mean (diamond), median (middle bar), 1st quartile (lower bar), 3rd quartile (upper bar), minimum observation above the lowest fence (lower whisker), and maximum observation below the upper fence (upper whisker) of the first PC of the acute local immune response data for each respiratory severity score value and by Shannon index level (defined as high vs. low based on being above or below the median). The fitted regression lines from corresponding linear regression models are also shown. The first PC of the acute local immune response was negatively correlated with all individual immune mediators. Definition of abbreviations: PC = Principal component.
Discussion:
In our study, we found that the URT microbiome during RSV ARI in infancy is associated with the acute local immune response, the disease severity, and the number of wheezing episodes in the 4th year of life. Our results also suggest complex URT bacterial-immune interactions that can impact the severity of the RSV ARI.
The acute URT immune response to RSV —a leading cause of bronchiolitis, hospitalizations, and health care costs in young children worldwide29— is multifaceted and depends on a fine balance between anti-viral, anti-inflammatory (e.g., Treg-related), pro-inflammatory (e.g., type 1- and 3-related), and pro-allergic (e.g., type 2-related) immune pathways.9, 30, 31 To our knowledge, our study is the first to comprehensively examine the associations of the URT microbiome with a wide spectrum of local immune mediators during RSV ARI in infancy. Other studies examining the impact of bacterial communities on immune responses to RSV have been conducted in vitro or in animal models, focused on the gut microbiome or individual URT taxa, only assessed the systemic immune response, or only measured a small number of immune mediators.5, 6 In a smaller study of a subset of infants with RSV-only ARIs included in the current study (n=105), we showed that the URT abundance of Haemophilus (most likely H. influenzae) was associated with a higher proportion of the levels of local immune mediators when compared to the one noted in the current study.6 This difference may be related to the individual study designs, populations of interest, and/or the statistical methods we used (as in the current study we took into account all taxa of the URT microbiome). Because the dilution effect of nasal washes leads to very low concentrations of analytes and a large number of missing data when using the usual standard curve-based data-processing methods, we opted to use MFIs instead of concentrations, as other studies have shown that this method increases the sensitivity and accuracy of high-throughput immunoassays.9, 18 Interestingly, we found several specific taxa-immune mediator associations. The abundances of well-known respiratory pathogens (e.g., Klebsiella, Haemophilus, Moraxella, or Streptococcus) were not consistently related to the levels of key anti-viral (e.g., IFN-α or IL-28A) or type 1 (e.g., IFN-γ, IL-2, or IL-12) cytokines. However, they were positively associated with the levels of multiple pleiotropic (e.g., TNF-α, IL-1β, IL-6, IL-8, MCP-1, MIP-1α) and type 3 (e.g., IL-17A) pro-inflammatory immune mediators. The majority of these have been shown to play a role in increased disease severity, although the effect of type 3 immune responses during RSV ARI in infancy remains controversial, with some studies showing heightened IL-17A production to be associated with impaired viral clearance, increased airway neutrophilic inflammation, and more abundant mucus production, whereas other studies have suggested that increased IL-17 levels could actually be beneficial.30 In regard to type 2 pathways, there was a positive association of the abundance of Klebsiella with the levels of IL-13 and of the abundance of Moraxella with the levels of IL-4, but not with other key type 2 cytokines (e.g., IL-5, IL-33, or TSLP). In contrast, the abundances of other genera (e.g., Methylobacterium, Ralstonia, and Cloacibacterium) appeared to have opposite effects to those described above. Taken together, our findings suggest that the balance between taxa of URT microbial ecosystem can potentially alter the delicate equilibrium between immune response pathways (e.g., type 1 vs. type 2 or type 3 vs. Treg9, 30, 31) and ultimately impact the pathophysiology of RSV ARI in infancy.
In line with our findings, other studies have shown that the URT microbiome can impact viral load,5 viral clearance,32 and host gene expression4 during an early-life RSV ARI, which collectively support an association of the URT microbiome with RSV ARI severity. One smaller study (n=58) also showed similar findings to ours.4 In contrast, 2 other pediatric studies found no clear relation between the URT microbiome and several markers of RSV ARI severity.3, 5 However, the latter 2 studies were limited to RSV-infected children requiring hospitalization, used other sample collection methods, focused on β-diversity-related metrics, and adjusted for a limited number of confounders, which may explain the discrepant results.
Few other studies have also suggested an effect of the early-life URT microbiome on the development of childhood wheezing illnesses, most of which have been conducted in the general pediatric population.7, 33, 34 In a prior study conducted on a subset of RSV-infected infants included in the current study (n=118), we found that an increased abundance of Lactobacillus in the URT reduced the odds of recurrent wheeze by age 2 years.7 One other study found the abundances of Haemophilus, Moraxella, and Klebsiella during RSV ARI to be positively associated with the onset of recurrent wheeze by age 3 years, although that study was smaller (n=74), focused on the lower airway microbiome, only included RSV-infected children requiring hospitalization, and did not adjust for potential confounders.35 In contrast, in our current study, we found no particular taxa of the URT microbiome to be associated with the risk of the number of wheezing episodes in the 4th year of life, although its α-diversity was associated with this clinical outcome. The prevalence of recurrent wheeze in the 2 prior studies was >35%, whereas that of any wheeze in the 4th year of life in our current study was ~20%, so it is possible that our statistical analyses were underpowered to detect individual associations (particularly of low-abundant taxa, such as Lactobacillus). It is also possible that childhood wheezing illnesses in the toddler and preschool-aged years represent distinctive phenotypes and that particular characteristics of the early-life upper and/or lower airway microbiome could impact these outcomes differently.
Unlike prior pediatric studies of the URT microbiome during RSV ARI that have largely focused on β-diversity-related metrics or only the top most abundant taxa,3, 32, 36, 37 we elected to include multiple microbial ecology metrics to better understand which of the URT microbiome characteristics (e.g., richness, α-diversity, β-diversity, and/or individual taxa abundance) are more relevant to this disease. Overall, metrics that take into account the community as a whole (and for β-diversity, those based in the presence vs. absence of taxa instead of their abundance) were more consistently associated with short- and long-term clinical outcomes, as well as with the acute local immune response. This suggests that low-abundant taxa may exert a strong effect on RSV-related outcomes, although it could also reflect low statistical power for certain measures and during taxa abundance testing.38
One other novel aspect of our study is the implementation of a causal mediation framework using a combination of next-generation sequencing, high-throughput immunoassays, and novel bioinformatics. The acute local immune response partly mediated the association between the URT microbiome and the severity of the RSV ARI in infancy, although this was only borderline significant. Our results also suggest complex URT bacterial-immune interactions that can impact RSV-related outcomes in infancy (with the URT microbiome having an effect on the acute local immune response and, in turn, the acute local immune response influencing the URT microbiome). This emphasizes the importance of study designs and statistical techniques that can examine these intricate relationships, even though these are certainly difficult to assess in pediatric studies. To our knowledge, prior studies in this field have only assessed exposure-outcome associations without taking into account mediation or interaction effects. In our study, most of the effect of the URT microbiome on RSV ARI severity in infancy was not explained by the acute local immune response, which implies that there could be other pathways underlying this association. For example, URT bacteria could have direct effects on respiratory cells, lead to epigenetic changes, or alter other microbial ecosystems (such as the host’s fungal or viral communities). Because we had to reduce the multi-dimensionality of both the URT microbiome and acute local immune response data to perform the mediation analyses, it is also possible that we may have lost some information and that the indirect effect is in fact higher than the one we found.
Our study has numerous additional strengths, including the population-based design, the large sample size, the close surveillance during the infants’ first winter viral season to capture their initial RSV ARI, the inclusion of the full spectrum of the RSV ARI severity and both short- and long-term clinical outcomes, and the use of statistical analysis taking into account the compositional nature of the 16S rRNA data, adjusting for potential confounders, and controlling for multiple comparisons. We should also acknowledge several limitations. First, as is the case with every observational study, there is always a possibility of selection bias or residual confounding. For example, we lacked data on introduction of solid foods or use of probiotics. We also only tested for co-infections with human rhinovirus or human enterovirus, so we could have missed other respiratory viruses. Likewise, although we used multiple methods to try to capture all RSV ARIs throughout the infancy period, only ~19% of infants in our study had ≥1 RSV ARI confirmed by RT-PCR, while previous studies have shown the prevalence of this disease to be ~50% in the first year of life.39 This discrepancy could be explained by us not capturing RSV ARIs among infants who were asymptomatic, had very mild symptoms, or that were missed or out of window for biospecimen collection. In this context, recent studies have shown that asymptomatic RSV ARIs are relatively common in the pediatric population.40, 41 Second, based on our hypothesis of interest, our study only included RSV-infected infants. We have previously shown that the URT microbiome and acute local immune response differ between RSV-infected infants and healthy infants or infants with ARIs due to other respiratory viruses (such as human rhinovirus).8, 14 Our findings are thus not generalizable to healthy infants or those with non-RSV ARIs, but are relevant to a group at a high risk of infant morbidity and of developing childhood asthma phenotypes.42, 43 Third, some of our statistical analyses had a cross-sectional design and we lacked infant nasal wash samples prior to the RSV ARI. Thus, there is a possibility of reverse causation for certain RSV-related outcomes (e.g., that it is actually the RSV ARI severity or the acute local immune response that impacts the URT microbiome and not the other way around). However, we found consistent results in our longitudinal analyses of the number of wheezing episodes in the 4th year of life, but —as suggested by our results— URT bacterial-immune interactions during RSV ARI are complex and likely multidirectional.44 Fourth, we used a rarefied microbiome dataset for some of the statistical analyses such that all samples had the same library size (the smallest total counts of all samples). Rarefying is a simple method to avoid bias in statistical analysis caused by the unbalanced library size, but it also tends to reduce power. To confirm our results were robust to the rarefying step, we repeated the statistical analyses using a non-rarefied dataset and rarefying to a higher library size (e.g., n=1,008 after discarding 19 samples with <1,000 sequences), obtaining similar results (data not shown). Fifth, the number of wheezing episodes at age 4 years was assessed by parental report and we lacked data on physician confirmation of the wheezing event, which could have led to a misclassification bias. However, unlike in the first few years of life, most episodes of wheezing at age 4 years are caused by a childhood wheezing phenotype, as the prevalence of other causes of early-life wheeze (such as bronchiolitis, airway malacia, reflux or aspiration, and other anatomical abnormalities) decreases after age 3 years. Lastly, we only had clinical outcomes until age 4 years, so whether the URT microbiome during RSV ARI in infancy influences childhood wheezing illnesses at later ages remains unknown. Nonetheless, our findings are particularly important as childhood wheezing illnesses in the preschool-age years remain a common cause of morbidity and health care utilization in the pediatric population.45
In summary, we found that the URT microbiome during RSV ARI in infancy is associated with short- and long-term clinical outcomes. Furthermore, our results suggest a complex interplay between the URT microbiome and the acute local immune impacting the severity of the RSV ARI in infancy. Our study adds new insights on URT bacterial-immune interactions during RSV ARI in infancy.
Supplementary Material
Key Messages:
The upper respiratory tract microbiome during acute respiratory infections due to respiratory syncytial virus in infancy is associated with the acute local immune response, the disease severity, and the number of wheezing episodes in the 4th year of life
Upper respiratory tract bacterial-immune interactions can impact the severity of acute respiratory infections due to respiratory syncytial virus in infancy
Declaration of All Sources of Funding:
This work was supported by funds from the National Institute of Allergy and Infectious Diseases (under award numbers U19AI095227, K24AI77930, HHSN272200900007C, R21AI142321, U19AI110819, R21AI154016, R21AI149262, and UG3OD023282); the National Heart, Lung, and Blood Institute (under award numbers K23HL148638 and R01HL146401); the National Institute of General Medical Sciences (under award number R01GM140464); the Parker B. Francis Fellowship Program; the Vanderbilt Institute for Clinical and Translational Research (grant support from the National Center for Advancing Translational Sciences under award number UL1TR000445); the Department of Pediatrics at Vanderbilt University Medical Center (grant support from the Eunice Kennedy Shriver National Institute of Child Health and Human Development under award number K12HD087023); the Vanderbilt Building Interdisciplinary Research Careers in Women’s Health K12 program (grant support from the Eunice Kennedy Shriver National Institute of Child Health and Human Development under award number K12HD04348318); and the Vanderbilt Technologies for Advanced Genomics Core (grant support from the National Institutes of Health under award numbers UL1RR024975, P30CA68485, P30EY08126, and G20RR030956).
Declaration of Conflict of Interests:
LJA has done paid consultancies on respiratory syncytial virus (RSV) vaccines for Bavarian Nordic, Novavax, Daiichi-Sankyo, ClearPath Development Company, and Pfizer. He receives funding from Pfizer through Emory University for laboratory surveillance studies of RSV infection in adults. He is a co-inventor on several Centers for Disease Control and Prevention patents on the RSV G protein and its CX3C chemokine motif relative to immune therapy and vaccine development. He is also co-inventor on a patent filing for the use of RSV platform virus-like particles with the F and G proteins for vaccines. The other authors do not have a commercial or other association that might pose a conflict of interest.
Abbreviations:
- ARI
Acute respiratory infection
- INSPIRE
Infant Susceptibility to Pulmonary Infections and Asthma Following Respiratory Syncytial Virus Infection in Infancy study
- IQR
Interquartile range
- MFI
Median fluorescence intensity
- PC
Principal component
- rRNA
Ribosomal ribonucleic acid
- RSS
Respiratory severity score
- RSV
Respiratory syncytial virus
- URT
Upper respiratory tract
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Notice of Prior Presentation:
This study was presented as an abstract at the American Thoracic Society Virtual International Conference 2020 (online presentation).
References:
- 1.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 2010;375:1545–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, Staat MA, et al. The burden of respiratory syncytial virus infection in young children. N Engl J Med 2009;360:588–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, et al. Nasopharyngeal Microbiota, Host Transcriptome, and Disease Severity in Children with Respiratory Syncytial Virus Infection. Am J Respir Crit Care Med 2016;194:1104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonawane AR, Tian L, Chu CY, Qiu X, Wang L, Holden-Wiltse J, et al. Microbiome-Transcriptome Interactions Related to Severity of Respiratory Syncytial Virus Infection. Sci Rep 2019;9:13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ederveen THA, Ferwerda G, Ahout IM, Vissers M, de Groot R, Boekhorst J, et al. Haemophilus is overrepresented in the nasopharynx of infants hospitalized with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses. Microbiome 2018;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shilts MH, Rosas-Salazar C, Turi KN, Rajan D, Rajagopala SV, Patterson MF, et al. Nasopharyngeal Haemophilus and local immune response during infant respiratory syncytial virus infection. J Allergy Clin Immunol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Schobel S, Chappell JD, Larkin EK, et al. Nasopharyngeal Lactobacillus is Associated with Childhood Wheezing Illnesses Following Respiratory Syncytial Virus Infection in Infancy. J Allergy Clin Immunol 2018;142:1447–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Chappell JD, Larkin EK, Nelson KE, et al. Nasopharyngeal Microbiome in Respiratory Syncytial Virus Resembles Profile Associated with Increased Childhood Asthma Risk. Am J Respir Crit Care Med 2016;193:1180–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turi KN, Shankar J, Anderson LJ, Rajan D, Gaston K, Gebretsadik T, et al. Infant Viral Respiratory Infection Nasal Immune-Response Patterns and Their Association with Subsequent Childhood Recurrent Wheeze. Am J Respir Crit Care Med 2018;198:1064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larkin EK, Gebretsadik T, Moore ML, Anderson LJ, Dupont WD, Chappell JD, et al. Objectives, design and enrollment results from the Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure Study (INSPIRE). BMC Pulm Med 2015;15:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.American Academy of Pediatrics. Respiratory Syncytial Virus. In: Kimberlin DW, Brady MT, Jackson MA, Long SS, eds. Red Book: 2018 Report of the Committee on Infectious Diseases. Elk Grove, IL: American Academy of Pediatrics; 2018. [Google Scholar]
- 12.Wu P, Dupont WD, Griffin MR, Carroll KN, Mitchel EF, Gebretsadik T, et al. Evidence of a causal role of winter virus infection during infancy in early childhood asthma. Am J Respir Crit Care Med 2008;178:1123–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kodani M, Yang G, Conklin LM, Travis TC, Whitney CG, Anderson LJ, et al. Application of TaqMan low-density arrays for simultaneous detection of multiple respiratory pathogens. J Clin Microbiol 2011;49:2175–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Schobel S, Chappell JD, Larkin EK, et al. Differences in the Nasopharyngeal Microbiome During Acute Respiratory Tract Infection With Human Rhinovirus and Respiratory Syncytial Virus in Infancy. J Infect Dis 2016;214:1924–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 2007;35:7188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018;6:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Won JH, Goldberger O, Shen-Orr SS, Davis MM, Olshen RA. Significance analysis of xMap cytokine bead arrays. Proc Natl Acad Sci U S A 2012;109:2848–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCallum GB, Morris PS, Wilson CC, Versteegh LA, Ward LM, Chatfield MD, et al. Severity scoring systems: are they internally valid, reliable and predictive of oxygen use in children with acute bronchiolitis? Pediatr Pulmonol 2013;48:797–803. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez H, Hartert TV, Gebretsadik T, Carroll KN, Larkin EK. A simple respiratory severity score that may be used in evaluation of acute respiratory infection. BMC Res Notes 2016;9:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2006. [Google Scholar]
- 22.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, et al. vegan: Community Ecology Package, 2014. Available from: http://CRAN.Rproject.org/package=vegan.
- 24.Lin W, Shi P, Feng R, Li H. Variable selection in regression with compositional covariates. Biometrika 2014;101:785–97. [Google Scholar]
- 25.Tang ZZ, Chen G, Alekseyenko AV, Li H. A general framework for association analysis of microbial communities on a taxonomic tree. Bioinformatics 2017;33:1278–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andabaka T, Nickerson JW, Rojas-Reyes MX, Rueda JD, Bacic Vrca V, Barsic B. Monoclonal antibody for reducing the risk of respiratory syncytial virus infection in children. Cochrane Database Syst Rev 2013:CD006602. [DOI] [PubMed] [Google Scholar]
- 27.Huang YT, Pan WC. Hypothesis test of mediation effect in causal mediation model with high-dimensional continuous mediators. Biometrics 2016;72:402–13. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological 1995;57:289–300. [Google Scholar]
- 29.Shi T, McAllister DA, O’Brien KL, Simoes EAF, Madhi SA, Gessner BD, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 2017;390:946–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangodt TC, Van Herck MA, Nullens S, Ramet J, De Dooy JJ, Jorens PG, et al. The role of Th17 and Treg responses in the pathogenesis of RSV infection. Pediatr Res 2015;78:483–91. [DOI] [PubMed] [Google Scholar]
- 31.Vazquez Y, Gonzalez L, Noguera L, Gonzalez PA, Riedel CA, Bertrand P, et al. Cytokines in the Respiratory Airway as Biomarkers of Severity and Prognosis for Respiratory Syncytial Virus Infection: An Update. Front Immunol 2019;10:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansbach JM, Hasegawa K, Piedra PA, Avadhanula V, Petrosino JF, Sullivan AF, et al. Haemophilus-Dominant Nasopharyngeal Microbiota Is Associated With Delayed Clearance of Respiratory Syncytial Virus in Infants Hospitalized for Bronchiolitis. J Infect Dis 2019;219:1804–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015;17:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teo SM, Tang HHF, Mok D, Judd LM, Watts SC, Pham K, et al. Airway Microbiota Dynamics Uncover a Critical Window for Interplay of Pathogenic Bacteria and Allergy in Childhood Respiratory Disease. Cell Host Microbe 2018;24:341–52 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Zhang X, Zhang N, Wang X, Sun L, Chen N, et al. Airway microbiome, host immune response and recurrent wheezing in infants with severe respiratory syncytial virus bronchiolitis. Pediatr Allergy Immunol 2020;31:281–9. [DOI] [PubMed] [Google Scholar]
- 36.Mansbach JM, Hasegawa K, Henke DM, Ajami NJ, Petrosino JF, Shaw CA, et al. Respiratory syncytial virus and rhinovirus severe bronchiolitis are associated with distinct nasopharyngeal microbiota. J Allergy Clin Immunol 2016;137:1909–13 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Man WH, Scheltema NM, Clerc M, van Houten MA, Nibbelke EE, Achten NB, et al. Infant respiratory syncytial virus prophylaxis and nasopharyngeal microbiota until 6 years of life: a subanalysis of the MAKI randomised controlled trial. Lancet Respir Med 2020. [DOI] [PubMed] [Google Scholar]
- 38.Tang ZZ, Chen G, Alekseyenko AV. PERMANOVA-S: association test for microbial community composition that accommodates confounders and multiple distances. Bioinformatics 2016;32:2618–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glezen WP, Taber LH, Frank AL, Kasel JA. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child 1986;140:543–6. [DOI] [PubMed] [Google Scholar]
- 40.Zylbersztejn A, Pembrey L, Goldstein H, Berbers G, Schepp R, van der Klis F, et al. Respiratory syncytial virus in young children: community cohort study integrating serological surveys, questionnaire and electronic health records, Born in Bradford cohort, England, 2008 to 2013. Euro Surveill 2021;26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munywoki PK, Koech DC, Agoti CN, Bett A, Cane PA, Medley GF, et al. Frequent Asymptomatic Respiratory Syncytial Virus Infections During an Epidemic in a Rural Kenyan Household Cohort. J Infect Dis 2015;212:1711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beigelman A, Rosas-Salazar C, Hartert TV. Childhood Asthma: Is It All About Bacteria and Not About Viruses? A Pro/Con Debate. J Allergy Clin Immunol Pract 2018;6:719–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abreo A, Gebretsadik T, Stone CA, Hartert TV. The impact of modifiable risk factor reduction on childhood asthma development. Clin Transl Med 2018;7:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol 2017;15:259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bisgaard H, Szefler S. Prevalence of asthma-like symptoms in young children. Pediatr Pulmonol 2007;42:723–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


